Abstract
Variants within non-coding genomic regions can greatly affect disease. In recent years, increasing focus has been given to these variants, and how they can alter regulatory elements, such as enhancers, transcription factor binding sites and DNA methylation regions. Such variants can be considered regulatory variants. Concurrently, much effort has been put into establishing international consortia to undertake large projects aimed at discovering regulatory elements in different tissues, cell lines and organisms, and probing the effects of genetic variants on regulation by measuring gene expression. Here, we describe methods and techniques for discovering disease-associated non-coding variants using sequencing technologies. We then explain the computational procedures that can be used for annotating these variants using the information from the aforementioned projects, and prediction of their putative effects, including potential pathogenicity, based on rule-based and machine learning approaches. We provide the details of techniques to validate these predictions, by mapping chromatin–chromatin and chromatin–protein interactions, and introduce Clustered Regularly Interspaced Short Palindromic Repeats-Associated Protein 9 (CRISPR-Cas9) technology, which has already been used in this field and is likely to have a big impact on its future evolution.
We also give examples of regulatory variants associated with multiple complex diseases. This review is aimed at bioinformaticians interested in the characterization of regulatory variants, molecular biologists and geneticists interested in understanding more about the nature and potential role of such variants from a functional point of views, and clinicians who may wish to learn about variants in non-coding genomic regions associated with a given disease and find out what to do next to uncover how they impact on the underlying mechanisms.
Keywords: variant analysis, regulatory variants, non-coding DNA, complex diseases, GWAS
Background
Thanks to improvements in sequencing technologies, it is now cheaper and easier than ever to sequence patient genomes with the aim of identifying variants associated with disease. Until recently, researchers were largely interested in variants that overlapped protein-coding regions of the genome. However, results from genome-wide association studies (GWAS) have found more than 88% of disease-associated variants to be in non-coding regions [1]. Accordingly, in the past 5 years, a great deal of research has been put into analysis of these regions, although their importance was already increasing thanks to initiatives such as the ENCODE project, whose pilot phase was released over 10 years ago [2, 3]. There are myriad ways by which variants in non-coding regions can affect disease, most of which involve the disruption of genomic elements that regulate gene expression, which we will refer to as regulatory elements [4]. These include cis- and trans-regulatory elements that bind transcription factors (TFs) and other proteins, such as enhancers or promoters [5], and transcribed non-coding regions with regulatory roles, such as micro RNAs (miRNAs) [6] and long non-coding RNAs (lncRNAs) [7]. We will refer to variants in non-coding regions that can affect regulation as regulatory variants.
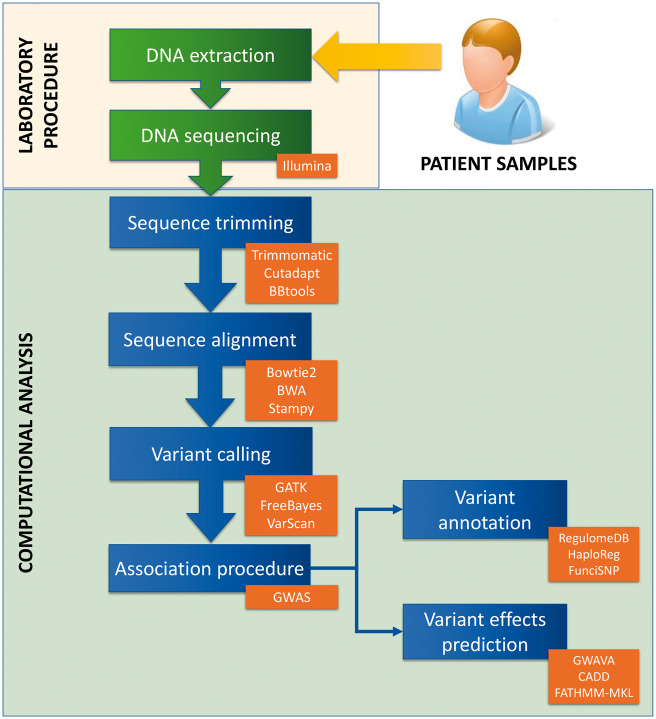
The procedure for identifying regulatory variants associated with disease is complex and involves both laboratory procedures and computational resources. Some steps are the same as for coding variant identification, such as genome-sequencing, computational analysis of the resultant data and identification of associated variants, for example by comparing allelic frequencies between individuals. For regulatory variant identification, the next steps involve annotation of overlapping regions and predicting their functional effects using a variety of software tools. There are currently around 20 tools available to perform these steps, each with their own advantages, disadvantages and peculiarities. Once regulatory variants have been identified using these tools, an experimental validation procedure is necessary to confirm their predicted impact. An overview of the different steps involved in the identification of regulatory variants associated with disease, from patient to confirmation, is shown in Figure 1.
Figure 1.
Description of the steps necessary to characterize regulatory variants. Following DNA extraction and sequencing of individual samples, a number of computational steps are performed. First, trimming and alignment of the sequences is necessary. Then a file with all the variants is obtained using the variant calling procedure. At this point, these variants must be associated with disease, using GWAS or other experimental designs such as trio analysis. Next the non-coding SNPs can be analysed and annotated putatively based on overlap with genomic functional elements (variant annotation) and their putative effects can be predicted (variant effects prediction). In orange boxes, there are some examples of tools or analysis types that can be used for each step, and in the case of variant annotation and variant effects prediction, we show examples of tools focused on regulatory variants.
In this review, we describe this entire process in detail, including how to detect variants using sequencing techniques, determining which of these are disease-associated, searching for overlap of these variants with regulatory elements and predicting their potential impact on the disease based on features of the overlapping region. We will also describe subsequent experimental approaches for validation. In addition, we will present previous studies that have identified and characterized regulatory variants associated with disease by following similar procedures to those described here, including for complex diseases such as coronary artery disease (CAD) [8], Crohn’s disease (CD) [9], schizophrenia [10] and cancer [11]. However, before we discuss these procedures, we must first introduce the main non-coding regulatory elements.
Key regulatory elements
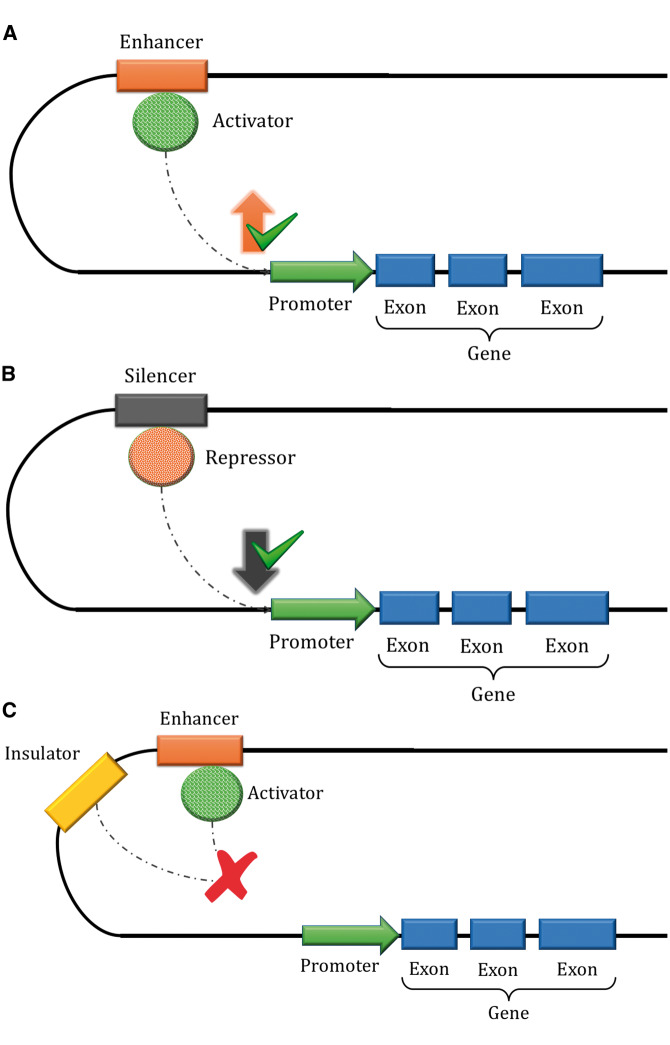
Regulatory elements coordinate the precise expression of genes in different cell types at the correct developmental stages and in response to changes in external conditions. Generally located within non-coding regions, they can exert their effects through various processes. An important class of element comprises specific regulatory sequences that affect transcription through binding with various proteins, generally known as TFs, including activators and repressors [4]. TFs can bind to cis-regulatory elements located in transcription start sites (TSS) such as promoters, as well as to distal elements including other regulatory regions that can be located thousands of base pairs away from the TSS, such as enhancers, silencers and insulators [12–14]. A representation of these cis-regulatory elements is shown in Figure 2. TFs also ensure the correct positioning of RNA polymerase II (RNAPII) to enable the correct formation of the transcription-initiation complex [15]. These regulatory elements are often located in conserved regions [16]. Key regulatory elements in this class include:
Figure 2.
