Abstract
The successful parasitisation of a plant by a phytophagous insect is dependent on the delivery of effector molecules into the host. Sedentary gall forming insects, such as grape phylloxera (Daktulosphaira vitifoliae Fitch, Phylloxeridae), secrete multiple effectors into host plant tissues that alter or modulate the cellular and molecular environment to the benefit of the insect. The identification and characterisation of effector proteins will provide insight into the host-phylloxera interaction specifically the gall-induction processes and potential mechanisms of plant resistance. Using proteomic mass spectrometry and in-silico secretory prediction, 420 putative effectors were determined from the salivary glands or the root-feeding D. vitifoliae larvae reared on Teleki 5C (V. berlandieri x V. riparia). Among them, 170 conserved effectors were shared between D. vitifoliae and fourteen phytophagous insect species. Quantitative RT-PCR analysis of five conserved effector candidates (protein disulfide-isomerase, peroxidoredoxin, peroxidase and a carboxypeptidase) revealed that their gene expression decreased, when larvae were starved for 24 h, supporting their assignment as effector molecules. The D. vitifoliae effectors identified here represent a functionally diverse group, comprising both conserved and unique proteins that provide new insight into the D. vitifoliae–Vitis spp. interaction and the potential mechanisms by which D. vitifoliae establishes the feeding site, suppresses plant defences and modulates nutrient uptake.
Introduction
The ability of phytophagous insects to feed from, or reproduce on plant hosts is dependent on the efficient modulation or evasion of plant defence systems. These defences may be constitutive or induced and can involve complex recognition and response systems that result in the release of defensive substances, targeted self-destruction of cells or even the attraction of predators to contend with the invading insect. Insects and particularly those that engage the plant host on cellular level, attempt to overcome these defences through a variety of strategies including the delivery of bioactive substances into the plant cellular environment [1]. Many of these substances, commonly referred to as effectors, are delivered via the insect saliva directly on- or into the host tissue and ultimately determine whether the prospected interaction is compatible or incompatible. Effectors of phytophagous insects are generally small proteins or molecules that alter host cell structures and/or biological functions once they are exposed to a suitable plant tissue. Thereby single effectors may interact with several plant protein networks and modulate various host physiological traits simultaneously [2, 3].
Identifying the effector repertoire of phytophagous insect species is essential to understand the underlying host-parasite interaction and potentially leads to the development of innovative pest control strategies reducing reliance on agrochemical application [4, 5]. The identification of effector molecules within plant tissues is challenging due to their low concentration and short ephemerality due to metabolic processing. An alternative strategy is to characterise the effector-enriched salivary gland tissue, which previously resulted in the discovery of multiple insect effector proteins released by aphids [6–8], hessian flies [9], planthoppers [10, 11], whiteflies [12, 13] and thrips [14]. Significant progress in protein-based mass spectrometry and next generation sequencing technologies (e.g. RNAseq) facilitated the large-scale identification of putative insect effectors lists within insect salivary glands or saliva [15–21]. Comparative In-silico analyses of predicted effector sets across phloem-feeding aphid species identified both conserved effectors used as part of shared infestation strategies aimed to modulate common host defensive and cellular processes and unique effectors employed to establish and maintain compatible interactions in a host-specific manner [22–24].
Historically grape phylloxera (Daktulosphaira vitifoliae Fitch; Phylloxeridae) was introduced from North America into Europe in the late 19th century with devastating economic consequences for the wine industry at the time. The sedentary insect infests the Vitis spp. root system by the formation of root gall tissues, thereby inducing changes in the water, mineral and assimilate transport affecting the host vine physiology and vigor [25–27]. In addition D. vitifoliae infestation promotes secondary soil-borne infections harming the root system of field-grown vines [28, 29]. The majority of commercial Vitis rootstocks tolerate minor phylloxera populations on the root system [30], however reports of phylloxera-caused root damages in vineyards increase worldwide [31–38]. Root gall formation by D. vitifoliae involves the regulation of various host plant physiological traits including the induction of structural root gall growth [39–43], interference with plant defence mechanisms [44–47] and alteration of systemic nutrient allocation [41, 48, 49].
Insect salivary glands are the principal organs for the production of effectors essential for the establishment and maintenance of compatible host-parasite interactions. In this study we aim to identify and characterise effector proteins of root-galling D. vitifoliae, a major pest of Vitis spp. hypothesising that salivary glands dissected from root-feeding D. vitifoliae larvae are enriched with effector proteins of diverse functional effector groups involved in the modulation of host physiological pathways and required for insect feeding.
Material and methods
Plant and insect material
Dormant single eye cuttings of the rootstock Teleki 5C (V. berlandieri x V. riparia, clone Gm 6–52) were propagated in Jiffy-7 pots (40 mm; Jiffy Products International AS, Norway) under controlled greenhouse conditions. In total 48 rooted plants were transplanted in four growth containers (38 x 28 x 20 cm) containing a mixed substrate of perlite and foamed clay (1:1). The growth containers were located in a climate chamber set to (25°C, 60% rH and 16 h photoperiod) and fertigated weekly (0.25‰ Ferty 3 MEGA) in quarantine conditions [50, 51]. Two hundred eggs of a D. vitifoliae single founder lineage were placed adjacent to the roots of each plant growth container for hatching and asexual reproduction.
Salivary gland collection
After 50 days feeding D. vitifoliae L3 larvae [52] were carefully from root galls and submerged in ice-cold phosphate buffered saline (PBS) with pH 7.0 (VWR, Radnor, USA) containing a protease inhibitor cocktail (0.2 mM, Complete Mini, Roche, Basel, CH). For each biological sample the salivary glands of 100 larvae were dissected under the binocular using two metallic needles and immediately snap-frozen in liquid N2. The salivary gland samples for the qRT-PCR analyses were collected likewise, but from L3 larvae submerged in a PBS buffer containing RNAlater (Qiagen, Hilden, DE). For the qRT-PCR analyses salivary glands were dissected from feeding (SG0) and starving D. vitifoliae larvae (SG24). For the starvation treatment, feeding L3 larvae were gently removed from the root galls and kept without plant contact on moist filter paper in isolated petri dishes for 24 h at 25±3°C in the dark simulating equal environmental conditions. For each treatment four or three biological samples were collected and stored at -80°C for the subsequent proteomic and transcriptomic analyses, respectively.
Protein extraction and purification
Salivary glands were homogenized with a motorised pestle in 150 μl lysis buffer (7 M urea, 2 M thiourea supplemented with a protease inhibitor cocktail, Roche, Complete Mini). Samples were briefly sonicated and centrifuged at 9000 g for 5 min to pellet the cellular debris. The supernatant was removed and purified using the 2D clean-up Kit (GE Heath Care, Chicago, USA) following the manufacturer’s instructions (procedure A) and the resulting protein pellet was resuspended in 50 μl of 6 M urea, 2 M thiourea, 0.1 M Tris-HCl, pH 8.0 and quantified using the Qubit Protein assay Kit (Thermo Scientific, Waltham, USA). 20 μl of this resuspension was removed for protein digestion and 50 mM ammonium bicarbonate was added to each sample. Proteins were reduced with 0.5 M dithiothreitol (DTT) at 56°C for 20 min and alkylated with 0.55 M iodoacetamide (IAA) at room temperature for 15 min, in the dark. 1 μl of a 1% w/v solution of Protease Max Surfactant Trypsin Enhancer (Promega) and 1 μg of Sequence Grade Trypsin (Promega) was added and the protein/trypsin mixture was incubated at 37°C for 18 hours. Digestion was terminated by adding 1 μl of 100% trifluoroacetic acid (Sigma Aldrich) and incubation at room temperature for 10 min. Samples were centrifuged for 10 min at 13,000 × g and a volume equivalent to 40 μg of pre-digested protein was removed and purified for mass spectrometry using ZipTips (Merck Millipore, Burlington USA) following the manufacturer’s instructions and the resulting eluant was concentrated with a SpeedyVac (Savant DNA120, Thermo Scientific Waltham, USA) and resuspended in acetonitrile (2% v/v); TFA (0.05% v/v) buffer.
