Abstract
The lipid aldehyde 4-oxo-2-nonenal (ONE) is a highly reactive protein crosslinker derived from peroxidation of n-6 polyunsaturated fatty acids and generated together with 4-hydroxynonenal (HNE). Lipid peroxidation product-mediated crosslinking of proteins in high-density lipoprotein (HDL) causes HDL dysfunction and contributes to atherogenesis. Although HNE is relatively well-studied, the role of ONE in atherosclerosis and in modifying HDL is unknown. Here, we found that individuals with familial hypercholesterolemia (FH) had significantly higher ONE-ketoamide (lysine) adducts in HDL (54.6 ± 33.8 pmol/mg) than healthy controls (15.3 ± 5.6 pmol/mg). ONE crosslinked apolipoprotein A-I (apoA-I) on HDL at a concentration of > 3 mol ONE per 10 mol apoA-I (0.3 eq), which was 100-fold lower than HNE, but comparable to the potent protein crosslinker isolevuglandin. ONE-modified HDL partially inhibited HDL's ability to protect against lipopolysaccharide (LPS)-induced tumor necrosis factor α (TNFα) and interleukin-1β (IL-1β) gene expression in murine macrophages. At 3 eq, ONE dramatically decreased apoA-I exchange from HDL, from ∼46.5 to ∼18.4% (p < 0.001). Surprisingly, ONE modification of HDL or apoA-I did not alter macrophage cholesterol efflux capacity. LC-MS/MS analysis revealed that Lys-12, Lys-23, Lys-96, and Lys-226 in apoA-I are modified by ONE ketoamide adducts. Compared with other dicarbonyl scavengers, pentylpyridoxamine (PPM) most efficaciously blocked ONE-induced protein crosslinking in HDL and also prevented HDL dysfunction in an in vitro model of inflammation. Our findings show that ONE-HDL adducts cause HDL dysfunction and are elevated in individuals with FH who have severe hypercholesterolemia.
Keywords: apolipoprotein, high-density lipoprotein (HDL), post-translational modification (PTM), lipid peroxidation, atherosclerosis, oxidative stress, mass spectrometry (MS), cardiovascular disease, apolipoprotein A-I, arterial plaque, hypercholesterolemia, metabolic disorder, reactive carbonyls, cardiovascular dysfunction
Introduction
Oxidative stress and increased net production of free radicals represent an important pathogenic mechanism in atherosclerosis. Free radicals react with unsaturated fatty acids in a chain reaction to yield lipid peroxides and secondary aldehyde products. These lipid aldehydes are highly reactive and selectively modify proteins or lipids to cause cellular and tissue damage. An important role for reactive aldehydes in the pathogenesis of atherosclerosis is suggested by increased aldehyde-protein adducts in plasma (1, 2), aortic atherosclerotic lesions (3–6), and lipoproteins. Adduction to apolipoprotein B in low-density lipoprotein (LDL)2 enhances their recognition and uptake by macrophages (7, 8) which converts macrophages to lipid-laden foam cells (9–11). HDL normally protects LDL from aldehyde adduction by serving as a “sink” for lipid peroxides and their reactive by-products (12), but when HDL becomes modified, it results in numerous dysfunctions of HDL (13, 14).
HNE is one of the most investigated aldehydic end products of oxidative breakdown of membrane n-6 polyunsaturated fatty acids. Its adduction to LDL accelerates its uptake by macrophages (15), and its adduction to HDL causes crosslinks of apoA-I and inhibits its ability to activate lecithin-cholesterol acyltransferase (LCAT) (14). However, the role of its more reactive 4-keto counterpart ONE in atherogenesis is not well-studied. Studies characterizing the side-chain modifying chemistry of ONE showed that ONE is a more reactive protein modifier and crosslinking agent than HNE (16–19). In this way, ONE is very similar to the isolevuglandins (IsoLGs), a family of 4-ketoaldehydes generated by peroxidation of arachidonic acid that potently crosslink HDL proteins (20). ONE is generated in parallel to HNE through the intermediate 4-hydroperoxy-2-nonenal (HPNE) and is a direct product of lipid oxidation (21, 22) (Fig. 1). Like HNE, ONE reacts rapidly with the side chains of Cys, His, and Lys residues in proteins via Michael addition, but unlike HNE, ONE also reacts with Arg. Kinetic experiments reveal that the reactivity of ONE toward amino acids of proteins occurs in the following order: Cys ≫ His > Lys > Arg (26). Although both HNE and ONE form Schiff bases with Lys, only ONE is capable of forming the 4-ketoamide adduct (24, 31).
Figure 1.
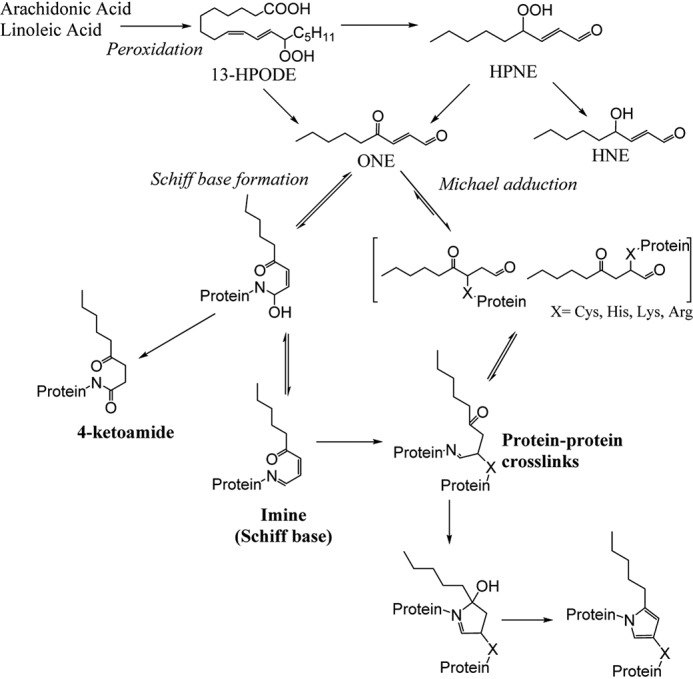
Formation of ONE from the peroxidation of arachidonic or linoleic acid and its adduction to proteins.
In vivo, HNE- and HPNE-specific epitopes exist in atherosclerotic plaques (23, 24). Studies using carnosine as an aldehyde scavenger show the presence of carnosine-ONE in oxidized LDL (25). However, no other studies to our knowledge have examined the biological consequences of ONE modification of lipoproteins in atherosclerosis. In this study, we utilized LC-MS/MS to measure ONE-lysine adducts in HDL derived from patients with familial hypercholesterolemia compared with control healthy age-matched subjects. We also identified the amino acid sites of apoA-I that ONE targets and determined the consequences of ONE modification on HDL function. We demonstrate for the first time that ONE-HDL adducts are elevated in atherosclerosis. Interestingly, ONE causes HDL dysfunction in terms of rendering HDL unable to protect against LPS-induced macrophage activation but does not alter its ability to promote macrophage cholesterol efflux.
Results
ONE-lysine adducts in human HDL are elevated in atherosclerosis
Levels of ONE protein adducts in HDL were determined in patients with familial hypercholesterolemia (FH) (n = 8) compared with healthy control volunteers (n = 9). Because ONE-induced crosslinking generates multiple chemical structures depending on the microenvironment (26–28), we focused instead on quantifying the lysine 4-ketoamide adduct. These monoadducts are irreversible, highly stable, and longer-lived (26). By LC-MS/MS, we found that the levels of ONE-ketoamide adducts were significantly higher (p = 0.013, unpaired t test with Welch's correction) in individuals diagnosed with familial hypercholesterolemia (54.6 ± 33.8 pmol/mg protein, mean ± S.E.) than in healthy controls (15.3 ± 5.6 pmol/mg protein) (Fig. 2A). Six of the eight study subjects were diagnosed with heterozygous FH and two were diagnosed with homozygous FH using the Dutch and the Simon Broome criteria for FH. Four subjects were genotyped. Detailed descriptions of individual demographics, disease scores, and LDL-C levels are found in Table S1. Of interest, the levels of ONE-ketoamide adducts in these samples were greater than the levels of IsoLG-lysine adducts measured in these same samples (15.6 ± 4.2, FH; 0.8 ± 0.1 pmol/mg protein, control; mean ± S.E.) (Fig. 2B). These results are first to demonstrate that ONE-protein adducts are elevated on human HDL in conditions of severe hypercholesterolemia associated with atherosclerosis.
Figure 2.
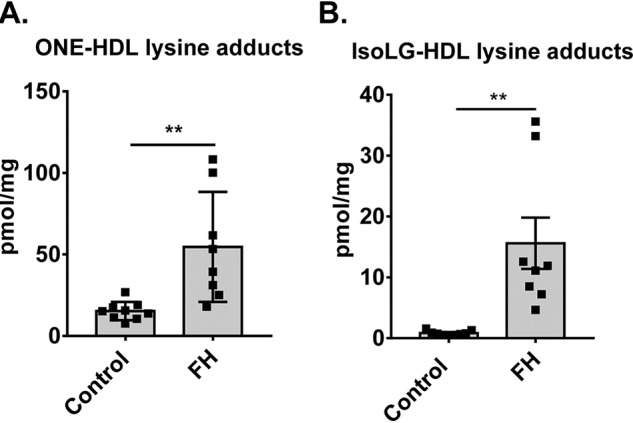
ONE-protein adducts are increased in HDL isolated from plasma of patients with FH and are higher than IsoLG-protein adducts measured from the same samples. Plasma was isolated from the blood of FH patients (n = 8) and healthy volunteers (n = 9) and subjected to DGUC to isolate the HDL. A and B, levels of (A) ONE-lysine ketoamide and (B) IsoLG-lysine adducts were determined by LC-MS/MS. Values are shown as mean ± S.D. Statistical significance was calculated by Student's t test. **, p < 0.01.
