Abstract
Color patterns in African cichlid fishes vary spectacularly. Although phylogenetic analysis showed already 30 years ago that many color patterns evolved repeatedly in these adaptive radiations, only recently have we begun to understand the genomic basis of color variation. Horizontal stripe patterns evolved and were lost several times independently across the adaptive radiations of Lake Victoria, Malawi, and Tanganyika and regulatory evolution of agouti-related peptide 2 (agrp2/asip2b) has been linked to this phenotypically labile trait. Here, we asked whether the agrp2 locus exhibits particular characteristics that facilitate divergence in color patterns. Based on comparative genomic analyses, we discovered several recent duplications, insertions, and deletions. Interestingly, one of these events resulted in a tandem duplication of the last exon of agrp2. The duplication likely precedes the East African radiations that started 8–12 Ma, is not fixed within any of the radiations, and is found to vary even within some species. Moreover, we also observed variation in copy number (two to five copies) and secondary loss of the duplication, illustrating a surprising dynamic at this locus that possibly promoted functional divergence of agrp2. Our work suggests that such instances of exon duplications are a neglected mechanism potentially involved in the repeated evolution and diversification that deserves more attention.
Keywords: Cichlidae, teleosts, pigmentation, coloration, agouti gene family, agrp2, asip2b
Introduction
The East African cichlid fish adaptive radiations, with their over 1,200 species, are one of the most prominent examples for repeated, convergent evolution (Meyer 1993; Stiassny and Meyer 1999; Muschick et al. 2012). In the large species flocks of Lakes Tanganyika, Malawi, and Victoria, body shapes, trophic morphologies, and color patterns evolved dozens of times independently (Kocher 2004; Muschick et al. 2012; Kratochwil et al. 2018; Salzburger 2018). This exuberant variation demanded the search for genomic explanations for the exceptional rates of diversification and repeated evolution (Brawand et al. 2014; Kratochwil and Meyer 2015; Salzburger 2018; Kratochwil 2019). Recent work from sticklebacks supports that genomic features such as DNA conformation can promote evolutionary adaptations and do so repeatedly (Xie et al. 2019). A meta-analysis of several cases of parallel evolution also suggests an influence of mutational biases on repeated adaptive evolution (Lynch et al. 2016; Stoltzfus and McCandlish 2017). Moreover, gene duplications are important molecular mechanisms of diversification (Ohno 1970; Van de Peer et al. 2009; Musilova et al. 2019) and convergence (Denoeud et al. 2014; Ishikawa et al. 2019).
Recently, it has been shown that the evolution of cichlid fish stripe color patterns is facilitated by independent regulatory evolution at the agrp2 locus (Kratochwil et al. 2018). Yet, it remains unclear what genomic features influenced the evolution of the underlying flexible gene regulatory architectures and the evolution of novel cis-regulatory elements that clearly account for diversification and repeated evolution in cichlids. Here, we report on an investigation of the agrp2 locus and asked whether particular genomic features including structural variation might affect the evolutionary dynamics of this important locus for cichlid color pattern diversification.
Materials and Methods
Pairwise Global Alignment
To perform pairwise global alignments, we extracted ∼200 kb (100 kb for the reference) intervals flanking the agrp2 gene from Ensembl 94 (Cunningham et al. 2019) and NCBI. The Maylandia zebra genome (Ensembl 94, GCA_000238955.5) was used as a reference. Before alignment, the reference was masked using the Tilapia repeat masker (Shirak et al. 2010). Nine genomes were aligned to the reference: Pundamilianyererei (Ensembl 94, GCA_000002035.4), Neolamprologus brichardi (Ensembl 94, GCA_000239395.1), Oreochromis niloticus (NCBI, MKQE02), Amphilophuscitrinellus (Ensembl 94, GCA_000751415.1), Xiphophorus maculatus (Ensembl 94, GCA_002775205.2), Gasterosteus aculeatus (Ensembl 94, BROAD S1), Scophthalmus maximus (Ensembl 94, GCA_003186165.1), Astyanax mexicanus (Ensembl 94, GCA_000372685.2), and Danio rerio (Ensembl 94, GCA_000002035.4). For P.nyererei, we used a curated version (description see below; fasta file available in the Supplementary Material online). For alignment, we used the Shuffle-LAGAN algorithm (Brudno et al. 2003) with a RankVISTA probability threshold of 0.5 and providing the following pairwise phylogenetic tree: ((((((((M. zebra P. nyererei) N. brichardi) O. niloticus) Amp. citrinellus) X. maculatus) G. aculeatus) S. maximus)(Ast. mexicanus D. rerio)). For the M. zebra reference, we used an annotation based on the ensemble annotation and manually annotated the previously described cis-regulatory region of agrp2 (Kratochwil et al. 2018). Finally, we extracted a ∼45 kb interval of the Shuffle-LAGAN alignment to be used in figure 1A. Genomic coordinates of agrp2 coding regions, cis-regulatory elements, and deletions in the different genomes are summarized in supplementary table S1, Supplementary Material online.
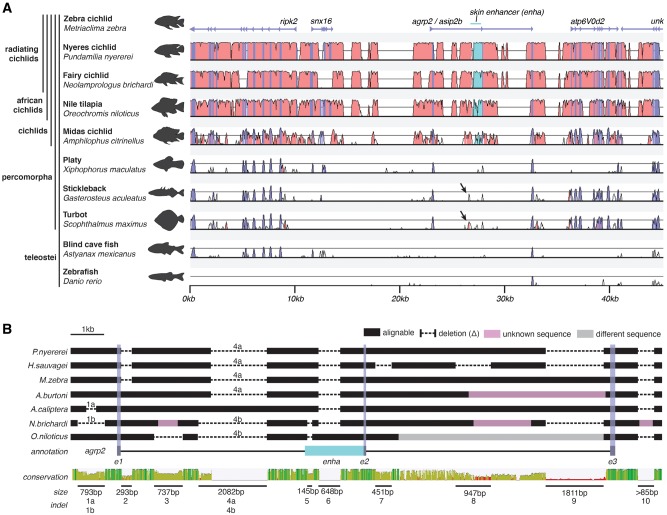
Fig. 1.
—Sequence conservation at the agrp2 locus. (A) Shuffle-LAGAN global pairwise alignment of genomic sequences from cichlids as well as representatives from the Order Percomorpha and non-Percomorpha outgroups. The used reference is the genome of the Lake Malawi cichlid Maylandia zebra. Although exonic regions of agrp2 and the neighboring genes ripk2, snx16, agrp2, atp6V0d2 and an unknown gene are largely conserved across the Order Percomorpha and partially seemingly even in all teleosts, there are only a few conserved noncoding elements. One of these partially conserved elements (with stickleback and turbot; divergence times 105–154 Ma; Kumar et al. 2017) is 5′ of a previously described regulatory element of agrp2, enhA (black arrow). (B) Multiple sequence alignments of available assemblies of the agrp2 locus indicate several larger (>50 bp) deletions (numbered from 1 to 10).
Cichlid Genome Alignments
For the cichlid genome alignments, we selected an 18 kb region encompassing the agrp2 gene. Sequences from the genomes of M. zebra, P. nyererei, and N.brichardi as well as Astatotilapiacalliptera (Ensembl 94, GCA_900246225.3) and Astatotilapiaburtoni (Ensembl 94, GCA_000239415.1). Additionally, we added a Sanger sequencing assembly of the locus from the Lake Victoria species Haplochromissauvagei (Kratochwil et al. 2018). Sequences were aligned using MAFFT 7 (Katoh et al. 2017) using automatic strategy choice, and standard settings. Conservation score and annotations were added using Geneious 2019. The alignment was visualized using Adobe Illustrator CC 2018. Dot plots were generated using MAFFT 7 (Katoh et al. 2017) with a threshold score of 39 (E = 8.4e-11).
