Abstract
Breathing rate and depth influence the concentration of carbon dioxide in the blood, altering cerebral blood flow and thus functional magnetic resonance imaging (fMRI) signals. Such respiratory fluctuations can have substantial influence in studies of fMRI signal covariance in subjects at rest, the so-called “resting state functional connectivity” technique. If respiration is monitored during fMRI scanning, it is typically done using a belt about the subject’s abdomen to record abdominal circumference. Several measures have been derived from these belt records, including the windowed envelope of the waveform (ENV), the windowed variance in the waveform (respiration variation, RV), and a measure of the amplitude of each breath divided by the cycle time of the breath (respiration volume per time, RVT). Any attempt to gauge respiratory contributions to fMRI signals requires a respiratory measure, but little is known about how these measures compare to each other, or how they perform beyond the small studies in which they were initially proposed. Here, we examine the properties of these measures in hundreds of healthy young adults scanned for an hour each at rest, a subset of the Human Connectome Project chosen for having high-quality physiological records. We find: 1) ENV, RV, and RVT are all correlated, and ENV and RV are more highly correlated to each other than to RVT; 2) respiratory events like deep breaths exhibit characteristic heart rate elevations, fMRI signal changes, head motions, and image quality abnormalities time-locked to large deflections in the belt traces; 3) all measures can “miss” deep breaths; 4) RVT “misses” deep breaths more than ENV or RV; 5) all respiratory measures change systematically over the course of a 14.4-minute scan. We discuss the implication of these findings for the literature and ways to move forward in modeling respiratory influences on fMRI scans.
Introduction:
Modulation of functional magnetic resonance imaging (fMRI) signals by respiration has come under increased scrutiny in recent years, largely due to the emergence of “resting state functional connectivity” approaches to signal analysis, which tend to examine signals in terms of covariance rather than in terms of pre-defined events modeled in time. Changes in breathing depth and rate modulate the amount of carbon dioxide (CO2) exhaled, and thereby alter the concentration of CO2 in the bloodstream (pCO2). Cerebral blood flow is controlled by homeostatic loops governed mainly by pCO2, such that increased arterial pCO2 causes increased cerebral blood flow (Hall, 2016). A typical baseline pCO2 is 35–45 mm Hg. Increases of ~5 mm Hg caused by decreased breathing rate or depth, or inhalation of CO2-enriched gas mixtures, cause cerebral blood flow increases on the order of 50%, which in turn cause large multiple-percent blood oxygen level dependent (BOLD) signals changes throughout the brain (Bright et al., 2009; Ito et al., 2003; Kastrup et al., 1999; Poulin et al., 1996). In this manner, breathing patterns can alter fMRI signals (and signal covariance).
Any effort to identify respiratory influences on fMRI signals – whether in task or task-free settings – must begin with a measure of respiration. In modern studies of respiratory physiology, it is common practice to take multiple kinds of measures at once, providing both unique and redundant information, and enabling high-confidence identification and characterization of respiratory phenomena. Typical noninvasive measures of respiration are optical or inductance respiratory plethysmography (Carry et al., 1997; Nierat et al., 2017), which involve simultaneous optical or mechanical measurement of thoracic and abdominal excursions (since either compartment can move air nearly independently). Such measures are often paired with nasal cannula or face-mask measures of air flow and end-tidal pCO2 (i.e., capnography) (Heinzer et al., 2015; Yumino and Bradley, 2008). These approaches can produce precise, quantitative measurement of air flow and blood gas changes, in addition to characterizing other respiratory parameters.
By contrast, in typical fMRI studies, if respiratory monitoring occurs at all, it is accomplished via a “respiratory bellows”, which is a single mechanical belt strapped about the subject’s abdomen that measures tension on the belt (see blue traces in Figure 1A). This belt has no back-up source of redundant information (e.g., a chest belt), and will index both respiratory motion and non-respiratory motions if they involve shifts of the abdomen and/or tightening of abdominal muscles. The abdominal belt does not permit quantitative measurement of air flow in the manner of calibrated plethysmography, but several semi-quantitative measurements have been proposed to index respiratory phenomena from the belt traces. These include respiration variation (RV), which is the standard deviation of the belt trace within a (6-second) window (Chang et al., 2009), the envelope of the respiratory trace (over a 10-second window, ENV) (Power et al., 2018), and the change in belt magnitude over a breath cycle (respiratory volume per time, RVT) (Birn et al., 2006).
Figure 1:
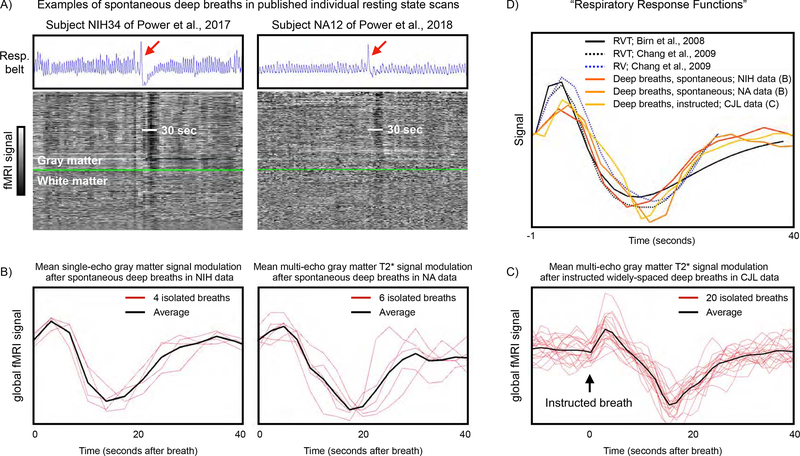
Examples of respiratory traces, effects of deep breaths, and respiratory response functions. A) For individual scans published in {Power, 2017} and {Power, 2018}, blue traces show respiratory belt records, and the gray-scale heat maps show all in-brain fMRI signals ordered by anatomic compartment, with gray matter above the bright green line and white matter below the bright green line. A single deep breath is evident in each scan (red arrow), and accompanying fMRI signal modulations last about 30 seconds in both scans. B) Mean gray matter signals in the 40 seconds after spontaneous, isolated deep breaths in subjects at rest in the above studies. NIH signals are single-echo mean gray-matter fMRI signals extracted from 4 subjects at times of 4 isolated deep breaths (modified from Fig. 6 of {Power, 2017}). NA signals are multi-echo mean gray-matter T2* estimates extracted from 4 subjects at times of 6 isolated deep breaths (modified from Fig. S4 of {Power, 2018}). C) Mean gray matter signals 10 seconds before until 40 seconds after 20 instructed widely-space deep breaths in a single subject. Signals are multi-echo mean gray-matter T2* estimates as in the NA data. D) Reproduction of published “respiratory response functions” alongside the signals shown in (B) and (C).
These semi-quantitative respiratory measures (RV, ENV, and RVT) have been used to model physiological contributions to fMRI signals, and their explanatory power has been greatest when such measures are convolved with “respiratory response functions” of the kind shown in Figure 1B–D (Birn et al., 2006; Birn et al., 2008; Chang et al., 2009; Chang and Glover, 2009; Power et al., 2017b). These respiratory response functions characterize the relatively slow fMRI signal changes that occur in the 30–40 seconds after a brief increase in ventilation. The functions are similar no matter whether they are derived from instructed deep breaths (Birn et al., 2008), from a deconvolution of multiple cardiac and pulmonary signals in task-free fMRI signals (Chang et al., 2009; Chang and Glover, 2009), or when estimated from widely spaced instructed deep breaths or isolated spontaneous deep breaths in subjects at rest (Power et al., 2019a; Power et al., 2018; Power et al., 2017b). In short, measures like ENV, RV, and RVT are intended to index respiration and respiratory “events”, and the respiratory response functions are intended to describe the consequent fMRI signal changes.
In this paper we explore the major features of respiration that these measures capture. We have used all of these measures in multiple datasets over many years, and have noticed particular features of certain measures that constrain their uses in certain ways. In particular, RVT, due to its definition, often “misses” deep breaths that are evident visually in respiratory traces. Deep breaths are prevalent in resting state fMRI scans and cause transient elevations in heart rate and also prolonged, prominent changes in fMRI signals, making them an important kind of respiratory event to capture. We first noticed this behavior years ago in an AFNI implementation of RVT (RetroTS.m), and in this paper we employ a “stripped down” version of the core RVT calculation (amplitude/time) to illustrate the issue clearly (the full implementation of RVT with resampling is also shown, with the same effect, in our own code and in AFNI and FSL implementations). More broadly, all of the measures (ENV, RV, and RVT) can “miss” an event depending on the characteristics of the respiratory belt trace. We illustrate these phenomena in Human Connectome Project (HCP) data, which are attractive for the sheer quantity of data (hundreds of subjects with an hour each of resting state fMRI with accompanying respiratory belt measurements), and also show examples from previous datasets in which we first noticed these properties. One unique advantage of the HCP dataset is that it contains four unusually long resting state scans, permitting us to assay for repeated, systematic changes in respiratory measures over scanning periods. We observe such changes, and link our systematic findings to those of other groups in the Discussion.
For readers unfamiliar with respiratory terminology, the following terms are used: tidal breathing is normal, cyclic breathing, tidal volume is the volume of air inspired in a normal breath, eupnea describes the state of normal, unlabored tidal breathing, hyperpnea and hypopnea describes increased and decreased ventilation, respectively, and apnea is an absence of ventilation. Readers desiring further information on respiratory physiology and terminology are directed to texts such as (Hall, 2016).
Methods
Data
The Human Connectome Project (HCP) data are publicly available and have been comprehensively described (Glasser et al., 2013; Van Essen et al., 2013). We give a brief overview here. The HCP dataset encompasses healthy young adults scanned in their third and fourth decades of life in the American Midwest. Subjects were all scanned on the same modified Siemens Skyra 3T scanner at Washington University in Saint Louis, over a 2-day period during which they underwent comprehensive neuropsychological testing as well as multiple scanning sessions including task fMRI, resting state fMRI, and anatomical and structural imaging protocols. During scanning, physiology data was acquired via the Siemens Physiology Monitoring Unit (PMU), which is standard equipment that accompanies the scanner for purposes of cardiac and physiological gating. The signals acquired were 400 Hz recordings of an abdominal belt and a finger pulse oximetry waveform. The Siemens respiratory record is obtained via a pressure hose connected to a respiratory cushion placed under an elastic belt strapped around the subject’s abdomen, and output is in arbitrary units. In this paper we focus on the resting state fMRI scans and the accompanying physiology data.
For the fMRI scans, subjects were scanned 4 times for 14.4 minutes (two scans per day on two days) at rest with a 32-channel head coil. Sequences were counterbalanced LR/RL phase encode sequences, with TR = 720 ms, TE = 33.1 ms, flip angle 52 degrees, multi-band factor 8, and 2 mm isotropic voxels with full brain coverage (FOV 208 × 180 mm). Realignment was calculated using the FSL MCFLIRT command. Data were registered to the MNI atlas space for volumetric analyses and to the Conte69 surface for surface-based analyses. Anatomical images were 3D MPRAGE sequences with 0.7 mm isotropic resolution (FOV 224 mm, matrix = 320, 256 sagittal slices in a single slab, TR = 2400 ms, TE = 2.14 ms, TI = 1000 ms, flip angle 8 degrees). A core mission of the HCP is to provide the public with registered, realigned data, as comprehensively described in (Glasser et al., 2013; Van Essen et al., 2013). These “ready to use” data are the data we accessed.
