Abstract
Eukaryotic ribosomal biogenesis is a high-energy–demanding and complex process that requires hundreds of trans-acting factors to dynamically build the highly-organized 40S and 60S subunits. Each ribonucleoprotein complex comprises specific rRNAs and ribosomal proteins that are organized into functional domains. The RNA exosome complex plays a crucial role as one of the pre-60S–processing factors, because it is the RNase responsible for processing the 7S pre-rRNA to the mature 5.8S rRNA. The yeast pre-60S assembly factor Nop53 has previously been shown to associate with the nucleoplasmic pre-60S in a region containing the “foot” structure assembled around the 3′ end of the 7S pre-rRNA. Nop53 interacts with 25S rRNA and with several 60S assembly factors, including the RNA exosome, specifically, with its catalytic subunit Rrp6 and with the exosome-associated RNA helicase Mtr4. Nop53 is therefore considered the adaptor responsible for recruiting the exosome complex for 7S processing. Here, using proteomics-based approaches in budding yeast to analyze the effects of Nop53 on the exosome interactome, we found that the exosome binds pre-ribosomal complexes early during the ribosome maturation pathway. We also identified interactions through which Nop53 modulates exosome activity in the context of 60S maturation and provide evidence that in addition to recruiting the exosome, Nop53 may also be important for positioning the exosome during 7S processing. On the basis of these findings, we propose that the exosome is recruited much earlier during ribosome assembly than previously thought, suggesting the existence of additional interactions that remain to be described.
Keywords: ribosome assembly, rRNA processing, precursor ribosomal RNA (pre-rRNA), exosome complex, protein–protein interaction, pre-60S particles
Introduction
The assembly of eukaryotic ribosomal subunits is a complex process that requires, in addition to the ribosomal proteins, more than 200 biogenesis factors (1). Synthesis of ribosomal subunits starts in the nucleolus, where the precursors rRNAs pre-35S and pre-5S are transcribed and bound by early processing and assembly factors (2). Early endonucleolytic cleavage reactions separate the pre-20S that is going to be part of the pre-40S subunit, from the pre-27SA2, which will give rise to the pre-rRNAs 7S and 26S, and subsequently to the mature rRNAs 5.8S and 25S (3).
In the pre-rRNA maturation pathway, the RNA exosome complex is responsible for the degradation of the spacer sequence 5′-external transcribed spacer (5′-ETS)3 after cleavage at A0 and for the 3′–5′-end processing of 7S pre-rRNA to the mature 5.8S rRNA (4, 5). In Saccharomyces cerevisiae, the exosome is composed of a nine-subunit core (Exo9) arranged as a heterohexameric ring (Rrp41, Rrp42, Rrp45, Rrp43, Mtr3, and Rrp46) capped by a heterotrimeric “cap” (Rrp40, Csl4, and Rrp4) that lacks catalytic activity. In the nucleus and cytoplasm, Exo9 interacts with Rrp44/Dis3 to form a 10-subunit complex (Exo10). Rrp44 is an RNase II family member and catalyzes both endoribonucleolytic and processive 3′–to–5′-exoribonucleolytic reactions (6–8). The yeast nuclear exosome (Exo11) contains an extra catalytic subunit, Rrp6, which is a distributive 3′–to–5′-exoribonuclease homolog of Escherichia coli RNase D (9), that binds to the trimeric cap and upper portion of the hexameric ring, opposite to the Rrp44-binding site. Although RRP6 is not essential, its deletion causes a slow-growth phenotype, temperature sensitivity, and RNA-processing defects (4, 10).
Nuclear and cytoplasmic forms of the exosome interact with distinct cofactors in these subcellular compartments (11). In the nucleus, Rrp47, Mpp6, and the RNA helicase Mtr4 associate with the exosome (Exo11) to form a 14-subunit complex (12). The exosome cofactor Mpp6 can associate with the Exo9 core through interactions with the “cap” subunit Rrp40 to recruit the other cofactor Mtr4 and to stimulate the activities of the exosome catalyzed by Rrp6 (13). Rrp47 is a nuclear exosome cofactor important for Rrp6 stability and activity (14). The N-terminal domains of Rrp6 and Rrp47 interact in an intertwined set of α-helices that form an interface for interaction with the N terminus of the RNA helicase Mtr4 (15, 16), a subunit of the TRAMP complex that bridges the exosome and the TRAMP (Trf4p/Air2p/Mtr4p polyadenylation complex) (17, 18) and that is also essential for the 7S pre-rRNA processing (19). The determination of Mtr4 structure revealed that it interacts with the other two subunits of the TRAMP complex through its DEXH domain (20), whereas its KOW domain is involved in the interaction with the pre-60S trans-acting factor Nop53 (17).
Nop53 is a pre-60S assembly factor (21, 22) that recently had its structure determined in the Nog2 intermediate (23–25). In that particle, Nop53 binds 25S rRNA and interacts with Nop7, Rpl7, and L27 in a region comprising the pre-60S foot structure assembled around the ITS2 (internal transcribed spacer 2) and 5.8S 3′ end (25, 26). Additionally, Nop53 has also been shown to interact with the exosome catalytic subunit Rrp6 and with the TRAMP subunits Trf4 and Mtr4 (27–30). Interestingly, depletion of Nop53 leads to the accumulation of 7S pre-rRNA (29), in agreement with its role as an adaptor protein that recruits the exosome associated with Mtr4 (23).
Endonucleolytic cleavage at the C2 site in ITS2 of pre-rRNA 27S separates pre-rRNAs 7S (5.8S+5′ region of ITS2) and 26S (3′ region of ITS2 plus 25S) (31), which undergo exonucleolytic processing by the exosome and Rat1/Rai1, respectively, to generate mature rRNAs 5.8S and 25S (31, 32).
Altogether, in the current model Nop53, Mtr4, and the RNA exosome are essential for the ITS2 processing of 7S after the cleavage at C2 (32), when the exosome subunit Rrp44 shortens 7S to the intermediate 5.8S+30, which is then handed over to Rrp6 that trims it to 6S pre-rRNA, which gives rise to the mature 5.8S rRNA after further processing in the cytoplasm (33).
Despite the available information on Nop53 interactions and structure, a complete understanding of its interplay with the exosome during pre-rRNA 7S processing is lacking. The elongated topology of Nop53 establishing several protein–protein interactions with pre-60S components (24) raised the possibility that it could also interact with other exosome subunits. In this work, we show evidence that Nop53 may be involved not only in the recruitment but also in positioning of the nuclear exosome on the pre-60S particle for processing of 7S pre-rRNA, based on the observation that, in addition to its direct interaction with Rrp6 and Mtr4, Nop53 also interacts with the core subunit Rrp45 and the exosome cofactor Mpp6. Furthermore, we show that Nop53 depletion affects the exosome interactome, particularly enriching components of early pre-ribosomal subunits.
Results
Nop53 can interact with the exosome core subunit Rrp45 and the cofactor Mpp6
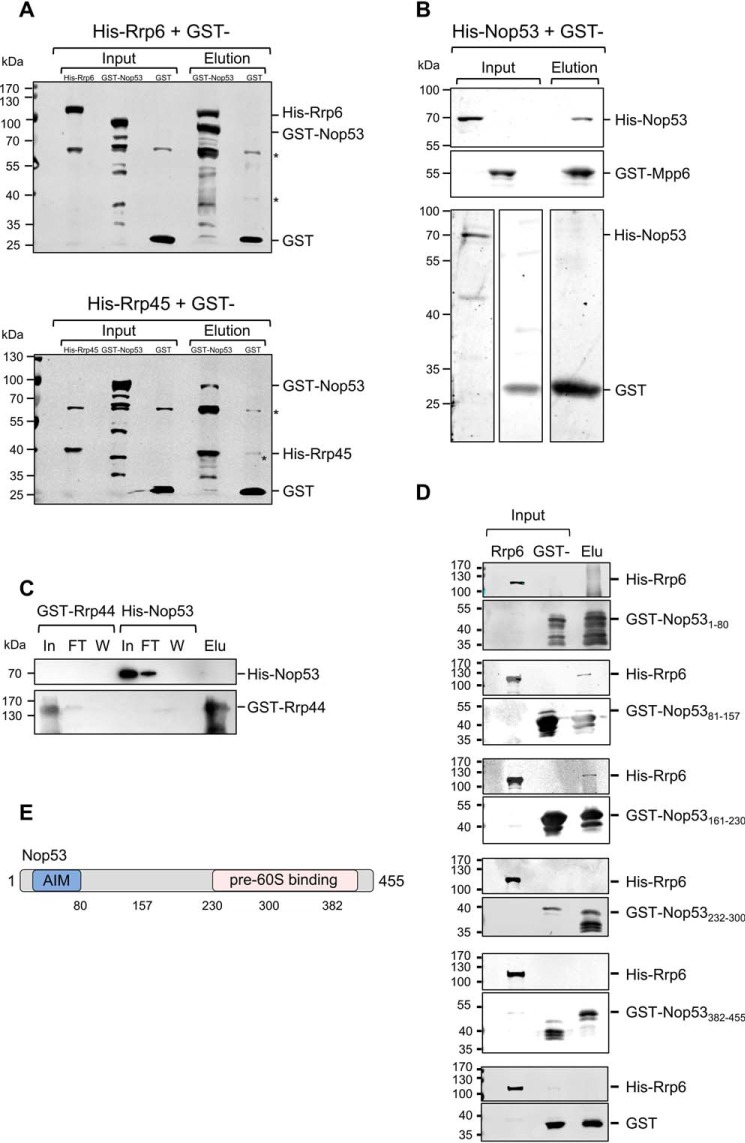
To investigate in more detail the role played by Nop53 in exosome recruitment to pre-60S for processing of the pre-rRNA 7S, we tested its interaction with core exosome subunits and with the exosome cofactor Mpp6. In addition to its interaction with Rrp6 (30), we show by pulldown assays that Nop53 also interacts with the exosome core subunit Rrp45 and with Mpp6, but not with Rrp44 (Fig. 1, A–C). Based on the available structure of the nuclear exosome (12) showing that Rrp45 and Mpp6 are exposed on the same side of the complex, it is conceivable to hypothesize that Nop53 could not only recruit the 14-subunit exosome but also be important for the positioning of the exosome on the pre-60S particle during 7S pre-rRNA processing. This hypothesis takes into account that although Nop53 interacts with Rrp6, Rrp45, Mtr4, and Mpp6, the complex subunit that process the 7S substrate is Rrp44 (4), positioned on the opposite side of the exosome (34).
