Abstract
Purpose
LncRNAs are important regulators in cancers. In this study, we investigated the role of lncRNA PCAT18 in gastric cancer (GC).
Patients and Methods
The level of PCAT18 in GC tissues and cells was determined by qRT-PCR. The cellular behaviors of GC cells with knockdown or overexpression of PCAT18 were respectively detected by CCK-8 assays, colony formation assays, flow cytometry and Western blot. A GC mice model was established by subcutaneous injection of MGC-803 and HGC-27 cells with the knockdown or overexpression of PCAT18. The tumor size and weight were measured, and IHC was performed to determine ki-67 level. Predicted by bioinformatics software and confirmed by dual-luciferase reporter assay, PCAT18 was involved in miR-107/PTEN axis, thus, the expression of and relationship among PCAT18, miR-107 and PTEN pathway were explored in clinical cases and GC cell lines. Rescue assay was performed in GC cells by co-transfection with miR-107 mimic or PCAT18. The PTEN/PI3K/AKT pathway was then detected by Western blot.
Results
PCAT18 was down-regulated in GC tissues and cells, and it had a significant diagnostic value for GC. The expression of PCAT18 was highly associated with tumor size, and PCAT18 was found to inhibit GC growth in vitro and in vivo. It was also found that PCAT18 was involved in PTEN/PI3K/AKT signaling pathway through targeting miR-107.
Conclusion
PCAT18 inhibits the progression of GC via miR-107/PTEN/PI3K/AKT signaling pathway. Additionally, PCAT18 is possibly a promising target for treatment of GC.
Keywords: gastric cancer, PCAT18, miR-107, PI3K/AKT signaling pathway
Introduction
Gastric cancer (GC) is a leading cause of cancer-related death worldwide. According to the cancer statistics provided by SEER (Surveillance, Epidemiology, and Results Program) from 2009 to 2015. GC has a poor 5-year survival rate, and the incidence of GC is the third highest in males and the fourth highest in females in China.1 Multiple factors, such as genetic, epigenetic and environmental risk factors, Helicobacter pylori (Helicobacter pylori) infection, diet, lifestyle, all lead to the occurrence and development of GC.2,3 GC is difficult to be diagnosed at early stage, due to the lack of evident clinical symptoms and diagnostic biomarkers.4
Long non-coding RNAs (lncRNAs) are non-protein coding transcripts longer than 200 nucleotides.5,6 Increasing evidence revealed that lncRNAs are involved in a series of biological processes in GC, such as tumorigenesis,6 proliferation,7 metastasis,8 chemoresistance and prognosis.9 However, the role of lncRNAs in GC has not been comprehensively studied. Prostate cancer-associated transcript 18 (PCAT18) is a less reported lncRNA, however, current researches on PCAT18 showed that the roles of PCAT18 vary in different cancers, for example, in metastatic prostate cancer, PCAT18 silencing inhibits cell migration and invasion,10 moreover, PCAT18 is considered as a tumor suppressor in the development of gastric tumors.11 Up to now, the molecular mechanism of PCAT18 in cancers, especially in GC, remains unclear, therefore, the current study aims to explore the role of PCAT18 in the progression of GC.
LncRNAs regulate critical pathways through competitively binding to microRNAs (miRNAs).12 MiRNAs are short nucleotides and are involved in regulating post-transcriptional gene expression level by miRNA-mRNA interaction. Previous studies showed that miR-107 could be used as a candidate biomarker, as its expression level is up-regulated at the early stage of GC.13 In addition, the down-regulation of miR-107 inhibits GC cell growth, promotes cell cycle arrest and apoptosis through regulating PTEN.14 MiR-107 also induces GC cell growth by targeting FOXO1.15 However, the role of miR-107 reported in GC researches still remains controversial, for example, one study also reported that miR-107 inversely inhibits GC progress by targeting PI3K/AKT pathway, and therefore functions as a tumor suppressor.16
In this study, we aim to investigate the functions of lncRNA PCAT18 and the possible mechanisms in GC, hoping to find a potential drug target for GC treatment.
Methods
GC Samples Collection
Sixty GC tissues and adjacent normal tissues were collected from GC patients who received resection surgeries at the Jiaozuo People’s hospital of Henan Province from May 2017 to May 2018. This study was approved by the Ethics Committee of Jiaozuo People’s Hospital of Henan Province, and all patients participated in our research signed the informed consent. The tissue samples were maintained in RNAlater (AM7020, ThermoFisher, USA) at −80°C. The clinical characteristics of GC patients were shown in Table 1.
Table 1.
The Relationship Between PCAT18 Expression Level in Cancer Tissues and Clinical Pathological Factors of Patients with Gastric cancer
| Parameters | Cases (n) | PCAT18 | χ2 | P Value | |
|---|---|---|---|---|---|
| High (n=25) | Low (n=35) | ||||
| Age (years) | 0.002 | 0.965 | |||
| <60 | 29 | 12 | 17 | ||
| ≥60 | 31 | 13 | 18 | ||
| Gender | 0.031 | 0.861 | |||
| Male | 32 | 13 | 19 | ||
| Female | 28 | 12 | 16 | ||
| Site of primary tumor | 0.429 | 0.807 | |||
| Gastric cardia | 10 | 4 | 6 | ||
| Antrum | 12 | 6 | 6 | ||
| Stomach | 38 | 15 | 23 | ||
| Tumor size (cm) | 14.343 | 0.000 | |||
| <5 | 26 | 18 | 8 | ||
| ≥5 | 34 | 7 | 27 | ||
| Histological grade | |||||
| 1,2 | 40 | 18 | 22 | 0.549 | 0.459 |
| 3,4 | 20 | 7 | 13 | ||
| Lymph node status | 0.571 | 0.450 | |||
| N0 | 45 | 20 | 25 | ||
| ≥N1 | 15 | 5 | 10 | ||
| Smoking status | 0.433 | 0.511 | |||
| Ever | 27 | 10 | 17 | ||
| Never | 33 | 15 | 18 | ||
Cell Culture
Human normal gastric epithelial cell line (GES-1, CBP60512) and human GC cell lines (AGS (CBP60476), MGC-803 (CBP60485), NCI-N87 (CBP60491), and HGC-27 (CBP60480)) were purchased from Cobioer Co., Ltd. The cells were cultured in RPMI1640 (61870044, ThermoFisher, USA) supplemented with 10% FBS (16140071, ThermoFisher, USA), 1% penicillin/streptomycin (15070063, ThermoFisher, USA) at 37°C in 5% CO2.
RNA Isolation and qRT-PCR
Total RNAs from tissues and cells were extracted using Qiagen RNeasy Plus Mini Kit (74134, Qiagen, Germany) according to the instructions. QRT-PCR assays were performed using 10 µL SYBR® Green PCR Master Mix (4312704, ABI, USA) on Bio-Rad CFX 96 Touch Real-Time PCR Detection System (1855196, Bio-Rad, China). One step miRNA RT kit (D1801, HaiGene, China) was used for reverse- transcription. Forward primer sequence 5ʹ-AGGAGACAGGCCCCAGATTT-3ʹ and reverse primer sequence 5ʹ-TGAAGTGCTGGGACAACGTA-3ʹ were used for PCAT18, while forward primer sequence 5ʹ-TGGATTCGACTTAGACTTGACCT-3ʹ and reverse primer sequence 5ʹ-GGTGGGTTATGGTCTTCAAAAGG-3ʹ were used for PTEN. Forward primer sequence 5ʹ- AAGGTGAAGGTCGGAGTCAAC-3ʹ and reverse primer sequence 5ʹ- GGGGTCATTGATGGCAACAATA-3ʹ were used for GAPDH. HaiGene SYBR Green miRNA qRT-PCR kit (AP01390, HaiGene, China) and HaiGene SYBR Green miRNA qRT-PCR kit (AP02055, HaiGene, China) were used to respectively determine the expressions of miR-107 and U6. U6 snRNA served an internal control. Parameters were set as follows: 95°C for 5 min, 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30s, and extension at 70°C for 10s. The relative expression level was determined by 2−∆∆CT.