Representation of the effects of cis-regulatory elements: enhancers (A), silencers (B) and insulators (C). In 2A, the enhancer region binds to a protein (activator) that joins to a specific transcription factor binding site (TFBS) in the promoter region, upregulating the target gene. In 2B, the silencer region binds to another protein (repressor) that binds to a specific TFBS in the promoter region, leading to reduced gene expression. Finally, in 2C, the insulator region interacts with the activator protein of an enhancer, blocking its binding to the promoter and inhibiting gene expression. These interactions are highly controlled and dynamic, and modifications to these elements can dysregulate expression and lead to disease.
Promoters: DNA sequences located in the 5’ region of genes that activate transcription via RNAPII. Through the action of various TFs, the RNAPII binds to a consensus sequence, the TATA box, forming the RNAPII transcription-initiation complex [17].
Enhancers: Short DNA regions that can be bound by DNA-binding proteins called activators and increase gene transcription through their interaction with RNAPII. They can be located thousands of base pairs away from the TSS, but through their interaction with activators, they often form DNA loops that bring them closer to the promoter region [18].
Silencers: Short DNA sequences that bind to DNA-binding proteins called repressors, causing a decrease in gene transcription by inhibiting other genomic elements such as promoters. Like enhancers, they can be located near the TSS or thousands of base pairs away from it, forming loops using DNA-binding and other proteins to get closer to the promoter regions [19].
Insulators: These regulatory sequences have a key role in chromatin state regulation, by preventing interactions between chromatin domains. Probably the most well-known is the transcriptional repressor CCCTC-binding factor (CTCF). By binding with target sequences, CTCF can act as an insulator by both blocking interactions between enhancers and promoters and preventing heterochromatin expansion, effectively acting as a chromatin barrier [20].
These functional elements often form clusters that regulate the expression of the same or different genes [21]. As a result, a single loss-of-function mutation in a cluster of enhancers will not necessarily alter gene expression leading to pathogenic effects, due to redundancy between the enhancers in the cluster [22]. However, a gain-of-function mutation is more likely to result in a pathological phenotype due to expanded enhancer activity [22].
Another important class of regulatory elements includes the non-coding RNAs (ncRNAs). They tend to influence regulation post-transcriptionally, by modifying the primary transcript or mature mRNA. This process can involve several mechanisms, including capping, splicing and polyadenylation [14, 23, 24], as well as binding to proteins. For example, ribonucleoproteins are RNA-containing proteins that take part in splicing processes, regulating mRNA maturation [25]. Post-transcriptional regulation can dramatically affect mRNA abundance and can be carried out via different types of ncRNAs, the most well-known of which include the following:
miRNA: Small, single-stranded RNA molecules (<25 nucleotides) that are involved in gene silencing processes. These small molecules are often found in non-coding intronic regions and are generated by the Dicer enzyme and the RNA-induced silencing complex (RISC). Dicer cuts the miRNA precursor molecule and the mature miRNA is then incorporated into the RISC. This allows the complex to recognize mRNAs complementary to the mature miRNA and cleave them, preventing the mRNA from being translated into protein [26].
lncRNA: Long non-coding RNA molecules (>200 nucleotides) that are thought to be involved in many types of gene regulation, not only post-transcriptionally. They can interact with TFs to modify their activity, such as those that regulate RNAPII [27]. They also bind mRNA molecules to affect post-transcriptional gene expression [28]. They can also play a role in the regulation of mRNA translation [29] and epigenetic modification by affecting histone methylation [30].
There are other types of ncRNAs that can influence regulation, including enhancer RNAs [31], piwi-interacting RNAs [32] and more [33]; the full details of which are outside the scope of this review.
Clinical genome sequencing for detecting variants in patients
It is now possible to sequence the genome of a patient and detect their variants in matter of hours using next-generation sequencing (NGS) [34]. Initial problems related to technical issues such as sequencing errors still persist but are being ameliorated by technological improvements and advances in bioinformatics techniques [35]. Although tools for prioritizing variants associated with coding regions and predicting their effects on protein structure and function are well established [36], predicting the function of regulatory variants is a relatively new area of research. As the majority of variants in non-coding regions have no obvious effect on disease, and despite multiple notable efforts such as the aforementioned ENCODE project, the characterization of all non-coding elements and how they can affect disease is still far from complete [37]. In the next section, we will describe the state of the art in sequencing platforms for clinical genomics and variant determination, including key methods for pre-processing, alignment and genomic variant calling. This procedure is largely the same for coding and regulatory variants, with some important distinctions that will be discussed.
Current sequencing platforms
There are two main considerations when selecting a sequencing platform for variant determination: (i) coverage per nucleotide: the number of reads that support a certain genomic position, better if the selected technology gives a larger number of sequences per run, and (ii) sequencing error rate: the average proportion of nucleotides not correctly sequenced. The most well-known sequencing platforms are Illumina, Roche 454, PacBio and Oxford Nanopore. Despite recent advances in the latter two platforms, Illumina remains the most popular and well-established sequencing technology for single-nucleotide variant determination. It allows the generation of a large number of short reads, ensuring a good coverage per nucleotide ratio [38], indispensable for the correct determination of this type of variant. Illumina can also be used for targeted gene sequencing [39], which can also be applied to the sequencing of predetermined regulatory regions, and has overtaken Roche 454 for clinical application [35]. PacBio and Oxford Nanopore, which generate long reads, look set to play a key role in the detection of large regulatory regions in the future, due to their ability to detect structural variants; however, they currently have a relatively low coverage, making them unsuitable for the routine detection of single-nucleotide variants at this moment in time [40, 41]. Therefore, we will focus on Illumina, who claim that their latest sequencing platform, HiSeq® X, can sequence one human genome per run in a single day for $1000, with a coverage of 30× [42].
Read pre-processing and alignment
Pre-processing
Illumina, like most sequencing platforms, produces a FASTQ format data file, consisting of the raw sequences (reads), derived from the patient genome, and their assigned quality scores, represented as a Phred score (Q), which shows the probability that a nucleotide is erroneously sequenced, expressed as . These scores should be used to remove poor-quality reads and adapters and detect other errors produced during the sequencing process [43, 44]. This can be performed using various tools such as Trimmomatic [45] or Cutadapt [46]. There are also various suites such as BBTools, which has a range of tools for these processes.
Alignment
Once the reads have been pre-processed, they must be aligned to a reference genome to allow us to identify areas where they differ, indicative of variation in the sequenced genome [47]. Good alignment tools must be both accurate and fast, as they are typically dealing with many millions of reads per sample; moreover, they must discriminate sequencing errors from true variants [48]. Some, such as Bowtie [49], and BWA [50], use the Burrows–Wheeler transform, while others, such as Stampy [51], use hash-based algorithms, which have been shown to give the most accurate results for Illumina sequences in terms of variant calling [48, 52].
These tools generate a Sequence Alignment Map (SAM) file [53], which contains details of the reads and their alignment positions. This is typically then transformed to a BAM (Binary Alignment Map) format, to save space [54]. They include a mapping quality (MAPQ) score, which can be used to decide the best position for a read that matches to the genome in multiple positions. These files are the starting point for the next analysis step: variant calling.
Variant calling
Variant calling tools determine the locations in which a sequenced genome differs from the reference using different statistical methods [52]. They attempt to avoid potential errors that have slipped through the sequencing and alignment processes, using the nucleotide quality scores and minimizing the false-positive variant call rate [55]. Two variant calling software use a Bayesian-approach algorithm for determining true variants from NGS data. The first one, the Genome Analysis Toolkit (GATK) [56], is a popular package with customizable modules not only for detecting different types of variants but also for determining whether these variants are likely correctly identified, which makes it one of the most-used tools for variant calling [57]. The second one, FreeBayes, is a framework specially developed for detecting single-nucleotide polymorphisms (SNPs) and short structural variants in haplotypes [58]. Other tools, such as VarScan [59], use statistical approaches to sort and score the alignments and reject those deemed ambiguous. Another tool, LoFreq, is based on a Poisson-binomial distribution and can be used to find rarer variants. In all cases, these tools return a variant call format (VCF) file containing variants, their positions, identifiers and quality scores, among other parameters [60]. Of these variant callers, it is recommended to use FreeBayes when variants with a variant allele frequency less than 0.10 are expected, and if higher than 0.20, the use of GATK is recommended [61].
Strategies for associating variants with disease
Once the VCF has been obtained, it is necessary to determine which variants are related to the disease (pathogenic) and those that have no effect (neutral). A common strategy is to compare allelic and genotypic frequencies between disease sufferers and controls, as is the case in GWAS. These studies are focused on identifying putative genomic variants, such as SNPs, that are associated with disease and population traits [62]. The canonical GWAS procedure compares variants from patients that share the same disease or phenotype against variants from healthy individuals from the same population, used as a control data set. Thus, disease-associated variants can be determined because of their relative frequency [63]. It is important to understand that SNPs associated with a disease are not necessarily causal, instead they may be inherited within the same haplotype block, potentially in linkage disequilibrium (LD) with the casual SNP [64]. This is particularly important when using SNP microarray-derived GWAS data, in which only a representative subset of variants is determined, which means that fine-mapping studies will usually be necessary. In relation to regulatory variants, this is a problem, as a haplotype block may contain several regulatory elements. Multiple testing correction is also critical to reduce the number of spurious associations [65], which can be exacerbated by the structure in the data related to LD [66]. Detecting associations is typically performed using tools such as PLINK, which can use multiple association models such as allelic or genotypic recessive [67]. Other tools include PRESTO [68] and PERMORY; the latter performs its statistical corrections through permutation tests and is optimized for sequencing data [69]. For further information about bioinformatics procedures related to GWAS, we highly recommend the work by Bush and Moore [63]. Higher order associations can also be examined using epistasis-based approaches [70].
