Abstract
The tnaC regulatory gene from the tna operon of Escherichia coli controls the transcription of its own operon through an attenuation mechanism relying on the accumulation of arrested ribosomes during inhibition of its own translation termination. This free l-Trp–dependent mechanism of inhibition of translation termination remains unclear. Here, we analyzed the inhibitory effects of l-Trp on the function of two known E. coli translation termination factors, RF1 and RF2. Using a series of reporter genes, we found that the in vivo l-Trp sensitivity of tnaC gene expression is influenced by the identity of its stop codon, with the UGA stop codon producing higher expression efficiency of the tnaA-lacZ gene construct than the UAG stop codon. In vitro TnaC-peptidyl-tRNA accumulation and toe-printing assays confirmed that in the presence of l-Trp, the UGA stop codon generates higher accumulation of both TnaC-peptidyl-tRNA and arrested ribosomes than does the UAG stop codon. RF-mediated hydrolysis assays corroborated that l-Trp blocks RF2 function more than that of RF1. Mutational analyses disclosed that amino acids substitutions at the 246 and 256 residue positions surrounding the RF2-GGQ functional motif reduce l-Trp–dependent expression of the tnaC(UGA) tnaA-lacZ construct and the ability of l-Trp to inhibit RF2-mediated cleavage of the TnaC-peptidyl-tRNA. Altogether, our results indicate that l-Trp preferentially blocks RF2 activity during translation termination of the tnaC gene. This inhibition depends on the identities of amino acid residues surrounding the RF2-GGQ functional motif.
Keywords: translation release factor, tryptophan, ribosome function, translation regulation, protein synthesis, gene regulation, translation control, GGQ tripeptide motif, peptidyl transferase center, regulatory nascent peptides, ribosomal arrest, tnaC, release factor, L-tryptophan, translation termination
Introduction
Translation termination in bacteria is initiated by a couple of protein paralogs named release factor 1 (RF1)5 and 2 (RF2). Once bound to the ribosome, these RF proteins promote hydrolysis of the resident peptidyl-tRNA by aiding in the accommodation of a molecule of water at the ribosome active site, also known as the peptidyl transferase center (PTC) (1, 2). RF1 or RF2 initiates translation termination by recognizing distinct stop codons (UAA/UAG for RF1; UAA/UGA for RF2) located at the small subunit ribosomal A-site decoding center (3). Once these release factors recognize their corresponding stop codons, they change their free-state compact conformation (closed conformation) to an extended conformation (open conformation) where the tip of RF1/RF2 domain 3 reaches the PTC (4). The tip of the domain 3 of both proteins contains a conserved GGQ tripeptide motif whose methylated glutamine residue presumably aids in the positioning of a molecule of water (1, 5–7). Accommodation of the tip of domain 3, suggested to be the rate-limiting step during hydrolysis of the peptidyl-tRNA, requires several structural changes in the release factor protein as well as in the PTC (8–10). RF2 promotes hydrolysis of peptidyl-tRNA three to six times more efficiently than RF1 (11). After hydrolysis of the peptidyl-tRNA, RF1 resides within the ribosomal complex until it dissociates from the ribosome with the help of release factor 3 (RF3), whereas RF2 can independently dissociate itself from the ribosomes (12). RF1 protein has shown longer residence time within the ribosome than RF2, explaining the need for the release action of RF3 (12), indicating that RF1 has stronger affinity for the ribosome than RF2 (13). Translation termination is completed when the ribosome release factor interacts at the empty A-site, and along with elongation factor G disassembles the translating ribosome separating the two subunits from the translated mRNA and the last deacylated tRNA (14).
The tnaCAB (tna) operon is a bacterial chromosomal unit whose function is related to the degradation of external l-tryptophan (l-Trp) and the formation of indole, a biofilm, and quorum-sensing regulatory metabolite (15). The tna operon is constituted by a 5′-regulatory leader region and two structural genes tnaA and tnaB, which express the tryptophanase enzyme and an l-Trp–specific transporter, respectively. The 5′-regulatory leader region of the tna operon contains a small ORF, tnaC, and a Rho-dependent transcription termination sequence. Transcription of the tna operon is induced primarily by the absence of a rich carbon source (16). Transcription coupled to translational expression of tnaC, which synthesizes the TnaC regulatory nascent peptide, allows RNA polymerase to continue synthesizing mRNA downstream of the tnaC gene. In Escherichia coli, under limiting concentrations of l-Trp, translation of the tnaC gene terminates at its UGA stop codon, releasing translating ribosomes from the nascent mRNA (17). The release of the ribosomes exposes a rut site that overlaps the tnaC stop codon, allowing Rho to terminate transcription before reaching the first structural gene, tnaA. Under activating concentrations of l-Trp, ribosomes translating tnaC are arrested at its UGA stop codon, blocking the rut site and action of Rho and allowing transcription of the structural genes (18, 19). Previous in vivo studies have shown that the expression of the tna structural genes is affected by the identity of the tnaC stop codon (20). Under the presence of activating concentrations of l-Trp, tnaC genes with either UAG or UAA codons induced less expression of tnaA'-'lacZ reporter genes than tnaC genes with UGA codons (20). Where 85 and 50% reporter gene expression reduction were observed after replacing the tnaC UGA stop codon with UAG and UAA stop codons, respectively (20). These observations suggest that the degree of inhibition of translation termination by l-Trp at the tnaC sequences may depend on the identity and kinetic characteristics of the release factor involved, with RF2, which recognizes the UGA stop codon, being more susceptible to l-Trp-induced inhibition of peptide release than RF1, which recognizes the UAG stop codon. Notably, these previous results were obtained in E. coli K12 strains that express a RF2 Thr-246 variant, a less active protein compared with the RF2 Ala-246 variant present in E. coli B strains (6). Unfortunately, there are no studies of the expression of the tna operon under the action of the more active versions of RF2.
Biochemical and structural analyses have shown that arrested ribosomes at the tnaC sequences contain TnaC-peptidyl-tRNAPro molecules in their P-sites (21, 22). These studies also indicate that the TnaC nascent peptide acquires a specific conformation, induced by its interactions with the exit tunnel and free l-Trp molecules (22–25). Once l-Trp is removed, TnaC-tRNAPro molecules within the arrested ribosomes become sensitive to the hydrolysis action of RF2, indicating that l-Trp is required for the inhibition of the activity of RF2 at the PTC (26). Several studies indicate that conformational changes in the PTC induced through the TnaC nascent peptide and the exit tunnel components are important for inhibiting RF2 (22). Cryo-EM structures of the ribosome in complex with the stalled TnaC nascent peptide show that the PTC nucleotide residues U2585 and A2602 acquire a conformational distribution that would clash with the amino acid residues at the tip of the domain 3 of RF2, likely hindering their proper positioning or accommodation within the ribosome (22, 25). Even though the structural studies revealed unfavorable conformational changes of PTC nucleotides, it remained unclear why the TnaC-mediated stalling could preferentially affect the function of RF2 over that of RF1, when their conserved GGQ motif acts in the same region of the ribosome during peptidyl-tRNA hydrolysis.
In this work, we aimed to determine why the function of RF2 in translation termination is more susceptible to the inhibitory action of the TnaC nascent peptide and l-Trp in the ribosomal termination complex as compared with the function of RF1. We performed genetic and biochemical assays to study the effects of RF1 and RF2 protein variants on the expression of the tna operon, accumulation of arrested ribosomes, and cleavage of TnaC-tRNAPro2 molecules in the presence of l-Trp. Our results indicate that distinct RF2 residues located at the PTC during translation termination make this protein more susceptible than RF1 to translation termination inhibition by the TnaC nascent peptide and l-Trp.
