Abstract
Lipopolysaccharide (LPS) from the Gram-negative bacterial outer membrane potently activates the human innate immune system. LPS is recognized by the Toll-like receptor 4/myeloid differentiation factor-2 (TLR4/MD2) complex, leading to the release of pro-inflammatory cytokines. Alkaline phosphatase (AP) is currently being investigated as an anti-inflammatory agent for detoxifying LPS through dephosphorylating lipid A, thus providing a potential treatment for managing both acute (sepsis) and chronic (metabolic endotoxemia) pathologies wherein aberrant TLR4/MD2 activation has been implicated. Endogenous LPS preparations are chemically heterogeneous, and little is known regarding the LPS chemotype substrate range of AP. Here, we investigated the activity of AP on a panel of structurally defined LPS chemotypes isolated from Escherichia coli and demonstrate that calf intestinal AP (cIAP) has only minimal activity against unmodified enteric LPS chemotypes. Pi was only released from a subset of LPS chemotypes harboring spontaneously labile phosphoethanolamine (PEtN) modifications connected through phosphoanhydride bonds. We demonstrate that the spontaneously hydrolyzed O-phosphorylethanolamine is the actual substrate for AP. We found that the 1- and 4′-lipid A phosphate groups critical in TLR4/MD2 signaling become susceptible to hydrolysis only after de-O-acylation of ester linked primary acyl chains on lipid A. Furthermore, PEtN modifications on lipid A specifically enhanced hTLR4 agonist activity of underacylated LPS preparations. Computational binding models are proposed to explain the limitation of AP substrate specificity imposed by the acylation state of lipid A, and the mechanism of PEtN in enhancing hTLR4/MD2 signaling.
Keywords: endotoxin, lipopolysaccharide (LPS), toll-like receptor (TLR), lipid A, phosphatase, intestinal alkaline phosphatase, intestinal flora, metabolic endotoxemia, metabolic syndrome, systemic inflammation
Introduction
Low-grade systemic inflammation associated with a number of metabolic syndromes has been proposed to originate from bacterial flora (1–7). Bacterial products (microbe-associated molecular patterns (MAMPs)2 or pathogen-associated molecular patterns (PAMPs)) from the intestinal flora are normally contained within the lumen by the intestinal epithelium layer. However, high-fat diets, obesity, and inflammatory bowel diseases help increase the permeability of the intestinal mucosal barrier, allowing luminal contents to breach containment and enter the bloodstream. MAMP engagement by pattern recognition receptors (PRR) on target cells triggers the release of pro-inflammatory cytokines, setting a low-grade systemic inflammatory tone (8, 9). Inflammation and oxidative stress is further accelerated by adipose tissue deposits, which can directly respond to MAMPs as well and secrete pro-inflammatory adipocytokines (10, 11). A central PRR-MAMP pair in these processes is the Gram-negative bacterial outer membrane component lipopolysaccharide (LPS or endotoxin) and the Toll-like receptor 4/myeloid differentiation factor-2 (TLR4/MD2) complex. Collectively termed metabolic endotoxemia (ME), elevated serum endotoxin levels and the ensuing aberrant TLR4/MD2 activity have been associated with the complications of metabolic diseases (6, 12). So far LPS constitutes the only known bacterial MAMP that can induce obesity as well as insulin resistance when infused subcutaneously into mice (2, 13). Thus, preventing ME through degradation and detoxification of LPS offers a potential therapeutic strategy for managing the onset, severity, and progression of ME (14).
According to the endotoxin principle, the 1- and 4′-lipid A phosphates of LPS are critical for biological activity as chemically dephosphorylated lipid A congeners weakly stimulate TLR4/MD2-directed pro-inflammatory cytokine release (15). Alkaline phosphatase (AP) has thus far received the most attention as a potential tool to detoxify LPS and attenuate inflammation through lipid A dephosphorylation (16–19). Recombinant AP is currently being examined in multiple clinical trials to treat a host of chronic (ME) or acute (sepsis) inflammatory conditions where endotoxin-induced inflammation is a suspected component of the underlying pathology (20–22). There are four AP-encoding genes in humans, three of which demonstrate tissue-specific expression (intestinal AP (IAP, ALPI), placental or embryonic AP (PLAP, ALPP) and germ cell AP (GCAP, ALPPL2)) along with a tissue-nonspecific AP (TNAP, ALPL) isoform (23, 24). IAP, being a native defense factor within the intestinal mucosal brush border, is a particularly promising therapeutic candidate as the anti-inflammatory mechanism of IAP has been attributed in part to the direct dephosphorylation of LPS (22, 25–30). Presumably, IAP hydrolyzes critical phosphates from lipid A involved in TLR4/MD2 recognition, although structural characterization of the IAP dephosphorylated LPS product has not hitherto been reported (31, 32). IAP activity to this point has instead been indirectly evaluated through the release of Pi using variants of the molybdate malachite green assay (33), in tandem with the apparent loss of biological activity. The potential LPS chemotype substrate specificity spectrum is unknown, as is the unanswered question of which of the many phosphate groups present on LPS are actually subject to dephosphorylation by IAP.
Given the complex and structurally heterogeneous nature of LPS (34, 35), we determined IAP substrate specificity using a panel of defined LPS chemotypes isolated from Escherichia coli. LPS from E. coli possesses among the highest endogenous endotoxin activity, and can be nonstoichiometrically modified with a variety of moieties. Variations in LPS acylation state, saccharide core, and phosphorylation patterns could conceivably affect IAP recognition and processing. Of note, E. coli converts the 1- and 4′-lipid A phosphate groups from phosphomonoesters into phosphodiesters upon addition of phosphoethanolamine (PEtN) and 4-amino-4-deoxy-l-arabinose (Ara4N) residues. PEtN and Ara4N addition are governed by a complex network of alternative transcription factors, two component systems, and regulatory RNAs (36). In Salmonella enterica, the regulators controlling LPS remodeling with PEtN/Ara4N are activated upon exposure to the mouse intestinal lumen environment (37). These regulatory systems could thus not only enable bacterial survival, but also block IAP-mediated LPS detoxification.
Herein, we report that IAP and other APs isozymes have minimal activity against unmodified LPS chemotypes (from hexa- to tetra-acylated lipid A) typical to the outer membrane of enteric bacteria such as E. coli. Unexpectedly, we have observed that Pi only appears to be released from LPS that has been modified with PEtN groups. We demonstrate that PEtN attached to LPS in a phosphoanhydride linkage spontaneously hydrolyzes at neutral to basic pH to release free PEtN, which in turn generates the actual substrate for IAP. Lipid A phosphate groups critical to TLR4/MD2 signaling only become susceptible to IAP activity after de-O-acylation of ester-linked primary acyl chains on lipid A. Importantly, LPS with PEtN covalently attached to lipid A on the 1- and 4′-lipid A phosphates through phosphoanhydride bonds stimulates TLR4/MD2 signaling activity compared with congeners lacking PEtN. The extent of TLR4/MD2 stimulation by PEtN modifications on lipid A is enhanced with decreasing acylation states, including for the otherwise antagonist tetra-acylated lipid IVA ligand. Computational binding models are applied to explain the substrate specificity of IAP, as well as the effects of PEtN modification on lipid IVA binding to human and murine TLR4/MD2 receptors. The apparent inability for IAP to dephosphorylate the critical lipid A backbone phosphates in fully acylated substrates suggests the physiological role of IAP in suppressing LPS-induced inflammation may be more complicated and invoke mechanisms beyond regulating TLR4/MD2 receptor activity.
Results
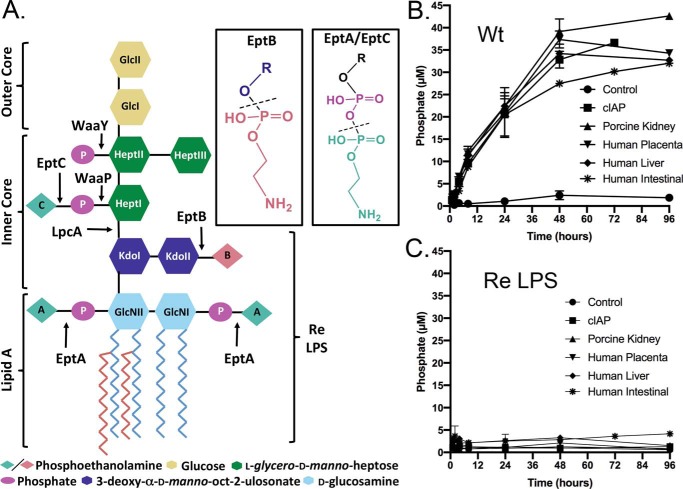
Inorganic phosphate is only released from WT E. coli B LPS chemotype
To study the effect of LPS modifications on processing by AP, a highly purified E. coli B LPS feedstock sample was first isolated using an extended protocol that included specific steps to remove phospholipid and nucleic acids contaminants that could contribute to background phosphate release. We chose E. coli B as the parent LPS strain source, first because of a native insertion sequence element within the outer saccharide core waaT gene, encoding for a UDP-galactose:(glucosyl) LPS α1,2-galactosyltransferase glycosyltransferase, which truncates the LPS to a structure of relatively low complexity (38, 39), and second for the high levels of endogenous PEtN and Ara4N modifications observed when this strain is grown in standard rich medium (Fig. 1A) (40). Analysis by MS confirmed a highly PEtN/Ara4N-substituted LPS (Fig. S1A), with an average total phosphate content of between 4 and 5 Pi equivalents per molecule of LPS (Table S2). We initially tested calf intestinal alkaline phosphatase (cIAP) with LPS preparations from WT E. coli B LPS for phosphate release using the Pi-specific malachite green assay (Fig. 1B). Although phosphate release was readily detected, the amount plateaued well short of the total LPS-associated phosphate input (∼25 μm LPS with 100–125 μm total phosphate). Because there are multiple tissue-specific AP isoforms (23, 24), a panel of commercially available APs was tested to determine whether a more robust phosphate release could be realized. However, all APs (including human intestinal alkaline phosphatase, hALPI) demonstrated a plateau similar to cIAP even after a prolonged 96-h incubation period (Fig. 1B). Varying reaction conditions by adding bile salts to act as detergent, BSA, 10% whole serum, or extensive pre-sonication of LPS vesicles did not appreciably enhance the amount of phosphate released (data not shown). Detection of Pi was completely dependent on inclusion of AP. This suggested that only a fraction of the total LPS phosphate content was subject to hydrolysis, irrespective of either the AP isoform or presence of de-aggregation agents.
Figure 1.
Pi released from LPS chemotypes in the presence of various alkaline phosphatases. A, schematic of hexa-acylated WT LPS chemotype from E. coli BL21 (DE3) with the nonstoichiometric phosphoanhydride (EptA/EptC) and phosphodiester (EptB)-linked PEtN modifications indicated. Ara4N modifications (not shown) share a common site of attachment with PEtN added by EptA to lipid A substrate. The structure of Re LPS resulting from lpcA deletion is indicated. LPS was extracted from either WT (Wt) (B) or unmodified Re LPS (TXM333 ΔlpcAΔeptAΔarnA) (C) producing strains and incubated with the indicated AP (100 μg/ml of substrate, 50 mm Tris-HCl (pH 8.25), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2 at 37 °C). Phosphate release was measured using the malachite green assay and the data plotted as the mean ± S.D. of three independent replicates.
To determine which LPS phosphate group(s) was being released, we repeated the assay using a structurally defined Re LPS chemotype as substrate. Re LPS extracted from E. coli TXM333 lacks all sugars but Kdo (3-deoxy-α-d-manno-oct-2-ulosonic acid) from the saccharide core due to deletion of the d-sedoheptulose 7-phosphate isomerase gene lpcA (gmhA) as well as all types of PEtN/Ara4N modifications on lipid A due to deletions in the respective biosynthetic genes eptA/arnA (Fig. 1A). Re LPS was extensively purified as described above for WT, and analysis by MS confirmed a nearly homogeneous population of the Re chemotype (Fig. S1B and Table S2). In striking contrast to WT LPS, no phosphate was liberated from the Re chemotype when incubated under identical conditions (Fig. 1C). This indicated inefficient cleavage of core lipid A phosphates by APs, and that the phosphate liberated from WT LPS must originate from either lipid A PEtN or saccharide core modifications attached distal to Kdo residues.
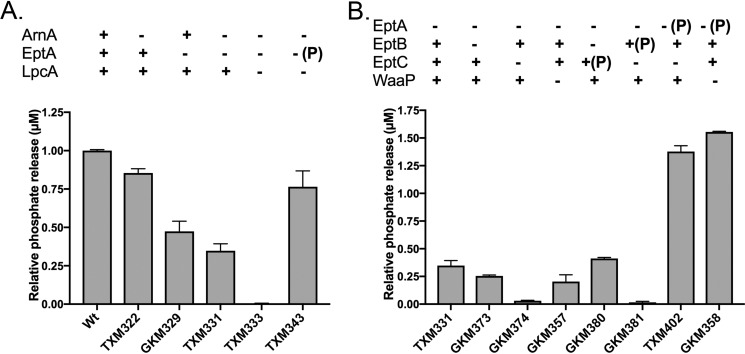
Released phosphate originates from PEtN added by EptA and EptC
PEtN groups are added in nonstoichiometric amounts to LPS at three distinct positions by a set of related membrane-bound transferases: (i) EptA onto either lipid A phosphate (41), (ii) EptB onto KdoII (42), or (iii) EptC onto phosphorylated HepI (l-glycero-d-manno-heptose) (43). We first constructed a panel of deletions in eptA, because this transferase can add PEtN to both phosphates of lipid A (Fig. 1A). Indeed, the amount of phosphate released was closely correlated with the presence of EptA, and plasmid-borne EptA alone restored phosphate release when testing Re LPS chemotype as substrate (Fig. 2A). Deletion of arnA, which modifies lipid A phosphate groups with Ara4N (44), alone or in tandem with eptA had minimal influence on phosphate release.
Figure 2.
Phosphate released during incubation of (A) BL21 (DE3) (Wt), TXM322 (ΔarnA), GKM329 (ΔeptA), TXM331 (ΔeptAΔarnA), TXM333 (Re LPS, ΔlpcAΔeptAΔarnA), TXM343 (Re LPS, ΔlpcAΔeptAΔarnA + pEptA), and (B) TXM331 (ΔeptAΔarnA), GKM373 (ΔeptAΔarnAΔeptB), GKM374 (ΔeptAΔarnAΔeptC), GKM357 (ΔeptAΔarnAΔwaaP), GKM380 (ΔeptAΔarnAΔeptB + pEptC), GKM381 (ΔeptAΔarnAΔeptC + pEptB), TXM402 (ΔeptAΔarnAΔeptC + pEptA), and GKM358 (ΔeptAΔarnAΔwaaP + EptA) with cIAP (100 μg/ml of substrate, 4 units/ml, 50 mm Tris-HCl (pH 8.25), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2 at 37 °C). Released phosphate was measured after 48 h of incubation using the malachite green assay and normalized to BL21 (DE3) (Wt). Data are representative of three independent experiments conducted in duplicates and the error bars show S.D.; ± indicates chromosomal genes; (P) denotes gene introduced on a plasmid.
