Abstract
Phosphatidylinositol-transfer proteins (PITPs) are key regulators of lipid signaling in eukaryotic cells. These proteins both potentiate the activities of phosphatidylinositol (PtdIns) 4-OH kinases and help channel production of specific pools of phosphatidylinositol 4-phosphate (PtdIns(4)P) dedicated to specific biological outcomes. In this manner, PITPs represent a major contributor to the mechanisms by which the biological outcomes of phosphoinositide are diversified. The two-ligand priming model proposes that the engine by which Sec14-like PITPs potentiate PtdIns kinase activities is a heterotypic lipid-exchange cycle where PtdIns is a common exchange substrate among the Sec14-like PITP family, but the second exchange ligand varies with the PITP. A major prediction of this model is that second-exchangeable ligand identity will vary from PITP to PITP. To address the heterogeneity in the second exchange ligand for Sec14-like PITPs, we used structural, computational, and biochemical approaches to probe the diversities of the lipid-binding cavity microenvironments of the yeast Sec14-like PITPs. The collective data report that yeast Sec14-like PITP lipid-binding pockets indeed define diverse chemical microenvironments that translate into differential ligand-binding specificities across this protein family.
Keywords: phosphoinositide, signaling, sterol, lipid metabolism, computational biology, cavity mapping, phosphatidylinositol transfer proteins, Sec14-domain, squalene
Introduction
Lipids play central roles in cell signaling in addition to their critical involvements in maintaining intracellular compartmentation. A significant aspect of lipid signaling involves the action and metabolism of phosphorylated derivatives of PtdIns.6 These phosphoinositides modulate an impressively-wide range of cellular processes, including regulation of G-protein–coupled receptor and receptor tyrosine kinase signaling at plasma membranes, actin dynamics, transcription, and membrane trafficking (1, 2). Although the basic involvements of specific lipid kinases in generating specific phosphoinositide species are well-appreciated, recent studies indicate that the regulation of phosphoinositide production for specific signaling events is far more complicated. PtdIns kinases, particularly the PtdIns 4-OH kinases, are biologically insufficient enzymes that by themselves cannot produce sufficient PtdIns(4)P to overcome the activities of negative regulators of PtdIns(4)P signaling to promote productive signaling events (3–6). Rather, these enzymes require the activities of Sec14-like PtdIns transfer proteins (PITPs) to potentiate the robust synthesis of PtdIns(4)P so that the actions of PtdIns(4)P-signaling antagonists are overcome in a precisely-regulated manner. The importance of PITPs in regulating phosphoinositide metabolism is amply demonstrated by the biological consequences associated with defects in individual PITPs in yeast (7–10), plant (11–13), insect (14, 15), and vertebrate systems (16–19).
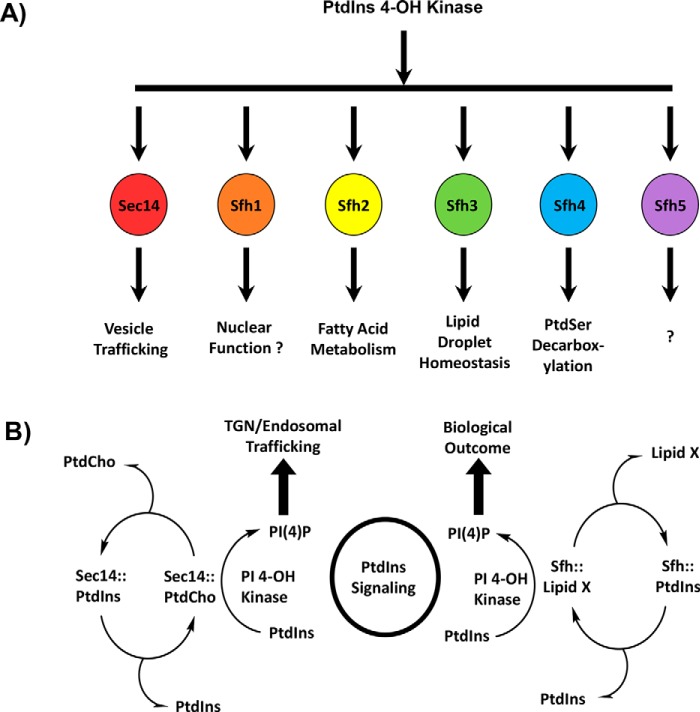
Sec14-like PITPs are founding members of the large Sec14 protein superfamily, and the common unifying element of this superfamily is a conserved Sec14 (or CRAL-TRIO) fold (3, 20–22). In that regard, whereas broad conservation of fold can provide indications of protein functional properties, functional diversification within a single fold class is manifested by virtue of finer structural features embedded within individual proteins. Such is the case within the Sec14 protein superfamily as exemplified by the fact that Sec14-like PITPs channel PtdIns 4-OH kinase activities to specific yet diverse biological outcomes (Fig. 1A) (3–6). The engine for doing so is proposed to be a heterotypic lipid-exchange cycle where exchange of a “second” ligand for PtdIns renders the PtdIns molecule a superior substrate for the lipid kinase (Fig. 1B). The available data project that this primed “PtdIns presentation” regime is a major mechanism for functionally specifying diverse biological outcomes for PtdIns(4)P signaling, even in simple eukaryotic cells such as yeast (10, 22–24).
Figure 1.
Sec14-like PITPs and diversification of PtdIns(4)P signaling. A illustrates the concept that the biological outcome of PtdIns(4)P signaling in yeast is primarily determined by the Sec14-like PITP that stimulates activity of a PtdIns 4-OH kinase to produce a functionally channeled PtdIns(4)P pool. This strategy is a major mechanism for diversifying PtdIns(4)P signaling. B depicts a heterotypic PtdIns/“second lipid” exchange cycle where the second lipid (lipid X) is a priming lipid that potentiates presentation of PtdIns to the lipid kinase, thereby rendering the PtdIns a superior substrate for the kinase resulting in stimulation of PtdIns(4)P production (3, 4, 6). At left is shown the concept for the Sec14 PtdIns/PtdCho-exchange cycle where PtdIns(4)P is channeled to promotion of membrane trafficking from the TGN/endosomal system. At right is shown the general case proposed for Sec14-like PITPs that fail to bind PtdCho and for whom identification of the exchangeable lipid X is sought.
The PtdIns presentation model for Sec14-like PITP function is founded upon structural data that project PtdIns binding as a conserved activity across the Sec14 superfamily, while the nature of the second ligand is highly diversified (3, 4). This marked diversity in ligand-binding specificities, together with the flexibility of the Sec14-fold, suggests that evolution has (i) taken advantage of the fundamental architecture of this fold to accommodate a large variety of lipid ligands and (ii) has translated that biochemical diversity into a diversity of biological outcomes. Thus, although the general notion of conservation of overall fold across the Sec14 superfamily is an interesting one, it is the specific and differential biological outcome associated with each of these proteins that is central to our understanding of how phosphoinositide signaling is spatially organized and functionally diversified in cells. These underlying mechanisms remain largely uninvestigated, however. It is in this context that integrating primary sequence comparisons with structural properties offers powerful opportunities to glean new insights into how functional diversity is incorporated into an otherwise hardwired Sec14-fold.
Herein, we report systematic investigation of the structural features of yeast Sec14-like PITPs to gain an understanding of molecular mechanisms that generate diversity and specificity for lipid substrates across the Sec14 superfamily. The data indicate that the chemical microenvironments of yeast Sec14-like PITP lipid-binding pockets are diverse. Using the Sfh2 and Sfh3 PITPs as experimental models, we show that these structural diversifications translate to differential “second ligand” lipid-binding specificities.
Results
Sec14 PtdIns/PtdCho-transfer proteins conserve both the general fold and protein surface electrostatics
Sec14 PtdIns- and PtdCho-transfer proteins from evolutionarily divergent fungal species (Candida albicans, Candida glabrata, Kluyveromyces lactis, Candida dubliniensis, Aspergillus nidulans, and Schizosaccharomyces pombe) share 53–87% primary sequence identity and 71–92% sequence similarity to Saccharomyces cerevisiae Sec14. The high-sequence homologies shared by these Sec14 proteins permitted the construction of 3D homology models using high-resolution crystal structures of PtdIns-bound Sfh1 (1.86 Å, PDB code, 3B7N (3)), PtdCho-bound Sfh1 (2.00 Å; PDB code, 3B7Q (3)), and β-octylglucoside–bound Sec14 (2.50 Å; PDB code, 1AUA (20)) as templates. As expected, all homology models conformed to a two-domain structure with an N-terminal tripod motif and a C-terminal lipid-binding domain (LBD), and an overall fold consisting of 11 α-helices, eight 310-helices, and six β-strands (Fig. 2A). An equally prominent feature of all of the Sec14 proteins was a β-sheet floor of the large hydrophobic lipid-binding cavity with a hybrid α/310-helix gating access to that cavity. A composite superimposition of all of these models emphasizes the structural conservation at the level of the α-carbon fold in this protein class (Fig. 2A).
Figure 2.
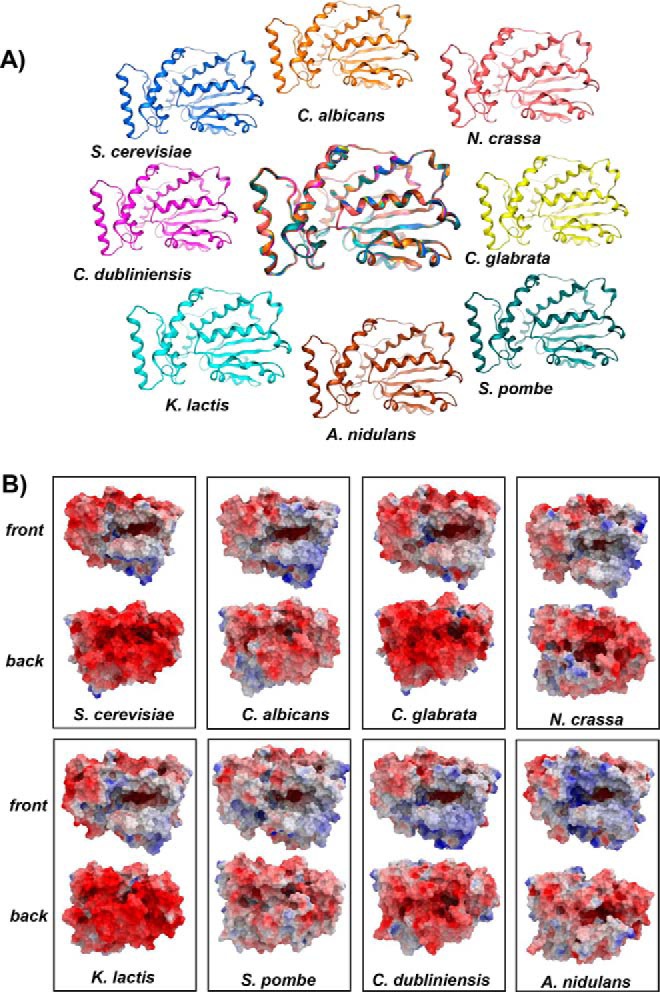
Structural features of Sec14 orthologs. A, α-carbon backbones of the basic folds of open conformers of Sec14 orthologs from the indicated fungal species: S. cerevisiae (blue); C. albicans (orange); Neurospora crassa (pink); C. dubliniensis (magenta); C. glabrata (yellow); K. lactis (cyan); A. nidulans (brown); and S. pombe (teal). At center is a structural overlay of the collective α-carbon backbones onto that of Sec14 that highlights the high overall conservation of the basic fold across large evolutionary distances. B, electrostatic potential surfaces of open conformers Sec14 and Sec14 orthologs from the indicated fungal species are shown. The en face orientation illustrates the open gating helices and the exposed lipid-binding cavities. Surfaces with electropositive potential are rendered blue, and electronegative surfaces are highlighted in red.
The impressive structural conservation among this protein class was further reflected in the general similarities of the molecular surface properties of these PITPs. Evaluation of the electrostatic surface maps of Sec14 and its orthologs from widely divergent fungi illustrated that these proteins are in effect dipoles. Whereas a significant portion of the exposed surface area of these Sec14 proteins is electronegative, conspicuous regions of electropositive potential are apparent at the solvent-exposed surfaces of substructures that form the helical gate and floor of the lipid-binding cavity in en face views of these molecules (Fig. 2B). Electronegative and electropositive charges are distributed similarly across those surfaces of the other Sec14s from widely divergent fungal species (Fig. 2B). These asymmetric surface charge distributions are most likely involved in orienting the Sec14 molecules in such a way that the electropositive regions are directed toward interactions with the electronegative membrane surface, thereby forecasting potential orientations by which Sec14 and its orthologs dock onto membranes.
Divergent Sec14 homologs conserve the general fold but not protein surface electrostatics
S. cerevisiae expresses five other Sec14-like PITPs in addition to Sec14 (23). These proteins are designated Sec fourteen homologs (Sfh) 1–5. The Sfh proteins share ∼25–78% primary sequence similarity to Sec14, and high-resolution crystal structures are available for PtdIns- and PtdCho-bound Sfh1 (PDB codes 3B7N, 3B7Q, and 3B7Z) (3) and for apo-Sfh3 (PDB codes 4J7Q and 4M8Z) (21, 22). Inspection of those structures, and of 3D homology models created by threading of the primary sequences of the Sfh2, Sfh4, and Sfh5 (i.e. proteins whose crystal structures are not yet solved) onto the structure of S. cerevisiae Sec14 (PDB code 1AUA) (20), demonstrated all of these Sfh proteins similarly shared a typical Sec14-fold with minor differences in detail (primarily in the loops; Fig. 3A).
