Abstract
Cancer stem cells (CSCs) are a subpopulation with the properties of extensive self‐renewal, capability to generate differentiated cancer cells and resistance to therapies. We have previously shown that malignant pleural effusions (MPEs) from patients with non‐small‐cell lung cancer (NSCLC) represent a valuable source of cancer cells that can be grown as three‐dimensional (3D) spheroids enriched for stem‐like features, which depend on the activation of the Yes‐associated protein‐transcriptional coactivator with PDZ‐binding motif (YAP‐TAZ)/Wnt‐βcatenin/stearoyl‐CoA desaturase 1 (SCD1) axis. Here, we describe a novel support, called CytoMatrix, for the characterization of limited amounts of cancer cells isolated from MPEs of patients with NSCLC. Our results show that this synthetic matrix allows an easy and fast characterization of several epithelial cellular markers. The use of CytoMatrix to study CSCs subpopulation confirms that SCD1 protein expression is enhanced in 3D spheroids when compared with 2D adherent cell cultures. YAP/TAZ nuclear‐cytoplasmic distribution analysed by CytoMatrix in 3D spheroids is highly heterogeneous and faithfully reproduces what is observed in tumour biopsies. Our results confirm and extend the robustness of our workflow for the isolation and phenotypic characterization of primary cancer cells derived from the lung MPEs and underscore the role of SCD1.
Keywords: cancer stem cells, CytoMatrix, lung cancer, MPEs, SCD1
Malignant pleural effusions (MPEs) are a good source of lung cancer cells and represent an excellent model to investigate tumour heterogeneity. MPE‐derived tumour cells can be grown as three‐dimensional (3D) spheroids enriched for stem‐like properties. CytoMatrix is a reliable support for the characterization of primary 2D and 3D MPE‐derived cell cultures using minimum amount of biological material.
Abbreviations
- ALDH
aldehyde dehydrogenases
- ALK
anaplastic lymphoma kinase
- CK7
cytokeratin 7
- CSC
cancer stem cell
- CTLA4
cytotoxic T‐lymphocyte antigen‐4
- EGFR
epidermal growth factor receptor
- ICC
immunocytochemistry
- ICI
immune checkpoint inhibitor
- IF
immunofluorescence
- IHC
immunohistochemistry
- MPE
malignant pleural effusion
- NSCLC
non‐small‐cell lung cancer
- PAN‐CK
Pan‐cytokeratin
- PDL1
programmed death‐ligand 1
- SCD1
stearoyl‐CoA desaturase 1
- TAZ
transcriptional coactivator with PDZ‐binding motif
- TTF‐1
thyroid transcription factor‐1
- YAP
Yes‐associated protein
1. INTRODUCTION
Lung cancer is the most common cause of cancer deaths. Approximately 85% of those cases are classified as non‐small‐cell lung cancer (NSCLC), of which adenocarcinoma is the most common histological subtype. Despite advances in anticancer therapies, the 5‐year survival rate (17.8%) for NSCLC is much lower than that of other leading cancers (Ettinger et al., 2017; Ferlay et al., 2015; Siegel, Miller, & Jemal, 2018).
For decades, the gold standard therapy for NSCLC patients in the first‐line setting has been platinum‐based doublet chemotherapy with targeted agents limited to a subset of patients with specific genetic alterations such as epidermal growth factor receptor mutations or anaplastic lymphoma kinase (ALK) and reactive oxygen species (ROS) translocations, and concomitant development of strategies for undruggable targets such as KRAS mutations (Acunzo et al., 2017; Reck et al., 2014). The recent introduction of immunotherapy with immune checkpoint inhibitors (ICI) directed against targets such as cytotoxic T‐lymphocyte antigen‐4 (CTLA‐4) and PD‐1/programmed death‐ligand 1 (PDL1) is rapidly changing standard treatments for NSCLC patients as well as for several other advanced malignancies (Callahan, Postow, & Wolchok, 2016; Herbst et al., 2016; Reck et al., 2016). However, it is important to point out that the use of both cisplatin and ICI therapy results in an unfavourable outcome for a substantial proportion of patients due to drug resistance (Goss & Tsvetkova, 2012; Ilie et al., 2017).
A growing body of evidence suggests that treatment failures in patients with NSCLC can be attributed to the presence of a subpopulation of cancer cells with distinctive features, defined as cancer stem cells (CSCs) (Lopez‐Ayllon et al., 2014; Zhang et al., 2016). These cells are characterized by the ability to self‐renew (Plaks, Kong, & Werb, 2015; Reya, Morrison, Clarke, & Weissman, 2001; Shackleton, Quintana, Fearon, & Morrison, 2009). CSCs, for their intrinsic stem‐like properties, seem to be responsible for the development of resistance to anticancer treatments such as conventional chemotherapy, radiation, and targeted therapies (Abdullah & Chow, 2013; Mihanfar et al., 2018; Pisanu et al., 2018; Yakisich, Azad, Kaushik, & Iyer, 2017). In contrast, growing evidence suggest that CSCs are by themselves weakly immunogenic and are not efficiently recognized and eliminated by the immune system. In addition, CSCs have been postulated to foster the creation of an immune‐evading microenvironment, which is emerging as a critical element in limiting the efficacy of ICI therapy (Codd, Kanaseki, Torigo, & Tabi, 2018; Sultan et al., 2017).
One of the main difficulties consists in the isolation of CSCs, given their poor abundance in the tumour tissue, their slow replication kinetics, their plasticity and phenotypic heterogeneity (Grimshaw et al., 2008; Ricci‐Vitiani et al., 2006). Even though several methods for isolating, propagating and characterizing CSCs have been developed (Collins, Berry, Hyde, Stower, & Maitland, 2005; Eramo et al., 2007; Grimshaw et al., 2008; Hemmati et al., 2003; Ricci‐Vitiani et al., 2006), there is still the need to develop robust and reproducible protocols. In recent years, we and others have reported that malignant pleural effusions (MPEs) could represent an excellent model to investigate tumour heterogeneity and to isolate putative CSCs, based on the rationale that it is an easily accessible form of metastatic malignancy (Basak et al., 2009; Giarnieri et al., 2015, 2013; Mancini et al., 2011; Roscilli et al., 2016; Tiran et al., 2017). MPEs, defined as pleural fluid containing malignant cells, are a complication caused by primary malignant pleural mesothelioma or by metastatic cancers originating, most commonly, from the lung and breast. MPE occurrence is observed in about 30% of lung cancer cases and is associated with poor prognosis (Penz, Watt, Hergott, Rahman, & Psallidas, 2017). Among several techniques proposed to isolate and enrich CSCs, many evidence support the concept that cell cultures in nonadherent conditions and in a serum‐free medium supplemented with growth factors promote the formation of spheroids with stem‐like properties (Eramo et al., 2007; Grimshaw et al., 2008; Mancini et al., 2011).
