Abstract
Background
Nontyphoidal Salmonella (NTS) is an important foodborne pathogen worldwide. We investigated a 2018 outbreak of highly antimicrobial-resistant Salmonella enterica serotype Goldcoast in northern Taiwan.
Methods
We collected 30 clinical isolates and 2 meat isolates from this outbreak in New Taipei and Taoyuan, Taiwan in 2018. The clinical manifestations and the treatment of the patients were reviewed. To trace the source, we examined NTS isolated from food samples collected from the markets in northern Taiwan. All of the isolates along with an additional human isolate from China were sequenced and compared with the sequences of Salmonella Goldcoast reported by other countries.
Results
The outbreak involved 14 pediatric patients (<5 years old) and 16 adults (36 to 83 years old). Nine patients with invasive or severe disease required carbapenem treatment. The MIC90 of ceftriaxone and ciprofloxacin for the outbreak isolates was >256 μg/mL and 1 μg/mL, respectively, and a conjugative 278-kilobase plasmid harboring blaCTX-M-55 and qnrS1 contributed towards the resistance. Whole-genome sequencing revealed a clonal relationship among the outbreak isolates and the 2 collected from the retail meats. The outbreak clone was phylogenetically close to that of Salmonella Goldcoast reported in the United Kingdom, Poland, and China, whereas similar resistance plasmids were found in China and Cambodia.
Conclusions
The clinical spectrum of the high-level cephalosporin-resistant Salmonella Goldcoast is similar to that of other NTS serotypes, but severe cases required carbapenem treatment. The study confirmed the emergence of a highly antimicrobial-resistant clone of Salmonella Goldcoast, highlighting the importance of surveillance for food safety.
Keywords: cephalosporin resistance, clone, conjugative plasmid, Salmonella Goldcoast, whole-genome sequencing
An outbreak caused by ceftriaxone- and ciprofloxacin-resistant Salmonella Goldcoast was investigated. A conjugative 278-kb plasmid harboring blaCTX-M-55 and qnrS1 contributed to the resistance. Whole-genome sequencing revealed a clonal relationship among the clinical isolates and another 2 from retail meats.
Nontyphoidal Salmonella (NTS) is a prevalent foodborne pathogen that usually causes gastroenteritis, but it can occasionally cause invasive diseases in humans [1, 2]. It has been estimated that there are more than 90 million cases of gastroenteritis due to NTS globally each year, with 85% of those cases being linked to foods consumed [3]. More than 2500 NTS serotypes have been identified, but only a few dozen, particularly Salmonella Typhimurium and Salmonella enteritidis, accounted for 50%–70% of NTS infections worldwide [4–6].
Salmonellosis caused by NTS is usually self-limiting, but when antimicrobial treatment is necessary, ampicillin and trimethoprim-sulfamethoxazole were the drugs of choice in the clinical setting. Antimicrobial-resistant NTS has emerged since the 1980s, in which the ACSSuT-resistant phenotype (defined as resistant to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline) was first identified in Salmonella Typhimurium DT104 in England [7, 8]. This penta-resistance pattern is mediated by a Class I integron, which is usually associated with the mosaic structure of the Salmonella genomic island 1 (SGI1) [9]. Due to the rapid dissemination of ACSSuT resistance in Salmonella, fluoroquinolones (FQs) and extended-spectrum cephalosporins are recommended for treating NTS infections in such settings. In 2017, the susceptible rate of Salmonella to ceftriaxone and ciprofloxacin in Taiwan was 92.3% and 78.7%, respectively [10].
Salmonella Goldcoast, a rare serotype for human infection, belongs to serogroup C2. To date, all reported foodborne outbreaks linked to Salmonella Goldcoast have occurred in Europe, with the contamination source being French pate, watercress, cheddar cheese, and raw fermented sausages [11–14]. The Salmonella Goldcoast strains recovered from these outbreaks were mostly sensitive to FQs and cephalosporins. In 2018, we experienced an Salmonella Goldcoast outbreak in Taiwan. This emerging Salmonella Goldcoast clone exhibited high-level resistance to extended-spectrum cephalosporins and concurrent resistance to FQs. Moreover, although the genomic analysis demonstrated that the outbreak isolates showed a close clonal relationship with strains isolated from the United Kingdom and China, the conjugative resistance plasmid was highly similar to the plasmids circulating in China and Cambodia. In this study, we report the clinical and microbiological investigation of this outbreak in northern Taiwan and highlight the importance of monitoring antimicrobial resistance (AMR) of foodborne pathogens.
