Abstract
Which comes first: tissue structure or cell differentiation? Although different cell types establish distinct structures delineating the inside and outside of an embryo, they progressively become specified by the blastocyst stage, when two types of cell lineages are formed: the inner cell mass (ICM) and the trophectoderm (TE). This inside–outside aspect can be experimentally converted by the isolation of the ICM from a blastocyst, leading to a posteriori externalization of the blastomeres composing the outermost layer of the ICM. Here, we investigated the totipotency of isolated mouse and bovine ICMs to determine whether they are competent for TE regeneration. Surprisingly, a calf was generated from the bovine isolated ICM with re-formed blastocoel (re-iICM), but no mouse re-iICMs developed to term. To further explore the cause of difference in developmental competency between the mouse and bovine re-iICMs, we investigated the SOX17 protein expression that is a representative molecular marker of primitive endoderm. The localization pattern of SOX17 was totally different between mouse and bovine embryos. Particularly, the ectopic SOX17 localization in the TE might be associated with lethality of mouse re-iICMs. Meanwhile, transcriptome sequencing revealed that some of the bovine re-iICMs showed transcriptional patterns of TE-specific genes similar to those of whole blastocysts. Our findings suggest that TE regeneration competency is maintained longer in bovine ICMs than in mouse ICMs and provide evidence that the ICM/TE cell fate decision is influenced by structural determinants, including positional information of each blastomere in mammalian embryos.
Keywords: embryo, gene expression, Hippo pathway, mouse, protein–protein interaction, Yes-associated protein (YAP), blastocyst, cattle, inner cell mass, trophectoderm
Introduction
Preimplantation development in all eutherians involves the blastocyst stage with a fluid-filled cavity (blastocoel), in which the first cell fate decision occurs. This decision is a fork in the road to either the pluripotent inner cell mass (ICM)2 or an extra-embryonic tissue, the trophectoderm (TE), during ontogenesis. Cell lineage asymmetry becomes specified in the blastomeres at different positions: inside and outside. Once blastomeres are allocated to the different positions, outer blastomeres asymmetrically or symmetrically divide, resulting in the generation of one inner and the other outer daughter blastomeres or both outer daughter blastomeres, respectively (1, 2). The blastomeres allocated to different positions are restricted to specific developmental fates. As blastomere allocation with cleavages proceeds, polarization begins (3, 4). The outer blastomeres retain the polarized apical features, whereas the inner ones become morphologically apolar. Positional allocation of blastomeres concomitant with polarization has been shown to influence the election to become either ICM or TE (5, 6), which has also been confirmed by numerous studies (7–10).
In mice, differences in blastomere position become apparent at the 16-cell stage; subsequently, lineage-specific transcriptional profiles are simultaneously composed and completed by the 32-cell stage, embryonic day (E) 3.0 (11). By E4.5 (128-cell stage), the primitive endoderm (PrE) is specified within the ICM to give rise to the yolk sac, which is the second cell fate decision after the first ICM/TE decision. The blastomeres other than PrE within the ICM are epiblast, which gives rise to the embryo proper. The PrE layer is arranged in the laminae adjacent to the blastocoel. After these blastomere specifications, three components are generated to construct a whole conceptus: epiblast, PrE, and TE. Thus, the different microenvironments provided by the inside and outside blastomeres lead to and ensure each blastomere specification through the accommodation of key differentiation markers to determine lineage-specific transcriptional profiles.
These microenvironments can be experimentally redesigned by isolating the ICM derived from a mouse blastocyst to explore the plasticity of cell fate decisions (12). The isolation of the ICM by TE removal leads to a posteriori externalization of the blastomeres composing the outermost layer of the ICM that was originally allocated to the inside beneath an epithelial TE layer. The isolated ICM (iICM) from mouse blastocyst has the ability to regenerate the TE because of the inactivation of Hippo signaling, which is essential for the conversion of information regarding blastomere position, polarity, and contact into cell lineage–specific transcriptional commands (12). The Hippo signaling effector YAP1 is a key regulator of cell lineage specificity in developing embryos. A nucleocytoplasmic shift in YAP1 localization characterizes the boundary between the ICM and TE within a blastocyst. Hippo signaling inactivation induces the nuclear localization of YAP1, resulting in CDX2 expression through interaction with TEAD4 in the TE (13). Interestingly, in mouse iICMs, CDX2, which commits cells to the TE lineage, as well as GATA4, an early marker for the PrE lineage, are expressed in the outer blastomeres, i.e. the “regenerated TE” (12). In contrast, the TE of a normal mouse blastocyst never expresses PrE markers, including SOX17 (14, 15). However, whether this “regenerated TE” observed in the iICM is equivalent to the “intrinsic TE” has not yet been determined.
Another important question is whether the iICM expressing lineage-specific transcription factors for three primary cell lineages is totipotent (defined as the developmental ability to form an entire individual). Expression of the complete set of lineage-specific markers does not ascertain totipotency. For example, although somatic cell nuclear transfer embryos express all the lineage-specific marker genes (16–19), the efficiency of normal birth remains extremely low (20, 21), suggesting that most of the somatic cell nuclear transfer embryos do not possess totipotency. Hence, cell totipotency and the potential for full-term development can only be determined by transferring embryos to recipient females, which would provide conclusive evidence that positional information of blastomeres is essential for cell fate decision in mammalian embryos.
The microenvironmental events associated with cell fate specification at the blastocyst stage described above have rarely been investigated in nonrodent species other than mice. However, crucial differences exist in the molecular mechanisms that regulate cell fate specification at the blastocyst stage among species (22–24). For example, OCT4, a representative basis of cell pluripotency, is not restricted to the ICM in human and bovine blastocysts, unlike that in mouse (25, 26), suggesting that diverse regulatory mechanisms are applied in a species-specific developmental manner. Comparing cell specifications through the microenvironment between rodent and nonrodent species may contribute to our understanding of diverse regulation patterns during preimplantation development across species.
We determined the TE regeneration competency of the outmost blastomeres within the iICM by investigating the totipotency of both mouse and bovine iICMs. Mouse iICMs could not develop to term, but a calf was born from bovine iICM with regenerated TE. This difference in maintenance of TE regeneration competency between the species might be attributed to species specificity, in part, in the spatiotemporal regulation of SOX17. The regeneration of TE in bovine as well as mouse iICM was mediated by Hippo signaling. Some bovine iICMs exhibited normal expression levels of bovine TE-specific genes, which was supported by the well-developed placenta of the newborn cattle derived from the iICM. Our findings suggest that the TE regeneration competency is maintained in bovine ICMs for relatively longer than in mouse ICMs, revealing the remarkable flexibility in cell differentiation during preimplantation development in mammals.
Results
Totipotency of the ICM with regeneration of TE in mice and cattle
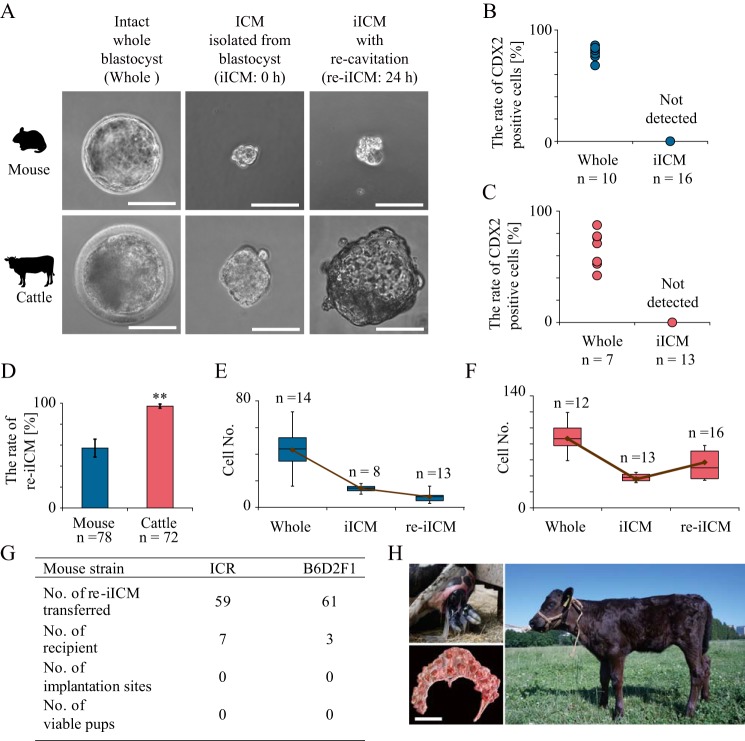
We assessed the competency of the iICMs to re-form a blastocoel (re-cavitation; Fig. 1A). The purity of the iICMs was confirmed on the basis of CDX2 fluorescent signals that characterize TE, and no TE carryover was noted in both mouse and bovine iICMs (Fig. 1, B and C). When the iICMs were cultured in the embryo-cultivation medium for 24 h, the formation of morphologically clear blastocoels was induced in both mouse and bovine iICMs (Fig. 1A). However, the size of the blastocoel in mouse iICMs with re-cavitation (re-iICMs) was markedly smaller than that in intact whole blastocysts. In contrast, the blastocoel of bovine re-iICMs was relatively well-developed, and the blastocoel size of some bovine re-iICMs was similar to that of intact whole blastocysts. The rate of re-iICMs to all the cultured iICMs in mice (57.1% ± 8.5) was remarkably lower than that in cattle (97.2% ± 2.0; Fig. 1D). For further comparison of re-iICMs between mice and cattle, we assessed the total cell number. Immediately after the isolation of ICMs, the cell numbers of both mouse and bovine iICMs decreased because of TE removal. The total cell number in mouse re-iICMs tended to decrease after 24 h of cultivation compared with that in iICMs immediately after isolation, both of which were considerably lower than that of intact whole blastocysts (Whole; Fig. 1E). In contrast, the total cell number in bovine re-iICM increased by ∼1.5-fold relative to that in the iICM immediately after isolation (Fig. 1F), which was equivalent to half of that in Whole. These results suggest that the competences of re-cavitation and cell proliferation in bovine re-iICM are relatively higher than those in mouse re-iICM.
Figure 1.