We previously reported a protocol to establish MPE‐derived tumour cell cultures from NSCLC patients. These cells can be grown in nonadherent conditions as three‐dimensional (3D) spheroids enriched for stem‐like properties, including upregulation of aldehyde dehydrogenase (ALDH) activity, nanog, oct4 and sox2 markers, and give rise efficiently to tumours, which reproduce the same histological features of the original human tumour, when implanted in recipient mice (Mancini et al., 2011). Lung cancer 3D spheroids overexpress genes involved in lipid metabolism. Among them we identified stearoyl‐CoA desaturase 1 (SCD1), the main biosynthetic enzyme responsible for the conversion of saturated fatty acids into monounsaturated fatty acids, as a key factor for lung CSCs and showed that its inhibition reverts resistance to cisplatin treatment acting synergistically with chemotherapy (Noto et al., 2013; Pisanu et al., 2017). At a mechanistic level, SCD1 inhibition inactivates β‐catenin and Yes‐associated protein/transcriptional coactivator with PDZ‐binding motif (YAP/TAZ) signalling pathways involved in the maintenance of the CSCs pool (Noto et al., 2017). We also used primary MPE‐derived tumour cultures as a tool for screening sensitivity to chemotherapeutic agents and for genetic profiling, to select the most effective therapy regimen (Roscilli et al., 2016).
Over the last 5 years we have been able to collect and analyze in our laboratory more than 300 MPE‐derived NSCLC samples. Of these, only ~15% are able to give rise to slowly proliferating cultures that can be propagated for at least four to five passages. Primary cultures may be difficult not only to establish, due to poor quality or abundance of biological material of origin, but also to maintain for a sufficient number of passages to perform complex analyses (Janik et al., 2016). Furthermore, MPEs contain a heterogeneous repertoire of cell types of which cancer cells represent only a small fraction because they secrete cytokines and chemokines that promote the recruitment of immunoregulatory cells, such as tumour‐associated macrophages, which are the major component of MPEs and are involved in cancer progression (Li et al., 2016). For all these reasons, it is important to subject MPE‐derived cell cultures to a thorough phenotypic characterization through the evaluation of a variety of markers. To avoid selection of subpopulation of cells with particular growth advantages and which may not faithfully represent the composition of the originating tumour, it is necessary to proceed to phenotypic characterization of isolated cultures when the number of available cells is often very low (Janik et al., 2016). Hence, there is a need to establish methods for the rapid characterization of cancer‐initiating cells soon after their isolation. In this paper, we describe the use of an innovative support, CytoMatrix, for the characterization of limited amounts of cancer cells isolated from MPEs of patients with NSCLC.
2. MATERIALS AND METHODS
2.1. MPE sample processing and cell cultures
Primary cell cultures were obtained from MPEs of patients (n = 3) with lung adenocarcinoma enroled by Istituti Fisioterapici Ospitalieri under an Institutional Research Ethics Committee approved protocol (Number 1032/17). All patients agreed to participate in the study and signed an informed consent form; additional information regarding subjects’ clinical history is reported in Table 1. Tumour cells were isolated from MPEs as previously described (Mancini et al., 2011). Briefly, pleural fluids (up to 1,000 ml) were obtained by thoracentesis and collected aseptically in nonheparinized bottles/tubes. After centrifugation at 1,200 rpm for 5 min at 4°C, the supernatant is decanted and the cell pellet resuspended in phosphate‐buffered saline (PBS). The procedure is repeated twice and then the cell suspension layered on Oncoquick gradient (Greiner Bio‐One) to partially purify tumour cells from the other MPE components. The cell suspension on the Oncoquick discontinuous gradient is then centrifuged at 800 g for 20 min at 4°C without brake. During the centrifugation step, the cells are separated according to their different buoyant densities: the denser fluid components such as erythrocytes and leucocytes migrate into the lower phase through the bottom of the tube, whereas the less‐dense cell fraction, including tumour cells, is enriched at the interphase layer. After the harvesting and washing step, tumour cells are cultured in RPMI‐1640 (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS; Sigma) to obtain 2D primary adherent cultures. The 3D spheroid cultures were obtained from adherent culture plating single cells at a clonal density in ultra‐low attachment flasks (Corning, NY) and in serum‐free Dulbecco's modified Eagle medium/F12 supplemented with basic fibroblast growth factor (20 ng/ml), epidermal growth factor (20 ng/ml), insulin (20 µg/ml), 0.5% glucose, heparin (5 µg/ml; Sigma, St. Louis, MO) and B27 (Gibco, Invitrogen, Carlsbad, CA).
Table 1.
Clinicopathological characteristics of three MPE‐cases included in the study
| Patient ID | Sex/age | Surgery location | Diagnosis | Grading | Diagnostic markers | NGS mutational analysis—biopsy | NGS mutational analysis—primary cell line | Therapy |
|---|---|---|---|---|---|---|---|---|
| BBIRE‐T238 | F/60 | Pleural biopsy | Adenocarcinoma | 3 | TTF‐1+, PDL1+, ALK−, ROS1− | EGFR (p.M766_A767 ins ASV (Level III/II) c.23_2309insCCAGCGTGG, EXON20): VAF 22% | EGFR (p.M766_A767 ins ASV (Level III/II) c.23_2309insCCAGCGTGG, EXON20): VAF 43% | CDDP/PEM |
| BBIRE‐T248 | M/74 | Pleural biopsy | Adenocarcinoma | 3 | TTF‐1+, PDL1+, ALK−, ROS1− | KRAS (p. G12V (Level III/II), c.35G>T, EXON2): VAF 36% | KRAS (p. G12V (Level III/II), c.35G>T, EXON2): VAF 86% | PEM |
| BBIRE‐T570 | F/46 | Pleural biopsy | Adenocarcinoma | 3 | TTF‐1+, PDL1+, ALK+, ROS1− | ALK‐rearrangment: Fusion EML4‐ALK.E17 ins30a20_V8a, gain of function | ALK‐rearrangment: Fusion EML4‐ALK.E17 ins30a20_V8a, gain of function | N/A |
| MYC‐amplification: CNV 20.4 | MYC‐amplification: CNV 26.44 |
Note: Patient and tumour features. Specific mutation sites are also reported from NGS mutational analysis on both biopsy and primary cell line.
Abbreviations: ALK, anaplastic lymphoma kinase; CDDP, cisplatin; CNV, copy number variants; EGFR, epidermal growth factor receptor; NGS, next‐generation sequencing; PDL1, programmed death‐ligand 1; PEM, pembrolizumab; ROS, reactive oxygen species; TTF‐1, thyroid transcription factor‐1; VAF, variant allele frequency.
Established human NSCLC cell line, NCI‐H460, was obtained from American Type Culture Collection (ATCC, Manassas, VA); 2D and 3D cultures were maintained as described for primary cells.
All cells were tested for mycoplasma contamination and subjected to short tandem repeat analysis by ATCC Cell Line Authentication Service.