Results
The identity of the tnaC stop codon is associated with the sensitivity to detect the inducer 1-methyl-l-Trp during the expression of the tna operon
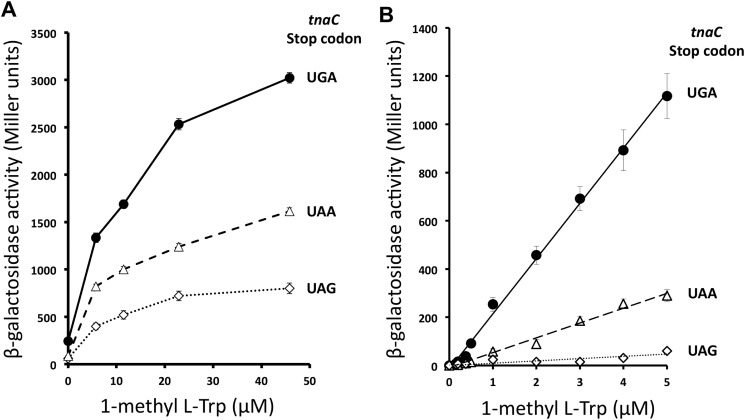
Previous experiments that compared the expression of tnaC tnaA'-'lacZ reporter genes containing variations in their tnaC stop codon (20) were performed under conditions where cellular l-Trp concentrations are affected by the expression of the tryptophanase enzyme (30), making it difficult to compare the sensory capacity of these reporter genes for free l-Trp. We decided to analyze the in vivo expression of these tnaC tnaA'-'lacZ reporter gene variants using 1M-l-Trp as an inducer, because this analog of l-Trp can induce the expression of the tna operon without being metabolized by tryptophanase (27). Bacterial cultures for each strain, SVS1144 (tnaC-UGA tnaA'-'lacZ) (28), VK800 (tnaC-UAG tnaA'-'lacZ) (20), and PDG1144 (tnaC-UAA tnaA'-'lacZ) (29), were grown under several concentrations of 1M-l-Trp. It is important to note that these are K12-derived bacterial strains, and they therefore express the RF2 Thr-246 protein variant (6). Bacterial pellets from the final cultures were used to determine β-gal activity (see “Experimental procedures”). Data are summarized in Fig. 1. We observed that the β-gal expression increased with increasing amounts of inducer in all three bacterial strains (Fig. 1A). At more than 40 μm 1M-l-Trp the SVS1144 (tnaC-UGA) strain generated the highest β-gal synthesis, two and five times more than the enzyme synthesis observed in the PDG1144 (tnaC-UAA) and VK800 (tnaC-UAG) strains, respectively. The graphs in Fig. 1A show a sharp increase in the enzyme expression between 0 and 5 μm 1M-l-Trp, which was previously observed with the SVS1144 strain (30). After testing the range between 0 and 5 μm 1M-l-Trp, we observed a linear correlation between the expression of the enzyme and the concentration of inducer (Fig. 1B). We calculated that the reporter gene of the SVS1144 (tnaC-UGA) strain produced an expression efficiency of 230 Miller units of β-gal activity per 1 μm 1M-l-Trp, four times greater than the expression efficiency of the reporter gene of the PDG1144 (tnaC-UAA) strain and 20 times higher than the expression efficiency of the reporter gene of the VK800 (tnaC-UAG) strain. These results indicate that the reporter gene with a tnaC-UGA sequence is more sensitive to 1M-l-Trp than the constructs containing a tnaC sequence with either a UAA or a UAG codon. These data also suggest that 1M-l-Trp could be more efficient in blocking translation termination at the RF2-specific UGA codon than the RF1/2 UAA and RF1-specific UAG codons.
Figure 1.
Expression of tnaC tnaA'-'lacZ reporter gene variants under several concentrations of 1-methyl-l-Trp. A and B, bacterial cells with tnaA'-'lacZ reporter genes controlled by tnaC genes with different stop codons (indicated in the figure) were grown in minimal media under different concentrations of 1-methyl-l-Trp, 0–45 μm (A) and 0–5 μm (B). β-gal activity was tested in the final cultures as indicated under “Experimental procedures.” The following strains were used in these experiments: SVS1144 strain carrying the tnaC(UGA) tnaA'-'lacZ construct (closed circles), PDG1158 strain with the tnaC(UAA) tnaA'-'lacZ construct (open triangles), and VK800 strain containing the tnaC(UAG) tnaA'-'lacZ construct (open diamonds). These results represent four independent experiments with a corresponding replica.
l-Trp preferentially induces accumulation of stalled ribosomes at the tnaC UGA codon
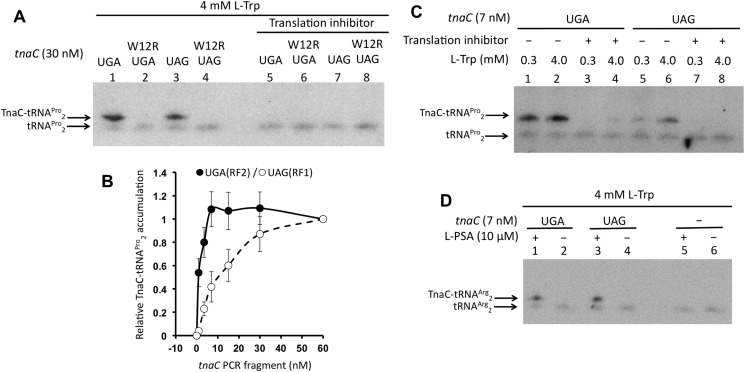
The results shown in Fig. 1 suggest that l-Trp–induced ribosome arrest at the tnaC sequences could be more efficient if the tnaC stop codon is UGA rather than UAA or UAG. To determine whether such an argument could be true, we decided to analyze the formation in vitro of l-Trp–arrested ribosomes within the tnaC sequences containing variations at their stop codon positions. We used a reconstituted protein synthesis system obtained from E. coli K12 strains where T7 RNA polymerase transcribes supplied DNA fragments (PURExpress from New England Biolabs). The reconstituted protein synthesis system contains the RF2 Thr-246 protein variant. Reactions were performed using PCR fragments that contained the tnaC gene variants, where transcription is controlled by a T7 RNA polymerase promoter, and translation is enhanced by an omega sequence located upstream of the tnaC-Shine-Dalgarno ribosome-binding sequence (30). Northern blotting assays were performed to detect the accumulation of TnaC-peptidyl-tRNAPro2 (see “Experimental procedures”). Accumulation of TnaC-peptidyl-tRNAPro2 indicates that translation termination is inhibited during the expression of tnaC, generating arrested ribosomes at its stop codon. As seen in Fig. 2A, in the presence of 4 mm l-Trp, both 30 nm UGA and UAG tnaC-PCR fragments induced accumulation of TnaC-peptidyl-tRNAPro2 molecules (see lanes 1 and 3). To confirm that the detected signal is a product of the inhibitory action of l-Trp and the regulatory TnaC nascent peptide we used tnaC-PCR fragments containing a change at the tnaC Trp-12 codon (W12R). This amino acid substitution generates a nonfunctional TnaC nascent peptide that cannot produce ribosome arrest under activating concentrations of l-Trp (18). We did not observe accumulation of TnaC-peptidyl-tRNAPro2 under the same conditions when the tnaC (W12R)-PCR fragments UGA and UAG were used for the reactions (compare lane 1 with 2 or lane 3 with 4), or when a translation inhibitor was added to the reactions (compare lanes 1 and 3 with lanes 5 and 7, respectively). These last sets of data suggest that such TnaC-peptidyl-tRNAPro2 accumulation depends on the amino acid composition of the TnaC nascent peptide and on the translation of both tnaC variants, UGA and UAG.
Figure 2.
In vitro accumulation of peptidyl-tRNA during the expression of tnaC variants. Northern blotting assays were performed on the resolved final products of in vitro reactions (see “Experimental procedures”). A, in vitro translation reactions were performed with the indicated tnaC PCR fragments in the presence of high l-Trp concentrations (4 mm). To reveal the TnaC-tRNAPro2 signals, 50 μm Borrelidin (translation inhibitor), which inhibits the synthesis of Threonyl-tRNA inducing translation arrest before reaching the tnaC Pro-24 codon, was added to the reactions 10 min prior the addition of the PCR fragments. B, plot showing relative accumulation of TnaC-peptidyl-tRNAPro2 with respect concentration of tnaC PCR fragments used in the reactions. Three independent experiments were performed with concentrations of tnaC PCR fragments 0, 3, 7, 15, 30, and 60 nm. The six reactions for each experiment were run and resolved in parallel, and their relative accumulation of TnaC-tRNAPro2 determined in the same membrane. The TnaC-tRNAPro2 band intensity obtained from the reaction using 60 nm of tnaC PCR fragment was considered for each experiment as maximum accumulation, or 1. Band intensity values obtained for each PCR concentrations were divided by the maximum accumulation value to obtain their relative accumulation value. Average of each relative accumulation value of three experiments and their error bars are shown in the graph. tnaC-UGA (closed circles) and tnaC-UAG (open circles) PCR fragments were used in these experiments. C, reactions were performed with low (0.3 mm) or high (4 mm) l-Trp with the indicated tnaC PCR fragments. 50 μm Borrelidin was added (+), or not (−), as indicated in panel A. D, reactions containing high concentrations of l-Trp were performed by adding (+) or not (−) l-PSA to inhibit the synthesis of prolyl-tRNA 10 min prior the addition of the indicated tnaC PCR fragments. As a control, reactions without addition of PCR fragments were run in parallel.