RNA Extraction and cDNA Synthesis
Fish were obtained from commercial breeders and euthanized with an overdose of MS-222. Experiments were performed in accordance with animal research regulations (Regierungspräsidium Freiburg, Baden Württemberg, Germany, Reference number: G-17/110). Skin, brain (both RNA-seq, cDNA sequencing), liver, eye, and muscle tissue (all cDNA sequencing) were dissected and kept in RNAlater (Invitrogen) at 4 °C overnight and transferred to −20 °C for long-term storage. RNAlater was removed prior to homogenization. Skin samples and appropriate amount of TRIzol (Invitrogen) (1 ml TRIzol per 0.1 g sample) were homogenized in 2 ml lysing matrix A tube (MP Biomedicals) using FastPrep-24 Classic Instrument (MP Biomedicals). RNA was extracted according to the manufacturer’s recommendations with additional 75% ethanol wash one time. Subsequent purification and on-column DNase treatment was performed with RNeasy Mini Kit (Qiagen) and RNase-Free DNase Set (Qiagen). The other organs were extracted using RNeasy Mini Kit (Qiagen). DNA was removed using RNase-Free DNase Set (Qiagen) according to the manufacture’s protocol. Following extraction and purification, RNA was quantified using the Qubit RNA HS Assay Kit (Invitrogen) with a Qubit Fluorometer (Life Technologies). First-strand cDNA was synthesized using 1 μg total RNA and the GoScript Reverse Transcription System (Promega).
5′ and 3′ Rapid Amplification cDNA Ends
5′ Rapid amplification of cDNA end (5′RACE) was performed as previously described (Scotto–Lavino et al. 2006b). We performed RT-PCR and nested PCR by two sets of gene specified primers: aR5 + aR4 +aR3 and aR4 + aR3 + aR2 (supplementary fig. S1, Supplementary Material online). To generate “5′ end” partial cDNA, reverse transcription was carried out using gene specified primers aR5 or aR4 by M-MLV Reverse Transcriptase [H–] (Promega) to generate first-strand product. Following the reverse transcription, a poly(A) tail was appended using terminal deoxynucleotidyl-transferase Tdt (Promega) and dATP. First-strand gene-specific cDNA with poly(A) tail was then purified by NucleoSpin Gel and PCR Clean-up (MACHEREY-NAGEL). Amplification was achieved by PCR using Qtotal and Qouter and gene specified primers, aR4 for the first-strand cDNA product of aR5, aR3 for the first-strand cDNA product of aR4. PCR was performed using DreamTaq DNA Polymerase (Thermo Fisher Scientific) in 50 μl reactions with the following program: initial denaturation at 98 °C for 3 min, annealing at 60 °C for 2 min, extension of cDNA at 72 °C for 40 min, followed by 30 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 3 min, final extension at 72 °C for 15 min. A second set of PCR cycles was carried out to increase the yield of specific product using nested primers Qinner and upstream gene specified primers, aR3 for the first-strand PCR product of aR4, aR2 for the first-strand PCR product of aR3. The second PCR was performed using the same condition as for the first-strand PCR without cDNA extension. After the second PCR, bands were purified using NucleoSpin Gel and PCR Clean-up (MACHEREY-NAGEL).
3′RACE was performed as previously described (Scotto–Lavino et al. 2006a). Total RNA was reverse transcribed to first-strand cDNA by M-MLV Reverse Transcriptase [H–] (Promega) using primer Qtotal in 20 μl reverse transcription reaction. To increase the specificity of the 3′RACE PCR, we performed a nested PCR by two sets of gene specified primers, aF1 + aF2 and aF2 + aF3 (supplementary fig. S2, Supplementary Material online). For the first RACE PCR, we used the outer primer Qouter with gene-specific primer aF1 or aF2. PCR was performed using DreamTaq DNA Polymerase (Thermo Fisher Scientific) as described for 5′RACE. For the nested PCR, inner primer Qinner was used in combination with gene-specific primers, aF2 for the first PCR product of Qouter and aF1, aF3 for the first PCR product of Qouter and aF2. The second PCR was performed using the same condition as for the first PCR without cDNA extension. Following the second PCR, we gel-purified bands using NucleoSpin Gel and PCR Clean-up (MACHEREY-NAGEL).
All DNA fragments from 3′RACE and 5′RACE were processed by Sanger sequencing using Applied Biosystems 3130 Genetic Analyzers. Sanger Sequencing data were analyzed in Geneious 2019. Primer aF3, aF5, aR5, and aR6 were used for sequencing of 3′RACE product. aF1, aF2, aR1, and aR2 were the sequencing primers of the 5′RACE products.
To confirm the sequencing result of 3′RACE and 5′RACE, we additionally performed RT-PCRs. For this, cDNA was synthesized as described earlier. Three forward primers (F1, F2, and F3) and one reverse primer (R1–R3) were used in PCR with DreamTaq DNA Polymerase (Thermo Fisher Scientific) with the following program: initial denaturation at 95 °C for 10 min, 30 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min 30 s, final extension at 72 °C for 7 min. Primers are summarized in supplementary table S2, Supplementary Material online.
Tandem Duplication-Specific PCRs
DNA was extracted from fin clips using ethanol precipitation. To identify species and individuals with and without duplication, we used two approaches (supplementary fig. S3A, Supplementary Material online). The first approach included PCRs with a forward primer in the 3′ end of exon 3a and a reverse primer in the 5′end of exon 3 b (supplementary figs. S3 and S4 and table S2, Supplementary Material online). This approach was more sensitive to detect duplications across species (as primer binding sites were inside coding regions). PCR was performed using DreamTaq DNA polymerase (Thermo Fisher Scientific) in a 20 µl reaction with the following program: initial denaturation at 95 °C for 5 min, 32 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min, final extension at 72 °C for 15 min.
The second approach was a long-range PCR that spanned the tandem duplication (supplementary fig. S5 and table S2, Supplementary Material online). Long-range PCRs failed in many species, likely because of binding site mutations and repeat regions that flank the duplication (supplementary table S4, Supplementary Material online). PCR was performed using DreamTaq DNA polymerase (Thermo Fisher Scientific) in a 20 µl reaction with the following program: initial denaturation at 95 °C for 5 min, 35 cycles of 95 °C for 30 s, 62 °C for 30 s, 72 °C for 7 min, final extension at 72 °C for 30 min.
TaqMan Probe Assay
TaqMan probe-based qPCR was used to determine the copy number of exon 1 (as a control) and exon 3 of agrp2. Genomic DNA was extracted from fin clips using DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s protocol. Following extraction, the purity of genomic DNA was checked by Colibri spectrometer (Berthold), the degradation was evaluated with 1% agarose gel and the concentration was quantified using Qubit DNA BR Assay Kit (Invitrogen) with Qubit Fluorometer (Life Technologies). qPCRs were performed with 10 ng genomic DNA, 1 µl of each forward and reverse primer (20 µM stock), 1 µl hydrolysis probe (5 µM stock), and GoTaq Probe qPCR Master Mix (Promega) with Nuclease-Free Water to make the final volume of 20 μl in a 96-well plate. Primers and probes for agrp2 exon 1 and exon 3 are listed in supplementary table S2, Supplementary Material online. We used 40 cycles of amplification on a CFX96 Real-Time PCR Detection System (Bio-Rad) with the program: polymerase activation at 95 °C for 2 min, 40 cycles of denaturation (95 °C for 15 s) and annealing/extension (60 °C for 1 min). Ct values were defined as the point at which fluorescence crossed a threshold adjusted manually to be the point at which fluorescence rose above the background level. We generated the standard curve by using different amounts (2.5 ng, 5 ng, 10 ng) of genomic DNA from H. sauvagei (mixture of three individuals) as DNA template. Copy number variation for each sample was detected based on the position of the Ct value on the standard curve. We assayed copy number variation using three technical replicates for each sample and three biological replicates for each species. Primers and probes are listed in supplementary table S2, Supplementary Material online.