Figure 3.
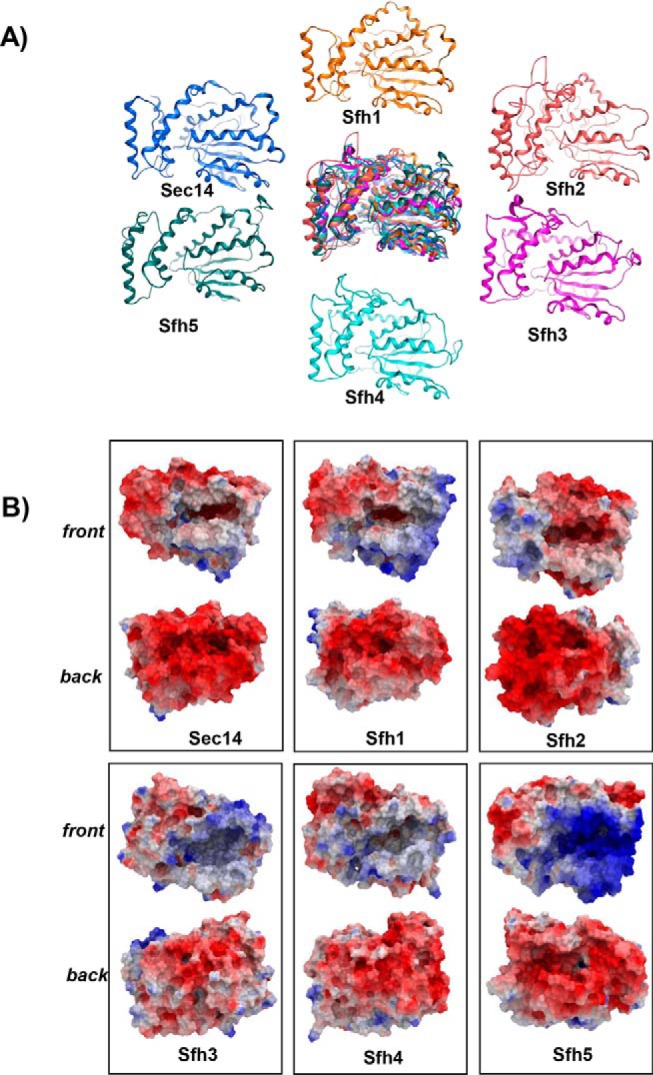
Structural features of Sec14 paralogs, the Sec14-like Sfh proteins. A, α-carbon backbones of the basic folds of open conformers of Sec14 and the Sec14-like Sfh PITPs from S. cerevisiae: Sec14 (blue); Sfh1 (orange); Sfh2 (pink); Sfh3 (magenta); Sfh4 (cyan); and Sfh5 (teal). At center is a structural overlay of the collective α-carbon backbones superimposed onto that of Sec14 that highlights the overall conservation of the basic Sec14-fold across the yeast Sfh1 family. B, electrostatic potential surfaces of open conformers Sec14 and indicated Sfh PITPs are shown. The en face orientation illustrates the open-gating helices and the exposed lipid-binding cavities and is labeled as “front,” and the opposite side of the molecule is identified as “back.” Surfaces with electropositive potential are rendered blue, and the electronegative surfaces are highlighted in red.
The conservation of fold notwithstanding, evaluation of the electrostatic surface maps of the Sfh proteins showed the protein-surface properties to be distinct from those of Sec14 and its orthologs. Although Sfh1 exhibited a Sec14-like dipole pattern of surface charge distribution, the electropositive charge was further expanded onto the protein surface covering the structural elements perpendicular to the side of the lipid-binding cavity (Fig. 3B). Sfh2 also showed electropositive character, but it was concentrated on the tripod motif surface rather than at the helical gate surface and the floor of the cavity. Sfh3 presented strong electropositive character, whereas Sfh4 exhibited more diffusely distributed areas of electropositive charge over its molecular surface (Fig. 3B). Sfh5 was an extreme case where the LBD surface was highly electropositive throughout. The distinct surface charge distributions and surface electrostatics of Sec14 and Sfh proteins likely contribute to the distinguishing molecular recognition determinants that help specify the diverse biological functions of these proteins.
Inferred heterogeneities in lipid-binding specificity
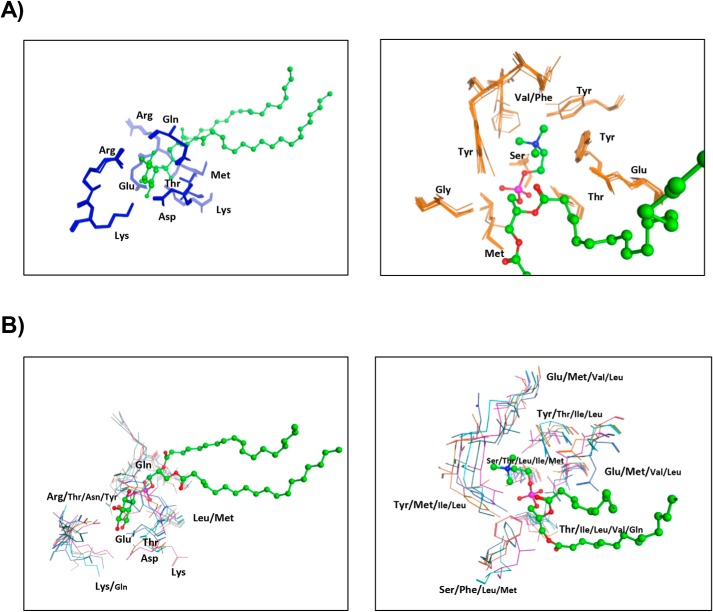
A second significant property proposed to contribute to functional diversification of Sec14-like PITPs is diversity in the nature of the second “ligand” bound by these proteins even with conserved PtdIns-binding activity. Evidence for this comes from sequence and structural alignments of divergent Sec14 orthologs that identify residues engaged in specific lipid headgroup binding (i.e. binding barcodes). Both primary sequence alignments and 3D homology models of fungal Sec14 orthologs show highly-conserved PtdIns- and PtdCho-binding barcodes (Fig. S1A; Fig. 4A, left panel). When compared with S. cerevisiae Sec14, the structural elements that coordinate binding of the inositol and choline headgroups are highly conserved as are the hydrophobic residues that lie within 4.5 Å of the fatty acyl chains. The PtdIns-binding barcode is composed of highly-conserved Arg, Lys, Glu, and Asp residues that engage in H-bond interactions with the headgroup inositol ring, and Pro, Thr, and Lys residues that coordinate the PtdIns phosphate moiety via H-bonds. In addition, an essentially invariant Thr within the LBD engages in H-bond interactions with the PtdIns glycerol backbone, whereas a conserved Arg residue contacts the PtdIns glycerol backbone and phosphate moieties via side-chain and backbone H-bonds, respectively.
Figure 4.
Structural barcodes for PtdIns- and PtdCho-binding in Sec14 orthologs and Sec14-like Sfh proteins. A, structural overlay of the PtdIns-binding (left panel) and PtdCho-binding barcode residues (right panel) from Sec14 orthologs superimposed onto those of Sec14. PtdIns-binding barcode residues are shown in blue stick model with PtdIns rendered in green ball and stick. The PtdCho-binding barcode residues are shown in orange stick model, and PtdCho is rendered in green ball and stick. Both barcodes are structurally preserved in Sec14 orthologs across large evolutionary distances. B, structural overlay of the PtdIns-binding (left panel) and PtdCho-binding barcode residues (right panel) from the indicated Sec14-like Sfh PITPs superimposed onto those of Sec14. PtdIns- and PtdCho-binding barcode residues are shown in a stick model using the color code of Fig. 3A to identify residues of individual Sfh proteins. PtdIns is rendered in green ball and stick. The PtdCho-binding barcode regions are shown using the color code of Fig. 3A to identify residues of individual Sfh proteins. PtdCho is rendered in green ball and stick. Although the PtdIns-binding barcodes are structurally well-preserved in the Sfh proteins, the PtdCho barcode is highly divergent (with the exception of Sfh1).
Similarly, the PtdCho barcode is also conserved in the 3D homology models of divergent fungal Sec14 orthologs. Those barcode residues coordinate PtdCho binding via a network of hydrogen bonds and water bridges and cation–π interactions between Tyr/Phe residues and the choline methyl groups. In Sec14, the PtdCho phosphate oxygen engages in a hydrogen bond network involving Tyr, Ser, and Thr residues to further stabilize the complex (Fig. S1A; Fig. 4A, right panel). Sec14 and Sfh1 share an identical PtdCho-binding barcode, and this conservation is of functional relevance. Both proteins bind/transfer PtdCho in vitro (3, 8, 25), and PtdCho binding is an essential property required for these proteins to potentiate PtdIns 4-OH kinase activity in vivo (3, 25).
With regard to the functionally-divergent Sfh proteins, the key PtdIns headgroup-binding barcode residues are conserved in the Sec14-like Sfh PITPs (Fig. S1B and Fig. 4B, left panel). This conservation reflects a functional property of these Sfh proteins as all exhibit PtdIns-binding/transfer activity in vitro (3, 4, 22, 23, 25).
However, there was a clear departure from the PtdCho-binding barcode in the other Sfh proteins as indicated by both primary sequence divergences (Fig. S1B) and by inspection of the analogous PtdCho headgroup coordinating regions of these proteins (Fig. 4B, right panel). For Sfh2, Sfh3, Sfh4, and Sfh5, the corresponding cavity microenvironment was distinct for each member, and the divergences in the amino acids corresponding to PtdCho-binding barcode residues are incompatible with PtdCho binding. Indeed, none of these Saccharomyces Sfh proteins bind/transfer PtdCho in vitro (23). Those sequence divergences forecast distinct ligand specificities that confer differential ligand-binding properties, and therefore distinct functional properties, to each Sfh protein (Fig. S1B and Fig. 4B, right panel).
Comparative PtdIns-binding topographies of Sec14 and Sfh lipid-binding cavities
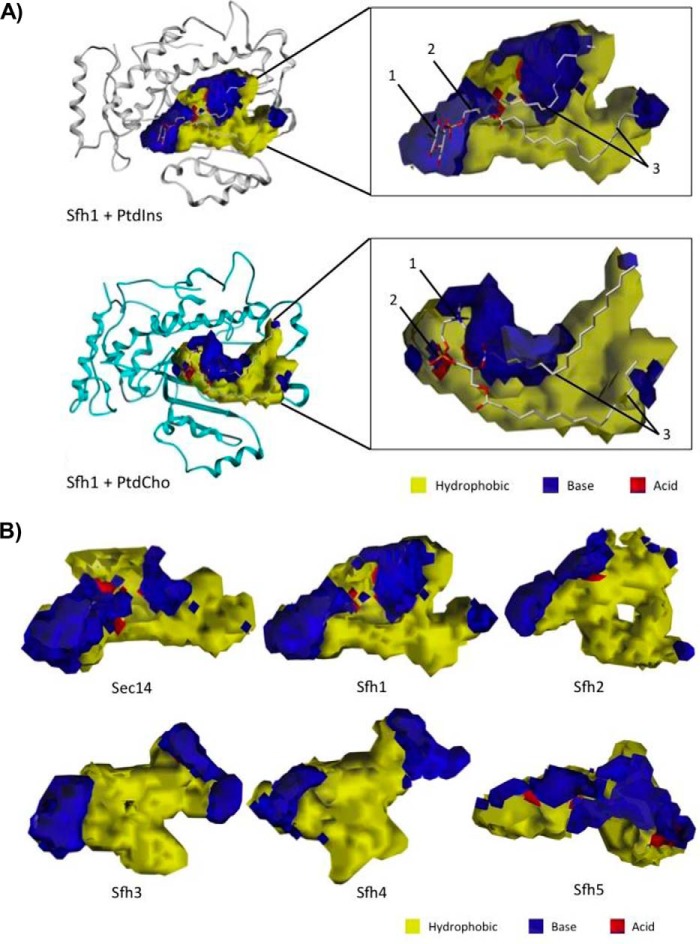
As the chemical landscape of the PITP lipid-binding cavity suggests a chemistry that governs ligand recognition/selectivity, a detailed structural and physiochemical characterization of each lipid-binding cavity was warranted. To that end, we determined the dimensions and extents of each cavity, and then we characterized each cavity with respect to its three-dimensional hydropathic properties. The first step involved application of the cavity search and characterization algorithm (vectorial identification of cavity extents (VICE)), which we described previously (26). VICE offers a complete set of metrics, including the cavity's volume, entrance opening size, and surface area. The resulting Boolean cavity extent maps were integrated with hydropathic “complement” property maps calculated by HINT (27, 28). The complement map represents the physical and chemical environments within the lipid-binding cavities in terms of the hydropathic properties of the ideal “complement” to a site, i.e. mapping where a complementary molecule should be hydrophobic, acidic, or basic (see under “Materials and methods”). We were thus able to obtain detailed fingerprints of Sec14/Sfh lipid-binding cavity microenvironments in terms of their polar and hydrophobic character (Fig. 5A). The integration of VICE and HINT maps, which is a point–by–point multiplication of the two maps, allowed extraction of unique information as to what the general features of a ligand might be, such that the extracted parameters of Sec14 and Sfh lipid-binding cavities could be translated to physical/chemical/spatial (rather than primary sequence) barcodes for ligand-binding activity. As PtdIns binding is a conserved property of all fungal Sec14 PtdIns/PtdCho-transfer proteins, and of the S. cerevisiae Sfh proteins, comparative analyses of the PtdIns-binding topographies of the various lipid-binding pockets served as test case for assessing the suitability of this general approach.
