Abstract
Background
To compare the prognostic utility of the new definition of difficult-to-treat resistance (DTR) vs established definitions in a cohort of patients with Gram-negative bloodstream infections (GNBSIs).
Methods
This was a retrospective single-center study of adult patients with monomicrobial GNBSI, hospitalized from 2013 to 2016. DTR was defined as isolate demonstrating intermediate or resistant phenotype to all reported agents in the carbapenem, beta-lactam, and fluoroquinolone classes. Carbapenem resistance (CR) was defined according to 2015 Centers for Disease Control and Prevention criteria. Each isolate was further classified according to the Magiorakos et al. criteria as non-multidrug-resistant (non-MDR), MDR, extensively drug-resistant (XDR), or pan-drug-resistant (PDR). The primary outcome was all-cause 30-day mortality.
Results
Overall, 1576 patients were analyzed. Enterobacteriaceae accounted for 88.7% of BSIs, with Escherichia coli (n = 941) and Klebsiella pneumoniae (n = 326) being the most common pathogens. Pseudomonas aeruginosa was the most common nonfermentative bacteria (n = 130, 8.2%). Overall, 11% of strains were defined as DTR and 13% as CR. Episodes were further classified as non-MDR (68.8%), MDR (21.9%), XDR (8.8%), and PDR (0.4%). The prevalence rates of DTR, CR, and XDR were similar among Enterobacteriaceae and Acinetobacter baumannii, whereas they differed in P. aeruginosa. All the analyzed resistance definitions significantly improved prediction of 30-day mortality when introduced into a baseline multivariate model, to a similar degree: 9%, 10%, and 11% for DTR, Magiorakos, and CR definitions, respectively.
Conclusions
DTR seems a promising tool to identify challenging GNBSIs, mainly those due to P. aeruginosa. With the availability of new agents for CR infections, further multicenter assessments of DTR are needed.
Keywords: bloodstream infection, Gram-negative, carbapenem resistance, difficult-to-treat resistance, all-cause 30-day mortality
Increasing rates of antibiotic resistance among Gram-negative bacteria have prompted investigators to analyze several issues concerning patients with severe infections due to these microorganisms.
In a majority of studies, the definition adopted for multidrug resistance was that proposed in 2008 by the US Centers for Disease Control and Prevention (CDC) and the European Centre for Disease Prevention and Control [1]. The authors defined 3 resistance phenotypes: multidrug resistance (MDR) as nonsusceptibility to ≥1 agent in ≥3 antimicrobial categories; extensive drug resistance (XDR) as susceptibility limited to ≤2 categories; and pan-drug resistance (PDR) as nonsusceptibility to all agents in all antimicrobial categories [1]. Although epidemiologically useful, this definition has the limitation of weighing all antibiotics equally and only considering their in vitro activity, regardless of their “real-life” effectiveness and toxicity, limiting the bedside applicability of MDR and XDR categories. Indeed, MDR and XDR infections were not consistently associated with poorer patient outcomes in some studies [2, 3]; initial appropriate therapy has failed to improve the outcomes of patients with such drug resistance categories [4]. These issues are pivotal in designing and evaluating clinical trials on the therapeutic management of MDR Gram-negative infections [5].
Indeed, in a white paper from the Infectious Diseases Society of America (IDSA) on the conduct of clinical trials for the treatment of drug-resistant bacteria, the authors proposed a new concept of “extreme drug resistance” as an alternative to the Magiorakos XDR class. They defined XDR organisms as those resistant to all Food and Drug Administration (FDA)–approved, systematically active antibacterial agents except for those known to be substantially more toxic than or less efficacious than alternative agents [6]. For Gram-negative bacteria, they reported the example of strains resistant to all FDA-approved agents except for aminoglycosides, tigecycline, or colistin. In accordance with this concept, a new definition of resistance for Gram-negative infections has been recently proposed by Kadri et al. [7]. The authors defined difficult-to-treat resistance (DTR) as a treatment-limiting resistance to all firstline agents including all beta-lactams and fluoroquinolones [7]. This definition should reflect the use of second-line agents, such as those mentioned in the IDSA white paper, which are characterized by poorer pharmacokinetic properties and increased risk of toxicity, resulting in a better prediction of poor outcome. The 5-year prevalence of DTR, the associated risk factors, and the impact on in-hospital mortality rates were analyzed using a very large US cohort of patients with Gram-negative bloodstream infections (GNBSIs) [7]. However, in this study, the prevalence of DTR was very low and administrative data were used.
