Abstract
Long noncoding RNAs (lncRNAs) are a group of transcripts that are longer than 200 nucleotides (nt) without coding potential. Over the past decade, tens of thousands of novel lncRNAs have been annotated in animal and plant genomes because of advanced high-throughput RNA sequencing technologies and with the aid of coding transcript classifiers. Further, a considerable number of reports have revealed the existence of stable, functional small peptides (also known as micropeptides), translated from lncRNAs. In this review, we discuss the methods of lncRNA classification, the investigations regarding their coding potential and the functional significance of the peptides they encode.
Keywords: long noncoding RNA (lncRNA), small ORF, small peptide, coding-potential prediction
Introduction
Long noncoding RNAs (lncRNAs) are a heterogeneous group of RNAs >200 nucleotides (nt) in length that lack coding potential, but their gene structures resemble those of RNA polymerase II products, such as mRNAs [1–3]. For a decade, since their discovery in the 1990s, lncRNAs were arguably considered to be junk or by-products of transcription [4]. In 2007, with the aid of high-throughput sequencing technologies, the ENCODE project unveiled an extensive set of noncoding elements with biochemical functions, which largely overlapped with the lncRNA gene loci from mammalian genomes [5]. Since then, researchers have been exploring these cryptic yet possibly functional noncoding transcripts from genomes. As lncRNAs are known to be expressed in specific cell types and developmental stages, early studies aimed at the computational identification of novel transcribed regions, using complementary DNA (cDNA); RNA sequencing (RNA-seq); chromatin immunoprecipitation followed by sequencing (ChIP-seq), 3P-seq and many other types of transcriptome data; and the transcriptome assembly of high-throughput short reads from different cell types and stages [6–13].
As only sequence and locus information were available for candidate noncoding transcripts, lncRNA classifications were initially based on the features that could be derived from sequences, such as predicted open reading frame (ORF) length, sequence conservation and sequence similarity to known coding genes [10–23]. However, the introduction of high-throughput sequencing of ribosome-protected fragments (Ribo-seq) helped us to examine the ribosome association of candidate transcripts in vivo [24]. Surprisingly, many studies repeatedly reported that some lncRNAs showed a strong association with ribosomes, although the association does not always imply that they are actively translated [9, 25–29]. To address whether the ribosomes associated with lncRNAs actively translate them, several studies attempted to detect either movement of the translating ribosome along the lncRNA transcripts, using Ribo-seq [26, 30–36] or peptides coded by lncRNAs, using mass spectrometry (MS), which is an analytical tool that ionizes peptides and measures their mass-to-charge ratio to identify their amino acid (aa) sequences [37–39].
Meanwhile, functional studies of a few well-conserved lncRNAs, such as XIST [40–42], OIP5-AS1 [7], NEAT1 [43, 44] and MALAT1 [45–47], and of cancer-related lncRNAs, such as GAS5, LUCAT1, HOTAIR and ANRIL [48–51], have shed light on the various regulatory roles of lncRNAs in cells. On investigating the functions of lncRNAs, a few studies confirmed that some lncRNAs indeed had small open reading frames (sORF, length <300 nt) that could code for a short peptide with key biological functions [52–63]. The presence of functional small peptides coded by the lncRNAs suggests that these lncRNAs could play dual roles, with both RNA and peptides, and therefore should be reclassified as bifunctional RNAs [64–66]. This review provides a brief overview of computational and combinatorial approaches for the classification of coding/noncoding RNAs and for the systematic identification of small peptides coded by these transcripts and summarizes functional small peptides encoded by invertebrate and vertebrate lncRNAs. Finally, this review discusses the clinical implications of these small peptides and their host lncRNAs.
Classification and annotation of coding and noncoding RNAs
Computational approaches for lncRNA classification
The advancement in RNA-seq and bioinformatics technologies led to the genome-wide identification of novel transcripts from plant and animal genomes. Although the assessment of coding potential was originally devised to detect novel protein-coding genes, the large number of novel transcripts sequenced with RNA-seq motivated researchers to apply it to distinguish protein-coding and noncoding RNAs. Early methods of estimating coding potential were intended to predict characteristics of translated RNAs from the sequence and locus information of novel transcripts. Intrinsic sequence features, including ORF length, sequence homology to known protein sequences, sequence conservation, nucleotide composition, substitution ratio and secondary structure, were invented and used for the calculation of coding potential (Table 1). ORF length is one of the most commonly used features. The use of ORF length is based on the premise that genuine protein-coding genes would include ORFs of sufficient lengths [10, 13–16, 19–22]. Other features include ORF integrity (whether the ORF includes start and stop codons to define its range) [22]. Protein homology is used to search for conserved segments among protein families and is often assessed by alignment with a protein database [14, 15, 20, 21, 67]. Conservation is considered to be a powerful feature because it is known that lncRNAs are less conserved than mRNAs [14, 18, 20, 23, 67]. Nucleotide composition refers to the frequency of certain k-mers or codon usage in coding or noncoding sequences [10–14, 16, 17, 19, 20, 22, 23]. The substitution ratio is the ratio between synonymous and nonsynonymous mutations in a given sequence and is used to assess whether the mutation profile of a given sequence is better explained by those of protein-coding sequence or those of noncoding sequence [17, 18, 67]. Secondary structures were applied to lncRNAs, as it was hypothesized that functional noncoding RNAs would have different secondary structures from mRNAs [12, 14, 23]. However, prediction algorithms available at the time did not consider the biological characteristics of lncRNAs, and, therefore, some features could be less accurate [12]. To train a computational coding-potential model without any biases, many tools adopted machine learning techniques using known coding and noncoding transcripts as training/test data sets. The most popular method, the coding potential calculator (CPC), takes advantage of BLAST-related features (sequence homology) and ORF-related features to train their model using a support vector machine (SVM) [15]. CPC is favored by many researchers, because of its robust performance, despite relatively long running times. Recently, an updated version of CPC, coding potential calculator 2 (CPC2), was introduced [22]. Unlike the original version, CPC2 excludes the time-consuming sequence alignment step and examines four intrinsic features: Fickett TESTCODE score [68], ORF length, ORF integrity and isoelectric point (the pH at which the peptide carries zero net charge), as implemented in other computational tools (Table 1). In addition to CPC and CPC2, other SVM-powered computational approaches, such as CONC, PORTRAIT, CNCI, iSeeRNA and PLEK, have also been developed with different intrinsic features (Table 1). Moreover, Linc-SF combines genetic algorithm and SVM (GA-SVM) techniques to optimize the classification model [12]. Other machine learning approaches, such as logistic regression [coding potential assessment tool (CPAT)], random forest (LncRNA-ID, COME), deep stacking network (lncRNA-MFDL), and the expectation–maximization (EM) algorithm (PhyloCSF), have also been applied to classify the coding/noncoding transcripts (Table 1). Although there have been a few other approaches, such as RNAcode [67], that avoid the training process to enable a more generic use of the algorithm, this trend toward using machine learning approaches continued after the ribosome profiling data were used in the field of classifying coding/noncoding transcripts.
Table 1.
