Summary
Awns, bristle‐like structures extending from grass lemmas, provide protection against predators, contribute to photosynthesis and aid in grain dispersal. In wheat, selection of awns with minimal extension, termed awnletted, has occurred during domestication by way of loci that dominantly inhibit awn development, such as Tipped1 (B1), Tipped2 (B2), and Hooded (Hd). Here we identify and characterize the B1 gene.
B1 was identified using bulked segregant RNA‐sequencing of an F2 durum wheat population and through deletion mapping of awned bread wheat mutants. Functional characterization was accomplished by gene overexpression while haplotype analyses assessed B1 polymorphisms and genetic variation.
Located on chromosome 5A, B1 is a C2H2 zinc finger encoding gene with ethylene‐responsive element binding factor‐associated amphiphilic repression (EAR) motifs. Constitutive overexpression of B1 in awned wheat produced an awnletted phenotype with pleiotropic effects on plant height and fertility. Transcriptome analysis of B1 overexpression plants suggests a role as transcriptional repressor, putatively targeting pathways involved in cell proliferation. Haplotype analysis revealed a conserved B1 coding region with proximal polymorphisms and supported the contention that B1 is mainly responsible for awnletted wheats globally.
B1, predominantly responsible for awn inhibition in wheat, encodes a C2H2 zinc finger protein with EAR motifs which putatively functions as a transcriptional repressor.
Keywords: awn, awnletted, B1 locus, C2H2, EAR motif, transcriptional repression, wheat, zinc finger
Introduction
Awns are bristle‐like structures extending from the lemma‐tip‐midvein in the Poaceae grasses including cereal crop species such as wheat (Triticum aestivum and T. durum), barley (Hordeum vulgare) and rice (Oryza sativa). As an extension of the lemma, awns have long been considered modified leaves; however, evidence suggests that the origin or homology of the lemma itself may be as a modified sepal or novel organ type with bract and sepal characteristics (Grundbacher, 1963; Fabien & Hitoshi, 2015; Schrager‐Lavelle et al., 2017). In wild species, the main function of awns is grain dispersal by way of hygroscopically propelling the seed dispersal unit on the ground and into the soil (Elbaum et al., 2007). Furthermore, awns protect the grain from predation by animals or birds, especially if the awns are barbed. The barbs also aid in dispersal through attachment to animal fur (Hua et al., 2015). The grain dispersal functionality of awns has been lost during domestication in cereals due to inclusion of the nonshattering trait which is controlled by, amongst others, the Tenacious glumes (Tg) locus and the Q allele in wheat or naked caryopsis (nud) and thresh‐1 genes in barley (Haas et al., 2019). Moreover, a general reduction in grain‐dispersal‐aiding appendages such as awns, barbs and hairs has accompanied domestication (Fuller & Allaby, 2009). In rice, breeding has artificially selected against awn presence or length because awns hinder harvesting and storage practices, and do not seem to contribute significantly to grain yield – likely related to the fact that rice awns lack chlorenchyma required for photosynthesis (Tatsumi & Kawano, 1972; Luo et al., 2013). In contrast to rice, wheat and barley awns have been retained to a great extent in domesticated cultivars, however the awns are shorter, thinner and lighter than those of their wild progenitors (Peleg et al., 2010; Haas et al., 2019). Awns in wheat and barley contribute to photosynthesis and can promote yield in warmer and drier rainfed environments ‐ as such awned wheats are prevalent in such areas as Australia, South and Central America, and the USA (Rebetzke et al., 2016). However, the absence of awns, often defined as awnletted due to minimal awn extension, also has been selected for particularly in wetter, more humid environments such as those in northern and central Europe where awns would not necessarily provide an adaptive advantage and may be a resource drain (Börner et al., 2005; Rebetzke et al., 2016).
Genes involved in awn development and elongation have been identified in rice and include: the basic helix‐loop‐helix transcription factor Awn‐1 (An‐1) whose expression at the apex of lemma primordia causes continuous cell division for long awn formation (Luo et al., 2013); Awn‐2 (An‐2) or LONG AND BARBED AWN1 (LABA1) encoding Lonely Guy Like 6 (OsLOGL6), which catalyzes the final step of cytokinin synthesis and promotes awn elongation and growth (Gu et al., 2015; Hua et al., 2015); the YABBY transcription factor DROOPING LEAF (DL), which promotes awn formation in a noncell‐autonomous manner; the auxin response factor OsETTIN whose expression is required for awn development (Toriba & Hirano, 2014); and REGULATOR OF AWN ELONGATION 2 (RAE2) or GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT1 (GAD1), belonging to the EPIDERMAL PATTERNING FACTOR‐LIKE family of secretory peptides, whose peptide is cleaved by SUBTILISIN‐LIKE PROTEASE 1 to induce awn elongation (Bessho‐Uehara et al., 2016; Jin et al., 2016). Genes suppressing awn formation also have been identified: the YABBY transcription factor TONGARI BOUSHI1 (TOB1) (Tanaka et al., 2012); and the mitogen‐activated protein kinase (MAPK) phosphatase, GRAIN LENGTH AND AWN 1 (GLA1) (T. Wang et al., 2018), which negatively regulates both grain size and awn length. Other rice genes promoting awn development also are associated with grain number or length. For example, wild‐type (WT) An‐1 or RAE2/GAD1 alleles produce long awns, longer grains and reduce grains per panicle, whereas WT An‐2/LABA1 alleles reduce grains per panicle. Similarly, in wheat and barley, awn presence or increases in awn length reduce grain number while increasing grain size (Schaller & Qualset, 1975; Rebetzke et al., 2016).
In barley, genes involved in brassinosteroid (BR) biosynthesis or signalling have been shown to affect awn length in addition to the multiple traits affected by BR including plant height, culm robustness and spike compactness (Dockter et al., 2014). The Lks2 gene, underlying the short awn 2 (lks2, for length 2) and allelic mutants unbranched style 4 and breviaristatum (ari‐d), encodes a SHORT INTERNODES family transcription factor regulating awn length and pistil morphology (Yuo et al., 2012). Similar to wheat, where awn absence seems suited to moist environments, Yuo et al. (2012) suggest that the short‐awned lks2 allele provides an adaptive advantage in the high‐precipitation areas of eastern Asia where natural variants of the allele originated. An intronic duplication in the homeobox gene HvKnox3 produces the Hooded phenotype in which ectopic meristems develop on the lemma to form inflorescence‐like structures instead of normal awns (Müller et al., 1995; Williams‐Carrier et al., 1997).
In wheat, three dominant loci inhibiting awn development, Tipped 1 (B1), Tipped 2 (B2) and Hooded (Hd), have been identified and localized to chromosome arms 5AL, 6BL and 4BS, respectively (Watkins & Ellerton, 1940; Kato et al., 1998; Sourdille et al., 2002; Yoshioka et al., 2017). Wheat with dominant B1 and B2 genotypes produce small outgrowths of awns from the tip of the lemma, hence the terminology Tipped, and often are referred to as awnletted. However, in B1 genotypes awns at the top of the head often reach 1 cm in length; while in B2 genotypes awn length is more consistent with awns showing a slight curvature. Wheat with dominant Hd genotypes produces awns of reduced length that are pronouncedly curved with an inflexion from the base; there can also be membranous lateral expansions near the tip of the lemma which can resemble the Hooded phenotype from barley (Watkins & Ellerton, 1940; Yoshioka et al., 2017). B1 is the most prevalent allele inhibiting awn development in both hexaploid and tetraploid wheats (Goncharov et al., 2003; Le Couviour et al., 2011; Mackay et al., 2014; Yoshioka et al., 2017). Despite the excellent progress in characterizing and mapping B1, B2 and Hd, the genes responsible for awn inhibition at these loci have not been identified. Here, we report the identification of the B1 awn inhibition gene through fine‐mapping using an F2 biparental durum wheat population, as well as a series of bread wheat mutants to delimit the genic region and identify the gene responsible for the awnletted trait. Furthermore, we characterize the identified B1 gene, TraesCS5A02G542800, annotated as a C2H2 zinc finger gene, through constitutive gene overexpression in both durum and bread wheat to suggest a role as transcriptional repressor. Finally, haplotype analyses revealed B1 diversity in wheat and were consistent with the assertion that B1 is the most prevalent gene for awn inhibition. Our identification of B1 parallels a companion paper in this issue by DeWitt et al. (2020), but whereas the present paper provides evidence for the repressor functionality of B1, DeWitt et al. (2020) detail the intricate relationship of B1 and awn inhibition to grain length and spikelets per spike.
Materials and Methods
Plant materials and growth conditions
Durum wheat
Wheat plants were grown in 10‐ or 15‐cm pots containing Sunshine 8 (Sun Gro® Horticulture, Agawam, MA, USA) mixed with a slow‐release fertilizer 14‐14‐14 in a sun room or glasshouse with approx. 16 h : 8 h, light : dark photoperiod and day/night temperatures of 22°C/18°C. To generate a durum wheat population segregating for awn presence, reciprocal crosses between the Canadian cultivar Strongfield (ST) and Australian Glossy Huguenot (GH, or AUS2499) were performed (Johnson et al., 1983; Clarke et al., 2005). F1 plants were self‐pollinated to produce an F2 population that consisted of 1936 plants, with 714 individuals grown in a glasshouse in 2016, 1135 grown in a glasshouse in 2017, and 87 F2 lines grown in a sunroom in 2016. F1 were also back‐crossed to ST to generate a BC1 population of 131 individuals in 2016. For awn phenotyping, F2 individuals were classified as fully awned or awnletted. Fully awned spikes had awn lengths > 1 cm throughout the entire spike whereas awnletted consisted of < 1 cm awns in the mid and basal spike regions; in some cases the apical part of an awnletted spike, especially the tip, had awn lengths of up to c. 3 cm. Additional phenotyping of the F2 population included measures of thousand kernel weight and grain size including area, length and width, performed using a Marvin seed analyzer (GTA Sensorik GmbH, Germany).
Hexaploid wheat
Spring wheat varieties Paragon and Cadenza are both awnless and carry the B1 awn suppressor on chromosome 5A. Awned mutants were selected from three populations: a population of 6500 mutants of Paragon derived by ethyl methane sulfonate (EMS) treatment to induce nucleotide substitutions and developed to the M6 generation; a population of 2000 mutants in the Paragon background derived by gamma irradiation to produce deletions in DNA and developed to M4, (both populations were generated under the Defra‐funded Wheat Genetic Improvement Network at the John Innes Centre, Norwich, UK); and a population of 1200 mutants of Cadenza derived following EMS treatment, which has been re‐sequenced by exome capture and contains an estimated 9.0 million mutations. In addition, 258 wheat accessions or varieties (Supporting Information Table S6a) were screened using the primer sets designed for the awn‐suppressor candidate as described in the Supporting Information. Selected lines showing awn characteristics inconsistent with the PCR results were further characterized by sequencing and/or gene expression.
Additional materials and methods
Details on the bulked segregant analysis, marker development and fine mapping, mutant deletion mapping, and transcriptome and haplotype analyses, with associated references can be found in Methods S1–S9 and Notes S1. The RNA‐seq data have been submitted to the Sequence Read Archive (SRA) at the NCBI with the accession numbers 11588058–11588082 (total RNA from Triticum durum and Triticum aestivum).
