Abstract
The SecYEG translocon constitutes the major protein transport channel in bacteria and transfers an enormous variety of different secretory and inner-membrane proteins. The minimal core of the SecYEG translocon consists of three inner-membrane proteins, SecY, SecE, and SecG, which, together with appropriate targeting factors, are sufficient for protein transport in vitro. However, in vivo the SecYEG translocon has been shown to associate with multiple partner proteins, likely allowing the SecYEG translocon to process its diverse substrates. To obtain a global view on SecYEG plasticity in Escherichia coli, here we performed a quantitative interaction proteomic analysis, which identified several known SecYEG-interacting proteins, verified the interaction of SecYEG with quality-control proteins, and revealed several previously unknown putative SecYEG-interacting proteins. Surprisingly, we found that the chaperone complex PpiD/YfgM is the most prominent interaction partner of SecYEG. Detailed analyses of the PpiD–SecY interaction by site-directed cross-linking revealed that PpiD and the established SecY partner protein YidC use almost completely-overlapping binding sites on SecY. Both PpiD and YidC contacted the lateral gate, the plug domain, and the periplasmic cavity of SecY. However, quantitative MS and cross-linking analyses revealed that despite having almost identical binding sites, their binding to SecY is noncompetitive. This observation suggests that the SecYEG translocon forms different substrate-independent subassemblies in which SecYEG either associates with YidC or with the PpiD/YfgM complex. In summary, the results of this study indicate that the PpiD/YfgM chaperone complex is a primary interaction partner of the SecYEG translocon.
Keywords: protein secretion, protein sorting, protein translocation, chaperone, proteomics, PpiD, protein cross-linking, quantitative affinity purification-mass spectrometry (qAP-MS), SecYEG translocon, YidC
Introduction
The universally-conserved Sec translocon orchestrates protein transport at the cytoplasmic membrane in prokaryotes and at the endoplasmic reticulum membrane in eukaryotes (1–6). It consists of the three core components SecY, SecE, and SecG in bacteria and Sec61α, Sec61γ and Sec61β in eukaryotes. The channel-forming subunit SecY/Sec61α is composed of 10 transmembrane domains (TMs)3 and has a unique structure composed of two water-filled cavities, which are separated by a central constriction in the middle of the membrane, called the pore ring (7). In Escherichia coli, the pore ring is composed of six isoleucine residues, which project their hydrophobic side chains radially into the lumen of the pore. The pore ring, together with a short helix on its periplasmic side, called the plug, is likely required for preventing uncontrolled ion movement across the membrane in the resting translocon (8, 9). On the cytosolic side of the membrane, extensions of the TMs protrude into the cytosol and provide the docking side for SecA, the signal-recognition particle (SRP) receptor FtsY, and the ribosome (10–13). SecA functions as a motor protein for the post-translational protein transport and inserts into the cytosolic cavity of SecY via the two-helix-finger domain (14, 15). In contrast, FtsY and the SRP–ribosome nascent chain complex form a transient quaternary complex with the SecYEG complex during co-translational insertion of bacterial membrane proteins (13, 16–19). In line with its role in both protein translocation across the membrane and protein insertion into the membrane, SecY not only opens toward the periplasmic side of the membrane but also toward the lipid phase. This is achieved by movements of TMs 2B, 3, 7 and 8 at the front of the SecY channel, which constitute a lateral gate that allows access of TMs to the lipid bilayer (20–23). The back of the SecY channel is encircled by SecE, which in E. coli consists of three TMs. SecE contributes to ribosome binding (24) and to the overall stability of the SecYEG translocon (25, 26). In the absence of SecE, SecY is rapidly degraded by the protease FtsH (27). In contrast to SecY and SecE, SecG is not essential for protein transport and is primarily required for the SecA-dependent translocation of secretory proteins (28). SecG contains two TMs and a connecting cytosolic loop and has only limiting contact to SecY and SecE. As SecA-dependent translocation is unique to bacteria and bacteria-derived chloroplasts, SecG is replaced by the nonhomologous Sec61β subunit in archaea and eukaryotes (29).
Reconstitution experiments have shown that SecYEG proteoliposomes are capable of transporting model protein substrates in the presence of the appropriate targeting factors, i.e. SecA for secretory proteins or SRP/FtsY for bacterial membrane proteins (30, 31). Thus, the SecYEG translocon represents the minimal membrane-embedded unit required for protein transport. However, multiple partner proteins of SecYEG have been identified, which either improve protein transport in general or facilitate the transport of particular substrates. One example is the trimeric SecDFYajC complex, which is suggested to function as a proton-driven membrane-integral chaperone that enhances both the translocation and the insertion of proteins (32–34). YidC, however, is specifically required during membrane protein insertion (35), where it facilitates the release of TMs from the SecY channel (36, 37) and supports TM folding (38) and the subsequent assembly of individual membrane proteins into oligomeric complexes (39). In addition to its SecYEG-associated role, YidC and its mitochondrial or chloroplast homologs Oxa1/Alb3 can also act as SecYEG-independent insertases for some membrane proteins (40, 41). In bacteria, these are primarily small membrane proteins and membrane proteins lacking extended periplasmic domains (31). The most complex SecYEG assembly described so far is a heptameric SecYEG/SecDFYajC/YidC complex that is referred to as a holo-translocon (42). The holo-translocon was shown to be more efficient in membrane protein insertion than the trimeric SecYEG complex, but it appears to be less efficient in SecA-dependent translocation of secretory proteins (42, 43).
In contrast to SecDFYajC and YidC, the function of other SecYEG-associated proteins is less defined. One example is the chaperone PpiD and its partner protein YfgM (44–46). PpiD is tethered to the bacterial membrane by a single transmembrane domain and contains an inactive peptidyl-prolyl isomerase motif within its large periplasmic domain (47). PpiD was shown to interact with proteins exiting the SecYEG translocon (45) and to enhance their release into the periplasm (48). Like PpiD, YfgM is a single-spanning membrane protein with an extended periplasmic domain. Both proteins have been identified as complex in the bacterial membrane (49) and in contact with the SecYEG translocon (44–46, 48). So far, little is known about the presence of SecYEG–PpiD/YfgM complexes in the E. coli membrane, but PpiD/YfgM was generally not considered to be major contact partners of the SecYEG complex.
For a global analysis of the SecYEG interactome in the E. coli membrane, we performed a quantitative affinity purification MS approach (qAP-MS) either with or without cross-linking and validated the most prominent interaction partners by site-directed cross-linking and functional assays. Unexpectedly, our data identified PpiD/YfgM as major partner proteins of the SecYEG complex. Our data furthermore show that PpiD and YidC share largely overlapping binding sites on SecY. Both proteins contact the lateral gate of SecY, but also deeply penetrate into the aqueous channel and the periplasmic cavity. Nevertheless, binding of YidC or PpiD to the SecYEG translocon appears to be noncompetitive, suggesting the presence of two distinct SecYEG populations, one interacting with YidC and a second interacting with PpiD/YfgM.
Results
Global identification of SecYEG-associated proteins in E. coli
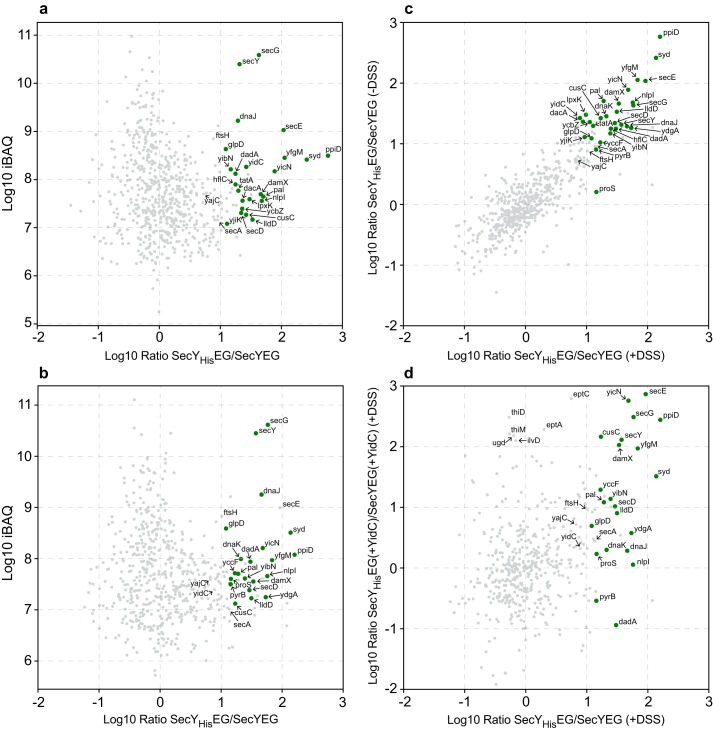
For identifying SecYEG interacting proteins on a global scale, a label-free qAP-MS approach was employed. SecYHisEG or SecYEG (serving as control) was expressed in E. coli from plasmid pTrc99a. After cell breakage and fractionation, the membrane fraction was solubilized with dodecyl maltoside, and the solubilized material was loaded onto a Talon metal-affinity chromatography (IMAC) column. For stabilizing potential protein–protein interactions of the SecYEG translocon, on-column cross-linking was performed on some samples, using the homobifunctional cross-linker disuccinimidyl suberate (DSS). The DSS-treated and -nontreated samples were then eluted from the column after several washing steps and subjected to digestion using trypsin followed by label-free quantitative MS. Only proteins that were reproducibly detected with ≥20% sequence coverage and at least two peptides were considered to be significant (Table S1). Significant interactors were determined from label-free protein intensity ratios (SecYHisEG versus SecYEG) by performing t tests across replicate measurements. This also takes into account that unspecific binding to the IMAC column might increase in the absence of a His-tagged protein. Resulting p values and fold changes are shown in volcano plots for both the nontreated (Fig. S1A) and the DSS-treated sample (Fig. S1B) visualizing significantly-enriched proteins. Enrichment ratios for proteins identified in both samples were plotted against the total iBAQ intensities (intensity-based absolute quantification) for both the nontreated sample (Fig. 1A) and the DSS-treated sample (Fig. 1B), highlighting significant interactors by green symbols. In both samples, SecY, SecE, and SecG were enriched, as expected. However, in the on-column cross-linking approach, the values for SecE were just below the defined threshold level (Fig. S1), and SecE was therefore not highlighted in Fig. 1B. This could be due to the presence of DMSO as a solvent for DSS. In general, the at least 10-fold–enriched proteins were almost identical between the DSS-treated and -nontreated sample (Pearsons's correlation coefficient r = 0.826; Fig. 1C; Table S1, and Fig. S1), suggesting that they are rather stable interaction partners of the SecYEG translocon. Among them are known SecYEG-interacting proteins (Syd, SecD, YajC, SecA, YidC, PpiD, and YfgM). We also identified TatA, the translocase of the twin-arginine–dependent protein translocation pathway (50), suggesting that the previously observed cooperation between the Sec and Tat pathways in Gram-positive bacteria (51) also exists in E. coli. The enrichment of proteins of the cellular quality control (FtsH, HflC, DnaK, DnaJ, HslU, and YcbZ) supports the previous assumption that protein transport via SecYEG is intrinsically linked to protein quality control (52, 53). However, the detection of these proteins could also result from the higher expression level of SecYEG in these experiments.
Figure 1.
