Abstract
Understanding the aspects of cell functionality that account for disease mechanisms or drug modes of action is a main challenge for precision medicine. Classical gene-based approaches ignore the modular nature of most human traits, whereas conventional pathway enrichment approaches produce only illustrative results of limited practical utility. Recently, a family of new methods has emerged that change the focus from the whole pathways to the definition of elementary subpathways within them that have any mechanistic significance and to the study of their activities. Thus, mechanistic pathway activity (MPA) methods constitute a new paradigm that allows recoding poorly informative genomic measurements into cell activity quantitative values and relate them to phenotypes. Here we provide a review on the MPA methods available and explain their contribution to systems medicine approaches for addressing challenges in the diagnostic and treatment of complex diseases.
Keywords: systems biology, mathematical models, signaling pathways, disease mechanism, transcriptomics, networks
Background
Since the beginning of the century, two technological waves (first microarray and subsequently massive sequencing) have been producing an increasingly vast amount of genomic data on gene variation and gene expression for different phenotypes, diseases and conditions [1]. As these methodologies gain in maturity, different statistical methods were used to derive progressively reliable and robust biomarkers and gene signatures [2] that, despite not being exempt of problems [3, 4], have contributed toward increasing the knowledge on the relationships between genotypes and phenotypes and have become mainstream in clinics [5, 6]. A major drawback from biomarkers and gene signatures is that they do not have an easy interpretation because they frequently lack any mechanistic link to the fundamental cellular processes that account for the phenotype studied. Actually, phenotypic differences are better understood as alterations in the operation of functional modules in the cell, which can be multiprotein complexes, a pathway or a single cellular or subcellular organelle [7]. Such alterations can be caused by different combinations of perturbations (mutations or gene expression changes) of functionally related genes [8, 9]. The interest in defining these biological modules and using them to interpret gene-based analyses resulted in early proposals of functional enrichment methods [10–12]. Such approaches consider functional modules just as plain lists of genes without taking into account either the underlying pathway network topology or the involved functional relations among genes. In the simplest approach, the over-representation analysis (ORA), the fraction of genes belonging to a particular gene set found within a predefined list of genes (e.g. genes showing significant changes in expression) is assessed [13, 14] (Figure 1B and C). A subsequent and more sensitive approach, the gene set enrichment analysis (GSEA), does not need a predefined list of genes but rather searches for functionally related gene sets constitutively up or down within all the genes studied, ranked by a relevance criterion (e.g. differential expression or association to a disease) [15–20]. Typical gene sets used in GSEA are gene ontology [21] categories, but other applications include gene sets sharing common regulation by transcription factors [22] or miRNAs [23], or even assess the impact of regulatory dynamics within functional gene sets, like the ROMA method [24]. However, similarly to ORA, GSEA methods ignore the complex network of functional relations among the proteins that compose the gene sets. Another generation of methods, generically known as pathway topology-based (PT-based) [11] or structure-based [25] methods, constitute different versions of GSEA for structured gene sets (pathways defined in the Kioto Encyclopedia of Genes and Genomes (KEGG) [26], Reactome [27], etc.). Contrarily to ORA and GSEA, these methods take into account the internal relationships among the proteins that compose the pathway (see Figure 1D and 1E for an example of the importance of internal gene–gene connections in the definitions of gene sets). Methods such as SPIA [28], CePa [29], NetGSA [30] or Pathifier [31] use different strategies to produce an enrichment value of the pathway, which is weighted according to the relationships among the activated constituent genes. Even with this approach, assessing and scoring whole pathway activities limits our understanding on the actual functionalities, triggered by specific subpathways of the pathway, carried out by the cell in the conditions studied. Actually, these methods ignore the obvious fact that many pathways are multifunctional entities composed of different subpathways that often may trigger very different and even opposite cell behavioral outcomes, depending on the dynamic progression of the signal propagation between the receptor and the effector proteins. For example, the apoptosis pathway may trigger cell survival or death depending on the specific receptor activated and how the signal is transduced within the pathway to reach a specific effector protein at the end of the signaling circuit. Actually, the most frequently used pathway descriptions (KEGG [26], Reactome [27], Pathway Commons [32], WikiPathways [33], etc.) are abstract concepts built around whole biological notions (e.g. apoptosis or cell cycle) and can be understood as compendiums of different possible cell activities that ultimately account for phenotypes (e.g. disease mechanisms or drug mechanisms of action—MoA). These cell activities are caused by the combined action of different genes that, in turn, are highly pleiotropic and often participate in more than one cell activity. This is the reason why gene activity by itself is often not descriptive of the cell functionality because it depends critically on the activity of other partner genes in the pathway to produce specific cell functionalities, as described in the map of interactions that is the pathway. On the other hand, the activity of the whole pathway does not provide a solution either. Activity measurement at the level of the whole pathway does not have a unique mechanistic consequence, and therefore, its use for the interpretation of genomic or transcriptomic experiments is purely indicative and of limited practical utility. Actually, methods like OCSANA [34] that seek minimal combinations of interventions to disrupt the subnetworks that connect source nodes and target nodes point to substructures within the pathway instead of whole pathways as the relevant entities to target. Moreover, evidences from the experimental side consisting of the prediction of patient survival based on the activity of the JNK subpathway provided a solid demonstration that subpathways can be considered real mechanistic biomarkers of clinical utility [35].
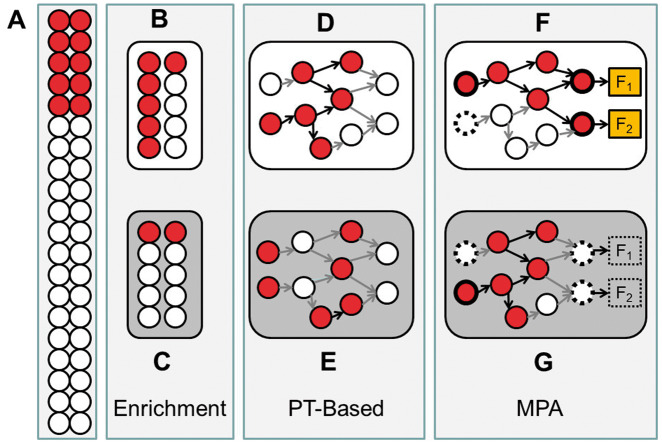
Figure 1.
Schematic representation of the three families of methods: enrichment analysis, PT-based analysis and MPA. The conventional enrichment analysis assumes the existence of a background (A) in which an observed percentage (25% in the example) of the genes differentially expressed (or mutated, associated to a trait, etc.). If gene sets are sampled based on some property shared by all the genes (e.g. they belong to a given pathway), a scenario (B) in which 60% of them are differentially expressed is found; the application of a simple test will evidence that this gene set is significantly enriched in differentially expressed genes, whereas in other scenarios (C), the gene set would not be different from a random sample of genes from the background. A PT-based algorithm takes into consideration the topology of the gene set, and a scenario (C) in which the differentially expressed genes are more connected among them would get a better score that an alternative scenario (D) in which the level of connection of the genes is lower. The significance of this data set would depend on the algorithm that estimates the score and the specific test applied. In MPA, there is more or less specific definition of circuits (subnetworks) within the pathway that should be related to cell activity in some way, and the connectivity of such circuits will determine the potential changes in cell activity. If circuits define subnetworks connecting receptor proteins to effector proteins in a signal transduction pathway, the same number of active genes could allow signal transduction (E) or being incompatible with the arrival of the signal to the current effector proteins (F), even in scenarios that would be significantly enriched in a conventional enrichment method.
