Abstract
As one of the few irreversible protein posttranslational modifications, proteolytic cleavage is involved in nearly all aspects of cellular activities, ranging from gene regulation to cell life-cycle regulation. Among the various protease-specific types of proteolytic cleavage, cleavages by casapses/granzyme B are considered as essential in the initiation and execution of programmed cell death and inflammation processes. Although a number of substrates for both types of proteolytic cleavage have been experimentally identified, the complete repertoire of caspases and granzyme B substrates remains to be fully characterized.
To tackle this issue and complement experimental efforts for substrate identification, systematic bioinformatics studies of known cleavage sites provide important insights into caspase/granzyme B substrate specificity, and facilitate the discovery of novel substrates. In this article, we review and benchmark 12 state-of-the-art sequence-based bioinformatics approaches and tools for caspases/granzyme B cleavage prediction. We evaluate and compare these methods in terms of their input/output, algorithms used, prediction performance, validation methods and software availability and utility. In addition, we construct independent data sets consisting of caspases/granzyme B substrates from different species and accordingly assess the predictive power of these different predictors for the identification of cleavage sites. We find that the prediction results are highly variable among different predictors. Furthermore, we experimentally validate the predictions of a case study by performing caspase cleavage assay. We anticipate that this comprehensive review and survey analysis will provide an insightful resource for biologists and bioinformaticians who are interested in using and/or developing tools for caspase/granzyme B cleavage prediction.
Keywords: caspase, cleavage sites, prediction tool
Introduction
Proteases are proteolytic enzymes that catalyze the breakdown of protein or peptide substrates by hydrolysis of peptide bonds [1–9]. They represent 2% (at least 500–600 proteases) of all gene products in human and are involved in the functional regulation of a large number of important physiological processes such as cell cycle [10, 11], cell proliferation [12], programmed cell death [13–15], DNA replication [16], tissue remodeling [17] and immune response [18, 19]. The members in this class of enzymes recognize specific substrate protein sequences and catalyze the hydrolysis of peptide bonds to activate or degrade the substrate proteins. The effects of the hydrolysis reactions are frequently amplified, resulting in a rapid and substantial change of the biological systems through modulating the balance of proteomic dynamics. Such highly orchestrated sequence of events is thus termed ‘proteolytic cascades’ [20].
Caspases are a family of proteases that can be subdivided functionally into those involved in either apoptosis or inflammation [21]. In 1993, researchers found that the Caenorhabditis elegans cell death gene, ced-3, has a remarkable sequence similarity to interleukin-1b-converting enzyme (caspase-1), a mammalian protease responsible for proteolytic maturation of pro-interleukin-1b [22]. This finding elucidated the first two members of the caspase family and provided evidence that these proteinases might play an essential role in apoptosis. Subsequent studies of these proteinases drove the identification of several other caspase family members important for apoptosis or inflammation.
Apoptosis, or programmed cell death, is a fundamental process that occurs in all tissues during development, homeostasis and disease [23–26]. On the other hand, the inflammatory response is triggered by innate immune sensors following cellular damage, infection or stress, and serves to clear the harmful stimulus and initiate healing [27, 28].
To date, at least 15 mammalian caspases have been identified [29], and they are categorized into three groups, based on their substrate specificities: Group I caspases (caspase-1, 4, 5 and 13) prefer bulky hydrophobic amino acids at the P4 site and cleave the peptide sequence (W/L)EHD, Group II caspases (caspase-2, 3 and 7) preferentially cleave the sequence motif DEXD, whereas Group III caspases (caspase-6, 8, 9 and 10) cleave the motif (I/V/L)E(H/T)D. In contrast to the caspases, granzyme B, another essential serine protease for apoptosis/inflammation, prefers to cleave the sequence motif IEXD [30].
For caspases not falling into Groups I, II and III, caspase-11 is considered as an orthologue of caspase-4 and 5 found in murine. Activation of caspase-11 leads to septic shock, pyroptosis and often organismal death. Caspase-12 is closely related to caspase-1, and the activating form of caspase-12 is only found in people of African descent in Homo sapiens. Caspase-14 is enriched in human skin and mainly expressed in the upper layers of the epidermis. The protein is mainly localized to the cytosol according to the Cell Atlas.
Caspases are essential to coordinating and integrating signals, which lead to apoptosis, inflammation and other forms of programmed death, including pyroptosis and necroptosis [31, 32]. This view is supported by observations that proteins involved in apoptosis and inflammation contain common conserved domains, including caspase-associated recruitment domains and death effector domains, which are also present in caspases. Recent findings have indicated that classically ‘apoptotic’ caspases have essential roles in initiating inflammation, both directly and via inflammatory cell death pathways [33].
The specificity of proteases like caspases depends primarily on their active sites, whose selectivity depends on preferences for a number of specific amino acids at defined positions. In addition to the primary amino acid sequence of the substrate, the substrate specificity of a protease is also influenced by the three-dimensional conformation of its substrates. In particular, proteases preferentially cleave substrates within extended loop regions, while residues that are buried within the interior of the protein substrate are usually inaccessible to the protease active site.
Identification of native substrates of caspases and granzyme B is the key to the understanding of their physiological roles, implicated in the pathological processes contributing to proteolytic cascades, and leading to apoptotic cell death. Identification of native substrates also means to find potential substrates that can serve as viable therapeutic targets. Although the application of advanced large-scale high-throughput proteomic techniques has significantly increased the number of experimentally verified caspase and granzyme B substrates, the complete repertoire of the native substrates remains to be discovered, and furthermore, many other cleavage sites within the known substrates are not fully experimentally identified. Moreover, experimental identification and characterization of protease substrates are often time-consuming, expensive and requiring extensively trained personnel. Therefore, bioinformatic prediction of caspase and granzyme B substrates may provide valuable and experimentally testable information regarding novel potential cleavage sites or putative substrates, i.e. ranking the candidate protein target list according to their likeliness, narrowing it down to a reasonable number to be validated in the test tube.
Sequence and structural analysis of substrates of caspases and granzyme B has enabled the development of computational approaches for prediction of potential cleavage sites and putative substrates from sequence alone [34–36] using techniques for analyzing protein sequences [37–40]. However, the rapid growth in prediction approaches since the last comprehensive comparison [41], which was reported almost 5 years ago, creates a need to critically assess and compare the expanding and diverse bundle of prediction methods. In this article, therefore, we present a comprehensive review of 12 sequence-based methods for caspases/granzyme B cleavage prediction, offering insights into the nature of different predictors and facilitating potential improvement of caspases/granzyme B cleavage prediction. All predictors are critically reviewed in terms of input/output, algorithm, prediction performance, validation method and software utility, i.e. whether a stand-alone software is available. To evaluate the performance of caspases/granzyme B cleavage predictors, we assembled independent testing data sets containing substrates of caspases/granzyme B of various species with carefully collected and curated data.
To address whether the predicted caspase substrate is really cleaved by caspase, we selected top-scoring sequences and test cleavage in vitro with a caspase assay. These sequences are top-ranked by prediction tools showing outstanding performances in our independent testing datasets.
Methods
Existing tools reviewed in this study
We briefly summarize the key aspects of the 12 tools evaluated for caspase/granzyme B cleavage prediction in Table 1. The tools included in the benchmarking analysis are GraBCas [48], CaSPredictor [49], PoPS [50], SitePrediction [51], Cascleave [52], Cascleave 2.0 [41], Pripper [53], PCSS [54], CASVM [55], CAT3 [56], PROSPER [57] and Blast [58].
Table 1.
