Abstract
BACKGROUND
Human polyomaviruses (HPyVs), like herpesviruses, cause persistent infection in a large part of the population. In immunocompromised and elderly patients, PyVs cause severe diseases such as nephropathy (BK polyomavirus [BKPyV]), progressive multifocal leukoencephalopathy (JC polyomavirus [JCPyV]), and skin cancer (Merkel cell polyomavirus [MCPyV]). Like cytomegalovirus, donor‐derived PyV can cause disease in kidney transplant recipients. Possibly blood components transmit PyVs as well. To study this possibility, as a first step we determined the presence of PyV DNA in Dutch blood donations.
STUDY DESIGN AND METHODS
Blood donor serum samples (n = 1016) were analyzed for the presence of DNA of 14 HPyVs using HPyV species‐specific quantitative polymerase chain reaction (PCR) procedures. PCR‐positive samples were subjected to confirmation by sequencing. Individual PCR findings were compared with the previously reported PyV serostatus.
RESULTS
MC polyomavirus DNA was detected in 39 donors (3.8%), JCPyV and TS polyomavirus (TSPyV) DNA in five donors (both 0.5%), and HPyV9 DNA in four donors (0.4%). BKPyV, WU polyomavirus (WUPyV), HPyV6, MW polyomavirus (MWPyV), and LI polyomavirus (LIPyV) DNA was detected in one or two donors. Amplicon sequencing confirmed the expected product for BKPyV, JCPyV, WUPyV, MCPyV, HPyV6, TSPyV, MWPyV, HPyV9, and LIPyV. For JCPyV a significant association was observed between detection of viral DNA and the level of specific IgG antibodies.
CONCLUSION
In 5.4% of Dutch blood donors PyV DNA was detected, including DNA from pathogenic PyVs such as JCPyV. As a next step, the infectivity of PyV in donor blood and transmission via blood components to immunocompromised recipients should be investigated.
ABBREVIATIONS
- BKPyV
BK polyomavirus
- Ct
cycle threshold
- HPyV(s)
human polyomaviruses
- JCPyV
JC polyomavirus
- LIPyV
LI polyomavirus
- KIPyV
KI polyomavirus
- MCPyV
Merkel cell polyomavirus
- MWPyV
MW polyomavirus
- NJPyV
NJ polyomavirus
- PML
progressive multifocal leukoencephalopathy
- PyV
polyomavirus
- qPCR
quantitative polymerase chain reaction
- STLPyV
STL polyomavirus
- TS
trichodysplasia spinulosa
- TSPyV
TS polyomavirus
- WUPyV
WU polyomavirus
Human polyomaviruses (HPyVs) cause asymptomatic persistent infection in healthy humans,1 whereas they can cause severe disease in immunocompromised patients and elderly persons. Latter groups increasingly receive blood components, although the presence of HPyVs in blood donors has not been studied extensively. Transfusion‐transmitted HPyV infection has not been reported, which can be explained by lack of such transmissions or by an erroneous assumption that HPyV‐related disease in immunocompromised patients always is caused by reactivation of their own, hitherto silent infection. In kidney transplant patients a substantial proportion of BK polyomavirus (BKPyV) infections and pathology is donor derived.2, 3
Polyomaviruses are ubiquitous viruses that frequently infect human beings. During childhood the seroprevalence of most HPyVs rapidly increases, sometimes reaching 100%.4, 5, 6, 7 PyVs can be detected in healthy persons, for example, in skin,8 urine,9 tonsillar tissue,10 and respiratory samples.11 Despite the persistence of these viruses, little is known about the occurrence of viremia in the healthy population, especially regarding the recently discovered HPyVs. In immunocompromised patients, HPyVs can be found also in blood and cerebrospinal fluid.12, 13 PyV‐associated diseases are increasingly relevant in the immunocompromised population. Two well‐known examples of PyV‐associated disease are BKPyV‐associated nephropathy14 and JC polyomavirus (JCPyV)‐associated progressive multifocal leukoencephalopathy (PML).15 Nowadays these severe conditions are primarily seen, respectively, in immunosuppressed kidney transplant recipients and patients on immunomodulatory drugs, such as multiple sclerosis patients taking natalizumab.15 In the past decade, with the identification of at least 10 novel HPyVs,8, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 the number of PyV‐associated diseases has increased and now includes Merkel cell carcinoma and trichodysplasia spinulosa (TS). Merkel cell carcinoma, caused by Merkel cell polyomavirus (MCPyV), is an aggressive, potentially lethal tumor that occurs in the elderly and in immunocompromised patients.16 TS, caused by TS polyomavirus (TSPyV), is a dysplastic and disfiguring skin disease that is especially found in solid organ transplant patients and lymphocytic leukemia patients.17 HPyV6 and ‐7 cause pruritic and dyskeratotic dermatoses in immunocompromised patients.28 KIPyV and WU polyomavirus (WUPyV) were first detected in human nasopharyngeal aspirates from patients with respiratory infection.18, 20 MWPyV and STLPyV were found in stool samples of healthy children.23, 27 HPyV9 was discovered in the serum of a kidney transplant patient.26 HPyV12, NJPyV, and LI polyomavirus (LIPyV) were all identified in human samples;21, 22, 24 however, seroprevalence of these viruses is low. Vaccination or proven effective antiviral therapy is not available for HPyVs.