Despite their widespread usage and popularity, GWAS have only been able to explain a small proportion of estimated disease heritability [71]. Moreover, the GWAS approach requires a large number of patients to obtain statistical significance in most cases [72]. Other alternative approaches exist for associating variants with disease. These include parent–child trio analysis, which consists of sequencing and comparing the genome of related individuals to distinguish between variants that are inherited from parents and de novo variants [73]. These de novo variants can be putatively associated with disease if the parents do not show the same phenotype as the child. Parent–child trio studies have been shown to decrease the type I error inherent to GWAS, as constructing parent–child relations avoids population stratification [74]. There are several tools to perform parent–child trio analysis using VCF files, such as Trio-SVM [75], which uses a support vector machine (SVM), TrioVis [76], a visualization tool that groups variants according to the Mendelian inheritance model, and trio [74], an R-Bioconductor package able to conduct multiple analyses. Another approach is to perform variant prioritization directly on a given genome, using annotation and other prioritization algorithms [36]. The annotation procedure for non-coding regions will be explained in the next few sections, along with how to prioritize potential regulatory variants in these regions. This remains an important challenge, as we are far from knowing the positions of all regulatory regions across all cell lines, tissues and conditions, and it is a rapidly evolving field.
Projects to detect regulatory elements and their usage to annotate and prioritize non-coding variants
Projects to annotate the non-coding genome
Regulatory variant annotation involves determining the regulatory elements with which variants overlaps. There are many tools available for this process, which use data from multiple resources, including TF binding sites (TFBSs), enhancers, promoters, DNA methylation sites, introns and splicing sites and others. Several consortia and international projects have been set up to produce such data, with the aim of identifying and characterizing all the regulatory elements in the non-coding genome, using multiple experimental techniques including Chromatin Immunoprecipitation Sequencing (ChIP-Seq) [4], chromosome conformation capture methods [77], DNase I hypersensitivity assays and DNase Sequencing (DNase-Seq) [78] and RNA Sequencing (RNA-Seq) [79]. Further information regarding regulatory sequence identification can be found here [80, 81]. Key international projects for the characterization of regulatory elements within the human genome include the following:
ENCODE (https://www.encodeproject.org/): The ENCyclopedia of DNA Elements project combines the efforts of many research groups to create a full catalogue of functional annotations of genomic elements [81, 82]. Through various high-throughput techniques, they have identified regulatory regions in diverse cell lines, providing information on regulatory features such as TFBSs and motifs, TSS, histone marks, DNA methylation sites and open chromatin regions, revealing genomic regulatory mechanisms that were previously unknown. Data can be accessed directly through the ENCODE Web portal or using the online resource University of California Santa Cruz (UCSC) Genome Browser (https://genome.ucsc.edu/) [3].
FANTOM5 (http://fantom.gsc.riken.jp/5/): The Functional Annotation of the Mammalian Genome (FANTOM) is an international consortium that undertakes large-scale complementary DNA (cDNA) sequencing projects to map and identify TSS and provide a comprehensive atlas of human gene expression [83], as well as functional annotation of mammalian genomes and expression profiles [84]. Through Cap Analysis of Gene Expression (CAGE), a technology that captures short 5′ ends of mRNA for producing short nucleotide sequences with NGS, they can determine genes, detect TSSs and recognize the activity of regulatory elements such as promoters, enhancers and others [85]. They have identified more than 180 000 promoters and almost 44 000 candidate enhancers in hundreds of primary cells from human and mouse genomes [83]. Data can be accessed through their website and have been used together with ENCODE to identify disease-associated variants within regulatory elements [86].
Roadmap Epigenomics Project (http://www.roadmapepigenomics.org/): This project aims to create a complete human epigenetics map, using NGS technology for analysing genome marks (DNA methylation, histone modifications, chromatin accessibility and RNA transcripts), in different cell types and tissues [87]. The website offers a genome browser and has a repository to download the latest data sets. Information can be used for identifying and annotating disease-associated variants that affect specific regulatory elements whose expression is different according to cell type, tissue or the stage of development [88].
GTEx (https://www.gtexportal.org/home/): The Genotype-Tissue Expression project encompasses a large community of research groups with the shared aim of demonstrating the relationship between variants and human traits or diseases by analysing changes in gene expression [89]. This project determines expression quantitative trait loci (eQTL) by combining genetic variation with gene expression in post-mortem tissues. It can be used to determine which pathways are affected in disease [90]. They provide an expression atlas for the identification of putative regulatory regions and determining eQTLs associated with disease [89].
Other projects include those attempting to unravel the epigenetic profiles of different human cell types, such as the International Human Epigenome Consortium (IHEC) [91] and BLUEPRINT [92]. We describe key resources in Table 1.
Table 1.
A selection of databases that contains annotations of regulatory elements
| Name | Regulatory elements | Database description | Reference |
|---|---|---|---|
| GTRD | TFBS | Stores TFBS information of ChIP-Seq experiments from different resources (including ENCODE) | [93] |
| TRANSFAC | TFBS | Contains experimental data of eukaryotic TFs, their binding sites, consensus sequences and regulated genes | [94] |
| JASPAR | TFBS | Includes curated and non-redundant experimentally determined TFBS in different eukaryote organisms | [95] |
| DENdb | Enhancers | Integrates predicted information of enhancers in different cell lines that overlap DNAse I HS and TFBS | [96] |
| Enhancer Atlas | Enhancers | Contains annotations of human enhancers from experimental data sets, including histone modifications, TFBS, DNAse I HS and additional information using the CAGE technique | [85] |
| dbSUPER | Super enhancers | Integrates ChIP-Seq signals of clusters of enhancers in different cell types of human and mouse | [97] |
| CTCFBSDB | Insulators | Contains information on CTCF binding sites, including experimentally determined and predicted | [98] |
| EPD | Promoters | Collects information on promoters recognized by the RNA polymerase II in eukaryotes | [99] |
| RNAcentral | ncRNAs | Integrates ncRNA information from high-quality resources | [100] |
| ncRNAdb | ncRNAs | Collects information on ncRNA sequences from various databases | [101] |
| NONCODE | lncRNAs | Contains a complete collection of lncRNA data from various resources (including lncRNAdb) for 16 different organisms | [102] |
TF: transcription factor; TFBS: transcription factor binding site; DNAse I HS: DNAse I hypersensitive site; ncRNA: non-coding RNA; lncRNA: long non-coding RNA.
Regulatory variant annotation tools
Various computational tools use the information generated by these projects to annotate regulatory variants. Such tools usually combine genomic information from multiple projects to determine the regulatory elements close to a query variant. Three of the main annotation tools for this process are described here. In addition, in Table 2, we provide details of, to the best of our knowledge, all freely available tools for this purpose.
Table 2.
Summary of the main variant annotation tools for non-coding DNA regions
| Name | Uses | Main data sources | Advantages | Limitations | Reference |
|---|---|---|---|---|---|
| RegulomeDB | Prioritization of functional variants, using a score based on the number of elements with which the variant overlaps | ENCODE, Roadmap Epigenomics Project | Includes information from numerous functional annotation sources | The scoring system can be difficult to interpret | [103] |
| HaploReg | Annotation of variants in LD, located within or next to regulatory elements | ENCODE, GTEx, Roadmap Epigenomics Project | Allows the identification and mining of causal variants in LD that affect regulatory sites | Functional annotations are not updated periodically | [106] |
| FunciSNP | Identification and prioritization of putative regulatory SNPs | ENCODE, Roadmap Epigenomics Project | Large data queries are fast to perform | A minimum knowledge of R is needed for its use | [107] |
| rVarBase | Annotation of regulatory variants that are involved in transcriptional and post-transcriptional regulation | ENCODE, Roadmap Epigenomics Project | Uses annotations of numerous regulatory features, easy to use, intuitive website | Results summary can be initially confusing, i.e. a SNP can appear annotated with both strong and weak transcription | [108] |
| FunSeq2 | Prioritization of cancer-associated SNVs in non-coding DNA | ENCODE | Can annotate and prioritize variants directly from BED or VCF files and the analysis can be customized | It is specifically designed to annotate cancer-associated variants but not for variants associated with other diseases | [109] |
| ENlight | Annotation of GWAS variants and analysing their putative effects through plot visualization | GWAS, ENCODE, GTEx | Plot system is useful to visually identify causal variants and the analysis can be customized | Functional annotations are not updated periodically | [110] |
| INFERNO | Characterization and prioritization of regulatory variants in different tissues | GTEx, FANTOM5, Roadmap Epigenomics Project | Prioritize variants by calculating an empirical p-value | Large Web queries take a long time to complete | [111] |
| Cepip | Prioritization of gene regulatory variants using tissue-expression data and predicted scores | GTEx, ENCODE, scores from different prediction tools | Integrates the effect of multiple chromatin states to identify and prioritize functional regulatory variants | A minimum knowledge of the command line is needed for its installation and use | [112] |
| GEMINI | Annotation of non-coding variants by integrating chromatin information for different cell types | ENCODE | Incorporates a workflow that automatically annotates variants from VCF or pedigree files | Requires command line use and lacks regulatory features in comparison with some other annotation tools | [113] |
| OncoCis | Prioritization and annotation of cis-regulatory somatic variants in cancer samples | ENCODE, Human Epigenome Atlas, Jaspar, FANTOM5 | The annotation procedure is more rigorous in comparison with other tools for identifying cis-regulatory mutations and it can be applied for identifying cell type-specific variants | It is specifically designed to annotate cancer-associated variants but not for variants associated with other diseases | [114] |
| SuRFR | R package that integrates annotations from different resources to prioritize functional regulatory SNPs | ENCODE, FANTOM5 | Short execution times and higher data confidentiality in comparison with Web-based tools | The user must be familiar with the R programming language | [115] |
RegulomeDB (http://regulomedb.org/): It includes data from resources such as ENCODE and the Roadmap Epigenomics Project for annotating variants within regulatory elements [103], including different regulatory features: TFBSs, chromatin states of different cell types and eQTL data. This information is combined to calculate a score for variant prioritization, useful when annotating GWAS variants associated with disease [104].