Although phosphate installed by EptA accounted for the bulk of the total released, significant amounts of phosphate (up to ∼30% of the total) were still liberated from LPS chemotypes isolated from ΔeptA strain backgrounds. This suggests some of the detected phosphate originated from the saccharide core as well. We thus tested phosphate liberation from LPS chemotypes produced by strains harboring various combinations of eptA/B/C (Fig. 2B). All strains were ΔarnA to limit any variability arising from substrate competition by ArnA with EptA. Because the native promoters of each of the PEtN transferases is subject to complex regulation, constructs with constitutive promoters were used to achieve comparable high-level PEtN modification levels as assessed by MS (Fig. S2). Next to ΔeptA, the least amount of phosphate release was detected from LPS extracted from strains lacking EptC and WaaP, the HepI kinase that forms the P-HepI acceptor substrate utilized by EptC (45). Overexpression of EptC, but not EptB, enhanced phosphate release although the amount remained well below EptA. The data collectively suggests EptA installs most of the total labile phosphate pool in E. coli WT LPS, with EptC making a minor contribution. The phosphate content added by EptB is stable to hydrolysis.
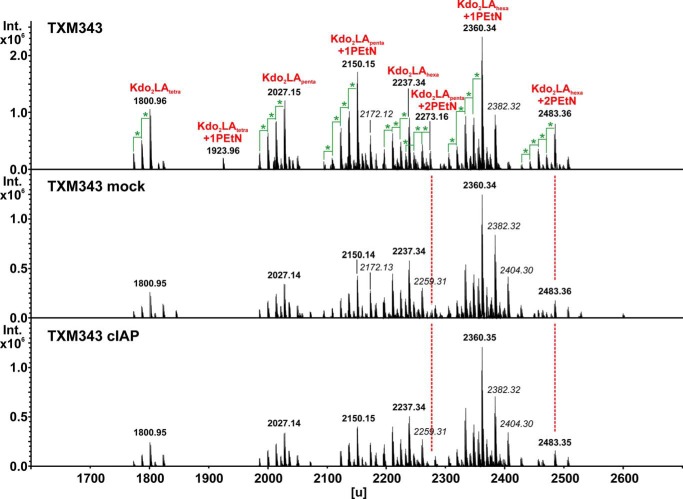
PEtN release from LPS is spontaneous
The PEtN groups attached by EptA and EptC are both connected by a phosphoanhydride bond, whereas EptB installs a typical phosphodiester bond at KdoII (Fig. 1A, inset). The data thus supported one of three scenarios, wherein: (i) cIAP specifically recognizes PEtN residues attached by EptA/EptC, (ii) cIAP only cleaves phosphoanhydride linked PEtN groups, or (iii) PEtN connected through a phosphoanhydride bond spontaneously hydrolyzes to free O-PEtN monoester that then becomes the actual substrate for cIAP. We initially suspected the third scenario, given the known instability of high-energy phosphoanhydride bonds and because the majority of characterized AP substrates are phosphomonoesters (23, 46). To test for nonenzymatic hydrolysis, Re LPS modified with phosphoanhydride-linked PEtN added by EptA (from TXM343) was incubated with or without cIAP, extracted, and analyzed by MS for doubly modified, singly modified, and unmodified Re LPS (Fig. 3). Reactions were performed within dialysis tubing and continuously dialyzed to prevent any potential product inhibition by liberated Pi. PEtN hydrolysis was monitored by a mass shift of Δm = 123 units, which corresponds to a single PEtN residue. Although the total PEtN content clearly decreased after incubation (buffer pH 7.1, 16 h) when compared with directly injected samples, the MS profile of mock-treated Re LPS was nearly indistinguishable from cIAP-treated samples (Fig. 3, middle and bottom panels).
Figure 3.
MS analysis of EptA-modified Re LPS. Re LPS was extracted from TXM343 (lpcA::gentR eptA::catR arnA::kanR (pSEVA434-eptA)) and analyzed by MS. Samples were either directly analyzed (top panel), or incubated for 16 h at 37 °C in buffer (100 μg/ml substrate, 50 mm Tris-HCl (pH 7.1), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2) alone (mock) or with cIAP (4 units/ml). Reactions were assembled within dialysis tubing and continuously dialyzed against buffer to remove potentially inhibitory Pi product. Masses in italic type represent sodium adducts (Δm = 22 units), whereas those marked with a green star account for a difference of Δm = 14 units, consistent with a methylene unit (-CH2-). Chemical composition assignments (red) for unmodified Re LPS species are listed in Table S2.
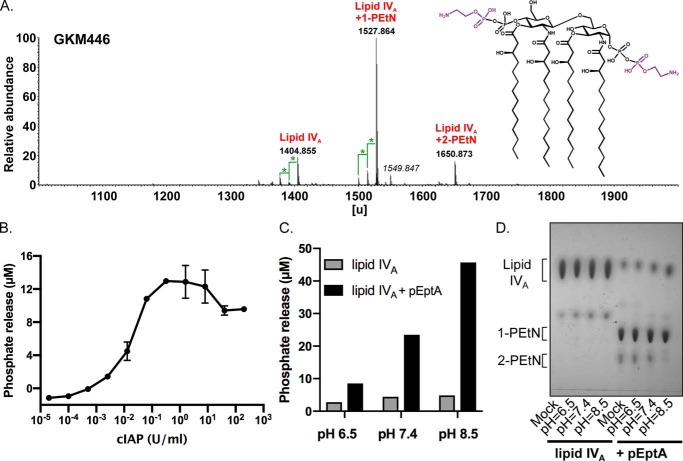
To confirm spontaneous (i.e. nonenzymatic) hydrolysis, we examined a second set of PEtN-modified chemotypes with a more homogenous composition because quantitative comparison of Re LPS MS peaks is complicated by multiple glycoforms within the same population. We previously had constructed a mutant E. coli strain that elaborates only lipid IVA, a chemotype lacking glycosylation with uniform 3-OH-C14:0 tetra-acylation, which remains viable due to suppressor mutations in LPS transport systems (47, 48). By introducing pEptA into this genetic background, we obtained PEtN-modified lipid IVA substrate preparations with 2-PEtN, 1-PEtN, or unmodified lipid IVA (Fig. 4A). When assayed for phosphate release, a nonlinear correlation between the amount of released Pi and the units of cIAP added was observed well before the total amount of input PEtN-linked lipid IVA phosphate should become rate-limiting (Fig. 4B). This is consistent with an initial slower, nonenzymatic hydrolytic step preceding cIAP catalysis, namely the putative dephosphorylation of free PEtN. To support this theory, the O-PEtN monoester was directly tested as a substrate for cIAP and hALPI (Fig. S3A). Typical Michaelis-Menten kinetics (Km of 173 ± 27 μm and Vmax of 1.09 ± 0.05 μm/min for cIAP and Km of 197 ± 56 μm and Vmax of 1.05 ± 0.1 μm/min for hALPI) were observed for both enzymes with O-PEtN as substrate. Using the crystal structure of the highly homologous rat IAP ortholog (∼70% identity with cIAP across 486 nongapped residues) as a model (49), O-PEtN can be readily accommodated within the substrate-binding pocket in the active site (Fig. S3B). This is in contrast to modeling of acylated lipid A as the putative IAP substrate (see below).
Figure 4.
Hydrolysis of phosphoanhydride-linked PEtN-lipid IVA is spontaneous and pH dependent. A, MS analysis in the negative anion mode of lipid IVA isolated from GKM446 (ΔeptAΔgutQΔkdsDΔlpxLΔlpxMΔlpxPΔpagP + pEptA). The structure of lipid IVA was modified with two PEtN residues (in purple) in nonstoichiometric amounts at C1-GlcNI and C4′-GlcNII when EptA is expressed. Masses in italic represent sodium adducts (Δm = 22 units), differences marked with a green star account for a difference of Δm = 14 units, consistent with a methylene unit (-CH2-). B, phosphate release was measured as a function of increasing concentrations of cIAP after incubation for 6 h at 37 °C (100 μg/ml of PEtN-lipid IVA substrate, 50 mm Tris-HCl (pH 8.25), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2) using PEtN-lipid IVA substrate isolated from GKM446. Phosphate was measured using the malachite green assay, and data are representative of two independent experiments conducted in triplicate with the error bars showing S.D. values. C, either lipid IVA alone (ClearColi® K-12 GKM445 ΔeptAΔgutQΔkdsDΔlpxLΔlpxMΔlpxPΔpagP) or with PEtN added by EptA (GKM446 ΔeptAΔgutQΔkdsDΔlpxLΔlpxMΔlpxPΔpagP + pEptA) was incubated for 48 h at 37 °C in MOPS-Tris buffer (100 μg/ml of substrate, 50 mm MOPS, 50 mm Tris (adjusted to pH 6.5, 7.4, or 8.5), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2), and then treated with cIAP (10 units/ml of cIAP) to release Pi from spontaneously hydrolyzed PEtN. Phosphate was quantified using the malachite green assay. D, lipid IVA species were hydrolyzed for 48 h in MOPS-Tris buffer at the indicated pH as described in C, except samples were isolated by extraction before separation by TLC. Total lipid was visualized by sulfuric acid charring.
Hydrolysis of phosphoanhydride-linked PEtN from LPS is pH-dependent
We next tested the stability of PEtN linkages attached by EptA on lipid IVA by incubation in buffer at defined pH (Fig. 4C). Liberated PEtN was indirectly quantified by adding excess cIAP at the end of the incubation period and measuring Pi levels using the malachite green assay. The extent of phosphate released increased as the pH of the preincubation buffer was raised, consistent with a base-labile phosphoanhydride bond. Hydrolysis was minimal at acidic pH. Interestingly, a mildly acidic environment naturally induces eptA expression and lipid A modification with PEtN in E. coli and S. enterica (50, 51). We repeated the incubation a second time, except the resulting lipid IVA population was extracted and separated by TLC before being visualized by sulfuric acid charring (Fig. 4D). Unmodified lipid IVA was stable across the entire pH range tested, whereas the dually modified 2-PEtN lipid IVA population disappeared with a concomitant increase in free lipid IVA as hydrolysis incubation conditions became more basic. Considering no cIAP was utilized, these data in combination with the qualitative MS results using PEtN-modified Re LPS (Fig. 3) supports a two-step mechanism whereby cIAP catalyzes phosphate release from spontaneously hydrolyzed O-PEtN that had initially been bound to LPS in a labile phosphoanhydride linkage.
cIAP directly releases Pi from de-O-acylated lipid A chemotypes
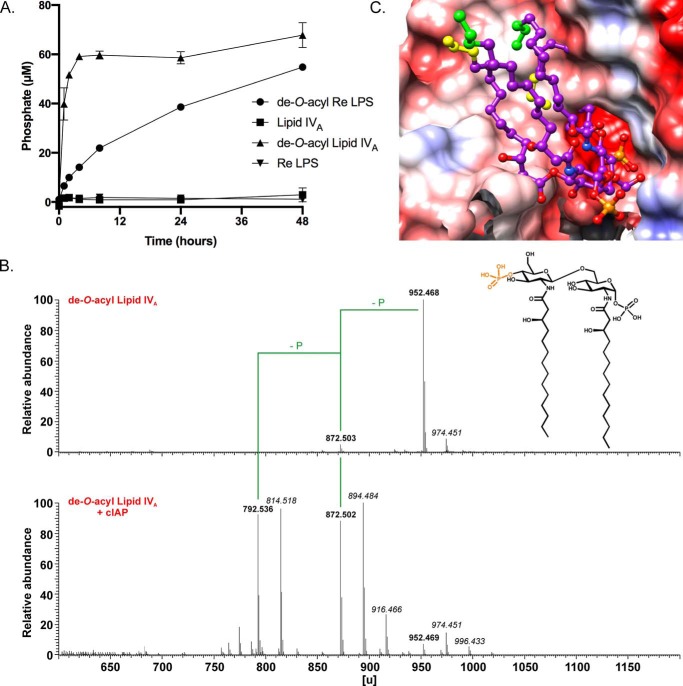
The recalcitrance of all LPS chemotypes thus far tested to being directly dephosphorylated by cIAP suggested that steric interference may prevent the AP active site from engaging target lipid A phosphate monoesters. To test this hypothesis, we removed all ester-linked acyl chains from lipid IVA and Re LPS to generate di-N-acyl de-O-acyl lipid A derivatives (Fig. 5). De-O-acyl lipid A (the N,N-diacylated) only contains amide-linked 3-OH-C14:0 acyl chains at C2 of GlcNI and C2′ of GlcNII. If steric hindrance is indeed problematic, this should facilitate enzyme access to the phosphate groups, particularly at C4′ of GlcNII, which is now adjacent to a free hydroxyl group compared with the steric bulk of a 3-OH-C14:0 acyl chain in lipid IVA. Furthermore, decreasing the acyl chain density on the lipid A backbone substrate also increases the conformational flexibility. Unlike the tetra-acylated lipid IVA parent, de-O-acyl lipid A was rapidly dephosphorylated in an initial phase that was followed by an extended period of slow phosphate release (Fig. 5A). De-O-acylated Re LPS, which has two Kdo residues attached to C6′ of GlcNII (Fig. S4A, inset), was likewise dephosphorylated in a biphasic manner albeit at a slower overall rate (Fig. 5A). MS analysis of cIAP-treated products revealed the entire population had lost at least one phosphate group from both de-O-acyl lipid A and Re LPS substrates (Fig. 5B and Fig. S4A). NMR analysis of the residual phosphate remaining after treatment of de-O-acyl lipid A with cIAP indicated the majority (∼77%) of the anomeric GlcNI phosphate group was retained under these conditions (Fig. S4B). In summary, the data are consistent with quantitative hydrolysis of a highly cIAP susceptible phosphate group on GlcNII followed by a slower second dephosphorylation event on GlcNI that remains incomplete even after a 48-h reaction period.
Figure 5.