HPyVs are non‐enveloped viruses, 40 to 50 nm in diameter, with circular double‐stranded DNA genomes. It can be expected that common pathogen reduction techniques used in blood banking have limited efficacy against HPyVs, because these viruses are non‐enveloped. HPyVs have been isolated from lymphocytes and hence leukoreduction of blood donations might decrease the presence of HPyVs in donated blood, but the extent of this reduction is unknown.29, 30, 31 It is uncertain whether higher levels of specific HPyV‐antibodies decrease potential infectivity by neutralization. On the one hand, kidney transplant recipients with a high antibody titer against BKPyV have a lower risk of developing BKPyV viremia compared to recipients with low antibody titers, but on the other hand kidney transplant patients have an increased risk of developing BKPyV viremia after receiving a kidney from a donor with high BKPyV antibody levels.3, 32 No group is fully protected and as such it seems likely that a seropositive transfusion recipient is not necessarily protected against PyV infection.
Since latent, persistent PyV infections bear a risk for the immunocompromised, one can wonder about the contribution of blood components as a vehicle for HPyV transmission. To start answering this question, we recently determined the seroprevalence of all known, thus far 14, HPyVs in a large group of blood donors and estimated that each blood donor is persistently infected with on average nine HPyVs.4 To further explore the risk from these potentially blood‐transmitted viruses, in this study we analyzed the same blood donor cohort by HPyV‐specific polymerase chain reaction (PCR) procedures for the presence of circulating genomic DNA of all currently known HPyVs.4
MATERIALS AND METHODS
DNA extraction
Nucleic acid extraction was performed on a nucleic acid purification instrument (MagNA Pure LC, Roche Diagnostics) using a large‐volume DNA isolation kit (MagNA Pure LC, Roche Diagnostics), according to the manufacturer's instructions, with an input volume of 1000 μL and an output volume of 65 μL. Extraction efficiency and PCR inhibition was controlled by adding a fixed concentration of phocine herpesvirus (PhHV) DNA to the lysis buffer that was added to each sample.33
PyV DNA detection
Each sample was analyzed for the presence of HPyV genomic DNA with the help of three real‐time multiplex quantitative PCR (qPCR) procedures (Multiplex 1, 2, and 3), developed to detect 14 PyVs (Table 1). The PCR procedures for BKPyV, HPyV6, HPyV7, TSPyV, and HPyV9 were previously designed and described.34, 35, 36 The PCR procedures for JCPyV, WUPyV, and MCPyV were developed by other research groups.37, 38, 39 Novel primers and probes were designed for KIPyV, MWPyV, STLPyV, HPyV12, NJPyV, and LIPyV using computer software (Geneious, Version 10.2.4, Biomatters; Table 1 and Table S1, available as supporting information in the online version of this paper). Multiplex 1 was developed to detect MCPyV, HPyV6, HPyV7, TSPyV, and HPyV9; Multiplex 2 to detect BKPyV, WUPyV, MWPyV, and NJPyV (and the internal control phocine herpesvirus); and Multiplex 3 to detect JCPyV, KIPyV, STLPyV, HPyV12, and LIPyV.
Table 1.
PyV PCR primers and probes
| Multiplex | Target species | Refseq no. | Gene | Product length (bp) | Sense primer sequence (5′‐3′) | Probe sequence (5′‐3′) | Antisense primer sequence (5′‐3′) |
|---|---|---|---|---|---|---|---|
| 1 | MCPyV | NC_010277 | LT | 149 | CCACAGCCAGAGCTCTTCCT | CY5‐TCCCAGGCTTCAGACTCCCA* | TGGTGGTCTCCTCTCTGCTACTG |
| 1 | HPyV6 | NC_014406 | VP1 | 150 | GTAGGGTATGCTGGTAAC | YAK‐CTCTCCTCTGTCTGAAGTGAACTCTAA | CAGGAATTGTCTAAACATCATATC |
| 1 | HPyV7 | NC_014407 | VP1 | 116 | GTGCTGATATGGTTGGAA | TXR‐AGCCTGTACTGTTCTCTGGTTACT | TCTGCAGTGGACTCTAAA |
| 1 | TSPyV | NC_014361 | VP1 | 104 | GAGTCTAAGGACAACTATGG | Q705‐CTTGTCCTGGTCACTGCTGTT | CTAGCTGTACTGTAGGTTG |
| 1 | HPyV9 | NC_015150 | VP1 | 109 | CCTGTAAGCTCTCTCCTTA | FAM‐CTTGTTCTCTGGTCTTATGCCTCA | CCTGATAAATTCTGACTTCTTC |
| 2 | BKPyV | NC_001538 | VP1 | 90 | GAAAAGGAGAGTGTCCAGGG | FAM‐CCAAAAAGCCAAAGGAACCC | GAACTTCTACTCCTCCTTTTATTAGT |
| 2 | WUPyV | NC_009539 | VP1 | 74 | AACCAGGAAGGTCACCAAGAAG | TXR‐CAACCCACAAGAGTGCAAAGCCTTCC | CTACCCCTCCTTTTCTGACTTGTTT |
| 2 | MWPyV | NC_018102 | VP1 | 86 | GACACCACAATGACAGTTGAG | CY5‐CCAAGGATGGGCAATGATGTAAAAACA | GGATCACTGTAGCCATACCAT |
| 2 | NJPyV | NC_024118 | VP1 | 135 | CCCACCAAGTAAAGTAAC | YAK‐AAGTGTCCTATACCTACTCCAGTGC | CAGAGTTCAATTTCAGTAGTA |
| 3 | JCPyV | NC_001699 | LT | 129 | GTCTCCCCATACCAACATTAGCTT | YAK‐TCTTTCCACTGCACAATCCTCTCATGAATG | GGTTTAGGCCAGTTGCTGACTT |
| 3 | KIPyV | NC_009238 | VP1 | 148 | AAGTTCCCCGGGTACAAACTC | TXR‐GGTAGAAGTACTAGCCGCAGTACCACTGT | CCATCCTGAGCAGCTGTTGTA |
| 3 | STLPyV | NC_020106 | VP1 | 101 | TTGAAAATGGCTCCAAAAAGAAAATCT | CY5‐ AGATGCACCTCACAGACATGTCCAATGGA | TGGCACGGATCATATTCACATCT |
| 3 | HPyV12 | NC_020890 | VP1 | 139 | AAGGGCTGTAAGAAATCC | FAM‐CCAGTATCTGCTCTCCTAACCAGT | CTCCAAACCCTCATATACC |
| 3 | LIPyV | NC_034253 | VP1 | 83 | TGACAGGTGACAATTCCCAGG | Q705‐AGAGGAAGTACGCGTCTATGATGGCAGAG | CCTTGGCAGATCTAACCCTCC |
Probe modified from original article, Goh S, Lindau C, Tiveljung‐Lindell A, et al. Emerg Infect Dis 2009;15:489–91.38
AS = antisense; LT = large T; S = sense; VP1 = viral protein 1.
