Abstract
We used single‐cell RNA sequencing (seq) on several human induced pluripotent stem (iPS) cell‐derived neural stem cell (NSC) lines and one fetal brain‐derived NSC line to study inherent cell type heterogeneity at proliferating neural stem cell stage and uncovered predisposed presence of neurogenic and gliogenic progenitors. We observed heterogeneity in neurogenic progenitors that differed between the iPS cell‐derived NSC lines and the fetal‐derived NSC line, and we also observed differences in spontaneous differentiation potential for inhibitory and excitatory neurons between the iPS cell‐derived NSC lines and the fetal‐derived NSC line. In addition, using a recently published glia patterning protocol we enriched for gliogenic progenitors and generated glial cells from an iPS cell‐derived NSC line.
Keywords: glia, induced pluripotent stem cells, modeling gliogenesis, modeling neurogenesis, neural stem cells, neurons, single cell RNA sequencing
Single‐cell RNA sequencing on neural stem cell (NSC) lines showed inherent cell type heterogeneity for neurogenic and gliogenic progenitors. Induced pluripotent stem (IPS) cell‐derived NSC lines and fetal‐derived NSC line showed differences in spontaneous differentiation outcome for inhibitory and excitatory neurons. iPS cell‐derived NSCs hold enrichment potential for gliogenic progenitors.

1. INTRODUCTION
The advent of iPS cell technology over a decade ago opened a new field of scientific study into biological processes governing development and establishment of cells. Importantly, iPS cell technology enables the study of human development in laboratory setting and the cell culture dish (Okano & Yamanaka, 2014; Takahashi et al., 2007). NSC lines derived from human iPS cells and NSC lines isolated from human fetal sources provide a robust platform for study of neuronal development and have demonstrated the potential for generating neurons, oligodendrocytes and astrocytes (Falk et al., 2012; Tailor et al., 2013). As cell models, NSC lines provide expandable and stable source of cells for the use of in vitro modeling of neurogenesis. The point of having access to robust neural stem cell sources is key for maintaining reproducible sets of source materials to set up and prove different hypothesis in the experimental setting (Gage & Temple, 2013; Mertens, Marchetto, Bardy, & Gage, 2016).
The cells of the developing mammalian neuroectoderm display gene expression coding for genetic pathways, biological function and cellular structures. In this perspective, reference genes can be used to identify and classify cell types along the temporal and spatial axis of embryonic central nervous system (CNS) development. The CNS developmental process can be modeled and mimicked with embryonic stem (ES) cells and iPS cells when induced toward NSC lines with above‐mentioned protocols (for reviews see Mertens et al., 2016; Shparberg, Glover, & Morris, 2019). Emerging functional cell types in the CNS can be classified by gene expression, for example, for the neurogenic lineage: DCX for neuroblasts (Gleeson, Lin, Flanagan, & Walsh, 1999), GAD2 for inhibitory neurons (McBain & Fisahn, 2001; Taniguchi et al., 2011), NPTX2 for synaptic priming of excitatory neurons (Pelkey et al., 2015), SLC17A6 (vGLUT2) for excitatory neurons (Fremeau, Voglmaier, Seal, & Edwards, 2004), LMX1B for dopaminergic neurons (Nakatani, Kumai, Mizuhara, Minaki, & Ono, 2010) and for the gliogenic lineage, PTPRZ1 for radial glia (Pollen et al., 2015) and FABP7 for astroglia (Ebrahimi et al., 2016). In light of the complexity and challenges presented when investigating cells of the emerging CNS, single‐cell RNA‐seq offers powerful precision and highly resourceful technology to resolve and expose subtle differences between seemingly similar cells. The adapted use of single‐cell RNA‐seq will have a lasting and permanent impact on the current and future study of biology (Linnarsson & Teichmann, 2016).
Here, we uncovered the presence of neurogenic progenitors and gliogenic progenitors in NSC lines by using single‐cell RNA‐seq and found inherent heterogeneity in established NSC lines used in studies modeling neurogenesis and gliogenesis by known differentiation protocols (Falk et al., 2012; Lam et al., 2019; Lundin et al., 2018; Tailor et al., 2013).
2. RESULTS
2.1. AF22 NES cell line inherently contains both neurogenic and gliogenic progenitors
To study NSC biology, we used AF22 neuroepithelial stem (NES) cells, an established human iPS cell‐derived NSC line with proven characteristics to model neural stem cell biology related to early fetal central nervous system (CNS) development. Derived from a robust and stable method, AF22 cells at NES stage are proliferating, are highly expandable and hold potential for multipotency of differentiation into neurons, oligodendrocytes and astrocytes (in this paper collectively called glia; Alvarez‐Buylla, Garcia‐Verdugo, & Tramontin, 2001; Falk et al., 2012).
We sorted and sequenced AF22 NES cells at passage 52 using RamDA‐seq (Hayashi et al., 2018) total single‐cell RNA protocol with deep sequencing at 20 million reads/cell to generate a super resolution data set of gene expression. We obtained 88 high‐quality single cells with observed median of ~5,647,922 million mapped exonic reads per cell and median 21,620 genes expressed per cell (Figure 1a). We used Seurat v3 (Butler, Hoffman, Smibert, Papalexi, & Satija, 2018) and carried out cell clustering and aggregation of gene expression profiles based on 5 k‐nearest neighbors (KNN) and observed 4 cell clusters on UMAP plot with percentage distribution of cell cluster 1 (33%), 2 (28.4%), 3 (19.3%) and 4 (19.3%; Figure 1b,c).
Figure 1.