The data studied are exactly the data studied in (Power et al., 2019b). The “900-subject” release of the Human Connectome Project data was obtained, with a focus on the following files in each subject: four resting state fMRI scans transformed to atlas space (in each subject’s /MNINonLinear/Results folder): {RUN}=REST1_LR, REST1_RL, REST2_LR, REST2_RL (this order is runs 1–4 in the text). rfMRI_{RUN}.nii.gz and rfMRI_{RUN}_hp2000_clean.nii.gz scans were obtained, representing minimally preprocessed data and data after FIX independent components analysis (FIX-ICA) denoising. For each of these four scans, the {RUN}_Physio_log.txt and Movement_Regressors_dt.txt files were also obtained. Structural scans transformed to atlas space were also obtained (in each subject’s /MNINonLinear/ folder): the T1w.nii.gz and the aparc+aseg.nii.gz files, representing the anatomical T1-weighted scan and its FreeSurfer segmentation.
Image Processing
The aparc+aseg.nii.gz file for each subject underwent a set of serial erosions within white matter and ventricle segments, exactly as in (Power et al., 2017b). The segmentation image had 0.7 mm isotropic resolution, and 1-voxel erosions were serially performed (four times for the white matter, two times for ventricles). Masks of cortical gray matter, the cerebellum, and subcortical nuclei were extracted, as were serially eroded layers of superficial, deep, and deepest (with respect to distance from gray matter) masks of the white matter (0–1, 2–3, and 4 erosions) and ventricles (0–1, and 2 erosions). These masks, together, include all in-brain voxels of these tissue types, and are used to order signals for “gray plots” (Power, 2017).
For the purpose of making useful gray plots, because of the considerable thermal noise in HCP scans, a within-mask 6 mm FWHM Gaussian kernel was applied to the data using the above masks (illustrated for HCP data in (Power, 2017)). This blurring does not mix tissue compartments due to the use of masks (beyond partial volume effects inherent to the voxels).
Parameter processing
Respiratory and cardiac measures:
Respiratory belt and pulse oximeter traces from the Siemens PMU (sampled at 400 Hz) first underwent visual inspection in their entirety to determine if the quality was sufficient for reliable peak detection, since traces are often partially or fully corrupted. Only subjects with traces deemed likely to successfully undergo peak detection in all runs were analyzed. Readers may view all physiology traces and decisions on quality in the supplemental movies and files of (Power, 2019)1. A list of the subjects with “good” data is in those materials. All further signal processing was performed using Matlab 2019a.
After selection, for respiratory traces, an outlier replacement filter was used to eliminate spurious spike artifacts (Matlab command: filloutliers(resp_trace, ‘linear’, ‘movmedian’, 100)) and the traces were then gently blurred to aid peak detection (Matlab command: smoothdata(resp_trace, ‘sgolay’, 400)) (a 1-second window for a 400 Hz signal). These traces were then z-scored since the records were in arbitrary units. These treated respiratory traces are the ones shown in Figures.
Following prior literature, several respiratory measures were derived from the treated respiratory belt trace. First, the envelope of the trace over a 10-second window (at 400 Hz) was calculated after (Power et al., 2018) (Matlab command: envelope(zscore_resp_trace, 4000, ‘rms’)). Second, the RV measure, defined as the standard deviation of the treated respiratory trace within a 6-second window, was calculated following (Chang and Glover, 2009) (Matlab command: movstd(zscore_resp_trace, 2400, ‘endpoints’, ‘shrink’)). These measures were termed ENV and RV in figures.
Calculation of ENV and RV is straightforward, but RVT can be complicated to calculate due to peakfinding and sampling steps. The core computation is one of amplitude/time, namely peak amplitude divided by time between peaks. This core computation can be ambiguous due to challenges in identifying “the right” peaks, as will be discussed at length in the article. Additionally, because this core computation is a ratio of amplitude and time, a deep breath occurring over a long time may yield a ratio much like a typical breath over a typical time, despite having different consequences physiologically and in fMRI timeseries. To make these properties plain, we calculated RVTcore simply as ((peak-prior trough)/(time between peaks)). Peak detection on the trace yielded peaks (and troughs, using the inverted trace) (Matlab command: (findpeaks(zscore_resp_trace, ‘minpeakdistance’, 800, ‘minpeakprominence’, .5)). The minimum peak distance presumes breaths occur more than 2 seconds apart. If a peak did not have a preceding trough after the previous peak, no value was scored at that peak (i.e., peaks and troughs were required to alternate). All traces and derived measures were visually checked to ensure that outliers and abnormalities would not drive results. This measure is termed RVTcore in Figures.
The original implementation of RVT described by (Birn et al., 2006; Birn et al., 2008) does not require alternation of peaks and troughs and has several additional steps of interpolation and sampling: after detection of peaks, linear interpolation creates a continuous peak trace; after detection of troughs, linear interpolation creates a continuous trough trace; cycle times (defined by peak-to-peak intervals) are placed at the mid-peak time, and linear interpolation between these mid-point cycle times creates a continuous cycle time trace; then at the time of each TR, the (peak-trough)/(cycle time) values are computed from the continuous traces. This procedure can smooth respiratory events to varying extents depending on the TR, obscuring the properties of the core calculation. This algorithm was also implemented in custom Matlab code, and is termed RVTBirn. The AFNI implementation of this algorithm, in RetroTS.m, is termed RVTAFNI (Birn), and the FSL implementation, via the “popp” command, is termed RVTFSL (Birn). A Matlab wrapper that feeds HCP files into each of these algorithms and returns RVTcore, RVTBirn, RVTAFNI (Birn), and RVTFSL (Birn) is available at this paper’s website2.
To most clearly illustrate why RVT “misses” deep breaths, the paper focuses on RVTcore, but the effects we note are present in RVTBirn, RVTAFNI (Birn), and RVTFSL (Birn). This commonality occurs because the misses originate via the core computation, not via sampling calculations. In the main text, simply for convenience and unless stated otherwise, reference to RVT values refers to RVTcore. Figures always label the version of RVT being displayed.
Pulse oximeter traces underwent z-scoring then peak detection (Matlab command: findpeaks(zscore_pulseox, ‘minpeakdistance’, 180, ‘minpeakprominence’, .5)). Heart rate was calculated from the interval between peaks. The minimum peak distance presumes heart rates are under 133 beats per minute. Cardiac traces are prone to transient disruptions when fingers move, and it is laborious to check and correct cardiac measures due to the large numbers of peaks. A limited number of cardiac records are therefore used in this report, but those select traces and their derived measures were visually checked and if necessary corrected to ensure accuracy.
In one supplemental figure, we illustrate convolution of “respiratory response functions” with different respiratory measures and regression of the convolved signal in fMRI timeseries. The respiratory response functions are shown in Figure 1D and can be downloaded at this paper’s website. To convolve such functions with a respiratory measure timeseries, the response function must be resampled at the respiratory measure frequency and then convolved. Example commands for such steps are (Matlab command: [resample_RRF_TR] = resample(ChangRV_value, ChangRV_time, (1/0.72)), for a 0.72 second TR; and [RV_conv_RRF] = conv(RV_TRsampling, resample_RRF_TR, ‘full’)).
Data quality measures:
The data quality measure DVARS was calculated after (Power et al., 2012; Smyser et al., 2010) as the root mean squared value in the brain at each timepoint of all voxel timeseries differentiated in time by backwards differences. DVARS by convention is 0 at the first timepoint. DVARS was computed in both the minimally preprocessed and FIX-ICA-denoised images.
Head position and head motion measures:
Head position was taken from the Movement_Regressors_dt.txt files. In Figures these position parameters are displayed after subtracting the first timepoint value from the timeseries (so that all traces start at zero). Head motion was represented by Framewise Displacement (FD) measures, following (Power et al., 2012), wherein all position measures were differentiated in time by backwards differences, rotational measures were converted to arc displacement at 5 cm radius, and the sum of the absolute value of these measures was calculated.
FD is typically calculated by backwards differences to the preceding timepoint (here 720 ms prior), but historically FD measures using sampling rates of 2–4 seconds were common. For comparison to such measures, FD was also calculated by backwards differences over 4 timepoints (4 * 720 ms = 2.88 seconds effective sampling rate) where indicated, using position estimates that had been filtered to suppress dominant respiratory frequencies (0.2–0.5 Hz stop band). This reformulation of FD is presented and discussed extensively in (Power et al., 2019b). In brief, in fast-TR data with sampling rates near or under 1000 ms, the head is in constant motion due to respiratory cycles. Additionally, large movements are subdivided by rapid sampling, and can “disappear” into the constant cyclic respiratory motion. To counter these two effects, respiratory frequencies can be suppressed in position estimates by frequency filtering, and position can be compared over multiple seconds (here, 4 timepoints), which permits large motions to “re-emerge” from the cyclic respiratory motion. This version of motion estimation, FDfiltered,4-TR, visually matches changes in position estimates and changes in fMRI signals as indexed by DVARS, making it useful for understanding events in gray plots. In gray plots, we show FDfiltered,1-TR in a faint light red, and FDfiltered,4-TR in bold bright red, corresponding to the versions of FD examined in (Power et al., 2019b).
Frequency content:
For identifying peak respiratory rates in early and late portions of scans, power spectral density estimates were generated via Welch’s method (e.g., for a respiratory trace, Matlab command: [pw pf] = pwelch(signal,[],[],[],400, ‘power’)). All spectra were visually checked to ensure correct peak frequency identification.
Accessory data
At times, we illustrate respiratory events in additional datasets to extend and contextualize the findings in the HCP data. The NIH and NA datasets are resting state fMRI data, and the CJL data are resting state fMRI data that include intermittent instructed breaths.
NIH dataset: N = 91 subjects scanned at rest for 8.2 minutes each at the NIH intramural campus in Bethesda, MD on a GE Signa 3T scanner using GE physiology recording equipment. These data were comprehensively described in (Gotts et al., 2012), and physiology data were presented in (Power et al., 2017b). These data were the first data in which we noticed differential performance of RVT and RV, using RVTAFNI (Birn). Instances of “missed” deep breaths, with the respiratory traces and measures from the 2017 paper (RVTR and RVTAFNI (Birn)) as well as measures of the present paper (ENV, RV, RVT, and RVTBirn), are shown in the supplemental materials as gray plots. RVTR denotes RV sampled at each TR. The examples of gray plots and global timeseries shown in Figure 1 are modified from (Power et al., 2017b), as are the gray plots shown in supplemental materials.
NA dataset: N = 12 subjects scanned at rest twice for 6.25 minutes at the NIH intramural campus in Bethesda, MD on a Siemens Skyra 3T scanner using Biopac physiology recording equipment. These data were presented in (Power et al., 2018), and the gray plots and global timeseries shown in Figure 1 are modified from that paper’s figures. Sequences were multiecho (TE = 12, 24.5, 37 ms), and fMRI signals underwent fitting of T2* parameters via tedana.py (and also congruent voxelwise monoexponential fits) to yield global signals as described in (Power et al., 2018).
