Abstract
Photoisomerization of the 11-cis-retinal chromophore of rod and cone visual pigments to an all-trans-configuration is the initiating event for vision in vertebrates. The regeneration of 11-cis-retinal, necessary for sustained visual function, is an endergonic process normally conducted by specialized enzyme systems. However, 11-cis-retinal also can be formed through reverse photoisomerization from all-trans-retinal. A nonvisual opsin known as retinal pigment epithelium (RPE)-retinal G-protein–coupled receptor (RGR) was previously shown to mediate visual chromophore regeneration in photic conditions, but conflicting results have cast doubt on its role as a photoisomerase. Here, we describe high-level production of 11-cis-retinal from RPE membranes stimulated by illumination at a narrow band of wavelengths. This activity was associated with RGR and enhanced by cellular retinaldehyde-binding protein (CRALBP), which binds the 11-cis-retinal produced by RGR and prevents its re-isomerization to all-trans-retinal. The activity was recapitulated with cells heterologously expressing RGR and with purified recombinant RGR. Using an RGR variant, K255A, we confirmed that a Schiff base linkage at Lys-255 is critical for substrate binding and isomerization. Single-cell RNA-Seq analysis of the retina and RPE tissue confirmed that RGR is expressed in human and bovine RPE and Müller glia, whereas mouse RGR is expressed in RPE but not in Müller glia. These results provide key insights into the mechanisms of physiological retinoid photoisomerization and suggest a novel mechanism by which RGR, in concert with CRALBP, regenerates the visual chromophore in the RPE under sustained light conditions.
Keywords: G-protein–coupled receptor (GPCR), retinol, retinoid-binding protein, rhodopsin, retina
Introduction
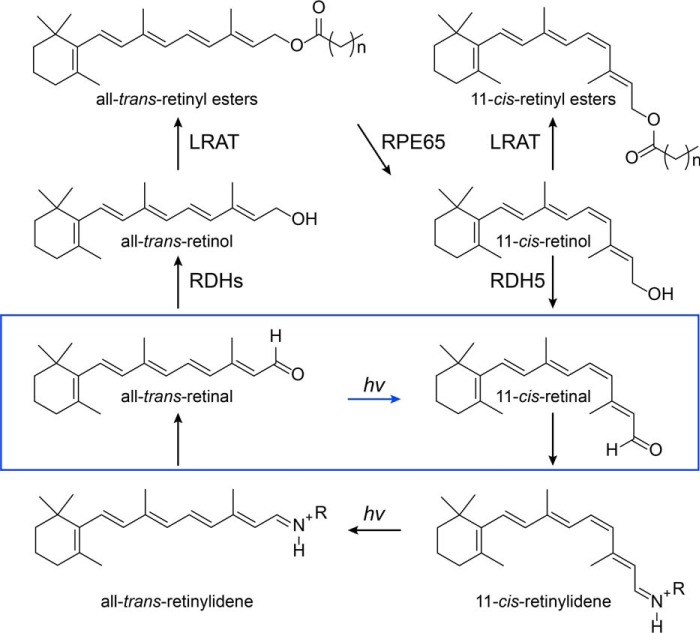
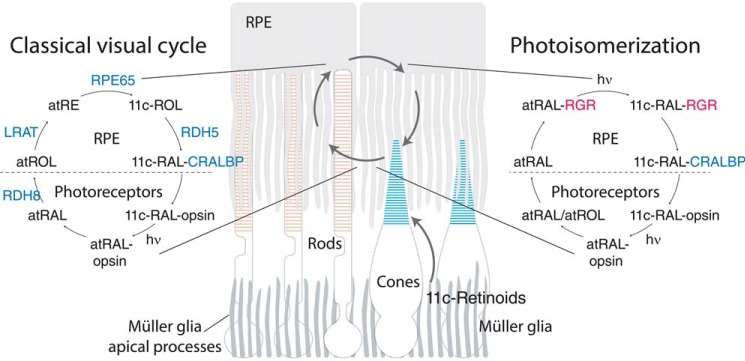
Our visual system is based on the photoisomerization of visual pigments in rod and cone photoreceptors (1–3). The forward bleaching reaction (cis-trans retinal isomerization) must be countered by trans-cis re-isomerization for continuous regeneration of the visual pigments. One source of regenerated 11-cis-retinal is a metabolic pathway referred to as the retinoid (visual) cycle (Fig. 1) (4) with a critical step that generates 11-cis-retinol from all-trans-retinyl esters via the action of a retinoid isomerase called RPE65 (5–7). This activity is consistent with an earlier proposal that light-independent isomerization of all-trans- to 11-cis-retinoids in the eye occurs at the alcohol oxidation state (8). It is also in agreement with genetic evidence of RPE65 playing a key role in 11-cis-retinal biosynthesis (9). An alternative light-independent pathway has also been described (10, 11), but the physiological relevance of its proposed enzyme activities has not been established (12). The overall rate of visual cycle activity can be estimated by electroretinography through dark adaptation measurements (13), in which visual sensitivity is monitored over time under dark conditions following light exposure (14–16) or by direct measurement of visual pigment levels (17–19). The dark adaptation process can be modulated by visual chromophore delivery to the photoreceptor outer segments or by the release of spent chromophore from opsins, which can be regulated by rhodopsin kinase (20).
Figure 1.
Schematic representation of retinoid transport and transformations in vision. Conversion of all-trans-retinal to 11-cis-retinal occurs through the classical visual cycle (black arrows) or the pathway described in this work, involving the RPE-retinal G-protein–coupled receptor (RGR) in the RPE under visible light (blue box).
Although the eye has mechanisms to limit retinal illuminance and visual pigment activation, for example by pupillary constriction, bright light exposure can result in high isomerization rates that may exceed the regeneration capacity of the visual cycle, particularly for foveal cones (21). This imbalance may be countered by the photochemical production of 11-cis-retinal under sustained light conditions. For example, it is known that exposure of all-trans-retinal–containing solutions to light results in the formation of a sizable fraction of 11-cis-retinal even though this isomer makes up a minority of the thermal equilibrium mixture (4). In vivo, it has been observed that blue light also can cause the regeneration of rhodopsin through direct photoreversal processes (9) or by photoisomerization of all-trans-retinal–phosphatidylethanolamine adducts (22, 23). However, because of the relatively low quantum yields of these photoisomerization reactions and the gradual loss of hydrolyzed all-trans-retinal (through reduction and trapping in the RPE3), these processes would serve only to slow the depletion of visual pigments rather than provide a means to sustain their levels in the face of high illumination. They also lack specificity and could generate nonphysiological toxic isomers such as 13-cis-retinoids, which are not normally observed in the eyes of healthy animals. Furthermore, in humans and other primates, the formation of cis-retinoids by blue light–dependent mechanisms is suppressed by the filtering of blue light by xanthophyll macular pigments before it can reach the photoreceptor and RPE cell layers (24).
Mollusks possess a well-characterized system for photic regeneration of 11-cis-retinal that consists of an opsin protein known as retinochrome and a retinoid-binding protein, which are phylogenetically related to RPE-retinal G-protein–coupled receptor (RGR)-opsin and to cellular retinaldehyde-binding protein (CRALBP) in vertebrates, respectively (25). RGR was originally reported to mediate a photic visual cycle in vertebrates (26), and activity was observed in Rpe65−/− but not Rpe65−/− Rgr−/− mice under light conditions (27). However, low 11-cis-retinal synthetic activity cast doubt on its physiological relevance (26, 28). The slightly higher 11-cis-retinal production at a shorter wavelength from the main Rgr absorbance maximum suggested nonenzymatic photochemical isomerization rather than RGR-dependent activity. Other studies have also indicated that RGR plays a more indirect role in 11-cis-retinal synthesis by mobilizing retinyl esters and enhancing RPE65 activity (29, 30), possibly through G-protein signaling. Furthermore, RGR inactivity or deficiency has been associated with recessive rod-cone dystrophy in humans, thus signifying its potential importance (31), although a cis-acting mutation in a nearby gene called CDHR1 likely contributes significantly to the phenotypes observed in these patients (32). Recent work on in mice on Rgr expressed in Müller cell using electrophysiological approaches has indicated that this protein, possibly working in concert with a retinol dehydrogenase enzyme (33, 34), makes a sizable contribution to cone visual pigment regeneration under sustained light conditions (35). However, robust Rgr-dependent 11-cis-retinal production has never been demonstrated at the biochemical level, and the function of RGR present in the RPE remains incompletely understood. Considering the importance of cone pigment regeneration in human visual physiology and pathophysiology, a more complete description of alternative visual cycle pathways is highly desirable. The goal of this study was to characterize a robust photoisomerase activity that we discovered in the microsomal fraction from RPE cells.
Results
Bovine RPE microsomes contain a high-level photoisomerase activity responsive to green light
The all-trans-retinal contained in isolated bovine RPE microsomes is not isomerized to its cis products if kept in the dark. However, when RPE microsomes were exposed to white light, we observed the formation of 11-cis-retinal, in amounts that fluctuated from experiment to experiment. To further explore this phenomenon and to minimize the variability of 11-cis-retinal production, we designed and built a precise illumination system capable of delivering narrow bandwidth light to the bovine RPE microsomal samples (Fig. S1a). The illumination was generated by LEDs with peak wavelengths ranging from 385 to 625 nm and bandwidths of about 50 nm or less, except for two diodes at long wavelength (Fig. S1b). The system covered most of the absorption spectra of cone and rod opsins (Fig. 2, a and b, and Fig. S1b) (36–38). For most experiments, the light power was kept below 250 μW. At such levels, temperature changes to the reaction solution were not evident. For experiments requiring variable temperature or higher light power, a temperature-controlled cuvette holder was utilized (see under “Experimental procedures” and Fig. S1c). Exogenous all-trans-retinal was added to the RPE microsomes at a molar excess of at least 10 times with respect to RPE65, the most abundant protein in the RPE (39), and recombinant human CRALBP was used to prevent any further nonphotoenzymatic isomerization events (40). The photoproducts were extracted with hexanes and analyzed by normal-phase HPLC (Fig. S2). The light-induced 11-cis-retinal production in the RPE was profiled according to three variables: (a) wavelength, (b) illumination intensity, and (c) length of light exposure (Fig. S3). With a fixed amount of RPE microsomes and the substrate under near-UV visible light (385–455 nm, overlapping with the absorbance of blue cone opsin, Fig. 2b), the maximum level of 11-cis-retinal was achieved in less than 20 min, and corresponded to ∼30% of the original, exogenous all-trans-retinal (Fig. S3, a–e). Increasing the near-UV light power did not increase 11-cis-retinal generation (Fig. S3, a–e). In contrast, medium wavelength light (490–565 nm, overlapping with the absorbance of human green and red cone opsin and rhodopsin, Fig. 2b (36–38)) converted all-trans-retinal into the 11-cis–configuration almost quantitatively, and it almost completely depleted the available exogenous substrate (Fig. S3, g–j). The reaction was efficient with well-tuned light power as low as 25 μW and illumination time of 60–120 min (Fig. S3, g–j). Further shift in the illumination wavelength toward the IR region resulted in a dramatic drop in photoisomerization efficiency. Generation of 11-cis-retinal became marginal when the 625-nm peak wavelength was used (Fig. S3l). Production of 11-cis-retinal was not observed in darkness (Fig. S2, a and b), whereas trace amounts of cis-retinals observed in the samples even before illumination (Fig. S2, c and d) were most likely derived from the incomplete depletion of retinoids in RPE microsomes pre-treated with intense UV light at 302 nm. In 395 nm light the amount of 9-cis-, 11-cis-, and 13-cis-retinal plateaued at 20 min, suggesting that photoisomerization had achieved equilibrium (Fig. S2a). In contrast, the accumulation of 11-cis-retinal under 530 nm light continued over the 60 min course of the experiment, accompanied by the gradual decrease of both 13-cis-retinal and all-trans-retinal (Fig. S2b). After 60 min of illumination, the amount of 11-cis-retinal reached 2,825 ± 189 pmol, more than 87% of the amount of the original all-trans-retinal, whereas the levels of the other retinoids were minimal (Fig. S2j). We found that under optimized conditions, the photic regeneration system can produce 11-cis-retinal at a rate of 1 nmol/min/mg crude protein, which is about 10 times greater than the rate of 11-cis-retinol formation by the RPE in the dark (41), indicating a substantial capacity of this system to support physiological visual pigment regeneration.
Figure 2.