We tested l-Trp–induced accumulation of TnaC-peptidyl-tRNAPro2 using several concentrations of tnaC-PCR fragments ranging from 0 to 60 nm to compare the capacity of both tnaC UGA and UAG variants for accumulating arrested ribosomes under high concentrations of l-Trp (4 mm). We observed that both tnaC variants generated maximum accumulation of TnaC-peptidyl-tRNAPro2 at DNA concentrations higher than 30 nm (Fig. 2B). We suspect that in the presence of 4 mm l-Trp, high concentrations of the tnaC PCR fragments could reduce the pool of tRNAPro2 limiting the production of stalled ribosomes, generating the plateau behavior observed in the curve obtained with the tnaC UGA PCR fragments (Fig. 2B, closed circles). Depletion of the cellular pool of tRNAPro2 has been seen in vivo during overexpression of the tnaC UGA gene (31). At concentrations lower than 30 nm, however, the tnaC-UAG DNA variant generated lesser amounts of TnaC-peptidyl-tRNAPro2 than the tnaC-UGA variant. The biggest differences between both variants were seen at concentrations lower than 7 nm, where the tnaC-UGA variant still reached maximum accumulation, whereas the tnaC-UAG variant produced roughly 40% of maximum accumulation. Therefore, we decided to do further in vitro tests using 7 nm for the two tnaC-PCR fragment variants. To determine the differences in l-Trp sensitivity of both constructs, we tested accumulation of TnaC-tRNAPro2 under two concentrations of l-Trp, 0.3 mm (low) and 4 mm (high), which have shown significant differences in in vitro accumulation of arrested ribosomes (27). We observed that at 7 nm concentrations both the tnaC-UGA and tnaC–UAG DNA variants accumulated TnaC-peptidyl-tRNAPro2 under both high and low l-Trp concentrations (Fig. 2C). At high l-Trp concentrations the tnaC-UGA DNA generated twice the amount of TnaC-peptidyl-tRNAPro2 molecules than at low l-Trp concentrations. Similarly, at high l-Trp concentrations the tnaC-UAG variant generated four times more TnaC-peptidyl-tRNAPro2 molecules than at low l-Trp concentrations. Both results indicate that each of these tnaC variants are still sensitive to changes in l-Trp concentrations. However, the tnaC-UGA DNA variant accumulates more TnaC-peptidyl-tRNAPro2 than the tnaC-UAG DNA variant at either l-Trp concentrations (Fig. 2C, compare lanes 1 and 2 with lanes 5 and 6). At high l-Trp concentrations, the tnaC-UGA DNA variant accumulates four times more TnaC-peptidyl-tRNAPro2 molecules than the tnaC-UAG DNA variant (Fig. 2C, compare lane 2 with lane 6). Meanwhile, at low l-Trp concentrations, the tnaC-UGA DNA variant accumulates eight times more TnaC-peptidyl-tRNAPro2 molecules than the tnaC-UAG DNA variant (Fig. 2C, compare lane 1 with lane 5). These results suggest that the tnaC-UGA variant is more efficient at accumulating arrested ribosomes than the tnaC-UAG variant at the l-Trp concentrations described above. To confirm that both tnaC DNA variants are transcribed and translated similarly, we induced the accumulation of TnaC-peptidyl-tRNAArg2 molecules at the tnaC Arg-23 codon position, the codon prior to the l-Trp–dependent ribosome-arresting position. We used an inhibitor of the l-prolyl-tRNA synthase, 5′-O-(N-(l-prolyl)-sulfamoyl)-adenosine (l-PSA), which action will reduce the pool of available prolyl-tRNA (32). As seen in Fig. 2D, the presence of l-PSA induced accumulation of TnaC-peptidyl-tRNAArg2 molecules using both tnaC DNA variants (compare lane 1 with lane 2, and lane 3 with lane 4). We did not observe significant differences in the amount of TnaC-peptidyl-tRNAArg2 accumulated at both tnaC DNA variants (compare lane 1 with lane 3), which suggests that these tnaC DNA variants have similar capacity for being transcribed and translated under our in vitro protein synthesis system. Therefore, we propose that the differences in accumulation of TnaC-peptidyl-tRNAPro2 observed between the tnaC-UGA and tnaC-UAG DNA variants are because of their sensitivity to l-Trp.
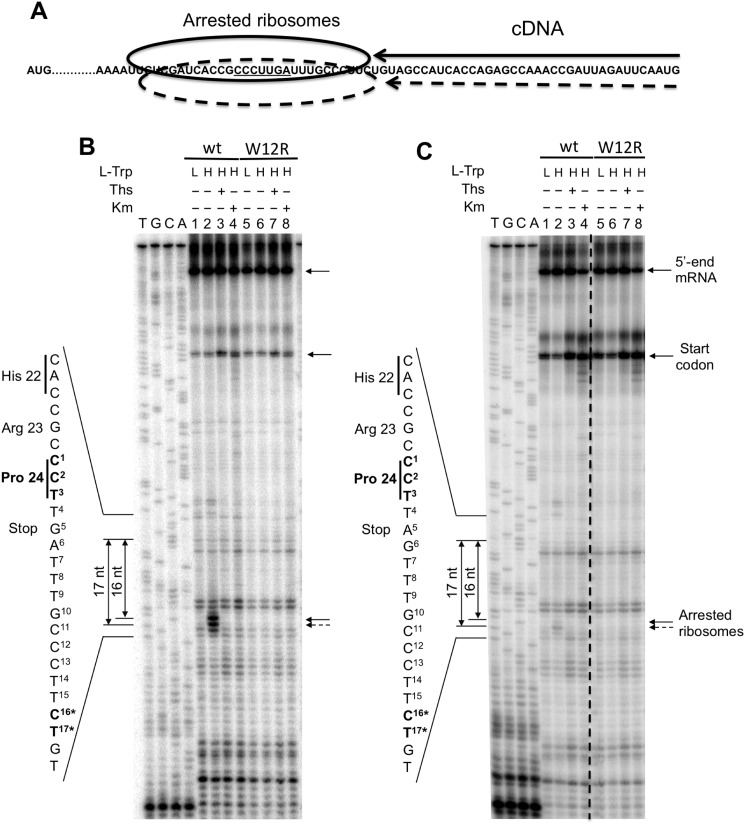
Toe-printing assays were used to confirm the accumulation of arrested ribosomes at the stop codon of the tnaC ORF (see “Experimental procedures”). In these assays, arrested ribosomes are detected during the production of cDNA where the progression of retrotranscriptase is inhibited by the stalled ribosomal complex. This allows for precise localization of the ribosomes at the translated mRNA (Fig. 3A). For these reactions, we used 5 nm of each tnaC DNA variants, a concentration that showed significant difference in TnaC-peptidyl-tRNAPro2 accumulation between both tnaC-UGA and tnaC-UAG DNA variants (Fig. 2B). We observed that the expression of the tnaC-UGA variant produced two toe-printing signals under the presence of high concentrations (4 mm) of l-Trp. These signals were located 16 and 17 nucleotides downstream of the last tnaC proline codon indicating the presence of arrested ribosomes at this tnaC codon. These arrested ribosome signals were not observed in reactions performed under low concentrations (0.3 mm) of l-Trp (Fig. 3B, compare lane 2 with lane 1), as well as in the reactions using nonfunctional mutant tnaC-UGA W12R variants under the presence of 4 mm l-Trp (Fig. 3B, compare lane 6 with lane 2). These data indicate that during in vitro expression of the tnaC-UGA variant, high concentrations of l-Trp induce accumulation of arrested ribosomes at the tnaC Pro-24 codon. These events depend on the nature of the TnaC nascent peptide, because replacements of its conserved Trp-12 residue abolished the accumulation of arrested ribosome signals. Similar experiments were performed with the tnaC-UAG variant (Fig. 3C). In this case, the accumulation of both arrested ribosome signals was also observed under 4 mm l-Trp but not under 0.3 mm l-Trp (Fig. 3C, compare lane 2 with lane 1). Accumulation of arrested ribosome signals was not observed in the corresponding tnaC-UAG variant containing the W12R replacement at both l-Trp concentrations (Fig. 3C, compare lane 6 with lane 2). However, using three independent in vitro experiments we calculated (see “Experimental procedures”) that under high concentrations of l-Trp the accumulation of arrested ribosomes produced with the tnaC-UGA variant was approximately four times higher than the accumulation observed with the tnaC-UAG variant. These results suggest that under high l-Trp concentrations the tnaC-UGA gene variant is more efficient at accumulating arrested ribosomes at their Pro-24 codon position than the tnaC-UAG gene variant.