SecYEG interactome of E. coli. A, SecYEG or SecYHisEG was expressed in E. coli, and subsequently, SecYEG/SecYHisEG was purified via metal-affinity chromatography and subjected to MS analyses as described under “Experimental procedures” (n = 4). The x axis displays the log10 intensity ratio of the respective protein in the SecYHisEG (His)/SecYEG (Ctl) pulldowns. The y axis displays the log10 of the total iBAQ value. Specific interactors of the SecYEG complex are marked by green dots. B, as in A but protein–protein interactions were stabilized by on-column cross-linking using the chemical cross-linker DSS. C, comparison of mean log10 ratios of DSS-treated (x axis) against the mean log10 ratios of nonDSS-treated samples (y axis). D, comparison of the mean log ratios of SecYEG-interacting proteins from cells without (x axis) or with (y axis) simultaneous YidC expression. The analyses were performed after on-column cross-linking. A complete list of all proteins is shown in Table S1.
In addition, several membrane proteins were enriched, although the physiological relevance of their interaction with SecYEG is currently unknown (Table 1). Among them are proteins, like Pal, that might be detected due to their high abundance, but also the outer membrane protein CusC, which is of very low abundance and linked to heavy metal export (54). One of the most enriched proteins was YicN, a predicted single-spanning membrane protein of unknown function.
Table 1.
SecYEG interactome in E. coli
Listed are the most abundant proteins that were either found co-purifying with SecYEG or were cross-linked to SecYEG on-column. Indicated is their localization (C, cytosolic; IMA, inner membrane-associated; IM, inner membrane; OM, outer membrane), the log10 ratio of SecYHisEG/SecYEG (His/Ctl), the known or predicted function, and the cellular copy number of the protein in E. coli, based on ribosome profiling (54). ↑ indicates that the copy number based on ribosome profiling is significantly higher than the copy number based on other methods (89).
| Protein | Localization | Log10 ratio (−DSS) | Log10 ratio (+DSS) | Function | Copy no. |
|---|---|---|---|---|---|
| Known interactors of the SecYEG translocon | |||||
| PpiD | IM | 2.76 | 2.20 | Part of the PpiD–YfgM chaperone complex | 3,717 |
| Syd | IMA | 2.42 | 2.14 | Regulator of SecY function | 928 |
| YfgM | IM | 2.05 | 1.84 | Part of the PpiD–YfgM chaperone complex | 2,852 |
| YidC | IM | 1.42 | 0.89 | SecYEG-associated insertase | 8,807 |
| SecD | IM | 1.34 | 1.46 | Part of the SecDFYajC complex | 2,699↑ |
| TatA | IM | 1.29 | 1.11 | Tat-translocase | 6,336 |
| SecA | IMA | 0.95 | 1.13 | SecYEG-associated ATPase | 3,987 |
| YajC | IM | 0.73 | 0.82 | Part of the SecDFYajC complex | 26,903↑ |
| Quality control proteins | |||||
| DnaK | C | 1.45 | 1.32 | Hsp70 chaperone | 44,581 |
| YcbZ | C | 1.36 | 1.05 | Putative ATP-dependent protease | 700 |
| DnaJ | C | 1.29 | 1.66 | DnaK co-chaperone | 5,688 |
| HflC | IM | 1.25 | 1.4 | Modulator of FtsH | 923 |
| HslU | C | 1.16 | 1.13 | Part of the HslVU protease complex | 9,329 |
| FtsH | IM | 0.88 | 1.01 | SecY-associated protease | 4,550 |
| Putative novel interactors of the SecYEG translocon | |||||
| YicN | IM | 1.89 | 1.68 | Unknown | 104 |
| Pal | OM | 1.7 | 1.28 | Peptidoglycan-associated lipoprotein | 64,020 |
| NlpI | IM | 1.68 | 1.76 | Lipoprotein, involved in cell division | 389 |
| DamX | IM | 1.66 | 1.53 | Cell division protein | 1,649 |
| LldD | IM | 1.53 | 1.50 | l-Lactate-dehydrogenase | 445 |
| LpxK | IM | 1.48 | 0.99 | Tetraacyldisaccharide 4′-kinase | 264 |
| CusC | OM | 1.42 | 1.23 | Cu/Ag export system | 5 |
| DacA | IM | 1.36 | 0.94 | d-Alanyl-d-alanine carboxypeptidase | 4,143 |
| YdgA | IM | 1.26 | 1.73 | Unknown, implicated in swarming | 1,875 |
| DadA | IM | 1.25 | 1.48 | d-Amino acid dehydrogenase | 1,586 |
| YibN | IM | 1.17 | 1.39 | Putative sulfur transferase | 8,459 |
| YjiK | IM | 1.10 | 0.968 | Unknown | 40 |
| GlpD | IM | 1.09 | 1.08 | Glycerol-3-phosphate dehydrogenase | 127 |
| YccF | IM | 1.02 | 1.23 | Unknown | 800 |
One of the best-characterized partner proteins of SecYEG is YidC, which was enriched about 10-fold in the SecYHisEG sample, and similar enrichments were observed for SecD, which together with SecF form the SecDFYajC complex. SecF was not detected in all MS analyses and is therefore not listed in Fig. 1 (Table S1). Surprisingly, the most abundant proteins in both the DSS-treated and -nontreated samples were the membrane-bound chaperone PpiD and its cognate partner protein YfgM. PpiD/YfgM have been previously identified as interaction partners of SecYEG (45, 48), and it was shown that both PpiD and YidC bind to the lateral gate of SecY (22, 46). Therefore, we tested by using the on-column cross-linking approach whether the simultaneous co-expression of YidC together withSecYHisEG would reduce the abundance of PpiD/YfgM. Both YidC and SecYEG are functional in this co-expression system and are detectable in the membrane as SecYEG/YidC complexes (55). However, the co-expression of YidC did not significantly influence the enrichment of PpiD/YfgM with the SecYEG translocon as detected by MS (Fig. 1D). This indicates that the interaction of PpiD/YfgM with SecY is not influenced by the YidC concentration, although both PpiD and YidC bind to the lateral gate of SecY (22, 46).
Noncompetitive binding of YidC and PpiD to the SecYEG translocon
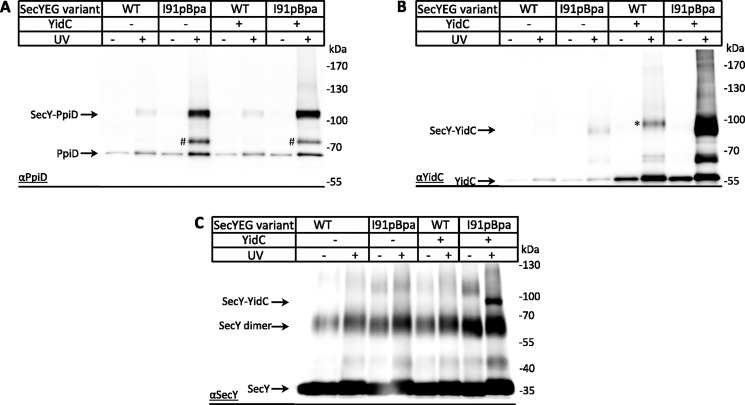
The potentially noncompetitive binding of PpiD and YidC to the SecYEG translocon was further analyzed by in vivo site-directed cross-linking with a SecY variant that had the UV-reactive phenylalanine derivative para-benzoyl-l-phenylalanine (pBpa) site-specifically inserted into position 91 within TM2b of the lateral gate. Upon UV exposure of E. coli cells expressing SecY(I91pBpa)EG and subsequent purification of the SecYEG complex and its cross-linked partner proteins, a 110-kDa cross-link product was detected by α-PpiD antibodies (Fig. 2A). This cross-link product had been observed before and was identified by MS as a SecY–PpiD cross-link product (46). The intensity of the SecY–PpiD cross-link did not significantly change when YidC was co-expressed together with SecYEG, further supporting that the SecY–PpiD contact is largely independent of the cellular YidC concentration. An additional UV-dependent band at ∼85 kDa was also recognized by α-PpiD antibodies, which likely reflects a SecY–YfgM cross-link, because the available polyclonal antibodies react with both PpiD and YfgM (Fig. S2), and an 85-kDa YfgM cross-link to the lateral gate had been observed before by MS (46).
Figure 2.
Noncompetitive binding of PpiD and YidC to the lateral gate of SecY. A, SecY–PpiD interaction was monitored in BL21 cells expressing either plasmid-borne WT SecYEG (WT) or SecY(I91pBpa)EG, carrying the UV-dependent cross-linker pBpa at position 91 within TM2a of the lateral gate. When indicated, YidC was co-expressed from the same plasmid. After in vivo UV exposure, the SecY was purified via a C-terminal His-tag using metal-affinity chromatography, and the purified sample was analyzed on SDS-PAGE followed by Western blotting and immunodetection using α-PpiD antibodies. As a control, SecY was also purified from samples that were not UV-exposed. The SecY–PpiD cross-linking product and PpiD co-purifying with SecY are indicated. A UV-dependent band migrating at ∼kDa (#) was also detected and could reflect a cross-link between SecY and YfgM, because the available antibodies against PpiD also recognize YfgM. B, same material analyzed in A was analyzed with α-YidC antibodies, and the SecY–YidC cross-linking product is indicated. The UV-dependent band observed in the absence of pBpa (*) could reflect UV-induced radical formation of aromatic amino acids, but this was not further analyzed. C, same material as in A was analyzed with α-SecY antibodies, but due to the low specificity of the antibody, only the SecY–YidC cross-link is detected in addition to the SecY dimer and SecY monomer.
When the same material was probed with antibodies against YidC, the SecY–YidC cross-link at ∼95 kDa was detected, which strongly increased when YidC was co-expressed with SecYEG (Fig. 2B). The strong SecY–YidC cross-link product was also visible when the blot was decorated with SecY antibodies (Fig. 2C), although the quality of the available SecY antibody did not allow for the detection of the SecY–PpiD cross-link. Thus, increasing the cellular YidC concentration increases the SecY–YidC contact without influencing the SecY–PpiD contact, which is in agreement with the quantitative MS analysis. In vivo UV exposure of cells co-expressing YidC with WT SecY, i.e. not containing pBpa, also resulted in a cross-linking product (Fig. 2B, *). This band was significantly weaker and migrated slightly above the SecY–YidC cross-link. This pBpa-independent cross-link is probably the result of UV-dependent radical formation of aromatic amino acids that favors nonspecific protein–protein and protein–nucleic acid cross-links (56, 57).
In summary, although both YidC and PpiD bind to the lateral gate of SecY, under the conditions tested here, they do not seem to compete with each other.
The on-column cross-linking did not reveal increased enrichment of YidC upon YidC co-expression (Fig. 1D). This was different for the pBpa cross-linking approach, which showed increased amounts of the SecY–YidC cross-linking product upon YidC co-expression (Fig. 2B). This probably reflects the different chemistry of both cross-linkers and differences in the experimental approaches. The in vivo pBpa cross-linking will also detect transient interactions that are probably lost in the detergent-solubilized and washed samples that were used for in vitro DSS cross-linking. In addition, the pBpa cross-linking approach is less dependent on the amino acid position of the partner protein, because it primarily reacts with C–H bonds (58). In contrast, DSS will form cross-links between primary amines, e.g. via the N-terminal methionine and via lysine residues and thus requires the stable proximity between specific residues.