ONE potently crosslinks HDL proteins but does not significantly alter HDL size distribution
We previously showed that IsoLGs crosslink HDL proteins even when added at only 0.3 molar equivalents (per mole of apoA-I), and that this level of modification by IsoLG is sufficient to reduce HDL's ability to inhibit LPS-stimulated inflammatory cytokine release (20). Given that ONE-Lys levels in FH HDL were greater than those of IsoLG-Lys levels, we performed studies to determine whether ONE could also crosslink HDL at similar concentrations as IsoLG. Incubation of ONE with HDL induced crosslinking of HDL proteins starting at only ∼0.1 eq (Fig. 3A), as indicated by SDS-PAGE and Coomassie Blue staining to visualize the proteins. Structural proteins apoA-I and apoA-II are crosslinked beginning at 0.1 eq (Fig. 3, B and C). This concentration is 10-fold lower than required for HNE (20). At 10 eq, the monomers of apoA-I and apoA-II begin to disappear, as they are crosslinked and form high-molecular-weight oligomers. The patterns of these high-molecular-weight oligomers appear different from that formed by IsoLG-crosslinking (20), indicating different reaction chemistries between the two dicarbonyls. Interestingly, 3 eq ONE was required to crosslink apoA-IV (Fig. 3D), another HDL protein that can activate LCAT (29) and has antioxidant properties (30, 31). Potentially, ONE targets apoA-I and apoA-II because of their greater abundance on HDL. The data indicate that ONE is a very reactive electrophile that potently crosslinks HDL proteins, making ONE equally as reactive as the IsoLGs.
Figure 3.
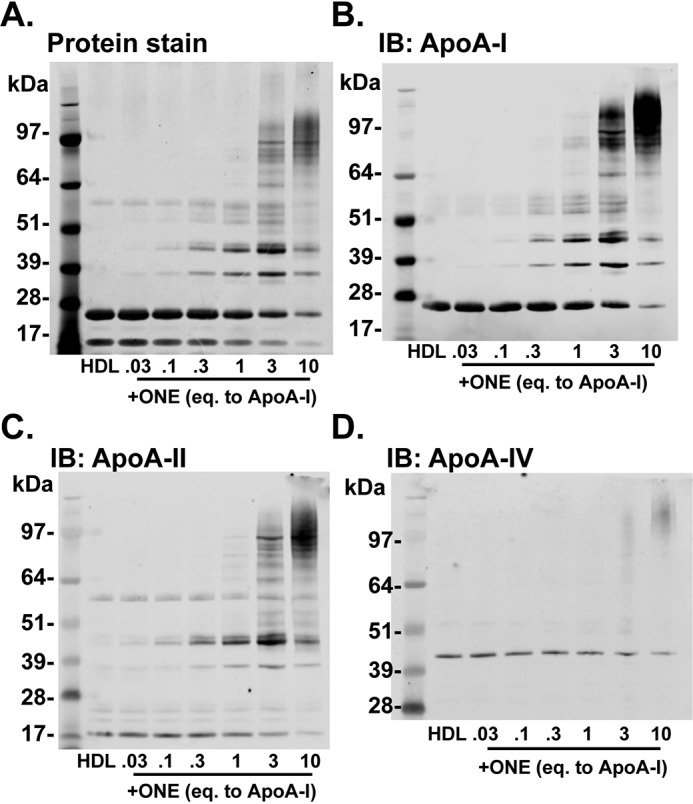
ONE crosslinks HDL proteins beginning at 0.1 molar eq to apoA-I. A–D, crosslinking of HDL proteins by ONE was determined by (A) Coomassie Blue staining and (B–D) immunoblotting (IB) against apoA-I, apoA-II, and apoA-IV. HDL isolated from normal healthy subjects by DGUC was subjected to ex vivo modification of ONE. SDS-PAGE and Western blots are representative of experiments performed three times.
Because IsoLGs crosslink HDL to produce a HDL subpopulation of larger size (20), we sought to determine the impact of ONE modification on HDL size distribution. Using fast protein LC (FPLC), we found that both unmodified and ONE-modified HDLs fractionate into two main subpopulations: spherical and lipid-poor HDL (Fig. 4A). However, ONE did not significantly change the size or distribution of HDL, suggesting that the particles were not fusing to form larger-sized HDL, unlike what it seen with IsoLG modification. Native PAGE gel electrophoresis of reconstituted HDL modified by ONE also shows no change in electrophoretic mobility of these particles (Fig. 4B). Together, the data would suggest that ONE mainly forms intramolecular crosslinks or crosslinks of proteins originally on the same HDL particles, rather than crosslinking proteins on adjacent particles that lead to fusion of the HDL particles.
Figure 4.
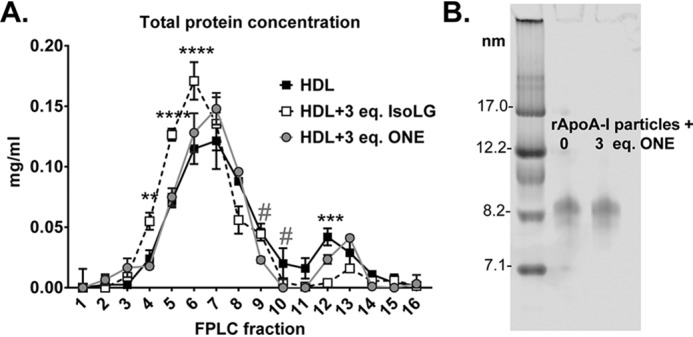
A and B, ONE does not significantly alter HDL size or distribution as indicated by (A) size-exclusion chromatography and by (B) Native PAGE gel electrophoresis. HDL was subjected to ex vivo modification of ONE under aqueous conditions 37 °C overnight. HDL was subjected to FPLC using the Superdex 200 column using A280 monitoring. Protein concentration of fractions was assessed using the Bradford assay. Values are represented as mean ± S.D. Statistical significance was calculated by 2-way ANOVA with Dunnett's multiple comparisons test compared with HDL. Asterisk represents statistical significance between HDL and IsoLG-HDL; # represents significance between HDL and ONE-HDL. #, p < 0.05; **, p < 0.01; ***, p < 0.005; ****, p < 0.001. Native PAGE gel electrophoresis was run under standard conditions and the gel was stained with Coomassie Blue G250 to visualize particles.
ONE-modified HDL has lower HDL–apoA-I exchange but no effect on total macrophage cholesterol efflux
Previously we showed that HDL highly crosslinked by IsoLG has reduced ability to exchange apoA-I, which correlates with reduced ability to promote macrophage cholesterol efflux (20). Because ONE also potently crosslinks HDL proteins, we sought to characterize HDL-apoA-I exchange in ONE-modified HDL. Using the method by Borja et al. (32), which utilizes EPR to detect conformational changes resulting from lipid binding of nitroxide-labeled apoA-I to HDL, we found that ONE modification of HDL dose-dependently reduced apoA-I exchangeability. Although unmodified HDL had an apoA-I exchange rate of 46.5 ± 5.6%, HDL exposed to 1 eq ONE yielded only 19.8 ± 5.7% exchange (p < 0.01) (Fig. 5).
Figure 5.
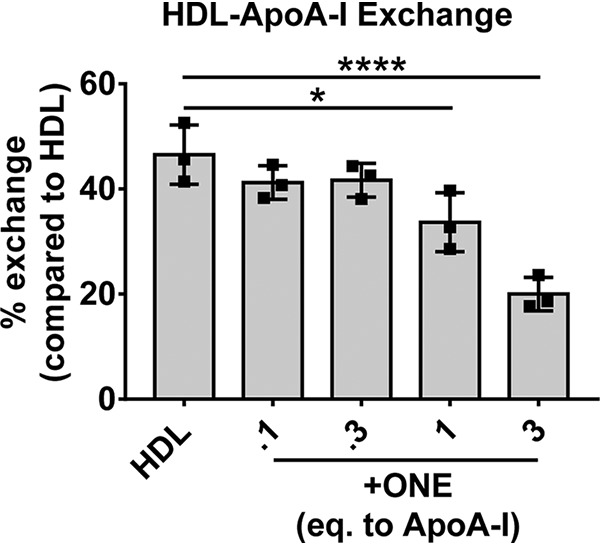
Modification of HDL by ONE inhibits the exchangeability of apoA-I on HDL. HDL-apoA-I exchange was analyzed by EPR as described in “Experimental procedures.” Reactions were performed at a constant apoA-I concentration of 1 mg/ml. Samples were assayed in triplicate. Results are plotted as mean ± S.D. Statistical significance was calculated by one-way ANOVA with Dunnett's multiple comparisons compared with unmodified HDL (control). *, p < 0.05; ****, p < 0.0001.
A decrease in apoA-I exchange often results in less efficient cholesterol mobilization from macrophages via ABCA1, so we examined the effect of ONE modification on cholesterol efflux as described previously (20). Using peritoneal murine macrophages isolated from apoE−/− mice (to exclude the effect of macrophage apoE) and loaded with acetylated LDL, we found that ONE modification did not affect the ability of HDL to efflux 3H-cholesterol even at concentrations up to 3 eq (Fig. 6). This showed that although ONE modification of HDL dramatically affected apoA-I exchange, the capacity of HDL to efflux cholesterol was unaffected.
Figure 6.
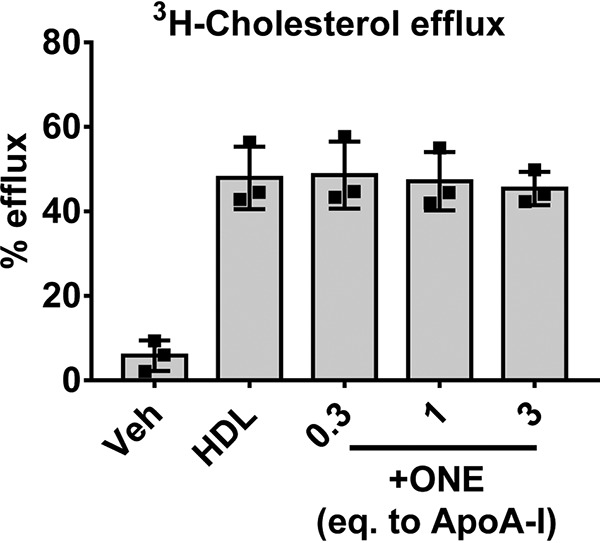
Modification of HDL by ONE does not affect cholesterol efflux from apoE-deficient murine macrophages. HDL isolated from normal healthy subjects by DGUC was subjected to ex vivo modification of ONE. Macrophage cholesterol efflux was assessed using thioglycollate-induced macrophages harvested from the peritoneum of apoE-deficient mice and loaded with 3H-cholesterol and acetylated LDL. Vehicle denotes cell culture media with no HDL added to the cells. Efflux of 3H to unmodified and modified HDL was calculated based on radioactive counts in the supernatant after 4 h and normalized to the HDL control. Results from three individual experiments with replicate wells per treatment are plotted as mean ± S.D.