Results
Fine‐mapping the B1 awn inhibitor in durum wheat
As a strategy to identify traits and genes associated with productivity under water stress, a biparental durum wheat population was developed from the cross of the Canadian cultivar Strongfield and the Australian Glossy Huguenot cultivar, which is further denoted as STxGH (Johnson et al., 1983; Clarke et al., 2005). The two cultivars differ in numerous traits including plant height, glaucousness, awn presence and grain size (Figs 1, S1). As demonstrated by the companion paper in this issue, DeWitt et al. (2020), and consistent with the previous studies in rice, barley and wheat discussed in the Introduction above, awn presence in STxGH promoted increases in grain size and in particular grain length (Fig. 1d,e). In terms of awn presence, Strongfield is fully awned whereas Glossy Huguenot is awnletted. In the F1 generation of the STxGH cross, the apical awns, around the top of the head, were longer in length suggesting that the awnletted trait is not completely dominant (Fig. 1a). However, for ease of phenotyping, phenotypes were grouped as fully awned or awnletted which included fully awnletted or awnletted awns of slightly longer length around the apical tip. With this phenotyping, plants segregated into awnletted : fully awned at a ratio of near 3 : 1 in the F2 population and 1 : 1 in the BC1 population, indicating that awn length is controlled by a single dominant inhibitor.
Figure 1.
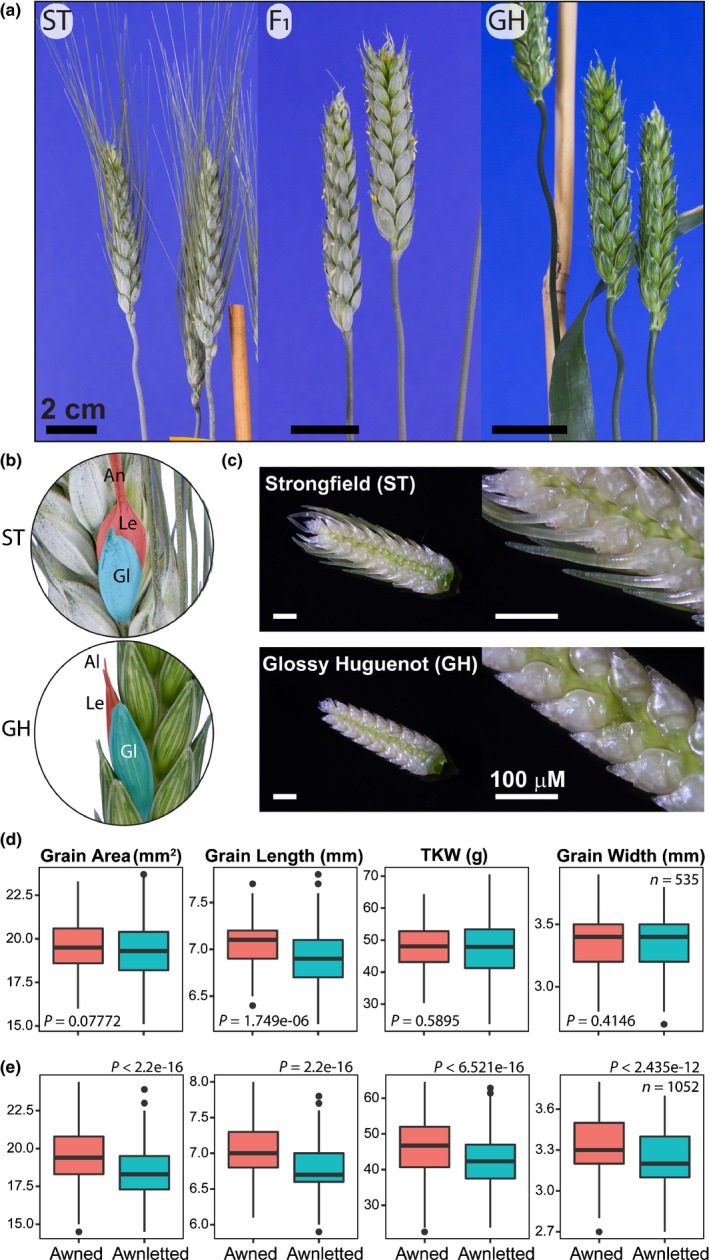
A durum wheat population segregating for awn presence. (a) Reciprocal crosses of the awned Strongfield (ST) and awnletted Glossy Huguenot (GH) cultivars were performed. All F1 plants were awnletted suggesting dominance over the awned trait. (b) The tip of the GH lemma (Le) was capped with an awnlet (Al), whereas the top of the glumes (Gl) were smooth and beakless. A predominant awn (An) extended from the tip of the lemma in ST whereas the top of the glume displayed a slight beak. (c) Awn growth was inhibited during early inflorescence development from the terminal spikelet stage during initial awn elongation. (d) In an initial glasshouse experiment in 15‐cm pots, awnletted lines of the STxGH F2 population were significantly reduced in grain length (P < 0.05). (e) In a second glasshouse experiment in 10‐cm pots, all measured grain traits including area, length, width and thousand kernel weight (TKW) were significantly reduced (P < 0.05) in awnletted lines of the STxGH F2 population. Boxplots of the grain trait data in (d, e) show the median (horizontal line), the interquartile range (boxes), 1.5‐times the interquartile range or maximum/minimum values (whiskers), and extreme values (dots). Red boxplots represent awned lines and blue boxplots represent awnletted lines of the GxH F2 population.
In order to fine‐map the awnletted trait bulked segregant analysis coupled with RNA sequencing (BSR‐seq) was performed using 714 F2 generation plants from the STxGH cross (Liu et al., 2012). The BSR‐seq pools, which consisted of c. 50–60 awned or awnletted lemmas or florets, were collected during swelling of the boot (Zadok stages 43–45). Following RNA sequencing and variant calling of the BSR‐seq pools, the greatest number of differential variants between the awned and awnletted pools were located on chromosome 5A and in particular the distal end of 5AL with a physical location between 680 and 705 million nt in the RefSeq 1.0 Chinese Spring genome reference (Figs 2, S2a,b; Table S1a) (International Wheat Genome Sequencing Consortium (IWGSC) et al., 2018). The dominant nature of the awnletted trait and 5A peak of differential variants between the awned‐awnletted pools is consistent with the location of the B1 inhibitor locus (Yoshioka et al., 2017).
Figure 2.
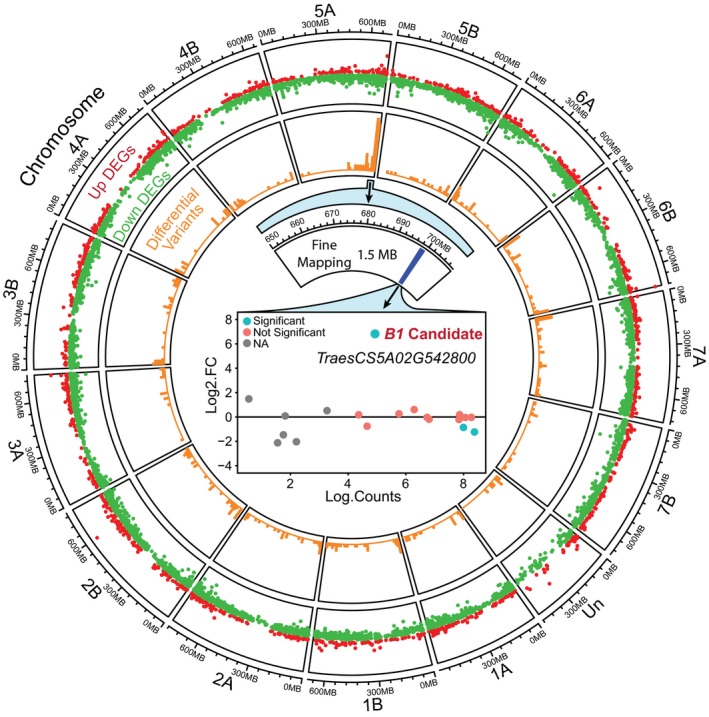
Identification of a candidate gene for the Tipped1 (B1) awn inhibitor in durum wheat using bulked segregant RNA‐sequencing (BSR‐seq) and fine mapping. Circular tracks display chromosome name and size (MB, megabases) followed by differentially expressed genes (DEGs) and differential variants from BSR‐seq. Analysis of awned and awnletted BSR‐seq pools identified 6890 DEGs (P‐adjusted < 0.05) with the 5560 of these being downregulated in the awnletted pool. Peak differential variants were located on the distal end of chromosome 5A between 680 and 705 million‐nt in the Chinese Spring RefSeq 1.0 genome. Fine‐mapping using KASP markers delineated the genomic region responsible for the awn inhibitor trait to a c. 1.5‐Mbp region beginning at 698 003 657‐nt. Within this 1.5‐Mbp region, there were three DEGs: TraesCS5A02G542600, TraesCS5A02G542800 and TraesCS5A02G543000. However, only TraesCS5A02G542800 was upregulated in the awnletted pool and thus became the chief candidate for the B1 awn inhibitor gene. Further, TraesCS5A02G542800 was only expressed in the awnletted pool which is consistent with the model of a dominant inhibitor.
Following the discovery that the distal end of chromosome 5AL is associated with awn growth inhibition, 11 KASP markers were designed spanning the 670–708 million nt region on 5A for further fine‐mapping. A 652 F2 subset of the STxGH population was genotyped with the 11 KASP markers, and the markers were separated and ordered based on linkage analysis to create a local genetic map (Meng et al., 2015). Quantitative trait loci (QTL) mapping was performed with three different algorithms: inclusive composite interval mapping (ICIM), logistic regression for binary traits (LRB) and linear mixed model (LMM) (Meng et al., 2015; Broman et al., 2019). Significant QTL peaks for the awn trait were detected with logarithm of the odds (LOD) scores ranging from 377 to 128 (Fig. S2c). The QTL peaks were between markers 5A_B1_4 and 5A_B1_2 for ICIM and centred on marker 5A_B1 for LRB and LMM mapping with a Bayes credible interval of 48.73–58.43 cM, which spans the region from markers 5A_B1_4 to 5A_B1_2 (Fig. S2c). Marker 5A_B1 is based on the same polymorphism [A/C] as the published B1 marker BobWhite_c8266_227 with different flanking sequences (Mackay et al., 2014). The single nucleotide polymorphism (SNP) markers 5A_B1_4 and 5A_B1_2 are physically located at 698 003 657 and 699 477 743 nt on chromosome 5A, which delimited the region containing the candidate gene for awn inhibition to c. 1.5 Mbp (Fig. 2).
In order to identify the gene responsible for awn inhibition on chromosome 5AL, the BSR‐seq data was analyzed for differentially expressed genes (DEGs, P‐adjusted < 0.05) between the awnletted and awned pools. There were 6890 DEGs (1330 up‐ and 5560 downregulated) between the two groups; however, there were only three DEGs, TraesCS5A02G542600, TraesCS5A02G542800 and TraesCS5A02G543000, in the 1.5‐Mbp region between markers 5A_B1_4 and 5A_B1_2 (Fig. 2; Table S1b,c; Notes S2). Of the three DEGs, only TraesCS5A02G542800 was upregulated in the awnletted pool and, moreover, showed no expression in the awned pool. The specific expression of TraesCS5A02G542800 in the awnletted pool is consistent with the model of a dominant inhibitor. Thus, TraesCS5A02G542800, annotated as a C2H2 zinc finger encoding gene, became the chief candidate gene for awn inhibition in Glossy Huguenot and a suggested candidate gene for the B1 awn inhibitor based on proximity to published markers for B1 including BobWhite_c8266_227 and RAC875_C8642_231 (Mackay et al., 2014).