Figure 1.
Nop53 interacts with the exosome and with exosome cofactors. GST pulldown assays were performed to test the interaction between Nop53 and the exosome components Rrp44 and Rrp45 and with the exosome cofactor Mpp6. A, GST–Nop53 immobilized in GSH-Sepharose beads pulled down His–Rrp6 and His–Rrp45. GST was used as negative control of interaction. *, E. coli proteins nonspecifically recognized by anti-GST antibody. B, GST–Mpp6, but not GST, also pulled down His–Nop53, whereas GST–Rrp44 did not (C). D, Rrp6 interacts with the N-terminal half of Nop53. The interaction between different GST-fused Nop53 truncation mutants was tested against His–Rrp6 by GST pulldown assay indicating that only the N-terminal region of Nop53 is able to interact with the exosome catalytic subunit Rrp6. GST was used as a negative control. Total extract of cells expressing His–Rrp6 was incubated either with purified GST or GST–Nop53 mutants. In, input; FT, flow-through; W, wash; Elu, elution. E, schematic representation of Nop53 with the relative positions of the amino acids indicated in the truncation mutants.
Because of its simultaneous interaction with many pre-60S proteins (25), we set out to determine the portions of Nop53 involved in the interaction with Rrp6. With this aim, deletion mutants of Nop53 were constructed, fused to GST, and used in pulldown experiments with His–Rrp6. His–Rrp6 interacted with mutants corresponding to the N-terminal portion of Nop53, namely Nop531–80, Nop5381–157, and Nop53161–230 (Fig. 1, D and E). The C-terminal half of Nop53 has been shown to be involved in anchoring this protein to pre-60S (25), and consistently, the corresponding mutants did not interact with Rrp6. Importantly, Nop53 interacts with Mtr4 through its N-terminal AIM motif (residues 59–64) (23). Because the mutant Nop53161–230, which does not contain the AIM motif, interacts with Rrp6, these results suggest that Nop53 may interact with Rrp6 and Mtr4 at the same time during pre-rRNA 7S processing.
Nop53 affects the exosome interactome but not the exosome complex stability
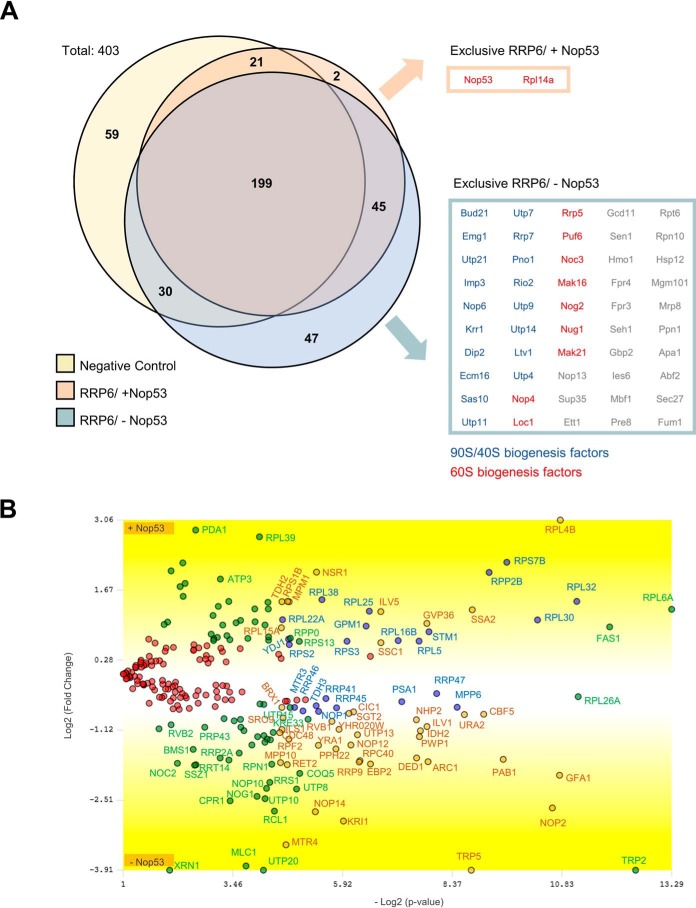
Considering the Nop53 direct interaction with the exosome, we set out to analyze in more detail the effects of Nop53 on the Rrp6 interactome. For this purpose, coimmunoprecipitation experiments were performed with Rrp6-TAP expressed in the conditional strain Δnop53/tetOff-GFP-NOP53 carrying the endogenously tagged Rrp6–TAP. In this strain, Nop53 expression was repressed by the addition of doxycycline to the medium. Proteins coimmunoprecipitated with Rrp6–TAP, in the presence or absence of Nop53, were identified by MS-based proteomics. Coimmunoprecipitation assays were also performed using the same conditional strain expressing only the TAP tag, used as control for unspecific binding to the resin. Biological triplicates (Fig. S1) of Rrp6–TAP and TAP tag coimmunoprecipitations were subjected to MS-based label-free quantitative proteomics. Interestingly, Nop53 depletion led to higher levels of ribosomal assembly factors being copurified with the exosome (Fig. 2), indicating that in addition to its recruitment function, Nop53 may also be required for the release of the exosome from pre-ribosomal particles.
Figure 2.
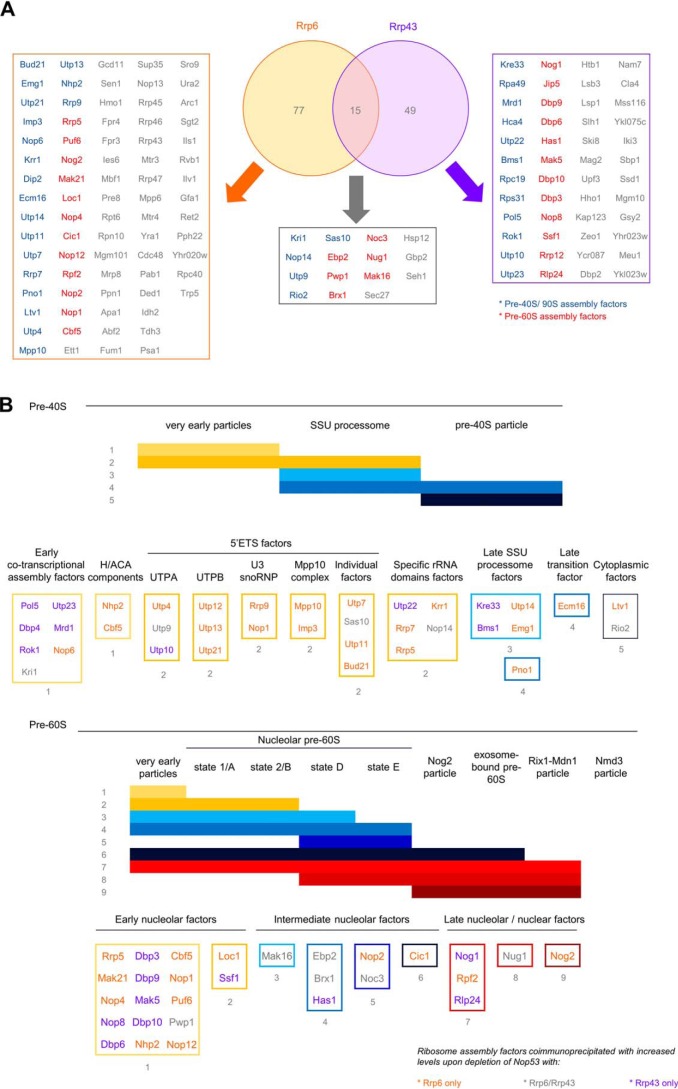
Depletion of Nop53 affects the Rrp6 interactome. To evaluate how Nop53 modulates the interaction of Rrp6 with other factors, a coimmunoprecipitation assay was performed both in the presence (−doxycycline) and upon depletion (+doxycycline) of Nop53 using the conditional strain Δnop53/tetOff::GFP-NOP53 carrying the endogenous Rrp6–TAP fusion. As a negative control, the same strain carrying only the TAP tag was employed. For each condition, the elution of biological triplicates was subjected to label-free quantitative analysis. Using the PatternLab software, the identified proteins in at least two biological replicates were grouped in a Venn diagram (A), showing that among a total of 403 proteins, two were exclusively identified in the presence of Nop53, and 47 uniquely upon depletion of Nop53. Three additional proteins were included in this latter group because, despite being present in the negative control, they were significantly enriched upon depletion of Nop53 (Fig. S3). Separately, the exclusively identified proteins in each condition are listed in boxes, highlighting the over-representation of pre-60S (red) and pre-90S/pre-40S (blue) assembly factors. B, proteins that were coimmunoprecipitated with Rrp6 both in the presence and absence of Nop53 (identified in at least two replicates per group) were compared through TFold analysis (benjamini-hochberg q-value; F-stringency (fold-change stringency parameter); L-stringency (stringency parameter for lowly-abundant proteins)) using PatternLab software (52). The top and bottom of the volcano plot display proteins, respectively, depleted and enriched upon depletion of Nop53. Among the 244 commonly identified proteins, 104 did not show a statistically significant change upon depletion of Nop53 (red dots), 74 met the fold change criteria but were not statistically significant (green dots), 43 are low abundance proteins that met both the fold change and statistical criteria (orange dots), and 23 proteins met both the fold change and statistical criteria (blue dots).
Using PatternLab for proteomics 4.1.1.4 software, the identified proteins in at least two biological replicates for each condition (presence and depletion of Nop53) were grouped and depicted in a Venn diagram (Fig. 2A; Table S1) along with the negative control. From the total 403 proteins, 47 were exclusively identified upon depletion of Nop53, and two were uniquely detected in the presence of Nop53. Two independent TFold analyses were performed (Rrp6–TAP against the Negative Control, Fig. 2A; Table S1) to classify the proteins that were also identified in the negative control. Only those with a statistically significant increase in the Rrp6–TAP group (in the presence or absence of Nop53) were considered. Applying this filter, the number of uniquely identified proteins coimmunoprecipitated with Rrp6 in the presence and absence of Nop53 were 2 and 50, respectively. Three additional proteins were included in this latter group, because, despite being present in the negative control, they were significantly enriched upon depletion of Nop53.