The PCR mix (total volume, 25 μL) consisted of a master mix kit (HotStarTaq, Qiagen), MgCl2, primers, probes (see Table 1 for concentrations) and 10 μL of input DNA isolate. Cycling conditions for the PCR procedures were as follows: 95°C for 15 minutes, followed by 45 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. qPCR procedures were performed on a real‐time PCR detection system (Model CFX96, Bio‐Rad Laboratories). Analysis of the qPCR data was performed with computer software (CFX Manager, Version 3.1, Bio‐Rad Laboratories). Baseline threshold values were determined separately for each target, and fluorescence drift correction was applied.
Polymerase chain reaction efficiency and analytical sensitivity of each PyV PCR were determined on replicates of serial dilution series of 10,000 to 1 copy per reaction of a plasmid containing a single cloned copy of the PyV target gene (VP1 or Large T‐antigen) and was defined as the ability of the assay to detect the target concentration with a probability higher than 95% in a number of replicates (Tables S2 and S3, available as supporting information in the online version of this paper).
PCR product sequencing
Polymerase chain reaction products amplified with a cycle threshold (Ct) value below 40 were analyzed by Sanger sequencing for confirmation, with a maximum of 10 positive samples (amplicons) per HPyV. The generated PCR products were run on a 2% agarose gel. Bands of the expected size (74‐150 bp) were isolated using a PCR and gel kit (Isolate II, Bioline Reagents), ligated, and cloned in Escherichia coli using a cloning kit (TOPO TA, Thermo Fisher Scientific), according to manufacturer's instructions. For each successful ligation, three colonies per plate were picked, and plasmid DNA was isolated with an isolation kit (NucleoSpin Plasmid EasyPure, Macherey‐Nagel). Sanger sequencing was performed on a DNA analyzer (ABI3730xl, Thermo Fisher Scientific) using M13 forward primer.
Study population
The study population consisted of 1050 serum samples from healthy Dutch blood donors. The samples were previously used for routine blood donor screening for human immunodeficiency virus, hepatitis B and C virus, and syphilis.4 Of 1050 samples, 34 were excluded due to insufficient volume for DNA extraction or due to inhibition in qPCR (Fig. 1). The presence of PyV antibodies in this sample set was determined previously. On average, a donor from this population is seropositive for nine different PyV species and seropositivity ranged from 5% to 100% depending on PyV species.4 Basic population demographics (age and sex) of the fully screened donor population are shown in Table 2. Samples from all regions of the Netherlands were included, as reported previously.4
Figure 1.
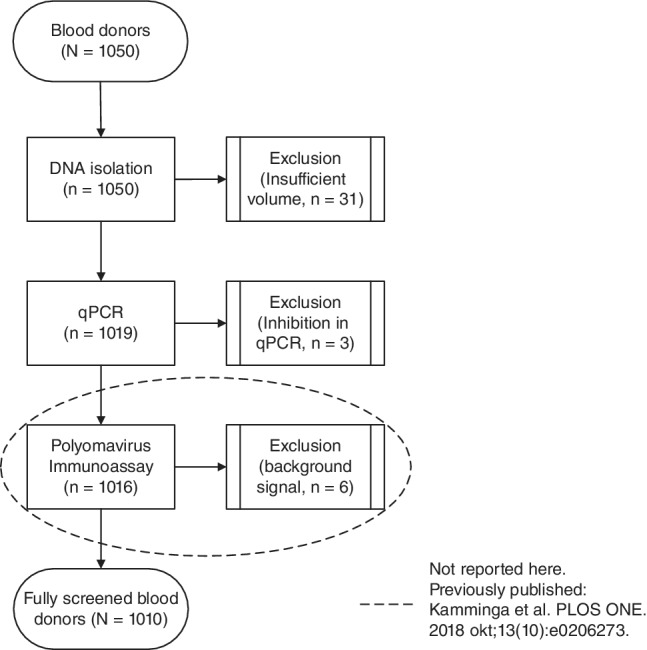
Flow chart for study population. Numbers in parentheses indicate serum samples that were successfully isolated, PCR amplified, and assessed with the immunoassay. Boxes on the right side of the figure state reasons for exclusion of samples.
Table 2.