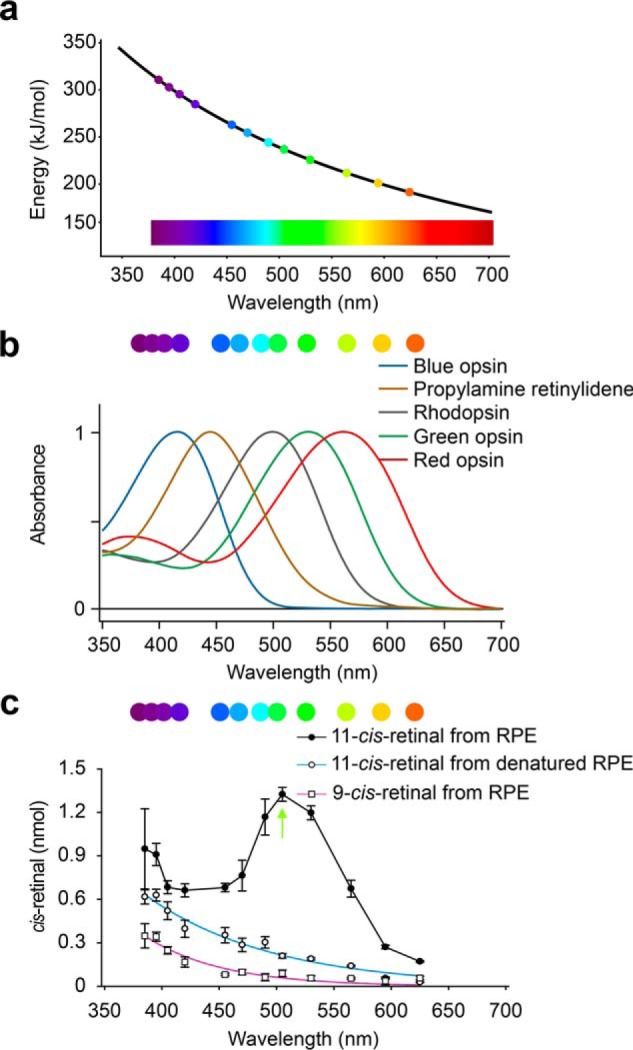
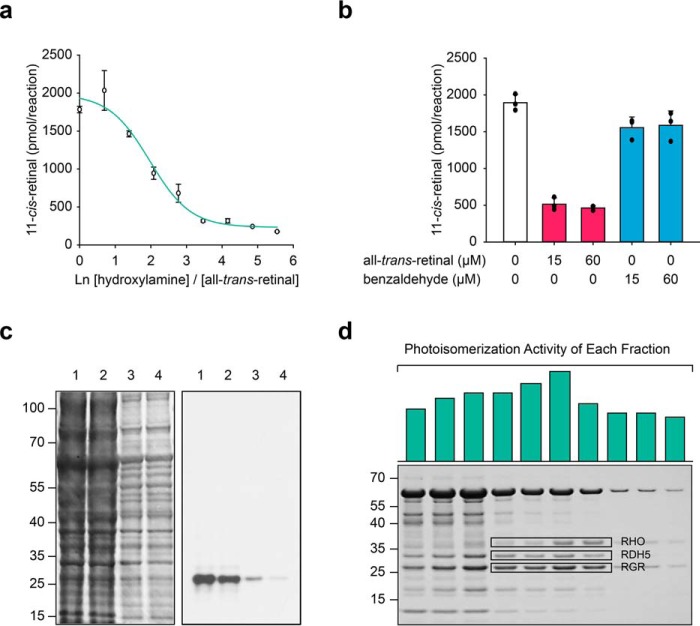
Enzymatic 11-cis-retinal generation depends on the wavelength of light. a, energy-wavelength relationship for the spectral range covered by the diodes used in this study. b, range of the wavelengths investigated covers the absorbance of human cone opsins and rhodopsin. c, graphs showing the 9-cis- and 11-cis-retinal generation rates during a 20-min light exposure with the given wavelength of light and a fixed power at 50 μW. ●, 11-cis-retinal generation with native RPE microsomes. The green arrow indicates the optimum wavelength for the enzymatic reaction. ○, 11-cis-retinal generation with denatured RPE microsomes. □, 9-cis-retinal generation with RPE microsomes. Data points are shown as means ± S.D. (n = 3).
Green light-stimulated photoisomerase activity in bovine RPE cells is mediated by protein rather than lipid adducts
A previous report showed that nonenzyme-associated photoisomerization of all-trans-retinal to cis-retinals was boosted by amines through Schiff base formation, independent of the RPE (42). We therefore compared the production of cis-retinals in native and denatured RPE samples illuminated by 12 different wavelengths. Under constant light power output (50 μW) and a set illumination time (20 min), the highest rate of 11-cis-retinal production by native RPE microsomes was observed at a wavelength of 505 nm, corresponding to the UV-visible spectrum of rhodopsin (Fig. 2c). However, upon an increase in the energy of the light source to 100 μW, the production of 11-cis-retinal was higher at a wavelength of 530 nm (Fig. S3, h and i). In contrast, generation of 11-cis-retinal in denatured RPE, as well as 9-cis-retinal in nondenatured RPE, was consistently low and correlated with the energy of the photons (Fig. 2c). The maximum amount of 11-cis-retinal generated upon green light illumination of native RPE suggested the presence of a wavelength-specific all-trans-retinal to 11-cis-retinal photoisomerase, activated optimally in the 505–530 nm range, prompting us to further explore this hypothesis.
Dependence of the photoisomerase activity on the retinaldehyde isomeric state
In subsequent RPE microsomal experiments, if not indicated otherwise, photoreactions were carried out using 530 nm light at 100 μW for 15 min. Under such conditions increased amounts of bovine RPE proportionally accelerated the generation of 11-cis-retinal (Fig. 3a). Similarly, higher concentrations of all-trans-retinal increased the production of 11-cis-retinal, but the effect was saturated above 15 μm (Fig. 3b). The RPE microsomes also converted both pure 13-cis-retinal, and to a lesser extent 9-cis-retinal, to the 11-cis conformer (Fig. 3c). The efficiency of 11-cis-retinal production with 13-cis-retinal as substrate (964 ± 88 pmol) was comparable with that of all-trans-retinal (1,363 ± 125 pmol) (Fig. 3c). These data are consistent with the thermal isomerization of 13-cis-retinal during photoisomerization, which exists in thermodynamic equilibrium ∼1:9 with all-trans-retinal (4). Interestingly, a more complex conversion of 9-cis- to 11-cis-retinal was also observed. Double isomerization is highly unlikely, so the conversion probably requires all-trans-retinal as an intermediate. The relatively low 11-cis-retinal yield in the presence of 9-cis-retinal therefore could be due to a lower amount of all-trans-retinal generated thermally.
Figure 3.
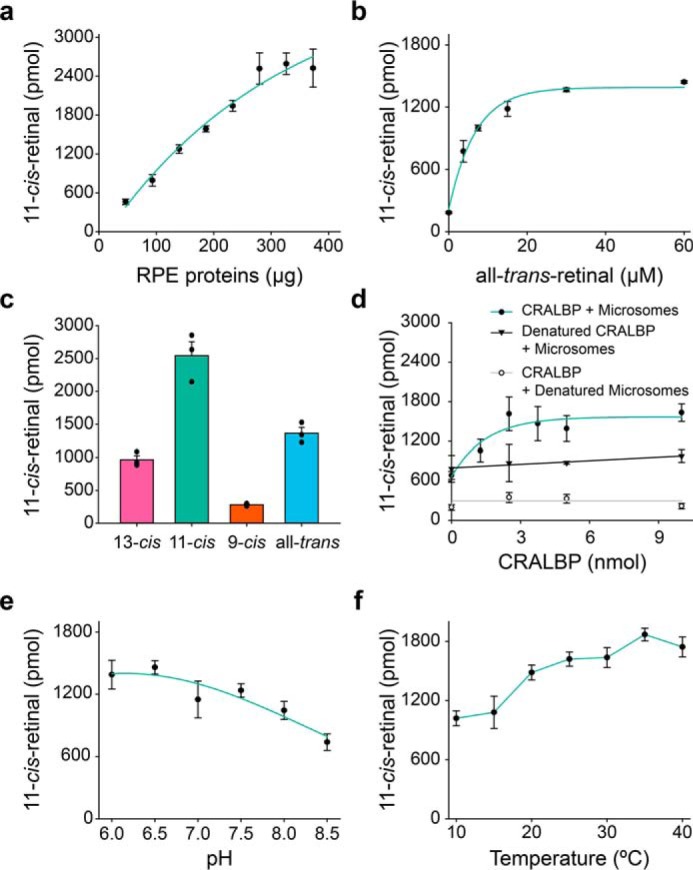
Optimization of the enzymatic production of 11-cis-retinal in RPE under green light. Photoreactions were carried out under 530 nm light (100 μW) for 15 min in the presence of CRALBP. a, production of 11-cis-retinal depends on the amount of RPE. b, all-trans-retinal serves as substrate for the photoisomerase. c, all-trans-retinal is the favored substrate of RGR as compared with 9-cis-retinal and 13-cis-retinal. The retinal isomer used as substrate in each case, at 15 μm concentration, is marked below each bar. d, production of 11-cis-retinal in the presence of CRALBP (green trace) or denatured CRALBP (black trace) in native RPE microsomes or denatured RPE microsomes (gray trace). e, photoisomerization is pH-sensitive. f, influence of temperature on photoisomerization. Data points are shown as means ± S.D. (n = 3–4).
Impact of CRALBP on the RPE photoisomerase activity
The fact that CRALBP has been shown to serve as a retinoid isomerase facilitating the conversion of 9-cis-retinal to 9,13-di-cis-retinal (43) prompted us to quantify its contribution to the generation of 11-cis-retinal in our experimental protocol. The 11-cis-retinal production was marginal in the presence of denatured RPE and nondenatured CRALBP, even with an excess of the latter (Fig. 3d). CRALBP added to nondenatured RPE improved the yield of 11-cis-retinal to ∼1,400 pmol, but the effect was saturated when the molar ratio of CRALBP exceeded that of all-trans-retinal. Yet, when CRALBP was boiled and mixed with nondenatured RPE, the generation of 11-cis-retinal in light fell to ∼800 pmol (Fig. 3d). These observations suggested that CRABLP facilitates the photoisomerization in RPE and protects the resulting 11-cis-retinal from re-isomerization to all-trans-retinal.
Photoisomerase activity in the RPE is not dependent on retinol dehydrogenase enzymes
Recent data have suggested that RGR in Müller glia works in concert with retinol dehydrogenase 10 to achieve photic production of 11-cis-retinol (35). To test whether RDH enzymes are involved in the photoisomerase activity of RGR in the RPE, we carried out photoisomerase assays in the presence of dinucleotide cofactors with RPE65 activity pharmacologically eliminated (Fig. S4). NAD+/NADP+ (oxidation) did not markedly affect the photoisomerase activity in RPE microsomes when compared with the dinucleotide-free controls. As expected, addition of NADH promoted 11-cis-retinol formation via the action of 11-cis-dehydrogenases (e.g. RDH5), but the total amount of 11-cis-retinoid was somewhat reduced compared with the nucleotide-free control. In contrast, NADPH reduced total cis-retinoid production likely because of contamination of the isolated bovine RPE with photoreceptor-derived RDH8, which stereospecifically reduces the all-trans-retinal substrate of RGR, converting it to all-trans-retinol. In summary, dinucleotides did not enhance photoisomerization activity, but they did affect the ratio of retinol/retinal as expected.
Impact of temperature and pH on the RPE photoisomerase activity
To further explore the properties and physiological relevance of the presumed photoisomerase, we tested the pH and temperature dependence of the 11-cis-retinal production. Maximal photoisomerization was observed at pH values in the physiological range, suggesting that the protonation to a Schiff base derived from retinal binding to the active site of the enzyme is critical to the production of 11-cis-retinal (Fig. 3e). We also utilized a variable temperature apparatus to test the effect of temperature on the enzymatic photoisomerization (see under “Experimental procedures” and Fig. S1c). The photoisomerization of all-trans-retinal by RPE microsomes was only moderately temperature-dependent in the range of 10–40 °C (Fig. 3f). The enzymatic activity at 10 °C remained surprisingly high and was equal to 54% of the value observed at 37 °C. The observed pH and temperature dependence patterns provided more support for the existence of a physiological photoisomerase of the receptor type rather than a classic enzyme. Many enzymes are acutely pH-sensitive due to the protonation of key residues in their active sites, and there is typically a 2-fold increase in enzymatic activity with every 10 °C increase of temperature up to the point of protein denaturation (44).
Light-dependent 11-cis-retinal production is independent of RPE65 and DES1
In the classical visual cycle, RPE65 isomerizes all-trans-retinyl esters to 11-cis-retinol without the assistance of light (Fig. 4a) (45). The subsequent oxidation of 11-cis-retinol to the opsin chromophore requires retinal dehydrogenase 5 (RDH5). DES1 may also contribute to 11-cis-retinol generation, which can be inhibited by fenretinide (11). To evaluate their relative roles in the regeneration of 11-cis-retinoids, both retinol and retinal were exposed to 530 nm light at 37 °C in RPE samples. The HPLC analysis of the isomerization products showed that the 11-cis-retinal yield was almost twice that of 11-cis-retinol when their respective all-trans substrates were added separately to the RPE (Fig. 4b), and the generation of 13-cis- or 9-cis-retinoids was negligible. The presence of both retinol and retinal in the RPE samples did not impair the photoisomerase activity, whereas the activity of RPE65 was greatly inhibited by all-trans-retinal (Fig. 4b). Emixustat, a visual cycle modulator that is currently in clinical trials for dry age-related macular degeneration (AMD), noncovalently binds to the active site of RPE65 and strongly inhibits 11-cis-retinol production, leading to undetectable levels of 11-cis-retinol (Fig. 4c). Here, the visual cycle modulator did not interfere with the activity of the photoisomerase (Fig. 4, b and c). Because of the strong inhibition of RPE65, adding all-trans-retinol did not increase the total amount of 11-cis-retinal, whereas an increase was evident in the absence of emixustat (Fig. 4, b and d). The presence of fenretinide (10 μm) in RPE samples demonstrated marginal effects on both the production of 11-cis-retinol and 11-cis-retinal, suggesting that RPE65 dominated the 11-cis-retinol production, and DES1 was irrelevant to the photoisomerization (Fig. 4d). Des1 is more likely involved in lipid metabolism and not retinoid transformations. In summary, enzymatic photoisomerization is the major pathway of 11-cis-retinal regeneration under illumination, independent of the classical visual cycle.