Importantly, Nop53 was identified among the exclusively coimmunoprecipitated proteins with Rrp6–TAP (Fig. 2A; Table S1), corroborating the pulldown assays (Fig. 1). The most striking result to emerge from these data, however, was that upon depletion of Nop53, several early nucleolar pre-60S transacting factors (e.g. Mak21, Loc1, and Mak16) as well as small subunit processome (SSU) processome factors (e.g. Utp7, Utp9, and Imp3) (35) appeared to become stably associated with the Rrp6 complex (Fig. 2A).
For the coimmunoprecipitated proteins identified in both conditions (in at least two biological replicates), a TFold analysis was performed, in which the more negative the fold change, the more enriched the protein is upon depletion of Nop53 (Fig. 2B; Fig. S2; Table S2). Similarly, the higher a positive fold change is, the more reduced the level of this protein is found upon depletion of Nop53. Altogether, the volcano plot shows that among the 244 commonly identified proteins, 104 satisfy neither the fold change cutoff nor the FDR cutoff α (red dots), 74 satisfy the fold change cutoff but not α (green dots), 43 satisfy both the fold change cutoff and α but are proteins of low abundance in the samples (orange dots), and 23 proteins satisfy all statistical filters (blue dots), being significantly enriched or reduced upon Nop53 depletion (Fig. 2B).
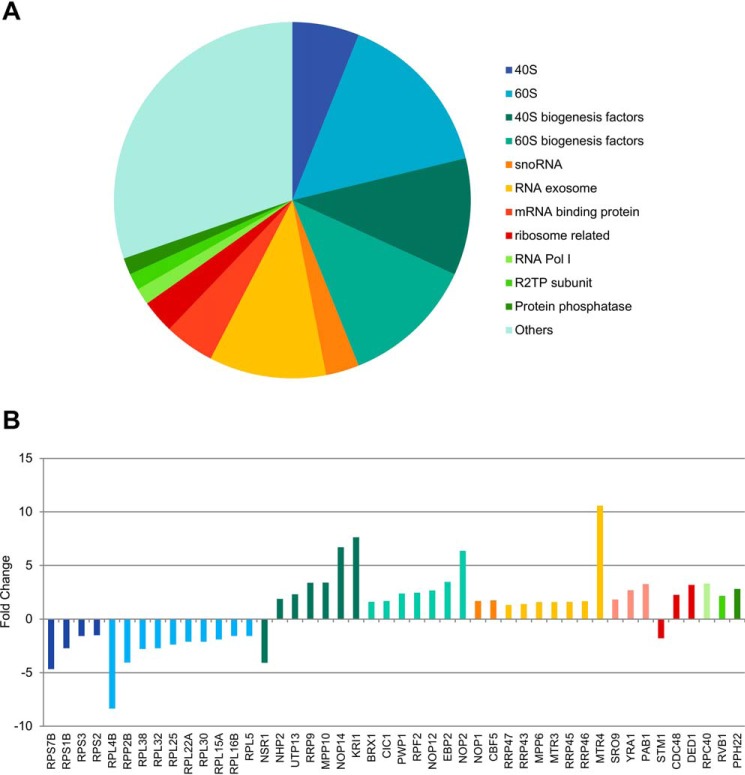
In Fig. 3, only the statistically significant differences (orange and blue dots) are depicted in a pie chart divided according to biological function. It is quite evident that the coimmunoprecipitated proteins whose levels were significantly affected by the Nop53 depletion are mainly related to ribosome biogenesis and the exosome complex. To evaluate in detail how these groups varied, each protein comprising each pie chart sector is shown with the respective fold change (Fig. 3B). (Positive fold change: increase upon Nop53 depletion; Negative fold change: decrease upon Nop53 depletion.) Surprisingly, one of the most significant increases of proteins associated with Rrp6–TAP was that of the RNA helicase Mtr4 (Fig. 3), with which Rrp6 directly interacts during 7S pre-rRNA processing. These results suggest that in the absence of Nop53, Rrp6 and Mtr4 are more tightly associated.
Figure 3.
Rrp6 remains associated with the core exosome complex and is retained in immature pre-ribosomal subunits upon depletion of Nop53. A, proteins that showed a statistically significant enrichment or depletion (both orange and blue dots in Fig. 4B) in the absence of Nop53 were categorized according to their biological function in a pie chart. Among the most enriched classes, both ribosome biogenesis and exosome complex stood out. B, same proteins, significantly affected by Nop53 depletion, are depicted with their respective fold changes. Positive and negative fold change values indicate, respectively, enrichment and decrease upon Nop53 depletion. Of note, several early pre-60S and pre-40S transacting factors were found coimmunoprecipitated with increased levels in the absence of Nop53. The exosome subunits and cofactors were slightly enriched, but not as much as the RNA helicase Mtr4.
When comparing both conditions, the statistically valid results with high fold changes were found mainly related to ribosomal maturation factors (Figs. 2 and 3). Considering the role of Nop53 in recruiting the exosome to the pre-60S (23), we expected that upon its depletion the exosome would be associated with pre-ribosomal assembly factors to a lesser extent. Intriguingly, however, the results indicate that the major effect of Nop53 depletion on the exosome interactome is its stronger association with early pre-60S particles, as well as with 90S and pre-40S particles. Among the possible interpretations, the exosome could be retained with earlier pre-ribosome intermediates or there could be an increased recruitment of the exosome to degrade defective pre-ribosomal subunits. In the first case, the exosome could already be associated with pre-60S even before the recruitment of Nop53 to the Nog2 pre-60S particle.
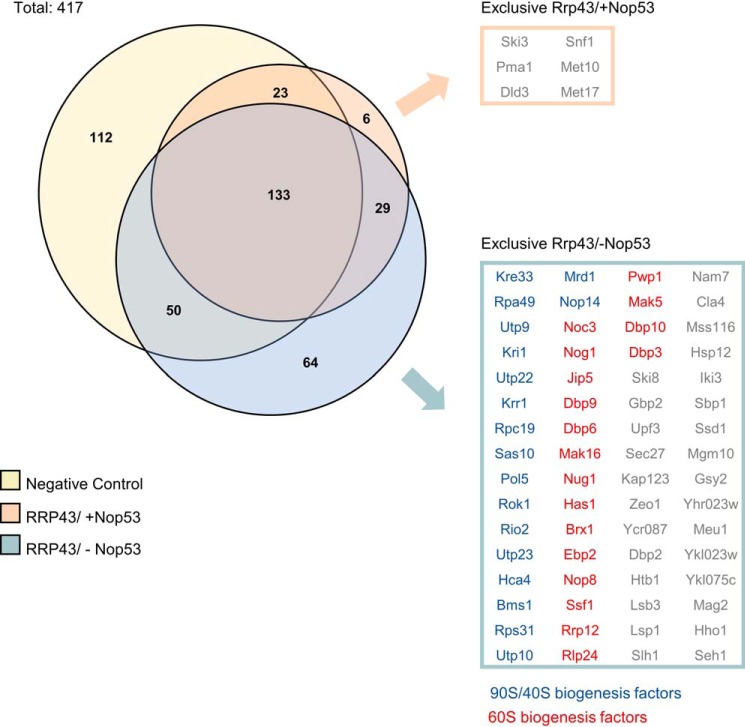
To verify whether Nop53 was affecting the association of only Rrp6 with pre-ribosomes or whether the whole exosome was being retained in pre-ribosomal complexes, similar experiments were performed with Δnop53/tetOff-GFP-NOP53 expressing Rrp43-TAP. The results show that the exosome core subunit Rrp43 also copurified more pre-60S, 90S, and pre-40S factors upon depletion of Nop53 (Fig. 4; Table S3), further indicating that the whole exosome could be retained in earlier pre-ribosomal particles.
Figure 4.
Depletion of Nop53 also affects the Rrp43 interactome. To analyze how Nop53 modulates the interaction of Rrp43 with other factors, a coimmunoprecipitation assay was performed both in the presence (−doxycycline) and upon depletion (+doxycycline) of Nop53 using the conditional strain Δnop53/tetOff::GFP-NOP53 containing RRP43-TAP fusion in a plasmid. As a negative control, the same strain carrying only the TAP tag was used. For each condition, the elution of biological triplicates was subjected to label-free quantitative analysis. Using the PatternLab software, the identified proteins in at least two biological replicates were grouped in a Venn diagram, showing that among a total of 417 proteins, six were exclusively identified in the presence of Nop53, and 64 uniquely upon depletion of Nop53. The exclusively identified proteins in each condition are listed in boxes, highlighting the over-representation of pre-60S (red) and pre-90S/pre-40S (blue) assembly factors.
Analysis of the protein components of pre-ribosomal particles 90S, 40S, and 60S that had their levels increased in association with the exosome upon depletion of Nop53 allowed us to pinpoint the specific stages of ribosome maturation that were more enriched (Fig. 5). Most of the identified 90S/pre-40S components corresponded to early cotranscriptional (e.g. Pol5, Kri1, and Nop6) and 5′ETS factors (e.g. Utp9, Mpp10, and Sas10), in agreement with the exosome role in the degradation of that upstream spacer. Interestingly, several SSU processome factors (e.g. Nop14, Krr1, Kre33, Pno1, and Emg1) were also found enriched in the samples, which could suggest an earlier association of the exosome, prior to the endonucleolytic cleavage that releases 5′ETS. The identification of two late 40S maturation factors (Ltv1 and Rio2), however, could be related to an increased recruitment of the exosome to degrade misassembled intermediates (Fig. 5B).
Figure 5.
Comparison of the proteins copurified with Rrp6 and Rrp43 that showed increased levels upon depletion of Nop53. A, Venn diagram depicting the identified proteins in at least two replicates coimmunoprecipitated with Rrp6 and Rrp43, whose levels were increased upon depletion of Nop53. B, schematics of pre-40S and pre-60S maturation pathways, depicting stages at which the identified proteins copurified with Rrp6 or Rrp43 associate with pre-ribosomes. Classification was based on Ref. 29. Proteins not classified are as follows: pre-40S proteins Rpa49 and Rpc19; pre-60S proteins Jip5 and Rrp12. No common proteins had their levels reduced upon depletion of Nop53.
