Abstract
Ribosome biogenesis is critical for proliferating cells and requires the coordinated activities of three eukaryotic RNA polymerases. We recently showed that the small ubiquitin-like modifier (SUMO) system controls the global level of RNA polymerase II (Pol II)–controlled transcription in mammalian cells by regulating cyclin-dependent kinase 9 activity. Here, we present evidence that the SUMO system also plays a critical role in the control of Pol I transcription. Using an siRNA-based knockdown approach, we found that multiple SUMO E3 ligases of the PIAS (protein inhibitor of activated STAT) family are involved in SUMO-mediated repression of ribosomal DNA (rDNA) gene transcription. We demonstrate that endogenous SUMO represses rDNA transcription primarily by repressing upstream-binding factor and proto-oncogene c-Myc expression and that ectopic overexpression of SUMO-associated enzymes additionally represses rDNA transcription via c-Myc SUMOylation and its subsequent degradation. The results of our study reveal a critical role of SUMOylation in the control of rDNA transcription, uncover the underlying mechanisms involved, and indicate that the SUMO system coordinates Pol I– and Pol II–mediated transcription in mammalian cells.
Keywords: SUMOylation, UBC9, ribosomal ribonucleic acid (rRNA) (ribosomal RNA), RNA polymerase I, transcription, transcription factor, c-Myc, PIAS, UBF
Introduction
Ribosome biogenesis is a major cellular process that occurs in specific nuclear compartments, the nucleoli. A rate-limiting step in this process is ribosomal DNA (rDNA)5 transcription by RNA polymerase (Pol) I, which accounts for up to 60% of cellular RNA synthesis in proliferating cells (1–3). Pol I transcription is initiated by binding of upstream-binding factor (UBF) and selectivity factor 1 (SL1) complex to the rDNA promoter. The UBF–SL1 complex in turn promotes the recruitment of a subpopulation of RRN3 (also known as TIF-1A)–associated Pol I to form a Pol I preinitiation complex at the rDNA promoter (4, 5). In mouse and human cells, ∼200 rDNA gene copies per haploid genome are distributed in 5 clusters on different chromosomes. Despite the need for a high level of rDNA transcription, typically only a fraction of the rDNA genes is transcriptionally active, and the remaining genes are epigenetically silenced (6, 7). Although much progress has been made in study of regulation of Pol I transcription by signaling pathways and epigenetic mechanisms (7–12), the less well-understood is the molecular mechanism(s) that coordinates the transcription by Pol I and transcription by Pol II. In this regard, transcription factor c-Myc has been shown to enhance Pol I transcription directly by binding to the rDNA promoter region (13, 14), and in a model of granulocyte differentiation, c-Myc also stimulates Pol II–dependent transcription of a cohort of factors associated with Pol I transcription (termed “Pol I regulon”) (15, 16).
Reversible post-translational modification with the small ubiquitin-related modifier SUMO (SUMOylation) is involved in essentially all fundamental cellular processes in eukaryotic cells (17, 18). SUMOylation is governed by a conserved cascade consisting of an E1-activating enzyme, an E2-conjugating enzyme UBC9, and multiple E3 ligases. By modification of a large number of transcription factors and regulatory proteins, SUMO has a broad role in regulation of transcription by Pol II (19–22). Furthermore, we recently reported that SUMO has a novel role in regulating the global level of Pol II transcription via inhibiting transcription elongation through CDK9 SUMOylation (23). SUMO has also been shown to regulate the stability of rDNA repeats in Saccharomyces cerevisiae (24) and rRNA processing and ribosome maturation in metazoan (25–28). However, it is surprisingly not known whether SUMO also regulates Pol I transcription.
In this study we investigated whether and how SUMO regulates Pol I transcription in mammalian cells. We show that similar to its role in repressing global Pol II transcription, SUMO also represses Pol I transcription. Furthermore, we present evidence that SUMO represses Pol I transcription indirectly, primarily through control of UBF and c-Myc expression.
Results
SUMO represses Pol I transcription
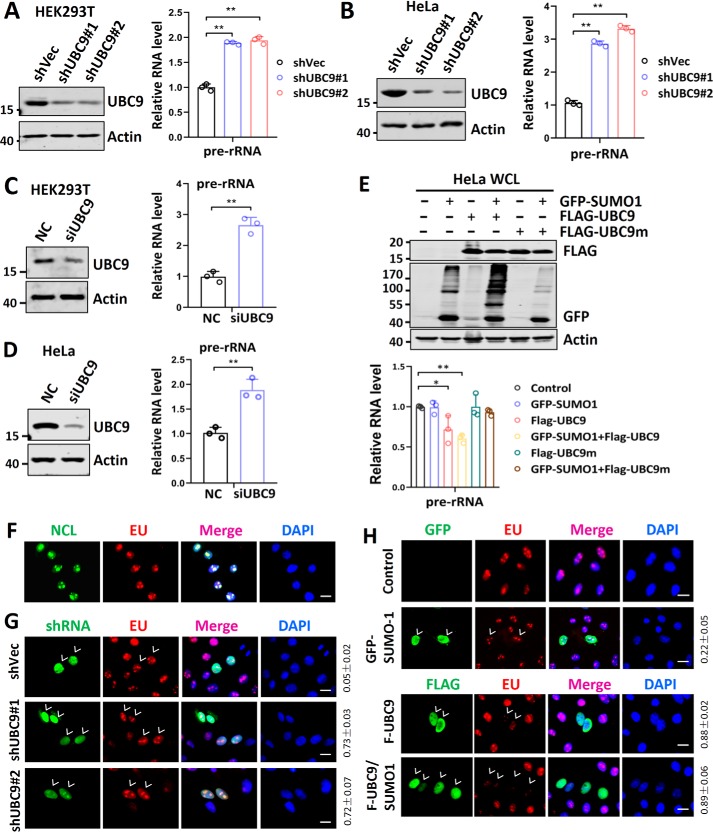
We recently showed that SUMO represses the global Pol II transcription by SUMOylation of CDK9 and consequently prevents the formation of P-TEFb complex (23) that is required for efficient transcriptional elongation of Pol II–transcribed genes. To determine whether SUMO also represses Pol I transcription, we knocked down UBC9, the sole SUMO E2-conjugating enzyme in HEK293T and HeLa cells by two distinct shRNAs (Fig. 1, A and B, left panel). Subsequent analysis of newly transcribed rDNA products, the 47S rRNA precursor, by quantitative RT-PCR (RT-qPCR) as reported (11, 12) showed that knockdown of UBC9 led to substantially increased levels of the rRNA precursor in both cell lines (Fig. 1, A and B, right panel). This observation was further confirmed by knockdown of UBC9 in these cells using a synthetic siRNA (Fig. 1, C and D). Thus, knockdown of UBC9 elevated the level of rDNA transcription, suggesting that the Pol I–mediated rDNA transcription is repressed by the endogenous SUMO system. In support of a repressive role of SUMO on Pol I transcription, we found that ectopic overexpression of FLAG-tagged SUMO E2 enzyme UBC9 inhibited rDNA transcription in HeLa cells, and this inhibition is further enhanced by co-expression of a GFP-tagged SUMO1 (Fig. 1E). However, no inhibition of rDNA transcription was observed when a SUMO E2-inactive UBC9 mutant (C93S) was expressed alone or together with GFP–SUMO1. Thus, UBC9 suppresses rDNA transcription in a SUMOylation-dependent manner.
