Abstract
Purpose
To explore the effect of miR-449a inhibits migration and invasion by targeting Notch1 and regulating epithelial–mesenchymal transition (EMT) in hepatocellular carcinoma (HCC), and further study on the molecular mechanism.
Patients and methods
The expression of miR-449a and Notch1 in HCC cells and tissues was detected by qRT-PCR. The HCC cell line HCCLM3 and SMMC-7721 were transfected with miR-449a. The invasion and migration of HCC cell lines were detected by transwell assay and wound healing assay. The Notch pathway and EMT related protein were detected with Western Blotting. The specific binding site of mir-449a on notch1 gene was detected by luciferase assay.
Results
We found the expression of miR-449a was related to short-term recurrence of hepatocellular carcinoma after hepatectomy and acted as independent risk factors of DFS and OS. The expression of miR-449a decreased in tumor tissues and HCC cell lines, but the expression of Notch1 increased. The overexpressed miR-449a promoted the invasiveness in vitro by regulating EMT via Notch pathway. Mechanically, miR-449a inhibited the translation of Notch1 protein by binding to 3ʹ UTR of its mRNA directly.
Conclusion
miR-449a is short-term recurrence-related miRNA and inhibits the invasion and metastasis ability of HCC cells by regulating EMT via Notch pathway. miR-449a may be a new effective therapeutic target for HCC.
Keywords: signal pathway, microRNA, epithelial-mesenchymal transition, bioinformatics
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignancy in the world and the second leading cause of cancer death.1 At present, the main treatment of HCC is surgical resection, transplantation, radiofrequency ablation (RFA) and transcatheter arterial chemoembolization (TACE). But the recurrence and metastasis rate is still high.2–4 Therefore, it is essential to study the molecular mechanisms that affect the invasion and migration of HCC. Related studies have shown that Epithelial–Mesenchymal Transition (EMT) is a key factor in tumor migration and invasion.5 The transition of tumor from epithelial to mesenchymal was accomplished by loss of cell adhesion and changed expression of mesenchymal markers such as E-cadherin and vimentin, and increased invasive ability of tumor cells.
MicroRNA binds to target mRNA by identifying specific site then exerting regulation of post-transcriptional level.6–8 The miR-449a which located in the first intron of cdc20b, chromosome 5q11, were decreased in a variety of tumor. It can inhibit invasion and migration of many tumors such as gastric cancer, lung cancer, nasopharyngeal cancer, etc.9–14 Related study found that the expression of miR-449a was decreased in HCC, and inhibited metastasis through targeted FOS and Met.11 In addition, it can induce apoptosis of HCC by targeting Calpain6 and POU2F1,15 and inhibit the proliferation, migration, and invasion of HCC by targeted ADAM10.16 However, the biological function of miR-449a which can inhibit invasion and migration of HCC cell by regulating Notch pathway has not been reported, and the molecular mechanism of miR-449a directly targeting Notch1 in HCC cell is not fully elucidated.
In this study, we demonstrate that miR-449a can inhibit invasion and migration of HCC cells in vitro and in vivo through inactivating Notch pathway by directly targeting notch1. Therefore, the data implicate miR-449a as a target of therapy for metastasis of HCC.
Materials and Methods
microRNA Sequencing
Total RNAs from 8 HCC tissue samples with short-term recurrence (STR, recurrence in 6 months) and 8 HCC samples without recurrence were pooled, respectively, and then sent to Beijing Genomics Institute (Shenzhen, China) for miRNAs sequencing. The analysis was made on Illumina HiSeq2000 platform (Illumina Inc., San Diego, CA, USA). Comparisons were made between the sequencing results and the Homo Sapiens microRNA precursor in miRbase database (if there was no microRNA precursor in the miRbase, the mature microRNA sequence was used) to obtain the quantity of microRNA in HCC samples.
Tissue Samples and Patient’s Follow-Up
A total of 50 consecutive patients with HCC, who underwent hepatectomy at the Affiliated Hospital of Qingdao University between January 2011 and December 2012, were enrolled in this study. All patients were followed-up every month during the first half-year and at least every 3–4 months thereafter. The mean follow-up period that ended on June 30th 2018 was 76.1 months. All tissue samples were immediately added with RNAlaterTM (Beyotime Biotechnology, Shanghai, China) after resection and stored at 80°C until required. This study was approved by the Research Ethics Committee of the Affiliated Hospital of Qingdao University, and written informed consent was obtained from all patients.
Cell Culture
HCC cell lines (SMMC-7721, QGY-7703, BEL-7402 and HCCLM3) and normal hepatic cell line HL-7702 were purchased from the cell resource center of the Shanghai Institutes for Sciences (Shanghai, China). SMMC-7721 cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco BRL, MD, USA) supplemented with 10% fetal bovine serum (FBS, Gibco BRL, MD, USA) and incubated in 5% CO2 at 37°C.
The Expression of miR-449a and Notch1 by Real-Time Quantitative PCR Assay
Total RNA was extracted from HCC cell lines and tissue samples using TRIZOL ® reagent (Takara, Otsu, Japan). For quantify miR-449a, cDNA was synthesized with the TaqMan MicroRNA reverse transcription kit (GenePharma, Jiangsu, China), and then was quantified using the QuantMir RT Kit (GenePharma, Jiangsu, China) under LightCycler® Instrument (Roche Diagnostics, Indiana, USA) with miR-449a (forward: 5ʹ-CTCGCTGGCAGTGTATTGTTAG-3ʹ;reverse:5ʹ-TATCGTTGTACTCCAGACCAAGAC-3ʹ) and U6 (forward: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; reverse: 5 ʹ-AACGCTTCACGAATTTGCGT-3ʹ). The expression of Notch1, RNA was reverse-transcribed into cDNA by a PrimscriptTM RT Reagent (Takara, Otsu, Japan), then was quantified using Real-time quantitative PCR Mixture Reagent (Takara, Otsu, Japan) under LightCycler® Instrument with Notch1 (forward: 5ʹ-CGGGTCCACCAGT TTGAATG-3ʹ; reverse: 5ʹ-GTTGTATTGGTTCGGCACCAT-3ʹ) and β-actin (forward: 5ʹ-CACCATGAAGATCAAGATCATTGC-3ʹ; reverse: 5ʹ-GGCCGGACTCATCGT ACTCCTGC-3‘). The relative expression of miR-449a was normalized to U6 and the expression of Notch1 was normalized to β-actin by using 2-△△Ct method.