In an attempt to gain a more precise knowledge on the functional consequences of the activity of pathways, methods that focus on mechanistic pathway activity (MPA) have emerged. MPA methods constitute a new paradigm by providing an alternative way to study the specific activities of internal elementary subpathways or circuits that compose the whole pathway. Circuits are subpathways, defined in different ways by different MPA methods, which are expected to be better descriptors of cell functional activity than whole pathways or single genes. Circuit definitions made by different MPA methods range from simple cliques or strings of connected nodes, that most likely account for unspecific activity within the pathway [36–39], to subpathways with a sound biological foundation (e.g. receptor-to-effector signaling circuits, which are defined from the chain of proteins that connect a specific receptor protein to a specific effector protein at the end of the signaling circuit) that are known to account for specific cell behavioral outcomes [40–43]. Under this mechanistic perspective, what is relevant is not the pure enrichment or a high connectivity by itself, but rather that the observed connectivity within the gene set will be compatible with a biological mechanism of the cell as, for example, signal transduction in a signaling pathway (Figure 1F and G). Most of the methods for computing circuit activity scores use gene expression values [36, 37, 40–42], but also methods that use mutations [44], have been proposed. The main difference with respect to PT-based methods is that MPA methods directly focus on pathway substructures and calculate the corresponding score for them rather than using such substructures to calculate a global score for the whole pathway.
MPA methods are likely to gain importance as systems medicine becomes increasingly relevant in understanding complex diseases and helping in their precise diagnosis and therapeutic decisions [45]. Initiatives such as Disease Maps (http://disease-maps.org/) have already provided detailed maps of complex gene relationships among genes in several diseases, such as cancer [46], Parkinson [47], Alzheimer [48] and influenza [49], and eight more are under development. These curated disease maps, along with the classical pathways, such as KEGG [26], Reactome [27], Pathway Commons [32], WikiPathways [33] and others [50], are detailed static diagrams whose dynamics and activity can be understood with MPA methods.
Despite the fact that numerous reviews of pathway enrichment methods can be found [10, 51], along with a few reviews on pathway-topology analysis methods [11, 25], to our knowledge, there are no reviews on specific methods that focus on testing independently the activity of pathway substructures (circuits). Here, we present a comprehensive review of MPA methods in which their relative advantages, drawbacks and performances are compared.
Methods
Methods analyzed
Contrarily to different versions of GSEA methods, such as PT-based [11] or structure-based [25] methods, in which pathway structures are taken into account to calculate a topology-weighted score for the whole gene set, MPA methods use pathway topologies to define subnetworks within, based on different criteria, which potentially constitute better descriptors of the real functional activity of the cell. MPA methods differ mainly in the way in which they define circuits within the pathways and in the manner that they estimate the activity of such circuits. A comprehensive collection of the available MPA methods is listed in Table 1. The most pervasively used pathway definition across all the methods is KEGG. Some methods use the KEGG pathway map to define circuits within with a biological rationale (e.g. receptor-to-effector circuits that effectively model signal transduction events), whereas others use the pathway topology to find subgraphs that behave differently among the compared conditions (Table 1). Scoring methods are also diverse, and in some cases, circuit activities can be calculated for individuals and then a test can be performed if a comparison is necessary (e.g. Pathiways [41, 42], HiPathia [40], developed by us, MinePath [52], TAPPA [39]), whereas other methods require of a comparison either to use differentially expressed genes (DEG) or to find subgraphs that explain the comparison. Another interesting aspect is that almost half of the methods do not distinguish between activations and inhibitions in the calculation of the score, despite the availability of this information and its potential importance (Table 1). Since circuit definition and scoring methods used will potentially have an impact on the relative performances of the methods, we have carried out a comparative benchmark to study their relative importance. From the methods listed in Table 1, nine were selected that satisfy three basic conditions: they can be applied to RNA-seq data, they have a common definition of pathway (KEGG pathways constituted the unique common pathway definition) and there is software available for running them.
Table 1.
List of mechanistic pathway activity methods
| Method | Date | Code | Pathway modeled | Circuit definition | Scoring method | Activation / inhibition | Input | Result | Scope |
|---|---|---|---|---|---|---|---|---|---|
| Hipathia [40] | 2017 | Web application | KEGG | Receptor-to-effector circuits | Propagation algorithm | Yes | MA, RNA-seq | P-value per circuit | Multiple analyses |
| http://hipathia.babelomics.org | |||||||||
| Hipathia R code | |||||||||
| MinePath [52] | 2016 | Web application | KEGG | All possible circuits | Discretized gene expression (GE) values with logical operators | Yes | MA RNA-seq | P-value per circuit | Multiple analyses |
| http://minepath.org/ | |||||||||
| subSPIA [53] | 2015 | R code | KEGG | Minimal spanning trees (MST) | Differentially expressed (DE) genes used to define the MST | No | MA RNA-seq | P-value of DE per circuit | Comparison |
| PathiVar [44] | 2015 | Web application | KEGG | Receptor-to-effector circuits | Probabilistic model | Yes | MA, VCF | P-value per circuit | Multiple analyses |
| http://pathivar.babelomics.org | |||||||||
| Pathome [54] | 2014 | NA | KEGG | Receptor-to-effector linear circuits | Correspondence between pattern activation/inhibition and co-expression in adjacent gene pairs | Yes | MA RNA-seq | P-value per circuit | Comparison |
| Pathiways [41, 42] | 2013 | Web application http://pathiways.babelomics.org | KEGG | Receptor-to-effector circuits | Probabilistic model | Yes | MA | P-value per circuit | Multiple analyses |
| DEAP [55] | 2013 | Python code | KEGG | Receptor-to-effector linear circuits | Running sum of discretized DE | Yes | MA RNA-seq | Maximally differential expressed pathway | Comparison |
| CLIPPER [37] and GraphiteWeb [56] | 2013 | R package | KEGG; Reactome | All possible circuits | Weighted sum of GE | No | MA RNA-seq | Most relevant circuit per pathway | Comparison |
| ToPASeq R package | |||||||||
| Web application: | |||||||||
| http://graphiteweb.bio.unipd.it/ | |||||||||
| TEAK [57] | 2013 | Code @ Google (Windows and Mac) | KEGG | Receptor-to-effector circuits | Fits a Bayesian network for circuit and uses the BIC | No | MA | Ranked circuits | Comparison |
| PRS [58] | 2012 | ToPASeq R package | KEGG | Trees of associated DE genes | Topologically weighted sum of DE | No | MA RNA-seq | Ranked subpathways | Comparison |
| DEGraph [36] | 2012 | R package | KEGG; User defined pathways | All possible circuits | Multivariate two-sample tests of means of DE genes within a subgraph. | No | MA RNA-seq | P-value of DE per circuit | Comparison |
| ToPASeq R package | |||||||||
| Rivera et al. [59] | 2012 | NA | NetPath | All possible circuits | Weighted Z-score of genes within the subgraph | No | MA | P-value of most perturbed circuit | Comparison |
| Chen et al. [60] | 2011 | NA | KEGG | Receptor-to-effector circuits | Euclidian distance | No | MA | P-value per circuit | Comparison |
| PWEA [61] | 2010 | ToPASeq R package | User defined pathways | All possible circuits | Mutual influence among gene expression within the circuit | No | MA RNA-seq | P-value of DE per pathway | Comparison |
| TopologyGSA [38] | 2010 | ToPASeq R package | User defined pathways | All possible circuits | Comparison of covariance matrices of genes in the circuit | Yes | MA RNA-seq | P-value of DE per pathway | Comparison |
| DEGAS [62] | 2010 | Java (Windows) | KEGG | All possible circuits | Heuristic to find the largest dysregulated circuit | No | MA | One circuit per pathway | Comparison |
| TAPPA [39] | 2007 | ToPASeq R package | KEGG | All possible circuits | Scores of co-expression that explain the compared conditions | No | MA RNA-seq | P-value of DE per pathway | Multiple analyses |
The first column (Method) contains the name or acronym of the method, if exists, otherwise, we refer to it as the first author of the publication. The second column (Date) contains the publication date. The third column (Code) informs on the availability of the code to run the method. The fourth column (Pathway modeled) indicates the database used for pathway definition used by the method. The fifth column (Circuit definition) is the type of circuit used by the method. The sixth column (scoring method) summarizes how the circuit activity is inferred in the method. The seventh column (Activation/inhibition) denotes whether the scoring method uses the information of activation or inhibition nodes. The eight column (Input) indicates the data type that inputs the method (MA: expression microarray; RNA-seq: counts of RNA-seq experiments; VCF: mutation files). The ninth column (Result) describes the results provided by the method. And the tenth column (Scope) indicates the type of analyses the method permits, which can be either only conventional two conditions comparison or a wide range of analyses if the method first recodes gene expression into circuit activities.
