Abstract
The purpose of this study was to investigate the synergistic and bactericidal effects of combinations of colistin with meropenem or amikacin in vitro and provide laboratory data needed for development of therapeutic strategies for the treatment of carbapenem-resistant Klebsiella pneumoniae (CRKP) infection. We found that minimum inhibitory concentration (MIC) of colistin, meropenem and amikacin were 2~32, 4~256, and 1~16384 μg/ml, respectively. The minimum bactericidal concentration of the antibiotics was either 1× or 2×MIC. Treatments of 6 CRKP isolates at 1 μg/ml colistin completely killed 2 of them and suppressed 4 others growth. 4 CRKP isolates at 16 μg/ml meropenem or amikacin completely killed and suppressed 2 others growth. 2 CRKP isolates showed synergic effects in all colistin combination and 3 CRKP isolates showed synergic effects in part of colistin combination. Our data suggest that colistin in combination with either meropenem or amikacin could be a valid therapeutic option against colistin-resistant CRKP isolates. Moreover, the combination of colistin-amikacin is less expensive to treat CRKP infections in Eastern Heilongjiang Province.
Keywords: carbapenem-resistant Klebsiella pneumoniae, colistin, combination therapy, synergistic effect, bactericidal effect
Introduction
The mergence and dissemination of carbapenem-resistant Klebsiella pneumoniae (CRKP) may poses serious threat to the elderly and sick persons. These organisms are associated with high mortality rates and have the potential to spread widely around the world (Patel et al., 2008; Grundmann et al., 2017; Zhang et al., 2017). It has been reported that the mortality rates can be as high as 40–50% in patients with CRKP infections (Patel et al., 2008). The pathogens often harbor multiple drug-resistant genes that cause the bacteria hetero-resistance to antibiotics in addition to β-bactams (Band et al., 2018, 2019), severely limiting alternative treatment options and increasing therapeutic cost (Huang et al., 2018; Santos and Secoli, 2019). The development of new antimicrobial agents to combat new CRKP isolates is extremely time-consuming, and therapeutic replacement with novel alternative agents may cause potential toxicities. Recently, Band et al. reported that two multidrug-resistant CRKP isolates exhibiting colistin hetero-resistance, a phenomenon in which only a subpopulation of genetically identical bacterial is resistant to the drug (Band et al., 2018, 2019). Therefore, the use of combination therapy of two or more antibiotics instead of monotherapy in CRKP-infected patients is a feasible option, and the efficacy of such combinations to kill the hetero-resistant CRKP isolates remains to be determined.
In our previous studies, we have demonstrated that carbapenem-non-susceptible Enterobacteriaceae (CNSE) isolates from the Eastern Heilongjiang Province of China carry carbapenem hydrolases encoded by the blaKPC-2, blaNDM-1, blaNDM-5, blaNDM-7, and blaIMP-4 genes. Most of these isolates harbor multiple drug-resistance genes related to extended-spectrum beta lactamase (ESBL) such as the blaSHV, blaTEM, blaCTX-M-15, and CTX-M-177 genes (Gong et al., 2018). The Carbapenemase-producing K. pneumoniae (KP) isolates are sometime dangerous because these pathogens are frequently co-resistant to multiple antibiotic classes, and only a few of available antibiotics remain effective. So far, several combinations such as colistin (COL), tigecycline (TGC), gentamicin (GEN), and fosfomycin (FM) have been reported to be effective against CRKP isolates as measured by in vitro checkerboard technique or time-kill assays (Morici et al., 2017). Clinical retrospective studies have also demonstrated favorable outcomes for the patients treated with combinations of COL and a carbapenem, TGC, FM, or an aminoglycoside (Tangden et al., 2014). Thus, COL has been recommended as the first line of option against CRKP isolates (Gibson et al., 2016). However, effectiveness of COL combinations with meropenem (MEM) or amikacin (AMK) against CRKP isolates emerged in the Heilongjiang Province of China has not been studies yet.
In this study, we investigated the synergistic and bactericidal effects of combinations of COL with either the most sensitive carbapenem antibiotic, MEM or the antibiotic with the lowest resistance rate in our region, AMK against the CRKP isolates resistant to COL or MEM alone. Our studies provided valuable laboratory data of antibiotic susceptibility for clinicians to develop therapeutic strategies for the treatment of drug-resistant CRKP-infections in the eastern Heilongjiang Province, China. In addition, combination therapy may also help to prevent the spread of the bacteria in this region as well as other parts of China.
Materials and Methods
Bacterial Isolates
Forty clinical CRKP isolates were collected from patients with infectious diseases at the First Affiliated Hospital of Jiamusi University from October 2015 to January 2019. The CRKP isolates were characterized by using the Vitek 2 system and the AST-GN card (bioMérieux, France). Routine multilocus sequence typing (MLST) and polymerase chain reaction (PCR) were performed to verify the presence of carbapenemase and ESBL genes (Table 1).
Table 1.