PpiD and YidC reach into the periplasmic cavity of the SecY channel
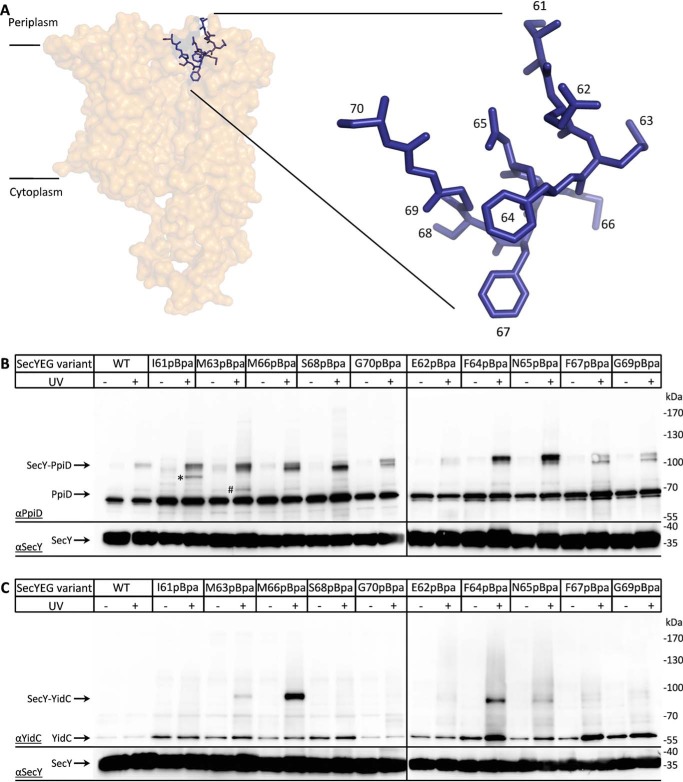
A common feature of both PpiD and YidC is that they both contain large periplasmic loops. Although the exact function of these loops is unknown, a part of YidC's periplasmic loop is involved in contacting the SecDFYajC complex (59), whereas the periplasmic loop of PpiD appears to facilitate the folding of outer membrane proteins (60). The close interaction of SecY with either YidC or PpiD prompted us to determine whether this not only involves the lateral gate but possibly also the periplasmic cavity of the SecY channel. Therefore, we generated several pBpa insertions in helix 2a, the plug domain of SecY, which shields the pore ring between the cytoplasmic and periplasmic cavities (Fig. 3A). E. coli cells expressing SecYEG with pBpa at different positions within the plug were UV-exposed and analyzed by immunodetection using both α-PpiD and α-YidC antibodies. The 110-kDa PpiD–SecY cross-link product was detectable for almost all plug residues that were analyzed, but it was most prominent for residues Ser-68, Phe-64, and Asn-65 (Fig. 3, A and B). In addition, Ile-61 showed an additional cross-linking product at ∼100 kDa (*) and Met-63 at ∼85 kDa (Fig. 3, #). Although the latter could correspond to the SecY–YfgM cross-linking product, the identity of the 100-kDa cross-link was not further analyzed.
Figure 3.
Plug domain of SecY makes multiple contacts to both YidC and PpiD. A, cartoon showing the structure of the SecYEG complex (PDB 4V6M) with the position of the plug helix (blue). Residues that were replaced with pBpa for in vivo cross-linking are indicated. B, BL21 cells expressing SecYEG variants containing pBpa at different positions of the plug domain were UV-exposed when indicated, and SecYEG was purified via the C-terminal His-tag on SecY. The purified samples were further analyzed by SDS-PAGE and Western blotting for identification of cross-linking products. The membrane was decorated with α-PpiD antibodies, and the SecY–PpiD cross-linking products as well as the co-purifying PpiD are indicated. The lower part of the membrane was also decorated with α-SecY antibodies for controlling that comparable amounts of SecY were loaded. However, because of the low quality of the α-SecY antibody, detection required longer exposure times. The SecY(I61pBpa) cross-linking product at ∼100 kDa (*) reflects an uncharacterized product, whereas the 85-kDa product of SecY(M63pBpa) (#) likely reflects the SecY–YfgM product. C, as in B, but the membrane was decorated with α-YidC antibodies, and the SecY–YidC cross-linking products as well as the co-purifying YidC are indicated.
When the same material was decorated with α-YidC antibodies, the 95-kDa YidC–SecY cross-link product was detectable for residues Met-63, Met-66, Phe-64, and Asn-65 and most pronounced for residue Met-66 (Fig. 3C). Thus, YidC and PpiD not only bind to the lateral gate via their respective TMs, but their large periplasmic domains also reach into the periplasmic cavity of SecY. Note that due to the lower quality of the SecY antibody compared with the α-PpiD and α-YidC antibodies, the lower parts of each gel panel required longer exposure.
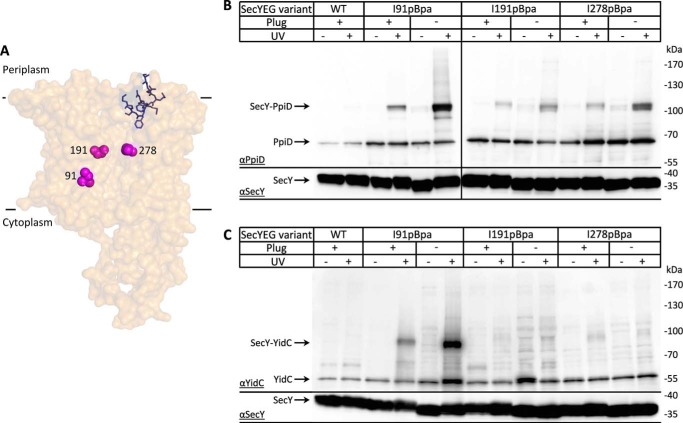
Although the plug domain is conserved in prokaryotic and eukaryotic SecY/Sec61α homologs, its deletion is tolerated (8, 9, 61–63), likely because additional residues move into the vacated area (64). Nevertheless, the deletion of the plug causes a prlA-like phenotype. The expression of prlA alleles of SecY lead to reduced subunit interactions within the SecYEG complex and to a lower dependence of protein translocation on functional signal sequences (63). Whether deleting the plug influences the interaction of SecY with either PpiD or YidC was analyzed by inserting pBpa into a plug deletion mutant. When pBpa was inserted at position 91 within TM2b of the lateral gate (Fig. 4A), the cross-link to both PpiD and YidC increased in the absence of the plug domain (Fig. 4, B and C). For the YidC–SecY interaction, this confirms previous data that had shown that a prlA phenotype is associated with an increased SecY–YidC interaction (22, 65). The same was now also observed for PpiD, suggesting that the absence of the plug enhances the interaction of SecYEG with both YidC and PpiD.
Figure 4.
Absence of the plug enhances the interaction of PpiD with the lateral gate and the SecY channel interior. A, cartoon showing the structure of the SecYEG complex (PDB 46VM). The positions where pBpa was inserted are indicated. B, in vivo cross-linking of BL21 cells expressing SecYEG with pBpa inserted at either position Ile-91 (TM2b, lateral gate), Ile-191 (TM5, channel), or Phe-278 (TM3, channel). When indicated, the plug domain of SecY was deleted. Samples were processed as described in the legend to Fig. 3 and decorated with either α-PpiD (B) or α-YidC antibodies (C). The lower part of the membrane was decorated with α-SecY antibodies.
This was further analyzed by inserting pBpa into positions Ile-191 (TM5) and Ile-278 (TM7), which are both part of the pore ring and located inside of the aqueous SecY channel. PpiD cross-linked to both residues within the SecY channel, demonstrating that PpiD has access to the channel interior (Fig. 4B). The amount of cross-linked material increased when the plug helix was deleted, indicating that access of PpiD to the channel interior is controlled by the plug domain. Upon UV-irradiation, we noticed a weak increase of PpiD co-purifying with WT SecY and the Ile-278 variant. However, the reason for this is unknown.
YidC contacts to the channel interior have been observed in vitro using purified inner membrane vesicles (INV) (65). However, in contrast to PpiD, YidC does not show significant cross-links to the SecY channel interior in vivo, and deleting the plug domain does not significantly influence this (Fig. 4C).
In summary, although both PpiD and YidC contact the periplasmic vestibule of the SecY channel, PpiD covers a larger surface area of the plug domain and contacts, at least transiently, also the SecY pore ring within the aqueous protein-conducting channel.
Contribution of YidC and PpiD to protein transport
The deep protrusion of PpiD and YidC into the periplasmic cavity of SecY, together with published data showing that PpiD interacts with nascent chains that are trapped within the SecYEG translocon (45, 48), would be in line with a role of PpiD and YidC in helping substrates to exit the SecYEG translocon. This could also explain the increased translocation activity of prlA translocons, because they show enhanced contact to YidC and PpiD (Fig. 4).
This was analyzed by using a molecular force sensor, a method that was established for monitoring the force that is exerted on a nascent chain during protein transport (66). In brief, leader peptidase (LepB) was fused to the Mannheimia succiniproducens SecM–stalling sequence, which is arrested within the ribosomal peptide tunnel during translation (Fig. 5A) (67, 68). This construct was expressed in WT cells in the presence of 35S-labeled methionine and -cysteine. After labeling for several minutes, the cells were denatured, and the arrested protein was detectable as a stalled protein of lower mass. However, if a pulling force is large enough to overcome the stalling of the SecM peptide within the ribosomal tunnel, then full-length LepB is synthesized. Thus, the fraction of full-length versus arrested LepB can serve as indicator for the exerted force.
Figure 5.
YidC but not PpiD/YfgM executes a pulling force on nascent SecY substrates. A, cartoon showing the LepB–SecM(Ms) force sensor used in this study. The arrest peptide (AP) is shown in red, and the distance between the arrest peptide and the C terminus is 23 amino acids. B, LepB–SecM(Ms) was expressed in vivo in WT E. coli strain MC4100 and labeled with [35S]methionine/cysteine. After the indicated time, whole cells were precipitated with ice-cold trichloroacetic acid (TCA), and samples were after centrifugation and denaturation in loading dye separated by SDS-PAGE and analyzed by phosphorimaging. Indicated are the full-length LepB–SecM(Ms) and the stalled version, lacking the C-terminal 23 amino acids. When indicated the protonophore CCCP was added for dissipating the proton-motive force. C, as in B, but the ΔppiD/yfgM double deletion strain and the conditional YidC-depletion strain JS7131 were used for expression. JS7131 was grown either in the presence of arabinose (YidC+) or in the presence of glucose (YidC−). The amounts of full-length and stalled LepB were quantified after phosphor-imaging using the ImageQuantTL/ImageJ software, and the fraction of the full-length(Ffl) is displayed. The values correspond to the mean of at least three independent replicates, and the standard deviation is indicated by error bars. D, amounts of YidC, SecY, PpiD, and YfgM in JS7131 cells analyzed in C were determined by Western blotting of TCA-precipitated cells. E, fraction of full-length LepB after 9 min of labeling was analyzed in the indicated strains in the presence or absence of CCCP for dissipating the pmf. Shown are the mean values of at least three independent experiments, and the standard deviations are indicated by the error bars. p values were calculated with an unpaired t test (n ≥3). ****, p value < 0.0001; **, p value < 0.01. n.s. is not significant. F, amounts of PpiD, YfgM, SecY, and YidC in the indicated single and double deletion strains were analyzed as in D.
When the LepB construct was expressed in E. coli WT cells (MC4100), an increase in full-length LepB was observed over time that was accompanied by a decrease of the arrested LepB (Fig. 5B). When the same experiment was performed with cells that had been treated with CCCP to collapse the proton-motive force (pmf), a significant portion of the truncated LepB was still detectable after 6 min of labeling. Thus, as shown before, this approach is suitable for detecting the force that is applied on a substrate during translocation across the SecYEG channel (66). The data also show that efficient translocation requires the proton-motive force.