METHODS
Patients and Setting
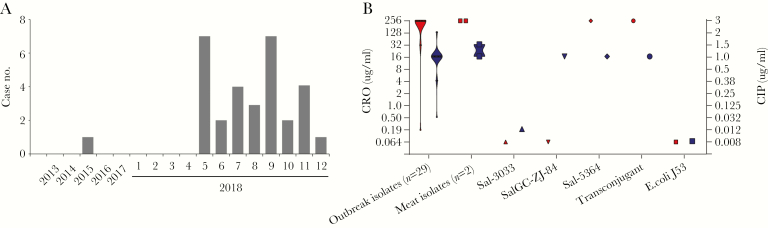
Chang Gung Memorial Hospital (CGMH) is a main referral hospital for cities in northern Taiwan, including Taipei, New Taipei, and Taoyuan. The population in this region is approximately 7 million. The Clinical Microbiology Laboratory has launched a program to monitor the serotypes of the NTS causing human infections since 2012. All Salmonella isolates derived from patients were collected and serotyped. Antimicrobial susceptibility testing was performed using the disc diffusion method specified in the Clinical and Laboratory Standards Institute (CLSI) guidelines. Salmonella Goldcoast is a relatively uncommonly recorded serotype for human infections. Before 2015, only 1 Salmonella Goldcoast isolate was recovered from the stool of a child with acute gastroenteritis. From 2016 to 2017, no Salmonella Goldcoast was further reported. However, in 2018, 30 culture-confirmed infections were found to have been caused by Salmonella Goldcoast (Figure 1A; Supplementary Table 1). This outbreak is still ongoing in 2019: there were already 20 cases of Salmonella Goldcoast infections by the end of July in 2019. Therefore, an outbreak investigation was conducted, including an analysis of the following: clinical manifestations, treatments, and patient outcomes; bacterial source tracing; and microbiological and genomic examinations of all isolates of Salmonella Goldcoast. Only 1 sample per patient was collected for downstream analyses. If both blood and stool isolates were available from the same patient, only the blood isolate was analyzed. Pulsed-field gel electrophoresis (PFGE) was performed by the Centers for Disease Control and Prevention in Taiwan, following the protocol as described [15]. The minimum inhibitory concentration (MIC) of ceftriaxone and ciprofloxacin to these isolates were determined by E-test and interpreted according to the recommendations given by CLSI [16]. The Student’s t test, the χ 2 test, and Fisher’s exact test or analysis of variance were used for comparisons whenever appropriate. All statistical analyses were 2-sided, and significance was set at P < .05.
Figure 1.
Salmonella Goldcoast infection in Taiwan and antimicrobial resistance of the isolates. (A) Number of the Salmonella Goldcoast cases in Chang Gung Memorial Hospital (CGMH). Between January 2013 and April 2018, only 1 case was reported, whereas between January 2018 and December 2018, 30 cases were identified. (B) Violin plot of the minimum inhibitory concentration (MIC) values of ceftriaxone ([CRO] in red, left y-axis) and ciprofloxacin ([CIP] in blue, right y-axis) for the studied isolates. Isolate Sal-3033 was the only one collected at CGMH before 2018. Isolate SalGC-ZJ-84 was collected from Zhejiang, China instead of Taiwan. The MICs for the donor strain, Sal-5364, the recipient strain, Escherichia coli J53, and the transconjugant obtained in the conjugation experiment demonstrate that the resistance was mediated by a conjugative plasmid.
To trace the source of Salmonella Goldcoast, we obtained Salmonella isolates from food samples collected from markets in Linko District, New Taipei City and Kweishan District, Taoyuan City. These 2 districts, with their high population densities, are 2 main areas of origin for the patients treated in CGMH. As with the clinical isolates, the Salmonella isolates obtained from food samples were further examined for their serotypes. Antimicrobial susceptibility to ceftriaxone and ciprofloxacin for these isolates were assessed using E-test strips.
Genome Sequencing and Assembly
In addition to the 31 Salmonella Goldcoast isolates collected from CGMH (30 isolates collected in the 2018 outbreak and 1 in 2015), 1 Salmonella Goldcoast isolate from the neighboring Zhejiang province, China (SalGC-ZJ-84), which was collected from the stool of an 80-year-old patient with acute gastroenteritis in June 2017, was also included in this study. All of these isolates were subjected to whole-genome sequencing (WGS).
Genomic deoxyribonucleic acid (DNA) of all the sequenced isolates was prepared using the QIAamp DNA Mini Kit (Qiagen, Düsseldorf, Germany) and then subjected to WGS using the Illumina Miseq platform (Illumina, San Diego, USA). The short reads generated were de novo assembled into contigs using SPAdes version 3.11.1 with the “--careful” option [17].
The isolate, Sal-5364, was subjected to sequencing using both Illumina Miseq sequencer and the long-read MinION Sequencer (Nanopore, Oxford, UK). A de novo hybrid assembly of the short Illumina and long MinION reads was performed using Unicycler version 0.4.79 under the “normal” mode [18]. The complete circular contigs generated were then corrected using Pilon version 2.14 10 with Illumina reads [19].
Genomic Analysis
The multilocus sequence typing (MLST) and prediction of the AMR genes were performed by analyzing the WGSs through BacWGSTdb [20]. To search for genetically close isolates from the Taiwan outbreak isolates, the global Salmonella Goldcoast isolates were downloaded from Enterobase (https://enterobase.warwick.ac.uk/) and NCBI GenBank database (Supplementary Table 2). An initially quick phylogeny construction was carried out using parsnp software [21].