2.2. Immunohistochemistry (IHC) and immunocytochemistry (ICC)
IHC on formalin‐fixed paraffin‐embedded (FFPE) tissue sections obtained from pleural biopsy of human lung adenocarcinomas and ICC on FFPE cellular sections obtained from NCI‐H460 cell line and MPE primary cell lines entrapped in CytoMatrix were performed on an automated immunostainer (Bond‐III; Leica, Biosystems, Italy). Briefly, paraffin sections were cut at 5 µm using a microtome LEICA SM 2000R (Advanced Research Systems, Inc., Macungie, PA). The slides were dewaxed in xylene and rehydrated through a series of graded ethanol solutions and stained with Gill's haematolylin (Bio‐Optica, Milan) and eosin (Bio‐Optica). A citrate buffer, pH 6 or pH 8 (for SCD1 and YAP/TAZ staining), was used to unmask the antigens. The primary antibodies used are listed in Table S1.
Images were obtained by using a light microscope (Nikon ECLIPSE 55i) equipped with a Digital Image Capture software (Leica Application Suite V4.8).
2.3. Immunofluorescence (IF) analysis and optical microscopy
IF was performed on FFPE NCI‐H460 cells entrapped in CytoMatrix. After solvent dewaxing with Clearify Agent (Dako; Agilent, Santa Clara) and antigen retrieval with a citrate buffer pH 8 in an automated immunostainer (Omnis, Dako; Agilent, Santa Clara, CA) IF was performed as previously described (Pisanu et al., 2018). Briefly, cells were fixed with 10% neutral buffered formalin, incubated 5 min in glycine 0.1 M and permeabilized in 0.2% Triton‐X (Sigma‐Aldrich, Milan, Italy). After washing two times with PBS and blocking with 3% FBS in PBS, the cells were stained with anti‐YAP/TAZ (Santa Cruz Biotechnologies, Dallas, TX) antibody (1:50 dilution) and incubated at 4°C overnight. Next day, cells were washed by PBS three times to remove unbound antibodies, then secondary antibody (1:200 dilution) was added in the dark and incubated at room temperature for 1 hr. Then, cells were stained with Hoechest 33342 (1:1,000 dilution) for 5 min in the dark. IF images and morphology observations of cell lines were performed on Axiocam Camera (Zeiss), a digital camera, coupled with Zeiss Axiovert optical microscope and analysed using ZEN core software (Zeiss).
2.4. Quantitative real‐time PCR analysis
For quantitative real‐time PCR (qRT‐PCR), total RNA was isolated by TRIzol Reagent (Life Technologies, Gaithersburg, MD) according to the manufacturer's guidelines. RNA was reverse transcribed into cDNA using High Capacity RNA‐to cDNA Kit (Applied Biosystems; Life Technologies). The qRT‐PCR was performed using SYBR green detection (Applied Biosystem; Life Technologies, Gaithersburg, MD) and the ∆∆C t method for relative quantification. The expression of Histone H3 or β‐actin were used as internal control, as specified. The primers used for individual genes are listed in Table S2.
2.5. Sequencing technologies
FFPE biopsy sections and primary cultures of tumour cells from MPEs of patient samples were processed by the Ion AmpliSeq Colon and Lung Cancer Panel (Life Technologies) and The Oncomine Focus Assay (Life Technologies).
For the Ion AmpliSeq Colon and Lung Cancer Panel, DNA was extracted using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions. For library preparation, 10 ng of DNA for each sample, obtained by using the Ion AmpliSeq Library 96LV Kit 2.0 (Life Technologies) and the Colon and Lung Cancer Panel (Life Technologies), were adopted. The panel provides 93 amplicons covering 504 mutational hotspot regions in 22 genes. The Ion Torrent Oncomine Focus Assay is a multi‐biomarker next‐generation sequencing (NGS) assay that permits immediate analysis of DNA and RNA, to simultaneously detect multiple types of variants in 52 genes. In detail, it enables simultaneous detection of 35 targeted hotspot mutations, 23 fusion genes, and 19 copy number variations (CNVs). A minimum of 10 ng of DNA and RNA were amplified using the NGS‐targeted panel Oncomine Focus Assay. On the Chef system (Life Technologies) was performed the template preparation by emulsion PCR (emPCR). In agreement with the manufacturer's instructions, library quality control was performed using the Ion Sphere Quality Control Kit, to obtain that 10–30% of template‐positive Ion Sphere particles (ISPs) were targeted in the emPCR reaction. At the end, sequencing primer and polymerase were added to the enriched ISPs and loaded onto Ion 520 (100 Mb output) chip. Each library was barcoded with the Ion Xpress Barcode Adapters 1–16 Kit (Life Technologies). Barcoded libraries were combined to a final concentration of 100 pM. Sequencing was performed on the Ion S5 (Life Technologies) platform. Data analysis was carried out with Torrent Suite Software V.3.2 (Life Technologies). The detected variants were annotated and filtered with the Ion Reporter software and reviewed with the Integrative Genomics Viewer v2.1 (Broad Institute, Cambridge, MA). Nucleotide variations detected by the Ion AmpliSeq Colon and Lung Cancer Panel, and the Oncomine Focus Assay with less than a 5% variant frequency and covered by <1,000 reads were not considered. Fusion genes were considered positive when ≥20 fusion amplicons were found. CNVs were annotated when confidence interval at 5% was ≥4 gene copies with ≤0.5 median of the absolute value of all pairwise differences quality control measure.
2.6. Statistical analysis
All experiments were performed at least three times and the values were calculated as mean ± standard deviation or standard error of the mean. Differences between two groups were determined using Student's t test and p < .05 was considered statistically significant.
3. RESULTS
3.1. CytoMatrix: A support to entrap limited amounts of cytological material from MPEs
CytoMatrix is an innovative support, originally designed to permit an easy and rational management of the biological material from needle aspirates. A detailed study of different biocompatible materials characterized by ion affinity for cell samples allowed to discover that chitosan derivatives increase the effectiveness of the process of retaining cell suspensions dispensed on a synthetic porous matrix. This study was conducted through a collaboration between the University Campus Bio‐Medico of Rome and the industrial partner UCS Diagnostic S.r.l., and led to the development and joint patenting of CytoMatrix (Pub No. WO2018083616. International Application No: PCT/IB2017/056812). The powerful feature of this support is the ability to permit a rapid and reliable diagnosis even in the absence of large amounts of material. In line with this concept, we thought that CytoMatrix could be a useful tool for the cytological evaluation of tumour cells directly isolated from lung MPEs, to obtain a rapid and complete phenotypic characterization. CytoMatrix's peculiarity is to efficiently restrain small amounts of biological material inside its 3D structure. The porous support is provided into a plastic bio‐cassette allowing to make easy the following steps of classical ICC technique, such as formalin fixation, paraffin inclusion and microtome cut (Figure 1a).
Figure 1.