Totipotency of re-cavitated ICM after isolation from mouse and bovine blastocysts. ICMs isolated from each blastocyst were prepared by detergent treatment. A, representative images of left panels: Whole, intact whole blastocyst; center panels: iICM, ICM isolated from blastocyst before in vitro cultivation (0 h); right panels: iICM that re-formed blastocoels after 24 h of cultivation(re-iICM). Bar, 50 μm. B and C, cell numbers with CDX2 fluorescent signals to characterize TE were counted in Wholes and iICMs in mice (B) and cattle (C). No CDX2-positive blastomeres were detected in iICMs in both species. D, proportions of re-iICMs with a blastocoel in cultivated iICMs. Dark blue and light red bars denote mean values in mice and cattle, respectively. **, p < 0.01. E, box plots represent the cell number in mouse whole blastocyst, iICM, and re-iICM: the two quartiles, 25th and 75th percentile, form the box with the media marked as a line; the maximum and minimum values form the whiskers within the acceptable range defined by the two quartiles. The brown line denotes the mean values. F, cell number in bovine whole blastocyst, iICM, and re-iICM is represented as box plots shown in G. G, table represents the full-term developmental ability in mouse iICMs. Donor oocytes were prepared from two strains of ICR and B6D2F1 for producing re-iICMs. Embryo transfer was repeated at least three times in each mouse strain. No mouse re-iICMs transferred showed implantation sites and developed to term at E19.5. H, viable calf was generated by embryo transfer of four re-iICMs after a 24-h cultivation (Japanese black beef cattle × Holstein) to three recipient Holstein cows, one of which was pregnant. On gestation day 282, a female individual with black coat was born vaginally on June 24, 2018 (upper left), and the afterbirth was morphologically normal with 92 cotyledons (lower left; bar, 30 cm). The bottom photograph shows the calf 1 month after birth. We named the calf “Matoryona” as it originated from a TE-decapsulated iICM resembling a “Matryoshka,” a Russian nesting doll that separates to reveal smaller figures of the same sort inside.
Subsequently, to determine the totipotency of the re-iICMs in mice and cattle, we evaluated the full-term development potential after transfer to the uteri of recipient females. First, we transferred 59 mouse re-iICMs derived from ICR (Institute of Cancer Research). None of the mouse re-iICMs developed to term (Fig. 1G). In general, the developmental competence of mouse embryos from closed colonies, including ICR, is considerably lower than that of embryos from hybrids, such as B6D2F1 (C57BL/6 × DBA/2). Therefore, we transferred 61 B6D2F1 re-iICMs; nevertheless, none of them showed implantation sites and full-term development. Like in mice, we transferred four bovine re-iICMs to three recipient cows. We used oocytes derived from Japanese black beef cattle for producing iICMs because Japanese black beef cattle have a distinct black coat-color phenotype, and neonates derived from them are distinguishable from recipient Holstein cows. Among the recipient cows, one was pregnant. Surprisingly, on day 282 (June 24, 2018) of gestation, a female calf with black coat was born vaginally (Fig. 1H). The placenta discharged from the uterus after birth (afterbirth) was morphologically normal with 92 fetal cotyledons. The birth weight was also normal (46 kg). The birth of the calf provides definitive evidence that some bovine iICMs can regenerate functional TE to support full-term development. In contrast, no mouse iICMs developed to term, indicating that species-specific differences exist in the maintenance of totipotency during preimplantation development. However, mouse iICMs could also regenerate TE with primary differentiation markers such as CDX2, which has been supported by the findings of one previous study, although whether the regenerated TE was functional was not determined (12).
Resemblance and difference in TE regeneration from iICMs between mice and cattle
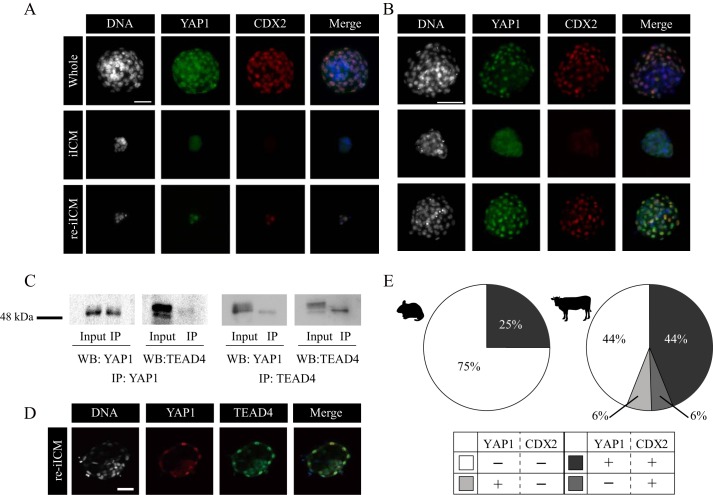
To explore how TE regeneration was achieved in mouse and bovine re-iICMs, we investigated CDX2 activation within the iICM by precisely observing the dynamics of three proteins: YAP1, TEAD4, and CDX2. In mouse normal embryos, the interaction between YAP1 and TEAD4 is believed to be an upstream mediator of CDX2 expression (13). YAP1 is shuttled between the nucleus and cytoplasm in a phosphorylation-dependent manner through the Hippo pathway, where the inside–outside spatial information translates into the transcriptional regulation of CDX2. We confirmed that YAP1 nuclear translocation in the outer blastomeres was coordinated with CDX2 expression during re-iICM formation in mice (Fig. 2A), which is consistent with the findings of a previous study (12).
Figure 2.
Localization patterns of YAP1 and CDX2 proteins in isolated ICMs derived from mouse and bovine blastocysts. A and B, dual immunofluorescence analysis of both YAP1 and CDX2 within Whole, iICM, and re-iICM in mice (A, bar, 50 μm) and cattle (B, bar, 100 μm). At least three embryos were analyzed for each experiment. YAP1 nuclear localization was absent in iICMs but present in the outer blastomeres in re-iICMs, regardless of species. C, immunoprecipitation (IP) with anti-YAP1 (left panel) or anti-TEAD4 (right panel) antibodies, followed by WB with anti-YAP1 and anti-TEAD4 antibodies by using bovine fetal fibroblasts. Input controls were nonimmunoprecipitated proteins and similarly used for WB with anti-YAP1 and anti-TEAD4 antibodies. D, localization patterns of YAP1 and TEAD4 proteins in re-iICM. TEAD4 was observed within all nuclei in all embryos. Bar, 50 μm. E, quantification of blastomeres with YAP1 nuclear localization and/or CDX2 expression in mouse (left) and bovine (right) re-iICMs. Light gray: YAP1-positive (+) and CDX2-negative (−) blastomeres; dark gray, YAP1+ and CDX2+ blastomeres; medium gray, YAP1− and CDX2+ blastomeres; white, YAP1− and CDX2− blastomeres. The values are represented as the percentage of the above four types of blastomeres to the total number of blastomeres.
However, little is known about this elegant regulatory mechanism of ICM/TE cell differentiation through the Hippo pathway during early bovine embryogenesis. To ensure the conserved molecular interaction in the Hippo pathway, we determined the localization patterns of both YAP1 and TEAD4 in intact bovine embryos from D5.0 to D6.5 (Fig. S1, A and B). Nuclear translocation of YAP1 in the outer cells began from D5.0 to D5.5 and was completed in the outermost blastomeres by the early blastocyst stage at D6.0 (Fig. S1A). In addition, TEAD4 was localized within the nuclei after D5.5 in almost all blastomeres composing an embryo (Fig. S1B). CDX2 started to localize within the nuclei in the outer blastomeres at D6.0 (Fig. S1B). To compare the localization patterns of YAP1 and CDX2 between bovine and mouse re-iICMs, we performed dual immunostaining (Fig. 2, A and B). In both mouse and bovine re-iICMs, YAP1 and CDX2 were colocalized within the nuclei in the outer blastomeres. Next, we double-checked the restricted localization of YAP1 and CDX2 to the outer blastomeres in bovine Whole and re-iICMs by using confocal microscopic images (Fig. S2). Moreover, because direct interaction between YAP1 and TEAD4 in species other than humans has not yet been confirmed (27), we performed immunoprecipitation for these proteins by using bovine fetal fibroblasts (Fig. 2C). We detected YAP1/TEAD4 proteins by using Western blotting and samples immunoprecipitated with YAP1/TEAD4 antibodies. Moreover, we confirmed the localization patterns of YAP1 and TEAD4 in bovine re-iICMs. YAP1 nuclear localization in the outermost blastomeres and TEAD4 nuclear localization in most blastomeres of bovine re-iICMs were similar to those observed in intact whole blastocysts after D6.0 (Fig. S1, A and B, and Fig. 2D). The spatiotemporal localization patterns of YAP1, TEAD4, and CDX2 in bovine embryos largely corresponded to those in mouse embryos; the interaction between YAP1 and TEAD4 has also been confirmed by immunoprecipitation experiments in human HEK293 cells (27). Thus, these results confirm that the underlining regulatory mechanism through the interaction between YAP1 and TEAD4 is conserved in bovine embryos.
To further explore the differences in the localization patterns of YAP1 and CDX2 between mouse and bovine re-iICMs, we assessed the proportion of blastomeres with or without YAP1 and CDX2 fluorescent signals in mouse and bovine re-iICMs (Fig. 2E). In mice, the proportion of YAP1+/CDX2+ and YAP1−/CDX2− blastomeres was 25.0 ± 25.0 and 75.0 ± 25.0%, respectively. Neither YAP1- nor CDX2-positive blastomeres (YAP1+/CDX2− and YAP1−/CDX2+) was detected in the analyzed mouse re-iICMs. Conversely, the proportion of YAP1+/CDX2+, YAP1+/CDX2−, YAP1−/CDX2+, and YAP1−/CDX2− blastomeres was 44.0 ± 11.1, 6.0 ± 2.4, 6.0 ± 2.8, and 44.0 ± 10.5% in bovine re-iICMs, respectively. Unlike in mouse re-iICMs, ∼50% of blastomeres exhibited CDX2 expression to characterize the TE. These results indicate that the competence of TE regeneration through YAP1 expression in bovine iICM is relatively higher than that in mouse iICM.