Mass spectrometry and functional annotation
A volume equivalent to 1 μg of tryptic peptides was loaded onto a QExactive high resolution accurate mass spectrometer (Thermo Scientific Waltham, USA) connected to a Dionex Ultimate 3000 chromatography system (RSLCnano, Thermo Scientific Waltham, USA). The peptides were separated by a 2% to 40% gradient of acetonitrile on a Biobasic C18 PicofritTM column (100 mm length, 75 mm ID), using a 120 minute reverse-phase gradient at a flow rate of 250 nL min-1. All data were acquired with the mass spectrometer operating in automatic data dependent switching mode. A full MS scan at 140,000 resolution and a range of 300–1700 m/z was followed by an MS/MS scan, resolution 17,500 and a range of 200–2000 m/z, selecting the 15 most intense ions prior to MS/MS.
Protein identification and LFQ normalisation of MS/MS data was performed using MaxQuant v1.5.6.5 (http://www.maxquant.org) to correlate data against the predicted protein set derived from the D. vitifoliae genome version 3.2 [53] produced and provided by the International Aphid Genomic Consortium (IAGC) (bipaa.genouest.org/is/aphidbase) and a contaminant sequence set provided by MaxQuant. The following search parameters were used: first search peptide tolerance of 20 ppm, second search peptide tolerance 4.5 ppm with cysteine carbamidomethylation as a fixed modification and N-acetylation of protein and oxidation of methionine as variable modifications and a maximum of two missed cleavage sites allowed. False Discovery Rates (FDR) were set to 1% for both peptides and proteins and the FDR was estimated following searches against a target-decoy database. Perseus v.1.5.5.3 (www.maxquant.org/) was used for data analysis, processing and visualisation. The data matrix was first filtered for the removal of contaminants and peptides identified by site. LFQ intensity values were log2 transformed, ranked on their average MS intensities and proteins not found in three of the four replicates were omitted from the analysis.
Functional annotation of the identified proteins and the assignment of gene ontology (GO) terms was conducted using Blast2GO v. 2.5.0 [54]. BlastP searches were conducted against the NCBI nr database (as of 12.11.2018) reporting matches with a HSP cut-off value ≥ 33, BLAST hits with an E≤10−3 value and > 90% sequence similarity were reported. GO terms for all identified proteins were categorized by molecular function (MF), biological process (BP) and cellular component (CC) to determine the functional component of the salivary gland proteome. Signal secretion peptides were identified using a combined In-silico pipeline consisting of: TMHMM Server 2 [55], PredGPI [56], SignalP Server 5.0 for Eukarya [57] and SecretomeP Server 2.0 with NN-score > 0.6 [58]. Local BlastP analyses against published salivary effector candidates of fourteen phytophagous insect species [11, 15, 16, 19, 24, 59–62] were performed with NCBI’s Blast+ tool. Effectors having an e-value < e-50 and a bit score >100 were considered as orthologous matches for the effector candidates of D. vitifoliae.
RNA extraction and qRT-PCR analysis
RNA was extracted using the NucleoSpin RNA XS kit (Macherey-Nagel, Düren, Germany) following manufacturer’s instructions yielding 100–250 ng total RNA per sample. Subsequent reverse transcription (Sensiscript RT kit, Qiagen, Hilden, Germany) and normalisation were conducted using 50 ng of cDNA. Out of six candidate genes, actin A1 (DV3000100) was chosen as a reference gene after optimisation tests [63] following the Normfinder procedure [64]. The primer pairs were designed based on the D. vitifoliae genome version 3.2 using the NCBI primer tool software (Table 1). Quantitative RT-PCR analyses were performed using the Rotor-Gene Cycler Q (Qiagen, Hilden, Germany) employing KAPA SYBR FAST qPCR Universal (Kapa Biosystems, Wilmington, US) as detector agent in technical duplicates (total reaction vol. 12 μl with 2 μl cDNA (1:7)). Primer efficiencies were determined by conducting standard curves with four step template dilutions. Cycling conditions were: one cycle for 5 min at 95°C, 45 cycles for 8 sec at 95°C, 20 sec at 60°C, 15 sec at 72°C, 3 sec at 75°C [48]. Relative gene expression levels were calculated using the EasyqpcR package (bioconductor) in R. Statistics were performed with Independent T tests (p < 0.05) in SPSS (IBM, v24) comparing the gene expression levels in salivary glands dissected from feeding larvae (SG0) versus starving larvae at 24 h (SG24).
Table 1. Analysed DvEffector genes and primers used for qRT-PCR.
| Gene accession ID (Dv 3.2) | Annotation | Forward sequence | Reverse sequence | R2 | Primer efficiency | |
|---|---|---|---|---|---|---|
| (5'-3') | (5'-3') | |||||
| Reference gene | ||||||
| DV3000100 | actA1 | Actin A1 | TGGGGTGGTAGTGGTGATGA | ACACGTCGGATGTAAACGACA | 0,981 | 1,10 |
| DvEffectors matched with effectors of Acyrthosiphon pisum | ||||||
| DV3009058.1 | Dv1 | Protein disulfide-isomerase A3 | TCATGGCTTAGTTGGACATCGT | TTACGCCAGTAGTTGGTTCCT | 0,997 | 0,98 |
| DV3001833 | Dv2 | Peroxiredoxin-2 | AAACCAGCCCCAGATTGGAA | TTGAACGCCAATATCTCAGTTGG | 0,999 | 0,94 |
| DV3017719 | Dv3 | Venom serine carboxypeptidase-like | CAGAGCTGCGTGCAAAGTGA | ACCAAAGCAAGACAGACGCA | 0,993 | 0,93 |
| DV3010201 | Dv4 | Peroxiredoxin-2 | TGTTTTAGTGGAACCTGATGGTGT | AGCCTGAACAAGCCGTAAGAC | 0,997 | 1,09 |
| DV3014676 | Dv5 | Peroxidase | TCTGCGACAACAGTGACGAT | TGTGGAGCAGCATGACCATA | 0,997 | 0,96 |
| False positives | ||||||
| DV3014415 | Fp1 | Muscle-specific 20-like | ATTGAATATGGCGTTCCTGATGTAG | AGGGCCTAAAAATGGACCTTTCC | 0,993 | 1,02 |
| DV3008102 | Fp2 | Myofilin isoform a | CAGATCCATACCATTCTGAGCCT | TCGGGCAGAGGTAGAGGTTT | 0,993 | 0,96 |
| DV3011193 | Fp3 | Serine arginine repetitive matrix 1 isoform X1 | AGTTCAACGTCCAGCGGAAA | ACCGGGAATCACTTTCTTCGT | 0,989 | 0,99 |
The presented genes coded for proteins identified within the salivary gland proteome of D. vitifoliae larvae feeding on root galls of Teleki 5C. The gene accession IDs refer to the D. vitifoliae genome v 3.2 provided by the International Aphid Genomic Consortium (IAGC). ‘Dv 1–5’ refers to five conserved DvEffectors, whereas ‘Fp1-3’ to false positives functionally not associated to D. vitifoliae feeding. The gene annotation corresponds to the A. pisum effector ortholog.