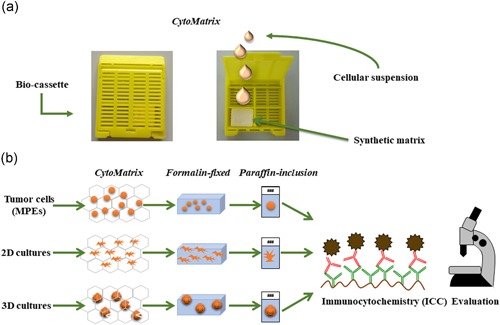
CytoMatrix: a support to entrap limited amounts of cytological material from MPEs. (a) Representative pictures of the plastic bio‐cassette containing CytoMatrix in the closed (left photo) and open (right photo) conformations. (b) The synthetic matrix restraining the biological material either derived from primary cancer cells directly isolated from MPEs and from established primary cell cultures grown in 2D or 3D conditions may be subjected to classical ICC steps: formalin fixation, paraffin inclusion, and microtome cut. 2D, two‐dimensional; MPE, malignant pleural effusions [Color figure can be viewed at wileyonlinelibrary.com]
In brief, a minimum of 100,000 2D adherent cells that roughly correspond to 250 3D spheroids are collected and centrifuged at 1,200 rpm for 5 min at 4°C. The supernatant is discarded and the cell pellet is resuspended in 1 ml of PBS and centrifuged at 1,200 rpm for 5 min at 4°C. After this washing step, the pellet is resuspended in a small volume of PBS (50–70 µl) and a single drop of cell suspension is laid on the matrix and trapped into its 3D structure. Then the bio‐cassette is closed and processed according to a classical ICC protocol. The sample is formalin‐fixed, blocked in paraffin and the cell block cut using a microtome into 5 μm sections, which are then mounted onto the slides. These slides are dewaxed in xylene and rehydrated through a series of graded ethanol solutions and stained with Gill's haematoxylin and eosin (H&E), to assess the presence and the morphology of the cells. Finally, the slides are subjected to unmasking process and incubation with the primary antibodies in an automated immunostainer (see Section 2). Starting from a limited number of cells it is possible to obtain from each single CytoMatrix several slides, to perform a panel of immunostainings for various markers, which result in an accurate characterization of cells isolated from the lung MPEs (Figure 1b).
In this context, it is important to underline the possibility to exploit the biological material trapped into CytoMatrix, also to perform molecular analyses by extraction of DNA and RNA or other investigations such as fluorescent in situ hybridization and IF, and how a single tool permits to take advantage of use of a minimal amount of precious biological material to perform a reliable diagnosis or research investigations also in the perspective of precision medicine.
3.2. Characterization of primary cells from MPEs using CytoMatrix
We first sought to set up the CytoMatrix‐based immunostaining protocol using a stable lung cancer cell line, namely NCI‐H460, obtained from ATCC. In brief, NCI‐H460 cells grown in 2D adherent conditions were harvested, resuspended in PBS and a drop of cellular suspension was trapped into CytoMatrix before formalin fixation and paraffin inclusion. Those samples were then evaluated for different markers by ICC. Our results, in line with ATCC data, confirmed that NCI‐H460 cells are characterized by a strong, positive immunostaining for Pan‐cytokeratin (PAN‐CK), a blend of cytokeratins (CKs), which are well‐known epithelial cell markers. Furthermore, those cells also presented positivity for vimentin, a mesenchymal marker that may be co‐expressed with CKs in a wide range of carcinomas. Of note, the pan leucocyte marker CD45 was used as a negative control (Figure 2a).
Figure 2.
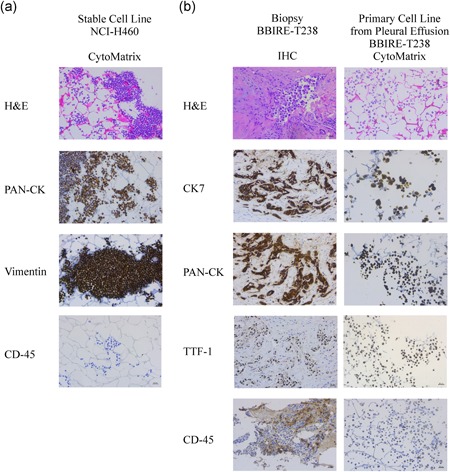
Characterization of primary cells from malignant pleural effusions (MPEs) using CytoMatrix. (a) CytoMatrix‐based ICC staining of haematoxylin and eosin, PAN‐CK, Vimentin, and CD45 on NCI‐H460 stable cell line. (b) Comparison between staining of different markers obtained from paraffin immunohistochemistry (IHC) and CytoMatrix‐based ICC samples of patient BBIRE‐T238. Original magnification = ×200. ICC, immunocytochemistry; PAN‐CK, Pan‐cytokeratin [Color figure can be viewed at wileyonlinelibrary.com]
Thereafter, we decided to use CytoMatrix for the ICC characterization of primary cultures derived from MPEs of patients with NSCLC. We describe here below the characterization of cell lines isolated from MPEs of three representative cases with adenocarcinoma of the lung. Detailed information regarding patients’ clinicopathological history is provided in Table 1. To confirm the epithelial origin of the isolated cells, we performed a panel of immunostaining routinely used for distinguishing lung adenocarcinoma cells, namely thyroid transcription factor‐1 (TTF‐1), PAN‐CK, and CK7 (Carney, Kraynie, & Roggli, 2015; Woo et al., 2017). Also, the pan leucocyte marker CD45 was evaluated to assess the purity of cell preparations. As shown in Figure 2b (and Figure S1), primary cells exhibited a marked positivity for cytoplasmic PAN‐CK and CK7, and a variable positivity for TTF‐1 depending on the cell line. As expected, CD45 marker showed no staining in all the three cell lines. Finally, we compared CytoMatrix results with those obtained with standard IHC performed on the solid biopsy from the same patients (Figures 2b and S1). We observed for all the three patients a complete correspondence of the results obtained with the two samples, thus confirming the sensitivity of the CytoMatrix approach and also the maintenance in our culture conditions of the phenotypic features of the original tumour. Of note, we have also observed a genotypic correspondence between pleural biopsy and low‐passage culture cells from the same patients, as showed by the analysis of mutational status reported in Table 1. It is important to point out that CytoMatrix also ensures the preservation of the cell morphology, tested by the routine H&E staining.
Overall, these results indicate that CytoMatrix constitutes a reliable support for the characterization with different cellular markers of primary cultures through ICC, taking advantages of using minimum amount of precious biological material compared with the classical approaches.
3.3. CytoMatrix for the study of MPE‐derived CSCs
Having documented the potential of CytoMatrix in the characterization of 2D adherent cell cultures established from MPEs of lung adenocarcinoma and given the evidence that MPEs represent an excellent model to isolate putative CSCs (Giarnieri et al., 2013; Mancini et al., 2011; Roscilli et al., 2016), we decided to evaluate whether CytoMatrix was a useful tool for the study of this subpopulation of cells. To test this hypothesis, we first set the conditions to enrich the cell population with stem‐like properties. When cultured in nonadherent conditions and in a serum‐free medium supplemented with growth factors, BBIRE‐T238 and BBIRE‐T248 MPE‐derived primary cell lines were able to give rise to 3D spheroids at variable efficiency. On average, 3D cultures were characterized by an enrichment of stem cell markers such as nanog, sox2 and oct4. The 3D spheroids from the stable lung cancer cell line, NCI‐H460, previously characterized (Noto et al., 2013; Pisanu et al., 2017), were used as control (Figures 3a,b and S2a,b).