De-O-acylated lipid A is rapidly dephosphorylated by cIAP. A, phosphate released by cIAP from either de-O-acylated lipid IVA or Re LPS (100 μg/ml substrate, 4 units/ml cIAP, 50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2) at 37 °C was quantified at the indicated times using the malachite green assay. The data are plotted as the mean ± S.D. of three independent replicates. B, MS analysis in the positive anion mode of de-O-acyl lipid IVA before (top panel, calculated mass 952.467 units; recorded in negative ion mode) and after treatment with cIAP (bottom panel, calculated masses of 872.501 and 792.535 units for monophosphoryl and nonphosphorylated products, respectively; recorded in positive ion mode). The highly cIAP susceptible phosphate at C4′-GlcNII is colored orange. Masses resulting from dephosphorylation events are indicated (-P), whereas masses in italic represent sodium adducts (Δm = 22 units). C, binding model of tetra-acylated lipid IVA with the C4′-GlcNII phosphate at the active site of cIAP. The phosphorylated catalytic serine covalent intermediate (bottom right, deeply buried in the cleft) is aligned with the C4′-GlcNII lipid IVA phosphate as reference (Fig. S3). Proximal atoms of the two O-acylated side chains (with the terminal Ω, Ω-1, Ω-2 carbon atoms colored in yellow) permeate the protein surface. The two N-acylated side chains can occupy the cleft in their entire length without any steric clashes (green caps). The anomeric 1-phosphate group of GlcNI lies to the front (bottom most, right) and is only slowly cleaved in de-O-acyl lipid A (N,N-di-acylated lipid IVA derivative, modeled in Fig. S5). Red, blue, and white surface colors indicate negative, positive, and neutral partial charges, respectively. Hydrogen atoms are not displayed.
We subsequently generated a model of the cIAP active site using the crystal structure of the highly similar rat IAP ortholog (∼70% identity with cIAP across 486 nongapped residues) as a structural template (49). Whereas de-O-acyl lipid A could be accommodated within the active site, lipid IVA could not be docked successfully (Fig. 5C). Detrimental van der Waals contacts arise between the protein surface with the two O-acyl side chains at all times when the GlcNII phosphate was positioned to occupy the active site. In particular, the 3′-O-acyl chain on GlcNII clashed with amino acid chains flanking the active site. During simulations, the N-acylated side chains, however, adopted conformations that could avoid steric clashes when docked with the cIAP susceptible de-O-acyl lipid A GlcNII phosphate orientated in the active site. Likewise, docking of the anomeric C1-GlcNI phosphate lipid IVA into the catalytic cleft resulted in multiple steric interferences (Fig. S5). The computed binding models are consistent with the nonanomeric 4′-phosphate of de-O-acyl lipid A being the preferred position for cIAP-mediated dephosphorylation.
PEtN modification enhances hTLR4 agonist activity of underacylated LPS chemotypes
Our data indicate a mechanistic model whereby PEtN spontaneously hydrolyzes from phosphoanhydride linkages on LPS to generate free O-PEtN, a monoester substrate that is processed by cIAP to liberate Pi. This interpretation would explain the apparent dependence of cIAP for detection of free Pi release from LPS under in vitro reaction conditions because the malachite green assay does not detect organic phosphate as in O-PEtN. Yet that alone does not account for the observed decrease in TLR4/MD2 activity after in vitro cIAP treatment considering the critical role lipid A phosphates at C1-GlcNI and C4′-GlcNII play during binding to TLR4/MD2 (52, 53). Our data indicates these key phosphate groups remain intact after exposure to cIAP unless primary O-ester acyl chains on lipid A have first been removed. Transforming lipid A into a good cIAP substrate through prior de-O-acylation would itself abrogate TLR4/MD2 activity, which argues against de-O-acyl glycoform dephosphorylation being relevant to endotoxin neutralization.
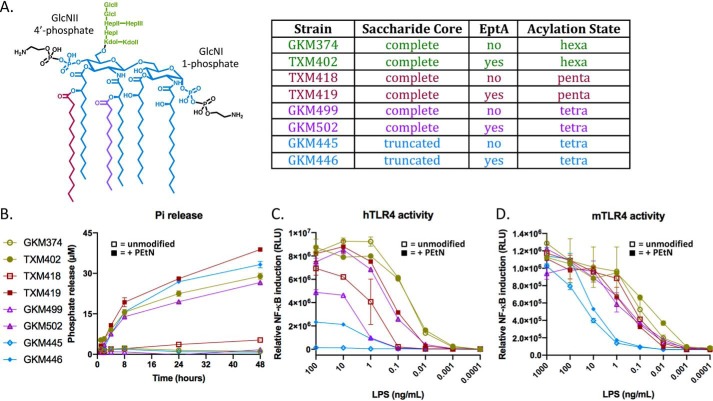
Previous studies have, however, demonstrated PEtN modifications of lipid A in Neisseria meningitidis (54–57) and Campylobacter jejuni (58) increase TLR4/MD2 signaling. Spontaneous hydrolysis of PEtN, which can only be detected by Pi assays with added AP, could account for the apparent decrease in biological activity. We therefore determined whether PEtN modification of E. coli lipid A, which has an asymetric lipid A acyl chain distribution unlike in N. meningitidis, affects TLR4/MD2 recognition in a similar fashion. We initially constructed a strain panel that synthesized lipid A glycoforms varying in acylation state and either with or without EptA-appended PEtN modifications (Fig. 6A). As expected, comparable amounts of Pi were only detected in the presence cIAP with LPS substrates that had been isolated from parent strains expressing EptA (Fig. 6B). We next directly compared TLR4/MD2 stimulation using a HEK293/hTLR4/MD2-CD14 whole cell NF-κB reporter assay (Fig. 6C). We utilized a luciferase-based reporter assay instead of the secreted embryonic AP (SEAP) HEK-BlueTM colorimetric reporter system, because there are reports that the PLAP isoform dephosphorylates LPS (29). Although PEtN addition to hexa-acylated lipid A had minimal impact, PEtN modifications of the penta- and tetra-acylated lipid A glycoforms containing a full saccharide core enhanced TLR4 signaling by ∼10-fold. Tetra-acylated lipid IVA glycoform did not stimulate hTLR4/MD2, consistent with lipid IVA being a known human TLR4 antagonist (59–62). Surprisingly, PEtN addition by EptA to lipid IVA imparted low but definite agonist activity (Fig. 6C). Restoration of hTLR4/MD2 activity by PEtN modification of lipid IVA was suppressed by preincubation in buffer in a pH-dependent manner (Fig. S6), as expected considering the labile nature of the lipid IVA-PEtN phosphoanhydride bond with increasing pH (Fig. 4, C and D). Collectively this suggests that PEtN addition to lipid IVA can convert an LPS-like hTLR4 antagonist into a weak agonist. The contribution of PEtN to hTLR4 activity is more determinant with suboptimal, underacylated E. coli LPS ligands, and agrees with observations made using the N. meningitidis lipid A scaffold (55).
Figure 6.
LPS modification with PEtN by EptA induces NF-κB activity through hTLR4/MD2 signaling. A, the structure of hexa-acylated lipid A modified with PEtN at both the C1-GlcNI and C4′-GlcNII phosphates. The major lipid A acylation chemotype being produced in paired pEptA−/+ E. coli constructs GKM374/TXM402 (ΔeptAΔarnAΔeptC), TXM418/TXM419 (ΔeptAΔarnAΔeptCΔlpxM), GKM499/TXM502 (ΔeptAΔarnAΔeptCΔlpxLΔlpxMΔpagP), and ClearColi® K-12 GKM445/GKM446 (ΔeptAΔgutQΔkdsDΔlpxLΔlpxMΔlpxPΔpagP) are indicated. B, the Pi released during incubation with cIAP (4 units/ml, 100 μg/ml substrate, 50 mm Tris-HCl (pH 8.25), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2) was measured using the malachite green assay. Data are representative of three independent experiments done in duplicates and the error bars show S.D. values. C, HEK293/hTLR4-MD2-CD14 NF-κB reporter cells were stimulated with the indicated LPS chemotypes and the luciferase activity was measured. D, the bioactivity of the LPS chemotypes with murine TLR4/MD2 receptor was re-tested using stably transfected HEK293/mTLR4-MD2-CD14 reporter cells. For both hTLR4 and mTLR4 assays, data are representative of three independent experiments conducted in duplicates with the error bars showing S.D. values.
Because key amino acid differences at the TLR4/MD2/LPS interface endow species-specific lipid IVA responses (63), we repeated the assay using the same panel of LPS chemotypes but with mouse TLR4/MD2 expressing NF-κB reporter cells (Fig. 6D). The pattern observed with hTLR4/MD2 was not replicated with the murine receptor complex, as PEtN attachment had minimal effect on signaling for any of the tested lipid A acylation states. This demonstrates that the enhanced signaling observed with hTLR4/MD2 is not an inherent biophysical property of PEtN-modified lipid A, but rather due to species-specific ligand recognition and engagement features unique to the respective TLR4/MD2 receptor complexes.
PEtN substitution of both lipid IVA phosphates is required for maximum hTLR4 agonism
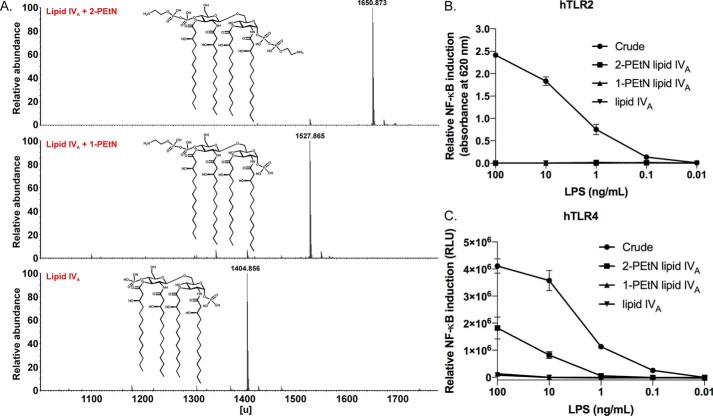
EptA can covalently add PEtN groups via phosphoanhydride bonds to either of the two lipid A phosphate groups, at C1 of GlcNI or C4′ of GlcNII. Hence we sought to determine whether both PEtN groups (2-PEtN) are required or if a single PEtN moiety (1-PEtN) is sufficient to trigger hTLR4/MD2 activity. To accomplish this, we utilized an E. coli B lipid IVA PEtN producing strain because this genetic background elaborates higher levels of PEtN-modified lipid A compared with the K-12 strain used in the prior experiments (40). We next developed a lipid IVA purification protocol to remove any contaminating lipoproteins and phospholipids, as well as to isolate 1-PEtN and 2-PEtN lipid IVA species to allow for more quantitative comparisons between glycoforms. Established purification methods used prior for LPS chemotypes containing at least part of the saccharide core failed when applied to PEtN-lipid IVA material extracted using the PCP method (see “Experimental procedures”). We thus optimized a pair of nonionic detergent-aided lipase pre-treatment steps to remove phospholipids and deacylate interfering lipoproteins, which improved the ensuing chromatographic separation of PEtN-lipid IVA species in the following step (Fig. S7). Using an adapted anion exchange chromatography protocol developed by Raetz and co-workers (64, 65) to separate Ara4N- and PEtN-modified chemotypes, we were able to isolate 2-PEtN and 1-PEtN lipid IVA species in high purity. A final purification step using HPLC reverse phase chromatography yielded homogeneous 2- and 1-PEtN lipid IVA samples that were essentially free of contaminating lipoproteins and chemically pure with respect to PEtN content as judged by MS analysis and TLR2 activation (Fig. 7, A and B). In addition, 31P NMR analysis confirmed PEtN substitution solely at C4′ of GlcNII in the purified 1-PEtN fraction (Fig. S8). When comparing purified fractions, the 2-PEtN fraction alone accounted for the bulk of the recovered hTLR4/MD2-stimulating activity (Fig. 7C). PEtN substitution at C1 of GlcNI on the lipid IVA scaffold (with four symmetrically distributed acyl chains) therefore constitutes a critical determinant of restoring agonistic character.
Figure 7.
MS analysis and TLR activity of purified 2-PEtN and 1-PEtN–modified lipid IVA samples. A, MS analysis of the purified PEtN-lipid IVA species isolated from E. coli strain TXM844 (msbA148ΔeptAΔgutQΔkdsDΔlpxLΔlpxMΔpagPΔlpxP + pEptA). B, relative NF-κB induction in HEKBlue hTLR2 reporter cells stimulated with crude LPS, 2-PEtN lipid IVA, 1-PEtN lipid IVA, and unmodified lipid IVA. Data are from experiments done in triplicate and the error bars show S.D. values. C, relative NF-κB induction in HEK293/hTLR4-MD2-CD14 reporter cells stimulated with crude PEtN lipid IVA (TXM844) and the purified 2-PEtN lipid IVA, 1-PEtN lipid IVA, and unmodified lipid IVA fractions. Data are from experiments conducted in triplicate and the error bars show S.D. values.
Molecular modeling suggests hTLR4/MD2 residues responsible for species-specific enhanced recognition of PEtN-modified lipid A
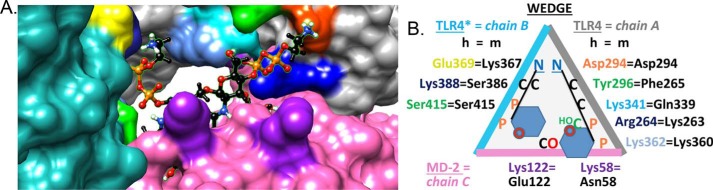
Signaling assays with TLR4/MD2 reporter cells unveiled that whereas lipid IVA is endotoxically inactive in human receptor complexes as expected, PEtN addition by EptA restores detectable activity (Figs. 6C and 7C). The contribution of PEtN to enhancing activity was more pronounced as the lipid A ligand became increasingly underacylated (Fig. 6). This trend, however, was not observed in murine receptor (mTLR4/MD2) reporter cells. MD2 is highly conserved between species, except for a few key residues (Fig. 8) (63). In the human receptor complex, a nonconserved cationic residue (hLys-122 versus anionic mGlu-122) on the rim of MD2 interacts with the negative charge of lipid A phosphate anions and causes the ligand to be buried more deeply within the MD2 cavity in an antagonistic pose. In contrast, mGlu-122 forces the ligand's phosphate groups to move away by charge repulsion into an agonist pose that is well-positioned for interactions with other subunits within the complex. Given the constant space in the MD2-binding cleft, underacylated lipid A congeners become more deeply buried than fully acylated ligands in hMD2 until eventually all agonist character is lost as with tetra-acylated lipid IVA. Our binding model suggest that as a direct consequence of PEtN substitution, ligand is sufficiently exposed to form contacts with the second TLR4* subunit, triggering dimerization between the [TLR4/MD2] and [TLR4*/MD2*] ectodomains and initiating downstream signaling (Fig. 8). The influence of PEtN groups is thus most evident when needed, i.e. for binding underacylated lipid A ligands in the hTLR4/MD2 complex. This effect is more muted in mTLR4/MD2 because mGlu-122 makes the binding contribution of PEtN substitution redundant as mGlu-122 already serves a similar function. In addition, there is more potential for favorable electrostatic interaction points between hTLR4 residues (e.g. hAsp-294 and hGlu-369) with the amino groups of PEtN (Fig. 8B), helping to bridge the space between TLR4 and TLR4*. In contrast, the murine receptor complex with mLys-367 in place of hGlu-369 is less favorable due to positive charge repulsion.