HaploReg (http://compbio.mit.edu/HaploReg): It combines information from ENCODE and the RoadMap Epigenomics Project [105] for annotating regulatory variants [106]. To help the user zero in on the casual SNP, the tool combines variants into haplotype blocks, so that SNPs correlated with the query SNP can also be examined. This is achieved using 1000 Genomes Project data, which provides sequence data for many individuals from different populations, for calculating the LD between different variants and producing haplotype blocks. Thus, HaploReg can predict the putative effects of GWAS-derived disease-associated variants and their impact on disease [105].
FunciSNP: The Functional Identification of SNPs is a bioinformatics tool developed in R/Bioconductor that uses genomic data from different resources, such as 1000 Genomes Project and ENCODE, to annotate disease-associated variants [107]. The tool integrates this information to identify SNPs in LD with functional genomic regions. Unlike RegulomeDB and HaploReg, it has no Web service, but is implemented in R.
Other regulatory variant annotation tools include rVarBase [108], FunSeq2 [109], ENlight [110], INFERNO [111], Cepip [112], GEMINI [113], OncoCis [114] and SuRFR [115], and we describe their characteristics, advantages and limitations in Table 2.
Prediction of the pathogenicity and putative effects of regulatory variants
Many variants overlap regulatory elements in the genome; however, they are not necessarily functionally important. Thus, to predict the potential impact of a putative disease-associated variant, several prediction algorithms have been developed that use positional information of the variant alongside genomic annotations to calculate the probability of this variant to affect regulatory motifs and lead to disease [116]. These algorithms use various classification methods, typically based on machine learning, with the aim of obtaining knowledge from large amounts of data. These include supervised methods, which learn the characteristics that correspond to pathogenic or benign variants using a training data set of known variants. To perform this learning step, functional annotations are used, including regulatory and conserved features from different resources, such as ENCODE. To validate the prediction results, they are compared with a testing data set, also consisting of true pathogenic and non-pathogenic (control) variants. Such variants are typically obtained from the Human Gene Mutation Database (HGMD) [117], ClinVar [118] or the 1000 Genomes Project for control variants [119]. First, the algorithm must be trained, to learn which features are able to distinguish pathogenic and control variants. Then, this learned information is used to classify new variants. We will describe some of the most commonly used methods for predicting the putative effects of regulatory variants:
CADD (The Combined Annotation Dependent Depletion, http://cadd.gs.washington.edu/): It is a framework for predicting the deleteriousness of coding and regulatory variants [120]. For predicting the impact of the second type of variants, it is trained on integrated regulatory element annotations, primarily obtained from ENCODE. With these annotations, CADD creates a C-score that is used to measure the variant’s effects. Through an SVM algorithm, CADD contrasts annotated alleles (considered as non-pathogenic) to simulated variants and calculates the lineal relationships between them. CADD classifies unknown variants, prioritizes them using their calculated C-score and quantifies their deleteriousness degree. In addition, pre-calculated C-scores are available to easily annotate GWAS experiments.
DANN (The Deleterious Annotation of genetic variants using Neural Networks tool): It uses a Deep Neural Network (DNN) algorithm that captures linear relationships among different annotations, including evolutionary features, to predict the impact of non-coding variants [121]. This tool was developed to improve the SVM algorithm results of CADD, using DNN it is able to capture a higher number of relationships between annotations. Using the same annotations and training data sets, it has been demonstrated that DANN outperforms CADD results [121].
GWAVA (The Genome-wide annotation of variants, https://www.sanger.ac.uk/sanger/StatGen_Gwava): It is specifically designed to predict the functional impact of regulatory variants and prioritize them, combining conserved features and regulatory annotations from ENCODE and GENCODE [122]. Using a modified random forest algorithm, specifically designed to address the class imbalance issue attached to combining genomic features, the training is performed in three rounds, with the same pathogenic variants set from the HGMD, against three control variant subsets from the 1000 Genomes. Then, it discriminates pathogenic variants using the annotations described before.
FATHMM-MKL (Functional Analysis through Hidden Markov Models, http://fathmm.biocompute.org.uk/): It is based on a machine learning algorithm that uses a multiple kernel (MK) learning method for predicting the putative effects of regulatory variants [123], using annotations from ENCODE. During training, it weights all the annotations, by means of their relevance, and creates matrices that will be used for an MK algorithm, with the aim of classifying input variants and finally predicting their putative effects. The gold-standard data set includes benign variants from the 1000 Genomes Projects and pathogenic variants from the HGMD. Predictions performed by FATHMM-MKL are given as p-values that can be used in other integrative studies. FATHMM-MKL recently improved it prediction system with the FATHMM-XF method, which trains a supervised machine learning approach with additional genetic and epigenetic features from ENCODE and the Roadmap Epigenomics Project, assigning a confidence score to all predictions [124]. FATHMM-XF was recently shown to outperform other predictors, including CADD and DANN [124].
LINSIGHT: It combines linear and probabilistic models with functional and evolutionary conservation data to calculate a fitness consequences score for predicting the putative effects of regulatory variants and ranking them [125]. LINSIGHT uses information for different genomic features from resources such as ENCODE and FANTOM5 to identify deleterious regulatory variants related to inherited diseases. This approach is used to infer the selective pressure on regulatory regions and is applied for evaluating the fitness consequences of regulatory variants and predicting their impact.
A recent comparison has demonstrated that FATHMM-MKL and GWAVA outperformed other tools such as CADD and DANN using the same benchmarking data set; however, despite all the advances in regulatory variants prediction, there are still issues related to our incomplete knowledge of regulatory regions and how base-changes can affect them [126]. Other tools described in Table 3 include deltaSVM [116], DeepSEA [127], Eigen [128], GenoCanyon [129], PRVCS [130], ARVIN [131] and DIVAN [132]. In addition, there are specific methods for predicting the impact of somatic and germline regulatory variants associated with cancer [133] and cis-regulatory variants in cancer gene regulatory networks [134] and for quantifying deleteriousness for a regulatory variant that affects enhancer TFBSs [135].
Table 3.