The aim of our study was to compare the prognostic utility of DTR vs established resistance definitions in a cohort of patients with GNBSIs hospitalized in a tertiary teaching hospital from an area with a high prevalence of antibiotic resistance.
METHODS
Study Design and Setting
We performed a retrospective cohort study of patients hospitalized at S. Orsola-Malpighi Hospital, a 1450-bed tertiary care university institute in Bologna, in the region of Emilia-Romagna (Northern Italy), from January 1, 2013, to December 31, 2016.
Patients were identified through microbiology databases. Clinical charts and hospital records were reviewed to gather study variables using a case report form (CRF) for up to 90 days after the index blood cultures (BCs). The accuracy of the data was systematically reviewed by a senior investigator before inclusion in the database.
Our ethics committee approved the study; informed consent was waived due to the retrospective noninterventional study design. Data were collected anonymously.
Participants
We included all adult (≥18 years) patients diagnosed with Gram-negative BSI, defined as ≥1 positive BC obtained from a patient suspected of having infection. Patients were considered only once at the time of the first episode (index BCs).
Patients were excluded from the analysis if they were found to have (i) a polymicrobial BSI, defined as growth of >1 micro-organism, excluding potential contaminants (ie, coagulase-negative staphylococci, Corynebacterium spp., Propionibacterium spp.); (ii) died within 72 hours of drawing the index BCs; or (iii) no clinical data available.
Variables and Definitions
For each GNBSI, we determined if the infection met the proposed definition for DTR according to Kadri et al. [7]. Specifically, DTR was defined as any GNBSI isolate demonstrating an intermediate or resistant phenotype to all reported agents in the carbapenem, beta-lactam, and fluoroquinolone categories (including additional agents when results were available). Stenotrophomonas maltophila, which showed resistance to all tested antimicrobials (including TMP/SMX, levofloxacin, and minocycline), was also considered DTR.
Isolates were further classified according to the Magiorakos et al. criteria [1] as non-MDR, MDR, XDR, or PDR.
In addition, we categorized antibiotic class resistance as carbapenem resistance (CR), extended-spectrum cephalosporin resistance (ECR), and fluoroquinolone resistance (FQR) based on Centers for Disease Control and Prevention (CDC) surveillance definitions (https://gis.cdc.gov/grasp/PSA/Downloads/AR-PhenotypeDefinitions.pdf). Finally, beta-lactam/betalactamase inhibitor resistance (BL/BLI-R) was assessed according to European Committee for Antimicrobial Susceptibility Testing (EUCAST) criteria.
The primary outcome used to assess the prognostic significance of each resistance definition was 30-day mortality, defined as all-cause mortality within 30 days of the index BC [8].
We analyzed patient risk factors for their association with 30-day mortality, including age, sex, and underlying disease severity according to the Charlson comorbidity index [9]. Immunosuppression included neutropenia (neutrophil count < 500/mm3), solid organ transplantation, hematopoietic stem cell transplantation, corticosteroid therapy at a dosage higher than or equivalent to prednisone 16 mg/da ≥15 days, and uncontrolled HIV infection (<200 CD4/mm3).
BSI was classified according to the site of acquisition into nosocomial, health care–associated, and community-acquired using Friedman’s criteria [10]. Clinical severity at infection onset was assessed according to updated sepsis definitions [11]. BSI sources were established according to CDC criteria [12]. In the absence of a recognized source, BSI was considered primary. BSI was defined as complicated when the infection source was not fully removable.
According to the causative species and susceptibility to carbapenems, etiologies were classified into 3 groups: (i) carbapenem-susceptible Enterobacteriaceae (CSE), (ii) carbapenem-resistant Enterobacteriaceae (CRE), and (iii) nonfermentative Gram-negative bacteria (NF-GNB).
Empirical therapy was defined as antibiotics administered before the susceptibility report was available. It was considered appropriate when at least 1 in vitro active drug (according to the susceptibility pattern of the isolate) was administered within 24 hours of drawing the index BC. Delayed or no active antibiotic administration within this period was considered inappropriate empirical therapy. Definitive antibiotic therapy was defined as antibiotic treatment administered according to susceptibility results. Combination therapy was defined as a regimen including >1 anti-Gram-negative drug irrespective of relative in vitro activity. Duration of antibiotic treatment was defined as the number of consecutive days during which the patient received an appropriate antibiotic regimen. Source control was defined as the removal of the infection source within 7 days of index BC, including the performance of nonsurgical or surgical procedures to treat an obstructive focus, collection, or abscess at any site, including, among others, the urinary tract, biliary tract, and surgical site, and the removal of any device deemed the source of the BSI.