Luciferase Reporter Assay
The wild type and mutant 3´untranslated region (3´UTR) of Notch1 were synthesized by GenePharma (Jiangsu, China), and inserted into downstream of the cytomegalovirus promoter in a pMIR-REPORT vector. For the luciferase reporter assay, SMMC-7721cells were cultured in the 96-well plate at 50–60% confluence before transfection. Co-transfections of Notch1 3´UTR plasmid with lenti-miR-449a or lenti-miR-control into SMMC-7721 cells were accomplished with Lipofectamine 2000 (Invitrogen, CA, USA). Luciferase activities were determined 48 hrs after transection using a Dual-luciferase Reporter Analysis System (Pro-mega, WI, USA).
Transfection
Human miR-449a mimic, negative control (NC mimic), miR-449a inhibitor and negative control (NC inhibitor) were synthesized by GenePharma (Jiangsu, China). The cells of SMMC-7721 and HCCLM3 were seeded in 12-well plate at 70% confluence and transfected with 20 pmol miR-449a per well using Lipofectamine 2000 (Invitrogen, CA, USA). Cells transfected with miRNA were harvested 48 hrs after transfection. Then, the efficiency of overexpression and knockdown of miR-449a was detected by real-time quantitative PCR.
Cell Invasion and Migration Assay
The invasion and migration of SMMC-7721 and HCCLM3 were assessed using 8.0 μm pores transwell polycarbonate membranes (Corning Inc., NY, USA). A total of 1×105 cells were plated in 12-well plated in DMEM containing 10% FBS and transfected with miRNA for 48 hrs. For invasion assay, SMMC-7721 cells (20×105 cells/well) and HCCLM3 cells (10 ×105 cells/well) were seeded on the transwell membranes precoated with Matrigel (BD, NJ, USA) using serum-free DMEM for 48 hrs and 24 hrs at 37°C. After that, the non-invading cells were gently removed with a cotton swab, while cells were fixed with paraformaldehyde for 15 min, and stained with 0.1% crystal violet for 15 mins. For migration assay, SMMC-7721 cells (20×105 cells/well) and HCCLM3 cells (10 ×105 cells/well) were seeded on the membranes without Matrigel using serum-free DMEM for 48 hrs and 24 hrs at 37°C. The cells were photographed and quantified by counting them in randomly six fields by a light microscope (Olympus; Olympus Corp., Tokyo, Japan) at ×200 magnification.
Western Blot Assay
Total protein of cell was extracted and protein concentration was measured using the BCA (bicinchoninic acid) protein quantitation kit (Thermo Fisher Scientific, MA, USA). A total of 20μg protein from each sample were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto 0.45 μm polyvinylidene difluoride membranes (Bio-Rad Laboratories, CA, USA). Then, the membranes were incubated with primary antibodies overnight at 4°C. The primary antibodies for Notch1 (D1E11; #3608s; 1:1000 dilution), E-cadherin (4A2; #14472s; 1:1000 dilution), Vimentin (D21H3; #5741s; 1:1000 dilution), Cleaved Notch1 (Val1744; D3B8; #4147; 1:1000 dilution), c-Myc (D3N8F; #13987; 1:1000 dilution) and β-actin (8H10D10; #3700s; 1:1000 dilution) were purchased from Cell Signaling Technology (Danvers, MA, USA). The membranes were incubated with secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse (ab6789, 1:10,000 dilution, Abcam, Cambridge, UK) and goat anti-rabbit antibodies (ab6721, 1:10,000 dilution, Abcam, Cambridge, UK) at room temperature for 2 hrs. The bands were detected using an enhanced chemiluminescence reagent and visualized with a Fusion FX7 System (Vilber Lourmat, France). ImageJ software was used to calculate the intensity (gray value) of each protein band, which was normalized to that for GAPDH.
Statistical Analysis
Data were analyzed using GraphPad Prism 6.0 software (San Diego, CA, USA). Quantitative variables were expressed as mean SD, and analyzed by Student´s t-test and one-way ANOVA. Experimental results are independently repeated three times and P<0.05 was considered statistically significant.
Results
Small RNA Sequencing
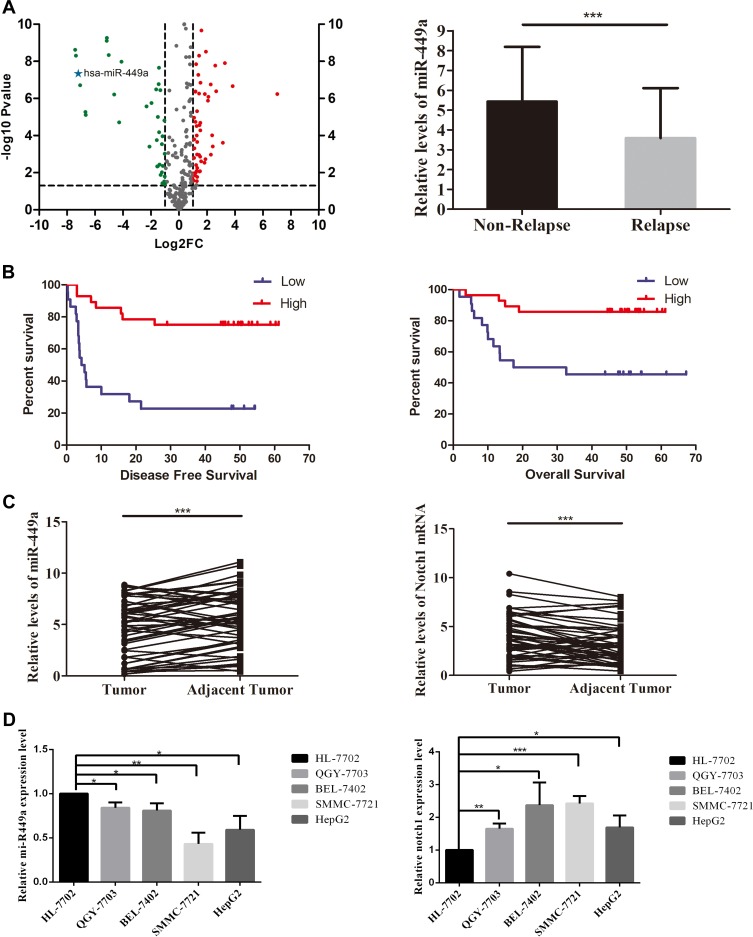
To explore the significance of miRNAs in HCC tissue with recurrence in 6 months, small RNA high-throughput sequencing was performed to compare the miRNA expression patterns between recurrence group and non-recurrence group. We found that miR-449a was one of the significantly downregulated miRNAs in HCC tissues with recurrence in 6 months (Figure 1A).