Summary of the main non-coding variant effect prediction tools
| Name | Description | Type | Advantages | Limitations | Reference |
|---|---|---|---|---|---|
| CADD | Framework designed to predict the impact of variants in coding and non-coding regions | SVM | Precomputes C-scores that can be used for other tools to prioritize variants | Limited performance due to SVM, as this cannot capture non-linear relationships between annotations | [120] |
| DANN | Software that predicts the deleteriousness of genetic variants, using the same data set as CADD with a different algorithm | DNN | DNN algorithm can capture non-linear relationships between annotations that SVM cannot | Does not have a website and the user requires Python knowledge to use it | [121] |
| GWAVA | Tool for predicting the impact of coding and non-coding variants using different annotations to prioritize those that are functional | Random forest | The random forest algorithm is modified to overcome the class imbalance issue when combining different genomic features | During the execution through the website, parameters to prioritize variants cannot be modified and it only supports a single region as a query | [122] |
| FATHMM-MKL | Software for predicting the impact of coding and non-coding SNVs using annotations grouped in categories | MK learning | They convert annotation groups into multiple kernels to perform the evaluation, and it outperforms other tools for non-coding SNV effects prediction | During the execution through the website, parameters to prioritize variants cannot be modified | [123] |
| FATHMM-XF | FATHMM-MKL improvement, with extended features gathered in a single-kernel data set | Supervised machine learning | In terms of accuracy, outperforms other prediction tools, including its predecessor, FATHMM-MKL, for non-coding SNV effect prediction | During the execution through the website, parameters to prioritize variants cannot be modified | [124] |
| LINSIGHT | Tool that determines the probability of negative selection in non-coding regions to predict the impact of variants that overlap them | Linear and probabilistic models | Combines both linear and probabilistic model for functional genomic and evolutionary data, respectively, to identify causal inherited non-coding variants in conservation sites | The user must be familiar with the use of the command line to install and execute the code | [125] |
| deltaSVM | Software that predicts and quantifies the impact of regulatory variants based on regulatory features whose role is cell-dependent | SVM | Uses a catalogue of DNase I hypersensitivity sites, histone modifications and TFBSs information to predict the effects of variants within enhancer regions with high accuracy | The user must be familiar with the use of the command line to install and execute the code and lacks genomic features in comparison with other tools | [116] |
| DeepSEA | Tool for predicting the functional impact of non-coding variation by evaluating molecular function | Deep learning | It combines de novo (predicted) information of chromatin effects and conservation sites to prioritize functional variants | Using the default threshold, DeepSEA returns a large amount of information, even for a small search using only a handful of variants | [127] |
| Eigen | Software for scoring, prioritising and predicting the impact of putative causal coding and non-coding variants using ENCODE annotations | Unsupervised spectral learning | In comparison with other score calculations, Eigen uses a more refined annotation data set to make its predictions and the unsupervised algorithm deals well with the class imbalance issue | To download and use of the tool, the user has must be familiar with the use of the command line and R | [128] |
| GenoCanyon | Tool that calculates a prediction score for each nucleotide of a given genomic region, using conservation data and epigenetic information | Unsupervised statistical learning | Predicts impact using conservation sites and molecular features, generating a plot that includes the analysed region and the prediction score for each nucleotide | Large regions cannot be analysed through their Web app and parameters other than those related to the plot cannot be changed | [129] |
| PRVCS | Software package that integrates functional annotations from different tools to predict and prioritize regulatory variants | Composite statistics model | Offers a database that integrates functional prediction scores for non-coding variants, computing a composite likelihood score to estimate if the variant is causal or not | The user must be familiar with the use of the command line to install and execute the code | [130] |
| ARVIN | Framework that predicts the impact of non-coding variants through network analysis | Random forest | Analyses an integrative gene regulatory network that identifies SNPs within enhancers in LD related to genes for predicting their impact | The user must be familiar with the use of the command line and R to install and execute the code | [131] |
| DIVAN | Framework designed to identify risk variants associated to specific diseases using epigenomic profiles of multiple cell types and genomic annotations | Decision tree learning | DIVAN take advantage of risk variants that are associated to a specific disease, instead of including a whole set of disease-associated variants | To date, it only presents 45 diseases to analyse the impact of functional variants in non-coding regions and the user must be familiar with the use of the command line/R | [132] |
SVM: support vector machine; DNN: deep neural network; MK: multiple kernel; SNV: single-nucleotide variant; TFBS: transcription factor binding site; LD: linkage disequilibrium.
Experimental methods to identify regulatory variants
As we have shown, there is a wide range of tools to predict the potential impact of variants in non-coding regions. However, it is crucial to validate these predictions, in terms of whether the variant overlaps a regulatory element, and how this affects gene expression. This is no easy task and is arguably the most important bottleneck in determining disease-associated regulatory variants. Many cis-regulatory elements (e.g. enhancers and insulators) can be located thousands of base pairs away from the genes that they regulate; moreover, they can regulate multiple genes. Therefore, confirming the impact of regulatory variation requires the use of multiple experimental techniques. These include determining the location of different regulatory elements through chromatin association methods and molecular cytogenetic techniques, and correlating variants with changes in gene expression. The genome editing technique, CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats-Associated Protein 9), is likely to be of high impact in this field.
Determining regulatory element location
Chromatin association methods
Once a variant has been predicted as pathogenic, multiple experimental techniques can be used to identify whether it overlaps a regulatory element. Chromatin association methods can be used for this purpose, by determining the physical connections between different loci, allowing us to visualize interactions between different areas of the genome; thus, we can explore whether variants affect these interactions. Multiple techniques are available, further described in Table 4, such as Hi-C, which had led to the development of full-genome interaction maps for different cell types and developmental stages [136]. However, although these methods can be used to analyse interactions between loci, they cannot identify interactions between loci and proteins. For this purpose, it is necessary to adapt the chromatin association methods and combine them with other techniques, such as molecular cytogenetic techniques.
Table 4.
Description of current experimental techniques for determining regulatory elements (adapted from [137])
| Technique name | Description | Advantages | Limitations |
|---|---|---|---|
| Chromosome conformation capture (3C) | Analyse chromatin structure by quantifyng interactions between two selected loci. | Reveal the role of particular regulatory elements in genes at high resolution | The region of interest must be previously specified and no dimensional information of the interactions is provided |
| Chromosome conformation capture-on-chip (4C) | Analyse the chromatin structure by quantifyng interactions between a specific locus and other loci. | High resolution for chromosome variants identification | Elements that interact must be close in the sample |
| Chromosome conformation capture carbon copy (5C) | Analyse the chromatin structure by quantifyng all possible interactions within different genomic regions. | Detects and quantifies a large number of DNA interactions at the same time | Expensive costs and a reference sample requirement |
| Hi-C | Analyse the genome-wide chromatin structure using high-throughput sequencing techniques | High resolution, useful to characterize the whole genome structure when no reference is available | Expensive costs, high workload and less resolution when fewer loci are analysed |
| Chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) | Combination of ChIP-based methods with 3C and sequencing for identifying chromatin interactions | Detection of protein–DNA interactions and long binding sites | Low resolution when identifying if a protein interacts with a given genomic element |
| Luciferase reporter assay | Quantitative technique for detecting the activity of genomic functional elements | Useful to detect changes in gene expression when the activity of regulatory elements involved in transcriptional regulation is dysregulated | Requires a construct and cell culture system |
| DNA fluorescence in situ hybridization (FISH) | Cytogenetic technique for locating specific DNA sequences within chromosomes | Allows the identification of gene position and detection of genetic aberrations for medical studies | Limited efficiency when loci has repeated DNA sequences |
Molecular cytogenetic techniques
These include DNA fluorescence in-situ hybridization (FISH) [138] and combined methods like chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) [139], which uses both chromatin immunoprecipitation and chromatin interaction-based techniques with NGS. Such methods are able to detect interactions between proteins and their biding sites with high resolution [140]. FISH has been used to study the three-dimensional organization of the chromosomes. This technique allows the visualization of interactions between loci located far away in individual cells [141] and has several applications. In combination with other chromatin-based techniques, for example with capture Hi-C, it can improve the resolution for detecting physical chromatin interactions and open chromatin conformations in large genomic regions. This has been useful to determine chromatin interactions in different risk loci where regulatory variants have been associated with colorectal cancer [142]. In addition, it can be used in combination with luciferase reporter assays to detect the activity of functional regulatory elements [143]. Moreover, it has recently been demonstrated that modifications of this technique, such as the single molecule FISH protocol (smFISH), can be used for studying transcriptional and post-transcriptional regulatory processes in different animal model tissues [144]. In the case of transcriptional regulation, this technique can quantify various TFBSs and molecules that bind them [145]. For post-transcriptional regulation, smFISH can be used to quantify the abundance and visually detect ncRNA molecules in different cellular compartments [146].
CRISPR-Cas9 method for evaluating variant effects
Genetic editing of specific loci can provide information about chromatin structure and interactions with other loci and proteins. A revolutionary technique for genome edition is the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) associated nuclease 9 (Cas9), CRISPR-Cas9 system, which can be used to mutate genomic regions of interest to study their effects [147]. For example, using this system it has been possible to target mutations to specific regulatory elements in experimental models: within the intronic CC(A/T)6GG (CArG) box in the promoter region that regulates the expression of CNN1 gene (whose loss of function has been associated with ovarian cancer [148]), and to later apply quantitative techniques to measure the functional impact [149]. CRISPR-Cas9 has also been used to delete TF binding motifs in the β-globin locus control regions and investigate their effects [150]. In addition, this technique can be applied to introduce variation within non-coding regions that are transcribed into ncRNAs, to study their implication in diseases like schizophrenia [151].
Regulatory variants associated with disease: case studies
Research of variants associated with disease in non-coding regions often exploits existing GWAS data to prioritize variants and annotate the genomic regions to which they map, followed by experimental validation of their putative effects. We will now provide some examples where variants in non-coding regions have been found to be involved in complex diseases. These diseases typically involve combinations of different factors, including environmental, genetics and lifestyle; as such, it is likely that regulatory variants will play an important role, as their influence is often context-dependent.