Figure 4.
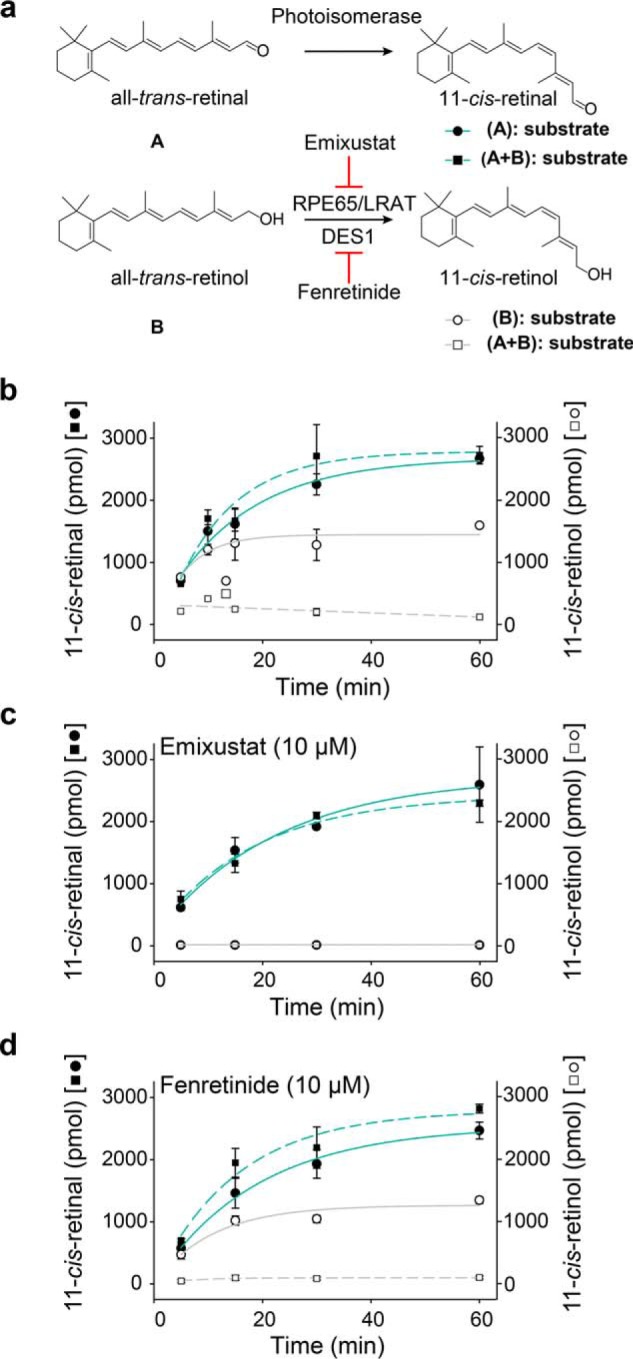
Light-dependent 11-cis-retinal generation is independent of the classical visual cycle. Retinal/retinol were incubated with RPE microsomes at 37 °C under 530 nm light (100 μW). a, schematic representation of two routes of isomerization generating 11-cis-retinoids. The 11-cis-retinal production was investigated with all-trans-retinal (15 μm) only (●) or both all-trans-retinal (15 μm) and all-trans-retinol (15 μm) (■) as the substrate; the 11-cis-retinol production was also studied with all-trans-retinol (15 μm) only (○) or both all-trans-retinal (15 μm) and all-trans-retinol (15 μm) (□) as substrates. b–d, time course of 11-cis-retinoid generation in RPE microsomes under 530 nm light (100 μW) at 37 °C without inhibitors (b), with 10 μm emixustat (c), or with 10 μm fenretinide (d). Data points are shown as means ± S.D. (n = 3).
Further biochemical properties of the RPE photoisomerase
Because the photoisomerase candidate appeared to be a membrane protein, solubilization was critical for its separation and identification. The washing of RPE microsomes with 3 m urea did not affect the activity of the isomerase and removed RPE constituent proteins associated with the membrane (Fig. S5a). Incubation with 1% trypsin for 30 min reduced the enzymatic photoisomerization (Fig. S5b). The presence of a high concentration of salts (Fig. S5c) and transition metal chelators such as EDTA and EGTA (Fig. S5d) did not alter the activity of the photoisomerase. In contrast, the isomerase was sensitive to most detergents (Fig. S5e), except for LMNG. With 10 mm LMNG, most membrane proteins were solubilized, and the isomerase retained at least 75% of its activity, even after being kept on ice for more than 12 h; however, it was deleterious to the function of RPE65 (Fig. S5f). The preincubation of RPE microsomes with phospholipase 2 altered the membrane composition and inhibited RPE65 function, but it did not impact photoisomerization (Fig. S5g) (46). The repeated freezing and thawing of the RPE microsomal suspension in 10 mm LMNG did not cause a substantial decrease in photoisomerization activity (Fig. S5h).
Identification of the RPE photoisomerase as RGR
The formation of a Schiff base at the active site of the putative photoisomerase has been proposed to be critical for substrate binding and retinal isomerization (47–49). Hydroxylamine, an aldehyde scavenger through stable Schiff base formation, competed with the binding of all-trans-retinal to the isomerase (Fig. 5a). The inhibition of isomerase activity required hydroxylamine at a concentration at least 32 times higher than the substrate, suggesting the specific binding of retinal to the protein also involved the noncovalent interaction between the retinoid backbone and the protein residues in the active site. The reduction of the retinylidene Schiff base with cyanoborohydride resulted in the formation of an irreversible bond with the Lys residue in the active site, which substantially inhibited photoisomerization (Fig. 5b). The substitution of all-trans-retinal with benzaldehyde abolished the inhibition, suggesting that benzaldehyde was incapable of fitting into the active site. The fluorography of RPE microsomal proteins incubated with radioactive all-trans-[3H]retinal revealed that all-trans-retinal specifically bound to a single protein migrating to band with an apparent molecular mass of around 27 kDa (Fig. 5c). To identify the all-trans-retinal–binding protein, detergent-solubilized RPE microsomes were subjected to cation-exchange chromatography followed by size-exclusion chromatography (SEC). SDS-PAGE of the SEC fractions containing isomerase activity again revealed a 27-kDa band (Fig. 5d) accompanied by substantial amounts of RPE65, rhodopsin, and RDH5 among other minor impurities. This band was further analyzed via MS, showing bovine RGR as its major component (over one-third of total peptide–to–spectrum matches), and absence of any significant alternative isomerase candidate (Table S1).
Figure 5.
Photoisomerization requires the binding of all-trans-retinal to RGR through a Schiff base formation. a, hydroxylamine competes with the photoisomerase to bind all-trans-retinal through a Schiff base formation. The x axis represents the natural logarithm of the hydroxylamine/all-trans-retinal molar ratio. Data points are shown as means ± S.D. (n = 3). b, 11-cis-retinal generation after RPE membranes had been reduced with sodium cyanoborohydride, in the presence of all-trans-retinal (red) or benzaldehyde (blue). Numbers below the graph denote the concentration of aldehyde used during the preincubation of the RPE samples, before conducting the 11-cis-retinal generation assay. Data points are shown as means ± S.D. (n = 3). c, RPE membranes incubated with all-trans-[3H]retinal were analyzed by SDS-PAGE and stained with Coomassie (left) or processed for fluorography (right). RPE microsomal proteins were labeled with 2 μm [3H]retinal (lane 1) or 2 μm [3H]retinal plus 12 μm nonradioactive retinal (lane 2) and then reduced with sodium cyanoborohydride. Samples in lanes 3 and 4 are 7.5 times dilutions of samples in lanes 1 and 2, respectively. d, SDS-polyacrylamide gel of solubilized RPE microsomes after ion-exchange and SEC. Green bars represent the relative photoisomerase activity in each SEC fraction.
Origin of the all-trans-retinal substrate of RGR in bovine RPE
The concentration of all-trans-retinal in the RPE is known to be very low, which raises the question of how RGR within the RPE gains access to its substrate. When RPE microsomes were treated with an excess of exogenous all-trans-retinol (30 μm) and emixustat (10 μm), the esterification of retinol by LRAT became much less favored (Fig. 6a), which resulted in a substantial elevation of the all-trans-retinol level, whereas 11-cis-retinol generation was marginal due to the inhibition of RPE65. After 30 min of light exposure (530 nm and 200 μW) at 37 °C, the observed small amount of 11-cis-retinal (108.2 ± 2.3 pmol) came mainly from the enzymatic photoisomerization of endogenous retinals in RPE microsomes. The 11-cis-retinal generation by RGR reached 532.1 ± 13.1 pmol if NAD+ (2 mm) was added to the reaction mixture (Fig. 6b). This boost of 11-cis-retinal production with NAD+ addition indicated that the all-trans-retinol escaping from the LRAT esterification could be oxidized by retinol dehydrogenase in the presence of the dinucleotide cofactor, and the product, all-trans-retinal, served as the substrate of photoisomerization.
Figure 6.
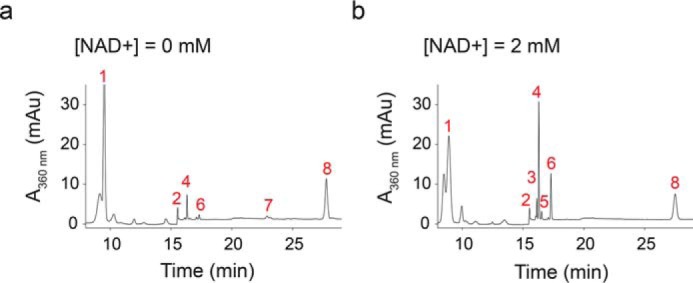
11-cis-Retinal in RPE also originates from the oxidation of all-trans-retinol and subsequent photoisomerization. RPE microsomes with exogenous all-trans-retinol (30 μm) were exposed to 530 nm light (200 μW) for 30 min at 37 °C. Emixustat (10 μm) was added to inhibit the isomerization by RPE65. The products were extracted with hexanes and analyzed with normal-phase HPLC. a and b, representative chromatographs of products extracted from reactions in the presence of 0 (a) or 2 mm (b) NAD+. Peak 1, retinyl esters; peak 2, solvent gradient change; peak 3, 13-cis-retinal; peak 4, 11-cis-retinal; peak 5, 9-cis-retinal; peak 6, all-trans-retinal; peak 7, 11-cis-retinol and 13-cis-retinol; and peak 8, all-trans-retinol. The trace amounts of 11-cis-retinoids in a originate from residual retinoid in RPE microsomes.
Heterologously expressed RGR exhibits spectral and biochemical properties matching those of the native RPE photoisomerase
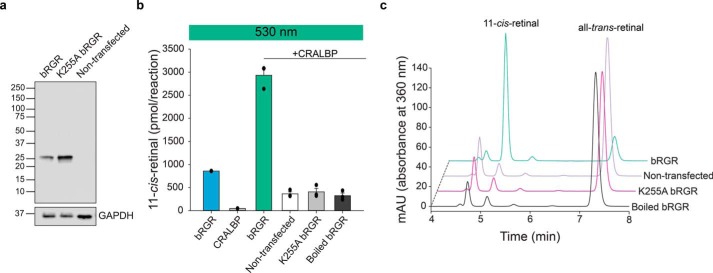
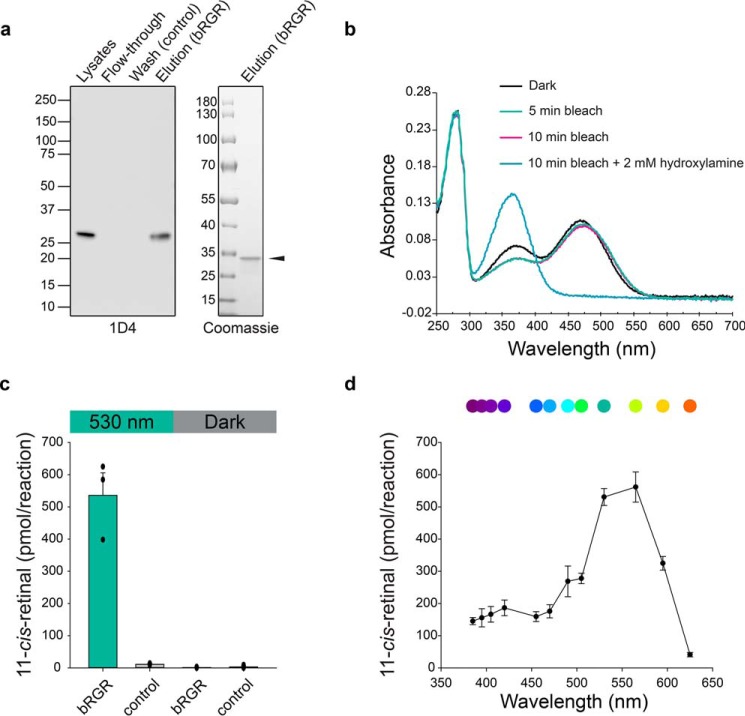
To further confirm that bovine RGR (bRGR) accounts for the isomerase activity found in bovine RPE microsomes, we heterologously expressed it in HEK293S GnTI− cells. Along with the wildtype (WT) bRGR, we also expressed a mutant bRGR that has an alanine residue instead of a Lys residue at position 255 in the active site. The Lys-255 residue is conserved from primates to zebrafish and is known to bind all-trans-retinal through a covalent Schiff base bond (50). Immunoblotting of cell lysates showed a comparable level of bRGR and K255A bRGR expression (Fig. 7a). Next, we incubated cell homogenates with all-trans-retinal in 530 nm light to measure photoisomerization activity. Accumulation of 11-cis-retinal was much more profound in bRGR cell homogenates in the presence of CRALBP than in its absence, reaching over a 7-fold excess compared with K255A bRGR or the nontransfected control (Fig. 7, b and c). Because LMNG has proven to preserve the photoisomerase activity of bovine RPE microsomes, we utilized this detergent to solubilize and purify recombinant bRGR. A one-step immunoaffinity chromatography procedure performed in the dark and in the presence of all-trans-retinal achieved significant enrichment of the target protein and limited nonspecific contaminants (Fig. 8a). The UV-visible light absorption spectrum of bRGR bound to all-trans-retinal showed a peak around 470 nm, as reported previously (47–49), with little change observed upon 530 nm light illumination. However, the 470 nm peak completely disappeared after 10 min of incubation with hydroxylamine (Fig. 8b). The HPLC analysis confirmed that all-trans-retinal bound to RGR was isomerized to 11-cis-retinal upon 530 nm light illumination, and that the incubation with hydroxylamine induced the formation of retinal oximes (Fig. S6a). In line with the previous cell lysate–based assays, the purified bRGR robustly photoisomerized exogenous all-trans-retinal to 11-cis-retinal upon 530 nm light illumination in the presence of CRALBP, yet 11-cis-retinal was almost absent in the negative control sample (Fig. 8c and Fig. S6b). The action spectrum for 11-cis-retinal production by purified bRGR, exhibited a maximum at ∼550 nm, in line with the action spectrum obtained from native bovine RPE microsomes, which corroborates the assignment of RGR as the photoisomerase in RPE microsomes (Figs. 2c and 8d). Together, these findings demonstrate that bRGR efficiently photoisomerizes all-trans-retinal to 11-cis-retinal upon 530–550 nm light exposure and that the Schiff base linkage at Lys-255 is critical for this process.