Figure 8.
Simulated hTLR4/MD2 binding of 2-PEtN–modified lipid IVA. A, computational model of hTLR4/MD2 with 2-PEtN–modified lipid IVA substrate bound at the hTLR4 homodimer interface. Surface color codes are gray for TLR4 (A chain, right), turquoise for the second TLR4* subunit (B chain, left), and pink for MD2 (C chain, bottom). Key substrate binding determinants on each subunit chain are indicated as follows. For MD2, hLys-122 and hLys-58 (bottom) are indicated by two purple patches. Patches on TLR4* to the left highlight hGlu-369 (yellow, equivalent to mLys-367), hLys-388 (blue, behind hGlu-369), and hSer-415 (green, below hGlu-369). Highlighted mid-section patches on TLR4 are hLys-362 (powder blue) and hLys-341 (cyan), whereas top right-side patches include hTyr-296 (green), hAsp-294 (orange), and hArg-264 (blue). Atom colors for the 2-PEtN–modified lipid IVA ligand: black, C-H; orange, P; red, O; blue, N; white, polar H. The GlcNII ring is visible in the cleft, whereas the GlcNI moiety is occluded by hLys-122. The two cationic H3N+ head groups of PEtN contact anionic hGlu-369 (anomeric C1-phosphate of GlcNI) and hAsp-294 (nonanomeric C4′-phosphate of GlcNII). The MD2 lipophilic cavity buries all four acyl chains of lipid IVA. Tyr-296 is positioned to contact the 6′-C-OH group or the 4′-pyrophosphate group of PEtN on GlcNII. B, binding map schematic highlighting critical residues that vary between the human (colored text to match A) and murine (black text) TLR4/MD2 receptor complex. To bind LPS-like ligands, the dimerized receptor complex provides a binding site contoured by TLR4*/MD2/TLR4. When projected onto a plane from a certain perspective, the three proteins (B/C/A chains) form a triangle (wedge). Amino acids potentially serving as favorable electrostatic contact points for the cationic amino head groups of PEtN moieties are noted, including anionic hAsp-294 (TLR4) and hGlu-369 (TLR4*) residues. Of note, the latter is replaced by a nonhomologous lysine (mLys-367) residue in the mTLR4 receptor. As in A, hTyr-296 (green) is interacting with the C6-OH group of GlcNII (green) or with the adjacent pyrophosphate group (orange). The amino acid residue numbering scheme has been described (63).
Discussion
The anti-inflammatory properties of IAP and APs in general have generated broad interest as potential therapeutics for treating a number of systemic and acute inflammatory conditions. Dissecting and isolating the relevant phosphorylated molecular targets involved, however, is a complicated endeavor due to the broad substrate specificity of IAP. IAP can dephosphorylate both bacterial derived MAMP/PAMPs as well as pro-inflammatory endogenous damage-associated molecular patterns (DAMPs) of host cell origin produced in response to LPS-mediated TLR4/MD2 inflammation. Along with LPS, flagellin, CpG DNA motifs and extracellular nucleotides have all been proposed as relevant IAP dephosphorylation targets (28). Flagellin and CpG motifs in bacterial DNA are MAMP/PAMP TLR ligands like LPS (66), whereas extracellular ATP nucleotide binds to purinergic receptors (67) and UDP to the P2Y6 pyrimidinergic receptor (68). The anti-inflammatory mechanism of IAP is further confounded by the difficulty in separating IAP-specific effects (i.e. the direct dephosphorylation of MAMP/PAMP/DAMP stimuli) from nonspecific ones resulting from general down-regulation of the inflammatory tone (17).
LPS has emerged as an often cited and most likely MAMP/PAMP-related molecular target for AP, due to its potent inflammatory potential when present at low concentrations. The conclusion that IAP dephosphorylates LPS in vitro predominantly rests on the observation that LPS-derived Pi is only detectable if AP is added when using either the molybdate complex or tissue histology-based assays as readouts. The corresponding LPS preparations treated with AP have diminished capacity to induce inflammation through TLR4/MD2 stimulation, as would be expected for hydrolysis of the lipid A backbone phosphate groups because these groups are central in TLR4/MD2 recognition. Direct evidence pertaining to which phosphate groups are indeed removed by AP is lacking, however. LPS preparations can also have multiple phosphate groups on the inner saccharide core in addition to those on lipid A. In E. coli, for instance, WaaP and WaaY heptose kinases phosphorylate HepI and HepII, respectively (45), whereas KdkA kinase modifies Kdo in bacteria with phosphorylated mono-Kdo residues such as Haemophilus influenzae (69). We thus sought to determine the origin of the Pi released by AP. Although our results are consistent with Pi release being contingent on AP activity for structurally heterogeneous LPS preparations, we found defined substrates such as Re LPS chemotype that only have lipid A phosphate monoester groups to be completely inert to dephosphorylation (Fig. 1, B and C). In previous studies where defined LPS chemotypes were tested for AP activity, a decrease in phosphate release has also been noted. Tuin et al. (70) recorded lower phosphate release when histology scoring rat liver sections were challenged with Re LPS from S. enterica sv. Minnesota, whereas Pettengill et al. (31) observed low phosphate release from S. enterica sv. Minnesota Re LPS and no measurable phosphate release from monophosphoryl lipid A (MPLA) substrate using recombinant TNAP. Re LPS lacks the Hep acceptor for the inner core PEtN transferase EptC, which we have now shown is an apparent source of cIAP-released phosphate along with the lipid A PEtN transferase EptA (Fig. 2). PEtN groups are common in many Gram-negative bacteria where they are added to both lipid A and the LPS inner saccharide core to increase the net positive charge (71). The increased positive charge imparts resistance to cationic peptides and enhances outer membrane integrity, particularly under more challenging growth environments. Both EptC and EptA enzymes add PEtN in a phosphoanhydride linkage using phosphatidylethanolamine phospholipid head groups as PEtN donors, forming a rather unique bond type in the process considering the extracellular location (Fig. 1A, inset). Whereas other bacterial surface phosphoanhydride bonds such as in bactoprenol glycosyl donors do exist, these are generally fleeting intermediates whose high energy character is used to drive the polymerization of peptidoglycan, O-antigen, and other cell surface polymer pathways as intracellular ATP is not available. Considering the inherent reactivity of phosphoanhydride bonds, we suspected spontaneous hydrolysis might play a role because phosphate was only released from LPS substrate containing phosphoanhydride-linked PEtN groups and not from phosphodiester-linked PEtN groups appended to Kdo by EptB (Figs. 1, 2, and 6B). Indeed, phosphodiester-linked PEtN remained stable across a wide pH range, whereas PEtN connected by phosphoanhydride bonds became labile in neutral to basic pH and was only stable under mildly acidic conditions (Fig. 4, C and D). Intriguingly, acidic environmental conditions induce EptA (72), suggesting that phosphoanhydride-linked PEtN may be hydrolytically susceptible by design so as to transiently tailor the LPS layer provided acidic conditions persist. Certain bacteria do not add phosphoanhydride-linked PEtN to lipid A, but rather hydrolyze lipid A phosphate first before transferring PEtN to form phosphodiester bound PEtN (73, 74). Presumably such PEtN phosphodiesters are more stable, as observed here for phosphodiester-bound Ara4N-modified lipid A and PEtN attached by EptB (Fig. 2). Apparently, cIAP is unable to directly release PEtN or phosphate from LPS, but does dephosphorylate spontaneously hydrolyzed PEtN phosphomonoesters (Figs. 3 and 4). Free O-PEtN is a characterized substrate for PLAP (75), and experiments here confirm it is a good substrate for the cIAP isozyme as well (Fig. S3).
In this study, we used commercially purified cIAP as AP enzyme source. In the intestinal lumen, IAP is enriched within lumenal vesicles secreted by enterocytes (76), a native environment that could enhance IAP catalytic activity toward LPS. When the apparent phosphate released from LPS by lumenal vesicles and purified IAP was directly compared, however, activities were comparable (77). A second consideration for any in vitro based LPS assay is substrate presentation. Highly aggregated LPS may not be bioavailable to IAP without accessory proteins to expose the lipid A phosphate groups. In serum, lipopolysaccharide-binding protein (LBP), soluble CD14 (sCD14), and albumin all have integral roles in disaggregating LPS and enhancing presentation to the TLR4/MD2 receptor complex (78, 79). Likewise, the activity of the LPS acyloxyacyl hydrolase, which cleaves secondary acyl chains on lipid A (80, 81), is stimulated up to 100-fold when substrate is presented by either sCD14 or LBP (82). Although we tested native preparations of AP enzymes extracted from tissue to capture relevant post-translational modifications (Fig. 1), glycosylation can influence activity in certain AP isozymes (83). It should be noted that even with tetra-acylated lipid IVA, which lacks secondary acyl chains and is more polar than hexa-acylated lipid A, there was no appreciable phosphate released by cIAP (Fig. 6). If LPS is not a relevant IAP substrate, then the bulk of the anti-inflammatory effects observed with AP would seem more likely to arise from dephosphorylation of ATP and UDP rather than through detoxification of LPS (18, 19, 84). IAP has been reported to shape the microbiota composition in a mouse IAP Akp-3 knockout model (85) and to directly inhibit E. coli growth in culture (86), so that an indirect role of IAP in modulating the composition of the endotoxin load present in the intestinal lumen need also be considered.
Although the exact structure of LPS and lipid IVA was unknown at the time, initial studies investigating AP activity also concluded that LPS was completely resistant to bacterial AP (87–89). However, ∼50% of the total phosphate could be released from “lipid A precursor” (i.e. later named lipid IVA) if first de-O-acylated by treatment under mild alkaline conditions (88, 89). The authors concluded that the C4′-GlcNII phosphate is recognized by AP, and that the C1-GlcNI anomeric phosphate remains intact. The resistance of C1 to hydrolysis was attributed to steric hindrance by the C2 amide-linked 3-OH-C14:0 acyl chain, which unlike the 3-OH-C14:0 ester acyl chain neighboring the C4′ phosphate of GlcNII, remains intact after base treatment. It was also suggested that de-O-acylation may decrease the aggregation state, facilitating enzyme access to substrate. Because these studies focused on an AP enzyme of bacterial origin with low sequence similarity, we replicated their study using cIAP with highly purified lipid IVA preparations (Fig. 5). As reported for bacterial AP, cIAP rapidly dephosphorylates de-O-acylated lipid A compared with tetra-acylated lipid IVA. Bacterial phosphate monoesterases (EC 3.1.3.1) such as E. coli alkaline phosphatase (EcAP) resemble mammalian IAP in catalytic site topology (catalytic residues, trimetallo core, zinc-binding and crown-like flap) (90, 91). Bacterial and mammalian mono-phosphoesterases with a trimetallo core (here called EcAP and rat IAP) share highly conserved amino acids for Zn2+ and Mg2+ complexation (49, 90). Computational docking of lipid IVA ligand into the active site of mammalian IAP (Fig. 5C) as well as bacterial AP (Fig. S9A) revealed steric hindrance by neighboring 3′-OH C14:0 acyl chains that prevent positioning of the C4′-phosphate of lipid A at the active site. Likewise, the approach of the anomeric phosphate appears hindered by the amide acyl chain at C2 of GlcNI (Figs. S5 and S9B, for mammalian and bacterial AP, respectively), in keeping with our experimental data for cIAP (Fig. 5) and data using bacterial AP (88, 89). The anomeric phosphate can be removed after de-O-acylation, as a small but completely dephosphorylated population is clearly observed by MS and NMR (Fig. 5 and Fig. S4). The increased conformational flexibility of the remaining N-acyl chains after de-O-acylation likely accounts for the appearance of weak phosphatase activity at C1-GlcNI. The presence of Kdo residues in de-O-acyl Re LPS substrate slowed but did not prevent dephosphorylation (Fig. 5A), indicating prior hydrolysis of the saccharide core is not necessarily required for cIAP activity.
Although de-O-acylated lipid A is rapidly dephosphorylated, the direct relevance in endotoxin detoxification is questionable given de-O-acylated lipid A is already a weak TLR4/MD2 agonist. However, many of the more populous gut-associated bacteria (as from bacteria in the order Bacteroidales) remodel their lipid A to underacylated chemotypes (92, 93). Underacylated chemotypes not only have lower intrinsic endotoxin activity, but also dampen TLR4/MD2 signaling and inflammation from more potent lipid A congeners with higher acylation states as found in Enterobacteriaceae through competition for TLR4/MD2 receptor. It is conceivable that endogenous underacylated chemotypes from gut-associated bacteria may be selectively subject to further detoxification by IAP, or that IAP may play an indirect role in lowering the endotoxin burden by processing the underacylated antagonistic lipid A population pool.
The influence of lipid A acylation state on attenuating hTLR4/MD2 activity has long been recognized. However, the importance of PEtN modification on lipid A is just beginning to be appreciated. Indeed, the structure-hTLR4/MD2 activity relationship of EptA-modified lipid A is nearly as equipotent as the removal/addition of secondary acyl chains (Fig. 6C). The PEtN effect is not unique to E. coli, as the potency of other lipid A scaffolds in N. meningitidis (54–57) and C. jejuni (58) is likewise enhanced by PEtN. The influence of PEtN-modified lipid A on TLR4/MD2 engagement demonstrates marked species-dependent effects, as the degree of mTLR4/MD2 stimulation was agnostic to the presence of EptA in the producing strain across all the tested lipid A acylation states (Fig. 6D). Of note, EptA expression in a lipid IVA producing strain restored some hTLR4/MD2 agonist character (Fig. 7). This raises the possibility that detoxification by acyloxyacyl hydrolase through removal of secondary acyl chains may be less effective when PEtN modified LPS is abundant. Our computational binding model suggests this is due to key residues specific to hTLR4/MD2 (Fig. 8). The weak activity of 2-PEtN lipid IVA is particularly interesting, considering this is a lipid A-based hTLR4/MD2 agonist with inherently self-limiting biological activity as the phosphoanhydride-linked PEtN groups are unstable at physiological pH (Fig. 4, C and D). These properties suggest a potential design for engineering safe next generation adjuvants. Utilizing lipid A analogs agonists to bolster immune responses to synthetic vaccines antigens without inducing toxicity is a major challenge (55, 94–96). Chemically unstable 2-PEtN lipid IVA type analogs will spontaneously convert over time from an agonist to antagonist under physiological conditions, offering an intriguing adjuvant strategy that warrants further investigation.