Results for the bla genotype, MLST, MICs, MBCs, and FICIs of colistin, meropenem, and amikacin against CRKP isolates.
| Isolate | Source | bla genotype | MLST | MIC (μg/ml) | MBC | FICI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| COL | MEM | AMK | COL | MEM | AMK | COL+MEM | COL+AMK | ||||
| 700603 | 4 | 1/4 | 1/2 | ||||||||
| CRKP1 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 64 | 8 | 0.25 | 1 | |||
| CRKP2 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 128 | 4 | 0.25 | 0.375 | |||
| CRKP3 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 128 | 8 | 0.25 | 0.75 | |||
| CRKP4 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 16 | 16 | 8 | 0.375 | 0.625 | |||
| CRKP5 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 8 | 0.375 | 0.75 | |||
| CRKP6 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 16 | 16 | 8 | 0.375 | 0.625 | |||
| CRKP7 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 8 | 1 | 0.25 | |||
| CRKP8 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 4 | 0.625 | 0.375 | |||
| CRKP9 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 4 | 0.625 | 0.375 | |||
| CRKP10 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 8 | 0.5 | 0.25 | |||
| CRKP11 | Sputum | KPC-2, SHV, CTX-M-15 | ST76 | 8 | 16 | 16 | 0.5 | 0.375 | |||
| CRKP12 | Sputum | KPC-2, NDM-5, SHV, TEM, CTX-M-177 | ST76 | 8 | 64 | 2 | 2× | 1× | 2× | 0.375 | 0.375 |
| CRKP13 | Blood | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 4 | 16 | 4 | 1× | 1× | 1× | 0.75 | 0.25 |
| CRKP14 | Blood | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 4 | 16 | 2 | 1× | 1× | 1× | 0.75 | 0.375 |
| CRKP15 | Blood | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 4 | 64 | 2 | 2× | 1× | 1× | 0.375 | 1 |
| CRKP16 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 16 | 16 | 8 | 0.25 | 0.625 | |||
| CRKP17 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 16 | 16 | 8 | 0.375 | 0.625 | |||
| CRKP18 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 16 | 16 | 16 | 0.375 | 0.25 | |||
| CRKP19 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 2 | 64 | 2 | 1× | 1× | 2× | 0.375 | 0.75 |
| CRKP20 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 4 | 16 | 2 | 2× | 2× | 2× | 0.75 | 0.625 |
| CRKP21 | Secretion | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 4 | 8 | 4 | 0.25 | 0.375 | |||
| CRKP22 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 8 | 8 | 0.625 | 0.375 | |||
| CRKP23 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 4 | 0.5 | 0.375 | |||
| CRKP24 | Blood | KPC-2, SHV | ST11 | 4 | 128 | 1 | 1× | 2× | 2× | 0.625 | 0.375 |
| CRKP25 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 2 | 1× | 2× | 1× | 0.5 | 1 |
| CRKP26 | Blood | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 32 | 32 | 8 | 0.25 | 0.375 | |||
| CRKP27 | Sputum | KPC-2, TEM, CTX-M-15 | ST76 | 8 | 32 | 4 | 0.25 | 0.375 | |||
| CRKP28 | Sputum | KPC-2, TEM, CTX-M-15 | ST76 | 8 | 16 | 4 | 0.5 | 0.375 | |||
| CRKP29 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 16 | 8 | 4 | 0.375 | 0.375 | |||
| CRKP30 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 8 | 4 | 0.75 | 0.375 | |||
| CRKP31 | Sputum | SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 4 | 0.75 | 0.375 | |||
| CRKP32 | Sputum | KPC-2, SHV, TEM, CTX-M-15 | ST375 | 2 | 8 | 1 | 1× | 1× | 1× | 1 | 0.625 |
| CRKP33 | other specimens | KPC-2, SHV, TEM, CTX-M-15 | ST76 | 8 | 16 | 4 | 0.5 | 0.5 | |||
| CRKP34 | Sputum | SHV, TEM | ST530 | 2 | 4 | 1 | 1× | 1× | 1× | 1 | 0.625 |
| CRKP35 | Sputum | SHV, TEM | ST3335 | 8 | 4 | 4 | 0.75 | 0.375 | |||
| CRKP36 | Blood | KPC-2, SHV, TEM, CTX-M-177 | ST11 | 8 | 256 | 16,384 | 0.625 | – | |||
| CRKP37 | Sputum | KPC-2, SHV, TEM, CTX-M-15, CTX-M-177 | ST290 | 8 | 16 | 2 | 0.375 | 0.25 | |||
| CRKP38 | Sputum | KPC-2, SHV, TEM, CTX-M-15, CTX-M-177 | ST290 | 8 | 16 | 2 | 0.375 | 0.25 | |||
| CRKP39 | Sputum | KPC-2, SHV, TEM, CTX-M-15, CTX-M-177 | ST290 | 8 | 16 | 2 | 0.375 | 0.25 | |||
| CRKP40 | Sputum | KPC-2, NDM-1, SHV, TEM, CTX-M-15, CTX-M-177 | ST290 | 8 | 128 | 1 | 0.25 | 0.375 | |||
MLST, multilocus sequence typing; MIC, minimum inhibitory concentrations; MBC, minimum bactericidal concentration; FICI, fractional inhibitory concentration index; COL, colistin; MEM, meropenem; AMK, amikacin; –, not tested.