Coronary artery disease
This pathology affects oxygen flood to the heart due to narrowing of blood vessels, leading to chest angina and cardiac arrest. Common causes include high levels of cholesterol in blood, high blood pressure, insulin resistance, sedentary habits, smoking and alcoholism. Through the use of GWAS, risk variants associated with five different vascular diseases, including CAD, were found in the non-coding regions of 6p24, specifically at the PHACTR1 gene locus [8]. Using expression data and regulatory annotations from the ENCODE and the Roadmap Epigenomics Project database (mainly histone marks), the authors revealed that one of these disease-associated variants, rs9349379, was located in an enhancer region that regulates the expression of the endothelin 1 (EDN1) gene in aorta, located 600 bp upstream of rs9349379 [8]. The role of EDN1 is essential for correct vascular tissue development because it codes for the peptide ET-1, which regulates the vascular tone of smooth muscle [152]. To validate the regulatory role of the annotated 6p24 region as an enhancer of EDN1, they performed a targeted deletion in this region using the CRISPR-Cas9 genome editing method, showing an increase in the expression of EDN1 gene and the peptide ET-1 production in vascular cells [8]. Then, they validated the role of rs9349379 variant using CRISPR-Cas9 to generate different allelic series of this single nucleotide, analysing and quantifying the effects of this variant with RNA-Seq. In addition, they measured the three-dimensional contact between EDN1 and rs9349379 using chromatin association methods [8].
Crohn’s disease
CD is a complex disorder that mainly affects the lower digestive system [153]. GWAS have found a large proportion of non-coding regions to be associated with the disease [154]. These variants are thought to affect the correct regulation of genes expressed in various tissues, including liver, brain and several immune system cells. One of these regulatory variants is a SNP located within the intronic region of the FOXO3A gene, which has been indirectly associated with CD and other diseases by affecting FOXO3A regulation [9]. In this study, two groups of patients were established accordingly to whether they showed aggressive or indolent CD effects, respectively. Then, they searched for variants that affected the genes involved in two pathways related to this disease, including their flanking regions, with the aim of finding which SNPs differentiated each group. With this information, they performed a GWAS re-analysis, and determined a non-coding SNP (rs12212067) within the intronic region of the FOXA3A gene, one allele of which was related to the indolent group of patients with CD, and the other allele to the aggressive group. Finally, they performed an experimental validation to confirm the association between this variant and both groups of patients, using ChIP-qPCR and luciferase assays [9].
Schizophrenia
Schizophrenia is highly complex in terms of genetics, and GWAS studies have revealed that more than a hundred loci are associated with this disease, containing thousands of non-coding risk variants [155]. Several GWAS have been performed for this disease, and most existing studies attempt to confirm the causality of the significantly associated variants to explain their mechanism of action. Such is the case for SNP rs1625579, located in the locus of the MIR137 gene that has been associated with neuropsychiatric disorders, including schizophrenia [10]. Using genotype data from the HapMap project, they found another variant, rs2660304, in strong LD with rs1625579. This rs2660304 SNP affects the internal promoter of the MIR137 gene, reducing its expression. The association of the variant with the activity of the promoter was confirmed by luciferase assays and its regulatory role was determined computationally using the annotation tool HaploReg [10].
Cancer
There is increasing interest in the study of regulatory variants in relation to cancer, both in terms of inherited and somatic mutations [156]. For example, different types of cancer have been associated with variants in the promoter region of the telomerase reverse transcriptase TERT gene [157–159]. In another example, overexpression of MDM2 was found to be associated with the progression of multiple cancers, related to the SNP rs2279744, which affects the gene promoter in both hereditary and de novo cancer [11]. This variant produces an increase in the affinity for the TF Sp1, inhibiting the p53 pathway. To confirm this finding, computational analysis was performed to predict whether this SNP could alter affinity for other TFs. A possible alteration of the E2F1 binding site was predicted due to this SNP, which led to the study of the E2F1-mediated alteration of the transcriptional activity of the MDM2 promoter. To confirm the relationship between rs2279744 and the MDM2 promoter, they performed experimental validation using ChIP-qPCR, luciferase assays and gene silencing assays [11].
Future directions
Recent technological advances, both experimental and computational, combined with the establishment of important projects for data generation and cataloguing, have made it more achievable than ever to find variants in the non-coding regions of the genome associated with disease, map these variants to regulatory elements and predict pathogenicity and validate these variants by investigating how they affect chromosome structure, protein binding and gene expression.
Most of these advances have taken place in the past few years, and they are likely to continue advancing in the near future. Concurrently, we expect new techniques for regulatory element and discovery to emerge. Sequencing technology developments are also likely to play a key role. Although Illumina is currently the most widely used sequencing platform for variant determination, it still has some obstacles to overcome for daily clinical use; nevertheless, we believe that sequencing of patients will become routine in the next decade or so. These data must be kept, alongside detailed phenotypic information on the patient and their symptoms. This influx of data will accelerate the discovery of disease-associated regulatory variants, most of which are likely to be in non-coding regions of the genome. It is therefore important that we complement the expected influx of genetic and phenotypic data by expanding projects to map regulatory regions, as well as eQTL-related projects such as GTEx, to investigate the effects of variants on gene expression. This way we can find not only associations but also start to piece together the regulatory processes involved. Of course, as we have made clear, validation is still the major hurdle in terms of definitively proving the mechanistic link between a variant and its effects; however, we also believe that the parallel development of CRISPR-Cas9-based technologies, alongside molecular techniques to investigate chromatin–protein interactions will help us unravel the full details of how variants cause diseases. Doing so is unquestionably the first step towards developing personalized therapies for patients who suffer from them.
Key Points
Most investigation of disease-associated variants has been of those that affect the structure and function of proteins. However, increasing importance is now being given to variants that affect expression levels. These regulatory variants usually overlap with regulatory elements.
We briefly describe the process of obtaining variants from sequencing data, as well as the bioinformatics procedures involved, dealing with the most widely used platforms and software.
We present important international projects aimed at characterizing regulatory elements throughout the non-coding regions of the genome, such as the GTEx project and ENCODE.
We describe the key bioinformatics tools for the annotation of regulatory variants and the prediction of their possible impact to classify them as neutral or pathogenic, using a variety of machine learning methods, and how they can be validated, using various experimental approaches.
Regulatory variants have been determined that affect the expression levels of different genes that give rise to disease. Here we present examples of such variants for complex diseases, including schizophrenia, certain types of cancer, CD and CAD.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness with European Regional Development Fund (SAF2016-78041-C2-1-R), the Andalusian Government with European Regional Development Fund (CTS-486) and the Institute of Health Carlos III of the Ministry of Economy and Competitiveness (grants cofunded by European Regional Development Fund, ERDF: PI12/00900, PI12/02247, PI12/02481, PI12/02529, Red de Reacciones Adversas a Alergenos y Farmacos RD12/0013/0001). The CIBERER is an initiative from the Institute of Health Carlos III. Elena Rojano is a researcher from the Plan de Formacion de Personal Investigador (FPI) supported by the Andalusian Government. James Richard Perkins is a researcher from the Sara Borrell Program (CD14/00242) of the Carlos III National Health Institute, Spanish Ministry of Economy and Competitiveness (grants cofunded by European Social Fund [ESF]: CP14/00034 and CP15/00103, respectively).
Elena Rojano is a Predoctoral Researcher at the Department of Molecular Biology and Biochemistry at the University of Malaga. Her research interests include the development of bioinformatics software for genotype–phenotype associations and regulatory variant analysis.
Pedro Seoane is a Postdoctoral Researcher at the Supercomputing and Bioinnovation Center (SCBI) in the University of Malaga. His research interests include the developing of automatized workflows and bioinformatics software for the analysis of next-generation sequencing data.
Juan Antonio García-Ranea is a Postdoctoral Researcher at the Department of Molecular Biology and Biochemistry in the University of Malaga. His research includes the application of systems biology to study the genetic mechanisms underlying complex and rare diseases.
James Richard Perkins is a Postdoctoral Researcher at the IBIMA Research Laboratory in the Regional University Hospital of Malaga. His research interests include the analysis of genetic and transcriptomic data related to immunological disorders, with a focus on the functional impact of variants in non-coding regions.