We cannot definitively rule out a direct LPS dephosphorylating activity for select AP-LPS chemotype pairs, as the E. coli LPS chemotypes-AP pairs studied here is a far from exhaustive list. The complement of respective intestinal AP isozymes varies widely among vertebrates, so that biological roles may be species specific. It has been proposed that the intestinal AP dephosphorylation activities have co-evolved to complement the substrates being produced by the particular resident microbiota (97). If hexa-acylated LPS is a bona fide substrate, however, there is clearly an integral missing component from the reconstituted in vitro system. Regardless, the labile nature of phosphoanhydride-linked PEtN modifications on LPS and the ensuing diminished TLR4/MD2 signaling precludes using phosphate release and decreased endotoxin activity as sole determinants of AP-mediated LPS detoxification. It also emphasizes the advantages of using chemically defined LPS chemotypes, given that the PEtN content will not only depend on the particular source strain, but will also vary according to both the growth media and the extraction and purification methods. Defined chemotypes will allow more meaningful comparisons between studies and ultimately a better understanding of the role of AP in ameliorating LPS-induced inflammation.
Experimental procedures
Reagents
cIAP was purchased from New England BioLabs. Human placenta (PLAP) and human liver alkaline phosphatases (TNAP) were ordered from Lee Biosolutions Inc., whereas porcine kidney alkaline phosphatase was from Sigma. Human ALPI was obtained from Sino Biological. All chemicals were purchased from Sigma Millipore unless noted otherwise.
Bacterial strain construction
Gene deletions were introduced into E. coli strains using the λ-Red recombinase system as described (98). Targeting cassettes were obtained by PCR amplification using P1–P2 primer pairs (Table S1). Each primer contained a 5′ 42-bp homology extension arm and a 3′ 18-bp sequence specific for the indicated antibiotic selection marker of the plasmid template. For the arnA::kanR and lpxM::kanR cassettes, genomic DNA was purified from the Coli Genetic Stock Center strain CGSC numbers 9813 and 9540 and used as a template to flank the antibiotic cassette with FRT sites for subsequent marker excision using the FLP recombinase plasmid pCP20 (99). Integration cassettes were purified and electroporated into recipient strains harboring the arabinose-inducible λ-Red recombinase plasmid pKD46 (98). Plasmids were cured by passaging at 37 °C, colonies were checked for loss of plasmid, and cassette insertion was confirmed using check primers (Table S1). For construction of tetra-acylated LPS strains containing a complete saccharide core (GKM499 and GKM502), plasmid pMMW52-msbA was first introduced to enhance LPS transport and improve fitness. In this background, deletion cassettes were then introduced by generalized transduction using P1vir. Strain genotypes are listed in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant genotype or phenotypea | Source or reference |
|---|---|---|
| E. coli strains | ||
| TXM319 | Wildtype BL21 (DE3); E. coli B F− ompT hsdSB (rB− mB−) gal dcm lon λ (DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Lab stock |
| TXM322 | BL21 (DE3) arnA::kanR; Kanr | This study |
| GKM329 | BL21 (DE3) eptA::catR; Catr | This study |
| TXM331 | BL21 (DE3) eptA::catR arnA::kanR; Catr Kanr | This study |
| TXM333 | BL21 (DE3) lpcA::gentR eptA::catR arnA::kanR; Gentr Catr Kanr | This study |
| TXM343 | BL21 (DE3) lpcA::gentR eptA::catR arnA::kanR [pSEVA434-eptA]; Gentr Catr Kanr Specr | This study |
| GKM357 | BL21 (DE3) eptA::catR arnA::kanR waaP::gentR; Catr Kanr Gentr | This study |
| GKM358 | BL21 (DE3) eptA::catR arnA::kanR waaP::gentR [pSEVA434-eptA]; Catr Kanr Gentr | This study |
| GKM373 | BL21 (DE3) eptA::catR arnA::kanR eptB::gentR; Catr Kanr Gentr | This study |
| GKM374 | BL21 (DE3) eptA::catR arnA::kanR eptC::gentR; Catr Kanr Gentr | This study |
| GKM380 | BL21 (DE3) eptA::catR arnA::kanR eptB::gentR [pSEVA434-eptC]; Catr Kanr Gentr Specr | This study |
| GKM381 | BL21 (DE3) eptA::catR arnA::kanR eptC::gentR [pSEVA434-eptB]; Catr Kanr Gentr Specr | This study |
| TXM402 | BL21 (DE3) eptA::catR arnA::kanR eptC::gentR [pSEVA434-eptA]; Catr Kanr Gentr Specr | This study |
| TXM418 | BL21 (DE3) eptA::catR arnA::FRT eptC::gentR lpxM::kanR; Catr Gentr Kanr | This study |
| TXM419 | BL21 (DE3) eptA::catR arnA::FRT eptC::gentR lpxM::kanR [pSEVA434-eptA]; Catr Gentr Kanr Specr | This study |
| GKM445 | ClearColi® K-12 F−, λ- ΔendA ΔrecA msbA52 frr181 ΔgutQΔkdsDΔlpxLΔlpxMΔlpxPΔeptA | Lucigen |
| GKM446 | GKM445 (pSEVA434-eptA); Specr | This study |
| GKM499 | BL21 (DE3) eptA::catR arnA::FRT eptC::gentR lpxM::kanR lpxL::aprR pagP::hygR [pMMW52-msbA]; Catr Gentr Kanr Aprr Hygr Carbr | This study |
| GKM502 | BL21 (DE3) eptA::catR arnA::FRT eptC::gentR lpxM::kanR lpxL::aprR pagP::hygR [pMMW52-msbA, pSEVA434-eptA]; Catr Gentr Kanr Aprr Hygr Carbr Specr | This study |
| TXM843 | ClearColi® BL21 (DE3) F− ompT hsdSB (rB− mB−) gal dcm lonλ (DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) msbA148 ΔgutQ ΔkdsD ΔlpxLΔlpxMΔpagPΔlpxPΔeptA | Lucigen |
| TXM844 | TXM843 [pSEVA434-eptA]; Specr | This study |
| Plasmids | ||
| pSEVA434 | pBBR1 ori lacIq-Ptrc Specr | (100) |
| pSEVA434-eptA | Ptrc eptA from E. coli BL21 (DE3) | This study |
| pSEVA434-eptB | Ptrc eptB from E. coli BL21 (DE3) | This study |
| pSEVA434-eptC | Ptrc eptC from E. coli BL21 (DE3) | This study |
| pMMW52-msbA | pMBL19 carrying a subcloned 3.5-kb insert with ycaÍ, msbA, and lpxK; Carbr | (48) |
| pKD3 | Catr template | (98) |
| pCP20 | FLP recombinase expression plasmid; Carbr Catr | (99) |
| pKD46 | λ-Red recombinase expression plasmid; Carbr | (98) |
| pEXG2 | Gentr template | (119) |
| pSET152 | Aprr template | (120) |
| pUC19-oriT-hyg | Hygr template | Lab stock |
a Kanr, kanamycin; Catr, chloramphenicol; Gentr, gentamycin; Specr, spectinomycin; Carbr, carbenicillin; Aprr, apramycin; Hygr, hygromycin.
Plasmids expressing EptA, EptB, or EptC were constructed using the InFusion Cloning kit (Clontech). PCR primer pairs (Table S1) were used to amplify inserts from E. coli BL21 (DE3) genomic DNA and then cloned into the vector pSEVA434 (100) that had been digested with EcoRI/BamHI. Plasmids were maintained with spectinomycin (50 μg/ml) and used without induction as basal expression was sufficient for phenotypic conversion for all three constructs.
LPS purification
Bacteria were harvested from stationary phase cultures grown at 37 °C in either Lysogeny Broth (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl per liter) or TB media (39) (10 g of tryptone, 5 g of yeast extract, 3 g of NH4Cl, 12 g of Na2HPO4, 6 g of KH2PO4, 0.5 g of Na2SO4, 7.5 g of glucose/liter) supplemented with the necessary antibiotics for plasmid selection. Dried bacterial cell biomass was obtained by sequentially stirring in ethanol overnight and then two rounds of acetone (12 h each) at 4 °C, collecting biomass between incubations by centrifugation (6,000 × g, 10 min, 4 °C). All LPS and lipid IVA chemotypes were initially extracted from cell powders via the phenol/chloroform/petroleum ether (PCP) method (101). Briefly, dried biomass was resuspended in PCP solution (90% phenol/chloroform/petroleum ether in 2:5:8 (v/v/v) ratio) and incubated for 1 h on a tube rotator. Biomass was pelleted and the supernatant collected, with the extraction being repeated twice. Pooled supernatant extract was rotavaped to remove chloroform and petroleum ether, and 100 μl of 3 m sodium acetate (pH 7.0) was added to the phenol phase. Here treatment of the samples varied depending on the chemotype. For LPS samples, precipitation was carried out via dropwise addition of water. Pelleted LPS was washed once with 80% phenol and then a second time with acetone. For lipid IVA samples, 5 volumes of acetone were added to precipitate lipid IVA from the phenol phase and then the pellet was washed once with acetone.
To remove co-extracting phospholipids from LPS, samples underwent a modified chloroform/methanol wash (102). Briefly, pellets were resuspended in chloroform, methanol, 3 m sodium acetate (pH 7.0) (85:15:1 (v/v/v)) and 2 to 3 volumes of methanol were added to precipitate LPS. Phospholipid containing supernatant was decanted and this process was repeated twice. Removal of contaminating lipoprotein was achieved via phenol/sodium deoxycholate extraction as described by Hirschfeld et al. (103). Lipid IVA samples could not be efficiently recovered using either the chloroform/methanol wash or the phenol/deoxycholate extraction, hence these steps were omitted.
All chemotypes underwent ultracentrifugation to remove any malachite green reactive nucleic acid material. LPS or lipid IVA pellets were resuspended in 3 ml of water and added to 30 ml of a Tris saline buffer (50 mm Tris-HCl, 100 mm NaCl, 1 mm MgCl2, pH 7.0). Samples were centrifuged at 100,000 × g for 4–6 h, resulting in a translucent pellet. The resultant pellet was quickly rinsed with buffer, resuspended in water, and dialyzed (0.5–1 kDa MWCO) against three 5-liter portions of water for 48 h (24 h for PEtN-modified chemotypes) at 4 °C. Desalted LPS chemotypes were lyophilized to a white powder, whereas lipid IVA chemotypes were further purified as described below.
Pi release and dephosphorylated LPS product assay
LPS samples (100 μg/ml) were incubated in alkaline phosphatase buffer (50 mm Tris-HCl at the indicated pH (pH 8.25, 7.4, or 7.1), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2) with cIAP (4 units/ml), human liver phosphatase (0.1 units/ml), human placenta phosphatase (1 unit/ml), human intestinal phosphatase (0.17 μg/ml), or porcine kidney phosphatase (1 unit/ml) at 37 °C. Aliquots were taken at different time points and phosphate release was measured with the malachite green assay as described previously (33). Briefly, 1 volume of malachite green solution (0.1% malachite green, 14% sulfuric acid, 1.5% ammonium molybdate, and 0.18% Tween 20) was mixed with 4 volumes of LPS solution and the mixture was incubated at room temperature for 10 min. Absorbance was read at 630 nm on a SpectraMax Plus 384 plate reader (Molecular Devices). Sodium phosphate was used for standard curve determination.
For direct assay of dephosphorylated LPS, 10-ml reactions were assembled as described above in dialysis tubing (0.5–1 kDa MWCO). Reactions were continuously dialyzed against 500 ml of buffer to remove any released Pi. LPS products were isolated by extensive dialysis against water and lyophilized before analysis by MS as detailed below.
Buffer composition for assays conducted at varying pH values was 50 mm MOPS, 50 mm Tris adjusted to pH 6.5, 7.4, or 8.5 along with 100 mm NaCl, 1 mm MgCl2, and 20 μm ZnCl2. In the experiments where 10 units of cIAP was added after preincubation in the buffer alone, samples were incubated for 30 min at 37 °C to release Pi from spontaneously hydrolyzed PEtN.
TLR4 stimulation assay
HEK293/hTLR4-MD2-CD14, HEK293/mTLR4-MD2-CD14, and parental HEK293/Null2 control cells were grown as specified by the supplier (InvivoGen). For the stimulation assay, the cells were plated at 50,000 cells per well in a white 96-well plate with clear bottom (CostarTM 3610, Corning Incorporated) in 200 μl of growth medium (Dulbecco's modified Eagle's medium, 2 mm l-glutamine, 10% heat-inactivated fetal bovine serum, 50 units/ml of penicillin, 50 μg/ml of streptomycin, 100 μg/ml of NormocinTM). The pNiFty-Luc plasmid (InvivoGen) encoding five NF-κB repeated transcription factor-binding sites in front of the luciferase reporter gene was mixed with transfection reagent LyoVecTM (InvivoGen) at a concentration of 1 μg of plasmid, 100 μl of LyoVecTM, and after incubation at room temperature for 20 min, the mixture (10 μl per well) was added to cells in a 96-well microplate. The next day the medium was removed and replaced with 180 μl of fresh growth medium. Various LPS chemotypes were added at different concentrations in a 20-μl volume per well. Endotoxin-free water was used as a negative control and tumor necrosis factor α (200 ng/well) was used as a positive control. PierceTM Firefly Luciferase One-Step Glow Assay Kit was used according to the manufacturer's instructions with luminescence being measured after 20 h of stimulation. All LPS and lipid IVA preparations were confirmed to be negative for NF-κB induction when challenging HEK293/Null2 control cells (InvivoGen) up to the highest tested LPS concentration (100 ng/ml, data not shown).
TLR2 stimulation assay
HEK-BlueTM hTLR2 cells (InvivoGen) were propagated as specified by the supplier. For the stimulation assay, the cells were plated at 25,000 cells/well in a 96-well plate in 180 μl of growth medium (Dulbecco's modified Eagle's medium, 2 mm l-glutamine, 10% heat-inactivated fetal bovine serum, 50 units/ml of penicillin, 50 μg/ml of streptomycin, 100 μg/ml of NormocinTM). LPS was added at different concentrations in a 20 μl/well volume. Endotoxin-free water was used as a negative control. QUANTI BlueTM (InvivoGen) reagent was used, according to manufacturer's instructions, 20 h later to detect NF-κB-dependent SEAP activity. Absorbance at 620 nm was read following incubation of the samples with QUANTI BlueTM substrate for 3 h at 37 °C.