Computational lncRNA classification
| Method | Machine learning technique | Feature |
Result |
Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF length | Protein homology | Conservation | Nucleotide composition | Substitution ratio (dN/dS) | Secondary structure | sORF detection | Coding/ noncoding prediction | P value | |||
| CONC | SVM | O | O | O | O | O | O | [14] | |||
| CPC | SVM | O | O | O | [15] | ||||||
| PORTRAIT | SVM | O | O | [16] | |||||||
| sORF finder | – | O | O | O | O | O | [17] | ||||
| PhyloCSF | EM | O | O | O | [18] | ||||||
| RNAcode | – | O | O | O | O | O | [67] | ||||
| CNCI | SVM | O | O | O | [10] | ||||||
| CPAT | Logistic regression | O | O | [19] | |||||||
| iSeeRNA | SVM | O | O | O | O | O | [20] | ||||
| PLEK | SVM | O | O | [11] | |||||||
| Linc-SF | GA-SVM | O | O | ? | ? | [12] | |||||
| LncRNA-ID | Balanced random forest | O | O | ? | [21] | ||||||
| lncRNA-MFDL | Deep stacking network | O | O | ? | ? | [13] | |||||
| CPC2 | SVM | O | O | O | O | [22] | |||||
| COME | Balanced random forest | O | O | O | O | [23] | |||||
Note: ‘?’ mark indicates that the corresponding information could not be found.
Classification of lncRNA using experimental data
Ribosome profiling, also known as ribosome footprinting, was introduced in 2009 by Ingolia and his colleagues [24]. Ribosome profiling is a technique that reads ribosome-protected RNA fragments (RPFs), which are obtained by stalling ribosomes on RNA with translation-inhibiting chemicals, applying RNase to eliminate unprotected RNAs and sequencing the remaining RNA molecules [24]. Ribosome profiling has enabled the observation of the global translation status and the computational analysis of in vivo translation. A short time after its introduction, Ribo-seq was applied to examine not only ribosome association but also ribosome dynamics during translation to classify coding/noncoding transcripts (Table 2). Ingolia and his colleagues first devised ribosome association as a measure of translation and later adjusted the value with the expression level of each genes, which was termed the translation efficiency (TE) [24, 69]. As an initial metric, TE was based on the amount of RPFs associated with a transcript, and it could not distinguish translating ribosomes from either nonspecific or nontranslating ribosome interactions. To address this issue, diverse derivatives of RPF coverage have been developed as extensions, and some were fed into machine learning algorithms in combinatorial methods. Shortly after the introduction of TE, a method that compares the RPF depth within ORFs to those in untranslated regions (UTRs) was introduced by two groups [26, 70]. Although the metrics they used to measure the features were similar, their conclusions were different with respect to the translation activity of lncRNAs. One study claimed that most lncRNAs are not actively translated [26], while the other claimed that some lncRNAs contain actively translating regions [70]. Many others also deduced conclusions similar to the latter study on implementing certain forms of RPF coverage or the coverage of RPFs with specific lengths [27, 30–33, 39, 71] (Table 2). To further emphasize the characteristics of active translation using RPFs, features representing ribosome dynamics were described in following studies. Bazzini et al. [30] suggested a new metric, ORFscore, which tests the presence of three-nucleotide periodicity. The periodicity originates from the codon-base translocation of ribosomes during translation along mRNAs, which is often represented by the coverage of ribosome reads mapped to the first, second and third nucleotide positions of a codon (also called sub-codon position), with the fraction of the mapped reads being skewed toward the first position [72]. To assign the mapped reads to a certain nucleotide position, it is essential to predict the position of the ribosome P-site on the reads. Normally, mammalian ribosome covers approximately 30 nt of RNA, and, therefore, the P-site is considered to be located at the 15th nucleotide from the 5′-end of the protected read. The three-nucleotide periodicity was adopted by most succeeding methods, such as RibORF classifier, riboHMM, SPECtre, RiboTaper, Rp-Bp and TERIUS (Table 2) [31–36]. As the P-site in Ribo-seq reads can vary according to the RPF read length, these methods should include the information for the P-site to calculate the three-nucleotide periodicity. For instance, RiboTaper requires users to manually detect the P-sites in reads with certain lengths [34], whereas Rp-Bp automatically infers P-sites by Bayesian inference [35].
Table 2.
Combinatorial lncRNA classification
| Method | Experimental data | Feature |
Result |
Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Three-nucleotide periodicity | RPF coverage | RPF length distribution | sORF detection | Coding/ noncoding prediction | P value | |||
| RRS | Ribo-seq | O | [26] | |||||
| TOC | Ribo-seq | O | ? | ? | ? | [70] | ||
| FLOSS | Ribo-seq | O | O | [27] | ||||
| ORFscore | Ribo-seq | O | O | [30] | ||||
| PROTEOFORMER | Ribo-seq, MS | O | O | O | [39] | |||
| ORF-RATER | Ribo-seq | O | O | O | [71] | |||
| RibORF | Ribo-seq | O | O | O | O | O | [31] | |
| riboHMM | Ribo-seq, RNA-seq | O | O | O | [32] | |||
| SPECtre | Ribo-seq | O | O | O | O | [33] | ||
| RiboTaper | Ribo-seq, RNA-seq | O | O | O | O | [34] | ||
| Rp-Bp | Ribo-seq | O | O | [35] | ||||
| TERIUS | Ribo-seq | O | O | [36] | ||||
Note: ‘?’ mark indicates that the corresponding information could not be found.
However, the type of experimental data used to detect translated ORFs is not limited to Ribo-seq. MS spectra and global translation initiation sequencing (GTI-seq) have also been incorporated into classification tools to add additional layers of evidence. GTI-seq is a technique that uses two translation inhibitors, lactimidomycin (LTM) and cycloheximide (CHX), to differentiate ribosome initiation from elongation. GTI-seq has the potential to identify translation initiation sites because CHX binds to all translating ribosomes, while LTM preferentially binds to initiating ribosomes with free E-sites. PROTEOFORMER uses both harringtonine- and LTM-treated Ribo-seqs to identify translated regions and translation initiation sites and integrates MS data for peptide identification [39]. Lee and his colleagues [73] developed a GTI-seq technique by combining Ribo-seq data, generated from samples treated with two different translation-inhibiting chemicals, to generate two types of Ribo-seq signal landscapes and to enhance the accuracy of annotating the translation initiation site.
Methods for detecting sORFs
The extent to which lncRNAs can produce small peptides is still debatable; however, it is now widely accepted that some lncRNAs can be translated [64, 66]. Although computational approaches that do not use Ribo-seq and/or MS data have been successful in detecting coding potential in RNAs, the majority of them lack the capability to detect sORFs that could encode small peptides. Most tools, including CONC, CPC, PORTRAIT, CNCI, CPAT, iseeRNA, LncRNA-ID, lncRNA-MFDL and CPC2, consider sORFs to be a feature of noncoding transcripts [10, 13–16, 19–22]. For instance, a group specified that the length distributions of ORFs originated from noncoding transcripts and those originating from coding transcripts were distinct and most clearly separated at approximately 300 nt [19]. Only sORF finder is capable of detecting sORFs in transcripts [17] (Table 1).