Figure 1.
rDNA transcription is repressed by endogenous SUMO system. A and B, HEK293T cells (A) or HeLa cells (B) were transfected with two different shRNAs targeting UBC9, and Western blotting and RT-qPCR analyses were performed to detect the levels of UBC9 proteins (left panel) and the relative levels of 47S rRNA precursor (right panel), respectively. C and D, HEK293T cells (C) and HeLa cells (D) were treated with siRNA targeting UBC9, and Western blotting and RT-qPCR analyses were performed to detect the levels of UBC9 proteins (left panel) and the relative levels of 47S rRNA precursor (right panel), respectively. Note that the level of pre-rRNA in scramble control siRNA (NC)-transfected cells was set as 1. E, ectopic overexpression of UBC9 and SUMO1 repressed rDNA transcription. HeLa cells were transfected with or without FLAG–UBC9 and/or GFP–SUMO1 as indicated. Expression of UBC9 and SUMO1 was confirmed by Western blotting analysis (upper panel). The levels of rDNA transcription were determined by RT-qPCR. Note that Western blotting with anti-GFP antibody revealed that co-transfection of FLAG–UBC9 but not FLAG–UBC9m(C93S) promoted global SUMOylation. F, fluorescent imaging showing complete co-localization of EU staining with nucleolin. HeLa cells were pulse-labeled with ethyneluridine for 30 min and processed for detection of RNA-incorporated EU by click chemistry and nucleolin by immunostaining. Scale bar, 20 μm. G, the EU incorporation assay showing a significantly increased EU incorporation in shUBC9-transfected cells (marked by arrowheads) compared with untransfected control cells. HeLa cells were transfected with two different shRNAs against UBC9 as indicated. Two days after transfection, the cells were pulse-labeled with EU for 30 min and then processed for imaging of EU. Scale bar, 20 μm. H, EU incorporation assay showing the effect of ectopic overexpression of UBC9 and/or SUMO1 on rDNA transcription. Note that the EU incorporation was drastically repressed in UBC9 and UBC9 plus SUMO1-transfected HeLa cells (marked by arrowheads). Scale bar, 20 μm. All Western blots were performed at least three times, and the data were highly reproducible. The immunofluorescent staining experiments were also repeated at least three times, with the rate of transfected cells with increased or reduced levels of EU incorporation counted from all three or more representative experiments (means ± S.D.). DAPI, 4′,6′-diamino-2-phenylindole.
To further evaluate the effect of SUMO on rDNA transcription, we used an assay based on incorporation of the uridine analog 5-ethynyluridine (EU) into newly synthesized RNAs (29). Because rRNA synthesis accounts for ∼60% of total transcription and is restricted in the nucleolus, imaging detection of incorporated EU after a 30-min pulse labeling reaction using a click chemistry (29) revealed unique bright EU spots that showed a perfect co-localization with a nucleolus resident protein NCL (nucleolin), thus reflecting the levels of rDNA transcription (Fig. 1F). Using this assay, we found that knockdown of UBC9 led to increased size and intensity of EU foci (Fig. 1G). In contrast, ectopic overexpression of UBC9 markedly diminished the size and intensity of EU foci, and this effect was further enhanced by co-expression of GFP–SUMO1 (Fig. 1H). Although increased Pol II transcription activity may also partially contribute to increased EU incorporation in UBC9-knockdown cells, marked reduction of EU foci in FLAG–UBC9–expressing or FLAG–UBC9 plus GFP–SUMO1–expressing cells (Fig. 1H) provided clear evidence that SUMO plays a role in repression of rDNA transcription.
Multiple PIAS family E3 ligases are involved in repression of rDNA transcription
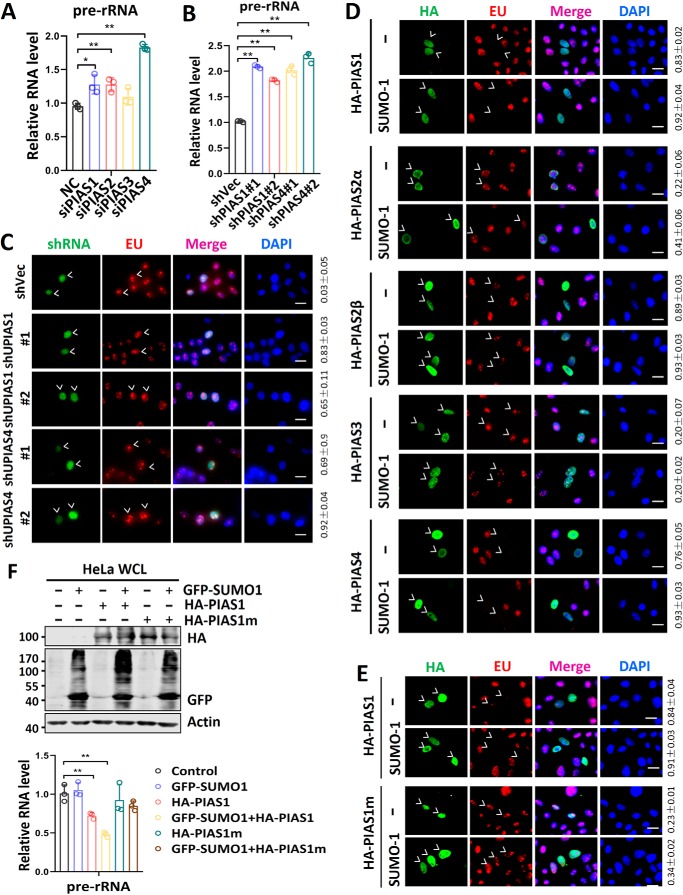
To further confirm a role of SUMO in repression of rDNA transcription, we tested whether the PIAS (protein inhibitor of activated STAT) family proteins, the major SUMO E3 ligases (30), are involved in this repression. We used smart pools of siRNAs against various PIAS mRNAs to treat HeLa cells for 2 days and measured the efficacy of knockdown by RT-qPCR analysis (Fig. S1A). Subsequent RT-qPCR analysis of 47S rRNA precursor showed that knockdown of PIAS1, PIAS2, and PIAS4 resulted in variously increased levels of rDNA transcription (Fig. 2A). To substantiate the siRNA-based observation, we designed two different shRNAs targeting PIAS1 and PIAS4, respectively. We validated by RT-qPCR analysis that transfection of HeLa cells with these shRNAs but not shVector (shVec) was able to down-regulate the levels of corresponding PIAS1 and PIAS4 mRNAs (Fig. S1B). Importantly, we found that knockdown of PIAS1 or PIAS4 by their specific shRNAs led to elevated levels of rDNA transcription in HeLa cells (Fig. 2B). Furthermore, by EU incorporation assay, we observed elevated levels of rDNA transcription in shPIAS1- or shPIAS4-transfected but not control shVect-transfected cells (Fig. 2C). Together, these loss-of-function assays suggest that multiple PIAS E3 ligases are likely involved in repression of rDNA genes by SUMO.
Figure 2.