Figure 5.
VICE/HINT mapping of the Sec14 and Sfh PITP lipid-binding cavities. A, VICE-calculated binding cavity of Sfh1 was mapped with HINT complementary maps, and the PtdIns (top) and PtdCho (bottom) headgroup-binding regions are illustrated. PtdIns is bound in the cavity at the interface between the LBD and tripod motifs. The inositol headgroup (1) and glycerol backbone-binding region (2) are localized in distinctive polar microenvironments of the cavity surface. The acyl chains (3) pack into the largely hydrophobic LBD. PtdCho binds the cavity at the interface between the LBD and tripod motifs. The choline headgroup (1) is located in a hydrophobic area in the near vicinity of an extensive polar region. The PtdCho phosphate moiety (2) is coordinated in a polar surface, and the acyl chains (3) pack into the largely hydrophobic LBD. The Sfh1 α-carbon backbone is rendered in ribbon style, and the lipid ligands are rendered with ball and stick. In HINT complementary maps, yellow contours identify hydrophobic regions, and blue and red contours identify polar electropositive and electronegative surfaces, respectively. B, lipid-binding cavity maps for Sec14 and the Sfh PITPs are shown. Binding cavities were mapped by the VICE cavity detection algorithm and HINT complementary maps. In HINT complementary maps, yellow contours depict hydrophobic regions, and blue and red contours identify polar electropositive and electronegative cavity surfaces, respectively.
The cavity properties of Sec14-like PITPs were characterized (i.e. volume, surface area, hydropathic character, entrance cross-sectional surface area, etc.; Fig. 5B). In these terms, the PtdIns-binding cavities were similar with respect to volume and surface area in all fungal Sec14s, even though PtdIns is a flexible molecule (Fig. S2 and Tables S1–S4). The PtdIns headgroup was consistently bound in these cavities at the interfaces of the LBD and tripod motifs. As a detailed PtdIns-binding fingerprint, the inositol headgroup and glycerol backbone-binding region (defined as the proximal end of the lipid-binding cavity) was landmarked by distinct polar patches ideal for engagement of these chemical moieties. As expected, the acyl chains were hosted by the hydrophobic LBD. However, the distal end of the PtdIns-binding pocket (toward helices α7–α8 and α9) was again landmarked by a patch of polar residues (Fig. 5A and Fig. S2).
Conservation of this general PtdIns-binding topography was evident for the Sfh proteins as well, and the Sec14-like PtdIns-binding microenvironment fingerprint was also nicely conserved in Sfh1, Sfh2, Sfh3, and Sfh4 (Fig. S3 and Tables S5–S8). By contrast, Sfh5 provided an exception in that it exhibited a rather polar and strikingly basic LBD environment, even though the fingerprint of the Sfh5 PtdIns headgroup–binding region conformed to the PtdIns-binding fingerprints calculated for Sec14 and the other Sfh proteins (Fig. 5B and Fig. S3). Taken together, these data validated the integrated VICE/HINT mapping approach as a reliable tool with which to produce spatial fingerprints of the physical/chemical parameters of individual Sec14/Sfh lipid-binding cavities for comparative analyses.
Comparative PtdCho-binding topographies of Sec14 and Sfh protein lipid-binding pockets
Sfh protein divergences from the PtdCho-binding primary sequence barcode are consistent with the idea that these proteins are able to prime PtdIns 4-OH kinase activity by heterotypic exchange of their distinct second ligands for PtdIns (3–6). The detailed fingerprinting of individual Sfh lipid-binding cavities, as described above, was applied to determine whether divergences in the primary sequence barcodes translated to unique physical/chemical properties of Sfh lipid-binding cavities. Of special interest were distinguishing polar microenvironments identified by HINT/VICE mapping within individual (and largely hydrophobic) ligand-binding cavities because it is those features that likely contribute to the second-ligand binding specificities of Sec14-like proteins.
Although the polar microenvironment associated with PtdIns headgroup binding was conserved throughout the Sec14 and Sfh protein cavity maps, the nature and distributions of other polar microenvironments in the lipid-binding cavities were unique to each Sfh protein (Fig. 5B and Fig. S3). Interestingly, very similar PtdCho headgroup-binding microenvironments were shared by the two PtdCho-binding members of this family (Sec14 and Sfh1), but the cavities of other Sfh proteins showed very different chemical properties in their corresponding microenvironments. Sfh2 exhibited unique polar character in the area of helix α9 and the loop connecting α9 to β-strand β4, whereas the large distal region of the lipid-binding cavity was hydrophobic, unlike the corresponding regions of the Sec14 and Sfh1 cavities (Fig. 5B and Fig. S3). The Sfh3 cavity was characterized by a deeply-buried cleft that was both larger and more hydrophobic than that of Sec14 or Sfh1, whereas the binding pocket shapes and topographies of Sfh4 and Sfh5 diverged significantly from those of Sec14, Sfh1, Sfh2, and Sfh3 (Fig. 5B and Fig. S3).
Sfh2 is a PtdIns- and squalene-exchange protein
The identification of second ligands for a Sec14-like PITP would afford an opportunity to relate the chemical/physical fingerprint of the corresponding lipid-binding cavity to binding of that second ligand. It is from this perspective that the remainder of this work focuses on the Sec14-like Sfh2 and Sfh3.
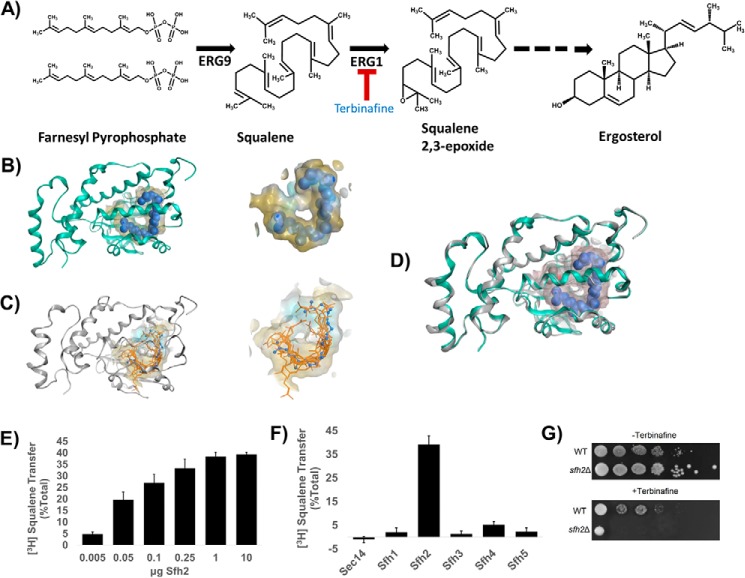
The most unexpected result from the VICE/HINT pocket mapping experiments was the altogether unique toroid-shaped topography of the Sfh2-binding pocket (Fig. 5B). Although Sfh2 is a PITP that shares 30–35% homology with Sec14 and Sfh1 (23), and the Sec14 and Sfh1 structures were used as templates to generate the Sfh2 homology model, the highly-divergent pocket topography projected Sfh2 bound an unusual second ligand. In that regard, the first clue was derived from the structural homology of Sfh2 with the mammalian Sec14L2 protein. This mammalian member of the Sec14 superfamily has been crystallized bound to squalene and 2,3-oxidosqualene (29). These two metabolites are sterol precursors, and squalene is converted to 2,3-oxidosqualene by the action of the terbinafine target squalene 2,3-epoxidase (ERG1 gene product; Fig. 6A). Indeed, structural overlay representations indicated that Sfh2 and Sec14L2 shared toroid-shaped and hydrophobic binding cavities within the LBDs (Fig. 6B). The data suggested these two Sec14-like proteins provide similar binding microenvironments for their respective ligands and strongly implicated squalene as a bona fide ligand for Sfh2.
Figure 6.
Sfh2 is squalene-binding/exchange PITP. A, squalene is a metabolic intermediate in the ergosterol biosynthesis pathway and is produced from farnesyl pyrophosphate by the action of the ERG9 gene product squalene synthase. Subsequently, squalene is converted to squalene 2,3-epoxide in a reaction catalyzed by the squalene 2,3-epoxidase (encoded by ERG1) that consumes molecular oxygen. Squalene 2,3-epoxidase is inhibited by the synthetic allylamine terbinafine. B, left, Sec14L2::squalene structural model is shown with the protein α-carbon backbone in ribbon style (green) with squalene as a space-filled model (blue) in a yellow translucent binding cavity. Right, close-up view of squalene within the Sec14L2-binding pocket is shown. The data were extracted from PDB code 4OMK (29). C, left shows the Sfh2 homology model (gray ribbon) with squalene (orange stick) docked into the lipid-binding cavity. The 10 highest scoring dock poses are shown. Right shows a close-up view of the model Sfh2 lipid-binding pocket with the 10 highest scoring squalene dock poses. The blue ball and stick model is a superimposition of the squalene pose extracted from Sec14L2 for comparison (PDB code 4OMK). D, structural overlay of the Sec14L2::squalene crystal structure (green ribbon, squalene rendered as a blue space-filled model) and a homology model of Sfh2 (gray ribbons). E, Sfh2-dependent [3H]squalene transfer. Squalene transfer is expressed as the fraction of total input of squalene transferred from donor liposomes to bovine heart mitochondria acceptor in 30 min at 37 °C after subtraction of background. [3H]Squalene input in these assays ranged from 21 to 27 × 103 cpm, and background ranged from 4 to 5 × 103 cpm. The mass amounts of Sfh2 assayed in these titration experiments are shown at the bottom. The values represent the averages of three independent determinations performed in triplicate. Error bars represent standard deviations. F, clamped amount (10 μg) of each of the indicated Sec14-like PITPs was assayed for squalene transfer as in E. Under those conditions, Sfh2 catalyzed ∼40% transfer of input [3H]squalene after subtraction of background. [3H]Squalene input in these assays ranged from 18 to 24 × 103 cpm and background ranged from 4 to 6 × 103 cpm. The values represent averages of triplicate determinations from three independent experiments. Error bars represent standard deviations. G, Sfh2-deficient yeast exhibit increased sensitivity to terbinafine. Isogenic WT and sfh2Δ yeast strains were spotted in 10-fold dilution series onto YPD plates without or with the squalene 2,3-epoxidase inhibitor terbinafine (250 μm), as indicated. Plates were incubated at 30 °C for 72 h.
To investigate in more detail the suitability of squalene as a putative second binding ligand for Sfh2, the molecule was computationally docked in the binding pocket of Sfh2, and the dock pose was optimized. The docked squalene models showed binding poses similar to that observed for squalene in the Sec14L2 lipid-binding cavity (Fig. 6C). Bound squalene, which describes an open hydrocarbon chain, was primarily coordinated by hydrophobic residues Phe268, Ile271, Trp272, Leu279, and Val283 positioned on the helical gate and lining the interior of the lipid-binding cavity. Residues comprising the floor of hydrophobic lipid-binding cavity also contributed to squalene binding, and these included Ile224, Phe226, Leu228, Leu256, Leu259, Leu260, and Ile261. Significant contributions for the hydrophobic microenvironment supporting squalene binding were also provided by residues on helix α9, which constitutes part of the cavity ceiling. Those residues included Pro240, Val241, Phe243, Leu244, Ile245, and Phe248.
We also note that, in addition to the largely hydrophobic character of the Sfh2 lipid-binding cavity, the distal and interior end of the cavity included hydrophilic residues Glu157, Thr158, Asn177, Arg185, His189, Gln194, Thr195, Glu198, and Lys201. Thus, the Sfh2 cavity exhibited significant amphipathic character. Interestingly, these features recapitulated the chemical landscape of the Sec14L2 lipid-binding cavity (Fig. 6D). Moreover, molecular modeling data further projected that the squalene binding space closely overlaps with that of the sn-2 acyl chain of PtdIns.
To directly determine whether squalene is an exchangeable ligand for Sfh2, an assay was developed to measure Sfh2-mediated transfer of [3H]squalene from donor liposomes to acceptor bovine heart mitochondria in vitro (see “Materials and methods”). Indeed, Sfh2 showed robust squalene transfer activity in this assay (Fig. 6E). A fixed amount of Sfh2 (10 μg) typically catalyzed transfer of ∼40% of the [3H]squalene from donor liposomes to acceptor membranes in a 30-min incubation at 37 °C. Those results came with a signal/background ratio of approximately 4, and squalene transfer exhibited Sfh2 concentration dependence (Fig. 6E). Moreover, Sfh2-mediated [3H]squalene transfer was a specific activity from the perspective that neither Sec14 nor any of the other Sfh proteins effected any significant squalene transfer above background (Fig. 6F). This level of transfer is notable as maximal achievable transfer is ∼50%. That squalene binding by Sfh2 is of physiological relevance is supported by our finding that Sfh2-deficient yeast cells exhibit enhanced sensitivity to terbinafine (Fig. 6G).