Cell Transfection
PcDNA3.1 and pcDNA3.1-PCAT18 were purchased from GenePharma Corporation, China. SiPCAT18 (siB160229100822-1-5), negative control siRNA (siN0000001-1-5), miR-107 mimic (miR10000104-1-5) and negative control mimic (miR1190315051351) were purchased from Ribobio Corporation, China. GC cells were grown in 6-well plates to 80% confluence. The plasmid, siRNA or mimic was transfected into the cells by using lipofectamine 2000 (11,668,019, Invitrogen, USA). After 4–6 h of incubation at 37ºC, the medium was replaced by fresh RPMI1640 medium containing 10% FBS. After incubation for 48 h at 37ºC, the cells were harvested for further analysis.
CCK-8 Assay
Cell Counting Kit-8 (70-CCK801, MultiSciences, China) was used to determine the cell viability. Briefly, the GC cells (5 ×103 per) were grown into a 96-well plate after the transfection. Next, 10 µL CCK-8 solution was added into each well and held for 24h, 48h and 72h at 37°C, respectively. The absorbance value was detected at the wavelength of 490nm by a microplate reader (PLUS 384, Molecular Devices, USA).
Colony Formation Assay
The cells (5 × 102 per well) were seeded in 6-well plates and incubated at 37°C for 14 days. The cells were then fixed by methanol and stained by 0.1% crystal violet (548-62-9, Aladdin, China) for 10min. Cell clones were observed under a microscope (TS100, Nikon, Japan).
Cell Apoptosis and Cell Cycle
Cell apoptosis detected using Annexin-V kit (70-AP101-100-AVF, MultiSciences, China). The cells were trypsinized at 37°C for 1min, then centrifuged at 450xg, 4°C for 5 min and washed by cold PBS and resuspended in 300µL binding buffer. After that, 5 µL Annexin-V-FITC solution was added into the cells and incubated at room temperature for 15 min away from light. 5 µL PI (BG-60910-00-200, Biogems, PeproTech, USA) was added to stain the cells for 5 min at room temperature. Finally, 200µL binding buffer was added, and cell apoptosis was determined using FACSCalibur flow cytometer (342973, BD Biosciences, USA). The apoptosis rate was determined by using BD FACSCanto™ system software v2.4 (646602, BD Biosciences, USA).
For detection of cell cycle, the cell were collected as described above, and fixed by 16% formaldehyde (1081-15-8, Aladdin, China) for 15min at room temperature and stained by PI (50 μg/mL). Cell cycle was measured and analyzed as described above.
Western Blot Analysis
The cells were washed by PBS twice and treated by RIPA protein extraction reagent (89901, Thermo Scientific, USA) at 4°C for 15 min. Then, the proteins were collected by centrifuging at 1000 xg, 4°C for 30 min. After denaturation at 100°C for 5 min, the proteins were mixed with loading buffer and separated on SDS/PAGE gels, then transferred onto a Polyvinylidene Fluoride membrane (LC2002, Invitrogen, Thermofisher, USA). The membrane was first sealed by non-fat milk (5%, PA201-01, BioMed, China) for 2 h at room temperature, and then incubated with primary antibodies (Anti-cyclinD1 (1:1000, ab134175, Abcam, UK), Anti-P27 KIP 1 (1:1000, ab32034, Abcam, UK), Anti-Caspase-3 (1:1000, ab13847, Abcam, UK), Anti-PTEN (1:1000, 9552, CST, USA), Anti-p-PI3K (1:1000, 4228, CST, USA), Anti-PI3K (1:1000, 4292, CST, USA), Anti-p-AKT (1:1000, 9271, CST, USA), Anti-AKT (1:1000, 9272, CST, USA), Anti-GAPDH (1:5000, ab8245, CST, USA)) at 4°C overnight. Next, the primary antibodies were removed by cold TBST, and HRP-linked anti-rabbit IgG antibody (1: 5000, 7074, Cell Signaling Technology, USA) was added to incubate with the membrane for 2 h at room temperature. The bound antibodies were detected using SignalFire™ ECL reagent (6883, Cell Signaling Technology, USA) and ImageQuant ECL Imager (28-9605-63, GE Healthcare, USA). GAPDH served as a control.
Dual-Luciferase Reporter Assay
The starBase (http://starbase.sysu.edu.cn/) and Targetscan7.2 (http://www.targetscan.org/vert_72/) were used to predict the interaction between PCAT18 and miRNA, and the interaction between miR-107 and its target mRNA, respectively, the results were further confirmed by dual-luciferase reporter assay. Briefly, the cells (5x104 cells/well) were seeded in 24-well plates and co-transfected with pmirGLO-PCAT18-WT and miR-107 mimic, or pmirGLO-PCAT18-MUT reporter and miR-107 mimic, with the cells transfected with reporter plasmid only (blank) served as control. After 48h of incubation, dual-luciferase assay system (E1910, Promega, USA) and Microplate Luminometer (11300010, Berthold, Germany) were used to determine the luciferase activity. The firefly luciferase activity was normalized against renilla luciferase activity and shown as mean ± SD.
Xenograft Assay
BALB/c Nude mice (4 weeks old) were obtained from Vitalriver, Co., ltd., China. The cells (107 cells) transfected with pcDNA PCAT18. siPCAT18, control plasmid or siRNA were resuspended in PBS/Matrigel mixture and then subcutaneously injected into the ventral side of the mice (with 3 mice in each group). To maintain the stability of the siRNA gene in mice, injections were given every four days for six times. The tumor volume was measured at day 0, 5, 10, 15, 20 and 25 after the injection. Finally, the mice were sacrificed and the tumors were isolated, weighted and stained with immunohistochemistry. Animal experiments were approved by the Institutional Animal Care and Use Committee of Jiaozuo People’s Hospital of Henan Province. All animal experiments were performed in accordance with the guidelines of the China Council on Animal Care and Use.
Immunohistochemistry
The tumors were fixed in 4% paraformaldehyde solution (30525-89-4, Aladdin, China) for 48 h at room temperature, embedded in paraffin and then sectioned into 4 μm thick. Slides were deparaffinized in a series of xylene (1330-20-7, Aladdin, China) and graded alcohols. After that, the sections were heated in citrate buffer (10mM, pH 6.0) in sub-boiling temperature for 10 min for antigen retrieval. Next, the sections were incubated with primary antibody (Ki-67, AF1738, 1:400, Beyotime, China), which was diluted in immunol staining blocking buffer (P0102, Beyotime, China) for 2h at 4°C. After that, the sections were washed by wash buffer (P0106, Beyotime, China) for 15min twice at room temperature and incubated with SignalStain® Boost IHC Detection Reagents (HRP, 8114S, CST, China) at room temperature for 30min. Then, the slices were observed under an ECLIPSE Ti2 microscope (Ti2-U, Nikon, Japan).
Data Analysis
Statistical analysis was performed using Graphad Prism 6. The significant differences among groups were analyzed by student’s t-test or one-way ANOVA followed by Tukey’s post-hoc test. Spearman correlation analyses between miR-107 and PCAT18 or PTEN expression levels were performed. P< 0.05 was considered as statistically significant.