Species-specific expression of SOX17 in mouse and bovine embryos
To obtain further insight into the transcriptional factor responsible for cell-fate decisions leading to the difference in the competence of TE regeneration among species, we focused on SOX17 expression during cell differentiation from ICM to PrE in mouse and bovine re-iICMs. In mouse embryos, SOX17 is a well-known PrE marker. However, because of insufficient knowledge on SOX17 expression pattern during bovine preimplantation development, we first determined the SOX17 dynamics in bovine embryos compared with those in mouse embryos (Fig. S3, A and B). SOX17 was absent in the mouse embryos at E3.0 (the late morula stage), and then was expressed in the ICM nuclei at E3.75 (the blastocyst stage). At E4.75 (the expanded blastocyst stage), SOX17 was clearly localized in the PrE nuclei of the ICM blastomeres facing the blastocoel (Fig. S3A), which is consistent with the findings of previous studies (14, 15, 28, 29). In contrast, bovine SOX17 was observed in both ICM and TE nuclei from day 5.5 (the late morula stage) and remained at D6.5 (the early blastocyst stage; Fig. S3B). SOX17 was localized in the ICM nuclei in the expanded bovine blastocyst at D8.0 as well as in the mouse blastocyst at E3.75. Although SOX17 is commonly localized in the ICM nuclei specifically by the late blastocyst stage in both species, it was characteristically expressed in both ICM and TE around the transition to the blastocyst stage in bovine embryos. These results indicate that SOX17 localization in the ICM nuclei is regulated in a species-specific manner.
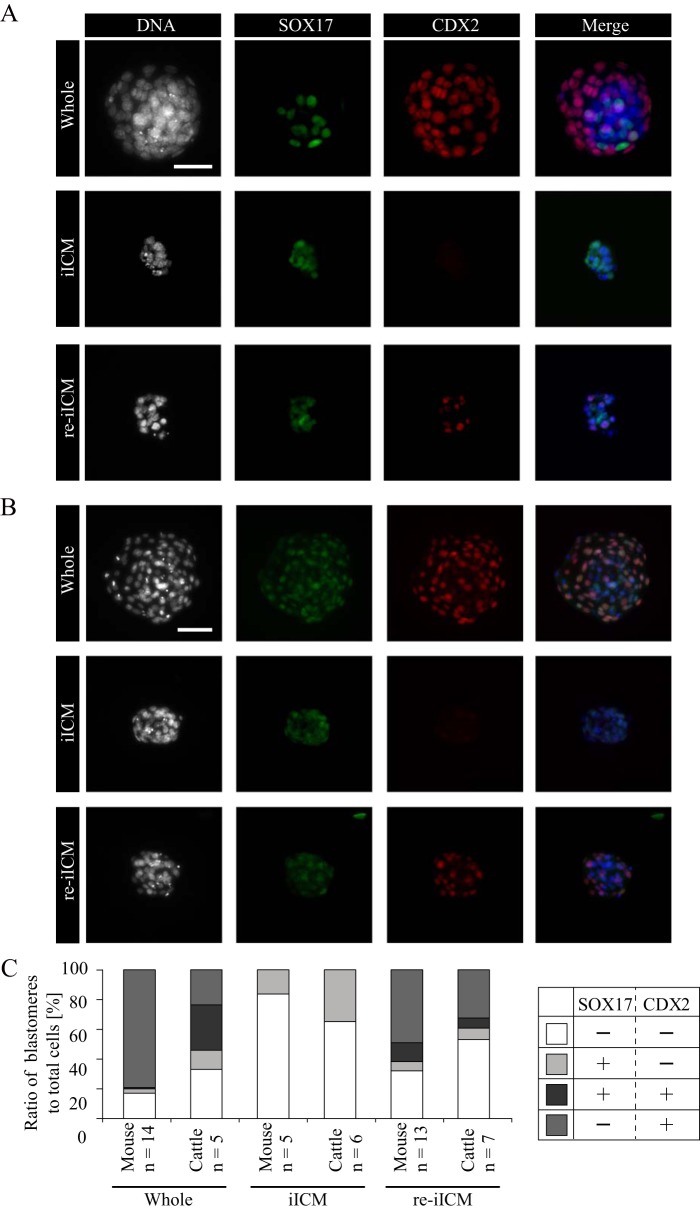
Based on the SOX17 localization pattern after the morula stage, we next assessed the localization patterns of both SOX17 and CDX2 by using Whole, iICM, and re-iICM of mice and cattle (Fig. 3). SOX17 remained localized in the ICM nuclei after TE removal in both the species. Interestingly, the two proteins were colocalized in the re-iICMs of both species. In particular, the co-localization of these proteins in mouse re-iICM was extraordinary because most of the intact mouse embryos did not show SOX17 expression in TE after the morula stage, as described above (Fig. S3A). In addition, SOX17 proteins were expressed in the TE of early bovine blastocysts at D6.5 (Fig. S3B). To quantitatively assess the SOX17 and CDX2 localization patterns, we investigated the proportion of blastomeres with or without SOX17 and CDX2 fluorescent signals by using Whole, iICM, and re-iICM of both species (Fig. 3C). In mice, SOX17+/CDX2+-coexpressing blastomeres were rarely observed in Whole (0.8 ± 0.4%), whereas they were markedly (12.7 ± 4.4%) noted in re-iICMs. In cattle, the proportion of SOX17+/CDX2+-coexpressing blastomeres in Whole and re-iICMs was 30.6 ± 7.6 and 6.9 ± 3.1%, respectively. These results show a species-specific expression pattern of SOX17 in mouse and bovine embryos, suggesting that the ectopic SOX17 expression in the TE of mouse re-iICM led to the disruption of TE function and failure of full-term development.
Figure 3.
Difference of SOX17 and CDX2 protein localizations between mouse and bovine iICMs. A and B, dual immunofluorescence analysis of SOX17 and CDX2 for whole blastocyst, iICM, and re-iICM in mice (A, bar, 50 μm) and cattle (B, bar, 100 μm). At least five embryos were analyzed for each experiment. C, quantification of blastomeres with SOX17 and/or CDX2 expression in whole, iICM, and re-iICM prepared from mouse and bovine blastocysts. Light gray: SOX17-positive (+) and CDX2-negative (−) blastomeres; dark gray, SOX17+ and CDX2+ blastomeres; medium gray, SOX17− and CDX2+ blastomeres; white, SOX17− and CDX2− blastomeres. The values are represented as the percentage of the above four types of blastomeres to the total number of blastomeres. Note that the bovine whole blastocysts affluently contained blastomeres positive for both proteins (dark gray), but mouse whole blastocysts rarely included such blastomeres. In contrast, in mouse re-iICM, both SOX17- and CDX2-positive blastomeres accounted for more than 12.7% of all the blastomeres.
Gene expression analyses of mouse and bovine iICMs after in vitro cultivation
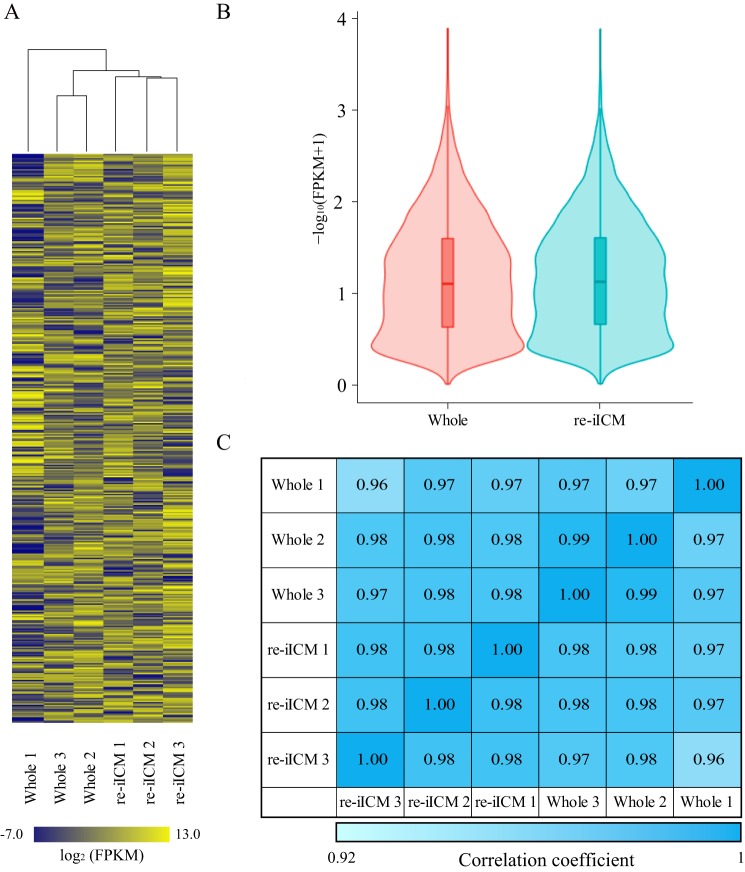
To investigate whether the transcriptional profile of the regenerated TE in bovine re-iICM is similar to that of Whole, we performed RNA-sequencing (RNA-seq) for bovine Whole and re-iICMs. Hierarchical clustering was used for all six samples (Whole 1–3 and re-iICM 1–3), and a dendrogram was constructed using the detected 9509 genes (Fig. 4A and Table S1). In the three re-iICM samples, normalization largely occurred in all the genes analyzed. Two Whole samples (Whole 2 and 3) and the three re-iICM samples were closely clustered. Furthermore, the violin plot showed that the global transcriptional profiles for Whole and re-iICM samples were obviously approximate, which was supported by the density plot and comparison of the Pearson correlation coefficient between samples (Fig. 4, B and C, and Fig. S4).
Figure 4.
Global gene expression analysis in bovine re-iICM using RNA-seq. A, heatmap representing the similarity of whole blastocyst and re-iICM in cattle. Hierarchical clustering alongside the heatmap shows the relation between inter-sample identities and similar expression patterns. Color scale represents the FPKM value from yellow (maximum) to blue (minimum). Gene expression levels are represented as FPKM values that are transformed by common logarithm with base 2. A total of 9509 genes (rows) were covered in each sample cluster (columns). B, violin plots of RNA-seq–based expression data for all 9509 genes in whole blastocysts and re-iICMs. Red and sky blue shapes show the distribution of the FPKM values for individual genes. Horizontal lines indicate group medians. C, to further confirm the similarity between Whole blastocyst and re-iICM gene expression patterns, we calculated the Pearson correlation coefficients for each sample. The values denote Pearson correlation coefficient (R2) of log2 (FPKM + 1) between samples. R2 >0.92 was considered “similarity” between two samples. Color scale represents the relative level of R2 from blue (high) to light blue (low).