Figure 1.
Small RNA sequencing and miR-449a expression in HCC tissue. (A) Left-Volcano plot for small RNA sequencing screening. The fold change was miRNA expression in short-term recurrence group (n=8) vs non-recurrence group (n=8). The miR-449a was one of the significantly downregulated miRNAs in HCC tissues with recurrence in 6 months. Right-The expression of miR-449a in non-relapse (n=34) and relapse (n=16) groups. (B) The expression of miR-449a was positively related to disease-free survival and overall survival of HCC patients. The mean miR-449a expression was used as cutoff value. The Kaplan–Meier analysis was used to estimate survival for the two groups. (C) The relative expression of mir-449a was significantly higher in HCC tissues than in adjacent nontumor tissues. Conversely, the Notch1 expression reduced in HCC tissues significantly. (D) The relative expression of mir-449a and Notch1 in HCC cell lines and hepatocyte cell line. *p<0.05, **p<0.01, ***p<0.001.
The Association of Clinicopathologic Characteristics and miR-449a Expression with STR
The basic characteristics of the patients in the STR and non-STR groups are shown in Table 117 that our team have reported it in a paper about miR-21 and STR. There was no significant difference between the two groups. But the relative expression of miR-449a was significantly lower in the STR group (≤6 months) than in non-STR group (3.59 ± 0.63 vs 5.43 ± 0.47, P < 0.05, Figure 1A).
Table 1.
Clinicopathological Characteristics of Patient in Short-Term Recurrence and Non-recurrence Groups
| Variables | Short Term Recurrence | ||||
|---|---|---|---|---|---|
| No (n=34) | Yes (n=16) | t-Test | χ2 | p-value | |
| Age (y) | 58.38±11.26 | 54.38±10.70 | 1.19 | 0.239 | |
| Gender (M/F) | 27/7 | 13/3 | 0.023 | 0.880 | |
| Tumor number | 1.03±0.17 | 1.06±0.25 | −0.547 | 0.587 | |
| Tumor size (cm) | 4.24±2.57 | 5.63±3.30 | −1.632 | 0.109 | |
| Tumor margin (cm) | 8.12±8.38 | 6.51±4.84 | 0.855 | 0.397 | |
| Pathologic differentiation(low/high) | 29/5 | 11/5 | 1.861 | 0.172 | |
| Capsule invasion | 8/34 | 7/16 | 2.118 | 0.146 | |
| Vascular invasion | 6/34 | 3/16 | 0.009 | 0.925 | |
| Liver cirrhosis | 31/34 | 13/16 | 1.015 | 0.314 | |
| HBV infection | 30/34 | 13/16 | 0.441 | 0.507 | |
| miR-449a | 3.59±0.63 | 5.43 ± 0.47 | 2.260 | 0.028 | |
Notes: Bold value represents statistically significant p<0.05. The data in this table were cited from a paper that our team have published.17
The Expression of miR-449a and Notch1 in HCC Cell Lines and Patient Samples
We found Notch1 was target gene of miR-449a with poorly conserved consequence by using the Target Scan Human Database (http://www.targetscan.org). To study the role of miR-449 and Notch1 in the development of HCC, the expression of miR-449a and Notch1 were analyzed in 50 matched HCC and adjacent nontumor tissues.
The 1-, 3-, and 5-year Disease-Free Survival (DFS) rates were 62.00%, 47.89%, and 45.43%, respectively. The overall median DFS time was 33.57 months [95% confidence interval (CI): 26.20–40.93], while the median DFS in the low expression group was 15.50 months (95% CI: 7.34–23.65) and 46.76 months (95% CI: 38.62–54.91) in the high expression group. A significant difference in DFS was detected between the low expression and high expression groups (χ2=17.570, P<0.001; Figure 1B).
The 1-, 3- and 5-year Over Survival (OS) rates were 82.00%, 68.00%, and 65.73%, respectively. The overall median OS time was 48.91months [95% confidence interval (CI): 41.66–56.16], while the median OS in the low expression group was 36.69 months (95% CI: 24.81–48.56) and 53.61 months (95% CI: 47.24–59.97) in the high expression group. A significant difference in OS was detected between the low expression and high expression groups (χ2=8.342, P=0.004; Figure 1B).
The expression of miR-449a decreased and the expression of Notch1 increased in HCC tissues compared with adjacent nontumor tissues (Figure 1C). Moreover, the expression of miR-449a in SMMC-7721 and HCCLM3 was comparatively lower than in hepatocyte and other HCC cell lines (Figure 1D). But the expression of Notch1 in SMMC-7721 and HCCLM3 was comparatively higher (Figure 1D). These results show that the expression of miR-449a and Notch1 in SMMC-7721 and HCCLM3 were consistent with HCC tissues.
Univariate and Multivariate Analysis of DFS and OS (Cox Risk Regression Model)
The factors that may influence HCC recurrence were individually included in the Cox proportional hazard model for univariate analysis (Table 2). The results showed that miR-449a expression (P< 0.001) and pathologic differentiation (P=0.016) significantly affected postoperative recurrence of HCC. However, miR-449a expression was the only independent risk factors for recurrence. (P=0.001)
Table 2.