ONE modification negates the ability to protect against LPS-stimulated macrophage inflammatory response
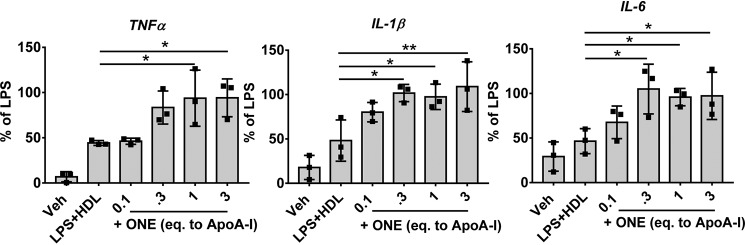
We next determined if ONE modification rendered HDL dysfunctional in protecting against LPS-induced proinflammatory cytokine expression in macrophages. Previously, we found that IsoLG-modified HDL not only rendered HDL ineffective at preventing LPS-induced inflammatory response but induced a greater response than LPS alone (20). In the current study, we found that although unmodified HDL inhibited the LPS-induced expression of Tnfα, IL-1β, and IL-6 by 77.0 ± 2.3%, 59.9 ± 2.7%, 37.3 ± 12.8%, respectively, modification with 0.1 eq ONE ablated its ability to prevent Tnfα, IL-1β, and IL-6 expression, resulting in 94.1 ± 20.9%, 108.9 ± 28.0%, 97.2 ± 26.5% of LPS, respectively, at 3 eq ONE (Fig. 7). However, these levels of cytokine expression are similar to LPS induction alone, indicating that ONE-modified HDL does not potentiate a greater inflammatory response by these macrophages.
Figure 7.
Modification of HDL by ONE renders HDL no longer protective against LPS-stimulated inflammatory cytokine expression. HDL isolated from normal healthy subjects by DGUC was subjected to ex vivo modification of ONE. Thioglycollate-elicited peritoneal murine macrophages were treated with LPS along with ONE modified HDL for 4 h. Results from three individual experiments with 3–4 wells per treatment are plotted as mean ± S.D. Statistical significance was determined by one-way ANOVA with Dunnett's multiple comparisons compared with LPS + HDL (control). *, p < 0.05; **, p < 0.01.
HDL isolated from FH subjects has reduced cholesterol efflux capacity and ability to protect against LPS-stimulated inflammation in macrophages compared with healthy controls
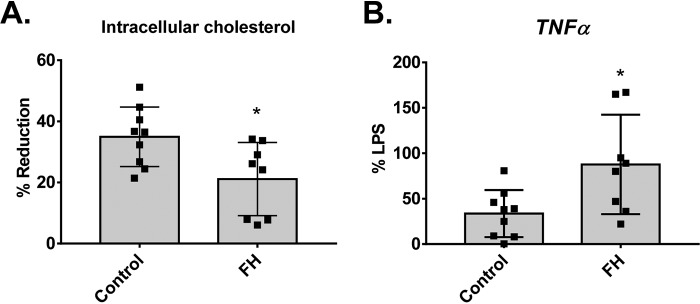
Because ONE modification of HDL does not affect macrophage cholesterol efflux capacity but renders HDL dysfunctional in protecting against LPS-induced macrophage activation, we tested the functions of HDL isolated from FH subjects compared with healthy controls. Interestingly, HDL isolated from FH subjects has lower cholesterol efflux capacity (as indicated as a % reduction in cellular cholesterol content of macrophages) (Fig. 8A) and reduced ability to protect against LPS-stimulated Tnfα expression in macrophages compared with healthy controls (Fig. 8B).
Figure 8.
HDL isolated from FH subjects have lower cholesterol efflux capacity and ability to protect against LPS-stimulated TNFα expression in macrophages compared with healthy controls. HDL-isolated human subjects by DGUC were subjected to ex vivo modification of ONE. Cholesterol efflux capacity was assessed by % reduction of cellular cholesterol in macrophages loaded with acetylated LDL. To determine capacity of HDL to protect against LPS-stimulated cytokine expression, macrophages were treated with LPS along with HDL samples for 4 h. Results are plotted as mean ± S.D. Statistical significance was determined by Student's t test. *, p < 0.05.
LC-MS/MS identification of ONE modified lysine residues in apoA-I
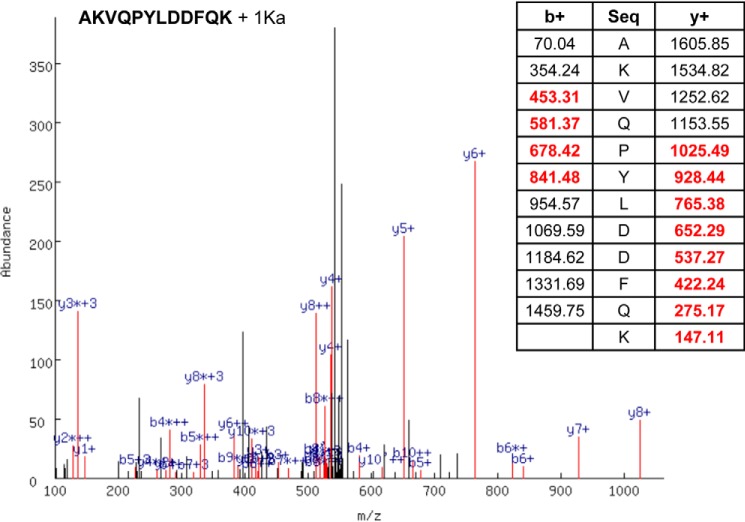
To identify the amino acid targets of ONE in apoA-I, we used reconstituted HDL generated using recombinant human apoA-I, phosphatidylcholine, and free cholesterol. Reconstituted HDL particles represent simplified systems that have been used for decades to study the HDL-like lipid bound state of apoA-I. Reconstituted HDL was modified with 3 eq ONE overnight. After reaction quenching, the particles were delipidated by a chloroform/methanol extraction and the resulting protein was exhaustively digested with trypsin. The resulting peptides were analyzed by LC-MS/MS and the data were interrogated for known mass adducts resulting from ONE chemical modifications. A representative MS/MS identification of the apoA-I peptide AKVQPYLDDFQK with a ONE ketoamide adduct on Lys-97 is shown in Fig. 9. The spectrum shows the MS/MS spectrum with b+ and y+ series ions highlighted in red and labeled in blue. The table shows the identified ions bolded. Table 1 summarizes all the identified peptides and ONE adducts resulting from three independent experiments. The identified residues include ketoamide adducts (+154.0994 atomic mass unit) on Lys-12, Lys-23, Lys-96, and Lys-226 in human apoA-I in triplicate experiments. One of the three experiments also identified ketoamide adducts on Lys-94 and Lys-118, whereas another experiment identified Michael adducts (+156.1200 atomic mass unit) on Lys-133 and Lys-226. A complete list of identified tryptic peptides and ONE modifications is provided in Table S2.
Figure 9.
Representative MS/MS identification of the apoA-I peptide AKVQPYLDDFQK with a ONE ketoamide adduct on Lys-97. The spectrum shows the MS/MS spectrum with b+ and y+ series ions highlighted in red and labeled in blue. The table shows the identified ions in bold red type.
Table 1.
Summary of all the identified peptides and ONE adducts resulting from three independent experiments
The identified residues include ketoamide adducts (+154.0994 atomic mass units) on Lys-12, Lys-23, Lys-96, and Lys-226 in human apoA-I in triplicate experiments. One of the three experiments also identified ketoamide adducts on Lys-94 and Lys-118, and another experiment identified Michael adducts (+156.1200 atomic mass units) on Lys-133 and Lys-226. Ka, ketoamide adduct; MA, Michael adduct. *, represents detection in a single experiment; ***, represents consistent detection in triplicate experiments.
| Lysine | Peptide + adduct | Frequency |
|---|---|---|
| Lys-12 | VKDLATVYVDVLK + 1Ka | *** |
| Lys-23 | DLATVYVDVLKDSGR + 1Ka | *** |
| Lys-94 | DLEEVKAK + 1Ka | * |
| Lys-96 | AKVQPYLDDFQK + 1Ka | *** |
| Lys-118 | QKVEPLR + 1Ka | * |
| Lys-133 | AELQEGARQK + 1MA | * |
| Lys-226 | QGLLPVLESFKVSFLSALEEYTK + 1Ka | *** |
| Lys-226 | QGLLPVLESFKVSFLSALEEYTK + 1MA | * |
Pentylpyridoxamine is more effective at scavenging ONE than other scavengers
2-Aminomethylphenols have been shown to scavenge ONE in situ, with PPM and chloro-salicylamine possessing the fastest second-order reaction rate (33). However, the effectiveness of these scavengers in preventing ONE from reacting with proteins embedded in a lipophilic environment of the HDL surface is unknown. Incubating the scavengers and ONE together with HDL overnight and assessing protein crosslinking by SDS-PAGE, we found that PPM was significantly more effective at scavenging ONE compared with salicylamine (2-hydroxybenzylamine) or its analogues (fluoro-salicylamine (F-SAM) and chloro-salicylamine (Cl-SAM)) (Fig. 10, A and B). Furthermore, PPM at 10 eq was able to prevent 1 eq ONE-induced HDL dysfunction in protecting against LPS-stimulated TNFα expression in macrophages, whereas its inactive analogue, pentylpyridoxine, did not (Fig. 10C). This demonstrates the effectiveness of PPM in preventing ONE-induced crosslinking as well as HDL dysfunction in an in vitro model of inflammation.