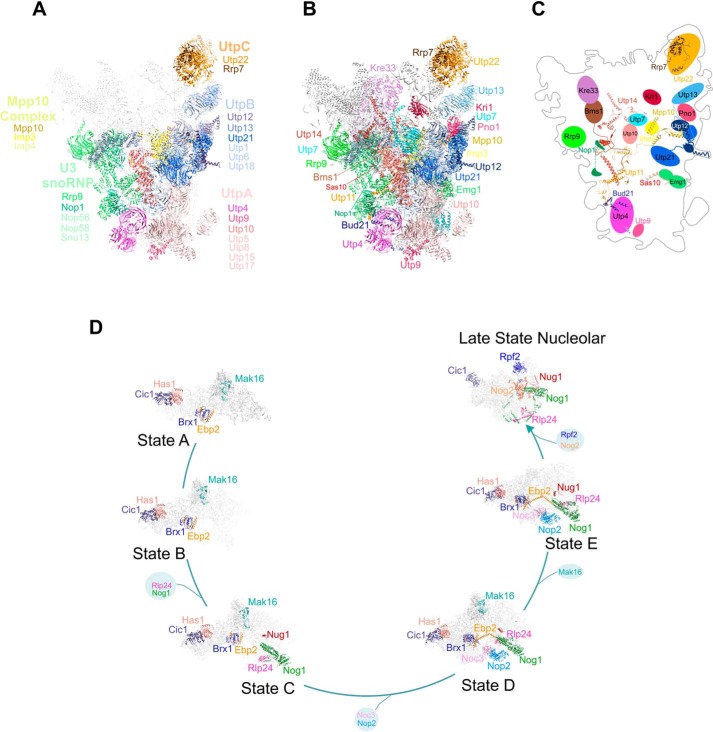
The most interesting observation, however, came from the analysis of the enriched pre-60S assembly factors coimmunoprecipitated with both Rrp6 and Rrp43 upon depletion of Nop53 (Fig. 5B). Not only were several late nucleolar factors constituents of the already described exosome-bound pre-60S particle detected (namely Cic1, Nog1, Nug1, Nog2, Rpf2, and Rlp24) (12, 25), but also earlier factors. Notably, along with several components of very early particles, one of the most enriched maturation stages was the state E, whose identified assembly factors Noc3, Brx1, Nop2, Ebp2, Has1, and Nop2 have to be released to allow the binding of Nog2, Rsa4, and Nop53 (25, 26, 36). This compositional remodeling coincides with the exit of the pre-60S from the nucleolus to the nucleoplasm (35). Late maturation factors, such as those found in Nmd3 particles, or cytoplasmic assembly factors were not enriched. Altogether, these results strongly converge to indicate an early association of the exosome both during 90S/pre-40S and pre-60S maturation. Interestingly, most of the 90S and pre-60S factors that were more abundantly associated with the exosome upon depletion of Nop53 interact with each other, are positioned in close proximity in these particles, or are on the same side of the particles (Fig. 6) (25, 36, 37).
Figure 6.
Proteins coimmunoprecipitated with the exosome in higher levels in the absence of Nop53 participate in different phases of ribosomal maturation. A, structural representation of 90S pre-ribosomes with the identified protein complexes are depicted in different colors as follows: pink, UTP-A; blue, UTP-B; orange, UTP-C; green, U3 snoRNP; yellow, Mpp10 complex. Proteins in bold letters are those indicated in Fig. 5B. The remaining parts of the particle are represented in light gray. B, individual proteins from the subcomplexes indicated in (A) are highlighted in different colors. C, schematics of the positions of the proteins in the 90S particle and their interactions within the particle. Note that all proteins interacting more efficiently with the exosome in the absence of Nop53 are exposed on the same face of 90S. Structure of the 90S particle was based on Ref. 37 (PDB 5WLC). D, representation of pre-60S maturation pathway, with the factors identified here highlighted in different colors. The exosome associates with various pre-60S intermediates in the absence of Nop53. Schematics show the interactions between the proteins identified here within the pre-60S. Structures of pre-60S particles were based on Refs. 25, 36. State A (PDB 6EM3), state B (PDB 6EM4), state C (PDB 6EM1), state D (PDB 6EM5), state E (PDB 6ELZ), and late nuclear states (PDB 3JCT) are shown.
The proteins that were copurified at increased levels with the exosome upon depletion of Nop53 were separated by functional complexes using the STRING database, which also allows the visualization of protein interactions (38). Fig. S3 shows the enriched 90S/pre-40S and pre-60S assembly factors copurified with Rrp6 and Rrp43 upon depletion of Nop53. The results summarized in this figure reinforce the conclusion that depletion of Nop53 leads to the retention of the exosome in pre-ribosomal complexes.
Regarding the exosome assembly, both Rrp6 and Rrp43 copurified all exosome subunits in both conditions (Fig. S4), showing that Nop53 did not significantly affect the assembly and/or stability of the exosome complex.
Depletion of Nop53 leads to more stable association of the exosome with pre-ribosomal particles
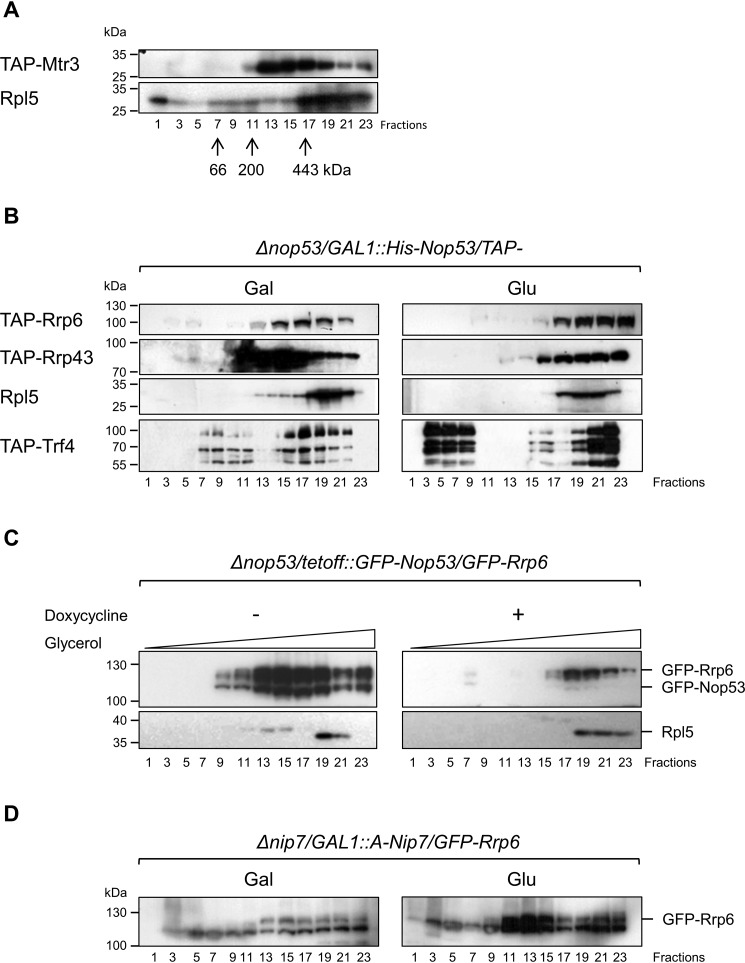
To confirm the hypothesis that Nop53 depletion affects the association of the exosome with pre-ribosomes, total extracts of different TAP-tagged strains were subjected to density gradient analysis thus allowing us to analyze the sedimentation of complexes. As a control, we used a strain expressing TAP–Mtr3 (representative of the exosome core). Despite being present in the bottom fractions, Mtr3 was mainly concentrated in fractions 13–17 (Fig. 7A), where complexes of ∼400 kDa sedimented (Fig. S5A) and could correspond to free exosome complexes. 60S ribosomal protein Rpl5 was clearly concentrated at the bottom of the glycerol gradient but was also present in higher fractions (Fig. 7A; Fig. S6A). In the strain Δnop53/GAL::NOP53 growing in galactose (presence of Nop53), Rrp43 showed the same profile as Mtr3 (Fig. 7B, Gal; Fig. S6B). Similar to the core exosome subunits Mtr3 and Rrp43, Rrp6 was present in fractions 13–17 but was more concentrated in fractions 15–19, denoting a larger complex (Fig. 7B; Fig. S6B). Interestingly, the TRAMP subunit Trf4 cosedimented with the exosome in the lower part of the gradient, and it was more concentrated in the same fractions as Rrp6 but was also present in upper fractions, probably corresponding to free TRAMP complex (Fig. 7B; Fig. S6B). Consistent with the results of protein coimmunoprecipitation described above, the sedimentation of the exosome subunits changed upon depletion of Nop53 (Glu), becoming more concentrated in the lower fractions of the gradient, where Rpl5 was also present (Fig. 7B, right panel; Fig. S6B). These results indicate that in the absence of Nop53, the exosome remains bound to pre-60S, suggesting a role for Nop53 in the release of the exosome from the nucleolar pre-ribosomes. Curiously, the peak of Trf4 was also shifted to lower fractions of the gradient upon depletion of Nop53, but unlike the exosome subunits, Trf4 was also present in higher fractions of the gradient, probably as part of free TRAMP complex (Fig. 7B; Fig. S6B). Control experiments for the analysis of RNAs extracted from the glycerol gradient fractions show that the bottom fractions contain early precursor rRNAs, whereas 7S and 6S intermediates sediment in the middle fractions (Fig. S5B). The sedimentation of the ribosomal protein Rpl5 did not change upon depletion of Nop53 (Fig. 7; Fig. S6B).
Figure 7.
Nop53 affects exosome association with pre-60S. The conditional strain Δnop53/GAL::NOP53 was transformed with plasmids expressing either the exosome subunits Rrp43 and Rrp6 or the TRAMP subunit Tfr4 fused to the TAP tag to monitor the association of these complexes to the pre-60S particle in the presence or absence of Nop53. A, cell extract from a strain expressing TAP–Mtr3 was subjected to centrifugation through the glycerol gradient for separation of soluble proteins from ribosomal particles. Rpl5 was used as a control for the 60S and pre-60S ribosomal subunit. Molecular mass markers were centrifuged in parallel, and their fractionation is indicated by arrows. B, fractionation of Rrp43, Rrp6, and Trf4 in the presence (Gal) or absence (Glu) of Nop53 shows their concentration in lower fractions of the gradient after depletion of Nop53. C, similar experiments with the conditional strain Δnop53/tetOff::GFP-NOP53-expressing GFP–Rrp6 show the same concentration of this exosome subunit in lower fractions after depletion of Nop53 (+doxycycline). Numbers of fractions from the gradient are indicated. D, conditional strain Δnip7/GAL::A-NIP7 expressing GFP-Rrp6 was used as a control for a pre-60S maturation factor and shows that its depletion (Glu) does not affect Rrp6 fractionation.