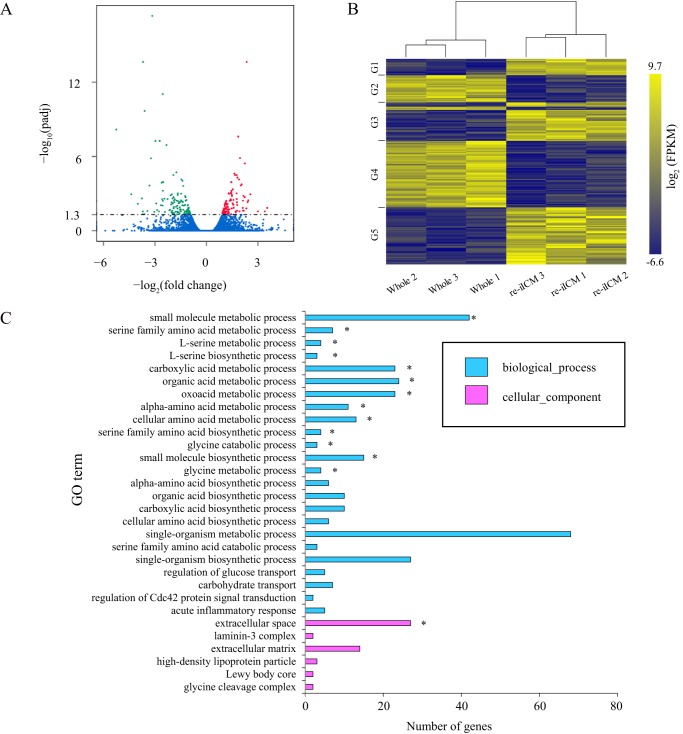
Although the global gene expression profiles between bovine re-iICM and Whole samples were generally similar, differentially-expressed genes (DEGs) were also noted, in which 99 and 123 genes were significantly (p < 0.05) up- and down-regulated in the re-iICMs compared with those in Whole, respectively (Fig. 5, A and B, and Tables S2 and S3). Next, we performed Gene Ontology (GO) enrichment analysis for these DEGs. The genes differentially expressed in the re-iICM were characterized in biological function, for which the GO terms associated with metabolism were output (Fig. 5C and Fig. S5). For example, small-molecule metabolic process, small-molecule biosynthetic process, single-organism metabolic process, organic acid metabolic process, and phosphorous metabolic process regulation were included in the list of GO terms that characterized the DEGs. In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis suggested that many DEGs were characterized in the metabolic pathways (Fig. S6). These results indicate that the global transcriptional profile of bovine re-iICM shares homology with that of Whole, sustaining the competence to develop to term; however, a portion of low-grade re-iICMs included in this analysis showed anomalous gene expression differing from those in intact blastocysts.
Figure 5.
Functional characterization of differentially-expressed genes in bovine re-iICM compared with those in Whole blastocyst. A, volcano plots show differentially-expressed genes in re-iICM compared with those in Whole blastocyst. Trimmed mean of M of each gene is plotted. The horizontal axis shows the fold-change in gene expression of re-iICM to that of Whole, and the vertical axis shows the p value after multiple comparison correction (Padj). Padj < 0.05 was considered statistically significant, the logarithmic value (=1.3) of which is indicated by a dashed line. Significantly up- and down-regulated genes are highlighted in red and green, respectively. Blue indicates genes showing no significant differential expression. B, heatmap representing the difference in whole blastocyst and re-iICM in cattle. Hierarchical clustering alongside the heatmap was performed using 188 genes that were differentially expressed and had raw FPKM >0 in the bovine re-iICMs (A). The gene sets corresponding to each group (G1 to 5) are shown in Table S3. C, GO enrichment analysis of differentially-expressed genes in re-iICM. The top 30 GO terms representing the functions known for these differentially-expressed genes are shown. The horizontal axis represents the number of differentially-expressed genes involved in a certain GO category, and the vertical axis shows GO terms enriched. The enriched GO terms are marked by an asterisk (p < 0.05).
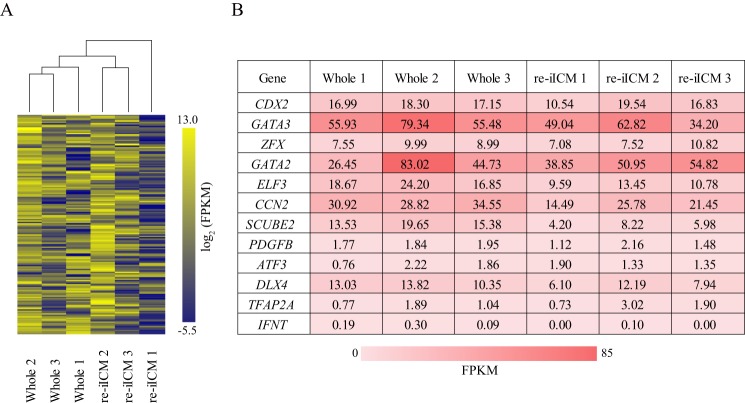
To gain a better understanding of the genes that were disturbed in the low-grade re-iICMs, we conducted hierarchical clustering analysis by focusing on 292 bovine TE-dominant genes that were determined in our previous study (30). Among the 9509 genes shown in Fig. 4A, 158 corresponded to the bovine TE-dominant genes (Table S4). We constructed a dendrogram representing the homology of the transcriptional profile among samples (Fig. 6A). The transcriptional profiles of re-iICM 2 and 3 were similar to those of Whole 1, 2, and 3. The transcriptional levels of TE-dominant genes in re-iICM 1 were markedly lower than those in the three Whole samples (Fig. 6A). Furthermore, we selected 12 especially significant TE- dominant genes related to transcription factors, growth factors, transporters, transcription activators, and the following oncogenes: CDX2, GATA3, ZFX, GATA2, ELF3, CCN2, SCUBE2, PDGFB, ATF3, DLX4, TFAP2A, and IFNT. Their fragments per kilobase of exon per million reads mapped (FPKM) values in re-iICMs (re-iICM 1, 2, and 3) were compared with those in Whole (Whole 1, 2, and 3; Fig. 6B). As shown in the dendrogram (Fig. 6A), the FPKM values for re-iICM 1 that did not include GATA2 and ATF3 were lower than those for each of the three Whole samples. In particular, the FPKM value for IFNT, which encodes a protein essential for maternal recognition and maintenance of pregnancy in ruminants, was not detected in re-iICM 1, as well as in another re-iICM sample (re-iICM 3). In contrast, the FPKM values detected in re-iICM 2 were similar to those in Whole samples, along with IFNT transcription. These results indicate that a part of bovine iICM is capable of regenerating TE normalized at the transcriptional level.
Figure 6.
Expression patterns of TE-predominant genes in bovine re-iICM using RNA-seq. A, hierarchical clustering analysis for 158 TE-predominant genes that were determined in our previous study (30). B, table represents FPKM values of each sample in 12 TE-specific genes that were identified as described in A. Color scale represents the FPKM value from yellow (maximum) to blue (minimum). Note that among the re-iICMs 1–3, IFNT expression was detected only in re-iICM 2, at a level equivalent to that in Whole 1–3.
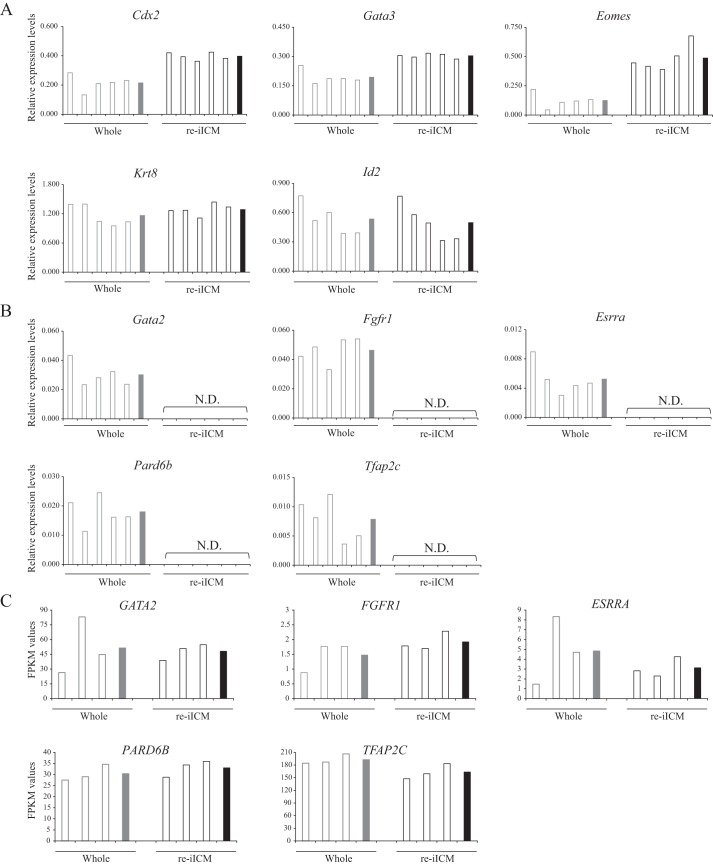
In addition, to explore the cause of insufficient TE regeneration in mouse embryos, we performed gene expression analysis of 10 primal TE-related genes, Cdx2, Gata2, Gata3, Eomes, Tfap2c, Fgfr1, Esrrα, Pard6b, Krt8, and Id2, that are critical for TE cell characterization (11, 31–38) in mouse Whole and re-iICMs using quantitative RT-PCR (qPCR; Fig. 7, A and B). Among these genes, Cdx2, Gata3, Eomes, Krt8, and Id2 were expressed in mouse re-iICMs (Fig. 7A). In contrast, the relative expression levels of Gata2, Tfap2c, Fgfr1, Erssα, and Pard6b were below detectable limits in re-iICMs, although they were stably expressed in whole samples (Fig. 7B). However, the 10 genes were expressed at similar levels in both bovine Whole and re-iICM samples (Fig. 7C). These results clearly showed that TE regeneration in mouse re-iICMs was grossly deficient at the transcriptional level, differing from that in bovine re-iICMs.
Figure 7.
Graphical representations of the expression of TE-related genes in Whole and re-iICM samples. The expression of 10 TE-related genes (Cdx2, Gata2, Gata3, Tfap2c, Eomes, Fgfr1, Erssa, Pard6b, Krt8, and Id2) in the Whole (gray) and re-iICM (black) samples were analyzed. A, genes with detectable expression levels were exhibited using qPCR data. B, genes with expression levels below detectable limits in the re-iICM samples were exhibited using qPCR data. The values shown in A and B represent the levels of expression relative to that of the reference gene (Gapdh). C, FPKM values of the genes examined in B from RNA-seq analysis of bovine re-iICM samples (Fig. 4). The values of single embryo samples are represented by white bars. The means of values are represented by gray bars (Whole) or black (re-iICM). N.D., not detected.