A summary of key features of each tool evaluated in this article
| Tools | SitePrediction | Cascleave | PoPS | Pripper |
|---|---|---|---|---|
| Species | Multispecies | Multispecies | Multispecies | Multispecies |
| Web server availability | http://www.dmbr.ugent.be/prx/bioit2-public/SitePrediction/ | http://sunflower.kuicr.kyoto-u.ac.jp/~sjn/Cascleave/ | http://pops.csse.monash.edu.au/ | No server |
| Algorithm | Combination of frequency score representing amino acids occurrence and position similarity | BEAA trained and tested support vector regression (SVR) model | PSSM matrix | Combination of SVM/random forest and J48 algorithm |
| Option of batch prediction | Yes | No | Yes | Yes |
| Adjustment of prediction thresholds | No | o | Yes | No |
| Standalone software availability | No | No | Yes | No |
| Language implemented | C++ | Perl | Java | Java |
| Dataset origin | Data from MEROPS | Multiple resources | Data from MEROPS | Data from EBI [42] |
| Ratio of positive to negative samples | – | 1:3 | – | 1: 1 |
| Sliding window size | – | 16 amino acids | – | 10 amino acids |
| Computing time for processing a sequence | Within a second | 5 min | Within a second | Within a second |
| Whether structural information considered | Secondary structure prediction, SA and PEST sequence occurrence considered | Secondary structure, SA and natively disordered regions considered | Secondary or tertiary structure of the substrate considered | Not considered |
| Types of caspases applicable | Specific training sets corresponding to caspases 1, 3, 6, 7, 8 | Mixed training sets for all caspases | Mixed training sets for all caspases | Mixed training sets for all caspases |
| Tools | CAT3 | PCSS | Blast | PROSPER |
| Species | Multispecies | Multispecies | N.A. | Multi-Species |
| Web server availability | No web server | http://salilab.org/peptide | N.A. | https://prosper.erc.monash.edu.au/webserver.html |
| Algorithm | PSSM matrix | SVM with radial basis function (RBF) kernel | N.A. | BEAA trained and tested SVR model with RBF kernel combined with MDGI feature selection |
| Option of batch prediction | Yes | Yes | N.A. | No |
| Adjustment of prediction thresholds | Yes | Yes | N.A. | No |
| Standalone software availability | Yes | No | N.A. | No |
| Language implemented | Perl | – | N.A. | Perl |
| Dataset origin | Data from PubMed [43] | Multiple resources | N.A. | Data from MEROPS, CutDB and PMAP [44] |
| Ratio of positive to negative samples | – | – | N.A. | 1:3 |
| Sliding window size | – | – | N.A. | Six amino acids |
| Computing time for processing a sequence | Within a second | A few minutes | N.A. | A few minutes |
| Whether Structure information considered | Not considered | Regular secondary structure considered | N.A. | Secondary structure, SA and native disorder considered |
| Types of caspases applicable | Training sets corresponding to caspases-3 | Separated training sets for caspases and granzyme B | N.A. | Mixed training sets for all caspases |
| Tools | GraBCas | CasPredictor | CASVM | Cascleave 2.0 |
| Species | Multispecies | Multispecies | Multispecies | Multispecies |
| Web server availability | http://wwwalt.med-rz.unikliniksaarland.de/med_fak/humangenetik/software/index.html (Not available) | http://icb.usp.br/ ∼farmaco/Jose/CaSpredictorfiles (Not available) | http://www.casbase.org/casvm/index.html (not available) | http://www.structbioinfor.org/cascleave2/ (not available) |
| Algorithm | Scoring matrices | BLOSUM 62 Substitution Matrix-based CCSearcher algorithm | SVM | Maximum relevance, minimum redundancy and forward feature selection techniques trained SVM model |
| Option of batch prediction | – | – | – | – |
| Adjustment of prediction thresholds | – | – | – | – |
| Standalone software availability | – | – | – | – |
| Language implemented | Java | Visual Basic | Perl | Java |
| Dataset origin | – | Various databases, including SwissProt [45], InterDom [46] and Pfam [47] | Various resources | MEROPS |
| Ratio of positive to negative samples | – | – | – | 1:1 |
| Sliding window size | – | – | Three scanning window sizes are available: P4P1, P4P2’ and P14P10’ | – |
| Computing time for processing a sequence | – | – | – | – |
| Whether Structure information considered | Not considered | Not considered | Not considered | Secondary structure, SA and natively disordered regions considered |
| Types of caspases applicable | Specific training sets corresponding to caspases-3 and granzyme B | Mixed training sets for all caspases | Mixed training sets for all caspases | Mixed training sets for all caspases |
Note: These features include applicable species, whether web server exists, algorithm used, whether the batch prediction option is available, whether threshold is adjustable, whether stand-alone software exists, programming language used to implement the program, the origins of training data set, ratio of positive and negative samples, sliding window size (if exists), computing time to process one sequence and whether SA and SS is considered. The ‘-’ option means not available or not mentioned in the original paper.
Model input
In machine learning, the data set is often divided into training and testing data sets. The training data sets are used to build a computational model to learn hidden patterns in the data. For caspase/granzyme B substrates, the data are usually collected from various databases, such as MEROPS peptidase database [59], which contains over 410 000 listed cleavage peptidases as well as 28 000 inhibitors (physiological and nonphysiological). Other databases are CutDB [60], CaMPDB [61], TopFIND [62] and Degrabase [63]. CutDB integrates 3070 proteolytic events for 470 different proteases captured from public archives. CaMPDB contains sequences of calpains, substrates and inhibitors as well as substrate cleavage sites, collected from the literature. TopFIND contains >290 000 N-/C-termini and >33 000 cleavage sites. Degrabase comprises about 8000 unique N termini from >3200 proteins directly identified in subtiligase-based positive enrichment mass spectrometry experiments in healthy and apoptotic human cell lines.
The issue of biased prediction often occurs when there is an extensive overlap between the training and testing data sets. To avoid such bias, tools such as Pripper constructed training data sets according to careful selection criteria of the data (including removing the sequence redundancy between the training and test data sets, controlling the ratio of positive data to negative data, as well as performing multiple rounds of randomization tests, e.g. 10 repeats of 10-fold cross-validation). Apart from the sequence overlap between the training data set and validation data set, the issue of data imbalance also needs to be addressed. Use of an unbalanced data set often leads to biased models that favor the prediction of the ‘majority’ class of samples. Most tools solve this issue by selecting the positive data set from experimental databases and manually generating/sampling the negative data set with different approaches. For example, Cascleave/Cascleave 2.0 address this issue by generating positive and negative data sets from substrate sequences using a local sliding window approach surrounding the experimentally verified cleavage sites and other residues that are found not to be cleaved by caspases, respectively. Cascleave also uses an undersampling approach by reducing the size of the overrepresented negative samples. Pripper creates the negative data set by selecting negative training sequences generated from the same substrate sequences that are used for positive sequences. Both Cascleave/Cascleave 2.0 and PROSPER set the ratio of the positive to negative data to approximately 1:3. Pripper sets the ratio of the positive data to negative data to approximately 1:1. Similar efforts to minimize the unbalanced data (such as controlling the ratio of positive and negative data), though not explained in detail, can be observed in the development of other prediction tools such as PoPS.
Models construction and development
Early tools for caspase/granzyme B cleavage site prediction predict caspase/granzyme B cleavage sites from sequence information only, while more sophisticated tools developed more recently consider additional information such as secondary structure (SS) information, hydrophilicity/hydrophobicity, as well as solvent accessibility (SA) and protein native disorder information. The prediction methods can be generally classified into two types, machine learning-based algorithms and statistical scoring method-based algorithms. Machine learning-based tools include CASVM, Pripper, PCSS, PROSPER, Cascleave and Cascleave 2.0. While Statistical scoring method-based tools include GraBCas, CaSPredictor, PoPS, SitePrediction, CAT3 and Blast.
GraBCas is a scoring method based on position-specific scoring matrices (PSSMs). The PSSM is constructed based on experimentally determined substrate specificities. For computing the score in PSSM, GraBCas screens for tetrapeptides with Asp (D) at their last position (P1) in a given amino acid sequence. Given the tetrapeptide A4A3A2D (P4P3P2P1) of a potential cleavage site, its cleavage score for a given endopeptidase is computed by multiplying the corresponding matrix entries of A2 at position P2, A3 at position P3 and A4 at position P4. To improve the performance, GraBCas analyzes the amino acid distribution of known granzyme B and caspase-3 cleavage sites at positions P6-P2’ [where P and P’ mean residues C-terminal to the cleavage site as prime (P’) site and N-terminal peptide residues as nonprime (P) site] taken from the literature. CaSPredictor, a tool published at the same time with GraBCas, developed a scoring algorithm named CCSearch (Caspase Cleavage Site searcher), which is based on three parameters. The first parameter is calculated from the BLOSUM62 Substitution Matrix. The second parameter is the relative frequency f(i) for each amino acid residue at position i (P4-P1) from annotated sequences. The last parameter is the PEST index, calculated by giving a value of 1 to the amino acids in the following set: Ser (S), Thr (T), Pro (P), Glu or Asp (E/D), Asn (N) and Gln (Q), which are the residues of PEST regions [64]. There is an evidence that PEST-like sequences, rich in the aforementioned amino acids, if located in the upstream or downstream of the cleavage site, may contribute to the specificity for at least 60% verified caspase substrates [64, 65]. PoPS is a tool based on a computational model built from three components. The first component is the number of subsites within the active site of the protease. The second component is the specificity profile of each subsite, assigning a value to each of the 20 amino acids based on the relative contribution of the amino acid at that subsite to the overall substrate specificity of the protease. The last component is the weight of the subsite [50]. SitePrediction is a tool based on the idea that besides the occurrences of fixed consensus cleavage sites in the substrate sequence, a second score is calculated to improve the performance. This score is based on the similarity of the potential cleavage sites to the known sites used. SitePrediction also makes use of extra features, including PEST sequences, SA and SS, which may also provide contribution to the prediction performance [51]. CAT3 is a PSSM matrix-based method developed in 2012 [56]. CAT3 exploits positional specific frequency matrices from the multiple sequence alignments of the relevant set of peptides. Each matrix consists of 14 rows, representing positions P9…P1P1’…P5’, where a D amino acid is at the position P1. The 20 columns of the matrix represent the frequencies of each amino acid. CAT3 also uses two weighting systems to correct the probability of overrepresented and underrepresented amino acids in the frequency matrices to establish the scoring matrices: Calculating log odd ratio and subtraction of negative control background, which also contributes to the accuracy of CAT3. Blast is an aligning tool ubiquitously used in proteomics. Assuming that target substrates share a similar sequence, it can be used as a rudimentary prediction tool [52]. For our assessment, the cleavage score of a query test protein corresponds to the highest Blast bit score (a normalized aligned value, independent from sequence length and database size) with the known substrates in the training set. PROSPERous [66] is a recently developed tool, which uses a combination of various scoring functions as the input, including nearest neighbor similarity (NNS), amino acid frequency (AAF), WebLogo-based Sequence conservation (WLS), BLOSUM62 Substitution Index (BSI) as well as pairs of these function, namely, AAF + NNS, WLS + BSI and NNS + WLS. More recently, an advanced version of PROSPER, termed iProt-Sub [67], was developed to provide optimized cleavage site prediction models with a larger coverage of more proteases (up to 4 major protease families and 38 different proteases). iProt-Sub uses 11 different sequence encoding schemes in combination with a two-step feature selection procedure to remove the redundant features and improve the accuracy [67].