The core genome MLST (cgMLST) was performed also using the BacWGSTdb service [20]. In brief, the cgMLST alleles were determined first, and then the allele matrix was put into GrapeTree software for construction of minimal spanning tree [22]. For single-nucleotide polymorphism (SNP) calling, the complete genome sequence of Sal-5364 was used as a reference; the query genomes were mapped against the reference genome by the nucmer program in MUMmer package version 3.22 [23]; the show-snps program was then used to call the SNPs, using the “-C” option to remove the SNPs from the alignments with ambiguous mapping. The 5-base pair adjacent SNPs were further filtered; then, Gubbins software was used to filter recombinant SNPs. An ML tree built using RAxML version 8.0.0 [24] was assessed with TempEst version 1.5.1 [25]. Given the strong temporal signal found in the data, we proceeded with time-calibrated Bayesian phylogenetic inference using BEAST version 1.10.4 [26]: the “HKY” substitution model, the “strict molecular clock” model, the “Coalescent: Constant size” tree prior model, and 10 000 000 Markov chain Monte Carlo (MCMC) chains were set for this analysis.
Similar plasmids to pSal-5364 were searched using the BacWGSTdb single-genome analysis module. In brief, BacWGSTdb collects all of the complete plasmid genomes and applies the MASH to compare them with the user uploaded sequences [27].
Conjugation Experiment
To investigate the transferability of the resistance plasmid identified in the genome sequencing, a conjugation assay was conducted using Escherichia coli J53 (a sodium azide-resistant strain) as the recipient. An outbreak Salmonella Goldcoast isolate (Sal-5364) was conjugated with the J53 at the donor-to-recipient ratio of 1:4 and coincubated for 12 hours on a 0.45-μm microporous membrane. Transconjugants were subsequently selected on Mueller-Hinton agar supplemented with sodium azide (200 μg/mL) and ceftriaxone (16 μg/mL). S1 nuclease-PFGE was performed to determine the plasmid profile. Genomic DNA preparations for J53, Sal-5364, and transconjugant strains were made in agarose plugs and digested with S1 nuclease, and then separated by PFGE. The conjugation efficiency was measured and calculated following the protocol in https://openwetware.org/wiki/conjugation.
Data Availability
The genome sequences sequenced in this study have been deposited in the NCBI GenBank database, and their accession numbers are listed in Supplementary Table 1.
RESULTS
Outbreak Description
Since May 2018, a sharp rise in infections caused by Salmonella Goldcoast has been observed; as of December 2018, 30 cases have been recorded (Figure 1A). All of the 14 pediatric patients were younger than 5 years of age, whereas the age of the adult patients ranged from 35 to 85 years (Supplementary Table 3). The most common symptoms noted in these patients were diarrhea, abdominal cramps, and fever (Supplementary Table 4). Most patients had only gastroenteritis, whereas 3 adults and 1 child were complicated with bacteremia, 1 adult with urinary tract infection, and 1 child with ileus. The extra-intestinal infection rate for Salmonella Goldcoast was 7.1% (1 of 14) for children less than 5 years of age and 25.0% (4 of 16) for adults. Pediatric patients showed a significantly longer hospital stay for treatment than did adults (P = .002) (Supplementary Table 4).
All isolates from this outbreak were 100% resistant to ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole. More importantly, all but 1 isolate were simultaneously resistant to ciprofloxacin and ceftriaxone. The MIC of ciprofloxacin and ceftriaxone for these isolates were mostly 1 μg/mL and >256 μg/mL (Figure 1B). Nine patients, including the 5 with extra-intestinal infections and the 1 with ileus, received carbapenem treatment, mainly with ertapenem.
Resistance Mechanism
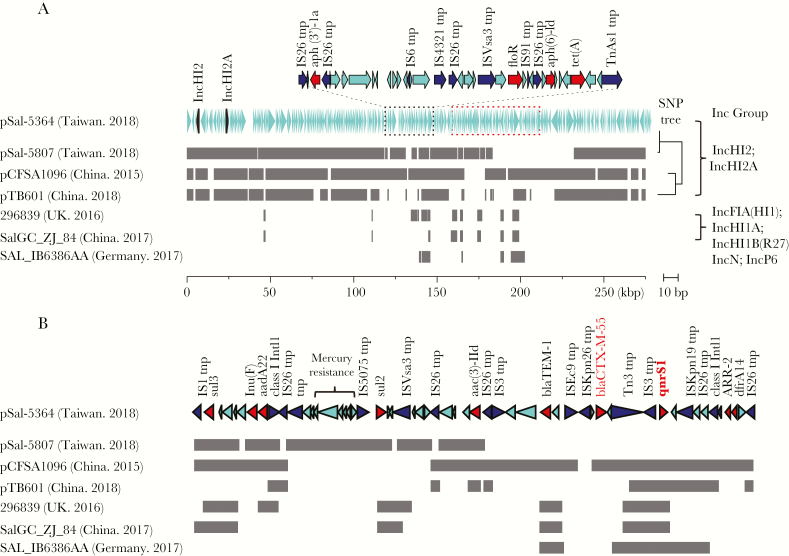
The Salmonella Goldcoast isolates were all subject to WGS. The resistant isolates invariably carried a 278-kilobase (kb) plasmid that belonged to the IncHI2/IncHI2A group. All resistance genes were clustered with 2 cassettes on the plasmid (Figure 2A). Insertion sequences (IS) and transposons were enriched in the 2 cassettes, indicative of their high frequency of lateral transfer. One of the 2 cassettes was bracketed by 2 integrases of the Class I integron and carried blaCTX-M-55 and qnrS1 simultaneously. This Class I integron-like element harbored sul2, sul3, lnu(F), aadA22, ARR-2, and dfrA14, and therefore conferred the ACSSuT phenotype to the bacteria. The only susceptible isolate from this outbreak, Sal-5807, also possessed this plasmid but lacked blaCTX-M-55 and qnrS1 (Figure 2B). An earlier isolate, Sal-5303, collected in 2015, was susceptible to ceftriaxone and ciprofloxacin due to its lack of the plasmid. The SGI1 was absent in all the Salmonella Goldcoast isolates.