In contrast, the combinatorial approaches that use Ribo-seq and MS data successfully detected sORFs in the UTRs of mRNAs and in noncoding RNAs. These approaches that rely on experimental data are free from length restrictions on ORFs, thereby enabling the detection of sORFs. Some groups aimed to design a tolerant classifier by implementing length normalization or additional translation signals independent of ORF length. For instance, translated ORF classifier (TOC) used several ribosome-protected read count per kilobase of ORF exons-based features, which were applied to a random forest classifier [70]. ORF-RATER is based on a nonnegative logistic regression of RPF coverage with harringtonine- and LTM-indicated translation start site data [71]. Although PROTEOFORMER takes similar steps as ORF-RATER, which analyzes translation start signals, it requires an RPF coverage of >85% of exons, hindering sORF detection [39]. In contrast, periodicity-based classification methods tend to be less susceptible to scarce ribosome coverage, although they could still suffer from a lack of a statistical significance arising from the scarce RPF coverage. For instance, ORFscore used a chi-square test to imply the significance of three-nucleotide periodicity and detected 190 sORFs coding for peptides of 20–100 aa in length when analyzing Ribo-seqs from zebrafish embryos [30]. RibORF calculates the maximum entropy value, which considers the fraction of RPF reads at the first and second nucleotides of codons to be a feature for building an SVM model [31]. Using RibORF, an 80-aa-ORF in the CEBPZOS lncRNA gene, along with other sORFs in upstream and downstream ORFs, were identified in a breast epithelial cell line [31]. riboHMM uses a hidden Markov model that considers nucleotide triplets associating with RPFs in all three possible frames to be emission probabilities and translated or untranslated states to be hidden states, resulting in a robust detection of sORFs in transcripts, even with a low RPF coverage [32]. In fact, more than half of the novel ORFs identified by riboHMM were shorter than 30 aa in length [32]. In addition, RiboTaper uses a Fourier transformation technique following a multitaper spectral density estimation of RPF signals at P-sites to detect the periodicity, allowing the discovery of multiple upstream ORFs in HEK293 cells [34]. Rp-Bp calculates the marginal likelihood ratios of the coding and noncoding profiles to determine which profile better describes observed data, leading to the detection of approximately 2500 sORFs in HEK293 cells [35]. In summary, computational methods can identify all possible ORFs, including those with low expression levels and without experimental data, but their results may include ORFs that are not translated. In contrast, combinatorial methods can identify ORFs that are actively translated, are non-canonical or are species specific. However, experimental data are needed to run combinatorial methods and often additional data are needed, such as matched RNA-seq; therefore, transcripts with low expression levels are likely to be neglected.
LncRNAs that encode small peptides
Numerous studies have identified translated ORFs from animal and plant lncRNAs using the previously mentioned approaches (Table 3), among which, RPFs, along with other sequence-related features, were most commonly used to detect sORFs in lncRNAs. For instance, a group profiled RPFs from breast epithelial cell and BJ fibroblast cells and found 1204 translated ORFs in 510 lncRNAs using the RibORF classifier [31]. Of 510 lncRNAs with translated ORFs, 412 encoded peptides <100 aa long, and 19 produced peptides <10-aa-long. Analyzing 93 human translated lncRNAs with orthologs in mice, they found that 41 encoded peptides are conserved in mice, presumably implicating their functional importance. Moreover, the translated lncRNAs were preferentially localized in the cytoplasm compared to other lncRNAs [31]. Crappé et al. [76] analyzed public RPFs, to find sORFs embedded in noncoding RNA (ncRNA) genes, and cryptic intergenic loci, using sORF finder, and found 528 and 226 sORFs, respectively, with supporting ribosome association with both ncRNAs and intergenic regions. Of the 528 sORFs found in ncRNAs, 514 were from lncRNAs (Table 3).
Table 3.
Studies that identified small ORFs and short peptides in lncRNA
| Species | Approacha | Method | Experimental data | Translated ORFs detected in lncRNAs | Translated sORFs detected in lncRNAs | MS evidence | Reference |
|---|---|---|---|---|---|---|---|
| Human | E | – | MS | – | – | 8 peptides | [74] |
| C+E | ORFscore | Ribo-seq | 261 from lncRNAs | 261 | – | [30] | |
| C+E | RibORF | Ribo-seq | 1204 from 510 lncRNAs | – | – | [31] | |
| C+E | PhyloCSF | Ribo-seq, MS | 354 from lncRNAs | 354 | 22 peptides | [75] | |
| C+E | Hexamer-based coding score | Ribo-seq | 143 from 390 lncRNAs | 99 | – | [28] | |
| C+E | RibORF | Ribo-seq, MS | 925 from 233 lncRNAs | 686 | 18 lncRNAs | [29] | |
| Mouse | C+E | sORF finder | Ribo-seq | 514 from lncRNAs | 514 | – | [76] |
| C+E | Hexamer-based coding score | Ribo-seq | 137 from 403 lncRNAs | 107s | – | [28] | |
| C+E | PhyloCSF | MS | 98 from lncRNAs | 98 | 11 peptides | [75] | |
| Zebrafish | C+E | ORFscore | Ribo-seq, MS | 535 from lncRNAs | 535 | 6 peptides | [30] |
| C+E | PhyloCSF | MS | 99 from lncRNAs | 99 | – | [75] | |
| C+E | Hexamer-based coding score | Ribo-seq | 379 from 726 lncRNAs | 155 | – | [28] | |
| Fruit fly | C+E | PhyloCSF | MS | 53 from lncRNAs | 53 | 2 peptides | [75] |
| C+E | Hexamer-based coding score | Ribo-seq | 7 from 22 lncRNAs | 7 | – | [28] | |
| Yeast | E | – | Ribo-seq, Polysome-seq | 47 from 331 lncRNAs | 47 | – | [77] |
| C+E | Hexamer-based coding score | Ribo-seq | 5 from 6 lncRNAs | 5 | – | [28] | |
| Worm | C+E | PhyloCSF | MS | 81 from lncRNAs | 81 | 1 peptide | [75] |
| Arabidopsis thaliana | C+E | Hexamer-based coding score | Ribo-seq | 43 from 93 lncRNAs | 43 | – | [28] |
Note: Approacha is denoted as E if the method is purely experimental, C if computational and C + E if combinatorial.
Although ribosome profiling successfully identified sORFs, ribosome occupancy does not guarantee an active translation signal that produces peptides. Therefore, several studies have adopted peptidomics that integrate RNA-seq, Ribo-seq and MS data to explore the peptide product from lncRNA sORFs (Table 3). For instance, Wang and colleagues identified actively translated sORFs using RibORF and detected 1332 ribosome-associated lncRNAs in eight human cell lines. Among those, 233 lncRNAs included 686 sORFs with RPF evidence, 18 of which were confirmed to express small peptides by MS data [29]. Conversely, Bazzini and colleagues [30] first analyzed Ribo-seq data using ORFscore in lncRNAs expressed in zebrafish, identifying 535 sORFs with ribosome association from lncRNAs. To verify the presence of peptides translated from sORFs, they used MS data from zebrafish embryos and confirmed the presence of peptides translated from six sORFs.
Many translated sORFs were also detected in invertebrate and metazoan lncRNAs, using Ribo-seq and/or MS data (Table 3). Smith et al. [77] identified 47 sORFs from 331 unannotated RNAs with ribosome occupancy, 20 of which were evolutionarily conserved in other yeasts. Mackowiak et al. [75] developed a computational pipeline that uses MS data to identify conserved sORFs in five vertebrate and invertebrate species. They predicted 2002 conserved sORFs in the UTRs of mRNAs or ncRNAs and validated them using MS spectra data from human cell lines, mouse cells and tissues, and whole animal zebrafish, fly and worm samples. As a result, a number of novel peptides were discovered in each species, including novel peptides from 36 lncRNAs (Table 3).
