Summary
The use of antimicrobials in human and veterinary medicine has coincided with a rise in antimicrobial resistance (AMR) in the food‐borne pathogens Campylobacter jejuni and Campylobacter coli. Faecal contamination from the main reservoir hosts (livestock, especially poultry) is the principal route of human infection but little is known about the spread of AMR among source and sink populations. In particular, questions remain about how Campylobacter resistomes interact between species and hosts, and the potential role of sewage as a conduit for the spread of AMR. Here, we investigate the genomic variation associated with AMR in 168 C. jejuni and 92 C. coli strains isolated from humans, livestock and urban effluents in Spain. AMR was tested in vitro and isolate genomes were sequenced and screened for putative AMR genes and alleles. Genes associated with resistance to multiple drug classes were observed in both species and were commonly present in multidrug‐resistant genomic islands (GIs), often located on plasmids or mobile elements. In many cases, these loci had alleles that were shared among C. jejuni and C. coli consistent with horizontal transfer. Our results suggest that specific antibiotic resistance genes have spread among Campylobacter isolated from humans, animals and the environment.
Introduction
Campylobacter is the leading cause of bacterial gastroenteritis in the European Union (EU) (Food and Authority, 2019). The most common pathogenic species, C. jejuni and C. coli, were responsible for over 245,658 cases of campylobacteriosis in the EU in 2016, surpassing disease caused by Escherichia coli, Salmonella and Listeria (Food and Authority, 2019). Campylobacter are a common constituent of the gut microbiota of livestock including poultry, ruminants and pigs (Sheppard et al., 2009a, 2011; Sproston et al., 2011), and are also found in wild birds (Sheppard et al., 2010a, 2010b; Griekspoor et al., 2013; Cody et al., 2015; Atterby et al., 2018) and environmental sources (Dingle et al., 2001; Colles et al., 2003; Sheppard et al., 2009a). Human infection is typically associated with the consumption of contaminated meat (Fravalo et al., 2009; Hermans et al., 2012; Guyard‐Nicodème et al., 2013) and causes acute gastroenteritis and is self‐limiting after 3–5 days. In severe cases, antibiotic treatment may be required with fluoroquinolones and macrolides being the drugs of choice (Acheson and Allos, 2001).
Despite the ban on the use of antibiotics as growth promoters in animal feed in 2006 in the EU (Castanon, 2007), antimicrobial resistance (AMR) is still common among bacteria of the gastrointestinal tract of farmed animals (Sheppard et al., 2009a,b; Sproston et al., 2011). According to the latest European Centre for Disease Prevention and Control (ECDC) report in 2017, C. jejuni and C. coli isolates of clinical and animal origin showed high levels of resistance to both ciprofloxacin and tetracycline (Food and Authority, 2019). Furthermore, C. coli from clinical and animal samples have displayed resistance to macrolides including erythromycin and the aminoglycoside streptomycin (Food and Authority, 2019). More worryingly, there is an apparent trend towards multidrug resistance (MDR), particularly among C. coli that regularly harbour different AMR genes simultaneously within the genome of a single isolate (Luangtongkum et al., 2009; Pascoe et al., 2017; Food and Authority, 2019).
Mechanisms of resistance are well documented for several drug classes including fluoroquinolones, tetracyclines, macrolides, aminoglycosides and β‐lactams. Fluoroquinolone treatment was traditionally the first line of defence against campylobacteriosis but resistance has rapidly increased among strains (Sproston et al., 2018), potentially because it requires only a single point mutation in the genome (in the gyrA gene; Luo et al., 2003; Gibreel, 2006; Payot et al., 2006; Luangtongkum et al., 2009). This has led to a shift in treatment in favour of erythromycin prescription (Nachamkin et al., 2000; Gibreel, 2006), where resistance arises from specific point mutations in 23S rRNA and develops relatively slowly (Lapierre et al., 2016). However, in 2014, erythromycin resistance was found in animal and clinical isolates that carried an rRNA methylating enzyme, the ermB gene (Qin et al., 2014; Wang et al., 2014). Two years later the ermB gene was detected in C. coli isolates from turkeys and chickens in Spain suggesting the mobilization of this gene through horizontal gene transfer (HGT; Florez‐Cuadrado et al., 2016, 2018). Tetracycline resistance, associated with the tetO gene encoding a ribosomal protection protein, has also been observed in Campylobacter since 1987 (Sougakoff et al., 1987) and new enzymes conferring resistance to aminoglycosides continue to be discovered in Campylobacter (Lambert et al., 1985; Iovine, 2013; Zhao et al., 2016). In addition to these emerging trends, Campylobacter is known to have ‘natural’ resistance to β‐lactams, such as penicillin, in large part due to the ubiquity of the bla OXA‐61 gene (Alfredson and Korolik, 2005; Griggs et al., 2009). As a result of the widespread resistance to multiple antibiotic classes, it is no surprise that Campylobacter is a high priority pathogen on the recently published World Health Organization (WHO) list of bacteria, for which new antibiotics are urgently needed (WHO, 2017).
Many studies have highlighted the potential for transmission of AMR bacteria between agricultural animals and humans following extended use of antibiotics (Boerlin and Reid‐Smith, 2008; Huttner et al., 2013). However, controversy surrounding evidence for a direct link is confounded by inconsistencies in interpreting what constitutes the spread of resistance. Broadly, the spread of AMR can be defined as a clonal transmission or gene pool transmission. In clonal transmission, bacteria that have acquired AMR in one niche are transmitted to another where they retain resistance, such as in the survival of resistant Campylobacter through the food production chain to infect humans (Yahara et al., 2017). In gene pool transmission, HGT facilitates the spread of resistance genes between strains and species and the movement of genes (rather than clones) into multiple genetic backgrounds can be seen to spread AMR. Efforts to reduce AMR and conserve the remaining efficacy of existing drugs are focussed on the judicious use of antibiotics in animals and humans. In this context, it is advantageous to consider gene pool transmission as this is directly influenced by the selection pressure to maintain resistance in a given environment.
Campylobacter jejuni and C. coli can evolve rapidly, accumulating large numbers of nucleotide substitutions through mutation and recombination (Wilson et al., 2009; Sheppard et al., 2010a, 2010b; Dearlove et al., 2016). This can lead to de novo development of AMR through point mutation as well as the acquisition of resistance elements from other bacteria through HGT (Yahara et al., 2014, 2016). HGT has a major role in the mobilization of AMR not only within bacterial species but even across species boundaries. For example, the tetO gene that confers resistance to tetracycline in Campylobacter (Taylor et al., 1983; Batchelor, 2004) is believed to have originated via HGT from a Gram‐positive bacterium, potentially mediated by plasmid transfer (Taylor et al., 1983; Taylor, 1986; Batchelor, 2004). Interspecies genetic exchange requires some degree of niche overlap or physical proximity of strains. However, while there is some understanding of host niche segregation and clonal transmission of particular Campylobacter lineages (Sheppard et al., 2009a, 2010a, 2010b, 2014), there is limited quantitative information about the transmission dynamics of AMR genes between human, animal and environmental gene pools (gene pool transmission) in this genus.
In this study, we sequence the genome of isolates from a survey of AMR Campylobacter from multiple sources in Spain. Multidrug resistance phenotypes are quantified in vitro and compared to putative genomic determinants identified from over 2,000 known AMR genes. The co‐localization of these genes within resistance islands is examined and the allelic variation is compared among isolates from different sample sources. These analyses provide a basis for considering the interaction of different AMR gene pools and the potential source/sink contribution of livestock, humans and sewage effluents to the Campylobacter resistome.
Results
Enhanced in vitro MDR in C. coli compared to C. jejuni
We collected 168 C. jejuni and 92 C. coli isolates of human, animal and sewage origin (Supporting Information Table S1). In vitro resistance to six antibiotics (ciprofloxacin, nalidixic acid, tetracycline, erythromycin, streptomycin and gentamicin) of isolates of animal origin (Table 1, Supporting Information Table S2) was compared to resistance profiles of isolates of human and sewage origin (Table 1, Supporting Information Table S2). All Campylobacter isolates that were resistant to both ciprofloxacin and nalidixic acid were referred to as ciprofloxacin resistant only because resistance is conferred by SNPs in the same gene. The highest proportion of AMR was to ciprofloxacin (146/163; 90.1% for C. jejuni and 86/91; 94.5% for C. coli) and tetracycline (149/163; 91.4% for C. jejuni and 86/91; 94.5% for C. coli), followed by streptomycin (24/163; 14.7% for C. jejuni and 58/91; 63.7% for C. coli), erythromycin (4/162; 2.5% for C. jejuni and 23/91; 25.3% for C. coli) and gentamicin (2/163; 1.2% for C. jejuni and 10/91; 11% for C. coli; Table 1, Supporting Information Table S2). Higher prevalence of resistance was observed in C. coli isolates to erythromycin, streptomycin and gentamicin compared to C. jejuni (Fisher's exact test; p < 0.001). Typically, an isolate is considered multidrug resistant when it is resistant to at least three different classes of antibiotics (European Centre for Disease Prevention and Control [ECDC] & European Food Safety Authority [EFSA], 2015). Based on this criterion, more C. coli isolates were MDR (49/91; 53.8%) than C. jejuni (27/163; 16.6%; Table 2). All C. coli isolates were resistant to at least one antibiotic. (Table 2). Six (out of 163; 3.7%) C. jejuni isolates were sensitive to all tested antibiotics. Most of the isolates tested were resistant to both ciprofloxacin and tetracycline (140/163 or 85.9% C. jejuni and 82/91 or 90.1% C. coli), of which 52 C. coli isolates (57.1%) were also resistant to streptomycin compared to 24 C. jejuni isolates (14.7%) and nine C. coli isolates (9.9%) were also resistant to gentamicin compared to two C. jejuni isolates (1.23%; Table 2).
Table 1.