Figure 2.
Alignment between the 278-kilobase plasmid and its closely related plasmids. The aligned regions are represented as gray blocks. (A) Sal-5364 and Sal-5807 were isolated from the outbreak; the former was resistant to ciprofloxacin and ceftriaxone, whereas the latter was susceptible to both. The replicon genes are marked in black. The resistance genes are concentrated within 2 cassettes (marked in dashed black and red boxes). The plasmids, pCFSA1096 (accession no. CP033347.1) and pTB601 (accession no. CP034832.1), revealed the same Inc group to the pSal-5364, but they did not originate from the Salmonella Goldcoast. To the right is the neighbor-joining tree built based on the single-nucleotide polymorphisms (SNPs) identified within the backbones of these plasmids. Isolate 296839 (accession no. SRR7204491) and isolate SAL_IB6386AA (accession no. ERR2984265) were the only European and American isolates that carried qnrS1 in the public database. Isolate SalGC_ZJ_84 from Zhejiang, China also carried qnrS1. These 3 plasmids belonged to different Inc groups from the others. (B) Gene structure of the red cassette (A).
To investigate the transferability of the resistance plasmid, conjugation experiments were carried out and revealed that the 278-kb plasmid was readily transferred into the recipient (Supplementary Figure 1), with a conjugation efficiency being 1.76 ± 1.02 × 10‒4. The transconjugant revealed the same level of resistance to ceftriaxone and ciprofloxacin as the donor strain and other outbreak isolates (Figure 1B), suggesting that the resistance was laterally transferrable between bacteria through plasmid conjugation.
Phylogeny Construction and Source Identification
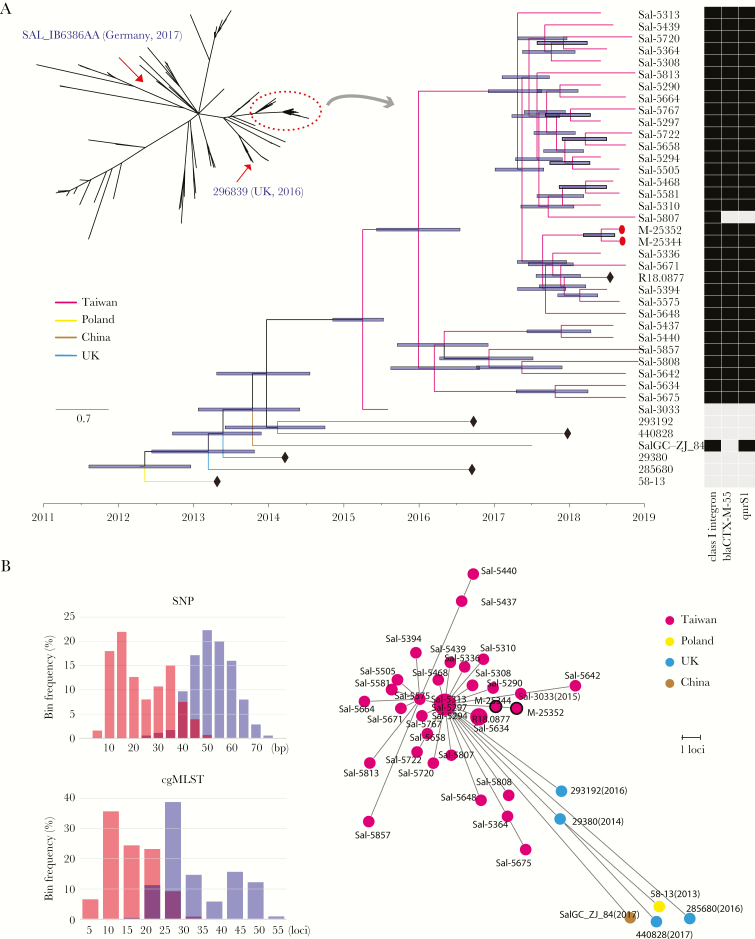
All isolates belonged to sequence type 358 (ST358) and had the same PFGE pattern. Whole-genome sequencing further demonstrated that they possessed a high level of similarity, with their chromosomal difference ranging from 7 to 46 SNPs (Figure 3). This result indicated that this outbreak had originated from the same contaminated foods. To trace the source of Salmonella Goldcoast, we obtained food samples from traditional markets in New Taipei and Taoyuan and identified 2 Salmonella Goldcoast isolates from retail meats, 1 being from pork and the other from chicken (Supplementary Table 5). Whole-genome sequencing showed that the isolates from the retail meats were closely connected with the clinical isolates (Figure 3A). The retail meat isolates harbored the same 278-kb plasmid and showed the same resistance profile. Taken together, the contaminated retail meats were considered a possible source of the outbreak.