Figure 3.
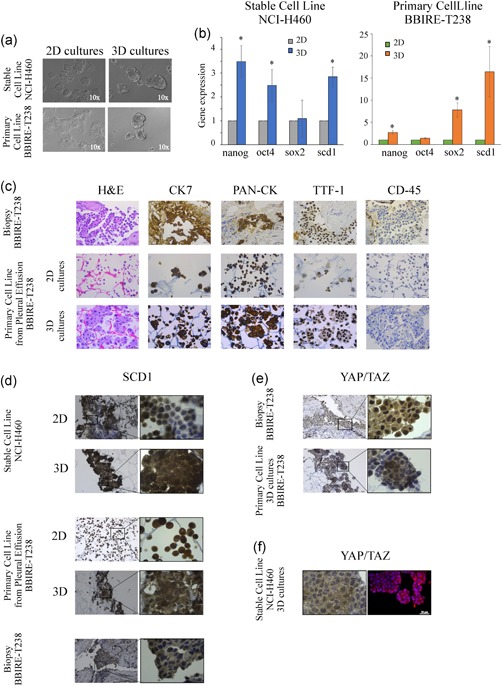
CytoMatrix for the study of MPE‐derived CSCs. (a) Representative images of a lung stable cell line NCI‐H460 and BBIRE‐T238‐derived primary cell line grown in adherent (2D) and in 3D conditions. The pictures were captured on Day 4. Original magnification = ×100. (b) Gene expression of stemness markers and scd1 performed by qRT‐PCR analyses on 3D and 2D cultures obtained from NCI‐H460 and the primary cell line isolated from MPEs of BBIRE‐T238 patient. The bar chart shows the fold change in the mRNA levels in 3D spheroids compared with 2D parental cells (histone H3 was used as housekeeping gene). Data represent the mean and standard deviation (SD) or standard error of mean (SEM) of at least three independent experiments and are statistically significant if *p < .05 (Student's t test). (c) Example of IHC staining of haematoxylin and eosin, CK7, PAN‐CK, CD45 and TTF‐1 performed on lung adenocarcinoma biopsy (upper panels), and of CytoMatrix‐based ICC staining of primary cell line derived from BBIRE‐T238 patient (middle and bottom panels). The results indicate a similar pattern of expression between biopsy and primary cell cultures. Original magnification = ×400. (d) SCD1 expression obtained from CytoMatrix‐based ICC staining of NCI‐H460 and BBIRE‐T238 primary cell line in 2D and 3D conditions, and from IHC staining of biopsy. Original magnification = ×200, detail ×1,000. (e) YAP/TAZ IHC (upper panel) and YAP/TAZ CytoMatrix‐based ICC (bottom panel) performed on samples obtained from BBIRE‐T238 patient. Original magnification = ×200, detail ×1,000. (f) Comparison between YAP/TAZ CytoMatrix‐based ICC (panel on the left) and YAP/TAZ CytoMatrix‐based IF (panel on the right) analyses obtained from NCI‐H460 grown in 3D conditions. Scale bar = 20 µm. 2D, two‐dimensional; CSC, cancer stem cells; ICC, immunocytochemistry;IF, immunofluorescence; IHC, immunohistochemistry; MPE, malignant pleural effusion; PAN‐CK, Pan‐cytokeratin; TAZ, transcriptional coactivator with PDZ‐binding motif; qRT‐PCR, quantitative reverse‐transcriptase polymerase chain reaction; YAP, Yes‐associated protein [Color figure can be viewed at wileyonlinelibrary.com]
The 3D primary spheroids were then subjected to the same phenotypic characterization performed on 2D adherent cells using CytoMatrix‐based immunostaining. First, H&E counterstains showed the power of this tool to preserve the 3D structure of the spheroids. Moreover, when compared with either 2D adherent culture and the solid biopsy from the same patient, the 3D spheroid culture showed the same pattern of expression for the markers evaluated (Figures 3c and S2c).
Recently, we and others have underlined the importance of lipid metabolism in controlling CSC maintenance (Igal, 2016; Li et al., 2017; Mancini et al., 2018). In line with previous studies where we demonstrated that SCD1 is a key factor for the propagation of lung CSCs (Mancini et al., 2011; Noto et al., 2013, 2017; Pisanu et al., 2017), we confirm here that 3D spheroids from both the NCI‐H460 NSCLC stable cell line and the BBIRE‐T238 primary culture show increased expression of SCD1 at mRNA level (Figure 3b). Furthermore, SCD1 expression was also assessed by performing CytoMatrix ICC analyses. As shown in Figure 3d, SCD1 staining shows increased expression of the protein in 3D spheroids compared with 2D adherent cell cultures. We also observed that SCD1 staining on MPE‐derived primary cultures is comparable with that obtained in the solid biopsy from the same patient, confirming that cancer cells isolated from MPEs maintain in our culture conditions the expression of SCD1 found in the primary tumour (Figures 3d and S2d). When we performed the same evaluation on 3D versus 2D cultures from BBIRE‐T248, we did not observe upregulation in the expression of SCD1 at the mRNA level (Figure S2b) as well as at the protein level (Figure S2d). This is probably due to patient heterogeneity as also stated by a very recent evidence that some cancer cell lines may activate an alternative fatty acid desaturation pathway to bypass the known pathway that is dependent on SCD1 (Vriens et al., 2019).
SCD1 expression was previously shown by our group to correlate with YAP/TAZ nuclear localization among different human lung adenocarcinoma samples (Noto et al., 2017). When we evaluated the expression of YAP/TAZ in BBIRE‐T238 and in BBIRE‐T248 patient biopsies, we observed that their subcellular localization is heterogeneous, and that only in some cells it is possible to detect a strong nuclear localization (Figure 3e, upper panels and Figure S2e, upper panels). As many evidence point to YAP/TAZ as key regulators of CSCs (Cordenonsi et al., 2011; Hao et al., 2014; Hayashi et al., 2015), we used CytoMatrix immunostaining for their evaluation in 3D primary spheroids. The staining reflects the result obtained from the patient's biopsies, with some cells that maintain cytoplasmic localization and some with a strong nuclear positivity, indicating that the 3D spheroid cell population is not homogeneous and reflects the heterogeneity found in the primary tumour (Figure 3e, lower panels and Figure S2e, lower panels). Finally, we evaluated the YAP/TAZ localization also in 3D spheroids from the NCI‐H460 cell line by setting up a CytoMatrix IF staining. Slides obtained after CytoMatrix processing were subjected to IF as described in Section 2. The results (Figure 3f) confirm also in this analysis a highly heterogeneous pattern of YAP/TAZ nuclear‐cytoplasmic localization. It has to be pointed out that Cytomatrix IF staining may suffer from unspecific binding of secondary antibodies to the matrix and, therefore, deserves further optimization (not shown).