MPA algorithms
TAPPA [39] was the first MPA method proposed by 2007. Although originally proposed as a PT-based algorithm, its implementation in the ToPASeq package [63] allows using it on not only whole pathways but also subpathways. The method is based on the concept of molecular connectivity, a well-known topological descriptor of chemical compounds in chemioinformatics [64]. In essence, the method calculates a score of gene co-expression, taking into account the adjacency in the topology of the pathway. Here, circuits are defined as all the possible subnetworks in the pathway. This method does not take into account activation/inhibition activities of the genes. Circuits with a molecular connectivity associated to the conditions compared by means of a Mann–Whitney test (for binary traits) or a Spearman correlation (for continuous traits) are considered to have a significant differential activity. Significant circuits that do not connect receptors to effector proteins are most probably not involved in triggering changes of cell functionality and consequently they will not be relevant to explain changes in cell behavior.
PWEA [61] was proposed in 2010. Similarly to TAPPA, this method was originally proposed as a PT-based algorithm, but the ToPASeq [63] implementation allows using it on subpathways. The method calculates for each gene its mutual influence with respect to the rest of genes within the pathway (or subpathway), obtained as a function of the correlation in their expression values. Then, for each pathway (or subpathway), it calculates a cumulative function based on all the genes within. The significance of this cumulative function is tested by means of a permutation test to compare the intrapathway with respect to the background (out of the pathway cumulative mutual influence). In these calculations, activation or inhibition gene activities are ignored. In the ToPASeq implementation, the circuits made out of KEGG pathways were defined as any subnetwork within the pathways, which again can define circuits not linked to the activation of cell functions.
CLIPPER [37], published in 2012, is a generalization of TopologyGSA [38]. The first step of the method converts pathways into gene–gene networks. Given that CLIPPER method only works on acyclic graphs, self-loops and cycles are solved by removing the weakest edge of the cycle based on expression data (with minimum expression profile correlation between nodes [65]). Then, the circuits are defined as receptor-to-effector subnetworks. Gene expression values are assigned to the nodes of the circuits, and the algorithm compares portions (any possible subnetwork) of the circuit between the conditions studied, with the aim of identifying subgroups of genes that appear to drive differences (deregulations) between them. CLIPPER does not distinguish between activation and inhibition activities in the proteins of the pathway. If the portion found as significant does totally connect receptors to effector proteins, the subnetworks detected could not have any relevance in terms of cell functionality.
PRS [58] was proposed in 2012. The algorithm maps discretized values of differential gene expression (a threshold of twofold is taken as differential expression and value is set to 1, otherwise to 0, without any further statistical assessment) at the nodes of the gene–gene directed network extracted from KEGG pathways, without considering their activation or inhibition roles. Then, the network is traversed with a depth-first search algorithm that essentially seeks for subnetworks of connected differentially expressed nodes by means of a normalized weighted sum of node values. A permutation test is used to assess the reliability of the subnetworks found. These significant subnetworks constitute the circuits defined by PRS, which, as in CLIPPER, could lack any mechanistic link with changes in cell functionality, if they do not connect receptors to effector proteins.
DEGraph [36] was proposed in 2012. This is a complex statistical approach in which first the topology of the graph representing the pathway undergoes Fourier analysis in which some additional information based on gene expression correlations is incorporated. Then, the method performs a Hotelling’s test in the graph–Fourier space for discovering non-homogeneous subgraphs within the whole pathway topology that exhibit a significant shift in means, by means of a pruning approach to reduce testing time and multiple testing. The way in which the algorithm defines circuits as significant subnets, extracted from the analysis of the graph–Fourier space, does not take into account activation/inhibition gene activities and do not specifically focus on circuits that can mechanistically explain cell activity.
DEAP [55] was proposed in 2013. The algorithm superimposes expression data onto the pathway graph. Every possible path that can be formed within the graph is independently examined by using a recursive function that calculates the differential expression for each path by adding or subtracting the discretized differential expression value of all downstream nodes with catalytic or inhibitory relationships, respectively. DEAP focuses on discovering the most differentially expressed portion of the pathway. Again, from a pure mechanistic point of view, it could be argued that the activity of some portions of the pathway does not necessarily involve a functional activity within the pathway.
SubSPIA [53] was proposed in 2015. SubSPIA maps the DEG into the pathway structure. Then, the method tries to connect all the possible DEG using the minimum possible number of non-DEGs by using a minimum spanning tree strategy. Activations and inhibitions are not taken into account. Circuits defined in this way do not necessarily involve a functional activity within the pathway.
MinePath [63] was proposed in 2016. The method first discretizes the expression values of the genes to active/inactive categories. Then, each pathway is decomposed in all the possible subgraphs, over which the gene expression categories are mapped. The functional status of any subgraphs is assessed by the Boolean algorithm that takes into account the activity of the involved genes and the type of relationship among them (activations and inhibitions). When the activity status changes between the conditions are compared, the subgraph is considered to be a differentially activated circuit. Again, the circuits found under this strategy do not necessarily involve a functional activity within the pathway.
HiPathia [40] was proposed by us in 2017. The method defines circuits as subgraphs that connect receptor proteins to effector proteins in the pathways. These types of circuits represent the logic of signal transduction in the cell and are related to the mechanistic of cell functionality. HiPathia uses the normalized gene expression values as proxies of protein activity and, taking into account the relationships among proteins (activation/inhibition) along the pathway, it uses a propagation algorithm to estimate the amount of signal that arrived to the effector protein from the receptor protein. Then, the profiles of circuit activity are compared by means of a conventional Wilcoxon test.
Data source
A total of 6246 samples, including 5647 tumor samples and 598 samples taken from the healthy reference tissue, belonging to 12 cancer types from The Cancer Genome Atlas (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga/) were used for the benchmarking. The following cancers were used: bladder urothelial carcinoma [66], breast invasive carcinoma [67], colon adenocarcinoma [68], head and neck squamous cell carcinoma [69], kidney renal clear cell carcinoma [70], kidney renal papillary cell carcinoma [71], liver hepatocellular carcinoma, lung adenocarcinoma [72], lung squamous cell carcinoma [73], prostate adenocarcinoma [74], thyroid carcinoma [75] and uterine corpus endometrial carcinoma [76].