Figure 3.
In vitro accumulation of arrested ribosomes during the expression of tnaC variants. A, diagram representing the final cDNA products produced by arrested ribosomes during toe-printing analysis. The arrows indicate the cDNA products that terminate once they encounter an arrested ribosome. B and C, toe-printing assays were performed using WT and mutant W12R tnaC DNA fragments which stop codon were either UGA (B) or UAG (C). Reactions were performed under 0.3 mm (L) or 4 mm (H) l-Trp, in the presence (+) or in the absence (−) of kanamycin (Km) or thiostreptone (Ths). Km and Ths are used to reveal signals that are dependent on the presence of arrested ribosomes. Final products were resolved as indicated under “Experimental procedures.” The dotted line on the image indicates a place where the original gel was spliced to deleted one lane containing repetitive data. Signals corresponding to the 5′-end of tnaC transcripts, arrested-ribosomes at the tnaC start codon, and arrested-ribosomes induced by l-Trp are indicated with arrows. The sequences of the 3′-end, the last three codons, and the stop codon of tnaC are shown in the left side of each panel. The nucleotide distances between the tnaC proline 24th codon and the l-Trp–dependent arrested signals are also indicated; nucleotide identities are indicated with asterisks in the left side sequence. These panels represent two independent experiments.
RF1 induces cleavage of TnaC-tRNAPro2 more efficiently compared with RF2 in the presence of l-Trp
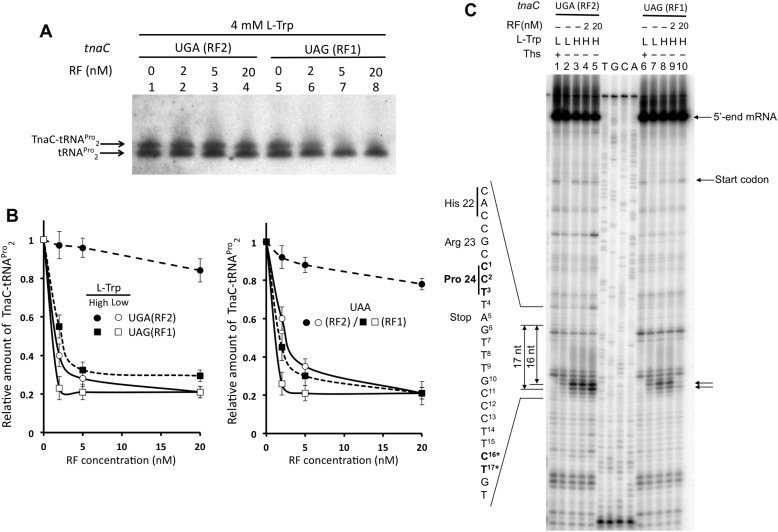
Data shown above (Fig. 2C) suggest that l-Trp has varying effects on the cleavage of TnaC-tRNAPro2, effects dependent upon whether the cleavage is induced by either RF1 or RF2. To determine such differences, we performed TnaC-tRNAPro2-cleavage assays using a reconstituted system nearly devoid of RF1, RF2 Thr-246, and RF3 proteins (PURExpress ΔRF123) to reduce background activity of these proteins. We initially allowed translation of tnaC (UGA or UAG) variants to proceed in the presence of high l-Trp concentrations to induce accumulation of arrested ribosomes and by consequence accumulation of TnaC-tRNAPro2 molecules. Reaction mixes were then supplemented with variable concentrations of either RF2 Thr-246 or RF1 (0, 2, 5, and 20 nm) along with 25 nm RF3. Final products of the reactions were then detected by Northern blotting (see “Experimental procedures”), an example can be seen at Fig. 4A. We observed that RF2 Thr-246 was less efficient than RF1 at inducing cleavage of accumulated TnaC-tRNAPro2 in the presence of l-Trp. 20 nm RF2 Thr-246 induced the cleavage of only 10% of the accumulated peptidyl-tRNA at the UGA stop codon under high l-Trp concentrations (Fig. 4B, left plot, closed circles). In contrast to the data obtained for RF2 Thr-246, accumulated TnaC-tRNAPro2 at the UAG stop codon was cleaved ∼45 and ∼70% using 2 and 20 nm RF1, respectively (Fig. 4B, check left plot, closed squares). As expected, accumulated TnaC-tRNAPro2 produced under low l-Trp concentrations at the UGA codon was 60% cleaved after adding 2 nm RF2 Thr-246, which increased up to ∼80% by adding higher RF2 Thr-246 concentrations (20 nm) (Fig. 4B, check left plot, open circles). Similar to the RF2 experiments, accumulated TnaC-tRNAPro2 produced under low l-Trp at the UAG codon was cleaved to a greater extent at lower and higher concentrations of RF1. We observed 80% cleavage using both 2 and 20 nm RF1 (Fig. 4B, check left plot, open squares). Similar results were obtained when a tnaC UAA variant was used, where both release factors should act to induce cleavage of the accumulated TnaC-tRNAPro2 (Fig. 4B, check right plot). These sets of data indicate that RF1 is more efficient at inducing the cleavage of TnaC-tRNAPro2 than RF2 Thr-246 under high l-Trp concentrations. Toe-printing assays were performed to determine whether the RFs activities on TnaC-tRNAPro2 translate into the release of stalled ribosomes from translating mRNAs. These assays were carried out using high l-Trp concentrations, because large differences between both RFs were observed with TnaC-tRNAPro2 cleavage assays (Fig. 4A) (23). We observed that both concentrations 2 and 20 nm RF2 Thr-246 did not reduce the arrested ribosome signals generated by the tnaC UGA mRNAs (Fig. 4C, compare lanes 4 and 5 with lane 3). That was not the case for the tnaC UAG mRNA and RF1, where the highest RF1 concentrations (20 nm) greatly reduced the intensity of the arrested ribosome signals to lower levels than those observed with low l-Trp concentrations (Fig. 4C, compare lane 10 with lane 7). These last sets of data suggest that high l-Trp concentrations affect ribosome release mediated by RF2 Thr-246 significantly more than the ribosome release mediated by RF1.
Figure 4.
Release factors have different efficiencies in inducing cleavage of TnaC-tRNAPro2 and releasing arrested ribosomes in the presence of l-Trp. A, Northern blotting assay performed as indicated in Fig. 2 using a cell-free extract nearly devoid of RF1, RF2 Thr-246, and RF3 proteins. Reactions containing either tnaC UGA or UAG DNA variants were performed with high concentrations of l-Trp and variable concentrations of the indicated RF. B, plots showing relative accumulation of TnaC-peptidyl-tRNAPro2 with respective concentrations of RF proteins used in the reactions. In vitro transcription translation assays were performed using tnaC DNA variants at their indicated stop codon and variable concentrations of the indicated RF proteins 0, 2, 5, and 20 nm. Northern blotting assays were performed and relative amounts of TnaC-tRNAPro2 were obtained as indicated in Fig. 2. The plots represent the average of three independent experiments. Reactions were performed with tnaC-UGA or tnaC-UAA PCR fragments and RF2 Thr-246 (RF2) proteins (circles), and tnaC-UAG or tnaC-UAA PCR fragments and RF1 proteins (squares) under 0.3 mm (closed symbols) or 4 mm (open symbols) l-Trp concentrations. C, toe-printing assays performed with the indicated tnaC DNA variants in the presence of 0.3 mm (L) or 4 mm (H) l-Trp. Reactions were performed with the addition or not (−) of the indicated concentrations of RF2 Thr-246 (RF2) or RF1 factors.