Mapping mutations of the B1 awn inhibitor in bread wheat
As part of the work to assess disease resistance between awned and awnletted wheat, mapping of mutations that caused an awned phenotype was used to identify the B1 candidate gene. Three awned mutants within a Cadenza‐EMS TILLING population (1378awn, 1636awn and 1978awn) were identified (Krasileva et al., 2017). Three fully awned lines (1533, 1695 and 1987) also were identified in a Paragon‐EMS population. Because a full wheat reference genome was not available at the time, primers for genes in the awn inhibitor region on chromosome 5A were designed based on available wheat sequences and synteny to rice, Brachypodium, and sorghum (IWGSC1 + popseq genome assembly in Ensembl Plants, http://mar2016-plants.ensembl.org/Triticum_aestivum/Info/Index). During primer design, wheat A, B and D homoeologs were compared to produce A genome‐specific PCR markers. When the gene‐based chromosome 5A markers were used to screen the six awned mutants, it became clear that all six lines had lost significant portions of chromosome 5A in the region of the B1 locus (Table 1). Awned Paragon line 1695 contained a smaller deletion than the other EMS‐treated awned lines. This delimited the awn‐suppressor to the region terminal to TraesCS5A02G542600 (annotated as a hexose carrier encoding gene). The presence of large deletions in both EMS populations led us to screen a Paragon gamma‐irradiated population to seek lines carrying overlapping deletions to further define the region responsible for B1 awn inhibition. In the Paragon‐Gamma population, the proportion of awned mutants was greater than in the EMS treated populations (eight awned lines among 2000 lines) as expected due to the nature of gamma irradiation, which tends to induce deletions > 10 kb (Morita et al., 2009). One awned mutant was identified from Paragon, line #10, which contained a small deletion spanning only two genes (Table 1). This Paragon awned mutant was backcrossed twice to the Paragon wild‐type (WT) to ensure that any additional mutations/deletions were removed and the mutation was consistent with the putative region for awn inhibition on chromosome 5AL. Similar to the durum crosses above in the STxGH population, it was evident that the awn inhibitor was dominant as the F1 plants all had short awns and plants segregated 3 : 1 for awnletted : fully awned in the F2 generation (Fig. 3a).
Table 1.
Mapping of deletions in bread wheat.
| IWGSC RefSeq v1.1 | Position | Original gene name | Paragon gamma | Paragon EMS | Cadenza EMS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | #10 | #11 | #1 | 1695 | 1533 | 1987 | 1378awn | 1636 awn | 1978 awn | ||
| TraesCS5A02G528400 | 688662668 | 688666371 | Traes_5AL_CC97471EB | nd | nd | nd | ||||||
| TraesCS5A02G529200 | 688782883 | 688785747 | Traes_5AL_1A8232916 | nd | nd | nd | nd | nd | nd | nd | ||
| TraesCS5A02G529700 | 688944988 | 688953005 | Traes_5AL_3524CED56 | nd | nd | nd | ||||||
| no gene annotated | 689721892 | 689722407 | Traes_5AL_33F0FB676 | nd | nd | nd | ||||||
| TraesCS5A02G529900 | 688973484 | 688976168 | Traes_5AL_74EEA8C04 | nd | nd | nd | nd | nd | nd | nd | ||
| TraesCS5A02G531200 | 689604419 | 689608548 | Traes_5AL_727C588D0 | |||||||||
| TraesCS5A02G531800 | 689896911 | 689912314 | Traes_5AL_D613D4F93 | |||||||||
| no gene annotated | 690356469 | 690356927 | Traes_5AL_F212D729D | |||||||||
| TraesCS5A02G534100 | 691436456 | 691441801 | Traes_5AL_9D148E259 | nd | nd | nd | nd | |||||
| TraesCS5A02G537000 | 693625968 | 693632648 | Traes_5AL_69D2D5116 | nd | nd | nd | nd | |||||
| TraesCS5A02G540400 | 697880912 | 697887556 | Traes_5AL_9F207C3A8 | nd | nd | nd | nd | |||||
| TraesCS5A02G541000 | 698010055 | 698012286 | Traes_5AL_34CE42B51 | |||||||||
| TraesCS5A02G542200 | 698448290 | 698451049 | Traes_5AL_CD47A37A6 | nd | nd | nd | nd | |||||
| TraesCS5A02G542600 | 698507247 | 698511217 | Traes_5AL_F49663738 | nd | nd | nd | nd | |||||
| TraesCS5A02G542700 | 698512064 | 698517752 | Traes_5AL_BDDA03F8B | nd | nd | nd | nd | |||||
| TraesCS5A02G542800 | 698528636 | 698529001 | TRIAE_CS42_5AL_TGACv1_374501_AA1201650 | nd | nd | nd | nd | |||||
| non coding sequence | 698581401 | 698582186 | non coding sequence | nd | nd | nd | nd | nd | ||||
| TraesCS5A02G542900 | 698629158 | 698637157 | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| no gene annotated | 698637620 | 698638706 | Traes_5AL_7F8E591BF | nd | nd | nd | nd | |||||
| TraesCS5A02G543100 | 698888618 | 698889913 | TRIAE_CS42_5AL_TGACv1_373973_AA1186000 | nd | nd | nd | nd | |||||
| TraesCS5A02G544900 | 700347633 | 700351845 | Traes_5AL_55669B33 | nd | nd | nd | nd | |||||
| TraesCS5A02G546600 | 700663097 | 700665985 | Traes_5AL_118352014 | nd | nd | nd | nd | |||||
| TraesCS5A02G547800 | 702132455 | 702136920 | Traes_5AL_14CC809DB | |||||||||
| TraesCS5A02G550800 | 704344835 | 704346995 | Traes_5AL_AB1566CF9 | nd | nd | nd | nd | |||||
| TraesCS5A02G551300 | 704832604 | 704835391 | Traes_5AL_14402DE4B | |||||||||
| TraesCS5A02G552700 | 705439246 | 705447278 | Traes_5AL_CFA8EDF16 | nd | nd | nd | ||||||
| TraesCS5A02G553300 | 705842223 | 705847913 | Traes_5AL_92DEAA86D | nd | nd | nd | ||||||
| TraesCS5A02G554100 | 706235563 | 706237619 | Traes_5AL_1BE4452BD | nd | nd | nd | nd | nd | nd | nd | ||
| TraesCS5A02G554600 | 706427023 | 706429950 | Traes_5AL_BBEF740F0 | nd | nd | nd | ||||||
| no gene annotated | 706436133 | 706438842 | Traes_5AL_BF079AF06 | nd | nd | nd | ||||||
Results of homoeologue specific PCR of genes on chromosome 5A. Red fill colour, gene amplicon absent; green fill colour, gene amplicon present; nd, not determined. The B1 gene, TraesCS5A02G542800, is denoted in bold type.IWGSC, International Wheat Genome Sequencing Consortium.
Figure 3.
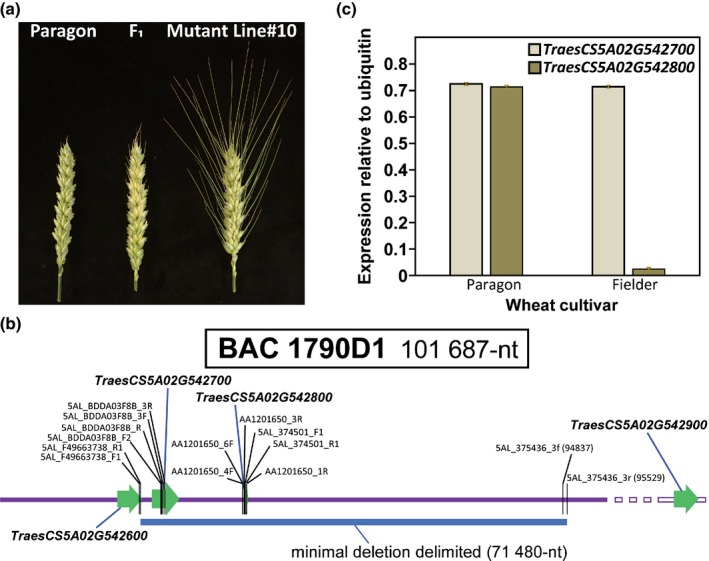
Utilizing deletion mutants in bread wheat to identify a candidate gene for the Tipped1 (B1) awn inhibitor. (a) Dominant awn inhibition in the F1 generation, following a backcross of Paragon mutant line#10 to wild‐type Paragon. (b) Analysis of BAC 1790D1 from the French awned cultivar Renan confirmed mutant deletions for awn inhibition were confined to two genes, TraesCS5A02G542700 and TraesCS5A02G542800. (c) Expression analyses of TraesCS5A02G542700 and TraesCS5A02G542800 revealed that only the expression of TraesCS5A02G542800 was limited to the awnletted cultivar Paragon and thus TraesCS5A02G542800 became the chief candidate for the B1 awn inhibitor gene.
A PCR designed from one of the potential candidate genes, TraesCS5A02G542800, was used to identify a bacterial artificial chromosome (BAC) clone (BAC 1790D1) from the French awned cultivar Renan that corresponded to the awn inhibitor sequence interval. The BAC sequence included three gene models TraesCS5A02G542600, TraesCS5A02G542700 and TraesCS5A02G542800. It was shown that the sequence extended beyond the deletion of the Paragon awned mutant line #10 using a PCR primer set (Noncoding Scaffold 37546 3f/3r) which locates near the end of the BAC (Fig. 3b). Mapping of the deletions and utilization of the BAC sequence revealed two potential candidate genes, TraesCS5A02G542700 and TraesCS5A02G542800, for awn inhibition. Next, RNA was isolated from developing spikes at the pre‐anthesis stage of the awned and awnletted varieties, Fielder and Paragon, respectively, and the expression of the two candidate genes was measured. The expression of TraesCS5A02G542800 was limited to the awnletted cultivar whereas TraesCS5A02G542700 was expressed to a similar level in both varieties (Fig. 3c). Thus, TraesCS5A02G542800 was the chief candidate for B1 in these UK bread wheats. Taken together with the BSR‐seq fine‐mapping of B1 in durum wheat, it is proposed that TraesCS5A02G542800, annotated as a C2H2 zinc finger encoding gene, is the B1 awn inhibitor gene – a statement also consistent with the companion paper by DeWitt et al. (2020).