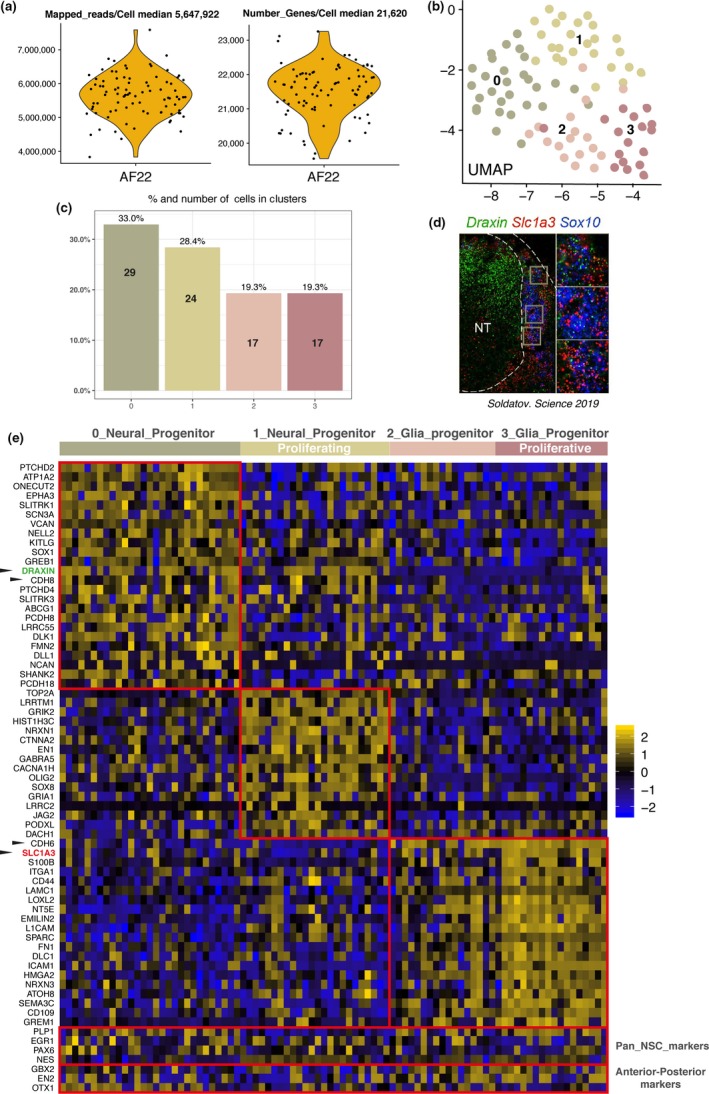
AF22 NES deep sequencing shows neurogenic and gliogenic progenitors. (a) Violin plot of mapped reads/cell and number of genes expressed/cell. (b) UMAP plot of 4 clusters over 88 AF22 NES single cells. (c) Barplot of percentage of AF22 NES cells in each of 4 clusters. (d) DRAXIN green (Neural tube) SLC1A3 red, in situ RNAScope E9.5 mouse developing spinal cord, from figure S2f Soldatov et al. (2019). (e) Heat map of genes enriched in cell clusters, highlighting neurogenic progenitor clustering division, selected by fold change
Our hypothesis relies on the developmental model concept that NES cells accurately represent a model of the developing neural ectoderm. In a recently published high impact study, we noted results describing neural tube and neural crest/glia cell identity during development by displaying Draxin + cells located in the neural tube region and Slc1a3 + cells located in the peripheral and outside region of the neural tube as glia progenitors/neural crest cells in mouse E9.5 developing spinal cord (Figure 1d; Soldatov et al., 2019).
We looked at gene expression enrichment for aggregating cell clusters in AF22 NES cells and observed a similar situation. Herein, DRAXIN‐expressing cells corresponding to proliferating/nonproliferating neurogenic progenitors co‐expressing NOTCH signaling markers DLL1, DLK1 and JAG2 and SLC1A3 expressing cells corresponding to gliogenic progenitors co‐expressing glia markers ITGA1, S100B and CD44. Exclusive expression of specific cadherins was observed, where CDH8 is expressed in neurogenic progenitors and CDH6 is expressed in gliogenic progenitors. We propose these cadherins might serve as cell surface markers for sorting out of progenitor populations. We also observed known pan neural stem cell markers in both neurogenic and gliogenic progenitors (e.g., NES, PAX6) and pan‐expression presence of anterior‐posterior GBX2 (hindbrain), EN2 (hindbrain/midbrain) and OTX1 (midbrain/forebrain) markers contained in AF22 NES cells (Figure 1e).
Our observations reveal pre‐existing progenitor heterogeneity residing inside AF22 NES cell line. This allows for explanation of a dual source of differentiation potential, the first source generating neurons from neurogenic progenitors (~60%) and the second source generating glia from gliogenic progenitors (~40%).
2.2. Fetal primary neural stem cells and iPS cell‐derived neural stem cells both contain neurogenic progenitors and gliogenic progenitors
Taking into account our observations from deep sequencing of AF22 NES cell line, we used C1 Fluidigm single‐cell capture system with STRT‐seq (Islam et al., 2014) to reexamine SAi2 NES passage 29 cell line, an established human fetal hindbrain‐derived primary NSC line, again the iPS cell‐derived AF22 NES passage 20 cell line and additionally the iPS cell‐derived Ctrl‐7 NES passage 12 cell line. The cells were captured in proliferating NES stage, and all three NSC lines have previously reported multipotency and differentiation capacity for generating neurons and glia (Falk et al., 2012; Lam et al., 2019; Tailor et al., 2013; Figure 2a).
Figure 2.