Drug resistance profiles of 254 Campylobacter isolates from humans, animals and sewage tested in the lab.
| Campylobacter jejuni | Campylobacter coli | |||||||
|---|---|---|---|---|---|---|---|---|
| Antibioticsa | Animals | Humans | Sewage | Total | Animals | Humans | Sewage | Total |
| Ciprofloxacin | 36/44 (81.8%) | 106/115 (88.7%) | 4/4 (100%) | 146/163 (90.12%) | 11/11 (100%) | 32/33 (97%) | 43/47 (91.5%) | 86/91 (94.5%) |
| Nalidixic acid | 35/44 (79.54%) | 78/115 (67.83%) | 3/4 (75%) | 116/163 (71.16%) | 11/11 (100%) | 30/33 (90.1%) | 43/47 (91.5%) | 84/91 (92.31%) |
| Tetracycline | 39/44 (88.6%) | 108/115 (93.91%) | 2/4 (50%) | 149/163 (91.41%) | 11/11 (100%) | 31/33 (94%) | 44/47 (93.6%) | 86/91 (94.5%) |
| Erythromycin | 3/44 (6.8%) | 1/115 (0.87%) | 0/4 (0%) | 4/163 (2.45%) | 10/11 (90.1%) | 6/33 (18.2%) | 7/47 (14.9%) | 23/91 (25.3%) |
| Streptomycin | 15/44 (34.1%) | 9/115 (7.83%) | 0/4 (0%) | 24/163 (14.72%) | 10/11 (90.1%) | 18/33 (54.5%) | 30/47 (63.8%) | 58/91 (63.7%) |
| Gentamicin | 0/44 (0%) | 2/115 (1.7%) | 0/4 (0%) | 2/163 (1.23%) | 4/11 (36.4%) | 2/33 (6.1%) | 4/47 (8.51%) | 10/91 (11%) |
| Total number of isolates | 44 | 115 | 4 | 163 | 11 | 33 | 47 | 91 |
Antibiotics resistance to: C, ciprofloxacin; T, tetracycline; E, erythromycin; S, streptomycin; G, gentamicin.
Table 2.
Multidrug resistant (in bold) and non‐multidrug resistant Campylobacter isolates (n = 254) from humans, animals and sewage.
| Campylobacter jejuni (n = 162) | Campylobacter coli (n = 91) | ||||||
|---|---|---|---|---|---|---|---|
| Antibioticsa | Animals | Humans | Sewage | Animals | Humans | Sewage | |
| CTESG | – | – | – | 4/11 (36.4%) | 1/33 (3%) | – | |
| CTES | – | – | – | 5/11 (45.5%) | 4/33 (12.1%) | 5/47 (10.6) | |
| Multiresistant | CTSG | – | 2/115 (1.7%) | – | – | 1/33 (3%) | 3/47 (6.4%) |
| CTS | 15/44 (34.1%) | 7/115 (6.9%) | – | 1/11 (9.1%) | 11/33 (33.3%) | 17/47 (36.2%) | |
| CTE | 2/44 (4.5%) | 1/115 (0/9%) | – | 1/11 (9.1%) | 1/33 (3%) | 2/47 (4.3%) | |
| CT | 16/44 (36.4%) | 95/115 (82.6%) | 2/4 (50%) | – | 12/33 (36.4%) | 13/47 (27.7%) | |
| CS | – | – | – | – | – | 1/47 (2.1%) | |
| TE | 1/44 (2.27%) | – | – | – | – | – | |
| Non‐multiresistant | TS | 1/44 (2.27%) | – | – | – | 1/33 (3%) | 4/47 (8.5%) |
| C | 3/44 (6.8%) | 1/115 (0.9%) | 1/4 (25%) | – | 2/33 (6.1%) | 2/47 (4.25%) | |
| T | 4/44 (11.4%) | 5/115 (4.4%) | – | – | – | – | |
| Non‐resistant | Sensitive | 2/44 (4.5%) | 4/115 (3.5%) | – | – | – | – |
| Total number of non‐multidrug resistant | 27/44 (61.36%) | 101/115 (8.69%) | 4/4 (100%) | – | 15/33 (45.45%) | 27/47 (57.44%) | |
| Total number of multidrug resistant | 17/44 (38.63%) | 10/115 (87.82%) | – | 11/11 (100%) | 18/33 (54.54%) | 20/47 (42.55%) | |
| Total number of isolates | 44 | 115 | 4 | 11 | 33 | 47 | |
Antibiotics resistance to: C, ciprofloxacin; T, tetracycline; E, erythromycin; S, streptomycin; G, gentamicin.
AMR isolates are distributed across highly structured populations
High levels of AMR observed in laboratory assays could indicate either an abundance of low diversity AMR clones or proliferation of AMR in multiple lineages. To investigate this, we analysed the population genomic structure of AMR isolates. The core genome phylogeny revealed that AMR isolates belonged to genome sequence clusters consistent with existing multilocus sequence typing (MLST) Sequence Type (ST) and clonal complex designations (Dingle et al., 2001; Miller, 2006; Fig. 1). Campylobacter jejuni isolates of chicken and cattle origin were mainly of host generalist (ST‐21, ST‐48, ST‐206 and ST‐45) clonal complexes (Sheppard et al., 2010a, 2010b, 2014; Fig. 1A, Supporting Information Table S1). Cattle isolates also belonged to ST‐61 and ST‐42 cattle associated clonal complexes, while human clinical isolates contained isolates of these generalist and cattle associated clonal complexes as well as additional generalist clonal complexes (ST‐22, ST‐52) and chicken associated clonal complexes (ST‐257, ST‐353, ST‐354, ST‐443, ST464, ST‐574 and ST‐658; Fig. 1A, Supporting Information Table S1). Campylobacter jejuni isolates from sewage belonged to ST‐362, a human associated complex and generalist ST‐22, ST‐45 and ST‐607 complexes (Fig. 1A, Supporting Information Table S1). Multidrug resistant C. jejuni isolates (27/167) were from generalist (ST‐21, ST‐206, ST‐45, ST‐52) complexes, chicken associated complexes (ST‐354, ST‐460 and ST‐464) and cattle associated complexes (ST‐42 and ST‐61; Fig. 1A, Supporting Information Table S1). Campylobacter coli isolates represented 28 different STs, all of which belonged to the ST‐828 clonal complex. The most abundant STs were 825 and 827, constituting 20.7% and 17.4% of all C. coli isolates (Fig. 1B, Supporting Information Table S1). The proportion of C. coli isolates displaying MDR (60.9%) was considerably higher than within C. jejuni (16.1%), nearly half of which were isolated from sewage highlighting the potential importance of urban effluents as reservoirs of AMR genes (Fig. 1B, Table 2). Clearly, diversity within this complex is lower than in agricultural/clinical C. jejuni and one might consider ST‐828 complex to be a single clone. However, as illustrated (Fig. 1B), AMR is found in divergent lineages within the ST‐828 complex and, importantly, is also absent in some closely related strains. This pattern is inconsistent with the proliferation of a clone that acquired AMR genes in a single ancestral acquisition event. Rather it suggests horizontal transfer of AMR genes among sublineages.
Figure 1.
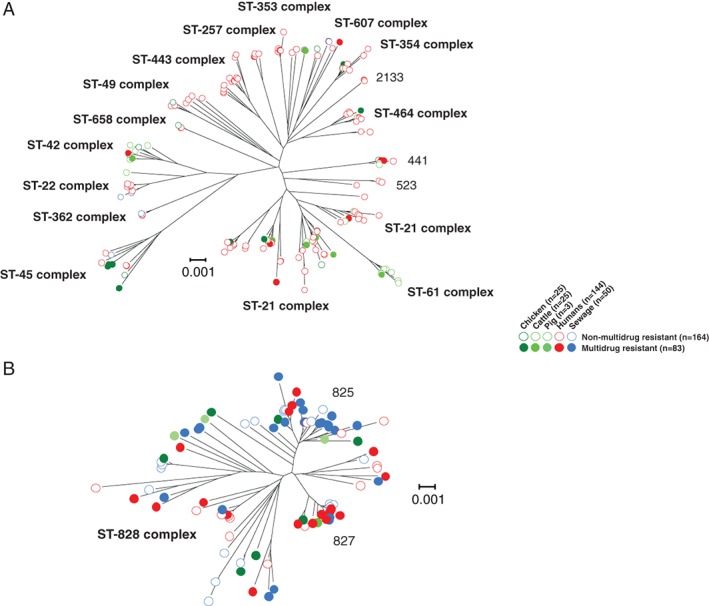
Phylogeny of antimicrobial resistant Campylobacter. Trees were reconstructed for 167 C. jejuni (A) and 92 C. coli (B) using concatenated gene‐by‐gene alignments of 595 core genes using the neighbour‐joining algorithm. Common sequence types and clonal complexes, defined by MLST, are indicated on the trees. Multidrug resistant isolates from chickens (dark green), cattle (intermediate green), pigs (light green), humans (red) and sewage (blue) are indicated with a filled circle, while the non‐multidrug resistant isolates are indicated with an open circle. The scale bars represent the number of substitutions per site. [Color figure can be viewed at http://wileyonlinelibrary.com]
Campylobacter coli genomes harbour more AMR genes than C. jejuni
The genome sequences of all Campylobacter isolates were compared to 2,158, 2,280 and 4,324 known antibiotic resistance genes and alleles from the Comprehensive Antibiotic Resistance Database (CARD; Cameron and Gaynor, 2014), ResFinder (Zankari et al., 2012) and the National Center for Biothechnology Information (NCBI) databases respectively. The analysis revealed the presence of 18 AMR genes including: cmeA, cmeB, cmeC, blaOXA‐61, tetO, ant‐like A, ant‐like B, ant(6)‐Ia, sat‐1, sat‐4, lnuC, ant(6)‐Ib, aad9, aph(3)‐IIIa, aph(2)‐IIIa, hpt, apmA and ermB (Fig. 2, Table 3; Trieu‐Cuot et al., 1985; Sougakoff et al., 1987; Achard et al., 2005; Alfredson and Korolik, 2005; Griggs et al., 2009; Qin et al., 2012; Toth et al., 2013; Cameron and Gaynor, 2014; Zhao et al., 2016; Florez‐Cuadrado et al., 2016; Olkkola et al., 2016; Yao et al., 2017). The cmeA, cmeB and cmeC genes, associated with efflux pump function, were present in all isolates. The bla OXA‐61 and tetO genes were common in resistant C. jejuni and C. coli isolates (Fig. 2, Table 3). The genes ant‐like A and ant‐like B have been described before as separate genes (Olkkola et al., 2016) and later revised as ant(6)‐Ie (Hormeño et al., 2018). To avoid the issues of gene duplication and gene paralogues they are considered as separate genes in this study. The bla OXA‐61 gene was significantly more prevalent in C. jejuni (64.8%) than C. coli isolates (51.1%; Fisher's exact test; p < 0.05), while the ant‐like A gene was more prevalent in C. coli (40.22% of C. coli and 1.19% of C. jejuni isolates, p < 0.001). The prevalence of the ant‐like A gene was also significantly higher in multidrug resistant isolates (33.7%) compared to non‐multidrug resistant isolates (6.7%; p < 0.001; Fig. 2, Table 3), and associated (p < 0.005) with isolates from humans (14.5%) and sewage (13.3%) compared to those from animals (1.2%; p < 0.005; Fig. 2, Table 3). In the case of non‐multidrug resistant isolates, the frequency difference of the ant‐like A gene can probably be attributed to the frequency of C. jejuni in human infection samples compared to the abundance of C. coli from sewage. Genes associated with aminoglycoside resistance (ant(6)‐Ia, sat‐4, ant(6)‐Ib, aad9, aph(3)‐IIIa, aph(2)‐IIIa, hpt and apmA) were mainly found in C. coli multidrug resistant isolates while sat‐1 was detected in only three C. jejuni strains from animals (Fig. 2, Table 3). Genes ant(6)‐Ia, sat‐4, ant(6)‐Ib and aph(3)‐IIIa were also found in C. jejuni isolates from animals (Fig. 2, Table 3). The lnuC gene, conferring resistance to lincosamides, was detected only in C. coli isolates and the ermB gene, which is not commonly found in Campylobacter, was detected in only one C. coli isolate from a chicken (Fig. 2, Table 3). A strong positive correlation (p < 0.001) between resistance phenotypes and genotypes was observed for tetracycline, streptomycin and gentamicin that were tested in vitro (Supporting Information Table S3). There was no correlation for erythromycin because the associated AMR gene ermB was only found in one isolate (Supporting Information Table S3). Concordance between putative resistance genotypes and laboratory phenotypes was lower than in some previous studies (Tyson et al., 2015; McDermott et al., 2016; Zhao et al., 2016). The main reason for this was that our study principally focused on the differential presence of AMR genes, to understand gene pool transmission, rather than resistance conferred by point mutation where it is more difficult to differentiate horizontal acquisition from de novo mutation. Other incongruences were observed between genotype prediction and laboratory phenotype. For example, not all isolates carrying aminoglycoside resistance genes were phenotypically resistant to streptomycin and gentamicin (Supporting Information Table S3). This is consistent with previous studies (Tyson et al., 2015; McDermott et al., 2016) and is potentially associated with variation in gene expression levels or synergistic effects among different resistance genes, warranting further study.