The B1 awn inhibitor TraesCS5A02G542800, a zinc finger protein with EAR motifs, belongs to a multigene family
The open reading frame (ORF) for the intron‐less B1 awn inhibitor gene resides from 698 528 636 to 698 529 001‐nt on chromosome 5A in the RefSeq 1.0 Chinese Spring genome reference. The 122‐aa B1 protein contains a C2H2‐type zinc finger domain (residues 25–47) and two leucine‐rich ethylene‐responsive element binding factor‐associated amphiphilic repression (EAR) motif‐like sequences in its N‐terminal (LDLSLSL; residues 7–14) and C‐terminal (LSLKL; residues 117–121) regions (Fig. 4a) (Ohta et al., 2001; Kagale et al., 2010). The C2H2 family of zinc finger proteins is a large class of transcriptional regulators involved in a number of processes including trichome and root hair formation, flower development, seed development, disease resistance and abiotic stress response (Luo et al., 1999; Kazan, 2006; Xiao et al., 2009; Yan et al., 2014; K. Wang et al., 2018). Furthermore, the EAR motif is the most prevalent form of transcriptional repression motif identified in plants (Kagale & Rozwadowski, 2011). EAR‐domain‐containing proteins have been demonstrated to interact physically with corepressors, TOPLESS and SIN3 ASSOCIATED POLYPEPTIDE 18 (SAP18), and recruit chromatin remodelling factors to form a transcriptional repression complex.
Figure 4.
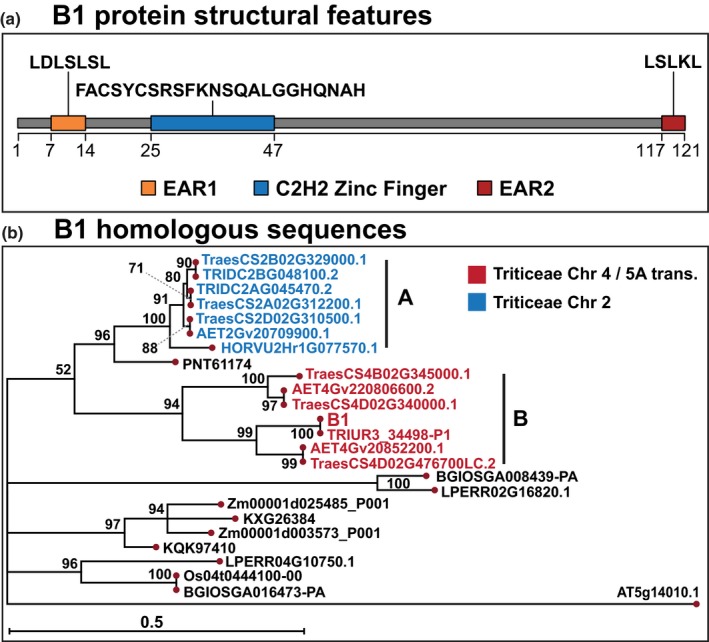
Tipped1 (B1) is a C2H2 zinc finger protein with ethylene‐responsive element binding factor‐associated amphiphilic repression (EAR) motifs and belongs to a multigene family. (a) Predicted structural features of the B1 protein including the location of a C2H2 zinc finger domain and two EAR motif sequences. (b) Maximum‐likelihood phylogeny of proteins with homology to B1. Triticeae species divide into two major grades: (A) B1‐related proteins from group‐2 chromosomes and (B) B1‐related proteins from group‐4 chromosomes. However, B1 has diverged from homoelogous proteins on chromosome 4B and 4D in sequence identity and may have undergone neo‐functionalization, perhaps due to the chromosome 4/5A translocation that occurred in the diploid progenitor of the A‐genome. TRIUR3_34498‐P1 from Triticum urartu was identical in sequence to B1; in wheat, the closest B1 homoelog was TraesCS4D02G476700LC with sequence identity of 74%. Protein sequences were downloaded from Ensembl Plants with identifiers retaining the isoform notation; full‐length proteins of TraesCS4D02G476700LC, TRIDC2AG045470, TRIDC2BG048100 and AET4Gv20806600 were identified as described (Supporting Information Fig. S3). Bootstrap support values from 1000 tests are shown; nodes were collapsed for bootstrap values < 50%; scale bar represents branch lengths as an estimation of the number of substitutions per amino acid. Species represented include: Aegilops tauschii (AET); Triticum aestivum (Traes); T. dicoccoides (TRIDC); T. urartu (TRIUR); Hordeum vulgare (HORVU); Sorghum bicolor (KXG); Zea mays (Zm); Setaria italica (KQK); Oryza sativa indica (BGIOSGA); O. sativa japonica (Os); Leersia perrieri (LPERR); and Arabidopsis thaliana (AT) (Methods S3).
In order to further characterize the B1 protein, Blast searches were performed of the Ensembl Plants database and ortholog, paralog, and homoeolog sequences in Ensembl were utilized to identify B1‐related amino acid sequences (Kersey et al., 2018). In addition, full‐length ORFs were identified from their respective genomes for the following genes: TraesCS4D02G476700LC, TRIDC2AG045470, TRIDC2BG048100 and AET4Gv20806600 (Avni et al., 2017; Luo et al., 2017; IWGSC et al., 2018). Following clustalw alignment, a maximum‐likelihood phylogeny was constructed to summarize relationships between B1 and sequences from Triticeae species Aegilops tauschii, barley, Triticum dicoccoides and T. uratu and additional members of the Class Liliopsida such as Brachypodium distachyon, Leersia perrieri, Zea mays, Setaria italica, O. sativa ssp. Indica and Japonica, and Sorghum bicolor (Figs 4b, S3) (Felsenstein, 1981; Jones et al., 1992; Thompson et al., 1994). Triticeae species divide into two major grades: (A) B1‐related proteins from group‐2 chromosomes and (B) B1‐related proteins from group‐4 chromosomes. The B1 awn inhibitor resides in the grade with group‐4 because the translocation T(4AL;5AL)1, a segmental interchange between chromosome arms 4AL and 5AL, occurred in the diploid progenitor of the wheat A subgenome (Dvorak et al., 2018; Grewal et al., 2018). The B1 protein shares important structural features with its homoeologs occurring on chromosome 4B (TraesCS4B02G345000) and chromosome 4D (TraesCS4D02G340000, TraesCS4D02G476700LC) but has diverged in sequence identity (58%, 60% and 74%, respectively) and promoter region, potentially leading to neo‐functionalization. Consistent with the companion paper by DeWitt et al. (2020), TraesCS4D02G476700LC shared the highest homology to B1 in wheat. However, TRIUR3_34498‐P1 from T. urartu was identical in sequence to B1. A paralog to B1, with 49% sequence identity, TraesCS2A02G312200, in the group‐2 chromosome grade, is orthologous to barley HORVU2Hr1G077570. HORVU2Hr1G077570 is upregulated during overexpression of FLOWERING LOCUS T3 (HvFT3); overexpression of HvFT3 accelerated the initiation of spikelet primordia and resulted in upregulation of auxin related‐genes (Mulki et al., 2018). The GenBank entry ADK55064 for rice Os04g0444100 denotes that the C2H2 zinc finger MALFORMED SPIKELET is involved in specifying rudimentary glume identity (Zhang et al., 2011). The presence of EAR motifs in TraesCS5A02G542800 advocates for a function of transcriptional repressor and evidence is provided below through constitutive overexpression of B1 that supports a repressor role.
B1 overexpression inhibits awn elongation with pleiotropic effects on plant development
Introduction of the 366‐nt ORF of B1 driven by the maize ubiquitin1 promoter into the durum cultivar Strongfield and bread wheat cultivar Bobwhite resulted in awn inhibition phenotypes (Figs 5, S4–S6; Table S2a–c). There were also additional pleiotropic phenotypes including reductions in plant height and floret fertility (Fig. S7; Table S2a–c). In some cases, T0 plants were totally infertile with no grain produced (Table S2a). Awn inhibition was heritable and was carried over into the T1 generation in both the Bobwhite and Strongfield backgrounds (Figs 5, S4).
Figure 5.
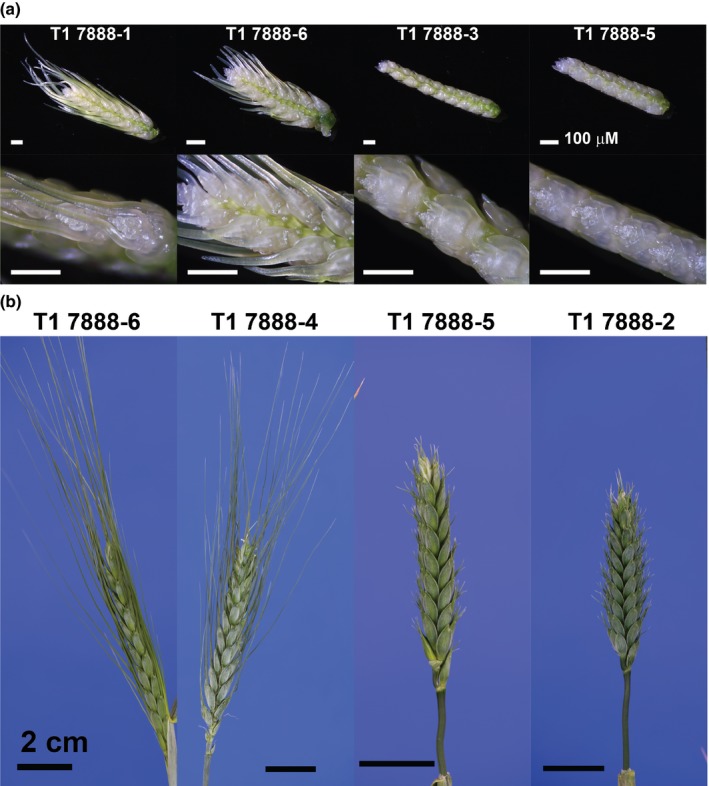
Tipped1 (B1) overexpression in durum wheat represses awn formation throughout inflorescence development. (a) Awn development is inhibited during early awn extension when floral tissues are forming in transgenic lines expressing B1 driven by the maize ubiquitin 1 promoter. In the T1 generation of line 7888, overexpression of B1 segregates: awn development is not inhibited in lines with low expression of B1 similar to wild‐type (WT) Strongfield and lines with high expression of B1 display awn inhibition (Supporting Information Table S3). Bars, 100 μm. (b) Once flower heads fully emerge from the boot, T1 lines expressing B1 displayed significant inhibition of awn growth with awnlets typically < 1 cm in length. Segregating T1 lines with low WT levels of B1 expression were fully awned. Additional awn phenotypes of B1 overexpression lines are shown in Figs S4–S7 for the durum wheat Strongfield and bread wheat Bobwhite backgrounds. Bars, 2 cm.
B1 overexpression causes repression of auxin and cell proliferation genes
Young inflorescences collected following the terminal spikelet stage during early awn extension in Strongfield and Bobwhite WT plants (awned) were compared to awn inhibition in B1 over‐expressing plants (awnletted) by performing RNA‐sequencing (Fig. 5a). Although overexpression through the ubiquitin promoter expanded the range of B1 expression, as observed by the pleiotropic phenotypes, it was hypothesized that comparisons of the transcriptomes of B1 over‐expressing plants would begin to reveal the molecular function of the B1 awn inhibitor gene, including the possible role of B1 in transcriptional repression. Three RNA‐sequencing experiments were compared: (1) awnletted vs awned segregants of T1 Strongfield line 7888 (ST‐T1), (2) T0 Strongfield to WT (ST‐T0), and (3) Bobwhite T0/T1 awnletted lines to awned T1 line 7933 and Bobwhite WT (BW‐OE). In terms of overexpression of B1, ST‐T1 displayed the greatest average difference with log2 fold‐change (FC) of 7.4 and the ST‐T0 and BW‐OE with FC of 6.8 and 5.5, respectively (Table S3).