Quantification of the fraction of full-length LepB (Ffl) in MC4100 cells and in a ΔppiD/yfgM double-deletion strain did not show a significant difference in the time-dependent accumulation of full-length LepB (Fig. 5C). This was different for the conditional YidC-depletion strain JS7131, which carries the yidC gene under the control of the arabinose promotor (40). When cells were grown in the presence of arabinose (YidC+), the accumulation of full-length LepB was comparable with MC4100 and ΔppiD/yfgM cells (Fig. 5C). However, when cells were grown in the absence of arabinose (YidC−), the amount of full-length LepB was significantly reduced (Fig. 5C). For excluding that secondary effects, like the reduction of the SecY content upon YidC-depletion, were responsible for the reduced transport of LepB, the cells used for the pulse-chase experiments were analyzed by Western blotting (Fig. 5D). Cells grown in the absence of YidC had only negligible amounts of YidC, whereas the levels of SecY, PpiD, and YfgM were unchanged (Fig. 5D).
Collapsing the pmf reduced the pulling force on LepB, and the effect of CCCP was also analyzed for the ΔppiD/yfgM strain and the conditional yidC mutant. The CCCP effect on the pulling force after 9 min of labeling was comparable between WT cells and the ΔppiD/yfgM cells or YidC-containing JS7131 cells. However, upon YidC depletion, CCCP did not further reduce the accumulation of full-length LepB (Fig. 5E). Immunodetection revealed that PpiD/YfgM were indeed absent in the corresponding single- or double deletion strains, without significantly affecting the SecY or YidC levels (Fig. 5F). Only the YfgM levels were slightly reduced in the absence of PpiD, further supporting the notion that both proteins form a complex in the E. coli membrane (44). These data indicate that PpiD/YfgM do not execute a detectable pulling force on LepB traversing the SecYEG translocon. This is different for YidC; in the absence of YidC, the pulling force is significantly reduced. However, it is difficult to determine whether this is a direct or indirect effect, because the depletion of YidC also reduces the pmf (69). Still, the deep penetration of YidC into the periplasmic vestibule of SecY would support a direct effect that needs to be further analyzed.
PpiD also interacts with YidC
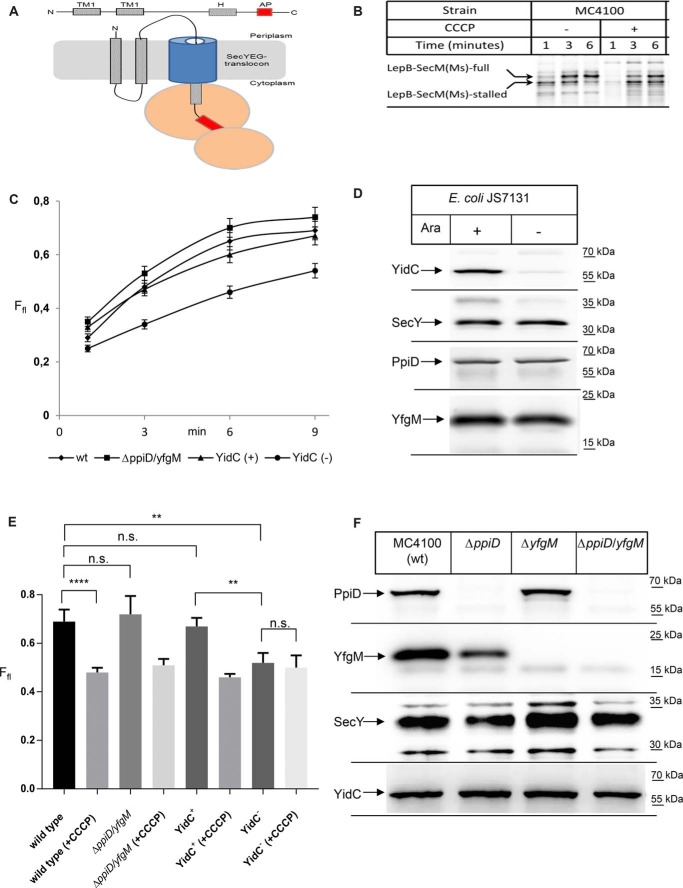
The SecYEG complex cooperates with multiple partner proteins, and this includes at least two periplasmic chaperones, the PpiD/YfgM complex as shown above and Skp (70). However, periplasmic partner proteins of the YidC insertase have not been described so far. We therefore analyzed whether YidC would also interact with periplasmic chaperones by using different cross-linking approaches in vivo and in vitro. In a first approach, a His-tagged YidC variant was expressed in WT E. coli cells and treated with the zero-length chemical cross-linker paraformaldehyde (PFA). After purification of YidC and its potential cross-linked partner proteins, the material was probed with α-PpiD antibodies, which detected multiple PFA-dependent bands. The most prominent PFA-dependent band was detected at ∼130 kDa, which would be in line with the predicted mass of a YidC–PpiD cross-link product (Fig. 6A). Noncross-linked PpiD was also present in the PFA-treated sample, which is probably the result of partial cleavage of the temperature-sensitive PFA-induced covalent bond (71).
Figure 6.
PpiD also interacts with YidC. A, in vivo p-formaldehyde (PFA) cross-linking was performed with BL21 cells expressing YidC. After PFA treatment, YidC was purified via metal-affinity chromatography, separated by SDS-PAGE, and analyzed by Western blotting with α-PpiD antibodies. PpiD and its potential cross-link to YidC are indicated. B, in vivo photocross-linking was performed with BL21 cells expressing either YidC WT or YidC with pBpa incorporated at position 399 within the cytosolic loop C1 of YidC. YidC was purified via metal-affinity chromatography and analyzed by SDS-PAGE and Western blotting with α-PpiD (top) or α-YidC (bottom) antibodies.
Previous data had shown that the cytosolic C1-loop of YidC and in particular residue Asp-399 is a hot spot for interactions with SRP, FtsY, and SecY (55). This residue was therefore also tested in vitro for interactions with PpiD. YidC(D399pBpa) was expressed in WT cells, and INV were isolated. After UV-exposure of INV, the material was analyzed by immunodetection with α-PpiD antibodies, which detected a cross-link at ∼130 kDa and a second UV-dependent band at ∼110 kDa (Fig. 6B). However, the latter was also present in WT INVs not containing the YidC(D399pBpa) variant. These data indicate that the cytosolically exposed N terminus of PpiD is in close proximity to the C1-loop of YidC.
pBpa was also inserted into position 249 of the periplasmic loop of YidC, and potential cross-links were analyzed by MS. This also revealed PpiD as a prominent contact partner of YidC (Table 2). In addition, we observed contacts to the periplasmic chaperone Skp, which like PpiD was suggested to be involved in releasing substrates from the SecYEG translocon (70), to the periplasmic chaperone FkbA and to leader peptidase LepB. In summary, PpiD is not only in close contact to the SecYEG translocon, but also to the YidC insertase, and this interaction involves both the cytosolically-exposed loops and the large periplasmic domain of YidC.
Table 2.
Interaction of YidC with periplasmic chaperones and leader peptidase
A non-UV–irradiated (not cross-linked, −UV) and a UV-irradiated (cross-linked, +UV) sample of YidC(K249pBpa) purified from whole cells was separated on a 5–15% SDS gel, and the proteins were visualized by Coomassie staining. The −UV and the +UV lanes were cut into equal slices followed by in-gel trypsin digestion and mass spectrometry. Shown are the quantification for PpiD, Skp, FkbA, and LepB.
| YidC(K249pBpa) in vivo cross-links | |||||
|---|---|---|---|---|---|
| Proteina | Molecular massb | Gel molecular massc | Relative intensity (−UV/+UV)d | Coveragee | Peptidesf |
| kDa | kDa | % | |||
| PpiD | 68.1 | 168 | 0.02 | 48.6 | 24 |
| Skp | 16 | 79 | 0.00 | 37.3 | 7 |
| FkbA | 28.9 | 116 | 0.00 | 3.7 | 1 |
| LepB | 35.9 | 133 | 0.00 | 36.7 | 13 |
a This is the protein identified.
b This is the calculated molecular mass.
c The molecular mass of the gel slice was determined by extrapolation.
d The relative intensity observed in gel slices from the control lane (−UV) compared with the +UV lane is shown.
e The sequence coverage of the total sequence was by detected peptides.
f This is the number of peptides detected.
Discussion
Dynamic protein assemblies facilitate the transport of a large variety of substrate proteins across biological membranes. This is exemplified by the Sec61 complex in the endoplasmic reticulum membrane or the SecYEG complex in cytoplasmic membrane of bacteria (1, 29). Both homologous complexes consist of three core proteins, which associate with multiple partner proteins. The best-characterized accessory proteins of the bacterial SecYEG complex are YidC (22, 40, 65, 72) and the SecDFYajC complex (32, 33, 73), which form together with SecYEG the so-called holo-translocon (42). Although additional accessory proteins of the SecYEG translocon have been identified and at least partially characterized (74–76), a global and quantitative assessment of the SecYEG interactome was still missing.
Unexpectedly, our analyses identified YicN, PpiD, and YfgM as dominant partner proteins of the SecYEG translocon in E. coli. YicN is an uncharacterized protein that is primarily present in enterobacterial genomes, but possible paralogs also exist in some eukaryotic parasites. Global transcriptome analyses suggest yicN induction upon heat stress (98), and the enrichment of YicN with the SecYEG translocon could be part of a stress response mechanism, which is currently under investigation.
The close proximity of PpiD/YfgM and SecYEG has been observed before by cross-linking (45, 46, 48); furthermore, PpiD/YfgM and SecYEG were shown to form a complex in the E. coli membrane (44, 49). A very recent E. coli membrane interactome study also detected PpiD and YfgM as partner proteins of SecY (77). However, PpiD/YfgM were generally considered to be minor SecYEG partner proteins, because no detectable protein transport defect was observed in their absence, and a ΔppiD deletion strain did not show a strong phenotype (46). However, because of the reported functional redundancy of the periplasmic chaperone systems, phenotypes are often only detectable when multiple periplasmic chaperones are deleted (78, 79). In addition, a recent study re-evaluated the effect of PpiD on protein secretion and found that at low SecYEG concentrations, PpiD stimulated the translocation of the outer membrane protein OmpA or the periplasmic protein SfmC in vitro and enhanced their detachment from the membrane (48). These observations could indicate that PpiD is involved in clearing the SecYEG translocon or in the subsequent release of substrates into the periplasm. This would be in line with the deep penetration of PpiD into the periplasmic cavity of the SecY channel, as observed here.
It was recently shown that during the translocon-mediated transport, pulling forces act on nascent substrates (66, 80, 81). These forces are proposed to be generated at least partially by the transmembrane electric potential and by interactions of charged residues with membrane or protein surfaces (66). By using the force sensor approach with an established arrest peptide, our data demonstrate that the absence of PpiD/YfgM does not reduce the force exerted on a translocating substrate. Thus, although PpiD was shown to bind to a nascent chain arrested within the SecYEG translocon (45), PpiD does not seem to apply a pulling force on its substrates. This is in line with the observation that PpiD contains an inactive peptidylprolyl-isomerase domain (47), which likely excludes force generation by substrate folding. PpiD could prevent back-sliding of the polypeptide chain and thus enhance the directionality of protein transport or facilitate the handover of substrates from the Sec machinery to the periplasmic chaperone network (82). Different from PpiD, the depletion of YidC reduces the pulling force exerted on a nascent protein. This could be due to the reduced pmf in the absence of YidC (69) and/or to a direct involvement of YidC in pulling. A direct involvement of YidC in substrate pulling might explain why the translocation of shorter periplasmic loops in membrane proteins is SecA-independent, whereas longer periplasmic loops (> ∼30 amino acids) require the ATPase activity of SecA (83, 84).