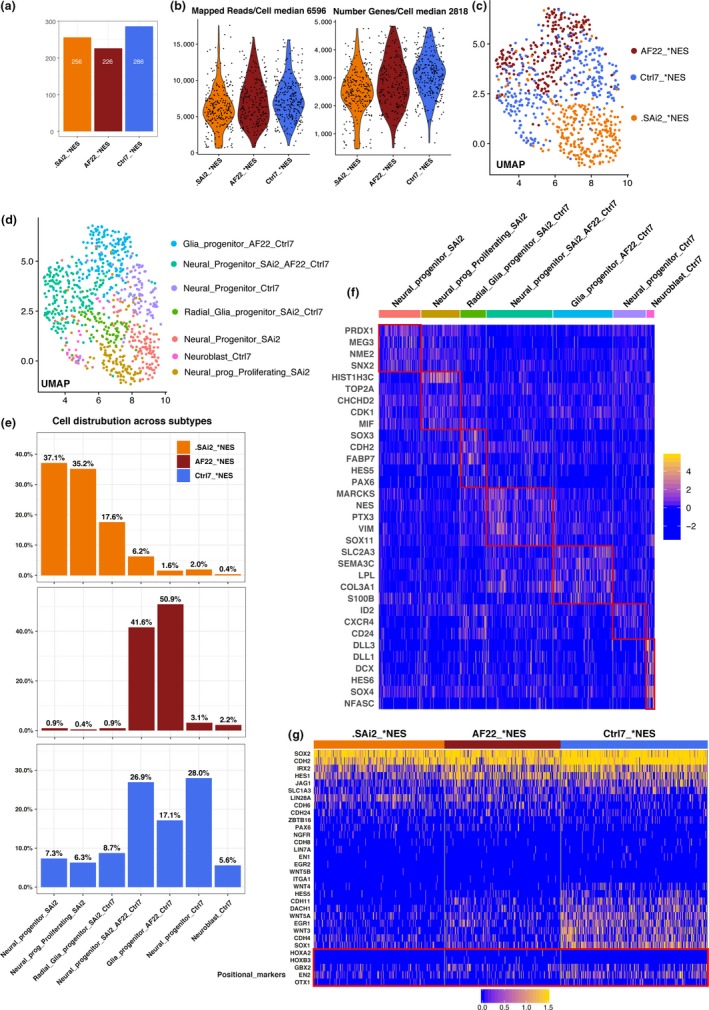
Characterizing heterogeneity of progenitors in fetal neural stem cells SAi2‐ and iPS‐derived neural stem cells AF22 and Ctrl7. (a) Barplot cell number across each neural stem cell lines. (b) Violin plots of mapped reads/cell and number of genes expressed/cell. (c) UMAP plot, location of neural stem cell line identity across cells. (d) UMAP plot, neurogenic and gliogenic progenitor identity distributed across cells. (e) Barplots percent contribution of each neural stem cell line to neurogenic and gliogenic progenitors. (f) Heat map genes enriched in neurogenic and gliogenic progenitors, adjusted p‐value. (g) Heat map of unclustered selected genes displays pluripotent, ectoderm, regional and positional identity across neural stem cell lines
We obtained high‐quality single cells across the NSC lines and estimated median 6,596 mapped exonic reads/cell and median 2,818 genes expressed/cell (Figure 2b). We used Seurat v3 (Butler et al., 2018) and carried out cell clustering and aggregation of gene expression profiles based on 10 KNN and plotted UMAP plot to visualize cell line identity and observed SAi2 NES and AF22 NES separated from each other while Ctrl7 NES localized between the other two lines (Figure 2c). Single cells clustered into 7 clusters, and we looked at distribution across clusters on UMAP plot and barplots in percent for each cell line to visualize overall cell type contribution profiles. Here, we observed AF22 NES mainly consisting of two main progenitor cell clusters at passage 20 (middle barplot Figure 2e); this was also observed at later passage 52 for RamDA‐seq (Figure 1c,e). Overall, we observed SAi2 and AF22 in majority contributing to nonoverlapping cell clusters while Ctrl7 contributed at a lesser extent to all clusters while simultaneously contributing its own unique cluster (Figure 2d,e). This observation suggests intrinsic cell line homogeneity that differs between AF22 NES and SAi2 NES based on overall gene expression profiles. In light of this observation, Ctrl7 NES holds a higher degree of heterogeneity at the NES cell stage when viewed side by side with AF22 NES and SAi2 NES cell lines.
Looking for overview of gene expression, we used heat map to visualize selected genes enriched in separate clusters and overlapping cells lines based in lowest adjusted p‐value as follows: Neural_progenitors_SAi2 (PRDX1, MEG3, NME2, SNX2), Neural_progenitors_proliferating_SAi2 (HIST1H3C, TOP2A, CHCHD2, CDK1, MIF), Radial_glia_progenitor_SAi2_Ctrl7 (SOX3, CDH2, FABP7, HES5, PAX6), Neural_progenitor_SAi2_AF22_Ctrl7 (MARCKS, NES, PTX3, VIM, SOX11), Glia_progenitor_AF22_Ctrl7 (SLC2A3, SEMA3C, LPL, COL3A1, S100B), neural_progenitor_Ctrl‐7 (ID2, CXCR4, CD24) and Neuroblast_Ctrl‐7 (DLL3, DLL1, DCX, HES6, SOX4, NFASC; Figure 2f).
We grouped the NSC lines separately and plotted selected genes across known developmental, regional and positional identity and observed an absence or decreased gene expression of pluripotency (LIN7A), neural crest (ITGA1), notochord (WNT5B) and spinal cord‐related (HOXA2, HOXB3) genes. Positive gene expression across cell lines demonstrated consensually known neural stem cell identity (i.e., SOX2, CDH2, HES1, JAG1) and spanning the presence of genes corresponding to hind‐ (GBX2), hind‐/mid‐ (EN2) and mid‐/forebrain (OTX1) positional regions. We also observed CDH6 and CDH24 preferentially enriched in SAi2 NES and AF22 NES. CDH4 and CDH11 were found enriched in Ctrl7 NES (Figure 2g). These observations again suggest cell adhesion molecules are related to important aspects of NSC identity and both neurogenic progenitors and gliogenic progenitors differentiation capacity. Overall, we found confirmation of inherent heterogeneity for both neurogenic progenitors and gliogenic progenitors in established NSC lines and our observation confirm previous publication describing character of Ctrl7 NES cell line (Lam et al., 2019).
2.3. Nondirected differentiation of neural stem cells allows for proliferation of neurogenic progenitors and differentiation of neurons
Next, we wanted to investigate the differentiation potential in SAi2, AF22, Ctrl7 NSC lines and captured neural stem cells in nondirected differentiation to neurons (DIFF) at 4 weeks by removal of growth factors. Again using C1 Fluidigm single‐cell capture system and STRT‐seq protocol, we obtained high‐quality cells with median 3,027 mapped exonic reads/cell and median 1,595 genes expressed/cell (Figure 3a,b).
Figure 3.