Gene ontology (GO) analysis was performed on the up‐ and downregulated DEGs from each awnletted‐awned comparison, ST‐T1, ST‐T0 and BW‐OE (Table S4). Common GO terms, for molecular function (MF), biological process (BP) and cellular compartment (CC), between the three comparisons were then selected (Table S4a). Significant GO terms (Fisher Classic, P < 0.05) for the upregulated DEGs included oxioreductase activity (MF), RNA glycosylase activity (MF), negative regulation of translation (BP) and extracellular region (CC). For the downregulated DEGs significant GO terms included: DNA binding transcription factor (TF) activity (MF), regulation of gene expression (BP), response to auxin (BP), and nucleus (CC). The effect of B1 overexpression on GO with enrichment of TF activity, regulation of gene expression, and nuclear localization in the downregulated DEGs is consistent with the annotated functionality of B1 as a transcriptional repressor.
In order to identify possible gene pathways that B1 might influence for awn inhibition and reduce possible artefacts from ubiquitous B1 overexpression, a series of analyses was performed on the gene expression data (Fig. 6; Tables S3, S5). Because B1 is annotated as a transcriptional repressor, which the GO analysis supported, the focus was on genes that were downregulated by B1 overexpression. First, an analysis was made of the functional annotations of the DEGs common in at least two of the three experiments that included ST‐T1, ST‐T0 or BW‐OE (Table S3d). Second, to further narrow the list of putative genes or transcription factors that B1 may regulate, linear regression was performed comparing the expression of genes to the expression of B1 in the over‐expression and control (WT) lines (Table S5a). Lastly, a clustering analysis was performed to identify genes expressed in an opposite pattern to B1 (Table S5b; Fig. S8). From these analyses, 49 DEGs from the DESeq2 expression analyses were compared to 609 genes from linear regression analyses which had an r 2 value of ≥ 0.5 and the top 100 oppositely expressed genes compared to B1 using the R/genefilter package (Fig. 6a; Table S5c) (Gentleman et al., 2018).
Figure 6.
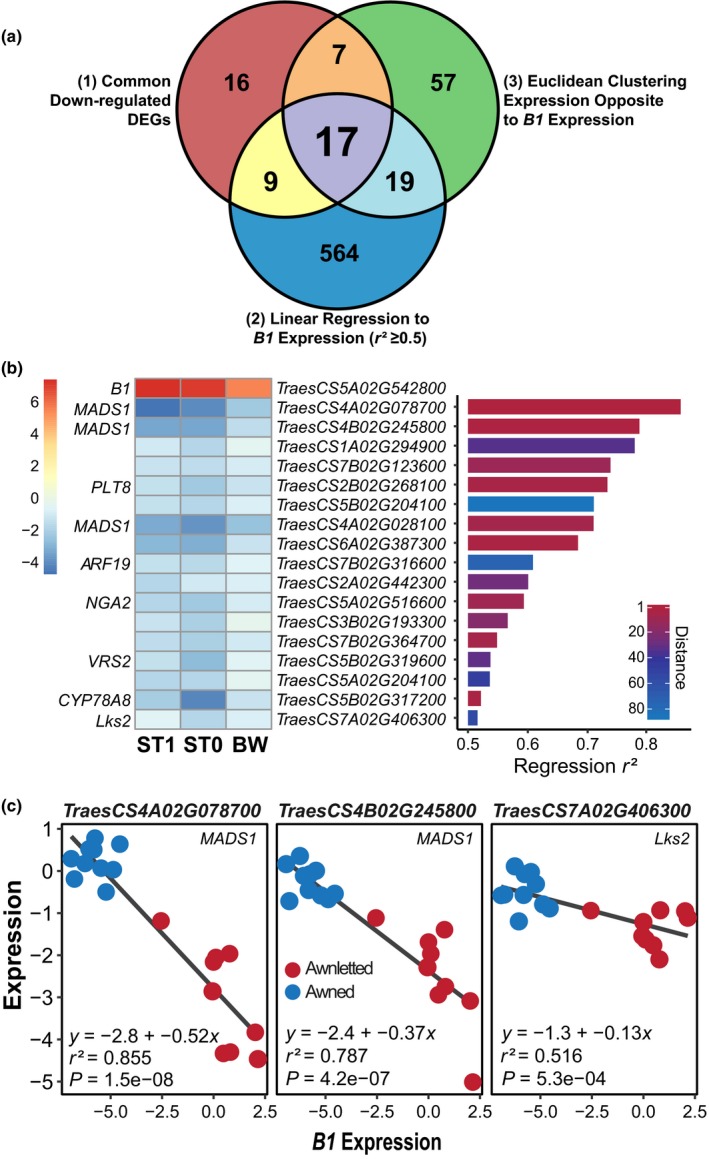
Tipped1 (B1) overexpression in durum and bread wheat represses auxin‐ and cell‐proliferation‐related genes. (a) Venn diagram representing the relationships between: (1) common downregulated differentially expressed genes (DEGs) in the B1 overexpression lines, (2) genes whose expression was linearly related to B1 with r 2 ≥ 0.5, and (3) genes which were expressed in an opposite manner to B1 as identified through Euclidean clustering of the additive inverse of log2‐normalized expression data (Supporting Information Fig. S8). (b) Seventeen genes were common to the three experimental analyses from (a). Within these, 17 were genes with orthology to OsMADS1, OsPLT8, OsARF19, AtNGA2, HvVRS2, AtCYP78A8, and HvLks2 (Os, Oryza sativa; At, Arabidopsis thaliana; Hv, Hordeum vulgare). Heatmap represents the log2 fold‐change in expression between awnletted and awned inflorescences of Strongfield T1 and T0 and Bobwhite (ST1, ST0 and BW). Bar graph displays the r 2 value from the linear regression comparing the expression of the gene to B1 expression; colours in the bar graph represent the Euclidean distance ranking from the clustering analysis. (c) Negative relationship of TraesCS4A02G078700, TraesCS4B02G245800 and TraesCS7A02G406300 expression to the expression of B1 using linear regression (Table S5a). Expression is displayed as 75th percentile normalized. P represents P‐value.
There were 17 genes common to the three analysis approaches used to identify potential pathways that could be regulated by B1 (Fig. 6a). These included genes associated with spike architecture, awn development and cell proliferation, and those participating in the auxin pathway (Figs 6b,c, S9). Within the gene set were MADS‐box genes TraesCS4A02G078700, TraesCS4B02G245800 and TraesCS4A02G028100 that have homology to rice OsMADS1, which is involved in meristem determinacy and differentiation and proliferation of the lemma and palea (Fig. S9a; Malcomber & Kellogg, 2004; Prasad et al., 2005). Transcription factors associated with auxin homeostasis included: (1) TraesCS5B02G319600 orthologous to the barley Six‐Rowed Spike gene VRS2, a SHORT INTERNODES (SHI) transcription factor that contributes to auxin and cytokinin gradients during spike development (Fig. S9b; Youssef et al., 2017); (2) TraesCS7B02G316600 which has homology to AUXIN RESPONSE FACTOR 19 (ARF19), whose mutant in rice has been associated with developmental abnormalities of the palea and lemma (Fig. S9c; Zhang et al., 2016); and (3) TraesCS2B02G268100, with homology to Arabidopsis thaliana (Arabidopsis) AINTEGUMENTA (ANT) and rice PLETHORA8 (PLT8). Members of the AINTEGUMENTA‐LIKE/PLETHORA (AIL/PLT) family in Arabidopsis are associated with auxin synthesis and signalling in both flower and root development (Krizek, 2011; Pinon et al., 2011; Mähönen et al., 2014). Also within the 17 genes common to the three analyses were: (1) TraesCS5A02G516600, a B3 domain transcription factor with homology to NGATHA2 (NGA2) from Arabidopsis; NGA genes have been shown to be important regulators of auxin accumulation and distribution in developing Arabidopsis gynoeciums (Alvarez et al., 2009; Martínez‐Fernández et al., 2014) and (2) TraesCS5B02G317200 with homology to the CYP78A class of cytochrome P450 genes such as CYP78A8‐9 of Arabidopsis involved in promotion of cell proliferation during floral organ growth (Sotelo‐Silveira et al., 2013). However, perhaps the most relevant gene common to the three analyses of B1 downregulated genes was TraesCS7A02G406300, the wheat gene orthologous to barley Short Awn 2 (Lks2) (Fig. S9d). In barley, Lks2 promotes awn elongation and has been proposed to affect auxin homeostasis (Yuo et al., 2012).
Overall, results of the gene expression analyses from the B1 overexpression experiments suggest a role for the C2H2 zinc finger as transcriptional repressor. Furthermore, although overexpression through the ubiquitin promoter expanded the range of B1 expression, and thus acted in tissues where it was not normally expressed, a common theme was the proportional downregulation of factors associated with cell proliferation and awn development.
Polymorphisms adjacent to a conserved B1 coding region are mostly predictive of B1 awn inhibition
In order to define possible polymorphisms important for B1 function, haplotype analyses were performed across accessions of wheat from the UK, Canada and Asia. Genomic alignment of the B1 region, 1000‐nt upstream and 850‐nt downstream of 698 528 636 to 698 529 001‐nt on chromosome 5A, identified SNPs, a 25‐nt deletion and a 1‐nt insertion that were present in genotypes with awn inhibition and revealed two major haplotype groups with a functional B1 inhibitor (A) compared to a nonfunction b1 (B) (Fig. 7a; Table S6). The polymorphisms were used to perform haplotype screens in diverse germplasm: (1) a PCR‐based screen to distinguish the 25‐nt polymorphism in a collection of 258 wheats including Watkins collection accessions (Wingen et al., 2014) and (2) KASP marker screening for select SNPs across a diverse collection of 562 wheats including Kyoto University accessions (Tanaka, 1983).
Figure 7.