Fourteen KEGG [26] pathways belonging to the subcategory of ‘Pathways in cancer’ were used to detect changes when cancers versus control comparisons were made. These are MAPK signaling pathway (hsa04010), Wnt signaling pathway (hsa04310), TGF-beta signaling pathway (hsa04350), VEGF signaling pathway (hsa04370), Jak-STAT signaling pathway (hsa04630), cAMP signaling pathway (hsa04024), PI3K-Akt signaling pathway (hsa04151), mTOR signaling pathway (hsa04150), cell cycle (hsa04110), apoptosis (hsa04210), p53 signaling pathway (hsa04115), focal adhesion (hsa04510), adherens junction (hsa04520) and PPAR signaling pathway (hsa03320).
Data processing
The COMBAT method [77] was used to remove non-biological experimental variations (batch effect) associated to a Genome Characterization Center (GCC) and plate ID from the RNA-seq data. Then, the trimmed mean of M-values normalization method (TMM) method [78] was used for data normalization. The resulting normalized values were used as input for the MPA methods.
Sensitivity of the methods
To estimate sensitivity or the true positive rate (TPR), we used the 12 cancer types mentioned earlier. In any of the 12 normal versus tumor comparison, we expect to detect changes of activity in the 14 cancer KEGG pathways. As different methods have different circuit definitions, the comparison of the methods was carried out at the level of the whole pathway definition. This is a quite liberal estimation because the detection of activity within a pathway by two methods does not mean that the activity detected really corresponds to the same cell behavioral outcome. However, possible overoptimistic TPRs obtained by this approximation will be compensated by low specificity determinations. Therefore, for each method, we estimated a rate of true positive as the number of cancer pathways in which the method discovered one or more differentially activated circuits divided by 12, the total number of cancer pathways. The distributions were ranked by TPR value and compared by means of a Wilcoxon test with Bonferroni correction to detect significant differences among them.
Specificity of the methods
The false positive rate (FPR) is estimated by comparing two groups composed by identical individuals for discovering differentially activated circuits in any of the cancer KEGG pathways. As the individuals compared belong to the same cancer type, any differentially activated circuit would be a false positive detection. For each comparison, we used two data sets of 25 tumor samples, each randomly chosen from each of the cancer types analyzed. Comparisons between both data sets were repeated 100 times per method and cancer. Similarly to the case of TPR, the FPR of any method is calculated as the number of cancer KEGG pathways in which the method finds one or more circuits significantly activated divided by the total number of pathways analyzed (14 pathways). Again, the distributions were ranked by FPR value and compared by means of a Wilcoxon test with Bonferroni correction to detect significant differences among them. If a method has a too liberal detection of active pathways, its high sensibility will be compensated with a low specificity.
Results
Data pre-processing
A large data set of RNA-seq counts for 12 cancer types analyzed was used for the benchmark provided in this review. The data were downloaded from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). The data were normalized using TMM [78] to account for RNA composition bias. Batch effect was corrected by the application of the COMBAT [77] method. Normalized data were used as input for the MPA methods.
Estimation of the sensitivity of the MPA methods
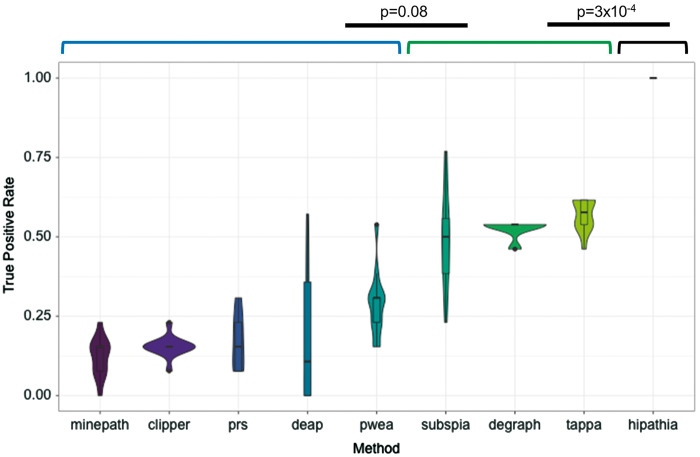
To estimate the relative TPR of the MPA methods analyzed, we compared cancer samples versus the corresponding healthy tissue for the 12 cancer types analyzed. In all these cancer versus control comparisons, statistically significant differences in the KEGG cancer-associated pathways are expected. Figure 2 shows how only HiPathia was able to detect changes in activity in circuits belonging to all the cancer pathways analyzed across the 12 cancer types analyzed. Other group of three methods (TAPPA, DEGraph and subSPIA), with a significantly different performance (P-value = 3 × 10−4), was able to detect between 50% and 75% of the cancer pathways used here. The rest of methods detected differential activity in less than 50% of the cancer pathways.
Figure 2.
TPR or sensitivity was computed as the number of significant cancer pathways found, when cancer samples are compared with samples of the tissue of reference, divided by the total number of cancer pathways (14 for HiPathia and DEAP and 13 for the rest of methods, because PPAR signaling pathway [hsa03320] was not implemented in them) per method and cancer. Violin plots obtained using 12 cancer types show for any method the mean TPR in the central dot, all possible results, with thickness indicating how common, in the outer shape and the layer inside, represents the values that occur 95% of the time. The figure shows the methods ranked by TPR value. A Wilcoxon test with Bonferroni correction was used to compare successive TPR distributions to detect significant differences among them. Black lines denote significant differences between consecutive methods. Brackets define groups of methods with no significant differences in their performances.
Estimation of the specificity of the MPA methods
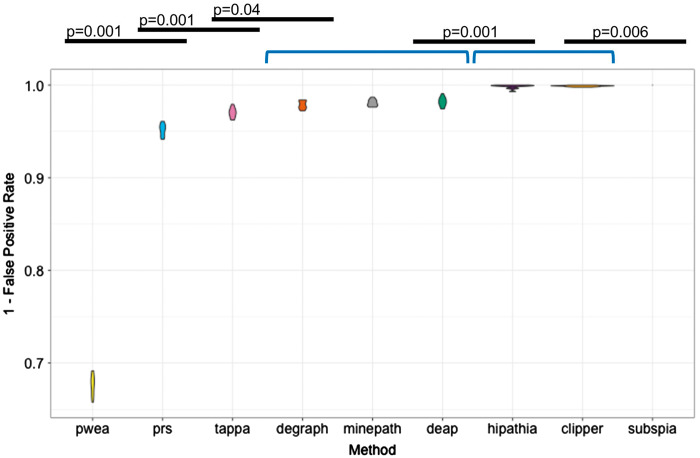
To check whether the high sensitivity of HiPathia, TAPPA, DEGraph and subSPIA is real or is only the consequence of a low specificity, we estimated the FPR for them. To achieve this, data sets of identical samples were compared and significant differences in circuit activity found by a particular method in the comparisons are considered false positives. A total of 10,800 comparisons (100 times × 9 methods × 12 cancer types) between pairs of data sets of 25 samples each, randomly sampled among cancer samples, were performed. The FPR was computed as the mean of the number of significant cancer pathways divided by the total number of cancer pathways per method and cancer. Figure 3 shows how most of the methods have a low FPR (below the conventional 5% P-value), except PWEA, which displays a high ratio of false positives (over 30%). The best performer is SubSPIA (P-value = 0.005), which, together with CLIPPER and HiPathia methods, showed the highest specificity (P-value = 0.001).
Figure 3.