CJL dataset: N = 1 subject scanned at rest 5 times on five consecutive days at the Citigroup Biomedical Imaging Center at Weill Cornell in New York City on a Siemens Prisma 3T scanner using Siemens PMU physiology recording equipment. These data were collected for other purposes but are included in this paper because a calibration procedure at the start of each scan elicited 4 widely-spaced instructed deep breaths (at 70-second intervals via visual cues), for a total of 20 instructed deep breaths. These data were multi-echo multi-band CMRR fMRI sequences (TE = 13.4, 31.1, 48.8, 66.5, 84.2 ms; TR = 1.355 sec; multi-band factor 6; 2.4 mm isotropic voxels) and underwent fitting of T2* parameters in the same manner as the NA data of (Power et al., 2018) to yield global T2* signals, shown in Figure 1. Estimates derived via multi-echo independent components analysis are shown, and the same result was obtained when checked via voxelwise monoexponential fits. Registration to surfaces was accomplished via the HCP preprocessing pipeline.
Results
Illustration of respiratory records and respiratory measures
Of the HCP “900 subject” release, only 440 subjects had full sets of fMRI data and full sets of physiologic data in which we believed signals were of sufficient quality that we could algorithmically obtain reliable peaks in cardiac and pulmonary traces. Only these subjects were analyzed further. Characteristics of the subjects were: age 28.6 ± 3.8 (range 22 – 36), 228 males and 212 females, BMI 26.5 ± 5.0 (range 16.5 – 43.9). Throughout the paper we will distinguish between “respiratory traces”, referring to the respiratory belt waveforms, and “respiratory measures”, meaning measures derived from the respiratory traces such as ENV, RV, and RVT. Unless stated otherwise, RVT in the text refers to RVTcore; figures will always label the version of RVT being shown.
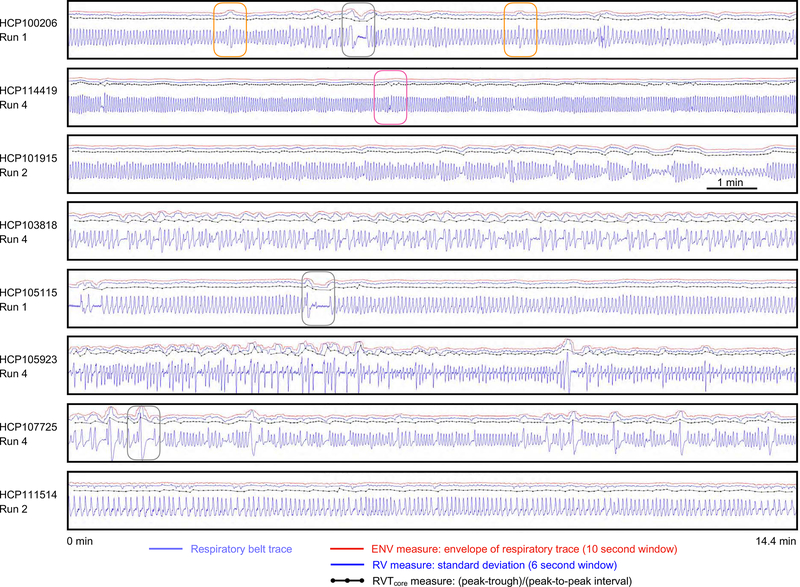
Examples of respiratory traces and associated ENV, RV, and RVT measures are shown for full runs of 8 subjects in Figure 2. Several points are noteworthy. First, the typical frequency of respiration varies by subject. Second, within a single run, there can be wide variety in respiratory frequency (e.g., 8th panel). Third, within a single run, there can be a wide variety in respiratory depth (e.g., 3rd panel). Fourth, there may be frank pauses in breathing (e.g., 1st, 5th, and 7th panels, gray boxes). Fifth, the ENV and RV waveforms tend to be quite similar and are both distinct from the RVT waveform (see Figure S1: across subjects, ENV and RV correlate at r = 0.85 ± 0.08, and ENV and RV correlate to RVT at r = 0.64 ± 0.20 and 0.66 ± 0.18; the difference between the first and the latter correlations are significant by two-sample t-test (p<10e-20 in each run)). Sixth, related to the fifth point, some abnormalities in respiratory traces are reflected mainly in ENV and RV (orange boxes in 1st panel) whereas others are more captured by the RVT measure (red box in 2nd panel). Such plots for all runs of all used subjects are shown in the supplemental movies3.
Figure 2:
Examples of respiratory traces and derived measures. For 8 scans from 8 subjects of the HCP dataset, respiratory traces and derived ENV, RV, and RVTcore measures are shown. Each scan lasts 14.4 minutes. Note the wide variety of periodicity within a scan and across subjects, as well as the varied shapes of the raw respiratory belt waveforms. Often the 3 derived measures capture similar properties of the respiratory trace, such as in the third panel. But in many instances there are discrepancies: the orange boxes in the 1st panel show ENV and RV capturing deep breaths, but RVTcore not capturing the same phenomena. Conversely, the red box in the 2nd panel shows ENV and RV not capturing a pause in breathing, which RVTcore captures. Gray boxes show several pauses in breathing.
Examination of respiration via gray plots demonstrates “missed” respiratory events
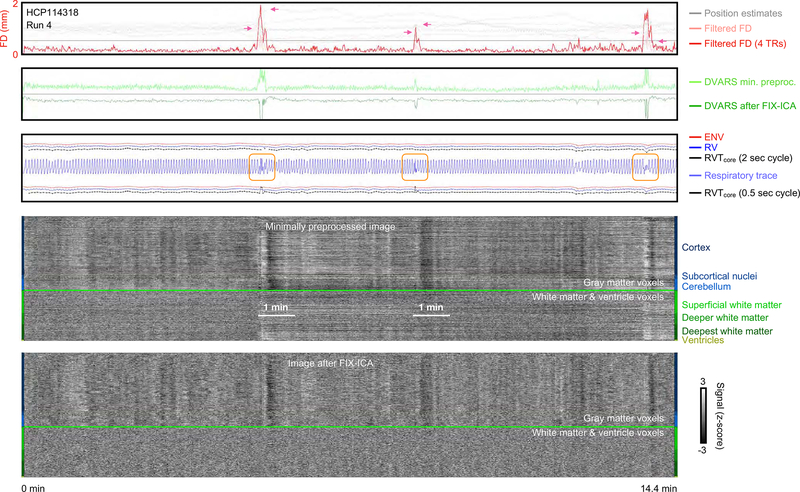
It is helpful to study respiratory traces in the context of other concurrent measures. The gray plot in Figure 3 shows a full run to orient the reader, and all gray plots in the paper follow the same scheme. The respiratory belt trace is shown in blue in the 3rd panel, and exhibits rather uniform periodicity and depth (i.e., eupnea) with a few exceptions, 3 of which are marked by orange boxes. The fact that something physiologically meaningful occurred at these times is evident from the fMRI timeseries heatmap below in the 4th panel, where vertical black bands representing pan-brain signal decreases follow each of the orange boxes. The heatmap shows signals from all voxels in the brain, ordered by anatomical compartment, with gray matter above the bright green line, and white matter below. The DVARS measures in the 2nd panel show at these boxed times spikes in the minimally preprocessed data (light green trace) and “DVARS dips” in the FIX-ICA-denoised data (dark green trace). These spikes and dips indicate that variance in these timepoints is changing abnormally rapidly in the minimally preprocessed image, and abnormally slowly in the FIX-ICA-denoised images. The explanation for the dips is that some ICA components act as pseudo-delta functions to remove large amounts of variance at specific time points during denoising. In our experience (Power et al., 2019b), DVARS dips occur nearly exclusively at times of major head motions (and may not occur during minor motions). Head position traces show two notable properties in the 1st panel (light gray traces). First, there are position zig-zags throughout the scan sharing the same periodicity as the respiratory trace, which reflects head motion and pseudomotion at the primary respiratory rate (Fair et al., 2018; Power et al., 2019c; Siegel et al., 2017). Second, at the three boxed times in question, there are sustained step changes in head position (pink arrows show starting and ending positions of a single position parameter). These position changes are captured in the FD measure of head motion, shown in bright red. Note that the FD shown here is a modified version of FD, namely one derived from comparisons of position traces across 4 TRs (2.88 seconds) that have had primary respiratory frequencies between 0.2–0.5 Hz filtered out. The rationale for these choices in constructing FD is detailed elsewhere (Power et al., 2019b) and summarized in the Methods. A 1-TR version of FD is shown in light red.
Figure 3:
Example of scan motion, image quality measures, respiratory measures, and fMRI timeseries before and after FIX-ICA denoising. In the first panel, unfiltered position estimates are shown in light gray, illustrating large trends in head position and also periodic respiratory motion. A bright red trace shows FD calculated in 4-TR intervals of the position estimates with respiratory frequencies filtered out, yielding motion estimates similar to those used in the literature in recent years (effective TR = 2.88 sec); the horizontal black line denotes FD = 0.5 mm. In the second panel, DVARS (DV) calculated in minimially preprocessed data and FIX-ICA denoised data. Note the correspondence of motion with DV spikes in minimally preprocessed data, and the “DV dips” after FIX-ICA, indicating that the ICA procedure acted as a kind of spike regressor at these times. In the third panel, respiratory traces and derived measures are shown. Note the circled abnormalities in the belt trace, which are not always captured by the derived measures. Two versions of RVTcore are shown (above and below the belt trace), the upper trace using standard peak finding settings and the lower trace using a setting permitting very rapid breaths. The bottom panels show “gray plots” of all timeseries in the image, organized by anatomical compartment. Such plots for all runs of all subjects are in the online movies. Figure S2 shows deep breaths in all runs in this subject for waveform comparison.
This scan illustrates important points about the respiratory measures derived from respiratory belt traces (i.e., the red ENV, blue RV, and black RVT traces immediately above the respiratory trace in the 3rd panel). First, as noted earlier, all measures are generally similar, especially ENV and RV. Second, respiratory events can be “missed” by any of the measures. In the present scan, all of the measures mark the third event with a dip, but there is little indication in the measures of the first and second boxed events. In this scan the “misses” are concordant across measures, but later figures will illustrate discordant situations where some measures mark an event but others do not.
The “misses” prompt some comments on respiratory measure definition and the qualities of the respiratory traces. Tautologically, ENV and RV “missed” events because the representations of the events in the belt trace produced little or no meaningful change in the waveform envelope or its windowed variance. But, unlike the other respiratory measures, RVT depends on peak detection (RVT = peak amplitude / time between peaks), and one might object that the “misses” occurred because RVT is not optimally calculated in this subject, for there are rapid oscillations in respiratory traces at the boxed times and RVT is not calculated for each of the peaks. However, as shown below, it is probably not possible to prevent “misses” by RVT in this scan and the challenge of dealing with such traces is precisely the reason this scan was chosen for illustration.