Figure 2.
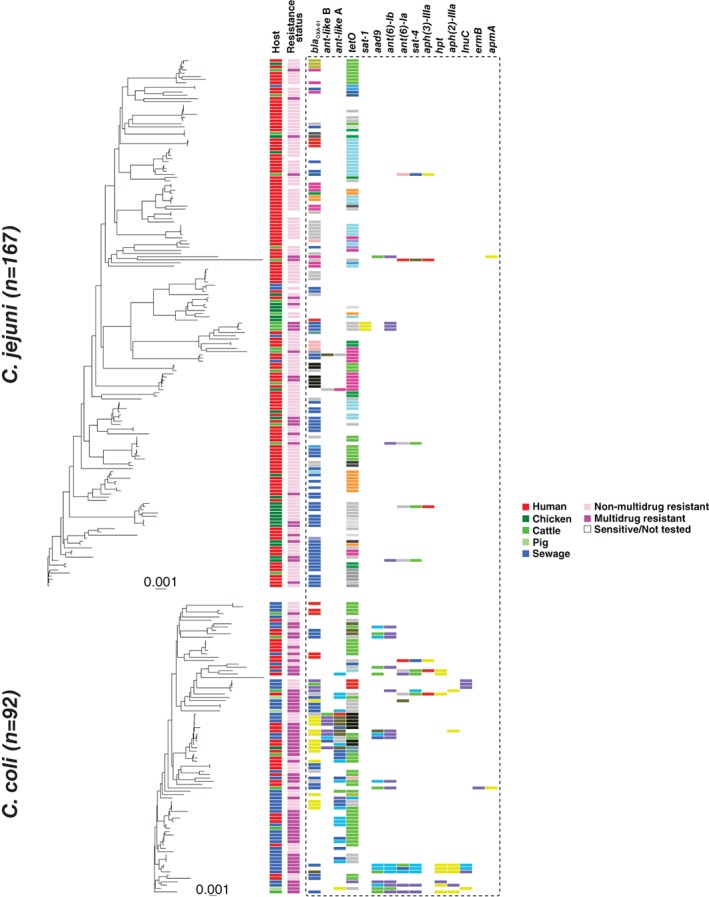
Presence and allelic diversity of 15 antimicrobial resistance genes in C. jejuni and C. coli genomes. Phylogenetic trees were reconstructed using gene‐by‐gene concatenated alignments of 595 core genes, and the neighbour‐joining algorithm for for 167 C. jejuni (A) and 92 C. coli (B). Isolate source is shown in the first column for chicken (dark green), cattle (green), pigs (light green), humans (red) and sewage (blue). The second column indicates the resistance status of each isolate as multidrug resistant (dark pink), non‐multidrug resistance (light pink) or not tested (white). Remaining columns indicate allelic variation at known resistance gene loci, with identical alleles coloured with the same colour. The scale represents the number of substitutions per site. [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 3.
Prevalence of 15 AMR genes in Campylobacter jejuni and C. coli isolates.a
| Multidrug resistant | Non‐multidrug resistant | Sensitive | Not tested | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Campylobacter jejuni (n = 27) | Campylobacter coli (n = 56) | Campylobacter jejuni (n = 129) | Campylobacter coli (n = 35) | Campylobacter jejuni (n = 7) | Campylobacter jejuni (n = 5)b | Campylobacter coli (n = 1)b | ||||||||||
| Animals (n = 17) | Humans (n = 10) | Animals (n = 11) | Humans (n = 18) | Sewage (n = 27) | Animals (n = 25) | Humans (n = 101) | Sewage (n = 3) | Humans (n = 15) | Sewage (n = 20) | Animals (n = 2) | Humans (n = 4) | Sewage (n = 1) | Humans (n = 3) | Sewage (n = 2) | Humans (n = 1) | |
| bla OXA‐61 | 15/17 (88.24%) | 6/10 (60.00%) | 8/11 (72.73%) | 9/18 (50.00%) | 9/27 (33.33%) | 16/25 (64.00%) | 65/101 (64.36%) | 1/3 (33.33%) | 8/15 (53.33%) | 13/20 (65.00%) | 1/2 (50.00%) | 3/4 (75.00%) | 0/1 (0%) | 1/3 (33.33%) | 0/2 (0.00%) | 0/1 (0.00%) |
| tet O | 14/17 (82.35%) | 4/10 (40.00%) | 8/11 (72.73%) | 16/18 (88.89%) | 23/27 (85.19%) | 20/25 (80.00%) | 80/101 (79.21%) | 1/3 (33.33%) | 12/15 (80.00%) | 16/20 (80.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 3/3 (100.00%) | 1/2 (50.00%) | 1/1 (100.00%) |
| ant‐like B | 0/17 (0.00%) | 0/10 (0.00%) | 1/11 (9.09%) | 3/18 (16.67%) | 2/27 (7.41%) | 1/25 (4.00%) | 1/101 (0.99%) | 0/3 (0.00%) | 0/15 (0.00%) | 3/20 (15.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| ant‐like A | 0/17 (0.00%) | 0/10 (0.00%) | 5/11 (45.45%) | 12/18 (66.67%) | 11/27 (40.74%) | 1/25 (4.00%) | 1/101 (0.99%) | 0/3 (0.00%) | 1/15 (6.67%) | 8/20 (40.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| ant(6) Ia | 4/17 (23.53%) | 0/10 (0.00%) | 1/11 (9.09%) | 3/18 (16.67%) | 6/27 (22.22%) | 1/25 (4.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 1/15 (6.67%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 1/1 (100.00%) |
| sat‐4 | 4/17 (23.53%) | 0/10 (0.00%) | 2/11 (18.18%) | 3/18 (16.67%) | 5/27 (18.52%) | 1/25 (4.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 1/15 (6.67%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 1/1 (100.00%) |
| lnuC | 0/17 (0.00%) | 0/10 (0.00%) | 1/11 (9.09%) | 0/18 (0.00%) | 3/27 (11.11%) | 0/25 (0.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 1/15 (6.67%) | 4/20 (20.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| ant(6)‐Ib | 5/17 (29.41%) | 1/10 (10.00%) | 5/11 (45.45%) | 4/18 (22.22%) | 8/27 (29.63%) | 0/25 (0.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 0/15 (0.00%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| aad9 | 0/17 (0.00%) | 1/10 (10.00%) | 4/11 (36.36%) | 5/18 (27.78%) | 8/27 (29.63%) | 0/25 (0.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 0/15 (0.00%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| aph(3)‐IIIa | 2/17 (11.76%) | 0/10 (0.00%) | 0/11 (0.00%) | 2/18 (11.11%) | 1/27 (3.7%) | 1/25 (4.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 1/15 (6.67%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 1/1 (100.00%) |
| aph(2)‐IIIa | 0/17 (0.00%) | 0/10 (0.00%) | 2/11 (18.18%) | 1/18 (5.56%) | 4/27 (14.81%) | 0/25 (0.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 0/15 (0.00%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| hyg | 0/17 (0.00%) | 0/10 (0.00%) | 1/11 (9.09%) | 2/18 (11.11%) | 6/27 (22.22%) | 0/25 (0.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 0/15 (0.00%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| apmA | 0/17 (0.00%) | 1/10 (10.00%) | 1/11 (9.09%) | 0/18 (0.00%) | 0/27 (0.00%) | 0/25 (0.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 0/15 (0.00%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| sat‐1 | 3/17 (17.65%) | 0/10 (0.00%) | 0/11 (0.00%) | 0/18 (0.00%) | 0/27 (0.00%) | 0/25 (0.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 0/15 (0.00%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
| ermB | 0/17 (0.00%) | 0/10 (0.00%) | 1/11 (9.09%) | 0/18 (0.00%) | 0/27 (0.00%) | 0/25 (0.00%) | 0/101 (0.00%) | 0/3 (0.00%) | 0/15 (0.00%) | 0/20 (0.00%) | 0/2 (0.00%) | 0/4 (0/00%) | 0/1 (0%) | 0/3 (0.00%) | 0/2 (0.00%) | 0/1 (0.00%) |
Isolates are separated as multidrug or non‐multidrug resistant based on their in vitro phenotypic profile.
Isolates id: 5087, 5093, 5111, 5095, 5100, 5215 were not tested for antibiotic resistant profile in vitro.