PpiD and YidC engage largely-identical contact sites on SecY and are in contact with the lateral gate, the plug, and the periplasmic vestibule. Thus, they basically align with the path a substrate protein takes during SecY-dependent transport. Despite using almost identical binding sites on SecY, our data indicate that PpiD and YidC show noncompetitive binding to SecY. This would point to the existence of different SecYEG subassemblies, in which SecYEG is either in contact with PpiD or with YidC. This is in line with the observation that YidC and PpiD were found in different SecYEG complexes on blue native–PAGE (44, 46). Furthermore, PpiD was not detected in the SecYEG–YidC–SecDF holo-translocon (43, 85).
The MS-based interactome analyses were performed in vitro with sucrose gradient–purified and solubilized INVs, whereas the pBpa cross-linking was performed in vivo. It therefore appears unlikely that the association of SecYEG with either PpiD or YidC is substrate-controlled, because the SecYEG translocon in INVs is largely in its resting state and does not contain bound substrates (86). An alternative explanation is that the association of SecYEG with either YidC or PpiD is controlled by additional partner proteins. A likely candidate is the SecDF complex, which was suggested to tether YidC to the SecYEG complex (33, 59). Although YidC binding to the lateral gate of SecY is observed even in the absence of the SecDF complex (22), the abundance of SecYEG/YidC complexes is reduced in the absence of SecDF (87). The exact copy number of the SecDF complex in E. coli is debated. Although biochemical characterizations indicate a very-low copy number (∼50 copies per E. coli cell (88, 89)), ribosome profiling data suggest more than 2,500 copies (54). If SecDF is indeed required for a stable YidC/YEG complex, SecDF would limit the amount of detectable SecYEG/YidC complexes if sub-stoichiometric to SecYEG. This would explain why the majority of SecYEG is in contact with PpiD/YfgM as observed here. It should be noted that the experiments described here are performed with a plasmid-encoded SecYEG copy and therefore at increased SecYEG concentrations. However, YidC (∼8,000 copies) and PpiD (∼4,000 copies) are much more abundant than the SecYEG complex (∼500 copies) (Table 1) (50). Thus, at the approximate 5–10-fold higher SecYEG levels as a consequence of the used expression system (55), the number of SecYEG molecules would approximately match the number of PpiD/YidC molecules.
In summary, our data have identified the PpiD/YfgM chaperone complex as a primary interaction partner of the SecYEG translocon. PpiD engages the same interaction surface on SecY as the established SecY partner protein YidC, yet their binding to SecYEG seems to be noncompetitive. This likely indicates that SecYEG exists in a at least two subassemblies, the SecYEG/YidC complex, probably also containing SecDF, and the SecYEG–PpiD/YfgM complex. These subassemblies might enhance the efficiency of protein transport, e.g. the SecYEG–SecDF–YidC would be optimized for inner membrane proteins and the SecYEG–PpiD/YfgM for secretory proteins/outer membrane proteins. This would be in line with the observation that the SecYEG–SecDF/YidC complex is less efficient in transporting secretory proteins (43), while the deletion of YfgM reduces the stability of the outer membrane (44, 90). Our data furthermore show that YidC also interacts with periplasmic chaperones, like PpiD, Skp, and FkbA. Thus YidC, like SecYEG, likely also acts in concert with accessory proteins during membrane protein insertion. The possible existence of different SecYEG subassemblies and heterooligomeric YidC complexes adds yet another level of complexity to the bacterial protein transport system that needs to be further explored.
Experimental procedures
Bacterial strains, plasmids, and bacterial growth
In this study, the following E. coli strains were used: BL21 (Merck, Darmstadt, Germany); Dh5α (91); MC4100 ΔppiD/yfgM::kan (44); JS7131 (40); BL21 pTRc99aSecYHisEG/pSup (22); BL21 pTRc99aSecYHisEG/pEVol or pTRc99aSecYHisEG-YidC/pEVol plasmids (55). For in vivo 35S-labeling, the plasmid pRS1 (ampR) was designed, containing the pBR322 origin and the coding sequence for Rop, which controls the plasmid copy number. It contains the T7 RNA polymerase under the control of the constitutive synthetic EM7 promoter, a lacI-controlled T7 promotor upstream of the multiple cloning site, and the T7 transcription terminator downstream of the multiple cloning site. This allows high level and IPTG-dependent in vivo expression. Custom cloning service for the plasmid, which was provided by VectorBuilder Inc.
The pRS1–LepB–SecM(Ms) plasmid was constructed by Gibson assembly using the LepB–SecM(Ms) insert amplified by PCR with the primer pING1-forward (5′-cagaagcagaaagaaggtaag-3′) and pING1-reverse (5′-tgccacctgacgtctaag-3′), and the pRS1 vector opened by reverse PCR with the primer pET-MCS anti-opening forward (5′-ctcgagtagcataaccccttggg-3′) and pET-MCS anti-opening reverse (5′-catggggtatatctccttcttaaag-3′). As template DNA for generation of the lepB-SecM(Ms) insert, the plasmid NB726 pING-Lep6LMSnoCys (kindly provided by G. von Heijne, University of Stockholm) (66) was used. All PCR steps were performed with PfuUltra II fusion HS DNA polymerase (Agilent) and the provided buffers according to the manufacturer's instructions. The insert-vector assembly was performed with Gibson Assembly® Cloning Kit (New England Biolabs) at a 1:3 ratio.
The SecY-pBpa plug variants (residues 61–71) and the plug deletion mutant were constructed by site-directed mutagenesis using the PfuUltra II fusion HS DNA polymerase kit (Agilent) inserting TAG stop codons at the indicated positions using pTrc99a-SecYHisEG as template. This plasmid was also used for removing residues 61–70 creating the plug-deletion variant of SecY. All primers used for TAG stop-codon insertion or plug removal are listed in the supporting Data.
In vivo and in vitro cross-linking
The in vivo paraformaldehyde cross-linking protocol was performed as described previously (55).
For site-directed cross-linking BL21 cells containing the pEVol plasmid together with SecY or YidC variants in pTrc99a plasmids were cultured overnight in LB medium at 37 °C. 10 ml of the overnight culture was further used for inoculation of 1 liter of LB medium supplemented with 1 ml of 0.5 m pBpa (final concentration, dissolved in 1 m NaOH), 50 μg/μl ampicillin, and 35 μg/μl chloramphenicol. The cultures were further incubated at 37 °C until they reached the early exponential growth phase (OD600 = 0.5–0.8) and were induced with 0.5 mm IPTG. After induction, the cultures were grown for 2 h at 37 °C, cooled down on ice for 10 min, and harvested by centrifugation at 3,738 × g in a JLA 9.1000 rotor for 10 min. The cell pellets were resuspended in 10 ml of PBS buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4 and 1.76 mm KH2PO4) and divided in two multiwell plates. One plate was exposed to UV light on ice for 20 min (UV chamber: BLX-365, from Vilber Lourmat), and the other plate was kept in the dark. After UV irradiation, the cell suspension was transferred to 50-ml Falcon tubes, and cells were collected by centrifugation at 4,500 × g for 10 min in an Eppendorf A-4-44 rotor. Each cell pellet was resuspended in 10 ml of resuspension buffer (50 mm Tris/HCl, pH 7.5, 300 mm NaCl, 10 mm Mg(Ac)2) for subsequent YidC purification or 20 mm Tris/HCl, pH 7.5, 300 mm NaCl, 5 mm MgCl2 for subsequent SecY purification. Next, the samples were lysed by French pressing, and the cell debris was removed by centrifugation at 30,000 × g for 30 min in an SS34 rotor. The supernatant was further centrifuged at 244,061 × g for 1.5 h in a TLA 50.2 rotor for membrane sedimentation. Next, the membrane pellets were solubilized in 1% (w/v) n-dodecyl β-d-maltoside (DDM) dissolved in resuspension buffer supplemented with 10% (w/v) glycerol for 1 h at 4 °C. YidC or SecY was purified via metal-affinity chromatography using TALON® metal-affinity resin (Clontech). The samples were incubated for 1 h at 4 °C with 1 ml/liter LB culture of pre-equilibrated TALON® metal-affinity resin, washed five times with 10 ml/sample of washing buffer (40 mm imidazole, 10% glycerol (w/v), 50 mm Tris/HCl, pH 7.5, 300 mm NaCl, 10 mm Mg(Ac)2, and 0.03% DDM for YidC purification or 5 mm imidazole, 10% (w/v) glycerol, 20 mm Tris/HCl, pH 7.5, 300 mm NaCl, 5 mm MgCl2, and 0.03% DDM for SecY purification), and eluted four times in a total volume of 2 ml of elution buffer (200 mm imidazole, 10% glycerol (w/v), 50 mm Tris/HCl, pH 7.5, 300 mm NaCl, 10 mm Mg(Ac)2, and 0.03% DDM for YidC purification or 200 mm imidazole, 10% (w/v) glycerol, 10 ml of 20 mm Tris/HCl, pH 7.5, 300 mm NaCl, 5 mm MgCl2, and 0.03% DDM for SecY purification). Next, the samples were precipitated with 1 volume of 10% trichloroacetic acid (TCA) and denatured at 56 °C for 10 min in 35 μl of TCA loading dye (prepared by mixing 1 part of solution III (1 m DTT) with 4 parts of solution II (6.33% SDS (w/v), 0.083 m Tris base, 30% glycerol, and 0.053% bromphenol blue) and 5 parts of solution I (0.2 m Tris/HCl, 0.02 m EDTA, pH 8). Samples were subsequently analyzed by Western blotting or by MS.
Preparation of material for mass spectrometric analyses
E. coli BL21 cells carrying plasmid pEVol and plasmid pTrc99a-SecYHisEG or pTRc99aSecYHisEG-YidC were cultured overnight at 37 °C in LB medium containing 50 μg/ml ampicillin and 30 μg/ml chloramphenicol. 10 ml of the overnight culture was further used for inoculation in 1 liter of minimal medium (1% w/v glycerol, 47.7 mm Na2HPO4, 22 mm KH2PO4, 8.6 mm NaCl, 18.7 mm NH4Cl, 0.153 mm CaCl2, 0.59 mm MgSO4, 2.07 μm Na2MoO4, 4.198 μm CoSO4, 37.49 μm MnSO4, 4.35 μm ZnSO4, 6.29 μm FeCl2, 16.17 μm H3BO3, 40 mg/liter l-leucine, 1 mg/ml d-biotin, 1 mg/ml thiamine, 7 mg/liter chloramphenicol, 10 mg/liter ampicillin) until the cells reached the early exponential growth phase (OD600 = 0.5–0.8), and protein expression was then induced with 0.5 mm IPTG. After induction, the cultures were grown for 4 h at 30 °C, cooled down on ice for 10 min, and harvested by centrifugation at 3,738 × g in a JLA 9.1000 rotor for 10 min. Each cell pellet was resuspended in 10 ml of resuspension buffer (20 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgCl2) to which 0.5 mm phenylmethylsulfonyl fluoride and 2 pills/liter cOmpleteTM protease inhibitor mixture (Roche Applied Science®, Mannheim, Germany) were added. Next, the samples were lysed by French pressing, and cell debris was removed by centrifugation at 30, 000 × g for 30 min in an SS34 rotor. The supernatant was further centrifuged at 244,061 × g for 1.5 h in a TLA 50.2 rotor for membrane sedimentation. Next, the membrane pellets were solubilized in solubilization buffer (20 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgCl2, 10% glycerol, 1% DDM) for 1 h at 4 °C. Nonsolubilized membranes were separated by centrifugation for 10 min at 18,407 × g in an Eppendorf FA-45-24-11 rotor at 4 °C. The supernatant was transferred onto TALON® metal-affinity resin, which was then washed twice with washing buffer (20 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgCl2, 10% glycerol, 0.03% DDM, 5 mm imidazole) and subsequently washed four times with imidazole-free washing buffer. Furthermore, the material was washed four times with imidazole-containing washing buffer and eluted in 2 ml of elution buffer (20 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgCl2, 10% glycerol, 0.03% DDM, 200 mm imidazole).