Coverage Analysis
To screen for coverage difference in whole genome resequencing data, we used previously published WGS data sets as summarized in supplementary table S3, Supplementary Material online (Valente et al. 2014; McGee et al. 2016; Meier et al. 2017; Malinsky et al. 2018). We downloaded the data, and trimmed Illumina adapters from raw fastq reads using picard v2.17.11. Reads were then aligned to a curated genome (see Supplementary Material online) of P.nyererei (Brawand et al. 2014) as agrp2 was split onto two scaffolds (exons 1 and 2 on scaffold_361/JH419567.1; exon 3 on scaffold_3/JH419209.1). The reverse complement of scaffold_361 was therefore concatenated with scaffold_3 (replacing scaffold_3 in the assembly). The original Scaffold_361 was then removed from the assembly. In addition, Scaffold_6781 (length: 1,095 bp) was removed from the assembly because it caused alignment problems as the nucleotide sequence was identical to a sequence 3′ of agrp2 exon 3 (but not overlapping with the duplication). The whole ∼25 kb agrp2 locus was replaced with a manually Sanger-sequencing-read-curated sequence previously published (Kratochwil et al. 2018). Using this approach, the assembly gap between the scaffolds was closed as well. We confirmed that the individual that has been used for Sanger Sequencing had no duplication of exon 3. Coverage was calculated for each position of the genome for each sample using samtools depth 1.9 (Li et al. 2009). For this, we counted only reads with a base quality >20 and a mapping quality >30. Further data analysis was performed in R (R Development Core Team 2019). Relative coverage was calculated by dividing the coverage at each position by the mean coverage of a 110 kb window around the agrp2 gene. Nonoverlapping sliding windows were calculated using the SlidingWindow function of the evobiR package (Blackmon and Adams 2015) with a window and step size of 100 bp. The following additional R packages were used: ggplot2 (Wickham et al. 2019) and stringR (Wickham 2019). Transposable element position and identity of the agrp2 locus are summarized in supplementary table S4, Supplementary Material online. We used both, the Tilapia repeat masker (Shirak et al. 2010) and the standard repeat masker (Smit et al. 2015).
Exon Duplicate-Specific Expression
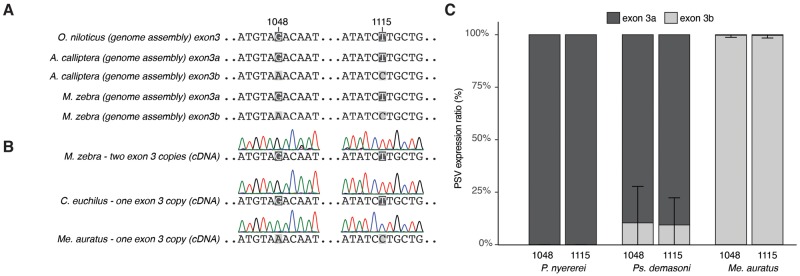
To determine the duplicate-specific expression of agrp2 exon 3a and 3 b, we first used the Pacbio assemblies of A. calliptera (GCA_900246225.3) and M. zebra (GCA_000238955.5) that resolved both exons, performed alignments using MAFFT and screened for paralogous sequence variants. Within the 3′-untranslated region (3′-UTR), we found two SNPs at position 1,048 and 1,115 of the agrp2 transcript (ENSHBUT00000024720) that are alternatively fixed between exon 3a and exon 3b of both species (fig. 3).
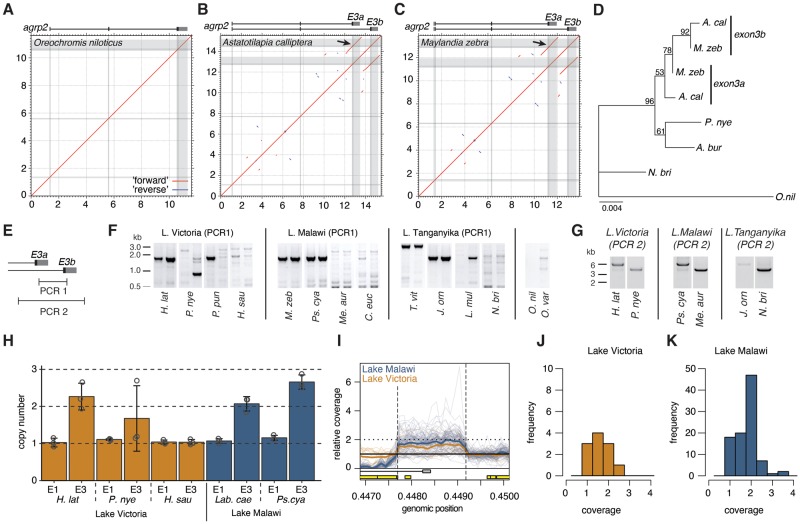
Fig. 3.
—The third exon of agrp2 was tandemly duplicated. (A) A syntenic dotplot of the agrp2 loci of O. niloticus against itself shows no evidence for duplicated sequences. (B) In contrast to that the genome of A. calliptera show multiple small duplications (red) and inverted duplications (blue) and a 1.6 kb tandem duplication that gives rise to a duplicated exon 3 (exon 3a and exon 3 b). (C) The genome of M. zebra shows a similar pattern of duplications, including a duplicated exon 3. (D) Maximum likelihood phylogeny of the genomic sequences for exon 3 (including 3a and 3 b) suggest a common origin of the duplication. (E) PCR design to confirm duplications of exon 3. (F) Short PCRs, amplifying a ∼1.6 kb sequence between the two copies confirms the duplication in several, but not all species. The presence also varies within populations (e.g., in P. nye and P. pun). (G) Long-range PCRs including sequence of both copies of exon 3 provide additional confirmation for the duplication. (H) TaqMan probe assay to quantify the copy number comparing exon 1 (no evidence for duplication) and exon 3. (I) Copy number quantification based on coverage of genome resequencing data. Thick lines indicate the average for Lake Malawi (blue) and Lake Victoria species (orange). Thin lines represent individuals. Relative coverage is increased in many individuals at a position corresponding to the estimated size of the duplication (dashed lines). Yellow boxes indicate repetitive elements that likely explain the coverage drop 5′ of the duplication in Lake Malawi. (I) Histogram representation of the Lake Victoria species’ mean relative read coverage within the duplicated region. Most species have one or two copies. (J) Same representation for Lake Malawi illustrating that the duplication is more common. Most species have two exon 3 copies, some even more than three. Abbreviations: A. bur, Astatotilapia burtoni; A. cal, Astatotilapia calliptera; C. euc, Cheilochromis euchilus; H. lat, Haplochromis latifasciatus; H. sau, Haplochromis sauvagei; J. orn, Julidochromis ornatus; L. mul, Lamprologus multifasciatus; Lab. cae, Labidochromis caeruleus; M. zeb, Maylandia zebra; Me. aur, Melanochromis auratus; N. bri, Neolamprologus brichardi; N. cau, Neolamprologus caudopunctatus; O. nil, Oreochromis niloticus; O. var, Oreochromis variabilis; P. nye, Pundamilia nyererei; P. pun, Pundamilia pundamilia; Ps. cya, Pseudotropheus cyaneorhabdos; T. vit, Telmatochromis vittatus.