Overview of HPyV PCR and sequence results of serum samples from 1016 blood donors
| PCR target | No. of PCR‐positive donors | PCR product sequencing | No. of positives per sex and age category | Viral load (copies/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Any Ct, n (%) | Ct < 40, n (%) | Successfully sequenced/total sequenced | HPyV confirmed/successfully sequenced (%) | Sex | Age category (years) | |||||||
| Male (n = 501) | Female (n = 509) | 18‐29 (n = 197) | 30‐39 (n = 201) | 40‐49 (n = 202) | 50‐59 (n = 206) | 60‐69 (n = 204) | ||||||
| BKPyV | 5 (0.5) | 1 (0.1) | 1/1 | 1/1 (100) | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 55 |
| JCPyV | 14 (1.4) | 5 (0.5) | 5/5 | 5/5 (100) | 2 | 3 | 0 | 1 | 1 | 2 | 1 | 9‐37 |
| KIPyV | 2 (0.2) | 1 (0.1) | 1/1 | 0/1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 46 |
| WUPyV | 2 (0.2) | 2 (0.2) | 2/2 | 1/2 (50) | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 8‐30 |
| MCPyV | 63 (6.2) | 39 (3.8) | 9/10 | 7/9 (78) | 23 | 16 | 5 | 5 | 12 | 7 | 10 | 24‐452 |
| HPyV6 | 4 (0.4) | 1 (0.1) | 1/1 | 1/1 (100) | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 13 |
| HPyV7 | 7 (0.7) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| TSPyV | 7 (0.7) | 5 (0.5) | 5/5 | 5/5 (100) | 4 | 1 | 0 | 2 | 2 | 1 | 0 | 9‐81 |
| HPyV9 | 6 (0.6) | 4 (0.4) | 3/4 | 1/3 (33) | 3 | 1 | 0 | 2 | 1 | 1 | 0 | 7‐68 |
| MWPyV | 3 (0.3) | 1 (0.1) | 1/1 | 1/1 (100) | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 15 |
| STLPyV | 1 (0.1) | 1 (0.1) | 1/1 | 0/1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 39 |
| HPyV12 | 11 (1.1) | 10 (1.0) | 10/10 | 0/10 | 6 | 4 | 0 | 2 | 3 | 4 | 1 | 2‐13 |
| NJPyV | 3 (0.3) | 1 (0.1) | 1/1 | 0/1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 123 |
| LIPyV | 6 (0.6) | 2 (0.2) | 2/2 | 2/2 (100) | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 13‐31 |
| Any PyV* | 111 (10.9) | 64 (6.3) | 55 (5.4)† | 45 | 28 | 6 | 16 | 21 | 17 | 13 | 55‡ | |
Codetection of multiple PyVs in a single donor counts as one for the total number of positive donors.
Total number of true‐positive donors based on positive PCR results with Ct value of less than 40 and sequence confirmation of at least one PCR product.
Median viral load.
Each blood donor gave permission to use residual blood samples for studies of blood‐borne agents. Hence, Sanquin's scientific board and the secretary of Sanquin's ethical advisory board decided that for this study additional permission from the ethical advisory board is not needed. The blood donors fulfilled all criteria for blood donation eligibility.
PyV serology
The PyV serostatus of all blood donors was determined and described in a previous study,4 using a multiplex immunoassay as previously described,40 employing a GST‐VP1 fusion protein for each PyV as antibody‐binding antigen.
Statistical analysis
Statistics were performed with computer software (SPSS Statistics, Version 23, IBM Corp.). Chi‐square tests were used to compare PCR results and seropositivity, age category, or sex. Mann‐Whitney U tests were used to compare seroreactivity results between samples positive or negative in qPCR analysis.
RESULTS
PyV PCR validation
The analytical sensitivity was 10 to 15 copies/reaction for all PCR procedures, except for the MCPyV PCR, which reliably detects 100 copies/reaction, although the dilution with 10 copies/reaction was detected in 90% of cases (Table S2, available as supporting information in the online version of this paper). High concentrations of nontarget PyV DNA with a Ct value between 25 and 30) did not inhibit the PCR (Table S4A‐N, available as supporting information in the online version of this paper). In addition, a panel of common double‐stranded DNA viruses containing herpes simplex virus 1 and 2, varicella zoster virus, cytomegalovirus, Epstein‐Barr virus, and adenovirus was tested negative in each HPyV PCR (data not shown). In short, all HPyV PCR procedures detect their target in a sensitive and specific manner.
Presence of PyV DNA in blood donors
Serum samples from 1016 blood donors were analyzed for the presence of HPyV DNA using three multiplex PCR procedures. In Table 2 the PCR results are summarized. MCPyV DNA was the most prevalent, detected in 39 of 1016 (3.8%) donors, with a viral load ranging between 24 and 452 genome equivalent copies/mL. Sequencing confirmed the presence of the MCPyV DNA in the PCR product in most samples (78%; Table 2). JCPyV, TSPyV, and HPyV9 were detected, respectively, in five (0.5%; range, 9‐37 copies/mL), five (0.5%; range, 9‐81 copies/mL), and four (0.4%; range, 7‐68 copies/mL) donors. Sequencing confirmed the presence of virus‐specific DNA in 100% of cases for JCPyV and TSPyV and in 33% for HPyV9. When sequencing was successful but HPyV‐specific sequences were not present, especially human genomic DNA and primer‐dimers were detected (Table 2). For example, the HPyV12‐positive findings in 10 donors with a low range of 2 to 13 copies/mL could not be not confirmed at all by sequencing. The other HPyVs were detected in only one or two donors, while HPyV7 was not detected at all. Summarizing, we found 64 donors to be HPyV PCR positive (6.3%), of which the detection of specific viral DNA was confirmed (in part) for BKPyV, JCPyV, WUPyV, MCPyV, HPyV6, TSPyV, HPyV9, MWPyV, and LIPyV in 55 blood donors (5.4%). HPyV codetection was observed in four donors (0.4%) and involved TSPyV, HPyV9, and LIPyV (all sequence‐verified); WUPyV, TSPyV, and HPyV9 (WUPyV and TSPyV sequence verified); TSPyV, KIPyV, and NJPyV (TSPyV sequence verified); and TSPyV and HPyV9 (TSPyV sequence verified), respectively. The distribution of PyV detection over sex and age category is summarized in Table 2. For none of the HPyVs a correlation was found between the detection in serum and sex or age category of the donor.