Microbiology
BCs were incubated using the BACTEC FX Automated Blood Culture System (Becton Dickinson, Franklin Lakes, NJ, USA). All positive BCs were processed with the Maldi Biotyper MALDI-TOF system (Bruker Daltonics, Bremen, Germany) for rapid and reliable species identification of microorganisms. Antimicrobial susceptibility testing of strains was performed using the Vitek 2 automated system (bioMerieux, Marcy l’Etoile, France). Enterobacteriaceae minimal inhibitory concentrations (MICs) were interpreted using EUCAST clinical breakpoints for all tested antibiotics.
Statistical Analysis
For the descriptive analysis, categorical variables were presented as absolute numbers and their relative frequencies. Continuous variables were presented as mean and standard deviation if normally distributed or as median and interquartile range (IQR) if non–normally distributed.
Univariate and multivariate analysis were performed to assess the relationship of study variables with 30-day all-cause mortality. First, categorical variables were compared using the χ 2 or Fisher exact test when appropriate, and continuous variables were compared using the Mann-Whitney U test. Then, significant and clinically relevant covariates identified in univariate analysis were introduced by a backward selection approach into a multivariable Cox regression survival model to ensure that all correlations between predictors were considered, using a P cutoff of .05. Patients were considered from the day of BSI onset (index BCs) until death or day 30. The discrimination and calibration of the Cox regression model were then analyzed without any variable defining a resistance category of the bloodstream isolate (baseline mortality model), vs the addition of 1 of the following categories: (i) Magiorakos et al. classifications (non-MDR, MDR, XDR, and PDR); (ii) DTR; and (iii) CR definitions, described previously. All analysis was performed with STATA IC 13.1 (Stata Corp., College Station, TX, USA) using the STCOXCAL package to compare model calibration. Model discrimination was assessed by the Harrel C statistic and Net Reclassification Index (NRI) of each resistance definition with the baseline survival model [13].
RESULTS
According to the study criteria (Supplementary Figure 1), 1576 patients with a first episode of monomicrobial GN-BSI during the study period were analyzed. The median age (IQR) was 72 (59–82) years, and 55.7% were male. The general characteristics of the study population are reported in Supplementary Table 1.
Enterobacteriaceae accounted for 88.7% of BSIs, with 1259 carbapenem-susceptible (CSE) and 140 carbapenem-resistant (CRE) pathogens. Escherichia coli was the most common causative microorganism (59.7%), followed by Klebsiella pneumoniae (20.7%). Pseudomonas aeruginosa was the most common nonfermentative bacteria (8.2%).
Overall, 11% of strains were defined as DTR. Distribution of resistance categories was as follows: non-MDR 68.8%, MDR 21.9%, XDR 8.8%, and PDR 0.4%. The distribution of antibiotic class resistance was: FQR 46.6%, BL/BLIR 44.7%, ESCR 36.1%, and CR 13.1%. The prevalence of resistance categories and classes of antibiotic resistance varied across pathogens, as shown in Table 1.
Table 1.
Prevalence of Resistance Among the Main Gram-Negative Species According to the Analyzed Definitions
| E. coli (n = 941), No. (%) | K. pneumoniae (n = 326), No. (%) | P. aeruginosa (n = 130), No. (%) | A. baumannii (n = 33), No. (%) | |
|---|---|---|---|---|
| Resistance categories | ||||
| Non-MDR | 742 (78.9) | 133 (40.8) | 111 (85.4) | 11 (33.3) |
| MDR | 197 (20.9) | 69 (21.2) | 19 (14.6) | 3 (9.1) |
| XDR | 2 (0.2) | 117 (35.9) | 0 | 19 (57.6) |
| PDR | 0 | 7 (2.1) | 0 | 0 |
| Antibiotic class resistance | ||||
| BL/BLIR | 371 (39.5) | 215 (66) | 37 (38.5) | NA |
| ECR | 296 (31.5) | 207 (63.5) | 26 (20) | NA |
| CR | 1 (0.1) | 140 (42.9) | 36 (27.7) | 22 (66.7) |
| FQR | 438 (46.5) | 198 (60.7) | 33 (25.4) | 22 (66.7) |
| New definition | ||||
| DTR | 1 (0.1) | 138 (42.3) | 10 (7.7) | 22 (66.7) |
Abbreviations: BL/BLIR, betalactam/betalactamase inhibitor resistance; CR, carbapenem resistance; DTR, difficult-to-treat resistance; ECR, extended-spectrum cephalosporin resistance; FQR, fluoroquinolone resistance; MDR, multidrug resistance; PDR, pandrug resistance, XDR, extensive drug resistance.