Antimicrobial Susceptibility Testing
Minimum inhibitory concentration (MIC) defined as the lowest compound concentration (μg/ml) required to stop bacterial growth was determined by using the microbroth dilution method according to the Clinical and Laboratory Standard Institutes (CLSI) recommendations (Clinical Laboratory Standards Institute, 2016). An ESBL-positive strain of KP ATCC700603 was used as a reference control. Three antimicrobial agents were tested: Colistin (COL, Sigma-Aldrich, St. Louis, MO), Meropenem (MEM, Haibin Pharmaceutical Co., Shenzhen, China), and amikacin (AMK, Sui Cheng Pharmaceutical Ltd., Tianjin, China). The concentration ranges of COL, MEM and AMK used in this study were 0.0625–64, 0.5–256, and 0.03125–16,384 μg/ml, respectively. The MEM and AMK results were interpreted per CLSI criteria (Clinical Laboratory Standards Institute, 2016), whereas the COL results were interpreted based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint recommendations (EUCAST, 2016). The MICs were determined by measuring optical density (OD) at 570 nm using a microplate reader.
Bactericidal/Static Determination Assay
After MIC tests, bacteria in each well (100 μL) corresponding to 1×, 2×, 4×, and 8× MIC were transferred and spread on Mueller-Hinton (MH) agar plates and cultured at 37°C overnight, followed by colony-forming units (CFUs) counting. Minimum bactericidal concentrations (MBCs) of the antibiotics were defined as the lowest antibiotic concentrations required to cause ≥99.99% cell death within 24 h. An antibiotic was classified as bactericidal if its MBC was between 1× to 4× MIC.
Checkerboard Technique
The synergistic effects of COL-MEM and COL-AMK combinations on CRKP isolates were tested using the checkerboard technique. The concentration ranges were determined based on MICs. Briefly, 50 μl of each antibiotic at five increasing (4-fold) concentrations (0.125× to 2× MIC) were used, and each well was inoculated with 100 μl of a 7.5 × 105 CFU/ml suspension of the test CRKP isolates in a final volume of 200 μl in duplicate. Results were measured by reading optical density (OD) after 24 h incubation at 37°C at 570 nm using a microplate reader. The effects of the antimicrobial combinations were defined according to the fractional inhibitory concentration index (FICI) as follows: FICI ≤ 0.5, synergism; 0.5 < FICI ≤ 1, additive; 1 < FICI ≤ 4, indifferent; or FICI > 4, antagonistic (Bai et al., 2015).
Antibacterial Time-Kill Assay
The antibacterial time-kill assay was performed on three individual antibiotics and two COL combinations against 6 distinct sequence types (STs) of the CRKP isolates. Antibiotic concentrations were calculated using the mean value of the steady-state concentrations of the non-protein-bound drug in humans as described previously (Clinical Laboratory Standards Institute, 1999). The concentrations of 250 and 1,000 μg/L COL, 4,000 and 16,000 μg/L MEM, and 8,000 and 16,000 μg/L AMK were used in both single-agent and combination studies (Kulengowski et al., 2018). Briefly, 5 × 105 CFU/ml of the tested organism were inoculated in 10 ml of the broth containing a single antibiotic or antibiotics combinations. Bacterial growth was quantified after 0, 2, 4, 8, 12, and 24 h incubation at 37°C by plating 10-fold dilutions on sheep blood agar, followed by counting CFU/ml. Time-kill curves were then constructed as a function of time, and the results are represented as the difference in log10 between the CFU/ml at 0 and 24 h. A decrease of ≥3 log10 in colony number as compared to the initial CFU/ml was considered as bactericidal effect while a decrease of <3 log10 CFU/ml over the initial CFU/ml was defined as bacteriostatic effect. An increase in colony count from the previous timepoint was thought as regrowth (Doern, 2014). A decrease of ≥2 log10 CFU/ml of combination cultures as compared to the most active single-drug broth at the same timepoint was defined as synergistic effect whereas an increase of >2 log10 in the combination cultures was interpreted as antagonism. Additivity and indifference were designated when the outcomes neither meet the criteria of synergy nor the criteria of antagonism (Petersen et al., 2006).
Statistical Analysis
The Kolmogorov–Smirnov test was used to analyze the data distribution. Student's t-test was used for treatment comparisons. SPSS 22.0 software was used for statistical analyses. P < 0.05 was considered as statistically significant.
Results
Characteristics of Bacterial Isolates
To characterize the isolates, we performed MLST and PCR to exam whether they carry carbapenemase and ESBL genes. We found that out of 40 CRKP isolates, 37 (92.5%) carried KPC-2 gene, which encodes carbapenemase class A, and 2 (13.2%) carried a gene that encodes New Delhi metallo-β-lactamase (NDM) carbapenemase. CRKP40 isolate carried blaNDM-1 gene while CRKP11 isolate harbored blaNDM-5 gene. Among total 6 ESBL genes amplified, blaSHV, bIaTEM, blaCTX-M-15, and blaCTX-M-177 were present in 38 (95.0%), 38 (95.0%), 35 (87.5%), and 6 (15.0%) of the CRKP isolates, respectively. These CRKP isolates were grouped into 6 distinct STs; ST76 was the most predominant clone (n = 30), followed by ST290 (n = 4), ST11 (n = 2), ST375 (n = 2), ST570 (n = 1), and ST3335 (n = 1) as shown in Table 1.