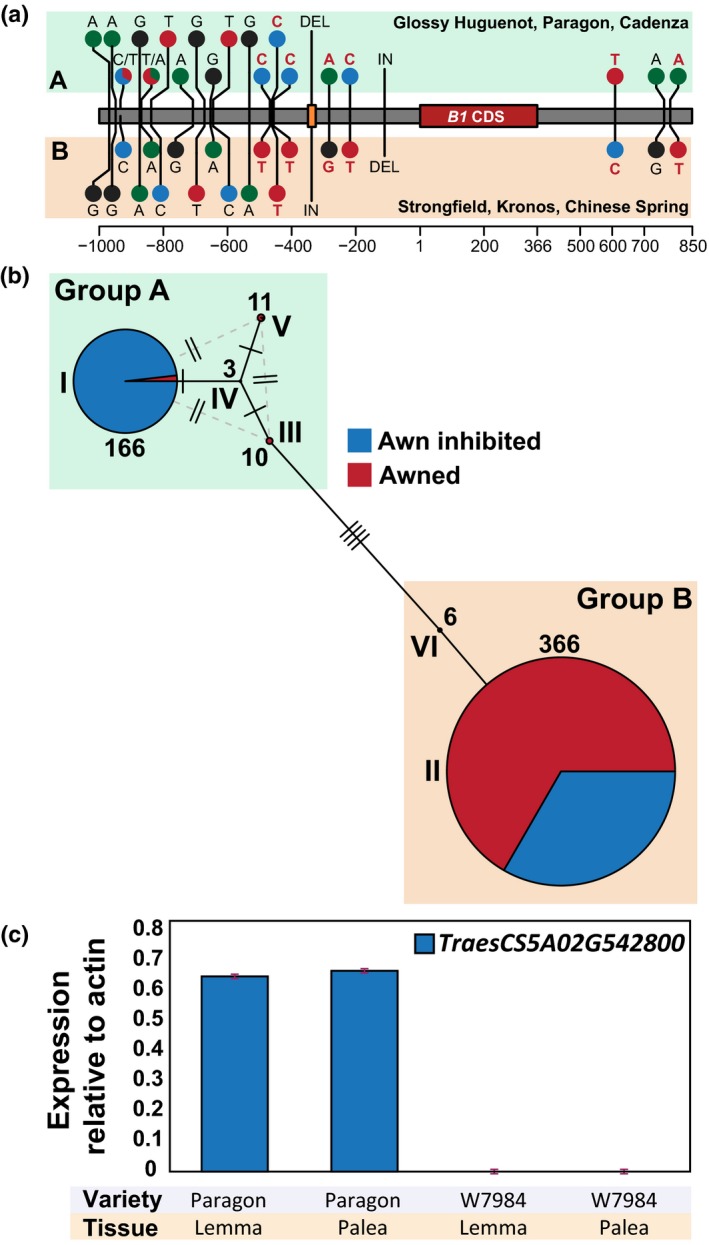
Haplotypes are mostly predictive of Tipped1 (B1) awn inhibition. (a) Initial single nucleotide polymorphism (SNP) and InDel identification in the B1 genomic region comparing awnletted‐B1 (A) to awned‐b1 (B) wheat accessions: Glossy Huguenot, Paragon and Cadenza to Strongfield, Kronos and Chinese Spring, respectively. There were no polymorphisms detected in the B1 coding region. Seven markers used for haplotype screening are noted in red letters (Supporting Information Table S7). (b) Bread wheat accessions surveyed with KASP markers contain six different haplotypes which can be divided into two subgroups: Group A, in which 87% of accessions are awn‐inhibited, and Group B, of which 67% of accessions are awned, with the resulting 33% presumably containing a different awn inhibitor gene such as Tipped2 (B2) or Hooded (Hd) (Table S6). In haplotype I, 98% of the accessions are awn‐inhibited indicating a functional B1 inhibitor; however, associated haplotypes III, IV and V are mostly awned (96%). Pie graphs represent awn presence, Roman numerals represent haplotype group, numbers represent accessions in each haplotype group, and each perpendicular hash‐mark represents a SNP that was screened with a KASP marker. (c) Within Group A haplotypes, the synthetic hexaploid wheat W7984 is awned but does not express B1 (Table S6).
In the 258‐line collection, the PCR‐based screening of the 25‐nt deletion between awn and awnletted genotypes was highly predictive of awn inhibition (98%) (Table S6a). However, there were five exceptions that included the awned synthetic hexaploid wheat W7984 and awnletted Chinese Spring cultivar. In the 562‐line collection, KASP marker screening identified two major groups of haplotypes consistent with the sequence alignment data (Fig. 7b; Table S6b). Haplotype Group A was predominantly awn‐inhibited with a functional B1 but contained a total of four haplotypes with the primary haplotype I containing 87% of the accessions. Ninety‐eight percent of haplotype I was awn‐inhibited, whereas minor haplotypes III, IV and V were 96% awned. Haplotype Group B was predominantly awned with a nonfunctional b1 and contained one dominant haplotype II in which 67% of accessions were awned with the remaining 33% awn‐inhibited due to B2, Hd, or other inhibitory mechanisms (Fig. 7b; Table S6b). For example, it is known that Chinese Spring contains the B2 and Hd awn inhibitors (Yoshioka et al., 2017). To confirm haplotypes identified from KASP marker screening in the 562‐line collection, a 1961‐nt B1 interval from 29 accessions was sequenced, 1027‐nt upstream to 934‐nt downstream of the start codon of the B1 gene (Table S6c). These B1 interval sequences together with available genomic sequences revealed a conserved B1 coding sequence and defined a total of nine haplotypes, six of which were within Haplotype Group A (Table S6c; Chapman et al., 2015; Avni et al., 2017; IWGSC et al., 2018; Clavijo et al., 2019). To further investigate the presence of awned accessions in Haplotype Group A, developing spikes from W7984 were isolated and it was found that B1 was not expressed consistent with the awned phenotype observed in W7984 (Fig. 7c). The presence of awned accessions in haplotypes III, IV and V is intriguing and suggests that the mechanism responsible for B1 expression is not present in these accessions. Sequencing analyses of the haplotype groups indicated that two polymorphisms are diagnostic for B1 awn inhibition: an A/G polymorphism 709‐nt upstream from the B1 start codon and an A/G polymorphism 761‐nt downstream of the start codon (Table S6c). However, the companion paper by DeWitt et al. (2020) suggests that no polymorphism within this region was diagnostic and that, rather, a 30‐nt deletion 4 kb downstream of the gene was the most (but not entirely) diagnostic for awn inhibition. Thus, the functional mechanism for B1 expression and subsequent awn inhibition remains enigmatic and further experiments are required. For example, further analyses of the awned accessions in Group A, with predominantly B1 haplotypes, are an important starting point for the discovery of the regulatory mechanisms for B1 expression and awn inhibition.
Discussion
Understanding the development of the inflorescence in cereal species is vital given the importance to yield and challenge for food security in the midst of climate change and population growth. In rice, numerous genes have been discovered and regulatory networks of inflorescence development have been defined (Zhang & Yuan, 2014; Chongloi et al., 2019). In barley and wheat, identification of regulators of inflorescence development is less mature but progressing given available genome sequences (Mascher et al., 2017; International Wheat Genome Sequencing Consortium (IWGSC) et al., 2018; Koppolu & Schnurbusch, 2019). Barley and wheat genes involved in inflorescence architecture have been discovered including: VRS genes involved in regulating two or six rows of grain in barley, wheat TEOSINTE BRANCHED1 (TB1) involved in regulating inflorescence architecture and paired spikelet formation, or Grain Number Increase 1 (GNI1) that contributes to wheat floret fertility (Komatsuda et al., 2007; Ramsay et al., 2011; Koppolu et al., 2013; Bull et al., 2017; Youssef et al., 2017; Dixon et al., 2018; Sakuma et al., 2019). The present study focused on the identification of the Tipped1 (B1) awn inhibitor gene. Awns are an important component of wheat spike architecture, with their presence facilitating ancestral grain dispersal and influencing grain productivity through contributions to photosynthesis. Analyses of B1, encoding a C2H2 zinc finger protein, advocate for a function as transcriptional repressor and we provide evidence that suggests B1 may inhibit awn development by repressing pathways related to auxin and cell proliferation.
Auxin is a central regulator for primordium formation and cell proliferation (Czesnick & Lenhard, 2015; Huang & Irish, 2016; Wang & Jiao, 2017). During awn formation, a primordium forms at the tip of the lemma and this region has been suggested to be a ‘quasi‐meristem’ (Girin et al., 2009; Toriba & Hirano, 2014). In rice, formation of awn primordia is driven by auxin and characterized by enhanced cell division and proliferation (Luo et al., 2013; Toriba & Hirano, 2014). With relevance to B1 function, although young inflorescences of constitutive overexpression plants were profiled and thus we cannot be certain of the precise genes B1 regulates during awn inhibition from the present results, there was an association of transcription factors related to auxin and cell proliferation. Furthermore, genes known to participate in awn and lemma development were among the genes downregulated in a proportional manner to B1 overexpression. For example, the wheat ortholog of length 2 (Lks2), a SHORT‐INTERNODES/STYLISH (SHI/STY) family transcription factor, was downregulated in unison with B1 overexpression. Mutants of Lks2 in barley are short‐awned and the lks2 mutation has been suggested to reduce cell divisions in developing awns by affecting auxin concentrations (Yuo et al., 2012).
The genes that displayed the highest negative correlation to B1 overexpression were orthologs of Oryza sativa (Os)MADS1, a key regulator of differentiation and proliferation of the lemma and palea in rice (Jeon et al., 2000; Prasad et al., 2005). The barley ortholog, MADS‐box 7 (HORVU4Hr1G067680), is specifically expressed in the lemma and palea at the awn primordium stage (Schmitz et al., 2000). Interestingly, expression of an alternatively spliced OsMADS1 is associated with increased grain length in rice (Liu et al., 2018). Liu et al. (2018) also demonstrated that OsMADS1 is a regulator of auxin synthesis, transport and response. Whether B1 regulates wheat MADS1 genes for awn inhibition is unknown, but nonetheless the present data suggest a working hypothesis that B1 expression inhibits genes involved in cell proliferation pathways required for awn development. As an analogy to this proposed B1 functionality, Arabidopsis KNUCKLES, a C2H2‐EAR‐motif protein with weak homology to B1, has been shown to repress the homeobox gene WUSCHEL to inhibit floral stem cell activity, helping to balance cell proliferation and differentiation in flower development (Sun et al., 2009).
Although it is suggested that B1 acts as a transcriptional repressor, targeting genes related to cell proliferation, questions remain as to the specific genes that B1 represses for awn inhibition. Analyses of transcriptomes of B1 knockdown plants with varied expression levels will contribute to defining the true B1 regulatory network. Furthermore, the tissues and cell layers, for example within the lemma, which express B1 in awned and awnletted wheat need to be established. Experiments examining the distribution of auxin, through reporter assays such as DR5::GUS, should be performed in awned and awnletted wheat (Ulmasov et al., 1997). Questions remain, but we nonetheless present the identification of the B1 awn inhibitor gene and its phylogeny and haplotype history. Furthermore, we provide evidence linking B1 to suppression of awn formation through transcriptional repression laying the groundwork to further explore B1 functionality including interacting protein partners and possible downstream targets that cause awn inhibition.
Author contributions
DH, AJC, CAM, PN and JAF designed the research; DH, QZ, TM, YB, DJFK, MM, MC, NMA, CC and AS performed the research; DH, FMY, SK, CAM, PN and JAF analyzed the data; and DH, SK, NMA, PN, CAM and JAF wrote the paper.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phenotypes of the parental cultivars and F1 offspring of Canadian cultivar Strongfield (ST) and the Australian cultivar Glossy Huguenot (GH).
Fig. S2 Fine‐mapping of awn trait in STxGH F2 population.
Fig. S3 clustalw alignment of B1 and related proteins.
Fig. S4 B1 overexpression in the bread wheat cultivar Bobwhite inhibits awn growth.
Fig. S5 B1 overexpression consistently represses awn growth in T0 plants.
Fig. S6 Inflorescence phenotypes of B1 overexpression lines at maturity.
Fig. S7 Reduction in plant height and awn length resulting from overexpression of B1 gene.
Fig. S8 Workflow to identify the top 100 B1 similarly and oppositely expressed genes in B1 overexpression lines.
Fig. S9 Amino acid alignments of wheat proteins with orthology to MADS1, VRS2 and Lks2, whose encoding genes were differentially regulated in B1 overexpression lines.
Fig. S10 Association of KASP markers 5A_AL7 and 5A_AL1 to awn inhibition.