Results
PCAT18 Was Down-Regulated in GC Tissues and Cells
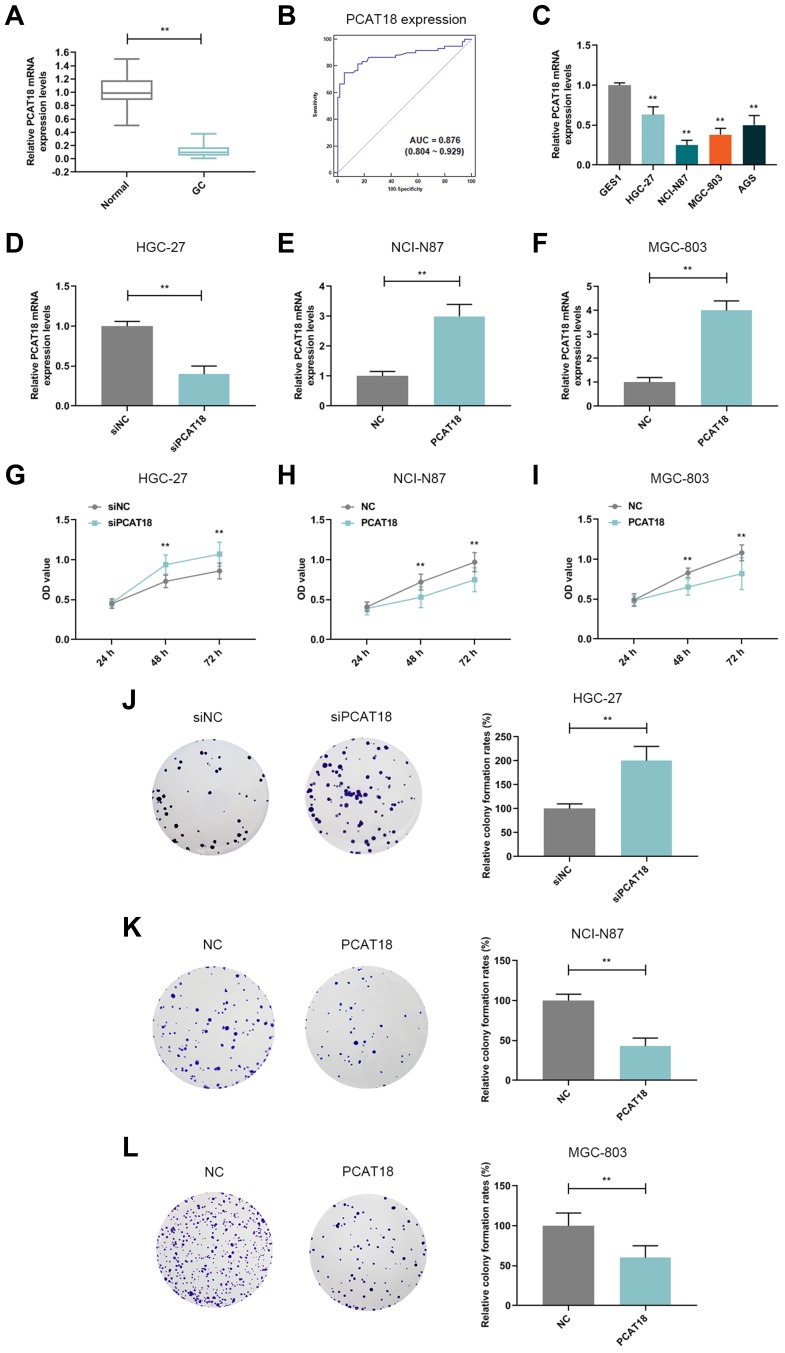
The expression of lncRNA PCAT18 in GC and adjacent normal tissues was determined by qRT-PCR, and we found that the level of PCAT18 was significantly down-regulated in GC tissues (Figure 1A, P<0.001). Receiver Operating Characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of lncRNA PCAT18 in GC tissues, and the area under the curve was 0.876 (95% confidence interval, 0.804–0.929; standard error, 0.02503) (Figure 1B). In addition, the expression of PCAT18 was significantly associated with tumor size (Table 1). Next, the expression level of PCAT18 in GC cell lines was detected, and the data showed that PCAT18 was down-regulated in GC cells (Figure 1C, P<0.001), and that the level of PCAT18 was relatively the highest in HGC-27 cells but lower in NCI-N87 and MGC-803 cell lines. Thus, the three cell lines were used for further experiments.
Figure 1.
PCAT18 was down-regulated in GC tissues and cells and it inhibited cell growth of GC cells. (A) The level of PCAT18 in GC and adjacent normal tissues was determined by qRT-PCR. n= 60. **P<0.001 vs. Normal. GC: Gastric Cancer. Normal: adjacent normal tissues. (B) ROC curve for the prediction of GC based on PCAT18 expression level, the AUC was 0.876 (95% CI 0.804‐0.929). (C) The level of PCAT18 in normal gastric epithelial cells and GC cell lines was measured by qRT-PCR. **P<0.001 vs. GSE1. (D,E) The efficiency of the knockdown (D) or overexpression (E and F) of PCAT18 in GC cells was measured by qRT-PCR. **P<0.001 vs. siNC or NC. (G–I) The cell viabilities of GC cells after transfection for 24h, 48h and 72h were determined by CCK-8 assays. **P<0.001 vs. siNC or NC. (J–L) The cell growth abilities of GC cells were determined by colony formation assays. **P<0.001 vs. siNC or NC.
Abbreviation: ROC curve, receiver operating characteristic curve.
Figure 2.
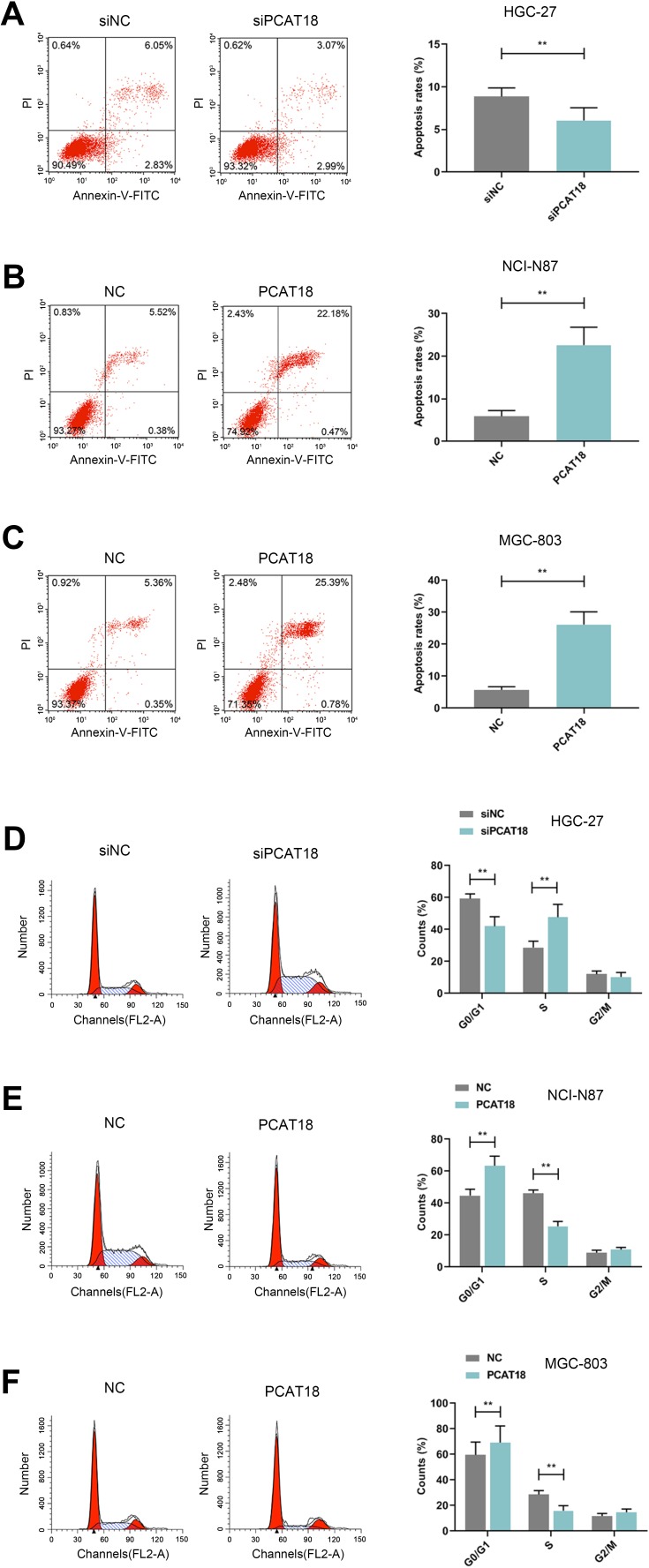
The effects of knockdown or overexpression of PCAT18 on cell apoptosis and cell cycle. (A–F) The cell apoptosis (A–C) and cell cycle (D–F) of GC cells were measured by flow cytometry. **P<0.001 vs. siNC or NC.