Multiple PIAS family proteins are involved in repression of rDNA transcription. A, effect of knockdown of PIAS family proteins on rDNA transcription. HeLa cells were transfected with siRNA against each of the PIAS family proteins, and 2 days after transfection the relative levels of 47S rRNA precursor were measured by RT-qPCR. B, effect of knockdown of PIAS1 and PIAS4 by specific shRNAs on rDNA transcription. HeLa cells were transfected with either control shVec or two different PIAS1 or PIAS4-specific shRNAs. Two days after transfection, the relative levels of 47S rRNA precursor were measured by RT-qPCR. C, the EU incorporation assay showing that knockdown of PIAS1 or PIAS4 in HeLa cells all led to increased rRNA synthesis. The shRNA-transfected cells are marked by arrowheads. Also shown on the right are the rates of transfected cells with increased levels of EU incorporation counted from three or more representative experiments (means ± S.D.). D, the EU incorporation assay showing the effect of ectopic overexpression of various PIAS proteins with or without SUMO1 on rDNA transcription in HeLa cells. Transfected cells are marked by arrowheads. Note that PIAS1, PIAS2b, and PIAS4 were able to repress rDNA transcription when they were ectopically overexpressed. Also shown on the right are the rates of transfected cells with decreased levels of EU incorporation counted from three or more representative experiments (means ± S.D.). Scale bar, 20 μm. E, the EU incorporation assay showing that ectopically expressed PIAS1 repressed rDNA transcription in HeLa cells in an E3 ligase activity-dependent manner. Transfected cells are marked by arrowheads. Also shown on the right are the rates of transfected cells with decreased levels of EU incorporation counted from three or more representative experiments (means ± S.D.). Scale bar, 20 μm. F, RT-qPCR analyses showing that ectopically expressed PIAS1 repressed rDNA transcription in HeLa cells in an E3 activity-dependent manner, and the repression was augmented by co-expressed SUMO1. DAPI, 4′,6′-diamino-2-phenylindole.
In agreement with the loss-of-function results by siRNA/shRNA knockdown, we found that ectopic overexpression of PIAS1, PIAS2β, and PIAS4 repressed rDNA transcription as revealed by EU incorporation assay, whereas ectopic overexpression of PIAS2α and PIAS3 failed to do so (Fig. 2D). Repression of rDNA transcription by PIAS1 depended on its SUMO E3 ligase activity, because a mutant PIAS1 defective in E3 ligase activity failed to repress rDNA transcription in EU incorporation assay (Fig. 2E). Furthermore, we confirmed by RT-qPCR that ectopic overexpression of PIAS1, but not its E3 ligase mutant, repressed rDNA transcription in HeLa cells (Fig. 2F) and HEK293T cells (Fig. S1C), and this repression was further enhanced by co-expression of GFP–SUMO1. Taken together, both loss- and gain-of-function assays provide evidence that multiple PIAS family E3 ligases are involved in repression of rDNA transcription, and experiments with PIAS1 indicate that PIASs are likely to repress rDNA transcription in a SUMO E3 activity-dependent manner.
Repression of rDNA by SUMO correlates with reduced levels of UBF and C-Myc proteins
Thus far, we have demonstrated that the SUMO system represses rDNA transcription. To define the mechanisms underlying SUMO-mediated rDNA transcriptional repression, we first examined whether key transcription factors required for Pol I transcription are sumoylated. Intriguingly, our extensive effort failed to detect significant SUMOylation of endogenous NCL, UBF, c-Myc, RRN3, and TAF1B, a subunit of the SL1 complex, even under the conditions with ectopically overexpressed UBC9 plus SUMO1 (Fig. S2A) or PIAS1 plus SUMO1 (Fig. S2B), with which rDNA transcription was markedly suppressed (Fig. 1E and Fig. 2F and Fig. S1C). In addition, we also failed to detect SUMOylation of RPA194, the large subunit of RNA Pol I (Fig. S2, A and B). The failure in detecting SUMOylation on these proteins was unlikely caused by technical problems, because an increase in SUMOylation was detected for endogenous HDAC1 and HDAC2 under the same condition (Fig. S2A).
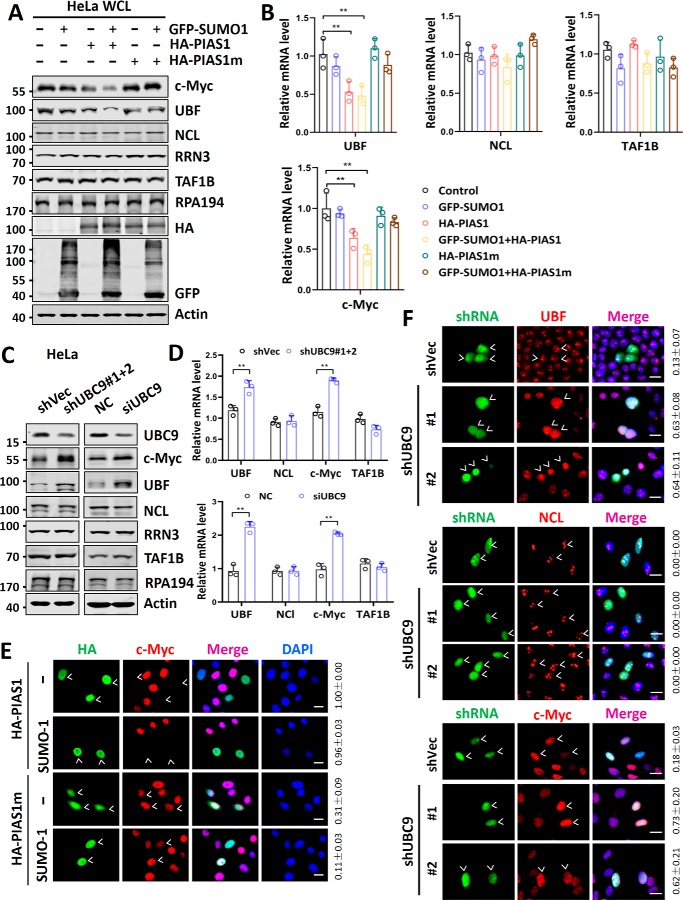
Although we could not detect SUMOylation on aforementioned Pol I regulatory proteins, we consistently observed a reduced level of UBF and c-Myc proteins upon overexpression of PIAS1 plus SUMO1 (Fig. 3A). Furthermore, the reduction of UBF and c-Myc proteins upon overexpression of PIAS1 plus SUMO1 depended on the E3 ligase activity of PIAS1 (Fig. 3A). On the other hand, ectopic overexpression of PIAS1 and SUMO1 did not significantly affect the levels of NCL, RRN3, TAF1B, and RPA194 proteins (Fig. 3A). Quantitative RT-PCR analysis revealed that ectopic overexpression of PIAS1 and SUMO1 also led to a substantial reduction of UBF and c-Myc mRNAs, but not that of NCL and TAF1B (Fig. 3B), suggesting that the reduction of UBF and c-Myc proteins upon overexpression of PIAS1 plus SUMO1 is likely due to repression of their transcription. Similarly, we found that ectopic overexpression of PIAS1 plus SUMO1 also reduced the levels of endogenous UBF and c-Myc proteins (Fig. S3A) and the levels of UBF and c-Myc mRNAs (Fig. S3B) in an E3 activity-dependent manner in HEK293T cells. Thus, repression of rDNA transcription by ectopically expressed UBC9 or PIAS1 plus SUMO1 correlates with repression of UBF and c-Myc expression. In contrast, we found that knockdown of UBC9 in HeLa cells led to an elevated level of UBF and c-Myc proteins (Fig. 3C) and elevated UBF and c-Myc mRNAs (Fig. 3D). Similarly, we found that knockdown of UBC9 in HEK293T cells by either shRNA or siRNA also led to an elevated level of UBF and c-Myc proteins (Fig. S3C). Thus, these data reveal a role of SUMO in repression of UBF and c-Myc expression.