Source control and appropriate empirical therapy were performed in 27.3% and 68% of cases, respectively. In both the empirical and definitive treatment cohorts, the antibiotic classes most commonly used were BL/BLI and carbapenems. Combination therapy was administered in 16.2% and 21.6% of empirical and definitive regimens, respectively (data shown in Supplementary Table 1).
All-cause 30-day mortality was 10.4%, with 5% of patients later presenting with a BSI relapse within 90 days after index BCs.
Compared with patients who were alive at day 30 (Table 2), nonsurviving patients exhibited higher Charlson index scores and were more likely to be in the ICU at BSI onset or have a hospital-acquired BSI. Nonsurviving patients also exhibited higher SOFA scores and higher rates of septic shock, nonurinary infection sources, and etiologies other than E. coli. In terms of therapeutic management, only appropriate empiric therapy was significantly associated with a lower mortality rate. All resistance definitions were associated with significantly higher 30-day mortality rates by univariate analysis (Supplementary Figure 2).
Table 2.
Univariate Analysis of Risk Factors for All-Cause 30-Day Mortality
| Survivors (n = 1412), No. (%) | Nonsurvivors (n = 164), No. (%) | p | |
|---|---|---|---|
| Demographics | |||
| Age, median (IQR), y | 72 (59–82) | 72 (62–83) | .20 |
| Male sex | 782 (55.4) | 96 (58.5) | .46 |
| Comorbidities | |||
| Charlson index, median (IQR) | 6 (4–8) | 6.6 (4.5–8.8) | .003 |
| Immunosuppression | 292 (20.7) | 40 (24.4) | .31 |
| Ward of admission | <.001 | ||
| Medical | 1136 (80.5) | 110 (67.1) | |
| Surgical | 181 (12.8) | 23 (14) | |
| ICU | 95 (6.7) | 31 (18.9) | |
| Site of BSI acquisition | <.001 | ||
| Community-acquired | 415 (29.4) | 22 (13.4) | |
| Health care–associated | 249 (17.6) | 32 (19.5) | |
| Hospital-acquired | 748 (53) | 110 (67.1) | |
| CRE carrier at BSI onset | 150 (10.6) | 33 (20.1) | .001 |
| Clinical severity at BSI onset | |||
| SOFA, median (IQR) | 3 (1–5) | 5 (3–7) | <.001 |
| Septic shock | 98 (6.9) | 45 (27.4) | <.001 |
| Source of BSI | |||
| Undefined | 265 (18.8) | 33 (20.1) | .75 |
| Urinary tract | 560 (39.7) | 33 (20.1) | <.001 |
| Biliary tract | 205 (14.5) | 19 (11.6) | .35 |
| Intra-abdominal | 170 (12) | 25 (15.2) | .26 |
| Lower respiratory tract | 99 (7) | 24 (14.6) | .001 |
| CVC-related | 71 (5) | 19 (11.6) | .001 |
| Complicated BSI | 358 (25.4) | 53 (32.3) | .06 |
| Etiology | |||
| Escherichia coli | 881 (62.4) | 60 (36.6) | <.001 |
| Klebsiella pneumoniae | 280 (19.8) | 46 (28) | .01 |
| Enterobacter spp. | 70 (5) | 7 (4.3) | .71 |
| Proteus spp. | 41 (2.9) | 14 (8.5) | .001 |
| Pseudomonas aeruginosa | 107 (7.6) | 23 (14) | .007 |
| Acinetobacter baumannii | 21 (1.6) | 10 (6.1) | .001 |
| Stenotrophomonas maltophilia | 10 (0.7) | 4 (2.4) | .05 |
| Etiology category | <.001 | ||
| CSE | 1163 (82.4) | 96 (58.5) | |
| CRE | 109 (7.7) | 31 (18.9) | |
| NF-GNB | 140 (9.9) | 37 (22.6) | |
| Resistance categoriesa | <.001 | ||
| Non-MDR | 1005 (71.2) | 80 (48.8) | |
| MDR | 295 (21) | 50 (30.5) | |
| XDR | 107 (7.6) | 32 (19.5) | |
| PDR | 5 (0.4) | 2 (1.2) | |
| Antibiotic class resistancea | |||
| ECR | 487 (34.5) | 82 (50) | <.001 |
| BL/BLIR | 612 (43.3) | 93 (56.7) | <.001 |
| CR | 154 (10.9) | 53 (32.3) | <.001 |
| FQR | 626 (44.3) | 109 (66.5) | <.001 |
| New definition | |||
| DTR | 129 (9.1) | 45 (27.4) | <.001 |
| Therapeutic management | |||
| Source control | 386 (27.3) | 45 (27.4) | 1 |
| Appropriate empirical therapy | 990 (70.1) | 85 (51.8) | <.001 |
Abbreviations: BL/BLIR, betalactam/betalactamase inhibitor resistance; BSI, bloodstream infection; CR, carbapenem resistance; CRE, carbapenem-resistant Enterobacteriaceae; CVC, central venous catheter; DTR, difficult-to-treat resistance; ECR, extended-spectrum cephalosporin resistance; FQR, fluoroquinolone resistance; ICU, intensive care unit; IQR, interquartile range; MDR, multidrug resistance; PDR, pandrug resistance; SOFA, sequential organ failure assessment; XDR, extensive drug resistance.