Univariate and Multivariate Analyses of Various Prognostic Parameters in Patients with HCC Cox-Regression Analysis for DFS
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| P-value | HR | 95% CI | p-value | HR | 95% CI | |
| miR-449a | <0.001* | 0.204 | 0.090–0.461 | 0.001* | 0.231 | 0.101–529 |
| Notch1 | 0.403 | 0.932 | 0.791–1.099 | |||
| Age (y) | 0.229 | 0.979 | 0.946–1.013 | |||
| Gender | 0.361 | 1.641 | 0.567–4.749 | |||
| AFP | 0.265 | 1.578 | 0.708–3.521 | |||
| Tumor number | 0.402 | 1.853 | 0.438–7.838 | |||
| Tumor size (cm) | 0.199 | 1.690 | 0.758–3.767 | |||
| Tumor margin (cm) | 0.332 | 0.346 | 0.151–0.795 | |||
| Pathologic differentiation | 0.016* | 2.532 | 1.186–5.406 | |||
| Vascular invasion | 0.708 | 1.189 | 0.479–2.950 | |||
| Capsule invasion | 0.074 | 2.021 | 0.933–4.379 | |||
| Liver cirrhosis | 0.396 | 0.631 | 0.218–1.827 | |||
| HBV infection | 0.365 | 0.638 | 0.241–1.687 | |||
Note: *p < 0.05, statistically significant.
Abbreviations: HCC, hepatocellular carcinoma; DFS, disease-free survival; HR, hazard ratio; CI, confidence interval.
The factors that influenced HCC survival were also individually included in the Cox proportional hazard model for univariate analysis (Table 3). The miR-449a expression (P= 0.008) and pathologic differentiation (P=0.033) significantly affected postoperative recurrence of HCC. However, miR-449a expression was the only independent risk factors for recurrence. (P=0.014)
Table 3.
Univariate and Multivariate Analyses of Various Prognostic Parameters in Patients with HCC Cox-regression Analysis for OS
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| P-value | HR | 95% CI | p-value | HR | 95% CI | |
| miR-449a | 0.008* | 0.241 | 0.085–0.687 | 0.014* | 0.265 | 0.092–0.762 |
| Notch1 | 0.943 | 0.966 | 0.372–2.504 | |||
| Age(y) | 0.317 | 0.611 | 0.232–1.606 | |||
| Gender | 0.339 | 2.054 | 0.469–8.987 | |||
| AFP | 0.298 | 1.697 | 0.626–4.596 | |||
| Tumor number | 0.082 | 3.725 | 0.848–16.371 | |||
| Tumor size (cm) | 0.537 | 1.389 | 0.489–3.946 | |||
| Tumor margin (cm) | 0.137 | 0.452 | 0.159–1.287 | |||
| Pathologic differentiation | 0.033* | 2.833 | 1.086–7.386 | |||
| Vascular invasion | 0.969 | 0.976 | 0.280–3.400 | |||
| Capsule invasion | 0.276 | 1.712 | 0.650–4.507 | |||
| Liver cirrhosis | 0.071 | 0.353 | 0.114–1.091 | |||
| HBV infection | 0.224 | 0.498 | 1.162–1.530 | |||
Note: *p < 0.05, statistically significant.
miR-449a Inhibits Migration and Invasion in HCC Cell Lines
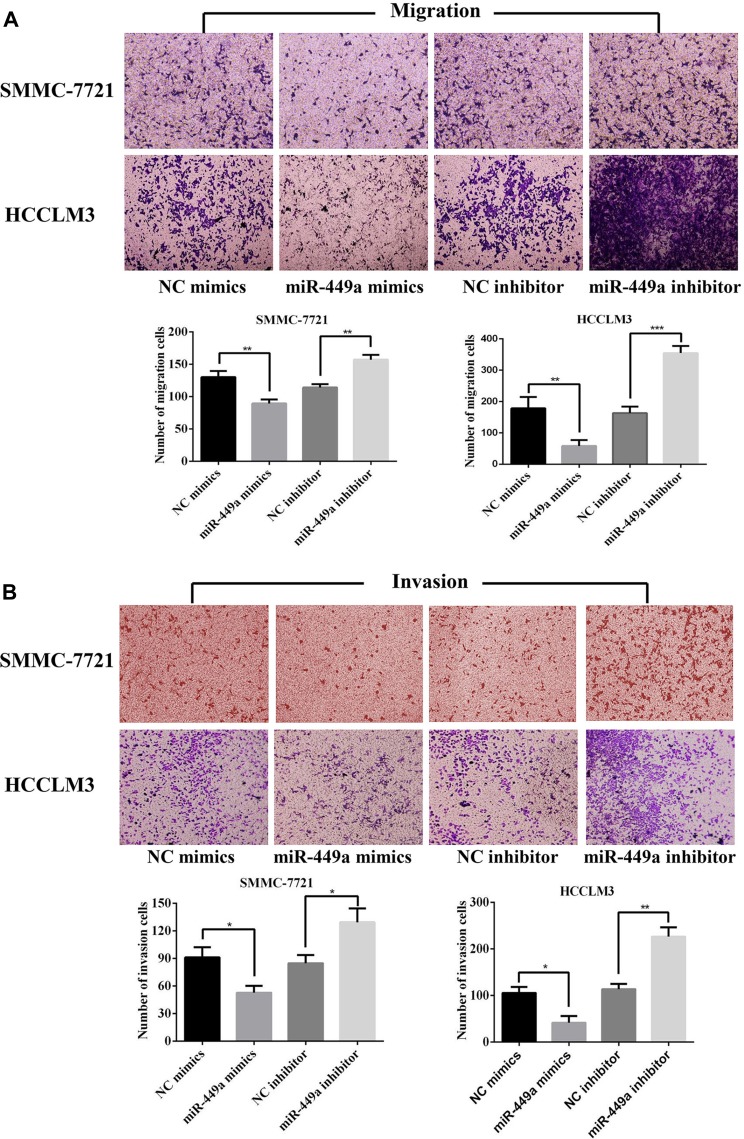
To determine whether miR-449a overexpression can suppress HCC cells migration and invasion, we transfected two HCC cell lines with miR-449a mimic and NC mimic. The metastasis ability of HCC cell lines was detected by transwell migration and invasion assay. The miR-449a up-regulation reduced SMMC-7721 and HCCLM3 cells invasion and migration compared with control group (P<0.05) (Figure 2).