Sfh3 is a PtdIns- and sterol-exchange protein
The second model for interrogation of lipid-exchange activity involved Sfh3, a PITP that localizes to a discrete subset of lipid droplets in sporulating yeast cells and functions to inhibit lipid mobilization from those structures (22). In a previous study of the role of sterol binding in activity of the yeast oxysterol–binding protein family member Kes1 in vivo (30), we serendipitously observed that Sfh3 (which was initially chosen as a presumptive negative control) showed appreciable cholesterol-binding activity. Moreover, an independent study using a permeabilized mammalian cell-based lipid-binding assay also suggested Sfh3 binds cholesterol (31). However, both sets of binding data were of limited information content as neither addressed the key issue of whether a sterol is a bona fide exchangeable second ligand. To build upon those preliminary observations, several variations of in vitro sterol transfer assays were performed with purified Sfh proteins using Sec14 and Kes1 as negative and positive sterol exchange protein controls, respectively. Dihydroergosterol (DHE), [3H]cholesterol, and cholestatrienol (CTL) were evaluated as sterol exchange substrates.
As DHE (an essentially “label-free” form of the major yeast sterol ergosterol) and CTL are both naturally fluorescent compounds, a fluorescence dequenching assay was used to measure transfer of these lipids between vesicle populations. Indeed, Sfh3 exhibited clear DHE exchange capability with an activity that was ∼75% of that measured for Kes1, a protein with known DHE exchange activity (32, 33). Sfh4 displayed a much reduced DHE exchange activity relative to Sfh3 (∼15% of the Kes1 control), whereas neither the negative Sec14 control nor the test Sfh1, Sfh2, or Sfh5 proteins exhibited detectable DHE exchange activities (Fig. 7A). Similar results were again obtained when [3H]cholesterol was used as a substrate in an assay measuring lipid transfer from liposomes to bovine heart mitochondria (Fig. 7C). When CTL was used as substrate in a fluorescence dequenching system, Sfh3 displayed the highest exchange activity with Kes1 exhibiting only some 30% of the Sfh3 activity (Fig. 7C). Neither Sec14 nor any other Sfh protein exhibited any detectable [3H]cholesterol or CTL transfer activity at all. As Sfh3 had been previously demonstrated to exchange PtdIns (22, 23), these data identified ergosterol as a likely second lipid exchange ligand for Sfh3.
Figure 7.
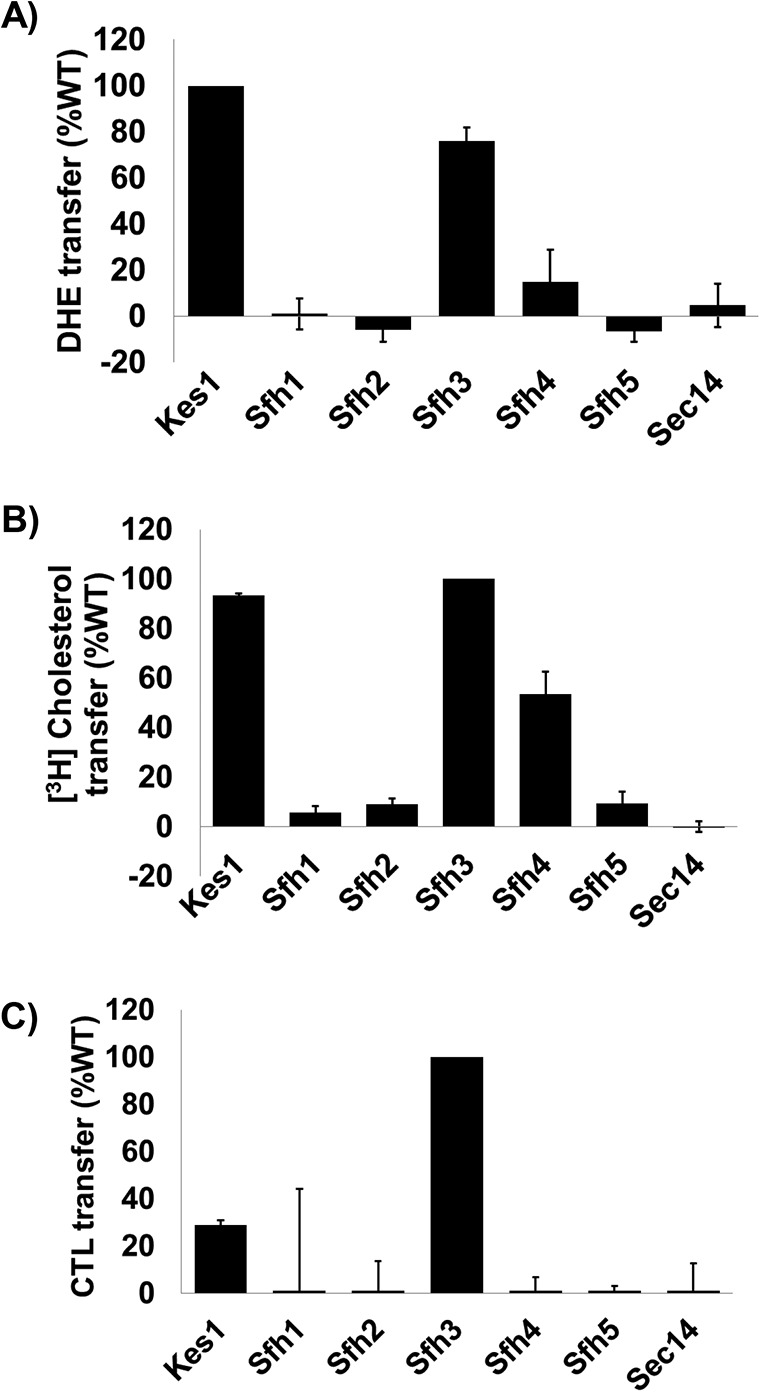
Sterol exchange activities for Sec14-like PITPs. In vitro transfer activities of the indicated Sec14-like PITPs were measured for DHE (A), [3H]cholesterol (B), and CTL (C) transfer. The sterol exchange protein Kes1 served as positive control and Sec14 as negative control. Assay incubations were for 10 min for the real-time fluorescence-based CTL and DHE assays and 30 min for [3H]cholesterol. All assays were performed at 37 °C. For CTL and DHE, values for spontaneous transfer (no protein) were measured at the same times indicated for protein assays, and spontaneous transfer backgrounds were subtracted from each measurement. Transfer activities were normalized to Kes1 (DHE) or Sfh3 (CTL). For all experiments, proteins were clamped 10 μg per assay. The averages of triplicate determinations from two independent experiments are shown for CTL and DHE, and averages of triplicate determinations from three independent experiments are shown for [3H]cholesterol transfer. Error bars represent standard deviations for the [3H]cholesterol assays and mean ± range for CTL and DHE assays.
Sterol binding pose within the Sfh3 lipid-binding pocket
The HINT hydropathic mapping of the Sfh3 lipid-binding cavity afforded an opportunity to further interrogate the recognition of sterols as Sfh3 ligands. To decipher the essential structural and functional features involved in sterol binding by Sfh3, several sterols (cholesterol, ergosterol, DHE, and CTL) were docked to the Sfh3 crystal structure (PDB code 4J7Q) (21). The lipid-binding cavity of the “open” Sfh3 conformer is solvent-accessible, is not well-defined, and with the flexible 210VPGNSKIP217 loop motif is much wider and draws a significantly larger surface area and volume relative to the cavities of the PtdIns/PtdCho-binding proteins Sec14 and Sfh1. To ensure sampling of all possible docking solutions and binding modes, we carried out unrestrained docking, which generated diverse solutions during the ligand-docking stage. Several independent docking runs were carried out to investigate the steric, electrostatic, and hydrophobic determinants for sterol binding within the Sfh3 hydrophobic cavity. The highest-scoring and most prominent binding modes were considered likely the most accurate estimations of an Sfh3-sterol complex (Fig. 8A).
Figure 8.
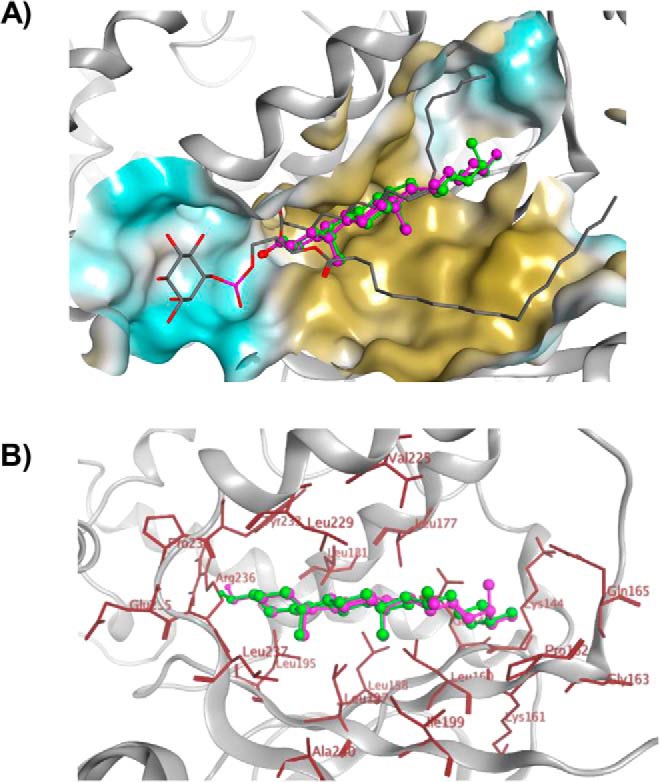
Sterol-binding model for Sfh3. A, image shows a binding model of ergosterol (green), cholesterol (magenta), and PtdIns (gray stick) with Sfh3 in open conformation. B, detailed depictions of the docked binding poses of ergosterol (green) and cholesterol (magenta) in the Sfh3 lipid-binding pocket are shown with coordinating residues highlighted in red.
Docking models for ergosterol, cholesterol, DHE, and CTL consistently indicated that the sterol-binding site was buried within the inner hydrophobic core region of the Sfh3 lipid-binding cavity far removed from the PtdIns-binding subregion. The hydrophobic sterol planar ring system was positioned in a narrow and highly-hydrophobic cleft of the pocket lined by Leu237 and Ala240 on β7; Leu195, Leu197, and Ile199 on β6; Leu158, Leu160, and Pro162 on β5; Leu177 and Leu181 on α6; and Val225, Leu229, Tyr233, and Pro234 on α7. By contrast, the hydroxyl oxygen at the 3-position of the sterol A-ring engaged in H-bond interactions with the backbone amide nitrogen of Leu237 on β7 and the hydroxyl hydrogen coordinating with the backbone oxygen of Pro234 (Fig. 8B).
Biochemical validation of in silico binding models
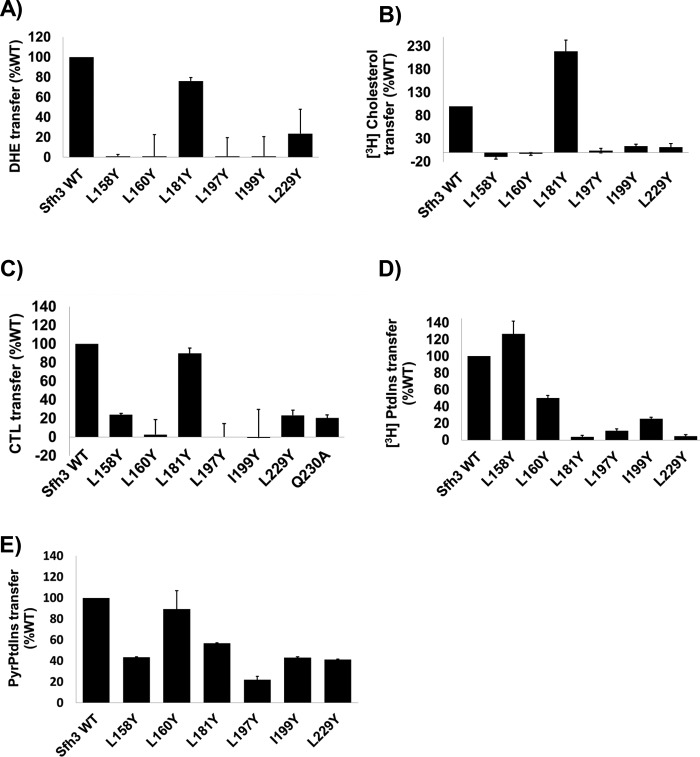
To experimentally test the quality of the Sfh3 sterol-binding model, the hydrophobic Ile/Leu residues padding the putative sterol-binding cavity were mutated to Tyr with the expectation that such substitutions would sterically occlude the binding pocket and further inhibit sterol binding by introducing polar character into an otherwise hydrophobic microenvironment. Thus, the model predicted that the Sfh3L158Y, Sfh3L160Y, Sfh3L181Y, Sfh3L197Y, Sfh3I199Y, and Sfh3L229Y proteins would all be compromised for sterol binding on the basis of steric and electrostatic considerations. That expectation was validated as all but one of those substitutions were incompatible with DHE, [3H]cholesterol, and CTL exchange activities (Fig. 9, A–C). Sfh3L181Y was the one exception, and this result is rationalized below.
Figure 9.