Most of the recently developed tools for predicting caspase/granzyme B cleavage sites are based on the support vector machine (SVM) algorithm. These tools include CASVM, Pripper, PCSS, Cascleave, Cascleave 2.0, PROSPER and iProt-Sub.
SVMs are classifiers that based on the maximization of the margin between classes. The data are considered as n-dimensional vectors, and the algorithm finds a hyperplane that separates vectors in different classes with a maximal margin. A kernel function can be used to map vectors of the original feature space to a higher-dimensional space in which the data can always be linearly separated. Note that the selection of training data greatly affects the performance of an SVM classifier; therefore, we will provide a detailed description of the way to select training data for each tool. CASVM is trained with sequences from a data set containing unique caspase cleavage sites, obtained from experimentally verified caspase substrates and an equal number of ‘non-cleavage’ sites, i.e. random tetrapeptide sequences extracted elsewhere on the same substrate. The tetrapeptide sequences are selected with the upstream 10 residues up to P14 position and downstream 10 residues up to P10’ position (i.e. the classifier is trained on a local window size of P14-P10’ sites) from the substrates. The SVM model of Pripper is trained on a balanced data set containing positive cleavage site samples gathered from 358 substrate proteins, and negative sequences generated from substrate sequences containing positive ones, specifically by selecting Asp (D) positions that have not been detected as caspase cleavage sites. Feature vectors consist of a fixed number of amino acids, encoded in a numerical form, incorporating both sides of the cleavage site. Each amino acid in the sequence is represented as an array of length 20 representing the 20 different amino acids. Only one element is set to 1, identifying the amino acid in question, while the rest is set to 0. PCSS model is based on single-cleavage sequences, each with eight features representing oligopeptides. To each residue, a feature number by the formula (n*20 + i) is assigned, where n represents the zero-based position in the peptide sequence of the residue, and i represents the position of the residue in line with a zero-based alphabetical ordering of all residues. In addition, PCSS also considers SS features, native disorder feature, SA feature calculated by DSSP [68], Disopred [69] and PSI-PRED [70], respectively. Cascleave uses a feature extraction method named binary encoding amino acid sequence profiles (BEAA) and its extension to include relevant structural features. In BEAA of which is encoded, substrate sequences are transformed into n-dimensional vectors using an orthonormal encoding scheme, in which each amino acid is represented by a 20-dimensional binary vector composed of either 0 or 1 elements. Similarly to PCSS, the structural information predicted by state-of-the-art algorithms, specifically, SSs, SA and natively unstructured regions are incorporated into the model to improve the performance. Cascleave also uses a novel approach named Bi-profile Bayesian signature, which is reported to significantly improve performance in methylation sites prediction [71]. Similarly to Cascleave, Cascleave 2.0 considers various structural information, including (but not limited to) SSs, solvent accessibility, disordered region and amino acid index (AAindex [72]). AAindex consists of a list of amino acid indices representing various physicochemical and biochemical properties. Cascleave 2.0 also involves an over- and underrepresented feature enrichment analysis. The rationale is that for each protein substrate, the set of various heterogeneous features generated above is highly dimensional, heterogeneous, noisy and redundant, and thus removing redundant features and using more relevant features might be useful for improving the predictive performance. Inclusion of noisy and redundant leads to a time-consuming practice to train classifiers, thereby resulting in possible biased model training and prediction. Cascleave 2.0 automatically estimates and eliminates noisy features. PROSPER, like Cascleave, is an SVM-based method; it can be applied to a broader range of proteases. Compared with Cascleave, PROSPER uses a feature selection method called mean decrease Gini index (MDGI) within the random forest algorithm, which can generate a score quantifying the importance and contribution of the individual element of a feature vector for correctly classifying a residue into a cleavage site or noncleavage site. The MGDI feature selection step has proven useful for improving the prediction accuracy [73] and is particularly useful for large training data sets.
Performance evaluation
To assess the performance of the compared methods, several cross-validation approaches are usually used, including N-fold, leave-one-out and leave-family-out. In addition, we look into prediction details by performing a case study. Cross-validation is typically exploited to avoid overfitting the training data set. Cross-validation consists in splitting the data set into N folds and combine N−1 folds as the training data set, while the left data set is regarded as test data set. Leave-one-out and leave-family-out are specific cases of N-fold cross-validation. Given a data set with D data samples, leave-one-out cross-validation (LOOCV) combines D−1 samples to form the training data set and leaves the remaining one sample as the test sample. By iteratively selecting test sample, each sample in the data set is used as a test sample once. On the other hand, in the leave-family- out cross-validation, if the data set is collected from different species/families, each subset from the same species/family is iteratively selected and regarded as test data sets once, while other subsets will be combined to form the training data set. As each sample/subset is iteratively selected as the test set, we need to perform the prediction many times with different combination of training datasets. We then average these results (usually accuracy) and acquire the final performance for cross-validation tests. Among the evaluated tools, Cascleave, Cascleave 2.0, PROSPER use the 5-fold cross-validation, while Cascleave 2.0 uses the LOOCV to assess their performance. Performing an independent test is another way to evaluate the performance of bioinformatics tools. In particular, it involves applying the algorithm to an independent test data set with a different data distribution, e.g. data obtained from other experiments. Finally, case study, or experimental validation of predictions, is another effective way to test the performance of a prediction tool in real-world applications, providing useful information of the scalability and usefulness of a tool on unknown data. Here, we perform both independent test and case study to assess and compare different methods.
Experimental validation
Recombinant target substrate proteins are incubated with active caspase-3 (Genetex) or active caspase-8 (Biovision) at 37°C for 2 h. After incubation, proteins are separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membrane. Transferred membrane is blocked with 4% blockace at 4°C for 2 h and incubated with anti-myc antibody (9E10) for 1:2000 in 0.2% blockace in tris-buffered saline (TBS) containing 0.02% tween 80 (TTBS). After incubation, membrane is incubated with peroxidase conjugated anti-mouse Ig for 1:5000 in 0.2% blockace in TBS-Tween (TTBS) for 1 h. Bound antibody is visualized by supersignal west pico (PIERCE) according to manufacturer’s instruction and LAS4000 mini (Fuji).
Predictor utility
An important consideration for developing practically useful predictors in the biological research community is to provide a user-friendly Web interface or a local software tool, to enable nonbioinformaticians to apply the model directly to their own data. The usefulness of bioinformatics tools depends on three main factors, i.e. the Web interface, the output and interpretation of prediction results and the availability of local executable software. A user-friendly interface can provide appropriate guidance and instructions for users to avoid making potential mistakes when exploiting the Web server. This is particularly important when parameter settings are required before conducting prediction tasks. Among the predictors we tested, SitePrediction, Cascleave, PCSS and Blast have implemented Web servers. All these tools require to provide parameters regarding penalty, prediction algorithm, error handling as well as e-mail address where the prediction results will be sent. Specifically, SitePrediction requires an input sequence to be submitted in the FASTA format, and the type of protease can be optionally selected, and multiple substrate protein sequences can be predicted at the same time. Cascleave requires users to input substrate sequences that need to be predicted, as well as algorithms used for performing the prediction. In addition, the user also needs to specify the e-mail address to acquire the prediction results. As Cascleave does not support batch submission, users can submit only one protein sequence to Cascleave each time. PCSS requires users to specify the training data as well as classifying data to perform the prediction, and also lists some pre-generated models for executing a quick prediction.
On the other hand, stand-alone software allows users to perform predictions for a large amount of sequences on local machines, offering an advantage over Web servers. However, there exists also a burden of installing (and perhaps compiling) the software locally, along with the dependent libraries. In addition, if the input sequence data are too large, there exists a possibility that the local resources may not be sufficient enough to run the program properly. Among the predictors we tested, Pripper and CAT3 are stand-alone software tools written using Java, whereas PoPS provides a JNLP file for downloading and local usage.
Result
Independent test and performance evaluation
In this section, to assess the performance of the reviewed tools in an objective and fair manner, we constructed independent test sets of caspase-1 and 3 substrates for H. sapiens. To evaluate the performance of these tools on other species, we also constructed independent test sets of caspase-1 and 3 substrates for Mus musculus and Escherichia coli.
Note that as some of the tools are not accessible (i.e. neither implemented as Web servers nor downloadable), we were forced to limit our assessment to the available ones: PoPS, SitePrediction, Cascleave, Pripper, PCSS, CAT3 and Blast.
Test data set construction
For each of the three species, we extracted all the fasta sequences from MEROPS of release 12.0 [59]. Training data sets for each tool and independent test data sets should have a minimum overlap, because a large overlap will likely result in an overestimation of the performance and biased prediction outcome. We therefore eliminated sequences that were overlapped in the training data sets of prediction tools, including Cascleave and Cascleave 2.0 from the independent test data sets. Both of these tools are recently developed, and thus, it is understandable that their training data set covered most of the extracted sequences (especially when compared with training datasets of tools developed in the early years). Our analysis showed that more than half of the extracted sequences were discarded for this reason, leaving 66 caspase-1 substrate sequences and 121 caspase-3 substrate sequences, respectively, in total, for all three species. For the negative data sets, we randomly selected proteins excluding those identified as substrates of caspase-1 or 3 of each species. To avoid biased performance evaluation, the size of negative data sets was set as the same as of positive data sets. These constructed independent test data sets are named as Cas1-all and Cas3-all, respectively.