Overall, these results underscore that CytoMatrix could be a useful tool not only for the phenotypic characterization of putative primary cancer cells isolated from lung MPEs but also for study of the key factors involved in the maintenance of stem‐like properties in 3D spheroids.
4. DISCUSSION
In this paper, we report the use of CytoMatrix, a porous support for trapping minimal amounts of cells, which allows cell fixation, slice cutting and analysis by ICC and IF. By presenting some examples of this procedure we show how CytoMatrix can easily allow the phenotypic characterization of small amounts of cancer cells derived from MPEs and assess that our culture conditions allow to faithfully preserve the features of the original tumour. Moreover, we have also observed by NGS analysis, identical mutational patterns between pleural biopsy and low‐passage culture cells from the same patients. CytoMatrix allows to generate from small amount of material, several slices, which can be subjected in theory to additional analyses such as electron microscopy, confocal microscopy, as well as DNA and RNA extraction for genomic studies.
Genomic analysis of CSC‐enriched 3D cultures from patients’ samples coupled with drug testing may allow to set up a workflow to predict drug sensitivity of patients to new drug combinations, and to facilitate the application of new concepts of personalized medicine (Amirouchene‐Angelozzi, Swanton, & Bardelli, 2017). In this respect, the technology described in this report has the advantage to allow an excellent characterization of the effects of drug testing on phenotypic changes of lung CSCs entrapped in CytoMatrix and biomarker analysis, and this can be in theory done in a rapid turnaround time of few days after CSCs isolation from MPEs.
Gaining importance is being attributed to the possibility to reproduce in vitro the complexity of the tumour microenvironment through the establishment of tumour organoids (or tumoroids) where tumour cells are embedded within a tumour stroma and interact with cancer‐associated fibroblasts and cells of the innate and acquired immune system (Lee et al., 2018; Nadkarni, Abed, & Draper, 2016). Tumoroids are believed to allow a better study of immune‐evasion mechanisms in cancer and to test in highly predictive systems the effect of immune‐enhancing therapeutic approaches. However, one of the major limitations of tumoroids is the limited amount of biological material and their slow propagation in the culture. In this regards, the possibility to analyse in depth the structure of tumoroids embedded into CytoMatrix in normal conditions and after drug testing will open up new promising perspectives.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Conception and design: G. C., S. Br., S. d. M., R. M., M. E. P., L. F. Development and patenting of CytoMatrix: M. T., A. Cr., A. S. Development of methodology: S. d. M., S. Br., V. L., S. Bu., E. T., C. D. V. Acquisition of data (acquired and managed patients, provided facilities, etc.): S. d. M., S. Br., V. L., S. Bu., P. V., G. A., F. F., C. N. Analysis and interpretation of data (e.g., statistical analysis, biostatistics): S. Br., M. E. P., L. F., S. d. M., G. C., R. M. Writing, review and/or revision of the manuscript: G. C., R. M., S. Br., L. F., M. E. P., S. d. M. Administrative, technical or material support: S. d. M., S. Br., V. L., E. T., S. Bu., C. D. V., A. Ce. Study supervision: G. C., R. M. All authors read and approved the final manuscript for publication.
DATA ACCESSIBILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGEMENTS
We thank the inventors of CytoMatrix and the researchers of Cyto+ project: Federica Cascone, Daniele Nicoletti, Chiara Taffon, Pamela Mozetic, Marco Costantini, Alberto Rainer, Sara Maria Giannitelli. “Porous material for the inclusion of cytologic preparations, process for obtaining the same and its use.” Pub No. WO2018083616. International Application No.: PCT/IB2017/056812. Publication Date: 2018 May 11 (https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2018083616&recNum=&maxRec=1000&office=&prevFilter=&sortOption=&queryString=&tab=PCTDescription). This study was supported by Italian Association for Cancer Research (AIRC; Grant Nos.: IG15216 and IG19865 to G. Ciliberto and IG17009 to R. Mancini); Lazio Innova (Grant No.: 2018 85–2017‐13750 to R. Mancini) and Fondo di Ricerca di Ateneo 2018 (Grant No.: B86C19001510005 to R. Mancini). This study was supported by the project “Incremento del TRL della tecnologia CytoMatrix‐Cyto+” funded in the framework of “INTESE: INnovazione e Trasferimento tEcnologico per Sostenere la fruizione dei risultati della ricerca sul territorio” (prot. FILAS‐RU‐2014‐1193, Lazio Region, Italy)”. All the information regarding CytoMatrix acquisition and applications are available on the online website: https://cytomatrix.it/.
Bruschini S, di Martino S, Pisanu ME, et al. CytoMatrix for a reliable and simple characterization of lung cancer stem cells from malignant pleural effusions. J Cell Physiol. 2019;235:1877–1887. 10.1002/jcp.29121
References
REFERENCES
- Abdullah, L. N. , & Chow, E. K. H. (2013). Mechanisms of chemoresistance in cancer stem cells. Clinical and Translational Medicine, 2(1), 3 10.1186/2001-1326-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acunzo, M. , Romano, G. , Nigita, G. , Veneziano, D. , Fattore, L. , Laganà, A. , … Croce, C. M. (2017). Selective targeting of point‐mutated KRAS through artificial microRNAs. Proceedings of the National Academy of Sciences of the United States of America, 114(21), E4203–E4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirouchene‐Angelozzi, N. , Swanton, C. , & Bardelli, A. (2017). Tumor evolution as a therapeutic target. Cancer Discovery, 7(8), 805–817. [DOI] [PubMed] [Google Scholar]
- Basak, S. K. , Veena, M. S. , Oh, S. , Huang, G. , Srivatsan, E. , Huang, M. , … Batra, R. K. (2009). The malignant pleural effusion as a model to investigate intratumoral heterogeneity in lung cancer. PLOS One, 4(6), e5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, M. K. , Postow, M. A. , & Wolchok, J. D. (2016). Targeting T cell co‐receptors for cancer therapy. Immunity, 44(5), 1069–1078. 10.1016/j.immuni.2016.04.023 [DOI] [PubMed] [Google Scholar]