To exclude the possibility that carbon source in the culture could influence the association of the exosome with other complexes in the cell, similar experiments were performed with strain Δnop53/tetOff::GFP-NOP53/GFP-Rrp6. In the presence of Nop53 (− doxycycline), Rrp6 was mainly present in the middle fractions of the gradient. After repression of Nop53 expression by the addition of doxycycline, Rrp6 sedimentation was shifted to the bottom of the gradient, cofractionating with Rpl5 (Fig. 7C, upper panels; Fig. S6C). These results strongly indicate that Nop53 plays a role in exosome release from pre-60S after processing of 7S pre-rRNA. The results of the coimmunoprecipitation of proteins with Rrp6–TAP shown above, however, indicate that Nop53 may influence the exosome release from even earlier pre-ribosome complexes. To determine whether the effect on the exosome sedimentation was due to the depletion of Nop53 or to a general inhibition of 60S maturation, Nip7, which is a pre-60S maturation factor and has been shown to interact with the exosome subunit Rrp43 (39), was also analyzed. The results show that depletion of Nip7 does not cause the retention of the exosome in larger complexes (Fig. 7D; Fig. S6D).
Rrp6–TAP copurifies early pre-rRNA intermediates
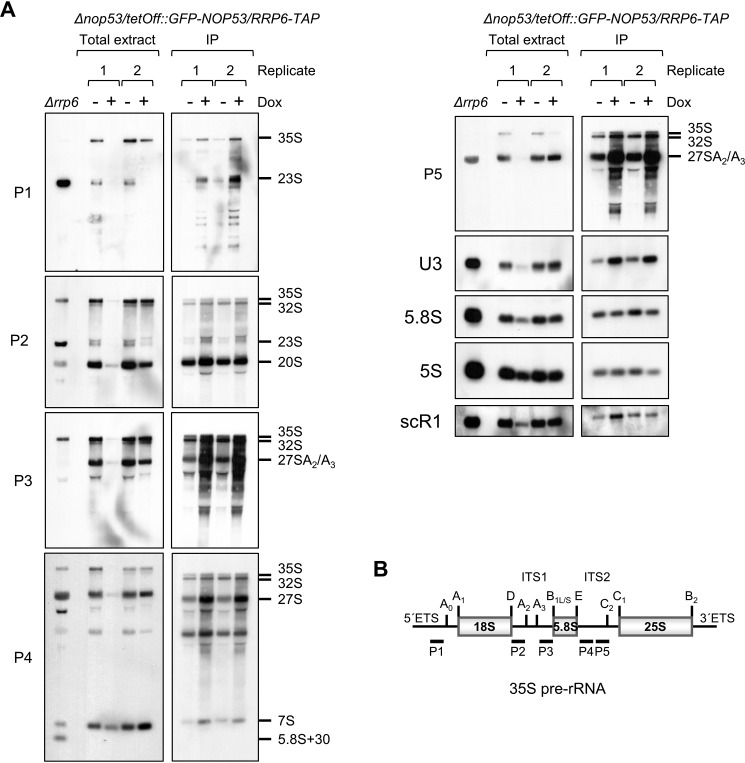
To further confirm the hypothesis that the depletion of Nop53 affects the exosome association with pre-ribosomes, RNA coimmunoprecipitation was performed using the strain Δnop53/tetOff::GFP-NOP53/RRP6-TAP. Corroborating the data presented above, Rrp6 copurified more pre-rRNA under depletion of Nop53 (Fig. 8, + dox). Interestingly, despite not leading to 23S accumulation in the cells, the absence of Nop53 led to more 23S coprecipitation with Rrp6 (Fig. 8A, probes P1 and P2). This dead-end processing intermediate is a substrate for degradation by the exosome (40–42), and its more efficient copurification with Rrp6 may suggest that absence of Nop53 affects earlier pre-rRNA processing steps. These results further confirm the more efficient copurification of pre-40S factors with Rrp6 upon depletion of Nop53. It remains to be defined whether the exosome found enriched in association with the 23S pre-rRNA targets it for degradation or waits for 5′ETS release. Further strengthening the hypothesis that Nop53 affects earlier pre-rRNA processing steps, 35S pre-rRNA was also more efficiently copurified with Rrp6 in the absence of Nop53 (Fig. 8A, probes P1–P5). Northern hybridization with different probes showed that Rrp6 not only co-purified more 35S and 23S pre-rRNAs upon depletion of Nop53, but also 32S (Fig. 8A, P2–P5), 27S (P3–P5) and 7S (P4) intermediates. More strikingly, Rrp6 copurified U3 small nuclear RNA and more efficiently in the absence of Nop53 (Fig. 8A, probe U3). 5S rRNA and scR1, which were nonspecifically copurified, did not show any enrichment in the absence of Nop53 (Fig. 8A; Fig. S7). Confirming the specificity of the RNAs being co-purified with Rrp6–TAP, mature 5.8S rRNAs was not enriched in the coimmunoprecipitated samples. These results strongly indicate that Nop53 affects the exosome release from pre-ribosomes and show that recruitment of the exosome may occur as early as the SSU processome is formed by the binding of U3 snoRNP.
Figure 8.
Rrp6–TAP co-purifies early precursor rRNAs. A, Δnop53/tetOff::GFP-NOP53/RRP6-TAP strain was used for RNA coimmunoprecipitation with Rrp6–TAP in the presence (−Dox (doxycycline )), or absence (+Dox) of Nop53. Total RNA extracted from Δrrp6 strain was used as a control of precursor rRNAs accumulating in the absence of Rrp6. Left panels show two biological replicates of RNAs extracted from aliquots of total cell extracts in the indicated conditions used in the coimmunoprecipitations as shown on the right panels. Precursor rRNAs are indicated on the right. 5S rRNA and scR1 were used as controls for nonspecific binding to the resin. Total RNA extracted from replicate 1 in the absence of Nop53 (+Dox) was lost during loading on the gel, but it was maintained in the figure because it clearly shows the accumulation of 7S pre-rRNA under this condition. B, schematic representation of the yeast 35S pre-rRNA indicating the hybridizing positions of the different probes.
In agreement with these observations, the analysis of Rrp6 subcellular localization, both in the presence and absence of Nop53, shows it is present in the nucleus, but particularly concentrated in the nucleolus (Fig. S8; supporting Experimental Procedure), further confirming its early binding to nucleolar pre-ribosome intermediates.
Discussion
The maturation factors involved in 60S ribosomal subunit formation, as well as the steps during which they bind and are released from the pre-60S, have been extensively studied in recent years (24, 33, 43), and Nop53 has been shown to bind pre-60S at the late nucleolar steps of maturation, at the same time as Rsa4 and Nog2, and to be released before the particle is transported to the cytoplasm (44). The binding and release of Nop53 coincide with the cleavage at site C2 in ITS2 and subsequent processing of 7S pre-rRNA by the exosome that will generate the intermediate 6S pre-rRNA (23). Depletion of Nop53 leads to the accumulation of the pre-rRNAs 27S and 7S (27–29), in agreement with its role in recruitment of the exosome. Nop53 has been shown to interact directly with the exosome catalytic subunit Rrp6 (30), and as shown here, this interaction is mediated by the N-terminal portion of Nop53, which includes a region encompassing the AIM motif required for the interaction with Mtr4 (45). The observation that partial deletions of the Nop53 AIM sequence cause the accumulation of 7S (a substrate for Rrp44) and of 5.8S + 30 (a substrate for Rrp6) (23) strongly suggests that the interaction of Nop53 with Rrp6 could be important during the 3′end handover from Rrp44 to Rrp6. Accordingly, it has recently been shown that during 7S pre-rRNA processing, Mtr4 directly binds the exosome subunit Rrp4, displacing the RNase domain of Rrp6, which becomes more exposed and available for other interactions (12). These observations, together with the results shown here, allow us to propose that Nop53 would be able to interact with Rrp6 concomitantly with the processing of 7S by Rrp44.
The recently resolved structure of the exosome-bound pre-60S particle containing 5.8S+30 pre-rRNA intermediate did not show clear cryo-EM density for Nop53 (12), indicating that Nop53 could be flexibly bound to pre-60S after processing by Rrp44 and formation of 5.8S+30. This observation is coherent with our postulation that Nop53 would be able to directly interact with different exosome subunits (Rrp6, Rrp45, Mtr4, and Mpp6) during 7S processing. Based on the position of the core exosome subunit Rrp45 and the cofactor Mpp6 on the same side of the complex (12), we can hypothesize that these interactions could play a role not only in the exosome recruitment to the pre-60S, but also in the proper positioning of the exosome during 7S processing by Rrp44 and even the hand over process to Rrp6 for trimming the remaining 5.8S+30.
In addition to recruiting and positioning the exosome, we show here that Nop53 also influences the release of the exosome from pre-ribosomal intermediates, because depletion of Nop53 causes the cosedimentation of the exosome with large mass complexes corresponding to pre-ribosomal particles. In the absence of Nop53, the exosome subunits Rrp43 and Rrp6, as well as the TRAMP subunit Trf4, were retained at the bottom of glycerol gradients as part of larger complexes. Confirming these results, we show that exosome subunits copurify more pre-ribosomal assembly factors upon depletion of Nop53 and, interestingly, earlier pre-rRNA processing factors than previously described.
The association of the exosome with pre-60S upon depletion of its recruiting factor Nop53 could be explained by the observation that Mtr4 binds pre-60S at an earlier stage than Nop53 (26), and it could recruit the exosome by its interaction with Rrp6. Importantly, Mtr4 binds ITS2 sequence on pre-60S (23), allowing this protein to recruit the exosome, despite not correctly positioning it for processing of the 7S pre-rRNA. The interaction of early precursor rRNAs with the exosome identified by RNA coimmunoprecipitation experiments further suggests that the exosome may be recruited by other factors in addition to Nop53. This early nucleolar exosome association could be related to a surveillance mechanism allowing rapid degradation of misassembled particles to ensure the recycling of assembly factors (46). It would also allow a rapid recruitment for 7S trimming and 5′ETS degradation.