Bactericidal/Static Determination and Antimicrobial Susceptibility
To determine whether COL, MEM, and AMK prevent bacterial growth or kill bacteria, we carried out bactericidal/static determination assays. Our genotyping and MLST mapping found that 40 strains carried 2–5 different bla genotypes, some of which have the same bla genotyping (Table 1). Thus, we selected 10 CRKP isolates out of 40 for bactericidal/static determination assays because these representative CRKP isolates included all bla genotypes. We found that COL, MEM and AMK were bactericidal since the MBC of COL, MEM or AMK was either 1× MIC or 2× MIC (Table 1). MICs against COL and MEM were ranged from 2 to 32 and 4 to 256 μg/ml, respectively. The CRKP's resistances to single COL and MEM treatment reached 92.5 and 100%, respectively. Of the CRKP isolates, 97.5% were sensitive to AMK. The MIC number of the CRKP isolates for AMK were high (range between 1 and 16 μg/ml) but the strains were sensitive except 1 CRKP isolate that had a MIC of 16,384 μg/ml (Tables 1, 2). We interpreted the susceptibility of AMK were: S (sensitive), ≤16 μg/ml; I (intermediate), 32 μg/ml; R (resistant), ≥64 μg/ml. The 50% minimum inhibitory concentrations (MIC50) of COL, MEM and AMK were found to be 8, 16, and 4 μg/ml, respectively. The 90% minimum inhibitory concentrations (MIC90) of COL, MEM and AMK were 16, 128, and 8 μg/ml, respectively (Table 2). To determine if the isolates display resistance to MEM and AMK, we performed disk diffusion assays. We found that the resistant rates of the CRKP isolates to MEM and AMK were 100 and 2.5%, respectively.
Table 2.
Antimicrobial resistance rates and MIC distributions of CRKP isolates.
| MIC (μg/ml) | ||||
|---|---|---|---|---|
| Cutoff value of resistance (%) | Range | MIC50 | MIC90 | |
| COL | >2 (92.5%) | 2~32 | 8 | 16 |
| MEM | ≥4 (100%) | 4~256 | 16 | 128 |
| AMK | ≥16 (97.5%) | 1~16384 | 4 | 8 |
MIC, minimum inhibitory concentration; AMK, amikacin; MIC50, MIC at which 50% of the isolates tested are inhibited; MIC90, MIC at which 90% of the isolates tested are inhibited; COL, colistin; MEM, meropenem.
Synergistic Effects of COL-MEM and COL-AMK Combinations
To determine whether treatment of CRKP isolates with a combination of either COL-MEM or COL-AMK exerts synergistic and additive effects, we performed the checkerboard experiments. We observed that FICI of the COL-MEM and COL-AMK combination was 0.484 ± 0.190 and 0.494 ± 0.269, respectively. Treatment of CRKP isolates with a combination of COL-MEM exhibited 65.0% synergistic and 35.0% additive effects while a combined treatment of COL-AMK resulted in 65.0% synergistic and 3.25% additive effects on 39 CRKP isolates tested (Table 1). The FICI value of 1 CRKP isolate was unable to be calculated because the high concentration of AMK did not meet the drug requirements of the combination (Table 1). It appeared that the treatment of COL-MEM showed better synergistic and additive effects than COL-AMK combination (P < 0.05). As expected, the combination of COL-MEM had better synergistic and additive effects on the COL-susceptible isolates than the COL-resistant isolates (P < 0.05; Table 3). In addition, the treatments of CRKP isolates with combinations of COL-MEM and COL-AMK decreased MICs of COL by 5.75- and 5.33-fold, respectively as compared to MIC of COL with COL treatment alone.
Table 3.
Combination against colistin-susceptible and colistin-resistant CRKP isolates.
| Comb | COL-S | COL-R | Impact | p-value | ||||
|---|---|---|---|---|---|---|---|---|
| Nb | Mean | SD | Nb | Mean | SD | |||
| COL+MEM | 3 | 0.542 | 0.144 | 37 | 0.48 | 0.194 | Synergistic+ Additive | <0.001 |
| COL+AMK | 3 | 0.875 | 0.572 | 36 | 0.462 | 0.212 | Synergistic+ Additive | 0.619 |
| p-value | 0.454 | 0.064 | ||||||
Comb, Combination; COL-S, colistin-susceptible; COL-R, colistin-resistant; COL, colistin; MEM, meropenem; AMK, amikacin.