PCAT18 Regulated Cell Growth, Apoptosis and Cell Cycle of GC Cells
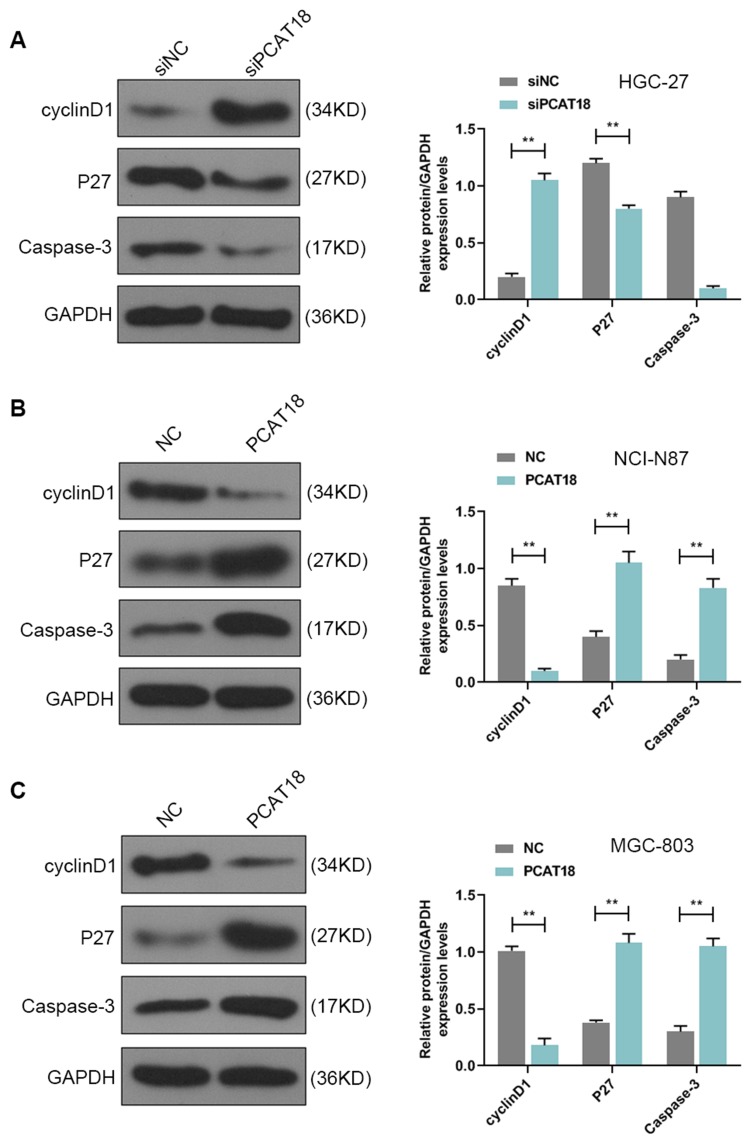
HGC-27 cells were successfully transfected with siPCAT18, while NCI-N87 and MGC-803 cells were transfected with overexpressed plasmid of PCAT18 (Figure 1D–F). CCK-8 assays were performed to detect the cell viability, and results showed that knockdown of PCAT18 increased cell viability, while the overexpression of PCAT18 produced the opposite effect (Figure 1G–I, P<0.001). We found that cell colony formation ability was promoted by siPCAT18 but decreased by overexpressed PCAT18 (Figure 1J–L, P<0.001). Furthermore, flow cytometry results showed that siPCAT18 decreased cell apoptosis rate, reduced the number of cells in G0/G1 phase, but increased those in S phase, however, overexpressed PCAT18 increased cell apoptosis rate, number of cells in G0/G1 phase but decrease those in S phase (Figure 2A–F, P<0.001). The levels of cell cycle-related proteins, cyclinD1, P27, cell apoptosis-associated protein, Caspase-3 were determined by Western blot, as the levels of cyclinD1 was up-regulated by siPCAT18, while P27 and Caspase-3 levels were down-regulated by siPCAT18 in HGC-27 cells (Figure 3A, P<0.001), the results from Western blot were consistent with the data from flow cytometry. The opposite results were found in NCI-N87 and MGC-803 cells (Figure 3B and C, P<0.001).
Figure 3.
The cell cycle and apoptosis-associated protein levels were regulated by PCAT18. (A–C) The levels of cyclinD1, P27, Caspase-3 in GC cells were measured by Western blot. **P<0.001 vs. siNC or NC.
PCAT18 Regulated GC Cell Growth in vivo
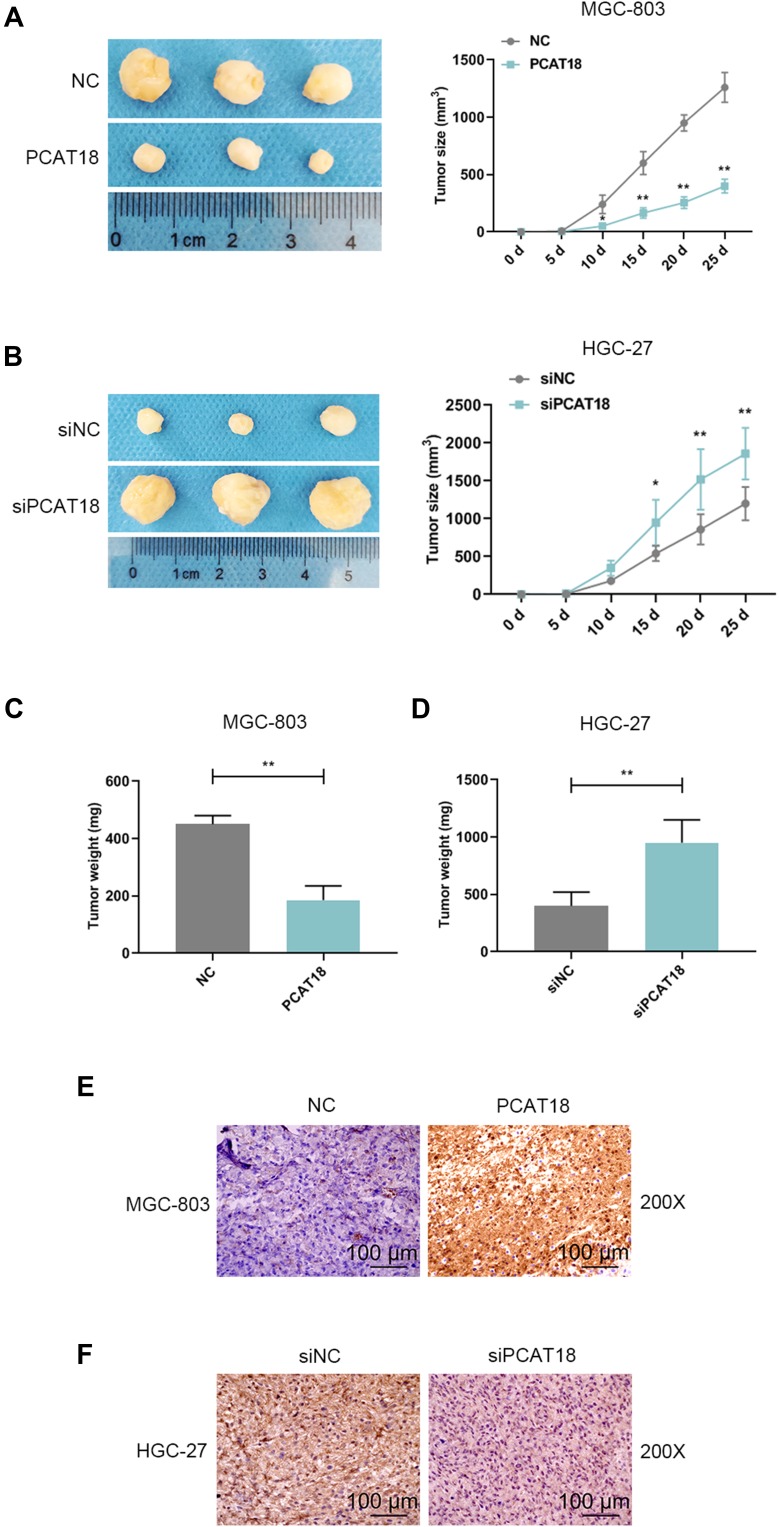
We explored the effect of PCAT18 on GC growth in vivo. A nude mice model of GC was constructed by subcutaneously injecting the GC cells into mice. Before the injection, MGC-803 cells were transfected with pcDNA-PCAT18 and control plasmid, while HGC-27 cells were transfected with siPCAT18 and control mimic. The tumors were monitored every 5 days after the injection. We observed that the tumors formed in the pcDNA-PCAT18 transfection group had smaller tumor sizes and lighter weights than those in control group (Figure 4A and C, P<0.001), while the tumors in the siPCAT18 transfection group had larger tumor sizes and heavier weights than those in control group (Figure 4B and D, P<0.001). Immunohistochemical staining of ki-67 further confirmed that ki-67 was down-regulated in pcDNA-PCAT18-treated group (Figure 4E, P<0.001), but was up-regulated in siPCAT18-treated group (Figure 4F, P<0.001). These data indicated that PCAT18 might act as a tumor suppressor gene to suppress GC cell growth.