Functional small peptides
Muscle-related small peptides
Despite the discovery of many small peptides coded by sORFs, the biological functions of only a handful of them have been described (Table 4). These peptides are usually conserved and are involved in a wide range of biological processes. Recent studies reported lncRNA-encoded small peptides related to specific muscle developmental processes in human and mouse, which participate in muscle regeneration and development (Table 4). Matsumoto and colleagues [59] used a peptidomics approach in human and mouse cell lines and tissues and identified an lncRNA encoding a peptide that is conserved in human and mouse. This small peptide, SPAR, is 90-aa-long in human and 75-aa-long in mouse, regulates mTORC1 activation and inhibits muscle regeneration. Zhang et al. [60] identified an 84-aa-long conserved peptide, Minion, which is involved in the regulation of muscle cell fusion in mouse. The human homologue of Minion was also encoded by a transcript previously annotated as an lncRNA and showed a similar function to its mouse counterpart. Similar functional small peptides related to muscle tissues were also discovered in other model organisms. One study identified a 46-aa-long evolutionarily conserved peptide from an lncRNA. This peptide, named myoregulin (MLN), interacts with the sarcoendoplasmic reticulum calcium transport ATPase (SERCA) calcium-ATPase and inhibits calcium reuptake into the sarcoplasmic reticulum [57]. The other peptide related to this function, DWORF, a 34-aa-long peptide, was also identified in mouse and was shown to regulate calcium reuptake [58]. DWORF enhanced the SERCA calcium-ATPase activity and calcium reuptake into the sarcoplasmic reticulum by displacing SERCA inhibitors, including phospholamban, sarcolipin and MLN. Invertebrates also have similar functional lncRNA-encoded peptides. In Drosophila, a member of the SERCA regulating family was initially identified as an ncRNA gene and encodes a transmembrane peptide, sarcolamban (Scl), of 28- or 29-aa [55]. Sarcolamban also inhibits the SERCA calcium-ATPase and regulates heart contractions.
Table 4.
Known functions of small peptides coded by lncRNAs
| Species | Peptide name | LncRNA | Peptide length (aa) | Function | Detailed function | Reference |
|---|---|---|---|---|---|---|
| Human | SPAR | ENSG00000235387 | 90 | Muscle and cancer-related (oncogenic) | Negatively regulates mTORC1 activation and inhibits muscle regeneration | [59] |
| Minion/myomixer | ENSG00000262179 | 84 | Muscle-related | Regulates muscle development and muscle cell fusion | [60] | |
| HOXB-AS3 | ENSG00000233101 | 53 | Cancer-related (tumor-suppressive) | Suppresses colon cancer aerobic glycolysis by inhibiting hnRNP A1-dependent PKM splicing | [62] | |
| NOBODY | ENSG00000204272 | 71 | Cancer-related and others | Involved in mRNA processing and negatively regulates P-body association | [63] | |
| Mouse | MLN | ENSMUSG00000019933 | 46 | Muscle-related | Interacts with SERCA (calcium-ATPase) and inhibits calcium reuptake into the sarcoplasmic reticulum | [57] |
| DWORF | ENSMUSG00000103476 | 34 | Muscle-related | Enhances SERCA activity and calcium reuptake into the sarcoplasmic reticulum | [58] | |
| SPAR | ENSMUSG0000002847 | 75 | Muscle and cancer-related (oncogenic) | Negatively regulates mTORC1 activation and inhibits muscle regeneration | [59] | |
| Minion/myomixer | ENSMUSG00000079471 | 84 | Muscle-related | Regulates muscle development and muscle cell fusion | [60, 61] | |
| Zebrafish | Toddler | ENSDARG00000094729 | 58 | Others | Activates G protein-coupled apelin receptor (APJ)/APJ signaling and promotes cell movement during gastrulation | [56] |
| Fruit Fly | Tarsal-less/tal | FBgn0087003 | 11 and 32 | Others | Activates the transcription factor responsible for cuticle formation | [53] |
| Scl | FBgn0266492 | 28 and 29 | Muscle-related | Regulates calcium transport and muscle contraction | [55] | |
| Pgc | FBgn0016053 | 71 | Others | Represses CTD2 serine phosphorylation in germline progenitor cells | [54] | |
| Soy bean | ENOD40 | GmENOD40 | 12 and 24 | Others | Interacts with sucrose synthase and is required for plant–bacteria symbiotic interactions | [52] |
Cancer-related small peptides
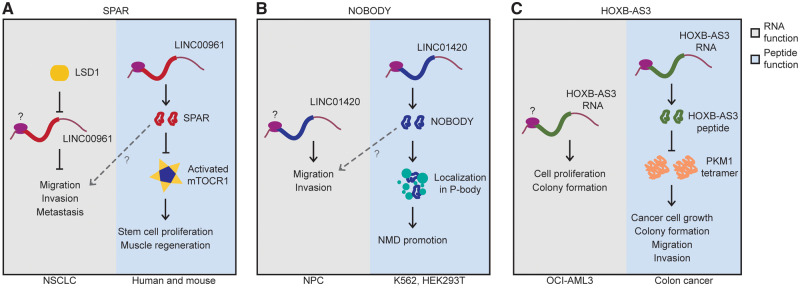
Recent studies found that small peptides are expressed or regulated during cancer progression, suggesting their roles in cancer development (Figure 1A and B). Matsumoto and his group [59] confirmed that the downregulation of the SPAR peptide resulted in the upregulation of mTORC1, even though the expression of SPAR RNA was unperturbed. Later, Jiang et al. [78] reported that the expression levels of the lncRNA encoding SPAR was inversely correlated with clinical outcomes in non-small cell lung cancer (NSCLC) (Figure 1A). The NOBODY peptide, a 71-aa-long peptide encoded by the lncRNA LINC01420, was also discovered before the reporting of a connection between its lncRNA and nasopharyngeal carcinoma (NPC) [79]. The lncRNA LINC01420 was negatively correlated with overall survival, and its knockdown by small interfering RNA reduced the migration and invasion of NPC cells. The authors observed that the expression of LINC01420 was elevated in both NPC cell lines and tissue samples and that NPC patients with high LINC01420 expression tended to show poor overall survival rates (Figure 1B). The authors, however, did not verify whether the molecule affecting cancer progression was the peptide or the lncRNA, leaving the function of the corresponding gene inconclusive. Huang et al. [62] reported the function of a 53-aa-long conserved peptide encoded in HOXB-AS3, which appeared to be downregulated in cancers. The expression of HOXB-AS3, previously annotated as an lncRNA, was downregulated in acute myeloid leukemia (AML) [80]. Ribo-seq implied that HOXB-AS3 could produce the encrypted peptide, which was also shown to be downregulated in cancer cells. In fact, HOXB-AS3 peptide, but not the RNA itself, suppressed cancer cell growth, colony formation, migration, invasion and tumorigenesis by inhibiting hnRNP A1-dependent PKM splicing [62] (Figure 1C).
Figure 1.
Cancer-related lncRNAs with functional peptides. Left side (gray box) of each figure shows the RNA function. (A)LINC00961 related to NSCLC. (B)LINC01420 related to NPC. (C)HOXB-AS3 transcript related to AML in OCI-AML3 cells. The right side (blue box) shows the functions for the peptides. (A) SPAR inhibiting mTORC1 activation. (B) NOBODY promoting NMD in K562 and HEK293T cells. (C) HOXB-AS3 peptide regulating PKM splicing and suppressing cancer growth.
Other functional small peptides
Other studies showed that small peptides from lncRNAs also participate in other biological processes (Table 4). The peptide NOBODY was identified through a proteomics approach in both K562 and HEK293T cell lines [63]. This peptide is thought to regulate mRNA processing by interacting with the mRNA decapping complex and to act as a negative regulator of P-body association. Pauli and colleagues [56] identified a zebrafish peptide of 58 aa, Toddler, from a transcript annotated as an lncRNA in zebrafish, mouse and human. Toddler binds to the apelin receptor (APJ) and induces G protein-coupled receptor signaling to promote cell movement during gastrulation in zebrafish. In soybean, using peptide mass fingerprinting, two small peptides (12- and 24-aa-long) translated from the ENOD40 transcript were identified to interact with sucrose synthase, which is required for plant symbiosis [52].