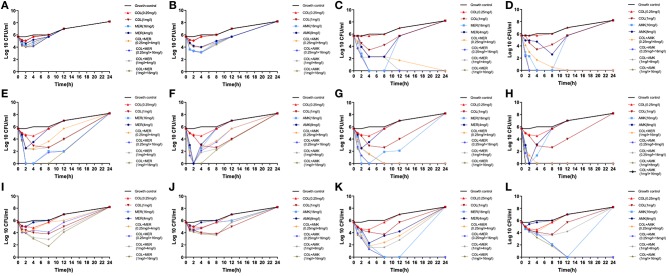
We found that the monotherapies exhibited considerable regrowth as determined by the time-kill assay (Figure 1). Treatment of CRKP 6 isolates (CRKP26, 32, 34, 35, 36, and 40) with COL at a dose of 0.25 mg/L did not kill the bacteria but exerted a bacteriostatic effect on them (Figure 1). Treatment of 6 CRKP isolates with COL at 1 mg/L displayed a bactericidal activity against 2 CRKP isolates (CRKP34 and 35) (Figures 1E–H). Treatment of 6 CRKP isolates with MEM at 4 mg/L exhibited a bacteriostatic effect on 2 out of (CRKP26 and 36) (Figures 1A,I) and bactericidal action on 4 others (CRKP32, 34, 35, and P40) (Figures 1C,E,G,K). Treatment of 6 CRKP isolate with AMK at 8 mg/L inhibited the growth of 3 CRKP isolates (CRKP26, 36 isolate and 40) (Figures 1B,J,L) and killed 3 others (CRKP32, 34 and 35) (Figures 1D,F,H). Treatment of 6 CRKP isolates with MEM or AMK at 16 mg/L killed 4 of them (CRKP32, 34, 35, and 40) (Figures 1C–H,K,L) and suppressed the growth of 2 others (CRKP26 and 36) (Figures 1A,B,I,J). Treatment of CRKP32 and 35 isolates with COL combinations either with MEM or AMK exhibited synergic actions and completely killed the bacteria without regrowth while mono-treatment with either of them failed to show bactericidal effects (Figures 1C,D,G,H). These combinations included 0.25 mg/L COL and 4 mg/L MEM, 0.25 mg/L COL and 16 mg/L MEM, 1 mg/L COL and 4 mg/L MEM, 1 mg/L COL and 16 mg/L MEM, 0.25 mg/L COL and 8 mg/L AMK, 0.25 mg/L COL and 16 mg/L AMK, 1 mg/L COL and 8 mg/L AMK, and 1 mg/L COL and 16 mg/L AMK. Treatment of CRKP34, 36, and 40 isolates with a combination of 1 mg/L COL and 16 mg/L MEM initially inhibited their growth but prolonged treatment and culture resulted in bacterial regrowth (Figures 1E,I,K). While treatment with a combination of 1 mg/L COL and 16 mg/L AMK showed synergic effects on CRKP 34 and 40 isolates, continuation of culture in in the presence of antibiotics caused bacterial regrowth (Figures 1F,L). Similarly, treatments of CRKP34 isolate with a combination of 0.25 mg/L COL and 8 mg/L AMK, or a combination of 0.25 mg/L COL and 16 mg/L AMK displayed a synergy, but long term treatment resulted in bacterial regrowth (Figure 1F).
Figure 1.
In vitro time-kill assays. Six CRKP isolates were treated with serum levels of colistin (COL), meropenem (MEM), or amikacin (AMK) alone or in a combination of COL MEM with either MEM or AMK. (A) CRKP26 isolate was treated with COL or MEM alone or in a combination of COL with MEM at the doses indicated. (B) CRKP26 isolate was treated with COL or AMK alone or in a com alone or a combination of COL with AMK at the doses indicated. (C) CRKP32 isolate was treated with COL or MEM alone or in a combination of COL with MEM at the doses indicated. (D) CRKP32 isolate was treated with COL or AMK alone or in a com alone or a combination of COL with AMK at the doses indicated. (E) CRKP34 isolate was treated with COL or MEM alone or in a combination of COL with MEM at the doses indicated. (F) CRKP34 isolate was treated with COL or AMK alone or in a com alone or a combination of COL with AMK at the doses indicated. (G) CRKP35 isolate was treated with COL or MEM alone or in a combination of COL with MEM at the doses indicated. (H) CRKP35 isolate was treated with COL or AMK alone or in a com alone or a combination of COL with AMK at the doses indicated. (I) CRKP36 isolate was treated with COL or MEM alone or in a combination of COL with MEM at the doses indicated. (J) CRKP36 isolate was treated with COL or AMK alone or in a com alone or a combination of COL with AMK at the doses indicated. (K) CRKP40 isolate was treated with COL or MEM alone or in a combination of COL with MEM at the doses indicated. (L) CRKP40 isolate was treated with COL or AMK alone or in a com alone or a combination of COL with AMK at the doses indicated.