In vitro lipid transfer activities of the predicted Sfh3 sterol-binding mutants. Transfer activities for the indicated Sfh3 variants were measured using DHE (A), [3H]cholesterol (B), CTL (C), [3H]PtdIns (D), and [Pyr]PtdIns (E) as transfer substrates. Assay incubations were for 10 min for the real-time fluorescence-based DHE and CTL assays and 30 min for [3H]cholesterol, [3H]PtdIns and [Pyr]PtdIns transfer assays. All assays were performed at 37 °C, and protein input was clamped at 10 μg for all experiments. For DHE, CTL, and [Pyr]PtdIns assays, values for spontaneous transfer (no protein) were determined, and spontaneous transfer backgrounds were subtracted from each measurement. Transfer activities were normalized to Sfh3 activity. The averages of triplicate determinations from two independent experiments for DHE, CTL, and [Pyr]PtdIns, and triplicate determinations from three independent experiments for [3H]cholesterol and [3H]PtdIns, are shown. Error bars represent standard deviations for the [3H]cholesterol and [3H]PtdIns assays and mean ± range for DHE, CTL, and [Pyr]PtdIns assays.
The mutant Sfh3 proteins were also evaluated for PtdIns transfer in parallel with [3H]PtdIns microsomal vesicle and [Pyr]PtdIns fluorescence dequenching assays. Whereas Sfh3L158Y, Sfh3L160Y, Sfh3L197Y, Sfh3I199Y, and Sfh3L229Y were all strongly compromised in terms of sterol-transfer activities, the effects on PtdIns transfer were generally milder than those recorded for sterol transfer, although most mutant proteins were measurably compromised for PtdIns-transfer activity as well regardless of whether the measurements employed [3H]PtdIns (Fig. 9D) or [Pyr]PtdIns (Fig. 9E) as transfer substrates.
Simulation of the sterol-binding pose within the closed Sfh3 lipid-binding pocket
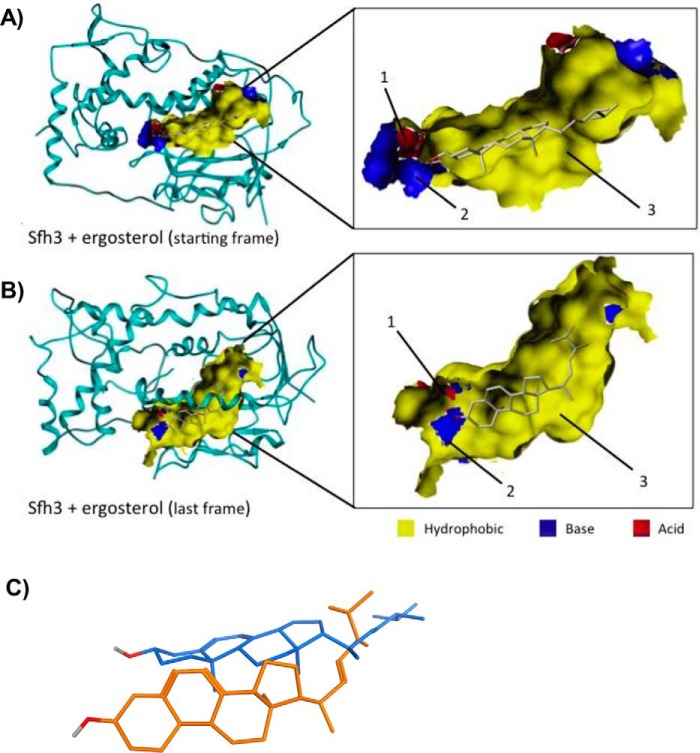
The sterol docking studies using the open Sfh3 conformer as the target-binding environment were incomplete in the sense that Sfh3 transitions to a closed conformation when ligand binds. Thus, to evaluate the sterol-binding pose in the closed Sfh3 conformer, a 50-ns MDS was run in a solvent box to model transition of the open sterol-bound conformer to a stable closed sterol-bound conformer. Consistent with expectations, the Sfh3-sterol complex converged to the “closed” conformer during the time course of the 50-ns simulation. Analysis of the simulation trajectories showed that the sterol molecule moved toward the PtdIns headgroup–binding region of the LBD, where its 3-hydroxyl group coordinated with Gln230 and Thr264 via a network of H-bond and water bridge interactions. In addition, Glu235 and Lys267 engaged sterol in the same manner. The rest of the planar sterol ring system was sandwiched between the ceiling and floor of the LBD's hydrophobic core. The primary lipid–Sfh3 interactions in the putative sterol-binding microenvironment involved dispersed van der Waals and hydrophobic interactions with Leu160, Leu197, Ile199, Leu226, Leu229, Leu237, Ala240, and Leu268 (Fig. 10).
Figure 10.
Ergosterol dynamics within the Sfh3-binding pocket as a function of transition from the open to the closed Sfh3 conformer. Models of the Sfh3::ergosterol complex in the open Sfh3 conformer (A) is compared with the projected ergosterol pose in the closed Sfh3 conformer (B). In the open conformer, the ergosterol headgroup is positioned close to the polar environment of the lower cavity and anchors via H-bond interactions with Pro234 (1) and Leu237 (2). The remainder of the ergosterol ring system is incorporated into the hydrophobic area of the Sfh3 lipid-binding pocket (3). B, Sfh3::ergosterol complex in the closed Sfh3 conformer. During the simulation, ergosterol moved toward the PtdIns headgroup-binding region of the lipid-binding cavity and engaged via H-bond interactions with residues Glu235 (1) and Lys267 (2). Complementarity maps depict hydrophobic regions in yellow. Electropositive and electronegative regions are depicted in blue and red, respectively. C, relative poses of ergosterol in the lipid-binding cavities of the open (from first frame of the MD simulation; blue stick model) and closed Sfh3 conformers (last frame of the MD simulation; orange stick model) are shown in superposition.
Moreover, MD simulation of the Sfh3-sterol complex predicted Sfh3T264A, Sfh3Q230A, Sfh3E235A, and Sfh3K267A would also show impaired sterol binding and transfer. The basis for this prediction was the bound sterol molecule engaged these residues via hydrogen bonding and water bridge interactions, as it shifted toward the more stabilizing PtdIns headgroup–binding region. The significance of this observation is detailed in the following section.
Attempts to uncouple Sfh3 PtdIns- and sterol-binding activities
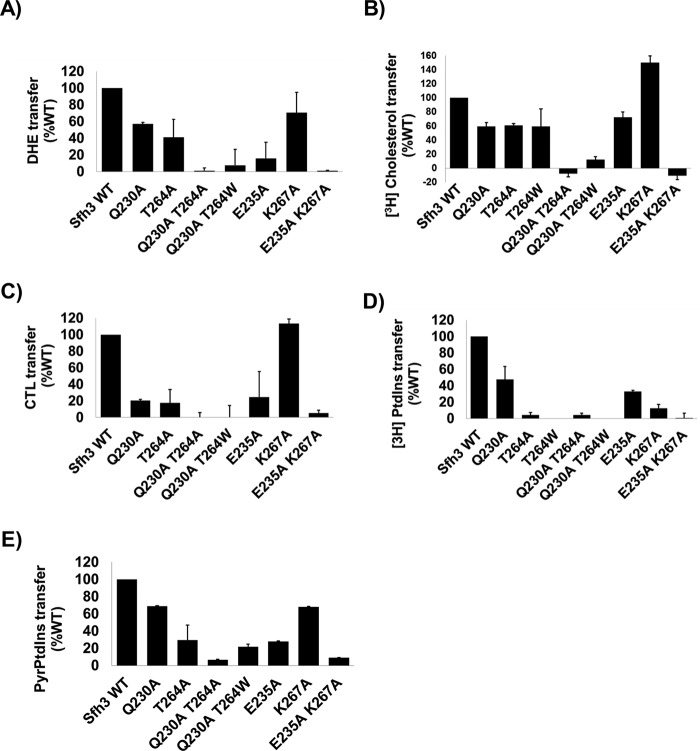
Sfh3 was previously shown to regulate lipid droplet homeostasis in vivo (22), and its activity is also reflected in the increased resistance of yeast to azole drugs when Sfh3 is overexpressed (31, 34). In those studies, PtdIns binding was concluded to be an essential activity required for Sfh3 to execute its biological functions. Those conclusions were based on demonstrations that compromise of PtdIns binding by mutation of PtdIns barcode residues (T264W in Ref. 22 and E235A and K267A in Ref. 31) ablated Sfh3 function in cells. However, the MD-simulated transition of the open Sfh3-sterol conformer to the closed conformer identified Gln230, Glu235, Thr264, and Lys267 as residues with which bound sterol was engaged in H-bond and water bridge interactions as the lipid orientation shifted toward the PtdIns headgroup–binding region (see previous section). Because Gln230 and Thr264 coordinate binding of the PtdIns PO4 moiety and Glu235 and Lys267 coordinate binding of the inositol headgroup (22), this raised the question of whether the Sfh3 PtdIns–binding barcode residues interacted only with PtdIns. This specificity concern was further highlighted by docking experiments that forecast proteins previously assumed to have PtdIns-specific binding defects (e.g. Sfh3T264A, Sfh3Q230A, Sfh3Q230A,T264A, and Sfh3Q230A,T264W) would also be impaired in sterol exchange.
Indeed, as predicted by the docking data, Sfh3Q230A, Sfh3T264A, and Sfh3E235A all showed significantly reduced DHE, [3H]cholesterol, and CTL exchange activities relative to Sfh3, whereas Sfh3K267A retained significant sterol exchange activity (Fig. 11, A–C). The Sfh3Q230A,T264A, Sfh3Q230A,T264W, and Sfh3E235A,K267A double mutant proteins had no measurable exchange activity for any of these sterol substrates (Fig. 11, A–C). These results closely mirrored data obtained from [3H]PtdIns transfer assays where Sfh3Q230A, Sfh3T264A, Sfh3E235A, and Sfh3K267A all showed reduced [3H]PtdIns transfer activities relative to Sfh3, whereas the Sfh3Q230A,T264A, Sfh3Q230A,T264W, and Sfh3E235A,K267A double mutant proteins were strongly defective in transfer of [3H]PtdIns (Fig. 11D) and [Pyr]PtdIns (Fig. 11E).
Figure 11.
In vitro lipid transfer activities of Sfh3 PtdIns-binding mutants. Transfer activities for the indicated Sfh3 variants were measured using DHE (A), [3H]cholesterol (B), CTL (C), [3H]PtdIns (D), and [Pyr]PtdIns (E) as transfer substrates. Assay incubations were for 10 min for the real-time fluorescence-based DHE and CTL assays and 30 min for [3H]cholesterol, [3H]PtdIns, and [Pyr]PtdIns transfer assays. All assays were performed at 37 °C, and protein input was clamped at 10 μg for all experiments. For DHE, CTL, and [Pyr]PtdIns assays, values for spontaneous transfer (no protein) were determined, and spontaneous transfer backgrounds were subtracted from each measurement. Transfer activities were normalized to Sfh3 activity. The averages of triplicate determinations from two independent experiments for DHE, CTL, and [Pyr]PtdIns, and triplicate determinations from three independent experiments for [3H]cholesterol and [3H]PtdIns, are shown. Error bars represent standard deviations for the [3H]cholesterol and [3H]PtdIns assays and mean ± range for DHE, CTL, and [Pyr]PtdIns assays.
However, as described above, the exceptional Sfh3L181Y mutant was the one case where the PtdIns/sterol-binding/transfer activities were successfully uncoupled, i.e. Sfh3L181Y unexpectedly retained sterol binding/transfer activity in the face of loss of PtdIns binding/transfer activity. Sterol-binding defects were predicted for Sfh3L181Y on the basis of initial docking models that placed Leu181 in proximity to both the bound sterol molecule and to the PtdIns acyl chains in the open Sfh3 conformer. However, MD simulation of the conformational transition of sterol-bound Sfh3 from the open to the closed conformer projected the sterol molecule to move toward the PtdIns headgroup–binding region, and away from Leu181, as the open sterol-bound Sfh3 conformer closed. The repositioning of the bound sterol during this conformational transition rationalizes the PtdIns-specific binding/transfer defects of Sfh3L181Y.
Discussion
Herein, we report a systematic investigation of structural features of Sec14-like PITPs, including their surface electrostatic properties, and the diverse shapes and chemical microenvironments of lipid-binding cavities, to gain an understanding of molecular mechanisms that generate diversity and specificity for lipid substrates across the Sec14 superfamily. Using integrated X-ray crystallographic and computationally-based approaches, the conservation and evolution of the diverse functionalities in Sec14-like PITPs were interrogated. Particular focus was trained on defining the comparative anatomies of the Sec14-like PITP LBDs in terms of the physical and chemical microenvironments that these structural elements provide for lipid binding. Using the functionally enigmatic Sfh2 and the lipid droplet–associated Sfh3 as experimental foci, we identify squalene and sterols as exchangeable second lipid-binding ligands for Sfh2 and Sfh3, respectively. Taken together, the collective data indicate that the Sec14-like PITP lipid-binding pockets define diverse chemical microenvironments, that these diversifications reflect differential ligand-binding specificities across this protein family, and that these ligand-binding specificities are associated with functional diversification.