Figure 10.
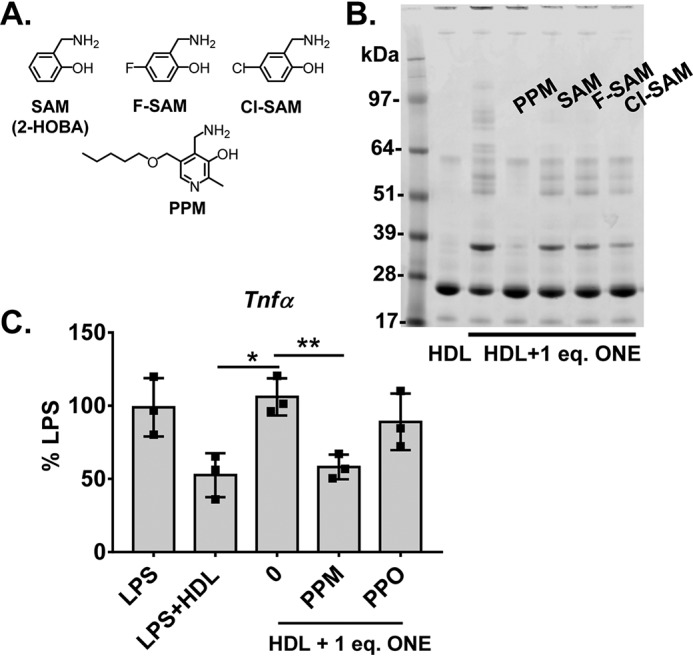
Effect of salicylamine analogues in scavenging ONE. A, molecular structures of salicylamine and various analogues (F-SAM, Cl-SAM, PPM). B, effect of analogues in preventing ONE-induced HDL protein crosslinking as shown by Coomassie Blue G250–stained SDS-PAGE gel of unmodified and 1 eq ONE-modified HDL. C, demonstration of PPM (10 eq) preventing ONE-induced HDL dysfunction in protecting against LPS-stimulated TNFα expression in thioglycollate-elicited peritoneal macrophages, whereas its inactive analogue, pentylpyridoxine (PPO), did not. Statistical significance was determined by one-way ANOVA with Dunnett's multiple comparisons compared with LPS + 1 eq ONE + HDL. Results from three individual experiments with three wells per treatment are plotted as mean ± S.D. *, p < 0.05; **, p < 0.01.
Discussion
Growing evidence suggests that modification of HDL by reactive lipid aldehydes renders HDL dysfunctional, which contributes to the pathogenesis of atherosclerotic disease. Although ONE is far more reactive than HNE, its role in atherosclerosis is unknown. In the present study, we show that ONE-protein adducts are elevated in human atherosclerosis. ONE crosslinks HDL proteins at low molar concentrations but does not alter HDL size. Although ONE modification alters HDL function in certain aspects (such as HDL–apoA-I exchange or anti-inflammation), it does not affect macrophage cholesterol efflux. ONE-modification of HDL contributes partly to the HDL dysfunctions observed in familial hypercholesterolemia. We also identify four lysines that ONE consistently targets in apoA-I. Finally, we show that the dicarbonyl scavenger, PPM, is most effective at scavenging ONE from crosslinking HDL proteins.
Because the 4-ketoamide adducts are known as the major long-lived protein adducts, we initially assayed for the ONE-ketoamide adducts in patient HDL. We found a significant elevation in ONE-ketoamide adducts in the HDLs of patients with familial hypercholesterolemia, somewhat greater than the levels of IsoLG adducts in these same samples.
Although ONE is structurally analogous to HNE, the ketone at the C4 position makes ONE much more reactive toward protein nucleophiles (16, 34). We demonstrate that ONE crosslinks the main structural proteins of HDL, apoA-I and apoA-II, at very low concentrations (0.1–0.3 eq to apoA-I), making ONE just as reactive as the potent isolevuglandins in protein crosslinking (20). Modification of HDL by above 1 eq ONE produces multimers of apoA-I and apoA-II that migrate as distinct oligomers through an SDS gel, which contrasts with the “smear” of oligomers created by isolevuglandin-induced crosslinking. This suggests that the proteins crosslinked by ONE differ to some extent from those crosslinked by IsoLGs, which may contribute to differences in the dysfunctions of HDL induced by these two reactive dicarbonyls.
The exchangeability of apoA-I in HDL governs many of HDL's functions, such as its ability to promote cholesterol efflux via ABCA1 (35). Decreases in HDL–apoA-I exchange were observed in both animal models of atherosclerosis and in human atherosclerosis (32). Decreases in HDL–apoA-I exchange also correlated with decreased cholesterol efflux capacity of IsoLG-modified HDL (20). When we measured the rate of exchange between spin-labeled apoA-I and ONE-modified HDL, we found a decrease in apoA-I exchangeability that depended on extent of ONE modification. This dose-dependent decrease is comparable to that of IsoLG-modified HDL. Crosslinking of HDL proteins may hinder the ability of HDL-bound apoA-I to exchange with exogenously added lipid-free apoA-I associated with the particles, thus resulting in decreased HDL–apoA-I exchange.
Although HDL–apoA-I exchange correlates with cholesterol efflux capacity, we find that ONE-modification of HDL or apoA-I did not alter the ability of HDL to induce cholesterol efflux from macrophages at any of the tested concentrations. This finding was initially surprising because ONE dramatically decreased HDL–apoA-I exchange as well as significantly crosslinked HDL proteins. Crosslinking of HDL proteins by IsoLGs correlated with the significant reduction of its cholesterol efflux capacity (20). However, size exclusion separation of spherical and lipid-poor HDL shows that ONE modification does not appear to alter HDL size or distribution of spherical versus lipid-poor particles. This contrasts with IsoLG modification, which increases HDL size (20) and decreases the population of lipid-poor particles. Because small lipid-poor HDL more efficiently promotes cholesterol efflux than larger spherical HDL via ABCA1 (36), it may be possible that ONE modification does not impact cholesterol efflux because of the lack of alteration of HDL size and population of the small particles. Additionally, not all endogenous crosslinkers of HDL proteins impair function. Peroxidase-generated tyrosyl radicals crosslink apoA-I and apoA-II in HDL but enhance its ability to promote cholesterol efflux (37, 38). We also found that HDL isolated from FH subjects has significantly decreased cholesterol efflux capacity compared with healthy controls, suggesting that ONE-modification does not contribute to this dysfunction.
Another function of HDL is to protect against inflammation partially by neutralizing LPS (39–42). Previously, we found that modification of HDL with IsoLGs not only completely blocks the ability of HDL to inhibit LPS-induced cytokine expression in macrophages but also further potentiates Il-1β and Il-6 expression induced by LPS. In this study, we find that ONE modification of HDL blocks its ability to inhibit LPS-induced Tnfα, Il-1β, and Il-6 expression beginning at 0.3 eq ONE. However, increasing ONE modification does not further potentiate expression of these cytokines by LPS. Of note, modification of HDL with HNE or succinaldehyde does not potentiate LPS expression either (20). These results suggest that the potentiation of cytokine expression induced by IsoLG modification of HDL results from a mechanism (e.g. formation of a receptor agonist) distinct from the mechanism whereby HDL inhibits LPS signaling (e.g. neutralization of LPS by apoAI). Potentiation of expression might result from IsoLG modification specifically forming receptor agonists whereas the block of inhibition might result from apoA-I modified by a wider array of aldehydes being unable to bind and neutralize LPS. In addition, HDL isolated from FH subjects has significantly decreased ability to protect against LPS-induced activation. A small subset of these HDLs further potentiates Tnfα expression over LPS. These data suggest that ONE modification likely contributes to the HDL dysfunction in protecting against endotoxin-mediated macrophage activation.
Analysis of proteolytic digests of apoA-I by LC-MS/MS revealed that ONE consistently formed ONE-ketoamide adducts on four lysine residues (Lys-12, Lys-23, Lys-96, Lys-226) in apoA-I. One of the three experiments also identified ketoamide adduct on Lys-118, whereas another experiment identified Michael adducts on Lys-133 and Lys-226. This may suggest that these sites are lower frequency targets. Unlike HNE (13), we saw no evidence of Michael adducts on any histidine residues nor on arginine residues. Interestingly, when superimposed onto the published crystal structure of an apoA-I dimer that has been reported to bear strong similarities to the lipid-bound form (43), the modified Lys residues are all highly solvent exposed (Fig. 11). In fact, Lys-96, Lys-118, and Lys-133 are pointing almost directly outward from the double belt structure that likely forms the basis of apoA-I's encapsulation of HDL lipids (44). Lys-12 was previously shown to be susceptible to glycation and important in apoA-I's anti-inflammatory function (43). Lys-23 resides in a region potentially significant in interactions with lipoprotein-binding protein (44), which plays an important role in not only facilitating transfer of LPS to soluble CD14 but also to neutralize these complexes via transfer to HDL (45). Lys-96 and Lys-133 reside in helix 3 and 5, respectively, and are potentially involved in LCAT activation (46) and also in interactions with lipoprotein-binding protein (44). Our finding that ONE modifies Lys implicated in the interaction with lipoprotein-binding protein suggests that the inability of ONE-modified HDL to inhibit LPS induced macrophage activation likely results from reduced ability to bind and neutralize LPS.
Figure 11.
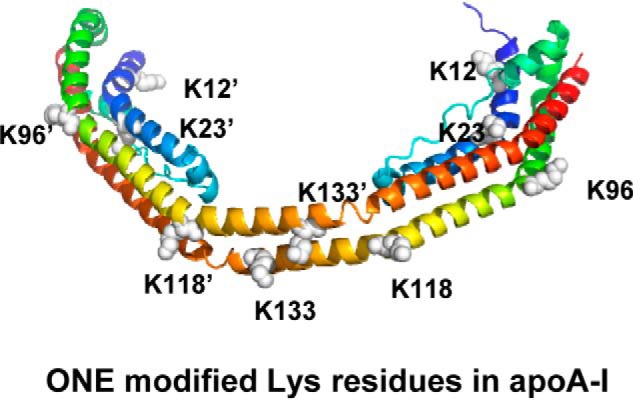
Superimposed ONE targeted lysines onto a published crystal structure of an apoA-I dimer that has been reported to bear strong similarities to the lipid-bound form.