To determine, which paralogous sequence is expressed across other cichlid species, we used Sanger sequencing of cDNA (description of extraction see above; primers see supplementary table S2, Supplementary Material online). Second, we used RNA-seq to determine paralog-specific expression by calculating variant ratios on the two variable positions. RNA-seq libraries were prepared using TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer’s protocol with first-strand cDNA synthesis by GoScript Reverse Transcriptase (Promega). The final libraries were amplified using 15 PCR cycles and quantified and quality-assessed by Agilent DNA 12000 Kit with 2100 Bioanalyzer (Agilent). Indexed DNA libraries were normalized then pooled in equal volumes. Libraries were sequenced on an Illumina HiSeq 2500 (P. nyererei skin) or HiSeq X Ten platform (Pseudotropheusdemasoni skin, Melanochromisauratus brain). Reads were mapped to the A. burtoni genome (as agrp2 is well annotated and the reference genome has only one copy of exon 3a) using STAR (Dobin et al. 2013). BAM files were extracted for the agrp2 transcript (ENSHBUT00000024720) and genotyped using FreeBayes with –pooled-continuous setting. Ratios between reference and alternative allele were calculated for the two positions using R (R Development Core Team 2019) and plotted as stacked bar plots.
Molecular Evolutionary Analyses
Maximum likelihood phylogenies of the agrp2 gene were based on genomic sequences (outgroups) and Sanger sequencing of agrp2 cDNA (synthesis of cDNA is described earlier; primers for amplification and Sanger sequencing are listed in supplementary table S2, Supplementary Material online). Trees were generated using PhyML (Guindon et al. 2010) with HKY85 substitution model, 100 bootstrap replicates, and Length/Rate optimization.
We tested for evidence of the evolution of the agrp2 coding sequence using codon-based models in PAML (Yang 2007). Different random site models (e.g., M0, M1a, M2a) were compared using log-likelihood ratio tests (LRT) to test for the presence of sites classes that differ in dN/dS ratio. Specifically, we first tested for evidence of two site classes (i.e., M1a/M0), one assumed to be evolving under purifying selection (dN/dS < 1) and a second class evolving under neutral selection (dN/dS = 1). Second, we tested for the presence of positively selected sites (i.e., M2a/M1a) (Yang 2014). Then, using clade model C (CmC) in PAML, we tested the hypothesis that the agrp2 gene is evolving divergently between the lineage where the duplication of the third exon occurred (e.g., the radiating cichlids from Lakes Tanganyika, Malawi, and Victoria) compared with lineages without this duplication. The significance of the CmC model was determined conducting a LRT against a null model that allowed for different site classes, but not for divergence among lineages in the alignment (M2a_rel) (Weadick and Chang 2012).
Single Breakpoint Recombination Analysis
Because the functional importance of the third exon of the agrp2 gene, the finding that this exon duplicated, and that this duplicated copy could occasionally become functional through the loss of exon 3a, it is possible that exon 3 shows phylogenetic discordance with the other two exons in the gene. To test this hypothesis, we conducted a single breakpoint recombination analysis (Kosakovsky Pond et al. 2006) as implemented in HYPHY (Pond and Frost 2005; Pond and Muse 2005). Because we found evidence for a recombination breakpoint between exons 1 and 2 and exon 3 (see results), we repeated the molecular evolution analyses described earlier but independently for an alignment including only exons 1 + 2 and one including only exon 3 of the agrp2 gene.
Results
Evolutionary History of the agrp2 Locus
The agrp2 locus has been previously implicated in regulating stripe patterns in the cichlid radiations of Lakes Victoria, Malawi, and Tanganyika. To gain a deeper understanding of the evolution of this locus, we investigate it in a comparative approach across ten teleost species (fig. 1A). The gene agrp2 is flanked by the ripk2 (Receptor interacting serine/threonine kinase 2) and snx16 (sorting nexin-16) on the 5′ side, and atp6V0d2 (v-type proton ATPase subunit d 2) and “unk,” an unknown gene 3′ of agrp2. Beyond cichlids, mainly coding sequences are conserved, with strong signals of conservation for ripk2, but also atp6V0d2 and agrp2. Interestingly, alignments show only low conservation 5′ of a skin-specific regulatory region (enha) that has been functionally linked to agrp2 expression (Kratochwil et al. 2018). This possibly suggests a larger regulatory module within this intron that might also contain elements that drive the brain-specific expression that shows greater evolutionary conservation (Zhang et al. 2010; Shainer et al. 2017). Within the family Cichlidae, we find several conserved blocks in the noncoding sequence in 5′ and 3′ as well as the intronic sequences including the enha regulatory sequence (fig. 1B).
Description of a Novel Noncoding Exon of agrp2
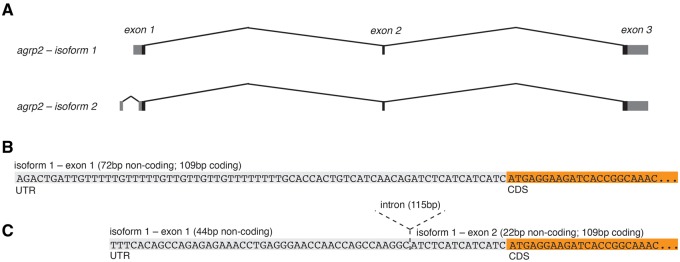
To be able to better interpret structural variation at the agrp2 locus within African cichlids, we first comprehensively described the transcript annotation of the agrp2 gene. Previously, it has been shown that agrp2 mRNA is mainly expressed in the brain and in the skin (Zhang et al. 2010; Shainer et al. 2017; Kratochwil et al. 2018). For the agouti signaling protein (asip), another member of the agouti family different tissue-specific isoforms have been reported (Mallarino et al. 2017). Therefore, we asked whether agrp2 also has different isoforms that may act—besides the described cis-regulatory variation—as an additional source of differential regulation between tissues (i.e., brain and skin) and species (i.e., striped and nonstriped species). As alternative splicing and the evolution of novel isoforms could be affected by structural variation in the agrp2 locus, we aimed to identify the full-length transcripts in an effort to discover potentially unknown exons. Based on 5′RACE, we amplified the full length 5′ sequence of agrp2 from both brain and skin tissue of the striped species H.sauvagei and nonstriped species P.nyererei. Sanger sequencing provided evidence for two isoforms (fig. 2A). Isoform 1 has three exons: the first exon consists of a 72 bp 5′-UTR and 109 bp of the coding sequence (fig. 2B and supplementary fig. S1, Supplementary Material online). Isoform 2 has an additional noncoding first exon with 44 bp of the 5′-UTR, the second exon has only a short 22 bp UTR and the 109 bp coding sequence (fig. 2C and supplementary fig. S1, Supplementary Material online). The isoforms can be found in both species and in both brain and skin tissue, as confirmed by isoform-specific PCR (supplementary fig. S1, Supplementary Material online).
Fig. 2.
—Isoforms of agrp2. (A) Using 5′- and 3′RACE as well as PCRs on cDNA, we characterized two isoforms of agrp2. Both have an identical coding sequences but differ in their 5′-UTR. (B) Isoform 1 has a 72 bp 5′-UTR. (C) Isoform 2 has an additional noncoding exon with 44 bp of the 5′-UTR, the second exon has only a short 22 bp UTR.
Additionally, we performed 3′ Rapid Amplification of cDNA Ends (3′RACE) to amplify the full length 3′-UTR of agrp2 (supplementary fig. S2, Supplementary Material online). Sequencing revealed a total 3′-UTR length of 616 bp in P. nyererei and 579 bp in H. sauvagei, respectively. Within the UTR, we found three potential cleavage sites with an AAUAAA or AUUAAA motif (supplementary fig. S2, Supplementary Material online). Sanger sequencing gave no indication for additional exons between exons 1 and 2 as well as exons 2 and 3 (supplementary figs. S1 and S2, Supplementary Material online) in any of the >30 analyzed species.