Figure 3.
Repression of rDNA transcription by SUMO correlates with transcriptional repression of UBF and c-Myc. A, Western blotting analysis showing the effect of ectopically overexpressed PIAS1 and/or SUMO on a panel of Pol I core transcription factors, regulators, and subunit RPA194. B, RT-qPCR analyses showing the effect of ectopically overexpressed PIAS1 and/or SUMO on the relative levels of mRNAs encoding Pol I core transcription factors and regulators. C, Western blotting analyses showing the effect of knockdown of UBC9 in HeLa cells by using either shRNA or siRNA on a panel of Pol I core transcription factors, regulators, and subunit RPA194. D, RT-qPCR analyses showing the effect of knockdown of UBC9 on the relative levels of mRNAs encoding Pol I core transcription factors and regulators. The level of each mRNA in shVec-transfected cells was set as 1. E, immunofluorescent staining assay showing that ectopically expressed PIAS1 proteins down-regulated the levels of c-Myc proteins in an E3 activity-dependent manner in HeLa cells. Also shown on the right are the rates of transfected cells with decreased levels of c-Myc proteins based on three or more representative experiments (means ± S.D.). Scale bar, 20 μm. F, immunofluorescent staining assay showing elevated levels of UBF and c-Myc proteins, but not NCL, in shUBC9-transfected HeLa cells. The rates of shUBC9-transfected cells with increased levels of UBF, NCL, or c-Myc are shown on the right, based on three representative experiments. Scale bar, 20 μm. All Western blots were performed at least three times, and the data were highly reproducible. DAPI, 4′,6′-diamino-2-phenylindole; WCL, whole cell lysate.
We also used immunofluorescence assay to examine whether SUMO represses UBF and c-Myc expression. First, we observed that ectopic co-expression of PIAS1 and SUMO1, but not PIAS1m and SUMO1, substantially reduced the levels of c-Myc (Fig. 3E) and UBF proteins (Fig. S4A), but not NCL and RPA194 proteins in HeLa cells (Fig. S4B). In contrast, knockdown of UBC9 by shRNAs resulted in elevated levels of UBF and c-Myc proteins, but not NCL in shUBC9-transfected cells (Fig. 3F). Together, these data suggest that SUMO may repress rDNA transcription through its ability to repress UBF and c-Myc expression. In support of this, we found that UBF and c-Myc are both important for rDNA transcription, because knockdown of either UBF (Fig. S5A) or c-Myc (Fig. S5C) led to reduced rDNA transcription in HeLa cells (Fig. S5, B and D), consistent with previous reports that UBF and c-Myc are essential for rDNA transcription (13, 14, 31, 32).
Endogenous SUMO represses rDNA transcription indirectly and mainly through repression of UBF and c-Myc expression
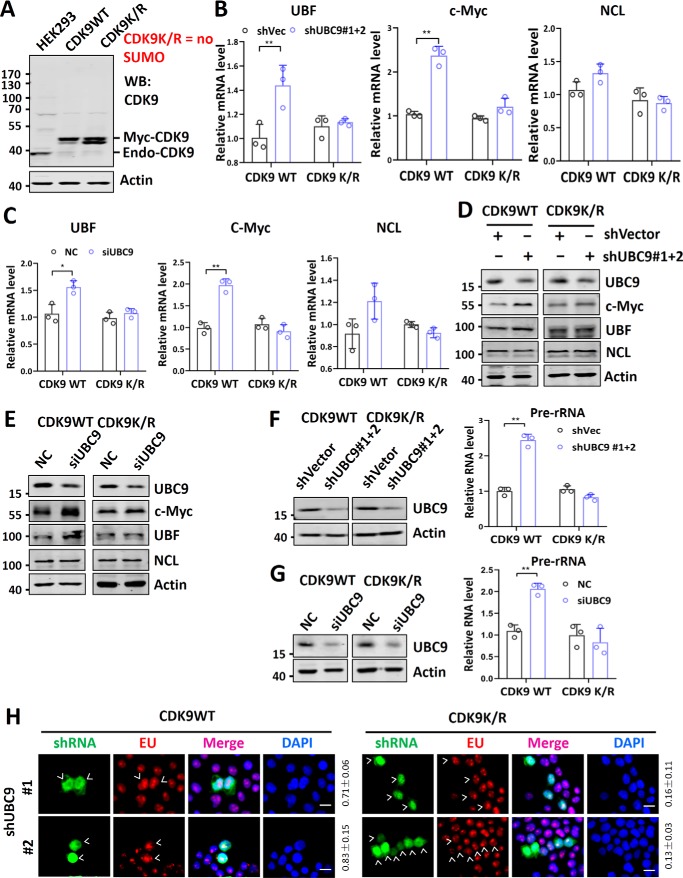
We recently showed that SUMO globally represses Pol II transcription through SUMOylation of CDK9 (23). SUMOylation of CDK9 blocks its interaction with cyclin T1/T2 and formation of a functionally active P-TEFb complex, thus controlling global Pol II transcriptional elongation and gene expression (23). Because repression of rDNA transcription correlates with reduced expression of UBF and c-Myc, we wished to test whether SUMO represses rDNA transcription through repression of UBF and c-Myc expression. To this end, we made use of two engineered HEK293T cell lines that were deleted of endogenous CDK9 genes by CRISPR-Cas9 and stably expressed either a Myc-tagged WT CDK9 (CDK9WT) or a SUMO-deficient CDK9 mutant (CDK9K/R) with all lysine residues mutated to arginine (Fig. 4A and Ref. 23). We showed previously that the CDK9K/R cell line was viable, whereas straight knockout of CDK9 in HEK293 cells was lethal (23). We found that although knockdown of UBC9 by either shRNA or siRNA in control CDK9WT cells led to elevated transcription of UBF and c-Myc, this was not observed in the CDK9K/R cell line (Fig. 4, B and C). We found by Western blotting analysis that although knockdown of UBC9 by either shRNA or siRNA led to elevated levels of UBF and c-Myc proteins, it failed to alter the levels of UBF and c-Myc proteins in the CDK9K/R cells (Fig. 4, D and E). We further confirmed by immunofluorescent staining assay that knockdown of UBC9 resulted in elevated levels of UBF and c-Myc in CDK9WT but not CDK9K/R cells (Fig. S6). Together, these data indicate that in the CDK9K/R cells the transcription of UBF and c-Myc genes and consequently their levels of proteins are not repressed by endogenous SUMO system.