Previously we analyzed every sample included in this study serologically for HPyV infection,4 which enabled us to compare the HPyV PCR findings with HPyV serostatus (seropositivity) and seroreactivity (the median seroresponse given as median fluorescence intensity value; Table 3). In case of BKPyV, JCPyV, KIPyV, WUPyV, MCPyV, HPyV6, TSPyV, and MWPyV, 77% to 100% of the positive PCR findings were obtained in donors seropositive for the detected HPyV. The HPyV9, STLPyV, HPyV12, NJPyV, and LIPyV DNA‐positive samples, however, were all from donors seronegative for the HPyV that was detected. No significant correlation was found between presence of PyV DNA and seropositivity. Seroreactivity was comparable between DNA‐positive and ‐negative samples for all HPyVs, except JCPyV where significantly higher seroresponses were measured in the JCPyV DNA–positive samples (Mann‐Whitney U test, p = 0.005; Table 3).
Table 3.
Overview of PyV serostatus and DNAemia in fully screened population (n = 1010)
| PyV | Seroprevalence* | Seropositives among PCR positives (%) | Median seroreactivity in MFI among PCR positives | Median seroreactivity in MFI among PCR negatives | p value† |
|---|---|---|---|---|---|
| BKPyV | 99 | 1/1 (100) | 22,519 | 18,936 | 0.635 |
| JCPyV | 62 | 5/5 (100) | 8,382 | 828 | 0.005 |
| KIPyV | 92 | 1/1 (100) | 11,956 | 9,755 | 0.775 |
| WUPyV | 99 | 2/2 (100) | 20,796 | 12,170 | 0.108 |
| MCPyV | 82 | 30/39 (77) | 9,799 | 6,297 | 0.239 |
| HPyV6 | 83 | 1/1 (100) | 3,022 | 8,140 | 0.575 |
| HPyV7 | 71 | ||||
| TSPyV | 79 | 4/5 (80) | 716 | 6,575 | 0.391 |
| HPyV9 | 19 | 0/4 (0) | −231 | −69 | 0.101 |
| MWPyV | 100 | 1/1 (100) | 11,023 | 10,425 | 0.909 |
| STLPyV | 65 | 0/1 (0) | −263 | 873 | 0.126 |
| HPyV12 | 4 | 0/10 (0) | −288 | −259 | 0.596 |
| NJPyV | 5 | 0/1 (0) | 367 | 204 | 0.466 |
| LIPyV | 6 | 0/2 (0) | −143 | −224 | 0.514 |
Overall percentage seroprevalence, as previously described by Kamminga et al. PLOS ONE. 2018 okt;13 (10):e0206273.
Mann‐Whitney U test (p value < 0.05 was considered significant).
MFI = median fluorescence intensity.
DISCUSSION
In this study we determined the presence of PyV DNA in serum samples taken from healthy Dutch blood donors. Our results show that the prevalence of PyV DNA varies from 0% to 3.8% depending on HPyV species. Importantly, we detected DNA from known pathogenic PyVs, BKPyV (0.1%), JCPyV (0.5%), MCPyV (3.8%), and TSPyV (0.5%), which suggests that these viruses may be present in blood components.
The prevalence of PyV DNA (5.4%) was based on PCR amplification (with Ct values <40) and sequence confirmation of at least one amplicon per HPyV, which was obtained for BKPyV, JCPyV, WUPyV, MCPyV, HPyV6, TSPyV, HPyV9, MWPyV, and LIPyV. Since blood donors in the Netherlands are selected on optimal health and minimal risk exposure (among others to infectious diseases), we believe that our estimation of HPyV presence is a minimum estimator of HPyV prevalences in the general adult Dutch population and probably in other western populations as well, as little differences in PyV seroprevalence are observed between these populations.4, 5, 6, 7
A strength of this study is the inclusion of many blood donors and all 14 currently described HPyVs. Except for MCPyV (see below), the most likely explanation for our findings is that the implicated blood donors were viremic at the time of blood collection. However, HPyV DNAemia as the result of disintegrating persistently infected cells cannot be excluded, which could be relevant for BKPyV and JCPyV that have been found in peripheral blood mononuclear cells from healthy persons.29, 30, 41 In a study of 400 plasma samples of American blood donors, BKPyV and JCPyV DNA was not detected,42 which might be explained by the smaller size of the study. Alternatively, technical or geographic differences could account for the negative outcome in that study.