AMR genes are co‐localized in the genome of multidrug resistant isolates
AMR genes are often found in close proximity in the genome. For example, aminoglycoside resistance genes can form localized clusters within the genome (Werner et al., 2003; Qin et al., 2012). The low numbers of apmA and ermB genes identified, excluded them from formal statistical comparison. Due to the high levels of resistance to fluoroquinolones and tetracycline, the presence of ant‐like A, ant(6)‐Ia, sat‐4, ant(6)‐Ib, aad9, aph(3)‐IIIa, aph(2)‐IIIa, sat‐1 and hpt genes, was by definition significantly associated with MDR (Fisher's exact test; p < 0.001), because this was defined as resistance to three or more antimicrobial classes (Table 3). There was a slight increasing trend in the presence of ant‐like A, ant‐like B, aad9, ant(6)‐Ia, sat‐4, ant(6)‐Ib and aph(3)‐IIIa genes from 2010 to 2015 (Supporting Information Fig. S1). Furthermore, the relative position of the 15 AMR genes (in contiguous sequence assemblies) detected in Campylobacter isolates revealed two types of genetic associations in animal, human and sewage isolates. The first was between ant(6)‐Ia, sat‐4 and aph(3)‐IIIa genes, which clustered together in three C. jejuni isolates (one from chicken and two from cattle) and in eight C. coli isolates (one from chicken, four from humans and three from sewage; Fig. 3). This cluster has been previously described with the three genes located on the same genomic island in C. coli (Qin et al., 2012). The further addition of the aph(2)‐IIIa gene to this genomic island was observed in two C. coli isolates from sewage (Fig. 3). The second type of genetic association involved the presence of tetO, aad9 and ant(6)‐Ib genes. These genes clustered together in six C. coli isolates (one from chicken, one from pig, one from human and three from sewage) but also in one C. jejuni isolate from a human patient (Fig. 3). The addition of the sat‐1, hpt, apmA and ermB genes was also observed in these two types of syntenic block (Fig. 3).
Figure 3.
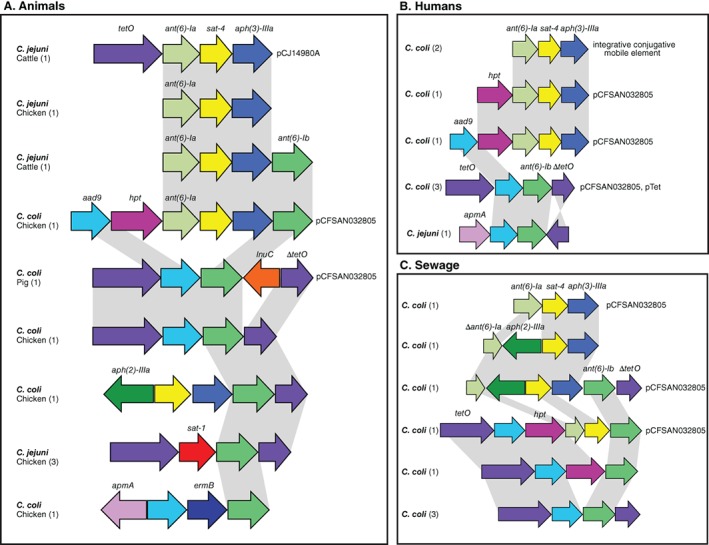
Comparative genetic organization of AMR GIs in Campyloabcter. The presence of each AMR gene, highlighted in different colours, is shown for representative C. jejuni and C. coli isolate genomes sampled from animals (A), humans (B) and sewage (C). The number of isolate genomes containing each genomic island arrangement is indicated in the parenthesis. Grey shading identifies sequence that shares > 95% nucleotide sequence identity. The name of the plasmid or mobile genetic element, associated with each genomic island, is indicated. [Color figure can be viewed at http://wileyonlinelibrary.com]
Evidence of gene pool transmission AMR genes
Evidence for HGT has been demonstrated for AMR genes in various bacteria, including Campylobacter (Sheppard et al., 2011, 2013; Wang et al., 2014; Sheppard and Maiden, 2015; Li et al., 2017), in some cases facilitated by mobile genetic elements including plasmids and transposons (Boerlin and Reid‐Smith, 2008). We identified one plasmid (pCFSAN032805; Accession: CP023546.1) in the genome sequences of eight C. coli isolates (one from chicken, one from a pig, three from humans and three from sewage; Fig. 3). Furthermore, a C. jejuni plasmid (pCJ14980A; Accession: CP017030.1) previously isolated from turkey faeces (Florez‐Cuadrado et al., 2017) was identified in a C. jejuni isolate from cattle in our study (Fig. 3). A pTet plasmid (Accession: CP002030.1) was also detected in one C. coli isolate of human origin (Fig. 3). A genomic region that was carrying the gene cluster ant(6)‐Ia, sat‐4 and aph(3)‐IIIa was highly similar to an integrative conjugative mobile element described in Erysipelothrix rhusiopathiae (Accession: MG812141.1) isolated from a pig farm. This region was also similar to sequences from other bacteria like Clostridium difficile, Staphylococcus aureus, S. pseudintermedius, Streptococcus suis and Enterococcus faecium. These findings are consistent with the circulation of genes, and more specifically alleles, not only between host microbiome gene pools but also between Campylobacter species. To investigate this further, we compared allelic diversity for the 15 identified AMR genes in C. jejuni and C. coli isolates.
The genes, bla OXA‐61 and tetO, had the highest diversity with 34 and 47 different alleles detected in C. jejuni and in C. coli isolates respectively (Fig. 3, Supporting Information Fig. S1). There were five bla OXA‐61 alleles, two of which were present in 16 and four C. jejuni and in 50 and five C. coli isolates, respectively (Fig. 3, Supporting Information Fig. S1). For the tetO gene, six alleles were present in more than five isolates each, with the most common allele present in 19 C. jejuni and in 35 C. coli. For the aad9 and ant(6)‐Ib gene, both of which had five alleles, the most common allele was present in both C. jejuni and C. coli isolates from multiple sources (Fig. 3, Supporting Information Fig. S1, Table S2). Finally, the sat‐4 gene shared two out of the six alleles between four C. jejuni and four C. coli isolates and the apmA gene had one allele which was shared by a C. jejuni of human origin and a C. coli isolated from a chicken (Fig. 3, Supporting Information Fig. S1, Table S2). Remaining alleles were detected exclusively in C. coli isolates.
Clonal descent is disrupted in AMR genes
The mean consistency index (CI) was significantly higher (Mann–Whitney test; U = 3307, p = 0.0214) among AMR genes (0.65581 ± 0.3531) compared with 595 core genes (0.4552 ± 0.05799; Fig. 4A). This provides evidence that the clonal mode of descent has been disrupted in AMR genes consistent with HGT. Furthermore, there was a significant decrease in the average allelic variation among AMR genes compared to core genes (Mann–Whitney test; U = 1004, p ≤ 0.0001; Fig. 4B). The average number of unique alleles per isolate was 0.03436 ± 0.05218 for the 15 AMR genes, compared with 0.1169 ± 0.05248 for 595 core genes. This is consistent with HGT facilitating the movement of AMR genes into multiple genetic backgrounds.
Figure 4.
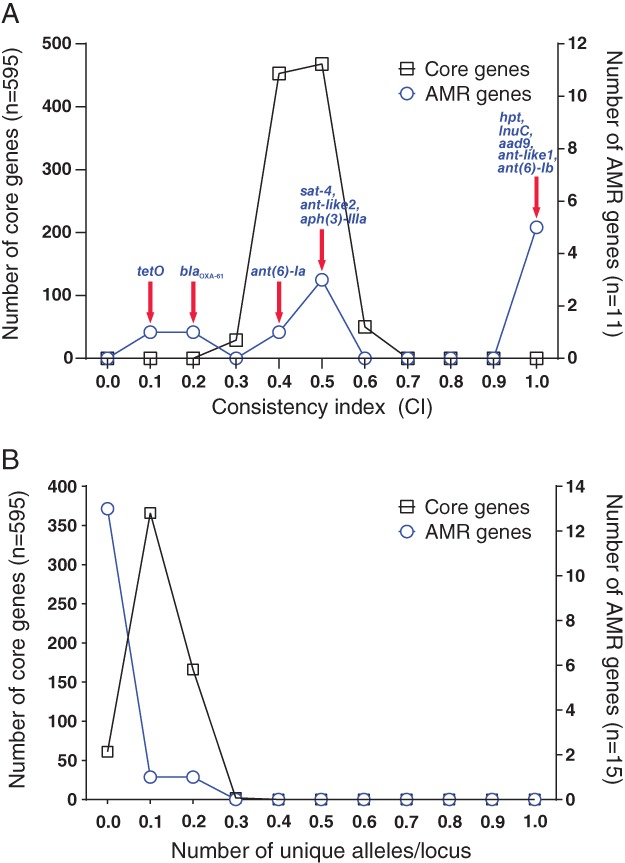
Comparison of consistency index and allelic variation between AMR and core genes. A. Consistency indices to a core phylogeny, were calculated for each gene alignment for AMR and core genes using the phangorn package in R. B. The number of alleles per locus. The left y‐axis indicates the number of core genes (black line), the right y‐axis indicates the number of AMR genes (blue line). For the consistency index, the two distributions were significantly different (two‐tailed Mann–Whitney test; p = 0.0214, Mann–Whitney U = 3307). For the number of alleles per locus, the two distributions were significantly different (two‐tailed Mann–Whitney test; p < 0.0001, Mann–Whitney U = 1004). [Color figure can be viewed at http://wileyonlinelibrary.com]
Among the AMR genes present in five or more isolates, the bla OXA‐61 and tetO alleles, associated with resistance to β‐lactams and tetracyclines respectively, were almost ubiquitous among C. jejuni and in C. coli from different sources. Two common bla OXA‐61 alleles were present in both Campylobacter species in all different hosts and sewage with other alleles shared only between human, chicken and sewage isolates (Fig. 5). A single tetO allele was present in the genomes of isolates from all different hosts and sewage except for C. jejuni from humans and C. coli cattle (Fig. 5), possibly due to low sample numbers (Supporting Information Table S1). Another tetO allele was shared between C. coli isolates from sewage and C. jejuni from chickens, cattle and humans (Fig. 5). In addition to evidence of frequent allele sharing between Campylobacter species from multiple sources, there were also several species‐specific alleles found in isolates from multiple sources (Fig. 5). AMR genes associated with aminoglycoside resistance had less allelic diversity compared to bla OXA‐61 and tetO (Fig. 2) and showed evidence of gene pool transmission between bacterial species and isolate source populations. Three alleles of the aad9, ant(6)‐Ib, sat‐4 genes were shared between C. jejuni and C. coli isolates. The ant(6)‐Ib allele was found in C. jejuni isolates from humans, cattle, chickens and in C. coli isolates from humans, chickens and sewage. The aad9 allele was found in human C. jejuni isolates and in C. coli isolates from humans, chickens, pigs and sewage. The sat‐4 allele was found in C. jejuni isolates from cattle and chicken and in C. coli isolates from human, chicken and sewage sources (Fig. 5). Alleles of other genes associated with aminoglycoside resistance (ant‐like A, aad9, ant(6)‐Ib, aph(3)‐IIIa, hpt and aph(2)‐IIIa) also showed evidence of transfer (allele sharing) between isolates sampled from different sources (Fig. 5).