Figure 4.
rDNA transcription is not repressed by endogenous SUMO in cells deficient in CDK9 SUMOylation. A, characterization of CDK9WT and CDK9K/R mutant cell lines. These HEK293T cell lines, with both endogenous CDK9 genes disrupted by CRISPR-Cas9, expressed either Myc-tagged WT CDK9 (CDK9WT) or a mutant CDK9 with all lysine residues converted to arginines (CDK9K/R). B, RT-qPCR analysis showing the effect of knockdown of UBC9 by shRNAs on the relative levels of UBF, c-Myc, and NCL mRNAs in both CDK9WT and CDK9K/R cells. Note that the level of mRNA for each protein in shVec-transfected cells was set as 1. C, RT-qPCR analysis showing the effect of knockdown of UBC9 by siRNA on the relative levels of UBF, c-Myc, and NCL mRNAs in both CDK9WT and CDK9K/R cells. Note that the level of mRNA for each protein in scramble control siRNA-transfected cells was set as 1. D, Western blotting analysis showing the effect of knockdown of UBC9 by shRNAs on the levels of c-Myc, UBF, and NCL proteins in both CDK9WT and CDK9K/R cells. E, Western blotting analyses showing the effect of knockdown of UBC9 by siRNA on the levels of c-Myc, UBF, and NCL in both CDK9WT and CDK9K/R cells. F, Western blotting and RT-qPCR analyses showing that knockdown of UBC9 by shRNAs led to elevated Pol I transcription in CDK9WT but not in CDK9K/R cells. Note that the level of pre-RNA in shVec-transfected cells was set as 1. G, Western blotting and RT-qPCR analyses showing that knockdown of UBC9 by siRNAs led to elevated Pol I transcription in CDK9WT but not in CDK9K/R cells. Note the level of pre-RNA in control scramble siRNA-transfected cells was set as 1. H, the EU incorporation assay showing that knockdown of UBC9 by transfected shRNAs elevated the level of rDNA transcription in CDK9WT but not in CDK9K/R cells. Transfected cells are marked by arrowheads. The rates of cells with increased EU incorporation versus counted transfected cells are shown on the right, based on three representative experiments. Scale bar, 20 μm. DAPI, 4′,6′-diamino-2-phenylindole.
We next examined whether rDNA transcription is repressed by SUMO in CDK9K/R cells. Importantly, we found that although knockdown of UBC9 by either shRNA or siRNA led to an elevated level of rDNA transcription in the CDK9WT cells, it failed to elevate rDNA transcription in the CDK9K/R cells (Fig. 4, F and G). By EU incorporation assay, we confirmed that knockdown of UBC9 led to an elevated level of rDNA transcription in CDK9WT but not in CDK9K/R cells (Fig. 4H). Together, these data indicate that the rDNA transcription in CDK9K/R cells is not repressed by endogenous SUMO, indicating that endogenous SUMO represses rDNA transcription primarily through its ability to repress transcription of UBF and c-Myc via CDK9 SUMOylation.
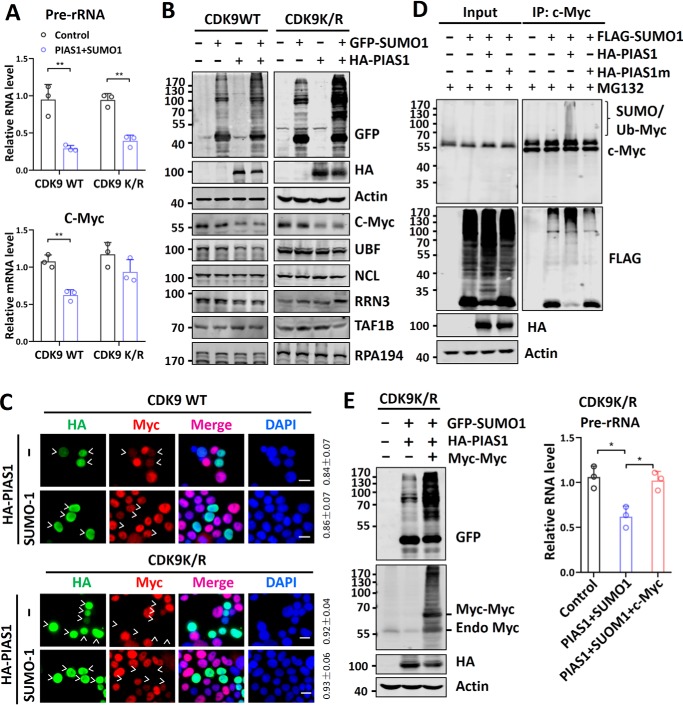
Ectopic overexpression of SUMO system components represses rDNA transcription through SUMOylation-induced c-Myc degradation
The above results suggest that endogenous SUMO represses rDNA transcription primarily in a CDK9 SUMOylation-dependent manner. To explore additional mechanisms that SUMO may repress rDNA transcription, we tested whether ectopic overexpression of the SUMO system is able to repress rDNA transcription in the CDK9K/R cells. RT-qPCR analysis revealed that overexpression of PIAS1 and SUMO1 was able to repress rDNA transcription in the CDK9K/R cells, albeit to a lesser extent compared with that in the CDK9WT cells (Fig. 5A, upper panel). We confirmed by EU incorporation assay that overexpression of PIAS1 and SUMO1 was able to repress rDNA transcription in both CDK9WT and CDK9K/R cells (Fig. S7A). However, overexpression of PIAS1 and SUMO1 did not significantly affect the transcription of c-Myc (Fig. 5A, lower panel) and UBF in the CDK9K/R cells (Fig. S7B), in agreement with their repression by SUMO being dependent on CDK9 SUMOylation.
Figure 5.
Ectopic overexpression of SUMO system components can repress rDNA transcription by down-regulation of c-Myc through SUMO-induced c-Myc degradation. A, RT-qPCR analysis showing that ectopic overexpression of PIAS1 and SUMO1 was able to repress rDNA transcription in both CDK9WT and CDK9K/R cells. Note that ectopic overexpression of PIAS1 and SUMO1 repressed c-Myc expression in CDK9WT but not in CDK9K/R cells. B, Western blotting analysis showing that ectopic overexpression of PIAS1 and SUMO1 down-regulated the levels of c-Myc proteins in both CDK9WT and CDK9K/R cells. Note that the levels of UBF proteins were down-regulated in CDK9WT but not in CDK9K/R cells. C, immunofluorescent staining assay showing that ectopic overexpression of PIAS1 and SUMO1 down-regulated the levels of c-Myc proteins in both CDK9WT and CDK9K/R cells. The rates of cells with reduced c-Myc versus counted transfected cells are shown on the right, based on analysis of three representative results. Scale bar, 20 μm. D, Western blotting analysis showing that addition of MG132 was able to block c-Myc down-regulation induced by ectopic co-expression of PIAS1 and SUMO1 and allowed the detection of sumoylated/ubiquitinated c-Myc proteins. E, ectopic overexpression of c-Myc abrogated PIAS1 and SUMO1-induced rDNA repression in CDK9K/R cells. The CDK9K/R cells were transfected with PIAS1/SUMO1, together with or without Myc-tagged c-Myc, as indicated. Two days after transfection, the cells were collected for Western blotting analysis (left panel) or RT-qPCR analysis of the 47S rRNA precursor (right panel). DAPI, 4′,6′-diamino-2-phenylindole; IP, immunoprecipitation.
To understand how overexpression of PIAS1 and SUMO1 repressed rDNA transcription in the CDK9K/R cells, we performed Western blotting analysis of Pol I regulatory proteins. We found that, similar to the CDK9WT cells, ectopic overexpression of PIAS1 plus SUMO1 in CDK9K/R cells resulted in down-regulation of c-Myc, whereas UBF was down-regulated only in the CDK9WT but not CDK9K/R cells (Fig. 5B). Consistent with the previous results, no change in NCL, RRN3, TAFIB, and RPA194 was observed (Fig. 5B). We confirmed by immunostaining assay that ectopic overexpression of PIAS1 and SUMO led to a reduced level of c-Myc proteins not only in the CDK9WT cells but also in CDK9K/R cells (Fig. 5C), whereas down-regulation of UBF was only observed in the CDK9WT cells (Fig. S6C). Thus, ectopic overexpression of SUMO system components is able to repress rDNA transcription in the CDK9 SUMOylation-resistant cells, possibly through down-regulation of c-Myc proteins.