Since HPyV infections are generally acquired during childhood,5, 6 HPyV‐detections probably result from persistent HPyV infections. Although primary infection is a possible explanation for PCR‐positive donors that are seronegative. Whether our findings result from continuous HPyV viremia, which occasionally exceeds the lower limit of PCR detection, or from an occasional viremic episode in the background of an otherwise latent infection cannot be deduced from our cross‐sectional data set. Furthermore, infectivity of the suspected HPyV, or loss of infectivity after nuclease treatment, to assess the presence of intact virions, will be difficult because of the detected low viral load levels (median, 55 copies/mL serum). Potential infectivity of blood components could be assessed by documented seroconversion or an increase in seroreactivity in the recipient after administration of blood components.
MC polyomavirus was detected in 3.8% of serum samples in our study, which is comparable to findings in other studies. For example, a study of 190 blood donors reported MCPyV in 2.6% of sera43 and another study of 621 sera from 394 elderly hospitalized patients older than 65 years of age found a prevalence of 9.9% for MCPyV,44 which suggests that the prevalence may increase with age. MCPyV has been detected in other blood compartments, for example, in 22% of buffy coats from blood donors.45 Interestingly, MCPyV was detected with whole‐genome sequencing as part of the blood virome46 and also with metagenomics in blood components eligible for transfusion.47, 48 KIPyV and WUPyV DNA have previously been reported in plasma from blood donors with prevalence ranging from 0.5% to 3.1% for KIPyV and 0.8% for WUPyV.49, 50 This is slightly higher than our finding of 0.1 and 0.2% in serum for KIPyV and WUPyV, respectively. Prevalence data in serum from healthy individuals for the other PyVs is currently lacking.
We consider it likely that a substantial part of the relatively high number of MCPyV PCR positives is explained by the high prevalence (>50%) of MCPyV (DNA) on skin of healthy individuals, as reported in several publications.51, 52, 53 During the hollow‐needle venipuncture, before the blood is actually collected, a small “biopsy” of skin tissue is punched that could act as a source of virus. For TSPyV, however, this scenario is unlikely, as it is barely found on the skin of asymptomatic immunocompetent and immunocompromised individuals.51 In addition to the potential “contamination” of donor blood through the skin punch, there is a theoretical risk of MCPyV contamination by blood bank and laboratory personnel, who carry MCPyV as well.
The seroprevalence of each PyV was determined within the same sample set in a previous study.4 Despite a high concordance (≥77%) between DNA positivity and seropositivity for most prevalent PyVs, we found no significant correlation between the two. This lack of association is likely caused by the low number of PCR positives among the generally high number of seropositives. The HPyV9‐, STLPyV‐, NJPyV‐, and LIPyV‐positive donors were seronegative for these PyVs, which could be explained by primary infection or a lack of productive infection. The latter seems likely for NJPyV and LIPyV as these viruses may not have humans as their primary host.4 For JCPyV we did observe an association between the height of the seroresponse and JCPyV DNA detections. An association between viral load and seroreactivity was previously observed for both BKPyVs in kidney transplant patients12, 32 and for JCPyV, where individuals with high seroreactivity had higher viral loads compared to individuals with low seroreactivity.54 JCPyV serology is also used as risk marker for PML.55 This suggests that there is an association between JCPyV viral load and JCPyV serology, both in healthy individuals and in patients at risk for PML.
Some limitations of this study include the chance of misclassification by sample contamination and the chance of erroneous detection. In addition, this study shows the presence of viral DNA, rather than the presence of encapsidated, infectious viral particles. The risk of lab contamination is reduced by storing and preparing reagents in separate rooms, using disposables and using no‐template controls. To further limit the chance of erroneous detection, prevalence calculations were based only on positive PCR results with a Ct value of less than 40. Furthermore, amplicon sequencing of samples (with a maximum of 10) with a Ct value below 40 was performed to check for presence of the expected product. For most PCR procedures the expected product was detected, although sometimes detection was difficult, for example, in case of codetection (TSPyV and HPyV9). Out of curiosity, we analyzed several very weak PCR‐positive samples and could confirm the presence of JCPyV‐, HPyV6‐, and TSPyV‐specific DNA in some of those samples (data not shown). For HPyV12, in all 10 PCR‐positive samples human genomic DNA was detected, which is probably amplified in a non‐specific manner, because of the absence of specific DNA template. Furthermore, the finding of a PyV similar to HPyV12 in shrews56 combined with a reported low seroprevalence4 suggests little circulation of this virus in humans.
In summary, DNA of HPyVs was detected in 5.4% of serum samples from a large cross‐section of Dutch blood donors. The detection of PyV DNA in these samples suggests that PyVs are present in blood components eligible for transfusion, which should be further investigated using infectivity assays and a donor–recipient transmission study.
CONFLICTS OF INTEREST
The authors have disclosed no conflicts of interest.
Supporting information
Table S1. Oligonucleotide and MgCl2 concentrations.
Table S2. PCR efficiency and limit of detection.
Table S3. Polyomavirus plasmids.
Table S4. Competition experiments
MF and HZ contributed equally.
Funded by Sanquin Blood Supply Foundation.