Figure 5.
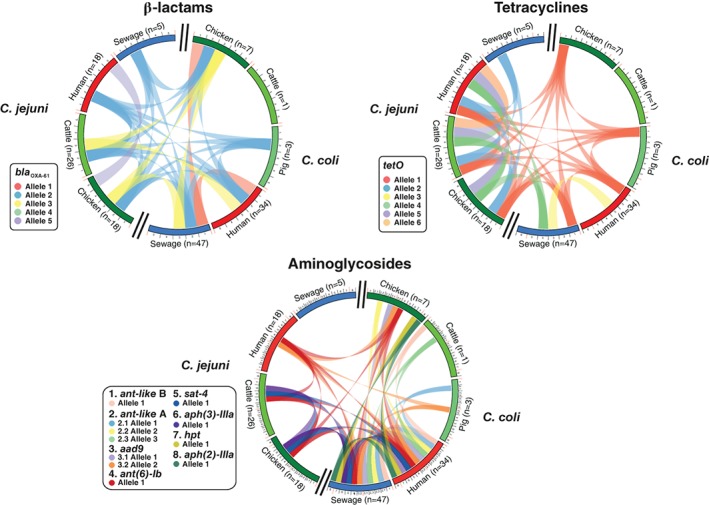
Distribution of AMR gene alleles among Campylobacter species and isolate source. Circus plots indicate the number of C. jejuni and C. coli isolates sampled from chickens (dark green), cattle (green), pigs (light green), humans (red) and sewage (blue) that contain genes associated with resistance to β‐lactam, tetracycline and aminoglycoside antimicrobials. Alleles present in > 5 isolate genomes are numbered around the perimeter. Exact matches between allele sequences are indicated by joining lines, coloured differently for different alleles. [Color figure can be viewed at http://wileyonlinelibrary.com]
Discussion
Forecasts of rising AMR in bacteria can make dramatic claims, such as an associated death toll of 10 million people by 2050 if no action is taken (Balouiri et al., 2016). However, for priority pathogens such as Campylobacter (WHO, 2017) it is not always clear where such action should be targeted. One reason for this is that zoonotic bacteria do not reside in a single host niche, therefore the source and sink dynamics of resistant strains may be poorly understood. Furthermore, the conduit for transmission between niches (in this case faeces) may also represent a reservoir of AMR. Here, by focussing analyses on comparison of gene pools, rather than individual resistant clones, we directly address if the alleles that confer resistance have spread between pathogenic Campylobacter species and the niches in which they reside.
Human infection is often a dead‐end for Campylobacter as disease is usually self‐limiting and human‐to‐human transmission is uncommon. As antibiotic treatment for campylobacteriosis is generally only given in acute or persistent cases, the heavy use of related antimicrobials in human and veterinary medicine (Schwarz et al., 2001; Teuber, 2001; Livermore, 2007) has raised concerns about how selection for resistance in livestock may lead to AMR in human pathogens. Despite the ban on the use of antibiotics as growth promoters in animals in 2006, quinolones and tetracyclines are still available for treatment of livestock all over the world (WHO, 2017). Consistent with trends in a recent ECDC report (Food and Authority, 2019), resistance to ciprofloxacin and tetracycline was seen in both Campylobacter species in our study, with resistance to streptomycin and gentamicin also frequent among sequenced C. coli isolates (Table 1). This may not be surprising as Spain has the highest sale of aminoglycosides for veterinary use in the EU (European Medicines Agency, 2018). Perhaps equally worrying was the isolation of C. coli resistant to erythromycin which is the drug of choice for antibiotic treatment of clinical campylobacteriosis (Acheson and Allos, 2001). The extent to which this level of resistance is a legacy of past use of fluoroquinolones, tetracyclines (Toth et al., 2013; Cameron and Gaynor, 2014) and other antimicrobials is not known but it is clear that Campylobacter harbour numerous resistance genes, potentially exacerbated by the carriage of similar genes among other components of the microbiota (van den Bogaard, 2000; Holmes et al., 2016).
AMR is widespread among Campylobacter isolated from livestock (Qin et al., 2014; Wang et al., 2014; Florez‐Cuadrado et al., 2016; Sproston et al., 2018), but the transmission dynamics are poorly understood. Where resistance is conferred by a single (or few) nucleotide substitution(s), such as in the gyrA gene (fluoroquinolone resistance; Engberg et al., 2001; Payot et al., 2006; Zhao et al., 2016), it is impossible to tell from sequence data if HGT or point mutation were responsible. For other classes of antibiotics, for example tetracyclines, there is evidence for the transfer of genes (e.g., tetO) between C. jejuni isolates, even in the absence antimicrobial selective pressure (Qin et al., 2012). In addition to tetO, our analyses identified 14 other accessory genes associated with Campylobacter resistance to other known antimicrobial classes (Supporting Information Table S4). These included aminoglycosides (10 genes), β‐lactams (bla OXA‐61) and macrolides (ermB) that have been variously used as treatments targeting Campylobacter and other infectious agents (or even as growth promoters; Engberg et al., 2001) in humans and animals (Lambert et al., 1985; Engberg et al., 2001; Griggs et al., 2009; Qin et al., 2012, 2014; Chen et al., 2013; Toth et al., 2013; Florez‐Cuadrado et al., 2016, 2017; Lapierre et al., 2016; Yao et al., 2017). Initial evidence of the importance of HGT in the transmission of these genes can be seen with inconsistent topology of individual AMR gene trees, compared to the Campylobacter core genome phylogeny (Supporting Information Fig. S1). Specifically, the CI varied for the 11 AMR genes, highlighting a disparity in the amount of inferred homoplasy in these genes, compared to genes in the core genome (Fig. 4B). Furthermore, the allelic variation in the AMR‐associated genes was significantly lower than the mean for core genes. Convergent genotypes may have evolved multiple times in different genetic backgrounds, however the most parsimonious explanation is the spread of AMR via HGT.
Perhaps the most compelling evidence for HGT is the identification of co‐localized clusters of genes that constitute GIs. Consistent with evidence of aminoglycoside resistance in Campylobacter (Lambert et al., 1985; Gibreel et al., 2004; Qin et al., 2012; Lapierre et al., 2016), all AMR genes detected in our study were found in MDR GIs, except for blaOXA‐61, ant‐like A and ant‐like B. There were multiple syntenic arrangements of genes with some GIs containing genes that confer resistance to more than one antimicrobial drug class (macrolides and aminoglycosides) as previously reported (Werner et al., 2003). Some of the MDR GIs are known from previous studies (ant(6)‐Ia, sat‐4 and aph(3)‐IIIa; Derbise et al., 1996, 1997), while others are reported here for the first time, such as the association between TetO, aad9 and ant(6)‐Ib genes. GI similarities provide evidence of transfer between C. jejuni and C. coli, and gene pool transmission among isolates from animals, humans and sewage. The transfer of GIs in Campylobacter can be via natural transformation (Qin et al., 2012), however several GIs were found on plasmids or integrative conjugative elements (Fig. 3) indicating the active mobilization of gene clusters. GIs containing the ant(6)‐Ia, sat‐4 and aph(3)‐IIIa cluster, and the tetO gene, have previously been described in staphylococci (Lambert et al., 1985; Derbise et al., 1996, 1997). Furthermore, the conjugative transposon found in C. coli was highly similar (~99.4% nucleotide identity over at least 60% of the sequence) to related sequence in other Gram‐positive bacteria. This is consistent with the circulation of AMR genes not only among Campylobacter species in different habitats but also HGT from other bacteria (Trieu‐Cuot et al., 1985; Zilhao et al., 1988).
An important finding in our study was that C. coli carry more combinations of AMR genes simultaneously than C. jejuni (Table 2). A simple explanation could be that C. coli ST‐828 complex isolates are more recombinogenic. There is evidence of the accumulation of C. jejuni DNA throughout the genome of this lineage (Sheppard et al., 2008, 2013) which could have led to the acquisition of multiple AMR genes. It is also possible that the dominance of this C. coli lineage (ST‐828 complex), that is much less diverse than C. jejuni as a whole, reflects a genetic bottleneck that favoured an ancestral AMR strain in, for example, the pig gut where C. coli (Thakur et al., 2006) and antimicrobial exposure (Aarestrup et al., 2000) are common. Whatever the reasons for differences in MDR between C. jejuni and C. coli, there is clear evidence for HGT and the transmission of AMR genes among bacterial species and host niche gene pools.
Contrasting evidence of HGT with quantitative information about the transmission of resistant bacteria between hosts would be extremely useful for understanding the dissemination of AMR among isolates from different habitats. In Campylobacter, studies have attempted to estimate the number of strains excreted into the environment by different animals (Ogden et al., 2009) and attribute the source of human infection to livestock (especially poultry) reservoirs (Sheppard et al., 2009b; Thépault et al., 2017, 2018). However, these large‐scale probabilistic studies are utterly underpowered for investigating the almost infinite number of possible transmission events, where the survival and proliferation of a single strain in a new niche could lead to the transfer of AMR genes between hosts and environments. A theoretical solution to the spread of AMR could be to use different drug classes in animals on the assumption that distinct antimicrobial selection pressures would sustain efficacy of drugs in humans. However, even if this was feasible, evidence from this study (and others (Hendriksen et al., 2019)) shows that multidrug resistant bacteria can be isolated and cultured from sewage, presenting a potential route for transmission of AMR in the environment. While the sources and implications of environmental contamination remain controversial (Rizzo et al., 2013; Munck et al., 2015), the evidence in our study is consistent with the horizontal transfer of AMR among Campylobacter isolated from livestock, humans and sewage. This suggests that judicious use of antimicrobials and monitoring of the amount of AMR Campylobacter entering the environment may be beneficial in combating the rise of resistance in this important zoonotic pathogen.