Discussion
Klebsiella pneumoniae strains are usually harmless. They often live in human's intestines without giving any problems. But the bacteria can be very dangerous if they invade into other parts of the body when people are sick, with weak immune systems. The germs can become “superbugs” and they can cause pneumonia or spread through blood, resulting in serious problems. Treatment of K. pneumoniae infections usually starts with broad-spectrum antibiotics such as cephalosporins (cefotaxime and ceftriaxone), β-lactams and β-lactamase inhibitors, and carbapenems (imipenem or cilastatin). However, the dissemination of CRKP strains has limited the therapeutic options. Thus, treatment with polymyxins such as colistin abandoned many years ago has been re-implicated as one of antibiotics of the last resort for CRKP infections. Recent studies reported that K. pneumoniae also became resistant to COL because of the prolonged or inappropriate treatments, and the monotherapy of colistin for CRKP infection caused some problems (Zhang et al., 2017; Huang et al., 2018). In our study, we found that the single antibiotic treatment of CRKP isolates from our hospital with exhibited considerable regrowth Treatment of CRKP isolates with COL at a dose of 250 μg/L did not kill the bacteria but exerted a bacteriostatic effect on 6 CRKP isolates. Treatment of the bacteria with MEM at 4,000 μg/L exhibited a bacteriostatic effect on 3 CRKP isolates and treatment of the bacteria with AMK at 8,000 μg/L inhibited the growth of 3 CRKP isolates. Our observed phenotype of the CRKP isolates was consistent with what has recently been reported in the United states, indicating the hetero-resistance of CRKP isolates to the drugs contributes to bacteriostatic effect and regrowth (Band et al., 2018, 2019). Therefore, combination therapy is needed not only to fight the multiple-drug resistant CRKP isolates but also to prevent the emergence of the resistance during the treatment.
Antibacterial action generally falls into four mechanisms, three of which involve the inhibition or regulation of enzymes involved in cell wall biosynthesis, nucleic acid synthesis and DNA repair, or protein synthesis, respectively (Kapoor et al., 2017; Reygaert, 2018). The fourth mechanism involves the disruption of membrane structure (Farkas et al., 2017; Rempe et al., 2017). In other hand, the bacteria have become smarter and along with the antibiotics, massive imprudent usage of antibiotics in clinical practice has caused resistance of bacteria to antimicrobial agents. The biochemical resistance mechanisms used by bacteria include antibiotic inactivation, target modification, altered permeability, and “bypass” of metabolic pathway. Colistin is a cationic antimicrobial peptide that targets bacterial lipopolysaccharide (LPS), causing cell membrane leakage (Hancock, 1997; Kwa et al., 2007; Yu et al., 2015). Previous studies have shown that the increased modification of LPS by the pmrHFIJKLM operon, and the two-component systems PhoPQ and PmrAB with connector PmrD changed the negative charge and reduced susceptibility to colistin in Enterobacteriaceae, therefore contributing to colistin resistance (Gunn et al., 2000; McPhee et al., 2006; Yan et al., 2007). Recent studies also demonstrated that the insertion, amino acid changes, or deletion of mgrB relieved the inhibition of PhoQ phosphorylation, leading to increased expression of pmrHFIJKLM transcript and reduced susceptibility to colistin in K. pneumonieae (Cannatelli et al., 2014; Olaitan et al., 2014; Cheng et al., 2015). In addition, the plasmid-mediated COL resistance gene, MCR-1, was discovered in 2015 (Liu et al., 2016).
Meropenem inhibits bacterial cell wall synthesis and is highly resistant to degradation by β-lactamases or cephalosporinases while the primary mechanism of amikacin action is to bind to bacterial 30S ribosomal subunits and interferes with mRNA binding and tRNA acceptor sites, interfering with bacterial growth by disrupting normal protein synthesis and producing non-functional or toxic peptides. Because the reduction in colistin permeability is the major mechanism of KP resistance to colistin, we hypothesized that COL combination with MEM or AMK would sensitize each other to inhibit biofilm formation and increase the membrane permeability by targeting multiple proteins via distinct mechanisms, therefore exerting synergistic effects on CRKP isolates (Kim et al., 2015; Su et al., 2019). As expected, our studies showed that COL-AMK had obvious synergistic and additive effects, and antibiotic's MICs in the combination were significantly lower than those of monotherapies. Though the COL combinations with AMK and MEM had the same antibacterial activity against the CRKP isolates of both COL-S and COL-R, the synergistic effect of COL-AMK was superior to the additive effect while the synergistic and additive effects of COL-MEM on the strains were same. Our data indicated that treatment of CRKP isolates with COL combination with aminoglycoside class antimicrobials resulted in a better synergistic effect than did the COL-carbapenems combination. In addition, we demonstrated that treatments of NDM-producing CRKP isolates with COL combinations lead to a better efficacy of antibacterial activity than KPC-producing CRKP strains.
In this study, we applied both the checkerboard techniques and the time-kill assays for the evaluation of the efficacies of the antibiotics in vitro and obtained a great agreement from these independent experiments. The COL combinations had synergistic or additive effects on all CRKP isolates in the checkerboard technique except one on which a higher AMK concentration was needed to obtain the same effect. The COL combinations were significantly synergistic against all distinct ST CRKP isolates except the ST76 in the time-kill assay. The difference in the outcomes of experiments was because the mean value for steady-state concentrations of non-protein-bound drugs in humans did not meet the single-drug MIC in vitro. It appears the results of the time-kill assay were more reliable and safer. In our study, we observed regrowth of some strains in the presence of antibiotic combinations. It is possible that the resistant subpopulations were selected through antimicrobial pressure in the time-kill assay (Gupta et al., 2014). Alternatively, the CRKP strains might adapt environmental stimuli by altering the outer membrane and became resistant to the combination after the prolonged treatment in vitro (Meletis et al., 2011).