aResistance categories were mutually exclusive, whereas antibiotic class resistances were not.
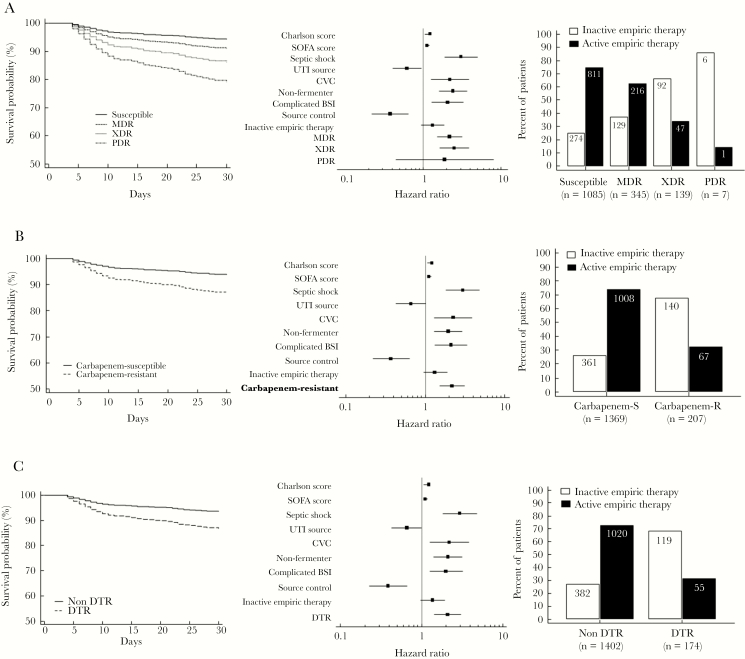
At multivariate analysis, the independent risk factors for all-cause 30-day mortality were Charlson index, SOFA score, septic shock, CVC-related BSI, BSI due to CRE or NF-GNB, and complicated BSI, whereas urinary source, source control, and active empiric therapy were protective factors (Table 3). The predicted impact of Magiorakos (non-MDR, MDR, XDR, and PDR), CR, and DTR definitions on 30-day survival adjusted for significant survival covariates is shown in Figure 1. The impact of DTR was also analyzed, including the 19 patients who died within 72 hours of index BC for whom clinical data were available, without observing different results (Supplementary Figure 3).
Table 3.
Multivariate Analysis of Risk Factors for All-Cause 30-Day Mortality
| Covariate | aHR (95% CI) | P |
|---|---|---|
| Charlson comorbidity score | 1.12 (1.06–1.18) | <.001 |
| Septic shock | 2.91 (1.81–4.70) | <.001 |
| SOFA score | 1.12 (1.07–1.18) | <.001 |
| Urinary tract source | 0.64 (0.42–0.96) | <.03 |
| CVC-associated infection | 2.19 (1.27–3.79) | .005 |
| Etiology category | ||
| CSE | Reference | |
| CRE | 1.95 (1.26–3.02) | .003 |
| Nonfermentative | 2.43 (1.59–3.73) | <.001 |
| Complicated BSI | 2.02 (1.26–3.24) | .003 |
| Source control | 0.38 (0.23–0.65) | <.001 |
| Active empiric therapy | 0.68 (0.49–0.95) | .02 |
Abbreviations: aHR, adjusted hazard ratio; BSI, bloodstream infection; CI, confidence interval; CSE, carbapenem-susceptible Enterobacteriaceae; CRE, carbapenem-resistant Enterobacteriaceae; CVC, central venous catheter; NFGN, nonfermentative Gram-negative; SOFA, sequential organ failure assessment.