Nonconserved residues at the tip of domain 3 of RF2 make the protein more sensitive to l-Trp inhibition
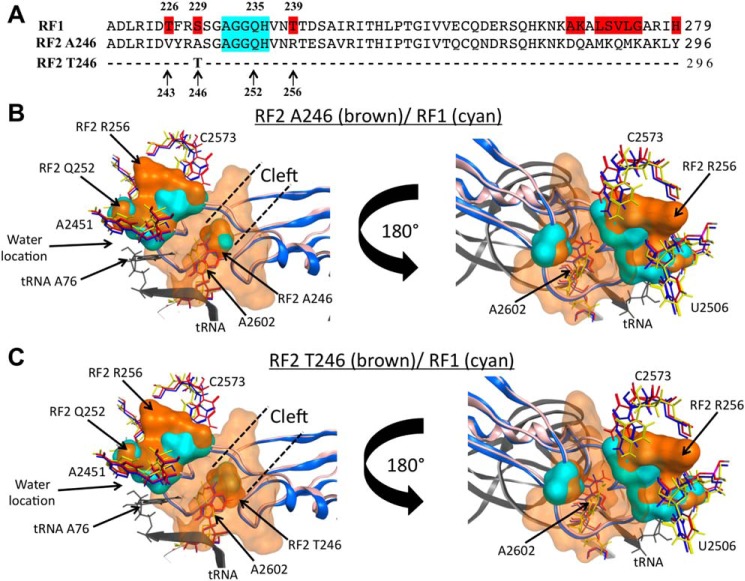
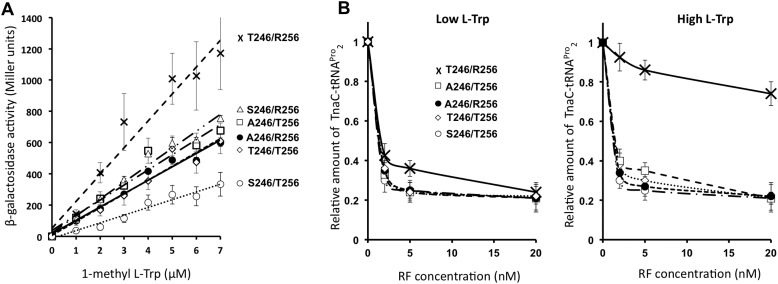
Inhibitory effects of TnaC and l-Trp are localized at the PTC region of the 50S ribosomal subunit (22, 26), where amino acid residues of the RF protein's domain 3 accommodate to assist in positioning reactive water at the PTC (33). The tips of the domain 3 of both RF2 and RF1 protein paralogs are nonconserved in three residue positions, which surround the functional GGQ motif (RF2/RF1: Val-243/Thr-226; Ala- or Thr-246/Ser-229; and Arg-256/Thr-239) (Fig. 5A). Furthermore, structural comparisons between isoforms of RF2, Ala-246 and Thr-246, and RF1 Ser-229 accommodated at the PTC have shown differences in their spatial distribution (Fig. 5, B and C). To determine whether these amino acid residues dictate the differences observed in the inhibition of the activity between RF2 and RF1, we expressed in vivo variants of RF2 with amino acid changes in both 246 and 256 residue positions to study their effects on the expression of the tnaC-UGA tnaA'-'lacZ reporter gene (Fig. 6). Importantly, the in vivo expression levels of the RF2 protein variants expressed from our plasmid constructs were highly comparable and their expression levels were ∼10 times higher than the protein produced from the chromosomal copy of the RF2 Thr-246/Arg-256 gene (Fig. S1). Cells transformed with these plasmids did not show significant differences in growth (Fig. S2) or in the expression of the lac gene from the lac operon (Fig. S3). Therefore, we expected that the overexpression of the RF2 protein variants would not alter the translation efficiency of the lacZ from our reporter gene, but rather, alter the action of tnaC sensing l-Trp. We observed that the expression efficiency of the reporter gene under the expression of the RF2 Thr-246/Arg-256 variant was 2-fold higher than the expression efficiency of the reporter gene under the expression of the RF2 Thr-246/Arg-256 protein variant (Fig. 6A, compare crosses with closed circles). These results were confirmed by comparing the expression of the reporter gene from E. coli K12 cells expressing chromosomal copies of the RF2 Thr-246/Arg-256 variant with cells expressing the RF2 Ala-246/Arg-256 variant (Fig. S4). These data indicate that the threonine residue at the 246 position of RF2 enhances the expression of the reporter gene under the presence of l-Trp. Interestingly, the expression efficiency of the reporter gene under the RF2 Ala-246/Thr-256 variant was similar to the expression efficiency observed under the RF2 Ala-246/Arg-256 variant (Fig. 6A compare open squares with closed circles). Indicating that the change R256T did not affect the expression of the reporter gene when RF2 holds an alanine residue at position 246. Surprisingly, the expression efficiency of the reporter gene under the RF2 Thr-246/Thr-256 protein variant was less than the expression observed under the RF2 Thr-246/Arg-256 variant (Fig. 6A, compare crosses with open diamonds), indicating that the R256T change in the RF2 protein prevents the enhancement in the expression of the reporter gene produced by the RF2 Thr-246 residue. These two last results indicate that the RF2 residue at position 256 also plays a role in the expression of the reporter gene, but it depends on the nature of the RF2 residue at position 246.
Figure 5.
Structural comparisons between RF2 and RF1 protein molecules. A, comparison of the primary sequences of the sections 220–279 and 237–296 of RF1 and two RF2 variants, respectively, which correspond to the tips of domain 3 that hold the GGQ functional motif (highlighted in cyan). Residues highlighted in red on the RF1 sequence are different between the three proteins. B and C, molecular modeling was performed as indicated under “Experimental procedures.” The structures show a view of overlapping molecular densities of the tips of domain 3 of (B) RF2 Ala-246/Arg-256 protein variant, and (C) Thr-246/Arg-256 (both in brown color), with RF1 (cyan color), all accommodated at the PTC A-site. The structural conformations of the A2602 nucleotide (sticks) observed in the presence of RF2 (red, PDB ID 5CZP) (7), RF1 (blue, PDB 5J30) (7), or with the TnaC peptide (yellow, PDB 6I0Y) (25) are also shown. The structural conformations of C2573 and U2506 acquired with RF2 (red), RF1 (blue), or TnaC (green) are shown as well. For both (B) and (C) panels, a tRNA located at the PTC P-site is shown in black. CCG MOE and Chimera were used to obtain the figures.
Figure 6.
The RF2 residues surrounding the functional GGQ motif contribute to the l-Trp-induced expression of the tna operon. A, cells containing plasmids that expressed the indicated RF2 variants at positions 246 and 256 were grown in minimal media supplemented with increasing concentrations of 1-methyl-l-Trp. Cultures at mid-log phase were used to determine β-gal activity (see “Experimental procedures”). These results represent four independent experiments; each experiment had a replica. B, plots showing relative amount of TnaC-peptidyl-tRNAPro2 with respect concentration of RF protein variants. Plots were obtained as indicated in Fig. 4. The following RF2 genes (A) and RF2 protein variants (B) were used in these experiments: Thr-246/Arg-256 (crosses), Ser-246/Arg-256 (open triangles), Ala-246/Thr-256 (open squares), Ala-246/Arg-256 (closed circles), Thr-246/Thr-256 (open diamonds), and Ser-246/Thr-256 (open circles).
Because RF1 is not efficiently inhibited by l-Trp, which leads to low expression of our reporter gene (Fig. 1A), we hypothesized that the RF1-like RF2 Ser-246/Thr-256 variant could reduce the l-Trp–mediated expression of the tnaA'-'lacZ reporter gene. As expected, the expression efficiency of the reporter gene under the RF2 Ser-246/Thr-256 variant was 2-fold lower than the expression efficiency observed with the RF2 Ala-246/Arg-256 variant (Fig. 6A, compare open circles with closed circles). This reduction effect depends on both 246 and 256 changes. We observed that the in vivo expression efficiency of the reporter gene under either the RF2 Ser-246/Arg-256 or Ala-246/Thr-256 variants was similar to the expression under the RF2 Ala-246/Arg-256 variant (Fig. 6A, compare open triangles and open squares with closed circles). These last results indicate that single changes A246S or R256T in the RF2 protein did not affect the l-Trp–inducible expression of the reporter gene, but the presence of both changes in the RF2 protein reduced the expression of the reporter gene. In conclusion, the RF2 Thr-246/Arg-256 variant allows for maximum expression of the reporter gene among all tested RF2 variants, whereas the RF1-like RF2 Ser-246/Thr-256 inefficiently promotes the l-Trp–mediated reporter gene expression.