We further divided these data sets according to the corresponding species these substrates belong to, resulting in another six data sets corresponding to caspase-1 and caspase-3 substrates of the three species. Each of these sets is named as Cas1-homo, Cas3-homo, Cas1-mus, Cas3-mus, Cas1-coli and Cas3-coli, respectively. We notice that for caspase-1 the sizes of cas1-mus and cas1-coli were too small to be used for an effective receiver operating characteristic (ROC) evaluation, and thus, we skipped these two data sets when drawing ROC curves. We also notice that CAT3 was designed only for predicting caspase-3 substrates, and thus, we only performed the evaluation of CAT3 on the sets that composed of caspase 1 substrates. The detailed description of the test sets used is shown in Table 2.
Table 2.
Detailed description of the eight test data sets used in this study
| Test set name | Positive or negative | Test set description |
|---|---|---|
| Cas1-all | Positive set | Combination of caspase-1 substrates from H. sapiens, M. musculus and Escherichia coli extracted from MEROPS |
| Negative set | Combination of protein from H. sapiens, M. musculus and E. coli excluding caspase-1 substrates. | |
| Cas3-all | Positive set | Combination of caspase-3 substrates from H. sapiens, M. musculus and E. coli extracted from MEROPS |
| Negative set | Combination of protein from H. sapiens, M. musculus and E. coli excluding caspase-3 substrates | |
| Cas1-homo | Positive set | Caspase-1 substrates from H. sapiens extracted from MEROPS |
| Negative set | Protein excluding caspase-1 substrates from H. sapiens | |
| Cas3-homo | Positive set | Caspase-3 substrates from H. sapiens extracted from MEROPS |
| Negative set | Protein excluding caspase-3 substrates from H. sapiens | |
| Cas1-mus | Positive set | Caspase-1 substrates from M. musculus extracted from MEROPS. |
| Negative set | Protein excluding caspase-1 substrates from M. musculus. | |
| Cas3-mus | Positive set | Caspase-3 substrates from M. musculus extracted from MEROPS |
| Negative set | Protein excluding caspase-3 substrates from M. musculus | |
| Cas1-coli | Positive set | Caspase-1 substrates from E. coli extracted from MEROPS |
| Negative set | Protein excluding caspase-1 substrates from E. coli | |
| Cas3-coli | Positive set | Caspase-3 substrates from E. coli extracted from MEROPS |
| Negative set | Protein excluding caspase-3 substrates from E. coli | |
Performance comparison
Among the reviewed predictors, as PoPS only has one parameter (threshold), we set the threshold as 0 to obtain more available results (as a lower threshold leads to a larger number of predicted potential cleavage sites [50]). For Cascleave, several prediction models (or the combination of models) such as BEAA, BPBAA and BPBDISO were tested. As the combination of BEAA, BPBAA and BPBDISO achieved the best in terms of the ROC curve, we chose this option for evaluation. For SitePrediction, different predefined training databases, corresponding to various species, exist. It is therefore possible to choose a particular database for each test set. For caspase-1 substrates prediction, we chose ‘caspase-1 for all species’ training database, and for caspase-3 substrates prediction, we chose the ‘caspase 3 for all species’ training database to perform the evaluation. For Pripper, as vote option gets the highest performance on the ROC curve testing, we chose the ‘vote’ option and cut option as ‘Full cut’. For PCSS, we chose the caspase option in the pre-generated model and selected the training iteration as 100. For Blast, we used the default parameters to perform the prediction. Then, we performed the prediction on each of the constructed independent test sets described in the test data set construction section, and for each set, we evaluated the performance using area under the curve (AUC) values.
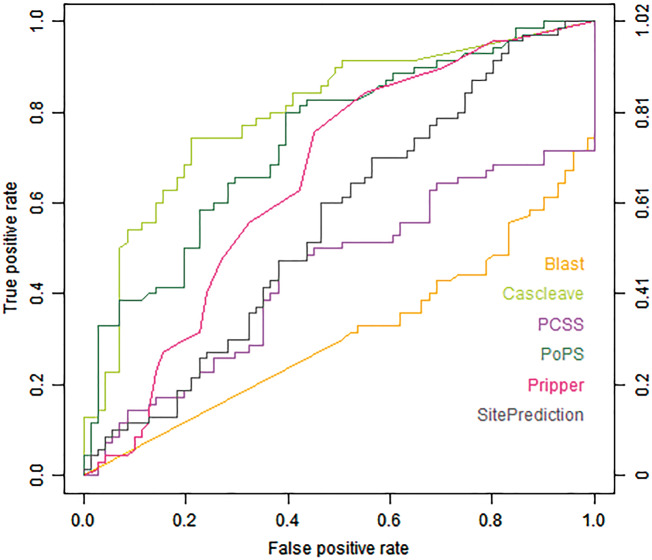
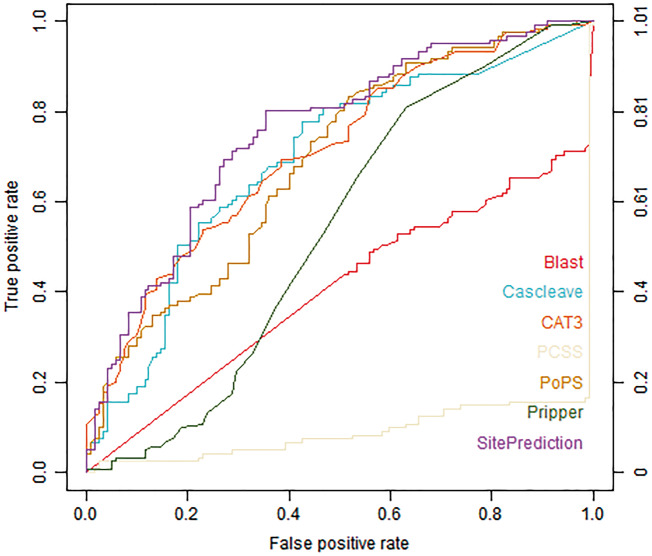
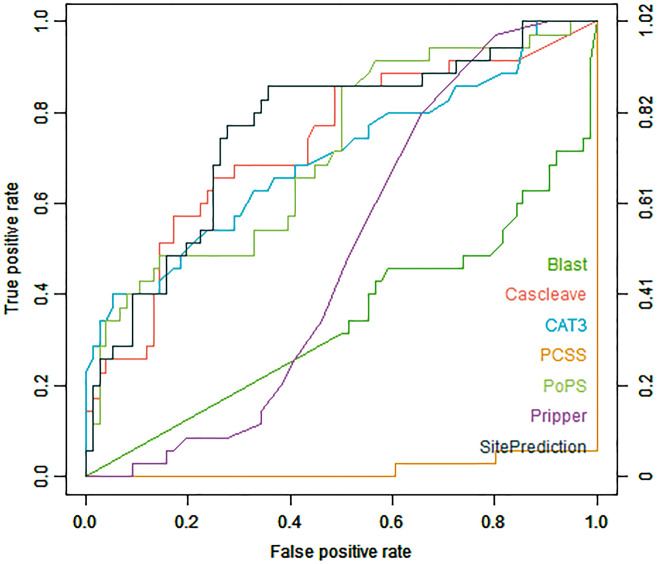
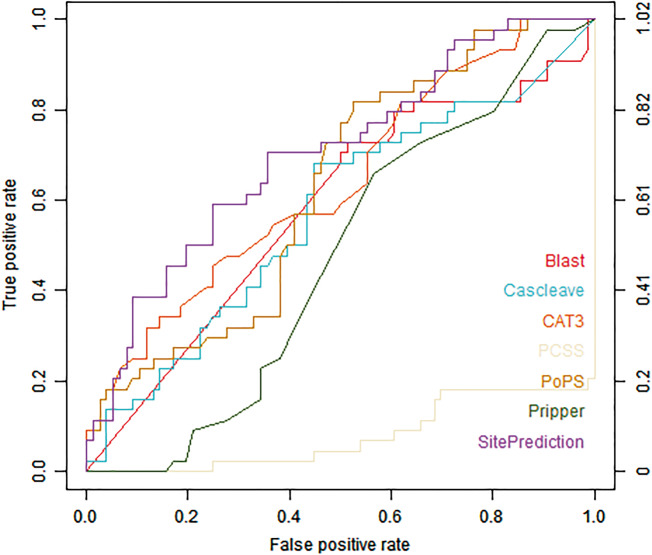
Figures 1 and 2 show the ROC curves of different tools assessed using the Cas1-all and Cas3-all test data sets, respectively. Cascleave, PoPS and Pripper outperformed other tools and achieved the best AUC values on the Cas1-all set (with an AUC value of 0.796 for Cascleave, 0.739 for PoPS and 0.655 for Pripper, respectively), while tools such as Blast, which depends on the sequence similarity, performed poorly in ROC performance. While on the Cas3-all set Cascleave, SitePrediction and CAT3 achieved the best AUC values (with an AUC value of 0.693 for Cascleave, 0.711 for CAT3 and 0.754 for SitePrediction, respectively).
Figure 1.
ROC curves of Blast, Cascleave, PCSS, PoPS, Pripper and SitePrediction on the Cas1-all set.
Figure 2.