Discussion
This study addresses the variable nature of blastomeres after allocation to the ICM and suggests that the species-specific molecular basis for ICM/TE cell lineages restricts the TE-regeneration capacity. We determined the totipotency of ICM isolated from the blastocyst-stage embryos in mice and cattle. Our data indicated the following. (i) iICMs form blastocoels with regenerated TE expressing CDX2 in mice and cattle. (ii) This TE regeneration observed in mouse and bovine re-iICMs was commonly mediated through the Hippo pathway in line with the YAP1–TEAD4–CDX2 axis to configure blastomeres to their fate via transcriptional modification. (iii) One bovine re-iICM showed full-term development, whereas none of the >100 mouse re-iICMs transferred to recipients developed to term or showed any implantation sites. (iv) Some bovine re-iICMs exhibited transcriptional patterns as those in the TE of bovine blastocysts. We also found that the PrE marker, SOX17, was ectopically expressed in the regenerated TE of mouse re-iICMs, although its localization was limited to the ICM nuclei in mouse normal blastocysts throughout development, providing one possible explanation for a species-specific molecular basis to determine and maintain cell-fate specification. Numerous studies have suggested that early ICM has the ability to differentiate into TE (39, 40). However, embryos are generally known to begin their ICM/TE cell-fate decision by the 16-cell stage, and restriction of the ICM to become TE cells is not observed until the 64-cell blastocyst stage in mice (41, 42). Our analysis of TE regeneration capacity in bovine blastocysts suggested that the developmental clock to provide such restriction of ICM to become TE is relatively slower in bovine embryos than in mouse embryos.
Our observation that blastomeres allocated to bovine ICM can develop to an individual organism with TE regeneration is, to our knowledge, the first evidence in mammalian embryos that the ICM possesses the ability to regenerate functionally-active TE to support full-term fetal development. Such a totipotential state could not be maintained in mouse embryos, likely because of the difference in the cell-fate decision process across species. To clarify the resemblance and difference in ICM/TE cell-fate decision between both the species, first, we investigated the manner in which the TE is regenerated from the iICM by analyzing YAP1 expression. In mouse embryos, YAP1 is an upstream mediator of CDX2 expression (13, 14, 43). YAP1 proteins localize into the nuclei in the outmost blastomeres, generating the TE and triggering the downstream elements of TEAD4 to induce CDX2 expression, which is essential for TE fate specification (13). In this study, we found direct interaction between YAP1 and TEAD4 in cattle (Fig. 2C), which, to our knowledge, is the first evidence in species other than human (27). Therefore, our data clearly show that the regulation of TE differentiation by the YAP1–TEAD4–CDX2 axis is conserved in bovine embryos. After the originally outer-positioned TE blastomeres of bovine blastocyst were stripped, new outer-positioned blastomeres re-expressed YAP1 and CDX2 in their nuclei; this was also noted in mouse blastocysts. Therefore, both mouse and bovine iICMs commonly possess the potency to convert to TE with CDX2 expression through the YAP1–TEAD4 interaction.
What led to the difference in the totipotential state of ICMs between both the species? The difference of cell fate determination via SOX17 might affect the cellular integrity of the regenerated TE. Interestingly, we found that mouse re-iICMs showed ectopic expression of SOX17, which was coexpressed with CDX2 in the regenerated TE (Fig. 3, A and C). SOX17 is a transcription factor belonging to the family of the sex-determining region Y–related high-mobility group box, namely SOX. It acts in various developmental processes (44–46). In addition, SOX17 has been proposed to function as a key regulator of endoderm formation and differentiation, a function that is conserved across vertebrates (29, 47–49). During mouse development, SOX17 protein localization within a blastocyst is coherently restricted to the PrE progenitors contained within the ICM (15, 50). Actually, before blastocyst formation, nuclear localization of SOX17 is never observed (Fig. S3A), which was consistent with the findings of a previous study (50). Mouse ICM blastomeres at E3.5 exclusively express either epiblast- or PrE-specific genes in a “salt–and–pepper” mosaic pattern before the appearance of the PrE layer (51, 52). Thus, the formation of epiblast and PrE is based on an initial mosaic of two lineage progenitors at E3.5. By E4.5, their sorting and appropriate positionings are established in the ICM. In this study, the SOX17 localization pattern of mouse iICM was also maintained in a mosaic pattern. In contrast, after 24 h of cultivation, 12.7% of blastomeres in the re-iICMs showed coexpression of both SOX17 and CDX2, which accounted for 20.6% of all CDX2-positive cells (Fig. 3C). The ectopic expression of SOX17 is commonly not observed in any developmental stage in mice; this might have induced disrupted transcription in the regenerated TE, leading to the defective TE regeneration and failure of full-term development in mouse re-iICMs.
As the SOX17 localization pattern in bovine embryos is not yet known, we investigated the SOX17 dynamics from the morula to blastocyst stages in cattle, matching the mouse developmental stages. Interestingly, unlike in mouse embryos, most of the bovine blastomeres at D5.5 showed nuclear localization of SOX17 (Fig. S3B). Eventually, nuclear SOX17 localization was limited to the blastomeres within the bovine ICM at D8.0; however, unlike the consistent restriction of SOX17 to the ICM nuclei in mouse embryos, SOX17 was localized to the nuclei in both the ICM and TE in bovine embryos until D6.5. Therefore, the nuclear SOX17 in the TE of bovine re-iICM might have facilitated preimplantation development, which was not observed in mouse re-iICM. Although which genes and how many genes are the targets of SOX17 in bovine embryos are not yet known, the nuclear localization of SOX17 in the TE was found to not disrupt embryo development in cattle, which is supported by the birth of a calf derived from the re-iICM. However, we cannot exclude the possibility that some transcriptional factors other than SOX17 are associated with failure of full-term development, because some TE-related genes were repressed in the mouse re-iICMs (Fig. 7, A and B).
To ensure the restoration of bovine re-iICM, we compared the global gene expression patterns of three re-iICM samples (re-iICM 1–3) with those of three Whole samples (Whole 1–3) by using RNA-seq analysis. The global comparisons targeted at all genes with detectable read counts (9509 genes) revealed that the gene expression patterns in the re-iICM samples approximated those in the Whole samples. Furthermore, the DEGs in the re-iICM samples were compared with the genes expressed in the Whole samples, and most biological functions were represented as GO terms associated with metabolism. This might reflect that some of the re-iICM samples contained regenerated TE with insufficient restoration because metabolism in TE is more active than that in the ICM (53); therefore, the disrupted TE in several re-iICMs might affect the transcription of the metabolism-associated genes. Therefore, we focused on the 158 TE-specific genes that were detected in our previous study (30), and we performed hierarchical clustering analysis for those genes in both the re-iICM and Whole samples. As expected, the TE-specific genes in one of the re-iICM samples (re-iICM 1) exhibited broadly lower expression levels than in the other re-iICM and Whole samples (re-iICM 2 and 3; Whole 1–3; Fig. 6A). Moreover, the expression level of the 12 core TE-specific genes in re-iICM 1 and 3 was considerably lower than that in re-iICM 2; furthermore, the IFNT gene was not detected in re-iICM 1 and 3 (Fig. 6B).
Only re-iICM 2 showed IFNT expression equivalent to that in Whole samples, indicating that, among the three re-iICM samples analyzed, re-iICM 2 showed a normal TE transcriptional profile. These results suggested that some bovine iICMs after TE removal are capable of regenerating TE with a transcriptional profile equivalent to that of the intact whole blastocyst. However, all the re-iICMs, including re-iICM 2, showed differential expression of genes compared with that in the Whole embryos (Fig. 5B). Therefore, elucidating which set of genes is critical for transcriptional restoration in bovine re-iICMs might further our understanding of the ICM → TE conversion to ensure embryo totipotency.
In conclusion, we found that the newly-externalized blastomeres from the originally internalized ones within a blastocyst could regenerate functional TE to support full-term development in cattle, i.e. bovine iICM could equally regenerate TE from both the side in contact with the original TE and the side in contact with the original blastocoel. Explaining this observation by a typical hypothesis, namely the inside–outside model (6), would be difficult because the microenvironment, including the positioning of the blastomeres on the side in contact with the original blastocoel, i.e. not in contact with the original TE, was probably not changed before and after TE removal. The conversion from the ICM to TE in this area would be assumed to be regulated by another mechanism that recasts the role of blastomeres allocated to the ICM. Furthermore, no mouse iICMs with re-cavitation developed to term, which may explain the diversity in TE lineage commitment of blastomeres allocated to the ICM among species, indicating the differences in the control of cell specification via key regulators such as SOX17. Transcription of lineage-specific genes has been shown to be driven by a developmental clock that is a temporal program to dictate correct gene expression patterns with progressive development in early embryos (54). Since the time course until direct communication with the maternal uterus is substantially distinct between mouse (around E4.5) and bovine embryos (around D19), the bovine developmental clock is most probably slower than that of the mouse. Taken together, the diversity in the totipotential state of the ICM probably reflects that developmental courses are continually changed during evolution and that the regulation of courses, including ICM/TE lineage specification, requires the momentous adjustability for these alterations.
Experimental procedures
Ethics approval
All experimental protocols were approved by the Regulatory Committee for the Care and Use of Laboratory Animals, Hokkaido University.
Embryo preparation in mice and cattle
Mouse and bovine preimplantation embryos for analyses in this study were produced using in vitro fertilization (IVF), as described in our previous studies (30, 55). In mice, female ICR or BDF1 (C57BL/6N × DBA/2N) mice were superovulated by injections of 5 IU PMSG (ASKA Pharmaceutical Co., Ltd., Tokyo, Japan) and 5 IU equine chorionic gonadotropin (ASKA Pharmaceutical Co., Ltd.), administered 48 h apart. Sperm was collected from the cauda epididymis of mature male ICR mice, suspended in a 200-μl drop of human tubal fluid (HTF) medium (56) in paraffin oil, and preincubated for 90 min in an atmosphere of 5% CO2 at 37 °C. Subsequently, oocytes at metaphase II were collected from the murine oviducts at 16 h after equine chorionic gonadotropin administration and transferred to a 100-μl drop of HTF medium in which the sperm concentration was adjusted to 0.5–1 × 106 cells/ml. At 4–6 h after insemination, the embryos were denuded of cumulus cells, washed with M2 medium (57), and transferred to a drop of M16 medium (58). The embryos were cultured in M16 until the blastocyst stage and used for subsequent experiments. Embryonic day 0 (E0) is defined as the time when the in vitro culture (IVC) was started.