REFERENCES
- 1. Moens U, Krumbholz A, Ehlers B, et al. Biology, evolution, and medical importance of polyomaviruses: an update. Infect Genet Evol 2017;54:18‐38. [DOI] [PubMed] [Google Scholar]
- 2. Wunderink HF, van der Meijden E, van der blij‐de Brouwer CS, et al. BK‐polyomavirus seroreactivity measured in kidney donors is strongly associated with incidence of viremia and nephropathy in their recipients. J Clin Virol 2015;70:S19‐20. [Google Scholar]
- 3. Bohl DL, Storch GA, Ryschkewitsch C, et al. Donor origin of BK virus in renal transplantation and role of HLA C7 in susceptibility to sustained BK viremia. Am J Transplant 2005;5:2213‐21. [DOI] [PubMed] [Google Scholar]
- 4. Kamminga S, van der Meijden E, MCW F, et al. Seroprevalence of fourteen human polyomaviruses determined in blood donors. PLoS One 2018;13:e0206273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van der Meijden E, Bialasiewicz S, Rockett RJ, et al. Different serologic behavior of MCPyV, TSPyV, HPyV6, HPyV7 and HPyV9 polyomaviruses found on the skin. PLoS One 2013;8:e81078 [cited 2015 Oct 5]. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3836759/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kean JM, Rao S, Wang M, et al. Seroepidemiology of human polyomaviruses. PLoS Pathog 2009;5:e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gossai A, Waterboer T, Nelson HH, et al. Seroepidemiology of human polyomaviruses in a US population. Am J Epidemiol 2016;183:61‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schowalter RM, Pastrana DV, Pumphrey KA, et al. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 2010;7:509‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ling PD, Lednicky JA, Keitel WA, et al. The dynamics of herpesvirus and polyomavirus reactivation and shedding in healthy adults: a 14‐month longitudinal study. J Infect Dis 2003;187:1571‐80. [DOI] [PubMed] [Google Scholar]
- 10. Sadeghi M, Wang Y, Ramqvist T, et al. Multiplex detection in tonsillar tissue of all known human polyomaviruses. BMC Infect Dis 2017;17:409 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5465560/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rockett RJ, Bialasiewicz S, Mhango L, et al. Acquisition of human polyomaviruses in the first 18 months of life. Emerg Infect Dis 2015;21:365‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wunderink HF, van der Meijden E, van der blij‐de Brouwer CS, et al. Stability of BK polyomavirus IgG seroreactivity and its correlation with preceding viremia. J Clin Virol 2017;90:46‐51. [DOI] [PubMed] [Google Scholar]
- 13. Ferretti F, Bestetti A, Yiannoutsos CT, et al. Diagnostic and prognostic value of JC virus DNA in plasma in progressive multifocal leukoencephalopathy. Clin Infect Dis 2018;67:65‐72. [DOI] [PubMed] [Google Scholar]
- 14. Hirsch HH, Steiger J. Polyomavirus BK. Lancet Infect Dis 2003;3:611‐23. [DOI] [PubMed] [Google Scholar]
- 15. Major EO. Progressive multifocal leukoencephalopathy in patients on immunomodulatory therapies. Annu Rev Med 2010;61:35‐47. [DOI] [PubMed] [Google Scholar]
- 16. Feng H, Shuda M, Chang Y, et al. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science 2008;319:1096‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van der Meijden E, Janssens RWA, Lauber C, et al. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog 2010;6:e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allander T, Andreasson K, Gupta S, et al. Identification of a third human polyomavirus. J Virol 2007;81:4130‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buck CB, Phan GQ, Raiji MT, et al. Complete genome sequence of a tenth human polyomavirus. J Virol 2012;86:10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaynor AM, Nissen MD, Whiley DM, et al. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog 2007;3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gheit T, Dutta S, Oliver J, et al. Isolation and characterization of a novel putative human polyomavirus. Virology 2017;506:45‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korup S, Rietscher J, Calvignac‐Spencer S, et al. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One 2013;8:e58021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lim ES, Reyes A, Antonio M, et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 2013;436:295‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mishra N, Pereira M, Rhodes RH, et al. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J Infect Dis 2014;210:1595‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sauvage V. Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg Infect Dis 2011;17:1364‐70 [cited 2015 May 26]. Available from: http://wwwnc.cdc.gov/eid/article/17/8/11-0278_article.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scuda N, Hofmann J, Calvignac‐Spencer S, et al. A novel human polyomavirus closely related to the African green monkey‐derived lymphotropic polyomavirus. J Virol 2011;85:4586‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siebrasse EA, Reyes A, Lim ES, et al. Identification of MW Polyomavirus, a Novel Polyomavirus in Human Stool. J Virol 2012. Oct 1;86:10321‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nguyen KD, Lee EE, Yue Y, et al. Human polyomavirus 6 and 7 are associated with pruritic and dyskeratotic dermatoses. J Am Acad Dermatol 2017;76:932‐940.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dolei A, Pietropaolo V, Gomes E, et al. Polyomavirus persistence in lymphocytes: prevalence in lymphocytes from blood donors and healthy personnel of a blood transfusion centre. J Gen Virol 2000;81:1967‐73. [DOI] [PubMed] [Google Scholar]
- 30. Delbue S, Tremolada S, Elia F, et al. Lymphotropic polyomavirus is detected in peripheral blood from immunocompromised and healthy subjects. J Clin Virol 2010;47:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frohman EM, Monaco MC, Remington G, et al. JC virus in CD34+ and CD19+ cells in patients with multiple sclerosis treated with natalizumab. JAMA Neurol 2014;71:596‐602. [DOI] [PubMed] [Google Scholar]