Discussion
The discovery of functional small peptides translated from lncRNAs has encouraged researchers to reexamine the roles of lncRNAs. The repertoire of biological processes that lncRNAs are involved in has grown rapidly, and lncRNAs serve as biomarkers and potential drug targets in many types of diseases [81]. However, the working mechanisms of disease-associated lncRNAs are largely unknown, and their coding potential under diseased conditions are rarely discussed, even for those that are known to harbor ORFs. For example, MALAT1 and KCNQ1OT1 have been associated with cardiovascular disease and are even used as biomarkers, but whether their functions are dependent on the lncRNA or the peptide is not clear [82]. The steroid receptor RNA activator (SRA) lncRNA produces SRA protein in breast cancer cells, and recently, the lncRNA showed a strong oncogenic property in cervical cancer; yet, the contribution of the SRA protein was not explored [83, 84]. The study of the translation status of these lncRNAs might shed light on their significance and clinical implications.
The existence of functional peptide products of lncRNAs emphasizes the need to thoroughly separate the RNA functions and peptide functions of lncRNAs. As in the case of HOXB-AS3 and SPAR, researchers that aim to investigate function of lncRNAs that encode a small peptide should clearly discriminate whether the acting molecule is the peptide, the lncRNA or both. In addition, when studying lncRNA function, the isoform that is responsible for the phenotype in question should be examined. This process is crucial, especially in cancer studies, where most isoform candidates are selected by differential expression. Several reports have shown that the major form of an effector lncRNA gene changes between alternative isoforms in cancer, without prominent differences in the gene expression level. It is also widely accepted that different isoforms may have different coding potentials and that changes in the expression levels of isoforms can affect the proteome of cells. Therefore, researchers should first define the exact effector and further inspect the behavior of the molecule to clarify its role.
Although researchers have focused on elucidating the functions of lncRNAs, studying the regulation of lncRNA expression is undoubtedly an equally crucial research focus. Given that most known functional small peptides are conserved in other species, the short, highly conserved regions in lncRNAs may be necessary for both producing peptides and the quality control of produced RNAs. Nonsense-mediated mRNA decay (NMD) is a surveillance mechanism that degrades erroneous mRNAs and reacts to spurious translation followed by premature stop codons. As lncRNAs that harbor sORFs are likely to be targeted by NMD, the extent of NMD targeting against lncRNAs should also be explored.
LncRNAs, or the peptides coded by them, are expected to be the missing pieces of many molecular mechanisms. Encouraged by the discovery of many novel sORFs residing in genomic locations that were previously thought to be noncoding, repositories of sORFs or small peptides identified by ribosome profiling and/or MS data, such as sORFs.org [85] and SmProt [86], have been developed. In the case of sORF.org, which stores all published sORFs from five species (human, mouse, rat fly, zebrafish and Caenorhabditiselegans), this database provides coding-potential evidence, such as PhyloCSF, FLOSS and ORFscore. SmProt incorporates small peptides from eight species (human, mouse, rat, zebrafish, fly, yeast, C. elegans and Escherichiacoli). However, the coding nature of lncRNAs is still largely unknown, and the group remains heterogeneous, thus far. More rigorous investigation of lncRNAs and the small peptides hidden within them would lead to a profound understanding and give new insights into numerous unsolved conundrums in the fields of biological and medical sciences.
Key Points
We presented computational and combinatorial approaches that use various experimental data to classify coding and noncoding RNAs and identify sORFs that encode small peptides.
Based on supporting experimental data, we demonstrated the challenges in identifying small peptides and the methods to improve their detection.
We summarized a list of functional peptides translated from lncRNAs, many of which are evolutionarily conserved in other species and some of which are known to be involved in muscle-related functions and cancer development.
Acknowledgements
The authors thank all BIG lab members for critical reading and comments.
Funding
This work was supported by the Bio and Medical Technology Development Program and the Basic Science Research Program through the National Research Foundation (NRF), funded by the Ministry of Science and ICT (grant numbers NRF-2017M3A9G8084539, 2018R1A2B2003782 and NRF-2014M3C9A3063541), and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), which was funded by the Ministry of Health and Welfare (grant number HI15C1578).
Box A.
Computational coding RNA classifier (related to Table 1)
Coding or noncoding
Coding or noncoding (CONC) [14] is one of the first SVM classifiers developed to detect protein-coding sequences. It uses various sequence-related features, secondary structure information and homologs from database searches to identify potential peptides. It is designed to compare input DNA/RNA sequences to confirmed coding cDNAs for eukaryotic proteins. CONC results predict the class (coding or noncoding) of an input sequence.
Coding potential calculator
Coding potential calculator (CPC) [15] is another SVM-based classifier that implements a protein database search for homology to known proteins. It also searches for possible ORFs and combines alignment-related features and ORF quality-related features to train the model. The protein database search makes CPC slow and reliant on the database, but it also improves the specificity greatly. CPC works with RNA sequences and provides predicted classes along with SVM scores. Recently, CPC was upgraded to CPC2 [22].
PORTRAIT
PORTRAIT [16] was developed to enable the coding classification of transcriptomes generated from poorly characterized species or from low sequence quality. It considers the scenario where most of the input sequences include truncated proteins with low conservation. PORTRAIT results provide coding and noncoding probabilities. PORTRAIT is also available online.
sORF finder
sORF finder [17] is specifically designed to detect sORFs with high coding potentials. It uses a hexamer frequency table to identify coding sequences, which means it requires prior knowledge. However, sORF finder currently provides hexamer table information for 11 organisms. sORF finder accepts sequence data and calculates the number and sequences of detected sORFs, the synonymous substitution ratio and the P value. A sORF finder Web server is also available.
PhyloCSF
PhyloCSF [18] attempted to overcome the limitations of previous classifiers that relied heavily on homology to known proteins. PhyloCSF examines evolutionary signatures in the form of synonymous or nonsynonymous codon substitution rates. PhyloCSF requires users to provide cross-species multiple sequence alignment results as inputs. It calculates a score that indicates the likelihood ratio of the protein-coding sequence evolution model. PhyloCSF also provides the start and end positions of the coding regions that it detects and the aa sequences corresponding to the regions.
RNAcode
RNAcode [67] implements evolutionary signatures regarding the reading frame, such as synonymous/conservative mutations, and conservation by gap scoring. However, it does not use species-specific characteristics or machine learning techniques to ensure general usage of the program. Like PhyloCSF, RNAcode also requires multiple sequence alignment results as inputs. RNAcode results include start and stop positions of the predicted coding region, coding potential scores and P values.
Coding–noncoding index
Coding–noncoding index (CNCI) [10] aims to classify input RNA sequences without known annotations. Its primary goal is to avoid false-positive and false-negative results issuing from the usage of evolutionary features. It is also tolerant of incomplete transcripts and sense–antisense pairs. Given input sequences, CNCI reports predicted ORFs and coding potential scores, based on adjoining nucleotide triplet usage frequency.