Lipase treatment of lipid IVA extracts
Crude lipid IVA (with or without PEtN) was treated with lipase via two sequential incubations. Both 12-h reactions were conducted at 45 °C in a 20 mm phosphate buffer (pH 7.0). Each reaction contained 40 mg of crude PCP-extracted lipid IVA, Thermomycyes lipase (TL, Sigma), and Novozyme® 51032 (Strem Chemicals, Newburyport, MA) at respective final concentrations of 0.1 mg/ml, 90 μg/ml, and 25 μg/ml. The initial reaction included 3.4 mm BIG CHAP (SolTec Bio Science, Beverly, MA) as a nonionic detergent additive. Immediately following this reaction, lipid IVA was recovered by conversion to a 2:2:1.8 (v/v/v) chloroform/methanol/water Bligh-Dyer biphasic mixture. Lipid IVA was isolated from the lower organic phase by rotary evaporation, resuspended in endotoxin-free water, and then lyophilized. Recovered lipid IVA was treated again in a second lipase reaction with 20 mm octyl β-d-glucopyranoside as the detergent additive, and re-isolated as described above. PEtN-modified lipid IVA was stored at −20 °C until further use.
Chromatographic purification of lipid IVA species
Ion exchange chromatography was performed using a 5-ml HiTrapTM SP HP cation exchange column connected in tandem to a 20-ml HiPrepTM DEAE FF 16/10 anion exchange column with the ÄKTATM Pure FPLC system. Lipase-treated PEtN lipid IVA prepared as described above was loaded in 15 ml of a 60% 1-propanol solution adjusted to pH 5 with acetic acid. The system was then washed with 10 ml of the same buffer, after which the cation exchange column was removed. The anion exchange column was subsequently washed with 4.5 column volumes of 60% 1-propanol (pH 5), before elution using a linear gradient of non-pH adjusted 0 to 80 mm ammonium acetate in 60% 1-propanol over 20 column volumes. To identify fractions containing nonvolatile organic compounds, 20 μl of each fraction was spotted onto an Analtech Silica Gel G TLC plate and visualized by charring using a 10% sulfuric acid/ethanol solution with heating at 160 °C for 10 min. Fractions containing organic material were further analyzed by spotting 20 μl on an Analtec Silica Gel H TLC plate, and developed using a pyridine/chloroform/formic acid/water (50:50:16:5, v/v/v/v) mobile phase before sulfuric acid charring (104). Fractions containing lipid IVA species were pooled, rotovaped to dryness, and subjected to two rounds of lyophilization to remove residual traces of ammonium acetate. The resulting powder was kept at −20 °C until further purification by reversed-phase HPLC (RP-HPLC). For this, lipid IVA samples were subjected to RP-HPLC essentially as described (105, 106), but with some modifications. A semi-preparative Kromasil C18 column (5 μm, 100 Å, 10 × 250 mm, MZ Analysentechnik, GmbH, Mainz, Germany) was used and samples (resuspended at 5 mg/ml in chloroform, methanol, 0.1 m acetic acid (8:2:1, v/v/v)) were eluted using a gradient consisting of methanol/chloroform/water (57:12:31, v/v/v) containing 10 mm ammonium acetate as mobile phase A and chloroform/methanol (70.2:29.8, v/v) with 50 mm ammonium acetate as mobile phase B. The initial solvent system consisted of 2% B and was maintained for 10 min, raised from 2 to 15% B (10–20 min), kept at 15% B for 20 min, raised from 15 to 25% B (40–50 min), kept at 25% B for 20 min, and raised from 25 to 100% B (70–100 min). The solvent was held at 100% B for 20 min, followed by re-equilibration of the column to 2% B for 10 min and held there for an additional 10 min prior to the next injection. The flow rate was 2 ml/min using a splitter between the evaporative light-scattering detector equipped with a low-flow nebulizer (Sedex model 75C ELSD, S.E.D.E.R.E., France). Nitrogen (purity 99.996%) was used as gas to nebulize the post-column flow stream at 3.5 bar into the detector at 50 °C setting the photomultiplier gain to 11. The detector signal was transferred to the Gilson HPLC Chemstation (Trilution LC, version 2.1, Gilson) for detection and integration of the ELSD signal.
De-O-acylation and dephosphorylation assays of lipid IVA and Re LPS with cIAP
Lipid IVA was dissolved (1 to 4 mg/ml) in a 1 m NaOH aqueous solution and incubated for 20 h at room temperature. Reactions were neutralized via addition of glacial acetic acid while stirring until pH 7.0. Neutralized reactions were extensively dialyzed against water (MWCO: 500–1000 Da) and lyophilized. Re LPS was likewise de-O-acylated, but was recovered by precipitation from neutralized solution using 5 volumes of ethanol. De-O-acylated products were further purified via anion exchange chromatography as described above. Fractions were pooled, concentrated by rotary evaporation, and dialyzed against water (MWCO: 500–1000 Da). Samples were lyophilized and stored at −20 °C.
Large scale (30 ml) cIAP reactions (100 μg/ml of de-O-acyl substrate, 4 units/ml of cIAP, 50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 1 mm MgCl2, 20 μm ZnCl2) were incubated for 24 to 48 h at 37 °C. Samples were recovered by dialysis against water (MWCO: 100–500 Da) followed by lyophilization.
Mass spectrometry
LPS samples were measured on a 7-tesla APEX Qe Electrospray Ionization Fourier Transform Ion Cyclotron Resonance (ESI-FT-ICR) mass spectrometer (Bruker Daltonics). Measurements were performed in negative ion mode. Samples (∼0.03 mg/ml) were dissolved in a water, 2-propanol/trimethylamine/acetic acid mixture (50:50:0.06:0.02, v/v/v/v). Spectra were acquired in broadband acquisition mode with nano-ESI using the Triversa Nanomate (Advion, Ithaca, NY) as ion source with a spray voltage set to −1.1 kV. Collision voltage was set to 5 V. Lipid IVA and de-O-acylated samples were measured on a Q Exactive Plus mass spectrometer (Thermo Scientific, Bremen, Germany) using a Triversa Nanomate (Advion, Ithaca, NY) as ion source. For negative ion mode, samples (∼0.05 mg/ml) were dissolved in either chloroform/methanol/water (60:35:4.5, v/v/v) or water, 2-propanol, 7 m trimethylamine, acetic acid mixture (50:50:0.06:0.02, v/v/v/v) and performed with a spray voltage set to −1.1 kV. For positive ion mode, samples were dissolved in water, 2-propanol, 30 mm ammonium acetate, acetic acid mixture (15:15:1:0.04, v/v/v/v) with a spray voltage set to +1.1 kV. Both mass spectrometers were calibrated externally with glycolipids of known structure. All mass spectra were charge deconvoluted and given mass values refer to the monoisotopic masses of the neutral molecules, if not indicated otherwise.
NMR spectroscopy
NMR spectroscopic measurement of PEtN-lipid IVA was performed in CDCl3/MeOH-d4/D2O (60:35:8, v/v/v) and de-O-acyl Re LPS after treatment with cIAP in CDCl3/MeOH-d4/D2O (2:3:1, v/v/v) (107), respectively, at 300 K on a Bruker AvanceIII 700 MHz (equipped with an inverse 5-mm quadruple-resonance Z-grad cryoprobe). Deuterated solvents were purchased from Deutero GmbH (Kastellaun, Germany). TMS was used as an external standard for calibration of 1H (δH 0.0) and 13C (δC 0.0) NMR spectra, and 85% of phosphoric acid was used as an external standard for calibration of 31P NMR spectra (δP 0.0). All data were acquired and processed using Bruker's TOPSPIN version 3.0 software. 1H NMR assignments were confirmed by 2D 1H,1H COSY and total correlation spectroscopy (TOCSY) experiments. 13C NMR assignments were indicated by 2D 1H,13C HSQC, based on the 1H NMR assignments. Inter-residue connectivity and further evidence for 13C assignment were obtained from 2D 1H,13C heteronuclear multiple bond correlation and 1H,13C HSQC-TOCSY. Connectivity of phosphate groups were assigned by 2D 1H,31P HMQC and 1H,31P HMQC-TOCSY.
Molecular modeling
Standard modeling tools and protocols were conducted according to published protocols (53, 63). Modeling software (Autodock 4.2 (108), Chimera 1.13.1 (109), SPDBV 4.10 (110), and VEGA ZZ 3.1.2 (111)) were licensed for academic use to generate and visualize three-dimensional model structures of lipid IVA, TLR4/MD2, and phosphatase enzymes, in addition to partial charges, electrostatic molecular surfaces, or fitted active site conformers (108, 109, 112, 113). The TLR4/MD2 docking protocol first presented by Meng et al. (53) and later adapted (63) using the hTLR4/MD2-E. coli LPS crystal structure (PDB 3FXI (114)) was utilized to model LPS-like congener binding. The large number (52) of freely rotatable bonds at the 4 side chains of lipid IVA had to be reduced and were limited to the first three chain members including the four H-O bonds as well as the two ester bonds, whereas excluding the two more rigid trans-amide bonds. To visualize the steric hindrance associated to nonbinders (no valid docking/scoring solutions found) we superimposed them onto the final docked poses of those ligand types with favorable side chain patterns for successful cavity binding, here called the binders (Fig. 5C). Whereas for nonbinders any of the calculated conformations under docking conditions would show detrimental van der Waals contacts, binders provide (some not all) conformational solutions for successful binding in the micromolar range. Lipid IVA (115) and all enzymes and receptors were retrieved from the PDB repository server (116) with the exception of hitherto structurally unknown cIAP that was generated by homology modeling using described methodology (117). The cIAP target (GenBank entry code AAA30571.1) shares more than 70% identity with rat IAP across 486 nongapped residues, including all active sites residues. Using the experimentally determined rat IAP crystal structures (PDB codes 4KJD and 4KJG) (49), a cIAP homodimer structure was generated under Swiss PDB Viewer (110). Multiple sequence alignments were carried out with built-in Clustal X under Vega ZZ (111). Of note, all phosphate groups were charged and modeled as monoanionic, e.g. bearing one -OH group. In some figures hydrogen atoms were not displayed for visual simplicity (118). All model figures were generated with Chimera (109).
Author contributions
G. K., M. M., R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. conceptualization; G. K., R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. resources; G. K., M. M., R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. data curation; G. K., M. M., R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. formal analysis; G. K., R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. supervision; G. K., M. M., R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. investigation; G. K., M. M., T. S., D. S., U. S., N. G., U. M., and T. C. M. methodology; G. K., M. M., R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. writing-original draft; G. K., D. S., U. S., N. G., U. M., and T. C. M. project administration; G. K., M. M., R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. writing-review and editing; R. W. W., T. S., D. S., U. S., N. G., U. M., and T. C. M. funding acquisition; T. S. software; T. S. visualization.
Supplementary Material
Acknowledgments
We thank Tania Laremore (Proteomics and Mass Spectrometry Core Facility, Penn State University) for help with experiments. We gratefully acknowledge Birte Buske, Dörte Grella, Manuel Hein, Heiko Käßner, and Brigitte Kunz (all Research Center Borstel) for excellent technical assistance.
This work was supported by National Institutes of Health NIAID Grant R21 AI138152 (to T. C. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S9 and Tables S1 and S2.
- MAMP
- microbe-associated molecular pattern
- hALPI
- human intestinal alkaline phosphatase
- AP
- alkaline phosphatase
- Ara4N
- 4-amino-4-deoxy-l-arabinose
- cIAP
- calf intestinal alkaline phosphatase
- DAMP
- damage-associated molecular pattern
- EcAP
- E. coli alkaline phosphatase
- Hep
- l-glycero-d-manno-heptose
- IAP
- intestinal alkaline phosphatase
- Kdo
- 3-deoxy-α-d-manno-oct-2-ulosonic acid
- LA
- lipid A
- LBP
- lipopolysaccharide-binding protein
- LPS
- lipopolysaccharide
- MD2
- myeloid differentiation factor-2
- ME
- metabolic endotoxemia
- PAMP
- pathogen-associated molecular pattern
- PCP
- phenol/chloroform/petroleum ether
- PEtN
- phosphoethanolamine
- PLAP
- placental/embryonic alkaline phosphatase
- PRR
- pattern recognition receptors
- sCD14
- soluble CD14
- SEAP
- secreted embryonic alkaline phosphatase
- TLR4
- Toll-like receptor 4
- TNAP
- tissue nonspecific alkaline phosphatase
- TOCSY
- total correlation spectroscopy
- HSQC
- heteronuclear single quantum coherence.