Results
Assessment and description of the salivary gland proteome of D. vitifoliae
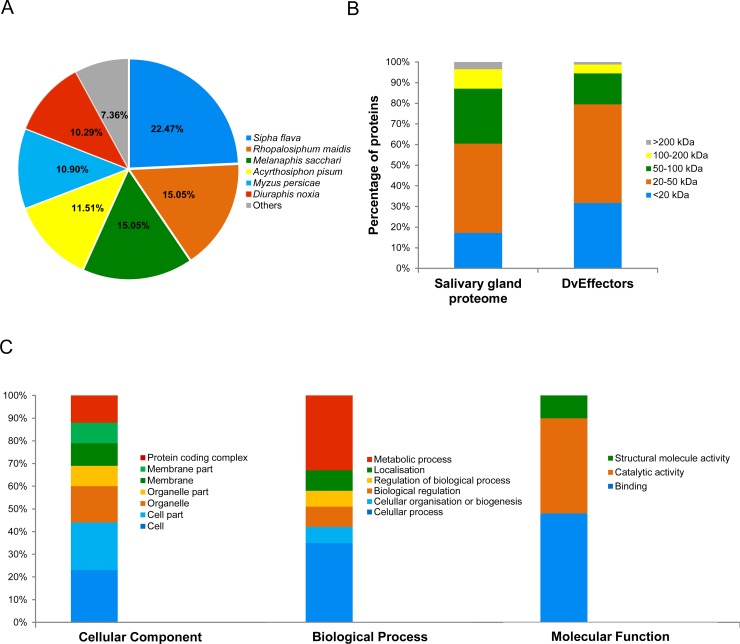
The dissection of the salivary glands from 100 feeding D. vitifoliae larvae, subsequent protein extraction and purification yielded 3.5–6.0 μg of total protein. In total 19,901 unique peptides representing 1511 proteins were identified (hereafter the salivary gland proteome) with log2 normalised LFQ intensities ranging between 21.6–35.9 (S1 Table). Comparative blast searches conducted with Blast2Go (as of 12.11.2018) revealed high similarities between the salivary gland proteome of D. vitifoliae and the aphid species. In total 92.64% of the identified proteins had top blast hits with proteins from: Sipha flava (22.47%), Rhopalosiphum maidis (15.05%), Melanaphis sacchari (15.05%), Acyrthosiphon pisum (11. 51%), Myzus persicae (10.90%) and Diuraphis noxia (10.29%) (Fig 1A). The majority of the identified salivary gland proteins, did not exceed 100 kDa in weight (87.09%) (Fig 1B). The smallest identified protein DV3009370 was annotated as cytochrome b-c1 complex subunit 9 (6.91 kDa, 59 AA), whereas the biggest protein DV3004164 was an annotated titin isoform X2 (1630.90 kDa, 14427 AA) (S1 Table). Of the top 100 most abundant proteins, determined by relative ranking based on average intensities, 36 were associated with structural components including contractile muscles, the cytoskeleton and the extracellular matrix. In addition, numerous primary metabolic enzymes (7) and mitochondrial associated proteins (14) were abundantly represented. To explore the functional composition of the salivary gland proteome in more detail, a Gene Ontology (GO) term analysis was conducted and terms were grouped into the categories: cellular component, molecular function and biological process (Fig 1C). For the cellular component category, 44% of the annotated proteins was associated to ‘cell’ or ‘cell part’, 25% to ‘organelle’ or ‘organelle part’, 19% to ‘membrane’ or ‘membrane part’ and 12% to ‘protein-containing complexes’. Within the biological process category the two most abundant groups were ‘cellular process’ 35% and ‘metabolic process’ 42%. The protein distribution within the category molecular function resulted in 42% of proteins with ‘binding’ properties, 40% with ‘catalytic activity’ and 10% with ‘structural molecule activity’ (Fig 1C).
Fig 1. Characterisation of the salivary gland proteome.
Gene ontology characterisation of the salivary gland proteome of D. vitifoliae larvae feeding on root galls of Teleki 5C (V. berlandieri x V. riparia). A) Protein top blast hit analysis ranked by species; B) Molecular weight distribution [kDa] of 1511 proteins of the salivary gland proteome and the secreted DvEffectors subset (420 proteins); C) GO enrichment analysis conducted with Blast2Go.
DvEffectors identified within the salivary glands of D. vitifoliae
A combined In-silico pipeline was employed to resolve proteins that are secreted with high probability among the D. vitifoliae salivary gland proteome (Table 2). The first step of the presented In-silico pipeline involved the screening for amino acid based structural retaining signals/motifs. Proteins with one or more transmembrane helices (TMHMMs) and/or glycosylphosphatidylinositol-anchors (GPI-anchors) were excluded as effectors due to their high probability of being retained by intra- and extracellular membranes. The remaining proteins without structural retaining signals were analysed for their likelihood to be secreted via classical (SignalP) or non-classical leaderless (SecretomeP, NN-score > 0.6) secretion pathway. Applying these criteria 420 D. vitifoliae proteins (27.80%) were detected to be secreted (Table 2) and therefore represented the putative D. vitifoliae effector set named ‘DvEffectors’ (S2 Table). The subsequent molecular weight analysis of the filtered DvEffectors showed a higher proportion of smaller proteins in comparison to the overall salivary gland proteome. Most of the identified DvEffectors (94.52%) did not exceed 100 kDa (Fig 1B).
Table 2. Protein secretion pipeline applied to the D. vitifoliae salivary gland proteome.
| Secretory prediction pipeline | Protein number | Percentage |
| Identified proteins | 1511 | 100,00% |
| TMHMMs | 240 | 15,88% |
| GPI-anchors | 23 | 1,52% |
| Proteins without structural retaining signals | 1259 | 82,32% |
| Classical secretion pathway | 151 | 10,19% |
| Non-classical secretion pathway | 354 | 23,43% |
| Secreted proteins | 420 | 27,80% |
The secretion pipeline used to identify secreted proteins within the salivary gland proteome of D. vitifoliae larvae feeding on root galls of Teleki 5C (V. berlandieri x V. riparia). The online tools TMHMM Server 2.0 (TMHMMs), PredGPI (GPI-anchors), SignalP Server 5.0 (classical secretion pathway) and SecretomeP Server 2.0 (non-classical secretion pathway) were used in a stepwise fashion to determine the likelihood of proteins to be secreted.