FPR or specificity was computed as the mean of the number of significant cancer pathways found, when cancer samples are compared with cancer samples, divided by the total number of KEGG cancer pathways along 100 bootstraps, per method and cancer. Violin plots show average values and distributions of the proportions of false discoveries made by any method. The figure shows the methods ranked by FPR value. A Wilcoxon test with Bonferroni correction was used to compare successive FPR distributions to detect significant differences among them. Black lines denote significant differences between consecutive methods. Brackets define groups of methods with no significant differences in their performances.
An example of MPA analysis of death and postmortem cold ischemia of blood transcriptome
Beyond the typical cancer study applications showed in all the MPA method articles, other biological systems can be studied at an unprecedented functional detail with the application of MPA methods. In a recent article [79], we studied the changes of blood transcriptome after death and subsequent postmortem cold ischemia and the functional consequences using the data available in the GTEx project [80]. Although changes in the transcriptome are expected as a response to the death, little is known about how death and the postmortem cold ischemia interval affect gene expression [81, 82]. In principle, gene expression measurements in postmortem samples will be affected both by biological responses to organism death and RNA degradation. MPA methods can help to understand the mechanisms of these effects and how they are dependent on the postmortem interval. This knowledge is essential for the proper use of postmortem gene expression measures as a proxy for antemortem physiological gene expression levels [83].
In the study, most of the changes in gene expression occur between 7 and 14 h after death, followed by a relative stabilization, between 14 and 24 h. The use of an enrichment method pointed to several processes affected by the gene expression changes, which include gene ontology terms related to immune response such as response to bacterium, response to other organisms, response to biotic stimulus, regulation of leukocyte migration, regulation of cytokine production and acute-phase response, to DNA replication, such as DNA packaging process, as well as some metabolic processes related to hypoxia, such as reactive oxygen process, peroxidase activity, oxidoreductase activity, carbohydrate binding and response to lipopolysaccharide [79]. All of them are very general processes and most of them can have an opposite outcome depending on the specific activity of the process (e.g. regulation of cytokine production can be positive or negative and there is no way of discovering the sense of the regulation from the pure enrichment of this ambiguous term).
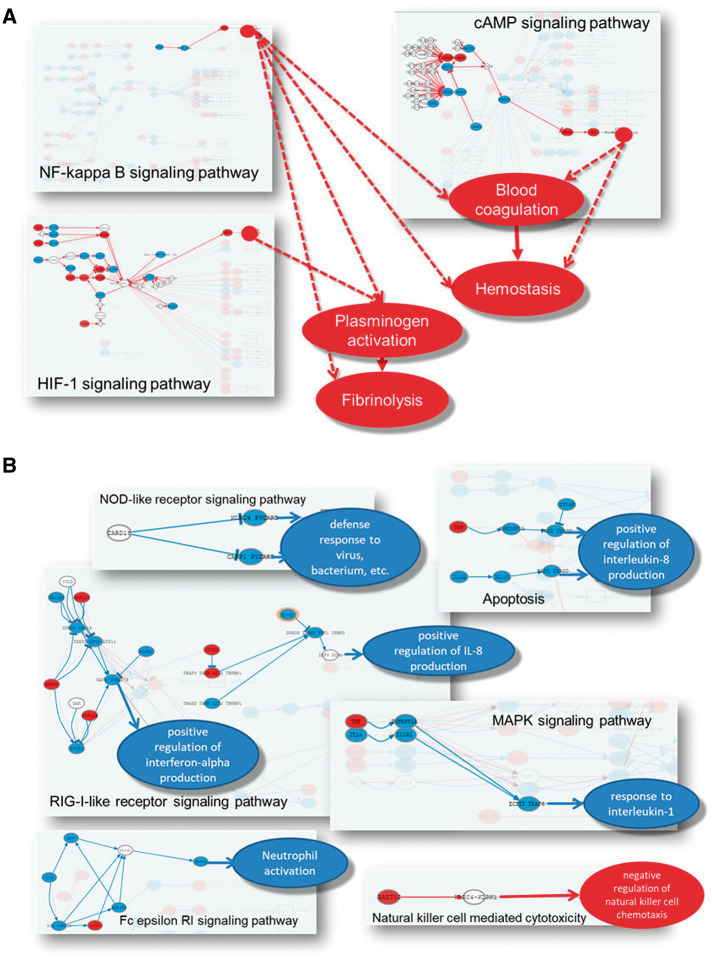
However, the application of the HiPathia [40] MPA method revealed significant changes in several relevant functional activities. Actually, HiPathia reveal a detailed picture on the ultimate mechanisms that cause the deactivation of the immune system. Regulation of interferon production is inhibited from several circuits of the apoptosis RIG-I-like receptor and the RIG-I-like receptor signaling pathways. Response to interleukin-1 is inhibited from the MAPK signaling pathway. Neutrophil activation is inhibited by a circuit of the Fc epsilon RI signaling pathway, whereas negative activity of natural killer cells is triggered from the natural killer cell-mediated cytotoxicity pathway (Figure 4A). As suggested by enrichment carbohydrate metabolism is affected, but MPA detects the mechanism behind, which involves a severe deactivation of the tricarboxylic acid cycle, while glycolysis is activated (FDR-adjusted P-value <10−27; [79] Figure 4D), and points to a major role in the initial pre- to postmortem transition for the hypoxia process (FDR-adjusted P-value 7.2 × 10−67), by the activation of HIF-1, mTOR, platelet activation and cGMP-PKG signaling pathways [79].
Figure 4.
Schema of the mechanisms behind the deactivation of the immune system (A) and the changes in the metabolism (B) caused by changes of blood transcriptome after death and subsequent postmortem cold ischemia [73].
Finally, activation of processes related to blood coagulation (FDR-adjusted P-value 1.52 × 10−54) and response to stress triggered by specific circuits in NF-kappa B, cAMP and HIF-1 signaling pathways (Figure 4B) are also detected by the MPA method.
Discussion
MPA, a new paradigm for the interpretation of genomic data
Transcriptomic experiments are affordable nowadays and provide a wealth of data that must be interpreted in the light of their biological consequences and implications. Different functional profiling methods have been proposed for this interpretation, such as over-representation methods or gene-set enrichment methods [15–18], that focus on the collective activity of genes within biologically relevant entities such as pathways. However, given that most pathways are multifunctional entities, these methods, even in their most sophisticated versions that consider the internal structure of the pathway, are simply illustrative and fail to provide real mechanistic information on specific behavioral outcomes of the cell. MPA methods provide an innovative, biologically inspired alternative for the interpretation of transcriptomic experiments. These methods use biological knowledge available on the interrelation among the genes that compose the pathways to provide hypothesis on how their perturbations ultimately cause diseases, responses to treatments or other complex phenotypes, such as the effects of death and postmortem cold ischemia on human tissues [78]. Therefore, MPA methods provide a link between gene perturbations (measured as gene expression changes) and disease mechanisms or drug MoAs [83, 84]. To understand how the different pathway definition and scoring strategies used by the different MPA methods capture biological information that account for cell behavior (such as signal transduction circuit activities) and relate them to phenotypic conditions, we have produced a benchmarking of nine MPA methods that could be compared in similar conditions.