Figure 3.
Phylogenetic analysis of the Salmonella Goldcoast outbreak isolates in Taiwan. (A) The phylogenetic relationship among global Salmonella Goldcoast isolates. The dendrogram built by parsnp software revealed that all of the Taiwanese isolates were clustered together with a few international isolates (marked in a red dashed circle). All isolates outside the red circle were in lack of blaCTX-M-55 and qnrS1, except the 2 isolates marked by the red arrows (see the detailed gene structure in Figure 2). A dated phylogeny was further built for the isolates within the red circle by the BEAST2 software. The branches are in different colors according to their geographic location. The black nodes in the diamond represent the data downloaded from public database, whereas the rest were sequenced in this study. The nodes in the form of red dots represent isolates of the food origin, whereas the rest were derived from the patients. The right heatmap represents the presence (in black) or absence (in gray) of key antimicrobial resistance genes. (B) Histograms of single-nucleotide polymorphism (SNP) distance and core genome multilocus sequence typing (cgMLST) distance. The pairwise distance within the Taiwanese outbreak isolates is marked in red, whereas that between the Taiwanese outbreak isolates and the international ones is in blue. (C) Minimal Spanning Tree built by GrapeTree software based on cgMLST alleles. The number in the parenthesis represents the collection year; isolates without this were all collected in 2018. Dots with black circles represent food isolates, whereas the others are clinical isolates.
One susceptible isolate, Sal-5303, which was collected earlier in 2015, was highly homologous to the outbreak isolates in genome sequence, with its distance from the other genomes ranging from 20 to 37 SNPs. A dated phylogeny dendrogram revealed that the isolate was at the root of the outbreak (Figure 3A).
Worldwide Dissemination of the Salmonella Goldcoast Clone
To detect the epidemiological and evolutionary signals from the global context, we compared the genomes of the Taiwan isolates with the 150 Salmonella Goldcoast genomes deposited in the GenBank, NCBI, including 44 from North America, 105 from Europe, and 1 from Taiwan. Because of the large geographic distances between the above isolates and the Taiwanese outbreak clone, we also collected and sequenced an Salmonella Goldcoast isolate from Zhejiang province, China, which was found 600 km across the strait from Taiwan.
As a result, a Taiwanese isolate in the public database, which had also been collected in 2018, was found nearly identical to the isolates studied (Figure 3B), suggesting that the outbreak caused by the Salmonella Goldcoast clone was not limited to northern Taiwan. It is interesting to note that the Zhejiang isolate sequenced, along with 4 British and 1 Polish isolates (collected in 2013–2016), were phylogenetically closely related to the Taiwanese outbreak clone, with a difference of 23–74 SNPs (Figure 3B). This distance range overlapped with that of the Taiwanese clone. The cgMLST presented similar results (Figure 3B and C). Thus, the Taiwanese isolates could not be distinguished from these international isolates purely by genetic distance, but they formed a topologically cohesive cluster. This close genetic distance demonstrated a short divergence history: the Taiwan isolates had not diverged from the international isolates until 2014 (Figure 3A).
The international Salmonella Goldcoast isolates were overall much more susceptible when compared with the Taiwan outbreak isolates. All of the international isolates lacked blaCTX-M-55 or other β-lactamase-encoding genes that can confer resistance to extended-spectrum cephalosporins. Almost all of them likewise lacked qnrS1 as well as effective mutations in their chromosomal quinolone resistance-determining regions, suggesting that they would be quinolone susceptible. Only the Zhejiang isolate as well as a British and a German isolate carried qnrS1. As with the Taiwanese clone, these isolates’ qnrS1 genes were carried by an IS3 transposase but belonged to different plasmid types, suggesting that the resistance genes were carried on an entirely different plasmid (Figure 2). The p.T57S mutation in parC was found in all Salmonella Goldcoast isolates, even those susceptible to FQs. No other point mutation was found. Thus, this mutation was merely a genetic marker for this serotype but did not contribute towards resistance. For other antimicrobial agents, the Class I integron was detected in the Zhejiang isolate and 12 European isolates from among the international isolates, indicating their potential to express the ACSSuT phenotype.
The conjugative nature of the 278-kb plasmid demonstrated its transmission route as being independent from that of the chromosome. We consequently developed an online service integrated in the BacWGSTdb database [20], which applies the MASH algorithm to compare the query sequence to all of the complete plasmid sequences deposited in the NCBI database. The closest plasmids to the 278-kb resistance plasmid of Salmonella Goldcoast were also from the Salmonella species isolated in China (Figure 2); the backbones of the plasmids were similar, and the differences were mainly distributed in mobile elements. In particular, the plasmid pCFSA1096 (accession no. CP033347.1), collected from the Hubei Province, China in 2015, also carried blaCTX-M-55 and qnrS1.