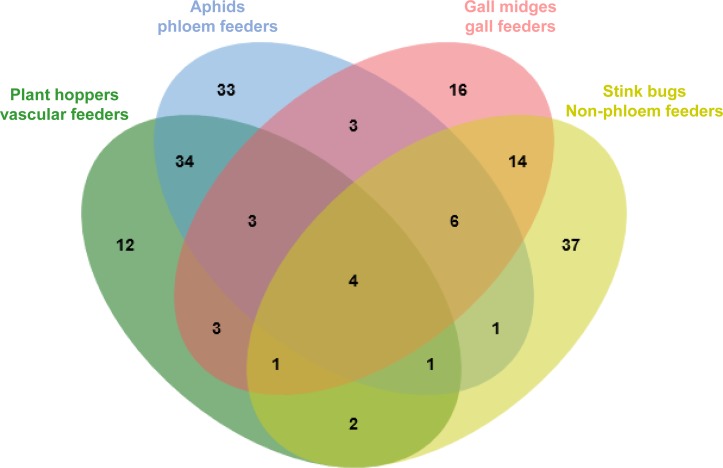
To identify potentially conserved effectors shared between D. vitifoliae and other phytophagous insect species, BlastP searches were conducted against salivary proteins of vascular (xylem and phloem) feeding plant hoppers: Nephotettix cincticeps [11], Nilaparvata lugens, Sogatella frucifera, Laodelphax striatellus [59]; phloem feeding aphids: Acyrthosiphon pisum [15, 16], Myzus persicae, Rhopalosiphum padi, Myzus cerasi [24], Sitobion avenae [19]; gall feeding gall midges: Mayetiola destructor [61], Mayetiola avenae, Mayetiola hordei [60] and non-phloem feeding stink bugs: Halyomorpha halys and Nezara viridula [62]. The conducted analysis (e-value < 1.0 e-50 and bit score >100) identified 170 conserved DvEffectors shared with salivary proteins of at least one analysed insect species (Fig 2, S3 Table). As one example 42 DvEffectors matched with predicted salivary effectors of the pea aphid Acyrthosiphon pisum (S4 Table, Table 3), being the species with most effector matches. Among them we identified numerous functional effector annotations such as esterases, glucose dehydrogenases, heat shock proteins, lysosomal alpha-mannosidases, maltases, peroxidases, peroxidoredoxins, protein disulfide-isomerases and carboxypeptidases (Table 3).
Fig 2. Conserved DvEffectors with phytophagous insects.
A summary of the BlastP search results of the DvEffectors against salivary gland proteins of phytophagous insects with different feeding behaviour: Plant hoppers–vascular feeders (green): Nephotettix cincticeps [11], Nilaparvata lugens, Sogatella frucifera, Laodelphax striatellus [59]; Aphids—phloem feeders (blue): Acyrthosiphon pisum [15, 16], Myzus persicae, Rhopalosiphum padi, Myzus cerasi [24], Sitobion avenae [19]; Gall midges—gall feeders (rosa): Mayetiola destructor [61], Mayetiola avenae, Mayetiola hordei [60] and Stink bugs—non-phloem feeders (yellow): Halyomorpha halys and Nezara viridula [62]. Numbers within the circles indicated the ortholog matches (-value < 1.0 e-50 and a bit score >100) between DvEffectors and the salivary gland proteins of at least one insect species within the insect feeding category.
Table 3. DvEffector matches with effectors of Acyrthosiphon pisum.
| Functional annotation | Acyrthosiphon pisum effector | Daktulosphaira vitifoliae ortholog | E-value | Bit score |
|---|---|---|---|---|
| Esterases | XP_001947304.2 | DV3004001.2 | 0.00E+00 | 569. |
| DV3013307.2 | 4.24E-97 | 302. | ||
| Glucose dehydrogenase | XP_001943395.1 | DV3014986 | 0.00E+00 | 905. |
| Heat shock protein | NP_001156420.1 | DV3001393 | 0.00E+00 | 1020 |
| Lysosomal alpha-mannosidase | XP_008187164.1 | DV3016723 | 0.00E+00 | 1390 |
| Maltase | XP_001943582.2 | DV3005150 | 0.00E+00 | 555. |
| Peroxidase | XP_001951217.2 | DV3006190 | 0.00E+00 | 1206 |
| DV3014676*.. | 1.71E-168 | 493. | ||
| Peroxidoredoxins | XP_001949571.1 | DV3001833*.. | 1.01E-156 | 430. |
| DV3010201*.. | 1.07E-91 | 265. | ||
| DV3001185.2 | 2.63E-92 | 265. | ||
| Protein disulfide-isomerase | XP_001950406.1 | DV3009058.1* | 0.00E+00 | 871. |
| XP_008183165.1 | DV3000226 | 0.00E+00 | 755. | |
| Carboxypeptidases | XP_001943316.2 | DV3017719*.. | 2.82E-152 | 434. |
| XP_008183303.1 | DV3004740.2 | 1.57E-70 | 239. |
Blast results between the DvEffectors set against effector lists of Acyrthosiphon pisum [15, 16] using the BlastP software of NCBI were presented. The match quality is determined by the e-value and bit score. The functional annotations are taken from the effector list of Acyrthosiphon pisum. Asterisks and bold protein IDs highlight effector candidates (Dv1-5) employed for the following qRT-PCR analysis.
Biosynthesis of D. vitifoliae effector candidates
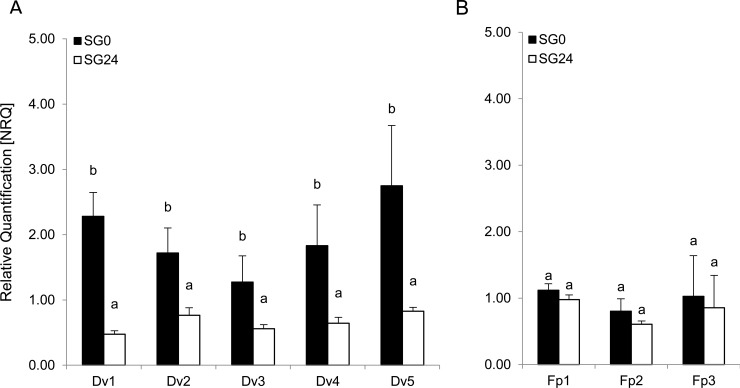
To increase the effector status of the predicted DvEffector candidates we challenged the hypothesis that the expression of the underlying DvEffector genes decreased when not required for feeding (= starved). In total five conserved effectors orthologs of Acyrthosiphon pisum (Dv1-5, Table 1) were chosen for a qRT-PCR analysis using RNA samples extracted from the salivary glands of feeding (SG0) and 24 h starving (SG24) D. vitifoliae larvae. In addition, three control metabolism genes, unrelated to insect feeding, were chosen (Fp1-3, Table 1) hypothesising that they their relative expression levels in salivary gland tissue remains unaffected by larval starvation. The results of the qRT-PCR analysis (Fig 3) showed significant gene expression levels for all tested candidate genes (Dv1-5 and Fp1-3) in salivary gland tissue of feeding and starving D. vitifoliae larvae (Fig 3A). Among them the gene expression level of Dv5 annotated as peroxidase was ranked as the highest with 2.75 NRQ in SG0. Comparing the SG0 and SG24 treatments, the analysis showed that the gene expression levels of all five DvEffectors decreased significantly, when insects were starved for 24 h without plant contact (Fig 3A), whereas the gene expression levels of Fp1-3 were statistically not affected by larval starvation (Fig 3B).
Fig 3. Relative gene expression levels of D. vitifoliae effector genes.