The chemical cross-linker DSS (Thermo Fisher Scientific, Langenselbold, Germany) was dissolved in DMSO and incubated with the samples for 30 min at 25 °C after the second imidazole-free washing step (on-column cross-linking) or after the elution step (solution cross-linking). A 100 mm DSS stock solution was freshly prepared, from which either 50 μl were added to a total of 5 ml or 20 μl to a total of 2 ml. The final DSS concentration was 1 mm. After the DSS incubation step, the reaction was quenched by adding Tris/HCl, pH 7.5, from a 1 m Tris/HCl stock solution to a final concentration of 20 mm.
Protein samples for MS analyses were precipitated with acetone at −20 °C overnight and subsequently centrifuged with 18,407 × g in an Eppendorf FA-45-24-11 rotor at 4 °C for 20 min. After the supernatant was removed, the protein samples were dried and denatured in 10 μl of 8 m urea (in 50 mm NH4HCO3). Then, 1.1 μl of 50 mm Tris(2-carboxyethyl)phosphine (in 50 mm NH4HCO3) was added, and the sample was incubated for 20 min at 37 °C with continuous shaking at 600 rpm. Subsequently, 1.23 μl of 500 mm iodoacetamide (in 50 mm NH4HCO3) was added and incubated for 20 min in the absence of light. Finally, 3 μl of 100 mm DTT (in 50 mm NH4HCO3) and subsequently 25 μl of 50 mm NH4HCO3 were added. The protein samples were digested overnight with 0.125 μg/μl trypsin at 37 °C and then acidified with 0.1% TFA and centrifuged for 6 min at 13,523 × g in an FA-45-24-11 rotor. In-gel digestion of proteins following site-directed cross-linking by UV irradiation of YidC(K249pBpa) cells was carried out after cutting the −UV and the +UV gel lanes into equal-sized slices as described previously (10).
Peptide mixtures were analyzed by LC-tandem MS (LC-MS/MS) using an Ultimate 3000 RSLCnano coupled to a Q Exactive Plus (Thermo Fisher Scientific) mass spectrometer as described (92) for SecYEG/SecYHisEG or an Ultimate inert HPLC system (Dionex) coupled to an LTQ-FT (Thermo Electron Corp.), as described previously (10) for YidC(K249pBpa). Proteins interacting with SecYHisEG were identified using the MaxQuant software (version 1.5.5.1 (93)) searching against the UniProt E. coli proteome set sequence database (version 2018_08, 4,344 entries) allowing a maximum of three missed cleavages of trypsin, a mass tolerance of 4.5 ppm for precursor, and 20 ppm for fragment ions. Methionine oxidation and hydrolyzed DSS at lysines were specified as variable modifications and carbamidomethylation of cysteine as fixed modification. The lists of both peptide and protein identifications were filtered applying a threshold for the false discovery rate of <0.01. Only proteins detected in at least two biological replicates, identified with ≥20% sequence coverage and a minimum of two peptides, were considered as reliably identified. The LFQ algorithm was used for label-free relative quantification with a minimum ratio count of 1. Intensity ratios (SecYHisEG versus SecYEG) were computed and, when needed because one value of a ratio pair was zero, missing values were replaced using data imputation to simulate signals of low abundant proteins near the detection limit of the instrument (94). Missing values were based on the mean and standard deviation of the distribution of all logarithmic intensities using a downshift of 2.1 and a width adjustment of 0.2. To determine significant interactors, logarithmic ratios were normalized for each replicate and subjected to two-sided t tests across all replicates. Threshold values for p value and ratio in volcano plots were defined using symmetric hyperbolic curves separating false-positive identifications on the left side (ratios <1) and a set of significant interactors on the right side (ratios >1) (95). Final parameters used to ensure a false-positive rate of <0.01 were a bend of 0.25, p value threshold of 0.05, and log10 ratio limit of 10. Relative protein abundances are best estimated by iBAQ intensities that correct protein intensities for the number of possible proteolytic peptides. For YidC(K249pBpa) in vivo cross-linking experiments, MS data analysis was carried out similarly, except that a maximum of two missed cleavages of trypsin was allowed, a mass tolerance of 0.5 Da for fragment ions was used, and no further filtering for the number of peptides or sequence coverage was applied. Only proteins having LFQ intensity gel lane profiles that showed a UV-dependent mass shift relative to their calculated molecular mass were accepted as specific interaction partners.
In vivo pulse labeling of LepB–SecM(Ms) ribosome-associated polypeptide chains
10 ml of LB medium were inoculated with either MC4100 or MC4100 ΔppiD + yfgM::kan carrying the pRS1-LepB–SecM(Ms) plasmid. 12.5 μg/ml kanamycin and 50 μg/ml ampicillin were used in the pre-culture of MC4100 ΔppiD + yfgM::kan strain, whereas for the MC4100 strain only ampicillin was used. The bacterial cultures were grown at 37 °C overnight, harvested, and resuspended after washing in 1 ml of M63 medium (20 g/liter glycerol, 13.6 g/liter KH2PO4, 2 g/liter (NH4)2SO4, 0.5 mg/liter FeSO4, pH 7.0, adjusted with KOH, 0.5 mg/ml thiamine, and 0.1 mm of the 18-amino acid mix (all essential amino acids with the exception of cysteine and methionine)). 200 μl of the cell suspension were used for inoculation of 20 ml of fresh M63 media supplemented with 50 μg/ml ampicillin in 100-ml Erlenmeyer flasks. The cultures were grown at 37 °C until they reached an OD600 of 0.5–0.9. Subsequently, 108 cells from each culture were collected and transferred to 2-ml Eppendorf tubes already containing 50 μg/mg rifampicin to inactivate the endogenous RNA polymerase for 10 min at 37 °C. The volume of each tube was adjusted to 2 ml with fresh M63 medium, and all cultures were incubated in a thermomixer for 10 min at 37 °C. Then 0.1 mm of the protonophore CCCP was added when indicated, and the cultures were incubated for 7.5 min at 37 °C. After the CCCP treatment, the production of LepB–SecM(Ms) ribosome-associated polypeptide chains was induced by simultaneous addition of 0.1 m IPTG and 2 μl of l-[35S]methionine and l-[35S]cysteine mix (7 mCi/ml, PerkinElmer Life Sciences). Subsequently, 100 μl of each sample were collected at the indicated time points and directly precipitated by addition of 10% TCA and incubation on ice for 30 min. The precipitated proteins were recovered as pellets by a 12-min centrifugation at 15,871 × g in an Eppendorf FA-45-24-11 rotor at 4 °C. Each pellet was denatured in 35 μl of TCA loading dye at 56 °C for 12 min with 1,400 rpm continuous shaking. The TCA loading dye is prepared by mixing 1 part of solution III (1 m DTT) with 4 parts of solution II (6.33% SDS (w/v), 0.083 m Tris base, 30% glycerol, and 0.053% bromphenol blue) and 5 parts of solution I (0.2 m Tris base, 0.02 m EDTA, pH 8). Subsequently, the samples were loaded onto a 15% SDS-PAGE and run overnight at 17 mA before the gel was dried and analyzed by phosphorimaging.
YidC depletion procedures
To deplete YidC, the E. coli strain JS7131 was used (40). The efficient depletion of YidC was initiated by inoculation of 10 ml of LB + 0.2% arabinose with JS7131 carrying the pRS1-LepB–SecM(Ms) plasmid, and cells were grown overnight at 37 °C. Spectinomycin and ampicillin were present at a final concentration of 50 μg/ml. 200 μl of the pre-culture were used for inoculation of 20 ml of LB culture containing 0.2% arabinose (for YidC expression), and the culture was grown at 37 °C until it reached an OD600 of 0.5. The cells were subsequently harvested at 4,500 × g for 10 min in an Eppendorf A-4-44 rotor at room temperature, washed with 10 ml of pre-warmed LB medium lacking arabinose, and collected again by centrifugation. The cell pellet was further resuspended in 1 ml of 37 °C fresh LB and transferred to a 19-ml pre-warmed LB medium lacking arabinose. Growth was monitored at OD600 until the OD600 doubled, and then 10 ml of the culture were replaced with 10 ml of pre-warmed LB medium, and the culture was grown again at 37 °C until the OD600 doubled. For complete depletion of YidC, five steps of LB medium exchange were required. In the fifth step the entire culture was centrifuged down and resuspended in 20 ml of M63 medium and further incubated at 37 °C for 30 min. 108 cells were collected for Western blotting with α-YidC antibodies or antibodies against SecY, PpiD, and YfgM, and the remaining of the culture was subjected to rifampicin treatment and radioactive labeling as described above.
Immunodetection and antibodies
For immunodetection of SDS-PAGE, proteins were electroblotted onto nitrocellulose membranes (GE Healthcare). Membranes were blocked with 5% milk powder in T-TBS buffer for at least 1 h. Polyclonal antibodies against YidC were generated in rabbits against the complete and SDS-denatured protein (96). α-YidC antibodies have been validated before (87). Antibodies against the SecY peptide MAKQPGLDFQSAKGGLGELKRRC were raised in rabbits by GenScript Biotech (Leiden, Netherlands) and have been validated before (30, 55, 87). Polyclonal antibodies against PpiD/YfgM were a gift of Dan Daley, University of Stockholm, and have been validated before (Fig. S2) (44). Peroxidase-coupled goat anti-rabbit antibodies (Caltag Laboratories, Burlingham, CA) were used as secondary antibodies with ECL (GE Healthcare).
Reproducibility statement and statistical analyses
All experiments were performed at least twice as independent biological replicates. Each replicate consisted of at least two technical replicates. Only representative Western blottings are displayed. Statistical significance between groups was established using the unpaired Student's t test. p < 0.05 was considered to be significant. Data analyses and presentation were performed in Excel 2002 (Microsoft Corp.) and GraphPad Prism for Windows version 7.00 (GraphPad Software, San Diego).
Author contributions
B. J., N.-A. P., F. D., L. F., I. S., and T. W. data curation; B. J., N.-A. P., F. D., L. F., I. S., T. W., and R. S. investigation; B. J., N.-A. P., F. D., and R. S. methodology; B. J., N.-A. P., F. D., I. S., R. S., B. W., and H.-G. K. writing-original draft; N.-A. P., F. D., I. S., B. W., and H.-G. K. formal analysis; F. D. validation; R. S. resources; B. W. and H.-G. K. conceptualization; B. W. and H.-G. K. supervision; B. W. and H.-G. K. funding acquisition; B. W. and H.-G. K. writing-review and editing; H.-G. K. project administration.
Supplementary Material
Acknowledgments
We thank Bettina Knapp for technical assistance in LC/MS analyses and Dan Daley and Gunnar von Heijne (University Stockholm) for antibodies, strains, and plasmids.
This work was supported by German Science Foundation (DFG) Grants KO2184/8-1 and KO2184/8-2 (to H. G. K.), Project-ID 403222702, SFB 1381 (to H. G. K. and B. W.), Excellence Strategy CIBBS-EXC-2189 Project ID 390939984 (to B. W.), the Else-Kröner-Fresenius-Foundation/Motivate Program of the University Freiburg Medical School, the Konrad-Adenauer-Foundation (to B. J.), and by an FF-Nord Fellowship (to R. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2, Table S1, and supporting Data.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (97) with the dataset identifier accession nos. PXD015985 and PXD015974.