Coding potential assessment tool
CPAT [19] is another popular method of coding RNA classification. Its alignment-free approach based on logistic regression takes little time while maintaining a robust performance that is comparable with those of alignment-based methods. The prediction model of CPAT is built with nucleotide sequence composition and codon usage bias. Therefore, CPAT requires prior knowledge of nucleotide composition information. The authors of CPAT provide this information for four species (human, mouse, zebrafish and fruit fly). However, CPAT includes a step that enables users to build their own logistic model. CPAT works with RNA sequence and provides mRNA size, ORF size, coding probability and two scores of its features (Fickett score and hexamer score). A Web application of CPAT is also available.
iSeeRNA
iSeeRNA [20] focuses on detecting lncRNA rather than protein-coding RNAs. Previous works of coding potential assessments were evaluated based on its performance for protein-coding RNAs and well-known noncoding RNAs. iSeeRNA exclusively chose lncRNAs as its target gene set to provide classifiers better suited for lncRNA identification. The feature set of iSeeRNA includes transcript conservation, ORF quality and nucleotide sequence composition. iSeeRNA accepts RNA sequences and gene annotation information as input and reports predicted classes and coding scores. iSeeRNA can also be accessed on the Web.
Predictor of lncRNAs and messenger RNAs based on an improved k-mer scheme
Predictor of lncRNAs and messenger RNAs based on an improved k-mer scheme (PLEK) [11] is another tool that is useful when dealing with an incomplete or erroneous transcriptome without reference genomes. PLEK implements an alignment-free approach by using k-mers and is not restricted by prior gene annotation. It is especially suitable for RNA-seq data with relatively higher error rates, such as those from PacBio. PLEK has been shown to be exceptionally tolerant of indel errors. PLEK works with input sequence data and reports predict classes. PLEK also enables users to build their own model of nonvertebrate species.
LncRNA-classifier based on selected features
LncRNA-classifier based on selected features (Linc-SF) [12] aims to distinguish lncRNAs from the others by using sequence and structure-related features with protein-coding potential features. It operates with a novel nucleotide composition feature selected by the GA-SVM algorithm and secondary structure-derived metrics. Linc-SF uses CPC to assess protein-coding features and, therefore, is expected to be much slower than other alignment-free methods. Linc-SF can be used when genome sequence data are available. The source code is not available online, so users will have to contact the authors.
Long noncoding RNA identification using balanced random forests
Long noncoding RNA identification using balanced random forests (LncRNA-ID) [21] is built with a model that can handle an imbalanced training data set, which is often the problem when constructing data sets consisting of lncRNA and protein-coding genes. It also works well with small training data sets. Another advantage of LncRNA-ID is that it uses a hidden Markov model, which enables fast and sensitive sequence comparison. Another interesting point regarding LncRNA-ID is that it attempts to infer potential ribosome interactions without experimental data. The authors provide source codes to extract features from RNA sequences, which are used as the inputs for LncRNA-ID.
lncRNA-MFDL
lncRNA-MFDL [13] is differentiated from other tools, as it implements deep learning to classify lncRNAs. It has been shown to have slightly better accuracy than tools that are based on the SVM model, such as CPC and CNCI. lncRNA-MFDL uses adjoining nucleotide triplet frequency, ORF quality, k-mer frequency and transcript secondary structure information. Its performance was tested on 11 species, including gorilla, lamprey and orangutan. The source code appears to be unavailable.
Coding potential calculator 2
CPC2 [22] is an upgraded version of CPC. The primary goal of CPC2 is to eliminate the time-consuming alignment step without compromising performance. It is a fast, species-neutral classifier and has added more information to the output files. Given RNA sequence data, CPC2 reports the putative peptide length, Fickett score, isoelectric point, ORF start position, the integrity of the ORF, coding probability and predicted class. CPC2 can be accessed on the Web.
Coding potential calculation tool based on multiple features
Coding potential calculation tool based on multiple features (COME) [23] is extraordinary in that it uses both experimental data and sequence-based features. Although it did not include ribosome profiling data, the authors of COME explored the impact of Ribo-seq on COME by comparing the results before and after adding the scores of combinatorial classifiers as features. COME uses small, polyA+/− RNA-seq and ChIP-seq (H3K36me3 and H3K4me3 signals) to predict coding potential. COME does not require the subjects to be conserved or fully assembled. The authors of COME stated in their paper that their model is focused only on canonical ncRNAs and therefore can only predict up to 70% of human lncRNAs. COME accepts gene annotation information and reports length, coding potential and predicted class for each transcript. A Web server version of COME is also available.
Box B.
Combinatorial coding RNA classifier (related to Table 2)
Ribosome release score
Ribosome release score (RRS) [26] is based on the discrepancy of ribosome occupancy between the ORF and non-ORF region. It is a metric defined as the ratio between the total reads in the coding region and the total reads in the 3′ UTR, normalized by the length of the regions and the ratio of RNA coverage. RRS requires ORF annotation, Ribo-seq, RNA-seq and chromosome size information as inputs. RRS reports the expression levels of coding sequence (CDS) and 3′ UTR, TE, as defined by Ingolia [69], and RRS score. RRS allows the detection of small coding regions smaller than Ribo-seq fragments (∼30 nt) and is robust to non-ribosomal proteins. However, RRS has the limitation that the 3′ UTR should have at least the length of the Ribo-seq fragment.
Translated ORF classifier
Translated ORF classifier (TOC) [70] is a random forest classifier trained on four metrics derived from a ribosome profile. It is based on translational efficiency, ribosome coverage in ORF and downstream of ORF and ORF fraction in the transcript. The training set of the classifier used a RefSeq gene set from zebrafish and a mouse genome assembly with a fragments per kilobase million (FPKM) >1. The source code appears to be unavailable.
Fragment length organization similarity score
Fragment length organization similarity score (FLOSS) [27] is a metric based on the calculation of RPF length distribution in a given transcript and a comparison with the reference coding region of the transcript. FLOSS discriminates true 80S ribosome-generated RPFs from a non-ribosomal background and measures the distribution discrepancy between protein-coding and true noncoding regions, with lower scores indicating that the given data are likely to contain true RPFs. It accepts Ribo-seq BAM files and gene annotation BED files as inputs to generate output files containing the distribution of RPF length and calculates a FLOSS score for each transcript.
ORFscore
ORFscore [30] is based on the three-nucleotide periodicity of ribosome occupancy. The score is calculated as a log-adjusted value of the difference between the number of RPF reads in each reading frame and the mean RPF reads across all three reading frames. This value is then normalized by the mean RPF reads. Given ORF annotation in each reading frame and Ribo-seq data, it quantifies the biased distribution of RPFs in the first reading frame. The threshold for ORFscore to classify a coding ORFs is recommended to be established empirically using known CDSs.
PROTEOFORMER
PROTEOFORMER [39] is a tool to visualize protein synthesis using Ribo-seq data. It maps RPFs and identifies translated transcripts and translation initiation sites and creates a protein sequence database that can be used for MS-based proteomics analysis. PROTEOFORMER takes two Ribo-seq data sets, an untreated or translation elongation inhibitor (CHX/emetine)-treated sample and a translation initiation inhibitor (LTM/puromycin/harringtonine)-treated sample. In addition, a species- and annotation version-specific SQlite Ensembl database should be provided as input [85]. The output of PROTEOFORMER is a FASTA file of nonredundant translation products. It also generates specific metrics regarding metagene classification, RPF abundance, transcripts with translation, single nucleotide polymorphism calling result and FLOSS score.
ORF-RATER
ORF-RATER [71] quantifies translation from an ORF, based on the concept that translated ORFs show similar ribosome occupancy patterns with annotated CDSs. ORF-RATER detects all NUG-starting ORFs and classifies all possible ORFs using linear regression and a random forest classifier trained with AUG-starting ORFs. It takes previously annotated CDSs as a positive set to determine true translation evidence. ORF-RATER works with FASTA files, gene annotation BED files and Ribo-seq BAM files and outputs ORFs with a high confidence of coding potential.