Expression levels [NRQ] of genes coding for A) conserved effectors of D. vitifoliae and A. pisum (Dv1-5) and B) false positives detected within the salivary gland proteome of D. vitifoliae but not associated to feeding (Fp1-3). RNA was extracted from salivary glands dissected from of feeding (SG0) and starving larvae (SG24). ActinA1 (DV3000100) was used as reference gene. Error bars indicate standard deviations of three independent biological replicates each consisting of salivary glands dissected from 100 individuals. Minor letters refer to significant differences obtained by Independent T-tests (p < 0.05).
Discussion
We present the first proteomic profile of salivary glands dissected from a root galling single founder lineage of D. vitifoliae feeding on root galls of Teleki 5C (V. berlandieri x V. riparia). Based on the salivary gland proteome and a subsequent In-silico secretory prediction 420 putative ‘DvEffectors’ were identified. Protein matches with salivary effector candidates of other phytophagous insects resulted in the identification of 170, conserved effectors. Quantitative RT-PCR analyses of five DvEffectors exemplarily complemented their potential roles as effectors since their expression within the salivary glands decreased by larval starvation.
The salivary gland proteome of D. vitifoliae is similar to aphids
In total 92.64% of identified D. vitifoliae proteins had a top blast hits with six aphid species (Fig 1A). Phylloxerids, in particular D. vitifoliae, are often described as ‘aphid-like’ or close aphid relatives because of their evolutionary relationship, similar modes of life and shared environmental niches [65, 66]. In fact Aphidoidea and Phylloxeroidea are suggested to share a common ancestor about 250 million years ago [67]. Recent genomic and transcriptomic studies confirm the phylogenetic proximity of D. vitifoliae and Aphidoidea [53, 68, 69]. Our results underlined these findings by showing a high degree of consistency between the proteomic profiles of D. vitifoliae salivary glands and aphids. Furthermore we showed a strong similarity in the effector profiles of root galling belowground D. vitifoliae and leaf feeding aphids as evidenced by the identification of conserved effector proteins (Fig 2 and S3 Table). In fact generalist and specialist insect species utilise conserved ‘core’ effectors involved in general plant infestation strategies [24, 70].
Conserved DvEffector groups
In total 420 DvEffectors of a D. vitifoliae single founder lineage feeding on root galls were identified based on their salivary gland proteome, among them 170 conserved effectors secreted by root feeding D. vitifoliae and other phytophagous insect species (Fig 2, S3 Table). Among them we detected numerous conserved DvEffectors associated to plant defence suppression such as peroxidoredoxins (DV3001185.2, DV3010201, DV3001833), peroxidases (DV3014676, DV3013176), superoxide dismutase (DV3003945) and serine protease (DV3001393) independently of the insect feeding category (S4 Table). For phloem feeders (aphids and plant hoppers) we found orthologous effector candidates potentially involved in stylet sheath formation (disulfide isomerases (DV3004088.2, DV3005250) and mucins (DV3010687.2, DV3001963) and DvEffector orthologs associated to cuticle functions proteins (e.g. DV3000905.2, DV3004090.2, DV3006395). Interestingly gall midges and stink bugs have an increased number of proteins associated to ribosomal processes (e.g. DV3005082.2, DV3007542.2, DV3007763.2). In the following we associated selected conserved effectors (Table 3) to functional effector groups and discuss their role for the compatible D. vitifoliae -Vitis spp. root interaction.
The first group is composed by DvEffectors involved in insect feeding site establishment. Protein disulfide isomerases (PDI) are commonly detected within insect salivary glands [16, 61, 71, 72]. Their abundancy across insect species with piercing-sucking mouthparts is explained by their catalytic role in the cross-linkage of cysteine bonds between mucin molecules essential for the stylet sheath formation [73, 74]. Other references suggest that phytophagous parasites secrete PDIs to detoxify host defensive reactive oxygen species [75, 76]. For D. vitifoliae we report the combined secretion of PDIs and mucins presuming their essential role for feeding site establishment by facilitating the stylet penetration into the root cortex and maintaining its mobility within the root gall tissue during insect feeding. Esterases and mannosidases, secreted by invading insects and phytopathogens, represent two groups of digestive enzymes assisting in the degradation of plant cell walls [77, 78]. Mobile D. vitifoliae larvae penetrate the cortexes of root tips or secondary lignified roots in order to establish the root gall feeding site [39, 41]. The secretion of enzymatic effectors contributing to the destabilisation of root cuticular layers and cell walls might facilitate the stylet penetration into the root cortex tissue. In addition increased cell wall flexibility could favour cell tissue expansion of root galls as shown exemplarily for the induction host expansin genes [43].
The second group of DvEffectors is composed by proteins primarily involved in the suppression of host plant defences. Glucose dehydrogenases (GDH) are among the most abundant proteins effectors detected within aphid saliva [79, 80]. Based on their structural similarity to glucose oxidases, GDHs are suggested to interfere with host plant defence mechanisms such as alterations of the salicylic and jasmonic acid signalling cascades, detoxification of reactive oxygen species, ethylene or other defensive compounds [81, 82], all of which might be part of the root infestation strategy of D. vitifoliae. Peroxidases and peroxidoredoxins catalyse the reduction of hydrogen peroxide (H2O2) known to be associated to incompatible host-parasite interactions such as hypersensitive cell death responses (HR) [83, 84]. Through the external delivery of detoxifying and scavenging enzymes into the root gall feeding site, D. vitifoliae might encounter the accumulation of defensive secondary metabolites to protect itself and/or to sustain the biological functionality of the feeding tissue; thus maintaining the compatible interaction despite activated host plant defences. Heat shock proteins (HSP) are evolutionary conserved key regulators known to interfere with stress signal cascades [85, 86]. In plants HSPs trigger the activation of host innate immunity traits against invading insect species [87], which would suggest a rather negative impact on the compatible D. vitifoliae–Vitis spp. interaction. On the opposite site HSPs trigger protective traits against a range of abiotic stresses [88], which would favour the sustainability of D. vitifoliae root galls in the soil. For instance in D. vitifoliae leaf galls HSPs are suggested to play a role for the suppression of host defences and the carbohydrate allocation [89].
D. vitifoliae feeds on the cell content withdrawn from parenchymal root cortex tissue marked by depleted levels of dissolved carbohydrates and elevated levels of free amino acids as compared to aphid species [90–92]. This implies the employment effectors assisting in and optimising the uptake of essential nutrients within the liquid diet. Maltases and GHDs are important enzymes for the endogenous sugar metabolism of insects [93, 94]. Both enzymes are previously detected in salivary glands of several aphids [16, 17, 19, 79, 80] thought to be involved in the extra oral digestion of carbohydrates increasing the digestibility of the sugar-rich phloem sap [14]. Root galls of D. vitifoliae contain abundant starch amounts due to redirected sucrose from the host plant’s primary metabolism [48]. It is not yet clear whether the accumulated starch serves as nutrient reservoir for the insect and its offspring or whether it represents a way to balance potentially phytotoxic sucrose levels. Nonetheless the secretion of sugar cleaving enzymes likely enables D. vitifoliae to exploit and feed from the accumulated carbohydrate reservoir within the root galls. Carboxypeptidases are involved in protein digestion processes aimed to provide peptides or amino acids for a balanced insect nutrition [95]. D. vitifoliae root galls and tuberosities contain elevated levels of essential amino acids such as glutamate, glutamic acid and asparagine and aspartic acid [41]. Secreted carboxypeptidases might be responsible for the protein or peptide cleavage explaining the accumulation of free amino acids in order to be dissolved and imbibed within the parenchymal sap by the insect. Additionally carboxypeptidases possess antimicrobial properties against bacterial and fungal pathogens [96, 97]. Alternatively carboxypeptidases may suppress feed-competitive or saprophytic microorganisms in the root tissue, which otherwise could contaminate the insect feed or degrade the host gall tissue [28].