Peak-finding settings must be chosen to operate reliably across a wide variety of respiratory waveforms, as evident in Figures 1 and 2. However, not only is there much variability in “true” respiratory waves, but the belt traces are also susceptible to abdominal contraction and motion, which can introduce rapid non-respiratory fluctuations into the trace of varying amplitude. Whereas large multi-second fluctuations of abdominal belts have face plausibility as respiratory phenomena, rapid fluctuations are less plausibly purely respiratory phenomena. For instance, amid several very small and rapid fluctuations, the first boxed event ostensibly contains two full breaths accomplished in under two seconds, an extremely rapid rate of breathing. Though there is doubtless a respiratory event at this time, it seems more likely that the subject breathed deeply but also moved and contracted the abdominal musculature to yield a rapidly fluctuating waveform, rather than suddenly breathing rapidly with odd variations in volume amidst an otherwise eupneic scan. Note that the black bands in the fMRI signals here span ~30–40 seconds, just as the deep breaths did in Figure 1. Additionally, as shown in Figure S2, each of the other runs in this subject contains clear deep breaths with similar-appearing fMRI signal changes, and it is noteworthy that in each of these runs, the deep breaths had irregularities in the peaks, as if the belt was slipping. Without another source of respiratory information, however, the truth of what occurred cannot be known with certainty.
Given the uncertainty in the “meaning” of rapid oscillations in the belt traces, it is prudent to choose peak-finding settings that minimize the production of “non-respiratory” values, especially the outliers that would likely be generated by brief cycle times. Nearly all scans have respiratory frequencies under 0.4 Hz, which is why a 2-second breath cycle is the standard setting in this paper. If more rapid peak detection is permitted, a greater number of high RVT values will be generated, and fewer low RVT values will be generated. Figure 3 illustrates this occurrence by re-presenting the RVT measure while permitting peaks to exist 0.5 seconds apart (see red, blue and black traces under the respiratory trace), now yielding prominent spikes in the RVT trace at the first two boxed events. Note, however, that the abnormality at the third boxed event initially seen in the “standard” RVT has now largely disappeared! As stated above, preventing “misses” in this scan due to peak-finding may not be possible.
Moreover - and this will be seen repeatedly in the paper - RVT can fail to flag a respiratory event even when there is no ambiguity about proper peak-finding. RVT is designed to index volume per time, and, if breath volume scales with breath time, changes in those parameters can cancel one another out. Clear examples are shown in Figure 4 and 5 of deep breaths being unremarkable in RVT simply because deeper-than-usual breaths occur over longer-than-usual time spans (e.g., HCP118528, HCP121618, etc.). Peak-finding plays no role in such “misses”. Neither are the misses a byproduct of our implementing only the “core” computation of RVT, for the same misses are also seen in the full, interpolated version of RVT, no matter whether in our own implementation (RVTBirn), in that of AFNI (RVTAFNI (Birn)), or that of FSL (RVTFSL (Birn)). See Figures S3 and S4 for reproductions of Figures 4 and 5 using RVTBirn; Figure S5, illustrating RVTcore, RVTBirn, and RVTAFNI (Birn), in NIH data from (Power et al., 2017b)); and Figure S6, illustrating RVTcore, RVTBirn, RVTAFNI (Birn), and RVTFSL (Birn), in entire runs of HCP data.
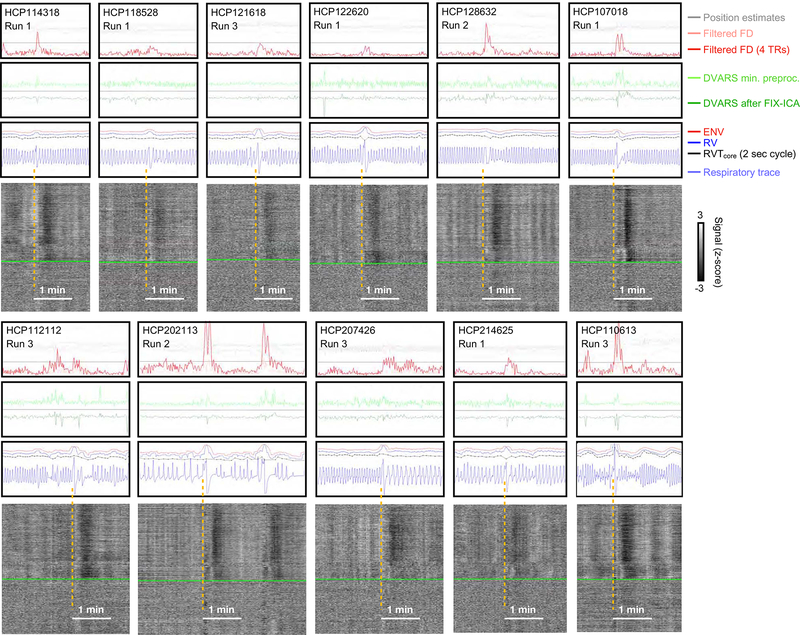
Figure 4:
Discordance of respiratory measures during “single deep breaths”. For 11 scans with relatively isolated deep breaths (see blue respiratory traces), excerpts of figures in the style of Figure 3 are shown. The gray plots are of minimally preprocessed data. Note that ENV (red trace) and RV (blue trace) often have bumps at the deep breaths, whereas RVTcore (black trace) may be flat, have a bump or spike, or may have a dip, depending on the shape of the breath. Breaths were selected by the respiratory trace and the presence of concomitant fMRI signal changes typical of deep breaths, similar to those shown in Figure 1, not on the basis of motion or DVARS or respiratory measures. Figure S3 reproduces this figure with RVTBirn added.
Figure 5:
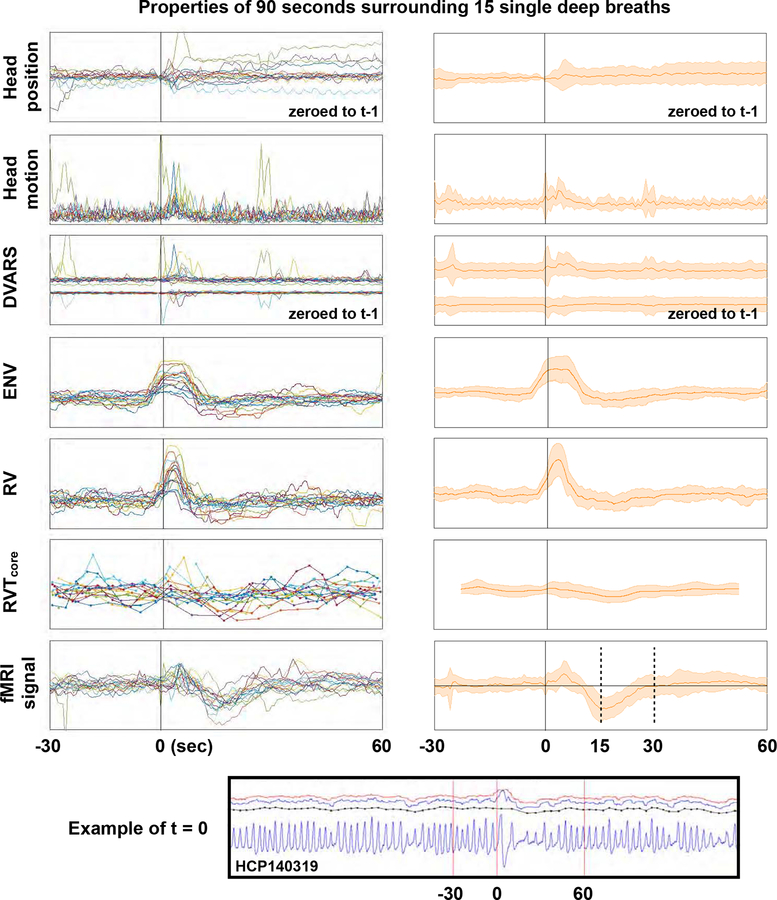
Summary of properties of 15 single deep breaths. Deep breaths were selected by respiratory traces and fMRI heat maps without knowledge of other properties, and full grayplots for all 15 scans are shown as a supplemental movie (these subjects are not in Figures 3 or 4). At bottom, an illustration of the respiratory trace and derived measures in a single subject, with vertical red lines indicating where t = 0 is set and the bounds 30 second prior to and 60 seconds after the breath. In the left column, data from each subject is shown in a different color, and traces for position, motion (FDoriginal), DVARS, ENV, RV, RVTcore and mean gray matter signals (in minimally preprocessed data) are plotted for 90 seconds surrounding deep breaths. In the right column, shaded plots show mean and standard deviations of the data. Axes are shifted slightly for the shaded head motion and DVARS plots to help visualization. Figure S4 reproduces the plot with RVTBirn added.
Thus, depending on the waveform and timing of a respiratory event, RVT may not mark an event, may mark it as a decrease, or may mark it as an increase. A corollary is that there is no universally satisfactory way to define peaks for RVT. Our standard settings will be to calculate RVT presuming respiratory cycles lasting 2 seconds or more. These settings may miss some rapid breaths, but will also avoid generating outliers in RVT of questionable accuracy. More generally – and this is an important point – there is no guarantee that a given respiratory event evident in the belt traces will be particularly noticeable in any of the derived respiratory measures. The measures are useful but imperfect indices of respiratory activity.
Discordance of respiratory measures during single deep breaths
Single deep breaths are useful illustrative respiratory phenomena, for they occur in most subjects and have been studied as both instructed and spontaneous breaths (Birn et al., 2006; Birn et al., 2008; Power et al., 2018; Power et al., 2017b). All animals with lungs sigh routinely to counter the natural collapse of lung compartments (i.e., atelectasis), which is one reason deep breaths are common in scans (Li and Yackle, 2017). To help convey the variety of respiratory waveforms denoting single deep breaths, 11 instances are shown in Figure 4 (there are thousands of examples in the online movies). In each of these instances, after an initial few seconds of moderate signal brightening, a black band in the gray plot reflects a pan-brain decrease in fMRI signals lasting until 20–30 seconds after the breath. There is almost always head motion associated with these breaths, often there is a DVARS spike in minimally preprocessed data, and there is sometimes, but sometimes not, a DVARS dip in FIX-ICA-denoised data. Note again that sometimes one or more respiratory measures do not capture the fact that something meaningful has occurred in the respiratory traces. Although ENV and RV often display bumps during the deep breath, RVT sometimes displays bumps, sometimes displays nothing, and sometimes displays a dip, consistent with our previous discussion.
A different set of 15 deep breaths in 15 different subjects was selected by examining respiratory belt traces and grayscale fMRI signal heat maps (in a chronological set of subjects, without knowledge of other properties like motion parameters, FD, DVARS, ENV, RVT, RV, etc.). Using only the traces and heatmaps let us to select isolated deep breaths that produced concomitant fMRI signal changes without actually knowing other properties of the scans. A gray plot for each of these 15 scans can be seen in an online movie4, with our markings of when the breaths began. For each breath, data from 30 seconds before and 60 seconds after the breath were extracted and are shown in Figure 5. Data for each of the different breaths are shown in a different color in the left column of Figure 5, and the right column summarizes the effects using mean and standard deviation shade plots. These events re-demonstrate the points above: 1) deep breaths cause ~30-second fMRI signal modulations, 2) deep breaths are marked by transient elevations of ENV and RV but often not RVT, 3) deep breaths often exhibit motion, often exhibit step changes in head position, often exhibit DVARS spikes, and less often exhibit DVARS dips. The scales of the left and right columns are changed slightly to facilitate visualization.