Bovine embryos were prepared using in vitro oocyte maturation (IVM), IVF, and subsequent IVC. Briefly, cumulus–oocyte complexes (COCs) collected from slaughterhouse-derived ovaries were matured by culturing in TCM-199 medium (Thermo Fisher Scientific, Inc., Waltham, MA) at 38.5 °C in a humidified atmosphere of 5% CO2 in air for 22–24 h. IVM oocytes were transferred to Brackett and Oliphant (B.O.) medium (59) containing 2.5 mm theophylline (Wako Pure Chemical Industries, Ltd., Osaka, Japan) and 7.5 μg/ml heparin sodium salt (Nacalai Tesque, Inc., Kyoto, Japan). Subsequently, frozen-thawed semen was centrifuged at 600 × g for 7 min in the B.O. medium, and the spermatozoa were added to the COCs at a final concentration of 5 × 106 cells/ml. After 18 h of incubation, the presumptive IVF zygotes were denuded by pipetting and cultured in mSOFai medium (60) at 38.5 °C in a humidified atmosphere of 5% CO2 and 5% O2 in air. The start of insemination was regarded as day 0 after IVC.
Isolation of ICM from mouse and bovine blastocysts
The ICM from blastocysts in both species was isolated as described previously (30). Briefly, the zona pellucida of the early blastocyst stage embryos was removed using acidic Tyrode's solution (pH 2.5) (61) or 0.05% (w/v) proteinase K (Wako Pure Chemical Industries, Ltd.) at E3.75 in mice and at D6.5 in cattle. The mouse and bovine blastocyst embryos were treated with PBS containing 0.1–0.125% (v/v) or 0.2% (v/v) Triton X-100 (Wako Pure Chemical Industries, Ltd.) and 0.2% (v/w) polyvinyl alcohol (Sigma) at room temperature (typically at 20–22 °C) for 3–10 s. The embryos were carefully washed, followed by gentle pipetting to collect pure ICMs without the TE cells. These iICMs were collected in the embryo culture medium corresponding to each species and cultured for another 24 h. The iICMs with an obvious cavity were defined as “re-cavitated iICM” (re-iICM).
Transfer of re-iICMs to recipient females in mice and cattle
The competence of full-term development in mouse and bovine re-iICMs was determined by transferring re-iICMs to recipient females, according to general embryo transfer techniques. Mouse re-iICMs were transferred to the uterine horns of recipient female ICR mice at 2.5 days of pseudo-pregnancy, as described previously (62). In cattle, a recipient cow was synchronized by CIDR synch (63) with a slight modification. The cow at the Hokkaido University was treated using an intravaginal progesterone-releasing device (CIDR; InterAg, Hamilton, New Zealand) for 7 days with 2 mg of estradiol benzoate and 25 mg of dinoprost at the times of insertion and withdrawal of the device, respectively. After the CIDR was withdrawn, we confirmed ovulation by ultrasonography daily (5 MHz, HS101V; Honda Electronics, Tokyo, Japan). Two bovine re-iICMs were transferred transcervically into a recipient cow synchronized at D6 after ovulation; this was repeated twice. Next, we inserted CIDR for 2 weeks from the day of re-iICM transfer, and pregnancy was diagnosed at the day of CIDR removal (∼26 days after ovulation).
Immunostaining
Immunofluorescence staining for embryos was performed as described previously (64). The re-iICMs were stained using the procedures identical to those for embryos. Briefly, zona pellucida–removed embryos were fixed and permeabilized. Subsequently, the embryos were blocked for 45 min with PBS containing 20% (v/v) Blocking One (Nacalai Tesque, Inc.) and 0.01% (v/v) Tween 20 (Wako Pure Chemical Industries, Ltd.). The following primary antibodies were used for analyses: anti-CDX2 (ab76541, rabbit monoclonal, 1:300; Abcam, Cambridge, UK); anti-SOX17 (AF1924, goat polyclonal, 1:1500; R&D Systems, Minneapolis); two types of anti-YAP1 (H00010413-M01, mouse monoclonal, 1:100; Novus Biologicals, Littleton, CO, and 4912S, rabbit polyclonal, 1:100; Cell Signaling Technology, Danvers, MA); and anti-TEAD4 (ab58310, mouse monoclonal, 1:1500; Abcam). The two types of anti-YAP1 were used properly according to species to avoid cross-staining for dual immunostaining. Temperature and incubation time for the primary antibody reaction varied depending on the target protein, i.e. overnight at 37 °C for CDX2, overnight at 4 °C for SOX17, overnight at room temperature for YAP1, and 2 h at room temperature for TEAD4. The following secondary antibodies were used: Alexa Fluor 488 donkey anti-rabbit IgG (A21206, polyclonal, 1:400; Invitrogen, Tokyo, Japan); Alexa Fluor 488 goat anti-mouse IgG (A11001, polyclonal, 1:400; Invitrogen); Alexa Fluor 488 donkey anti-goat IgG (ab150129, polyclonal, 1:400; Abcam); and Alexa Fluor 555 goat anti-rabbit IgG (A21428, polyclonal, 1:400; Invitrogen). All the antibodies were diluted in PBS containing 5% (v/v) Blocking One and 0.01% (v/v) Tween 20. For reaction with the secondary antibodies, the embryos were incubated for 30 min at room temperature. DNA was counterstained with 25 μg/ml Hoechst 33342 (Sigma) for 5 min at room temperature. Fluorescence signals were visualized using a TCS SP5 confocal laser-scanning microscope (Leica, Tokyo, Japan) or a LAS X with DMi8 fluorescence microscope (Leica). The proportion of marker protein (CDX2, YAP1, or SOX17)-positive blastomeres to all the blastomeres within an embryo was analyzed by manually counting the blastomeres from immunofluorescence images. For each protein, blastomeres in which nuclear fluorescence signals were obviously stronger than those within the cytoplasm were regarded as positive fluorescence signals. The total cell number was determined using Hoechst staining as described above.
Coimmunoprecipitation
Immunoprecipitation (IP) was performed using the Pierce coimmunoprecipitation kit (26149; Thermo Fisher Scientific) according to the manufacturer's instructions. Briefly, bovine fetal fibroblast cells were cultured in Dulbecco's modified Eagle's medium (Sigma) containing 10% (v/v) fetal bovine serum (PAA, The Cell Culture Co., Pasching, Austria) in an atmosphere of 5% CO2 at 38.5 °C until the cells became 80% confluent. Next, the cells were detached and centrifuged to obtain a pellet. Subsequently, 50 mg of cell pellet was lysed in 500 μl of IP Lysis/Wash Buffer. For immunoprecipitation, 45–50 mg of pre-cleared lysate was used. Incubations with 10 μg of anti-YAP (H00010413-M01) and 7 μg of anti-TEAD4 (ab58310) antibodies were performed initially at 37 °C for 2 h and subsequently at room temperature overnight. Immunoprecipitated samples were washed seven times in IP Lysis/Wash Buffer and eluted in sample buffer containing 62.5 mm Tris-HCl, 10% (v/v) glycerol, 5% (v/v) 2-mercaptoethanol, 2.5% (w/v) SDS, and 0.005% (w/v) bromphenol blue. The samples were treated at 70 °C for 15 min before electrophoresis. For Western blot analysis, 6 μl of precipitated sample per lane was run on an 8% (w/v) polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Bio-Rad). The membrane was blocked with PBS containing 4% (w/v) skim milk (Nacalai Tesque) and 0.1% (v/v) Tween 20 and incubated overnight with the following antibodies: anti-YAP1 (H00010413-M01, 1:500) at room temperature and anti-TEAD4 (ab58310, 1:500) at 4 °C. Subsequently, the membrane was incubated for 50 min at room temperature with horseradish peroxidase–conjugated donkey anti-rabbit IgG (NA934, monoclonal, 1:5000; GE Healthcare, UK) and horseradish peroxidase–conjugated sheep anti-mouse IgG (NA931, monoclonal, 1:5000; GE Healthcare). The images of the protein bands were obtained using a Bio-Rad ChemiDocTM EQ densitometer (Bio-Rad).
RNA-seq library construction
Total RNA from zona pellucida–removed whole blastocysts and re-iICMs after 24 h culture was extracted using ReliaPrep RNA Cell Miniprep System (Promega, Madison, WI). In each replication, 15 embryos and re-iICMs each were used per sample. The cDNA was then synthesized and amplified using SMART-Seq version 4 Ultra Low Input RNA kit for sequencing (Takara Bio, Shiga, Japan) and purified using Agencourt AMPure XP Kit (Beckman Coulter). RNA integrity was checked using Agilent 2100 Bioanalyzer (Agilent, Tokyo, Japan). Libraries were constructed using Nextera DNA Library Preparation Kit (Illumina, Tokyo, Japan). Sequencing was performed in 150-bp paired-end format on an Illumina HiSeq 2500 system (Illumina).
RNA-seq data analysis
Sequencing reads were mapped to the July, 2018, assembly of the bovine genome (UMD3.1) by using TopHat2 software. The FPKM values were calculated for each gene and used in the subsequent analyses. The logarithm of the >0 raw FPKM value was considered for graphical representation. Genes whose FPKM of ≥1 were considered as expressed. DEG analysis was performed using DESeq package by normalizing the expression values to Trimmed mean of M. Hierarchical clustering analysis was performed using Euclidean distance for similarity between samples, and Ward's method was used for distance measurement among clusters. The “TE-specific genes” (Fig. 6A) were the expressed (whose FPKM ≥1) genes included in the list of those predominantly expressed in the TE in our previously published microarray data (30). GO (http://www.geneontology.org/)3 analyses were performed using R package GOseq (66, 67). The enrichment of each GO term was determined using the hypergeometric distribution test, and the statistical index value was represented as the p value. In GO enrichment analysis (Fig. S5), the background denotes whole genes annotated by any term. Pathway enrichment analysis was performed based on the KEGG pathway database (https://www.genome.jp/kegg/pathway.html).3
Quantitative RT-PCR
qPCR for mouse embryos was performed using a LightCycler® 96 (Roche Diagnostics, Basel, Switzerland) as described previously (64). Total RNA from zona pellucida–removed single whole blastocysts and re-iICMs after 24 h culture was extracted using Arcturus® PicoPure® RNA isolation kit (Thermo Fisher Scientific). cDNA synthesis was conducted using ReverTra Ace qPCR RT Master Mix (Toyobo, Osaka, Japan). Quantitative PCR was performed after preparing the reaction mixtures in THUNDERBIRD SYBR qPCR Mix (Toyobo). The primer sets used for qPCR analysis are listed in Table S5. Thermal cycling conditions consisted of one cycle at 95 °C for 30 s (denaturation), followed by 50 cycles at 95 °C for 10 s (denaturation), and annealing temperature corresponded to each primer set for 15 s (primer annealing) and 72 °C for 30 s (extension). Five independent embryo samples were used for each experiment. Relative mRNA abundance was determined three times and calculated using the ΔΔCt method, with Gapdh as the reference gene (65) in each sample.