References
- 1. Edwards SL, Beesley J, French JD, et al. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 2013;93(5):779–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447(7146):799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosenbloom KR, Dreszer TR, Long JC, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res 2012;40:D912–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Narlikar L, Ovcharenko I.. Identifying regulatory elements in eukaryotic genomes. Brief Funct Genomics Proteomics 2009;8(4):215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mora A, Sandve GK, Gabrielsen OS, et al. In the loop: promoter–enhancer interactions and bioinformatics. Brief Bioinform 2016;17:980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu B, Li J, Cairns MJ.. Identifying miRNAs, targets and functions. Brief Bioinform 2014;15(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo X, Gao L, Wang Y, et al. Advances in long noncoding RNAs: identification, structure prediction and function annotation. Brief Funct Genomics 2016;15(1):38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta RM, Hadaya J, Trehan A, et al. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell 2017;170:522–33.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JC, Espéli M, Anderson CA, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell 2013;155(1):57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Warburton A, Breen G, Bubb VJ, et al. A GWAS SNP for schizophrenia is linked to the internal mir137 promoter and supports differential allele-specific expression. Schizophr. Bull 2016;42(4):1003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang ZH, Zhou CL, Zhu H, et al. A functional SNP in the MDM2 promoter mediates E2F1 affinity to modulate cyclin D1 expression in tumor cell proliferation. Asian Pacific J Cancer Prev 2014;15(8):3817–23. [DOI] [PubMed] [Google Scholar]
- 12. Kolovos P, Knoch TA, Grosveld FG, et al. Enhancers and silencers: an integrated and simple model for their function. Epigenetics Chromatin 2012;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghirlando R, Giles K, Gowher H, et al. Chromatin domains, insulators, and the regulation of gene expression. Biochim Biophys Acta 2012;1819(7):644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alberts B, Johnson A, Lewis J, et al. Control of gene expression In: Molecular Biology of the Cell, 4th edn New York: Garland Science, 2002. [Google Scholar]
- 15. Lodish H, Berk A, Zipursky L, et al. Regulation of transcription initiation In: Molecular Cell Biology, 4th edn New York: W. H. Freeman, 2000. [Google Scholar]
- 16. Liu Y, Liu XS, Wei L, et al. Eukaryotic regulatory element conservation analysis and identification using comparative genomics. Genome Res 2004;14(3):451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boettiger AN, Ralph PL, Evans SN.. Transcriptional regulation: effects of promoter proximal pausing on speed, synchrony and reliability. PLoS Comput Biol 2011;7(5):e1001136.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cooper GM, Hausman RE.. RNA synthesis and processing In: The Cell: A Molecular Approach, 2nd edn Sinauer Association, 2007. [Google Scholar]
- 19. Maston G. a, Evans SK, Green MR.. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet 2006;7(1):29–59. [DOI] [PubMed] [Google Scholar]
- 20. Ong C-T, Corces VG.. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 2014;15(4):234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moorthy SD, Davidson S, Shchuka VM, et al. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 2016;27(2):246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osterwalder M, Barozzi I, Tissiéres V, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 2018;554(7691):239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nelson CE. Gene regulation: a eukaryotic perspective. fifth edition. BIOS advanced text. By David S Latchman. Q Rev Biol 2007;82(1):48. [Google Scholar]
- 24. Braun KA, Young ET.. Coupling mRNA synthesis and decay. Mol Cell Biol 2014;34(22):4078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008;582(14):1977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tijsterman M, Plasterk RHA.. Dicers at RISC: the mechanism of RNAi. Cell 2004;117(1):1–3. [DOI] [PubMed] [Google Scholar]
- 27. Goodrich J. a, Kugel JF.. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol 2006; 7:612–16. [DOI] [PubMed] [Google Scholar]
- 28. Yoon J-H, Abdelmohsen K, Gorospe M.. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 2013;425(19):3723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karapetyan AR, Buiting C, Kuiper RA, et al. Regulatory roles for long ncRNA and mRNA. Cancers 2013;5(2):462–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joh RI, Palmieri CM, Hill IT, et al. Regulation of histone methylation by noncoding RNAs. Biochim Biophys Acta 2014;1839(12):1385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lam MTY, Li W, Rosenfeld MG, et al. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 2014;39(4):170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Teixeira FK, Okuniewska M, Malone CD, et al. PiRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 2017;552(7684):268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alonso CR. Post-transcriptional gene regulation via RNA control. Brief Funct Genomics 2013;12:1–2. [DOI] [PubMed] [Google Scholar]
- 34. Middha S, Baheti S, Hart SN, et al. From days to hours: reporting clinically actionable variants from whole genome sequencing. PLoS One 2014;9(2):e86803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pabinger S, Dander A, Fischer M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform 2014;15(2):256–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eilbeck K, Quinlan A, Yandell M.. Settling the score: variant prioritization and Mendelian disease. Nat Rev Genet 2017;18:599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward LD, Kellis M.. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol 2012;30:1095–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goodwin S, Mcpherson JD, Mccombie WR.. Coming of age : ten years of next- generation sequencing technologies. Nat Rev Genet 2016;17(6):333–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Munchel S, Hoang Y, Zhao Y, et al. Targeted or whole genome sequencing of formalin fixed tissue samples : potential applications in cancer genomics. Oncotarget 2015;6(28):25943–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weirather JL, de Cesare M, Wang Y, et al. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Res 2017;6:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rhoads A, Au KF.. PacBio Sequencing and Its Applications. Genomics Proteomics Bioinform 2015;13(5):278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Illumina. Whole-genome sequencing power. https://www.illumina.com/systems/sequencing-platfo
- 43. Chen S, Huang T, Zhou Y, et al. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics 2017;18:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Del Fabbro C, Scalabrin S, Morgante M, et al. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One 2013;8(12):e85024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011;17(1):10. [Google Scholar]
- 47. Goodwin S, McPherson JD, McCombie WR.. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17(6):333–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nielsen R, Paul JS, Albrechtsen A, et al. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 2011;12(6):443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lunter G, Goodson M.. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 2011;21(6):936–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shang J, Zhu F, Vongsangnak W, et al. Evaluation and comparison of multiple aligners for next-generation sequencing data analysis. Biomed Res Int 2014;2014:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cock PJA, Bonfield JK, Chevreux B, et al. SAM/BAM format v1.5 extensions for de novo assemblies. bioRxiv 2015, in press. doi: 10.1101/020024. [Google Scholar]
- 55. Guo Y, Ye F, Sheng Q, et al. Three-stage quality control strategies for DNA re-sequencing data. Brief Bioinform 2014;15(6):879–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pirooznia M, Kramer M, Parla J, et al. Validation and assessment of variant calling pipelines for next-generation sequencing. Hum Genomics 2014;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garrison E, Marth G.. Haplotype-based variant detection from short-read sequencing. arXiv 2012, in press. [Google Scholar]
- 59. Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009;25(17):2283–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics 2011;27(15):2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sandmann S, de Graaf AO, Karimi M, et al. Evaluating variant calling tools for non-matched next-generation sequencing data. Sci Rep 2017;7:43169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Visscher PM, Brown MA, McCarthy MI, et al. Five years of GWAS discovery. Am J Hum Genet 2012;90(1):7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bush WS, Moore JH.. Chapter 11: genome-wide association studies. PLoS Comput Biol 2012;8(12):e1002822.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Takeuchi F, Yanai K, Morii T, et al. Linkage disequilibrium grouping of single nucleotide polymorphisms (SNPs) reflecting haplotype phytogeny for efficient selection of tag SNPs. Genetics 2005;170(1):291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Duggal P, Gillanders EM, Holmes TN, et al. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics 2008;9:516.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Johnson RC, Nelson GW, Troyer JL, et al. Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genomics 2010;11(1):724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Browning BL. PRESTO: rapid calculation of order statistic distributions and multiple-testing adjusted P-values via permutation for one and two-stage genetic association studies. BMC Bioinformatics 2008;9(1):309.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pahl R, Schäfer H.. PERMORY: an LD-exploiting permutation test algorithm for powerful genome-wide association testing. Bioinformatics 2010;26(17):2093–100. [DOI] [PubMed] [Google Scholar]
- 70. Upton A, Trelles O, Cornejo-García JA, et al. Review: high-performance computing to detect epistasis in genome scale data sets. Brief Bioinform 2016;17(3):368–79. [DOI] [PubMed] [Google Scholar]
- 71. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009;461(7265):747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hong EP, Park JW.. Sample size and statistical power calculation in genetic association studies. Genomics Inform 2012;10(2):117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Goldstein DB, Allen A, Keebler J, et al. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet 2013;14(7):460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schwender H, Li Q, Neumann C, et al. Detecting disease variants in case-parent trio studies using the bioconductor software package trio. Genet Epidemiol 2014;38(6):516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lu AT, Cantor RM.. Identifying rare-variant associations in parent-child trios using a Gaussian support vector machine. BMC Proc 2014;8(Suppl 1):S98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sakai R, Sifrim A, Vande Moere A, et al. TrioVis: a visualization approach for filtering genomic variants of parent-child trios. Bioinformatics 2013;29(14):1801–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Davies JOJ, Oudelaar AM, Higgs DR, et al. How best to identify chromosomal interactions: a comparison of approaches. Nat Methods 2017;14(2):125–34. [DOI] [PubMed] [Google Scholar]