Methods S1 Bulked segregant analysis with RNA sequencing (BSR‐seq) in durum wheat.
Methods S2 Gene overexpression in durum and bread wheat.
Methods S3 Statistical analyses, data mining and data visualization.
Methods S4 Marker screening and QTL mapping in durum wheat.
Methods S5 Mapping B1 in hexaploid awned mutants.
Methods S6 BAC library screening.
Methods S7 Gene expression studies by quantitative PCR.
Methods S8 Characterizing wheat varieties for the TraesCS5A02G542800 haplotypes.
Methods S9 B1 mapping in hexaploid wheat biparental RIL population of BW278 and AC Foremost.
Notes S1 Association of two KASP markers to awn inhibition in durum and bread wheat.
Notes S2 Differentially expressed genes from BSR‐seq demonstrate awns are a photosynthetic organ.
Table S1 Bulked segregant RNA‐sequencing (BSR‐seq) analyses.
Table S2 Phenotypes of B1 overexpression plants.
Table S3 Differentially expressed genes in B1 overexpression plants.
Table S4 Gene ontology analyses of differentially expressed genes in B1 overexpression lines.
Table S5 Identification of genes with expression profiles similar or opposite to B1 expression.
Table S6 Haplotype analysis of B1 genomic region in diverse wheat germplasm.
Table S7 Primers used in the present study.
Acknowledgements
We thank the following individuals and organizations: Yuefeng Ruan and Ron Knox from Agriculture and Agri‐Food Canada for providing grain of the Strongfield cultivar; Sally Norton from the Australian Grains Genebank and Brett Lobsey from the New South Wales Department of Primary Industries who provided the Glossy Huguenot (AUS2499) grain; at the NRC, Joe Hammerlindl and Allan Kolenovsky for wheat transformation; NRC‐Saskatoon Genomics Facility for RNA sequencing; and the NRC‐Saskatoon Plant Growth Facilities team, especially Theo Maatman; Luzie Wingen and Brande Wulff of JIC for assistance with bioinformatics, and Marianna Pasquariello of JIC for screening of the BAC library. We thank Rainer Melzer, University College Dublin and an anonymous reviewer for helping improve the manuscript. Work at NRC was supported by the Canadian Wheat Improvement program of the National Research Council of Canada. Work at JIC was supported by grants BB/J004588/1, BB/P013511/1 and BB/P016855/1 from BBSRC, the International Wheat Yield Partnership consortium (project no. IWYP76), and the John Innes Foundation.
References
- Alvarez JP, Goldshmidt A, Efroni I, Bowman JL, Eshed Y. 2009. The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell 21: 1373–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K et al 2017. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. [DOI] [PubMed] [Google Scholar]
- Bessho‐Uehara K, Wang DR, Furuta T, Minami A, Nagai K, Gamuyao R, Asano K, Angeles‐Shim RB, Shimizu Y, Ayano M et al 2016. Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proceedings of the National Academy of Sciences, USA 113: 8969–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner A, Schäfer M, Schmidt A, Grau M, Vorwald J. 2005. Associations between geographical origin and morphological characters in bread wheat (Triticum aestivum L.). Plant Genetic Resources 3: 360–372. [Google Scholar]
- Broman KW, Gatti DM, Simecek P, Furlotte NA, Prins P, Sen Ś, Yandell BS, Churchill GA. 2019. R/qtl2: Software for mapping quantitative trait loci with high‐dimensional data and multiparent populations. Genetics 211: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull H, Casao MC, Zwirek M, Flavell AJ, Thomas WTB, Guo W, Zhang R, Rapazote‐Flores P, Kyriakidis S, Russell J et al 2017. Barley SIX‐ROWED SPIKE3 encodes a putative Jumonji C‐type H3K9me2/me3 demethylase that represses lateral spikelet fertility. Nature Communications 8: 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, Mascher M, Buluç A, Barry K, Georganas E, Session A, Strnadova V, Jenkins J, Sehgal S, Oliker L et al 2015. A whole‐genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biology 16: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chongloi GL, Prakash S, Vijayraghavan U. 2019. Rice shoot and floral meristem development: an overview of developmental regulators of meristem maintenance and organ identity. Journal of Experimental Botany 70: 1719–1736. [DOI] [PubMed] [Google Scholar]
- Clarke JM, McCaig TN, DePauw RM, Knox RE, Clarke FR, Fernandez MR, Ames NP. 2005. Strongfield durum wheat. Canadian Journal of Plant Science 85: 651–654. [Google Scholar]
- Clavijo B et al 2019. Sequencing the wheat genome. 10 + Genome Project . [WWW document] URL https://opendata.earlham.ac.uk/opendata/data/ [accessed 31 March 2019].
- Czesnick H, Lenhard M. 2015. Size control in plants – lessons from leaves and flowers. Cold Spring Harbor Perspectives in Biology 7: a019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt N, Guedira M, Lauer E, Sarinelli M, Tyagi P, Fu D, Hao Q, Murphy JP, Marshall D, Akhunova A et al 2020. Sequence based mapping identifies a candidate transcription repressor underlying awn suppression at the B1 locus in wheat. New Phytologist 225: 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Greenwood JR, Bencivenga S, Zhang P, Cockram J, Mellers G, Ramm K, Cavanagh C, Swain SM, Boden SA. 2018. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum). Plant Cell 30: 563–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockter C, Gruszka D, Braumann I, Druka A, Druka I, Franckowiak J, Gough SP, Janeczko A, Kurowska M, Lundqvist J et al 2014. Induced variations in brassinosteroid genes define barley height and sturdiness, and expand the green revolution genetic toolkit. Plant Physiology 166: 1912–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, Wang L, Zhu T, Jorgensen CM, Luo MC, Deal KR, Gu YQ, Gill BS, Distelfeld A, Devos KM et al 2018. Reassessment of the evolution of wheat chromosomes 4A, 5A, and 7B. Theoretical and Applied Genetics 131: 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum R, Zaltzman L, Burgert I, Fratzl P. 2007. The role of wheat awns in the seed dispersal unit. Science 316: 884–886. [DOI] [PubMed] [Google Scholar]
- Fabien L, Hitoshi Y. 2015. Interpreting lemma and palea homologies: a point of view from rice floral mutants. Frontiers in Plant Science 6: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution 17: 368–376. [DOI] [PubMed] [Google Scholar]
- Fuller DQ, Allaby R. 2009. Seed dispersal and crop domestication: shattering, germination and seasonality in evolution under cultivation In: Østergaard L, ed. Fruit development and seed dispersal. Oxford, UK: Blackwell Publishing, 238–295. [Google Scholar]
- Gentleman R, Carey V, Huber W, Hahne F. 2018. Genefilter: methods for filtering genes from high‐throughput experiments, R package v.1.64.0 . [WWW document] URL https://bioconductor.org/packages/genefilter/ [accessed 17 February 2019].
- Girin T, Sorefan K, Ostergaard L. 2009. Meristematic sculpting in fruit development. Journal of Experimental Botany 60: 1493–1502. [DOI] [PubMed] [Google Scholar]
- Goncharov NP, Mitina RL, Anfilova NA. 2003. Inheritance of awnlessness in tetraploid wheat species. Russian Journal of Genetics 39: 463–466. [PubMed] [Google Scholar]
- Grewal S, Hubbart‐Edwards S, Yang C, Scholefield D, Ashling S, Burridge A, Wilkinson PA, King IP, King J. 2018. Detection of T. urartu introgressions in wheat and development of a panel of interspecific introgression lines. Frontiers in Plant Science 9: 1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundbacher FJ. 1963. The physiological function of the cereal awn. Botanical Review 29: 366–381. [Google Scholar]
- Gu B, Zhou T, Luo J, Liu H, Wang Y, Shangguan Y, Zhu J, Li Y, Sang T, Wang Z et al 2015. An‐2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Molecular Plant 8: 1635–1650. [DOI] [PubMed] [Google Scholar]
- Haas M, Schreiber M, Mascher M. 2019. Domestication and crop evolution of wheat and barley: genes, genomics, and future directions. Journal of Integrative Plant Biology 61: 204–225. [DOI] [PubMed] [Google Scholar]
- Hua L, Wang DR, Tan L, Fu Y, Liu F, Xiao L, Zhu Z, Fu Q, Sun X, Gu P et al 2015. LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27: 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Irish VF. 2016. Gene networks controlling petal organogenesis. Journal of Experimental Botany 67: 61–68. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) , Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A et al 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG et al 2000. leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Hua L, Zhu Z, Tan L, Zhao X, Zhang W, Liu F, Fu Y, Cai H, Sun X et al 2016. GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell 28: 2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Richards RA, Turner NC. 1983. Yield, water relations, gas exchange, and surface reflectances of near‐isogenic wheat lines differing in glaucousness. Crop Science 23: 318–325. [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. 2010. Genome‐wide analysis of ethylene‐responsive element binding factor‐associated amphiphilic repression motif‐containing transcriptional regulators in Arabidopsis. Plant Physiology 152: 1109–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. 2011. EAR motif‐mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Miura H, Akiyama M, Kuroshima M, Sawada S. 1998. RFLP mapping of the three major genes, Vrn1, Q and B1, on the long arm of chromosome 5A of wheat. Euphytica 101: 91–95. [Google Scholar]
- Kazan K. 2006. Negative regulation of defence and stress genes by EAR‐motif‐containing repressors. Trends in Plant Science 11: 109–112. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, Carvalho‐Silva D, Christensen M, Davis P, Grabmueller C et al 2018. Ensembl Genomes 2018: an integrated omics infrastructure for non‐vertebrate species. Nucleic Acids Research 46: D802–D808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A et al 2007. Six‐rowed barley originated from a mutation in a homeodomain‐leucine zipper I‐class homeobox gene. Proceedings of the National Academy of Sciences, USA 104: 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppolu R, Anwar N, Sakuma S, Tagiri A, Lundqvist U, Pourkheirandish M, Rutten T, Seiler C, Himmelbach A, Ariyadasa R et al 2013. Six‐rowed spike4 (Vrs4) controls spikelet determinacy and row‐type in barley. Proceedings of the National Academy of Sciences, USA 110: 13198–13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppolu R, Schnurbusch T. 2019. Developmental pathways for shaping spike inflorescence architecture in barley and wheat. Journal of Integrative Plant Biology 61: 278–295. [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Vasquez‐Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, Simmonds J, Ramirez‐Gonzalez RH, Wang X, Borrill P et al 2017. Uncovering hidden variation in polyploid wheat. Proceedings of the National Academy of Sciences, USA 114: E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA. 2011. Auxin regulation of Arabidopsis flower development involves members of the AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) family. Journal of Experimental Botany 62: 3311–3319. [DOI] [PubMed] [Google Scholar]
- Le Couviour F, Faure S, Poupard B, Flodrops Y, Dubreuil P, Praud S. 2011. Analysis of genetic structure in a panel of elite wheat varieties and relevance for association mapping. Theoretical and Applied Genetics 123: 715–727. [DOI] [PubMed] [Google Scholar]