Characterization of Insertions and Deletion of the agrp2 Locus
To more comprehensively describe structural variation (i.e., larger insertions and deletions; indels or duplications), we aligned available cichlid fish genomic segments of the agrp2 locus (Brawand et al. 2014; Conte et al. 2017, 2019; Kratochwil et al. 2018) including seven African cichlid species: the geographically widespread O.niloticus, the Lake Tanganyika endemic N.brichardi, nonendemic of the Lake Tanganyika region A.burtoni, nonendemic of the Lake Malawi region A.calliptera, the Lake Malawi endemic M.zebra, and the Lake Victoria endemics P. nyererei and H. sauvagei. In our analyses, we found 12 larger indels (>50 bp) localized in intronic sequence as well as directly upstream and downstream of the agrp2 gene in this set of African cichlid fishes (fig. 1B). Based on maximum parsimony most of the indels within the analyzed 10–15 kb locus (10/12) likely arose within the radiating cichlid lineages (supplementary fig. S6, Supplementary Material online). Based on previous studies that performed genome-wide characterizations of indels and structural variations (Fan and Meyer 2014; Dolfin et al. 2018), density of indels at the agrp2 locus is four to five times higher than genome-wide average.
Several of the indels overlap with putatively functional DNA regions. Two independent deletions (nos. 1a and 1 b; compared with the outgroup O. niloticus) are located within the promoter region of agrp2, one in the Lake Malawi species A. calliptera (296 bp deletion; 418 bp 5′ of the start codon) and one in the Lake Tanganyika species N. brichardi (793 bp deletion; 157 bp 5′ of the start codon) (fig. 1B). Two further indels (nos. 5 and 6) overlap with a region within intron 1 that has been previously characterized as a cis-regulatory element of agrp2 containing several alternatively fixed alleles associated with stripe presence/absence in Lake Victoria cichlids (Kratochwil et al. 2018) (fig. 1B). Element number 5 is a 145 bp short interspersed nuclear element retrotransposon-derived insertion (supplementary tables S1 and S4, Supplementary Material online) specific to haplochromine cichlids (and therefore missing in the genomes of N. brichardi and O. niloticus) (fig. 1B). The 648 bp element number 6 is only present in O. niloticus (and more distantly related species, compared with the Lake Victoria and L. Malawi species, such as the neotropical cichlid Amp.citrinellus; BLAST E value: 7e-66) therefore suggest that it constitutes a deletion specific to the radiating cichlids including Lake Tanganyika and Haplochromine cichlids (fig. 1B).
Two of the deletions (451 and 947 bp) were variable even within the extremely young Lake Victoria radiation (∼10,000 years; Johnson et al. 1996; Elmer et al. 2009) and are of particular interest as they were specific to the striped species H. sauvagei (fig. 1B). To confirm a potential association with the stripe phenotype, we conducted PCRs to assay the variation in length. Indeed, amplicons of H. sauvagei were shorter than those of P. nyererei, also those of Haplochromischilotes but other striped species such as Haplochromisserranus and Haplochromisthereuterion as well as all nonstriped species showed no evidence for a deletion (supplementary fig. S7, Supplementary Material online). The lack of association is in line with the previous hypothesis that variation in stripe patterns of Lake Victoria cichlids is mainly associated with the regulatory element in the first intron of agrp2 (Kratochwil et al. 2018).
Tandem Exon Duplication of agrp2 in Several African Cichlids
The largest insertion (1,811 bp) is directly upstream of exon 3 and could be only found in the two long-read chromosome-level assemblies of M. zebra and A. calliptera (fig. 1B), yet sequences contained gaps in several of the short-read genomes suggesting assembly problems. To visualize repeats that could cause such assembly problems and to gain insights into the origin of the insertion, we generated syntenic dot plots of the available long-read assemblies (O. niloticus, M. zebra, and A. calliptera). The alignments provide evidence for several duplicated regions in the M. zebra and A. calliptera genomes that are unique in the O. niloticus genome (fig. 3A–C). Although most duplications are small (<50 bp), we found one ∼1.6 kb tandem duplication that included exon 3 of agrp2. Using PhyML, we generated a maximum likelihood phylogeny of exon 3 and the two copies (exon 3a and exon 3 b; fig. 3D and E) that were found in M. zebra and A. calliptera. Based on the phylogeny, parsimony suggests only one duplication event that gave rise to exon 3 b (bootstrap support 92%). In addition, the phylogeny provides no evidence of gene conversion or concerted evolution.
To test if the duplication is common across the radiation of Lake Malawi (where M. zebra and A. calliptera are from) but potentially also Lakes Victoria and Tanganyika, we used three complementary approaches: 1) a PCR-based approach, 2) a TaqMan probe assay, and 3) reanalysis of available genome-resequencing data.
In an effort to identify the phylogenetic timing when the exon 3 tandem duplication occurred, we first designed primers that specifically detect the duplication, both by designing primers that amplify the region between the duplicates (fig. 3E and F) or by amplifying the whole fragment containing the duplications (fig. 3E and G). The results of the PCR approach uncover a surprising degree of standing genetic variation within species as well as substantial variation between species of the flocks of Lakes Victoria, Malawi, and Tanganyika (supplementary table S3, Supplementary Material online and fig. 3F and G). In total, we found 16 species without duplication and 25 species with a duplication; and in 2 species from Lake Victoria (P. nyererei and Pundamiliapundamilia), we found individuals with and without duplication suggesting standing genetic variation in some species. More interestingly, duplications could be found across cichlid radiations from Lake Tanganyika, Malawi, and Victoria.
Second, as the PCR assay was not sensitive enough to detect heterozygote individuals, or individuals with more than two duplicates, we used a TaqMan probe assay on a subset of species to confirm the results and to determine the exact number of repeats (fig. 3H). The assay was performed using probes for exon 1 (that had no evidence of duplication in species for which long-read sequence data are available and thereby served as a control) and exon 3. Our analysis confirmed the PCR results and support tandem exon duplications in several species of the Lake Victoria radiation including Haplochromislatifasciatus, P. nyererei (1/3 individuals), endemic to Lake Malawi Labeotropheus caeruleus and Pseudotropheus cyaneorhabdos. There is some support for more than two copies in P. nyererei and Pse. cyaneorhabdos (fig. 3H).
Third, as a further independent approach, we realigned available cichlid genomes (Poletto et al. 2010; McGee et al. 2016; Meier et al. 2017; Malinsky et al. 2018) to the P. nyererei reference genome, a genome without the duplication (Brawand et al. 2014) and analyzed the relative coverage across the agrp2 region (supplementary table S3, Supplementary Material online). Here, we would expect to find a relative coverage of ∼1 for individuals without duplication, a relative coverage of ∼2 for individuals with a homozygous duplication, ∼1.5 for heterozygous individuals and >2 for individuals with more than two copies. Indeed, using this approach, we find support for tandem exon duplications of exon 3 (fig. 3I), with some individuals being likely heterozygote (e.g., Haplochromispaucidens and Haplochromisvittatus, both endemics to Lake Victoria) or having even more than two duplications (e.g., Thoracochromis pharyngalis) (supplementary table S3, Supplementary Material online). Generally, duplications and more than two tandem duplicates seem to be more prevalent in the Lake Malawi cichlid radiation (fig. 3J and K).