Experimental procedures
Culture and antimicrobial susceptibility testing
As part of routine Campylobacter surveillance in Spain, isolates were sampled and cultured on blood agar plates (bioMérieux) and incubated for 48 h at 37°C under microaerophilic conditions using Campygen atmosphere generation system packs (Oxoid, Basingstoke, U.K.). Subcultured colonies were harvested and suspended in sterile water to a standardized cell density (0.5 McFarland turbidity). Fifty microliters of this suspension was added to 11 ml of Mueller‐Hinton broth (TREK Diagnostics Systems, Waltham, MA) supplemented with 5.5% lysed horse blood (Oxoid). The solution was poured onto EUCAMP2 microdilution plates (TREK Diagnostics Systems) which were incubated under microaerophilic conditions for 48 h at 37°C as previously described (Florez‐Cuadrado et al., 2017). The interpretation of the quantitative data was performed according to the European Committee of Antimicrobial Susceptibility Testing, EUCAST (http://www.eucast.org/; last accessed: 06/2017).
DNA extraction, genome sequencing and archiving
A total of 260 Campylobacter isolates (167 C. jejuni and 92 C. coli) that displayed MDR phenotypes were chosen for genome sequencing. These represented strains sampled from humans, livestock and urban effluents in Spain. Of these, 55 isolates originated in animals (44 C. jejuni and 11 C. coli) including broiler chickens (18 C. jejuni and 7 C. coli), cattle (26 C. jejuni and 1 C. coli) and pigs (3 C. coli) and were collected from abattoirs in Spain (2008–2011) as part of the Spanish Veterinary Antimicrobial Resistance Surveillance (VAV) Network (Supporting Information Table S1). The isolates were chosen on the basis of resistance profiles (susceptible to resistant) to five different antibiotics (Table 1). Human samples (n = 152; 118 C. jejuni and 34 C. coli) were associated with campylobacteriosis cases in hospitals in the regions of Castilla y Leon, Extremadura and Andalucía between 2013 and 2016. Campylobacter isolates of urban effluent origin (n = 53; 6 C. jejuni and 47 C. coli) were collected from the wastewater treatment plants in the city of Madrid (Spain) between 2011 and 2013 (Ugarte‐Ruiz et al., 2015). All isolates were obtained using culture based methods (Moreno et al., 2000; Ugarte‐Ruiz et al., 2015; Hormeño et al., 2016) and speciated as C. jejuni or C. coli using a conventional multiplex PCR as previously described (Ugarte‐Ruiz et al., 2012).
For genome sequencing, isolates stored at −80°C in 1% protease peptone and 10% glycerol broth were cultured onto blood agar plates (bioMérieux) in microaerophilic conditions at 42°C for 48 h as previously described (Florez‐Cuadrado et al., 2017). Genomic DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN, Crawley, U.K.), according to manufacturer's instructions. Nucleic acid content was quantified on a Nanodrop spectrophotometer prior to normalization and sequencing. Libraries were prepared with Nextera XT kits (v2) and high‐throughput sequencing was performed using an Illumina MiSeq sequencer (Illumina, San Diego, CA; v3 technology, 300 bp paired‐end). Short reads were assembled de novo using SPAdes (version 3.8.0). All genomes used in this study were archived on the BIGSdb web‐based database platform (Jolley and Maiden, 2010) and given a unique identification number (BIGSid; Supporting Information Table S1).
Phylogenetic analysis
A pangenome was created for all isolate genomes in our collection as the sum of core genes, shared by all isolates, and accessory genes, present in at least one isolate. Genomes with a total assembly length > 1.9 Mbp, > 500 contigs or an N95 < 800 bp were considered poor quality and were excluded from the phylogenetic analyses. Whole genome multiple sequence alignments were obtained using MAFFT (Katoh, 2002) following a gene‐by‐gene approach as previously described (Méric et al., 2014). Phylogenetic trees, based on gene‐by‐gene alignments of core genes (Méric et al., 2014) or single gene sequences, were reconstructed using the Neighbour joining clustering method (Saitou and Nei, 1987).
Screening for AMR genes
AMR genes were identified in all Campylobacter genomes by comparison with the CARD (Jia et al., 2017; last assessed: 03/06/2017), the ResFinder (Zankari et al., 2012) and the NCBI databases using the BLAST algorithm (Sheppard et al., 2012; Maiden et al., 2013). A locus match was defined when genes had > 70% nucleotide identity over > 50% of the sequence length, and a matrix was generated that contained presence/absence information for each card gene and the allelic variation at that locus for every genome. Following the identification of isolate genomes harbouring one or more AMR genes, contigs were screened for upstream and downstream open reading frames (ORFs) to characterize the location of AMR relative to adjacent genes, using SnapGene software (GSL Biotech; available at http://snapgene.com). A second confirmatory analysis was performed, in which contigs were compared to NCBI database to identify whether they are associated with known plasmid or mobile elements. Sequence matches with > 95% nucleotide identity over > 50% of the sequence length were considered positive hits. A bivariate analysis was performed, in Stata version 14.0 (StataCorp, College Station, TX), to determine the relationship between phenotypes and genotypes for the presence of resistance using the Fisher's exact test. Associations were considered significant when p < 0.05.
HGT among infection‐associated genes
Population genetic analyses were undertaken to compare molecular variation among AMR genes to investigate patterns of HGT between species and isolates sampled form different niches. Genes where AMR is mediated by single nucleotide polymorphisms (SNPs), for example gyrA in fluroquinolone resistance (Sproston et al., 2018), were excluded from this analysis because of the inability to distinguish de novo mutation from homologous recombination of similar sequence. The allelic variation was calculated at loci associated with AMR genes (n = 15) and compared to variation at core loci (n = 595 genes). For both groups, the number of alleles at each locus (determined using a whole‐genome multilocus sequence typing, MLST, approach (Sheppard et al., 2012) and CI) were calculated. The consistency of a phylogenetic tree to patterns of variation in sequence alignments was determined for each gene of interest, and constituted an inference of the minimum amount of homoplasy in these genes, as implied by the tree (Kluge and Farris, 1969). The CI function from the R Phangorn package (Schliep, 2011) was used to calculate consistency indices for every single‐gene alignment of the 15 AMR genes to a phylogeny constructed from a concatenated gene‐by‐gene alignment of 595 core genes shared by all 259 isolates. The average CI of these shared genes was compared to that of the AMR genes.
Supporting information
Table S1. Details of isolates used in this study.
Table S2. Isolates and their MIC against different antibiotics used in this study.
Table S3. Resistance phenotype–genotype correlations among Campylobacter isolates.
Table S4. Antibiotic drug classes: mechanism of action/resistance and AMR genes.
Table S5. Genomic and phenotypic details of all isolates used in this study.
Fig. S1. Individual AMR gene trees. 14 single‐gene trees highlighting the allelic diversity in AMR genes found in C.jejuni (grey) and C.coli (black) isolates shown in the first column. The resistance status of each isolate is highlighted in the second column for multidrug resistant (dark pink), non‐multidrug resistance (light pink) or not tested (white). The host of every isolate is shown in the third column for chickens (dark green), cattle (green), pigs (light green), humans (red) and sewage (blue). The scale bars represent the number of substitutions per site.
Fig. S2. Prevalence of AMR genes over time. Graphs illustrate the presence of 15 putative AMR genes in isolate genomes sampled at each year in the study. Prevalence (%) was calculated by dividing the number of samples that had the AMR gene by the total number of samples in that year.
Acknowledgements
S.K.S., B.P. and S.C.B. were supported by grants from the Medical Research Council (MR/L015080/1), the Wellcome Trust (088786/C/09/Z), the Food Standards Agency (FS246004) and the Biotechnology and Biological Sciences Research Council (BB/I02464X/1). E.M. received a University of Bath Faculty of Science URSA studentship. D.F.C. is supported by the FPI program (BES‐2013‐065003) from the Spanish Ministry of Economy and Competitiveness. J.K.C. is supported by a BBSRC KTN PhD studentship (BB/P504750/1). All high performance computing was conducted with MRC CLIMB. We wish to thank our technicians María García, Estefanía Rivero and Nisrin Maasoumi for their excellent technical assistance.
Data Availability
All sequence data are linked to NCBI BioProject PRJNA528879. The bacterial genomes are available in GenBank under accession codes SRX5575129 to SRX5587545.