Selective conservation of physical properties across the Sec14-protein superfamily
A remarkable feature of Sec14-like proteins in yeast is their ability to diversify the biological outcomes of PtdIns(4)P signaling in cells. These available data suggest that Sec14-like PITPs instruct biological outcomes for PtdIns 4-OH kinase activity, and current ideas are that these PITPs do so via a regulated metabolic channeling of PtdIns(4)P production to dedicated effectors of PtdIns(4)P signaling (4, 12). In that regard, we find that, although the structural fold of Sec14-like PITPs (as defined by the α-carbon backbone) was highly conserved across widely-diverged fungal species (S. cerevisiae and S. pombe diverged from each other ∼350 million years ago (35)), proteins of this family differed in at least two general respects. The first involved protein surface chemistry where the electrostatic surface properties of Sec14 orthologs were impressively preserved across wide evolutionary distances. These PITPs showed highly-asymmetric distributions of negative and positive surface charge with the basic regions concentrated in the vicinity of the helical gate element that controls access to the lipid-binding pocket. By contrast, four of the five Sec14 homologs of S. cerevisiae presented very different surface charge distributions. Sfh1, the protein most similar in primary sequence to Sec14, was the single exception.
The second general area of divergence involved the nature of the lipid-binding cavity. An integrated VICE/HINT cavity mapping approach indicated that the dimensions and the chemical environments of the lipid-binding cavities of Sec14, its homologs from distant fungal species, and of Sfh1, were again highly conserved, as were the ligand-binding/exchange properties of those PITPs. The Sec14 proteins from S. cerevisiae and S. pombe are PtdIns/PtdCho-transfer proteins in vitro as is Sfh1 (3, 25, 36). By contrast, not only did the PtdCho-binding barcode regions deviate from those of Sec14 in the other four yeast Sfh proteins, but the general shapes and chemical fingerprints of their corresponding lipid-binding cavities were also distinct from those of Sec14 and from each other. Those features further reinforce the concept that Sec14-like proteins have diverged to accommodate the binding of a wide variety of lipids/lipophilic molecules while preserving the features critical for PtdIns-binding/exchange.
Squalene as second ligand for a Sec14-family protein
The most unusual yeast Sec14-like PITPs from the standpoint of the VICE/HINT analyses were Sfh2 and Sfh5. Whereas Sfh5 exhibited distinct electropositive character, Sfh2 was characterized by its unique toroid-shaped hydrophobic cavity. This property of a toroid-shaped lipid-binding cavity is also shared by the mammalian Sec14-like squalene-binding protein Sec14L2, and we demonstrate that Sfh2 is a robust PtdIns and squalene exchange protein, thereby identifying squalene as a bona fide second ligand for this Sec14-like PITP.
There are two satisfying aspects of those results. First, the fact that the unique topology of the Sfh2 lipid-binding cavity translated to a specific ability to bind and exchange squalene validated the basic premise of using the VICE/HINT approach to predict ligand-binding diversification in the yeast Sec14-like PITPs. Second, the data promise to inform downstream analyses of Sfh2 function in vivo. Sfh2 is a functionally enigmatic PITP that has been indirectly implicated, on the basis of genetic interactions, to be involved in a number of in vivo activities such as regulation of oxidative stress, hypoxia-related fatty acid synthesis, DNA replication stress, etc. (37–40). Our collective data, including the demonstration that Sfh2-deficient cells are sensitized to terbinafine challenge, a condition expected to elevate intracellular squalene levels, now suggest that Sfh2 activity is functionally linked either to squalene homeostasis or in potentiating a squalene-induced stress response.
Sterols as second ligands for Sec14-family proteins
Of the yeast Sec14-like PITPs, Sfh3 was notable for both the general hydrophobicity and the shallowness of its lipid-binding cavity, and those unique properties also translated to lipid ligand specificity. Sfh3 was the only one of the S. cerevisiae Sec14-like proteins that exhibited robust sterol transfer activity, and it did so regardless of whether DHE, cholesterol, or CTL was offered as binding/exchange substrates. Indeed, Sfh3 compared favorably with the known DHE-transfer protein Kes1 in its ability to transfer each of these sterol ligands between membranes in vitro. On the basis of such data, we conclude that the most likely binding ligand for Sfh3, in addition to PtdIns, is the major yeast sterol ergosterol. These data further demonstrate that Sec14-like PITPs, as a class of proteins, are not limited to phospholipids as binding/exchange ligands but are able to bind/exchange sterols or sterol-like molecules as well. This biochemical property is interesting given Sfh3 associates with a subset of lipid droplets enriched in sterol hydrolase activities in yeast cells undergoing meiosis, and that this PITP functions in inhibiting lipid mobilization from lipid droplets (22).
The closest yeast homolog to Sfh3 is Sfh4, and Sfh3 shares no obvious functional redundancy with Sfh4 in vivo (10, 22). Consistent with the functional divergence of Sfh3 and Sfh4, VICE/HINT cavity mapping data showed that, irrespective of the general shape similarity of the chemical profiles of the Sfh3 and Sfh4 lipid-binding cavities, the corresponding microenvironment profiles were quite different. Moreover, as was evident from direct biochemical assays, Sfh4 was a poor DHE exchange protein for which we measured no significant cholesterol or CTL exchange activity. Thus, whereas Sfh4 might also bind a sterol-like molecule as a second exchangeable ligand, we project that this ligand will be distinct from ergosterol (possibly a late-stage intermediate in the ergosterol biosynthetic pathway?).
Implications for interpretation and exploitation of lipid-binding barcodes
High-resolution crystal structures of closed Sfh1 conformers individually bound to PtdIns or PtdCho provided the startling demonstration that the motifs that coordinate headgroup binding for these phospholipids were physically and spatially distinct in Sec14-like PtdIns/PtdCho-transfer proteins (3). Those data permitted the recognition of physically discrete PtdIns- and PtdCho headgroup-specific binding barcodes at the level of the primary sequence (3–6). The definition of PtdIns- and PtdCho-binding barcodes, in turn, provide useful guides for generating headgroup-specific binding mutants for as yet poorly characterized Sec14-like proteins. This is particularly true for the PtdIns-binding barcode as it is conserved across the Sec14 protein superfamily. The ability to generate specific headgroup-binding mutants in Sec14-like PITPs that do not exchange PtdCho suggests experimental avenues for rigorous interrogation of the “second” ligand model for priming PITP-dependent presentation of PtdIns to the PtdIns 4-OH kinase (3, 10, 13, 22–24).
The results reported herein, however, discourage tacit extrapolation of the biochemical specificities of defects that come with altered PtdIns-binding barcodes for Sec14-like PITPs where the second ligand remains unknown. Two recent studies of Sfh3 function in vivo provide a case in point. Both studies assumed that perturbation of the Sfh3 PtdIns-binding barcode would result in specific inactivation of PtdIns-binding/exchange activity (22, 31). One of those studies directly confirmed that PtdIns-binding/exchange activity and the ability to stimulate PtdIns 4-OH kinase activity in vivo were indeed ablated by such perturbations (22), and both studies found that such perturbations completely abolished Sfh3 biological function. Those phenotypic data were taken as evidence that PtdIns-binding/exchange is an essential functional property of Sfh3 (22, 31). Although that conclusion may yet prove correct, the data reported herein demonstrate that neither of the PtdIns-binding mutants used in those studies inactivated Sfh3 PtdIns-binding activity specifically. Rather, both PtdIns- and sterol-exchange were compromised.
Unlike the case of the PtdIns and PtdCho binding barcodes, which might yet prove exceptional in their discreteness, the Sfh3 data indicate substantial overlap of lipid-binding barcodes might prove a common scenario in Sec14-like PITPs. Indeed, this also appears to be the case for Sfh2. The squalene binding space in the Sfh2 lipid-binding cavity overlaps extensively with that of PtdIns. Consistent with such an assignment, we have to date failed to generate mutants selectively ablated for squalene binding/exchange that retain uncompromised PtdIns-binding/exchange activity. The Sfh3L181Y mutant, where the sterol- and PtdIns-binding/transfer activities are cleanly uncoupled, is encouraging in that it suggests that specific headgroup-binding mutants will be forthcoming for Sfh3. Unfortunately, the utility of Sfh3L181Y is limited in this regard as the mutant protein is unstable when expressed in yeast (data not shown).
Taken together, the data herein clearly demonstrate that the hydrophobic lipid-binding cavities of Sec14-like PITPs present chemically diverse landscapes that translate to functional diversities for “second-ligand” binding. This property fulfills one prediction of the “second-ligand priming” model for how Sec14-like PITPs stimulate the activities of PtdIns 4-OH kinases and identifies Sec14-like PITPs as particularly well-suited to contribute to the remarkable diversification of the biological outcomes of phosphoinositide signaling. In that regard, these studies identify Sfh5 as a genuinely unique Sec14-like protein given its hydrophilic and highly-basic surface and cavity properties. Those unusual properties suggest the interesting possibility that the ligand-binding properties of Sfh5 and other Sec14-like proteins extend beyond lipids.
Materials and methods
Protein preparation and model building
The X-ray crystal structures of Sec14, Sfh1, Sfh3, and other mammalian proteins were obtained from PDB repository. The protein models were prepared using the Protein Preparation Wizard panel in the Schrödinger suite. The Prime program (41) was used to predict any missing loops, and the resulting complete structures were optimized with the OPLS_2005 force field in the Schrödinger suite to relieve all atom and bond strains found after adding all missing side chains and/or atoms. The small molecule model structures for cholesterol and ergosterol were prepared and energy-minimized in MOE (42), and the lowest energy conformations of each were selected for docking.
Sfh3 model building
The 1.55 Å X-ray crystal structure of the Sfh3 homodimer (PDB code 4J7Q) was used in this study. The protein model was prepared using the Protein Preparation Wizard Panel in Schrodinger Suite. As interpretable electron density for the N-terminal half of helix A7 (210VPGNSKIP217) was missing from the electron density map, Prime was used to predict the missing loop, and the complete structure was optimized to relieve all atom and bond constraints after adding all side chains and missing atoms. Structures of cholesterol and ergosterol were prepared in MOE, and the lowest energy conformation was selected for docking.
Homology modeling
All sequence and structural alignments were carried out within the MOE software package version 2013.08. Homology models of Sec14 for the fungal species were generated with the MOE 2013.08 modeling package. The Sec14 and homolog sequences to be modeled were aligned to the Sec14 (target) crystal structure sequences in the following: (i) its open conformation (PDB code 1AUA; 2.50 Å resolution); (ii) Sfh1 bound with PtdIns (PDB code 3B7N; 1.86 Å resolution); and (iii) Sfh1 bound with PtdCho (PDB code 3B7Q; 2.03 Å resolution). The latter two Sfh1 crystal structures, with the PtdIns and PtdCho ligands, were included in the environment to facilitate generating induced-fit homology models of Sec14-PtdIns/PtdCho complexes. By default, 10 independent intermediate models were generated. These different intermediate homology models were generated as a result of permutational selection of different loop candidates and side-chain rotamers. The intermediate model, which scored best according to the Amber99 force field, was chosen as the final model and was then subjected to further optimization.
Docking studies
Computational docking was carried out using the genetic algorithm-based ligand docking program GOLD 5.2 (43). GOLD 5.2 includes algorithms that provide limited flexibility to protein side chains with hydroxyl groups by reorienting the hydrogen bond donor and acceptor groups. The active site was defined by manually generating a centroid in the LBD of Sfh3 surrounded by a volume with a radius of 10 Å around that centroid created with the GOLD cavity detection algorithm. GOLD docking was carried out without using any constraints or biases to explore all possible solutions. In this study, we carried out three separate 10, 100, and 1000 GOLD genetic algorithm runs with early termination turned off. To explore all the possible binding modes, docking was carried out so as to generate diverse solutions. All other parameters were as defaults. To evaluate and validate GOLD performance, the co-crystallized substrate PtdIns was extracted and docked. GOLD, using our protocol, accurately reproduced the experimentally observed binding mode of PtdIns in Sfh3 and squalene in Sec14L2. Cholesterol and ergosterol were then docked to Sfh3, and squalene was docked to homology model of Sfh2 and scored using the CHEMPLP-scoring function within GOLD. CHEMPLP was used because it gives the highest success rates for both pose prediction and virtual screening experiments against diverse validation test sets (44).
Surface electrostatics
ICM-Pro was used to calculate the electrostatic potentials of the molecules in this study using the boundary element algorithm (45), and from that we generated a 3D surface skin model colored by potential. The ICM-Pro Rapid Exact-Boundary Electrostatics (REBEL) methodology allows solutions of the Poisson equation for a molecule without a grid and with exact positions of electric charges (46). The energies so calculated consisted of terms representing the intramolecular Coulomb energy and solvation energy, and those terms were plotted as colors on the surface elements according to potentials from red (most negative) to blue (most positive).
Cavity identification
Binding pockets of Sec14 and Sec14-like homologs were detected using the VICE algorithm (26), which is an option in HINT (28), implemented as a local module within SYBYL 8.1. The VICE algorithm is a grid-based method that propels vectors in directions that are defined by integer numbers of grid cubes, e.g. (0, 0, 1), (0, 1, 0), (1, 0, 0), (0, 1, 1), (1, 0, 1), (1, 1, 0), (1, 1, 1), (1, 1, 2), (1, 2, 1) (2, 1, 1), etc., rather than in directions defined by angles. These vectors are projected until they either hit a cube occupied by a molecule (putatively a cavity wall) or the edge of a grid box. For each grid cube, the fraction of vectors finding a cavity wall defines it “cavityness”; cubes with cavityness of > ∼0.5 are thus inside a cavity. Two clean-up phases finish the algorithm: 1) cubes with few adjacent neighbors also within the cavity are discarded as small crannies; 2) closed surfaces with volumes smaller than a default are also discarded. VICE was used to search for pockets within the LBDs of Sec14 and Sfh proteins in PtdIns/PtdCho complexes, where the LBD was defined to include all amino acid residues within 10 Å of the bound ligand. The grid resolution was set at 0.5 Å, and the minimum closed contour value was set to be 60 Å3. The default cavity definition was set to 0.45, and the contour value was set to 0.4. All other variables were kept at their default values.