Collectively, these observations demonstrate a disparity in the way various respiratory measures mark single deep breaths. The disparity arises chiefly because RVT is dependent on the timings and temporal characteristics of respiratory cycles: a larger-than-normal breath transpiring over a longer-than-normal time may appear quantitatively just like a typical breath occurring over a typical time. In contrast, the other measures are relatively insensitive to the exact temporal characteristics of the signal and are more sensitive to the amplitudes of the signal in the sampling window, which is why they routinely mark deep breaths. In a related study (Power et al., 2019b), we have found that the tidal breathing rates of HCP data tend to peak around 0.3 Hz, with nearly all scans of all subjects falling within 0.2–0.4 Hz; by contrast, deep breaths in HCP data tend to occur around 0.1–0.15 Hz.
As a final way to describe the tendencies of respiratory measure to identify deep breaths, the deep breaths of the first 10 and second 10 chronologically numbered subjects (total 20 hours of scanning) were visually identified in the belt traces, and each respiratory measure was scored as either clearly showing an abnormality (i.e., a bump or dip) or not clearly showing an abnormality at the times of clear deep breaths. In the first 10 subjects, in 21 clear deep breaths, ENV and RV marked 15 clearly (71%), and RVT marked 11 clearly (53%). In the next 10 subjects, of 48 clear deep breaths, 45 were marked clearly by ENV and RV (94%) and 24 by RVT (50%).
The decisions underpinning these numbers are subjective but the statistics conform to the characteristics described above. The bottom line is that ENV and RV “miss” modest fractions of physiologically meaningful respiratory events, and RVT misses a larger fraction of those same events.
Concordance of respiratory measures during changes in breathing depth
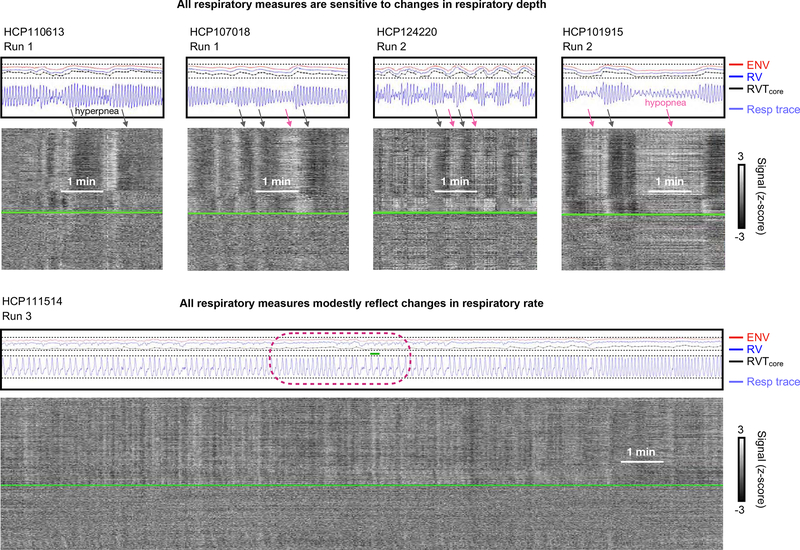
Respiratory depth can have a major effect on all respiratory measures, and the effect is concordant. While maintaining regular, periodic breathing, subjects can breathe deeply, increasing exhalation of CO2, lowering pCO2, and thereby decreasing cerebral blood flow and causing BOLD signal decreases (hyperpnea: black arrows of Figure 6). Periods of shallow breathing cause complementary phenomena (hypopnea: pink arrows of Figure 6). In the setting of respiratory depth modulation, high concordance is typically seen between ENV, RV, and RVT. The concordance follows from the sensitivity of all measures to changes in respiratory trace amplitude in the setting of relatively constant timings of cycles of breathing.
Figure 6:
Examples of concordance between respiratory measures during sustained changes in respiratory depth and respiratory rate. At top, segments are shown from four subjects with substantial variation in respiratory depth, demonstrating large, concordant changes in respiratory measures in all scans. At bottom, a subject with much variation in tidal respiratory rate (among the most variable we observed), with concordant but relatively modest modulation of respiratory measures by respiratory rate.
Considerations about breathing rate
Breathing rate has a less obvious influence on respiratory measures than breathing depth, for one principal reason: although many subjects exhibit marked modulation of tidal breathing depth, relatively few subjects show marked variation in tidal respiratory rate within or across scans (beyond the intermittent occurrence of slow deep breaths) (Power et al., 2019b). The bottom row of Figure 6 illustrates one of the most variable subjects we encountered in terms of tidal rate (the same subject shown at the bottom of Figure 2). Toward the end of the scan there is a clear increase in breathing rate, and there is also an increase in all respiratory measures comparing the first few minutes to the final few minutes (compare via dotted lines). However, the belt amplitude has actually also increased (compare via dotted lines). The most informative comparison is seen in the middle of the scan (maroon circle), where the respiratory rate falls almost by half while preserving belt amplitude: a decrease of respiratory measures with lowered respiratory rate is seen, but this decrease is much less than the decreases seen above (in the top panels of the figure) with respiratory depth modulation.
Certain properties of the respiratory traces only become relevant at low respiratory rates. When occurring sufficiently rapidly, respiratory cycles are continuous oscillations (akin to sinusoidal forms), seen at the top of Figure 6 and in several other figures. However, when respiratory rates drop, this sinusoidal form does not hold, and there may be actual pauses in the respiratory cycle near the functional residual capacity (the resting point of the lung where elastic recoil is balanced between chest expansion and lung contraction), rather than the continuous cycle of inspiration and expiration. This phenomenon is seen in the encircled portion of the bottom scan in Figure 6 where transient plateaus occur in the belt trace. When respiratory rates fall this low, distinctions among the respiratory measures emerge. Though RVT is sensitive to respiratory rates, precisely what happens in the waveform between the troughs and peaks is irrelevant (a sinusoidal cycle would have the exact same value as a flat line with a trough and a peak inserted at the appropriate times). In contrast, RV and ENV are windowed measures, and if the respiratory trace essentially pauses for some time, this pause can cause dramatic drops in the respiratory measure, depending on the relation of the pause to the windowing time. This sensitivity is seen in the RV trace in Figure 6, which exhibits marked dips during the pauses. Interestingly, the ENV measure is anticorrelated with RV at these times, displaying bumps during the plateaus. This bump is because of windowing – the ENV measure has 10 second windows (compared to 6 seconds for RV), and the respiratory rate is so low that at the midpoint between breaths the ENV window is encompassing one extra breath relative to when it is centered on a peak (small green line shows 10 second span). In contrast, the RV window only ever encompasses a single cycle at respiratory rates this low.
The subject in the lower panel of Figure 6 was specifically chosen for demonstrating potential influences of tidal respiratory rate. However, though differences in respiratory measures during especially slow breathing can be seen, this situation occurs infrequently (beyond intermittent deep breaths), and tidal respiratory rate modulation, when it occurs in HCP data, most often occurs at considerably higher frequencies than the low ones illustrated here (Power et al., 2019b), rendering the above considerations related to low rates largely academic, especially in comparison to the much more influential modulations of tidal respiratory depth that are commonly seen in subjects.
Systematic effects in respiratory measures over time
We turn now from properties of the respiratory measures in individual scans to properties at the group level. The HCP data are unusual in the length of scans (~15 minutes), in the size of the cohort (hundreds), and in the fact that four scans per subject are obtained, permitting investigation of systematic changes in respiration seen repeatedly in scans.
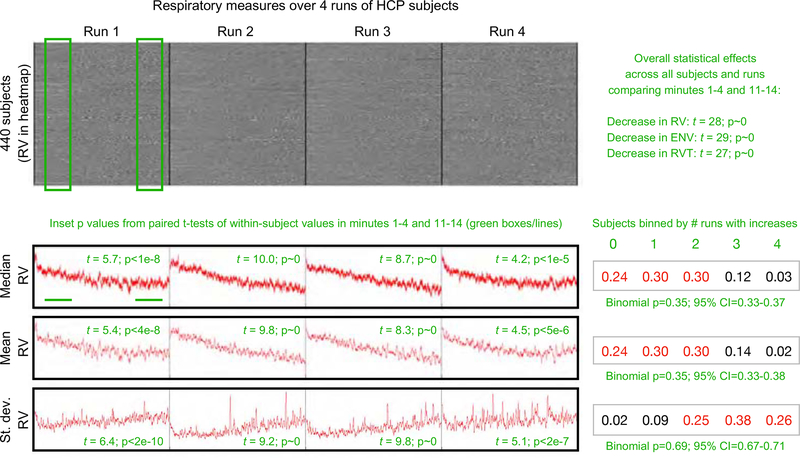
When ENV, RV, and RVT measures are examined over runs, a pattern emerges. Figure 7 shows the pattern for the RV measure (the same pattern is found in ENV and RV measures, see Figure S7). Nothing is easily seen in heatmaps of respiratory measures. But numerically, as shown in the red traces, the medians and means of respiratory measures across subjects decline over the course of each run, and the variance in those measures across subjects increases over the course of each run. To quantify these effects, values of these measures in minutes 1–4 and 11–14 were compared in each run by paired t-test (sampling once every 10 seconds to preserve independence in measures), yielding significant differences over time (green inset p values). To quantify the proportion of subjects displaying such effects, subjects were binned by the number of runs (0–4) in which they displayed increases in the median, mean, and standard deviation within these time windows, and these proportions were fitted to binomial distributions, yielding a probability of 65% per run (95% C.I. 63%−67%) that measures would decrease and a 69% chance (95% C.I. 67%−71%) that variance would increase. Overall statistical effects for each of the respiratory measures are shown in Figure 7, contrasting values in the early and late periods across all runs. The significance of this finding is that one should often expect systematic respiratory changes, and thus systematic BOLD respiratory effects, as fMRI runs progress.
Figure 7:
Respiratory measures systematically change during fMRI scans. At top, a heat map of the RV measures for all subjects with a complete set of 4 runs of fMRI data and acceptable physiologic measures. Median and mean RV values across subjects are plotted, both showing decreases over time in each run. Standard deviation calculations across subjects show variability increasing over time in each run. To assess within-run changes, the mean, median, and standard deviation of RV was calculated in minutes 1–4 and 11–14 of each subject’s trace, in each run (see green boxes and underscores, samples were taken every 10 seconds). The inset green numbers by traces indicate the p values of paired t-tests of within-run comparisons of mean, median, and standard deviation in early and late time periods. At right of traces, a binning of subjects by the number of runs in which their within-run comparisons yielded increases is shown, and each distribution is fit to a binomial distribution, with the probability and 95% confidence intervals shown in green. All intervals indicate an approximately 66% chance per run that a subject will show a decrease in RV and an increase in variance over each run. Similar findings are found using RVTcore and ENV measures (Figure S7). Statistical effects are found for all respiratory measures, shown in Figure S7 and listed at upper right.
Based on the properties noted above, decreases in breathing depth or rate could produce decreases in respiratory measure values over time. To test these possibilities, we calculated peak frequencies in minutes 1–4 and 11–14 of each run, and compared them within-subject by paired t-test. In each run, peak respiratory frequencies decreased in the later minutes compared to the earlier minutes, with small but significant effect sizes (p = 0.023, 3.0e-6, 0.002, and 6.6e-6 for the four runs). In the four runs, 39–46% showed a decrease in rate, 18–25% of subjects showed an increase in rate, and 34–42% showed no change in rate. We also examined the amplitude of the peaks that defined RVT within these early and late windows, and found that respiratory depth also decreased over each run (p = 1.4e-4, 3.4e-14, 4.6e-11, and 5.1e-5 for the four runs). In the four runs, 58–66% of subjects in each run exhibited decreases in amplitude. Thus, both reduced rate and reduced depth of breathing could contribute to decreases in respiratory measures over time during these resting state scans, though depth likely has the greater influence.