Figure 7.
Bovine RGR expressed in HEK293S GnTI− cells photoisomerizes all-trans-retinal to 11-cis-retinal. a, immunoblot of bRGR and K255A bRGR expressed in HEK293S GnTI− cells. Sample loading was normalized based on the signal for GAPDH. b, amounts of 11-cis-retinal photoisomerized from all-trans-retinal in cell homogenates exposed to 530 nm light. Homogenates of HEK293S GnTI− cells transfected with a plasmid encoding bRGR produced a significant amount of 11-cis-retinal. The presence of CRALBP increased the amount of 11-cis-retinal, whereas the mutation K255A within bRGR nearly abolished its photoisomerase activity. n = 3 for each group. Data are shown as mean ± S.E. c, representative chromatograms showing the amount of 11-cis-retinal and all-trans-retinal from different cell homogenates when exposed to 530 nm light.
Figure 8.
Photoisomerase activity of immunopurified bovine RGR expressed in HEK293S GnTI− cells. a, immunoblot of purified bRGR detected by 1D4 antibody (left). Coomassie-stained SDS-polyacrylamide gel of purified bRGR (right). b, absorption spectra for purified bRGR bound to all-trans-retinal. The spectrum of bRGR was measured in the dark and after 5 and 10 min of illumination with 530 nm light. Excess hydroxylamine was then added, and the spectrum of the sample was measured again. c, amounts of 11-cis-retinal produced from all-trans-retinal by purified bRGR in a photoisomerization assay. Control sample consists of the immunochromatography elution buffer. n = 3 for each group. Data are shown as mean ± S.E. d, action spectrum showing the amounts of 11-cis-retinal photoisomerized from all-trans-retinal by purified bRGR during 30 min of light exposure with the given wavelength and a fixed intensity at 400 μW. n = 4 for the 530 and 565 nm, n = 3 for the other wavelengths. Data are shown as mean ± S.E.
Analysis of RGR photoisomerase activity in other vertebrates
To test the green light-dependent photoisomerization potential of RGR in other species, we first prepared microsomal fractions with the same amount of protein from porcine and chicken RPE. Both were able to produce 11-cis-retinal upon 530 nm light illumination, although the efficiency of the latter was about 25% that of bovine or porcine RPE (Fig. S5i). Microsomal fractions derived from bovine rod outer segments, mouse brain, and mouse liver were incapable of generating 11-cis-retinal, confirming that RPE is the specific tissue contributing to the photoisomerase activity (Fig. S5i). To further explore the evolutionary conservation of this phenomenon, we heterologously expressed and tested chicken and mouse RGR, in a fashion similar to bRGR. In concert with the bovine data, cRGR cell lysates exhibited robust 11-cis-retinal production (Fig. S7, a and b). In contrast, photoisomerase activity of mRGR was much lower than that of cRGR even though the expression of mRGR and cRGR was comparable (Fig. S7, a and b).
Impact of human RGR splicing variation on photoisomerase activity
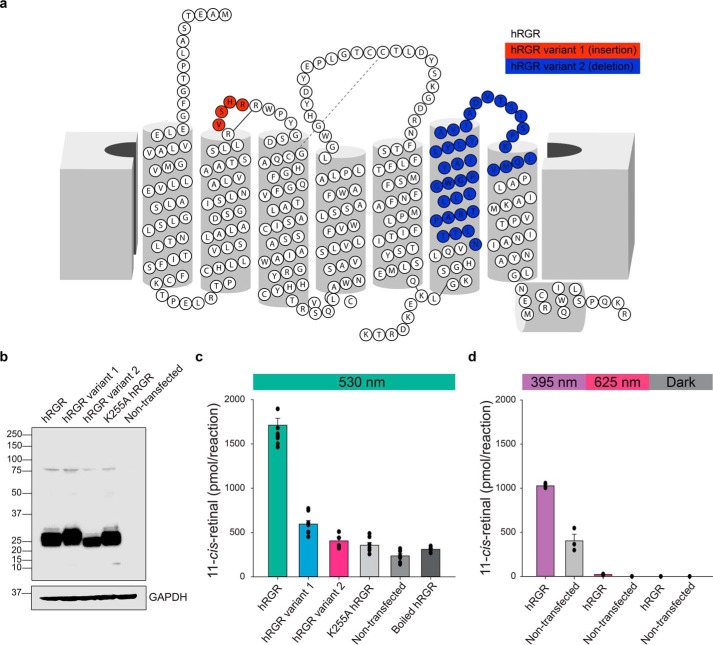
The notion that human RGR (hRGR) contributes to chromophore regeneration has been controversial, partly due to its low activity (26). Moreover, because multiple splice variants of the hRGR gene have been reported (Fig. 9a), our next goal was to quantify the relative abundance of these variants in the human eye and assess their photoisomerization capacity to produce 11-cis-retinal. Using available human retina and RPE RNA-seq datasets (51), we confirmed the existence of an alternative splice acceptor site of hRGR intron 2 leading to an insertion of four additional in-frame codons (amino acids 79–82, hRGR variant 1) in a small (<5%) fraction of the total transcripts. Another more common variant of hRGR transcripts (10–20% abundance) originates from the deletion of exon 6, encompassing 114 nucleotides (amino acids 211–248, hRGR variant 2), which removes the transmembrane domain 6 from the encoded protein. Next, we heterologously expressed the hRGR together with its two minor splice variants in HEK293S GnTI− cells to compare their photoisomerization activity. Immunoblotting analysis of cell lysates demonstrated efficient and comparable expression of all transcripts (Fig. 9b). The WT hRGR transcript generated the greatest amount of 11-cis-retinal upon 530 nm light exposure in the presence of CRALBP. Consistent with the findings from our bovine studies, the K255A substitution basically abolished this activity (Fig. 9c). hRGR variant 1, containing the nonconserved 4-amino acid insertion, exhibited much lower photoisomerization activity, whereas variant 2 was comparable with the K255A mutant. The level of 11-cis-retinal produced upon 625 nm light exposure was minimal across all test and control samples, whereas hRGR still produced significant amounts of 11-cis-retinal upon 395 nm light exposure compared with the nontransfected samples (Fig. 9d). Taken together, the assay demonstrates that hRGR can robustly photoisomerize all-trans-retinal to 11-cis-retinal.
Figure 9.
Photoisomerase activity of human RGR isoforms. a, putative two-dimensional model of the human RGR variants. Variant 1 has an insertion of 4 amino acids (red), and variant 2 has a deletion of 38 amino acids (blue) due to the deletion of exon 6. b, immunoblot of WT hRGR, hRGR variant 1, hRGR variant 2, and K255A hRGR expressed in HEK293S GnTI− cells. c, light at 395 nm produced a lower level of photoisomerization of all-trans-retinal by hRGR than 530 nm light, and light at 625 nm did not cause photoisomerization. n = 3–10 for each group. Data are shown as mean ± S.E. d, light at 395 nm produced a lower level of photoisomerization of all-trans-retinal by hRGR than 530 nm light, whereas light at 625 did not cause photoisomerization. n = 3 for each group. Data are shown as mean ± S.E.
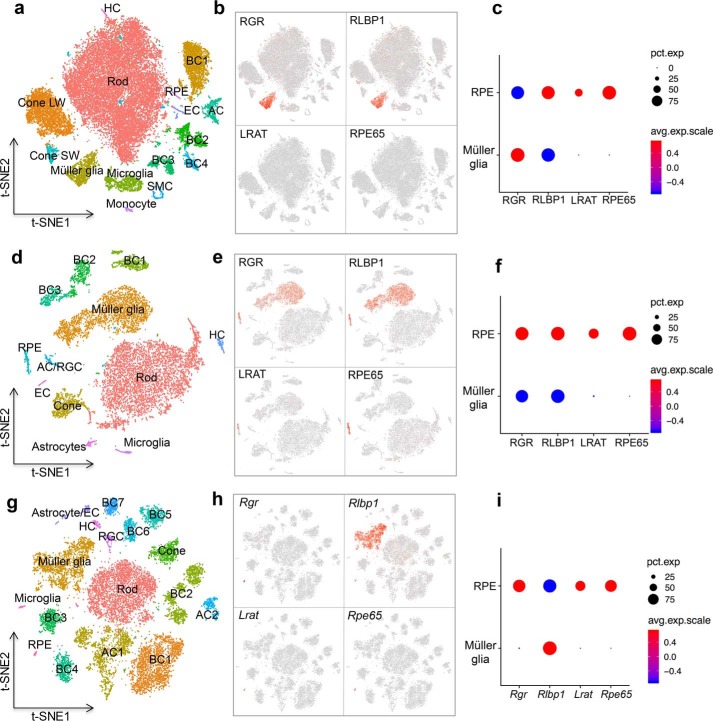
RNA-Seq analysis revealed species-specific differences in RGR and RLBP1 co-expression
Because our finding suggest that the co-presence of CRALBP is necessary for efficient RGR-dependent photoisomerization of all-trans-retinal, we examined the expression pattern of both RLBP1 and RGR transcripts in different retinal cell types and the RPE through a single-cell sequencing analysis. We performed a single-cell RNA-Seq of retina and RPE tissue from mouse, bovine, and human, and we analyzed the data with t-SNE (52) to visualize clusters corresponding to distinct cell types (Fig. 10, a, d, and g). In human and bovine samples, both RGR and RLBP1 were simultaneously expressed in Müller cells and RPE, supporting a possible interaction between the two proteins during RGR-dependent photoisomerization. (Fig. 10, b and e). In mice, Rlbp1 was expressed in both Müller cells and RPE (as observed in bovine and human), whereas Rgr was expressed only in RPE, but only barely detectable in Müller cells (Fig. 10h). Furthermore, the single-cell RNA-Seq allowed us to compare the expression level of RGR with other genes involved in the classical visual cycle. We found that RGR is expressed at a relatively lower level compared with that of RPE65 in RPE from bovine and human (Fig. 10, c and f). Interestingly, the expression level of Rgr in RPE from the mouse was comparable to that of Rpe65 (Fig. 10i). These data show that RGR and RLBP1 are co-expressed in RPE and Müller cells of bovine and human, but in the mouse eye the co-expression of Rgr was far higher in RPE, with only minimal expression detected in Müller cells.
Figure 10.
Single-cell RNAseq analysis of RGR expression in the bovine, human, and mouse retina. a, d, and g, analysis of the retinal cells and RPE cells isolated from bovine (a), human (d), and mouse (g) origins formed clusters representing each cell type. b, e, and h, expression of RGR, RLBP1, LRAT, and RPE65 at the single-cell level in samples from bovine (b), human (e), and mouse (h) tissues. c, f, and i, relative expression levels of RGR, RLBP1, LRAT, and RPE65 in Müller cells and RPE in samples from bovine (c), human (f), and mouse (i).
Discussion
The need for efficient chromophore regeneration in moderate to bright light conditions appears to exceed that supplied by the classical visual cycle (53, 54). One explanation for sustained visual function in vertebrates under these conditions could be the presence of a complementary photic visual cycle that responds to increasing light exposure, thus balancing the bleaching of cone visual pigments with increased 11-cis-retinal production. We have demonstrated in this study a pathway located in the RPE that may serve this function. We have found that 1) the RPE has the ability to respond to incident visible light with a robust photic response to regenerate 11-cis-retinal; 2) RGR is the enzymatic origin of this visible light-dependent 11-cis-retinal regeneration in the RPE; 3) both native and recombinant RGR show peak activity upon stimulation with green light (∼530 nm) with lower activity at other wavelengths; and 4) the RGR-dependent photic response appears to have a higher capacity for 11-cis-retinal production compared with the classical visual cycle.