Statistical analysis
Statistical significance was analyzed using Student's t test or analysis of variance, followed by the Tukey's post hoc tests. The data are presented as mean ± S.E. The percentage data were subjected to an arcsin transformation. R software (Comprehensive R Archive Network) was used for statistical analysis. p < 0.05 was considered statistically significant.
Author contributions
N. K. and M. Kawahara data curation; N. K. and M. Kawahara formal analysis; N. K. and M. Kawahara funding acquisition; N. K. and M. Kawahara validation; N. K., H. A., S. I., Y. Y., M. Komatsu, M. Kawai, M. N., and M. Kawahara investigation; N. K., H. B., and M. Kawahara visualization; N. K., H. A., and M. Kawahara writing-original draft; H. B., M. T., and M. Kawahara resources; H. B. and M. Kawahara writing-review and editing; M. Kawahara conceptualization; M. Kawahara supervision; M. Kawahara project administration.
Supplementary Material
Acknowledgments
We thank Dr. Tomohiro Mitani and the staff at the Field Science Center for Northern Biosphere, Hokkaido University, Sapporo, Japan, for taking care of the recipient cow. We thank Prof. Fumio Nakamura, Laboratory of Animal By-Product Science, Research Faculty of Agriculture, Hokkaido University, and Dr. Ken Kobayashi, Laboratory of Cell and Tissue Biology, Research Faculty of Agriculture, Hokkaido University, for critical discussions related to this work. We are grateful to Koki Shiina, Hikaru Takuma, and Fumi Yokoi, Laboratory of Animal Genetics and Reproduction, Research Faculty of Agriculture, Hokkaido University, for their support on mouse embryo transfer. We thank Genetics Hokkaido Association for the donation of frozen bull spermatozoa and the Hokkaido Hayakita meat inspection center for providing bovine ovaries.
This work was supported by Grant-in-aid for Scientific Research (B) 18H02321 (to M. K.) and Grant-in-aid for JSPS Fellows from the Japan Society for the Promotion of Science 17J03585 (to N. K.). The authors declare that they have no conflicts of interest with the contents of this article.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) DRA008645.
This article contains Figs. S1–S6 and Tables S1–S5.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ICM
- inner cell mass
- TE
- trophectoderm
- re-iICM
- isolated ICM with re-formed blastocoel
- E
- embryonic day
- PrE
- primitive endoderm
- iICM
- isolated ICM
- Whole
- intact whole blastocyst
- IP
- immunoprecipitation
- WB
- Western blotting
- DEG
- differentially-expressed gene
- GO
- gene ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- FPKM
- fragments/kilobase of exon per million reads mapped
- IVF
- in vitro fertilization
- HTF
- human tubal fluid
- IVC
- in vitro culture
- IVM
- in vitro oocyte maturation
- COCs
- cumulus–oocyte complexes
- qPCR
- quantitative PCR
- B.O.
- Brackett and Oliphant
- RNA-seq
- RNA-sequencing
- D
- day
- HTF
- human tubal fluid
- CIDR
- a controlled intravaginal progesterone-releasing device.
References
- 1. Anani S., Bhat S., Honma-Yamanaka N., Krawchuk D., and Yamanaka Y. (2014) Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 141, 2813–2824 10.1242/dev.107276 [DOI] [PubMed] [Google Scholar]
- 2. Hirate Y., Hirahara S., Inoue K., Kiyonari H., Niwa H., and Sasaki H. (2015) Par-aPKC-dependent and -independent mechanisms cooperatively control cell polarity, Hippo signaling, and cell positioning in 16-cell stage mouse embryos. Dev. Growth Differ. 57, 544–556 10.1111/dgd.12235 [DOI] [PubMed] [Google Scholar]
- 3. Hyafil F., Morello D., Babinet C., and Jacob F. (1980) A cell surface glycoprotein involved in the compaction of embryonal carcinoma cells and cleavage stage embryos. Cell 21, 927–934 10.1016/0092-8674(80)90456-0 [DOI] [PubMed] [Google Scholar]
- 4. Shirayoshi Y., Okada T. S., and Takeichi M. (1983) The calcium-dependent cell–cell adhesion system regulates inner cell mass formation and cell surface polarization in early mouse development. Cell 35, 631–638 10.1016/0092-8674(83)90095-8 [DOI] [PubMed] [Google Scholar]
- 5. Johnson M. H., and Ziomek C. A. (1983) Cell interactions influence the fate of mouse blastomeres undergoing the transition from the 16- to the 32-cell stage. Dev. Biol. 95, 211–218 10.1016/0012-1606(83)90019-2 [DOI] [PubMed] [Google Scholar]
- 6. Tarkowski A. K., and Wróblewska J. (1967) Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J. Embryol. Exp. Morphol. 18, 155–180 [PubMed] [Google Scholar]
- 7. Lorthongpanich C., and Issaragrisil S. (2015) Emerging role of the Hippo signaling pathway in position sensing and lineage specification in mammalian preimplantation embryos. Biol. Reprod. 92, 143 10.1095/biolreprod.114.127803 [DOI] [PubMed] [Google Scholar]
- 8. Sasaki H. (2017) Roles and regulations of Hippo signaling during preimplantation mouse development. Dev. Growth Differ. 59, 12–20 10.1111/dgd.12335 [DOI] [PubMed] [Google Scholar]
- 9. Boroviak T., and Nichols J. (2014) The birth of embryonic pluripotency. Philos. Trans. R. Soc. B Biol. Sci. 369, 20130541 10.1098/rstb.2013.0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitalipov S., and Wolf D. (2009) Totipotency, pluripotency and nuclear reprogramming. Adv. Biochem. Eng. Biotechnol. 114, 185–199 10.1007/10_2008_45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo G., Huss M., Tong G. Q., Wang C., Li Sun L., Clarke N. D., and Robson P. (2010) Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685 10.1016/j.devcel.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 12. Wigger M., Kisielewska K., Filimonow K., Plusa B., Maleszewski M., and Suwińska A. (2017) Plasticity of the inner cell mass in mouse blastocyst is restricted by the activity of FGF/MAPK pathway. Sci. Rep. 7, 15136 10.1038/s41598-017-15427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N., Makita R., Kurihara H., Morin-Kensicki E. M., Nojima H., Rossant J., et al. (2009) The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 14. Lorthongpanich C., Messerschmidt D. M., Chan S. W., Hong W., Knowles B. B., and Solter D. (2013) Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev. 27, 1441–1446 10.1101/gad.219618.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krawchuk D., Honma-Yamanaka N., Anani S., and Yamanaka Y. (2013) FGF4 is a limiting factor controlling the proportions of primitive endoderm and epiblast in the ICM of the mouse blastocyst. Dev. Biol. 384, 65–71 10.1016/j.ydbio.2013.09.023 [DOI] [PubMed] [Google Scholar]
- 16. Mallol A., Santaló J., and Ibáñez E. (2015) Improved development of somatic cell cloned mouse embryos by vitamin C and latrunculin A. PLoS ONE 10, e0120033 10.1371/journal.pone.0120033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Isaji Y., Murata M., Takaguchi N., Mukai T., Tajima Y., Imai H., and Yamada M. (2013) Valproic acid treatment from the 4-cell stage improves Oct4 expression and nuclear distribution of histone H3K27me3 in mouse cloned blastocysts. J. Reprod. Dev. 59, 196–204 10.1262/jrd.2012-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kishigami S., Hikichi T., Van Thuan N., Ohta H., Wakayama S., Bui H. T., Mizutani E., and Wakayama T. (2006) Normal specification of the extraembryonic lineage after somatic nuclear transfer. FEBS Lett. 580, 1801–1806 10.1016/j.febslet.2006.02.036 [DOI] [PubMed] [Google Scholar]
- 19. Balbach S. T., Esteves T. C., Brink T., Gentile L., McLaughlin K. J., Adjaye J. A., and Boiani M. (2010) Governing cell lineage formation in cloned mouse embryos. Dev. Biol. 343, 71–83 10.1016/j.ydbio.2010.04.012 [DOI] [PubMed] [Google Scholar]
- 20. Ogura A., Inoue K., and Wakayama T. (2013) Recent advancements in cloning by somatic cell nuclear transfer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110329 10.1098/rstb.2011.0329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Whitworth K. M., and Prather R. S. (2010) Somatic cell nuclear transfer efficiency: how can it be improved through nuclear remodeling and reprogramming? Mol. Reprod. Dev. 77, 1001–1015 10.1002/mrd.21242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simmet K., Zakhartchenko V., and Wolf E. (2018) Comparative aspects of early lineage specification events in mammalian embryos–insights from reverse genetics studies. Cell Cycle 17, 1688–1695 10.1080/15384101.2018.1496747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simmet K., Zakhartchenko V., Philippou-Massier J., Blum H., Klymiuk N., and Wolf E. (2018) OCT4/POU5F1 is required for NANOG expression in bovine blastocysts. Proc. Natl. Acad. Sci. U.S.A. 115, 2770–2775 10.1073/pnas.1718833115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuijk E. W., Du Puy L., Van Tol H. T., Oei C. H., Haagsman H. P., Colenbrander B., and Roelen B. A. (2008) Differences in early lineage segregation between mammals. Dev. Dyn. 237, 918–927 10.1002/dvdy.21480 [DOI] [PubMed] [Google Scholar]
- 25. Berg D. K., Smith C. S., Pearton D. J., Wells D. N., Broadhurst R., Donnison M., and Pfeffer P. L. (2011) Trophectoderm lineage determination in cattle. Dev. Cell 20, 244–255 10.1016/j.devcel.2011.01.003 [DOI] [PubMed] [Google Scholar]
- 26. Fogarty N. M. E., McCarthy A., Snijders K. E., Powell B. E., Kubikova N., Blakeley P., Lea R., Elder K., Wamaitha S. E., Kim D., Maciulyte V., Kleinjung J., Kim J. S., Wells D., Vallier L., et al. (2017) Genome editing reveals a role for OCT4 in human embryogenesis. Nature 550, 67–73 10.1038/nature24033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jang J. W., Kim M. K., Lee Y. S., Lee J. W., Kim D. M., Song S. H., Lee J. Y., Choi B. Y., Min B., Chi X. Z., and Bae S. C. (2017) RAC-LATS1/2 signaling regulates YAP activity by switching between the YAP-binding partners TEAD4 and RUNX3. Oncogene 36, 999–1011 10.1038/onc.2016.266 [DOI] [PubMed] [Google Scholar]