- 32. Wunderink HF, van der Meijden E, van der blij‐de Brouwer CS, et al. Pretransplantation donor‐recipient pair seroreactivity against BK polyomavirus predicts viremia and nephropathy after kidney transplantation. Am J Transplant 2017;17:161‐72. [DOI] [PubMed] [Google Scholar]
- 33. Niesters HGM. Quantitation of viral load using real‐time amplification techniques. Methods 2001;25:419‐29. [DOI] [PubMed] [Google Scholar]
- 34. van der Meijden E, Wunderink HF, van der blij‐de Brouwer CS, et al. Human polyomavirus 9 infection in kidney transplant patients. Emerging Infect Dis 2014;20:991‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Purdie KJ, Proby CM, Rizvi H, et al. The role of human papillomaviruses and polyomaviruses in BRAF‐inhibitor induced cutaneous squamous cell carcinoma and benign squamoproliferative lesions. Front Microbiol 2018;9:1806 [cited 2019 Apr 4]. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01806/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van der Meijden E, Horváth B, Nijland M, et al. Primary polyomavirus infection, not reactivation, as the cause of trichodysplasia spinulosa in immunocompromised patients. J Infect Dis 2017;215:1080‐4 jiw403. [DOI] [PubMed] [Google Scholar]
- 37. Pal A, Sirota L, Maudru T, et al. Real‐time, quantitative PCR assays for the detection of virus‐specific DNA in samples with mixed populations of polyomaviruses. J Virol Methods 2006. Jul;135:32‐42. [DOI] [PubMed] [Google Scholar]
- 38. Goh S, Lindau C, Tiveljung‐Lindell A, et al. Merkel cell polyomavirus in respiratory tract secretions. Emerg Infect Dis 2009;15:489‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rao S, Garcea RL, Robinson CC, et al. WU and KI polyomavirus infections in pediatric hematology/oncology patients with acute respiratory tract illness. J Clin Virol 2011;52:28‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kamminga S, van der Meijden E, Wunderink HF, et al. Development and evaluation of a broad bead‐based multiplex immunoassay to measure IgG seroreactivity against human polyomaviruses. J Clin Microbiol 2018;56:e01566‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bialasiewicz S, Whiley DM, Lambert SB, et al. Detection of BK, JC, WU, or KI polyomaviruses in faecal, urine, blood, cerebrospinal fluid and respiratory samples. J Clin Virol 2009;45:249‐54. [DOI] [PubMed] [Google Scholar]
- 42. Egli A, Infanti L, Dumoulin A, et al. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis 2009;199:837‐46. [DOI] [PubMed] [Google Scholar]
- 43. Mazzoni E, Rotondo JC, Marracino L, et al. Detection of Merkel cell polyomavirus DNA in serum samples of healthy blood donors. Front Oncol 2017;7:294 [cited 2017 Dec 19]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5712532/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sadeghi M, Aronen M, Chen T, et al. Merkel cell polyomavirus and trichodysplasia spinulosa‐associated polyomavirus DNAs and antibodies in blood among the elderly. BMC Infect Dis 2012;12:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pancaldi C, Corazzari V, Maniero S, et al. Merkel cell polyomavirus DNA sequences in the buffy coats of healthy blood donors. Blood 2011;117:7099‐101. [DOI] [PubMed] [Google Scholar]
- 46. Moustafa A, Xie C, Kirkness E, et al. The blood DNA virome in 8,000 humans. PLoS Pathog 2017;13:e1006292 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5378407/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lau P, Cordey S, Brito F, et al. Metagenomics analysis of red blood cell and fresh‐frozen plasma units. Transfusion. 2017;57:1787‐800. [DOI] [PubMed] [Google Scholar]
- 48. Brito F, Cordey S, Delwart E, et al. Metagenomics analysis of the virome of 300 concentrates from a Swiss platelet bank. Vox Sang 2018;113:601‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Babakir‐Mina M, Ciccozzi M, Farchi F, et al. KI and WU Polyomaviruses and CD4+ cell counts in HIV‐1–infected patients, Italy. Emerg Infect Dis 2010;16:1482‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Touinssi M, Galicher V, de Micco P, et al. Molecular epidemiology of KI and WU polyomaviruses in healthy blood donors, south‐eastern France. J Med Virol 2013;85:1444‐6. [DOI] [PubMed] [Google Scholar]
- 51. Hampras SS, Giuliano AR, Lin H‐Y, et al. Natural history of polyomaviruses in men: the HPV infection in men (HIM) study. J Infect Dis 2015;211:1437‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wieland U, Mauch C, Kreuter A, et al. Merkel cell polyomavirus DNA in persons without Merkel cell carcinoma. Emerg Infect Dis 2009. Sep;15:1496‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hashida Y, Nakajima K, Nakajima H, et al. High load of Merkel cell polyomavirus DNA detected in the normal skin of Japanese patients with Merkel cell carcinoma. J Clin Virol 2016;82:101‐7. [DOI] [PubMed] [Google Scholar]
- 54. Berger JR, Miller CS, Danaher RJ, et al. Distribution and quantity of sites of John Cunningham virus persistence in immunologically healthy patients: correlation with John Cunningham virus antibody and urine John Cunningham virus DNA. JAMA Neurol 2017;74:437‐44. [DOI] [PubMed] [Google Scholar]
- 55. White MK, Sariyer IK, Gordon J, et al. Diagnostic assays for polyomavirus JC and progressive multifocal leukoencephalopathy. Rev Med Virol 2016;26:102‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gedvilaite A, Tryland M, Ulrich RG, et al. Novel polyomaviruses in shrews (Soricidae) with close similarity to human polyomavirus 12. J Gen Virol 2017; 98: 3060‐3067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Oligonucleotide and MgCl2 concentrations.
Table S2. PCR efficiency and limit of detection.
Table S3. Polyomavirus plasmids.
Table S4. Competition experiments