Cavity mapping
HINT 3D complement maps were calculated for the cavity regions within the region of interest nominally defined as the molecular (dimensional) extent of the substrate within the binding cavity. This HINT algorithm projects the hydropathic character and intensity of the atoms adjacent to unoccupied grid cubes to hypothesize the character and intensity of an “ideal” atom occupying that space, i.e. a Lewis acid would be ideal for interacting with Lewis base neighbors, etc. (26–28). Its distance to these other atoms mediates its hydropathic intensity. In practice, this is accomplished with map pairs: a hydrophobic/polar map where positive field values represent hydrophobic regions of space, and negative field values represent polar (hydrophilic) regions. The second map, acid/base, depicts the acidic polar and basic polar regions of space with positive and negative field values, respectively. The VICE cavity map, represented as a Boolean (1 = inside cavity; 0 = outside cavity), was then multiplied by each of the complement maps in the map pair to focus the representation within the cavity. The final overall display of these contoured maps showed regions of the active site most hospitable to hydrophobic groups (green contours) and differentiates between the two types of polar regions (Lewis acid-like in red and Lewis base-like in blue). Generally, contour values were chosen such that their values were ∼50% of the maximum (or minimum) field value.
Molecular dynamics simulations
All atom molecular dynamics simulations were carried out using the Desmond MD simulation program (47), and analyses were performed using the Schrodinger Software package. Structures of Sfh3-sterol complex, Sfh3 mutants, and apo-Sfh3 were used as the starting structures for an all-atom molecular dynamics simulation in solvent boxes with explicit water molecules. All the parameters were kept the same for these simulations to maintain consistency. OPLS2005 force field as implemented in the Desmond MD package was used to describe all the atoms in the system. Periodic boundary conditions were set with an orthorhombic box with a 12 Å buffer to the box edge. The complex was solvated with TIP3P water model that is compatible with OPLS parameterization (48, 49). Physiological salt concentration was fixed to 150 mm by adding custom Na+ and Cl− counter ions to neutralize the total charge on the system. The system was set up at 300 K temperatures and at 1 bar of atmospheric pressure. A series of pre-defined energy minimization and equilibration steps was carried out to thoroughly relax and equilibrate the system before the final production run. A 50-ns production run was then carried out at NPT ensemble conditions. Post-MD simulation analyses were carried out with the simulation interaction diagram protocol as implemented in Schrodinger package.
Molecular graphics and chemical drawing
Molecular graphics and analyses were performed with the Schrodinger and MOE software suites. Marvin was used for drawing, displaying, and characterizing chemical structures, substructures, and reactions (Marvin 5.10.0 and 5.11.4, 2012; ChemAxon. Chemical drawings were also produced using ChemBioDraw Ultra, CambridgeSoft, version 11.01.1.
Lipids and liposomes
l-α-Phosphatidylcholine, (chicken) egg PtdCho, l-α-phosphatidylinositol (soy), and l-α-phosphatidic acid (chicken) egg of highest available quality were purchased from Avanti Polar Lipids (Alabaster, AL) and used without further purification. 1-Palmitoyl-2-decapyrenyl-sn-glycero-3-phosphocholine, [Pyr]PtdCho, and 1-palmitoyl-2-decapyrenyl-sn-glycero-3-phosphoinositol ([Pyr]PtdIns), were generous gifts from Dr. Pentti Somerharju (Helsinki University, Helsinki, Finland), and synthesis of these lipids was described previously (50, 51). 2,4,6-Trinitrophenyl-phosphatidylethanolamine (TNP-PtdEtn) was prepared from phosphoethanolamine as described by Gordesky and Marinetti (52) and purified by silica gel column chromatography. CTL was a generous gift from Dr. J. Peter Slotte (Åbo Akademi University, Turku, Finland). [3H]Cholesterol (1 mCi/ml) was obtained from American Radiolabeled Chemicals (St. Louis, MO). The concentrations of all phospholipid solutions were determined by Rouser et al. (53). [Pyr]PtdCho and [Pyr]PtdIns concentrations were determined spectroscopically by using ϵ = 42,000 and DHE and CTL by using ϵ = 13,000 and ϵ = 11,250, respectively.
For all fluorescence transfer assays, donor and acceptor vesicle solutions were prepared as follows. Donor vesicles were made from egg PtdCho, fluorescent probe ([Pyr]PtdIns, [Pyr]PtdCho, DHE, or CTL), and TNP-PtdEtn mixed together from stock solution in a 4:0.5:0.5 nmol ratio ([Pyr]PtdCho/[Pyr]PtdIns) or 20:2.5:2.5 nmol ratio (DHE/CTL). Solvent was evaporated under a stream of nitrogen, and the lipid film was resuspended in 10 μl of EtOH. For the acceptor vesicles, stock solutions of egg PtdCho and phosphatidic acid were mixed in a 291:9 nmol (97/3 mol %) ratio and dried under a stream of nitrogen. The resulting lipid film was then hydrated with 2 ml of low phosphate buffer and sonicated on ice for 10 min.
[3H]Squalene transfer assays
Donor liposomes were composed of PtdCho/[3H]squalene (99/1 weight %). For each assay, 40 μg of lipid was dried under a stream of N2 gas and resuspended by vortexing in 50 μl of phosphate buffer (300 mm NaCl, 25 mm Na2HPO4, 5 mm β-mercaptoethanol, 1 mm NaN3, pH 7.5). Small unilamellar vesicles were then generated from the suspension by sonication in an ice bath. Squalene transfer assays were then conducted using bovine heart mitochondria as acceptor membranes as described previously for PtdCho-transfer assays (3, 11, 13). [12,13-3H]Squalene (0.1 mCi/ml) was obtained from American Radiolabeled Chemicals (St. Louis, MO).
Fluorescence-based lipid transfer assays
The [Pyr]PtdCho and [Pyr]PtdIns binding and transfer measurements were performed as described by Somerharju et al. (54) with modification. For [Pyr]PtdIns/[Pyr]PtdCho transfer measurements, 10 μl of the donor vesicles were injected into 2 ml of acceptor vesicle solution. After donor vesicle addition, the solution was incubated for 5–10 min at 37 °C after which the fluorescence intensity was recorded at 378 nm (excitation 346 nm) as a function of time using a Horiba Fluorolog 3 fluorimeter (Horiba Ltd., Kyoto, Japan). To initiate protein-mediated lipid transfer, 1 μg of protein was injected every 250 s for a total of four injections followed by a final 5-μg injection at t = 1250 s to achieve binding-site saturation.
For the DHE and CTL transfer measurements, the method was adjusted to consider the rapid spontaneous transfer of sterol between membranes (t½ ∼10 min for CTL and DHE compared with t½ ∼24 h for [Pyr]PtdIns and [Pyr]PtdCho). 2 ml of acceptor vesicles were incubated for 5 min with 10 μg of protein after which 10 μl of donor vesicles were added to the solution, and the data collection was started immediately. Data were collected for a total of 10 min to reach equilibrium (excitation 338 nm/emission 395 nm for DHE and excitation 325 nm/emission 395 nm for CTL). For each prepared batch of donor and acceptor vesicles, a “no protein” measurement was also performed to record the amount of spontaneous transfer, and those data points were then subtracted from the measurements containing protein. All measurements were normalized for the value at t = 0.
[3H]PtdIns and [3H]cholesterol transfer assays
To measure lipid and sterol transfer using natural ligands (PtdIns and cholesterol), end-point radioactive transfer assays were performed. [3H]PtdIns transport was measured from rat liver microsomes to liposomes (98 mol % PtdCho and 2 mol % PtdIns) and [3H]cholesterol from liposomes (98 mol % PtdCho and 2 mol % cholesterol) to bovine heart mitochondria as described (8, 55).
Expression and purification of recombinant proteins
The SFH3 gene was amplified from the yeast genome and integrated into a pET28B-His8 protein expression plasmid. All mutations were confirmed by DNA sequencing outsourced to the Eton Biosciences service. Recombinant His8Sfh3 and mutant variants were purified as described by Ren et al. (22) using an E. coli BL21 (RIL/DE3; New England Biolabs Inc., Ipswich, MA) expression system.
Author contributions
A. T., E. M., A. J. O., M. G. L., M. L., D. K., K. G. S., C. J. M., G. E. K., and V. A. B. conceptualization; A. T., E. M., A. J. O., M. G. L., M. L., K. G. S., G. E. K., and V. A. B. formal analysis; A. T., E. M., M. G. L., M. L., D. K., K. G. S., G. E. K., and V. A. B. validation; A. T., E. M., A. J. O., M. G. L., M. L., D. K., K. G. S., C. J. M., G. E. K., and V. A. B. investigation; A. T., E. M., A. J. O., M. G. L., M. L., D. K., G. E. K., and V. A. B. visualization; A. T., E. M., A. J. O., M. G. L., M. L., D. K., K. G. S., G. E. K., and V. A. B. methodology; A. T., E. M., A. J. O., M. G. L., M. L., D. K., K. G. S., G. E. K., and V. A. B. writing-original draft; A. T., E. M., M. G. L., M. L., D. K., K. G. S., C. J. M., G. E. K., and V. A. B. writing-review and editing; A. T., E. M., and V. A. B. resources; A. T., A. J. O., and G. E. K. software; G. E. K. data curation; A. T., A. J. O., G. E. K. and V. A. B. supervision; G. E. K. and V. A. B. funding acquisition; G. E. K. and V. A. B. project administration.
Supplementary Material
Acknowledgments
The Laboratory for Molecular Simulation and High Performance Research Computing at Texas A&M University provided software, support, and computer time. We thank Kaitlyn McGrath for help with lipid transfer assays, and Pentti Somerharju (Helsinki, Finland) and J. Peter Slotte (Turku, Finland) for their generous gifts of fluorescent lipids.
This work was supported by National Institutes of Health Grants R35 GM131804 and Robert A. Welch Foundation Grant BE-0017 (to V. A. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Tables S1–S8.
- PtdIns
- phosphatidylinositol
- PtdIns(4)P
- phosphatidylinositol 4-phosphate
- DHE
- dehydroergosterol
- CTL
- cholestatrienol
- Pyr
- pyroglutamic acid
- MD
- molecular dynamics
- MDS
- molecular dynamics simulation
- PITP
- phosphatidylinositol-transfer protein
- LBD
- lipid-binding domain
- PDB
- Protein Data Bank
- PtdCho
- phosphatidylcholine
- VICE
- vectorial identification of cavity extent
- [Pyr]PtdIns
- 1-palmitoyl-2-decapyrenyl-sn-glycero-3-phosphoinositol
- TNP-PtdEtn
- 2,4,6-trinitrophenyl-phosphatidylethanolamine.