Figure 4.
The effects of knockdown or overexpression of PCAT18 on GC growth in vivo. (A,B) Xenograft assays were performed by subcutaneously injected of GC cells with the knockdown and overexpression of PCAT18 in mice. The tumors were shown in the left, and tumor size was recorded and shown in the right. *P<0.05 and **P<0.001 vs. NC or siRNA NC. (3 mice in each group). (C,D) The tumors were weighed (mg). **P<0.001 vs. siNC or NC. (E,F) The tumor sections under IHC staining of ki-67. bar=100µm.
Abbreviation: IHC, immunohistochemistry.
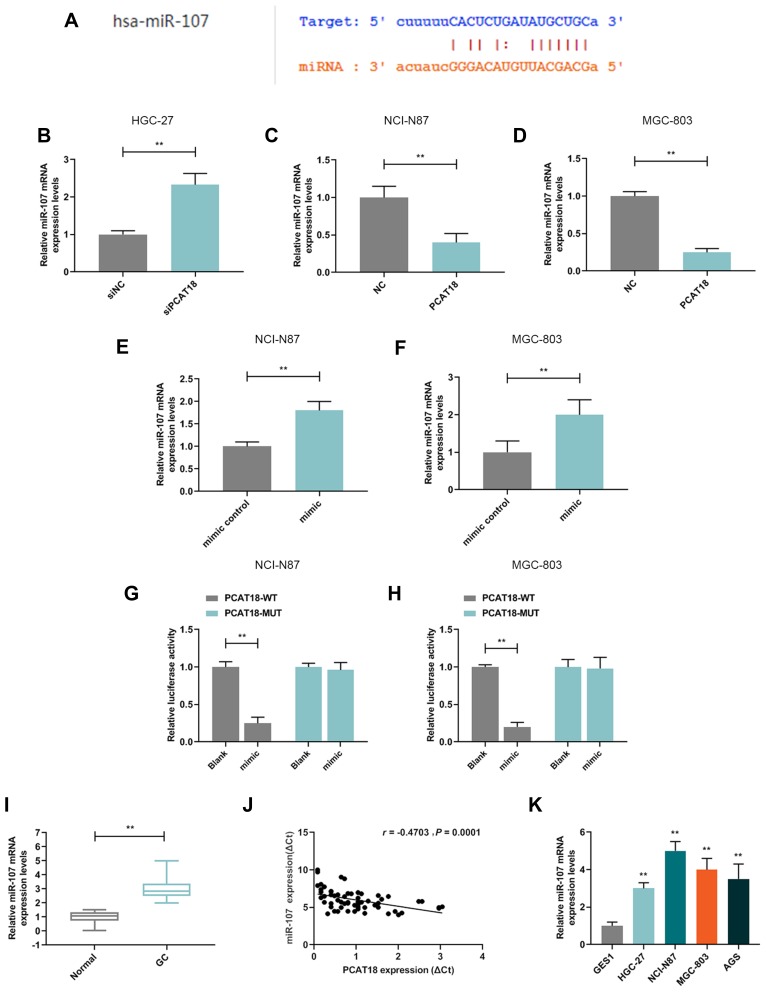
PCAT18 Targeted and Regulated miR-107 in GC
A potential binding site between miR-107 and PCAT18 was predicted by starBase (Figure 5A). QRT-PCR results showed that the level of miR-107 was up-regulated by siPCAT18 in HGC-27 cells but was down-regulated by overexpressed PCAT18 in NCI-N87 and MGC-803 cells (Figure 5B–D, P<0.001). In order to further confirm the relationship between PCAT18 and miR-107, miR-107 mimic and inhibitor were respectively transfected into NCI-N87 and MGC-803 cells (Figure 5E and F, P<0.001). The results of dual-luciferase reporter assays showed that miR-107 mimic decreased the luciferase activity in GC cells after transfection with the PCAT18-WT reporter plasmid (Figure 5G and H, P<0.001). QRT-PCR showed that the expression level of miR-107 in GC tissues was significantly up-regulated (Figure 5I, P<0.001). Correlation analysis also showed that miR-107 expression was negatively correlated with PCAT18 (r=−0.4703, P=0.0001, Figure 5J). The up-regulation of miR-107 in GC cell lines was further confirmed by qRT-PCR (Figure 5K, P<0.001).
Figure 5.
MiR-107 was up-regulated in GC tissues, cells, and directly regulated by PCAT18. (A) MiR-107 had a binding site at the PCAT18 predicted by starBase. (B–D) The level of miR-107 in GC cells was detected by qRT-PCR. **P<0.001 vs. siNC or NC. (E,F) The efficiencies of overexpression of miR-107 in GC cells by transfection with miR-107 mimic or mimic control were measured by qRT-PCR. **P<0.001 vs. mimic control. (G,H) Relative luciferase activity was analyzed in GC cells. **P<0.001 vs. blank. (I) The level of miR-107 in GC and adjacent normal tissues was determined by qRT-PCR. n= 60. **P<0.001 vs. Normal. GC: Gastric Cancer. Normal: adjacent normal tissues. (J) Correlation analysis between PCAT18 and miR-107 expression levels in GC tissues (n=60). r=−0.4703, P=0.0001. (K) The level of miR-107 in normal gastric epithelial cells and GC cell lines measured by qRT-PCR. **P<0.001 vs. GSE1.