Figure 1.
Survival curves for different resistance categories and forest plots according to Cox multivariate analysis of risk factors for all-cause 30-day mortality. The baseline model included the following resistance definitions: Magiorakos criteria (A); carbapenem resistance (B); difficult-to-treat resistance (C). Numbers and rates of active and inactive therapy for each category according to each resistance definition are shown. Abbreviations: BSI, bloodstream infection; CVC, central venous catheter; DTR, difficult-to-treat resistance; MDR, multidrug resistance; PDR, pandrug resistance; SOFA, sequential organ failure assessment; UTI, urinary tract infection; XDR, extensive drug resistance.
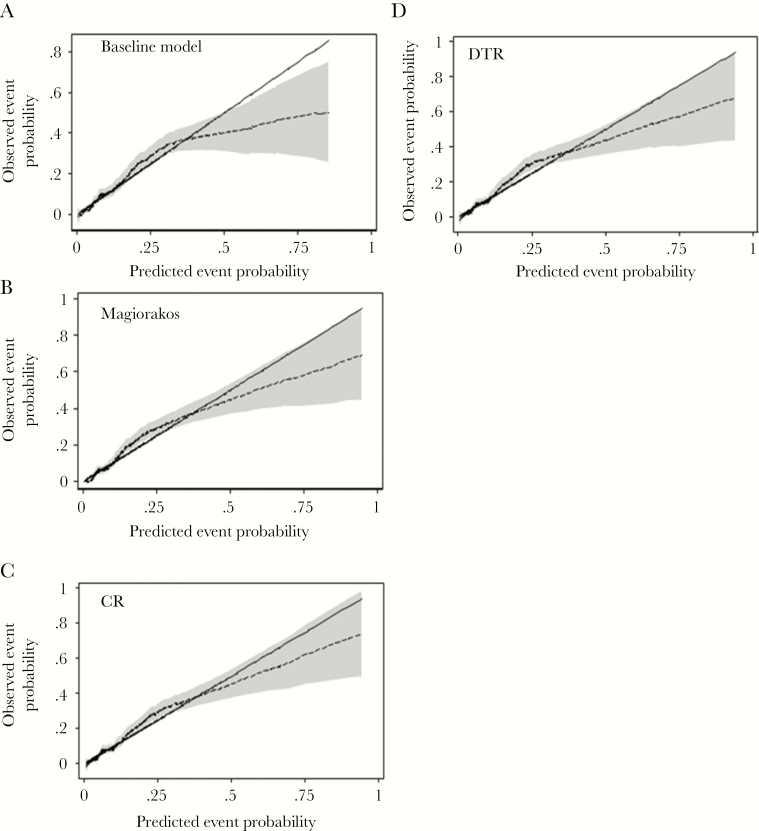
Incorporation of the resistance definitions into the baseline mortality model significantly improved discrimination of the multivariate model for predicting 30-day mortality; the net reclassification improvement was 9%, 10%, and 11% for DTR, Magiorakos et al., and CR definitions, respectively (Supplementary Table 2). Similarly, calibration of the baseline mortality risk model was improved with inclusion of each resistance definition, particularly for predicted 30-day mortality risk >20%, as shown in Figure 2.
Figure 2.
Calibration of 30-day mortality risk model by resistance definition. Smoothed pseudo-values (dashed lines) with pointwise 95% confidence intervals (shaded area) are plotted against predicted 30-day mortality probabilities. The solid line is the line of identity, denoting perfect calibration. Some miscalibration is evident with all models at predicted probabilities >0.5. A, Baseline mortality model without susceptibility categories. B, Magiorakos et al. definitions. C, Carbapenem resistance. D, Difficult-to-treat resistance. Abbreviations: CR, carbapenem resistance; DTR, difficult-to-treat resistance;
Discussion
We analyzed the prevalence of the new proposed definition of antibiotic resistance for Gram-negative bacteria and difficult-to-treat resistance in a cohort of 1576 patients with monomicrobial GN-BSI. In addition, we compared DTR with the previously proposed definitions of Magiorakos et al. and CR according to the 2015 CDC criteria. In our study, the prevalence of DTR was 11%. It varied across species and was highest among K. pneumoniae and A. baumannii BSIs. In these pathogens, CR and DTR rates were comparable, whereas they differed in P. aeruginosa. Specifically, DTR seemed to identify better than CR and XDR categories the cases of P. aeruginosa with limited treatment options. All the analyzed definitions significantly improved the prediction of 30-day mortality to a similar degree when introduced into a baseline mortality prediction model.