Interestingly, Lys-226 was one of the Lys residues found to be highly modified by ONE. Lys-226 is also a major target of acrolein (47) and malondialdehyde (13). Lys-226 appears to be important in cholesterol efflux (48). Deletions of helix 10 where Lys-226 resides (49) or helix 9 + 10 (50) greatly reduce cholesterol efflux and a synthetic 9/10 helix mediates high-affinity cholesterol efflux (51). Yet in our studies, ONE modification of HDL did not affect its macrophage cholesterol efflux capacity. As such, our study extends the findings of Shao et al. (13) showing that individual lipid aldehydes varied in their ability to impair specific HDL functions and suggests that specific structural features of various Lys-226 adducts determine whether there is an effect on cholesterol efflux. Elucidating the specific structural features that account for these differences will require additional studies. One important difference between the lysine adducts of IsoLG and ONE may be that the IsoLG adducts include a negatively charged carboxylate. IsoLGs could also potentially target different lysine residues.
We previously demonstrated the potential of using dicarbonyl scavengers with a 2-aminomethylphenol moiety (e.g. PPM) to block the ability of IsoLGs to modifying HDL and thus preserve its function. Measurements of the second-order rate constants for the reaction of various scavengers with ONE in vitro showed that PPM is much more reactive than SAM and Et-SAM but only slightly more reactive than Cl-SAM (33). This study did not test the ability of the scavengers to react with ONE in biological systems. We found that PPM is effective at preventing ONE-induced protein crosslinking in HDL, whereas SAM-based scavengers show little efficacy. In addition, PPM was also able to prevent ONE-induced HDL dysfunction in an in vitro model of inflammation. These results combined with our previous study (20) suggest that PPM may have potential use as a therapeutic to protect HDL in vivo.
In conclusion, we show that ONE modifies HDL in humans with FH, who have severe hypercholesterolemia and atherosclerotic disease, and exerts important biological effects; yet, ONE differs significantly in its effects from IsoLGs, which are similar in terms of reactivity and ability to crosslink proteins. Importantly, ONE crosslinking of HDL proteins does not alter the size or distribution of HDL particles. ONE modification reduces apoA-I exchange but does not alter macrophage cholesterol efflux. ONE modification blocks HDL inhibition of LPS-induced macrophage activation but does not further potentiate cytokine expression. It will be of interest for future studies to determine whether ONE impairs other functions of HDL and its relevance in LDL modifications in atherosclerosis.
Experimental procedures
Materials
Reagents for SDS-PAGE and immunoblotting were from Novex by Life Technologies (Carlsbad, CA). ApoA-I mouse/human (5F4) mAb was purchased from Cell Signaling Technology (Danvers, MA). ApoA-II human (EPR2913) mAb was purchased from Abcam (Cambridge, MA). Materials used for cell culture were from Gibco by Life Technologies (Grand Island, NY). [1,2-3H(N)]cholesterol was purchased from Perkin-Elmer Life Sciences. eBioscience LPS solution was purchased from Thermo Fisher Scientific. Acetylated LDL derived from human plasma was purchased from Alfa Aesar (Haverhill, MA) RNeasy Mini Kit was purchased from Qiagen (Hilden, Germany). iQ SYBR Green Supermix and iScript cDNA Synthesis Kit were purchased from Bio-Rad.
HDL from FH patients and healthy controls
Blood from eight patients diagnosed with FH and nine healthy control volunteers was collected through peripheral vein phlebotomy into purple top EDTA tubes (5 ml each) and placed on wet ice. Within a few hours, plasma was collected after centrifugation for 20 min (2000 rpm) at 4 °C. Then plasma samples were stored at −80 °C until lipoprotein extraction. HDL was isolated from thawed plasma by density-gradient ultra-centrifugation (DGUC) (1.061–1.21 g/ml), 330,000 × g followed by extensive dialysis at 4 °C in 1× PBS. These studies abide by the Declaration of Helsinki principles. They were approved by the Vanderbilt University Institutional Review Board (IRB), and all participants gave their written informed consent.
Animals
All procedures were approved by the Vanderbilt University Institutional Animal Care and Use Committee (IACUC). Breeding pairs of homozygous apoE−/− mice (C57BL/6J background, strain 002052) age 12 weeks were purchased from The Jackson Laboratory (Bar Harbor, ME). The animals were acclimated and housed in the Vanderbilt University animal facility in a 12-h light/12-h dark cycle, were maintained on standard rodent chow (LabDiet 5001), and were given free access to water. Progeny of the breeding pairs were at least 8 weeks of age before harvest of macrophages (described below).
Chemical synthesis of ONE and scavengers
ONE was synthesized as reported previously (52) and prepared as a 1 m solution in acetonitrile. ONE was diluted as a 10 mm stock solution in DMSO in aliquots and stored in −80 °C until use. Acetic acid salts of SAM, Cl-SAM, and F-SAM and the hydrochloride salt of PPM were synthesized as described (53, 54). Working solutions were prepared fresh before each assay and diluted in water to appropriate concentrations.
Measurement of ONE and IsoLG adducts
Quantitation of lysine modification of HDL by ONE (ONE-Lys) and IsoLG (IsoLG-Lys) was performed by subjecting an aliquot of HDL to proteolysis with Pronase and aminopeptidase M. The released ONE-Lys and IsoLG-Lys (the 4-ketoamide adduct for ONE and the lysyl-lactam adduct for IsoLG) were then measured using a slight modification of the stable isotope dilution LC-MS/MS assay previously used to measure IsoLG-Lys (55). As with IsoLG-Lys, two internal standards for ONE-Lys were used, with all four internal standards added to samples. The primary internal standard, [2H3]ONE-protein, was generated by reaction of [2H3]ONE (Cayman Chemical) with BSA. This internal standard requires proteolyis to release the [2H3]ONE-Lys measured in the LC-MS/MS assay, just as ONE-modified proteins in the sample that must undergo proteolysis to release the measured ONE-Lys. The second internal standard, ONE-[13C6]Lys, was made by coupling the N-hydroxysuccinimide ester of 4-oxononanoic acid to Fmoc-protected [13C6]Lys under basic conditions. The product was deprotected with piperidine and purified by flash chromatography on silica gel. The ONE-[13C6]Lys was used to quantify the amount of [2H3]ONE-Lys released when varying amounts of [2H3]ONE-protein was subjected to proteolytic digestion. It was also added to samples to confirm inter-sample consistency of protein digestion by measuring ratio of ONE-[13C6]Lys to [2H3]ONE-Lys in each sample. The original method of solid phase extraction for IsoLG-Lys was slightly revised to eliminate the 15% methanol wash step because some ONE-Lys was lost in this wash step. The LC-MS/MS assay previously used to measure IsoLG-Lys (55) was then modified to include three additional multiple reaction monitoring transitions: m/z 301 → m/z 84 (ONE-Lys), m/z 307 → m/z 89 (ONE-[13C6]Lys), and m/z 304 → m/z 84 ([2H3]ONE-Lys) and amount of ONE-Lys quantified by ratio of the ONE-Lys peak area to [2H3]ONE-Lys peak area.
ONE modification of HDL and the use of scavengers
HDL was exposed to various concentrations of ONE at 37 °C overnight to guarantee a complete reaction to form a stable end product. Control HDL was treated similarly in the absence of dicarbonyls. For experiments involving the use of scavengers, scavengers solubilized in water were incubated with HDL for 30 min at 37 °C before the addition of ONE.
Characterization of apolipoprotein crosslinking of modified HDL
HDL apolipoprotein crosslinking was assessed by SDS-PAGE performed under reducing conditions with Invitrogen's gel electrophoresis and transfer system. 4–20% Tris gradient gels were used. Western blotting analyses were carried out using polyclonal antibodies specific for human apoA-I, apoA-II, and apoA-IV.
Characterization of HDL size by FPLC and Native gel electrophoresis
To quantify HDL size by its distribution of subpopulations, HDL samples were diluted to 500 μl in size-exclusion chromatography (SEC) running buffer (10 mm Tris-HCl, 0.15 m NaCl, 0.2% NaN3) and injected an ÄKTA SEC system (GE Healthcare) with three in-series Superdex 200 Increase gel filtration columns (10/300 GL; GE Healthcare). Samples were applied to the column with a flow rate of 0.3 ml/min at room temperature and eluate collected as 72 × 1.5 ml fractions using a F9-C 96-well plate fraction collector (GE Healthcare). Each fraction was analyzed for total protein using the Bradford assay. Native gel electrophoresis was performed using the Invitrogen gel electrophoresis system using a Native PAGE 4–16% Bis-Tris gel. Reconstituted HDL was synthesized by a modified sodium cholate dialysis method (56, 57) from recombinant apoA-I, POPC, and cholesterol at a ratio of 1:80:5. Particles were subjected to ONE modification at 37 °C overnight. 1-μg samples and the lipoprotein marker (Amersham Biosciences HMW Calibration Kit for Native Electrophoresis, GE Healthcare) were mixed with Native loading buffer before being subjected to Native gel electrophoresis at 150 volts for 90 min. The gel was washed three times with deionized water before staining with Coomassie Blue G250 (Bio-Rad) to visualize the HDL. The lipoprotein standard contained the following proteins: porcine thyroglobulin (Mr = 669 kDa, Rs = 17 nm), equine spleen ferritin (Mr = 440 kDa, Rs = 12.2 nm), bovine heart lactate dehydrogenase (Mr = 232 kDa, Rs = 8.2 nm), BSA (Mr = 66 kDa, Rs = 7.1 nm).