DvEffectors involved in structural root gall formation
Among the set of DvEffectors, 65 (15.48%) do not have a functional annotation (‘not annotated’ or ‘uncharacterized protein’) and might represent novel D. vitifoliae-specific or even biotype-specific candidates involved in the establishment of a compatible root interaction. In particular, the identity and function of insect effectors that modulate plant physiological pathways resulting in structural root gall formation are yet unknown. However, through comparisons to identified effectors in other phytophagous organisms the potential function of newly identified effectors can be made. Although the feeding sites of root galling nematode species such as Heterodera sp., Globodera sp. [98] and Meloidogyne sp. [99] are structurally different from D. vitifoliae root galls [39, 41], effectors involved in root galling could be expected to have evolutionary conserved functions [100]. Based on experimental evidence Hassan (et al. 2010) [101] characterised effectors of root-galling nematodes with stringent biological functions involved in cell wall degradation, suppression of host plant defences, feeding site initiation, development and maintenance. Comparing the nematode effectors associated with structural root gall formation with the DvEffectors, numerous proteins are common to both. These include calreticulins (DV3007536, DV3007818.2) and proteins associated to ubiquitination (DV3014649, DV3013030, DV3002279, DV3001602, DV3004034, DV3012664) as well as nuclear DNA/RNA modifications (e.g. DV3010515, DV3009903.2, DV3016166, DV3010607 DV3002380, DV3001693.2, DV3017446 DV3019764, DV3008705, DV3003913, DV3000194) and could be the effector candidates associated with root gall formation described for an insect species.
Insect starvation supports effector status
In order to provide further evidence for the effector status of the identified DvEffector set, the biosynthesis of five selected effector candidates Dv1-5 (Table 1) was investigated on transcriptional level. A decrease in expression was observed for all five target genes in D. vitifoliae larvae that were removed from the gall and starved for 24 h (Fig 3). D. vitifoliae as a sedentary insect is in continuous contact with the root gall tissue and offers the possibility to assess temporarily well-defined starvation treatments [44, 48, 97]. When dealing with insect starvation, one key factor is the intensity and duration of the prospected treatment. A starvation regime that is too abrupt may affect overall insect metabolism and behaviour [102], whereas a targeted and mild starvation, as conducted in the present study, merely affects the metabolic processes related to insect feeding [103]. Here we show that the expression of effectors genes within the salivary glands of root-galling D. vitifoliae larvae decreased, when insects were starved for 24 h confirming the validity of our approach to provide improved characterisation and methodological complementation of the presented DvEffector set.
Supporting information
Proteins IDs were based on the D. vitifoliae genome v. 3.2. Annotations and protein length were obtained by BlastP searches against the NCBI nr database (as of 12.11.2018).
(XLSX)
Proteins IDs were based on the D. vitifoliae genome v. 3.2. Annotations and protein length were obtained by BlastP searches against the NCBI nr database (as of 12.11.2018).
(XLSX)
The first table provided information about the salivary gland protein sets used for the BlastP analysis referring to the feeding category, insect species and the literature reference. The second table presented conserved DvEffectors shared with plants hoppers (green), aphids (blue), gall midges (rosa) or stink bugs (yellow). Proteins IDs were based on the D. vitifoliae genome v. 3.2. (as of 12.11.2018).
(XLSX)
Proteins IDs were based on the D. vitifoliae genome v. 3.2. The table presented the BlastP results between the identified DvEffector set against effector lists of Acyrthosiphon pisum with a cut-off e-value < e-50 and bit score > 100. Protein annotations were taken from the effector lists of Acyrthosiphon pisum [15, 16].
(XLSX)
Acknowledgments
We thank Dr. S. Savoi (SupAgro Montpellier), Dr. Harald Berger (BOKU, Vienna) and N. Douglas (Maynooth University) for fruitful discussions and computational help. We greatly acknowledge the International Aphid Genomics Consortium (IAGC) for providing the genome of D. vitifoliae and effector gene annotation namely Prof. Dr. D. Tagu (INRA Rennes, F), Dr. C. Rispe (INRA Nantes, F), Prof. Dr. P. Nabity and Dr. C. Zhao (UC, Riverside, USA). This work contributes in part to the doctoral thesis of M. W. Eitle.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The presented project was supported by a DOC research fellowship for M.W. Eitle provided by the Austrian Academy of Sciences (ÖAW) and with the Schlumberger award 2018 provided by the Robert Schlumberger foundation. Funding for Daktulosphaira vitifoliae clone Pcf genomic sequencing was provided by INRA (AIP Bioressources), BGI Biotech and i5k initiatives. Parts of the transcriptomic resources were obtained within the 1KITE projects. The Maynooth University Q-Exactive Quantitative Mass Spectrometer was funded under the SFI Research Infrastructure Call 2012; Grant Number: 12/RI/2346 (3). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Giron D, Huguet E, Stone GN, Body M. Insect-induced effects on plants and possible effectors used by galling and leaf-mining insects to manipulate their host-plant. J Insect Physiol. 2016;84:70–89. 10.1016/j.jinsphys.2015.12.009 [DOI] [PubMed] [Google Scholar]
- 2.Win J, Chaparro-Garcia A, Belhaj K, Saunders D, Yoshida K, Dong S, et al. , editors. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb Symp Quant Biol. 2012;77:235–247. 10.1101/sqb.2012.77.015933 [DOI] [PubMed] [Google Scholar]
- 3.Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact. 2009;22(2):115–122. 10.1094/MPMI-22-2-0115 [DOI] [PubMed] [Google Scholar]
- 4.Robinson AS, Franz G, Atkinson PW. Insect transgenesis and its potential role in agriculture and human health. Insect Biochem Mol Biol. 2004;34(2):113–120. 10.1016/j.ibmb.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 5.Yin K, Qiu J-L. Genome editing for plant disease resistance: applications and perspectives. Philos Trans R Soc Lond B Biol Sci. 2019;374(1767):20180322 10.1098/rstb.2018.0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atamian HS, Chaudhary R, Cin VD, Bao E, Girke T, Kaloshian I. In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol Plant Microbe Interact. 2013;26(1):67–74. 10.1094/MPMI-06-12-0144-FI [DOI] [PubMed] [Google Scholar]