One might wonder if automatic gain control in the Siemens physiology equipment could contribute to or cause these effects. We see no possibility for gain modulation to affect breathing rate estimation, for good belt signals are present throughout all used scans and systematic changes in amplitude are subtle rather than marked. Though we cannot definitively exclude the possibility that dynamic gain contributes to amplitude changes, it is difficult to see how a systematic decrease in peak amplitude would result without it being a response to a change in breathing. If breathing depth truly decreased, one might expect gain to dynamically increase and cancel out the change, which did not happen. If one posits that the gain is gradually dynamically reduced, presumably this would be due to increases over time in detected breathing amplitude, which could either be increased tidal depth or increased intermittent deep breaths. In any event, a respiratory change would underlie the “observed” change (and, anecdotally, we see no evidence that deep breaths occur later in scans, they seem to occur at any time in a scan). As we will discuss below, there are also reasons to anticipate modest reductions in breathing rate and depth in these resting state scans. Taken together, and barring contradictory evidence, we interpret the reductions in respiratory depth and rate as true phenomena.
Discussion
In this paper we examined the characteristics of respiratory belt traces and of several respiratory measures used to index respiration and to flag respiratory events in fMRI studies. We used the HCP dataset to leverage its large number of subjects, the large amount of scan time per subject, and because this dataset has become a reference dataset for the field. Examination of hundreds of individual scans yielded three principal findings: 1) respiratory measures ENV, RV, and RVT were all correlated, especially RV and ENV, and all measures were prominently and similarly modulated by the depth of breathing; 2) although any measure could “miss” a deep breath, RVT tended to miss more deep breaths than the other measures; and 3) all respiratory measures changed systematically across resting state scans. The principal implications of these findings are twofold and will be discussed below: 1) respiratory measures, to the extent they are insensitive to deep breaths, may fail to link relevant phenomena to respiration; 2) respiratory modulation of pCO2 ought to cause systematic BOLD changes over time in resting state scans.
Implications of “missed” deep breaths
Deep breaths in resting state fMRI scans are sighs. Sighs are unusually deep breaths that interrupt tidal breathing, and are well-studied phenomena that occur for a variety of reasons (Li and Yackle, 2017). “Physiological” sighs are universal among humans. They are estimated to occur several times per hour in upright adults, and are more frequent in the supine positions used in most fMRI scanning. The purpose of sighs seems to be to reinflate collapsed alveoli, and to counter slight changes in blood gases like hypoxia or hypercapnia (Li et al., 2016). Sighs often include brief central apneas (i.e., breathing pauses) following the sigh, just as is seen in many of the deep breaths during HCP scans. “Emotional” sighs (e.g., indicating disappointment) probably occur infrequently in resting state scans. Sighs can also be seen in the transition to sleep, especially in exaggerated form as yawns. Some deep breaths in HCP resting state scans are likely yawns, since subjects stared at a black screen for a quarter of an hour in a warm, dark environment. We know of no way to discriminate between causes of deep breaths in the HCP data.
Deep breaths can be “missed” by any of the respiratory measures (Figure 3), but are more often “missed” by RVT than by RV or ENV (Figure 5). The significance of these misses is that if deep breaths are not recognized by a respiratory measure, then a model using those measures will fail to assign “deep breath variance” to its proper respiratory cause, and may either leave this variance unassigned or assign it improperly to a variable collinear with deep breaths. Several versions of this scenario will now be discussed.
The simplest model would include only respiratory variables, for example, for denoising purposes. Investigators creating physiological models of fMRI signals tend to record respiration, to derive a measure like RVT or RV to “index” respiration, and then to convolve that index with a kernel approximating the fMRI respiratory response function (RRF). Several examples of such kernels were shown in Figure 1D. During convolution, flat shapes in the index timeseries will fail to invoke the RRF, whereas a spike will invoke the RRF, placing the approximate fMRI respiratory response shape at the appropriate segment of the regressor timeseries. In a situation where a deep breath creates a bump in ENV or RV but leaves RVT flat, convolution will place the RRF via ENV*RRF or RV*RRF, but will fail to place it via RVT*RRF. In such situations, regression should largely remove respiratory variance via ENV*RRF or RV*RRF approaches, but largely leave respiratory variance in place via RVT*RRF approaches. Numerous examples of this situation are evident in the HCP data at times of discordance between RVT and RV or ENV, several of which are shown in Figure S8. Stated plainly, missed deep breaths cause the models to “overlook” respiratory variance. In the setting of denoising, this variance then remains available to subsequent models.
A different scenario occurs when variables collinear with deep breaths are modeled either without respiratory variables or with ineffective respiratory variables. In this scenario, missed breaths will cause the model to fail to assign deep breath variance to a respiratory cause, and will instead assign it to anything modeled that co-occurs with deep breaths. Multiple physiological and mechanical changes accompany deep breaths, providing ample opportunity for misattributions.
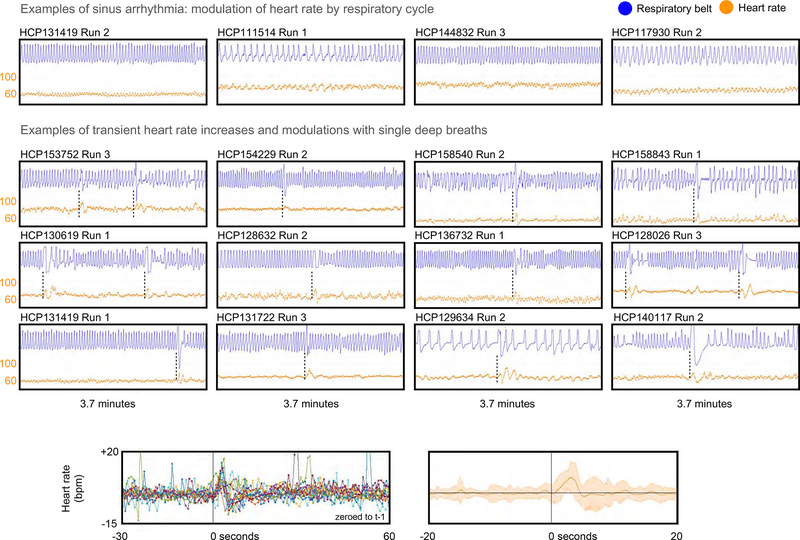
A version of this scenario could occur with variables describing heart rate. Breathing is intimately related with heart rate via physical forces in the chest as well as by autonomic feedback loops (Hall, 2016). An example is the ongoing modulation of heart rate by breathing cycle seen in all humans, a phenomenon termed sinus arrhythmia, illustrated in the top row of Figure 8. With regard to deep breaths specifically, when subjects take deep breaths, it is common for the heart rate to increase transiently by 5–15 beats per minute before settling back to baseline or transient below-baseline values several seconds later. We routinely observe this phenomenon in HCP scans, and have observed it previously in other datasets (Power et al., 2017b). Figure 8 shows twelve individual examples of heart rate increasing during and just after deep breaths, and also shows the effect in summary form for the set of deep breaths examined in Figure 5 (those breaths were chosen without any knowledge of cardiac properties). Figure S2, which illustrated deep breaths in each run of the single subject shown in Figure 3, also routinely shows such elevations of heart rate at deep breaths. To the extent that a respiratory variable “overlooks” deep breaths, by the logic discussed above, a study could assign deep breath fMRI signal changes to variables for heart rate or heart rate variability. This situation is most likely to occur with RVT respiratory measures, but will occur even with ENV and RV to the extent they “miss” individual events. Such misassignment of respiratory variance can happen for any number of physiologic modulations that co-occur with deep breaths (e.g., pulse pressure, (Cannesson et al., 2005; Natalini et al., 2006)).
Figure 8:
Examples of heart rate changes driven by respiration. At top, examples of sinus arrhythmia; heart rate is in orange in beats per minute (bpm). Middle, examples of single deep breaths causing transient elevations and modulations of heart rate. At bottom, heart rate from the 90 seconds around the 15 deep breaths examined in Figure 5 (only 13 subjects had usable cardiac records at the relevant time segment).
A different version of the scenario could occur with head motion, for deep breaths are also often accompanied by head motion. Multiple forms of respiratory-associated motion can be found in fMRI datasets, including both true motion and pseudomotion (an apparent but false shift of the brain caused by magnetic field instabilities when the lungs expand, occurring prominently in the phase encode direction (Bollmann et al., 2017; Brosch et al., 2002; Fair et al., 2018; Raj et al., 2001)). Both real motion and pseudomotion accompany deep breaths, and pseudomotion should be most marked during deep breaths. In HCP data, a uniquely slow motion during (slow) deep breaths is apparent, but it appears only in the phase encode parameter, and not in other directions, representing pseudomotion during deep breaths (this effect is demonstrated in several datasets, including the HCP, in (Power et al., 2019b)). Such results indicate that, at least in HCP-style datasets at 3T, pseudomotion is prominent only in the phase encode direction. Figure S3 reproduces Figure 4, using FD estimates that entirely exclude the phase encode direction, thereby eliminating the contribution of such pseudomotion to the motion estimate. All breaths continue to display motion, and many breaths display very large motions even after excluding phase encode pseudomotion. If one attempts to model “events” in timeseries, deep breath fMRI signal modulations spanning ~30 seconds could be assigned to “motion” if no respiratory cause is considered or if the respiratory variable is insensitive to deep breaths. This assignment will occur whether one is measuring true motion, pseudomotion, or both phenomena, since they co-occur. Such logic can explain why Byrge and colleagues (Byrge and Kennedy, 2018) find a 30-second “motion” signal following the larger motions of the HCP data with rather modest association to RVT and yet a waveform very like the respiratory response functions in Figure 1D.
This scenario can also affect attempts to classify signals, such as the temporal independent component analysis classification scheme proposed in (Glasser et al., 2018). That study used associations with RVT, with motion (via DVARS, not FD), and with sleepiness to help categorize temporal components. The decision to use DVARS dips as a measure of motion, rather than the actual motion estimates, was motivated by concern over the influence of pseudomotion, as described above. We view the substitution of DVARS for FD with reservations, for while DVARS deflections do routinely mark large motions, they do not so reliably mark smaller motions. For instance, Figure 4 shows several instances of deep breaths that not only fail to cause deflections in RVT, but which also have little or no representation in DVARS traces (e.g., HCP118528, HCP121618, HCP 128632, HCP207426, etc.) despite having modest or major motion (in non-phase-encode directions, see Figure S3). The relative insensitivity of RVT to deep breaths – and of DVARS dips to subtler motions - could help explain why Glasser and colleagues (Glasser et al., 2018), even while attempting to index respiratory influences (via RVT) found only 3 of 84 components to be associated with both respiration and motion, despite several components displaying spatial distributions that are associated with respiration as well as across-scan signal amplitude changes paralleling those of the respiratory measures in Figure 7. We expand on this topic below.