Transcription Bias and Resurrection of a Nontranscribed Exon 3 Copy
To assess potential functional effects, we first tested whether both or only one of the copies of exon 3 are expressed. Using the genomes of M. zebra and A. calliptera, we found copy-specific, paralogous sequence variants at position 1,048 and 1,115 within the UTR region of agrp2 (fig. 4A). We synthesized cDNA from RNA extractions from the skin of 33 species. Sanger sequencing shows that 32 of the 33 species, independent of the number of duplicates, showed the exon 3a-specific paralogous sequence variants in the chromatogram trace (fig. 4B). This suggests that in species with a duplicated third exon, only the 5′ exon 3 (exon 3a) is transcribed. Interestingly, one species, Mel.auratus, a species with a single copy had the paralogous sequence variant specific for exon 3 b. This suggests a secondary loss of exon 3a and a resurrection of the usually nonexpressed exon 3 b. To confirm the results with a more sensitive approach, we used RNA-seq data for three species (P. nyererei [n = 4], Pse.demasoni [n = 5], and Mel. auratus [n = 4]) and estimated the ratio of the paralogous sequence variants at the cDNA positions 1,048 and 1,115 that are located within the 3′-UTR (fig. 4C). The analysis confirmed that in both P. nyererei and Pse. demasoni exon 3a is predominantly expressed. In Pse. demasoni, two individuals showed some expression of exon 3 b (22% and 25%). We found no sequence variation (i.e., lack of a 3′-UTR cleavage site) that could explain these interindividual differences. In Mel. auratus, only exon 3 b is expressed (as exon 3a has been lost). If we assume that also in Mel. auratus exon 3a was initially the expressed exon, this suggest a resurrection of the nontranscribed exon 3 b.
Fig. 4.
—Expression bias toward exon 3a and resurrection of exon 3 b (A) Two paralogous sequence variants identify the two exon copies exon 3a and 3 b. The variant of exon 3a corresponds to the ancestral alleles as also found in Oreochromis niloticus. (B) In species with two copies (here, Maylandia zebra), exon 3a is mainly expressed as indicated by the lack of the exon 3 b-specific chromatogram trace. Species that lost (or never had) exon 3 b have the exon 3a variant. One exception is Melanochromis auratus that only has one copy, but in this species the sequence corresponds to exon 3 b, suggesting a secondary loss of exon 3a. (C) RNA-seq data confirm the expression bias toward exon 3a (no evidence of exon 3a in Pundamilia nyererei; some weak expression in Pseudotropheus demasoni) and the loss of the exon 3a paralogous sequence variants in Mel. auratus. Expression ratio was estimated based on two paralog-specific SNPs in the 3′-UTR. Error bars indicate standard deviation.
Signals of Selection
Such a scenario where a nontranscribed, likely neutrally evolving exon copy (exon 3 b), could occasionally become functional again through the loss of exon 3a (as shown for Mel. auratus), but also the observed evolutionary dynamics at this locus could promote the accumulation of genetic variation that positive selection could act upon. Therefore, we analyzed the molecular evolution of agrp2 (always using all three exons of the transcribed sequence across species) to determine if variation is neutral or if it had evolved adaptively (figs. 5A and 6). We found evidence that many codons in the gene (∼33%) are evolving under neutrality (dN/dS ≈1; LRTM1a/M0: X2(1)= 60.27, P < 0.0001), yet, we found no evidence for the presence of positively selected sites (LRTM2a/M1a: X2(2)= 0, P = 1). In addition, we did not find evidence that the agrp2 gene is evolving under divergent selection pressures in the cichlid lineage with the exon 3 duplication compared with other lineages in our alignment (LRTCmC-Great Lakes/M2a_rel: X2(1)= 0.46, P = 0.50). Yet, this analysis does not exclude that in some instances amino acid changes contributed to adaptive or nonadaptive pigmentation differences (figs. 5B–J and 6).
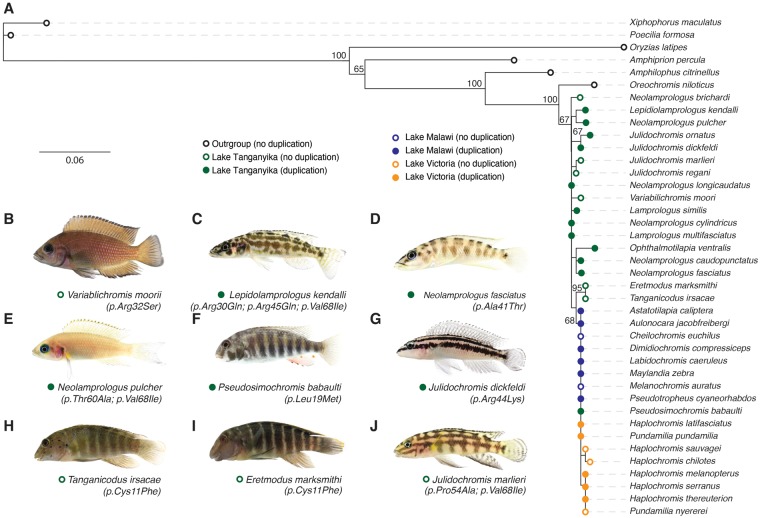
Fig. 5.
—Phylogeny of the agrp2 gene across cichlids. (A) Maximum likelihood phylogeny using agrp2 cDNA. Points connote the presence/absence of the duplication and origin. Numbers indicate bootstrap values >60. (B–J) Photographs of cichlid fishes studied in this research with information about the duplication and nonsynonymous mutations (see fig. 6).
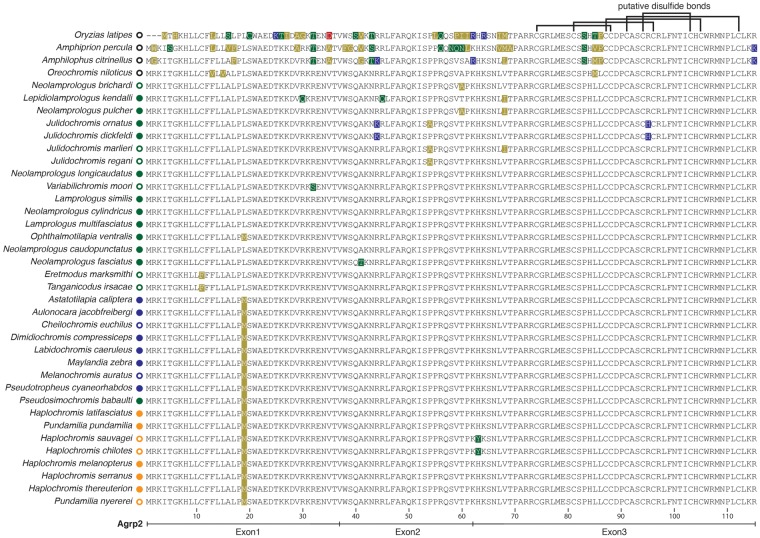
Fig. 6.
—Protein sequence evolution of agrp2 across cichlid fishes. Protein alignments show several amino acid substitutions across cichlids (at 12 positions in Lake Tanganyika cichlids, at 2 positions in Lake Malawi and Victoria cichlids). Putative disulfide bonds that are a characteristic of agouti proteins are indicated at the top. The involved cysteines are highly conserved.