DISCUSSION
Raw food products are popular in Taiwan. In combination with the location’s optimum temperature range for bacterial growth, NTS infections have long been rampant on this island. The present study reported an outbreak of highly antimicrobial-resistant Salmonella Goldcoast and confirmed that the offending Salmonella was from local retail meats. The entire meat production and sale chain consists of the following 4 stages: breeding and farming, slaughter, distribution, and sale. Although a much larger body of literature has focused on isolation rates on farms and in slaughter houses, contamination may occur more commonly at the distribution and sale phase [28]. In this study, we attributed the present outbreak to the contamination occurring at the distribution and sale phase rather than during farming and slaughter. We also speculated, based on the following epidemiological and genomic evidence, that this contamination had taken place for years. First, the sampled chicken and pork came from different farms but were sold at the same market. Second, the early Salmonella Goldcoast isolate collected in 2015 in the same geographic region belonged to the same clone as the outbreak isolates. Third, the genomes of the outbreak isolates were not exactly identical but differed from each other by a limited number of SNPs, indicating the presence of a regional source rather than point source pollution. One limitation of this study is that we did not conduct tests upon local farms, although the farms raising pigs and chicken were likely the source of the outbreak clone. This hypothesis could not be totally rejected. We propose that actions such as transportation by cold chain, fresh meat preservation, and the improvement of sanitation in traditional markets should be undertaken to reduce the contamination and thereby avoid the occurrence of subsequent foodborne infection.
Phylogenetically, genome comparison has revealed an unexpectedly limited genetic distance between the Salmonella Goldcoast isolates of Taiwan and other countries in Asia and Europe. The SNP distance within the Taiwan isolates overlapped with that between Taiwan and other international isolates, making it difficult to distinguish them merely by SNP distance. This situation also applies to the cgMLST strategy. Furthermore, the genetic distance between the Taiwanese and Chinese Salmonella Goldcoast isolates was not shorter than that between Taiwan and British isolates; this disproportion between the SNP and spatial distances strongly suggests the involvement of human activity, such as increases in global travel and food trade, upon the transmission of the infectious disease. The ancestor for all Taiwanese Salmonella Goldcoast isolates can be traced to as late as 2014. However, few cases, even sporadic ones, caused by this rare serotype, have been reported in Taiwan before 2014, whereas all previously reported cases of Salmonella Goldcoast have been concentrated in Europe. The global dissemination of a real bacterial clone, rather than a certain PFGE type or ST, has been reported for a number of other bacterial pathogens [29]. Therefore, increased awareness, timely global communication, and collaboration for food safety are essential towards containing foodborne disease outbreaks.
Salmonellosis is usually a self-limiting disease that does not require antimicrobial therapy, but in young children and the elderly population, if the offending Salmonella causes an invasive illness, antimicrobial therapy becomes necessary. Because the ACSSuT penta-resistance has reached 14.5%, 72.7%, and 28.8% in the United States, Taiwan, and China [30], respectively, FQs and extended-spectrum cephalosporins became the antimicrobials of choice for the treatment of salmonellosis. Before 2003, resistance to FQ was mainly attributed to an accumulation of mutations in such bacterial genes as gyrA and parC targeted by the drug. For example, the Salmonella Choleraesuis outbreak isolates in Taiwan in 2000 were highly resistant to FQ (>8 μg/mL) by their simultaneous carriage of F83S and N87D mutations in gyrA [31]. A single T57S mutation in an entire Salmonella Goldcoast lineage is not sufficient for a strain to achieve clinically significant resistance, but 1 additional mutation could result in highly FQ resistance. Meanwhile, during the past 15 years, there have been increasing reports of FQ-resistant Enterobacteriaceae encoding qnrA, qnrB, qnrD, aac(6’)-Ib-cr, and oqxAB, as well as qnrS1 [32], the gene responsible for the FQ resistance in Salmonella Goldcoast. These plasmid-mediated quinolone resistance genes generally facilitate low-level FQ resistance, but their potential to be laterally transferred promotes their rapid dissemination.
Growing resistance of NTS to extended-spectrum cephalosporins is usually linked to the spread of plasmid-mediated AmpC or extended-spectrum β-lactamase (ESBL) genes. The most common global contributor is blaCMY-2, followed by blaCTX-M-15 [33–36]. As a variant of blaCTX-M-15, blaCTX-M-55 increases the catalytic efficiency of the ESBL to cephalosporins, leading to a higher degree of resistance than blaCTX-M-15. The blaCTX- M-55 gene has already become prevalent in China, Thailand, and Cambodia [30, 37–42], but the present study is the first case reported in Taiwan. The 2 resistance genes, namely, blaCTX-M-55 and qnrS1, were both carried by a 278-kb conjugative plasmid in Salmonella Goldcoast outbreak strains. In this regard, surveillance and monitoring of the plasmid’s circulation among the Enterobacteriaceae species is crucial. Nowadays, a few databases, such as Enterobase (https://enterobase.warwick.ac.uk) and BacWGSTdb (http://bacdb.org/BacWGSTdb), have been constructed with the aim of investigating genetic relatedness based on genomic sequences and, accordingly, establishing epidemiological links between bacterial pathogens. Few databases, on the other hand, have been developed for plasmids. To help address this concern, we developed an online service for plasmid tracing. We have discovered highly similar plasmids from Salmonella isolates collected in Hubei province in 2015 and Zhejiang province in 2018 in China. We also noticed that an IncHI2 plasmid carrying blaCTX-M-55 and qnrS1 was reported in Cambodia in 2016 [40]; this plasmid was identical or highly similar to the 278-kb one found in Salmonella Goldcoast. Therefore, it is very likely that the 278-kb conjugative plasmid was transferred to Taiwan from other Asian countries.