The wavelength selectivity associated with the photoisomerase activity in native microsomal membranes and recombinant RGR is similar to the absorbance spectra of rhodopsin and green cone opsin (55). This correspondence is expected to maximize the efficiency of the system for visual chromophore production when it is most needed. It is interesting that the wavelength range (510–550 nm) optimally stimulating RGR photoisomerase activity only partially overlaps with the UV-visible absorption spectrum of purified RGR (Fig. 8b) (47). This observation is similar to the photoisomerase activity of retinochrome, which is most efficiently stimulated with yellow light (>530 nm) despite the absorbance maximum of the purified protein occurring at a shorter wavelength (496 nm) (56, 57). Differences between the absorption and action spectra likely arise from the multitude of competing photoisomerization reactions that occur in the complex mixture of proteins and membranes. For example, nonspecific retinal–protein Schiff base adducts may exist predominantly in the trans-configuration at equilibrium during illumination with 440 nm light. Such a reaction would counteract the activity of RGR resulting in a red shift of the action spectrum.
The rate at which 11-cis-retinal was regenerated via photoisomerization in the RPE depended on the isomeric state of the retinal substrate with the following order of substrate selectivity: all-trans-retinal > 13-cis-retinal > 9-cis-retinal (Fig. 3c). This is opposite to the order of rod and cone opsin affinity for retinals. Despite the lower specificity for the 13-cis-retinal and 9-cis-retinal isomers, enzymatic photoisomerization in the RPE could play a role in clearing these compounds that are produced nonspecifically during light exposure. Such a functional role is consistent with prior studies of Rgr−/− mice, which exhibit an accumulation of 9- and 13-cis-retinoids following exposure to short light flashes (50).
Prior work indicated that RGR can stimulate RPE65 activity independent of light (29) and increase all-trans-retinyl ester hydrolase activity (30). Further work is needed to reconcile these findings in the context of our results. Considering all reports, it is likely that RGR has a multifunctional role in visual physiology. Although direct signal transduction activity has not yet been demonstrated for RGR, it is interesting to note that the protein has conserved molecular features of G-protein–coupled receptors, including the (modified) (E/D)RY and NPXXY sequence motifs (58). Additionally, RGR physically associates with RDH5 and RPE65 (46) in the RPE, suggesting a mechanism by which light-driven conformational changes in RGR could be transmitted to these key components of the classical visual cycle. Despite the apparent 11-cis-stereospecificity of RPE65 in vivo, this enzyme can generate substantial amounts of 13-cis-retinol (59–61). Moreover, substantial 13-cis-retinoid accumulation occurs in vivo in the eyes of Rdh5−/− mice during dark adaptation (62) or in Rgr−/− mice after bleaching (50). These data suggest that RGR, working in conjunction with RDH5, also may function to remove the 13-cis-retinol by-product of the classical visual cycle in vivo.
Peropsin orthologs constitute a sister group to the RGR/retinochrome photoisomerase subfamily (63). Based on this close phylogenetic relationship and the fact that peropsin is expressed in the RPE (64), we considered the possibility that peropsin could contribute to the observed photoisomerase activity. However, MS analysis aiming to identify the RPE microsome component that bound all-trans-retinal did not show the presence of peropsin (Table S1). Additionally, a recent study on peropsin function indicated that peropsin plays a role in vitamin A transit between the retina and RPE, rather than acting as a visual chromophore-producing photoisomerase (65).
CRALBP moderately stimulated photic 11-cis-retinal production from bovine microsomes (Fig. 3d). However, our experiments using heterologously-expressed RGR or purified bovine RGR clearly demonstrated the importance of CRALBP in facilitating RGR-dependent 11-cis-retinal formation. It is possible that residual CRALBP present in the RPE microsome samples could have partially masked the CRALBP dependence of the reaction in the native system. Several lines of evidence have demonstrated that cephalopod retinal-binding protein is required to effectively bind 11-cis-retinal produced from retinochrome and act as a shuttle in the rhodopsin–retinochrome pathway (66, 67). Our data indicate a similar complementary relationship between RGR and CRALBP in vertebrates. Moreover, the co-localization of RGR and CRALBP in the same cell types of human and bovine eyes supports their potential interaction during the regeneration of chromophore under sustained photic conditions. Rod opsin was also present in the photoisomerase-active fractions, due to rod membrane contamination of the RPE microsomal preparation, and we considered the possibly that it could also act as an 11-cis-retinal sink stimulating 11-cis-retinal production through RGR by mass action. However, the excess of CRALBP over rod opsin suggests the latter would play only a minor role in binding 11-cis-retinal. Moreover, it is known from prior studies that detergent-purified rod opsin binds 9- or 11-cis-retinal poorly (68, 69). Finally, photoisomerization reactions carried out in the presence and absence of bleached ROS membranes yielded similar production rates and quantities of 11-cis-retinal. Therefore, the presence of native CRALBP in the purified photoisomerase-active samples is the most likely explanation for the weaker CRALBP dependence of the reactions carried out using material from native RPE microsomes.
RGR is a sink for all-trans-retinal, as we demonstrate in this work, and can be isolated with bound retinal even after extensive washing during column chromatography. All-trans-retinol is esterified by LRAT, and the esters can be hydrolyzed (70). The conversion of all-trans-retinol to all-trans-retinal is governed by the reducing power of the cell, and the reaction is reversible. From a thermodynamic point of view, there will always be some amount of all-trans-retinal in RPE cells. To support this view, the experiments described here (Fig. 6) demonstrate that 1) RPE microsomes generate all-trans-retinal from retinyl esters or retinol that is present in the RPE at a high concentration in the presence of NAD+, and 2) in the light, all-trans-retinal is selectively converted to 11-cis-retinal (Fig. 6). A second source of all-trans-retinal is derived directly from photoreceptors under strong light conditions. Note that retinal transport is not known to be protein-mediated. 11-cis-Retinal diffuses from the RPE to photoreceptors despite the high concentration of CRALBP in the RPE (with nanomolar affinity for retinal). This occurs because opsin is a sink for 11-cis-retinal. Likewise, RGR must act as a sink for all-trans-retinal.
The mechanism we describe depends on the availability of all-trans-retinal in RPE to serve as a substrate for RGR. Prior studies have shown that all-trans-retinal accumulates in the retina during periods of light exposure when RGR could act as a photoisomerase (18, 71). This retinal can likely transfer between the photoreceptor outer segments and the RPE. For example, Rando and Bangerter (72) demonstrated that the intermembranous rates of retinoid (all-trans-retinol(al) and 11-cis-retinol) transfer from vesicle to vesicle and vesicle to erythrocyte were exceedingly rapid, whereas all-trans-retinyl palmitate did not undergo transfer at an appreciable rate. Fex and Johannesson (73, 74) showed that retinol, similar to unesterified cholesterol and long-chain fatty acids but unlike phospholipids, can transfer rapidly and spontaneously between liposomal membranes. Noy et al. (75) suggested that retinoid transfer between different cell types in vivo may depend on the geometry of cellular surfaces, which are highly elaborated in the RPE apical membrane. These results suggest that retinoids may not require active transport or binding protein–mediated transfer between membranous compartments of ROS and the RPE. However, this work does not exclude the possibility that a yet unidentified active transport mechanism exists in the mammalian retina.
This study also highlights the importance of RGR sequence variations in determining photoisomerase activity. The alternative splicing of human RGR generates at least three different transcript isoforms, which were identified in post-mortem donor retinas (48, 49, 76). The dominant isoform 1, referred to as WT hRGR, is highly conserved, whereas variant 1 has a 4-amino acid–long insertion that is not observed in other species (Fig. 9a) (49). The second splicing variant lacks the nucleotide sequence encoding transmembrane domain 6, which likely renders the translated protein nonfunctional (76). Here, we confirmed that WT hRGR is the most abundant isoform in both human RPE and Müller glia and exhibits the highest photoisomerase activity, comparable with recombinant bovine and chicken RGRs.
The photoisomerase activity of recombinant mouse RGR was much lower than that of the human, chicken, and bovine orthologs, despite high overall sequence conservation (81% identity with human RGR), robust heterologous expression, and identical assay conditions. Single-cell RNA-Seq analysis of mouse retina and RPE showed a different pattern of RGR expression compared with the other species, with mouse RGR transcripts occurring solely in the RPE cells. These findings are consistent with prior immunohistochemistry data showing negligible RGR expression in mouse retinal tissue harvested at postnatal day 16 (77). Conversely, RGR expression in human and bovine Müller glia was readily detectable with immunostaining (78, 79). Considering that the RGR knockout mouse exhibits a relatively mild visual phenotype (50, 80), it is possible that the contribution of the RGR photoisomerase activity to the visual cycle differs considerably between the nocturnal mouse and diurnal animals such as human, cow, and chicken, although this hypothesis will need to be verified in additional nocturnal animals. This fundamental difference in retinal physiology is possibly why it has been difficult to observe contributions of RGR to light-dependent visual pigment regeneration in mice (29, 30, 50).
Our results indicate that the RGR isomerization pathway works in a complementary fashion to the classical visual cycle to regenerate bleached visual pigments via retinoid transport between photoreceptor outer segments and the RPE (Fig. 11). In the case of rods, which desensitize in relatively dim light, it is likely that the classical visual cycle is of greater relevance to their function. However, it was recently shown that rods can maintain some function under photopic conditions and that the RGR system may be involved in providing the chromophore under these conditions (81). Previous studies have shown that the classical visual cycle plays a significant role in maintaining responses from cones but that cones can continue to function in circumstances of profound RPE65 inhibition (54, 82). It has been speculated that 11-cis-retinal bound to CRALBP in Müller cells could act as a chromophore reserve to sustain cone function (54). The presence of RGR in Müller cells of diurnal animals suggests that the complementary visual cycle may be at work in the Müller cells as well, which in turn may explain the source of 11-cis-retinoids bound to CRALBP in these cells. The results described here indicate that the RGR system in the RPE adds another, potentially more significant source of 11-cis-retinal that could allow for long-term maintenance of cone function in the face of sustained bright light exposure.
Figure 11.
Proposed mechanism for photic chromophore regeneration mediated by RGR. A key step in visual pigment regeneration is the isomerization of all-trans-retinal to 11-cis-retinal. Whereas the classical visual cycle regenerates 11-cis-retinal through a series of enzymatic steps independently of light, RGR further contributes to regeneration of 11-cis-retinal in photic conditions. Absorption of a photon by rhodopsin and cone opsins isomerizes 11-cis-retinal to all-trans-retinal in photoreceptors. All-trans-retinal can diffuse directly to the RPE or be reduced to form all-trans-retinol, which is subsequently oxidized to all-trans-retinal in the RPE. RGR photoisomerizes all-trans-retinal formed by either route. CRALBP then binds 11-cis-retinal and shields it from re-isomerization. The resulting 11-cis-retinal is used as the chromophore to regenerate rhodopsin and cone opsins.
Experimental procedures
Animals
All experiments were approved by the Institutional Animal Care and Use Committees at the University of California, Irvine, and were conducted in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Visual Research.
Spectral illumination
To provide controlled spectral illumination, all components, including LEDs and cuvette holders, were rigidly mounted on solid aluminum optical breadboards (Fig. S1a). Light from LEDs, M385F1, M395F3, M405FP1, M420F2, M455F1, M470F3, M490F3, M505F1, M530F2, M565F3, M595F2, and M625F2 was guided via a multimode optical fiber. The fiber was fastened into a CP02 cage plate (Thorlabs, Newton, NJ) with the SM1SMA fiber adapter plate (Thorlabs). To ensure consistent placement between the cuvette and the light-emitting fiber-end, we used a cage-mounted customized CVH100 cuvette holder (Thorlabs) and customized t2Sport cuvette holder with Peltier-driven temperature control (Quantum Northwest, Liberty Lake, WA). M395L4, M530L3, and M625L4 LEDs (Thorlabs) with collimators were used for photoisomerase activity assays of recombinant RGR. Light intensity was adjusted using a DC4100 LED driver (Thorlabs), and light power was measured with the optical power meter PM100D equipped with S120C and S120VC light sensors (Thorlabs).
Reagents
Solvents and reagents were purchased from Sigma unless otherwise stated. All-trans-retinal was obtained from Toronto Research Chemicals, Inc. (Toronto, Canada), or Sigma. 11-cis-Retinal was obtained by illuminating all-trans-retinal in acetonitrile and separating the retinal isomers by normal-phase HPLC on a silica column (10 μm, 21.2 × 250 mm, Phenomenex, Torrance, CA) with a mobile phase of 10% ethyl acetate in hexanes flowing at a rate of 5 ml·min−1 (83).
RPE microsomal preparations
Bovine RPE microsomes were isolated from RPE homogenates by differential centrifugation as described previously (84). The resulting microsomal pellet was resuspended in 10 mm Bis-tris propane/HCl, pH 7.4 (BTP buffer), to achieve a total protein concentration of ∼5 mg·ml−1. The mixture then was placed in a quartz cuvette and irradiated with UV light for 6 min at 4 °C with a ChromatoUVB transilluminator (model TM-15; UVP, Upland, CA) to eliminate residual retinoids. After irradiation, 5 mm DTT was added to the RPE microsomal mixture.