Possible factors that determine the relative performance of MPA methods
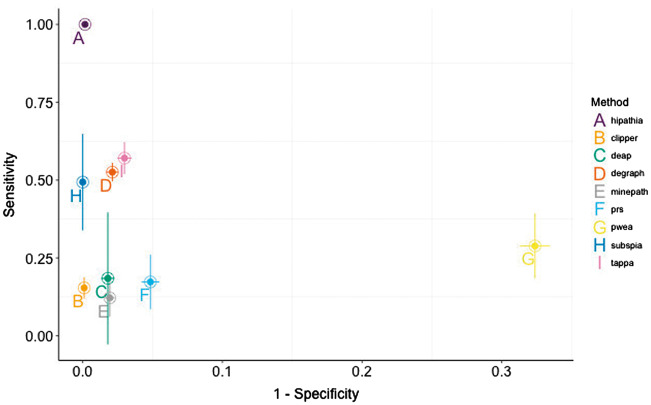
Figure 5 offers a combined view of both specificity and sensitivity of the methods analyzed in this review. Whereas most of the MPA methods show an excellent specificity (below the typical 5% conventional P-value threshold, with the exception of PWEA), the differences in terms of sensitivity are more pronounced. According to their relative sensitivity and specificity, we could distinguish four groups of methods. The first group of high sensitive and specific methods would include HiPathia, the only one able to detect differences in the activities of all the cancer pathways across all the cancer types analyzed while maintaining a high specificity as well. A second group of methods with medium sensitivity but still high specificity, which includes TAPPA, DEGraph and SubSPIA, detects changes in only 75%–50% of the cases (P-value = 0.08). A third group of methods, with low sensitivity but still high specificity, which includes CLIPPER, DEAP, MinePath and PRS, shows poorer performance, detecting changes in circuit activities in less than 25% of the cases. Finally, the conceptually most different method, PWEA, does not only present a low specificity but also a low sensitivity.
Figure 5.
Simultaneous comparison of sensitivities and specificities of the different MPA methods. The results obtained in the 12 cancers are used to obtain a mean value and an error. The x-axis represents 1 − the FPR. Horizontal bars represent in each point 1 SD of the FPR for the corresponding method.
It is difficult to attribute the relative performance of the methods to a unique factor and it rather seems to be a combination of several of them. Apparently, the use of receptor-to-effector definition of circuit and the distinction between activations and deactivations seem to be important factors that differentiate HiPathia from the rest of methods in terms of sensitivity. MinePath and DEAP also use activation/inhibition information to calculate the score and DEAP uses receptor-to-effector circuit definitions, but in both cases, the scoring algorithm uses discretized values of differential gene expression, which seem to reduce drastically the sensitivity. The most representative feature of the second group of methods seems to be the use of differential gene expression or co-expression to obtain scores for circuits. These circuits that can effectively separate the conditions compared are chosen as differentially activated. In the third group, showing poorer sensitivity (below 25%), the discretization of differential gene expression values seems to represent a hurdle for obtaining a better sensitivity for two of the methods (DEAP and MinePath). The case of CLIPPER and PRS is probably related to a combination between the scoring strategy and the circuit definition. Finally, the PWEA presents, in addition, a low specificity. Probably, the PWEA case is a combination of the circuit definition and a scoring algorithm, based on mutual influence among genes, which is not capturing properly the underlying biology of the pathway activity. It must be stated that all the methods included in the three first groups discover differentially activated circuits efficiently and with a low rate of false positives, thus providing a more informative interpretation than the simple description of differential gene expression. Moreover, all the methods in their original publications demonstrated to be more sensitive than the conventional ORA and GSEA methods [36, 37, 39, 40, 52, 53, 55, 58, 61].
Receptor-to-effector subpathways are relevant circuit definitions from a biological standpoint, as they represent the possible routes taken from the beginning of a pathway, where the signal is originated, to its end, where a function is triggered. Within the context of signaling pathways, such circuit definitions effectively model signal transduction events. MPA methods implementing these circuit definitions model more realistically biological events and consequently produce better results. In addition, an interesting feature of the methods that use receptor-to-effector circuits is that the changes in the activity of such circuits can be easily associated to cell functionalities triggered by the effector protein [40]. Contrarily, given the fact that many genes and subnetworks can be shared by several pathways, pathway definitions based in subnetworks are, in principle, more prone to false positives.
Most of the MPA methods are based on the use of differential gene expression or on any type of comparison-derived scoring system, which restricts its analytic use only to comparison scenarios. An interesting feature for a MPA method is the possibility of producing circuit values for each individual sample. This allows transforming an uninformative matrix of [samples × gene] expression values into a more informative matrix [samples × circuit activities] that can be used for detecting differentially active circuits, but also for many other types of analysis such as clustering, predictors, time-series analysis, survival analysis, patient stratification, etc. For example, the Pathiways method [41, 42] was used to predict cell response to different drugs [85].
Because most of the methods only accept KEGG pathways as input (Table 1), it is not possible to test potential biases of the different methods under different pathway definitions. In principle, given the way that the methods tested work, the two most important factors prone to cause any bias would be the length of the pathways and the existence of more or less loops. In principle, longer pathways would affect the performance in terms of runtime and, in some cases, could introduce a multiple testing issue in methods that decompose the pathway in all the possible subpathways. TAPPA and MinePath will probably be affected in their runtimes, and methods such as PRS of DEGraph even more and could even be impracticable for large pathway definitions, given the algorithm behind the definition of circuits. On the other hand, CLIPPER accuracy could be compromised by pathways with many loops, given that this algorithm breaks the loops to linearize the circuits.
Mechanistic biomarkers and systems medicine
Despite its unquestionable clinical utility, single-gene biomarkers and gene signatures are not exempt of problems [3, 4] due to the lack of any mechanistic link to the fundamental cellular processes that account for the mechanism of the disease [84]. Today’s empirical methods of diagnosis and therapeutic decision-making need to be transformed in ways that consider important facts such as the heterogeneity of the patients [86] and the modular nature of diseases [7], which are overlooked today. To achieve this transformation, the current method of classifying diseases needs to be modified from a phenotypic description to one that incorporates the different molecular drivers that created the observed phenotype. This can only be achieved through a deeper, systems-based understanding of these disease drivers [84]. Within this context, a mechanistic biomarker would be a multigenic biomarker that uses genes involved in the mechanism of a specific phenotype to estimate an activity value for cell functionalities related to, or directly accounting for, such a mechanism. The prediction of a complex phenotype such as patient survival based on a mechanism that is conceptualized as a mathematical model of the activity of the JNK signaling subpathway represents an example of a mechanistic biomarker [35]. MPA methods open the door to the systematic definition and use of mechanistic biomarkers [35]. Moreover, mechanistic biomarkers are not only diagnostic but also can be actionable entities that can efficiently and realistically address the problem of patient heterogeneity [86]. In fact, the concept of actionability within a systems medicine context is related to the activity of the mechanistic biomarker, rather than with the specific activity of a particular gene, and could require distinct interventions in different patients [35, 40]. Thus, MPA methods can be used to suggest and predict the effect of interventions (Knock Outs (KOs), drug inhibitions, over-expressions, etc.) on specific genes within circuits so as to find suitable clinical targets, predict side effects, speculate off-target activities, etc. This is the case of the PathAct application [87] that uses the HiPathia algorithm [40] to explore the effects of interventions that are simulated by directly changing the expression level of the genes, which can be considered an in silico equivalent to KOs, drug inhibitions or gene over-expressions, and can be carried out as individual interventions or in combinations. The use of actionable mechanistic biomarkers can pave the way for personalized and individualized therapies, especially in cancer, where many targeted therapies are already available. Another obvious area of application is drug discovery [83], where patient heterogeneity is behind the failure of many drugs [88].