- 78. Song L, Crawford GE.. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc 2010;2010(2):pdb.prot5384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Castel SE, Levy-Moonshine A, Mohammadi P, et al. Tools and best practices for data processing in allelic expression analysis. Genome Biol 2015;16:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Geertz M, Maerkl SJ.. Experimental strategies for studying transcription factor-DNA binding specificities. Brief Funct Genomics 2010;9(5-6):362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sheffield NC, Furey TS.. Identifying and characterizing regulatory sequences in the human genome with chromatin accessibility assays. Genes 2012;3(4):651–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bernstein BE, Birney E, Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Forrest ARR, Kawaji H, Rehli M, et al. A promoter-level mammalian expression atlas. Nature 2014;507(7493):462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lizio M, Harshbarger J, Shimoji H, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 2015;16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature 2014;507(7493):455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ma M, Ru Y, Chuang L-S, et al. Disease-associated variants in different categories of disease located in distinct regulatory elements. BMC Genomics 2015;16(Suppl 8):S3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 2010;28:1045–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Eggers SDZ, Horn AKE, Roeber S, et al. Epigenomic annotation of genetic variants using the Roadmap EpiGenome Browser. Nat Biotechnol 2016;1343:113–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Aguet F, Ardlie KG, Cummings BB, et al. Genetic effects on gene expression across human tissues. Nature 2017;550(7675):204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Stunnenberg HG, Abrignani S, Adams D, et al. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell 2016;167(5):1145–9. [DOI] [PubMed] [Google Scholar]
- 92. Adams D, Altucci L, Antonarakis SE, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotech 2012;30(3):224–6. [DOI] [PubMed] [Google Scholar]
- 93. Yevshin I, Sharipov R, Valeev T, et al. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res 2017;45(D1):D61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Matys V, Fricke E, Geffers R, et al. TRANSFAC®: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 2003;31(1):374–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Khan A, Fornes O, Stigliani A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 2018;46:D260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ashoor H, Kleftogiannis D, Radovanovic A, et al. DENdb: database of integrated human enhancers. Database 2015. pii: bav085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Khan A, Zhang X.. DbSUPER: a database of Super-enhancers in mouse and human genome. Nucleic Acids Res 2016;44(D1):D164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ziebarth JD, Bhattacharya A, Cui Y.. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res 2012;41(D1):D188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dreos R, Ambrosini G, Périer RC, et al. The eukaryotic promoter database: expansion of EPDNew and new promoter analysis tools. Nucleic Acids Res 2015;43(D1):D92–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. The RNAcentral Consortium. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res 2017;45:D128–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Szymanski M, Erdmann VA, Barciszewski J.. Noncoding RNAs database (ncRNAdb). Nucleic Acids Res 2007;35:D162.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhao Y, Li H, Fang S, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 2016;44(D1):D203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22(9):1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Liao X, Lan C, Liao D, et al. Exploration and detection of potential regulatory variants in refractive error GWAS. Sci Rep 2016;6:33090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ward LD, Kellis M.. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 2015;44:D877–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ward LD, Kellis M.. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40(D1):D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Coetzee SG, Rhie SK, Berman BP, et al. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res 2012;40:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Guo L, Du Y, Qu S, et al. rVarBase: an updated database for regulatory features of human variants. Nucleic Acids Res 2016;44(D1):D888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Fu Y, Liu Z, Lou S, et al. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol 2014; 15:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Guo Y, Conti DV, Wang K.. Enlight: web-based integration of GWAS results with biological annotations. Bioinformatics 2015;31(2):275–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Amlie-Wolf A, Tang M, Mlynarski EE, et al. INFERNO - INFERring the molecular mechanisms of NOncoding genetic variants. bioRxiv 2017, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Li MJ, Li M, Liu Z, et al. Cepip: context-dependent epigenomic weighting for prioritization of regulatory variants and disease-associated genes. Genome Biol 2017;18(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Paila U, Chapman BA, Kirchner R, et al. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol 2013;9(7):e1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Perera D, Chacon D, Thoms JA, et al. Oncocis: annotation of cis-regulatory mutations in cancer. Genome Biol 2014;15(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ryan NM, Morris SW, Porteous DJ, et al. SuRFing the genomics wave: an R package for prioritising SNPs by functionality. Genome Med 2014;6(10):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lee D, Gorkin DU, Baker M, et al. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet 2015;47(8):955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014;133(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016;44(D1):D862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46(3):310–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Quang D, Chen Y, Xie X.. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 2015;31(5):761–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ritchie GRS, Dunham I, Zeggini E, et al. Functional annotation of noncoding sequence variants. Nat Methods 2014;11:294–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Shihab H. a, Rogers MF, Gough J, et al. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 2015;31(10):1536–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Rogers MF, Shihab HA, Mort M, et al. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 2018;34(3):511–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Huang YF, Gulko B, Siepel A.. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet 2017;49(4):618–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Drubay D, Gautheret D, Michiels S, et al. A benchmark study of scoring methods for non-coding mutations. Bioinformatics 2018;34(10):1635–41. [DOI] [PubMed] [Google Scholar]
- 127. Zhou J, Troyanskaya OG.. Predicting effects of noncoding variants with deep learning– based sequence model. Nat Methods 2015;12:931–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ionita-Laza I, Mccallum K, Xu B, et al. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet 2016;48(2):214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lu Q, Hu Y, Sun J, et al. A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci Rep 2015;5:10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Li MJ, Pan Z, Liu Z, et al. Predicting regulatory variants with composite statistic. Bioinformatics 2016;32(18):2729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gao L, Uzun Y, Gao P, et al. Identifying noncoding risk variants using disease-relevant gene regulatory networks. Nat Commun 2018;9(1):702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chen L, Jin P, Qin ZS, et al. DIVAN: accurate identification of non-coding disease-specific risk variants using multi-omics profiles. Genome Biol 2016; 17:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Li J, Poursat M-A, Drubay D, et al. A dual model for prioritizing cancer mutations in the non-coding genome based on germline and somatic events. PLoS Comput Biol 2015;11(11):e1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kalender Atak Z, Imrichova H, Svetlichnyy D, et al. Identification of cis-regulatory mutations generating de novo edges in personalized cancer gene regulatory networks. Genome Med 2017;9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Li S, Alvarez RV, Sharan R, et al. Quantifying deleterious effects of regulatory variants. Nucleic Acids Res 2017;45:2307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hu M, Deng K, Qin Z, et al. Understanding spatial organizations of chromosomes via statistical analysis of Hi-C data. Quant Biol 2013;1(2):156–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Sati S, Cavalli G.. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma 2017;126(1):33–44. [DOI] [PubMed] [Google Scholar]
- 138. Cui C, Shu W, Li P.. Fluorescence in situ hybridization: cell-based genetic diagnostic and research applications. Front Cell Dev Biol 2016;4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Fullwood MJ, Ruan Y.. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem 2009;107(1):30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Heidari N, Phanstiel DH, He C, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res 2014;24(12):1905–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Le Scouarnec S, Gribble SM.. Characterising chromosome rearrangements: recent technical advances in molecular cytogenetics. Heredity 2012;108(1):75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Jäger R, Migliorini G, Henrion M, et al. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun 2015;6(1):6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Whitfield TW, Wang J, Collins PJ, et al. Functional analysis of transcription factor binding sites in human promoters. Genome Biol 2012;13(9):R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yang L, Titlow J, Ennis D, et al. Single molecule fluorescence in situ hybridisation for quantitating post-transcriptional regulation in Drosophila brains. Methods 2017;126:166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Xu H, Sepúlveda LA, Figard L, et al. Combining protein and mRNA quantification to decipher transcriptional regulation. Nat Methods 2015;12(8):739–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Fok ET, Scholefield J, Fanucchi S, et al. The emerging molecular biology toolbox for the study of long noncoding RNA biology. Epigenomics 2017;9(10):1317–27. [DOI] [PubMed] [Google Scholar]
- 147. Sander JD, Joung JK.. CRISPR-Cas systems for genome editing, regulation and targeting. Nat Biotechnol 2014;32(4):347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wang K-H, Chuv S-C, Chu T-Y.. Loss of calponin h1 confers anoikis resistance and tumor progression in the development of high-grade serous carcinoma originating from the fallopian tube epithelium. Oncotarget 2017;8:61133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Han Y, Slivano OJ, Christie CK, et al. CRISPR-Cas9 genome editing of a single regulatory element nearly abolishes target gene expression in mice—Brief report. Arterioscler Thromb Vasc Biol 2015;35(2):312–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kim YW, Kim A.. Deletion of transcription factor binding motifs using the CRISPR/spCas9 system in the β-globin LCR. Biosci Rep 2017;37(4):BSR20170976.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhuo C, Hou W, Hu L, et al. Genomic editing of non-coding RNA genes with CRISPR/Cas9 ushers in a potential novel approach to study and treat schizophrenia. Front Mol Neurosci 2017;10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Davenport AP, Hyndman KA, Dhaun N, et al. Endothelin. Pharmacol Rev 2016;68(2):357–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ha F, Khalil H.. Crohn’s disease: a clinical update. Therap Adv Gastroenterol 2015;8(6):352–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Jostins L, Ripke S, Weersma RK, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491(7422):119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Flint J, Munafò M.. Schizophrenia: genesis of a complex disease. Nature 2014;511(7510):412–13. [DOI] [PubMed] [Google Scholar]
- 156. Mansur Y, Rojano E, Ranea JA, et al. Analyzing the effects of genetic variation in noncoding genomic regions In: Deigner Hans-Peter, Kohl Matthias (eds) Precision Medicine Tools and Quantitative Approaches, 1st edn. 2018, 374. [Google Scholar]
- 157. Vinagre J, Almeida A, Pópulo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun 2013;4:2185. [DOI] [PubMed] [Google Scholar]
- 158. Heidenreich B, Rachakonda PS, Hemminki K, et al. TERT promoter mutations in cancer development. Curr Opin Genet Dev 2014;24:30–7. [DOI] [PubMed] [Google Scholar]
- 159. Cuykendall TN, Rubin MA, Khurana E.. Non-coding genetic variation in cancer. Curr Opin Syst Biol 2017;1:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]