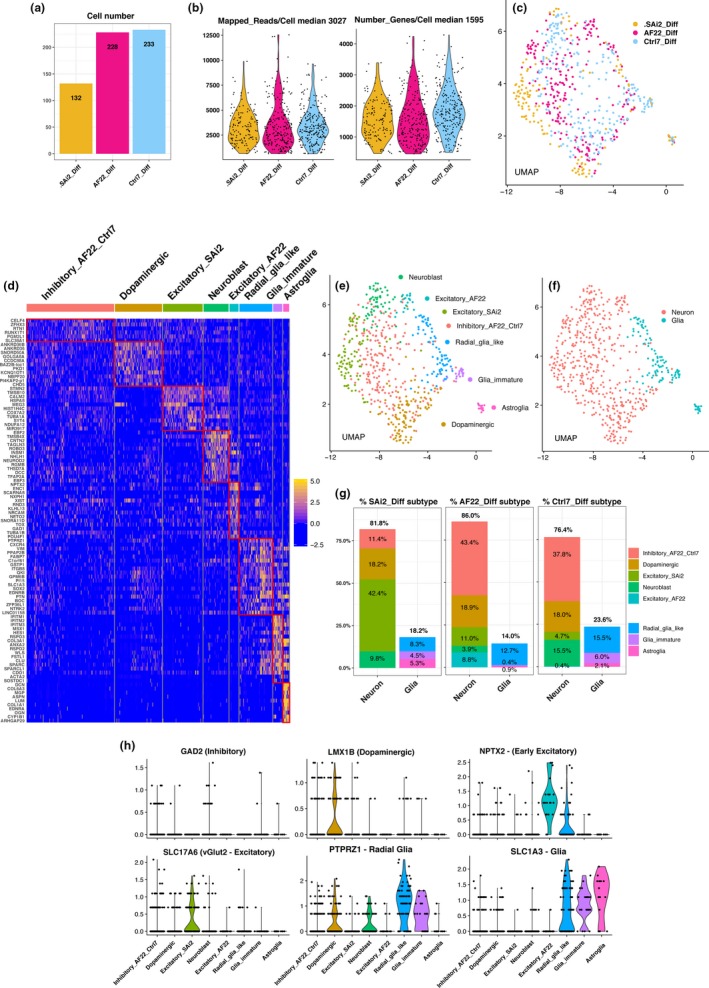
SAi2, AF22 and Ctrl‐7 NSC lines differentiated to neurons and glia show heterogeneity in neuronal subtypes. (a) Barplot cell number across differentiated cells. (b) Violin plots of mapped reads/cell and number of genes expressed/cell. (c) UMAP plot, differentiated cells across cell lines. (d) Heat map genes enriched across neuronal and glial subtypes, fold change. (e) UMAP plot neuronal and glial subtypes. (f) UMAP plot, neurons and glia. (g) Barplots percent of cell subtypes distributed across neurons and glia over cell lines. (h) Cell subtype markers, inhibitory neurons (GAD2), dopaminergic neurons (LMX1B), early excitatory neurons (NPTX2), excitatory neurons (SLC17A6), radial glia (PTPRZ1) and glia (SLC1A3)
We used Seurat v3 (Butler et al., 2018) to perform cell clustering and aggregation of gene expression profiles based on 10 KNN. We plotted single cells onto UMAP plot for visualization and observed intermingling cells across all cell lines (Figure 3c).
We looked at selected enriched gene expression based on fold change in cell clusters and annotated the cell clusters as inhibitory neurons enriched in AF22 DIFF and Ctrl7 DIFF (Inhibitory_AF22_Ctrl7), dopaminergic neurons enriched in all differentiated NSC lines (Dopaminergic_All), excitatory neurons enriched in SAi2 (Excitatory_SAi2), neuroblast, early excitatory neurons enriched in AF22 (Excitatory_AF22) and radial glia like, immature glia and astroglia (Figure 3d). Again, we used UMAP plot to visualize cell identity by overlaying cell subtype and neuron or glia identity and observed a clear separation between the cell subtypes and also between neuron and glia (Figure 3e,f).
Looking at the contribution of cell subtypes spanning across the nondirected differentiated NSC lines, we observed an overall approximate outcome of differentiation for neurons (80%) and glia (20%). We observed that SAi2 DIFF cells differentiate in majority to excitatory neurons (42.4%) while AF22 DIFF and Ctrl‐7 DIFF shared a similar preferential nondirected differentiation outcome of inhibitory neurons (43.4% and 37.8%, respectively). All NSC lines demonstrated differentiation capacity for dopaminergic neurons (~18% across all lines) and radial glia (SAi2 8.3%, AF22 12.7%, Ctrl7 15.5%). SAi2 DIFF and Ctrl‐7 DIFF showed the presence of retained differentiation capacity with the presence of neuroblast (9.8% and 15.5%, respectively) while AF22 DIFF contained cell cluster of early excitatory neurons (8.8%; Figure 3g).
Using violin plots to show the distribution and probability density of the data, we present neuronal subtype‐enriched genes for inhibitory neurons (GAD2), dopaminergic (LMX1B), early excitatory neurons (NPTX2), excitatory neurons (SLC17A6 or vesicular glutamate transporter vGlut2), radial glia (PTPRZ1) and glia lineage (SLC1A3; Figure 3h).
In conclusion, we observed nondirected differentiation of NES cells generating a wide variety of neuronal and non‐neuronal cells. Furthermore, combined observations from cell cluster analysis of NES cells and DIFF cells suggest heterogeneous neurogenic progenitors being important for neuronal differentiation and more specifically, in the predestined generation of neuronal subtypes.
2.4. Pattering on neural stem cells allows for enrichment of gliogenic progenitors and differentiation of astroglia
Inspired to investigate our observation of gliogenic progenitors in NES cells (Figure 1e), we used recently published NES‐Astro patterning protocol to study the potential of gliogenic progenitors derived from C9 NES cells (Lundin et al., 2018).
We captured C9 NES cells at 5 time points across the 4 weeks NES‐Astro patterning protocol and obtained high‐quality single cells with median 2,524 mapped exonic reads/cell and median 1,278 genes expressed/cell (Figure 4a).
Figure 4.