The 278-kb resistance plasmid harbored a high density of ISs and transposons that encompass not only blaCTX-M-55 and qnrS1 but also a wide variety of other AMR genes. Further dissemination of the 278-kb conjugative plasmid into other Salmonella serotypes or even other Enterobacteriaceae species would make ciprofloxacin and ceftriaxone no longer effective for the treatment of infections and lead to a more frequent use of carbapenems, a drug of last resort. The use of antimicrobials in clinical practice and, more importantly, in food source livestock, would create a selective pressure that pushes AMR genes-carrying ISs and transposons to enter the plasmid, or for the entire conjugative plasmid to be transferred into susceptible bacteria.
CONCLUSIONS
In this study, we have observed that Salmonella Goldcoast isolates in Europe and North America were much more susceptible than those in Taiwan and China because the isolates generally lack of ISs, transposons, or plasmids. The difference may be due to a more stringent control of antimicrobial use in patients as well as in food source livestock in Europe and North America.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Author contributions. Y.-J. C., S.-H. F., L.-H. S., H.-C. L., H.-P. Y., M.-J. Y., and C.-H. C. collected clinical and epidemiological data of the outbreak. H.-C. L., H.-P. Y., and M.-J. Y. carried out serotyping, antimicrobial susceptibility testing, and conjugation experiments. Y. F. and C.-H. C. coordinated the microbiological and genomic analysis. Y. F. and C.-H. C. analyzed the whole-genome sequencing data. Y. F. and C.-H. C. contributed to first drafts, and all authors reviewed and edited the final manuscript.
Financial support. The study was funded by Grant 31670132 from the National Natural Science Foundation, China, Grants CRRPG3F0083, CMRPG3D1721-3, CMRPG3G1451-3, CMRPG3E1371-3, and CMRPG3A1111-3 from Chang Gung Memorial Hospital, Taiwan, and Grants MOHW106- CDC-C-114-113702, MOHW107-CDC-C-114-123505, and MOHW108-CDC-C-114-133505 from Ministry of Health and Welfare, Taiwan.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Chiou CS, Hong YP, Liao YS, et al. New multidrug-resistant Salmonella enterica Serovar Anatum Clone, Taiwan, 2015-2017. Emerg Infect Dis 2019; 25:144–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Y, Zhang A, Yang Y, et al. Emergence of Salmonella enterica serovar Indiana and California isolates with concurrent resistance to cefotaxime, amikacin and ciprofloxacin from chickens in China. Int J Food Microbiol 2017; 262:23–30. [DOI] [PubMed] [Google Scholar]
- 3. Majowicz SE, Musto J, Scallan E, et al. ; International Collaboration on Enteric Disease ‘Burden of Illness’ Studies The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 2010; 50:882–9. [DOI] [PubMed] [Google Scholar]
- 4. Li Y, Xie X, Xu X, et al. Nontyphoidal salmonella infection in children with acute gastroenteritis: prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis 2014; 11:200–6. [DOI] [PubMed] [Google Scholar]
- 5. Saravanan S, Purushothaman V, Murthy TR, et al. Molecular epidemiology of nontyphoidal salmonella in poultry and poultry products in India: implications for human health. Indian J Microbiol 2015; 55:319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wei ZQ, Chang HL, Li YF, et al. Clinical epidemiology and antimicrobial resistance of nontyphoidal Salmonella enteric infections in children: 2012-2014. Zhonghua Er Ke Za Zhi 2016; 54:489–95. [DOI] [PubMed] [Google Scholar]
- 7. Lauderdale TL, Aarestrup FM, Chen PC, et al. ; TSAR hospitals Multidrug resistance among different serotypes of clinical Salmonella isolates in Taiwan. Diagn Microbiol Infect Dis 2006; 55:149–55. [DOI] [PubMed] [Google Scholar]
- 8. Yu CY, Chou SJ, Yeh CM, et al. Prevalence and characterization of multidrug-resistant (type ACSSuT) Salmonella enterica serovar Typhimurium strains in isolates from four gosling farms and a hatchery farm. J Clin Microbiol 2008; 46:522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boyd D, Peters GA, Cloeckaert A, et al. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 2001; 183:5725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jean SS, Lu MC, Shi ZY, et al. In vitro activity of ceftazidime-avibactam, ceftolozane-tazobactam, and other comparable agents against clinically important Gram-negative bacilli: results from the 2017 Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART). Infect Drug Resist 2018; 11:1983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bremer V, Leitmeyer K, Jensen E, et al. Outbreak of Salmonella Goldcoast infections linked to consumption of fermented sausage, Germany 2001. Epidemiol Infect 2004; 132:881–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horváth JK, Mengel M, Krisztalovics K, et al. Investigation into an unusual increase of human cases of Salmonella Goldcoast infection in Hungary in 2009. Euro Surveill 2013; 18:20422. [DOI] [PubMed] [Google Scholar]
- 13. Inns T, Beasley G, Lane C, et al. Outbreak of Salmonella enterica Goldcoast infection associated with whelk consumption, England, June to October 2013. Euro Surveill 2013; 18:20654. [DOI] [PubMed] [Google Scholar]