In addition, the systems biology perspective provided by MPA approaches can be used to address other biological problems with an unprecedented mechanistic detail. An interesting problem is the assumption that the use of postmortem samples for transcriptomic studies provides a good representation of the antemortem transcriptome [81], despite the fact that changes in gene expression levels are expectable and actually little is known about how death and the postmortem cold ischemia can affect gene expression measurements [81, 82]. A recent study using data available from the GTEx project [80] revealed important changes of blood transcriptome after death and subsequent postmortem cold ischemia with relevant functional consequences such as deactivation of the immune system, metabolic changes as response to hypoxia, DNA synthesis arrest, diverse responses to stress and the activation of blood coagulation [79].
Conclusion
MPA methods constitute an evolution of pathway analysis methods in which pathways are decomposed into elementary subpathways or circuits that potentially account for cell outcomes that can help to explain mechanistic features of phenotypes (disease mechanism, drug MoA, etc.). Here we provide a review of MPA methods that include a limited benchmarking of sensitivity and specificity. From this comparison we concluded that, although most of the methods were highly specific, they presented remarkable differences in terms of sensitivity. From their relative performances it can be concluded that a biologically realistic definition of the circuits within the pathways analyzed is a major determinant of the success of the method. However, the scoring methodology, which accounts for the activity of the circuit, must also be representative of the biological activity of the cell. Thus, the propagation method used by HiPathia seems to be the most efficient solution, followed by scores based on differential gene expression, implemented by subSPIA, DEGraph and TAPPA. In any case, MPA methods have demonstrated to be more sensitive than the conventional functional analysis (ORA or GSEA) and represent a promising alternative for the interpretation of genomic measurements. Actually, the increasing importance of systems medicine to face the challenges of diagnosis and treatment of complex diseases [45] will increase the relevance and make more mainstream the use of approaches such as MPA methods.
Key Points
MPA methods focus on the activity of pathway substructures defined in different ways.
Biologically inspired pathway substructures like receptor-to-effector circuits and methods to score their activity are relevant for the performance of MPA methods.
MPA can help to discover actionable mechanistic biomarkers.
Funding
This work was supported by grants SAF2017-88908-R from the Spanish Ministry of Economy and Competitiveness and “Plataforma de Recursos Biomoleculares y Bioinformáticos” PT13/0001/0007 from the ISCIII, both co-funded with European Regional Development Funds (ERDF), and EU H2020-INFRADEV-1-2015-1 ELIXIR-EXCELERATE (ref. 676559).
Biographies
Alicia Amadoz is a postdoctoral fellow in the Bioinformatics Department of the Igenomix, with a background in biology. She currently carries out genomic data analysis for the company.
Marta R. Hidalgo is a postdoctoral fellow in the Clinical Bioinformatics Area of the Fundación Progreso y Salud, with a background in mathematics and computing sciences. Her current research interests include mathematical modeling of biological systems.
Cankut Çubuk is a PhD student at the Clinical Bioinformatics Area of the Fundación Progreso y Salud, with a background in molecular biology and bioinformatics. His current research interest includes mathematical modeling of pathways.
José Carbonell-Caballero is a researcher at the Center for Genomic Regulation, with a background in scientific computing and bioinformatics. His current research interest includes mathematical modeling of pathways.
Joaquin Dopazo is the Director of the Clinical Bioinformatics Area of the Fundación Progreso y Salud, with a background in genomic data analysis, systems biology and machine learning. His current research interests include medical genomics, systems medicine and mathematical modeling.
References
- 1. Kahvejian A, Quackenbush J, Thompson JF.. What would you do if you could sequence everything? Nat Biotechnol 2008;26(10):1125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi L, Campbell G, Jones WD, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol 2010;28(8):827–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iwamoto T, Pusztai L.. Predicting prognosis of breast cancer with gene signatures: are we lost in a sea of data? Genome Med 2010;2(11):81.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ein-Dor L, Zuk O, Domany E.. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA 2006;103(15):5923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;5(1):530–6. [DOI] [PubMed] [Google Scholar]
- 6. Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov 2012;2(1):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oti M, Brunner HG.. The modular nature of genetic diseases. Clin Genet 2007;71(1):1–11. [DOI] [PubMed] [Google Scholar]
- 8. Barabasi AL, Gulbahce N, Loscalzo J.. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12(1):56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barabasi AL, Oltvai ZN.. Network biology: understanding the cell's functional organization. Nat Rev Genet 2004;5(2):101–13. [DOI] [PubMed] [Google Scholar]
- 10. Dopazo J. Formulating and testing hypotheses in functional genomics. Artif Intell Med 2009;45(2–3):97–107. [DOI] [PubMed] [Google Scholar]
- 11. Khatri P, Sirota M, Butte AJ.. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 2012;8(2):e1002375.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jin L, Zuo X-Y, Su W-Y, et al. Pathway-based analysis tools for complex diseases: a review. Genomics Proteomics Bioinform 2014;12(5):210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Al-Shahrour F, Diaz-Uriarte R, Dopazo J.. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 2004;20(4):578–80. [DOI] [PubMed] [Google Scholar]
- 14. Draghici S, Khatri P, Bhavsar P, et al. Onto-Tools, the toolkit of the modern biologist: onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res 2003;31(13):3775–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al-Shahrour F, Diaz-Uriarte R, Dopazo J.. Discovering molecular functions significantly related to phenotypes by combining gene expression data and biological information. Bioinformatics 2005;21(13):2988–93. [DOI] [PubMed] [Google Scholar]
- 17. Goeman JJ, van de Geer SA, de Kort F, et al. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics 2004;20(1):93–9. [DOI] [PubMed] [Google Scholar]
- 18. Montaner D, Dopazo J.. Multidimensional gene set analysis of genomic data. PLoS One 2010;5(4):e10348.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Medina I, Montaner D, Bonifaci N, et al. Gene set-based analysis of polymorphisms: finding pathways or biological processes associated to traits in genome-wide association studies. Nucleic Acids Res 2009;37:W340–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Al-Shahrour F, Arbiza L, Dopazo H, et al. From genes to functional classes in the study of biological systems. BMC Bioinformatics 2007;8(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Falco MM, Bleda M, Carbonell-Caballero J, et al. The pan-cancer pathological regulatory landscape. Sci Rep 2016;6(1):39709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bleda M, Medina I, Alonso R, et al. Inferring the regulatory network behind a gene expression experiment. Nucleic Acids Res 2012;40(W1):W168–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martignetti L, Calzone L, Bonnet E, et al. ROMA: representation and quantification of module activity from target expression data. Front Genet 2016;7:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jaakkola MK, Elo LL.. Empirical comparison of structure-based pathway methods. Brief Bioinform 2016;17(2):336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kotera M, Hirakawa M, Tokimatsu T, et al. The KEGG databases and tools facilitating omics analysis: latest developments involving human diseases and pharmaceuticals. Methods Mol Biol 2012;802:19–39. [DOI] [PubMed] [Google Scholar]
- 27. Croft D, Mundo AF, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res 2014;42(D1):D472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics 2009;25(1):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gu Z, Liu J, Cao K, et al. Centrality-based pathway enrichment: a systematic approach for finding significant pathways dominated by key genes. BMC Syst Biol 2012;6(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shojaie A, Michailidis G.. Network enrichment analysis in complex experiments. Stat Appl Genet Mol Biol 2010;9:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drier Y, Sheffer M, Domany E.. Pathway-based personalized analysis of cancer. Proc Natl Acad Sci USA 2013;110(16):6388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cerami EG, Gross BE, Demir E, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res 2011;39:D685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slenter DN, Kutmon M, Hanspers K, et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res 2018;46:D661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vera-Licona P, Bonnet E, Barillot E, et al. OCSANA: optimal combinations of interventions from network analysis. Bioinformatics 2013;29(12):1571–3. [DOI] [PubMed] [Google Scholar]