ROC curves of Blast, Cascleave, PCSS, PoPS, Pripper, CAT3 and SitePrediction on the Cas3-all set.
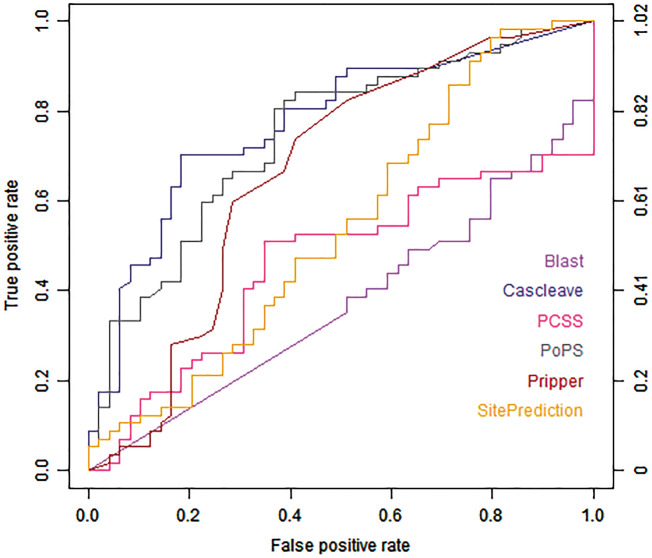
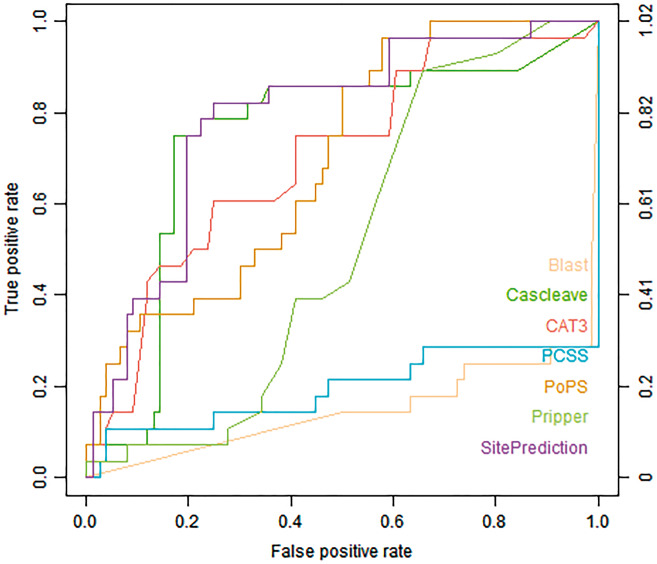
Figure 3 shows the ROC curves of different tools on the Cas1-homo set. PoPS, Cascleave and Pripper achieved the highest AUC values (PoPS with AUC value of 0.744, Cascleave with AUC value of 0.771 and Pripper with AUC value of 0.663, respectively). Figure 4 shows the ROC curves for the prediction result of the Cas3-homo set, for H. sapiens. SitePrediction, CAT3 and Cascleave achieved the best AUC values (SitePrediction with AUC value of 0.787, CAT3 with AUC value of 0.703 and Cascleave with AUC value of 0.745, respectively).
Figure 3.
ROC curves of Blast, Cascleave, PCSS, PoPS, Pripper and SitePrediction on the Cas1-homo set.
Figure 4.
ROC curves of Blast, Cascleave, PCSS, PoPS, Pripper, CAT3 and SitePrediction on the Cas3-homo set.
Figure 5 shows the ROC curves on the Cas3mus set. The ROC curves show that for the Cas3-mus set, SitePrediction, PoPS and Cascleave achieved the best performance in terms of AUC value, each with AUC value of 0.760, 0.712 and 0.729.
Figure 5.
ROC curves of Blast, Cascleave, PCSS, PoPS, Pripper, CAT3, SitePrediction on the Cas3-mus set.
Figure 6 shows the ROC curves on the Cas3coli set. We can see from the ROC curves that SitePrediction, PoPS and CAT3 achieved the best AUC value (SitePrediction with AUC value of 0.702, PoPS with AUC value of 0.627 and CAT3 with AUC value of 0.638, respectively).
Figure 6.
ROC curves of Blast, Cascleave, PCSS, PoPS, Pripper, CAT3, SitePrediction on the Cas3-coli set.
Combining the tool evaluation results in Table 1 and performance benchmarking results, we can draw the following conclusions:
SitePrediction achieves a better performance for general prediction (i.e. it provides a better performance for predicting caspase substrates from species excluding H. sapiens). This is perhaps because of the separation of the training sets provided by SitePrediction. The performance results in turn show that it is better to construct independent training sets for each species than to mix all sequences into a single training set. Considering the faster computing speed of SitePrediction, users are recommended to use SitePrecition to predict species excluding H. sapiens.
While SitePrediction possesses such a merit, it is also flawed when it has to address predictions on H. sapiens. From Table 3, we can see that while SitePrediction achieves the highest performance for caspase-3 substrate prediction, it performs poorly for caspase-1 substrate prediction. This indicates that it is generally better to use Cascleave to achieve the best performance on caspase-1 substrate prediction.
Table 3.
Summary of the top three tools that achieved the highest performance of AUC values for each set evaluated
| Data set | Top three tools of the highest performance of AUC values | ||
|---|---|---|---|
| Cas1-all | Cascleave (0.796) | PoPS (0.739) | Pripper (0.655) |
| Cas3-all | SitePrediction (0.754) | CAT3 (0.711) | Cascleave (0.693) |
| Cas1-homo | Cascleave (0.771) | PoPS (0.744) | Pripper (0.663) |
| Cas3-homo | SitePrediction (0.787) | Cascleave (0.745) | CAT3 (0.703) |
| Cas3-mus | SitePrediction (0.760) | Cascleave (0.729) | PoPS (0.712) |
| Cas3-coli | SitePrediction (0.702) | CAT3 (0.638) | PoPS (0.627) |
Note: The data sets used include are Cas1-all, Cas3-all, Cas1-homo, Cas3-homo, Cas3-mus and Cas3-coli.
Although Cascleave provides the best performance on caspase-1 substrate prediction and an acceptable performance on caspase-3 prediction, the computational cost for Cascleave is a little higher compared with other tools. Moreover, in addition to the requirement of submitting fasta files, Cascleave only allows the submission of one sequence to the server each time, thereby limiting the batch prediction for Cascleave.
If users want to perform a caspase-specific (i.e. the types of the caspase substrate is limited) substrate prediction offline, they are advised to use CAT3 for caspase-3 substrate prediction and Pripper for caspase-1 substrate prediction, as these two tools come with an implemented local package, while the other tools with better performance such as Cascleave and SitePrediction can only be used online.
We also notice that Blast performs poorly for almost all the training set, and this is perhaps because that the Blast predictions are not aiming at identifying specific cleavage sites, but in general work by identifying homologous protein sequences as a whole that are similar to known cleavable sequences. The result of Blast is specifically generated according to the following steps: for a given caspase, a positive set and a test protein, the test protein is aligned using Blast against all the proteins in the positive set, and then the best E-value is taken as the Blast prediction score, based on which Blast can predict the test protein to be a target substrate for the given caspase according to the highest similarity to the known targets.
On the other hand, the Blast calculation steps also indicate that Blast may perform better than specialized algorithms in certain cases (Figure 6); however, in terms of false-positive rates, it cannot identify the cleavage site. Furthermore, even if not designed for identifying small motifs, Blast can find an overall sequence similarity, which might result from a common ancestry, and therefore share a common function, such as the whole protein being a cleavage target for caspases.
The best AUC values for each prediction tool on each test set are summarized in Table 3. In summary, performance comparison analysis on the independent test indicates that SitePrediction and Cascleave are the two best-performing tools and generally provide an overall best performance among all the tools compared.
Although the AUC value shows that the state-of-the-art tools are still limited in terms of their predictive performance, there is room to further improve the performance by including newly discovered caspase substrates into training sets, integrating new informative features and developing novel models.
Case study: Caspase-3 and 8 substrate cleavage prediction
Selection of potential caspase substrates
To further evaluate each tool described in this research, we selected a number of potential substrates with high scores predicted by most tools in the human proteome, and experimentally validated these potential substrates for caspase-3 and caspase-8 by performing caspase assay experiments. In the independent test step, the AUC values of caspase-1 and caspase-3 for H. sapiens showed that among all the tools tested Cascleave, PoPS and SitePrediction achieved better performances than other tools (Table 3). Among these three tools, Cascleave and SitePrediction achieved the best AUC values. We further notice that while on the Cas3-homo set SitePrediction achieved the best AUC value but performed poorly on the Cas1-homo set (even not included in the top three tools), suggesting that SitePrediction might be more suitable for predicting caspase-3 substrates.