References
- 1. Boroni Moreira A. P., and de Cassia Goncalves Alfenas R. (2012) The influence of endotoxemia on the molecular mechanisms of insulin resistance. Nutr. Hosp. 27, 382–390 [DOI] [PubMed] [Google Scholar]
- 2. Cani P. D., Amar J., Iglesias M. A., Poggi M., Knauf C., Bastelica D., Neyrinck A. M., Fava F., Tuohy K. M., Chabo C., Waget A., Delmée E., Cousin B., Sulpice T., Chamontin B., et al. (2007) Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- 3. Duseja A., and Chawla Y. K. (2014) Obesity and NAFLD: the role of bacteria and microbiota. Clin. Liver Dis. 18, 59–71 10.1016/j.cld.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 4. Miura K., and Ohnishi H. (2014) Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World J. Gastroenterol. 20, 7381–7391 10.3748/wjg.v20.i23.7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moreno-Indias I., Cardona F., Tinahones F. J., and Queipo-Ortuño M. I. (2014) Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front. Microbiol. 5, 190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neves A. L., Coelho J., Couto L., Leite-Moreira A., and Roncon-Albuquerque R. Jr. (2013) Metabolic endotoxemia: a molecular link between obesity and cardiovascular risk. J. Mol. Endocrinol. 51, R51–R64 10.1530/JME-13-0079 [DOI] [PubMed] [Google Scholar]
- 7. Tilg H., and Moschen A. R. (2014) Microbiota and diabetes: an evolving relationship. Gut 63, 1513–1521 10.1136/gutjnl-2014-306928 [DOI] [PubMed] [Google Scholar]
- 8. Moreira A. P., Texeira T. F., Ferreira A. B., Peluzio Mdo C., and Alfenas Rde C. (2012) Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br. J. Nutr. 108, 801–809 10.1017/S0007114512001213 [DOI] [PubMed] [Google Scholar]
- 9. Tlaskalová-Hogenová H., Stěpánková R., Kozáková H., Hudcovic T., Vannucci L., Tučková L., Rossmann P., Hrnčiř T., Kverka M., Zakostelská Z., Klimešová K., Přibylová J., Bártová J., Sanchez D., Fundová P., et al. (2011) The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol. Immunol. 8, 110–120 10.1038/cmi.2010.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bès-Houtmann S., Roche R., Hoareau L., Gonthier M. P., Festy F., Caillens H., Gasque P., Lefebvre d'Hellencourt C., and Cesari M. (2007) Presence of functional TLR2 and TLR4 on human adipocytes. Histochem. Cell Biol. 127, 131–137 10.1007/s00418-006-0230-1 [DOI] [PubMed] [Google Scholar]
- 11. Ouchi N., Ohashi K., Shibata R., and Murohara T. (2012) Adipocytokines and obesity-linked disorders. Nagoya J. Med. Sci. 74, 19–30 [PMC free article] [PubMed] [Google Scholar]
- 12. Piya M. K., Harte A. L., and McTernan P. G. (2013) Metabolic endotoxaemia: is it more than just a gut feeling? Curr. Opin. Lipidol. 24, 78–85 10.1097/MOL.0b013e32835b4431 [DOI] [PubMed] [Google Scholar]
- 13. Fei N., and Zhao L. (2013) An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 7, 880–884 10.1038/ismej.2012.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cani P. D., and Delzenne N. M. (2011) The gut microbiome as therapeutic target. Pharmacol. Ther. 130, 202–212 10.1016/j.pharmthera.2011.01.012 [DOI] [PubMed] [Google Scholar]
- 15. Schromm A. B., Brandenburg K., Loppnow H., Zähringer U., Rietschel E. T., Carroll S. F., Koch M. H., Kusumoto S., and Seydel U. (1998) The charge of endotoxin molecules influences their conformation and IL-6-inducing capacity. J. Immunol. 161, 5464–5471 [PubMed] [Google Scholar]
- 16. Kaliannan K., Hamarneh S. R., Economopoulos K. P., Nasrin Alam S., Moaven O., Patel P., Malo N. S., Ray M., Abtahi S. M., Muhammad N., Raychowdhury A., Teshager A., Mohamed M. M., Moss A. K., Ahmed R., et al. (2013) Intestinal alkaline phosphatase prevents metabolic syndrome in mice. Proc. Natl. Acad. Sci. U.S.A. 110, 7003–7008 10.1073/pnas.1220180110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lallès J. P. (2014) Intestinal alkaline phosphatase: novel functions and protective effects. Nutr. Rev. 72, 82–94 10.1111/nure.12082 [DOI] [PubMed] [Google Scholar]
- 18. Malo M. S., Moaven O., Muhammad N., Biswas B., Alam S. N., Economopoulos K. P., Gul S. S., Hamarneh S. R., Malo N. S., Teshager A., Mohamed M. M., Tao Q., Narisawa S., Millán J. L., Hohmann E. L., et al. (2014) Intestinal alkaline phosphatase promotes gut bacterial growth by reducing the concentration of luminal nucleotide triphosphates. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G826–G838 10.1152/ajpgi.00357.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peters E., Geraci S., Heemskerk S., Wilmer M. J., Bilos A., Kraenzlin B., Gretz N., Pickkers P., and Masereeuw R. (2015) Alkaline phosphatase protects against renal inflammation through dephosphorylation of lipopolysaccharide and adenosine triphosphate. Br. J. Pharmacol. 172, 4932–4945 10.1111/bph.13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bilski J., Mazur-Bialy A., Wojcik D., Zahradnik-Bilska J., Brzozowski B., Magierowski M., Mach T., Magierowska K., and Brzozowski T. (2017) The role of intestinal alkaline phosphatase in inflammatory disorders of gastrointestinal tract. Mediators Inflamm. 2017, 9074601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fawley J., and Gourlay D. M. (2016) Intestinal alkaline phosphatase: a summary of its role in clinical disease. J. Surg. Res. 202, 225–234 10.1016/j.jss.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kiffer-Moreira T., Sheen C. R., Gasque K. C., Bolean M., Ciancaglini P., van Elsas A., Hoylaerts M. F., and Millán J. L. (2014) Catalytic signature of a heat-stable, chimeric human alkaline phosphatase with therapeutic potential. PLoS ONE 9, e89374 10.1371/journal.pone.0089374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buchet R., Millán J. L., and Magne D. (2013) Multisystemic functions of alkaline phosphatases. Methods Mol. Biol. 1053, 27–51 10.1007/978-1-62703-562-0_3 [DOI] [PubMed] [Google Scholar]
- 24. Rader B. A. (2017) Alkaline phosphatase, an unconventional immune protein. Front. Immunol. 8, 897 10.3389/fimmu.2017.00897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koyama I., Matsunaga T., Harada T., Hokari S., and Komoda T. (2002) Alkaline phosphatases reduce toxicity of lipopolysaccharides in vivo and in vitro through dephosphorylation. Clin. Biochem. 35, 455–461 10.1016/S0009-9120(02)00330-2 [DOI] [PubMed] [Google Scholar]
- 26. Poelstra K., Bakker W. W., Klok P. A., Kamps J. A., Hardonk M. J., and Meijer D. K. (1997) Dephosphorylation of endotoxin by alkaline phosphatase in vivo. Am. J. Pathol. 151, 1163–1169 [PMC free article] [PubMed] [Google Scholar]
- 27. Bates J. M., Akerlund J., Mittge E., and Guillemin K. (2007) Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382 10.1016/j.chom.2007.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen K. T., Malo M. S., Moss A. K., Zeller S., Johnson P., Ebrahimi F., Mostafa G., Alam S. N., Ramasamy S., Warren H. S., Hohmann E. L., and Hodin R. A. (2010) Identification of specific targets for the gut mucosal defense factor intestinal alkaline phosphatase. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G467–G475 10.1152/ajpgi.00364.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bentala H., Verweij W. R., Huizinga-Van der Vlag A., van Loenen-Weemaes A. M., Meijer D. K., and Poelstra K. (2002) Removal of phosphate from lipid A as a strategy to detoxify lipopolysaccharide. Shock 18, 561–566 10.1097/00024382-200212000-00013 [DOI] [PubMed] [Google Scholar]
- 30. Goldberg R. F., Austen W. G. Jr., Zhang X., Munene G., Mostafa G., Biswas S., McCormack M., Eberlin K. R., Nguyen J. T., Tatlidede H. S., Warren H. S., Narisawa S., Millán J. L., and Hodin R. A. (2008) Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc. Natl. Acad. Sci. U.S.A. 105, 3551–3556 10.1073/pnas.0712140105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pettengill M., Matute J. D., Tresenriter M., Hibbert J., Burgner D., Richmond P., Millan J. L., Ozonoff A., Strunk T., Currie A., and Levy O. (2017) Human alkaline phosphatase dephosphorylates microbial products and is elevated in preterm neonates with a history of late-onset sepsis. PLoS ONE 12, e0175936 10.1371/journal.pone.0175936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang W. H., Heithoff D. M., Aziz P. V., Haslund-Gourley B., Westman J. S., Narisawa S., Pinkerton A. B., Millán J. L., Nizet V., Mahan M. J., and Marth J. D. (2018) Accelerated aging and clearance of host anti-inflammatory enzymes by discrete pathogens fuels sepsis. Cell Host Microbe 24, 500–513.e5 10.1016/j.chom.2018.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baykov A. A., Evtushenko O. A., and Avaeva S. M. (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 171, 266–270 10.1016/0003-2697(88)90484-8 [DOI] [PubMed] [Google Scholar]
- 34. Maldonado R. F., Sá-Correia I., and Valvano M. A. (2016) Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 40, 480–493 10.1093/femsre/fuw007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raetz C. R., Reynolds C. M., Trent M. S., and Bishop R. E. (2007) Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 10.1146/annurev.biochem.76.010307.145803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klein G., and Raina S. (2019) Regulated assembly of LPS, its structural alterations and cellular response to LPS defects. Int. J. Mol. Sci. 20, e356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Merighi M., Ellermeier C. D., Slauch J. M., and Gunn J. S. (2005) Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J. Bacteriol. 187, 7407–7416 10.1128/JB.187.21.7407-7416.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klein G., Lindner B., Brade H., and Raina S. (2011) Molecular basis of lipopolysaccharide heterogeneity in Escherichia coli: envelope stress-responsive regulators control the incorporation of glycoforms with a third 3-deoxy-α-d-manno-oct-2-ulosonic acid and rhamnose. J. Biol. Chem. 286, 42787–42807 10.1074/jbc.M111.291799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jansson P. E., Lindberg A. A., Lindberg B., and Wollin R. (1981) Structural studies on the hexose region of the core in lipopolysaccharides from Enterobacteriaceae. Eur. J. Biochem. 115, 571–577 [DOI] [PubMed] [Google Scholar]
- 40. Mamat U., Wilke K., Bramhill D., Schromm A. B., Lindner B., Kohl T. A., Corchero J. L., Villaverde A., Schaffer L., Head S. R., Souvignier C., Meredith T. C., and Woodard R. W. (2015) Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb. Cell Fact. 14, 57 10.1186/s12934-015-0241-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee H., Hsu F. F., Turk J., and Groisman E. A. (2004) The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186, 4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reynolds C. M., Kalb S. R., Cotter R. J., and Raetz C. R. (2005) A phosphoethanolamine transferase specific for the outer 3-deoxy-d-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide: identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J. Biol. Chem. 280, 21202–21211 10.1074/jbc.M500964200 [DOI] [PubMed] [Google Scholar]
- 43. Klein G., Müller-Loennies S., Lindner B., Kobylak N., Brade H., and Raina S. (2013) Molecular and structural basis of inner core lipopolysaccharide alterations in Escherichia coli: incorporation of glucuronic acid and phosphoethanolamine in the heptose region. J. Biol. Chem. 288, 8111–8127 10.1074/jbc.M112.445981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Breazeale S. D., Ribeiro A. A., McClerren A. L., and Raetz C. R. (2005) A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-l-arabinose: identification and function of UDP-4-deoxy-4-formamido-l-arabinose. J. Biol. Chem. 280, 14154–14167 10.1074/jbc.M414265200 [DOI] [PubMed] [Google Scholar]
- 45. Yethon J. A., Heinrichs D. E., Monteiro M. A., Perry M. B., and Whitfield C. (1998) Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273, 26310–26316 [DOI] [PubMed] [Google Scholar]
- 46. Millán J. L. (2006) Alkaline phosphatases: structure, substrate specificity and functional relatedness to other members of a large superfamily of enzymes. Purinergic Signal. 2, 335–341 10.1007/s11302-005-5435-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mamat U., Meredith T. C., Aggarwal P., Kühl A., Kirchhoff P., Lindner B., Hanuszkiewicz A., Sun J., Holst O., and Woodard R. W. (2008) Single amino acid substitutions in either YhjD or MsbA confer viability to 3-deoxy-d-manno-oct-2-ulosonic acid-depleted Escherichia coli. Mol. Microbiol. 67, 633–648 [DOI] [PubMed] [Google Scholar]
- 48. Meredith T. C., Aggarwal P., Mamat U., Lindner B., and Woodard R. W. (2006) Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 1, 33–42 10.1021/cb0500015 [DOI] [PubMed] [Google Scholar]
- 49. Ghosh K., Mazumder Tagore D., Anumula R., Lakshmaiah B., Kumar P. P., Singaram S., Matan T., Kallipatti S., Selvam S., Krishnamurthy P., and Ramarao M. (2013) Crystal structure of rat intestinal alkaline phosphatase: role of crown domain in mammalian alkaline phosphatases. J. Struct. Biol. 184, 182–192 10.1016/j.jsb.2013.09.017 [DOI] [PubMed] [Google Scholar]
- 50. Perez J. C., and Groisman E. A. (2007) Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 63, 283–293 10.1111/j.1365-2958.2006.05512.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Soncini F. C., and Groisman E. A. (1996) Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178, 6796–6801 10.1128/jb.178.23.6796-6801.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meng J., Gong M., Björkbacka H., and Golenbock D. T. (2011) Genome-wide expression profiling and mutagenesis studies reveal that lipopolysaccharide responsiveness appears to be absolutely dependent on TLR4 and MD-2 expression and is dependent upon intermolecular ionic interactions. J. Immunol. 187, 3683–3693 10.4049/jimmunol.1101397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meng J., Lien E., and Golenbock D. T. (2010) MD-2-mediated ionic interactions between lipid A and TLR4 are essential for receptor activation. J. Biol. Chem. 285, 8695–8702 10.1074/jbc.M109.075127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. John C. M., Liu M., Phillips N. J., Yang Z., Funk C. R., Zimmerman L. I., Griffiss J. M., Stein D. C., and Jarvis G. A. (2012) Lack of lipid A pyrophosphorylation and functional lptA reduces inflammation by Neisseria commensals. Infect. Immun. 80, 4014–4026 10.1128/IAI.00506-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zariri A., Pupo E., van Riet E., van Putten J. P., and van der Ley P. (2016) Modulating endotoxin activity by combinatorial bioengineering of meningococcal lipopolysaccharide. Sci. Rep. 6, 36575 10.1038/srep36575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu M., John C. M., and Jarvis G. A. (2010) Phosphoryl moieties of lipid A from Neisseria meningitidis and N. gonorrhoeae lipooligosaccharides play an important role in activation of both MyD88- and TRIF-dependent TLR4-MD-2 signaling pathways. J. Immunol. 185, 6974–6984 10.4049/jimmunol.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Packiam M., Yedery R. D., Begum A. A., Carlson R. W., Ganguly J., Sempowski G. D., Ventevogel M. S., Shafer W. M., and Jerse A. E. (2014) Phosphoethanolamine decoration of Neisseria gonorrhoeae lipid A plays a dual immunostimulatory and protective role during experimental genital tract infection. Infect. Immun. 82, 2170–2179 10.1128/IAI.01504-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cullen T. W., O'Brien J. P., Hendrixson D. R., Giles D. K., Hobb R. I., Thompson S. A., Brodbelt J. S., and Trent M. S. (2013) EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infect. Immun. 81, 430–440 10.1128/IAI.01046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Loppnow H., Brade H., Dürrbaum I., Dinarello C. A., Kusumoto S., Rietschel E. T., and Flad H. D. (1989) IL-1 induction-capacity of defined lipopolysaccharide partial structures. J. Immunol. 142, 3229–3238 [PubMed] [Google Scholar]