A clarification on how we use physiologic records
A theme evident throughout this paper is that, with respect to physiological records, the devil is in the details. We caution against performing “physiological” analyses without inspecting records. Inspecting records both guards against poor quality and alerts investigators to phenomena or relationships that are evident in single scans but which could easily be missed in group-level analyses (e.g., because a respiratory measure frequently failed to index a respiratory event, or, because it depended on cross-subject or cross-day factors like subject hydration, etc.). We raise this point because neither of the just-mentioned studies manually inspected physiology records, and, in fact, both studies cited our own work to justify not looking at records (e.g., “following (Power et al., 2017b) the respiratory belt traces were preprocessed as-provided, without manual inspection for artifacts” in (Byrge and Kennedy, 2018) and “[the investigators] were unable to manually review and quality assure [the physiology data]… we rely on prior work… that did manual quality control… (Power et al., 2017b)” in (Glasser et al., 2018)). To be clear: we did manually review all physiology data in (Power et al., 2017b), and do so in all of our papers. We think it unwise to use physiology data that has not been manually inspected, for there are routinely substantial corruptions of the traces, as evidenced by the fact that we discarded over half of the HCP subjects considered for this study due to partially or fully corrupted physiology records. Even that estimate of usability turned out to be optimistic, for when we checked pulse oximetry peaks manually for the specific heart rate data illustrated in this paper, we could only use 13/15 instances of cardiac records in deep breaths in Figure 8, and we had to eliminate 1 of the 4 runs of pulse oximetry data of the subject in Figure S2, all because we were too frequently uncertain about the “true” peaks in the traces to calculate heart rate. What we said in (Power et al., 2017b) was that whereas we “intervened” in pulse oximetry traces to manually correct peaks for heart rate calculation (as we did in this paper), we did not intervene in the respiratory traces, for they are often so much more irregular than the cardiac traces (as the present paper illustrates). If this statement misled readers into thinking we did not inspect the respiratory data, we regret the misimpression. Many of the subjects we discarded from the HCP data were discarded due to poor respiratory records.
When a deep breath occurs, many things happen: the chest expands, the heart rate changes, the pulse pressure changes, the head can move, there are modulations of pCO2 and therefore cerebral blood flow, there are neural circuits detecting and guiding those changes, and there may be a variety of “triggers” of the deep breath, some cognitive (e.g., frustration, tiredness, etc.). To the extent that one wishes to disambiguate such effects or limit such influences in a study, one must know that a deep breath occurred in the first place. Our view is that, at least at this point in time, detecting deep breaths or other physiological occurrences with confidence requires personal attention to the data.
Implications of systematic changes in respiratory measures over time
Over the course of each 14.4-minute resting state scan of the HCP data, respiratory rates tended to decrease, respiratory depth tended to decrease, respiratory measures tended to decrease, and variance in respiratory measures tended to increase. Collectively, these observations indicate that investigators ought to expect pCO2 and thus BOLD signals to change systematically over a scan, at least at the group level.
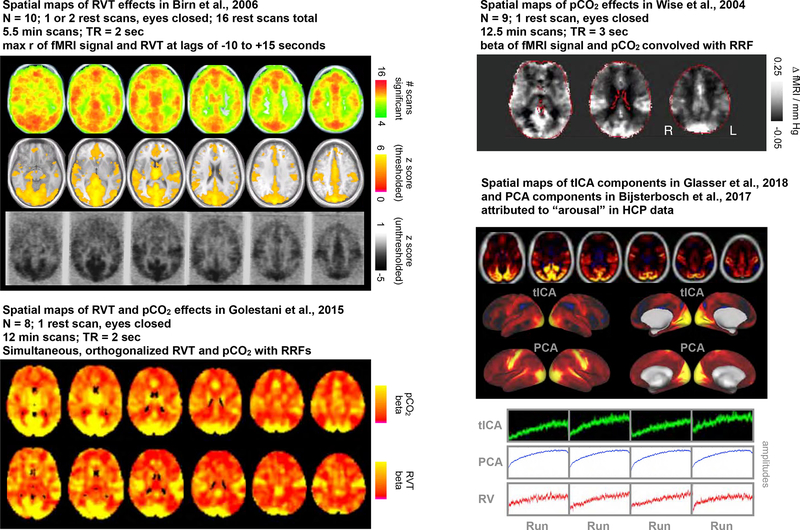
Many studies have investigated changes in fMRI signals contingent on pCO2 or respiratory belt records, including studies using spontaneous breathing records and instructed breathing paradigms (e.g., breath holds, single deep breaths, paced breaths, etc.). Several examples of published images are shown in Figure 9 (from (Birn et al., 2006; Golestani et al., 2015; Wise et al., 2004)), consistently obtaining modulation across all gray matter which is most pronounced in a “sensorimotor” distribution, emphasizing occipital, posterior temporal, and peri-Rolandic gray matter. If BOLD signals were to change systematically over a scan due to changes in respiration, it would likely occur in this spatial pattern.
Figure 9:
Spatial patterns of respiratory belt and end-tidal gas variable regressions in fMRI data, from published papers. Using different methods, Birn et al., 2006, Wise et al.,2004, and Golestani et al., 2015 all obtain rather similar spatial patterns of where respiratory variables explain the most variance: positively everywhere in gray matter but especially in occipital, posterior temporal, and peri-Rolandic cortex, a “sensorimotor” distribution. Separately, in HCP data, several papers have reported signals whose amplitude increases systematically in each run of the data, and these signals share a “sensorimotor distribution”. Amplitudes are plotted for these components (replicated for the PCA signal, which was the mean of the 4 runs). The (inverted) median RV respiratory signal from Figure 7 is placed for comparison. Figures modified with permission from {Birn, 2006; Wise, 2004; Golestani, 2015; Bijsterbosch, 2017; Glasser, 2018}.
Several reports in recent years have documented in HCP data a set of structured signals with amplitudes that increase gradually across each of the resting state runs, examples of which are also shown in Figure 9 (signals obtained by spatial ICA and/or PCA in (Bijsterbosch et al., 2017) and temporal ICA in (Glasser et al., 2018)). These reports explained such effects as effects of arousal, but a competing explanation of the component properties would be that they are consequences of respiratory phenomena in the scans: systematic signal amplitude changes across the scan resemble systematic respiratory measure changes, and the signal spatial pattern resembles where respiratory measures most strongly influence fMRI signals in the literature. Although it is possible these structured signals are direct signatures of arousal, these components may alternatively be signatures of arousal only insofar as states of arousal cause or correlate with altered respiratory properties (Power, 2019; Power et al., 2017a).
The reverse argument could potentially be made, that respiratory studies were actually identifying neural signatures of arousal, via systematic changes in respiration. This argument is somewhat undermined by the similarity of the spatial effects (r ~ 0.77) in rest and instructed breathing conditions (Birn et al., 2006; Birn et al., 2008), but such comparisons have been reported only for few subjects and with stimulus confounds, and further studies will be needed to convincingly disambiguate these possibilities. Because it is of potential interest, we illustrate in Figure S9 the spatial patterning of the instructed widely-spaced breaths shown in Figure 1C. These data were calibration data and were not acquired for the purpose of isolating the deep breath response, and each trial includes a 10-second visual instruction to inhale. Already in the Introduction we noted that the duration of the instructed respiratory responses is very like those of spontaneous breaths (Figures 1B–D; see also {Power, 2019}). These instructed breaths are of interest because the breaths are externally imposed, not internally triggered, and there is little obvious role for arousal to influence such responses. There is spatial topography to both maxima and minima in the respiratory response function, and the difference maps substantially conform to the sensorimotor distribution noted above. Because cue and response elements are intertwined, and because the data are only from one subject, these images should clearly be viewed as preliminary descriptors. Nevertheless, these preliminary data are compatible with a structured sensorimotor topography of respiratory responses. A more targeted study isolating deep breath from cue effects is now underway.
It will likely be challenging to clearly disambiguate – at rest - respiratory signals from “neural” signatures of arousal, for a large literature documents an intimate association of arousal and respiration. Studies in typical human adults indicate that the transition from wakefulness to non-rapid-eye-movement sleep is characterized by a mild reduction (~10%) in respiratory rate and a mild reduction in tidal volume (~5%) (Berssenbrugge et al., 1983), which would accord with our reduced rates and amplitudes over time. It is of interest that a large study that monitored sleep state during resting state fMRI scanning reported that approximately two thirds of subjects showed evidence of sleep or decreased arousal as a scan progressed (Tagliazucchi and Laufs, 2014). Two thirds is also the fraction of the HCP subjects examined here who exhibited decreased respiratory measures and increased respiratory variance over their scans. Studies that simultaneously monitored EEG alpha power and respiration and fMRI signals have noted links between all three phenomena (Yuan et al., 2013). Several studies of sleep have also recovered a “sensorimotor” set of signal changes that denote changes in arousal and sleep state (Horovitz et al., 2008; Tagliazucchi and Laufs, 2014; Tagliazucchi et al., 2013). These fMRI signatures of sleep may in part be respiratory signals. Given the expected association of respiratory properties with arousal (not only via changes in tidal parameters, but also yawns), one curious aspect of the study by Glasser and colleagues is that though several components were associated with “sleep”, none of them were also associated with RVT (illustrated in Figure S10). Potentially, this lack of association may have been due to insensitivity of RVT to discrete events; all component amplitudes increase over each run, and each component has strong “sensorimotor” loading. Our point in drawing comparisons between respiratory effects and signals and patterns attributed to arousal is not to dispute an association with arousal, but to note that the extent to which respiratory effects are contributing to these signals and patterns is unclear. Further study will be needed, and we advocate attention to effects in single scans in addition to group-level approaches.
On monitoring respiration
The one respiratory measure (RVT) that attempts to incorporate proxies of standard physiologic respiratory measures (e.g., inspired volume, cycle time) performs relatively poorly at identifying events like deep breaths compared to measures that are simply ad hoc descriptions of a waveform (RV and ENV). This poorer performance is partially explained by practical considerations about respiratory belt records. RV and ENV are relatively insensitive to the exact timings of wave modulations by virtue of their windowed calculation, and so brief non-respiratory motions or other artifactual disruptions of the belt record are often less problematic for those measures than for RVT, which strongly depends on times between peaks in the trace. But even absent such factors, the parameter indexed by RVT – air moved in a cycle time – is by definition relatively insensitive to outlier breath volumes so long as they scale with breath times. Deep breaths in the HCP data routinely occur more slowly than tidal breaths (Power et al., 2019b), and thus, RVT, by definition, is going to be less sensitive to such phenomena. RVT, by virtue of its rate dependence, may be especially sensitive to quick changes in breathing rate in the setting of preserved breathing depth, which may have particular utility in certain contexts. Our view is that of the present parameters, ENV and RV are similar in practice, are easy and unambiguous to compute, and usually “flag” noticeable events in the respiratory timeseries. There is room for improvement in all measures, given that all of them can fail to note deep breaths, but it seems that a substantial limiting factor in deriving quality respiratory measures is a lack of reliable and comprehensive respiratory records to begin with. For these and other reasons, we have begun to collect a longitudinal multi-echo, multi-band fMRI dataset with comprehensive physiologic recording that will eventually be made available to the public, including simultaneous capnography, inductance plethysmography, electrocardiography, pulse oximetry with gas saturations, galvanic skin responses, and pupillometry and eye tracking. With such data in hand, one hopes there would be little obstacle to creating precise physiological models, at least from a reliability or comprehensiveness standpoint.