References
- 1. Strahl T., and Thorner J. (2007) Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771, 353–404 10.1016/j.bbalip.2007.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schaaf G., Ortlund E. A., Tyeryar K. R., Mousley C. J., Ile K. E., Garrett T. A., Ren J., Woolls M. J., Raetz C. R., Redinbo M. R., and Bankaitis V. A. (2008) The functional anatomy of PL binding and regulation of phosphoinositide homeostasis by proteins of the Sec14-superfamily. Mol. Cell 29, 191–206 10.1016/j.molcel.2007.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bankaitis V. A., Mousley C. J., and Schaaf G. (2010) The Sec14-superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci 35, 150–160 10.1016/j.tibs.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nile A. H., Bankaitis V. A., and Grabon A. (2010) Mammalian diseases of phosphatidylinositol transfer proteins and their homologs. Clin. Lipidol. 5, 867–897 10.2217/clp.10.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grabon A., Khan D., and Bankaitis V. A. (2015) Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology. Biochim. Biophys. Acta 1851, 724–735 10.1016/j.bbalip.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bankaitis V. A., Malehorn D. E., Emr S. D., and Greene R. (1989) The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 108, 1271–1281 10.1083/jcb.108.4.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bankaitis V. A., Aitken J. R., Cleves A. E., and Dowhan W. (1990) An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347, 561–562 10.1038/347561a0 [DOI] [PubMed] [Google Scholar]
- 9. Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., Dowhan W., Goebl M., and Bankaitis V. A. (1991) Mutations in the CDP-choline pathway for PL biosynthesis bypass the requirement for an essential PL transfer protein. Cell 64, 789–800 10.1016/0092-8674(91)90508-V [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu W.-I., Routt S., Bankaitis V. A., and Voelker D. R. (2000) A new gene involved in transport-dependent metabolism of phosphatidylserine, PSTB2/PDR17, shares sequence similarity with the gene encoding the PI-/PC-TP, Sec14p. J. Biol. Chem. 275, 14446–14456 10.1074/jbc.275.19.14446 [DOI] [PubMed] [Google Scholar]
- 11. Vincent P., Chua M., Nogue F., Fairbrother A., Mekeel H., Xu Y., Allen N., Bibikova T. N., Gilroy S., and Bankaitis V. A. (2005) A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J. Cell Biol. 168, 801–812 10.1083/jcb.200412074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghosh R., de Campos M. K., Huang J., Hur S. K., Orlowski A., Yang Y., Tripathi A., Nile A., Lee H.-C., Schäfer H., Dynowski M., Róg T., Lete M. G., Ahyayauch H., Alonso A., et al. (2015) Sec14-nodulin proteins and the patterning of phosphoinositide landmarks for developmental control of membrane morphogenesis. Mol. Biol. Cell 26, 1764–1781 10.1091/mbc.E14-10-1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang J., Ghosh R., Tripathi A., Lönnfors M., Somerharju P., and Bankaitis V. A. (2016) Two-ligand priming mechanism for potentiated phosphoinositide synthesis is an evolutionarily conserved feature of Sec14-like phosphatidylinositol and phosphatidylcholine exchange proteins. Mol. Biol. Cell 27, 2317–2330 10.1091/mbc.E16-04-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Milligan S. C., Alb J. G. Jr., Elagina R. B., Bankaitis V. A., and Hyde D. R. (1997) The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration protein B is required for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 139, 351–363 10.1083/jcb.139.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giansanti M. G., Bonaccorsi S., Kurek R., Farkas R. M., Dimitri P., Fuller M. T., and Gatti M. (2006) The class I PITP giotto is required for Drosophila cytokinesis. Curr. Biol. 16, 195–201 10.1016/j.cub.2005.12.011 [DOI] [PubMed] [Google Scholar]
- 16. Hamilton B. A., Smith D. J., Mueller K. L., Kerrebrock A. W., Bronson R. T., van Berkel V., Daly M. J., Kruglyak L., Reeve M. P., Nemhauser J. L., Hawkins T. L., Rubin E. M., and Lander E. S. (1997) The vibrator mutation causes neurodegeneration via reduced expression of PITPα: positional complementation cloning and extragenic suppression. Neuron 18, 711–722 10.1016/S0896-6273(00)80312-8 [DOI] [PubMed] [Google Scholar]
- 17. Alb J. G. Jr, Cortese J. D., Phillips S. E., Albin R. L., Nagy T. R., Hamilton B. A., and Bankaitis V. A. (2003) Mice lacking phosphatidylinositol transfer protein-α exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J. Biol. Chem. 278, 33501–33518 10.1074/jbc.M303591200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alb J. G. Jr., Phillips S. E., Wilfley L. R., Philpot B. D., and Bankaitis V. A. (2007) The pathologies associated with functional titration of phosphatidylinositol transfer protein α activity in mice. J. Lipid Res. 48, 1857–1872 10.1194/jlr.M700145-JLR200 [DOI] [PubMed] [Google Scholar]
- 19. Ile K. E., Kassen S., Cao C., Vihtehlic T., Shah S. D., Mousley C. J., Alb J. G. Jr., Huijbregts R. P., Stearns G. W., Brockerhoff S. E., Hyde D. R., and Bankaitis V. A. (2010) The zebrafish class 1 phosphatidylinositol transfer protein family: PITPβ isoforms and double cone cell outer segment integrity in retina. Traffic 11, 1151–1167 10.1111/j.1600-0854.2010.01085.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sha B., Phillips S. E., Bankaitis V. A., and Luo M. (1998) Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391, 506–510 10.1038/35179 [DOI] [PubMed] [Google Scholar]
- 21. Yang H., Tong J., Leonard T. A., and Im Y. J. (2013) Structural determinants for phosphatidylinositol recognition by Sfh3 and substrate-induced dimer–monomer transition during lipid transfer cycles. FEBS Lett. 587, 1610–1616 10.1016/j.febslet.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 22. Ren J., Pei-Chen Lin C., Pathak M. C., Temple B. R., Nile A. H., Mousley C. J., Duncan M. C., Eckert D. M., Leiker T. J., Ivanova P. T., Myers D. S., Murphy R. C., Brown H. A., Verdaasdonk J., Bloom K. S., et al. (2014) A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress-induced membrane biogenesis. Mol. Biol. Cell 25, 712–727 10.1091/mbc.e13-11-0634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X., Routt S. M., Xie Z., Cui X., Fang M., Kearns M. A., Bard M., Kirsch D. R., and Bankaitis V. A. (2000) Identification of a novel family of nonclassical yeast PITPs whose function modulates activation of phospholipase D and Sec14p-independent cell growth. Mol. Biol. Cell 11, 1989–2005 10.1091/mbc.11.6.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Routt S. M., Ryan M. M., Tyeryar K., Rizzieri K. E., Mousley C., Roumanie O, Brennwald P. J., and Bankaitis V. A. (2005) Nonclassical PITPs activate phospholipase D via an Stt4p-dependent pathway and modulate function of late stages of the secretory pathway in vegetative yeast cells. Traffic 6, 1157–1172 10.1111/j.1600-0854.2005.00350.x [DOI] [PubMed] [Google Scholar]
- 25. Schaaf G., Dynowski M., Mousley C. J., Shah S. D., Yuan P., Winklbauer E. M., de Campos M. K., Trettin K., Quinones M. C., Smirnova T. I., Yanagisawa L. L., Ortlund E. A., and Bankaitis V. A. (2011) Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Mol. Biol. Cell 22, 892–905 10.1091/mbc.e10-11-0903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tripathi A., and Kellogg G. E. (2010) A novel and efficient tool for locating and characterizing protein cavities and binding sites. Proteins 78, 825–842 10.1002/prot.22608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tripathi A., Surface J. A., and Kellogg G. E. (2011) Using active site mapping and receptor-based pharmacophore tools: prelude to docking and de novo/fragment-based ligand design. Methods Mol. Biol. 716, 39–54 10.1007/978-1-61779-012-6_3 [DOI] [PubMed] [Google Scholar]
- 28. Eugene Kellogg G., and Abraham D. J. (2000) Hydrophobicity: is LogP(o/w) more than the sum of its parts? Eur. J. Med. Chem. 35, 651–661 10.1016/S0223-5234(00)00167-7 [DOI] [PubMed] [Google Scholar]
- 29. Christen M., Marcaida M. J., Lamprakis C., Aeschimann W., Vaithilingam J., Schneider P., Hilbert M., Schneider G., Cascella M., and Stocker A. (2015) Structural insights on cholesterol endosynthesis: binding of squalene and 2,3-oxidosqualene to supernatant protein factor. J. Struct. Biol. 190, 261–270 10.1016/j.jsb.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 30. Mousley C. J., Yuan P., Gaur N. A., Trettin K. D., Nile A. H., Deminoff S. J., Dewar B. J., Wolpert M., Macdonald J. M., Herman P. K., Hinnebusch A. G., and Bankaitis V. A. (2012) A sterol binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell 148, 702–715 10.1016/j.cell.2011.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holič R., Simová Z., Ashlin T., Pevala V., Poloncová K., Tahotná D., Kutejová E., Cockcroft S., and Griač P. (2014) Phosphatidylinositol binding of Saccharomyces cerevisiae Pdr16p represents an essential feature of this lipid transfer protein to provide protection against azole antifungals. Biochim. Biophys. Acta 1842, 1483–1490 10.1016/j.bbalip.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., and Drin G. (2011) Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195, 965–978 10.1083/jcb.201104062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang J., Mousley C. J., Dacquay L., Maitra N., Drin G., He C., Ridgway N. D., Tripathi A., Kennedy M., Kennedy B. K., Liu W., Baetz K., Polymenis M., and Bankaitis V. A. (2018) A lipid transfer protein signaling axis exerts dual control of cell-cycle and membrane trafficking systems. Dev. Cell 44, 378–391.e5 10.1016/j.devcel.2017.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van den Hazel H. B., Pichler H., do Valle Matta M. A., Leitner E., Goffeau A., and Daum G. (1999) PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem. 274, 1934–1941 10.1074/jbc.274.4.1934 [DOI] [PubMed] [Google Scholar]
- 35. Hoffman C. S., Wood V., and Fantes P. A. (2015) An ancient yeast for young geneticists: a primer on the Schizosaccharomyces pombe model system. Genetics 201, 403–423 10.1534/genetics.115.181503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakase Y., Nakamura T., Hirata A., Routt S. M., Skinner H. B., Bankaitis V. A., and Shimoda C. (2001) The S. pombe spo20+ gene encoding a homologue of S. cerevisiae Sec14p plays an important role in forespore membrane formation. Mol. Biol. Cell 12, 901–917 10.1091/mbc.12.4.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cha M. K., Hong S. K., Oh Y. M., and Kim I. H. (2003) The protein interaction of Saccharomyces cerevisiae cytoplasmic thiol peroxidase II with SFH2p and its in vivo function. J. Biol. Chem. 278, 34952–34958 10.1074/jbc.M301819200 [DOI] [PubMed] [Google Scholar]
- 38. Desfougères T., Ferreira T., Bergès T., and Régnacq M. (2008) SFH2 regulates fatty acid synthase activity in the yeast Saccharomyces cerevisiae and is critical to prevent saturated fatty acid accumulation in response to haem and oleic acid depletion. Biochem. J. 409, 299–309 10.1042/BJ20071028 [DOI] [PubMed] [Google Scholar]
- 39. Régnacq M., Ferreira T., Puard J., and Bergès T. (2002) SUT1 suppresses sec14-1 through upregulation of CSR1 in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 216, 165–170 10.1111/j.1574-6968.2002.tb11431.x [DOI] [PubMed] [Google Scholar]
- 40. Tkach J. M., Yimit A., Lee A. Y., Riffle M., Costanzo M., Jaschob D., Hendry J. A., Ou J., Moffat J., Boone C., Davis T. N., Nislow C., and Brown G. W. (2012) Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14, 966–976 10.1038/ncb2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jacobson M. P., Friesner R. A., Xiang Z., and Honig B. (2002) On the role of crystal packing forces in determining protein sidechain conformations. J. Mol. Biol. 320, 597–608 10.1016/S0022-2836(02)00470-9 [DOI] [PubMed] [Google Scholar]
- 42. Molecular Operating Environment (MOE) (2018) Chemical Computing Group ULC, Montreal, Quebec, Canada [Google Scholar]
- 43. Jones G., Willett P., Glen R. C., Leach A. R., and Taylor R. (1997) Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 10.1006/jmbi.1996.0897 [DOI] [PubMed] [Google Scholar]
- 44. Liebeschuetz J. W., Cole J. C., and Korb O. (2012) Pose prediction and virtual screening performance of GOLD scoring functions in a standardized test. J. Comput. Aided Mol. Des. 26, 737–748 10.1007/s10822-012-9551-4 [DOI] [PubMed] [Google Scholar]
- 45. Abagyan R. A., Totrov M. M., and Kuznetsov D. N. (1994) ICM–a new method for protein modeling and design. Applications to docking and structure prediction from the distorted native conformation. J. Comp. Chem. 15, 488–506 10.1002/jcc.540150503 [DOI] [Google Scholar]
- 46. Totrov M., and Abagyan R. (2001) Rapid boundary element solvation electrostatics calculations in folding simulations: successful folding of a 23-residue peptide. Biopolymers 60, 124–133 [DOI] [PubMed] [Google Scholar]
- 47. Bowers K. J., Chow E., Xu H., Dror R. O., Eastwood M. P., Gregersen B. A., Klepeis J. L., Kolossvary I., Moraes M. A., and Sacerdoti F. D. (November 11–17, 2006) in Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (SC06), Tampa, Florida [Google Scholar]
- 48. Jorgensen W. L., and Tirado-Rives J. (1988) The OPLS (optimized potentials for liquid simulations) potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 110, 1657–1666 10.1021/ja00214a001 [DOI] [PubMed] [Google Scholar]
- 49. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926 10.1063/1.445869 [DOI] [Google Scholar]
- 50. Gupta C. M., Radhakrishnan R., and Khorana H. G. (1977) GlyceroPL synthesis: improved general method and new analogs containing photoactivable groups. Proc. Natl. Acad. Sci. U.S.A. 74, 4315–4319 10.1073/pnas.74.10.4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Somerharju P., and Wirtz K. W. A. (1982) Semisynthesis and properties of a fluorescent phosphatidylinositol analogue containing a cis-parinaoyl moiety. Chem. Phys. Lipids 30, 81–91 10.1016/0009-3084(82)90009-3 [DOI] [Google Scholar]
- 52. Gordesky S. E., and Marinetti G. V. (1973) The asymmetric arrangement of PLs in the human erythrocyte membrane. Biochem. Biophys. Res. Commun. 50, 1027–1031 10.1016/0006-291X(73)91509-X [DOI] [PubMed] [Google Scholar]
- 53. Rouser G., Fkeischer S., and Yamamoto A. (1970) A two dimensional thin layer chromatographic separation of polar lipids and determination of PLs by phosphorus analysis of spots. Lipids 5, 494–496 10.1007/BF02531316 [DOI] [PubMed] [Google Scholar]
- 54. Somerharju P. J., van Loon D., and Wirtz K. W. (1987) Determination of the acyl chain specificity of the bovine liver phosphatidylcholine transfer protein. Application of pyrene-labeled phosphatidylcholine species. Biochemistry 26, 7193–7199 10.1021/bi00396a048 [DOI] [PubMed] [Google Scholar]
- 55. Khan D., McGrath K. R., Dorosheva O., Bankaitis V. A., and Tripathi A. (2016) Structural elements that govern Sec14-like phosphatidylinositol transfer protein sensitivities to potent small molecule inhibitors. J. Lipid Res. 57, 650–662 10.1194/jlr.M066381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.