HDL–ApoA-I exchange
HDL samples prepared by adding 15 μl 3 mg/ml spin-labeled apoA-I probe to 45 μl 1 mg/ml HDL and drawn into an EPR-compatible borosilicate capillary tube (VWR Life Science) (32). EPR measurements were performed using a Bruker eScan EPR spectrometer outfitted with temperature controller (Noxygen). Samples were incubated for 15 min at 37 °C and then scanned at 37 °C. The peak amplitude of the nitroxide signal from the apoA-I probe in the sample (3462–3470 gauss) was compared with the peak amplitude of a proprietary internal standard (3507–3515 gauss) provided by Bruker. The internal standard is contained within the eScan spectrometer cavity and does not contact the sample. Because the y axis of the EPR spectrometer is measured in arbitrary units, measuring the sample against a fixed internal standard facilitates normalization of the response. HDL–apoA-I exchange (HAE) activity represents the sample: internal standard signal ratio at 37 °C. The maximal % HAE activity was calculated by comparing HAE activity to a standard curve ranging in the degree of probe lipid-associated signal. Experiments were repeated two separate times. All samples were read in triplicate and averaged.
Cell culture
Male and female apoE−/− mice (C57/BL genetic background) were injected intraperitoneally with 3% thioglycollate and the macrophages were harvested by peritoneal lavage after 4 days. Cells were maintained in 24-well plates in DMEM with 10% (v/v) FBS and penicillin-streptomycin at 100 units/ml and 100 μg/ml, respectively.
Cholesterol efflux
Efflux was assessed by the isotopic method (58). Loading medium was prepared to consist of DMEM containing 100 μg/ml acetylated LDL with 6 μCi 3H-cholesterol/ml. After equilibration for 30 min at 37 °C, loading medium was added to cells for 48 h. After 48 h, the cells were incubated for 1 h with DMEM containing 0.1% BSA so that surface-bound acetylated LDL was internalized and processed. Cells were washed and incubated with efflux medium, which contained DMEM with 35 μg/ml HDL samples. After 24 h incubation, supernatants were collected, vacuum filtered, and prepared for β-scintillation counting. To measure the capacity of the FH versus control HDL to reduce macrophage cholesterol, apoE−/− macrophages were cholesterol-enriched by incubation for 48 h in DMEM containing 100 μg protein/ml of acetylated LDL. The cells were then washed and incubated for 24 h in DMEM alone or with 50 μg HDL protein/ml. Cellular cholesterol was measured before and after incubation with HDL using an enzymatic cholesterol assay as described (59).
Macrophage inflammation
Cells derived from female mice were incubated overnight in DMEM containing 0.5% FBS and 1% penicillin-streptomycin. The cells were washed two times with Hanks' Balanced Salt Solution and then incubated for 4 h with DMEM alone or containing 100 ng/ml LPS with or without the HDL preparations (50 μg/ml). The cells were lysed, mRNA harvested, and the cDNA synthesized. qPCR was performed with the following primer pairs: Tnf forward, 5′-CCATTCCTGAGTTCTGCAAAG-3′; Tnf reverse, 5′-GCAAATATAAATAGAGGGGGGC-3′; Il-1β forward, 5′-TCCAGGATGAGGACATGAGCA-3′; Il-1β reverse, 5′-GAACGTCACACACCAGCA-3′; Il-6 forward, 5′-TAGTCCTTCCTACCCCAATTTCC-3′; Il-6 reverse, 5′-TTGGTCCTTAGCCACTCCTTCC-3′.
Sample digestion, preparation, and LC-MS/MS analysis of ONE-modified peptides
Reconstituted HDL was synthesized by the cholate dialysis method (60) from recombinant human apoA-I, phosphatidylcholine, and free cholesterol. Particle size homogeneity was checked by Native gel electrophoresis. HDL was then dialyzed into PBS before modifying with 3 eq ONE at 37 °C overnight. HDL samples were dialyzed into 50 mm ammonium bicarbonate buffer. In the Davidson lab, the particles were lyophilized to dryness then the lipids were extracted in 1 ml ice cold 2:1 (v/v) chloroform/methanol (61) and the precipitated protein was resolubilized in 80% ammonium bicarbonate buffer and 20% methanol. Samples were reduced by addition of DTT to a final concentration of 10 mm and incubation for 30 min at 42 °C. Reduced protein was carbamidomethylated with iodoacetamide at a final concentration of 40 mm and incubated in the dark at room temperature for 30 min. 20 μg of each sample was digested by adding 1 μg of sequencing grade trypsin (Promega) for 16 h at 37 °C. An additional 1 μg of trypsin was added the following day and incubated for an additional 2 h at 37 °C. Samples were lyophilized to dryness using a SpeedVac and stored at −20 °C until ready for MS analysis.
Dried peptides were reconstituted in 15 μl of 0.1% formic acid in water and 5 μl of sample was applied to an ACQUITY UPLC C18 reverse phase column (Waters) maintained at 40 °C using an Infinity 1290 Autosampler and HPLC (Agilent). Peptides were eluted at 0.1 ml/min using a varying mobile phase gradient from 95% phase A (FA/H2O 0.1/99.9, v/v) to 32% phase B (FA/ACN 0.1/99.9 v/v) for 120 min followed by 32% B to 50% B for 2 min. Column cleaning was performed by varying the mobile phase gradient to 90% B for 10 min and the column was re-equilibrated at 95% A for 10 min. Peptides were introduced to the mass spectrometer using a Jet Stream source (Agilent) as described previously (62). Spectra were acquired using an iFunnel Q-Tof (Agilent) operating in positive ion mode. Precursors were limited to acquisition of ions with a charge states of 2+ and 3+ and required a minimum of 1500 counts. Each cycle acquired the 20 most intense precursors which were fragmented with a variable collision energy (CE) dependent on the precursor mass-to-charge (m/z) ratio: CE = k* (m/z) + b, with a slope (k) of 3 and an offset (b) of 2 for 2+ ions and −2 for 3+ ions. MS/MS spectra were acquired until at least 45,000 total counts were collected or a maximum accumulation time of 0.33 s. MGF files were generated using MassHunter Qualitative Analysis Software (v. B.07.00, Agilent). MS/MS peaks were limited to the top 150 peaks by height and precursors were limited to a maximum assigned charge state of 3+. MS/MS data were analyzed by MassMatrix Suite 3.10 using settings of peptide mass tolerance of 10 ppm, peptide length of 3–40 amino acids, 1 posttranslational modification per peptide, 2 missed tryptic cleavages, and minimum pp score of 5.0. Mass additions for a ONE-ketoamide (C9H14O2, 154.099 Da) or a ONE-Michael adduct (C9H14O2, 156.115) were searched for both Lys and His residues.
Data availability
Raw mass spectrometry data is publicly accessible through Figshare online repository: https://figshare.com/articles/Zhang_et_al_JBC_MS_Data_10_13_2019/9975560 under project IDs Zhang Set 1 Run 1 3 eq ONE, Zhang Set 1 Run 2 3 eq ONE 2A, Zhang Set 1 Run 2 3 eq ONE 2B, and Zhang Set 2 3 eq ONE.3
Author contributions
L. S. M.-Z., P. G. Y., W. S. D., M. F. L., and S. S. D. conceptualization; L. S. M.-Z., W. S. D., and S. S. D. data curation; L. S. M.-Z., W. S. D., and S. S. D. formal analysis; L. S. M.-Z., M. F. L., and S. S. D. funding acquisition; L. S. M.-Z. and P. G. Y. validation; L. S. M.-Z., V. Y., J. T. M., J. M., K. A. T., M. S. B., T. P., V. A., W. S., W. S. D., and S. S. D. investigation; L. S. M.-Z., P. G. Y., W. S. D., and S. S. D. visualization; L. S. M.-Z., V. Y., W. S. D., and S. S. D. methodology; L. S. M.-Z. writing-original draft; L. S. M.-Z., V. Y., K. A. T., M. S. B., V. A., P. G. Y., W. S. D., M. F. L., and S. S. D. writing-review and editing; P. G. Y., W. S. D., M. F. L., and S. S. D. resources; W. S. D., M. F. L., and S. S. D. supervision; S. S. D. project administration.
Supplementary Material
Acknowledgments
We acknowledge the Lipoprotein Analysis and HDL Function Core for providing the purified human HDL. We also thank Dr. James Galligan, Dr. Ryan Allen, Dr. Allison Cooke, Dr. Olivier Boutard, and Taneem Amin for their technical assistance.
This work was supported by National Institutes of Health Grants HL116263 (to M. F. L. and S. S. D.), HL138745 (to L. S. M.-Z.) and P01HL128203 (to W. S. D.). Drs. Amarnath, Davies, and Linton are inventors on a patent application for the use of PPM and related dicarbonyl scavengers for the treatment of cardiovascular disease. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- LDL
- low-density lipoprotein
- ONE
- 4-oxo-2-nonenal
- HNE
- 4-hydroxynonenal
- HDL
- high-density lipoprotein
- FH
- familial hypercholesterolemia
- apo
- apolipoprotein
- LPS
- lipopolysaccharide
- TNFα
- tumor necrosis factor α
- PPM
- pentylpyridoxamine
- LCAT
- lecithin-cholesterol acyltransferase
- IsoLG
- isolevuglandin
- HPNE
- 4-hydroperoxy-2-nonenal
- eq
- molar equivalence
- F-SAM
- fluoro-salicylamine
- Cl-SAM
- chloro-salicylamine
- DGUC
- density-gradient ultra-centrifugation
- HAE
- HDL-apoA-I exchange
- ANOVA
- analysis of variance.