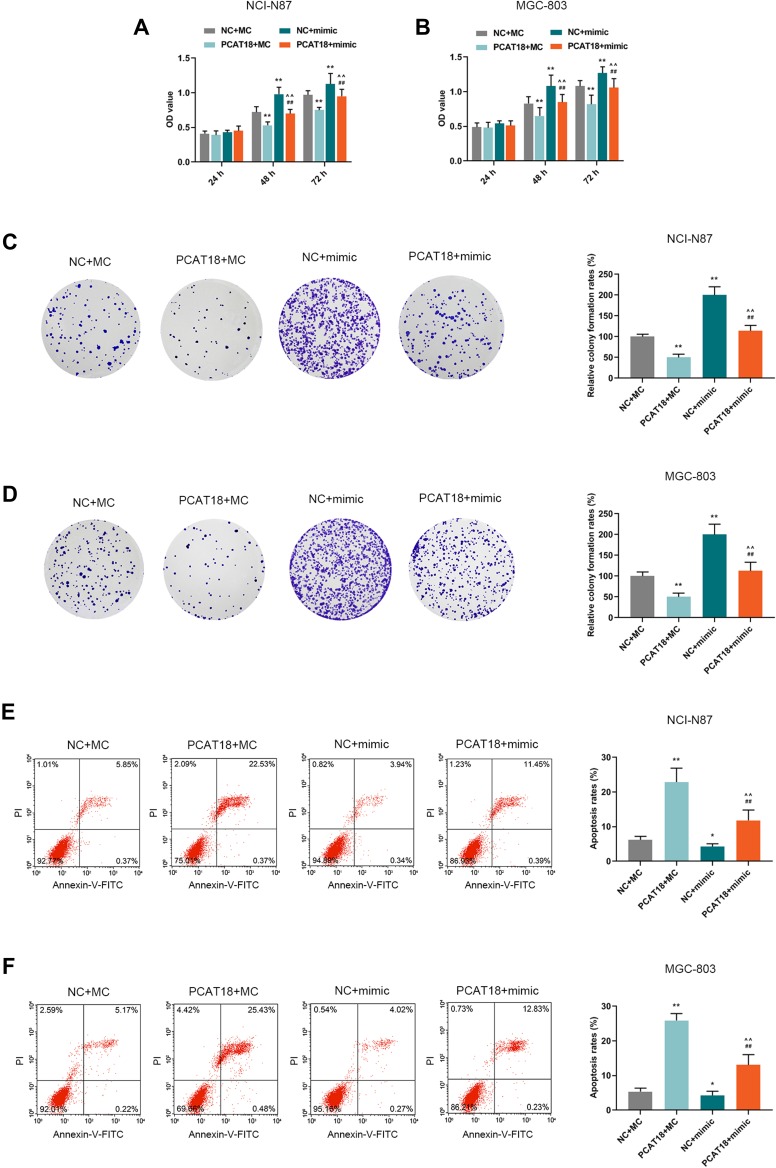
MiR-107 Mimic Rescued Suppressive Effects Induced by PCAT18 on GC Cells
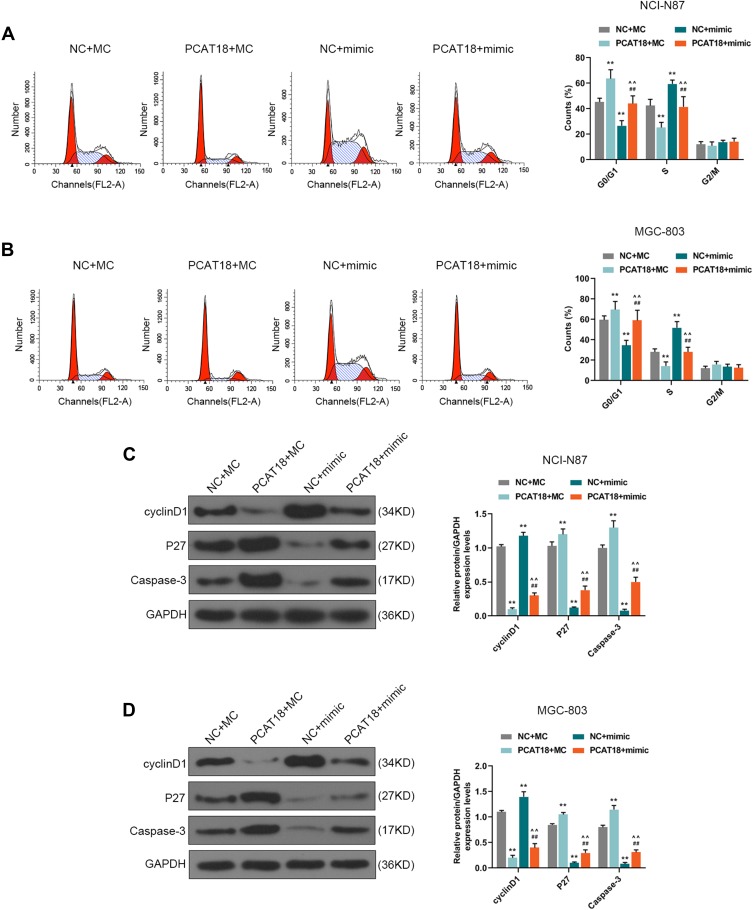
MiR-107 mimics were co-transfected with PCAT18 in GC cells to further confirm the mutual regulation between PCAT18 and miR-107. We found that co-transfection with PCAT18 and miR-107 mimic could reverse the reduced cell viability, colony formation ability, the increase of cell apoptosis and arrest of G0/G1 cell cycle caused by overexpression of PCAT18 (Figures 6A–F and 7A, B, P<0.001). The cell cycle-associated protein cyclinD1 was reduced by PCAT18 overexpression, while the expression of P27KIP1 (cyclin-dependent kinase inhibitor 1B) and Caspase-3 was increased by PCAT18 overexpression. However, these effects on cell cycle and apoptosis-associated protein expressions could be reversed by miR-107 mimic (Figure 7C and D, P<0.001).
Figure 6.
PCAT18 regulated cell viability, colony formation ability, and cell apoptosis by sponging miR-107 in GC cells. (A,B) The cell viabilities of GC cells after transfection for 24, 48and 72 h were determined by CCK-8 assays. **P<0.001 vs. NC+MC. ##P<0.001 vs. PCAT18+MC. ^^P<0.001 vs. NC+mimic. (C,D) The cell growth of GC cells was determined by colony formation assays. **P<0.001 vs. NC+MC. ##P<0.001 vs. PCAT18+MC. ^^P<0.001 vs. NC+mimic. (E,F) The cell apoptosis of GC cells was measured by flow cytometry. *P<0.05 and **P<0.001 vs. NC+MC. ##P<0.001 vs. PCAT18+MC. ^^P<0.001 vs. NC+mimic.
Figure 7.
PCAT18 regulated cell cycle and cell cycle/apoptosis-associated proteins expressions by sponging miR-107 in GC cells. (A,B) The cell cycles of GC cells were measured by flow cytometry. **P<0.001 vs. NC+MC. ##P<0.001 vs. PCAT18+MC. ^^P<0.001 vs. NC+mimic. (C,D) The levels of cyclinD1, P27, Caspase-3 in GC cells were measured by Western blot. **P<0.001 vs. NC+MC. ##P<0.001 vs. PCAT18+MC. ^^P<0.001 vs. NC+mimic.