- TM
- transmembrane domain
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazine
- DSS
- disuccinimidyl suberate
- iBAQ
- intensity-based absolute quantification
- IMAC
- immobilized metal-affinity chromatography
- INV
- inner membrane vesicles
- pBpa
- para-benzoyl-l-phenylalanine
- pmf
- proton-motive force
- PFA
- paraformaldehyde
- SRP
- signal-recognition particle
- PDB
- Protein Data Bank
- qAP-MS
- quantitative affinity purification-MS
- pmf
- proton-motive force
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- DDM
- n-dodecyl β-d-maltoside.
References
- 1. Denks K., Vogt A., Sachelaru I., Petriman N. A., Kudva R., and Koch H. G. (2014) The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 31, 58–84 10.3109/09687688.2014.907455 [DOI] [PubMed] [Google Scholar]
- 2. Knyazev D. G., Kuttner R., Zimmermann M., Sobakinskaya E., and Pohl P. (2018) Driving forces of translocation through bacterial translocon SecYEG. J. Membr. Biol. 251, 329–343 10.1007/s00232-017-0012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lycklama A., and Driessen A. J. (2012) The bacterial Sec-translocase: structure and mechanism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1016–1028 10.1098/rstb.2011.0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collinson I., Corey R. A., and Allen W. J. (2015) Channel crossing: how are proteins shipped across the bacterial plasma membrane? Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150025 10.1098/rstb.2015.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park E., and Rapoport T. A. (2012) Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 10.1146/annurev-biophys-050511-102312 [DOI] [PubMed] [Google Scholar]
- 6. Becker T., Bhushan S., Jarasch A., Armache J. P., Funes S., Jossinet F., Gumbart J., Mielke T., Berninghausen O., Schulten K., Westhof E., Gilmore R., Mandon E. C., and Beckmann R. (2009) Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 10.1126/science.1178535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van den Berg B., Clemons W. M. Jr, Collinson I., Modis Y., Hartmann E., Harrison S. C., and Rapoport T. A. (2004) X-ray structure of a protein-conducting channel. Nature 427, 36–44 10.1038/nature02218 [DOI] [PubMed] [Google Scholar]
- 8. Li W., Schulman S., Boyd D., Erlandson K., Beckwith J., and Rapoport T. A. (2007) The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol. Cell 26, 511–521 10.1016/j.molcel.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 9. Saparov S. M., Erlandson K., Cannon K., Schaletzky J., Schulman S., Rapoport T. A., and Pohl P. (2007) Determining the conductance of the SecY protein translocation channel for small molecules. Mol. Cell 26, 501–509 10.1016/j.molcel.2007.03.022 [DOI] [PubMed] [Google Scholar]
- 10. Kuhn P., Weiche B., Sturm L., Sommer E., Drepper F., Warscheid B., Sourjik V., and Koch H. G. (2011) The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic 12, 563–578 10.1111/j.1600-0854.2011.01167.x [DOI] [PubMed] [Google Scholar]
- 11. Angelini S., Deitermann S., and Koch H. G. (2005) FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 6, 476–481 10.1038/sj.embor.7400385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Keyzer J., Regeling A., and Driessen A. J. (2007) Arginine 357 of SecY is needed for SecA-dependent initiation of preprotein translocation. FEBS Lett. 581, 1859–1864 10.1016/j.febslet.2007.03.081 [DOI] [PubMed] [Google Scholar]
- 13. Kuhn P., Draycheva A., Vogt A., Petriman N. A., Sturm L., Drepper F., Warscheid B., Wintermeyer W., and Koch H. G. (2015) Ribosome binding induces repositioning of the signal recognition particle receptor on the translocon. J. Cell Biol. 211, 91–104 10.1083/jcb.201502103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banerjee T., Zheng Z., Abolafia J., Harper S., and Oliver D. (2017) The SecA protein deeply penetrates into the SecYEG channel during insertion, contacting most channel transmembrane helices and periplasmic regions. J. Biol. Chem. 292, 19693–19707 10.1074/jbc.RA117.000130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zimmer J., Nam Y., and Rapoport T. A. (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 10.1038/nature07335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Draycheva A., Bornemann T., Ryazanov S., Lakomek N. A., and Wintermeyer W. (2016) The bacterial SRP receptor, FtsY, is activated on binding to the translocon. Mol. Microbiol. 102, 152–167 10.1111/mmi.13452 [DOI] [PubMed] [Google Scholar]
- 17. Estrozi L. F., Boehringer D., Shan S. O., Ban N., and Schaffitzel C. (2011) Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat. Struct. Mol. Biol. 18, 88–90 10.1038/nsmb.1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jomaa A., Fu Y. H., Boehringer D., Leibundgut M., Shan S. O., and Ban N. (2017) Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome. Nat. Commun. 8, 15470 10.1038/ncomms15470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akopian D., Dalal K., Shen K., Duong F., and Shan S. O. (2013) SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J. Cell Biol. 200, 397–405 10.1083/jcb.201208045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. du Plessis D. J., Berrelkamp G., Nouwen N., and Driessen A. J. (2009) The lateral gate of SecYEG opens during protein translocation. J. Biol. Chem. 284, 15805–15814 10.1074/jbc.M901855200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gogala M., Becker T., Beatrix B., Armache J. P., Barrio-Garcia C., Berninghausen O., and Beckmann R. (2014) Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506, 107–110 10.1038/nature12950 [DOI] [PubMed] [Google Scholar]
- 22. Sachelaru I., Petriman N. A., Kudva R., Kuhn P., Welte T., Knapp B., Drepper F., Warscheid B., and Koch H.-G. (2013) YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J. Biol. Chem. 288, 16295–16307 10.1074/jbc.M112.446583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ge Y., Draycheva A., Bornemann T., Rodnina M. V., and Wintermeyer W. (2014) Lateral opening of the bacterial translocon on ribosome binding and signal peptide insertion. Nat. Commun. 5, 5263 10.1038/ncomms6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frauenfeld J., Gumbart J., Sluis E. O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., and Beckmann R. (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 10.1038/nsmb.2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaufmann A., Manting E. H., Veenendaal A. K., Driessen A. J., and van der Does C. (1999) Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry 38, 9115–9125 10.1021/bi990539d [DOI] [PubMed] [Google Scholar]
- 26. Lycklama a Nijeholt J. A., de Keyzer J., Prabudiansyah I., and Driessen A. J. (2013) Characterization of the supporting role of SecE in protein translocation. FEBS Lett. 587, 3083–3088 10.1016/j.febslet.2013.07.046 [DOI] [PubMed] [Google Scholar]
- 27. Akiyama Y., Kihara A., Tokuda H., and Ito K. (1996) FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 271, 31196–31201 10.1074/jbc.271.49.31196 [DOI] [PubMed] [Google Scholar]
- 28. Sugai R., Takemae K., Tokuda H., and Nishiyama K. (2007) Topology inversion of SecG is essential for cytosolic SecA-dependent stimulation of protein translocation. J. Biol. Chem. 282, 29540–29548 10.1074/jbc.M704716200 [DOI] [PubMed] [Google Scholar]
- 29. Zimmermann R., Eyrisch S., Ahmad M., and Helms V. (2011) Protein translocation across the ER membrane. Biochim. Biophys. Acta 1808, 912–924 10.1016/j.bbamem.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 30. Braig D., Mircheva M., Sachelaru I., van der Sluis E. O., Sturm L., Beckmann R., and Koch H. G. (2011) Signal sequence-independent SRP–SR complex formation at the membrane suggests an alternative targeting pathway within the SRP cycle. Mol. Biol. Cell 22, 2309–2323 10.1091/mbc.e11-02-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Welte T., Kudva R., Kuhn P., Sturm L., Braig D., Müller M., Warscheid B., Drepper F., and Koch H. G. (2012) Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell 23, 464–479 10.1091/mbc.e11-07-0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duong F., and Wickner W. (1997) Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 16, 2756–2768 10.1093/emboj/16.10.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nouwen N., and Driessen A. J. (2002) SecDFyajC forms a heterotetrameric complex with YidC. Mol. Microbiol. 44, 1397–1405 10.1046/j.1365-2958.2002.02972.x [DOI] [PubMed] [Google Scholar]
- 34. Pogliano J. A., and Beckwith J. (1994) SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 13, 554–561 10.1002/j.1460-2075.1994.tb06293.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dalbey R., Koch H. G., and Kuhn A. (2017) Targeting and insertion of membrane proteins. EcoSalPlus 2017, 7, 1–28 10.1128/ecosalplus.ESP-0012-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beck K., Eisner G., Trescher D., Dalbey R. E., Brunner J., and Müller M. (2001) YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2, 709–714 10.1093/embo-reports/kve154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Houben E. N., ten Hagen-Jongman C. M., Brunner J., Oudega B., and Luirink J. (2004) The two membrane segments of leader peptidase partition one by one into the lipid bilayer via a Sec/YidC interface. EMBO Rep. 5, 970–975 10.1038/sj.embor.7400261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu L., Kaback H. R., and Dalbey R. E. (2013) YidC protein, a molecular chaperone for LacY protein folding via the SecYEG protein machinery. J. Biol. Chem. 288, 28180–28194 10.1074/jbc.M113.491613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wagner S., Pop O., Haan G. J., Baars L., Koningstein G., Klepsch M. M., Genevaux P., Luirink J., and de Gier J. W. (2008) Biogenesis of MalF and the MalFGK(2) maltose transport complex in Escherichia coli requires YidC. J. Biol. Chem. 283, 17881–17890 10.1074/jbc.M801481200 [DOI] [PubMed] [Google Scholar]
- 40. Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., and Dalbey R. E. (2000) YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 10.1038/35020586 [DOI] [PubMed] [Google Scholar]
- 41. Facey S. J., Neugebauer S. A., Krauss S., and Kuhn A. (2007) The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J. Mol. Biol. 365, 995–1004 10.1016/j.jmb.2006.10.083 [DOI] [PubMed] [Google Scholar]
- 42. Komar J., Alvira S., Schulze R. J., Martin R., Lycklama A., Nijeholt J. A., Lee S., Dafforn T. R., Deckers-Hebestreit G., Berger I., Schaffitzel C., and Collinson I. (2016) Membrane protein insertion and assembly by the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Biochem. J. 473, 3341–3354 10.1042/BCJ20160545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schulze R. J., Komar J., Botte M., Allen W. J., Whitehouse S., Gold V. A., Lycklama A., Huard K., Berger I., Schaffitzel C., and Collinson I. (2014) Membrane protein insertion and proton-motive force-dependent secretion through the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Proc. Natl. Acad. Sci. U.S.A. 111, 4844–4849 10.1073/pnas.1315901111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Götzke H., Palombo I., Muheim C., Perrody E., Genevaux P., Kudva R., Müller M., and Daley D. O. (2014) YfgM is an ancillary subunit of the SecYEG translocon in Escherichia coli. J. Biol. Chem. 289, 19089–19097 10.1074/jbc.M113.541672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Antonoaea R., Fürst M., Nishiyama K., and Müller M. (2008) The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry 47, 5649–5656 10.1021/bi800233w [DOI] [PubMed] [Google Scholar]
- 46. Sachelaru I., Petriman N.-A., Kudva R., and Koch H.-G. (2014) Dynamic interaction of the Sec translocon with the chaperone PpiD. J. Biol. Chem. 289, 21706–21715 10.1074/jbc.M114.577916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weininger U., Jakob R. P., Kovermann M., Balbach J., and Schmid F. X. (2010) The prolyl isomerase domain of PpiD from Escherichia coli shows a parvulin fold but is devoid of catalytic activity. Protein Sci. 19, 6–18 10.1002/pro.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fürst M., Zhou Y., Merfort J., and Müller M. (2018) Involvement of PpiD in Sec-dependent protein translocation. Biochim. Biophys. Acta Mol. Cell Res. 1865, 273–280 10.1016/j.bbamcr.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 49. Maddalo G., Stenberg-Bruzell F., Götzke H., Toddo S., Björkholm P., Eriksson H., Chovanec P., Genevaux P., Lehtiö J., Ilag L. L., and Daley D. O. (2011) Systematic analysis of native membrane protein complexes in Escherichia coli. J. Proteome Res. 10, 1848–1859 10.1021/pr101105c [DOI] [PubMed] [Google Scholar]