RPE65 retinoid isomerase activity assay
RPE65 isomerase assays were initiated by addition of all-trans-retinol in dimethylformamide (DMF) to a final concentration of 15 μm in BTP buffer, to a suspension containing 150 μg of RPE microsomal proteins, 1% bovine serum albumin (BSA), 2 mm disodium pyrophosphate, and 25 μm human apo-CRALBP (85). The resulting mixture was incubated at 37 °C for 15 min to 1 h. The reaction was quenched by adding 400 μl of methanol (Thermo Fisher Scientific, Fair Lawn, NJ), and the products were extracted with 400 μl of hexanes. Production of 11-cis-retinol was quantified by normal-phase HPLC using a Zorbax Rx-SIL column (5 μm, 4.6 × 250 mm, Agilent, Santa Clara, CA) with 10% (v/v) ethyl acetate in hexanes as the eluent at a flow rate of 1.4 ml·min−1. Retinoids were detected by monitoring their absorbance at 325 nm and quantified based on a standard curve representing the relationship between the amount of 11-cis-retinol and the area under the corresponding chromatographic peak.
Photoisomerization assay
All-trans-retinal in DMF was added to a final concentration of 15 μm in BTP buffer, containing 150 μg of bovine RPE microsomal proteins, 1% BSA, 2 mm disodium pyrophosphate, and 25 μm CRALBP. The resulting mixture was subsequently transferred to quartz cuvettes (Starna Cells, Atascadero, CA, path length 1 mm) and illuminated with LED lights for 5 min to 2 h. The reaction was quenched by turning off the light source. Methanol (400 μl) then was added to the reaction mixture, and retinoid products were extracted with hexanes (400 μl). Production of 11-cis-retinal was quantified by normal-phase HPLC with 10% (v/v) ethyl acetate in hexanes as the eluent at a flow rate of 1.4 ml·min−1. Retinoids were detected by monitoring their absorbance at 360 nm and quantified based on a standard curve representing the relationship between the amount of 11-cis-retinal and the area under the corresponding chromatographic peak (46). All isomerization reactions were performed and analyzed in a darkroom.
Binding of all-trans-retinal to RPE microsomal protein
RPE microsomes were resuspended in 300 μl of BTP buffer, containing 2 μm all-trans-[3H]retinal with or without 12 μm nonradioactive all-trans-retinal. The resulting mixture was incubated at 4 °C for 1 h, and then 7.5 mg of sodium cyanoborohydride was added. The resulting samples were loaded onto a BisTris polyacrylamide gel (4–12%) (Thermo Fisher Scientific, Carlsbad, CA). After electrophoresis, the gel was washed with water, dehydrated with dimethyl sulfoxide (DMSO), and soaked in a 2,5-diphenyloxazole solution (20% in DMSO) for 1 h. The gel was subsequently washed with water, dried under vacuum at 60 °C for 1 h, and exposed to Kodak BioMax XAR film (Sigma) at −80 °C for 10 h.
Ion-exchange and size-exclusion chromatography of urea-washed RPE microsomal proteins
Bovine RPE microsomes were suspended in 6 ml of urea (3 m, pH 8) over ice. The mixture was centrifuged at 100,000 × g for 30 min, and the resulting pellet was gently solubilized with 10 mm LMNG in BTP buffer at 4 °C. After centrifugation at 100,000 × g for 30 min, 3.5 ml of the resulting supernatant was loaded onto a glass column containing 9.5 ml of DEAE-Sepharose (GE Healthcare, Uppsala, Sweden). The column was then washed with 20 ml of BTP buffer containing 0.25 mm LMNG. Bound proteins were eluted from the column with a 20-ml linear gradient from 0 to 1 m NaCl. Fractions exhibiting photoisomerase activity (as described above) were combined, concentrated to 2.5 ml, and loaded onto a 125-ml Superdex 200 size-exclusion column (GE Healthcare), and the proteins were separated at a flow rate of 0.5 ml·min−1 in BTP buffer containing 0.25 mm LMNG and 100 mm NaCl. The photoisomerase activities of the fractions were evaluated as described above.
Mass spectrometry
Fractions collected from SEC were analyzed by SDS-PAGE, and the gel was stained with Coomassie Blue. The visible bands in the lane with higher photoisomerase activity were individually excised, washed, and destained in 50% ethanol, 5% acetic acid, and then dehydrated in acetonitrile. The samples were then reduced with DTT and alkylated with iodoacetamide prior to in-gel digestion by addition of 50 ng of trypsin in 50 mm ammonium bicarbonate followed by overnight incubation at room temperature. Peptides were extracted from the polyacrylamide gel slices in two aliquots of 30 μl of 50% acetonitrile, 5% formic acid. These extracts were combined and evaporated to <10 μl using a Speedvac concentrator (Thermo Fisher Scientific) and then resuspended in 1% acetic acid to a final volume of ∼30 μl for analysis using an LTQ-Orbitrap Elite hybrid mass spectrometer system (Thermo Fisher Scientific). Peptides were separated on an Acclaim Pepmap C18 column (15 cm × 75 μm, inner diameter 2 μm, 100 Å reversed-phase capillary chromatography column, Thermo Fisher Scientific) with a 0–100% acetonitrile in 0.1% formic acid gradient at a flow rate of 0.3 μl·min−1, and the eluate was introduced into the source of the mass spectrometer online. The nano-electrospray ion source was operated at 1.9 kV. The digest was analyzed using the data-dependent multitask capability of the instrument acquiring full-scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans. The data were analyzed using all collision-induced dissociation spectra collected in the experiment to search against the UniProtKB bovine protein sequence database using Sequest and Mascot software (86, 87).
Protein expression and isomerase activity assay
Plasmids encoding hRGR and two splicing variants with a C-terminal FLAG-tag were purchased from Origene (Rockville, MD). pcDNA3.1-derived plasmids encoding cRGR and bRGR with the C-terminal His8- and 1D4-tags were produced by Gene Universal (Newark, DE). The nucleotide sequence for cRGR was codon-optimized for expression in mammalian cells. The RGR mutants were generated by site-directed mutagenesis using the Q5 kit (New England Biolabs, Ipswich, MA) according to the manufacturer's instructions with the primers listed in Table S2 and using the respective WT RGR plasmids as templates. All RGR variants were expressed in HEK293S GnTI− cells (ATCC, Manassas, VA) cultured in suspension in FreeStyle 293 medium (Thermo Fisher Scientific) with 2% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) and 1% penicillin–streptomycin (Thermo Fisher Scientific). Cells were transfected using polyethyleneimine Max (Polysciences, Warrington, PA) at 3:1 polyethyleneimine to DNA ratio following previously established protocols (88) and cultured for 48 h at 37 °C, 8% CO2 in a shaking incubator. For the retinal isomerization assay, 8 × 107 HEK293S GnTI− cells washed in phosphate-buffered saline (PBS, 10 mm sodium phosphate, 150 mm sodium chloride, pH 7.4) were suspended in the 10 mm BTP buffer and homogenized in a glass tissue grinder (Kimble Chase, Rockwood, TN). Cell lysates were complemented with 1 mm sodium pyrophosphate, 75 μm BSA, 30 μm CRALBP (85), and 12 μm all-trans-retinal and were incubated for 30 min in quartz cuvettes in the dark or attached to mounted 395:530:625 nm LEDs matched for 560 μW optical power output. Retinal isomerization products were extracted from the 600 μl of reaction mixture by vigorous shaking with 400 μl of methanol and 400 μl of hexanes and centrifuged for 5 min at 16,000 × g. Then, 100 μl of extracted retinoids were injected for the HPLC analysis. The retinoids in the organic phase were quantified as described above.
Purification of recombinant bRGR
The pellet from a 2.1-liter culture of HEK293S GnTI− cells expressing bRGR, which was C-terminally tagged with His8- and 1D4 antibody tags (TETSQVAPA), was resuspended in 160 ml of 20 mm HEPES, pH 7.4, plus protease inhibitor mixture (Roche Diagnostics GmbH, Mannheim, Germany), 1 mm MgCl2, and benzonase nuclease (New England Biolabs), homogenized with a glass tissue grinder, and centrifuged for 30 min at 50,000 × g. The membrane pellet (25 g) was solubilized in 4.5 mm LMNG, 50 mm HEPES, pH 7.4, 0.25 m NaCl, 1 mm MgCl2, plus protease inhibitors, benzonase, and 30 μm all-trans-retinal (final volume 160 ml) and then centrifuged for 50 min at 50,000 × g. The supernatant was incubated for 1 h on batch with 6 ml of 1D4 antibody immobilized in Sepharose beads (CNBr-activated SepharoseTM 4B, GE Healthcare) and then washed on a column with 200 mm HEPES, pH 7.4, with 0.15 m NaCl, and 0.2 mm LMNG. Finally, bRGR was eluted with washing buffer containing 0.8 mg/ml competing peptide. The purified protein was then used directly for UV-visible spectrometry and for the photoisomerase activity assay as described above. The purity of the fractions was assessed by SDS-PAGE on Coomassie Blue-stained BisTris (4–12%) polyacrylamide gels (Thermo Fisher Scientific).
Immunoblotting
Total protein fractions were extracted from HEK293S GnTI− cells using RIPA lysis buffer (Millipore, Burlington, MA), separated by SDS-PAGE on Tris-glycine (4–20%) polyacrylamide gels (Bio-Rad), and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked in 20 mm Tris, pH 7.5, with 0.5 m NaCl, 0.1% Tween 20, 5% nonfat dry milk (RPI, Mount Prospect, IL), and proteins were detected with the following primary antibodies: mouse anti-FLAG M2 (1:1000, Sigma), mouse anti-rhodopsin 1D4 (1:1000, produced in-house (89)), and rabbit anti-GAPDH (1:2000, Abcam, Cambridge, MA). Horseradish peroxidase-conjugated secondary antibodies were visualized on the membranes using SuperSignal West Pico Plus Chemiluminescent Substrate (Thermo Fisher Scientific). Images were collected with an Odyssey Fc Imaging System (LI-COR, Lincoln, NE).
Single-cell RNA-Seq analysis
A human eye globe from an 86-year-old Caucasian female, who died of a myocardial infarction and had no known ocular disease other than cataracts, was obtained from the Alabama Eye Bank (Birmingham, AL) and processed within 3.3 h after death. The study was approved by The Johns Hopkins Institutional Review Board. The neural retina and RPE/choroid were dissected from the globe in ice-cold PBS. First, a circular incision was made on the sclera, behind the limbus, to remove the anterior parts, lens, and vitreous body. The neural retina was then peeled from the eyecup, and retinal cells were dissociated using the Papain Dissociation System (Worthington Biochemical, Lakewood, NJ) following the manufacturer's instructions. RPE cells were dissociated from the eyecup by incubating with 2 ml of 0.05% trypsin-EDTA (Thermo Fisher Scientific) for 20 min at 37 °C. Dissociated cells were resuspended in ice-cold PBS, 0.04% BSA/ and 0.5 units/μl of RNase inhibitors. Cells were then filtered through a 50-μm cell strainer. CD1 mice around 2 months of age were purchased from Charles River Laboratories (Wilmington, MA). All experimental procedures were pre-approved by the Institutional Animal Care and Use Committee (IACUC) of The Johns Hopkins University School of Medicine. Mice were euthanized, and eye globes were removed and incubated in ice-cold PBS. The neural retinas were dissected, and cells were dissociated using the Papain Dissociation System as mentioned above. In total, four biological replicates were used for the mouse scRNA study. Each replicate contained four retinas from each of two male and two female mice. Dissociated cells were resuspended and filtered as above. Cell count and viability were assessed by trypan blue staining. Fresh bovine eyes were obtained from a local slaughterhouse. The bovine eyes were hemisected, and the neural retina was peeled off from the eyecups. The retinal cells were dissociated using the Papain Dissociation System as mentioned above. The RPE layer was detached from the eyecup by incubating with 2 ml of 0.05% trypsin-EDTA for 20 min at 37 °C. The detached RPE cells were resuspended in 8 ml of DMEM/F-12 medium (Thermo Fisher Scientific) containing 1% fetal bovine serum (Thermo Fisher Scientific) and 1% penicillin–streptomycin (Thermo Fisher Scientific). The RPE cells were pelleted through centrifugation for 10 min at 300 × g. Then, the pellet was triturated with 1 ml of Dulbecco's PBS (Thermo Fisher Scientific) and filtered by a 50-μm cell strainer to remove aggregates. The dissociated bovine retinal cells and RPE cells were washed with 1 ml of Dulbecco's PBS twice and fixed with 100 μl of PBS and 900 μl of methanol. The methanol-fixed bovine samples were rehydrated and processed by following the protocols provided from 10× Genomics (Pleasanton, CA). Briefly, the methanol-fixed cells were centrifuged at 3000 × g for 10 min at 4 °C and washed three times in an ice-cold resuspension buffer containing PBS, 1% BSA, and 0.5 units/μl RNase inhibitors. The dissociated cells were resuspended in the buffer and filtered through a 50-μm cell strainer. For human and mouse samples, freshly dissociated cells (∼10,000) were loaded into a 10× Genomics Chromium Single Cell system using v2 chemistry following the manufacturer's instruction. The bovine samples were processed using 10× Genomics v3 chemistry. Libraries were pooled and sequenced on Illumina NextSeq with ∼200 million reads per library. Sequencing results were processed through the Cell Ranger 3.0.1 pipeline (10× Genomics) with default parameters. Seurat version 2.3.4 (90) was used to perform downstream analysis following the standard pipeline using cells with more than 200 genes and 1000 UMI counts, resulting in 16,659 mouse cells, 14,286 human cells, and 34,881 bovine cells. Samples were aggregated, and cell clusters were annotated based on previous literature. A t-SNE dimension reduction was performed on the top principal components learned from high variance genes. Gene expression of each cell cluster was calculated using the average expression function of Seurat.