Utp18, a 90S pre-ribosomal particles subunit, has also been shown to interact with Mtr4 through an AIM domain (23), being important for the exosome recruitment to 90S particles. Although Rrp6 co-purifies more pre-rRNAs 35S and 23S upon depletion of Nop53, we did not find any significant change in the levels of Utp18 copurifying with Rrp6 in the presence or absence of Nop53. However, in addition to Utp18, other 90S subunits such as Sas10 and Lcp5 interact with the exosome (35) and could recruit the exosome to these particles. Interestingly, Sas10 and other UTP-B subunits were found enriched among the proteins copurified with Rrp6 and Rrp43 in the absence of Nop53. Together with the increased copurification of 23S pre-rRNA, these observations strengthen the hypothesis that upon depletion of Nop53 the exosome could be found associated with the SSU processome, being recruited by Sas10 and other factors.
The retention of the exosome on nucleolar pre-60S particles upon depletion of Nop53 complements previous in vitro results showing that when ITS2 processing is blocked, assembly factors comprising the foot structure of the pre-60S remain bound to these particles (33). Based on the results presented here, we propose a model in which the exosome may be correctly positioned on the pre-60S by the Nop53 interactions with Rrp6, Rrp45, and Mpp6. Furthermore, the identification of 90S, pre-40S, and early pre-60S assembly factors enriched with two of the exosome subunits, led us also to propose that the exosome is recruited in much earlier stages than previously predicted, opening the possibility for new interactors among the early assembly factors yet to be described.
Experimental procedures
Yeast strains manipulation
The maintenance and growth of yeast strains were carried out in minimal media (YNB) supplemented with required amino acids and were performed according to Ref. 47. The lithium acetate method was employed for yeast transformation (48). To deplete Nop53 expression, carbon source conditional-strains were shifted from galactose to glucose-containing medium for 18 h, whereas conditional-strains regulated by the Tet-Off system (49) were grown in glucose-containing media and transferred to the same media supplemented with doxycycline (1.5 μg/ml) for 18 h. The full list of yeast strains used in this study is described in Table 1.
Table 1.
Yeast strains used in this work
| Strain | Genotype | Refs. |
|---|---|---|
| Δnop53 | n MET15 his3Δ1 leu2Δ0 ura3Δ0 NOP53::kanMX4 | 29 |
| Δnop53/GAL::His-NOP53 | Δnop53, YCp111GAL::His-NOP53 | 29 |
| Δnop53/tetOff-GFP-NOP53 | Δnop53, ptetOff-GFP-NOP53 | This study |
| Δnop53/tetOff-GFP-NOP53/ RRP6-TAP | Δnop53/tetOff-GFP-NOP53/YCp111-RRP6::TAP (HIS3) | This study |
| Δnop53/tetOff-GFP-NOP53/ RRP43-TAP | Δnop53/tetOff-GFP-NOP53/YCp111-RRP43::TAP (HIS3) | This study |
| Δnop53/GAL::His-NOP53/TAP-RRP6 | Δnop53, YCp111GAL::His-NOP53, pMET-TAP-RRP6 | This study |
| Δnop53/GAL::His-NOP53/TAP-RRP43 | Δnop53, YCp111GAL::His-NOP53, pMET-TAP-RRP43 | This study |
| Δnop53/GAL::His-NOP53/TAP-TRF4 | Δnop53, YCp111GAL::His-NOP53, pMET-TAP-TRF4 | This study |
| Δnop53/TAP-NOP53 | Δnop53, pMET-TAP-NOP53 | This study |
| Δnop53/tetOff-GFP-NOP53/GFP-Rrp6 | Δnop53, ptetOff-GFP-NOP53, pUG34-Rrp6 | This study |
| Mtr3-TAP | MATa; ura3–52; leu2–3,112; YGR158c::TAP-KlURA3 | Euroscarf® |
| Δnip7/GAL::A-NIP7 | MATa; ade2-1 leu2-3,112 trp1-1 ura3-1 nip7::HIS3 p[LEU2 ARS1 GAL1::NIP7] | 59 |
Plasmids
Plasmids constructed in this work were obtained either by restriction cloning or by using InFusion HD (Clontech) recombination cloning system. The full list of plasmids used in this study is described in Table 2.
Table 2.
List of plasmids used in this work
| Plasmid | Description | Refs. |
|---|---|---|
| YCp111GAL::His-NOP53 | GAL1::His-NOP53, LEU2, CEN4 | 29 |
| yCP111 tetoff-GFP-NOP53 | tetO-CYC1::GFP-NOP53, LEU2, CEN4 | This study |
| ptetOff-GFP-NOP53 | tetO-CYC1::GFP-NOP53, URA3, CEN4 | This study |
| pMET-TAP-Rrp43 | MET25::TAP-RRP43, HIS3, CEN6 | 50 |
| pMET-TAP-Rrp6 | MET25::TAP-RRP6, HIS3, CEN6 | This study |
| pMET-TAP-Nop53 | MET25::TAP-NOP53, HIS3, CEN6 | This study |
| pMET-TAP-Trf4 | MET25::TAP-TRF4, HIS3, CEN6 | This study |
| pUG34-Rrp6 | MET25::GFP-RRP6, HIS3, CEN6 | 60 |
| pUG34-Rrp44 | MET25::GFP-RRP44, HIS3, CEN6 | This study |
| pET28a-Nop53 | 6xHis::NOP53, KanR | 30 |
| pET-Rrp45 | 6xHis::RRP45, KanR | 61 |
| pET-Rrp40 | 6xHis::RRP40, KanR | 61 |
| pET-Rrp6 | 6xHis::RRP6, KanR | This study |
| pGEX4T1 | GST, AmpR | GE Healthcare |
| pGEX4T1-Nop53 | GST::Nop53, AmpR | This study |
| pGEX4T1-Nop53(1–80) | GST::Nop53 1–80, AmpR | This study |
| pGEX4T1-Nop53(81–157) | GST::Nop53 81–157, AmpR | This study |
| pGEX4T1-Nop53(161–230) | GST::Nop53 161–230, AmpR | This study |
| pGEX4T1-Nop53(1–300) | GST::Nop53 1–300, AmpR | This study |
| pGEX4T1-Nop53(301–380) | GST::Nop53 301–380, AmpR | This study |
| pGEX4T1-Nop53(382–455) | GST::Nop53 382–455, AmpR | This study |
| pGEX4T1-Mpp6 | GST::Mpp6, AmpR | This study |
| pGEX4T1-Rrp44 | GST::Rrp44, AmpR | This study |
| pGEX4T1-Rrp6 | GST::Rrp6, AmpR | 30 |
Heterologous protein expression and GST-pulldown assay
GST and His6-tag fusion proteins respectively encoded by pGEX4T1(GE Healthcare) and pET28(Novagen) vectors were expressed in BL21 CodonPlus(DE3)–RIL E. coli strain (Agilent Technologies), as described previously (30). 500 ml of induced culture were harvested, suspended in lysis buffer (A: 20 mm Hepes, pH 8.0, 150 mm NaCl, 5 mm MgCl2, 1 mm PMSF; or B: 20 mm Tris-HCl, pH 8.0; 150 mm NaCl, 1 mm EDTA, pH 8.0; 0.8% Nonidet, 1 mm PMSF, 1 mm DTT), and lysed using a French press. Buffer A was used in the interaction assay between GST–Nop53 and His-tagged exosome subunits, and buffer B was employed both in the interaction assay between His–Rrp6 and GST–Nop53 truncation mutants and in the pulldown assay between His–Nop53 and GST-fused exosome subunits. Total cellular extract was cleared by 17,000 rpm centrifugation for 30 min at 4 °C. GST–Nop53 truncation mutants were purified through affinity, followed by size-exclusion chromatography in buffer B. GST fusion proteins were incubated with 50 μl of pre-equilibrated GSH–Sepharose beads (GE Healthcare) for 1 h at 4 °C. After extensive washing with lysis buffer, the cleared total extract containing the His6-tag fusion protein was added to the beads and incubated for 1 h at 4 °C. Extensive washing of the bound proteins with lysis buffer with higher ionic strength (250 mm NaCl) was performed before elution with 10 mm reduced GSH, 50 mm Tris-HCl, pH 8.0. As a negative control for unspecific interaction, GST-covered beads were incubated with the extract of His-tagged protein of study. Fractions of flow-through and wash were collected after incubation with both cellular extracts, and together with total extract and elution were resolved through SDS-PAGE separation and analyzed by Western blotting with anti-GST (Sigma) and anti-His (Sigma).
Protein coimmunoprecipitation
Rrp6–TAP, Rrp43–TAP, and TAP (tandem affinity purification) tag separately expressed in the carbon-source conditional strain Δnop53/tetOff::GFP-NOP53 grown in glucose-containing medium in the absence or presence of doxycycline (expression or depletion of Nop53, respectively) were used in coimmunoprecipitation experiments. The affinity purification procedure was carried out as described previously (50). For both conditions, 5 liters of cells grown to an OD600 of 1.0 were harvested by centrifugation, resuspended in resuspension buffer (50 mm Tris-HCl, pH 8.0, 200 mm KCl, 5 mm MgCl2, 5% glycerol, 1 mm PMSF) and flash-frozen in liquid N2. The total cellular extract that was obtained by grinding in a Ball Mill device (Retsch, Mixer Mill MM 200) was cleared by centrifugation at 40,000 rpm for 1 h at 4 °C. The cleared supernatant was then incubated for 2 h at 4 °C with IgG-Sepharose 6 Fast Flow (GE Healthcare) previously equilibrated with resuspension buffer. After extensive washing with the same buffer, elution of the bound proteins was performed by incubation with 500 mm ammonium hydroxide for 20 min (51).
Sample preparation
The protein content was quantified by Bradford assay. 75 μg of protein were reduced with 5 mm of DTT at 56 °C for 25 min. All samples were cooled to room temperature and incubated in the dark with 14 mm of iodoacetamide for 30 min. Afterward, the samples were digested for 16 h with sequence grade modified trypsin at a 1:50 (E:S) ratio at 37 °C. Following digestion, all reactions were acidified with 10% (v/v) TFA (0.4% v/v final concentration) to stop proteolysis. Desalting was carried out using SepPak C18 cartridges (Waters).