Given the evolutionary history and characteristics of the third exon of agrp2 in African cichlids (e.g., exon duplication, resurrection of nontranscribed exons, different functional importance of exon 3 compared with exons 1 and 2), it could be expected that this exon has evolved under divergent selection pressures relative to the other exons in the gene, potentially resulting in phylogenetic discordance among exons (Beltrán et al. 2002). We found some support for this prediction. The single breakpoint recombination analysis performed in HYPHY, found evidence for a breakpoint at base-pair position 185 (at the border between exons 2 and 3; ΔAIC = 19.78, Model support =1; although the signal was lost when considering cAIC). Thus, we conducted exon-specific analyses of nucleotide substitutions for the agrp2 gene. As for the whole gene, we found evidence for some codons neutrally evolving, both in exons 1 + 2 (LRTM1a/M0: X2(1)= 11.48, P = 0.0007) and in exon 3 (LRTM1a/M0: X2(1)= 40.73, P < 0.0001), but no evidence for positive selection (both P > 0.5). Interestingly, however, we found the proportion of sites evolving neutrally to be larger for exons 1 + 2 (40%) than for exon 3 (9%), suggesting that the functional importance of this exon might result in purifying selection across most codons. We found no evidence that exons 1 + 2 (LRTCmC-Great Lakes/M2arel: X2(1)= 0.55, P = 0.46) or exon 3 (LRTCmC-Great Lakes/M2arel: X2(1)= 1.49, P = 0.22) are evolving divergently in radiating African cichlids compared with lineages where the duplication of the third exon was not found (i.e., there is no evidence of codons evolving under different selection pressures in the compared lineages).
Discussion
It was Susumu Ohno who postulated 50 years ago how gene duplication might serve as a source of evolutionary novelties (Ohno 1970). As one copy is freed from stabilizing selection, the other can accumulate mutations “more freely” and evolve a novel function (neofunctionalization) as one possible consequence. Although there are many examples for the dynamics and evolutionary significance of gene duplications (Perry et al. 2007; Vonk et al. 2013; Denoeud et al. 2014; Ishikawa et al. 2019; Musilova et al. 2019), exon duplications have received considerably less attention (Kondrashov and Koonin 2001; Letunic et al. 2002; Keren et al. 2010; Lambert et al. 2015; Rogers et al. 2017). Especially tandem duplications of the terminal exon have been barely investigated in an evolutionary context. A lone exception is a recent study that described a duplication of the terminal exon of the alternative oxidase in oysters that has been involved in adaptation to environmental stress (Liu and Guo 2017).
Here, we found a tandem duplication of the third exon of agrp2 that likely occurred >8–12 Ma early in the evolution of the African cichlid fish radiations before the oldest of these, that from Lake Tanganyika, diversified (fig. 7). The duplication is not fixed, as its presence varies across the radiations of the Lakes Tanganyika, Malawi, and Victoria and even is polymorphic within some species. Furthermore, we found evidence for copy number variation (up to four or five copies) (fig. 7). In previous short-read assembly genomes (Brawand et al. 2014), this duplication was not resolved, illustrating that such instances might have been systematically overlooked as well as their evolutionary and phenotypic importance went unrecognized. In contrast, long-read assemblies (e.g., using PacBio or Nanopore technologies) have now the power to uncover such duplications (fig. 2B and C). Such assemblies therefore permit studying their evolutionary relevance. Despite not finding any evidence for diversifying selection, evolution of novel isoforms or a link to particular color patterns (fig. 5B–J), we discover a surprising dynamic of gains and losses of exon 3 that also includes (at least in one instance) the resurrection of the nonfunctional exon 3 b (fig. 7). As we only looked at a small selection of the over 1,200 species of East African cichlids, the dynamics of exon duplication and loss might indeed contribute to the diversity of cichlid color patterns in some instances. Clearly, an increasing availability of RNA-seq data and long-read genome data will permit much more comprehensive analyses in the future.
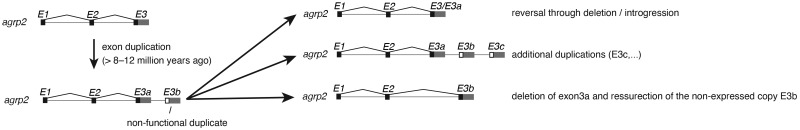
Fig. 7.
—Summary of the evolutionary history of the agrp2 gene. Exon 3 was duplicated before the radiations of Lake Tanganyika cichlids >8–12 Ma giving rise to exon 3a and 3 b. Exon 3 b was partially either lost again or the nonduplicated version was partially maintained through incomplete lineage sorting. Additional duplications occurred generating more copies of exon 3. At least in one species (Melanochromis auratus), exon 3a was subsequently lost.
Regulatory variation of agrp2 has been previously shown to be linked to absence and presence of stripe patterns across African cichlids (Kratochwil et al. 2018). High expression of agrp2 represses stripe patterns, while low levels lead to their formation. Moreover, also CRISPR-Cas9 knockouts of agrp2 (which as such constitutes an experimentally introduced coding variation) cause the reappearance of stripe patterns in nonstriped cichlids. This suggests that coding variation might be equally potent in driving diversification in stripe patterns. Loss- but also gain-of-function mutations in both the coding sequences and 3′-UTR of agrp2 could accumulate in the untranslated copy exon 3 b, and be resurrected through subsequent deletion of exon 3a. This, in turn, might lead to phenotypic diversification. Interestingly, the C-terminus encoded by exon 3 shows high evolutionary conservation across all agouti family members (agrp2/asip2b, agrp1, asip1, asip2/asip2a) as it is the part that binds to and antagonizes melanocortin receptor function (Kaelin et al. 2008). Mutations in this exon are therefore likely to have direct effects on the melanocortin pathway and thereby color pattern formation.
Our RNA-seq analysis suggests that only the first copy, exon 3a is expressed. This is in contrast to previous work that has shown alternative splicing of terminal exon copies, that can be even regulated through environmental factors (Liu and Guo 2017). As two out of the five Pse. demasoni individuals that we analyzed for relative expression of exon 3a/3b showed a low level of expression of exon 3b, it is possible that cryptic genetic variation or environmental factors might trigger a stronger expression of exon 3 b.
As copy number variations occur frequently through unequal crossing over, potentially leading to loss or gain of tandem copies (Hastings et al. 2009) such a tandem duplication could facilitate neofunctionalization or even loss-of-function of agrp2, especially if populations experience strong bottlenecks (e.g., during colonization events). Such a scenario would have implications similar to recent work from sticklebacks (Kratochwil and Meyer 2019; Xie et al. 2019). This study describes how repeated loss-of-function deletions of regulatory elements of pitx1 cause the independent loss of pelvic fin spines in sticklebacks. These recurrent deletions are driven by repeats that result in a more fragile, double-strand breaking DNA-formation (Z-DNA) that in turn results in an elevated mutation rate at this locus. This repeatedly led to loss-of-function mutations of the regulatory element necessary for pelvic fin spine development.
In summary, we conclude that the tandem duplication of agrp2 exon 3, or more generally tandem duplications at genomic hotspots are important loci to analyze in light of their evolutionary importance as they might facilitate diversification and adaptation in a similar manner as gene duplications. This analysis crucially depends on state-of-the-art technologies including PacBio and Nanopore sequencing, the so called third revolution in sequencing technology (van Dijk et al. 2018), sensitive enough to detect such variations. In the future, beyond the characterization of structural variation, insertions, and duplication, functional experiments have to more specifically address the role and phenotypic impact of such mutational events. Many of those will be difficult to assess through comparative approaches, but will need genetic manipulations, either through hybridization experiments or functional approaches including CRISPR-Cas9 genome editing that have become amenable in nonmodel organisms as cichlid fishes (Juntti et al. 2016; Kratochwil et al. 2018) and that steadily become more precise (Anzalone et al. 2019).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Baden-Württemberg Foundation (to C.F.K.), the Deutsche Forschungsgemeinschaft (DFG, KR 4670/2-1, KR 4670/4-1, TO914/2-1, ME 1725/20-1, ME 1725/21-1 to C.F.K., J.T.D., and A.M.) a stipend from the China Scholarship Council (CSC, to Y.L.), a bridge funding from the International Max Planck Research School (IMPRS) for Organismal Biology (to S.U.), and an ERC advanced grant (no. 293700 GenAdapt; to A.M.).