- 7.Elzinga DA, De Vos M, Jander G. Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol Plant Microbe Interact. 2014;27(7):747–756. 10.1094/MPMI-01-14-0018-R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mutti NS, Louis J, Pappan LK, Pappan K, Begum K, Chen M-S, et al. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc Natl Acad Sci. 2008;105(29):9965–9969. 10.1073/pnas.0708958105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao C, Shukle R, Navarro-Escalante L, Chen M, Richards S, Stuart JJ. Avirulence gene mapping in the Hessian fly (Mayetiola destructor) reveals a protein phosphatase 2C effector gene family. J Insect Physiol. 2016;84:22–31. 10.1016/j.jinsphys.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 10.Ji R, Ye W, Chen H, Zeng J, Li H, Yu H, et al. A salivary endo-β-1, 4-glucanase acts as an effector that enables the brown planthopper to feed on rice. Plant Physiol. 2017;173(3):1920–1932. 10.1104/pp.16.01493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto Y, Hattori M. The green rice leafhopper, Nephotettix cincticeps (Hemiptera: Cicadellidae), salivary protein NcSP75 is a key effector for successful phloem ingestion. PloS One. 2018;13(9):e0202492 10.1371/journal.pone.0202492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H-X, Qian L-X, Wang X-W, Shao R-X, Hong Y, Liu S-S, et al. A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway Proc Natl Acad Sci. 2019;116(2):490–495. 10.1073/pnas.1714990116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang N, Zhao P, Ma Y, Yao X, Sun Y, Huang X, et al. A whitefly effector Bsp9 targets host immunity regulator WRKY33 to promote performance. Philos Trans R Soc Lond B Biol Sci. 2019;374(1767):20180313 10.1098/rstb.2018.0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stafford-Banks CA, Rotenberg D, Johnson BR, Whitfield AE, Ullman DE. Analysis of the salivary gland transcriptome of Frankliniella occidentalis. PloS One. 2014;9(4):e94447 10.1371/journal.pone.0094447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulain H, Legeai F, Guy E, Morlière S, Douglas NE, Oh J, et al. Fast evolution and lineage-specific gene family expansions of aphid salivary effectors driven by interactions with host-plants. Genome Biol Evol. 2018;10(6):1554–1572. 10.1093/gbe/evy097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carolan JC, Caragea D, Reardon KT, Mutti NS, Dittmer N, Pappan K, et al. Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): A dual transcriptomic/proteomic approach. J Proteome Res. 2011;10(4):1505–1518. 10.1021/pr100881q [DOI] [PubMed] [Google Scholar]
- 17.Ji R, Yu H, Fu Q, Chen H, Ye W, Li S, et al. Comparative transcriptome analysis of salivary glands of two populations of rice brown planthopper, Nilaparvata lugens, that differ in virulence. PloS One. 2013;8(11):e79612 10.1371/journal.pone.0079612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Y-L, Li J-M, Li M, Luan J-B, Ye X-D, Wang X-W, et al. Transcriptomic analysis of the salivary glands of an invasive whitefly. PLoS One. 2012;7(6):e39303 10.1371/journal.pone.0039303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Fan J, Sun J, Francis F, Chen J. Transcriptome analysis of the salivary glands of the grain aphid, Sitobion avenae. Sci Rep. 2017;7(1):15911 10.1038/s41598-017-16092-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao C, Escalante LN, Chen H, Benatti TR, Qu J, Chellapilla S, et al. A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr Biol. 2015;25(5):613–620. 10.1016/j.cub.2014.12.057 [DOI] [PubMed] [Google Scholar]
- 21.Bos JI, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 2010;6(11):e1001216 10.1371/journal.pgen.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori K. Insect secretions and their effect on plant growth, with special reference to hemipterans Biology of insect-induced plant galls Oxford University Press, New York: 1992:157–170. [Google Scholar]
- 23.Stuart J. Insect effectors and gene-for-gene interactions with host plants. Curr Opin Insect Sci. 2015;9:56–61. [DOI] [PubMed] [Google Scholar]
- 24.Thorpe P, Cock PJ, Bos J. Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genomics. 2016;17(1):172–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wieczorek K, Hensler S, Forneck A, editors. Displaying sink-source flux in phylloxerated grapevines by microscopic techniques. Acta Hortic. 2013;1045:29–32. [Google Scholar]
- 26.Porten M, Huber L. An assessment method for the quantification of Daktulosphaira vitifoliae (Fitch)(Hem., Phylloxeridae) populations in the field. J Appl Entomol. 2003;127(3):157–162. [Google Scholar]
- 27.Powell K, Cooper P, Forneck A. The biology, physiology and host-plant interactions of grape phylloxera Daktulosphaira vitifoliae. Adv Insect Physiol. 2013;45:159–218. [Google Scholar]
- 28.Huber L. Schaderrerger im Wurzelraum von Reben (Vitis spp.)—Vorkommen, Wirkung, Interaktionen—und Möglichkeiten zu deren Kontrolle durch Maßnahmen des Integrated Pest Managements (IPM). Dissertation of the Johannes Gutenberg University Mainz. 2007. [Google Scholar]
- 29.Omer A, Granett J, De Benedictis J, Walker M. Effects of fungal root infections on the vigor of grapevines infested by root-feeding grape phylloxera. Vitis. 1995;34(3):165–170. [Google Scholar]
- 30.Powell K, Korosi G, editors. Taking the strain—selecting the right rootstock to protect against endemic phylloxera strains. Acta Hortic. 2013;1045:99–107. [Google Scholar]
- 31.Forneck A, Walker M, Blaich R. Ecological and genetic aspects of grape phylloxera Daktulosphaira vitifoliae (Hemiptera: Phylloxeridae) performance on rootstock hosts. Bull Entomol Res. 2001;91(06):445–451. [PubMed] [Google Scholar]
- 32.Forneck A, Powell KS, Walker MA. Scientific opinion: Improving the definition of grape phylloxera biotypes and standardizing biotype screening protocols. Am J Enol Vitic. 2016;67(4):371–376. [Google Scholar]
- 33.Granett J, Timper P, Lider L. Grape phylloxera (Daktulosphaira vitifoliae)(Homoptera: Phylloxeridae) biotypes in California. J Econ Entomol. 1985;78(6):1463–1467. [Google Scholar]
- 34.Kocsis L, Granett J, Walker M, Lin H, Omer A. Grape phylloxera populations adapted to Vitis berlandieri x V. riparia rootstocks. Am J Enol Vitic. 1999;50(1):101–106. [Google Scholar]
- 35.Korosi G, Powell K, Clingeleffer P, Smith B, Walker R, Wood J, editors. New hybrid rootstock resistance screening for phylloxera under laboratory conditions. Acta Hortic. 2013;904:53–58. [Google Scholar]
- 36.Riaz S, Lund KT, Granett J, Walker MA. Population Diversity of Grape Phylloxera in California and Evidence for Sexual Reproduction. Am J Enol Vitic. 2017;68(2):218–227. [Google Scholar]
- 37.Song G-C, Granett J. Grape phylloxera (Homoptera: Phylloxeridae) biotypes in France. J Econ Entomol. 1990;83(2):489–493. [Google Scholar]
- 38.Sun Q, Du Y, Wang Z, Di H. Research progress on biotypes and genetic diversity of grape phylloxera. J Fruit Sci. 2012;1:125–129. [Google Scholar]