- Liu Q, Han R, Wu K, Zhang J, Ye Y, Wang S, Chen J, Pan Y, Li Q, Xu X et al 2018. G‐protein βγ subunits determine grain size through interaction with MADS‐domain transcription factors in rice. Nature Communications 9: 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Tang HM, Nettleton D, Schnable PS. 2012. Gene mapping via bulked segregant RNA‐Seq (BSR‐Seq). PLoS ONE 7: e36406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Liu H, Zhou T, Gu B, Huang X, Shangguan Y, Zhu J, Li Y, Zhao Y, Wang Y et al 2013. An‐1 encodes a basic helix‐loop‐helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25: 3360–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. 1999. Genes controlling fertilization‐independent seed development in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 96: 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo N, Zhu T, Wang L, Wang Y et al 2017. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii . Nature 551: 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IJ, Bansept‐Basler P, Barber T, Bentley AR, Cockram J, Gosman N, Greenland AJ, Horsnell R, Howells R, O'Sullivan DM et al 2014. An eight‐parent multiparent advanced generation inter‐cross population for winter‐sown wheat: creation, properties, and validation. G3 4: 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz‐Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. 2004. Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16: 1692–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Fernández I, Sanchís S, Marini N, Balanzá V, Ballester P, Navarrete‐Gómez M, Oliveira AC, Colombo L, Ferrándiz C. 2014. The effect of NGATHA altered activity on auxin signaling pathways within the Arabidopsis gynoecium. Frontiers in Plant Science 5: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J et al 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433. [DOI] [PubMed] [Google Scholar]
- Meng L, Li H, Zhang L, Wang J. 2015. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop Journal 3: 269–283. [Google Scholar]
- Morita R, Kusaba M, Iida S, Yamaguchi H, Nishio T, Nishimura M. 2009. Molecular characterization of mutations induced by gamma irradiation in rice. Genes & Genetic Systems 84: 361–370. [DOI] [PubMed] [Google Scholar]
- Mulki MA, Bi X, von Korff M. 2018. FLOWERING LOCUS T3 controls spikelet initiation but not floral development. Plant Physiology 178: 1170–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller KJ, Romano N, Gerstner O, Garcia‐Maroto F, Pozzi C, Salamini F, Rohde W. 1995. The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374: 727–730. [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme‐Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z, Saranga Y, Fahima T, Aharoni A, Elbaum R. 2010. Genetic control over silica deposition in wheat awns. Physiologia Plantarum 140: 10–20. [DOI] [PubMed] [Google Scholar]
- Pinon V, Prasad K, Grigg SP, Sanchez‐Perez GF, Scheres B. 2011. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 110: 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U. 2005. OsMADS1, a rice MADS‐box factor, controls differentiation of specific cell types in the lemma and palea and is an early‐acting regulator of inner floral organs. The Plant Journal 43: 915–928. [DOI] [PubMed] [Google Scholar]
- Ramsay L, Comadran J, Druka A, Marshall DF, Thomas WT, Macaulay M, MacKenzie K, Simpson C, Fuller J, Bonar N et al 2011. INTERMEDIUM‐C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1 . Nature Genetics 43: 169–172. [DOI] [PubMed] [Google Scholar]
- Rebetzke GJ, Bonnett DG, Reynolds MP. 2016. Awns reduce grain number to increase grain size and harvestable yield in irrigated and rainfed spring wheat. Journal of Experimental Botany 67: 2573–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma S, Golan G, Guo Z, Ogawa T, Tagiri A, Sugimoto K, Bernhardt N, Brassac J, Mascher M, Hensel G, et al. 2019. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proceedings of the National Academy of Sciences, USA 116: 5182–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller CW, Qualset CO. 1975. Isogenic analysis of productivity in barley: interaction of genes affecting awn length and leaf‐spotting. Crop Science 15: 378–382. [Google Scholar]
- Schmitz J, Franzen R, Ngyuen TH, Garcia‐Maroto F, Pozzi C, Salamini F, Rohde W. 2000. Cloning, mapping and expression analysis of barley MADS‐box genes. Plant Molecular Biology 42: 899–913. [DOI] [PubMed] [Google Scholar]
- Schrager‐Lavelle A, Klein H, Fisher A, Bartlett M. 2017. Grass flowers: an untapped resource for floral evo‐devo. Journal of Systematics and Evolution 55: 525–541. [Google Scholar]
- Sotelo‐Silveira M, Cucinotta M, Chauvin AL, Chávez Montes RA, Colombo L, Marsch‐Martínez N, de Folter S. 2013. Cytochrome P450 CYP78A9 is involved in Arabidopsis reproductive development. Plant Physiology 162: 779–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourdille P, Cadalen T, Gay G, Gill B, Bernard M. 2002. Molecular and physical mapping of genes affecting awning in wheat. Plant Breeding 121: 320–324. [Google Scholar]
- Sun B, Xu Y, Ng KH, Ito T. 2009. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes and Development 23: 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M. 1983. Catalogue of Aegilops‐Triticum germ‐plasm preserved in Kyoto University. Kyoto, Japan: Plant Germ‐Plasm Institute, Faculty of Agriculture, Kyoto University. [Google Scholar]
- Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama‐Tsuchida T, Ichikawa H, Mitsuda N, Ohme‐Takagi M, Hirano HY. 2012. The YABBY gene TONGARI‐BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell 24: 80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi J, Kawano K. 1972. Suitou no noge ni tsuite (about the awn in rice). Nippon sakumotsu‐gakkai toukai‐shibu kenkyu‐happyou kougai (Research Reports of Tokai Branch of Crop Science Society of Japan) 56: 11–15. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T, Hirano HY. 2014. The DROOPING LEAF and OsETTIN2 genes promote awn development in rice. The Plant Journal 77: 616–626. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Ding Y, Cai C, Chen Z, Zhu C. 2018. The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiologia Plantarum 165: 690–700. [DOI] [PubMed] [Google Scholar]
- Wang T, Zou T, He Z, Yuan G, Luo T, Zhu J, Liang Y, Deng Q, Wang S, Zheng A et al 2018. GRAIN LENGTH AND AWN 1 negatively regulates grain size in rice. Journal of Integrative Plant Biology 61: 1036–1042. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiao Y. 2017. Auxin and above‐ground meristems. Journal of Experimental Botany 69: 147–154. [DOI] [PubMed] [Google Scholar]
- Watkins AE, Ellerton S. 1940. Variation and genetics of the awn in Triticum . Journal of Genetics 40: 243–270. [Google Scholar]
- Williams‐Carrier RE, Lie YS, Hake S, Lemaux PG. 1997. Ectopic expression of the maize kn1 gene phenocopies the Hooded mutant of barley. Development 124: 3737–3745. [DOI] [PubMed] [Google Scholar]
- Wingen LU, Orford S, Goram R, Leverington‐Waite M, Bilham L, Patsiou TS, Ambrose M, Dicks J, Griffiths S. 2014. Establishing the A. E. Watkins landrace cultivar collection as a resource for systematic gene discovery in bread wheat. Theoretical and Applied Genetics 127: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Tang J, Li Y, Wang W, Li X, Jin L, Xie R, Luo H, Zhao X, Meng Z et al 2009. STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. The Plant Journal 59: 789–801. [DOI] [PubMed] [Google Scholar]
- Yan A, Wu M, Zhao Y, Zhang A, Liu B, Schiefelbein J, Gan Y. 2014. Involvement of C2H2 zinc finger proteins in the regulation of epidermal cell fate determination in Arabidopsis. Journal of Integrative Plant Biology 56: 1112–1117. [DOI] [PubMed] [Google Scholar]
- Yoshioka M, Iehisa JCM, Ohno R, Kimura T, Enoki H, Nishimura S, Nasuda S, Takumi S. 2017. Three dominant awnless genes in common wheat: fine mapping, interaction and contribution to diversity in awn shape and length. PLoS ONE 12: e0176148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef HM, Eggert K, Koppolu R, Alqudah AM, Poursarebani N, Fazeli A, Sakuma S, Tagiri A, Rutten T, Govind G et al 2017. VRS2 regulates hormone‐mediated inflorescence patterning in barley. Nature Genetics 49: 157–161. [DOI] [PubMed] [Google Scholar]
- Yuo T, Yamashita Y, Kanamori H, Matsumoto T, Lundqvist U, Sato K, Ichii M, Jobling SA, Taketa S. 2012. A SHORT INTERNODES (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. Journal of Experimental Botany 63: 5223–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yuan Z. 2014. Molecular control of grass inflorescence development. Annual Review of Plant Biology 65: 553–578. [DOI] [PubMed] [Google Scholar]
- Zhang S, Wu T, Liu S, Xi L, Ling J, Wan J. 2016. Disruption of OsARF19 is critical for floral organ development and plant architecture in rice (Oryza sativa L.). Plant Molecular Biology Reporter 34: 748–760. [Google Scholar]
- Zhang Y, Xie H, Qian Q, Fang X, Chen F. 2011. GenBank: ADK55064.1, C2H2 zinc finger malformed spikelet [Oryza sativa Japonica Group] . [WWW document] URL https://www.ncbi.nlm.nih.gov/protein/301068493 [accessed 03 March 2019].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phenotypes of the parental cultivars and F1 offspring of Canadian cultivar Strongfield (ST) and the Australian cultivar Glossy Huguenot (GH).
Fig. S2 Fine‐mapping of awn trait in STxGH F2 population.
Fig. S3 clustalw alignment of B1 and related proteins.
Fig. S4 B1 overexpression in the bread wheat cultivar Bobwhite inhibits awn growth.
Fig. S5 B1 overexpression consistently represses awn growth in T0 plants.
Fig. S6 Inflorescence phenotypes of B1 overexpression lines at maturity.
Fig. S7 Reduction in plant height and awn length resulting from overexpression of B1 gene.
Fig. S8 Workflow to identify the top 100 B1 similarly and oppositely expressed genes in B1 overexpression lines.
Fig. S9 Amino acid alignments of wheat proteins with orthology to MADS1, VRS2 and Lks2, whose encoding genes were differentially regulated in B1 overexpression lines.
Fig. S10 Association of KASP markers 5A_AL7 and 5A_AL1 to awn inhibition.
Methods S1 Bulked segregant analysis with RNA sequencing (BSR‐seq) in durum wheat.
Methods S2 Gene overexpression in durum and bread wheat.
Methods S3 Statistical analyses, data mining and data visualization.
Methods S4 Marker screening and QTL mapping in durum wheat.
Methods S5 Mapping B1 in hexaploid awned mutants.
Methods S6 BAC library screening.
Methods S7 Gene expression studies by quantitative PCR.
Methods S8 Characterizing wheat varieties for the TraesCS5A02G542800 haplotypes.
Methods S9 B1 mapping in hexaploid wheat biparental RIL population of BW278 and AC Foremost.
Notes S1 Association of two KASP markers to awn inhibition in durum and bread wheat.
Notes S2 Differentially expressed genes from BSR‐seq demonstrate awns are a photosynthetic organ.
Table S1 Bulked segregant RNA‐sequencing (BSR‐seq) analyses.
Table S2 Phenotypes of B1 overexpression plants.
Table S3 Differentially expressed genes in B1 overexpression plants.
Table S4 Gene ontology analyses of differentially expressed genes in B1 overexpression lines.
Table S5 Identification of genes with expression profiles similar or opposite to B1 expression.
Table S6 Haplotype analysis of B1 genomic region in diverse wheat germplasm.
Table S7 Primers used in the present study.