To confirm that the differences in expression of the reporter gene produced by the RF2 variants are because of differences in the l-Trp-inhibition of the RF2-mediated TnaC-peptidyl-tRNA hydrolysis, we performed in vitro cleavage assays as shown in Fig. 4. For each RF2 variant, we tested different concentrations of pure RF protein on their ability to hydrolyze TnaC-tRNAPro2 under low and high concentrations of l-Trp. As seen in the left plot of Fig. 6B, under low concentrations of l-Trp mostly all RF2 variants have the same efficiency of promoting hydrolysis of TnaC-peptidyl-tRNAPro2 at 20 nm concentrations. The RF2 Thr-246/Arg-256 variant was less efficient at concentration ranges of 0 to 5 nm (Fig. 6B, left plot compare the curve with crosses with other curves). At high concentrations of l-Trp, the RF2 Thr-246/Arg-256 efficiency of hydrolysis was significantly reduced compared with all other variants (Fig. 6B, right plot, compare the curve with crosses with other curves). At 20 nm concentration, the RF2 Thr-246/Arg-256 protein induced just 20% hydrolysis of the TnaC-tRNAPro2 molecules when all other RF2 variants induced 80% hydrolysis of this peptidyl-tRNA. As seen in vivo, the RF2 Thr-246/Thr-256 variant did not behave like the RF2 Thr-246/Arg-256 variant in these experiments as well, where the RF2 Thr-246/Thr-256 variant was 4-fold more efficient promoting hydrolysis of TnaC-tRNAPro2 than the RF2 Thr-246/Arg-256 variant under high concentration of l-Trp (Fig. 6B, right plot, compare crosses with open diamonds). We conclude with these last data that the replacement R256T counteracts the effects of the Thr-246 residue during the inhibition of hydrolysis produced by l-Trp. Altogether, these results confirm that the nature of the RF2 residues at position 246 and 256 is important for the induction of the tna operon and for the l-Trp-inhibition of RF2-mediated hydrolysis of peptidyl-tRNA observed at the tnaC gene.
Discussion
In this work, we show that the regulatory TnaC nascent peptide bound to l-Trp at the ribosomal exit tunnel preferentially blocks the function of RF2 within the ribosome. In vivo analyses using reporter genes indicate that the identity of the tnaC stop codon determines the efficiency of l-Trp induction of the expression of the tna operon in E. coli; specifically, tnaC sequences with UGA stop codons better induce the expression of the reporter gene under activating concentrations of l-Trp than those sequences with UAA and UAG stop codons (Fig. 1). In vitro assays show that under l-Trp, tnaC constructs accumulate the most stalled ribosomes and TnaC-tRNAPro2 when its stop codon is UGA comparing with UAG constructs (Figs. 2 and 3). In vitro analyses indicate that l-Trp blocks better the RF2-mediated hydrolysis of TnaC-tRNAPro2 than RF1, and as a consequence the release of translating ribosomes from tnaC genes containing UGA stop codons (Fig. 4). Finally, mutational analyses indicated that the preferential inhibition of RF2 on our system depends on the nature of its residues at positions 246 and 256 surrounding the GGQ motif. The presence of a threonine residue at the RF2 246 position makes RF2 very sensitive to the inhibition action of l-Trp; changes in the RF2 arginine residue at position 256 reduce the inhibitory effects observed for the RF2 Thr-246 protein (Fig. 6).
l-Trp modifications targeting the RF2 246 residue
Genetic assays in concordance with cryo-EM models suggest that TnaC bound to l-Trp induces structural changes on nucleotides at the A-site of the PTC (22). It has been suggested that the TnaC peptide and free l-Trp molecules establish interactions with ribosomal nucleotides at the exit tunnel, causing the A2602 nucleotide to hold a spatial conformation that would generate an unproductive accommodation of the release factor (22, 34). A2602 is important for the action of release factor proteins (35, 36). The RF2 residue at position 246 is part of a cleft that holds the A2602 nucleotide once the release factor is located at the PTC (Fig. 5, B and C). RF2 proteins with the Thr-246 residue are less active in termination than RF2 proteins with the Ala-246 (37) or Ser-246 (38) residues. The structural comparisons shown at Fig. 5, B and C indicates to us that the RF2 Thr-246 residue makes a narrower cleft (Fig. 5C) compared with the RF2 Ala-246 residue (Fig. 5B) or with the RF1 Ser-229 residue suggesting that the RF2 Thr-246 protein might have slower kinetics of accommodation of the A2602 residue at its cleft than the RF2 Ala-246 or RF1 protein. Despite the observation that all RF2 and RF1 proteins could be inhibited by TnaC and l-Trp (Figs. 1 and 2), our results show that the RF2 Thr-246 protein is a better target for inhibition than the RF2 Ala-246, RF2 Ser-246 and RF1 protein (Fig. 6). Therefore, we suggest that TnaC and l-Trp alter transiently the A-site structural conformation of the PTC, which gives the opportunity for factors, such as RF2 Ala-246, RF2 Ser-246 or RF1, with perhaps faster accommodation kinetics than RF2 Thr-246 to induce hydrolysis of the TnaC-tRNAPro2 before the inhibitory conformation is acquired again. Therefore we are expecting any further studies to show that RF2 Thr-246 has slower accommodation kinetics than the kinetics of acquiring the inhibitory conformation.
The RF2 256 residue and its structural relationship with the 246 residue
We have seen that the interaction of l-Trp with the TnaC-ribosome complex alters the dimethyl sulfate reactivity (exposure to water) of several nucleotides of the PTC, and among them is A2572 (26). As a result, these changes could alter the conformation of neighboring residues such as C2573 which is in close proximity to the RF2 Arg-256 residue (Fig. 5, B and C) (22). The PTC nucleotides A2572 and C2753 are suggested to be part of an accommodation gate that controls the entrance of the acceptor arm of aminoacyl-tRNAs and the tip of domain 3 of the release factors at the PTC (39). Mutational changes in these two nucleotides impair RF2 activity but not aminoacyl-tRNA accommodation at the PTC and peptide bond formation (8). These observations suggest that the TnaC nascent peptide and l-Trp are affecting functional interactions between the nucleotides C2573 and/or A2572 with elements of the RF2 protein disturbing accommodation of domain 3 of RF2 at the PTC. As seen in Fig. 5, B and C, the RF2 Arg-256 residue is in close vicinity with the C2573 and 2506 nucleotides. Our data indicate that changes in this RF2 residue, specifically a threonine in the 256 position, overcome the inhibition induced by l-Trp (Fig. 4, C and D). Therefore, we suggest that the interactions of the Arg-256 residue with the PTC C2573 and 2506 nucleotides are important for blocking accommodation of RF2 at the PTC.
In conclusion, the evidence shown in this work indicates that the regulatory TnaC nascent peptide, in conjunction with l-Trp, inhibits better RF2- than RF1-mediated peptidyl-RNA hydrolysis and ribosome release action. These differences in inhibition depend on the identity of two nonconserved residues located at the tip of domain 3 of the RF2 protein, 246 and 256 positions. We suggest that changes in these two residue positions could affect the accommodation dynamics of the RF2 protein in the PTC; our claims could be supported by further kinetic analyses using our current RF2 variants.
Experimental procedures
Bacterial strains, plasmids, growth media, and mutagenesis
E. coli K12 bacteria strains and plasmids used in this work are indicated in Table S1. M9 media (140 mm disodium phosphate, 20 mm potassium monophosphate, 8 mm sodium chloride and 18 mm ammonium chloride, 2 mm magnesium sulfate, and 0.1 mm calcium chloride), supplemented with 0.2% glycerol and 0.05% casamino acids, was used to grow bacterial strains for β-gal assays. 100 μg/ml ampicillin and 50 μg/ml chloramphenicol were added when required for maintaining working plasmids. Luria Broth (LB) media supplemented with 100 μg/ml ampicillin and 50 μg/ml chloramphenicol was used to grow cultures for protein purification.
Mutagenesis and protein purification
Site-directed mutagenesis was performed on the pETDuet plasmids harboring the prfB and the prmC genes (7) using the QuikChange II Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Plasmids were Sanger-sequenced to confirm the presence of the mutations using the following primer: 5′-TAATACGACTCACTATAGG-3′. RF2 protein variants were purified as follows: cell cultures were grown in 20 ml LB supplemented with ampicillin and chloramphenicol at 37 °C for 3 h. Later, 0.01 mm l-arabinose (Sigma-Aldrich) and 0.1 mm isopropyl β-d-1 thiogalactopyranoside (IPTG) (Fisher Scientific) was added to induce the expression of the T7 RNA polymerase from the pTARA plasmid (40), which directs expression of the RF2 protein variants from the pETDuet plasmid variants. Cultures were incubated at 37 °C for 4 more hours. Cells for each culture were harvested by centrifugation at 4000 × g for 10 min at 4 °C, and later resuspended in 1 ml of binding buffer (20 mm sodium phosphate, 500 mm sodium chloride, and 20 mm imidazole, pH 7.4). 1 μl of 29 kilo units/μl of lysozyme (Epicenter), 1 μl of 1 units/μl DNase (RQ1, Promega), and 40 μl of 1 m magnesium chloride was added to the cell suspension. The final mix was incubated at room temperature for 30 min to allow cell disruption. The suspension was later sonicated and centrifuged for a minute at 13,000 × g to separate cell-free extract from cellular debris. RF2 proteins were purified by passing the cell-free extracts through a pretreated HisTrap Niquel column (GE Healthcare). After a couple of washings, the pure RF2 proteins were eluted from the columns with 200 μl of elution buffer (20 mm sodium phosphate, 500 mm sodium chloride, and 500 mm imidazole, pH 7.4). The final collected solutions were dialyzed against 137 mm sodium chloride, 2.7 mm potassium chloride, 8 mm disodium phosphate, and 2 mm potassium phosphate with 20% glycerol. Samples were stored at −80 °C. Degree of purification was assessed by gel electrophoresis. Concentration of proteins was determined using Bradford assays.