In this paper we focused on respiratory data from a single site, captured via the Siemens Physiologic Monitoring Unit. One might wonder if the properties noted here extend to other recording schemes (e.g., GE equipment, Biopac equipment). We actually first noticed the effects described in this paper years ago in data acquired on a GE scanner, using GE equipment for respiratory monitoring (Power et al., 2017b). Some of the data from that paper are reproduced in Figure S5, illustrating deep breaths captured by RV but missed by RVTAFNI (Birn) (calculated in 2016 via AFNI’s RetroTS.m). Differences in monitoring equipment could affect respiratory measure calculations via gain nonlinearities or differences in dynamic adjustment strategies, but our observations are likely of general relevance, as they hold under multiple recording schemes and in multiple datasets.
Conclusions
Respiration can cause a variety of effects in fMRI data, from alterations of cerebral blood flow and BOLD signal to real head motions to pseudomotion. Breathing is also yoked to cardiovascular parameters via the physics of the chest and by autonomic loops. There are thus many reasons an investigator might wish to monitor respiration during fMRI scans. However, making use of respiratory records is not trivial, and the different respiratory measures that can be derived from respiratory belt records can perform quite differently at indexing respiration and respiratory events depending on the kinds of breathing in a scan. We studied task-free scans of young adults here; similar analyses in other age ranges, in clinical populations, and in task settings may be of interest. The field would benefit from more robust and reliable measurement of respiration, both in terms of physiological monitoring at the scanner and in terms of deriving variables from the physiological records.
Supplementary Material
Highlights:
Data are resting state fMRI scans in healthy young adults
Examines respiratory belt records and derived measures ENV, RV, and RVT
All respiratory measures “miss” deep breaths, RVT more than others
Deep breaths exhibit characteristic heart rate and fMRI signal changes
All respiratory measures change systematically over 14.4 minute scans
Acknowledgments
This work was supported by Simons Foundation Grant 528440, a gift from the Mortimer D. Sackler, M.D. family, and the Intramural Research Program, National Institute of Mental Health/NIH (ZIAMH002920; ). We thank Kevin Tran and the NIH Helix/Biowulf staff for computing support. We thank Rasmus Birn for discussion of RVT calculations and Catie Chang for assistance with respiratory response functions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors declare no conflicts of interest with respect to this report.
References
- Berssenbrugge A, Dempsey J, Iber C, Skatrud J, Wilson P, 1983. Mechanisms of hypoxiainduced periodic breathing during sleep in humans. J Physiol 343, 507–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijsterbosch J, Harrison S, Duff E, Alfaro-Almagro F, Woolrich M, Smith S, 2017. Investigations into within- and between-subject resting-state amplitude variations. Neuroimage 159, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA, 2006. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage 31, 1536–1548. [DOI] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA, 2008. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage 40, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollmann S, Kasper L, Vannesjo SJ, Diaconescu AO, Dietrich BE, Gross S, Stephan KE, Pruessmann KP, 2017. Analysis and correction of field fluctuations in fMRI data using field monitoring. Neuroimage 154, 92–105. [DOI] [PubMed] [Google Scholar]
- Bright MG, Bulte DP, Jezzard P, Duyn JH, 2009. Characterization of regional heterogeneity in cerebrovascular reactivity dynamics using novel hypocapnia task and BOLD fMRI. Neuroimage 48, 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch JR, Talavage TM, Ulmer JL, Nyenhuis JA, 2002. Simulation of human respiration in fMRI with a mechanical model. IEEE Trans Biomed Eng 49, 700–707. [DOI] [PubMed] [Google Scholar]
- Byrge L, Kennedy DP, 2018. Identifying and characterizing systematic temporally-lagged BOLD artifacts. Neuroimage 171, 376–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannesson M, Besnard C, Durand PG, Bohe J, Jacques D, 2005. Relation between respiratory variations in pulse oximetry plethysmographic waveform amplitude and arterial pulse pressure in ventilated patients. Crit Care 9, R562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carry PY, Baconnier P, Eberhard A, Cotte P, Benchetrit G, 1997. Evaluation of respiratory inductive plethysmography: accuracy for analysis of respiratory waveforms. Chest 111, 910–915. [DOI] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH, 2009. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage 44, 857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH, 2009. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage 47, 1381–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Miranda-Dominguez O, Snyder AZ, Perrone AA, Earl EA, Van AN, Koller JM, Feczko E, Klein RL, Mirro AE, Hampton JM, Adeyemo B, Laumann TO, Gratton C, Greene DJ, Schlaggar B, Hagler D, Watts R, Garavan H, Barch DM, Nigg JT, Petersen SE, Dale A, Feldstein-Ewing SW, Nagel BJ, Dosenbach NUF, 2018. Correction of respiratory artifacts in MRI head motion estimates. bioRxiv, 337360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Bijsterbosch JD, Harrison SJ, Harms MP, Anticevic A, Van Essen DC, Smith SM, 2018. Using temporal ICA to selectively remove global noise while preserving global signal in functional MRI data. Neuroimage 181, 692–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, Consortium WU-MH, 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golestani AM, Chang C, Kwinta JB, Khatamian YB, Jean Chen J, 2015. Mapping the end-tidal CO2 response function in the resting-state BOLD fMRI signal: spatial specificity, test-retest reliability and effect of fMRI sampling rate. Neuroimage 104, 266–277. [DOI] [PubMed] [Google Scholar]
- Gotts SJ, Simmons WK, Milbury LA, Wallace GL, Cox RW, Martin A, 2012. Fractionation of social brain circuits in autism spectrum disorders. Brain 135, 2711–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JE, 2016. Guyton and Hall textbook of medical physiology, 13th edition ed. Elsevier, Philadelphia, PA. [Google Scholar]
- Heinzer R, Vat S, Marques-Vidal P, Marti-Soler H, Andries D, Tobback N, Mooser V, Preisig M, Malhotra A, Waeber G, Vollenweider P, Tafti M, Haba-Rubio J, 2015. Prevalence of sleep-disordered breathing in the general population: the HypnoLaus study. Lancet Respir Med 3, 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Fukunaga M, de Zwart JA, van Gelderen P, Fulton SC, Balkin TJ, Duyn JH, 2008. Low frequency BOLD fluctuations during resting wakefulness and light sleep: a simultaneous EEG-fMRI study. Hum Brain Mapp 29, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Kanno I, Ibaraki M, Hatazawa J, Miura S, 2003. Changes in human cerebral blood flow and cerebral blood volume during hypercapnia and hypocapnia measured by positron emission tomography. J Cereb Blood Flow Metab 23, 665–670. [DOI] [PubMed] [Google Scholar]
- Kastrup A, Kruger G, Glover GH, Neumann-Haefelin T, Moseley ME, 1999. Regional variability of cerebral blood oxygenation response to hypercapnia. Neuroimage 10, 675–681. [DOI] [PubMed] [Google Scholar]
- Li P, Janczewski WA, Yackle K, Kam K, Pagliardini S, Krasnow MA, Feldman JL, 2016. The peptidergic control circuit for sighing. Nature 530, 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Yackle K, 2017. Sighing. Curr Biol 27, R88–R89. [DOI] [PubMed] [Google Scholar]
- Natalini G, Rosano A, Franceschetti ME, Facchetti P, Bernardini A, 2006. Variations in arterial blood pressure and photoplethysmography during mechanical ventilation. Anesth Analg 103, 1182–1188. [DOI] [PubMed] [Google Scholar]
- Nierat MC, Dube BP, Llontop C, Bellocq A, Layachi Ben Mohamed L, Rivals I, Straus C, Similowski T, Laveneziana P, 2017. Measuring Ventilatory Activity with Structured Light Plethysmography (SLP) Reduces Instrumental Observer Effect and Preserves Tidal Breathing Variability in Healthy and COPD. Front Physiol 8, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin MJ, Liang PJ, Robbins PA, 1996. Dynamics of the cerebral blood flow response to step changes in end-tidal PCO2 and PO2 in humans. J Appl Physiol (1985) 81, 1084–1095. [DOI] [PubMed] [Google Scholar]
- Power JD, 2017. A simple but useful way to assess fMRI scan qualities. Neuroimage 154, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, 2019. Temporal ICA has not properly separated global fMRI signals: A comment on Glasser et al. (2018). Neuroimage. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE, 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Laumann TO, Plitt M, Martin A, Petersen SE, 2017a. On Global fMRI Signals and Simulations. Trends Cogn Sci. [DOI] [PubMed] [Google Scholar]
- Power JD, Lynch CJ, Gilmore AW, Gotts SJ, Martin A, 2019a. Reply to Spreng et al. : Multiecho fMRI denoising does not remove global motion-associated respiratory signals. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Lynch CJ, Silver BM, Dubin MJ, Martin A, Jones RM, 2019b. Distinctions among real and apparent respiratory motions in human fMRI data. Neuroimage 201, 116041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Plitt M, Gotts SJ, Kundu P, Voon V, Bandettini PA, Martin A, 2018. Ridding fMRI data of motion-related influences: Removal of signals with distinct spatial and physical bases in multiecho data. Proc Natl Acad Sci U S A 115, E2105–E2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Plitt M, Laumann TO, Martin A, 2017b. Sources and implications of whole-brain fMRI signals in humans. Neuroimage 146, 609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Silver BM, Silverman MR, Ajodan EL, Bos DJ, Jones RM, 2019c. Customized head molds reduce motion during resting state fMRI scans. Neuroimage 189, 141–149. [DOI] [PubMed] [Google Scholar]
- Raj D, Anderson AW, Gore JC, 2001. Respiratory effects in human functional magnetic resonance imaging due to bulk susceptibility changes. Phys Med Biol 46, 3331–3340. [DOI] [PubMed] [Google Scholar]
- Siegel JS, Mitra A, Laumann TO, Seitzman BA, Raichle M, Corbetta M, Snyder AZ, 2017. Data Quality Influences Observed Links Between Functional Connectivity and Behavior. Cereb Cortex 27, 4492–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyser CD, Inder TE, Shimony JS, Hill JE, Degnan AJ, Snyder AZ, Neil JJ, 2010. Longitudinal analysis of neural network development in preterm infants. Cereb Cortex 20, 2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H, 2014. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron 82, 695–708. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, von Wegner F, Morzelewski A, Brodbeck V, Jahnke K, Laufs H, 2013. Breakdown of long-range temporal dependence in default mode and attention networks during deep sleep. Proc Natl Acad Sci U S A 110, 15419–15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, Consortium WU-MH, 2013. The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I, 2004. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage 21, 1652–1664. [DOI] [PubMed] [Google Scholar]
- Yuan H, Zotev V, Phillips R, Bodurka J, 2013. Correlated slow fluctuations in respiration, EEG, and BOLD fMRI. Neuroimage 79, 81–93. [DOI] [PubMed] [Google Scholar]
- Yumino D, Bradley TD, 2008. Central sleep apnea and Cheyne-Stokes respiration. Proc Am Thorac Soc 5, 226–236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.