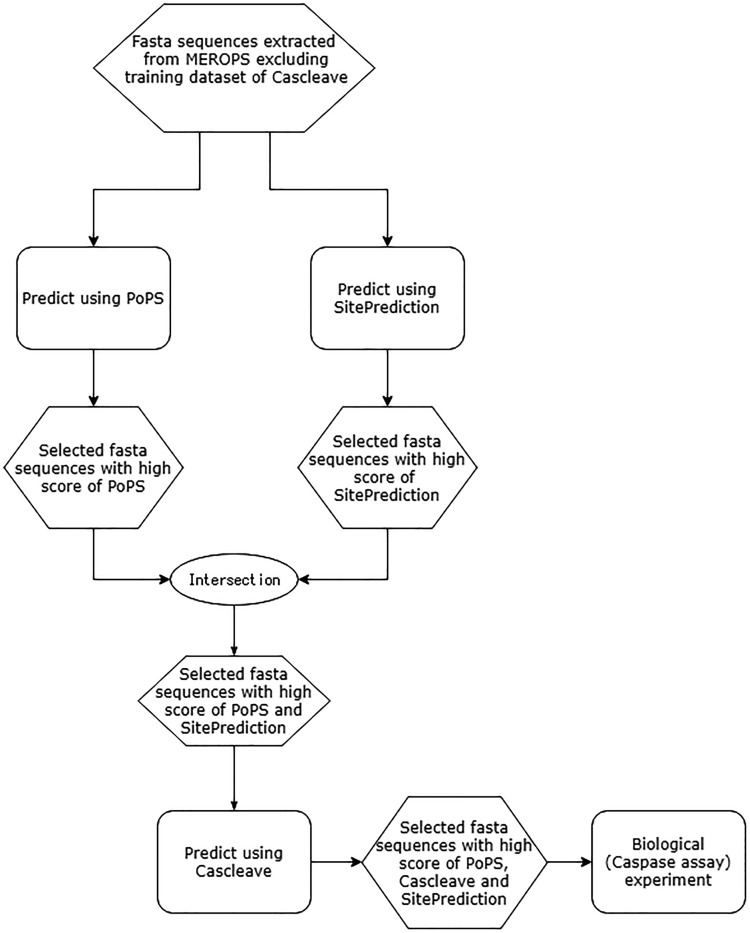
We first made a consensus-based decision for caspase prediction using the predictors that are capable of discriminating caspase substrates from non-caspase substrate based on the result described above. The predictors used for this purpose were SitePrediction, Cascleave and PoPS. During the prediction, we notice that it took a longer time for Cascleave to complete the prediction process for a single protein sequence; thus, it is reasonable to use SitePrediction and PoPS first to perform a rough discrimination of the sequences in the proteome of H. sapiens and apply Cascleave to predict specific cleavage sites within the proteins selected out. The detailed procedures can be found in Figure 7.
Figure 7.
A flowchart of the procedures for caspase-3 and caspase-8 substrate cleavage site prediction of the human proteome.
Considering the fact that Cascleave performed best in discriminating caspase substrates from non-caspase substrates, we sorted the final predicted caspase substrates based on predicted cleavage probability the score of Cascleave, and then performed the caspase cleavage assay experiments to validate these potential caspase substrates.
Caspase cleavage assay
The caspase cleavage assay is conducted to experimentally validate the potential substrates selected out in the previous section.
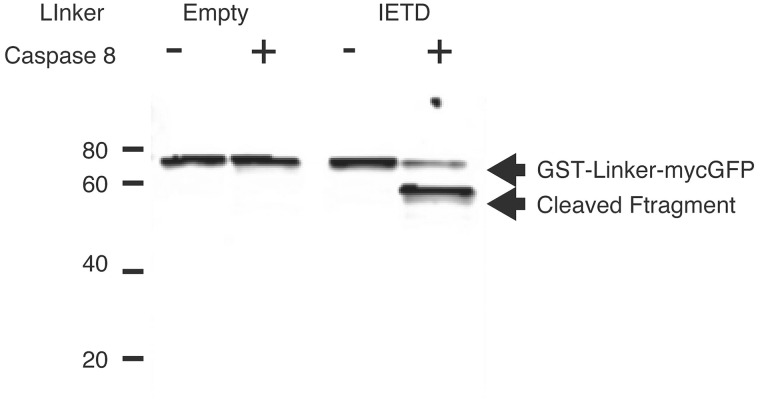
Figure 8 illustrates the caspase substrate cleavage results based on western blotting. Recombinant proteins encoding GST-myc-GFP with IETD linker between GST and GFP (Right) or without linker (Left) were digested by active caspase-8 (+) or control (−). After digestion, proteins were analyzed by western blotting using anti-myc. Recombinant GST-mycGFP, 75 kDa band, was detected in both conditions with and without caspase-8 treatment. In the recombinant GST-IETD-mycGFP protein case, a 75 kDa band was detected in caspase-8 nontreatment condition, and in contrast, a 50 kDa protein band was detected in caspase-8 treatment recombinant GSTIETD-mycGFP, indicating that IETD linker was cleaved by caspase-8.
Figure 8.
Western blotting of caspase assay analysis. Recombinant GST-mycGFP, 75 kDa band, was detected in both conditions with and without caspase-8 protein treatment. In the recombinant GST-IETD-mycGFP protein case, a 75 kDa band was detected in caspase-8 nontreatment condition, while in contrast a 50 kDa protein band was detected in caspase-8 treatment recombinant GSTIETD-mycGFP, indicating that the IETD linker was cleaved by caspase-8.
Caspase assay result discussion
Caspase cleavage assay results are summarized in Tables 4 and 5. As we can see, the experimental results clearly show that the majority of predicted potential substrates were cleaved. These results indicate that the tools tested in the evaluation step demonstrate an excellent performance for predicting both caspase-3 and 8 target substrates. In addition, there also exist a number of cleavage sites, which cannot be identified by most of the tools, highlighting that it remains a challenging task to develop more reliable and accurate caspase cleavage site prediction methods. See Supplemental Information for details of protein expression.
Table 4.
The caspase cleavage assay results of predicted potential caspase-3 substrates by PoPS, SitePrediction and cascleave
| Predicted caspase-3 substrate cleavage site | PoPS score | SitePrediction score | Cascleave score | Experimental result | Corresponding annotations in MEROPS |
|---|---|---|---|---|---|
| DVVD—GADT | 21.32 | 1560.39 | 1.578 | ◯ | – |
| EEVD—GSSP | 20.08 | 1515.922 | 1.461 | ◯ | – |
| EEVD—GSQG | 20.08 | 1515.922 | 1.461 | ◯ | C14 homologue |
| DETD—SGAG | 21.77 | 3400.88 | 1.345 | ◯ | C14.003: caspase-3, C14.005: caspase-6 |
| EEVD—GAPR | 20.08 | 1888.277 | 1.307 | ◯ | C14.005: caspase-6 |
| DSVD—GSLT | 21.26 | 1909.074 | 1.21 | ◯ | – |
| DDTD—GLTP | 17.79 | 791.345 | 1.157 | ◯ | C14.005: caspase-6, C14.006: caspase-2 |
| AEVD—GVDE | 19.93 | 295.25 | 1.061 | × | C14 homologue |
| DDPD—SAYL | 18.08 | 680.822 | 1.058 | ◯ | – |
| SEVD—GNDS | 20.05 | 449.294 | 1.039 | ◯ | C14.004: caspase-7, C14.006: caspase-2 |
| AEVD—GATP | 19.94 | 623.306 | 1.034 | ◯ | – |
| EEPD—GGFR | 16.97 | 414.973 | 0.969 | ◯ | – |
| TEPD—SPSP | Non-cleavage | Non-cleavage | 0.961 | × | – |
| SEID—GLKG | 18.7 | 220.873 | 0.911 | ◯ | – |
| EEPD—SANS | 17.14 | 761.635 | 0.82 | ◯ | C14.005: caspase-6, C14.006: caspase-2, C14 homologue |
| NEVD—GSNE | 20.01 | 223.501 | 0.766 | ◯ | – |
| EETD—GLDP | 16.86 | 886.89 | 0.747 | ◯ | C14.001: caspase-1, C14.005: caspase-6, C14.006: caspase-2, C14 homologue |
| EETD—GLHE | 16.86 | 886.89 | 0.747 | ◯ | – |
| GEVD—GKAI | 19.85 | 271.729 | 0.691 | ◯ | – |
| TEMD—SETL | Non-cleavage | Non-cleavage | 0.632 | × | – |
| LESD—SESL | Non-cleavage | Non-cleavage | 0.585 | × | – |
Note: ‘◯’ indicates the sequence is cleaved in the cleavage assay experiment, while ‘×’ indicates the sequence is not cleaved in the cleavage assay experiment.
Table 5.