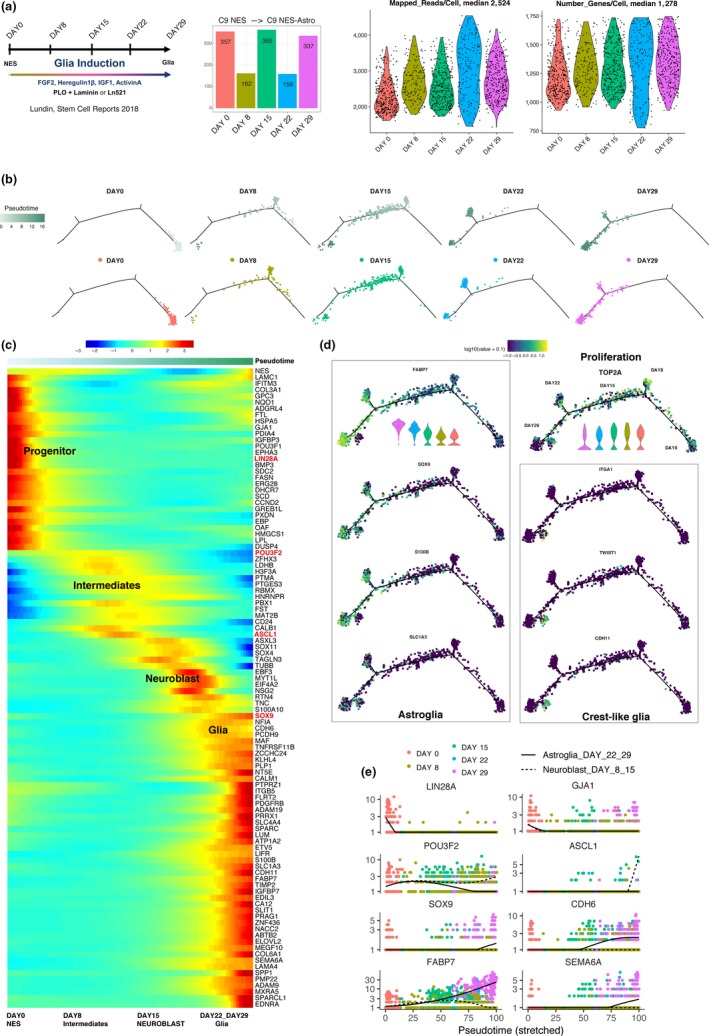
Gliogenic progenitor enrichment and glia differentiation from C9 NES cell line. (a) Barplot number of cells across cell lines and violin plots of mapped reads/cell and number of genes expressed/cell. (b) Pseudotime trajectory and day time points separated by day time points. (c) Heat map cells ordered by pseudotime display selected genes expressed by neural stem cells DAY0, intermediates (DAY8), neuroblast (DAY15) and astroglia (DAY22, DAY29), adjusted p‐value. (d) Gene expression profiles over trajectory, proliferation (TOP2A), astroglia (SOX9, FABP7, S100B, SLC1A3), crest‐like glia (ITGA1, TWIST1, CDH11), guide violin plots for FABP7 and TOP2A expression over days (DAY29, DAY22, DAY15, DAY8, Day0) in log10 scale. (e) Gene expression over time points for neural stem cells (LIN28A, GJA1), branching for spontaneous neuronal differentiation (POU3F2, ASCL1) and glial enrichment (SOX9, CDH6, FABP7, SEMA6A)
We used Monocle 2 (Qiu et al., 2017) to model a pseudotemporal developmental trajectory and matched time points of sampling with convergent correspondence between pseudotime and sampling time points across the single‐cell data set (Figure 4b).
We used heat map of patterned cells ordered by pseudotime, visualized selected genes and observed NES cells at DAY0 transitioning through intermediates at DAY8, neuroblast at DAY15 and glia cells emerging at DAY22 to DAY29 (Figure 4c).
Looking at genes defining gliogenic progenitor potential and glial cells, we observed TOP2A expression in actively proliferating cells up until DAY29 when emerging glia cells gradually slow down and halt in proliferation. We observed branching off at specific time points suggesting cells at distinct stages during patterning will acquire distinct gene expression profiles unique for the time of capture (Figure 4d). Further, we observed increasing gene expression corresponding to glia cells (SOX9, FABP7, S100B, SLC1A3) and crest‐like glia (ITGA1, TWIST1, CDH11; Figure 4d).
At DAY 8 and DAY15, we observed neurogenic genes suggesting nondirected neuronal differentiation (POU3F2, ASCL1) and ramping up of gene expression related to glial enrichment (SOX9, FABP7, CDH6, SEMA6A; Figure 4e).
All together, combined observations reveal successful enrichment of gliogenic progenitors and patterning toward glia. All the while, simultaneously nondirected neuronal differentiation will occur originating from subset of neurogenic progenitors.
3. DISCUSSION
In our study, we shed light on the definition and genes that govern the concept of a “neural stem cell.” Our observations set the perspective on an inclusive interpretation that committed neurogenic and gliogenic progenitors already co‐exist inside the cell population of established NSC lines and with proper cell culture handling persist over long‐term culture. We propose a summary model of occurring events for both neuronal differentiation and glial differentiation in Figure 5. This added perspective supports interpretation of defining progenitor populations residing in established NSC lines in past, current and future studies in the field of in vitro modeling of neurogenesis and gliogenesis.
Figure 5.