Figure 2.
miR-449a inhibited the migration and invasion levels of HCC cells. (A) The results of migration assays for SMMC-7721 and HCCLM4 after transfected. (B) The results of invasion assays for SMMC-7721 and HCCLM3 after transfected. *p<0.05, **p<0.01 and ***p<0.001 compared with control group.
To determine the role of miR-449a knockdown in the HCC cells in vitro, we transfected two HCC cell lines with miR-449a inhibitor and NC inhibitor. The cell migration and invasion analysis using a transwell assay suggested that miR-449a depletion in SMMC-7721 and HCCLM3 cells increased cell migration and invasion (Figure 2).
miR-449a Targets Notch1 via Binding to Its 3´UTR
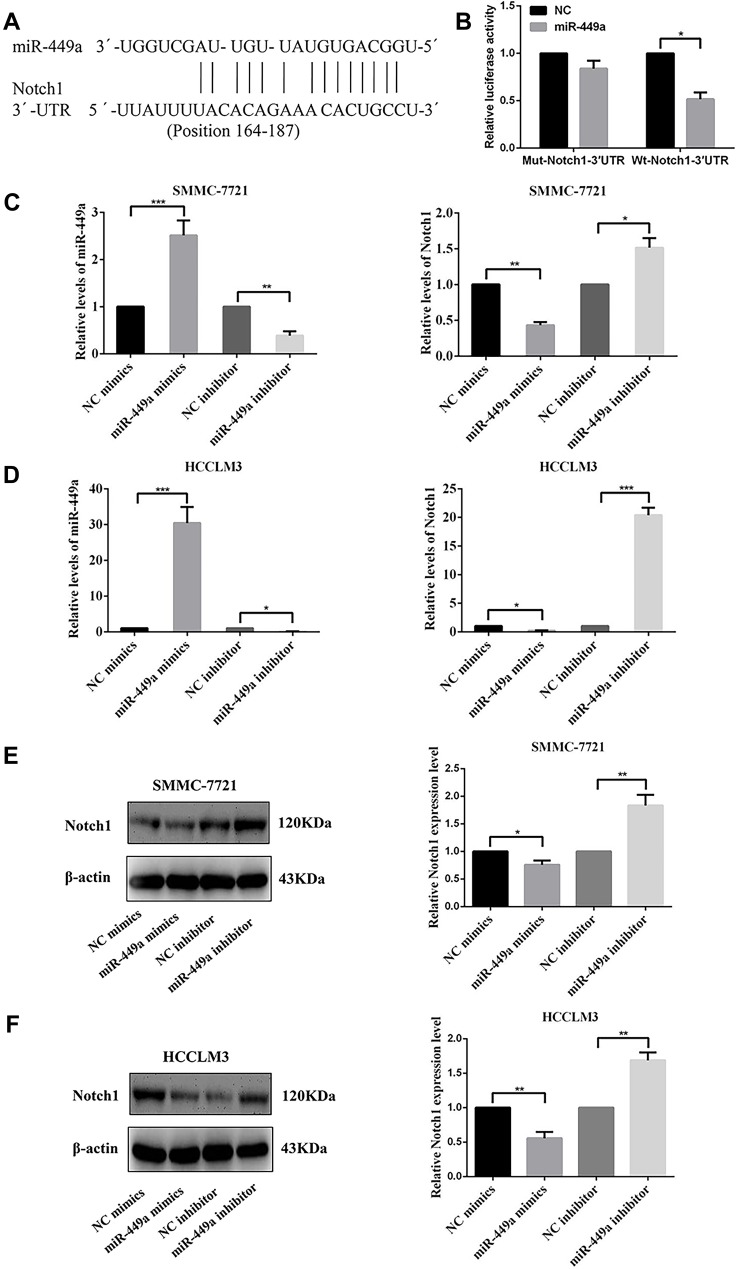
We predicted the possible downstream target genes of miR-449a with two bioinformatics algorithms (TargetScan 7.0 and miRanda) and found that Notch1 fit our criteria and 3´UTR of Notch1 contains a conserved binding site for miR-449a (Figure 3A). The Notch1 as a direct target of miR-449a was validated by luciferase reporter assay in SMMC-7721 cells. Overexpression of miR-449a in SMMC-7721 cells caused a significant decrease in luciferase activity transfected with the reporter plasmid with Wt-Notch1-3´UTR, but not Mut-Notch1-3´UTR (Figure 3B).
Figure 3.
miR-449a mimics and miR-449a inhibitor had influenced the gene expression both in SMMC-7721 and HCCLM3 cell lines. (A) The putative sequences of miR-449a and Notch1 with one binding site. (B) miR-449a significantly inhibited the luciferase activity of the wild-type reporter for Notch1; however, miR-449a did not inhibit the luciferase activity of the reporter vector containing the mutant binding sites of Notch1 in SMMC-7721. (C) The relative levels of miR-449a were up-regulated after transfected with miR-449a mimics while was down-regulated after transfected with miR-449a inhibitor compared with their own NC vector in both cell lines. (D) The relative levels of Notch1 mRNA were up-regulated after being transfected with miR-449a inhibitor and were down-regulated in miR-449a mimics group compared with their own NC vector in both cell lines. (E, F) The expression levels of Notch1 protein were up-regulated after being transfected with miR-449a inhibitor while was down-regulated in miR-449a mimics group compared with their own NC vector in SMMC-7721(E) and HCCLM3(F). *p<0.05, **p<0.01, and ***p<0.001 compared with the control group.