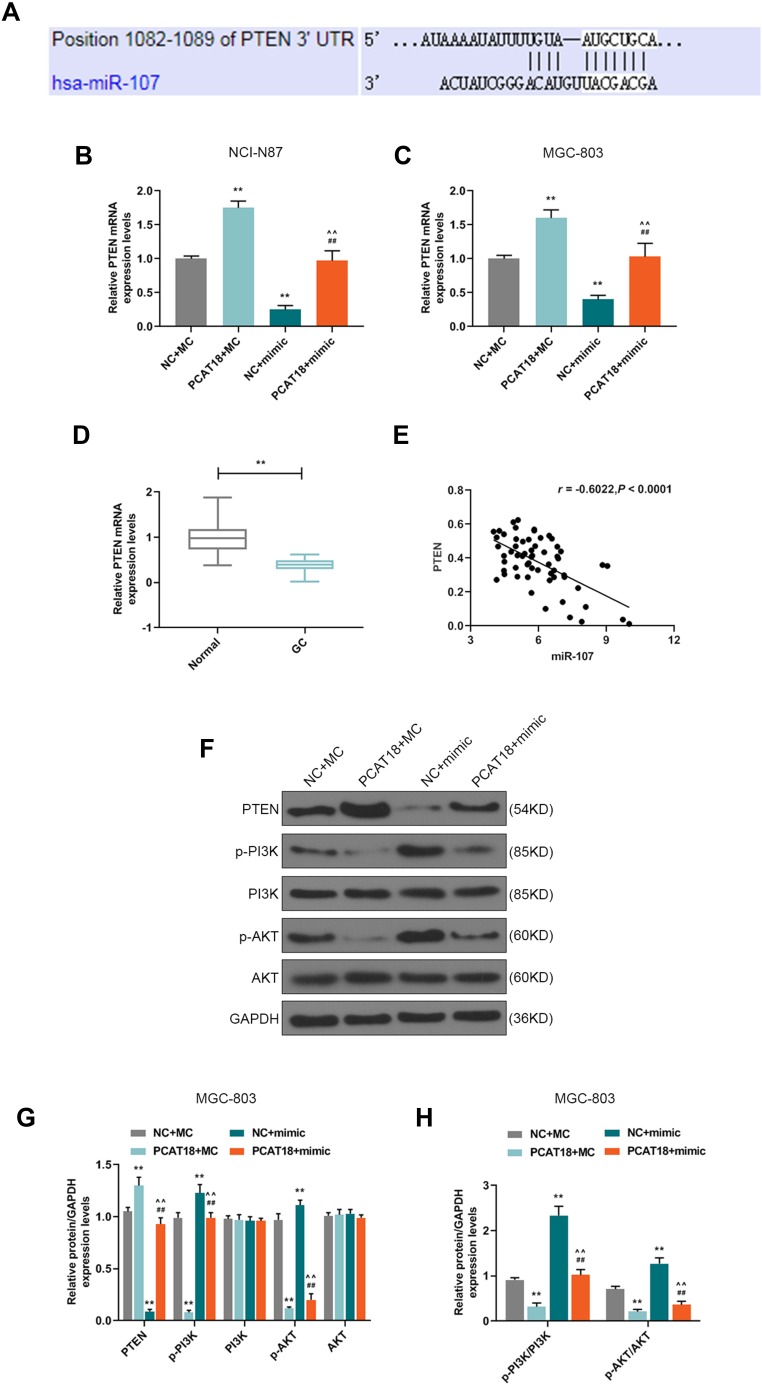
PCAT18 Regulated the Cell Viability of GC Cells via miR-107/PTEN/PI3K/AKT Signaling Pathway
Targetscan predicted that miR-107 had a binding site on the position 1082–1089 of PTEN 3ʹUTR (Figure 8A). The level of PTEN was negatively regulated by miR-107 in NCI-N87 and MGC-803 cells (Figure 8B and C, P<0.001). QRT-PCR also showed that the level of PTEN in GC was down-regulated compared with adjacent normal tissues (Figure 8D, P<0.001), and we also found that the mRNA level of PTEN was negatively correlated with miR-107 (Figure 8E, Pearson r = −0.6022, P < 0.0001), suggesting that PTEN might be a target gene for miR-107 in GC. Therefore, the effect of PCAT18/miR-107 axis on PI3K/AKT pathway was measured by transfection with PCAT18, miR-107, or co-transfection with PCAT18 and miR-107. Western blot results showed that the overexpression of PCAT18 down-regulated the level of phosphorylated-PI3K, which could be reversed by miR-107 mimic. However, the down-regulation of phosphorylated-AKT caused by overexpression of PCAT18 could be partly reversed by miR-107 mimic, and no changes were observed at the protein levels of PI3K, AKT (Figure 8F–G). Additionally, the expression ratio of p-PI3K to PI3K was also decreased by overexpression of PCAT18, which, however, can be blocked by miR-107 mimic, while the decreased ratio of p-AKT to AKT could be partly reversed by miR-107 mimic (Figure 8H).
Figure 8.
PCAT18 regulated cell viability of GC cells by miR-107/PTEN/PI3K/AKT signaling pathway. (A) MiR-107 has a binding site on the PTEN 3ʹUTR predicted by Targetscan7.2. (B,C) qRT-PCR was performed to detect PTEN expression level in GC cells. (D) The level of PTEN in GC and adjacent normal tissues was determined by qRT-PCR. n= 60. **P<0.001 vs. Normal. (E) Correlation analysis between PTEN and miR-107 in GC tissues (n=60). r=−0.6022, P<0.0001. (F,G) The levels of PTEN, p-PI3K, PI3K, p-AKT, AKT were measured by Western blot in MGC-803. (H) The expression ratios of p-PI3K to PI3K, p-AKT to AKT were calculated in MGC-803. **P<0.001 vs. NC+MC. ##P<0.001 vs. PCAT18+MC. ^^P<0.001 vs. NC+mimic.
Discussion
GC is a leading cause of cancer mortality. Evidence showed that lncRNAs play critical roles in multiple diseases, including in cancer progression.17 Though many long non-coding RNAs (ncRNAs) have been identified in cancer,18,19 the role and functions of most lncRNAs are not fully understood. In this study, lncRNA PCAT18 was down-regulated in GC and overexpression of PCAT18 inhibited GC cells growth in vitro and in vivo. The mechanism analysis results further found that PCAT18 was involved in PTEN/PI3K/AKT signaling pathway through targeting miR-107. To the best of our knowledge, our research was the first to report the molecular mechanism of PCAT18 in cancer.
PCAT18 is a prostate cancer-associated lncRNA, however, its molecular mechanism still remains unknown. In this study, we found that PCAT18 was down-regulated in GC and has a diagnostic value for GC, and such a finding was consistent with a previous study on GC.11 Furthermore, PCAT18 was confirmed to regulate GC cell growth and cell apoptosis in vitro and tumor growth in vivo. Interestingly, the role of PCAT18 in metastatic prostate cancer is different from that in GC. A study reported that lncRNA H19 plays critical roles in EMT and MET by sponging different miRNAs.20 Thus, we speculated that the participation of PCAT18 in different pathways could explain these controversial results.
LncRNA, as a ceRNAs, regulates miRNA and its related pathways. For example, lncRNA NORAD promotes the growth of GC cells via modulating the miR-608/FOXO6 pathway.21 In GC, lncRNA HOTAIR regulates HER2 by sponging miR-331-3p.22 In our current study, PCAT18 could function as miRNA sponge that directly interacts with miR-107, according to dual-luciferase reporter assay. Furthermore, data indicated that PCAT18 interacts with miR-107 to regulate the progress of GC. Though the role of miR-107 is controversial in GC, our study proved that miR-107 was up-regulated in GC and was negatively correlated with the expression of PCAT18. Multiple miRNAs has been found to regulate PTEN/PI3K/Akt pathway in GC,23 similar to a previous study, our further data indicated that PCAT18 regulated GC growth via modulating PI3K/AKT signaling pathway through sponging miR-107.
The phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling promotes cell growth and survival. The activated PI3K phosphorylates phosphatidylinositol-4,5-bisphosphate (PIP2) to form phosphatidylinositol-3,4,5-trisphosphate (PIP3). Akt is the downstream of PIP3, in which Akt is phosphorylated and stimulated by other kinases. PTEN (phosphatase and tensin homolog) primarily dephosphorylates phosphatidylinositol-3,4,5-trisphosphate (PIP3) and is a negative regulator of PI3K/AKT signaling. The PTEN/PI3K/AKT axis is a crucial signaling pathway in various of tumor biological processes,24–27 and molecules targeting PI3K pathway is widely used in GC therapies.28,29 PIK3CA and PTEN mutations are common events in genetic mutations in cancer progression.30–32 Previous study indicates that PIK3CA mutations are closely associated with survival time of patients with glioblastoma.32 PTEN loss is associated with resistance to trastuzumab-based therapy.33 Therefore, further understanding of the PTEN/PI3K/AKT pathways and their upstream signaling in GC is helpful in finding effective targets for GC therapies. Accumulating evidence indicates that lncRNAs participate in PI3K/AKT signaling pathway and thereby regulate tumor growth. For example, a study showed that LOC101928316 regulates the progression of GC via modulating PI3K-Akt-mTOR signaling pathway.34 A novel lncRNA TUBA4B can inhibit GC growth through regulating miR-214/miR-216a/b-PTEN axis.35 Research also reported that lncRNA SEMA3B-AS1 regulates the proliferation of hepatocellular carcinoma cells by regulating miR-718/PETN axis.36 In this study, a PCAT18/miR-107/PI3K/PTEN/AKT signaling axis was found in GC, and we showed that PCAT18 is a new target associated with PTEN/PI3K/AKT pathway and could be explored to be used in GC treatment.