- 14. Scavia G, Ciaravino G, Luzzi I, et al. A multistate epidemic outbreak of Salmonella Goldcoast infection in humans, June 2009 to March 2010: the investigation in Italy. Euro Surveill 2013; 18:20424. [DOI] [PubMed] [Google Scholar]
- 15. Ribot EM, Fair MA, Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 2006; 3:59–67. [DOI] [PubMed] [Google Scholar]
- 16. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-First Informational Supplement. CLSI document M100-S28. Wayne, PA; Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 17. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13:e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruan Z, Feng Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res 2016; 44:D682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 2014; 15:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Z, Alikhan NF, Sergeant MJ, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res 2018; 28:1395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol 2004; 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2016; 2:vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suchard MA, Lemey P, Baele G, et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 2018; 4:vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ondov BD, Treangen TJ, Melsted P, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 2016; 17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li WW, Bai L, Zhang XL, et al. Prevalence and antimicrobial susceptibility of Salmonella isolated from broiler whole production process in four provinces of China. Zhonghua Yu Fang Yi Xue Za Zhi 2018; 52:352–7. [DOI] [PubMed] [Google Scholar]
- 29. Ruan Z, Yu Y, Feng Y. The global dissemination of bacterial infections necessitates the study of reverse genomic epidemiology. Brief Bioinform 2019. https://doi.org/10.1093/bib/bbz010 [DOI] [PubMed] [Google Scholar]
- 30. Ma Y, Li M, Xu X, et al. High-levels of resistance to quinolone and cephalosporin antibiotics in MDR-ACSSuT Salmonella enterica serovar Enteritidis mainly isolated from patients and foods in Shanghai, China. Int J Food Microbiol 2018; 286:190–6. [DOI] [PubMed] [Google Scholar]
- 31. Chiu CH, Wu TL, Su LH, et al. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype choleraesuis. N Engl J Med 2002; 346:413–9. [DOI] [PubMed] [Google Scholar]
- 32. Rodríguez-Martínez JM, Machuca J, Cano ME, et al. Plasmid-mediated quinolone resistance: two decades on. Drug Resist Updat 2016; 29:13–29. [DOI] [PubMed] [Google Scholar]
- 33. Noda T, Murakami K, Etoh Y, et al. Increase in resistance to extended-spectrum cephalosporins in Salmonella isolated from retail chicken products in Japan. PLoS One 2015; 10:e0116927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shigemura H, Matsui M, Sekizuka T, et al. Decrease in the prevalence of extended-spectrum cephalosporin-resistant Salmonella following cessation of ceftiofur use by the Japanese poultry industry. Int J Food Microbiol 2018; 274:45–51. [DOI] [PubMed] [Google Scholar]
- 35. Su LH, Wu TL, Chia JH, et al. Increasing ceftriaxone resistance in Salmonella isolates from a university hospital in Taiwan. J Antimicrob Chemother 2005; 55:846–52. [DOI] [PubMed] [Google Scholar]
- 36. Yu F, Chen Q, Yu X, et al. High prevalence of plasmid-mediated quinolone resistance determinant aac(6’)-Ib-cr amongst Salmonella enterica serotype Typhimurium isolates from hospitalised paediatric patients with diarrhoea in China. Int J Antimicrob Agents 2011; 37:152–5. [DOI] [PubMed] [Google Scholar]
- 37. Zhao X, Yang J, Zhang B, et al. Characterization of integrons and resistance genes in Salmonella isolates from farm animals in Shandong Province, China. Front Microbiol 2017; 8:1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang L, Fu Y, Xiong Z, et al. Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front Microbiol 2018; 9:2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei Z, Xu X, Yan M, et al. Salmonella typhimurium and Salmonella enteritidis infections in sporadic diarrhea in children: source tracing and resistance to third-generation cephalosporins and ciprofloxacin. Foodborne Pathog Dis 2019; 16:244–55. [DOI] [PubMed] [Google Scholar]
- 40. Nadimpalli M, Fabre L, Yith V, et al. ; BIRDY study group CTX-M-55-type ESBL-producing Salmonella enterica are emerging among retail meats in Phnom Penh, Cambodia. J Antimicrob Chemother 2019; 74:342–8. [DOI] [PubMed] [Google Scholar]
- 41. Luk-In S, Chatsuwan T, Pulsrikarn C, et al. High prevalence of ceftriaxone resistance among invasive Salmonella enterica serotype choleraesuis isolates in Thailand: the emergence and increase of CTX-M-55 in ciprofloxacin-resistant S. choleraesuis isolates. Int J Med Microbiol 2018; 308:447–53. [DOI] [PubMed] [Google Scholar]
- 42. Li S, Zhou Y, Miao Z. Prevalence and antibiotic resistance of non-typhoidal Salmonella isolated from raw chicken carcasses of commercial broilers and spent hens in Tai’an, China. Front Microbiol 2017; 8:2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences sequenced in this study have been deposited in the NCBI GenBank database, and their accession numbers are listed in Supplementary Table 1.