References
- 1. Salomon R. G., Kaur K., Podrez E., Hoff H. F., Krushinsky A. V., and Sayre L. M. (2000) HNE-derived 2-pentylpyrroles are generated during oxidation of LDL, are more prevalent in blood plasma from patients with renal disease or atherosclerosis, and are present in atherosclerotic plaques. Chem. Res. Toxicol. 13, 557–564 10.1021/tx000007u [DOI] [PubMed] [Google Scholar]
- 2. Salomon R. G., Batyreva E., Kaur K., Sprecher D. L., Schreiber M. J., Crabb J. W., Penn M. S., DiCorletoe A. M., Hazen S. L., and Podrez E. A. (2000) Isolevuglandin-protein adducts in humans: Products of free radical-induced lipid oxidation through the isoprostane pathway. Biochim. Biophys. Acta 1485, 225–235 10.1016/S1388-1981(00)00038-X [DOI] [PubMed] [Google Scholar]
- 3. Nakamura J., Shimomoto T., Collins L. B., Holley D. W., Zhang Z., Barbee J. M., Sharma V., Tian X., Kondo T., Uchida K., Yi X., Perkins D. O., Willis M. S., Gold A., and Bultman S. J. (2017) Evidence that endogenous formaldehyde produces immunogenic and atherogenic adduct epitopes. Sci. Rep. 7, 10787 10.1038/s41598-017-11289-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palinski W., Rosenfeld M. E., Ylä-Herttuala S., Gurtner G. C., Socher S. S., Butler S. W., Parthasarathy S., Carew T. E., Steinberg D., and Witztum J. L. (1989) Low density lipoprotein undergoes oxidative modification in vivo. Proc. Natl. Acad. Sci. U.S.A. 86, 1372–1376 10.1073/pnas.86.4.1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenfeld M. E., Palinski W., Ylä-Herttuala S., Butler S., and Witztum J. L. (1990) Distribution of oxidation specific lipid-protein adducts and apolipoprotein B in atherosclerotic lesions of varying severity from WHHL rabbits. Arteriosclerosis 10, 336–349 10.1161/01.ATV.10.3.336 [DOI] [PubMed] [Google Scholar]
- 6. Hazen S. L., Gaut J. P., Crowley J. R., Hsu F. F., and Heinecke J. W. (2000) Elevated levels of protein-bound p-hydroxyphenylacetaldehyde, an amino-acid-derived aldehyde generated by myeloperoxidase, are present in human fatty streaks, intermediate lesions and advanced atherosclerotic lesions. Biochem. J. 352, 693–699 [PMC free article] [PubMed] [Google Scholar]
- 7. Kawamura M., Heinecke J. W., and Chait A. (2000) Increased uptake of α-hydroxy aldehyde-modified low density lipoprotein by macrophage scavenger receptors. J. Lipid Res. 41, 1054–1059 [PubMed] [Google Scholar]
- 8. Hoppe G., Subbanagounder G., O'Neil J., Salomon R. G., and Hoff H. F. (1997) Macrophage recognition of LDL modified by levuglandin E2, an oxidation product of arachidonic acid. Biochim. Biophys. Acta 1344, 1–5 10.1016/S0005-2760(96)00160-9 [DOI] [PubMed] [Google Scholar]
- 9. Jinnouchi Y., Sano H., Nagai R., Hakamata H., Kodama T., Suzuki H., Yoshida M., Ueda S., and Horiuchi S. (1998) Glycolaldehyde-modified low density lipoprotein leads macrophages to foam cells via the macrophage scavenger receptor. J. Biochem. 123, 1208–1217 10.1093/oxfordjournals.jbchem.a022062 [DOI] [PubMed] [Google Scholar]
- 10. Watanabe K., Nakazato Y., Saiki R., Igarashi K., Kitada M., and Ishii I. (2013) Acrolein-conjugated low-density lipoprotein induces macrophage foam cell formation. Atherosclerosis 227, 51–57 10.1016/j.atherosclerosis.2012.12.020 [DOI] [PubMed] [Google Scholar]
- 11. Hoff H. F., O'Neil J., Chisolm G. M. 3rd, Cole T. B., Quehenberger O., Esterbauer H., and Jürgens G. (1989) Modification of low density lipoprotein with 4-hydroxynonenal induces uptake by macrophages. Arteriosclerosis 9, 538–549 10.1161/01.ATV.9.4.538 [DOI] [PubMed] [Google Scholar]
- 12. Bowry V. W., Stanley K. K., and Stocker R. (1992) High density lipoprotein is the major carrier of lipid hydroperoxides in human blood plasma from fasting donors. Proc. Natl. Acad. Sci. U.S.A. 89, 10316–10320 10.1073/pnas.89.21.10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shao B., Pennathur S., Pagani I., Oda M. N., Witztum J. L., Oram J. F., and Heinecke J. W. (2010) Modifying apolipoprotein A-I by malondialdehyde, but not by an array of other reactive carbonyls, blocks cholesterol efflux by the ABCA1 pathway. J. Biol. Chem. 285, 18473–18484 10.1074/jbc.M110.118182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCall M. R., Tang J. Y., Bielicki J. K., and Forte T. M. (1995) Inhibition of lecithin-cholesterol acyltransferase and modification of HDL apolipoproteins by aldehydes. Arterioscler. Thromb. Vasc. Biol. 15, 1599–1606 10.1161/01.ATV.15.10.1599 [DOI] [PubMed] [Google Scholar]
- 15. Kumano-Kuramochi M., Shimozu Y., Wakita C., Ohnishi-Kameyama M., Shibata T., Matsunaga S., Takano-Ishikawa Y., Watanabe J., Goto M., Xie Q., Komba S., Uchida K., and Machida S. (2012) Identification of 4-hydroxy-2-nonenal-histidine adducts that serve as ligands for human lectin-like oxidized LDL receptor-1. Biochem. J. 442, 171–180 10.1042/BJ20111029 [DOI] [PubMed] [Google Scholar]
- 16. Doorn J. A., and Petersen D. R. (2002) Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem. Res. Toxicol. 15, 1445–1450 10.1021/tx025590o [DOI] [PubMed] [Google Scholar]
- 17. Oe T., Lee S. H., Silva Elipe M. V., Arison B. H., and Blair I. A. (2003) A novel lipid hydroperoxide-derived modification to arginine. Chem. Res. Toxicol. 16, 1598–1605 10.1021/tx034178l [DOI] [PubMed] [Google Scholar]
- 18. Oe T., Arora J. S., Lee S. H., and Blair I. A. (2003) A novel lipid hydroperoxide-derived cyclic covalent modification to histone H4. J. Biol. Chem. 278, 42098–42105 10.1074/jbc.M308167200 [DOI] [PubMed] [Google Scholar]
- 19. Stein D. B., Linne U., and Marahiel M. A. (2005) Utility of epimerization domains for the redesign of nonribosomal peptide synthetases. FEBS J. 272, 4506–4520 10.1111/j.1742-4658.2005.04871.x [DOI] [PubMed] [Google Scholar]
- 20. May-Zhang L. S., Yermalitsky V., Huang J., Pleasent T., Borja M. S., Oda M. N., Jerome W. G., Yancey P. G., Linton M. F., and Davies S. S. (2018) Modification by isolevuglandins, highly reactive γ-ketoaldehydes, deleteriously alters high-density lipoprotein structure and function. J. Biol. Chem. 293, 9176–9187 10.1074/jbc.RA117.001099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee S. H., and Blair I. A. (2000) Characterization of 4-oxo-2-nonenal as a novel product of lipid peroxidation. Chem. Res. Toxicol. 13, 698–702 10.1021/tx000101a [DOI] [PubMed] [Google Scholar]
- 22. Spiteller P., Kern W., Reiner J., and Spiteller G. (2001) Aldehydic lipid peroxidation products derived from linoleic acid. Biochim. Biophys. Acta 1531, 188–208 10.1016/S1388-1981(01)00100-7 [DOI] [PubMed] [Google Scholar]
- 23. Shimozu Y., Hirano K., Shibata T., Shibata N., and Uchida K. (2011) 4-Hydroperoxy-2-nonenal is not just an intermediate but a reactive molecule that covalently modifies proteins to generate unique intramolecular oxidation products. J. Biol. Chem. 286, 29313–29324 10.1074/jbc.M111.255737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumagai T., Matsukawa N., Kaneko Y., Kusumi Y., Mitsumata M., and Uchida K. (2004) A lipid peroxidation-derived inflammatory mediator: Identification of 4-hydroxy-2-nonenal as a potential inducer of cyclooxygenase-2 in macrophages. J. Biol. Chem. 279, 48389–48396 10.1074/jbc.M409935200 [DOI] [PubMed] [Google Scholar]
- 25. Barski O. A., Xie Z., Baba S. P., Sithu S. D., Agarwal A., Cai J., Bhatnagar A., and Srivastava S. (2013) Dietary carnosine prevents early atherosclerotic lesion formation in apolipoprotein E-null mice. Arterioscler. Thromb. Vasc. Biol. 33, 1162–1170 10.1161/ATVBAHA.112.300572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu X., and Sayre L. M. (2007) Long-lived 4-oxo-2-enal-derived apparent lysine Michael adducts are actually the isomeric 4-ketoamides. Chem. Res. Toxicol. 20, 165–170 10.1021/tx600295j [DOI] [PubMed] [Google Scholar]
- 27. Zhu X., and Sayre L. M. (2007) Mass spectrometric evidence for long-lived protein adducts of 4-oxo-2-nonenal. Redox Rep. 12, 45–49 10.1179/135100007X162176 [DOI] [PubMed] [Google Scholar]
- 28. Sayre L. M., Lin D., Yuan Q., Zhu X., and Tang X. (2006) Protein adducts generated from products of lipid oxidation: Focus on HNE and one. Drug Metab. Rev. 38, 651–675 10.1080/03602530600959508 [DOI] [PubMed] [Google Scholar]
- 29. Steinmetz A., and Utermann G. (1985) Activation of lecithin: Cholesterol acyltransferase by human apolipoprotein A-IV. J. Biol. Chem. 260, 2258–2264 [PubMed] [Google Scholar]
- 30. Ostos M. A., Conconi M., Vergnes L., Baroukh N., Ribalta J., Girona J., Caillaud J. M., Ochoa A., and Zakin M. M. (2001) Antioxidative and antiatherosclerotic effects of human apolipoprotein A-IV in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 21, 1023–1028 10.1161/01.ATV.21.6.1023 [DOI] [PubMed] [Google Scholar]
- 31. Qin X., Swertfeger D. K., Zheng S., Hui D. Y., and Tso P. (1998) Apolipoprotein AIV: A potent endogenous inhibitor of lipid oxidation. Am. J. Physiol. 274, H1836–H1840 [DOI] [PubMed] [Google Scholar]