- 35. Fey D, Halasz M, Dreidax D, et al. Signaling pathway models as biomarkers: patient-specific simulations of JNK activity predict the survival of neuroblastoma patients. Sci Signal 2015;8(408):ra130. [DOI] [PubMed] [Google Scholar]
- 36. Jacob L, Neuvial P, Dudoit S.. More power via graph-structured tests for differential expression of gene networks. Ann Appl Stat 2012;6(2):561–600. [Google Scholar]
- 37. Martini P, Sales G, Massa MS, et al. Along signal paths: an empirical gene set approach exploiting pathway topology. Nucleic Acids Res 2013;41(1):e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Massa MS, Chiogna M, Romualdi C.. Gene set analysis exploiting the topology of a pathway. BMC Syst Biol 2010;4:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao S, Wang X.. TAPPA: topological analysis of pathway phenotype association. Bioinformatics 2007;23(22):3100–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hidalgo MR, Cubuk C, Amadoz A, et al. High throughput estimation of functional cell activities reveals disease mechanisms and predicts relevant clinical outcomes. Oncotarget 2017;8(3):5160–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sebastian-Leon P, Carbonell J, Salavert F, et al. Inferring the functional effect of gene expression changes in signaling pathways. Nucleic Acids Res 2013;41(W1):W213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sebastian-Leon P, Vidal E, Minguez P, et al. Understanding disease mechanisms with models of signaling pathway activities. BMC Syst Biol 2014;8(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nam S, Park T.. Pathway-based evaluation in early onset colorectal cancer suggests focal adhesion and immunosuppression along with epithelial-mesenchymal transition. PLoS One 2012;7(4):e31685.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hernansaiz-Ballesteros RD, Salavert F, Sebastian LP, et al. Assessing the impact of mutations found in next generation sequencing data over human signaling pathways. Nucleic Acids Res 2015;43(W1):W270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gustafsson M, Nestor CE, Zhang H, et al. Modules, networks and systems medicine for understanding disease and aiding diagnosis. Genome Med 2014;6(10):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuperstein I, Bonnet E, Nguyen H, et al. Atlas of Cancer Signalling Network: a systems biology resource for integrative analysis of cancer data with Google Maps. Oncogenesis 2015;4(7):e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fujita KA, Ostaszewski M, Matsuoka Y, et al. Integrating pathways of Parkinson's disease in a molecular interaction map. Mol Neurobiol 2014;49(1):88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogishima S, Mizuno S, Kikuchi M, et al. AlzPathway, an updated map of curated signaling pathways: towards deciphering Alzheimer’s disease pathogenesis. Syst Biol Alzheimers Dis 2016;1303:423–32. [DOI] [PubMed] [Google Scholar]
- 49. Watanabe T, Kawakami E, Shoemaker JE, et al. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 2014;16(6):795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bader GD, Cary MP, Sander C.. Pathguide: a pathway resource list. Nucleic Acids Res 2006;34(90001):D504–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goeman JJ, Buhlmann P.. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics 2007;23(8):980–7. [DOI] [PubMed] [Google Scholar]
- 52. Koumakis L, Kanterakis A, Kartsaki E, et al. MinePath: mining for phenotype differential sub-paths in molecular pathways. PLoS Comput Biol 2016;12(11):e1005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li X, Shen L, Shang X, et al. Subpathway analysis based on signaling-pathway impact analysis of signaling pathway. PLoS One 2015;10(7):e0132813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nam S, Chang HR, Kim KT, et al. PATHOME: an algorithm for accurately detecting differentially expressed subpathways. Oncogene 2014;33(41):4941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haynes WA, Higdon R, Stanberry L, et al. Differential expression analysis for pathways. PLoS Comput Biol 2013;9(3):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sales G, Calura E, Martini P, et al. Graphite Web: web tool for gene set analysis exploiting pathway topology. Nucleic Acids Res 2013;41(W1):W89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Judeh T, Johnson C, Kumar A, et al. TEAK: topology enrichment analysis framework for detecting activated biological subpathways. Nucleic Acids Res 2013;41(3):1425–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ibrahim MA, Jassim S, Cawthorne MA, et al. A topology-based score for pathway enrichment. J Comput Biol 2012;19(5):563–73. [DOI] [PubMed] [Google Scholar]
- 59. Rivera CG, Tyler BM, Murali TM.. Sensitive detection of pathway perturbations in cancers. BMC Bioinformatics 2012;13(Suppl 3):S9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen X, Xu J, Huang B, et al. A sub-pathway-based approach for identifying drug response principal network. Bioinformatics 2011;27(5):649–54. [DOI] [PubMed] [Google Scholar]
- 61. Hung JH, Whitfield TW, Yang TH, et al. Identification of functional modules that correlate with phenotypic difference: the influence of network topology. Genome Biol 2010;11(2):R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ulitsky I, Krishnamurthy A, Karp RM, et al. DEGAS: de novo discovery of dysregulated pathways in human diseases. PLoS One 2010;5(10):e13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ihnatova I, Budinska E.. ToPASeq: an R package for topology-based pathway analysis of microarray and RNA-Seq data. BMC Bioinformatics 2015;16:350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hu Q-N, Liang Y-Z, Fang K-T.. The matrix expression, topological index and atomic attribute of molecular topological structure. J Data Sci 2003;1:361–89. [Google Scholar]
- 65. Edwards D, Wang L, Sørensen P.. Network-enabled gene expression analysis. BMC Bioinformatics 2012;13:167.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. The Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499(7456):43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Linehan WM, Spellman PT, Ricketts CJ, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 2016;374(2):135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. The Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. The Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014;159:676–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Johnson WE, Li C, Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8(1):118–27. [DOI] [PubMed] [Google Scholar]
- 78. Robinson MD, Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ferreira PG, Muñoz-Aguirre M, Reverter F, et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat Commun 2018;9(1):490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lonsdale J, Thomas J, Salvatore M, et al. The genotype-tissue expression (GTEx) project. Nat Genet 2013;45:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee J, Hever A, Willhite D, et al. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J 2005;19(10):1356–8. [DOI] [PubMed] [Google Scholar]
- 82. Heinrich M, Matt K, Lutz-Bonengel S, et al. Successful RNA extraction from various human postmortem tissues. Int J Legal Med 2007;121(2):136–42. [DOI] [PubMed] [Google Scholar]
- 83. Dopazo J. Genomics and transcriptomics in drug discovery. Drug Discov Today 2014;19(2):126–32. [DOI] [PubMed] [Google Scholar]
- 84. Fryburg DA, Song DH, Laifenfeld D, et al. Systems diagnostics: anticipating the next generation of diagnostic tests based on mechanistic insight into disease. Drug Discov Today 2014;19(2):108–12. [DOI] [PubMed] [Google Scholar]
- 85. Amadoz A, Sebastian-Leon P, Vidal E, et al. Using activation status of signaling pathways as mechanism-based biomarkers to predict drug sensitivity. Sci Rep 2016;5(1):18494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tian Q, Price ND, Hood L.. Systems cancer medicine: towards realization of predictive, preventive, personalized and participatory (P4) medicine. J Intern Med 2012;271(2):111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Salavert F, Hidago MR, Amadoz A, et al. Actionable pathways: interactive discovery of therapeutic targets using signaling pathway models. Nucleic Acids Res 2016;44(W1):W212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dugger SA, Platt A, Goldstein DB.. Drug development in the era of precision medicine. Nat Rev Drug Discov 2017;17(3):183.. [DOI] [PMC free article] [PubMed] [Google Scholar]