- Carney, J. M. , Kraynie, A. M. , & Roggli, V. L. (2015). Immunostaining in lung cancer for the clinician: Commonly used markers for differentiating primary and metastatic pulmonary tumors. Annals ATS, 12(3), 429–435. 10.1513/AnnalsATS.201501-004FR [DOI] [PubMed] [Google Scholar]
- Codd, A. S. , Kanaseki, T. , Torigo, T. , & Tabi, Z. (2018). Cancer stem cells as targets for immunotherapy. Immunology, 153(3), 304–314. 10.1111/imm.12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, A. T. , Berry, P. A. , Hyde, C. , Stower, M. J. , & Maitland, N. J. (2005). Prospective identification of tumorigenic prostate cancer stem cells. Cancer Research, 65(23), 10946–10951. [DOI] [PubMed] [Google Scholar]
- Cordenonsi, M. , Zanconato, F. , Azzolin, L. , Forcato, M. , Rosato, A. , Frasson, C. , … Piccolo, S. (2011). The hippo transducer TAZ confers cancer stem cell‐related traits on breast cancer cells. Cell, 147(4), 759–772. 10.1016/j.cell.2011.09.048 [DOI] [PubMed] [Google Scholar]
- Eramo, A. , Lotti, F. , Sette, G. , Pilozzi, E. , Biffoni, M. , Di Virgilio, A. , … De Maria, R. (2007). Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death & Differentiation, 15, 504–514. [DOI] [PubMed] [Google Scholar]
- Ettinger, D. S. , Wood, D. E. , Aisner, D. L. , Akerley, W. , Bauman, J. , Chirieac, L. R. , … Hughes, M. (2017). Non small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. Journal of the National Comprehensive Cancer Network, 15(4), 504–535. 10.6004/jnccn.2017.0050 [DOI] [PubMed] [Google Scholar]
- Ferlay, J. , Soerjomataram, I. , Dikshit, R. , Eser, S. , Mathers, C. , Rebelo, M. , … Bray, F. (2015). Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer, 136(5), E359–E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Giarnieri, E. , Bellipanni, G. , Macaluso, M. , Mancini, R. , Holstein, A. C. , Milanese, C. , … Russo, G. (2015). Review: Cell dynamics in malignant pleural effusions. Journal of Cellular Physiology, 230(2), 272–277. 10.1002/jcp.24806 [DOI] [PubMed] [Google Scholar]
- Giarnieri, E. , De Vitis, C. , Noto, A. , Roscilli, G. , Salerno, G. , Mariotta, S. , … Mancini, R. (2013). EMT markers in lung adenocarcinoma pleural effusion spheroid cells. Journal of Cellular Physiology, 228(8), 1720–1726. 10.1002/jcp.24300 [DOI] [PubMed] [Google Scholar]
- Goss, G. D. , & Tsvetkova, E. (2012). Drug resistance and its significance for treatment decisions in non‐small‐cell lung cancer. Current Oncology, 19, S45–S51. 10.3747/Co.19.1113. Personalized Medicine in Metastatic NSCLC: A Canadian PerspectiveDO ‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimshaw, M. J. , Cooper, L. , Papazisis, K. , Coleman, J. A. , Bohnenkamp, H. R. , Chiapero‐Stanke, L. , … Burchell, J. M. (2008). Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Research, 10(3), R52 10.1186/bcr2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, J. , Zhang, Y. , Jing, D. , Li, Y. , Li, J. , & Zhao, Z. (2014). Role of hippo signaling in cancer stem cells. Journal of Cellular Physiology, 229(3), 266–270. 10.1002/jcp.24455 [DOI] [PubMed] [Google Scholar]
- Hayashi, H. , Higashi, T. , Yokoyama, N. , Kaida, T. , Sakamoto, K. , Fukushima, Y. , … Baba, H. (2015). An imbalance in TAZ and YAP expression in hepatocellular carcinoma confers cancer stem cell‐like behaviors contributing to disease progression. Cancer Research, 75(22), 4985–4997. 10.1158/0008-5472.CAN-15-0291 [DOI] [PubMed] [Google Scholar]
- Hemmati, H. D. , Nakano, I. , Lazareff, J. A. , Masterman‐Smith, M. , Geschwind, D. H. , Bronner‐Fraser, M. , & Kornblum, H. I. (2003). Cancerous stem cells can arise from pediatric brain tumors. Proceedings of the National Academy of Sciences, 100(25), 15178–15183. 10.1073/pnas.2036535100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst, R. S. , Baas, P. , Kim, D. W. , Felip, E. , Pérez‐Gracia, J.,L. , Han, J. Y. , … Garon, E. B. (2016). Pembrolizumab versus docetaxel for previously treated, PD‐L1‐positive, advanced non‐small‐cell lung cancer (KEYNOTE‐010): A randomised controlled trial. The Lancet, 387(10027), 1540–1550. 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- Igal, R. A. (2016). Stearoyl CoA desaturase‐1: New insights into a central regulator of cancer metabolism. Biochimica Et Biophysica Acta (BBA)—Molecular and Cell Biology of Lipids, 1861(12), 1865–1880. 10.1016/j.bbalip.2016.09.009 [DOI] [PubMed] [Google Scholar]
- Ilie, M. , Benzaquen, J. , Hofman, V. , Lassalle, S. , Yazbeck, N. , Leroy, S. , & C‐H, M. A. (2017). Immunotherapy in non‐small cell lung cancer: Biological principles and future opportunities. Current Molecular Medicine, 17(8), 527–540. 10.2174/1566524018666180222114038 [DOI] [PubMed] [Google Scholar]
- Janik, K. , Popeda, M. , Peciak, J. , Rosiak, K. , Smolarz, M. , Treda, C. , … Ksiazkiewicz, M. (2016). Efficient and simple approach to in vitro culture of primary epithelial cancer cells. Bioscience Reports, 36(6), 10.1042/BSR20160208. pii: e00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. H. , Hu, W. , Matulay, J. T. , Silva, M. V. , Owczarek, T. B. , Kim, K. , … Shen, M. M. (2018). Tumor evolution and drug response in patient‐derived organoid models of bladder cancer. Cell, 173(2), 515–528. 10.1016/j.cell.2018.03.017. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Condello, S. , Thomes‐Pepin, J. , Ma, X. , Xia, Y. , Hurley, T. D. , … Cheng, J. X. (2017). Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell, 20(3), 303–314. 10.1016/j.stem.2016.11.004. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Yang, L. , Wang, L. , Wang, F. , Zhang, Z. , Li, J. , … Zhang, Y. (2016). Impaired T cell function in malignant pleural effusion is caused by TGF‐β derived predominantly from macrophages: Impaired T cell function in malignant pleural effusion. International Journal of Cancer, 139(10), 2261–2269. 10.1002/ijc.30289 [DOI] [PubMed] [Google Scholar]