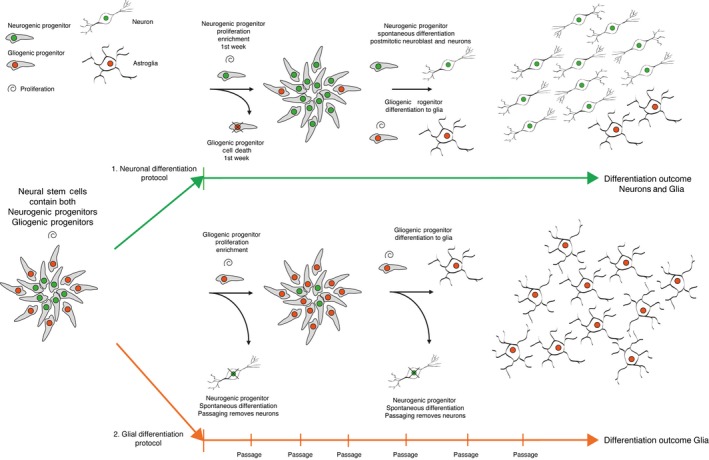
Summary model of occurring events in neurogenic and gliogenic differentiation. Neural stem cells inherently contain both neurogenic progenitors (~60%) and gliogenic progenitors (~40%). Upon differentiation with two separate protocols, in each protocol parallel set of events occurs. (1) For neuronal differentiation, in first few days neurogenic progenitors will continue to proliferate and a portion of gliogenic progenitors die off, then neurogenic progenitors undergo spontaneous differentiation to neuroblasts and further to neurons (80%). Remaining surviving gliogenic progenitors differentiate to glia (20%). (2) For glial differentiation, gliogenic progenitors continue to proliferate and neurogenic progenitors spontaneously differentiate to neuroblast and neurons, the cell culture remains in a proliferative state and the cells are repeatedly passaged to maintain optimal condition for cell culture. Repeated passage removes spontaneously differentiating postmitotic neurons and enriches gliogenic progenitors differentiating to glia
In this setting, nonoverlapping neurogenic progenitor identities of SAi2 NES and AF22 NES could help to predict outcome of neuronal subtype differentiation (Figure 2e). This meaning the nondirected neuronal differentiation protocol that involves removal of growth factors FGF2 and EGF will in the end result in different neuron subtypes based on the preset identity of the neurogenic progenitor. From our experience, observing NES cells over the first week of nondirected differentiation, three sequential events occur. First, NES cells continue to proliferate for the first 2–3 days, second, massive cell death occurs for the next 1–2 days and third, visible neuroblast emerge around day 7. These observed events suggest proliferating neurogenic progenitors thrive in the nondirected differentiation culture condition for the first few days and gliogenic progenitors do not, as seen in the massive cell death (1. neuronal differentiation protocol in Figure 5). The outcome of 80% neurons and 20% glia at 4 weeks of differentiation suggests higher overall survival of neurogenic progenitors over gliogenic progenitors during initiation of differentiation (Figure 3g).
NES‐Astro patterning protocol starting with NES cells with expression of LIN28A, (reported neural progenitor marker by Yang et al. (2015)) with gliogenic progenitor enrichment results in the differentiation of glial cells. During 4 weeks of patterning, NES‐Astro cells will continue to proliferate and in cell culture handling become split and reseeded for maintenance of optimal growth conditions. This action will inadvertently remove spontaneously differentiated postmitotic neurons originating from neurogenic progenitors expressing POU3F2 and ASCL1 (factors together with MYT1L used for direct reprogramming of fibroblast to neurons by Pang et al. (2011)) and dilute out the neurogenic subpopulation in the patterned NES‐Astro cell culture and enrich for SOX9 expressing gliogenic progenitors (reported glial marker by Kang et al. (2012); Pseudotime heat map Figure 4c; 2. glial differentiation protocol Figure 5).
Our combined observations demonstrate the power of analysis in single‐cell sequencing and in delivering high‐resolution perspective needed to uncover previously unattained observations in the study of biology. The proposed model of events in Figure 5 is perceived by the combined observations across single‐cell RNA‐seq analysis and summarizes the differentiation potential of established NSC lines. Uncovering the presence of neurogenic and gliogenic progenitors in NSC lines show clarity in the interpretation of definition for neural stem cell potential in the in vitro setting.
Speculatively, the proliferating neurogenic progenitors could represent the “true” neural stem cell and in the setting of modeling the developing neural tube corresponds to cells lining the inner wall of the neural cavity. Although our investigation adds information to understanding the usage of cell models for concept and action of modeling neurodevelopment, we acknowledge that further study will be needed to uncover additional aspects regarding the true neural stem cell.
4. EXPERIMENTAL PROCEDURES
4.1. Neural stem cell culture
Fetal‐derived neural stem cell line SAi2 (Tailor et al., 2013), iPSCs‐derived NSC line AF22 (Falk et al., 2012), iPSCs‐derived NSC line Ctrl‐7 (Lam et al., 2019) and iPSCs‐derived NSC line C9 (Lundin et al., 2018) were cultured as adherent cells at a substrate of 20 μg/ml polyornithine (Sigma) and 1 μg/ml Laminin2020 (Engelbreth‐Holm‐Swarm murine sarcoma, Sigma). NES culture medium contained DMEM/F12+GlutaMax (Gibco), supplemented with 10 μl/ml N‐2‐supplement (100×, Thermo Fisher Scientific), 10 μl/ml Penicillin‐Streptomycin (10,000 U/ml, Thermo Fisher Scientific), 1 μl/ml B27‐supplement (50×, Thermo Fisher Scientific), 10 ng/ml of bFGF (Life Technologies) and 10 ng/ml of FGF (PeproTech). The culture medium was replaced every second day. The NES cells were passaged enzymatically when reaching 100% confluency using Trypsin‐EDTA (0.025%, Thermo Fisher Scientific), the enzymatic reaction was inhibited by defined trypsin inhibitor (Thermo Fisher Scientific) at equal volume. NES cells were seeded at a density of 40,000 cells/cm2.
4.2. Differentiation of neural stem cells
Neuroepithelial stem cell medium was replaced by differentiation medium containing DMEM/F12+GlutaMax (Gibco), supplemented with 10 μl/ml B27‐supplement (50×, Thermo Fisher Scientific), 10 μl/ml N‐2‐supplement (100×, Thermo Fisher Scientific) and 10 μl/ml Penicillin‐Streptomycin (10,000 U/ml, Thermo Fisher Scientific). Differentiation medium was replaced every second to third days, and 1 μg/ml Laminin (Engelbreth‐Holm‐Swarm murine sarcoma, Sigma) was added to the differentiation medium from day 14 and onwards throughout the differentiation. The cells were passaged once between days 3 and 5 of differentiation.