- 28. Le Bin G. C., Muñoz-Descalzo S., Kurowski A., Leitch H., Lou X., Mansfield W., Etienne-Dumeau C., Grabole N., Mulas C., Niwa H., Hadjantonakis A.-K., and Nichols J. (2014) Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development 141, 1001–1010 10.1242/dev.096875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Artus J., Piliszek A., and Hadjantonakis A. K. (2011) The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev. Biol. 350, 393–404 10.1016/j.ydbio.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagatomo H., Kagawa S., Kishi Y., Takuma T., Sada A., Yamanaka K., Abe Y., Wada Y., Takahashi M., Kono T., and Kawahara M. (2013) Transcriptional wiring for establishing cell lineage specification at the blastocyst stage in cattle. Biol. Reprod. 88, 158 10.1095/biolreprod.113.108993 [DOI] [PubMed] [Google Scholar]
- 31. Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., and Rossant J. (2005) Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 10.1242/dev.01801 [DOI] [PubMed] [Google Scholar]
- 32. Home P., Kumar R. P., Ganguly A., Saha B., Milano-Foster J., Bhattacharya B., Ray S., Gunewardena S., Paul A., Camper S. A., Fields P. E., and Paul S. (2017) Genetic redundancy of GATA factors in the extraembryonic trophoblast lineage ensures the progression of preimplantation and postimplantation mammalian development. Development 144, 876–888 10.1242/dev.145318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ralston A., Cox B. J., Nishioka N., Sasaki H., Chea E., Rugg-Gunn P., Guo G., Robson P., Draper J. S., and Rossant J. (2010) Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 10.1242/dev.038828 [DOI] [PubMed] [Google Scholar]
- 34. Russ A. P., Wattler S., Colledge W. H., Aparicio S. A., Carlton M. B., Pearce J. J., Barton S. C., Surani M. A., Ryan K., Nehls M. C., Wilson V., and Evans M. J. (2000) Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95–99 10.1038/35003601 [DOI] [PubMed] [Google Scholar]
- 35. Cao Z., Carey T. S., Ganguly A., Wilson C. A., Paul S., and Knott J. G. (2015) Transcription factor AP-2γ induces early Cdx2 expression and represses HIPPO signaling to specify the trophectoderm lineage. Development 142, 1606–1615 10.1242/dev.120238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kurowski A., Molotkov A., and Soriano P. (2019) FGFR1 regulates trophectoderm development and facilitates blastocyst implantation. Dev. Biol. 446, 94–101 10.1016/j.ydbio.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alarcon V. B. (2010) Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol. Reprod. 83, 347–358 10.1095/biolreprod.110.084400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng X., Xu S., Song C., He L., Lian X., Liu Y., Wei J., Pang L., and Wang S. (2016) Roles of ERα during mouse trophectoderm lineage differentiation: revealed by antagonist and agonist of ERα. Dev. Growth Differ. 58, 327–338 10.1111/dgd.12276 [DOI] [PubMed] [Google Scholar]
- 39. Pierce G. B., Arechaga J., Muro C., and Wells R. S. (1988) Differentiation of ICM cells into trophectoderm. Am. J. Pathol. 132, 356–364 [PMC free article] [PubMed] [Google Scholar]
- 40. Rossant J., and Lis W. T. (1979) Potential of isolated mouse inner cell masses to form trophectoderm derivatives in vivo. Dev. Biol. 70, 255–261 10.1016/0012-1606(79)90022-8 [DOI] [PubMed] [Google Scholar]
- 41. Zernicka-Goetz M., Morris S. A., and Bruce A. W. (2009) Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 10, 467–477 10.1038/nrg2564 [DOI] [PubMed] [Google Scholar]
- 42. Rossant J. (2018) Genetic control of early cell lineages in the mammalian embryo. Annu. Rev. Genet. 52, 185–201 10.1146/annurev-genet-120116-024544 [DOI] [PubMed] [Google Scholar]
- 43. Cockburn K., Biechele S., Garner J., and Rossant J. (2013) The hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23, 1195–1201 10.1016/j.cub.2013.05.044 [DOI] [PubMed] [Google Scholar]
- 44. Kim I., Saunders T. L., and Morrison S. J. (2007) Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130, 470–483 10.1016/j.cell.2007.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Viotti M., Nowotschin S., and Hadjantonakis A. K. (2014) SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat. Cell Biol. 16, 1146–1156 10.1038/ncb3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lilly A. J., Lacaud G., and Kouskoff V. (2017) SOXF transcription factors in cardiovascular development. Semin. Cell Dev. Biol. 63, 50–57 10.1016/j.semcdb.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 47. Alexander J., and Stainier D. Y. (1999) A molecular pathway leading to endoderm formation in zebrafish. Curr. Biol. 9, 1147–1157 10.1016/S0960-9822(00)80016-0 [DOI] [PubMed] [Google Scholar]
- 48. Clements D., and Woodland H. R. (2000) Changes in embryonic cell fate produced by expression of an endodermal transcription factor, Xsox17. Mech. Dev. 99, 65–70 10.1016/S0925-4773(00)00476-7 [DOI] [PubMed] [Google Scholar]
- 49. Hudson C., Clements D., Friday R. V., Stott D., and Woodland H. R. (1997) Xsox17α and -β mediate endoderm formation in Xenopus. Cell 91, 397–405 10.1016/S0092-8674(00)80423-7 [DOI] [PubMed] [Google Scholar]
- 50. Morris S. A., Teo R. T., Li H., Robson P., Glover D. M., and Zernicka-Goetz M. (2010) Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl. Acad. Sci. U.S.A. 107, 6364–6369 10.1073/pnas.0915063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chazaud C., Yamanaka Y., Pawson T., and Rossant J. (2006) Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 10, 615–624 10.1016/j.devcel.2006.02.020 [DOI] [PubMed] [Google Scholar]
- 52. Gerbe F., Cox B., Rossant J., and Chazaud C. (2008) Dynamic expression of Lrp2 pathway members reveals progressive epithelial differentiation of primitive endoderm in mouse blastocyst. Dev. Biol. 313, 594–602 10.1016/j.ydbio.2007.10.048 [DOI] [PubMed] [Google Scholar]
- 53. Houghton F. D. (2006) Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation 74, 11–18 10.1111/j.1432-0436.2006.00052.x [DOI] [PubMed] [Google Scholar]
- 54. Lorthongpanich C., Doris T. P., Limviphuvadh V., Knowles B. B., and Solter D. (2012) Developmental fate and lineage commitment of singled mouse blastomeres. Development 139, 3722–3731 10.1242/dev.086454 [DOI] [PubMed] [Google Scholar]
- 55. Kawahara M., Koyama S., Iimura S., Yamazaki W., Tanaka A., Kohri N., Sasaki K., and Takahashi M. (2015) Preimplantation death of xenomitochondrial mouse embryo harbouring bovine mitochondria. Sci. Rep. 5, 14512 10.1038/srep14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Quinn P., Kerin J. F., and Warnes G. M. (1985) Improved pregnancy rate in human in vitro fertilization with the use of a medium based on the composition of human tubal fluid. Fertil. Steril. 44, 493–498 10.1016/S0015-0282(16)48918-1 [DOI] [PubMed] [Google Scholar]
- 57. Quinn P., Barros C., and Whittingham D. G. (1982) Preservation of hamster oocytes to assay the fertilizing capacity of human spermatozoa. J. Reprod. Fertil. 66, 161–168 [DOI] [PubMed] [Google Scholar]
- 58. Whitten W. K. (1956) Culture of tubal mouse ova. Nature 12, 260–274 [DOI] [PubMed] [Google Scholar]
- 59. Brackett B. G., and Oliphant G. (1975) Capacitation of rabbit spermatozoa in vitro. Biol. Reprod. 12, 260–274 10.1095/biolreprod12.2.260 [DOI] [PubMed] [Google Scholar]
- 60. Aono A., Nagatomo H., Takuma T., Nonaka R., Ono Y., Wada Y., Abe Y., Takahashi M., Watanabe T., and Kawahara M. (2013) Dynamics of intracellular phospholipid membrane organization during oocyte maturation and successful vitrification of immature oocytes retrieved by ovum pick-up in cattle. Theriogenology 79, 1146–1152.e1 10.1016/j.theriogenology.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 61. Gwatkin R. B. (1964) Effect of enzymes and acidity on the zona pellucida of the mouse egg before and after fertilization. J. Reprod. Fertil. 7, 99–105 10.1530/jrf.0.0070099 [DOI] [PubMed] [Google Scholar]
- 62. Kawahara M., Wu Q., Takahashi N., Morita S., Yamada K., Ito M., Ferguson-Smith A. C., and Kono T. (2007) High-frequency generation of viable mice from engineered bi-maternal embryos. Nat. Biotechnol. 25, 1045–1050 10.1038/nbt1331 [DOI] [PubMed] [Google Scholar]
- 63. Katagiri S., and Takahashi Y. (2006) Potential relationship between normalization of endometrial epidermal growth factor profile and restoration of fertility in repeat breeder cows. Anim. Reprod. Sci. 95, 54–66 10.1016/j.anireprosci.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 64. Akizawa H., Kobayashi K., Bai H., Takahashi M., Kagawa S., Nagatomo H., and Kawahara M. (2018) Reciprocal regulation of TEAD4 and CCN2 for the trophectoderm development of the bovine blastocyst. Reproduction 155, 563–571 10.1530/REP-18-0043 [DOI] [PubMed] [Google Scholar]
- 65. Yamazaki W., Amano T., Bai H., Takahashi M., and Kawahara M. (2016) The influence of polyploidy and genome composition on genomic imprinting in mice. J. Biol. Chem. 291, 20924–20931 10.1074/jbc.M116.744144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., et al. (2000) Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. The Gene Ontology Consortium (2017) Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.