- 32. Borja M. S., Zhao L., Hammerson B., Tang C., Yang R., Carson N., Fernando G., Liu X., Budamagunta M. S., Genest J., Shearer G. C., Duclos F., and Oda M. N. (2013) HDL-apoA-I exchange: rapid detection and association with atherosclerosis. PLoS One 8, e71541 10.1371/journal.pone.0071541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amarnath V., and Amarnath K. (2015) Scavenging 4-oxo-2-nonenal. Chem. Res. Toxicol. 28, 1888–1890 10.1021/acs.chemrestox.5b00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Doorn J. A., and Petersen D. R. (2003) Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chem. Biol. Interact. 143–144, 93–100 10.1016/s0009-2797(02)00178-3 [DOI] [PubMed] [Google Scholar]
- 35. Cavigiolio G., Geier E. G., Shao B., Heinecke J. W., and Oda M. N. (2010) Exchange of apolipoprotein A-I between lipid-associated and lipid-free states: A potential target for oxidative generation of dysfunctional high density lipoproteins. J. Biol. Chem. 285, 18847–18857 10.1074/jbc.M109.098434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du X. M., Kim M. J., Hou L., Le Goff W., Chapman M. J., Van Eck M., Curtiss L. K., Burnett J. R., Cartland S. P., Quinn C. M., Kockx M., Kontush A., Rye K. A., Kritharides L., and Jessup W. (2015) HDL particle size is a critical determinant of ABCA1-mediated macrophage cellular cholesterol export. Circ. Res. 116, 1133–1142 10.1161/CIRCRESAHA.116.305485 [DOI] [PubMed] [Google Scholar]
- 37. Francis G. A., Mendez A. J., Bierman E. L., and Heinecke J. W. (1993) Oxidative tyrosylation of high density lipoprotein by peroxidase enhances cholesterol removal from cultured fibroblasts and macrophage foam cells. Proc. Natl. Acad. Sci. U.S.A. 90, 6631–6635 10.1073/pnas.90.14.6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang W. Q., Merriam D. L., Moses A. S., and Francis G. A. (1998) Enhanced cholesterol efflux by tyrosyl radical-oxidized high density lipoprotein is mediated by apolipoprotein AI-AII heterodimers. J. Biol. Chem. 273, 17391–17398 10.1074/jbc.273.28.17391 [DOI] [PubMed] [Google Scholar]
- 39. Cavaillon J. M., Fitting C., Haeffner-Cavaillon N., Kirsch S. J., and Warren H. S. (1990) Cytokine response by monocytes and macrophages to free and lipoprotein-bound lipopolysaccharide. Infect. Immun. 58, 2375–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ulevitch R. J., Johnston A. R., and Weinstein D. B. (1979) New function for high density lipoproteins. Their participation in intravascular reactions of bacterial lipopolysaccharides. J. Clin. Invest. 64, 1516–1524 10.1172/JCI109610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ulevitch R. J., Johnston A. R., and Weinstein D. B. (1981) New function for high density lipoproteins. Isolation and characterization of a bacterial lipopolysaccharide-high density lipoprotein complex formed in rabbit plasma. J. Clin. Invest. 67, 827–837 10.1172/JCI110100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Levine D. M., Parker T. S., Donnelly T. M., Walsh A., and Rubin A. L. (1993) In vivo protection against endotoxin by plasma high density lipoprotein. Proc. Natl. Acad. Sci. U.S.A. 90, 12040–12044 10.1073/pnas.90.24.12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu D., Ji L., Zhao M., Wang Y., Guo Y., Li L., Zhang D., Xu L., Pan B., Su J., Xiang S., Pennathur S., Li J., Gao J., Liu P., Willard B., and Zheng L. (2018) Lysine glycation of apolipoprotein A-I impairs its anti-inflammatory function in type 2 diabetes mellitus. J. Mol. Cell. Cardiol. 122, 47–57 10.1016/j.yjmcc.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 44. Massamiri T., Tobias P. S., and Curtiss L. K. (1997) Structural determinants for the interaction of lipopolysaccharide binding protein with purified high density lipoproteins: Role of apolipoprotein A-I. J. Lipid Res. 38, 516–525 [PubMed] [Google Scholar]
- 45. Wurfel M. M., Hailman E., and Wright S. D. (1995) Soluble CD14 acts as a shuttle in the neutralization of lipopolysaccharide (LPS) by LPS-binding protein and reconstituted high density lipoprotein. J. Exp. Med. 181, 1743–1754 10.1084/jem.181.5.1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uboldi P., Spoladore M., Fantappiè S., Marcovina S., and Catapano A. L. (1996) Localization of apolipoprotein A-I epitopes involved in the activation of lecithin:cholesterol acyltransferase. J. Lipid Res. 37, 2557–2568 [PubMed] [Google Scholar]
- 47. Shao B., Fu X., McDonald T. O., Green P. S., Uchida K., O'Brien K. D., Oram J. F., and Heinecke J. W. (2005) Acrolein impairs ATP binding cassette transporter A1-dependent cholesterol export from cells through site-specific modification of apolipoprotein A-I. J. Biol. Chem. 280, 36386–36396 10.1074/jbc.M508169200 [DOI] [PubMed] [Google Scholar]
- 48. Shao B. (2012) Site-specific oxidation of apolipoprotein A-I impairs cholesterol export by ABCA1, a key cardioprotective function of HDL. Biochim. Biophys. Acta 1821, 490–501 10.1016/j.bbalip.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chroni A., Liu T., Gorshkova I., Kan H. Y., Uehara Y., Von Eckardstein A., and Zannis V. I. (2003) The central helices of ApoA-I can promote ATP-binding cassette transporter A1 (ABCA1)-mediated lipid efflux. Amino acid residues 220–231 of the wild-type ApoA-I are required for lipid efflux in vitro and high density lipoprotein formation in vivo. J. Biol. Chem. 278, 6719–6730 10.1074/jbc.M205232200 [DOI] [PubMed] [Google Scholar]
- 50. Panagotopulos S. E., Witting S. R., Horace E. M., Hui D. Y., Maiorano J. N., and Davidson W. S. (2002) The role of apolipoprotein A-I helix 10 in apolipoprotein-mediated cholesterol efflux via the ATP-binding cassette transporter ABCA1. J. Biol. Chem. 277, 39477–39484 10.1074/jbc.M207005200 [DOI] [PubMed] [Google Scholar]
- 51. Natarajan P., Forte T. M., Chu B., Phillips M. C., Oram J. F., and Bielicki J. K. (2004) Identification of an apolipoprotein A-I structural element that mediates cellular cholesterol efflux and stabilizes ATP binding cassette transporter A1. J. Biol. Chem. 279, 24044–24052 10.1074/jbc.M400561200 [DOI] [PubMed] [Google Scholar]
- 52. Zhang W. H., Liu J., Xu G., Yuan Q., and Sayre L. M. (2003) Model studies on protein side chain modification by 4-oxo-2-nonenal. Chem. Res. Toxicol. 16, 512–523 10.1021/tx020105a [DOI] [PubMed] [Google Scholar]
- 53. Amarnath V., Amarnath K., Avance J., Stec D. F., and Voziyan P. (2015) 5′-O-alkylpyridoxamines: Lipophilic analogues of pyridoxamine are potent scavengers of 1,2-dicarbonyls. Chem. Res. Toxicol. 28, 1469–1475 10.1021/acs.chemrestox.5b00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zagol-Ikapitte I., Amarnath V., Bala M., Roberts L. J. 2nd, Oates J. A., and Boutaud O. (2010) Characterization of scavengers of γ-ketoaldehydes that do not inhibit prostaglandin biosynthesis. Chem. Res. Toxicol. 23, 240–250 10.1021/tx900407a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yermalitsky V. N., Matafonova E., Tallman K., Li Z., Zackert W., Roberts L. J. 2nd, Amarnath V., and Davies S. S. (2019) Simplified LC/MS assay for the measurement of isolevuglandin protein adducts in plasma and tissue samples. Anal. Biochem. 566, 89–101 10.1016/j.ab.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 56. Bonomo E. A., and Swaney J. B. (1988) A rapid method for the synthesis of protein-lipid complexes using adsorption chromatography. J. Lipid Res. 29, 380–384 [PubMed] [Google Scholar]
- 57. Cooke A. L., Morris J., Melchior J. T., Street S. E., Jerome W. G., Huang R., Herr A. B., Smith L. E., Segrest J. P., Remaley A. T., Shah A. S., Thompson T. B., and Davidson W. S. (2018) A thumbwheel mechanism for APOA1 activation of LCAT activity in HDL. J. Lipid Res. 59, 1244–1255 10.1194/jlr.M085332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Low H., Hoang A., and Sviridov D. (2012) Cholesterol efflux assay. J. Vis. Exp., 61, e3810 10.3791/3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Robinet P., Wang Z., Hazen S. L., and Smith J. D. (2010) A simple and sensitive enzymatic method for cholesterol quantification in macrophages and foam cells. J. Lipid Res. 51, 3364–3369 10.1194/jlr.D007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lerch P. G., Förtsch V., Hodler G., and Bolli R. (1996) Production and characterization of a reconstituted high density lipoprotein for therapeutic applications. Vox Sang. 71, 155–164 10.1046/j.1423-0410.1996.7130155.x [DOI] [PubMed] [Google Scholar]
- 61. Melchior J. T., Street S. E., Andraski A. B., Furtado J. D., Sacks F. M., Shute R. L., Greve E. I., Swertfeger D. K., Li H., Shah A. S., Lu L. J., and Davidson W. S. (2017) Apolipoprotein A-II alters the proteome of human lipoproteins and enhances cholesterol efflux from ABCA1. J. Lipid Res. 58, 1374–1385 10.1194/jlr.M075382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. González Fernández-Niño S. M., Smith-Moritz A. M., Chan L. J., Adams P. D., Heazlewood J. L., and Petzold C. J. (2015) Standard flow liquid chromatography for shotgun proteomics in bioenergy research. Front. Bioeng. Biotechnol. 3, 44 10.3389/fbioe.2015.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry data is publicly accessible through Figshare online repository: https://figshare.com/articles/Zhang_et_al_JBC_MS_Data_10_13_2019/9975560 under project IDs Zhang Set 1 Run 1 3 eq ONE, Zhang Set 1 Run 2 3 eq ONE 2A, Zhang Set 1 Run 2 3 eq ONE 2B, and Zhang Set 2 3 eq ONE.3