- Lopez‐Ayllon, B. D. , Moncho‐Amor, V. , Abarrategi, A. , de Cáceres, I. I. , Castro‐Carpeño, J. , Belda‐Iniesta, C. , … Sastre, L. (2014). Cancer stem cells and cisplatin‐resistant cells isolated from non‐small‐lung cancer cell lines constitute related cell populations. Cancer Medicine, 3(5), 1099–1111. 10.1002/cam4.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini, R. , Giarnieri, E. , De Vitis, C. , Malanga, D. , Roscilli, G. , Noto, A. , … Ciliberto, G. (2011). Spheres derived from lung adenocarcinoma pleural effusions: Molecular characterization and tumor engraftment. PLOS One, 6(7), 10.1371/journal.pone.0021320. e21320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini, R. , Noto, A. , Pisanu, M. E. , De Vitis, C. , Maugeri‐Saccà, M. , & Ciliberto, G. (2018). Metabolic features of cancer stem cells: The emerging role of lipid metabolism. Oncogene, 37(18), 2367–2378. 10.1038/s41388-018-0141-3 [DOI] [PubMed] [Google Scholar]
- Mihanfar, A. , Aghazadeh Attari, J. , Mohebbi, I. , Majidinia, M. , Kaviani, M. , Yousefi, M. , & Yousefi, B. (2018). Ovarian cancer stem cell: A potential therapeutic target for overcoming multidrug resistance. Journal of Cellular Physiology, 234(4), 3238–3253. 10.1002/jcp.26768 [DOI] [PubMed] [Google Scholar]
- Nadkarni, R. R. , Abed, S. , & Draper, J. S. (2016). Organoids as a model system for studying human lung development and disease. Biochemical and Biophysical Research Communications, 473(3), 675–682. 10.1016/j.bbrc.2015.12.091 [DOI] [PubMed] [Google Scholar]
- Noto, A. , Raffa, S. , De Vitis, C. , Roscilli, G. , Malpicci, D. , Coluccia, P. , … Mancini, R. (2013). Stearoyl‐CoA desaturase‐1 is a key factor for lung cancer‐initiating cells. Cell Death & Disease, 4(12), 947 10.1038/cddis.2013.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto, A. , de Vitis, C. , Pisanu, M. E. , Roscilli, G. , Ricci, G. , Catizone, A. , … Mancini, R. (2017). Stearoyl‐CoA‐desaturase 1 regulates lung cancer stemness via stabilization and nuclear localization of YAP/TAZ. Oncogene, 36, 4573–4584. 10.1038/onc.2017.75 [DOI] [PubMed] [Google Scholar]
- Penz, E. , Watt, K. N. , Hergott, C. A. , Rahman, N. M. , & Psallidas, I. (2017). Management of malignant pleural effusion: Challenges and solutions. Cancer Management and Research, 9, 229–241. 10.2147/CMAR.S95663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanu, M. E. , Maugeri‐Saccà, M. , Fattore, L. , Bruschini, S. , De Vitis, C. , Tabbì, E. , … Mancini, R. (2018). Inhibition of stearoyl‐CoA desaturase 1 reverts BRAF and MEK inhibition‐induced selection of cancer stem cells in BRAF‐mutated melanoma. Journal of Experimental & Clinical Cancer Research, 37(1), 318 10.1186/s13046-018-0989-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanu, M. E. , Noto, A. , De Vitis, C. , Morrone, S. , Scognamiglio, G. , Botti, G. , … Mancini, R. (2017). Blockade of stearoyl‐CoA‐desaturase 1 activity reverts resistance to cisplatin in lung cancer stem cells. Cancer Letters, 406, 93–104. https://doi.org/S0304-3835(17)30463-9. [pii]. [DOI] [PubMed] [Google Scholar]
- Plaks, V. , Kong, N. , & Werb, Z. (2015). The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell, 16(3), 225–238. 10.1016/j.stem.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck, M. , Reinmuth, N. , De Ruysscher, D. , Kerr, K. M. , & Peters, S. On Belhalf of ESMO Guidelines Working Group . (2014). Metastatic non‐small‐cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Annals of Oncology, 25, iii27–iii39. [DOI] [PubMed] [Google Scholar]
- Reck, M. , Rodríguez‐Abreu, D. , Robinson, A. G. , Hui, R. , Csőszi, T. , Fülöp, A. , … Brahmer, J. R. (2016). Pembrolizumab versus chemotherapy for PD‐L1 Positive non small‐cell lung cancer. New England Journal of Medicine, 375(19), 1823–1833. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- Reya, T. , Morrison, S. J. , Clarke, M. F. , & Weissman, I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature, 414, 105–111. [DOI] [PubMed] [Google Scholar]
- Ricci‐Vitiani, L. , Lombardi, D. G. , Pilozzi, E. , Biffoni, M. , Todaro, M. , Peschle, C. , & De Maria, R. (2006). Identification and expansion of human colon‐cancer‐initiating cells. Nature, 445, 111–115. [DOI] [PubMed] [Google Scholar]
- Roscilli, G. , De Vitis, C. , Ferrara, F. F. , Noto, A. , Cherubini, E. , Ricci, A. , … Mancini, R. (2016). Human lung adenocarcinoma cell cultures derived from malignant pleural effusions as model system to predict patients chemosensitivity. Journal of Translational Medicine, 14, 61 10.1186/s12967-016-0816-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton, M. , Quintana, E. , Fearon, E. R. , & Morrison, S. J. (2009). Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell, 138(5), 822–829. 10.1016/j.cell.2009.08.017 [DOI] [PubMed] [Google Scholar]
- Siegel, R. L. , Miller, K. D. , & Jemal, A. (2018). Cancer Statistics, 2018. CA: A Cancer Journal for Clinicians, 68(1), 7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- Sultan, M. , Coyle, K. M. , Vidovic, D. , Thomas, M. L. , Gujar, S. , & Marcato, P. (2017). Hide‐and‐seek: The interplay between cancer stem cells and the immune system. Carcinogenesis, 38(2), 107–118. [DOI] [PubMed] [Google Scholar]
- Tiran, V. , Stanzer, S. , Heitzer, E. , Meilinger, M. , Rossmann, C. , Lax, S. , … Balic, M. (2017). Genetic profiling of putative breast cancer stem cells from malignant pleural effusions. PLOS One, 12(4):e0175223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriens, K. , Christen, S. , Parik, S. , Broekaert, D. , Yoshinaga, K. , Talebi, A. , … Fendt, S. M. (2019). Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature, 566(7744), 403–406. 10.1038/s41586-019-0904-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, J. S. , Reddy, O. L. , Koo, M. , Xiong, Y. , Li, F. , & Xu, H. (2017). Application of immunohistochemistry in the diagnosis of pulmonary and pleural neoplasms. Archives of Pathology & Laboratory Medicine, 141(9), 1195–1213. 10.5858/arpa.2016-0550-RA [DOI] [PubMed] [Google Scholar]
- Yakisich, J. S. , Azad, N. , Kaushik, V. , & Iyer, A. K. V. (2017). Cancer cell plasticity: Rapid reversal of chemosensitivity and expression of stemness markers in lung and breast cancer tumorspheres. Journal of Cellular Physiology, 232(9), 2280–2286. 10.1002/jcp.25725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Duan, S. , Tsai, Y. , Keng, P. C. , Chen, Y. , Lee, S. O. , & Chen, Y. (2016). Cisplatin treatment increases stemness through upregulation of hypoxia‐inducible factors by interleukin‐6 in non‐small cell lung cancer. Cancer Science, 107(6), 746–754. 10.1111/cas.12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