β-gal assays
For the experiments shown in Figs. 1; 4, C and D; and 6, A and B, bacterial cultures in M9 media with increasing concentrations of 1M-l-Trp (Sigma-Aldrich) and 0.1 mm IPTG were grown with aeration at 37 °C until they reached 0.6 A600. No l-arabinose was required for this experiment because the background expression of T7 RNA polymerase induced ∼10 more the expression of the RF2 protein variants than the expression of the endogenous RF2 protein (Fig. S1); however, we added 0.1 mm IPTG to block the action of the Lac I repressor from the pETDuet plasmids. Final cultures were kept on ice and small amounts from each culture were used for β-gal assays as described previously (24). The products of reactions were detected using a microplate reader (Bio-TEK, powerwave-HT). The normalized values were reported in Miller units. Replicas of each independent experiment were performed.
Determination of in vitro accumulation and cleavage of peptidyl-tRNA
Northern blotting assays were performed to determine in vitro accumulation of TnaC-peptidyl-tRNAs (Fig. 2) as indicated previously (26). Translation assays using the PURExpress protein synthesis system were performed as follows: 7 μl reaction containing 2.5 μl of solution A, 2 μl of solution B, 5 units of SUPERase-In (Ambion), and variable concentrations of l-Trp and tnaC PCR fragment variants were incubated for 30 min at 37 °C. In the case of Fig. 2D, the mix was pre-incubated with 5′-O-(N-(l-prolyl)-sulfamoyl)adenosine (l-PSA) (provided by Karin Musier-Forsyth) for 10 min at room temperature prior to the addition of tnaC PCR fragments. Reactions were stopped by the addition of 1 volume of loading buffer (4% SDS, 12% glycerol, 50 mm Tris-HCl, pH 6.8, 2% 2-mercaptoethanol, 1% bromphenol-blue) and resolved in 10% tris-tricine polyacrylamide gels containing a 5% stacking layer. The resolved proteins were transferred to N+ Nylon membranes (Hybond). Membranes were exposed to UV for 5 min to fix RNA molecules to the membrane, blocked and hybridized as indicated previously (26) using the following 5′-end labeled [32P]oligodeoxynucleotides: anti-tRNAPro2, 5′-CCTCCGACCCCCGACACCCCAT-3′; anti-tRNAArg2, 5′-CCTCCGACCGCTCGGTTCG-3′ (41). Membranes were developed using X-ray films (Kodak) and bands relative intensities were determined using ImageJ2 (42).
Toe-printing assays
Toe-printing assays were performed as described previously (23), using customized PURExpress protein synthesis systems (E6800, New England Biolabs) (Fig. 3) and ΔRF123, which are nearly devoid of the RF1, RF2 Thr-246, and RF3 proteins (E6850, New England Biolabs) (Fig. 4C). DNA fragments containing WT and mutant alleles of the tnaC gene were obtained by PCR using previously described primers (23). 5 μl of reactions were carried out using 5 to 30 × 10−3 pmol/μl tnaC DNA fragments, 0.3 mm all amino acids and variable concentrations of l-Trp at 37 °C for 20 min. For Fig. 4C the indicated pure release factors (supplemented by New England Biolabs for the ΔRF123 system) were added after 20 min of reaction, and the final mixtures incubated 5 more minutes. cDNA was synthesized in all cases by adding 4 units of AMV reverse transcriptase enzyme (Roche) and 0.5 μl per reaction of 20 pmol/μl reverse primer labeled with [32P] to the reactions as described previously (23). Reactions were then incubated at 37 °C for 15 min. cDNA products were resolved by electrophoresis using 6% urea-polyacrylamide gels. Final gels were exposed to a storage phosphor screen for 1 h and scanned using Typhoon Imager 9410. Band intensities were quantified using ImageJ2 (42). To compare each lane, the intensities of the arrested ribosome signals were normalized with the intensity of an internal control signal. We selected internal control signals that are located below the arrested bands (because their intensities are not affected by the signals of the arrested ribosomes) and whose presence is independent of the translation of the mRNAs (they remain present despite the addition of translation inhibitors).
Molecular modeling
Separate structure files for the TnaC (PDB ID: 6I0Y) (25), RF1, and RF2-bound ribosome (PDB IDs: 5J30 and 5CZP) (7) were structure corrected, protonated, and partially charged in CCG MOE 2019.0101 (Chemical Computing Group). Additionally, the cryo-EM density map coefficients for RF1 and RF2 were aligned with their respective structures; RF1 residues Ser-229, His-236, Thr-240, and Ser-242, and RF2 residues Ala-246, His-253, Arg-256, Thr-257, and Ser-259 were manually fitted to their corresponding cryo-EM density maps. All structures were then imported to UCSF Chimera 1.13.1 (43) for superimposition of all three structures using the MatchMaker tool. After superimposition, the structures were combined into one PDB and exported back to MOE 2019.0101 for comparison of the molecular surface of residues and nucleotides of interest. To generate the molecular surface model shown in Fig. 5, B and C, a solid molecular surface was generated for RF2 residues Ala-246 through Ser-259, Asn-277, and Arg-278, and for the RF1 spatially corresponding residues. The conformation of the RF2 Thr-246 residue was generated by the replacement A246T, and then by aligning the hydroxyl group of Thr-246 with Asn-1 of RF2-A2602. Fig. 5, B and C were rendered with POV-Ray (Persistence of Vision Pty. Ltd. version 3.6).
Author contributions
J. S. E., A. S., and E. R. G. conceptualization; J. S. E., A. S., E. R. G., J. T. N., and L. R. C.-V. data curation; J. S. E. and L. R. C.-V. formal analysis; J. S. E. and L. R. C.-V. supervision; J. S. E., A. S., and L. R. C.-V. validation; J. S. E., A. S., E. R. G., J. T. N., and L. R. C.-V. investigation; J. S. E., A. S., E. R. G., J. T. N., and L. R. C.-V. methodology; J. S. E., A. S., and L. R. C.-V. writing-original draft; J. S. E. and L. R. C.-V. project administration; J. S. E., A. S., E. R. G., J. T. N., and L. R. C.-V. writing-review and editing.
Supplementary Material
Acknowledgments
We are grateful to Hani Zaher for the kind provision of the pETDuet plasmids and Karin Musier-Forsyth for providing us with l-PSA used in this study. We also thank Valérie Heurgué-Hamard for providing us with the VH1125 strain; Nora Vazquez-Laslop, Gabriel Guarneros, Frances Yap, and Tanya Sysoeva for valuable critiques regarding this version of the manuscript; and Yan Zhang and Carol Gross for helpful discussion about data on the expression of the tna operon under the expression of the RF2 Ala-246 protein variant. We also thank previous anonymous reviewers for their observations that led to improving the scientific content of this article.
This work was supported by National Science Foundation Grant MCB-1158271 (to L. R. C.-V.) and UAH RCEU program and Alabama Space Grant Consortium (to J. T. N.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Table S1.
- RF
- release factor
- 1M-l-Trp
- 1-methyl-l-Trp
- IPTG
- isopropyl β-d-1 thiogalactopyranoside
- l-PSA
- 5′-O-(N-(l-prolyl)-sulfamoyl)-adenosine
- l-Trp
- l-tryptophan
- PTC
- peptidyl transferase center.