To further confirm that miR-449a targets Notch1, NC mimics, miR-449a mimics, NC inhibitor and miR-449a inhibitor were transfected into SMMC-7721 and HCCLM3 cells. Compared with cells transfected with NC mimics, the expression of miR-449a was significantly higher in SMMC-7721 and HCCLM3 transfected with miR-449a mimics. On the contrary, the expression of miR-449a in SMMC-7721 and HCCLM3 transfected with miR-449a inhibitor was lower than cells transfected with NC inhibitor (Figure 3C). Meanwhile, the expression of Notch1 in SMMC-7721 and HCCLM3 transfected with miR-449a mimics was lower than that with NC mimics. And the expression of Notch1 in SMMC-7721 and HCCLM3 transfected with miR-449a inhibitor increased significantly compared with NC inhibitor group (Figure 3D and E).
miR-449a Affected Migration and Invasion of HCC Cell Lines by Regulating EMT
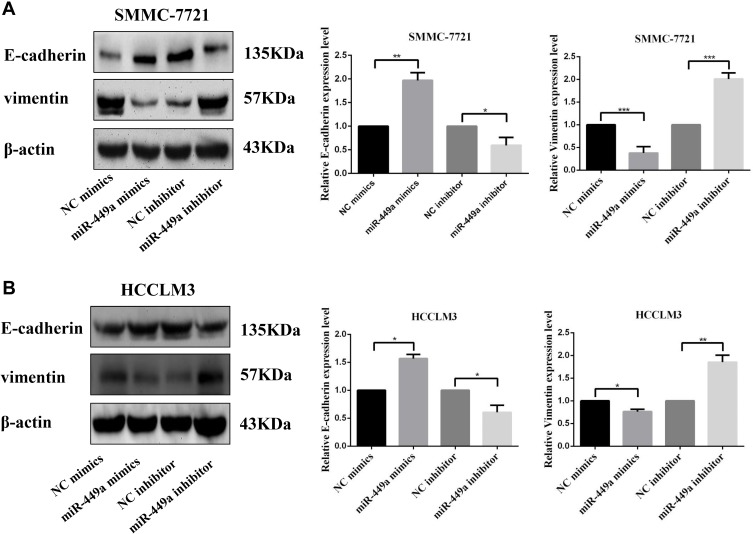
To further study the role of miR-449a in HCC migration and invasion, we detected the E-cadherin and vimentin which are classical EMT related protein. According to the Western blot analyses, the overexpressed miR-449a in SMMC-7721 and HCCLM3 cells significantly enhanced the expression of E-cadherin and reduced vimentin compared with NC mimic. On the contrary, knockdown of miR-449a significantly reduced the expression of vimentin and activated E-cadherin compared to NC inhibitor (Figure 4). These results suggested that miR-449a inhibits the migration and invasion by regulating the EMT proteins.
Figure 4.
miR-449a affected migration and invasion of HCC cell lines by regulating EMT. (A) Western Blot assays for E-cadherin and Vimentin expression in SMMC-7721 after transfection. (B) Western Blot assays for E-cadherin and Vimentin expression in HCCLM3 after transfection. *p<0.05, **p<0.01, and ***p<0.001 compared with control group.
miR-449a Affected Notch Signaling Pathway in HCC Cells
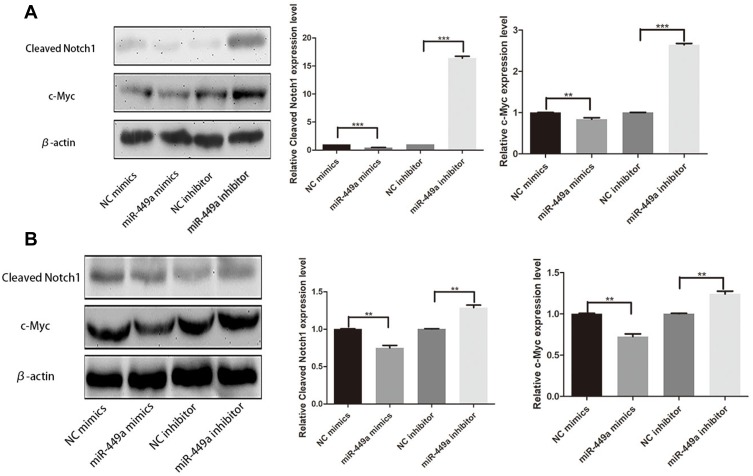
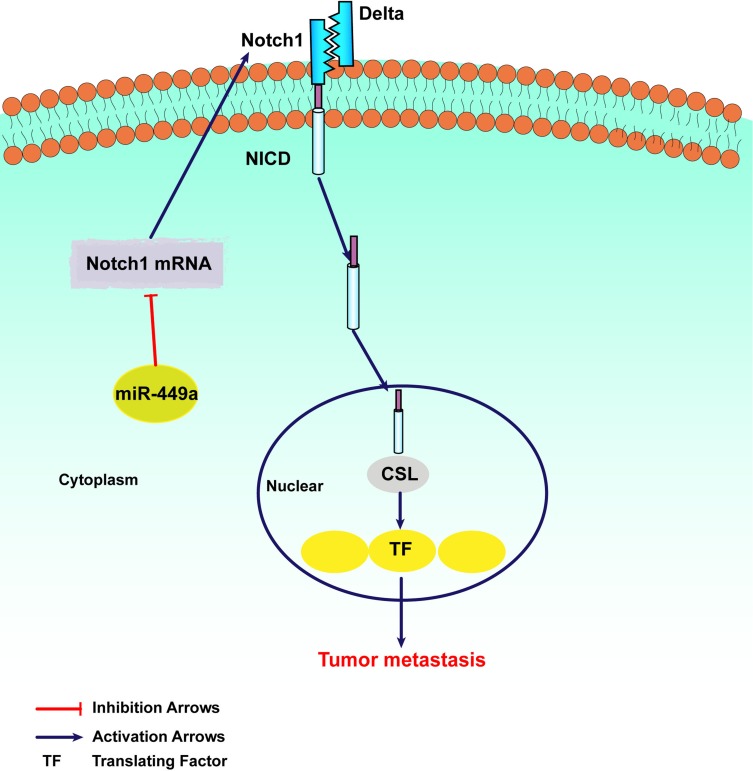
As shown in Figure 5, in order to uncover the mechanism of miR-449a and EMT, Cleaved Notch1 and c-Myc were detected by Western blot assays. In SMMC-7721 and HCCLM3, the assay showed miR-449a mimic significantly decreased the expression of Cleaved Notch1 and c-Myc, while miR-449a inhibitor promoted them (Figure 5). Together these data suggest that miR-449a plays a crucial role in regulating Notch1 in hepatocellular carcinoma, as illustrated in Figure 6.