Literature Cited
- Anzalone AV, et al. 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán M, et al. 2002. Phylogenetic discordance at the species boundary: comparative gene genealogies among rapidly radiating Heliconius butterflies. Mol Biol Evol. 19(12):2176–2190. [DOI] [PubMed] [Google Scholar]
- Blackmon H, Adams RH. 2015. R Package ‘evobiR’ v.1.1. Cran R.
- Brawand D, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513(7518):375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, et al. 2003. Glocal alignment: finding rearrangements during alignment. Bioinformatics 19(Suppl 1):i54–i62. [DOI] [PubMed] [Google Scholar]
- Conte MA, et al. 2019. Chromosome-scale assemblies reveal the structural evolution of African cichlid genomes. Gigascience 8(4):giz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MA, Gammerdinger WJ, Bartie KL, Penman DJ, Kocher TD. 2017. A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 18(1):341.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, et al. 2019. Ensembl 2019. Nucleic Acids Res. 47(D1):D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, et al. 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345:1181–1184. [DOI] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfin LP, Man A, Haerty W, Di-Palma F. 2018. Analysis of structural variants in four African cichlids highlights an association with developmental and immune related genes. BioRxiv 473710; doi: https://doi.org/10.1101/473710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, et al. 2009. Pleistocene desiccation in East Africa bottlenecked but did not extirpate the adaptive radiation of Lake Victoria haplochromine cichlid fishes. Proc Natl Acad Sci U S A. 106(32):13404–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Meyer A. 2014. Evolution of genomic structural variation and genomic architecture in the adaptive radiations of African cichlid fishes. Front Genet. 5:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. 2009. Mechanisms of change in gene copy number. Nat Rev Genet. 10(8):551.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, et al. 2019. A key metabolic gene for recurrent freshwater colonization and radiation in fishes. Science 364(6443):886–889. [DOI] [PubMed] [Google Scholar]
- Johnson TC, et al. 1996. Late Pleistocene desiccation of Lake Victoria and rapid evolution of cichlid fishes. Science 273(5278):1091–1093. [DOI] [PubMed] [Google Scholar]
- Juntti SA, et al. 2016. A neural basis for control of cichlid female reproductive behavior by prostaglandin F2α. Curr Biol. 26(7):943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin C, et al. 2008. New ligands for melanocortin receptors. Int J Obes. 32(S7):S19.. [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinformatics 20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 11(5):345.. [DOI] [PubMed] [Google Scholar]
- Kocher TD. 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 5(4):288.. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. 2001. Origin of alternative splicing by tandem exon duplication. Hum Mol Genet. 10(23):2661–2669. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 23(10):1891–1901. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF. 2019. Molecular mechanisms of convergent color pattern evolution. Zoology 134:66–68. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, et al. 2018. Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362(6413):457–460. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, Meyer A. 2015. Closing the genotype-phenotype gap: emerging technologies for evolutionary genetics in ecological model vertebrate systems. Bioessays 37(2):213–226. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, Meyer A. 2019. Fragile DNA contributes to repeated evolution. Genome Biol. 20(1):39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Lambert MJ, Cochran WO, Wilde BM, Olsen KG, Cooper CD. 2015. Evidence for widespread subfunctionalization of splice forms in vertebrate genomes. Genome Res. 25(5):624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Bork P. 2002. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet. 11(13):1561–1567. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Guo X. 2017. A novel and stress adaptive alternative oxidase derived from alternative splicing of duplicated exon in oyster Crassostrea virginica. Sci Rep. 7(1):10785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, et al. 2016. Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet. 17:704–714. [DOI] [PubMed]
- Malinsky M, et al. 2018. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat Ecol Evol. 2:1940.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallarino R, Linden TA, Linnen CR, Hoekstra HE. 2017. The role of isoforms in the evolution of cryptic coloration in Peromyscus mice. Mol Ecol. 26(1):245–258. [DOI] [PubMed] [Google Scholar]
- McGee MD, Neches RY, Seehausen O. 2016. Evaluating genomic divergence and parallelism in replicate ecomorphs from young and old cichlid adaptive radiations. Mol Ecol. 25(1):260–268. [DOI] [PubMed] [Google Scholar]
- Meier JI, et al. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat Commun. 8(1):14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. 1993. Phylogenetic relationships and evolutionary processes in East African cichlid fishes. Trends Ecol Evol. 8(8):279–284. [DOI] [PubMed] [Google Scholar]
- Muschick M, Indermaur A, Salzburger W. 2012. Convergent evolution within an adaptive radiation of cichlid fishes. Curr Biol. 22(24):2362–2368. [DOI] [PubMed] [Google Scholar]
- Musilova Z, et al. 2019. Vision using multiple distinct rod opsins in deep-sea fishes. Science 364(6440):588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin: Springer-Verlag. [Google Scholar]
- Perry GH, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 39(10):1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poletto AB, Ferreira IA, Martins C. 2010. The B chromosomes of the African cichlid fish Haplochromis obliquidens harbour 18S rRNA gene copies. BMC Genet. 11(1):1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21(10):2531–2533. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. In: Nielsen R, ed. Statistical methods in molecular evolution: New York, NY: Springer; p. 125–181. [Google Scholar]
- R Development Core Team. 2019. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Rogers RL, Shao L, Thornton KR. 2017. Tandem duplications lead to novel expression patterns through exon shuffling in Drosophila yakuba. PLoS Genet. 13(5):e1006795.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W. 2018. Understanding explosive diversification through cichlid fish genomics. Nat Rev Genet. 19(11):705–717. [DOI] [PubMed] [Google Scholar]
- Scotto–Lavino E, Du G, Frohman MA. 2006a. 3′ end cDNA amplification using classic RACE. Nat Protoc. 1:2742–2745. [DOI] [PubMed] [Google Scholar]
- Scotto–Lavino E, Du G, Frohman MA. 2006b. 5′ end cDNA amplification using classic RACE. Nat Protoc. 1:2555.. [DOI] [PubMed] [Google Scholar]
- Shainer I, et al. 2017. Novel hypophysiotropic AgRP2 neurons and pineal cells revealed by BAC transgenesis in zebrafish. Sci Rep. 7(1):44777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirak A, et al. 2010. Identification of repetitive elements in the genome of Oreochromis niloticus: tilapia repeat masker. Mar Biotechnol. 12(2):121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2015. RepeatMasker Open-4.0. Available from: http://www.repeatmasker.org; last accessed December 6, 2019.
- Stiassny MLJ, Meyer A. 1999. Cichlids of the Rift lakes. Sci Am. 280(2):64–69. [Google Scholar]
- Stoltzfus A, McCandlish DM. 2017. Mutational biases influence parallel adaptation. Mol Biol Evol. 34(9):2163–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente GT, et al. 2014. Origin and evolution of B chromosomes in the cichlid fish Astatotilapia latifasciata based on integrated genomic analyses. Mol Biol Evol. 31(8):2061–2072. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. 2009. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 10(10):725.. [DOI] [PubMed] [Google Scholar]
- van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. 2018. The third revolution in sequencing technology. Trends Genet. 34(9):666.. [DOI] [PubMed] [Google Scholar]
- Vonk FJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 110:20651–20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weadick CJ, Chang BS. 2012. An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol Biol Evol. 29(5):1297–1300. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2019. R Package ‘stringR’ v.1.4.0. Cran R.
- Wickham H, et al. 2019. R Package ‘ggplot2’ v.3.1.1. Cran R.
- Xie KT, et al. 2019. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 363(6422):81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2014. Molecular evolution: a statistical approach. Oxford, UK: Oxford University Press. [Google Scholar]
- Zhang C, et al. 2010. Pineal-specific agouti protein regulates teleost background adaptation. Proc Natl Acad Sci U S A. 107(47):20164–20171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.