The caspase cleavage assay results of predicted potential caspase-8 substrates by PoPS, SitePrediction and Cascleave
| Predicted caspase-8 substrate cleavage site | PoPS score | SitePrediction score | Cascleave score | Experimental result | Corresponding annotations in MEROPS |
|---|---|---|---|---|---|
| DVVD—GADT | 17.9 | 206.97 | 1.578 | ◯ | – |
| EEVD—GSSP | 21.24 | 771.98 | 1.461 | ◯ | – |
| EEVD—GSQG | 21.24 | 771.98 | 1.461 | ◯ | C14 homologue |
| DETD—SGAG | 17.77 | 3194.444 | 1.345 | ◯ | C14.003: caspase-3, C14.005: caspase-6 |
| EEVD—GAPR | 21.24 | 1621.17 | 1.307 | ◯ | C14.005: caspase-6 |
| DEVD—GAND | 22.46 | 3371.648 | 1.261 | ◯ | – |
| DETD—SPTV | 21.14 | 4921.875 | 1.236 | ◯ | C14.005: caspase-6, C14.006: caspase-2 |
| DSVD—GSLT | 17.59 | 525.68 | 1.21 | ◯ | C14 homologue |
| AEVD—GVDE | 22.46 | 1010.936 | 1.061 | ◯ | – |
| SEVD—GNDS | Non-cleavage | Non-cleavage | 1.039 | ◯ | C14.004: caspase-7, C14.006: caspase-2 |
| AEVD—GATP | 21.21 | 1268.26 | 1.034 | ◯ | – |
| TETD—SVGT | 20.01 | 854.701 | 0.999 | ◯ | – |
| EEPD—GGFR | Non-cleavage | Non-cleavage | 0.969 | ◯ | – |
| TEPD—SPSP | 17.38 | 92.307 | 0.961 | × | – |
| LEMD—SVLK | 19.27 | 412.088 | 0.935 | ◯ | C14.005: caspase-6, C14.006: caspase-2, C14 homologue |
| EEPD—SANS | Non-cleavage | Non-cleavage | 0.82 | ◯ | – |
| EETD—GLDP | 22.17 | 559.69 | 0.747 | ◯ | C14.001: caspase-1, C14.005: caspase-6, C14.006: caspase-2, C14 homologue |
| EETD—GLHE | 22.17 | 559.69 | 0.747 | ◯ | – |
| TEED—SVSV | 18.61 | 275.71 | 0.714 | ◯ | – |
| TEMD—SETL | 19.27 | 167.993 | 0.632 | × | – |
| LESD—SESL | 18.58 | 526.556 | 0.585 | × | – |
Note: ‘◯’ indicates the sequence is cleaved in the cleavage assay experiment while ‘×’ indicates the sequence is not cleaved in the cleavage assay experiment.
From the result, we can also see that some predicted substrates are both cleaved by caspases in the Group II and Group III, such as DETD—SGAG (both cleaved by caspase-3 and caspase-6) and DDTD—GLTP (both cleaved by caspase-2 and caspase-6). This is the reason why many substrates cleaved by caspase-3 are also cleaved by caspase-8. The result also indicates that there is a tendency that PoPS, SitePrediction and Cascleave prefer to predict substrates cleaved by caspases from the Groups II and III rather than Group I.
Conclusion
Owing to the functional significance of caspase substrate identification problem, computational biologists are motivated to develop more accurate and reliable predictors for caspase substrate prediction. Aiming at providing a comprehensive review of the status quo of caspase substrate predictors to nonbioinformaticians, this article describes and compares a number of widely used caspase substrate predictors in terms of their input/output, model construction and development, model performance evaluation as well as predictor utility. Benchmarking analysis on the independent test data sets revealed that Cascleave and SitePrediction achieve the overall highest AUC value when used for predicting caspase substrates in different species, especially for H. sapiens. In particular, SitePrediction achieved the highest AUC value when used for predicting caspase substrates for species other than H. sapiens. Detailed case studies of 21 caspase-3 substrate sequences and 21 caspase-8 substrate sequences demonstrate that while Cascleave, PoPS and SitePrediction achieved acceptable performance, there still exist some sequences that most currently available tools failed to predict. We conclude that caspase substrate prediction remains a challenging task, and we expect that more powerful next-generation algorithms with improved prediction performance will emerge with the increasing availability of caspase substrate cleavage data that can be used as high-quality training data for constructing the prediction models.
Key Points
We tested and evaluated 12 state-of-the-art tools for caspase cleavage site prediction according to their input/output, model construction strategies, predictive performance and predictor utility.
We constructed a series of testing data sets and performed independent data set testing.
We performed a case study and experimentally validated the cleavage prediction results by conducting caspase assay to verify whether sequences with high predicted scores can be cleaved in vitro.
Supplementary Data
Supplementary data are available online at https://academic.oup.com/bib.
Supplementary Material
Acknowledgements
The authors would like to thank the three anonymous reviewers for their helpful and constructive comments that have greatly helped improve the presentation of this article. Availability: All the independent test data sets are available at http://sunflower.kuicr.kyoto-u.ac.jp/~houu/Caspase_tool_review/Original_input.zip.
Funding
T.A. and H.S. are supported by JSPS, Japan (grant-in-aid #26240034). J.S. is supported by grants from the National Health and Medical Research Council of Australia (NHMRC) (grant number 490989), the Australian Research Council (ARC) (grant numbers LP110200333 and DP120104460), the National Institute of Allergy and Infectious Diseases of the National Institute of Health (grant number R01 AI111965), a Major Inter-Disciplinary Research (IDR) project (grant number 201402) awarded by Monash University and the Collaborative Research Program of Institute for Chemical Research, Kyoto University (grant number 2018-28).
Biographies
Yu Bao is currently a PhD candidate in the Bioinformatics Center, Institute Chemical Research at Kyoto University. His main research interests are machine learning in bioinformatics, algorithms on biological networks and biology.
Simone Marini is a research investigator at the University of Michigan. His main research interests are data integration and machine learning in bioinformatics, medicine and biology.
Takeyuki Tamura received PhD degree in Informatics from Kyoto University, Japan, in 2006. Currently, he works as an associate professor for Bioinformatics Center, Institute for Chemical Research, Kyoto University.
Mayumi Kamada has PhD in Informatics from Kyoto University and is an associate professor at Kyoto University. She currently works on developing and validating computational methods for genomic medicine.
Shingo Maegawa received his PhD degree in Molecular Biology in 2002 from Nara Institute of Science and Technology, Japan. His research interests include developmental biology and behavioral genetics.
Hiroshi Hosokawa received his PhD degree in Human and environmental studies in 1998 from Kyoto University, Japan. He is a senior lecture in Graduate School of Informatics, Kyoto University, Japan. His research interests include molecular biology, biochemistry and pharmacology.
Jiangning Song received his BEng and DEng degrees from Jiangnan University, China. He is currently a senior research fellow and group leader in the Biomedicine Discovery Institute, Monash University, Melbourne, Australia. He is affiliated with the Monash Centre for Data Science and ARC Centre of Excellence in Advanced Molecular Imaging, Monash University. His research interests include bioinformatics, computational biology, machine learning, data mining and pattern recognition.
Tatsuya Akutsu received his Dr. Eng. degree in Information Engineering in 1989 from University of Tokyo, Japan. Since 2001, he has been a professor in Bioinformatics Center, Institute for Chemical Research, Kyoto University, Japan. His research interests include bioinformatics and discrete algorithms.
References
- 1. Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer 2004;4(5):349–60. [DOI] [PubMed] [Google Scholar]
- 2. Adam Z. Protein stability and degradation in chloroplasts. Plant Mol Biol 1996;32(5):773–83. [DOI] [PubMed] [Google Scholar]
- 3. Anwar A, Saleemuddin M.. Regulation of digestive proteolytic activity in the larvae of Spilosoma obliqua (Lep., Arctiidae). J Appl Entomol 2001;125(9–10):577–82. [Google Scholar]
- 4. Nicholson DW, Thornberry NA.. Caspases: killer proteases. Trends Biochem Sci 1997;22(8):299–306. [DOI] [PubMed] [Google Scholar]
- 5. Bonfil RD, Cher ML.. The role of proteolytic enzymes in metastatic bone disease. IBMS Bonekey 2011;8(1):16–36. [Google Scholar]
- 6. duVerle DA, Mamitsuka H.. A review of statistical methods for prediction of proteolytic cleavage. Brief Bioinform 2012;13(3):337–49. [DOI] [PubMed] [Google Scholar]
- 7. Lohmüller T, Wenzler D, Hagemann S, et al. Toward computer-based cleavage site prediction of cysteine endopeptidases. Biol Chem 2003;384(6):899–909. [DOI] [PubMed] [Google Scholar]
- 8. Mizianty MJ, Kurgan L.. Sequence-based prediction of protein crystallization, purification and production propensity. Bioinformatics 2011;27(13):i24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang H, Feng L, Zhang Z, et al. Crysalis: an integrated server for computational analysis and design of protein crystallization. Sci Rep 2016;6(1):21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li SJ, Hochstrasser M.. A new protease required for cell-cycle progression in yeast. Nature 1999;398(6724):246–51. [DOI] [PubMed] [Google Scholar]
- 11. Jenal U, Fuchs T.. An essential protease involved in bacterial cell-cycle control. EMBO J 1998;17(19):5658–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerdes J, Li L, Schlueter C, et al. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol 1991;138(4):867–73. [PMC free article] [PubMed] [Google Scholar]
- 13. Cardone MH, Roy N, Stennicke HR, et al. Regulation of cell death protease caspase-9 by phosphorylation. Science 1998;282(5392):1318–21. [DOI] [PubMed] [Google Scholar]
- 14. Boldin MP, Goncharov TM, Goltseve YV, et al. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1-and TNF receptor–induced cell death. Cell 1996;85(6):803–15. [DOI] [PubMed] [Google Scholar]
- 15. Krajewska M, Wang HG, Krajewski S, et al. Immunohistochemical analysis of in vivo patterns of expression of CPP32 (Caspase-3), a cell death protease. Cancer Res 1997;57(8):1605–13. [PubMed] [Google Scholar]
- 16. McGarry TJ, Kirschner MW.. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998;93(6):1043–53. [DOI] [PubMed] [Google Scholar]