Figure 5.
miR-449a affected Notch Signaling Pathway. (A) Western Blot assays for Cleaved Notch1 and c-Myc expression in SMMC-7721 after transfection. (B) Western Blot assays for Cleaved Notch1 and c-Myc expression in HCCLM3 after transfection. **p<0.01, and ***p<0.001 compared with control group.
Figure 6.
The mechanism of miR-449a in regulating Notch Signaling Pathway.
Discussion
In this study, we have screened a group of miRNAs related to postoperative STR by RNA sequencing and miR-449a was one of them. The clinicopathological characteristic factors that related to prognosis of HCC in STR and non-STR groups have no significant difference. There is a strong possibility that the differentially expressed HCC-related genes in the STR group lead to increased invasiveness of tumors. Moreover, we found that MiR-449a expression was also significantly lower in tumor tissue than in adjacent tumor tissue. Importantly, miR-449a was independent risk factor for DFS and OS of HCC. Furthermore, overexpression of miR-449a promoted the invasiveness in vitro by regulating EMT via Notch pathway. Mechanically, miR-449a inhibited the translation of Notch1 protein by binding to 3ʹ UTR of its mRNA directly. This study demonstrated that miR-449a affected prognosis of HCC and inhibited invasion by regulating Notch1 expression.
The miR-449a were highly expressed in human normal tissues, such as lungs,1 testis, and trachea18 and usually decreased in various malignant tumors, such as prostate cancer,19 gastric cancer,20 and bladder cancer.21 We found that the expression of miR-449a in HCC tissue was significantly lower than that in normal liver tissue, and its expression level in HCC cell line was also lower than that in normal hepatocytes. The low-level expression of miR-449a was reported to be associated with short disease-free survival of patients with HCC and independent prognostic factor for poor survival of patients with HCC.11 In this study, we observed that miR-449a was not only an independent risk factor for poor prognosis of HCC patients but also a short-term recurrence-related miRNA. The result suggested that the HCC cells with low expression of miR-449a probably have quite high malignancy degree that leads to recurrence within 6 months after surgery.
The migration and invasion ability of HCC are the main factors that affect recurrence and prognosis after surgical resection.22–24 Some research found that miR-449a can inhibit the migration and invasion of HCC cells.11,16,25 We also found that overexpressed miR-449a inhibited the migration and invasion of HCC cells and miR-449a knockdown promoted migration and invasion. Our data are in line with the previous reports that miR-449a has such a similar ability.
In HCC, there are some target genes regulated by miR-449a such as FOS, C-MET, and ADAM10.11,16 We made a prediction of Notch1 as a potential target gene of miR-449a with miRNA Database. However, there is no report about mechanism study on miR-449a-Notch1 signal axis in the migration and invasion of HCC cells. The notch family is composed of a single transmembrane receptor, after interacting with ligands DLL1, 3, 4 and Jagged1-2, the proteins are hydrolyzed, dissected and translocated into the nucleus to activate target gene transcription in mammals.26 Based on the prediction of the binding sites of miR-449a and Notch1 genes, we performed a luciferase reporter assay which demonstrated that miR-449a can bind specifically to the wild-type Notch1 gene. The results showed that miR-449a could inhibit the expression of Notch1 protein by regulate Notch1 mRNA directly. Moreover, we also found that the expression of notch1 in HCC tissues and HCC cell lines was higher than that in normal liver tissues and hepatocytes. These results suggest that there may be a negative correlation between miR-449a and Notch1 in HCC.
Notch1 is involved in a variety of tumor processes, including cell proliferation, apoptosis, metastasis, EMT and angiogenesis.27–29 In addition, the EMT and Notch1 signaling activations mediated by CD24 were elucidated as potential mechanisms of HCC promotion.30 Our previous study also found that the activation of Notch pathway can promote invasion and migration of HCC by regulating EMT.31 In the present study, we found that the expression of E-cadherin increased and the expression of Vimentin reduced in miR-449a group. On the contrary, the expression of Vimentin was increased and E-cadherin reduced in miR-449a inhibitor group. Both E-cadherin and vimentin were pivotal EMT related proteins.
c-Myc, a conventional transcriptional factor, plays an important role in the regulation of many biological processes including cell proliferation, apoptosis, and cell metabolism. In our study, we have Western blot assays to confirm that miR-449a could suppress Cleaved Notch1 and c-Myc. The results above suggest that miR-449a-Notch1 signaling pathway affected the migration and invasion by regulating EMT in HCC cells.
In conclusion, our finding shows for the first time that miR-449a is short-term recurrence-related miRNA which reflects the malignancy degree of HCC. We also found a novel miRNA regulating mechanism for migration and invasion in HCC via Notch pathway. These results suggest that miR-449a may be a new effective therapeutic target for HCC patients with short-term recurrence after hepatectomy in the future.