References
- Aarestrup, F.M. , Kruse, H. , Tast, E. , Hammerum, A.M. , and Jensen, L.B. (2000) Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb Drug Resist 6: 63–70. [DOI] [PubMed] [Google Scholar]
- Achard, A. , Villers, C. , Pichereau, V. , and Leclercq, R. (2005) New lnu(C) gene conferring resistance to Lincomycin by Nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob Agents Chemother 49: 2716–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson, D. , and Allos, B.M. (2001) Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis 32: 1201–1206. [DOI] [PubMed] [Google Scholar]
- Alfredson, D.A. , and Korolik, V. (2005) Isolation and expression of a novel molecular class D‐lactamase, OXA‐61, from Campylobacter jejuni . Antimicrob Agents Chemother 49: 2515–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atterby, C. , Mourkas, E. , Méric, G. , Pascoe, B. , Wang, H. , Waldenström, J. , et al (2018) The potential of isolation source to predict colonization in avian hosts: a case study in Campylobacter jejuni strains from three bird species. Front Microbiol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balouiri, M. , Sadiki, M. , and Ibnsouda, S.K. (2016) Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal 6: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor, R.A. (2004) Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150: 3507–3517. [DOI] [PubMed] [Google Scholar]
- Boerlin, P. , and Reid‐Smith, R.J. (2008) Antimicrobial resistance: its emergence and transmission. Anim Health Res Rev 9: 115–126. [DOI] [PubMed] [Google Scholar]
- van den Bogaard, A. (2000) Epidemiology of resistance to antibiotics links between animals and humans. Int J Antimicrob Agents 14: 327–335. [DOI] [PubMed] [Google Scholar]
- Cameron, A. , and Gaynor, E.C. (2014) Hygromycin B and Apramycin antibiotic resistance cassettes for use in Campylobacter jejuni . PLoS One 9: e95084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon, J.I.R. (2007) History of the use of antibiotic as growth promoters in European poultry feeds. Poult Sci 86: 2466–2471. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Mukherjee, S. , Hoffmann, M. , Kotewicz, M.L. , Young, S. , Abbott, J. , et al (2013) Whole‐genome sequencing of gentamicin‐resistant campylobacter coli isolated from U.S. retail meats reveals novel plasmid‐mediated aminoglycoside resistance genes. Antimicrob Agents Chemother 57: 5398–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody, A.J. , McCarthy, N.D. , Bray, J.E. , Wimalarathna, H.M.L. , Colles, F.M. , Jansen van Rensburg, M.J. , et al (2015) Wild bird‐associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environ Microbiol Rep 7: 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colles, F.M. , Jones, K. , Harding, R.M. , and Maiden, M.C.J. (2003) Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl Environ Microbiol 69: 7409–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearlove, B.L. , Cody, A.J. , Pascoe, B. , Méric, G. , Wilson, D.J. , and Sheppard, S.K. (2016) Rapid host switching in generalist Campylobacter strains erodes the signal for tracing human infections. ISME J 10: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbise, A. , de Cespedes, G. , and El Solh, N. (1997) Nucleotide sequence of the Staphylococcus aureus transposon, Tn5405, carrying aminoglycosides resistance genes. J Basic Microbiol 37: 379–384. [DOI] [PubMed] [Google Scholar]
- Derbise, A. , Dyke, K.G.H. , and El Solh, N. (1996) Characterization of a Staphylococcus aureus transposon, Tn5405, Located within Tn5404and carrying the aminoglycoside resistance Genes,aphA‐3andaadE. Plasmid 35: 174–188. [DOI] [PubMed] [Google Scholar]
- Dingle, K.E. , Colles, F.M. , Wareing, D.R.A. , Ure, R. , Fox, A.J. , Bolton, F.E. , et al (2001) Multilocus sequence typing system for Campylobacter jejuni . J Clin Microbiol 39: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg, J. , Aarestrup, F.M. , Taylor, D.E. , Gerner‐Smidt, P. , and Nachamkin, I. (2001) Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis 7: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control [ECDC] & European Food Safety Authority [EFSA] . (2015) EU summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. EFSA J 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency (2018) Reflection paper on use of aminoglycosides in animals in the European Union: development of resistance and impact on human and animal health. 44: 1–42. [Google Scholar]
- Florez‐Cuadrado, D. , Moreno, M.A. , Ugarte‐Ruíz, M. , and Domínguez, L. (2018) Antimicrobial resistance in the food chain in the European Union. Adv Food Nutr Res 86: 115–136. [DOI] [PubMed] [Google Scholar]
- Florez‐Cuadrado, D. , Ugarte‐Ruiz, M. , Meric, G. , Quesada, A. , Porrero, M.C. , Pascoe, B. , et al (2017) Genome comparison of erythromycin resistant Campylobacter from turkeys identifies hosts and pathways for horizontal spread of erm(B) genes. Front Microbiol 8: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez‐Cuadrado, D. , Ugarte‐Ruiz, M. , Quesada, A. , Palomo, G. , Domínguez, L. , and Porrero, M.C. (2016) Description of an erm (B)‐carrying Campylobacter coli isolate in Europe. J Antimicrob Chemother 71: 841–843. [DOI] [PubMed] [Google Scholar]
- Food, E. , and Authority, S. (2019) The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J 17: 1–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fravalo, P. , Laisney, M.‐J. , Gillard, M.‐O. , Salvat, G. , and Chemaly, M. (2009) Campylobacter transfer from naturally contaminated chicken thighs to cutting boards is inversely related to initial load. J Food Prot 72: 1836–1840. [DOI] [PubMed] [Google Scholar]
- Gibreel, A. (2006) Macrolide resistance in Campylobacter jejuni and Campylobacter coli . J Antimicrob Chemother 58: 243–255. [DOI] [PubMed] [Google Scholar]
- Gibreel, A. , Sköld, O. , and Taylor, D.E. (2004) Characterization of plasmid‐mediated aphA – 3 kanamycin resistance in Campylobacter jejuni . Microb Drug Resist 10: 98–105. [DOI] [PubMed] [Google Scholar]
- Griekspoor, P. , Colles, F.M. , McCarthy, N.D. , Hansbro, P.M. , Ashhurst‐Smith, C. , Olsen, B. , et al (2013) Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol Ecol 22: 1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs, D.J. , Peake, L. , Johnson, M.M. , Ghori, S. , Mott, A. , and Piddock, L.J.V. (2009) Lactamase‐mediated β‐lactam resistance in Campylobacter species: prevalence of Cj0299 (blaOXA‐61) and evidence for a novel β‐lactamase in C. jejuni . Antimicrob Agents Chemother 53: 3357–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyard‐Nicodème, M. , Tresse, O. , Houard, E. , Jugiau, F. , Courtillon, C. , El Manaa, K. , et al (2013) Characterization of Campylobacter spp. transferred from naturally contaminated chicken legs to cooked chicken slices via a cutting board. Int J Food Microbiol 164: 7–14. [DOI] [PubMed] [Google Scholar]
- Hendriksen, R.S. , Munk, P. , Njage, P. , van Bunnik, B. , McNally, L. , Lukjancenko, O. , et al (2019) Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat Commun 10: 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans, D. , Pasmans, F. , Messens, W. , Martel, A. , Van Immerseel, F. , Rasschaert, G. , et al (2012) Poultry as a host for the zoonotic pathogen Campylobacter jejuni . Vector Borne Zoonotic Dis 12: 89–98. [DOI] [PubMed] [Google Scholar]
- Holmes, A.H. , Moore, L.S.P. , Sundsfjord, A. , Steinbakk, M. , Regmi, S. , Karkey, A. , et al (2016) Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387: 176–187. [DOI] [PubMed] [Google Scholar]
- Hormeño, L. , Palomo, G. , Ugarte‐Ruiz, M. , Porrero, M.C. , Borge, C. , Vadillo, S. , et al (2016) Identification of the main quinolone resistance determinant in Campylobacter jejuni and Campylobacter coli by MAMA‐DEG PCR. Diagn Microbiol Infect Dis 84: 236–239. [DOI] [PubMed] [Google Scholar]
- Hormeño, L. , Ugarte‐Ruiz, M. , Palomo, G. , Borge, C. , Florez‐Cuadrado, D. , Vadillo, S. , et al (2018) Ant(6)‐I genes encoding aminoglycoside O‐Nucleotidyltransferases are widely spread among streptomycin resistant strains of Campylobacter jejuni and Campylobacter coli . Front Microbiol 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner, A. , Harbarth, S. , Carlet, J. , Cosgrove, S. , Goossens, H. , Holmes, A. , et al (2013) Antimicrobial resistance: a global view from the 2013 world healthcare‐associated infections forum. Antimicrob Resist Infect Control 2: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine, N.M. (2013) Resistance mechanisms in Campylobacter jejuni . Virulence 4: 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, B. , Raphenya, A.R. , Alcock, B. , Waglechner, N. , Guo, P. , Tsang, K.K. , et al (2017) CARD 2017: expansion and model‐centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45: D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley, K.A. , and Maiden, M.C. (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge, A.G. , and Farris, J.S. (1969) Quantitative Phyletics and the evolution of anurans. Syst Biol 18: 1–32. [Google Scholar]
- Lambert, T. , Gerbaud, G. , Trieu‐Cuot, P. , and Courvalin, P. (1985) Structural relationship between the genes encoding 3′‐aminoglycoside phosphotransferases in Campylobacter and in gram‐positive cocci. Ann l'Institut Pasteur/Microbiol 136: 135–150. [DOI] [PubMed] [Google Scholar]
- Lapierre, L. , Arias, M.L. , and Fernández, H. (2016) Antimicrobial resistance in Campylobacter spp In Campylobacter Spp. and Related Organisms in Poultry. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- Li, J. , Jiang, N. , Ke, Y. , Feßler, A.T. , Wang, Y. , Schwarz, S. , and Wu, C. (2017) Characterization of pig‐associated methicillin‐resistant Staphylococcus aureus . Vet Microbiol 201: 183–187. [DOI] [PubMed] [Google Scholar]
- Livermore, D.M. (2007) Introduction: the challenge of multiresistance. Int J Antimicrob Agents 29: S1–S7. [DOI] [PubMed] [Google Scholar]
- Luangtongkum, T. , Jeon, B. , Han, J. , Plummer, P. , Logue, C.M. , and Zhang, Q. (2009) Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol 4: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, N. , Sahin, O. , Lin, J. , Michel, L.O. , and Zhang, Q. (2003) In vivo selection of Campylobacter isolates with high levels of Fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother 47: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden, M.C.J. , van Rensburg, M.J.J. , Bray, J.E. , Earle, S.G. , Ford, S.A. , Jolley, K.A. , and McCarthy, N.D. (2013) MLST revisited: the gene‐by‐gene approach to bacterial genomics. Nat Rev Microbiol 11: 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott, P.F. , Tyson, G.H. , Kabera, C. , Chen, Y. , Li, C. , Folster, J.P. , et al (2016) Whole‐genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella . Antimicrob Agents Chemother 60: 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric, G. , Yahara, K. , Mageiros, L. , Pascoe, B. , Maiden, M.C.J. , Jolley, K.A. , and Sheppard, S.K. (2014) A reference pan‐genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic Campylobacter . PLoS One 9: e92798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W.G. (2006) Identification of host‐associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152: 245–255. [DOI] [PubMed] [Google Scholar]
- Moreno, M.A. , Domínguez, L. , Teshager, T. , Herrero, I.a. , and Porrero, M.C. (2000) Antibiotic resistance monitoring: the Spanish programme. The VAV network. Red de Vigilancia de Resistencias Antibióticas en Bacterias de Origen Veterinario. Int J Antimicrob Agents 14: 285–290. [DOI] [PubMed] [Google Scholar]
- Munck, C. , Albertsen, M. , Telke, A. , Ellabaan, M. , Nielsen, P.H. , and Sommer, M.O.A. (2015) Limited dissemination of the wastewater treatment plant core resistome. Nat Commun 6: 8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachamkin, I. , Engberg, J. , and Aarestrup, F. (2000). In Diagnosis and Antimicrobial Susceptibility of Campylobacter Species, Nachamkin I., and Blaser M.J. (eds). Washington DC, USA: ASM Press. [Google Scholar]