- 17. Pellettieri J, Fitzgerald P, Watanabe S, et al. Cell death and tissue remodeling in planarian regeneration. Dev Biol 2010;338(1):76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muruve DA, Pétrilli V, Zaiss AK, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 2008;452(7183):103–7. [DOI] [PubMed] [Google Scholar]
- 19. Franchi L, Eigenbrod T, Muñoz-Planillo R, et al. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol 2009;10(3):241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cerenius L, Kawabata S, Lee BL, et al. Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem Sci 2010;35(10):575–83. [DOI] [PubMed] [Google Scholar]
- 21. Cohen GM. Caspases: the executioners of apoptosis. Biochem J 1997;326(Pt 1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuan J, Shaham S, Ledoux S, et al. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell 1993;75(4):641–52. [DOI] [PubMed] [Google Scholar]
- 23. Ashkenazi A, Dixit VM, et al. Death receptors: signaling and modulation. Science 1998;281(5381):1305–8. [DOI] [PubMed] [Google Scholar]
- 24. Barry M, Bleackley RC.. Cytotoxic T lymphocytes: all roads lead to death. Nat Rev Immunol 2002;2(6):401–9. [DOI] [PubMed] [Google Scholar]
- 25. Bortner CD, Oldenburg NB, Cidlowski JA.. The role of DNA fragmentation in apoptosis. Trends Cell Biol 1995;5(1):21–6. [DOI] [PubMed] [Google Scholar]
- 26. Rathmell JC, Thompson CB.. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell 2002;109(2):S97–107. [DOI] [PubMed] [Google Scholar]
- 27. Everett H, McFadden G.. Apoptosis: an innate immune response to virus infection. Trends Microbiol 1999;7(4):160–5. [DOI] [PubMed] [Google Scholar]
- 28. Dostert C, Pétrilli V, Van Bruggen R, et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 2008;320(5876):674–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Earnshaw WC, Martins LM, Kaufmann SH.. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 1999;68(1):383–424. [DOI] [PubMed] [Google Scholar]
- 30. Thornberry NA, Rano TA, Peterson EP, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B Functional relationships established for key mediators of apoptosis. J Biol Chem 1997;272(29):17907–11. [DOI] [PubMed] [Google Scholar]
- 31. Lauber K, Bohn E, Kröber SM, et al. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell 2003;113(6):717–30. [DOI] [PubMed] [Google Scholar]
- 32. Salvesen GS, Dixit VM.. Caspase activation: the induced-proximity model. Proc Natl Acad Sci USA 1999;96(20):10964–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Creagh EM, Conroy H, Martin SJ.. Caspase-activation pathways in apoptosis and immunity. Immunol Rev 2003;193(1):10–21. [DOI] [PubMed] [Google Scholar]
- 34. Song J, Tan H, Boyd SE, et al. Bioinformatic approaches for predicting substrates of proteases. J Bioinform Comput Biol 2011;09(01):149–78. [DOI] [PubMed] [Google Scholar]
- 35. Wee LJ, Tong JC, Tan TW, et al. A multi-factor model for caspase degradome prediction. BMC Genomics 2009;10(Suppl 3):S6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wee LJ, Tan TW, Ranganathan S.. SVM-based prediction of caspase substrate cleavage sites. BMC Bioinformatics 2006;7(Suppl 5):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhasin M, Raghava G.. Pcleavage: an SVM based method for prediction of constitutive proteasome and immunoproteasome cleavage sites in antigenic sequences. Nucleic Acids Res 2005;33:W202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suresh MX, Gromiha MM, Suwa M.. Development of a machine learning method to predict membrane protein-ligand binding residues using basic sequence information. Adv Bioinform 2015;2015:843030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chauhan JS, Mishra NK, Raghava GP.. Prediction of GTP interacting residues, dipeptides and tripeptides in a protein from its evolutionary information. BMC Bioinformatics 2010;11(1):301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gromiha MM, Ou YY.. Bioinformatics approaches for functional annotation of membrane proteins. Brief Bioinform 2014;15(2):155–68. [DOI] [PubMed] [Google Scholar]
- 41. Wang M, Zhao XM, Tan H, et al. Cascleave 2.0, a new approach for predicting caspase and granzyme cleavage targets. Bioinformatics 2014;30(1):71–80. [DOI] [PubMed] [Google Scholar]
- 42. Apweiler R, Biswas M, Fleischmann W, et al. Proteome analysis database: online application of interPro and CluSTr for the functional classification of proteins in whole genomes. Nucleic Acids Res 2001;29(1):44–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Acland A, Agarwala R, Barrett T, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res 2014;42(D1):D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Igarashi Y, Heureux E, Doctor KS, et al. PMAP: databases for analyzing proteolytic events and pathways. Nucleic Acids Res 2009;37(Suppl 1):D611–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boeckmann B, Bairoch A, Apweiler R, et al. The SWISS-PROT protein knowledgebase and its supplement Trembl in 2003. Nucleic Acids Res 2003;31(1):365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ng SK, Zhang Z, Tan SH, et al. InterDom: a database of putative interacting protein domains for validating predicted protein interactions and complexes. Nucleic Acids Res 2003;31(1):251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bateman A, Coin L, Durbin R, et al. The Pfam protein families database. Nucleic Acids Res 2004;32(90001):D138–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Backes C, Kuentzer J, Lenhof HP, et al. GraBCas: a bioinformatics tool for score-based prediction of Caspase-and Granzyme B-cleavage sites in protein sequences. Nucleic Acids Res 2005;33:W208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garay-Malpartida HM, Occhiucci JM, Alves J, et al. CaSPredictor: a new computer-based tool for caspase substrate prediction. Bioinformatics 2005;21(Suppl 1):i169–76. [DOI] [PubMed] [Google Scholar]
- 50. Boyd SE, Pike RN, Rudy GB, et al. PoPS: a computational tool for modeling and predicting protease specificity. J Bioinform Comput Biol 2005;03(03):551–85. [DOI] [PubMed] [Google Scholar]
- 51. Verspurten J, Gevaert K, Declercq W, et al. SitePredicting the cleavage of proteinase substrates. Trends Biochem Sci 2009;34 (7):319–23. [DOI] [PubMed] [Google Scholar]
- 52. Song J, Tan H, Shen H, et al. Cascleave: towards more accurate prediction of caspase substrate cleavage sites. Bioinformatics 2010;26(6):752–60. [DOI] [PubMed] [Google Scholar]
- 53. Piippo M, Lietzén N, Nevalainen OS, et al. Pripper: prediction of caspase cleavage sites from whole proteomes. BMC Bioinformatics 2010;11 (1):320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barkan DT, Hostetter DR, Mahrus S, et al. Prediction of protease substrates using sequence and structure features. Bioinformatics 2010;26 (14):1714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wee LJ, Tan TW, Ranganathan S.. CASVM: web server for SVM-based prediction of caspase substrates cleavage sites. Bioinformatics 2007;23(23):3241–3. [DOI] [PubMed] [Google Scholar]
- 56. Ayyash M, Tamimi H, Ashhab Y.. Developing a powerful in silico tool for the discovery of novel caspase-3 substrates: a preliminary screening of the human proteome. BMC Bioinformatics 2012;13(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Song J, Tan H, Perry AJ, et al. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS One 2012;7(11):e50300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rawlings ND, Waller M, Barrett AJ, et al. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 2014;42(D1):D503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Igarashi Y, Eroshkin A, Gramatikova S, et al. CutDB: a proteolytic event database. Nucleic Acids Res 2007;35:D546–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. duVerle D, Takigawa I, Ono Y, et al. CaMPDB: a resource for calpain and modulatory proteolysis. In: Genome Informatics. International Conference on Genome Informatics, Vol. 22. Singapore: World Scientific Publishing, 2010, 202–13. [PubMed]
- 62. Lange PF, Overall CM.. TopFIND, a knowledgebase linking protein termini with function. Nat Methods 2011;8(9):703–4. [DOI] [PubMed] [Google Scholar]
- 63. Crawford ED, Seaman JE, Agard N, et al. The DegraBase: a database of proteolysis in healthy and apoptotic human cells. Mol Cell Proteomics 2013;12(3):813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rogers S, Wells R, Rechsteiner M.. Amino acid sequences common to rapidly degrade proteins: the PEST hypothesis. Science 1986;234(4774):364–9. [DOI] [PubMed] [Google Scholar]
- 65. Rechsteiner M, Rogers SW.. PEST sequences and regulation by proteolysis. Trends Biochem Sci 1996;21(7):267–71. [PubMed] [Google Scholar]
- 66. Song J, Li F, Leier A, et al. PROSPERous: high-throughput prediction of substrate cleavage sites for 90 proteases with improved accuracy. Bioinformatics 2018;34(4):684–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Song J, Wang Y, Li F, et al. iProt-sub: a comprehensive tool for accurately mapping and predicting protease-specific substrates and cleavage sites. Brief Bioinform 2018, doi: 10.1093/bib/bby028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kabsch W, Sander C.. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983;22(12):2577–637. [DOI] [PubMed] [Google Scholar]
- 69. Ward JJ, McGuffin LJ, Bryson K, et al. The DISOPRED server for the prediction of protein disorder. Bioinformatics 2004;20(13):2138–9. [DOI] [PubMed] [Google Scholar]
- 70. Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 1999;292 (2):195–202. [DOI] [PubMed] [Google Scholar]
- 71. Shao J, Xu D, Tsai SN, et al. Computational identification of protein methylation sites through bi-profile Bayes feature extraction. PLoS One 2009;4(3):e4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kawashima S, Kanehisa M.. AAindex: amino acid index database. Nucleic Acids Res 2000;28(1):374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ebina T, Toh H, Kuroda Y.. DROP: an SVM domain linker predictor trained with optimal features selected by random forest. Bioinformatics 2011;27(4):487–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.