References
- 1. Korostelev A., Asahara H., Lancaster L., Laurberg M., Hirschi A., Zhu J., Trakhanov S., Scott W. G., and Noller H. F. (2008) Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. U.S.A. 105, 19684–19689 10.1073/pnas.0810953105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weixlbaumer A., Jin H., Neubauer C., Voorhees R. M., Petry S., Kelley A. C., and Ramakrishnan V. (2008) Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956 10.1126/science.1164840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scolnick E., Tompkins R., Caskey T., and Nirenberg M. (1968) Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. U.S.A. 61, 768–774 10.1073/pnas.61.2.768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trappl K., and Joseph S. (2016) Ribosome induces a closed to open conformational change in release factor 1. J. Mol. Biol. 428, 1333–1344 10.1016/j.jmb.2016.01.021 [DOI] [PubMed] [Google Scholar]
- 5. Shaw J. J., and Green R. (2007) Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol. Cell 28, 458–467 10.1016/j.molcel.2007.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mora L., Heurgue-Hamard V., de Zamaroczy M., Kervestin S., and Buckingham R. H. (2007) Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 282, 35638–35645 10.1074/jbc.M706076200 [DOI] [PubMed] [Google Scholar]
- 7. Pierson W. E., Hoffer E. D., Keedy H. E., Simms C. L., Dunham C. M., and Zaher H. S. (2016) Uniformity of peptide release is maintained by methylation of release factors. Cell Rep. 17, 11–18 10.1016/j.celrep.2016.08.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burakovsky D. E., Sergiev P. V., Steblyanko M. A., Kubarenko A. V., Konevega A. L., Bogdanov A. A., Rodnina M. V., and Dontsova O. A. (2010) Mutations at the accommodation gate of the ribosome impair RF2-dependent translation termination. RNA 16, 1848–1853 10.1261/rna.2185710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Svidritskiy E., and Korostelev A. A. (2018) Conformational control of translation termination on the 70S ribosome. Structure 26, 821–828.e823 10.1016/j.str.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Korostelev A., Zhu J., Asahara H., and Noller H. F. (2010) Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 29, 2577–2585 10.1038/emboj.2010.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freistroffer D. V., Kwiatkowski M., Buckingham R. H., and Ehrenberg M. (2000) The accuracy of codon recognition by polypeptide release factors. Proc. Natl. Acad. Sci. U.S.A. 97, 2046–2051 10.1073/pnas.030541097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adio S., Sharma H., Senyushkina T., Karki P., Maracci C., Wohlgemuth I., Holtkamp W., Peske F., and Rodnina M. V. (2018) Dynamics of ribosomes and release factors during translation termination in E. coli. Elife 7, e34252 10.7554/eLife.34252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zavialov A. V., Mora L., Buckingham R. H., and Ehrenberg M. (2002) Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell 10, 789–798 10.1016/S1097-2765(02)00691-3 [DOI] [PubMed] [Google Scholar]
- 14. Pavlov M. Y., Freistroffer D. V., MacDougall J., Buckingham R. H., and Ehrenberg M. (1997) Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J. 16, 4134–4141 10.1093/emboj/16.13.4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wood T. K., Hong S. H., and Ma Q. (2011) Engineering biofilm formation and dispersal. Trends Biotechnol. 29, 87–94 10.1016/j.tibtech.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deeley M. C., and Yanofsky C. (1982) Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J. Bacteriol. 151, 942–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gong F., and Yanofsky C. (2001) Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 276, 1974–1983 10.1074/jbc.M008892200 [DOI] [PubMed] [Google Scholar]
- 18. Stewart V., and Yanofsky C. (1986) Role of leader peptide synthesis in tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 167, 383–386 10.1128/jb.167.1.383-386.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gong F., Ito K., Nakamura Y., and Yanofsky C. (2001) The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro). Proc. Natl. Acad. Sci. U.S.A. 98, 8997–9001 10.1073/pnas.171299298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Konan K. V., and Yanofsky C. (1999) Role of ribosome release in regulation of tna operon expression in Escherichia coli. J. Bacteriol. 181, 1530–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cruz-Vera L. R., Rajagopal S., Squires C., and Yanofsky C. (2005) Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell 19, 333–343 10.1016/j.molcel.2005.06.013 [DOI] [PubMed] [Google Scholar]
- 22. Bischoff L., Berninghausen O., and Beckmann R. (2014) Molecular basis for the ribosome functioning as an L-tryptophan sensor. Cell Rep. 9, 469–475 10.1016/j.celrep.2014.09.011 [DOI] [PubMed] [Google Scholar]
- 23. Martínez A. K., Gordon E., Sengupta A., Shirole N., Klepacki D., Martinez-Garriga B., Brown L. M., Benedik M. J., Yanofsky C., Mankin A. S., Vazquez-Laslop N., Sachs M. S., and Cruz-Vera L. R. (2014) Interactions of the TnaC nascent peptide with rRNA in the exit tunnel enable the ribosome to respond to free tryptophan. Nucleic Acids Res. 42, 1245–1256 10.1093/nar/gkt923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martínez A. K., Shirole N. H., Murakami S., Benedik M. J., Sachs M. S., and Cruz-Vera L. R. (2012) Crucial elements that maintain the interactions between the regulatory TnaC peptide and the ribosome exit tunnel responsible for Trp inhibition of ribosome function. Nucleic Acids Res. 40, 2247–2257 10.1093/nar/gkr1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian P., Steward A., Kudva R., Su T., Shilling P. J., Nickson A. A., Hollins J. J., Beckmann R., von Heijne G., Clarke J., and Best R. B. (2018) Folding pathway of an Ig domain is conserved on and off the ribosome. Proc. Natl. Acad. Sci. U.S.A. 115, E11284–E11293 10.1073/pnas.1810523115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cruz-Vera L. R., Gong M., and Yanofsky C. (2006) Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. U.S.A. 103, 3598–3603 10.1073/pnas.0600082103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yanofsky C., Horn V., and Gollnick P. (1991) Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 173, 6009–6017 10.1128/jb.173.19.6009-6017.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stewart V., and Yanofsky C. (1985) Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 164, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gollnick P., and Yanofsky C. (1990) tRNA(Trp) translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J. Bacteriol. 172, 3100–3107 10.1128/jb.172.6.3100-3107.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vazquez-Laslop N., Thum C., and Mankin A. S. (2008) Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell 30, 190–202 10.1016/j.molcel.2008.02.026 [DOI] [PubMed] [Google Scholar]
- 31. Gong M., Gong F., and Yanofsky C. (2006) Overexpression of tnaC of Escherichia coli inhibits growth by depleting tRNA2Pro availability. J. Bacteriol. 188, 1892–1898 10.1128/JB.188.5.1892-1898.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heacock D., Forsyth C. J., Shiba K., and Musier-Forsyth K. (1996) Synthesis and aminoacyl-tRNA synthetase activity of prolyl adenylate analogs. Bioorg. Chem. 24, 273–289 10.1006/bioo.1996.0025 [DOI] [Google Scholar]
- 33. Jin H., Kelley A. C., Loakes D., and Ramakrishnan V. (2010) Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc. Natl. Acad. Sci. U.S.A. 107, 8593–8598 10.1073/pnas.1003995107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang R., Cruz-Vera L. R., and Yanofsky C. (2009) 23S rRNA nucleotides in the peptidyl transferase center are essential for tryptophanase operon induction. J. Bacteriol. 191, 3445–3450 10.1128/JB.00096-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Polacek N., Gomez M. J., Ito K., Xiong L., Nakamura Y., and Mankin A. (2003) The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol. Cell 11, 103–112 10.1016/S1097-2765(02)00825-0 [DOI] [PubMed] [Google Scholar]
- 36. Youngman E. M., Brunelle J. L., Kochaniak A. B., and Green R. (2004) The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 10.1016/S0092-8674(04)00411-8 [DOI] [PubMed] [Google Scholar]
- 37. Dinçbas-Renqvist V., Engström A., Mora L., Heurgué-Hamard V., Buckingham R., and Ehrenberg M. (2000) A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19, 6900–6907 10.1093/emboj/19.24.6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heurgué-Hamard V., Champ S., Engström A., Ehrenberg M., and Buckingham R. H. (2002) The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21, 769–778 10.1093/emboj/21.4.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanbonmatsu K. Y., Joseph S., and Tung C. S. (2005) Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. U.S.A. 102, 15854–15859 10.1073/pnas.0503456102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wycuff D. R., and Matthews K. S. (2000) Generation of an araC-araBAD promoter-regulated T7 expression system. Anal. Biochem. 277, 67–73 10.1006/abio.1999.4385 [DOI] [PubMed] [Google Scholar]
- 41. Dong H., Nilsson L., and Kurland C. G. (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 10.1006/jmbi.1996.0428 [DOI] [PubMed] [Google Scholar]
- 42. Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T., and Eliceiri K. W. (2017) ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 18, 529 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.