Conclusion
In conclusion, we demonstrated that AMK was a better antibacterial agent against all CRKP isolates than MEM and COL in vitro. Treatments of 6 CRKP isolates at 16 μg/ml MEM or AMK in addition to 2 μg/ml COL completely killed 4 of them and suppressed the growth of 2 others (ST76 and ST11) while monotherapy showed no bactericidal effects. Our data suggest that COL could be a valid therapeutic option against COL-resistant CRKP isolates when used in combination with MEM or AMK. There are a few limitations in this study. First, while our in vitro experiments showed antimicrobial activity of two antibiotic combinations, and the experiments yielded statistical significances in the checkerboard technique and time-kill assays, the findings from this study need to be validated in vivo and in clinical trials. The isolates tested in the time kill assay should be further studies with the E-test for Colistin, Amikacin and Meropenem to confirm if the isolates exhibit phenotypic signs of hetero-resistance. Second, we only tested 6 CRKP isolates from a Healthcare Center with combination treatments. More tests are needed to determine if the combination treatment of two antibiotics is effective to other 34 CRKP isolates that have not been tested in this study as well as the CRKP isolates from other medical hospitals. Third, a pharmacokinetic evaluation of the antibiotics is also required to optimize the antibiotic combination regimens.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
XZ conceived and designed the study. LY and JSZ wrote this paper and contributed equally to this work. LY, JSZ, YF, and YZ performed the experiments. YW, JZ, YG, and CL analyzed the data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by Excellent Team of Young Teachers Foundation of Heilongjiang Province (2018-KYYWF-0916), and Heilongjiang Provincial Health and Family Planning Commission on Scientific Research Project (2018-290 and 2018-291) and Jiamusi University President Innovation and Entrepreneurship Fund Project (XZYF2018-43).
References
- Bai Y., Liu B., Wang T., Cai Y., Liang B., Wang R., et al. (2015). In vitro activities of combinations of rifampin with other antimicrobials against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 59, 1466–1471. 10.1128/AAC.04089-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band V. I., Hufnagel D. A., Jaggavarapu S., Sherman E. X., Wozniak J. E., Satola S. W., et al. (2019). Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat. Microbiol. 4, 1627–1635. 10.1038/s41564-019-0480-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band V. I., Satola S. W., Burd E. M., Farley M. M., Jacob J. T., Weiss D. S. (2018). Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. MBio 9:e02448-17. 10.1128/mBio.02448-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A., Giani T. M. M, D'Andrea V., Di Pilato Arena F., Conte V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. 10.1128/AAC.03110-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. H., Lin T. L., Pan Y. J., Wang Y. P., Lin Y. T., Wang J. T. (2015). Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob. Agents Chemother. 59, 2909–2913. 10.1128/AAC.04763-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (1999). Methods for Determining Bactericidal Activity of Antimicrobial Agents. Document M26-A. Wayne, PA: CLSI; IOP Publishing. [Google Scholar]
- Clinical and Laboratory Standards Institute (2016). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Sixth Informational Supplement. Document M100-S26. Wayne, PA: CLSI; IOP Publishing. [Google Scholar]
- Doern C. D. (2014). When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J. Clin. Microbiol. 52, 4124–4128. 10.1128/JCM.01121-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2016). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 6.0. Bristol, UK: IOP Publishing. [Google Scholar]
- Farkas A., Maroti G., Kereszt A., Kondorosi E. (2017). Comparative analysis of the bacterial membrane disruption effect of two natural plant antimicrobial peptides. Front. Microbiol. 8:51. 10.3389/fmicb.2017.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. A., Bauer S. R., Neuner E. A., Bass S. N., Lam S. W. (2016). Influence of colistin dose on global cure in patients with bacteremia due to carbapenem-resistant gram-negative Bacilli. Antimicrob. Agents Chemother. 60, 431–436. 10.1128/AAC.01414-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X., Zhang J., Su S., Fu Y., Bao M., Wang Y., et al. (2018). Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the Eastern region of Heilongjiang Province, China. BMC Infect. Dis. 18:417. 10.1186/s12879-018-3294-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H., Glasner C., Albiger B., Aanensen D. M., Tomlinson C. T., Andrasevic A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163. 10.1016/S1473-3099(16)30257-2 [DOI] [PubMed] [Google Scholar]