In the daily practice, DTR and CR definitions offer some important advantages over Magiorakos criteria as (i) being easier to establish; (ii) providing more descriptive information that enhances pathogen-directed treatment; and (iii) capturing excess mortality attributable to both discordant empirical regimens and subsequent reliance on less effective and/or more toxic compounds (eg, colistin, tigecycline, and aminoglycosides).
Some authors have observed that CR, when appropriately applied, encompasses most DTR Gram-negative infections, providing useful information for guiding therapy [14]. This was confirmed in our study for Enterobacteriaceae and A. baumannii but not for P. aeruginosa. Unfortunately, the low number of P. aeruginosa BSIs limited our ability to analyze the prognostic significance of DTR in this subgroup. In addition, our epidemiology and the therapeutic approach to CR infections during the study period could have influenced our results. Indeed, with the introduction of new drugs for CR infections, the predictive value of CR has been changing [15]. This fact underlines a strength of a new definition: “DTR is not a fixed phenotype but rather a flexible framework” [16]. Indeed, the authors who proposed this definition recognized the need to periodically revise the rubric of firstline, high-efficacy, and low-toxicity agents in order to continue to capture, with the DTR definition, how resistance is perceived and confronted at the bedside.
The extreme drug resistance and DTR concepts were primarily developed to design clinical trials on new antibacterial agents for drug-resistant infections [6]. Resistance to all firstline drugs should reflect excess mortality attributable not only to initial inappropriate therapy, but also to the use of alternative drugs with suboptimal pharmacokinetic/pharmacodynamic (PK/PD) profiles and greater toxicity [17]. Indeed, in our basic model for mortality prediction, active empiric therapy was an independent protective factor, along with source control. However, when drug resistance categories were added to the model, the association between initial appropriate therapy and mortality was no longer significant, whereas source control remained a strong protective factor. It is worth noting that our multivariate analysis focused solely on appropriate empiric therapy; that is, it is still possible to receive inappropriate empiric therapy even in patients infected with susceptible isolates. Indeed, in our analysis, we found that nearly 25% of patients with “susceptible” Gram-negative pathogens did not receive appropriate empirical therapy, thus providing one explanation of how MDR or DTR resistance definitions could be retained simultaneously in a multivariate model adjusted for inappropriate therapy. Another possible explanation is that both resistance categories and in vitro active therapy do not take into account eventual drug exposure in real life. These considerations underline the need to determine local microbiology and optimize dosing schedules to improve the rates of appropriate empiric therapy and patient survival.
Our study has several limitations. The single-center design could limit the generalizability of our results. However, this is the first validation of the DTR definition in a Southern European country, where the prevalence and impact on mortality of antibiotic resistance are much higher than in the population used to develop the definition [18]. In addition, in US studies, both urban and rural hospitals were included, diluting the prevalence and impact of DTR. Our cohort is from a large tertiary teaching hospital, reflecting the complexity and epidemiology of patients managed in similar institutions from our area. The retrospective collection of patient and microbiological data could have limited integrity and accuracy. However, a senior investigator revised all CRFs and reconciled data reports and missing data with medical records before including information in the database. This approach ensured that patient-level data were accurate and of high quality, whereas in prior studies clinical information was mainly obtained from administrative data [7, 19]. Most antibiotic susceptibility data were generated by an automated system (Vitek 2) that could have over- or underestimated MICs for some antibiotics in some episodes. However, this reflects real life, as in most hospitals physicians establish treatment on the basis of laboratory results generated using similar methods. Finally, clinical competency can contribute to the outcomes of patients with drug-resistant GN-BSI. However, this was not systematically assessed in our retrospective analysis.