RibORF
RibORF [31] is an SVM classifier to identify translated ORFs based on ribosomal A-site alignment, three-nucleotide periodicity and the distribution of RPFs across codons. Annotated CDSs were used as the positive set, while off-frame ORFs of protein-coding transcripts and ORFs of small noncoding RNA were used as the negative set. RibORF takes Ribo-seq, gene annotation files and candidate ORF files as inputs, and read lengths with offset distances should be manually checked and given in parameter files. RibORF reports the translation probability and number of RPFs, percentage of maximum entropy score and P value for each transcript.
riboHMM
riboHMM [32] is a model to identify translated ORFs using abundance and three-nucleotide periodicity of RPFs. riboHMM takes Ribo-seq data, annotation files, genome FASTA files and RNA-seq data as inputs and outputs translated sequences for each transcript. riboHMM also provides ORF annotation and an opportunity to identify novel coding ORFs. However, riboHMM cannot discriminate RPFs arising from different isoforms.
SPECtre
SPECtre [33] classifies actively translated regions based on three-nucleotide periodicity. SPECtre does not require matched RNA-seq data and takes Ribo-seq data, isoform.fpkm_tracking files from Cufflinks (transcript abundance calculated from Ribo-seq or RNA-seq) and annotation files as inputs. SPECtre outputs a metric containing the translational status for each transcript. Users can customize parameters regarding false discovery rate, window size, RPF abundance cutoff and step size of the window to optimize the performance. Application of FLOSS and ORFscore are available in the pipeline.
RiboTaper
RiboTaper [34] identifies actively translated regions based on the three-nucleotide periodicity of Ribo-seq data. RiboTaper creates every annotated exonic regions and applies a multitaper approach and Fourier transformation to determine the significance of the exonic periodic profiles. RiboTaper takes genome FASTA files, gene annotation files, Ribo-seq and RNA-seq data as inputs. Read lengths and offset distances should be manually checked and provided as parameters. It outputs translated ORFs, translated aa sequences, quality control plots and summaries of translated ORFs. RiboTaper can detect translation regardless of expression level, but it only considers AUG-initiated ORFs and does not account for frame shifting.
Ribosome profiling with Bayesian predictions
Ribosome profiling with Bayesian predictions (Rp-Bp) [35] predicts the likelihood of ORF translation based on the three-nucleotide periodicity of RPFs. Rp-Bp detects all ORFs with a three-nucleotide periodicity pattern, regardless of how many ORFs are present in the same transcript. It contains two phases, ORF profile construction and translation prediction. In ORF profile construction, it constructs a profile for each ORF and infers the P-site offset automatically by Bayesian inference. In the next phase, translation is predicted by calculating the likelihood ratio for the determination of which profile fits the given data. Apart from the actual prediction, creating reference genome indices is mandatory. Rp-Bp takes reference gene annotation files, genome FASTA files, ribosomal RNA FASTA files and Ribo-seq data as inputs and runs STAR to align RPF reads [87]. Rp-Bp provides ORF profiles and final prediction sets, including sequences of ORFs, DNA and proteins.
Translation-dependent ensemble classifier with ribosome and UPF1 association score
Translation-dependent ensemble classifier with ribosome and UPF1 association score (TERIUS) [36] consists of a two-step identification of lncRNAs. The first step counts for Ribo-seq reads mapped to each frame of transcripts and calculates their coding probabilities based on Bayesian inference. It does not search for ORFs in transcripts, to avoid false-negatives arising from erroneous ORFs. Transcripts without coding potential are further filtered with UPF1 association to eliminate possible mRNA fragments. TERIUS requires gene annotation, Ribo-seq, RNA-seq and CLIP-seq data and reports predicted class, predicted coding frame and noncoding probability for each transcript.
Biographies
Seo-Won Choi is a PhD candidate in the Department of Life Science at Hanyang University, and has studied and developed lncRNA classification algorithms.
Hyun-Woo Kim is an undergraduate student at Hanyang University, who is working with lncRNAs encoding functional peptides in cancer.
Jin-Wu Nam is an associate professor at Hanyang University. He received a PhD in Bioinformatics from Seoul National University and studied computational biology at the Whitehead Institute for Biomedical Research, affiliated with MIT, as a postdoctoral associate.
References
- 1. Rinn JL, Chang HY.. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Batista PJ, Chang HY.. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152(6):1298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ulitsky I, Bartel DP.. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154(1):26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kung JT, Colognori D, Lee JT.. Long noncoding RNAs: past, present, and future. Genetics 2013;193(3):651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447(7146):799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jia H, Osak M, Bogu GK, et al. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 2010;16(8):1478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ulitsky I, Shkumatava A, Jan CH, et al. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011;147(7):1537–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pauli A, Valen E, Lin MF, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 2012;22(3):577–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nam JW, Bartel DP.. Long noncoding RNAs in C. elegans. Genome Res 2012;22(12):2529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun L, Luo H, Bu D, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 2013;41(17):e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li A, Zhang J, Zhou Z.. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 2014;15:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Li Y, Wang Q, et al. Computational identification of human long intergenic non-coding RNAs using a GA-SVM algorithm. Gene 2014;533(1):94–9. [DOI] [PubMed] [Google Scholar]
- 13. Fan XN, Zhang SW.. lncRNA-MFDL: identification of human long non-coding RNAs by fusing multiple features and using deep learning. Mol Biosyst 2015;11(3):892–7. [DOI] [PubMed] [Google Scholar]
- 14. Liu J, Gough J, Rost B.. Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet 2006;2(4):e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kong L, Zhang Y, Ye ZQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 2007;35(Suppl 2):W345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arrial RT, Togawa RC, Brigido Mde M.. Screening non-coding RNAs in transcriptomes from neglected species using PORTRAIT: case study of the pathogenic fungus Paracoccidioides brasiliensis. BMC Bioinformatics 2009;10(1):239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanada K, Akiyama K, Sakurai T, et al. sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics 2010;26(3):399–400. [DOI] [PubMed] [Google Scholar]
- 18. Lin MF, Jungreis I, Kellis M.. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011;27(13):i275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L, Park HJ, Dasari S, et al. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res 2013;41(6):e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun K, Chen X, Jiang P, et al. iSeeRNA: identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC Genomics 2013;14(Suppl 2):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Achawanantakun R, Chen J, Sun Y, et al. LncRNA-ID: long non-coding RNA IDentification using balanced random forests. Bioinformatics 2015;31(24):3897–905. [DOI] [PubMed] [Google Scholar]
- 22. Kang YJ, Yang DC, Kong L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res 2017;45(W1):W12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu L, Xu Z, Hu B, et al. COME: a robust coding potential calculation tool for lncRNA identification and characterization based on multiple features. Nucleic Acids Res 2017;45(1):e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ingolia NT, Ghaemmaghami S, Newman JR, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009;324(5924):218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ingolia NT, Brar GA, Rouskin S, et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 2012;7(8):1534–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guttman M, Russell P, Ingolia NT, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 2013;154(1):240–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ingolia NT, Brar GA, Stern-Ginossar N, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 2014;8(5):1365–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruiz-Orera J, Messeguer X, Subirana JA, et al. Long non-coding RNAs as a source of new peptides. Elife 2014;3:e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang H, Wang Y, Xie S, et al. Global and cell-type specific properties of lincRNAs with ribosome occupancy. Nucleic Acids Res 2017;45(5):2786–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bazzini AA, Johnstone TG, Christiano R, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. Embo J 2014;33(9):981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ji Z, Song R, Regev A, et al. Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 2015;4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raj A, Wang SH, Shim H, et al. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. Elife 2016;5:e13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chun SY, Rodriguez CM, Todd PK, et al. SPECtre: a spectral coherence–based classifier of actively translated transcripts from ribosome profiling sequence data. BMC Bioinformatics 2016;17(1):482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Calviello L, Mukherjee N, Wyler E, et al. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods 2016;13(2):165–70. [DOI] [PubMed] [Google Scholar]