It was reported that SUMOylation of c-Myc can lead to subsequent ubiquitination and proteasome degradation (33, 34). We detected c-Myc SUMOylation when HeLa cells were overexpressed with PIAS1 and SUMO1, and ubiquitin-dependent protein degradation was blocked by MG132 (Fig. 5D). Thus, consistent with previous studies (33, 34), ectopically expressed SUMO is likely to reduce the level of c-Myc proteins through SUMOylation-induced, ubiquitin-dependent degradation. In support of this, we found that ectopic overexpression of PIAS1 and SUMO1 reduced the half-life of c-Myc proteins in CDK9K/R cells (Fig. S8).
To test whether reduced c-Myc proteins were responsible for reduced rDNA transcription in the CDK9K/R cells, we tested whether ectopic overexpression of c-Myc could relieve PIAS1 and SUMO1-induced rDNA repression. We observed that ectopic overexpression of Myc-tagged c-Myc proteins (Fig. 5E, left panel) was able to abrogate rDNA repression instigated by ectopically overexpressed PIAS1 and SUMO1 in the CDK9K/R cells (Fig. 5E, right panel). Thus, ectopic overexpression of the SUMO system components can additionally repress rDNA transcription through a SUMOylation-mediated degradation of c-Myc proteins.
Discussion
In this study we unravel a critical role for SUMO in regulation of rDNA transcription. Knockdown of SUMO E2 enzyme UBC9 or E3 PIAS proteins in various cells significantly elevated rDNA transcription, whereas ectopic overexpression of SUMO system markedly repressed rDNA transcription. We present evidence that the endogenous SUMO system represses rDNA transcription primarily through regulating the expression of transcription factors UBF and c-Myc, whereas ectopic overexpression of the SUMO system components can additionally repress rDNA transcription via SUMOylation-induced c-Myc degradation.
In principle, SUMO is a dynamic modification and regulates substrate proteins through its effect on protein–protein interaction, subcellular localization, activity, and stability (17, 18). Because transcription factors and co-regulators are among the most frequently identified and characterized SUMO substrates, it is not surprising that SUMO has been shown to play a broad role in regulation of transcription by Pol II (20, 21, 35, 36). Interestingly, SUMO in general is involved in transcriptional repression, and multiple mechanisms have been proposed to explain the roles of SUMOylation in transcriptional repression (21, 37). We recently uncovered a novel role of SUMO in repression of the global level of transcription via control of P-TEFb complex formation by SUMOylation of CDK9 (23). This finding provides a novel mechanism for transcriptional repression by SUMO. In contrast to the well-recognized role of SUMO in regulation of Pol II transcription and to the best of our knowledge, it is surprisingly not known prior to this study whether SUMO regulates rDNA transcription by Pol I. Because knockdown of UBC9 or multiple PIAS proteins all led to increased transcription of rDNA genes (Figs. 1 and 2), SUMO has a role in repression of Pol I transcription. In this regard, the EU incorporation assay, although not entirely quantitative, provides a sensitive measurement for the levels of newly synthesized rRNAs in nucleoli. Because rDNA transcription accounts for up to 60% RNA synthesis and is restricted to nucleoli compartments, our findings that knockdown or overexpression of SUMO system components led to elevated or diminished EU foci staining in nucleoli (Figs. 1 and 2) provide compelling evidence for repression of rDNA transcription by SUMO.
In search of the potential mechanisms by which SUMO represses rDNA transcription, we initially focused our attention on SUMOylation of key transcription factors required for rDNA transcription. However, no significant SUMOylation of endogenous UBF, NCL, RRN3, TAF1B, and c-Myc was observed under the regular culture condition, despite the previous reports on SUMOylation of nucleolin (38) and c-Myc proteins (33), both required for efficient rDNA transcription (13, 14, 39). Even with overexpression of UBC9 or PIAS1 and SUMO1, we could not observe significant SUMOylation on these proteins (Fig. S1). Instead, we observed elevated or reduced levels of UBF and c-Myc proteins and mRNAs upon UBC9 knockdown or overexpression of the SUMO system components, respectively (Figs. 1–3). These observations point to an alternative mechanism that SUMO represses rDNA transcription indirectly through regulation of UBF and c-Myc transcription. In support of this idea, we find that rDNA transcription is no longer repressed by the endogenous SUMO system in the CDK9K/R cell line in which SUMO does not repress UBF and c-Myc transcription (Fig. 4). Thus, endogenous SUMO most likely represses rDNA transcription through repression of UBF and c-Myc transcription, although we could not exclude the possibility that SUMOylation of the Pol I regulatory proteins may also partially contribute to repression of rDNA transcription.
Although our study suggests that endogenous SUMO represses rDNA transcription via repression of UBF and c-Myc expression, we show that ectopic overexpression of the SUMO system components can repress rDNA transcription through SUMOylation-induced degradation of c-Myc proteins (Fig. 5), in agreement with previous studies (33, 34). Indeed, c-Myc SUMOylation can be detected upon overexpression of PIAS1 and SUMO1 and in the presence of the proteasome inhibitor MG132 (Fig. 5D).
In sum, our study demonstrates that, similar to transcription by Pol II, rDNA transcription by Pol I is also repressed by SUMO. We uncover two mechanisms for SUMO-mediated transcriptional repression of rDNA genes, an indirect one by repression of UBF and c-Myc transcription via CDK9 SUMOylation that appears to be the major repression force by the endogenous SUMO system and the other one by SUMO-mediated c-Myc degradation that may dominate under an overexpression condition. However, these two mechanisms are not mutually exclusive and may act together to control rDNA transcription and cell proliferation. Furthermore, both mechanisms converge on the control of c-Myc protein levels, in good agreement with c-Myc as a master regulator of rDNA transcription and cell proliferation. Given the critical role of c-Myc in regulating global as well as specific programs of Pol II transcription (15, 40, 41), our study suggests that SUMO can coordinate Pol I and Pol II transcription by controlling c-Myc expression. In light of reported regulation of Pol III transcription by SUMO (42, 43), it is tempting to propose that SUMO has a role in coordinating transcription by all three RNA polymerases.