4.3. NSCs‐derived astroglia differentiation
Neural stem cell line C9 cells passage 18 were plated at 60,000 cells/cm2 on 2 mg/cm2 poly‐l‐ornithine and 0.2 mg/cm2 laminin (PLO‐Laminin; Sigma) double‐coated culture vessels in FHIA differentiation medium; DMEM/F12, N2 supplement (1:100; Invitrogen), B27 (1:100; Invitrogen), FGF2 (8 ng/ml; PeproTech), heregulin1b (10 ng/ml; Sigma), IGF1 (200 ng/ml; Sigma) and activin A (10 ng/ml; PeproTech). The medium was changed every other day, and cells were passaged once they reached 80% confluency; 7–9 passages during the differentiation protocol of 28 days (Lundin et al., 2018).
4.4. RamDA‐seq
AF22 NES cells at passage 52 were sorted into 96‐well plate and subjected to RamDA‐seq total RNA sequencing protocol (Hayashi et al., 2018) and processed through deep sequencing at 20 million reads per cell. For analysis, high‐quality single cells were selected through cutoff to fit between 400,000 and 800,000 mapped reads/cell.
4.5. STRT‐seq for neural stem cells and differentiated neurons
Sai2 NES cells passage 29, AF22 NES cells passage 20 and Ctrl‐7 NES cells passage 12 were harvested as neural stem cells at day 0 and Sai2 DIFF passage 27, AF22 DIFF passage 16 and 23, Ctrl‐7 DIFF passage 12 as differentiated neurons at day 28 of differentiation.
The cells were dissociated as described under neural stem cell culture. The dissociated cells were passaged through a cell strainer (40 μm, VWR) and diluted to 2,000 cells/ml in NES medium with addition of 5% DNaseI (2,000 U/ml, Qiagen) and 1% BSA (Sigma). Cells were placed on ice until further processed. The cells were loaded according to manufacturers protocol on C1 Single‐Cell AutoPrep IFC microfluidic chip (for cell size 10–17 μm) and processed on a Fluidigm C1 instrument. This chip contains 96 wells for single‐cell capture.
Each chamber of the chip was optically inspected by automated microscope (Nikon TE2000E), empty chambers, cell clumps and dying cells were discarded from further analyze. Lysis of the cells and cDNA production were carried out according to the Linnarsson's laboratory protocol (Islam et al., 2014). For analysis, high‐quality single cells were selected through cutoff to fit between 500 and 5,000 genes/cell.
4.6. Droplet‐based single cells for NES‐Astro data set
Cells undergoing NES‐Astro protocol were detached and resuspended to a single‐cell suspension. Cell count, viability and aggregation level were assessed using the Cedex instrument. The Illumina® Bio‐Rad® SureCell WTA 3′ Library Prep Kit for the ddSEQ System (Illumina) was used for preparation of the single‐cell suspension and library workflow. All libraries were quantified with the Bioanalyzer using the High Sensitivity DNA kit (Agilent Technologies). Libraries were pooled and quantified using the Qubit instrument, DNA HS kit (Thermo Fisher Scientific), and the library pool was further diluted to 1.7 pM. Pair‐end sequencing was carried out using a High Output Kit v2 (150 cycles) on an Illumina NextSeq500.
For analysis, high‐quality single cells were selected through cutoff to fit between 800 and 1,800 genes/cell.
4.7. Data analysis
Seurat 3 (version 3.0.2) standard analysis pipeline was used for analysis of AF22 NES RAMDA‐seq (20 million reads/cell), AF22 STRT‐seq (NES and DIFF) and SAi2 STRT‐seq (NES and DIFF).
Monocle 2 (version 2.12.0) standard analysis pipeline was used for pseudotemporal ordering and trajectory projection for C9 NES‐Astro single cells.
AUTHOR CONTRIBUTIONS
Matti Lam conducted the single‐cell analysis investigation, created all figures and wrote the manuscript. Tsukasa Sanosaka, Anders Lundin, Kent Imaizumi, Hideyuki Okano and Anna Falk reviewed and proposed suggestions of improvement for manuscript. Okano laboratory members at Keio University generated data used for analysis in creation of Figure 1. Falk laboratory members at Karolinska Institute generated data used for analysis in creation of Figures 2 and 3. AstraZeneca employees generated data used in analysis for creation of Figure 4.
ACKNOWLEDGMENTS
Authors thank the iPS Core (ipscore.se) and ESCG (escg.se) facilities at Karolinska Institute. Anders Lundin is part of AstraZeneca's Post‐Doc Program. Anders Lundin, Damla Etal, Fredrik H. Karlsson, Maryam Clausen and Jonathan Cairns are employees of AstraZeneca.
Lam M, Sanosaka T, Lundin A, et al. Single‐cell study of neural stem cells derived from human iPSCs reveals distinct progenitor populations with neurogenic and gliogenic potential. Genes Cells. 2019;24:836–847. 10.1111/gtc.12731
Communicated by: Takashi Tada
DATA AVAILABILITY STATEMENT
Figure 1 raw data and processed data sets are available at NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136611). Figures 2, 3 and 4 processed data sets are available at Array Express (E‐MTAB‐8363, E‐MTAB‐8383 and E‐MTAB‐8379).