- 35. Malone B, Atanassov I, Aeschimann F, et al. Bayesian prediction of RNA translation from ribosome profiling. Nucleic Acids Res 2017;45(6):2960–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi SW, Nam JW.. TERIUS: accurate prediction of lncRNA via high-throughput sequencing data representing RNA-binding protein association. BMC Bioinformatics 2018;19(S1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koch A, Gawron D, Steyaert S, et al. A proteogenomics approach integrating proteomics and ribosome profiling increases the efficiency of protein identification and enables the discovery of alternative translation start sites. Proteomics 2014;14(23–24):2688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun H, Chen C, Shi M, et al. Integration of mass spectrometry and RNA-Seq data to confirm human ab initio predicted genes and lncRNAs. Proteomics 2014;14(23–24):2760–8. [DOI] [PubMed] [Google Scholar]
- 39. Crappe J, Ndah E, Koch A, et al. PROTEOFORMER: deep proteome coverage through ribosome profiling and MS integration. Nucleic Acids Res 2015;43(5):e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brockdorff N, Ashworth A, Kay GF, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992;71(3):515–26. [DOI] [PubMed] [Google Scholar]
- 41. Herzing LB, Romer JT, Horn JM, et al. Xist has properties of the X-chromosome inactivation centre. Nature 1997;386(6622):272–5. [DOI] [PubMed] [Google Scholar]
- 42. Engreitz JM, Pandya-Jones A, McDonel P, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013;341(6147):1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009;33(6):717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. West JA, Davis CP, Sunwoo H, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 2014;55(5):791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22(39):8031–41. [DOI] [PubMed] [Google Scholar]
- 46. Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39(6):925–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013;73(3):1180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mourtada-Maarabouni M, Pickard MR, Hedge VL, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009;28(2):195–208. [DOI] [PubMed] [Google Scholar]
- 49. Sun Y, Jin SD, Zhu Q, et al. Long non-coding RNA LUCAT1 is associated with poor prognosis in human non-small lung cancer and regulates cell proliferation via epigenetically repressing p21 and p57 expression. Oncotarget 2017;8(17):28297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464(7291):1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang EB, Kong R, Yin DD, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget 2014;5(8):2276–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rohrig H, Schmidt J, Miklashevichs E, et al. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci USA 2002;99(4):1915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kondo T, Hashimoto Y, Kato K, et al. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 2007;9(6):660–5. [DOI] [PubMed] [Google Scholar]
- 54. Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, et al. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 2008;451(7179):730–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Magny EG, Pueyo JI, Pearl FM, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 2013;341(6150):1116–20. [DOI] [PubMed] [Google Scholar]
- 56. Pauli A, Norris ML, Valen E, et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 2014;343(6172):1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015;160(4):595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nelson BR, Makarewich CA, Anderson DM, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016;351(6270):271–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matsumoto A, Pasut A, Matsumoto M, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017;541(7636):228–32. [DOI] [PubMed] [Google Scholar]
- 60. Zhang Q, Vashisht AA, O'Rourke J, et al. The microprotein Minion controls cell fusion and muscle formation. Nat Commun 2017;8:15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bi P, Ramirez-Martinez A, Li H, et al. Control of muscle formation by the fusogenic micropeptide myomixer. Science 2017;356(6335):323–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang JZ, Chen M, Chen D, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell 2017;68(1):171–84.e6. [DOI] [PubMed] [Google Scholar]
- 63. D'Lima NG, Ma J, Winkler L, et al. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol 2017;13(2):174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ulveling D, Francastel C, Hube F.. When one is better than two: rNA with dual functions. Biochimie 2011;93(4):633–44. [DOI] [PubMed] [Google Scholar]
- 65. Ulveling D, Francastel C, Hube F.. Identification of potentially new bifunctional RNA based on genome-wide data-mining of alternative splicing events. Biochimie 2011;93(11):2024–7. [DOI] [PubMed] [Google Scholar]
- 66. Nam JW, Choi SW, You BH.. Incredible RNA: dual functions of coding and noncoding. Mol Cells 2016;39(5):367–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Washietl S, Findeiss S, Muller SA, et al. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA 2011;17(4):578–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fickett JW. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res 1982;10(17):5303–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ingolia NT, Lareau LF, Weissman JS.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011;147(4):789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chew GL, Pauli A, Rinn JL, et al. Ribosome profiling reveals resemblance between long non-coding RNAs and 5' leaders of coding RNAs. Development 2013;140(13):2828–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fields AP, Rodriguez EH, Jovanovic M, et al. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell 2015;60(5):816–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Guo H, Ingolia NT, Weissman JS, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010;466(7308):835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lee S, Liu B, Lee S, et al. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA 2012;109(37):E2424–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Slavoff SA, Mitchell AJ, Schwaid AG, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol 2013;9(1):59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mackowiak SD, Zauber H, Bielow C, et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol 2015;16:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Crappe J, Van Criekinge W, Trooskens G, et al. Combining in silico prediction and ribosome profiling in a genome-wide search for novel putatively coding sORFs. BMC Genomics 2013;14(1):648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smith JE, Alvarez-Dominguez JR, Kline N, et al. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep 2014;7(6):1858–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jiang B, Liu J, Zhang YH, et al. Long noncoding RNA LINC00961 inhibits cell invasion and metastasis in human non-small cell lung cancer. Biomed Pharmacother 2018;97:1311–18. [DOI] [PubMed] [Google Scholar]
- 79. Yang L, Tang Y, He Y, et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer 2017;8(1):97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Papaioannou D, Petri A, Thrue CA, et al. HOXB-AS3 regulates cell cycle progression and interacts with the Drosophila Splicing Human Behavior (DSHB) complex in NPM1-mutated acute myeloid leukemia. Blood 2016;128:1514. [Google Scholar]
- 81. Prabhakar B, Zhong XB, Rasmussen TP.. Exploiting long noncoding RNAs as pharmacological targets to modulate epigenetic diseases. Yale J Biol Med 2017;90(1):73–86. [PMC free article] [PubMed] [Google Scholar]
- 82. Wu T, Du Y.. LncRNAs: from basic research to medical application. Int J Biol Sci 2017;13(3):295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chooniedass-Kothari S, Emberley E, Hamedani MK, et al. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett 2004;566(1–3):43–7. [DOI] [PubMed] [Google Scholar]
- 84. Eoh KJ, Paek J, Kim SW, et al. Long non-coding RNA, steroid receptor RNA activator (SRA), induces tumor proliferation and invasion through the NOTCH pathway in cervical cancer cell lines. Oncol Rep 2017;38:3481–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Olexiouk V, Crappe J, Verbruggen S, et al. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 2016;44(D1):D324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hao Y, Zhang L, Niu Y, et al. SmProt: a database of small proteins encoded by annotated coding and non-coding RNA loci. Brief Bioinform 2017, pii: bbx005. doi: 10.1093/bib/bbx005. [DOI] [PubMed] [Google Scholar]
- 87. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]