To conclude, this is the first validation of DTR in a large non-US cohort of patients with GN-BSI. DTR seems a promising tool to identify challenging cases, mainly among patients with P. aeruginosa BSI. However, due to the high prevalence of CR in our study, mainly among K. pneumoniae and A. baumannii, the CR category was associated with the highest net reclassification improvement in predicting mortality. Further studies assessing the value of DTR in multinational European cohorts, considering also newly available drugs for CR infections, are encouraged.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support. No external funding was received for the present study.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. M.G.: study design, data analysis, and drafting the manuscript; L.B.: data collection and support in data analysis and drafting the manuscript; R.P.: revision of collected data; M.B.: revision of collected data; M.M.: data collection; L.P.: data collection; A.T.: data collection; G.F.: data collection; L.M.: support in data collection and analysis; S.A.: support in collection and revision of microbiological data; R.L.: data analysis and drafting the manuscript; P.V.: study design and manuscript revision.
References
- 1. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18:268–81. [DOI] [PubMed] [Google Scholar]
- 2. Tabah A, Koulenti D, Laupland K, et al. Characteristics and determinants of outcome of hospital-acquired bloodstream infections in intensive care units: the EUROBACT International Cohort Study. Intensive Care Med 2012; 38:1930–45. [DOI] [PubMed] [Google Scholar]
- 3. Zilberberg MD, Nathanson BH, Sulham K, et al. Multidrug resistance, inappropriate empiric therapy, and hospital mortality in Acinetobacter baumannii pneumonia and sepsis. Crit Care 2016; 20:221–31 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zak-Doron Y, Dishon Benattar Y, Pfeffer I, et al. ; AIDA Study Group The association between empirical antibiotic treatment and mortality in severe infections caused by carbapenem-resistant gram-negative bacteria: a prospective study. Clin Infect Dis 2018; 67:1815–23. [DOI] [PubMed] [Google Scholar]
- 5. de Kraker MEA, Sommer H, de Velde F, et al. ; COMBACTE-NET Consortium Optimizing the design and analysis of clinical trials for antibacterials against multidrug-resistant organisms: a white paper from COMBACTE’s STAT-Net. Clin Infect Dis 2018; 67:1922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. White paper: recommendations on the conduct of superiority and organism-specific clinical trials of antibacterial agents for the treatment of infections caused by drug-resistant bacterial pathogens. Clin Infect Dis 2012; 55:1031–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kadri SS, Adjemian J, Lai YL, et al. ; National Institutes of Health Antimicrobial Resistance Outcomes Research Initiative (NIH–ARORI) Difficult-to-treat resistance in Gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin Infect Dis 2018; 67:1803–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harris PNA, McNamara JF, Lye DC, et al. Proposed primary endpoints for use in clinical trials that compare treatment options for bloodstream infection in adults: a consensus definition. Clin Microbiol Infect 2017; 23:533–41. [DOI] [PubMed] [Google Scholar]
- 9. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 10. Friedman ND, Kaye KS, Stout JE, et al. Health care–associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 2002; 137:791–7. [DOI] [PubMed] [Google Scholar]
- 11. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016; 315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 2008; 36:309–32. [DOI] [PubMed] [Google Scholar]
- 13. Sundström J, Byberg L, Gedeborg R, et al. Useful tests of usefulness of new risk factors: tools for assessing reclassification and discrimination. Scand J Public Health 2011; 39:439–41. [DOI] [PubMed] [Google Scholar]
- 14. Echols RM, Tillotson GS. Difficult to treat: do we need a new definition? Clin Infect Dis 2019; 69:1641–2. [DOI] [PubMed] [Google Scholar]
- 15. Tumbarello M, Trecarichi EM, Corona A, et al. Efficacy of ceftazidime-avibactam salvage therapy in patients with infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Clin Infect Dis 2019; 68:355–64. [DOI] [PubMed] [Google Scholar]
- 16. Kadri SS, Danner RL. Reply to Raoult and Rolain, and to Echols and Tillotson. Clin Infect Dis. 2019; 69:1642–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pea F, Viale P. The antimicrobial therapy puzzle: could pharmacokinetic-pharmacodynamic relationships be helpful in addressing the issue of appropriate pneumonia treatment in critically ill patients? Clin Infect Dis 2006; 42:1764–71. [DOI] [PubMed] [Google Scholar]
- 18. Cassini A, Hogberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019; 19:129–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kadri SS, Lai YLE, Ricotta EE, et al. External validation of difficult-to-treat resistance prevalence and mortality risk in Gram-negative bloodstream infection using electronic health record data from 140 US hospitals. Open Forum Infect Dis 2019; 6:ofz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.