REFERENCES
- Alvarez‐Buylla, A. , Garcia‐Verdugo, J. M. , & Tramontin, A. D. (2001). A unified hypothesis on the lineage of neural stem cells. Nature Reviews Neuroscience, 2, 287–293. 10.1038/35067582 [DOI] [PubMed] [Google Scholar]
- Butler, A. , Hoffman, P. , Smibert, P. , Papalexi, E. , & Satija, R. (2018). Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology, 36, 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi, M. , Yamamoto, Y. , Sharifi, K. , Kida, H. , Kagawa, Y. , Yasumoto, Y. , … Owada, Y. (2016). Astrocyte‐expressed FABP7 regulates dendritic morphology and excitatory synaptic function of cortical neurons. Glia, 64, 48–62. 10.1002/glia.22902 [DOI] [PubMed] [Google Scholar]
- Falk, A. , Koch, P. , Kesavan, J. , Takashima, Y. , Ladewig, J. , Alexander, M. , … Brüstle, O. (2012). Capture of neuroepithelial‐like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS ONE, 7, e29597 10.1371/journal.pone.0029597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremeau, R. T. Jr , Voglmaier, S. , Seal, R. P. , & Edwards, R. H. (2004). VGLUTs define subsets of excitatory neurons and suggest novel roles for glutamate. Trends in Neurosciences, 27, 98–103. 10.1016/j.tins.2003.11.005 [DOI] [PubMed] [Google Scholar]
- Gage, F. H. , & Temple, S. (2013). Neural stem cells: Generating and regenerating the brain. Neuron, 80, 588–601. 10.1016/j.neuron.2013.10.037 [DOI] [PubMed] [Google Scholar]
- Gleeson, J. G. , Lin, P. T. , Flanagan, L. A. , & Walsh, C. A. (1999). Doublecortin is a microtubule‐associated protein and is expressed widely by migrating neurons. Neuron, 23, 257–271. 10.1016/S0896-6273(00)80778-3 [DOI] [PubMed] [Google Scholar]
- Hayashi, T. , Ozaki, H. , Sasagawa, Y. , Umeda, M. , Danno, H. , & Nikaido, I. (2018). Single‐cell full‐length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nature Communications, 9, 619 10.1038/s41467-018-02866-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, S. , Zeisel, A. , Joost, S. , La Manno, G. , Zajac, P. , Kasper, M. , … Linnarsson, S. (2014). Quantitative single‐cell RNA‐seq with unique molecular identifiers. Nature Methods, 11, 163–166. 10.1038/nmeth.2772 [DOI] [PubMed] [Google Scholar]
- Kang, P. , Lee, H. K. , Glasgow, S. M. , Finley, M. , Donti, T. , Gaber, Z. B. , … Deneen, B. (2012). Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron, 74, 79–94. 10.1016/j.neuron.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, M. , Moslem, M. , Bryois, J. , Pronk, R. J. , Uhlin, E. , Ellström, I. D. , … Falk, A. (2019). Single cell analysis of autism patient with bi‐allelic NRXN1‐alpha deletion reveals skewed fate choice in neural progenitors and impaired neuronal functionality. Experimental Cell Research, 383(1), 111469– 10.1016/j.yexcr.2019.06.014 [DOI] [PubMed] [Google Scholar]
- Linnarsson, S. , & Teichmann, S. A. (2016). Single‐cell genomics: Coming of age. Genome Biology, 17, 97 10.1186/s13059-016-0960-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin, A. , Delsing, L. , Clausen, M. , Ricchiuto, P. , Sanchez, J. , Sabirsh, A. , … Falk, A. (2018). Human iPS‐derived astroglia from a stable neural precursor state show improved functionality compared with conventional astrocytic models. Stem Cell Reports, 10, 1030–1045. 10.1016/j.stemcr.2018.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain, C. J. , & Fisahn, A. (2001). Interneurons unbound. Nature Reviews Neuroscience, 2, 11–23. 10.1038/35049047 [DOI] [PubMed] [Google Scholar]
- Mertens, J. , Marchetto, M. C. , Bardy, C. , & Gage, F. H. (2016). Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nature Reviews Neuroscience, 17, 424–437. 10.1038/nrn.2016.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani, T. , Kumai, M. , Mizuhara, E. , Minaki, Y. , & Ono, Y. (2010). Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Developmental Biology, 339, 101–113. 10.1016/j.ydbio.2009.12.017 [DOI] [PubMed] [Google Scholar]
- Okano, H. , & Yamanaka, S. (2014). iPS cell technologies: Significance and applications to CNS regeneration and disease. Molecular Brain, 7, 22 10.1186/1756-6606-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, Z. P. , Yang, N. , Vierbuchen, T. , Ostermeier, A. , Fuentes, D. R. , Yang, T. Q. , … Wernig, M. (2011). Induction of human neuronal cells by defined transcription factors. Nature, 476, 220–223. 10.1038/nature10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkey, K. A. , Barksdale, E. , Craig, M. T. , Yuan, X. , Sukumaran, M. , Vargish, G. A. , … McBain, C. J. (2015). Pentraxins coordinate excitatory synapse maturation and circuit integration of parvalbumin interneurons. Neuron, 85, 1257–1272. 10.1016/j.neuron.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen, A. A. , Nowakowski, T. J. , Chen, J. , Retallack, H. , Sandoval‐Espinosa, C. , Nicholas, C. R. , … Kriegstein, A. R. (2015). Molecular identity of human outer radial glia during cortical development. Cell, 163, 55–67. 10.1016/j.cell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X. , Mao, Q. , Tang, Y. , Wang, L. , Chawla, R. , Pliner, H. A. , & Trapnell, C. (2017). Reversed graph embedding resolves complex single‐cell trajectories. Nature Methods, 14, 979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shparberg, R. A. , Glover, H. J. , & Morris, M. B. (2019). Modeling mammalian commitment to the neural lineage using embryos and embryonic stem cells. Frontiers in Physiology, 10, 705 10.3389/fphys.2019.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov, R. , Kaucka, M. , Kastriti, M. E. , Petersen, J. , Chontorotzea, T. , Englmaier, L. , … Adameyko, I. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science, 364(6444), eaas9536 10.1126/science.aas9536 [DOI] [PubMed] [Google Scholar]
- Tailor, J. , Kittappa, R. , Leto, K. , Gates, M. , Borel, M. , Paulsen, O. , … Smith, A. (2013). Stem cells expanded from the human embryonic hindbrain stably retain regional specification and high neurogenic potency. Journal of Neuroscience, 33, 12407–12422. 10.1523/JNEUROSCI.0130-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K. , Tanabe, K. , Ohnuki, M. , Narita, M. , Ichisaka, T. , Tomoda, K. , & Yamanaka, S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Taniguchi, H. , He, M. , Wu, P. , Kim, S. , Paik, R. , Sugino, K. , … Huang, Z. J. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron, 71, 995–1013. 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Yang, S.‐L. , Herrlinger, S. , Liang, C. , Dzieciatkowska, M. , Hansen, K. C. , … Chen, J.‐F. (2015). Lin28 promotes the proliferative capacity of neural progenitor cells in brain development. Development, 142, 1616–1627. 10.1242/dev.120543 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Figure 1 raw data and processed data sets are available at NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136611). Figures 2, 3 and 4 processed data sets are available at Array Express (E‐MTAB‐8363, E‐MTAB‐8383 and E‐MTAB‐8379).