Statistical analyses
Data are presented as the means ± S.D. or means ± S.E. for the results of at least three independent experiments. Significant differences between groups were assessed by Student's t test or one-way analysis of variance with the Bonferroni post-test for comparisons of more than two groups. Sigma Plot 11.0 (Systat Software) was used to perform the statistical analyses.
Author contributions
J. Z., E. H. C., D. S., H. L., C. L. S., T. V. H., and G. P. investigation; J. Z., E. H. C., A. T., D. S., G. P., P. D. K., and K. P. writing-original draft; J. Z., E. H. C., A. T., D. S., H. L., C. L. S., S. B., G. P., P. D. K., and K. P. writing-review and editing; E. H. C., A. T., P. D. K., and K. P. conceptualization; E. H. C., A. T., D. S., T. V. H., and K. P. data curation; E. H. C. and S. B. formal analysis; E. H. C., A. T., D. S., and S. B. methodology; A. T., J. T. H., G. P., and K. P. resources; G. P. and K. P. funding acquisition; P. D. K. and K. P. supervision.
Supplementary Material
Acknowledgments
We thank Drs. H. Jin and W. Sun for assistance with 1D4 antibody production and tissue culture studies. We thank Dr. S. Gulati and A. Sears for providing rod outer segments and interphotoreceptor retinoid-binding protein, respectively. We thank Drs. B. Willard and L. Li from Cleveland Clinic for assistance with MS. We are also grateful to members of the Palczewski laboratory for their helpful comments regarding this project. The Department of Ophthalmology, University of California, Irvine, was the recipient of an RPB unrestricted grant.
This work was supported in part by National Institutes of Health Grants EY009339, EY027283, EY025451, and EY019312 (to K. P.), Department of Veterans Affairs Grant IK2BX002683 (to P. D. K.), and National Institutes of Health Grants T32GM007250 and T32GM008803 (to E. H. C.). K. P. is Chief Scientific Officer at Polgenix, Inc. G. P. is an employee of Polgenix, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Identification of RGR accession no. 10.5281/zenodo.3519404 has been deposited to Zenodo.
This article contains Figs. S1–S7 and Tables S1 and S2.
- RPE
- retinal pigment epithelium
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- BTP
- Bistris propane
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- CRALBP
- cellular retinaldehyde-binding protein
- DES1
- sphingolipid Δ4-desaturase 1
- DMF
- dimethylformamide
- LED
- light-emitting diode
- LMNG
- lauryl maltose neopentyl glycol
- LRAT
- lecithin:retinol acyltransferase
- RDH
- retinol dehydrogenase
- RGR
- RPE-retinal G-protein-coupled receptor
- μW
- microwatt
- SEC
- size-exclusion chromatography
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- b
- bovine
- h
- human
- m
- mouse
- c
- chicken
- RNA-Seq
- RNA-sequencing
- ROS
- rod outer segment
- t-SNE
- t-distributed stochastic neighbor embedding.
References
- 1. Palczewski K. (2006) G protein-coupled receptor rhodopsin. Annu. Rev. Biochem. 75, 743–767 10.1146/annurev.biochem.75.103004.142743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schoenlein R. W., Peteanu L. A., Mathies R. A., and Shank C. V. (1991) The first step in vision: femtosecond isomerization of rhodopsin. Science 254, 412–415 10.1126/science.1925597 [DOI] [PubMed] [Google Scholar]
- 3. Barry B., and Mathies R. (1982) Resonance Raman microscopy of rod and cone photoreceptors. J. Cell Biol. 94, 479–482 10.1083/jcb.94.2.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kiser P. D., Golczak M., and Palczewski K. (2014) Chemistry of the retinoid (visual) cycle. Chem. Rev. 114, 194–232 10.1021/cr400107q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin M., Li S., Moghrabi W. N., Sun H., and Travis G. H. (2005) Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell 122, 449–459 10.1016/j.cell.2005.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moiseyev G., Chen Y., Takahashi Y., Wu B. X., and Ma J. X. (2005) RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc. Natl. Acad. Sci. U.S.A. 102, 12413–12418 10.1073/pnas.0503460102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Redmond T. M., Poliakov E., Yu S., Tsai J. Y., Lu Z., and Gentleman S. (2005) Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. U.S.A. 102, 13658–13663 10.1073/pnas.0504167102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernstein P. S., and Rando R. R. (1986) In vivo isomerization of all-trans- to 11-cis-retinoids in the eye occurs at the alcohol oxidation state. Biochemistry 25, 6473–6478 10.1021/bi00369a020 [DOI] [PubMed] [Google Scholar]
- 9. Grimm C., Wenzel A., Hafezi F., Yu S., Redmond T. M., and Remé C. E. (2000) Protection of Rpe65-deficient mice identifies rhodopsin as a mediator of light-induced retinal degeneration. Nat. Genet. 25, 63–66 10.1038/75614 [DOI] [PubMed] [Google Scholar]
- 10. Mata N. L., Radu R. A., Clemmons R. C., and Travis G. H. (2002) Isomerization and oxidation of vitamin a in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight. Neuron 36, 69–80 10.1016/S0896-6273(02)00912-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaylor J. J., Yuan Q., Cook J., Sarfare S., Makshanoff J., Miu A., Kim A., Kim P., Habib S., Roybal C. N., Xu T., Nusinowitz S., and Travis G. H. (2013) Identification of DES1 as a vitamin A isomerase in Muller glial cells of the retina. Nat. Chem. Biol. 9, 30–36 10.1038/nchembio.1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kiser P. D., Kolesnikov A. V., Kiser J. Z., Dong Z., Chaurasia B., Wang L., Summers S. A., Hoang T., Blackshaw S., Peachey N. S., Kefalov V. J., and Palczewski K. (2019) Conditional deletion of Des1 in the mouse retina does not impair the visual cycle in cones. FASEB J. 33, 5782–5792 10.1096/fj.201802493R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lamb T. D., and Pugh E. N. Jr. (2004) Dark adaptation and the retinoid cycle of vision. Prog. Retin. Eye Res. 23, 307–380 10.1016/j.preteyeres.2004.03.001 [DOI] [PubMed] [Google Scholar]
- 14. Rushton W. A. (1977) Visual adaptation. Biophys. Struct. Mech. 3, 159–162 10.1007/BF00535812 [DOI] [PubMed] [Google Scholar]
- 15. Hecht S. (1937) The instantaneous visual threshold after light adaptation. Proc. Natl. Acad. Sci. U.S.A. 23, 227–233 10.1073/pnas.23.4.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahroo O. A., and Lamb T. D. (2004) Recovery of the human photopic electroretinogram after bleaching exposures: estimation of pigment regeneration kinetics. J. Physiol. 554, 417–437 10.1113/jphysiol.2003.051250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wenzel A., Reme C. E., Williams T. P., Hafezi F., and Grimm C. (2001) The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J. Neurosci. 21, 53–58 10.1523/JNEUROSCI.21-01-00053.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saari J. C., Garwin G. G., Van Hooser J. P., and Palczewski K. (1998) Reduction of all-trans-retinal limits regeneration of visual pigment in mice. Vision Res. 38, 1325–1333 10.1016/S0042-6989(97)00198-3 [DOI] [PubMed] [Google Scholar]
- 19. Nymark S., Frederiksen R., Woodruff M. L., Cornwall M. C., and Fain G. L. (2012) Bleaching of mouse rods: microspectrophotometry and suction-electrode recording. J. Physiol. 590, 2353–2364 10.1113/jphysiol.2012.228627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frederiksen R., Nymark S., Kolesnikov A. V., Berry J. D., Adler L. 4th., Koutalos Y., Kefalov V. J., and Cornwall M. C. (2016) Rhodopsin kinase and arrestin binding control the decay of photoactivated rhodopsin and dark adaptation of mouse rods. J. Gen. Physiol. 148, 1–11 10.1085/jgp.201511538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodieck R. W. (1998) The First Steps in Seeing, Sinauer Associates, Sunderland, MA [Google Scholar]
- 22. Kaylor J. J., Xu T., Ingram N. T., Tsan A., Hakobyan H., Fain G. L., and Travis G. H. (2017) Blue light regenerates functional visual pigments in mammals through a retinyl-phospholipid intermediate. Nat. Commun. 8, 16 10.1038/s41467-017-00018-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shichi H., and Somers R. L. (1974) Possible involvement of retinylidene phospholipid in photoisomerization of all-trans-retinal to 11-cis-retinal. J. Biol. Chem. 249, 6570–6577 [PubMed] [Google Scholar]
- 24. Widjaja-Adhi M. A. K., Ramkumar S., and von Lintig J. (2018) Protective role of carotenoids in the visual cycle. FASEB J. 2018, fj201800467R 10.1096/fj.201800467R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Albalat R. (2012) Evolution of the genetic machinery of the visual cycle: a novelty of the vertebrate eye? Mol. Biol. Evol. 29, 1461–1469 10.1093/molbev/msr313 [DOI] [PubMed] [Google Scholar]
- 26. Yang M., and Fong H. K. (2002) Synthesis of the all-trans-retinal chromophore of retinal G protein-coupled receptor opsin in cultured pigment epithelial cells. J. Biol. Chem. 277, 3318–3324 10.1074/jbc.M108946200 [DOI] [PubMed] [Google Scholar]
- 27. Van Hooser J. P., Liang Y., Maeda T., Kuksa V., Jang G. F., He Y. G., Rieke F., Fong H. K., Detwiler P. B., and Palczewski K. (2002) Recovery of visual functions in a mouse model of Leber congenital amaurosis. J. Biol. Chem. 277, 19173–19182 10.1074/jbc.M112384200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hao W., and Fong H. K. (1999) The endogenous chromophore of retinal G protein-coupled receptor opsin from the pigment epithelium. J. Biol. Chem. 274, 6085–6090 10.1074/jbc.274.10.6085 [DOI] [PubMed] [Google Scholar]
- 29. Wenzel A., Oberhauser V., Pugh E. N. Jr., Lamb T. D., Grimm C., Samardzija M., Fahl E., Seeliger M. W., Remé C. E., and von Lintig J. (2005) The retinal G protein-coupled receptor (RGR) enhances isomerohydrolase activity independent of light. J. Biol. Chem. 280, 29874–29884 10.1074/jbc.M503603200 [DOI] [PubMed] [Google Scholar]
- 30. Radu R. A., Hu J., Peng J., Bok D., Mata N. L., and Travis G. H. (2008) Retinal pigment epithelium-retinal G protein receptor-opsin mediates light-dependent translocation of all-trans-retinyl esters for synthesis of visual chromophore in retinal pigment epithelial cells. J. Biol. Chem. 283, 19730–19738 10.1074/jbc.M801288200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morimura H., Saindelle-Ribeaudeau F., Berson E. L., and Dryja T. P. (1999) Mutations in RGR, encoding a light-sensitive opsin homologue, in patients with retinitis pigmentosa. Nat. Genet. 23, 393–394 10.1038/70496 [DOI] [PubMed] [Google Scholar]
- 32. Arno G., Hull S., Carss K., Dev-Borman A., Chakarova C., Bujakowska K., van den Born L. I., Robson A. G., Holder G. E., Michaelides M., Cremers F. P., Pierce E., Raymond F. L., Moore A. T., and Webster A. R. (2016) Reevaluation of the retinal dystrophy due to recessive alleles of RGR with the discovery of a cis-acting mutation in CDHR1. Invest. Ophthalmol. Vis. Sci. 57, 4806–4813 10.1167/iovs.16-19687 [DOI] [PubMed] [Google Scholar]
- 33. Xue Y., Sato S., Razafsky D., Sahu B., Shen S. Q., Potter C., Sandell L. L., Corbo J. C., Palczewski K., Maeda A., Hodzic D., and Kefalov V. J. (2017) The role of retinol dehydrogenase 10 in the cone visual cycle. Sci. Rep. 7, 2390 10.1038/s41598-017-02549-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sahu B., Sun W., Perusek L., Parmar V., Le Y. Z., Griswold M. D., Palczewski K., and Maeda A. (2015) Conditional ablation of retinol dehydrogenase 10 in the retinal pigmented epithelium causes delayed dark adaption in mice. J. Biol. Chem. 290, 27239–27247 10.1074/jbc.M115.682096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morshedian A., Kaylor J. J., Ng S. Y., Tsan A., Frederiksen R., Xu T., Yuan L., Sampath A. P., Radu R. A., Fain G. L., and Travis G. H. (2019) Light-driven regeneration of cone visual pigments through a mechanism involving RGR opsin in Muller glial cells. Neuron 102, 1172–1183.e5 10.1016/j.neuron.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oprian D. D., Asenjo A. B., Lee N., and Pelletier S. L. (1991) Design, chemical synthesis, and expression of genes for the three human color vision pigments. Biochemistry 30, 11367–11372 10.1021/bi00112a002 [DOI] [PubMed] [Google Scholar]
- 37. Filipek S., Teller D. C., Palczewski K., and Stenkamp R. (2003) The crystallographic model of rhodopsin and its use in studies of other G protein-coupled receptors. Annu. Rev. Biophys. Biomol. Struct. 32, 375–397 10.1146/annurev.biophys.32.110601.142520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Filipek S., Stenkamp R. E., Teller D. C., and Palczewski K. (2003) G protein-coupled receptor rhodopsin: a prospectus. Annu. Rev. Physiol. 65, 851–879 10.1146/annurev.physiol.65.092101.142611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Båvik C. O., Busch C., and Eriksson U. (1992) Characterization of a plasma retinol-binding protein membrane receptor expressed in the retinal pigment epithelium. J. Biol. Chem. 267, 23035–23042 [PubMed] [Google Scholar]