Conclusion
In conclusion, PCAT18 is a tumor suppressor gene in GC. Additionally, PCAT18 might regulate the PTEN/PI3K/AKT sgnalling pathway through sponging miR-107, and thereby regulating tumor growth. Therefore, PCAT18 could be potentially used as a therapeutic target for treatment of GC.
Disclosure
The authors declare no conflicts of interest.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Karim-Kos HE, Coebergh JW, et al. Recent trends in incidence of five common cancers in 26 European countries since 1988: analysis of the European Cancer Observatory. Eur J Cancer. 2015;51(9):1164–1187. doi: 10.1016/j.ejca.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 3.Chmiela M, Karwowska Z, Gonciarz W, Allushi B, Staczek P. Host pathogen interactions in Helicobacter pylori related gastric cancer. World J Gastroenterol. 2017;23(9):1521–1540. doi: 10.3748/wjg.v23.i9.1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saka M, Morita S, Fukagawa T, Katai H. Present and future status of gastric cancer surgery. Jpn J Clin Oncol. 2011;41(3):307–313. doi: 10.1093/jjco/hyq240 [DOI] [PubMed] [Google Scholar]
- 5.Ye N, Wang B, Quan ZF, et al. Functional roles of long non-coding RNA in human breast cancer. Asian Pac J Cancer Prev. 2014;15(15):5993–5997. doi: 10.7314/apjcp.2014.15.15.5993 [DOI] [PubMed] [Google Scholar]
- 6.Zhang E, He X, Zhang C, et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YW, Xia R, Lu K, et al. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16(1):39. doi: 10.1186/s12943-017-0588-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuo W, Liu Y, Li S, et al. Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of Ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019;156(3):676–91.e11. doi: 10.1053/j.gastro.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 9.Lin LY, Yang L, Zeng Q, et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17(1):84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crea F, Watahiki A, Quagliata L, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foroughi K, Amini M, Atashi A, Mahmoodzadeh H, Hamann U, Manoochehri M. Tissue-specific down-regulation of the long non-coding RNAs PCAT18 and LINC01133 in gastric cancer development. Int J Mol Sci. 2018;19:12. doi: 10.3390/ijms19123881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G, Li S, Lu J, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17(1):87. doi: 10.1186/s12943-018-0829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang J, Min BH, Jang J, et al. MicroRNA expression profiles in gastric carcinogenesis. Sci Rep. 2018;8(1):14393. doi: 10.1038/s41598-018-32782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T, Liu S, Xu Y, et al. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res Treat. 2018;50(4):1396–1417. doi: 10.4143/crt.2017.537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Li F, Liu B, Gao Y, et al. Upregulation of microRNA-107 induces proliferation in human gastric cancer cells by targeting the transcription factor FOXO1. FEBS Lett. 2014;588(4):538–544. doi: 10.1016/j.febslet.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Cheng F, Yang Z, Huang F, Yin L, Yan G, Gong G. microRNA-107 inhibits gastric cancer cell proliferation and metastasis by targeting PI3K/AKT pathway. Microb Pathog. 2018;121:110–114. doi: 10.1016/j.micpath.2018.04.060. [DOI] [PubMed] [Google Scholar]
- 17.Tam C, Wong JH, Tsui SKW, Zuo T, Chan TF, Ng TB. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: updates in recent years. Appl Microbiol Biotechnol. 2019;103(12):4649–4677. doi: 10.1007/s00253-019-09837-5. [DOI] [PubMed] [Google Scholar]
- 18.Ding Y, Yang DZ, Zhai YN, et al. Microarray expression profiling of long non-coding RNAs in epithelial ovarian cancer. Oncol Lett. 2017;14(2):2523–2530. doi: 10.3892/ol.2017.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 20.Zhou W, Ye XL, Xu J, et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10:483. doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 21.Miao Z, Guo X, Tian L. The long noncoding RNA NORAD promotes the growth of gastric cancer cells by sponging miR-608. Gene. 2019;687:116–124. doi: 10.1016/j.gene.2018.11.052. [DOI] [PubMed] [Google Scholar]
- 22.Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu M, Zhu S, Xiong S, Xue X, Zhou X. MicroRN As and the PTEN/PI3K/Akt pathway in gastric cancer (Review). Oncol Rep. 2019;41(3):1439–1454. doi: 10.3892/or.2019.6962. [DOI] [PubMed] [Google Scholar]
- 24.Tsai CY, Wu JCC, Fang C, Chang AYW. PTEN, a negative regulator of PI3K/Akt signaling, sustains brain stem cardiovascular regulation during mevinphos intoxication. Neuropharmacology. 2017;123:175–185. doi: 10.1016/j.neuropharm.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/Mtor Signaling Pathway in Ovarian Cancer: Biological and Therapeutic Significance. Semin Cancer Biol 2019.. [DOI] [PubMed] [Google Scholar]
- 26.Wang LL, Hao S, Zhang S, et al. PTEN/PI3K/AKT protein expression is related to clinicopathological features and prognosis in breast cancer with axillary lymph node metastases. Hum Pathol. 2017;61:49–57. doi: 10.1016/j.humpath.2016.07.040. [DOI] [PubMed] [Google Scholar]
- 27.Patel K, Pagel JM. Exploring a future for PI3K Inhibitors in chronic lymphocytic leukemia. Curr Hematol Malig Rep. 2019;14(4):292–301. doi: 10.1007/s11899-019-00525-9. [DOI] [PubMed] [Google Scholar]
- 28.Farran B, Muller S, Montenegro RC. Gastric cancer management: kinases as a target therapy. Clin Exp Pharmacol Physiol. 2017;44(6):613–622. doi: 10.1111/1440-1681.12743. [DOI] [PubMed] [Google Scholar]
- 29.Haddadi N, Lin Y, Travis G, Simpson AM, Nassif NT, McGowan EM. PTEN/PTENP1: ‘Regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy. Mol Cancer. 2018;17(1):37. doi: 10.1186/s12943-018-0803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavagnari MAV, Silva TD, Pereira MAH, et al. Impact of genetic mutations and nutritional status on the survival of patients with colorectal cancer. BMC Cancer. 2019;19(1):644. doi: 10.1186/s12885-019-5837-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu W, Li X, Wang T, Zheng S. Association mining of mutated cancer genes in different clinical stages across 11 cancer types. Oncotarget. 2016;7(42):68270–68277. doi: 10.18632/oncotarget.11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao H, Wang N, Shi LX, Wang Z, Song WB. Overexpression of mircoRNA-137 inhibits cervical cancer cell invasion, migration and epithelial-mesenchymal transition by suppressing the TGF-beta/smad pathway via binding to GREM1. Cancer Cell Int. 2019;19:147. doi: 10.1186/s12935-019-0852-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deguchi Y, Okabe H, Oshima N, et al. PTEN loss is associated with a poor response to trastuzumab in HER2-overexpressing gastroesophageal adenocarcinoma. Gastric Cancer. 2017;20(3):416–427. doi: 10.1007/s10120-016-0627-z. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Liang G, Yang S, et al. LncRNA-LOC101928316 contributes to gastric cancer progression through regulating PI3K-Akt-mTOR signaling pathway. Cancer Med. 2019;8(9):4428–4440. doi: 10.1002/cam4.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo J, Li Y, Duan H, Yuan L. LncRNA TUBA4B functions as a competitive endogenous RNA to inhibit gastric cancer progression by elevating PTEN via sponging miR-214 and miR-216a/b. Cancer Cell Int. 2019;19:156. doi: 10.1186/s12935-019-0879-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Zhong Y, Li Y, Song T, Zhang D. MiR-718 mediates the indirect interaction between lncRNA SEMA3B-AS1 and PTEN to regulate the proliferation of hepatocellular carcinoma cells. Physiol Genomics. 2019;51(10):500–505. doi: 10.1152/physiolgenomics.00019.2019. [DOI] [PubMed] [Google Scholar]