MS/MS analysis
Peptide samples were suspended in 0.1% formic acid and analyzed two times. The setup used a Proxeon EASY-nLC II coupled online with an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). The peptide mixture was loaded in a 100-μm inner diameter × 2-cm trap column (Dr. Maish-ReproSil-Pur C18-AQ, 5-μm beads) and was separated using a New Objective PicoFrit® column (25 cm length × 75-μm inner diameter) packed in-house with ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH HPLC). The flow rate was 200 nl/min, and we applied a 90-min gradient using the steps 95% of mobile phase A (5% acetonitrile in 0.1% formic acid) to 40% of mobile phase B (95% acetonitrile in 0.1% formic acid) for 75 min, 40–95% of B for 5 min, and 95% of B for 10 min. After each run, the trap column and column were equilibrated with mobile phase A. The LTQ-Orbitrap Velos instrument was set to data-dependent acquisition to automatically switch between full-scan MS and MS/MS acquisition with a dynamic exclusion of 90 s. Survey scans (375–2,000 m/z) were acquired in the Orbitrap system with a resolution of 60,000 at m/z 400. The 10 most intense ions, excluding unassigned and 1+ charge state, were sequentially isolated and fragmented using collision-induced dissociation with a normalized collision energy of 30. The fragment ions were analyzed in the LTQ.
Peptide spectrum matching (PSM)
The data analysis was performed with the PatternLab for proteomics 4.1.1.4 software that is freely available at http://www.patternlabforproteomics.org (52). The sequences from S. cerevisiae were downloaded on May 7th, 2019, from the UniProtKB/Swiss-Prot, and then a target-decoy database was generated to include a reversed version of each sequence plus those from 127 common MS contaminants. The number of entries in the database was 12,344 (6049 forward sequences, 6172 decoys, and 123 contaminant sequences). The Comet 2016.01 revision 3 search engine was used for identifying the mass spectra (53). The following search parameters were considered: fully and semi-tryptic peptide candidates with masses between 550 and 5500 Da, up to two missed cleavages, 40 ppm for precursor mass, and bins of 1.0005 m/z for MS/MS. The modifications were carbamidomethylation of cysteine and oxidation of methionine as fixed and variable, respectively. Mass spectrometry raw data is shown in Table S4.
Validation of PSMs
The validity of the peptide spectrum matches was assessed using Search Engine Processor (SEPro) (54, 55). The identifications were grouped by charge state (2+ and ≥3+), and then by tryptic status, resulting in four distinct subgroups. For each group, the XCorr, DeltaCN, DeltaPPM, and Peaks Matches values were used to generate a Bayesian discriminator. The identifications were sorted in nondecreasing order according to the discriminator score. A cutoff score was accepting an FDR of 1% at the peptide level based on the number of decoys. This procedure was independently performed on each data subset, resulting in an FDR that was independent of charge state or tryptic status. Additionally, a minimum sequence length of five amino acid residues and a protein score greater than 3 were imposed. Finally, identifications deviating by more than 10 ppm from the theoretical mass were discarded. This last filter led to FDRs, now at the protein level, to be lower than 1% for all search results (56).
Relative quantitation of proteins
A label-free protein quantitative analysis was performed with PatternLab's XIC (57) module allowing up to 20 ppm deviation from the precursor. Differentially abundant proteins were pinpointed using PatternLab's TFold module (58). The differential proteomic comparison only considered proteins identified with two or more unique peptides. Only proteins present in at least two technical replicates were considered for the TFold analysis. The PatternLab's Venn diagram module was used to pinpoint proteins uniquely identified to a condition; likewise, only proteins identified in two replicates were considered.
Immunoblot
Protein samples resuspended in loading buffer were resolved by SDS-PAGE (in Tris-Glycine Buffer) and transferred to polyvinylidene difluoride membranes (GE Healthcare). Western blotting assays were carried out using as primary antibodies anti-GST (Sigma), anti-GFP (Sigma), anti-CBP (Millipore), anti-polyHis (Sigma), and anti-Rpl5 (gift from Dr. C. F. Zanelli). The secondary antibodies employed, anti-rabbit IgG (GE Healthcare®) and anti-mouse IgG (GE Healthcare), were both conjugated to horseradish peroxidase (HRP). Protein detection was carried out using Immobilon Western Chemiluminescent HRP Substrate (Millipore).
Glycerol gradient sedimentation
Cleared total yeast extracts obtained from 2-liter cultures by grinding with a Ball Mill device (Retsch, Mixer Mill MM 200) were quantified for protein content with bicinchoninic acid (BCA, Sigma) method. Approximately 10 mg of total protein were loaded on the top of a 12-ml linear glycerol gradient 10–30% (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1.5 mm MgCl2, 10% glycerol, 1 mm PMSF) previously obtained using a Gradient Master device (Biocomp®). The samples were subjected to a 40,000 rpm centrifugation for 16 h at 4 °C in a Hitachi ultracentrifuge. Using an EconoSystem (Bio-Rad), each gradient was aliquoted in 24,500-μl fractions, whereas the sedimentation profile was monitored by an A260 measurement. The gradient fractions were then either precipitated with 10% trichloroacetic acid (TCA) before SDS-PAGE separation for protein detection by immunoblot or subjected to RNA extraction for Northern blot analysis.
Coimmunoprecipitation of RNAs
Total cellular extracts were prepared from strain Δnop53/tetOff::GFP-NOP53/RRP6-TAP growing in the presence or absence of doxycycline (depletion or expression of Nop53, respectively) and added to IgG-Sepharose beads (Amersham Biosciences). Immunoprecipitation was performed at 4 °C for 2 h. IgG-Sepharose beads were washed with buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1.5 mm MgCl2, 5% glycerol, 1 mm PMSF), and RNA was isolated from bound fractions by adding phenol directly to the beads. After precipitation, the recovered RNA was denatured and separated by electrophoresis on 1.5% agarose gels and transferred to nylon membranes. For comparison, 20 μg of RNA from total extract was loaded on the gel. Hybridization was performed using biotin-labeled probes specific to rRNAs. 5S rRNA and scR1 were used as controls for nonspecific binding to the resin. Quantification of bands from Northern hybridizations were performed with ImageJ software.
Author contributions
L. P. P. C., F. F. B., and C. C. O. conceptualization; L. P. P. C., F. F. B., R. M. S., M. D. S., F. C. N., and C. C. O. formal analysis; F. C. N. and C. C. O. supervision; C. C. O. funding acquisition; L. P. P. C., F. F. B., and R. M. S. validation; L. P. P. C., F. F. B., and R. M. S. investigation; L. P. P. C., F. F. B., R. M. S., M. D. S., F. C. N., and C. C. O. writing-original draft; C. C. O. project administration; M. D. S. software; L. P. P. C., F. F. B., and C. C. O. visualization.
Supplementary Material
Acknowledgments
We thank Dr. Cleslei F. Zanelli (UNESP, Araraquara, SP, Brazil) for the kind gift of antibodies and Frederico Gueiros Filho for the use of the fluorescence microscope.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grant 15/06477-9 (to C. C. O.), by FAPESP fellowship 2015/05776-2 (to F. F. M. B.), and by fellowships from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) (to L. P. C.).
This article contains Figs. S1–S8 and Tables S1–S4.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier accession no. PXD014187.
- 5′-ETS
- 5′-external transcribed spacer
- FDR
- false-discovery rate
- PDB
- Protein Data Bank
- PMSF
- phenylmethylsulfonyl fluoride
- snoRNP
- small nucleolar ribonucleoprotein
- HRP
- horseradish peroxidase
- PSM
- peptide spectrum match
- LTQ
- linear trap quadropole orbitrap velos mass spectrometer
- TAP
- tandem affinity purification.
References
- 1. de la Cruz J., Karbstein K., and Woolford J. L. Jr. (2015) Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 84, 93–129 10.1146/annurev-biochem-060614-033917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kressler D., Hurt E., and Bassler J. (2017) A puzzle of life: crafting ribosomal subunits. Trends Biochem. Sci. 42, 640–654 10.1016/j.tibs.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 3. Fatica A., and Tollervey D. (2002) Making ribosomes. Curr. Opin Cell Biol. 14, 313–318 10.1016/S0955-0674(02)00336-8 [DOI] [PubMed] [Google Scholar]
- 4. Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., and Tollervey D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18, 5399–5410 10.1093/emboj/18.19.5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitchell P., Petfalski E., Shevchenko A., Mann M., and Tollervey D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91, 457–466 10.1016/S0092-8674(00)80432-8 [DOI] [PubMed] [Google Scholar]
- 6. Liu Q., Greimann J. C., and Lima C. D. (2006) Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127, 1223–1237 10.1016/j.cell.2006.10.037 [DOI] [PubMed] [Google Scholar]
- 7. Dziembowski A., Lorentzen E., Conti E., and Séraphin B. (2007) A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14, 15–22 10.1038/nsmb1184 [DOI] [PubMed] [Google Scholar]
- 8. Lebreton A., Tomecki R., Dziembowski A., and Séraphin B. (2008) Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456, 993–996 10.1038/nature07480 [DOI] [PubMed] [Google Scholar]
- 9. Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., and Mitchell P. (1999) The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 13, 2148–2158 10.1101/gad.13.16.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Briggs M. W., Burkard K. T., and Butler J. S. (1998) Rrp6p, the yeast homologue of the human PM–Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273, 13255–13263 10.1074/jbc.273.21.13255 [DOI] [PubMed] [Google Scholar]
- 11. Zinder J. C., and Lima C. D. (2017) Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 31, 88–100 10.1101/gad.294769.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schuller J. M., Falk S., Fromm L., Hurt E., and Conti E. (2018) Structure of the nuclear exosome captured on a maturing preribosome. Science 360, 219–222 10.1126/science.aar5428 [DOI] [PubMed] [Google Scholar]
- 13. Wasmuth E. V., Zinder J. C., Zattas D., Das M., and Lima C. D. (2017) Structure and reconstitution of yeast Mpp6–nuclear exosome complexes reveals that Mpp6 stimulates RNA decay and recruits the Mtr4 helicase. Elife 6, e29062 10.7554/eLife.29062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feigenbutz M., Garland W., Turner M., and Mitchell P. (2013) The exosome cofactor Rrp47 is critical for the stability and normal expression of its associated exoribonuclease Rrp6 in Saccharomyces cerevisiae. PLoS ONE 8, e80752 10.1371/journal.pone.0080752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makino D. L., Schuch B., Stegmann E., Baumgärtner M., Basquin C., and Conti E. (2015) RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 524, 54–58 10.1038/nature14865 [DOI] [PubMed] [Google Scholar]