- Gunn J. S., Ryan S. S., Van Velkinburgh J. C., Ernst R. K., Miller S. I. (2000). Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect. Immun. 68, 6139–6146. 10.1128/IAI.68.11.6139-6146.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K., Padmanabhan B. R., Diene S. M. R, Lopez-Rojas Kempf M., Landraud L., et al. (2014). ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220. 10.1128/AAC.01310-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E. (1997). Peptide antibiotics. Lancet 349, 418–422. 10.1016/S0140-6736(97)80051-7 [DOI] [PubMed] [Google Scholar]
- Huang W., Qiao F., Zhang Y., Huang J., Deng Y., Li J., et al. (2018). In-hospital medical costs of infections caused by carbapenem-resistant Klebsiella pneumoniae. Clin. Infect. Dis. 67, S225–S230. 10.1093/cid/ciy642 [DOI] [PubMed] [Google Scholar]
- Kapoor G., Saigal S., Elongavan A. (2017). Action and resistance mechanisms of antibiotics: a guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 33, 300–305. 10.4103/joacp.JOACP_349_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Kim T., Pan J. G. (2015). In vitro evaluation of ciclopirox as an adjuvant for polymyxin B against gram-negative bacteria. J. Antibiot. 68, 395–398. 10.1038/ja.2014.164 [DOI] [PubMed] [Google Scholar]
- Kulengowski B., Rutter W. C., Campion J. J., Lee G. C., Feola D. J., Burgess D. S. (2018). Effect of increasing meropenem MIC on the killing activity of meropenem in combination with amikacin or polymyxin B against MBL- and KPC-producing Enterobacter cloacae. Diagn. Microbiol. Infect. Dis. 92, 262–266. 10.1016/j.diagmicrobio.2018.06.013 [DOI] [PubMed] [Google Scholar]
- Kwa A., Kasiakou S. K., Tam V. H., Falagas M. E. (2007). Polymyxin B: similarities to and differences from colistin (polymyxin E). Expert. Rev. AntiInfect. Ther. 5, 811–821. 10.1586/14787210.5.5.811 [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- McPhee J. B., Bains M., Winsor G., Lewenza S., Kwasnicka A., Brazas M. D., et al. (2006). Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188, 3995–4006. 10.1128/JB.00053-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis G., Tzampaz E., Sianou E., Tzavaras I., Sofianou D. (2011). Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 66, 946–947. 10.1093/jac/dkr007 [DOI] [PubMed] [Google Scholar]
- Morici P., Florio W., Rizzato C., Ghelardi E., Tavanti A., Rossolini G. M., et al. (2017). Synergistic activity of synthetic N-terminal peptide of human lactoferrin in combination with various antibiotics against carbapenem-resistant Klebsiella pneumoniae strains. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1739–1748. 10.1007/s10096-017-2987-7 [DOI] [PubMed] [Google Scholar]
- Olaitan A. O., Diene S. M., Kempf M., Berrazeg M., Bakour S., Gupta S. K., et al. (2014). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. 10.1016/j.ijantimicag.2014.07.020 [DOI] [PubMed] [Google Scholar]
- Patel G., Huprikar S., Factor S. H., Jenkins S. G., Calfee D. P. (2008). Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect. Control Hosp. Epidemiol. 29, 1099–1106. 10.1086/592412 [DOI] [PubMed] [Google Scholar]
- Petersen P. J., Labthavikul P., Jones C. H., Bradford P. A. (2006). In vitro antibacterial activities of tigecycline in combination with other antimicrobial agents determined by chequerboard and time-kill kinetic analysis. J. Antimicrob. Chemother. 57, 573–576. 10.1093/jac/dki477 [DOI] [PubMed] [Google Scholar]
- Rempe C. S., Burris K. P., Lenaghan S. C., Stewart C. N., Jr. (2017). The potential of systems biology to discover antibacterial mechanisms of plant phenolics. Front. Microbiol. 8:422. 10.3389/fmicb.2017.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reygaert W. C. (2018). An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 4, 482–501. 10.3934/microbiol.2018.3.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos W. M. D., Secoli S. R. (2019). Economic burden of inpatients infected with Klebsiella pneumoniae carbapenemase. Einstein 17:eGS4444. 10.31744/einstein_journal/2019GS4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Li D., Guo Q., Guo Y., Zheng Y., Xu X. (2019). In vitro bactericidal activity of trimethoprim-sulfamethoxazole/colistin combination against carbapenem-resistant Klebsiella pneumoniae clinical isolates. Microb. Drug Resist. 25, 152–156. 10.1089/mdr.2018.0085 [DOI] [PubMed] [Google Scholar]
- Tangden T., Hickman R. A., Forsberg P., Lagerback P., Giske C. G., Cars O. (2014). Evaluation of double- and triple-antibiotic combinations for VIM- and NDM-producing Klebsiella pneumoniae by in vitro time-kill experiments. Antimicrob. Agents Chemother. 58, 1757–1762. 10.1128/AAC.00741-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A., Guan Z., Raetz C. R. (2007). An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J. Biol. Chem. 282, 36077–36089. 10.1074/jbc.M706172200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Qin W., Lin J., Fang S., Qiu J. (2015). Antibacterial mechanisms of polymyxin and bacterial resistance. Biomed. Res. Int. 2015:679109. 10.1155/2015/679109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Chan E. W., Zhou H., Chen S. (2017). Prevalence and genetic characteristics of carbapenem-resistant Enterobacteriaceae strains in China. Lancet Infect. Dis. 17, 256–257. 10.1016/S1473-3099(17)30072-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.