- 50. Kudva R., Denks K., Kuhn P., Vogt A., Müller M., and Koch H. G. (2013) Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534 10.1016/j.resmic.2013.03.016 [DOI] [PubMed] [Google Scholar]
- 51. Keller R., de Keyzer J., Driessen A. J., and Palmer T. (2012) Co-operation between different targeting pathways during integration of a membrane protein. J. Cell Biol. 199, 303–315 10.1083/jcb.201204149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ito K., and Akiyama Y. (2005) Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59, 211–231 10.1146/annurev.micro.59.030804.121316 [DOI] [PubMed] [Google Scholar]
- 53. Castanié-Cornet M. P., Bruel N., and Genevaux P. (2014) Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim. Biophys. Acta 1843, 1442–1456 10.1016/j.bbamcr.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 54. Li G. W., Burkhardt D., Gross C., and Weissman J. S. (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 10.1016/j.cell.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Petriman N. A., Jauss B., Hufnagel A., Franz L., Sachelaru I., Drepper F., Warscheid B., and Koch H. G. (2018) The interaction network of the YidC insertase with the SecYEG translocon, SRP and the SRP receptor FtsY. Sci. Rep. 8, 578 10.1038/s41598-017-19019-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Neves-Petersen M. T., Klitgaard S., Pascher T., Skovsen E., Polivka T., Yartsev A., Sundström V., and Petersen S. B. (2009) Flash photolysis of cutinase: identification and decay kinetics of transient intermediates formed upon UV excitation of aromatic residues. Biophys. J. 97, 211–226 10.1016/j.bpj.2009.01.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harris M. E., and Christian E. L. (2009) RNA crosslinking methods. Methods Enzymol. 468, 127–146 10.1016/S0076-6879(09)68007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dormán G., and Prestwich G. D. (1994) Benzophenone photophores in biochemistry. Biochemistry 33, 5661–5673 10.1021/bi00185a001 [DOI] [PubMed] [Google Scholar]
- 59. Xie K., Kiefer D., Nagler G., Dalbey R. E., and Kuhn A. (2006) Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45, 13401–13408 10.1021/bi060826z [DOI] [PubMed] [Google Scholar]
- 60. Dartigalongue C., and Raina S. (1998) A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17, 3968–3980 10.1093/emboj/17.14.3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Junne T., Schwede T., Goder V., and Spiess M. (2006) The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol. Biol. Cell 17, 4063–4068 10.1091/mbc.e06-03-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Erlandson K. J., Or E., Osborne A. R., and Rapoport T. A. (2008) Analysis of polypeptide movement in the SecY channel during SecA-mediated protein translocation. J. Biol. Chem. 283, 15709–15715 10.1074/jbc.M710356200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Maillard A. P., Lalani S., Silva F., Belin D., and Duong F. (2007) Deregulation of the SecYEG translocation channel upon removal of the plug domain. J. Biol. Chem. 282, 1281–1287 10.1074/jbc.M610060200 [DOI] [PubMed] [Google Scholar]
- 64. Park E., and Rapoport T. A. (2011) Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 473, 239–242 10.1038/nature10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sachelaru I., Winter L., Knyazev D. G., Zimmermann M., Vogt A., Kuttner R., Ollinger N., Siligan C., Pohl P., and Koch H.-G. (2017) YidC and SecYEG form a heterotetrameric protein translocation channel. Sci. Rep. 7, 101 10.1038/s41598-017-00109-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ismail N., Hedman R., Schiller N., and von Heijne G. (2012) A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat. Struct. Mol. Biol. 19, 1018–1022 10.1038/nsmb.2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ito K., Chiba S., and Pogliano K. (2010) Divergent stalling sequences sense and control cellular physiology. Biochem. Biophys. Res. Commun. 393, 1–5 10.1016/j.bbrc.2010.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cymer F., Hedman R., Ismail N., and von Heijne G. (2015) Exploration of the arrest peptide sequence space reveals arrest-enhanced variants. J. Biol. Chem. 290, 10208–10215 10.1074/jbc.M115.641555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van der Laan M., Urbanus M. L., Ten Hagen-Jongman C. M., Nouwen N., Oudega B., Harms N., Driessen A. J., and Luirink J. (2003) A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. U.S.A. 100, 5801–5806 10.1073/pnas.0636761100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schäfer U., Beck K., and Müller M. (1999) Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274, 24567–24574 10.1074/jbc.274.35.24567 [DOI] [PubMed] [Google Scholar]
- 71. Lohmeyer E., Schröder S., Pawlik G., Trasnea P. I., Peters A., Daldal F., and Koch H. G. (2012) The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb(3)-type cytochrome oxidase in Rhodobacter capsulatus. Biochim. Biophys. Acta 1817, 2005–2015 10.1016/j.bbabio.2012.06.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Scotti P. A., Urbanus M. L., Brunner J., de Gier J. W., von Heijne G., van der Does C., Driessen A. J., Oudega B., and Luirink J. (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 10.1093/emboj/19.4.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tsukazaki T., Mori H., Echizen Y., Ishitani R., Fukai S., Tanaka T., Perederina A., Vassylyev D. G., Kohno T., Maturana A. D., Ito K., and Nureki O. (2011) Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235–238 10.1038/nature09980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chorev D. S., Baker L. A., Wu D., Beilsten-Edmands V., Rouse S. L., Zeev-Ben-Mordehai T., Jiko C., Samsudin F., Gerle C., Khalid S., Stewart A. G., Matthews S. J., Grünewald K., and Robinson C. V. (2018) Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science 362, 829–834 10.1126/science.aau0976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang X. X., Chan C. S., Bao H., Fang Y., Foster L. J., and Duong F. (2012) Nanodiscs and SILAC-based mass spectrometry to identify a membrane protein interactome. J. Proteome Res. 11, 1454–1459 10.1021/pr200846y [DOI] [PubMed] [Google Scholar]
- 76. Stenberg F., Chovanec P., Maslen S. L., Robinson C. V., Ilag L. L., von Heijne G., and Daley D. O. (2005) Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280, 34409–34419 10.1074/jbc.M506479200 [DOI] [PubMed] [Google Scholar]
- 77. Carlson M. L., Stacey R. G., Young J. W., Wason I. S., Zhao Z., Rattray D. G., Scott N., Kerr C. H., Babu M., Foster L. J., and Duong Van Hoa F. (2019) Profiling the E. coli membrane interactome captured in peptidisc libraries. Elife 8, e46615 10.7554/eLife.46615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stull F., Betton J. M., and Bardwell J. C. A. (2018) Periplasmic chaperones and prolyl isomerases. EcoSal. Plus 2018, 8, 10.1128/ecosalplus.ESP-0005-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Matern Y., Barion B., and Behrens-Kneip S. (2010) PpiD is a player in the network of periplasmic chaperones in Escherichia coli. BMC Microbiol. 10, 251–267 10.1186/1471-2180-10-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cymer F., Ismail N., and von Heijne G. (2014) Weak pulling forces exerted on Nin-orientated transmembrane segments during co-translational insertion into the inner membrane of Escherichia coli. FEBS Lett. 588, 1930–1934 10.1016/j.febslet.2014.03.050 [DOI] [PubMed] [Google Scholar]
- 81. Ismail N., Hedman R., Lindén M., and von Heijne G. (2015) Charge-driven dynamics of nascent-chain movement through the SecYEG translocon. Nat. Struct. Mol. Biol. 22, 145–149 10.1038/nsmb.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Merdanovic M., Clausen T., Kaiser M., Huber R., and Ehrmann M. (2011) Protein quality control in the bacterial periplasm. Annu. Rev. Microbiol. 65, 149–168 10.1146/annurev-micro-090110-102925 [DOI] [PubMed] [Google Scholar]
- 83. Andersson H., and von Heijne G. (1993) Sec dependent and sec independent assembly of E. coli inner membrane proteins: the topological rules depend on chain length. EMBO J. 12, 683–691 10.1002/j.1460-2075.1993.tb05702.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Neumann-Haefelin C., Schäfer U., Müller M., and Koch H. G. (2000) SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 19, 6419–6426 10.1093/emboj/19.23.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Botte M., Zaccai N. R., Nijeholt J. L., Martin R., Knoops K., Papai G., Zou J., Deniaud A., Karuppasamy M., Jiang Q., Roy A. S., Schulten K., Schultz P., Rappsilber J., Zaccai G., et al. (2016) A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion. Sci. Rep. 6, 38399 10.1038/srep38399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Koch H. G., and Müller M. (2000) Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. J. Cell Biol. 150, 689–694 10.1083/jcb.150.3.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Boy D., and Koch H. G. (2009) Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol. Biol. Cell 20, 1804–1815 10.1091/mbc.e08-08-0886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Matsuyama S., Fujita Y., Sagara K., and Mizushima S. (1992) Overproduction, purification and characterization of SecD and SecF, integral membrane components of the protein translocation machinery of Escherichia coli. Biochim. Biophys. Acta 1122, 77–84 10.1016/0167-4838(92)90130-6 [DOI] [PubMed] [Google Scholar]
- 89. Pogliano K. J., and Beckwith J. (1994) Genetic and molecular characterization of the Escherichia coli secD operon and its products. J. Bacteriol. 176, 804–814 10.1128/jb.176.3.804-814.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Götzke H., Muheim C., Altelaar A. F., Heck A. J., Maddalo G., and Daley D. O. (2015) Identification of putative substrates for the periplasmic chaperone YfgM in Escherichia coli using quantitative proteomics. Mol. Cell. Proteomics 14, 216–226 10.1074/mcp.M114.043216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 92. Chan A., Schummer A., Fischer S., Schröter T., Cruz-Zaragoza L. D., Bender J., Drepper F., Oeljeklaus S., Kunau W. H., Girzalsky W., Warscheid B., and Erdmann R. (2016) Pex17p-dependent assembly of Pex14p/Dyn2p-subcomplexes of the peroxisomal protein import machinery. Eur. J. Cell Biol. 95, 585–597 10.1016/j.ejcb.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 93. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- 94. Deeb S. J., D'Souza R. C., Cox J., Schmidt-Supprian M., and Mann M. (2012) Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell. Proteomics 11, 77–89 10.1074/mcp.M111.015362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., and Mann M. (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 10.1083/jcb.200911091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Koch H. G., Moser M., Schimz K. L., and Muller M. (2002) The integration of YidC into the cytoplasmic membrane of Escherichia coli requires the signal recognition particle, SecA and SecYEG. J. Biol. Chem. 277, 5715–5718 10.1074/jbc.C100683200 [DOI] [PubMed] [Google Scholar]
- 97. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Santos-Zavaleta A., Salgado H., Gama-Castro S., Sanches-Perez M., Gomez-Romero L., Ledezma-Tejeida D., Garcia-Sotelo J. S., Alquicira-Hernandez K., Muniz-Rascado L. J., Pena-Loredo P., Ishada-Gutierrez C., Velazquez-Ramirez D. A., Del Moral-Chavez V., Bonavides-Martinez C., Mendez-Cruz C. F., et al. (2018) RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K12. Nucleic Acid Research 47, D212–D220 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.