- 16. Schuch B., Feigenbutz M., Makino D. L., Falk S., Basquin C., Mitchell P., and Conti E. (2014) The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 33, 2829–2846 10.15252/embj.201488757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Falk S., Weir J. R., Hentschel J., Reichelt P., Bonneau F., and Conti E. (2014) The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities. Mol. Cell 55, 856–867 10.1016/j.molcel.2014.07.020 [DOI] [PubMed] [Google Scholar]
- 18. LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., and Tollervey D. (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724 10.1016/j.cell.2005.04.029 [DOI] [PubMed] [Google Scholar]
- 19. de la Cruz J., Kressler D., Tollervey D., and Linder P. (1998) Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 17, 1128–1140 10.1093/emboj/17.4.1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weir J. R., Bonneau F., Hentschel J., and Conti E. (2010) Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc. Natl. Acad. Sci. U.S.A. 107, 12139–12144 10.1073/pnas.1004953107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nissan T. A., Bassler J., Petfalski E., Tollervey D., and Hurt E. (2002) 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539–5547 10.1093/emboj/cdf547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ohmayer U., Gamalinda M., Sauert M., Ossowski J., Pöll G., Linnemann J., Hierlmeier T., Perez-Fernandez J., Kumcuoglu B., Leger-Silvestre I., Faubladier M., Griesenbeck J., Woolford J., Tschochner H., and Milkereit P. (2013) Studies on the assembly characteristics of large subunit ribosomal proteins in Saccharomyces cerevisiae. PLoS ONE 8, e68412 10.1371/journal.pone.0068412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thoms M., Thomson E., Bassler J., Gnädig M., Griesel S., and Hurt E. (2015) The exosome is recruited to RNA substrates through specific adaptor proteins. Cell 162, 1029–1038 10.1016/j.cell.2015.07.060 [DOI] [PubMed] [Google Scholar]
- 24. Wu S., Tan D., Woolford J. L. Jr, Dong M. Q., and Gao N. (2017) Atomic modeling of the ITS2 ribosome assembly subcomplex from cryo-EM together with mass spectrometry-identified protein–protein crosslinks. Protein Sci. 26, 103–112 10.1002/pro.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu S., Tutuncuoglu B., Yan K., Brown H., Zhang Y., Tan D., Gamalinda M., Yuan Y., Li Z., Jakovljevic J., Ma C., Lei J., Dong M. Q., Woolford J. L. Jr., and Gao N. (2016) Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534, 133–137 10.1038/nature17942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Biedka S., Micic J., Wilson D., Brown H., Diorio-Toth L., and Woolford J. L. Jr. (2018) Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J. Cell Biol. 217, 2503–2518 10.1083/jcb.201711037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sydorskyy Y., Dilworth D. J., Halloran B., Yi E. C., Makhnevych T., Wozniak R. W., and Aitchison J. D. (2005) Nop53p is a novel nucleolar 60S ribosomal subunit biogenesis protein. Biochem. J. 388, 819–826 10.1042/BJ20041297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomson E., and Tollervey D. (2005) Nop53p is required for late 60S ribosome subunit maturation and nuclear export in yeast. RNA 11, 1215–1224 10.1261/rna.2720205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Granato D. C., Gonzales F. A., Luz J. S., Cassiola F., Machado-Santelli G. M., and Oliveira C. C. (2005) Nop53p, an essential nucleolar protein that interacts with Nop17p and Nip7p, is required for pre-rRNA processing in Saccharomyces cerevisiae. FEBS J. 272, 4450–4463 10.1111/j.1742-4658.2005.04861.x [DOI] [PubMed] [Google Scholar]
- 30. Granato D. C., Machado-Santelli G. M., and Oliveira C. C. (2008) Nop53p interacts with 5.8S rRNA cotranscriptionally, and regulates processing of pre-rRNA by the exosome. FEBS J. 275, 4164–4178 10.1111/j.1742-4658.2008.06565.x [DOI] [PubMed] [Google Scholar]
- 31. Gasse L., Flemming D., and Hurt E. (2015) Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol. Cell 60, 808–815 10.1016/j.molcel.2015.10.021 [DOI] [PubMed] [Google Scholar]
- 32. Konikkat S., and Woolford J. L. Jr. (2017) Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem. J. 474, 195–214 10.1042/BCJ20160516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fromm L., Falk S., Flemming D., Schuller J. M., Thoms M., Conti E., and Hurt E. (2017) Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat. Commun. 8, 1787 10.1038/s41467-017-01786-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Januszyk K., and Lima C. D. (2014) The eukaryotic RNA exosome. Curr. Opin Struct. Biol. 24, 132–140 10.1016/j.sbi.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klinge S., and Woolford J. L. Jr. (2019) Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20, 116–131 10.1038/s41580-018-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kater L., Thoms M., Barrio-Garcia C., Cheng J., Ismail S., Ahmed Y. L., Bange G., Kressler D., Berninghausen O., Sinning I., Hurt E., and Beckmann R. (2017) Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171, 1599–1610.e14 10.1016/j.cell.2017.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barandun J., Chaker-Margot M., Hunziker M., Molloy K. R., Chait B. T., and Klinge S. (2017) The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 24, 944–953 10.1038/nsmb.3472 [DOI] [PubMed] [Google Scholar]
- 38. Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., and Mering C. V. (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zanchin N. I., and Goldfarb D. S. (1999) The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 27, 1283–1288 10.1093/nar/27.5.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Callahan K. P., and Butler J. S. (2008) Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 36, 6645–6655 10.1093/nar/gkn743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oliveira C. C., Gonzales F. A., and Zanchin N. I. (2002) Temperature-sensitive mutants of the exosome subunit Rrp43p show a deficiency in mRNA degradation and no longer interact with the exosome. Nucleic Acids Res. 30, 4186–4198 10.1093/nar/gkf545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Choque E., Schneider C., Gadal O., and Dez C. (2018) Turnover of aberrant pre-40S pre-ribosomal particles is initiated by a novel endonucleolytic decay pathway. Nucleic Acids Res. 46, 4699–4714 10.1093/nar/gky116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanghai Z. A., Miller L., Molloy K. R., Barandun J., Hunziker M., Chaker-Margot M., Wang J., Chait B. T., and Klinge S. (2018) Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 556, 126–129 10.1038/nature26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Biedka S., Wu S., LaPeruta A. J., Gao N., and Woolford J. L. Jr. (2017) Insights into remodeling events during eukaryotic large ribosomal subunit assembly provided by high resolution cryo-EM structures. RNA Biol. 14, 1306–1313 10.1080/15476286.2017.1297914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Falk S., Tants J. N., Basquin J., Thoms M., Hurt E., Sattler M., and Conti E. (2017) Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the pre-ribosomal protein Nop53. RNA 23, 1780–1787 10.1261/rna.062901.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Koš-Braun I., Koš M., Thoms M., Berninghausen O., Beckmann R., and Hurt E. (2016) Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166, 380–393 10.1016/j.cell.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 47. (1991) Guide to yeast genetics and molecular biology. Methods Enzymol. 194, 1–863 [PubMed] [Google Scholar]
- 48. Chen D. C., Yang B. C., and Kuo T. T. (1992) One-step transformation of yeast in stationary phase. Curr. Genet 21, 83–84 10.1007/BF00318659 [DOI] [PubMed] [Google Scholar]
- 49. Gossen M., and Bujard H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5551 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lourenço R. F., Leme A. F., and Oliveira C. C. (2013) Proteomic analysis of yeast mutant RNA exosome complexes. J. Proteome Res. 12, 5912–5922 10.1021/pr400972x [DOI] [PubMed] [Google Scholar]
- 51. Ohmayer U., Gil-Hernández Á., Sauert M., Martín-Marcos P., Tamame M., Tschochner H., Griesenbeck J., and Milkereit P. (2015) Studies on the coordination of ribosomal Protein assembly events involved in processing and stabilization of yeast early large ribosomal subunit precursors. PLoS ONE 10, e0143768 10.1371/journal.pone.0143768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carvalho P. C., Lima D. B., Leprevost F. V., Santos M. D., Fischer J. S., Aquino P. F., Moresco J. J., Yates J. R. 3rd., and Barbosa V. C. (2016) Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nat. Protoc. 11, 102–117 10.1038/nprot.2015.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eng J. K., Hoopmann M. R., Jahan T. A., Egertson J. D., Noble W. S., and MacCoss M. J. (2015) A deeper look into Comet–implementation and features. J. Am. Soc. Mass. Spectrom. 26, 1865–1874 10.1007/s13361-015-1179-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carvalho P. C., Fischer J. S., Xu T., Cociorva D., Balbuena T. S., Valente R. H., Perales J., Yates J. R. 3rd., and Barbosa V. C. (2012) Search engine processor: filtering and organizing peptide spectrum matches. Proteomics 12, 944–949 10.1002/pmic.201100529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yates J. R. 3rd., Park S. K., Delahunty C. M., Xu T., Savas J. N., Cociorva D., and Carvalho P. C. (2012) Toward objective evaluation of proteomic algorithms. Nat. Methods 9, 455–456 10.1038/nmeth.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barboza R., Cociorva D., Xu T., Barbosa V. C., Perales J., Valente R. H., França F. M., Yates J. R. 3rd., and Carvalho P. C. (2011) Can the false-discovery rate be misleading? Proteomics 11, 4105–4108 10.1002/pmic.201100297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Neilson K. A., Ali N. A., Muralidharan S., Mirzaei M., Mariani M., Assadourian G., Lee A., van Sluyter S. C., and Haynes P. A. (2011) Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11, 535–553 10.1002/pmic.201000553 [DOI] [PubMed] [Google Scholar]
- 58. Carvalho P. C., Yates J. R. 3rd., and Barbosa V. C. (2012) Improving the TFold test for differential shotgun proteomics. Bioinformatics 28, 1652–1654 10.1093/bioinformatics/bts247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zanchin N. I., Roberts P., DeSilva A., Sherman F., and Goldfarb D. S. (1997) Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell. Biol. 17, 5001–5015 10.1128/MCB.17.9.5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gonzales-Zubiate F. A., Okuda E. K., Da Cunha J. P. C., and Oliveira C. C. (2017) Identification of karyopherins involved in the nuclear import of RNA exosome subunit Rrp6 in Saccharomyces cerevisiae. J. Biol. Chem. 292, 12267–12284 10.1074/jbc.M116.772376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Luz J. S., Tavares J. R., Gonzales F. A., Santos M. C., and Oliveira C. C. (2007) Analysis of the Saccharomyces cerevisiae exosome architecture and of the RNA binding activity of Rrp40p. Biochimie 89, 686–691 10.1016/j.biochi.2007.01.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.