Experimental procedures
Cell lines, plasmids, and antibodies
HeLa and HEK293T cells were cultured in Dulbecco's modified Eagle's Medium (Gibco, Thermo Fisher) with 10% fetal bovine serum (Gibco) in a humidified incubator at 37 °C with 5% CO2. Plasmids encoding UBC9, UBC9m (C93S), SUMO1, CDK9, and CDK9K/R mutant were constructed as previously described (23). The CDK9 knockout cell lines with expression of Myc-CDK9 or Myc-CDK9K/R mutant were generated as described (23). The plasmids for pCDNA3.0-HA-PIAS1m(C346SC351SC356S) was derived from pCDNA3.0-HA-PIAS1 by site-directed mutagenesis. Various PIAS expression constructs were generated by subcloning the corresponding coding sequences into the pCDNA3.0-HA vector. The antibodies used in this study were listed as follows: rabbit anti-UBC9 (CST catalog no. 4918, dilution 1:5000 for WB), rabbit anti–c-Myc (Abcam catalog no. ab32072, dilution 1:5000 for WB and 1:1000 for IF, 0.5 μg for immunoprecipitation), mouse anti-UBF (Santa Cruz catalog no. sc-13125, dilution 1:1000 for WB and 1:100 for IF), rabbit anti-NCL (homemade polyclonal antibody, dilution 1:10000 for WB and 1:2000 for IF), anti-RRN3 (Abcam catalog no. ab112052, dilution 1:5000 for WB), rabbit anti-TAF1B (Absci catalog no. AB44736, dilution 1:1000 for WB), mouse anti-RPA194 (Santa Cruz catalog no. sc-48385, dilution 1:1000 for WB and 1:100 for IF), rabbit anti-CDK9 (Abclonal catalog no. A1564 dilution 1:1000 for WB), rabbit anti-HDAC1 (Abclonal catalog no. A0238, dilution 1:5000 for WB), rabbit anti-HDAC2 (Abclonal catalog no. A2084, dilution 1:5000 for WB), mouse anti-HA (Abmart catalog no. M2003, dilution 1:5000 for WB and 1:1000 for IF), rabbit anti-HA (Santa Cruz catalog no. sc-805 dilution 1:1000 for WB and 1:200 for IF), mouse anti-GFP (Abmart catalog no. M2004L dilution 1:5000 for WB and 1:1000 for IF), mouse anti-FLAG (Sigma catalog no. F7425 dilution 1:5000 for WB and 1:1000 for IF), and mouse anti–β-actin (Sigma catalog no. A5316, dilution 1:5000 for WB).
Transfections, Western blotting analysis, immunoprecipitation, and immunofluorescence staining
DNA and siRNA transient transfection were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Western blotting, immunoprecipitation, and immunofluorescence staining were performed essentially as described (23), using the antibodies as indicated. Images were obtained at 40× magnification using Olympus IX73P2F microscope and processed using ImageJ. Images in Figs. 1F and 2 (B and C) were taken with a Leica DM4000BLED microscope and processed using ImageJ.
Knockdown with siRNAs or shRNAs and quantitative RT-PCR
The sequence information for siRNAs targeting human UBC9, PIAS1, PIAS2, PIAS3, and PIAS4 were provided in Table S1. All siRNAs were synthesized by Genepharma. The sequences for shRNA targeting human UBC9, c-Myc, and UBF are also listed in Table S1. The vector for shRNAs was pLKO.1. For RT-qPCR analysis, an equal number of cells for each sample was collected, and preparation of total RNA was carried out using the RNAiso Plus kit (Takara catalog no. D9108A). All cDNAs were prepared using the TransScript one-step gDNA removal and cDNA synthesis SuperMix (TransGen Biotech catalog no. AT311) according to the manufacturer's instructions. Quantitative PCR analysis was performed using CFX96 touch real-time PCR detection system (Bio-Rad). The following cycle conditions were applied for PCR analysis: initial denaturation at 94 °C for 10 min, followed by 40 cycles at 94 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s. The ΔCt obtained was used to find the relative expression of genes according to the formula: relative expression n = 2−ΔΔCt, where ΔΔCt = (ΔCt of respective genes in experimental groups) − (ΔCt of the same genes in control group). GAPDH was used as an internal control. GraphPad Prism 8 software was used for plotting. The primer sequences used in the RT-qPCR are listed in Table S1.
EU labeling assay
The EU incorporation assay was carried out essentially as described (29). Briefly, 48 h after initial transfection, EU was added to the complete culture medium at a final concentration of 1 mm, and cells were incubated at 37 °C for 30 min. After EU labeling, the cells were washed with PBS and fixed in 125 mm Pipes, pH 6.8, 10 mm EGTA, 1 mm magnesium chloride, 0.2% Triton X-100, and 3.7% formaldehyde for 30 min at room temperature. The cells were then washed with TBS and stained with 10 μm Alexa 594–azide.
Data analysis
Statistical analyses were performed using IBM SPSS statistics 23. The data are presented as mean of two or three independent experiments. Statistical relevance was determined using the unpaired Student's test for RT-qPCR. The error bars represent ± S.D. Differences of p < 0.05 were considered significant. *, 0.01 < p < 0.05; **, p < 0.01.
Author contributions
Y. P., Zhiqiang Wang, and F. Y. data curation; Y. P., Zhenxing Wang, Zhiqiang Wang, and F. Y. investigation; Y. P. writing-original draft; Zhenxing Wang validation; F. Y. methodology; J. L. and J. W. supervision; J. L. and J. W. project administration; J. W. conceptualization; J. W. funding acquisition; J. W. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Cheng-Ming Chiang (University of Texas Southwestern Medical Center) for critical reading of the manuscript. We also thank members of Wong's lab for valuable discussion.
This work is supported by Ministry of Science and Technology of China Grant 2017YFA054201 (to J. W.) and National Natural Science Foundation of China Grants 31730048, 81530078, and 31571325 (to J. W.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S8.
- rDNA
- ribosomal DNA
- Pol
- polymerase
- SUMO
- small ubiquitin-like modifier
- CDK
- cyclin-dependent kinase
- UBF
- upstream-binding factor
- SL1
- selectivity factor 1
- RT-qPCR
- quantitative RT-PCR
- EU
- 5-ethynyluridine
- WB
- Western blotting
- IF
- immunofluorescence
- HA
- hemagglutinin
- shVec
- shVector.