- Ogden, I.D. , Dallas, J.F. , MacRae, M. , Rotariu, O. , Reay, K.W. , Leitch, M. , et al (2009) Campylobacter excreted into the environment by animal sources: prevalence, concentration shed, and host association. Foodborne Pathog Dis 6: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkola, S. , Culebro, A. , Juntunen, P. , Hänninen, M.‐L. , and Rossi, M. (2016) Functional genomics in Campylobacter coli identified a novel streptomycin resistance gene located in a hypervariable genomic region. Microbiology 162: 1157–1166. [DOI] [PubMed] [Google Scholar]
- Pascoe, B. , Méric, G. , Yahara, K. , Wimalarathna, H. , Murray, S. , Hitchings, M.D. , et al (2017) Local genes for local bacteria: evidence of allopatry in the genomes of transatlantic Campylobacter populations. Mol Ecol 26: 4497–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payot, S. , Bolla, J.‐M. , Corcoran, D. , Fanning, S. , Mégraud, F. , and Zhang, Q. (2006) Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect 8: 1967–1971. [DOI] [PubMed] [Google Scholar]
- Qin, S. , Wang, Y. , Zhang, Q. , Chen, X. , Shen, Z. , Deng, F. , et al (2012) Identification of a novel Genomic Island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli . Antimicrob Agents Chemother 56: 5332–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, S. , Wang, Y. , Zhang, Q. , Zhang, M. , Deng, F. , Shen, Z. , et al (2014) Report of ribosomal RNA methylase gene erm(B) in multidrug‐resistant Campylobacter coli . J Antimicrob Chemother 69: 964–968. [DOI] [PubMed] [Google Scholar]
- Rizzo, L. , Manaia, C. , Merlin, C. , Schwartz, T. , Dagot, C. , Ploy, M.C. , et al (2013) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447: 345–360. [DOI] [PubMed] [Google Scholar]
- Saitou, N. , and Nei, M. (1987) The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schliep, K.P. (2011) Phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, S. , Kehrenberg, C. , and Walsh, T.R. (2001) Use of antimicrobial agents in veterinary medicine and food animal production. Int J Antimicrob Agents 17: 431–437. [DOI] [PubMed] [Google Scholar]
- Sheppard, S.K. , Cheng, L. , Méric, G. , de Haan, C.P.A. , Llarena, A.‐K. , Marttinen, P. , et al (2014) Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol Ecol 23: 2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , Colles, F. , Richardson, J. , Cody, A.J. , Elson, R. , Lawson, A. , et al (2010a) Host association of Campylobacter genotypes transcends geographic variation. Appl Environ Microbiol 76: 5269–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , Colles, F.M. , McCarthy, N.D. , Strachan, N.J.C. , Ogden, I.D. , Forbes, K.J. , et al (2011) Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol Ecol 20: 3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , Dallas, J.F. , MacRae, M. , McCarthy, N.D. , Sproston, E.L. , Gormley, F.J. , et al (2009a) Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int J Food Microbiol 134: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , Dallas, J.F. , Strachan, N.J.C. , MacRae, M. , McCarthy, N.D. , Wilson, D.J. , et al (2009b) Campylobacter genotyping to determine the source of human infection. Clin Infect Dis 48: 1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , Dallas, J.F. , Wilson, D.J. , Strachan, N.J.C. , McCarthy, N.D. , Jolley, K.A. , et al (2010b) Evolution of an agriculture‐associated disease causing Campylobacter coli clade: evidence from National Surveillance Data in Scotland. PLoS One 5: e15708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , Didelot, X. , Jolley, K.A. , Darling, A.E. , Pascoe, B. , Meric, G. , et al (2013) Progressive genome‐wide introgression in agricultural Campylobacter coli . Mol Ecol 22: 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , Jolley, K.A. , and Maiden, M.C.J. (2012) A gene‐by‐gene approach to bacterial population genomics: whole genome MLST of Campylobacter . Genes 3: 261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , and Maiden, M.C.J. (2015) The evolution of Campylobacter jejuni and Campylobacter coli . Cold Spring Harb Perspect Biol 7: a018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, S.K. , McCarthy, N.D. , Falush, D. , and Maiden, M.C.J. (2008) Convergence of Campylobacter species: implications for bacterial evolution. Science 320: 237–239. [DOI] [PubMed] [Google Scholar]
- Sougakoff, W. , Papadopoulou, B. , Nordmann, P. , and Courvalin, P. (1987) Nucleotide sequence and distribution of gene tetO encoding tetracycline resistance in Campylobacter coli . FEMS Microbiol Lett 44: 153–159. [Google Scholar]
- Sproston, E.L. , Ogden, I.D. , MacRae, M. , Dallas, J.F. , Sheppard, S.K. , Cody, A.J. , et al (2011) Temporal variation and host association in the Campylobacter population in a longitudinal ruminant farm study. Appl Environ Microbiol 77: 6579–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproston, E.L. , Wimalarathna, H.M.L. , and Sheppard, S.K. (2018) Trends in fluoroquinolone resistance in Campylobacter . Microb Genomics 4: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D.E. (1986) Plasmid‐mediated tetracycline resistance in Campylobacter jejuni: expression in Escherichia coli and identification of homology with streptococcal class M determinant. J Bacteriol 165: 1037–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D.E. , Garner, R.S. , and Allan, B.J. (1983) Characterization of tetracycline resistance plasmids from Campylobacter jejuni and Campylobacter coli . Antimicrob Agents Chemother 24: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuber, M. (2001) Veterinary use and antibiotic resistance. Curr Opin Microbiol 4: 493–499. [DOI] [PubMed] [Google Scholar]
- Thakur, S. , Morrow, W.E.M. , Funk, J.A. , Bahnson, P.B. , and Gebreyes, W.A. (2006) Molecular epidemiologic investigation of Campylobacter coli in swine production systems, using multilocus sequence typing. Appl Environ Microbiol 72: 5666–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thépault, A. , Méric, G. , Rivoal, K. , Pascoe, B. , Mageiros, L. , Touzain, F. , et al (2017) Genome‐wide identification of host‐segregating epidemiological markers for source attribution in Campylobacter jejuni . Appl Environ Microbiol 83: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thépault, A. , Rose, V. , Quesne, S. , Poezevara, T. , Béven, V. , Hirchaud, E. , et al (2018) Ruminant and chicken: important sources of campylobacteriosis in France despite a variation of source attribution in 2009 and 2015. Sci Rep 8: 9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, M. , Frase, H. , Antunes, N.T. , and Vakulenko, S.B. (2013) Novel aminoglycoside 2″‐phosphotransferase identified in a gram‐negative pathogen. Antimicrob Agents Chemother 57: 452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu‐Cuot, P. , Gerbaud, G. , Lambert, T. , and Courvalin, P. (1985) In vivo transfer of genetic information between gram‐positive and gram‐negative bacteria. EMBO J 4: 3583–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson, G.H. , McDermott, P.F. , Li, C. , Chen, Y. , Tadesse, D.A. , Mukherjee, S. , et al (2015) WGS accurately predicts antimicrobial resistance in Escherichia coli . J Antimicrob Chemother 70: 2763–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte‐Ruiz, M. , Florez‐Cuadrado, D. , Wassenaar, T. , Porrero, M. , and Domínguez, L. (2015) Method comparison for enhanced recovery, isolation and qualitative detection of C. jejuni and C. coli from wastewater effluent samples. Int J Environ Res Public Health 12: 2749–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte‐Ruiz, M. , Gómez‐Barrero, S. , Porrero, M.C. , Álvarez, J. , García, M. , Comerón, M.C. , et al (2012) Evaluation of four protocols for the detection and isolation of thermophilic Campylobacter from different matrices. J Appl Microbiol 113: 200–208. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Zhang, M. , Deng, F. , Shen, Z. , Wu, C. , Zhang, J. , et al (2014) Emergence of multidrug‐resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob Agents Chemother 58: 5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, G. , Hildebrandt, B. , and Witte, W. (2003) Linkage of erm (B) and aadE‐sat4‐aphA‐3 in multiple‐resistant Enterococcus faecium isolates of different ecological origins. Microb Drug Resist 9: 9–16. [DOI] [PubMed] [Google Scholar]
- WHO . (2017) WHO publishes list of bacteria for which new antibiotics are urgently needed. WHO Media Cent. [Google Scholar]
- Wilson, D.J. , Gabriel, E. , Leatherbarrow, A.J.H. , Cheesbrough, J. , Gee, S. , Bolton, E. , et al (2009) Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni . Mol Biol Evol 26: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara, K. , Didelot, X. , Ansari, M.A. , Sheppard, S.K. , and Falush, D. (2014) Efficient inference of recombination hot regions in bacterial genomes. Mol Biol Evol 31: 1593–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara, K. , Didelot, X. , Jolley, K.A. , Kobayashi, I. , Maiden, M.C.J. , Sheppard, S.K. , and Falush, D. (2016) The landscape of realized homologous recombination in pathogenic bacteria. Mol Biol Evol 33: 456–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara, K. , Méric, G. , Taylor, A.J. , de Vries, S.P.W. , Murray, S. , Pascoe, B. , et al (2017) Genome‐wide association of functional traits linked with Campylobacter jejuni survival from farm to fork. Environ Microbiol 19: 361–380. [DOI] [PubMed] [Google Scholar]
- Yao, H. , Liu, D. , Wang, Y. , Zhang, Q. , and Shen, Z. (2017) High prevalence and predominance of the aph(2″)‐if gene conferring aminoglycoside resistance in Campylobacter . Antimicrob Agents Chemother 61: e00112‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari, E. , Hasman, H. , Cosentino, S. , Vestergaard, M. , Rasmussen, S. , Lund, O. , et al (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67: 2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S. , Tyson, G.H. , Chen, Y. , Li, C. , Mukherjee, S. , Young, S. , et al (2016) Whole‐genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 82: 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilhao, R. , Papadopoulou, B. , and Courvalin, P. (1988) Occurrence of the Campylobacter resistance gene tetO in Enterococcus and Streptococcus spp. Antimicrob Agents Chemother 32: 1793–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Details of isolates used in this study.
Table S2. Isolates and their MIC against different antibiotics used in this study.
Table S3. Resistance phenotype–genotype correlations among Campylobacter isolates.
Table S4. Antibiotic drug classes: mechanism of action/resistance and AMR genes.
Table S5. Genomic and phenotypic details of all isolates used in this study.
Fig. S1. Individual AMR gene trees. 14 single‐gene trees highlighting the allelic diversity in AMR genes found in C.jejuni (grey) and C.coli (black) isolates shown in the first column. The resistance status of each isolate is highlighted in the second column for multidrug resistant (dark pink), non‐multidrug resistance (light pink) or not tested (white). The host of every isolate is shown in the third column for chickens (dark green), cattle (green), pigs (light green), humans (red) and sewage (blue). The scale bars represent the number of substitutions per site.
Fig. S2. Prevalence of AMR genes over time. Graphs illustrate the presence of 15 putative AMR genes in isolate genomes sampled at each year in the study. Prevalence (%) was calculated by dividing the number of samples that had the AMR gene by the total number of samples in that year.
Data Availability Statement
All sequence data are linked to NCBI BioProject PRJNA528879. The bacterial genomes are available in GenBank under accession codes SRX5575129 to SRX5587545.