Acknowledgments
This research is supported by ‘Clinical Medicine +X’ Project of Medical Department of Qingdao University, the Science and Technology Plan of Qingdao City Shinan District (grant no. 2018-4-018-YY), the Science and Technology for People’s Livelihood Project of Qingdao (grant no. 18-6-1-89-nsh).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2.Mazzola A, Costantino A, Petta S, et al. Recurrence of hepatocellular carcinoma after liver transplantation: an update. Future Oncol. 2015;11(21):2923–2936. doi: 10.2217/fon.15.239 [DOI] [PubMed] [Google Scholar]
- 3.Morise Z, Kawabe N, Tomishige H, et al. Recent advances in the surgical treatment of hepatocellular carcinoma. World J Gastroenterol. 2014;20(39):14381–14392. doi: 10.3748/wjg.v20.i39.14381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu LN, Li DD, Xu HX, Zheng SG, Zhang XP. Role of microRNAs in hepatocellular carcinoma. Front Biosci (Landmark Ed). 2015;20:1056–1067. doi: 10.2741/4358 [DOI] [PubMed] [Google Scholar]
- 5.O’Brien-Ball C, Biddle A. Reprogramming to developmental plasticity in cancer stem cells. Dev Biol. 2017;430(2):266–274. doi: 10.1016/j.ydbio.2017.07.025 [DOI] [PubMed] [Google Scholar]
- 6.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-Specific upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joyce BT, Zheng Y, Zhang Z, et al. miRNA-processing gene methylation and cancer risk. Cancer Epidemiol Biomarker Prev. 2018;27(5):550–557. doi: 10.1158/1055-9965.EPI-17-0849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 9.Shi J, Liu Y, Liu J, Zhou J. Hsa-miR-449a genetic variant is associated with risk of gastric cancer in a Chinese population. Int J Clin Exp Pathol. 2015;8(10):13387–13392. [PMC free article] [PubMed] [Google Scholar]
- 10.You J, Zhang Y, Li Y, et al. MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am J Cancer Res. 2015;5(9):2730–2744. [PMC free article] [PubMed] [Google Scholar]
- 11.Chen SP, Liu BX, Xu J, et al. MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer. 2015;15:706. doi: 10.1186/s12885-015-1738-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar P, Sharad S, Petrovics G, et al. Loss of miR-449a in ERG-associated prostate cancer promotes the invasive phenotype by inducing SIRT1. Oncotarget. 2016;7(16):22791–22806. doi: 10.18632/oncotarget.v7i16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu M, Gao D, Wen Q, et al. MiR-29c regulates the expression of miR-34c and miR-449a by targeting DNA methyltransferase 3a and 3b in nasopharyngeal carcinoma. BMC Cancer. 2016;16:218. doi: 10.1186/s12885-016-2253-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Peng J, Li X, Leng A, Liu T. miR-449a targets Flot2 and inhibits gastric cancer invasion by inhibiting TGF-beta-mediated EMT. Diagn Pathol. 2015;10:202. doi: 10.1186/s13000-015-0435-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Wang Y, Sun X, et al. miR-449a promotes liver cancer cell apoptosis by downregulation of Calpain 6 and POU2F1. Oncotarget. 2016;7(12):13491–13501. doi: 10.18632/oncotarget.4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Liu K, Zhang W, et al. miR-449a inhibits proliferation and invasion by regulating ADAM10 in hepatocellular carcinoma. Am J Transl Res. 2016;8(6):2609–2619. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Zhu C, Zhang M, Hu J, et al. Prognostic effect of IL-6/JAK2/STAT3 signal-induced microRNA-21-5p expression on short term recurrence of hepatocellular carcinoma after hepatectomy. Int J Clin Exp Pathol. 2018;11(8):4169–4178. [PMC free article] [PubMed] [Google Scholar]
- 18.Lize M, Pilarski S, Dobbelstein M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2010;17(3):452–458. doi: 10.1038/cdd.2009.188 [DOI] [PubMed] [Google Scholar]
- 19.Noonan EJ, Place RF, Pookot D, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28(14):1714–1724. doi: 10.1038/onc.2009.19 [DOI] [PubMed] [Google Scholar]
- 20.Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, et al. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Lin YW, Mao YQ, et al. MicroRNA-449a acts as a tumor suppressor in human bladder cancer through the regulation of pocket proteins. Cancer Lett. 2012;320(1):40–47. doi: 10.1016/j.canlet.2012.01.027 [DOI] [PubMed] [Google Scholar]
- 22.Mao B, Wang G. MicroRNAs involved with hepatocellular carcinoma (Review). Oncol Rep. 2015;34(6):2811–2820. doi: 10.3892/or.2015.4275 [DOI] [PubMed] [Google Scholar]
- 23.Mansoori B, Mohammadi A, Shirjang S, Baradaran B. MicroRNAs in the diagnosis and treatment of cancer. Immunol Invest. 2017;46(8):880–897. doi: 10.1080/08820139.2017.1377407 [DOI] [PubMed] [Google Scholar]
- 24.Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol. 2015;21(34):9853–9862. doi: 10.3748/wjg.v21.i34.9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandbothe M, Buurman R, Reich N, et al. The microRNA-449 family inhibits TGF-beta-mediated liver cancer cell migration by targeting SOX4. J Hepatol. 2017;66(5):1012–1021. doi: 10.1016/j.jhep.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 26.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770 [DOI] [PubMed] [Google Scholar]
- 27.Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28(3):339–363. doi: 10.1210/er.2006-0046 [DOI] [PubMed] [Google Scholar]
- 28.Xu P, Qiu M, Zhang Z, et al. The oncogenic roles of Notch1 in astrocytic gliomas in vitro and in vivo. J Neurooncol. 2010;97(1):41–51. doi: 10.1007/s11060-009-0007-1 [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Zhang J, Gan X, et al. LGR5 promotes epithelial ovarian cancer proliferation, metastasis, and epithelial-mesenchymal transition through the Notch1 signaling pathway. Cancer Med. 2018;7(7):3132–3142. doi: 10.1002/cam4.2018.7.issue-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan X, Cheng C, Shao Q, Lin Z, Lu S, Chen Y. CD24 promotes HCC progression via triggering Notch-related EMT and modulation of tumor microenvironment. Tumour Biol J Int Soc Oncodev Biol Med. 2016;37(5):6073–6084. doi: 10.1007/s13277-015-4442-7 [DOI] [PubMed] [Google Scholar]
- 31.Han B, Liu SH, Guo WD, et al. Notch1 downregulation combined with interleukin-24 inhibits invasion and migration of hepatocellular carcinoma cells. World J Gastroenterol. 2015;21(33):9727–9735. doi: 10.3748/wjg.v21.i33.9727 [DOI] [PMC free article] [PubMed] [Google Scholar]