References
- 1. Ide S., Miyazaki T., Maki H., and Kobayashi T. (2010) Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327, 693–696 10.1126/science.1179044 [DOI] [PubMed] [Google Scholar]
- 2. Moss T., Langlois F., Gagnon-Kugler T., and Stefanovsky V. (2007) A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 64, 29–49 10.1007/s00018-006-6278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schneider D. A. (2012) RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene 493, 176–184 10.1016/j.gene.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drygin D., Rice W. G., and Grummt I. (2010) The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 50, 131–156 10.1146/annurev.pharmtox.010909.105844 [DOI] [PubMed] [Google Scholar]
- 5. Goodfellow S. J., and Zomerdijk J. C. (2013) Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell. Biochem. 61, 211–236 10.1007/978-94-007-4525-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gagnon-Kugler T., Langlois F., Stefanovsky V., Lessard F., and Moss T. (2009) Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell 35, 414–425 10.1016/j.molcel.2009.07.008 [DOI] [PubMed] [Google Scholar]
- 7. Grummt I., and Längst G. (2013) Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim. Biophys. Acta 1829, 393–404 10.1016/j.bbagrm.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 8. Quin J. E., Devlin J. R., Cameron D., Hannan K. M., Pearson R. B., and Hannan R. D. (2014) Targeting the nucleolus for cancer intervention. Biochim. Biophys. Acta 1842, 802–816 10.1016/j.bbadis.2013.12.009 [DOI] [PubMed] [Google Scholar]
- 9. Kusnadi E. P., Hannan K. M., Hicks R. J., Hannan R. D., Pearson R. B., and Kang J. (2015) Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene 556, 27–34 10.1016/j.gene.2014.11.010 [DOI] [PubMed] [Google Scholar]
- 10. Diesch J., Hannan R. D., and Sanij E. (2014) Perturbations at the ribosomal genes loci are at the centre of cellular dysfunction and human disease. Cell Biosci. 4, 43 10.1186/2045-3701-4-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cong R., Das S., Douet J., Wong J., Buschbeck M., Mongelard F., and Bouvet P. (2014) macroH2A1 histone variant represses rDNA transcription. Nucleic Acids Res. 42, 181–192 10.1093/nar/gkt863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi G., Wu M., Fang L., Yu F., Cheng S., Li J., Du J. X., and Wong J. (2014) PHD finger protein 2 (PHF2) represses ribosomal RNA gene transcription by antagonizing PHF finger protein 8 (PHF8) and recruiting methyltransferase SUV39H1. J. Biol. Chem. 289, 29691–29700 10.1074/jbc.M114.571653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arabi A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., Larsson L. G., and Wright A. P. (2005) c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7, 303–310 10.1038/ncb1225 [DOI] [PubMed] [Google Scholar]
- 14. Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., and White R. J. (2005) c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell. Biol. 7, 311–318 10.1038/ncb1224 [DOI] [PubMed] [Google Scholar]
- 15. Poortinga G., Wall M., Sanij E., Siwicki K., Ellul J., Brown D., Holloway T. P., Hannan R. D., and McArthur G. A. (2011) c-Myc coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 39, 3267–3281 10.1093/nar/gkq1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poortinga G., Hannan K. M., Snelling H., Walkley C. R., Jenkins A., Sharkey K., Wall M., Brandenburger Y., Palatsides M., Pearson R. B., McArthur G. A., and Hannan R. D. (2004) MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 23, 3325–3335 10.1038/sj.emboj.7600335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hay R. T. (2013) Decoding the SUMO signal. Biochem. Soc. Trans. 41, 463–473 10.1042/BST20130015 [DOI] [PubMed] [Google Scholar]
- 18. Gareau J. R., and Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ouyang J., and Gill G. (2009) SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics 4, 440–444 10.4161/epi.4.7.9807 [DOI] [PubMed] [Google Scholar]
- 20. Hendriks I. A., and Vertegaal A. C. (2016) A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595 10.1038/nrm.2016.81 [DOI] [PubMed] [Google Scholar]
- 21. Chymkowitch P., Nguéa P. A., and Enserink J. M. (2015) SUMO-regulated transcription: challenging the dogma. BioEssays 37, 1095–1105 10.1002/bies.201500065 [DOI] [PubMed] [Google Scholar]
- 22. Srikumar T., Lewicki M. C., Costanzo M., Tkach J. M., van Bakel H., Tsui K., Johnson E. S., Brown G. W., Andrews B. J., Boone C., Giaever G., Nislow C., and Raught B. (2013) Global analysis of SUMO chain function reveals multiple roles in chromatin regulation. J. Cell Biol. 201, 145–163 10.1083/jcb.201210019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu F., Shi G., Cheng S., Chen J., Wu S. Y., Wang Z., Xia N., Zhai Y., Wang Z., Peng Y., Wang D., Du J. X., Liao L., Duan S. Z., Shi T., et al. (2018) SUMO suppresses and MYC amplifies transcription globally by regulating CDK9 SUMOylation. Cell Res. 28, 670–685 10.1038/s41422-018-0023-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., Reid R., Jentsch S., Rothstein R., Aragón L., and Lisby M. (2007) The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9, 923–931 10.1038/ncb1619 [DOI] [PubMed] [Google Scholar]
- 25. Finkbeiner E., Haindl M., and Muller S. (2011) The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 30, 1067–1078 10.1038/emboj.2011.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Finkbeiner E., Haindl M., Raman N., and Muller S. (2011) SUMO routes ribosome maturation. Nucleus 2, 527–532 10.4161/nucl.2.6.17604 [DOI] [PubMed] [Google Scholar]
- 27. Liang J., Singh N., Carlson C. R., Albuquerque C. P., Corbett K. D., and Zhou H. (2017) Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity. Genes Dev. 31, 802–815 10.1101/gad.296145.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haindl M., Harasim T., Eick D., and Muller S. (2008) The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 9, 273–279 10.1038/embor.2008.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jao C. Y., and Salic A. (2008) Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 105, 15779–15784 10.1073/pnas.0808480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt D., and Müller S. (2003) PIAS/SUMO: new partners in transcriptional regulation. Cell. Mol. Life Sci. 60, 2561–2574 10.1007/s00018-003-3129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Learned R. M., Learned T. K., Haltiner M. M., and Tjian R. T. (1986) Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell 45, 847–857 10.1016/0092-8674(86)90559-3 [DOI] [PubMed] [Google Scholar]
- 32. Stefanovsky V. Y., Pelletier G., Hannan R., Gagnon-Kugler T., Rothblum L. I., and Moss T. (2001) An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8, 1063–1073 10.1016/S1097-2765(01)00384-7 [DOI] [PubMed] [Google Scholar]
- 33. González-Prieto R., Cuijpers S. A., Kumar R., Hendriks I. A., and Vertegaal A. C. (2015) c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 14, 1859–1872 10.1080/15384101.2015.1040965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun X. X., Chen Y., Su Y., Wang X., Chauhan K. M., Liang J., Daniel C. J., Sears R. C., and Dai M. S. (2018) SUMO protease SENP1 deSUMOylates and stabilizes c-Myc. Proc. Natl. Acad. Sci. U.S.A. 115, 10983–10988 10.1073/pnas.1802932115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang X. J., and Chiang C. M. (2013) SUMOylation in gene regulation, human disease, and therapeutic action. F1000prime Rep. 5, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raman N., Nayak A., and Muller S. (2013) The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma 122, 475–485 10.1007/s00412-013-0429-6 [DOI] [PubMed] [Google Scholar]
- 37. Gill G. (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18, 2046–2059 10.1101/gad.1214604 [DOI] [PubMed] [Google Scholar]
- 38. Zhang D., Liang Y., Xie Q., Gao G., Wei J., Huang H., Li J., Gao J., and Huang C. (2015) A novel post-translational modification of nucleolin, SUMOylation at Lys-294, mediates arsenite-induced cell death by regulating gadd45alpha mRNA stability. J. Biol. Chem. 290, 4784–4800 10.1074/jbc.M114.598219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cong R., Das S., Ugrinova I., Kumar S., Mongelard F., Wong J., and Bouvet P. (2012) Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 40, 9441–9454 10.1093/nar/gks720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D. R., Tessarollo L., Casellas R., Zhao K., and Levens D. (2012) c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 10.1016/j.cell.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sabo A., Kress T. R., Pelizzola M., de Pretis S., Gorski M. M., Tesi A., Morelli M. J., Bora P., Doni M., Verrecchia A., Tonelli C., Fagà G., Bianchi V., Ronchi A., Low D., et al. (2014) Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 10.1038/nature13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rohira A. D., Chen C. Y., Allen J. R., and Johnson D. L. (2013) Covalent small ubiquitin-like modifier (SUMO) modification of Maf1 protein controls RNA polymerase III-dependent transcription repression. J. Biol. Chem. 288, 19288–19295 10.1074/jbc.M113.473744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chymkowitch P., Nguéa P. A., Aanes H., Robertson J., Klungland A., and Enserink J. M. (2017) TORC1-dependent SUMOylation of Rpc82 promotes RNA polymerase III assembly and activity. Proc. Natl. Acad. Sci. U.S.A. 114, 1039–1044 10.1073/pnas.1615093114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.