- 60. Nogare A. R., and Yarbrough W. C. Jr. (1990) A comparison of the effects of intact and deacylated lipopolysaccharide on human polymorphonuclear leukocytes. J. Immunol. 144, 1404–1410 [PubMed] [Google Scholar]
- 61. Pohlman T. H., Munford R. S., and Harlan J. M. (1987) Deacylated lipopolysaccharide inhibits neutrophil adherence to endothelium induced by lipopolysaccharide in vitro. J. Exp. Med. 165, 1393–1402 10.1084/jem.165.5.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang M. H., Flad H. D., Feist W., Brade H., Kusumoto S., Rietschel E. T., and Ulmer A. J. (1991) Inhibition of endotoxin-induced interleukin-6 production by synthetic lipid A partial structures in human peripheral blood mononuclear cells. Infect. Immun. 59, 4655–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scior T., Alexander C., and Zaehringer U. (2013) Reviewing and identifying amino acids of human, murine, canine and equine TLR4/MD-2 receptor complexes conferring endotoxic innate immunity activation by LPS/lipid A, or antagonistic effects by Eritoran, in contrast to species-dependent modulation by lipid IVa. Comput. Struct. Biotechnol. J 5, e201302012 10.5936/csbj.201302012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Raetz C. R., Purcell S., Meyer M. V., Qureshi N., and Takayama K. (1985) Isolation and characterization of eight lipid A precursors from a 3-deoxy-d-manno-octylosonic acid-deficient mutant of Salmonella typhimurium. J. Biol. Chem. 260, 16080–16088 [PubMed] [Google Scholar]
- 65. Strain S. M., Armitage I. M., Anderson L., Takayama K., Qureshi N., and Raetz C. R. (1985) Location of polar substituents and fatty acyl chains on lipid A precursors from a 3-deoxy-d-manno-octulosonic acid-deficient mutant of Salmonella typhimurium: studies by 1H, 13C, and 31P nuclear magnetic resonance. J. Biol. Chem. 260, 16089–16098 [PubMed] [Google Scholar]
- 66. McClure R., and Massari P. (2014) TLR-dependent human mucosal epithelial cell responses to microbial pathogens. Front. Immunol. 5, 386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Faas M. M., Sáez T., and de Vos P. (2017) Extracellular ATP and adenosine: the Yin and Yang in immune responses? Mol. Aspects Med. 55, 9–19 10.1016/j.mam.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 68. Grbic D. M., Degagné E., Langlois C., Dupuis A. A., and Gendron F. P. (2008) Intestinal inflammation increases the expression of the P2Y6 receptor on epithelial cells and the release of CXC chemokine ligand 8 by UDP. J. Immunol. 180, 2659–2668 10.4049/jimmunol.180.4.2659 [DOI] [PubMed] [Google Scholar]
- 69. White K. A., Lin S., Cotter R. J., and Raetz C. R. (1999) A Haemophilus influenzae gene that encodes a membrane bound 3-deoxy-d-manno-octulosonic acid (Kdo) kinase: possible involvement of Kdo phosphorylation in bacterial virulence. J. Biol. Chem. 274, 31391–31400 10.1074/jbc.274.44.31391 [DOI] [PubMed] [Google Scholar]
- 70. Tuin A., Huizinga-Van der Vlag A., van Loenen-Weemaes A. M., Meijer D. K., and Poelstra K. (2006) On the role and fate of LPS-dephosphorylating activity in the rat liver. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G377–385 10.1152/ajpgi.00147.2005 [DOI] [PubMed] [Google Scholar]
- 71. Needham B. D., and Trent M. S. (2013) Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 11, 467–481 10.1038/nrmicro3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen H. D., and Groisman E. A. (2013) The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 67, 83–112 10.1146/annurev-micro-092412-155751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cullen T. W., Giles D. K., Wolf L. N., Ecobichon C., Boneca I. G., and Trent M. S. (2011) Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 7, e1002454 10.1371/journal.ppat.1002454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Renzi F., Zähringer U., Chandler C. E., Ernst R. K., Cornelis G. R., and Ittig S. J. (2016) Modification of the 1-phosphate group during biosynthesis of Capnocytophaga canimorsus lipid A. Infect. Immun. 84, 550–561 10.1128/IAI.01006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Harkness D. R. (1968) Studies on human placental alkaline phosphatase: II. kinetic properties and studies on the apoenzyme. Arch. Biochem. Biophys. 126, 513–523 10.1016/0003-9861(68)90436-0 [DOI] [PubMed] [Google Scholar]
- 76. McConnell R. E., Higginbotham J. N., Shifrin D. A. Jr., Tabb D. L., Coffey R. J., and Tyska M. J. (2009) The enterocyte microvillus is a vesicle-generating organelle. J. Cell Biol. 185, 1285–1298 10.1083/jcb.200902147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shifrin D. A. Jr, McConnell R. E., Nambiar R., Higginbotham J. N., Coffey R. J., and Tyska M. J. (2012) Enterocyte microvillus-derived vesicles detoxify bacterial products and regulate epithelial-microbial interactions. Curr. Biol. 22, 627–631 10.1016/j.cub.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Esparza G. A., Teghanemt A., Zhang D., Gioannini T. L., and Weiss J. P. (2012) Endotoxin·albumin complexes transfer endotoxin monomers to MD-2 resulting in activation of TLR4. Innate Immun. 18, 478–491 10.1177/1753425911422723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gioannini T. L., Zhang D., Teghanemt A., and Weiss J. P. (2002) An essential role for albumin in the interaction of endotoxin with lipopolysaccharide-binding protein and sCD14 and resultant cell activation. J. Biol. Chem. 277, 47818–47825 10.1074/jbc.M206404200 [DOI] [PubMed] [Google Scholar]
- 80. Gorelik A., Illes K., and Nagar B. (2018) Crystal structure of the mammalian lipopolysaccharide detoxifier. Proc. Natl. Acad. Sci. U.S.A. 115, E896–E905 10.1073/pnas.1719834115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hall C. L., and Munford R. S. (1983) Enzymatic deacylation of the lipid A moiety of Salmonella typhimurium lipopolysaccharides by human neutrophils. Proc. Natl. Acad. Sci. U.S.A. 80, 6671–6675 10.1073/pnas.80.21.6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gioannini T. L., Teghanemt A., Zhang D., Prohinar P., Levis E. N., Munford R. S., and Weiss J. P. (2007) Endotoxin-binding proteins modulate the susceptibility of bacterial endotoxin to deacylation by acyloxyacyl hydrolase. J. Biol. Chem. 282, 7877–7884 10.1074/jbc.M605031200 [DOI] [PubMed] [Google Scholar]
- 83. Halling Linder C., Narisawa S., Millán J. L., and Magnusson P. (2009) Glycosylation differences contribute to distinct catalytic properties among bone alkaline phosphatase isoforms. Bone 45, 987–993 10.1016/j.bone.2009.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Moss A. K., Hamarneh S. R., Mohamed M. M., Ramasamy S., Yammine H., Patel P., Kaliannan K., Alam S. N., Muhammad N., Moaven O., Teshager A., Malo N. S., Narisawa S., Millán J. L., Warren H. S., et al. (2013) Intestinal alkaline phosphatase inhibits the proinflammatory nucleotide uridine diphosphate. Am. J. Physiol. Gastrointest. Liver Physiol. 304, G597–G604 10.1152/ajpgi.00455.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Malo M. S., Alam S. N., Mostafa G., Zeller S. J., Johnson P. V., Mohammad N., Chen K. T., Moss A. K., Ramasamy S., Faruqui A., Hodin S., Malo P. S., Ebrahimi F., Biswas B., Narisawa S., et al. (2010) Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut 59, 1476–1484 10.1136/gut.2010.211706 [DOI] [PubMed] [Google Scholar]
- 86. Campbell E. L., MacManus C. F., Kominsky D. J., Keely S., Glover L. E., Bowers B. E., Scully M., Bruyninckx W. J., and Colgan S. P. (2010) Resolvin E1-induced intestinal alkaline phosphatase promotes resolution of inflammation through LPS detoxification. Proc. Natl. Acad. Sci. U.S.A. 107, 14298–14303 10.1073/pnas.0914730107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lehrer S., and Nowotny A. (1972) Isolation and purification of endotoxin by hydrolytic enzymes. Infect. Immun. 6, 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rick P. D., Fung L. W., Ho C., and Osborn M. J. (1977) Lipid A mutants of Salmonella typhimurium: purification and characterization of a lipid A precursor produced by a mutant in 3-deoxy-d-manno-octulosonate-8-phosphate synthetase. J. Biol. Chem. 252, 4904–4912 [PubMed] [Google Scholar]
- 89. Rosner M. R., Tang J., Barzilay I., and Khorana H. G. (1979) Structure of the lipopolysaccharide from an Escherichia coli heptose-less mutant: I. chemical degradations and identification of products. J. Biol. Chem. 254, 5906–5917 [PubMed] [Google Scholar]
- 90. Hulett F. M., Kim E. E., Bookstein C., Kapp N. V., Edwards C. W., and Wyckoff H. W. (1991) Bacillus subtilis alkaline phosphatases III and IV: cloning, sequencing, and comparisons of deduced amino acid sequence with Escherichia coli alkaline phosphatase three-dimensional structure. J. Biol. Chem. 266, 1077–1084 [PubMed] [Google Scholar]
- 91. Sunden F., AlSadhan I., Lyubimov A. Y., Ressl S., Wiersma-Koch H., Borland J., Brown C. L. Jr., Johnson T. A., Singh Z., and Herschlag D. (2016) Mechanistic and evolutionary insights from comparative enzymology of phosphomonoesterases and phosphodiesterases across the alkaline phosphatase superfamily. J. Am. Chem. Soc. 138, 14273–14287 10.1021/jacs.6b06186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. d'Hennezel E., Abubucker S., Murphy L. O., and Cullen T. W. (2017) Total lipopolysaccharide from the human gut microbiome silences Toll-like receptor signaling. mSystems 2, e00046–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Vatanen T., Kostic A. D., d'Hennezel E., Siljander H., Franzosa E. A., Yassour M., Kolde R., Vlamakis H., Arthur T. D., Hämäläinen A. M., Peet A., Tillmann V., Uibo R., Mokurov S., Dorshakova N., et al. (2016) Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 10.1016/j.cell.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gregg K. A., Harberts E., Gardner F. M., Pelletier M. R., Cayatte C., Yu L., McCarthy M. P., Marshall J. D., and Ernst R. K. (2017) Rationally designed TLR4 ligands for vaccine adjuvant discovery. MBio 8, e00046–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Needham B. D., Carroll S. M., Giles D. K., Georgiou G., Whiteley M., and Trent M. S. (2013) Modulating the innate immune response by combinatorial engineering of endotoxin. Proc. Natl. Acad. Sci. U.S.A. 110, 1464–1469 10.1073/pnas.1218080110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zariri A., and van der Ley P. (2015) Biosynthetically engineered lipopolysaccharide as vaccine adjuvant. Expert Rev. Vaccines 14, 861–876 10.1586/14760584.2015.1026808 [DOI] [PubMed] [Google Scholar]
- 97. Yang Y., Wandler A. M., Postlethwait J. H., and Guillemin K. (2012) Dynamic evolution of the LPS-detoxifying enzyme intestinal alkaline phosphatase in zebrafish and other vertebrates. Front. Immunol. 3, 314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cherepanov P. P., and Wackernagel W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 10.1016/0378-1119(95)00193-A [DOI] [PubMed] [Google Scholar]
- 100. Silva-Rocha R., Martínez-García E., Calles B., Chavarría M., Arce-Rodríguez A., de Las Heras A., Páez-Espino A. D., Durante-Rodríguez G., Kim J., Nikel P. I., Platero R., and de Lorenzo V. (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675 10.1093/nar/gks1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Galanos C., Lüderitz O., and Westphal O. (1969) A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9, 245–249 10.1111/j.1432-1033.1969.tb00601.x [DOI] [PubMed] [Google Scholar]
- 102. Strain S. M., Fesik S. W., and Armitage I. M. (1983) Characterization of lipopolysaccharide from a heptoseless mutant of Escherichia coli by carbon 13 nuclear magnetic resonance. J. Biol. Chem. 258, 2906–2910 [PubMed] [Google Scholar]
- 103. Hirschfeld M., Ma Y., Weis J. H., Vogel S. N., and Weis J. J. (2000) Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine Toll-like receptor 2. J. Immunol. 165, 618–622 10.4049/jimmunol.165.2.618 [DOI] [PubMed] [Google Scholar]
- 104. Henderson J. C., O'Brien J. P., Brodbelt J. S., and Trent M. S. (2013) Isolation and chemical characterization of lipid A from Gram-negative bacteria. J. Vis. Exp. 79, e50623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kutschera A., Dawid C., Gisch N., Schmid C., Raasch L., Gerster T., Schäffer M., Smakowska-Luzan E., Belkhadir Y., Vlot A. C., Chandler C. E., Schellenberger R., Schwudke D., Ernst R. K., Dorey S., et al. (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364, 178–181 [DOI] [PubMed] [Google Scholar]
- 106. Ranf S., Gisch N., Schäffer M., Illig T., Westphal L., Knirel Y. A., Sánchez-Carballo P. M., Zähringer U., Hückelhoven R., Lee J., and Scheel D. (2015) A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 16, 426–433 [DOI] [PubMed] [Google Scholar]
- 107. Zhou Z., Ribeiro A. A., and Raetz C. R. (2000) High-resolution NMR spectroscopy of lipid A molecules containing 4-amino-4-deoxy-l-arabinose and phosphoethanolamine substituents. Different attachment sites on lipid A molecules from NH4VO3-treated Escherichia coli versus kdsA mutants of Salmonella typhimurium. J. Biol. Chem. 275, 13542–13551 10.1074/jbc.275.18.13542 [DOI] [PubMed] [Google Scholar]
- 108. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 110. Guex N., and Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- 111. Pedretti A., Villa L., and Vistoli G. (2004) VEGA: an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Aided Mol. Des. 18, 167–173 10.1023/B:JCAM.0000035186.90683.f2 [DOI] [PubMed] [Google Scholar]
- 112. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., and Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [DOI] [Google Scholar]
- 113. Thompson M. A. (Mar-Apr, 2004) Molecular docking using ArgusLab, an efficient shape-based search algorithm and the AScore scoring function. in Proceedings of the ACS Meeting, Vol. 172, CINF 42, Philadelphia, PA [Google Scholar]
- 114. Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., and Lee J. O. (2009) The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 10.1038/nature07830 [DOI] [PubMed] [Google Scholar]
- 115. Ohto U., Fukase K., Miyake K., and Satow Y. (2007) Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science 316, 1632–1634 10.1126/science.1139111 [DOI] [PubMed] [Google Scholar]
- 116. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., and Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Scior T., and Wahab H. A. (2007) Structure prediction of proteins with very low homology: a comprehensive introduction and a case study on aminopeptidase (Kaplan S., ed) Drug Design Research Perspectives, pp. 675–708, Nova Science Publishers, Hauppauge, NY [Google Scholar]
- 118. Gasteiger J., and Marsili M. (1981) Prediction of proton magnetic-resonance shifts: the dependence on hydrogen charges obtained by iterative partial equalization of orbital electronegativity. Organic Magnetic Resonance 15, 353–360 10.1002/mrc.1270150408 [DOI] [Google Scholar]
- 119. Rietsch A., Vallet-Gely I., Dove S. L., and Mekalanos J. J. (2005) ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102, 8006–8011 10.1073/pnas.0503005102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Bierman M., Logan R., O'Brien K., Seno E. T., Rao R. N., and Schoner B. E. (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 10.1016/0378-1119(92)90627-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.