- 40. McBee J. K., Van Hooser J. P., Jang G. F., and Palczewski K. (2001) Isomerization of 11-cis-retinoids to all-trans-retinoids in vitro and in vivo. J. Biol. Chem. 276, 48483–48493 10.1074/jbc.M105840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang J., Kiser P. D., Badiee M., Palczewska G., Dong Z., Golczak M., Tochtrop G. P., and Palczewski K. (2015) Molecular pharmacodynamics of emixustat in protection against retinal degeneration. J. Clin. Invest. 125, 2781–2794 10.1172/JCI80950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alchalel A., Honig B., Ottolenghi M., and Rosenfeld T. (1975) Triplet-sensitized cis-trans isomerization of the protonated Schiff base of retinal isomers. J. Am. Chem. Soc. 97, 2161–2166 10.1021/ja00841a029 [DOI] [PubMed] [Google Scholar]
- 43. Bolze C. S., Helbling R. E., Owen R. L., Pearson A. R., Pompidor G., Dworkowski F., Fuchs M. R., Furrer J., Golczak M., Palczewski K., Cascella M., and Stocker A. (2014) Human cellular retinaldehyde-binding protein has secondary thermal 9-cis-retinal isomerase activity. J. Am. Chem. Soc. 136, 137–146 10.1021/ja411366w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chance B. (1999) Classics in enzymology: the kinetics of the enzyme-substrate compound of peroxidase (reprinted from J. Biol. Chem. 151, 553–573). Adv. Enzymol Relat. Areas Mol. Biol. 73, 3–23 [PubMed] [Google Scholar]
- 45. Kiser P. D., and Palczewski K. (2016) Retinoids and retinal diseases. Annu. Rev. Vis. Sci. 2, 197–234 10.1146/annurev-vision-111815-114407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Golczak M., Kiser P. D., Lodowski D. T., Maeda A., and Palczewski K. (2010) Importance of membrane structural integrity for RPE65 retinoid isomerization activity. J. Biol. Chem. 285, 9667–9682 10.1074/jbc.M109.063941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hao W., and Fong H. K. (1996) Blue and ultraviolet light-absorbing opsin from the retinal pigment epithelium. Biochemistry 35, 6251–6256 10.1021/bi952420k [DOI] [PubMed] [Google Scholar]
- 48. Shen P., Canoll P. D., Sap J., and Musacchio J. M. (1999) Expression of a truncated receptor protein tyrosine phosphatase κ in the brain of an adult transgenic mouse. Brain Res. 826, 157–171 10.1016/S0006-8993(99)01179-8 [DOI] [PubMed] [Google Scholar]
- 49. Shen D., Jiang M., Hao W., Tao L., Salazar M., and Fong H. K. (1994) A human opsin-related gene that encodes a retinaldehyde-binding protein. Biochemistry 33, 13117–13125 10.1021/bi00248a022 [DOI] [PubMed] [Google Scholar]
- 50. Maeda T., Van Hooser J. P., Driessen C. A., Filipek S., Janssen J. J., and Palczewski K. (2003) Evaluation of the role of the retinal G protein-coupled receptor (RGR) in the vertebrate retina in vivo. J. Neurochem. 85, 944–956 10.1046/j.1471-4159.2003.01741.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim E. J., Grant G. R., Bowman A. S., Haider N., Gudiseva H. V., and Chavali V. R. M. (2018) Complete transcriptome profiling of normal and age-related macular degeneration eye tissues reveals dysregulation of anti-sense transcription. Sci. Rep. 8, 3040 10.1038/s41598-018-21104-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van der Maaten L., and Hinton G. (2008) Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 [Google Scholar]
- 53. Lyubarsky A. L., Savchenko A. B., Morocco S. B., Daniele L. L., Redmond T. M., and Pugh E. N. Jr. (2005) Mole quantity of RPE65 and its productivity in the generation of 11-cis-retinal from retinyl esters in the living mouse eye. Biochemistry 44, 9880–9888 10.1021/bi0505363 [DOI] [PubMed] [Google Scholar]
- 54. Kiser P. D., Zhang J., Sharma A., Angueyra J. M., Kolesnikov A. V., Badiee M., Tochtrop G. P., Kinoshita J., Peachey N. S., Li W., Kefalov V. J., and Palczewski K. (2018) Retinoid isomerase inhibitors impair but do not block mammalian cone photoreceptor function. J. Gen. Physiol. 150, 571–590 10.1085/jgp.201711815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hofmann L., and Palczewski K. (2015) Advances in understanding the molecular basis of the first steps in color vision. Prog. Retin. Eye Res. 49, 46–66 10.1016/j.preteyeres.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hara T., and Hara R. (1973) Isomerization of retinal catalysed by retinochrome in the light. Nat. New Biol. 242, 39–43 10.1038/newbio242039a0 [DOI] [PubMed] [Google Scholar]
- 57. Ozaki K., Hara R., Hara T., and Kakitani T. (1983) Squid retinochrome. Configurational changes of the retinal chromophore. Biophys. J. 44, 127–137 10.1016/S0006-3495(83)84285-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Batten M. L., Imanishi Y., Maeda T., Tu D. C., Moise A. R., Bronson D., Possin D., Van Gelder R. N., Baehr W., and Palczewski K. (2004) Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem. 279, 10422–10432 10.1074/jbc.M312410200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McBee J. K., Kuksa V., Alvarez R., de Lera A. R., Prezhdo O., Haeseleer F., Sokal I., and Palczewski K. (2000) Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: dependence on the specificity of retinoid-binding proteins. Biochemistry 39, 11370–11380 10.1021/bi001061c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Redmond T. M., Poliakov E., Kuo S., Chander P., and Gentleman S. (2010) RPE65, visual cycle retinol isomerase, is not inherently 11-cis-specific: support for a carbocation mechanism of retinol isomerization. J. Biol. Chem. 285, 1919–1927 10.1074/jbc.M109.027458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maeda A., Maeda T., Imanishi Y., Golczak M., Moise A. R., and Palczewski K. (2006) Aberrant metabolites in mouse models of congenital blinding diseases: formation and storage of retinyl esters. Biochemistry 45, 4210–4219 10.1021/bi052382x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miyagawa Y., Ohguro H., Odagiri H., Maruyama I., Maeda T., Maeda A., Sasaki M., and Nakazawa M. (2003) Aberrantly expressed recoverin is functionally associated with G-protein–coupled receptor kinases in cancer cell lines. Biochem. Biophys. Res. Commun. 300, 669–673 10.1016/S0006-291X(02)02888-7 [DOI] [PubMed] [Google Scholar]
- 63. Terakita A. (2005) The opsins. Genome Biol. 6, 213 10.1186/gb-2005-6-3-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun H., Gilbert D. J., Copeland N. G., Jenkins N. A., and Nathans J. (1997) Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc. Natl. Acad. Sci. U.S.A. 94, 9893–9898 10.1073/pnas.94.18.9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cook J. D., Ng S. Y., Lloyd M., Eddington S., Sun H., Nathans J., Bok D., Radu R. A., and Travis G. H. (2017) Peropsin modulates transit of vitamin A from retina to retinal pigment epithelium. J. Biol. Chem. 292, 21407–21416 10.1074/jbc.M117.812701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Terakita A., Hara R., and Hara T. (1989) Retinal-binding protein as a shuttle for retinal in the rhodopsin-retinochrome system of the squid visual cells. Vision Res. 29, 639–652 10.1016/0042-6989(89)90026-6 [DOI] [PubMed] [Google Scholar]
- 67. Ozaki K., Terakita A., Hara R., and Hara T. (1987) Isolation and characterization of a retinal-binding protein from the squid retina. Vision Res. 27, 1057–1070 10.1016/0042-6989(87)90020-4 [DOI] [PubMed] [Google Scholar]
- 68. Chen Y., Chen Y., Jastrzebska B., Golczak M., Gulati S., Tang H., Seibel W., Li X., Jin H., Han Y., Gao S., Zhang J., Liu X., Heidari-Torkabadi H., Stewart P. L., et al. (2018) A novel small molecule chaperone of rod opsin and its potential therapy for retinal degeneration. Nat. Commun. 9, 1976 10.1038/s41467-018-04261-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ortega J. T., Parmar T., and Jastrzebska B. (2019) Flavonoids enhance rod opsin stability, folding, and self-association by directly binding to ligand-free opsin and modulating its conformation. J. Biol. Chem. 294, 8101–8122 10.1074/jbc.RA119.007808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Isken A., Golczak M., Oberhauser V., Hunzelmann S., Driever W., Imanishi Y., Palczewski K., and von Lintig J. (2008) RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 7, 258–268 10.1016/j.cmet.2008.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee K. A., Nawrot M., Garwin G. G., Saari J. C., and Hurley J. B. (2010) Relationships among visual cycle retinoids, rhodopsin phosphorylation, and phototransduction in mouse eyes during light and dark adaptation. Biochemistry 49, 2454–2463 10.1021/bi1001085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rando R. R., and Bangerter F. W. (1982) The rapid intermembranous transfer of retinoids. Biochem. Biophys. Res. Commun. 104, 430–436 10.1016/0006-291X(82)90655-6 [DOI] [PubMed] [Google Scholar]
- 73. Fex G., and Johannesson G. (1988) Retinol transfer across and between phospholipid bilayer membranes. Biochim. Biophys. Acta 944, 249–255 10.1016/0005-2736(88)90438-5 [DOI] [PubMed] [Google Scholar]
- 74. Fex G., and Johannesson G. (1987) Studies of the spontaneous transfer of retinol from the retinol:retinol-binding protein complex to unilamellar liposomes. Biochim. Biophys. Acta 901, 255–264 10.1016/0005-2736(87)90122-2 [DOI] [PubMed] [Google Scholar]
- 75. Noy N., Kelleher D. J., and Scotto A. W. (1995) Interactions of retinol with lipid bilayers: studies with vesicles of different radii. J. Lipid Res. 36, 375–382 [PubMed] [Google Scholar]
- 76. Jiang M., Shen D., Tao L., Pandey S., Heller K., and Fong H. K. (1995) Alternative splicing in human retinal mRNA transcripts of an opsin-related protein. Exp. Eye Res. 60, 401–406 10.1016/S0014-4835(05)80096-X [DOI] [PubMed] [Google Scholar]
- 77. Tao L., Shen D., Pandey S., Hao W., Rich K. A., and Fong H. K. (1998) Structure and developmental expression of the mouse RGR opsin gene. Mol. Vis. 4, 25 [PubMed] [Google Scholar]
- 78. Pandey S., Blanks J. C., Spee C., Jiang M., and Fong H. K. (1994) Cytoplasmic retinal localization of an evolutionary homolog of the visual pigments. Exp. Eye Res. 58, 605–613 10.1006/exer.1994.1055 [DOI] [PubMed] [Google Scholar]
- 79. Jiang M., Pandey S., and Fong H. K. (1993) An opsin homologue in the retina and pigment epithelium. Invest. Ophthalmol. Vis. Sci. 34, 3669–3678 [PubMed] [Google Scholar]
- 80. Chen P., Hao W., Rife L., Wang X. P., Shen D., Chen J., Ogden T., Van Boemel G. B., Wu L., Yang M., and Fong H. K. (2001) A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat. Genet. 28, 256–260 10.1038/90089 [DOI] [PubMed] [Google Scholar]
- 81. Tikidji-Hamburyan A., Reinhard K., Storchi R., Dietter J., Seitter H., Davis K. E., Idrees S., Mutter M., Walmsley L., Bedford R. A., Ueffing M., Ala-Laurila P., Brown T. M., Lucas R. J., and Münch T. A. (2017) Rods progressively escape saturation to drive visual responses in daylight conditions. Nat. Commun. 8, 1813 10.1038/s41467-017-01816-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jacobson S. G., Aleman T. S., Cideciyan A. V., Heon E., Golczak M., Beltran W. A., Sumaroka A., Schwartz S. B., Roman A. J., Windsor E. A., Wilson J. M., Aguirre G. D., Stone E. M., and Palczewski K. (2007) Human cone photoreceptor dependence on RPE65 isomerase. Proc. Natl. Acad. Sci. U.S.A. 104, 15123–15128 10.1073/pnas.0706367104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. He M., Du W., Du Q., Zhang Y., Li B., Ke C., Ye Y., and Du Q. (2013) Isolation of the retinal isomers from the isomerization of all-trans-retinal by flash countercurrent chromatography. J. Chromatogr. A 1271, 67–70 10.1016/j.chroma.2012.11.022 [DOI] [PubMed] [Google Scholar]
- 84. Stecher H., and Palczewski K. (2000) Multienzyme analysis of visual cycle. Methods Enzymol. 316, 330–344 10.1016/S0076-6879(00)16733-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Crabb J. W., Chen Y., Goldflam S., West K., and Kapron J. (1998) Methods for producing recombinant human cellular retinaldehyde-binding protein. Methods Mol. Biol. 89, 91–104 10.1385/0-89603-438-0:91 [DOI] [PubMed] [Google Scholar]
- 86. Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- 87. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 88. Longo P. A., Kavran J. M., Kim M. S., and Leahy D. J. (2013) Transient mammalian cell transfection with polyethyleneimine (PEI). Methods Enzymol. 529, 227–240 10.1016/B978-0-12-418687-3.00018-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Molday L. L., and Molday R. S. (2014) 1D4: a versatile epitope tag for the purification and characterization of expressed membrane and soluble proteins. Methods Mol. Biol. 1177, 1–15 10.1007/978-1-4939-1034-2_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R. (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.