- 39.Forneck A, Kleinmann S, Blaich R, Anvari S. Histochemistry and anatomy of phylloxera (Daktulosphaira vitifoliae) nodosities on young roots of grapevine (Vitis spp). Vitis. 2002;41(2):93–98. [Google Scholar]
- 40.Hofmann E. Die Histologie der Nodositäten verschiedener Rebensorten bei Reblausbefall. Vitis. 1957;1:66–81. [Google Scholar]
- 41.Kellow AV, Sedgley M, Van Heeswijck R. Interaction between Vitis vinifera and grape phylloxera: changes in root tissue during nodosity formation. Ann Bot. 2004;93(5):581–590. 10.1093/aob/mch082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niklowitz W. Histologische Untersuchungen an Reblausgallen. Phytopath Z. 1955;24:299–340. [Google Scholar]
- 43.Lawo N, Griesser M, Forneck A. Expression of putative expansin genes in phylloxera (Daktulosphaira vitifoliae Fitch) induced root galls of Vitis spp. Eur J Plant Pathol. 2013;136(2):383–391. 10.1007/s10658-013-0173-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eitle MW, Loacker J, Meng-Reiterer J, Schuhmacher R, Griesser M, Forneck A. Polyphenolic profiling of roots (Vitis spp.) under grape phylloxera (D. vitifoliae Fitch) attack. Plant Physiol Biochem. 2019;135:174–181. 10.1016/j.plaphy.2018.12.004 [DOI] [PubMed] [Google Scholar]
- 45.Eitle MW, Griesser M, Vankova R, Dobrev P, Aberer S, Forneck A. Grape phylloxera (D. vitifoliae) manipulates SA/JA concentrations and signalling pathways in root galls of Vitis spp. Plant Physiol Biochem. 2019;144:85–91. 10.1016/j.plaphy.2019.09.024 [DOI] [PubMed] [Google Scholar]
- 46.Lawo NC, Weingart GJ, Schuhmacher R, Forneck A. The volatile metabolome of grapevine roots: First insights into the metabolic response upon phylloxera attack. Plant Physiol Biochem. 2011;49(9):1059–1063. 10.1016/j.plaphy.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabity PD, Haus MJ, Berenbaum MR, DeLucia EH. Leaf-galling phylloxera on grapes reprograms host metabolism and morphology. Proc Natl Acad Sci. 2013;110(41):16663–16668. 10.1073/pnas.1220219110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griesser M, Lawo NC, Crespo-Martinez S, Schoedl-Hummel K, Wieczorek K, Gorecka M, et al. Phylloxera (Daktulosphaira vitifoliae Fitch) alters the carbohydrate metabolism in root galls to allowing the compatible interaction with grapevine (Vitis ssp.) roots. Plant Sci. 2015;234:38–49. 10.1016/j.plantsci.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan F, Omer A, Aung L, Granett J. Effects of infestation by grape phylloxera on sugars, free amino acids, and starch of grapevine roots. Vitis. 2000;39(4):175–176. [Google Scholar]
- 50.Forneck A, Walker M, Blaich R. An in vitro assessment of phylloxera (Daktulosphaira vitifoliae Fitch)(Hom., Phylloxeridae) life cycle. J Appl Entomol. 2001;125(8):443–447. [Google Scholar]
- 51.Harsch M. Detektion differenziell exprimierter Gensequenzen bei Galleninduktion von Rebläusen (D. vitifoliae) an Reben (Vitis ssp.) [Diploma]: Universität Hohenheim, Stuttgart; 2004. [Google Scholar]
- 52.Forneck A, Huber L. (A) sexual reproduction–a review of life cycles of grape phylloxera, Daktulosphaira vitifoliae. Entomol Exp Appl. 2009;131(1):1–10. [Google Scholar]
- 53.Rispe C, Legeai F, Jaquiéry J, Breteaudeau A, Tagu D, Powell K, et al. , editors. The grape phylloxera genome sequencing project. Acta Hortic. 2013;1045:15–19. [Google Scholar]
- 54.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. 10.1093/nar/gkn176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 56.Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9(1):392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- 58.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17(4):349–356. 10.1093/protein/gzh037 [DOI] [PubMed] [Google Scholar]
- 59.Huang H-J, Lu J-B, Li Q, Bao Y-Y, Zhang C-X. Combined transcriptomic/proteomic analysis of salivary gland and secreted saliva in three planthopper species. J Proteomics. 2018;172:25–35. 10.1016/j.jprot.2017.11.003 [DOI] [PubMed] [Google Scholar]
- 60.Al-jbory Z, Anderson KM, Harris MO, Mittapalli O, Whitworth RJ, Chen M-S. Transcriptomic analyses of secreted proteins from the salivary glands of wheat midge larvae. J Insect Sci. 2018;18(1):17–22. 10.1093/jisesa/iex103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen M-S, Zhao H-X, Zhu YC, Scheffler B, Liu X, Liu X, et al. Analysis of transcripts and proteins expressed in the salivary glands of Hessian fly (Mayetiola destructor) larvae. J Insect Physiol. 2008;54(1):1–16. 10.1016/j.jinsphys.2007.07.007 [DOI] [PubMed] [Google Scholar]
- 62.Serteyn L, Francis F. Insight into salivary gland proteomes of two polyphagous stink bugs: Nezara viridula L. and Halyomorpha halys Stål. Proteomics. 2019;19(7):1800436. [DOI] [PubMed] [Google Scholar]
- 63.Teng X, Zhang Z, He G, Yang L, Li F. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. J Insect Sci. 2012;12(60):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 65.Davis GK. Cyclical parthenogenesis and viviparity in aphids as evolutionary novelties. J Exp Zool B Mol Dev Evol. 2012;318(6):448–459. 10.1002/jez.b.22441 [DOI] [PubMed] [Google Scholar]
- 66.Favret C, Blackman RL, Miller GL, Victor B. Catalog of the phylloxerids of the world (Hemiptera, Phylloxeridae). Zookeys. 2016;(629):83–101. 10.3897/zookeys.629.10709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heie O. Paleontology and phylogeny. Aphids: Their biology, natural enemies and control. 1987;2a:367–391. [Google Scholar]
- 68.Ortiz-Rivas B, Martínez-Torres D. Combination of molecular data support the existence of three main lineages in the phylogeny of aphids (Hemiptera: Aphididae) and the basal position of the subfamily Lachninae. Mol Phylogenet Evol. 2010;55(1):305–317. 10.1016/j.ympev.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 69.Rispe C, Legeai F, Papura D, Bretaudeau A, Hudaverdian S, Le Trionnaire G, et al. De novo transcriptome assembly of the grapevine phylloxera allows identification of genes differentially expressed between leaf-and root-feeding forms. BMC Genomics. 2016;17(1):219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guiguet A, Dubreuil G, Harris MO, Appel HM, Schultz JC, Pereira MH, et al. Shared weapons of blood-and plant-feeding insects: surprising commonalities for manipulating hosts. J Insect Physiol. 2016;84:4–21. 10.1016/j.jinsphys.2015.12.006 [DOI] [PubMed] [Google Scholar]
- 71.Konishi H, Noda H, Tamura Y, Hattori M. Proteomic analysis of the salivary glands of the rice brown planthopper, Nilaparvata lugens (Stål) (Homoptera: Delphacidae). Appl Entomol Zool. 2009;44(4):525–534. [Google Scholar]
- 72.Miao Y-T, Deng Y, Jia H-K, Liu Y-D, Hou M-L. Proteomic analysis of watery saliva secreted by white-backed planthopper, Sogatella furcifera. PloS One. 2018;13(5):e0193831 10.1371/journal.pone.0193831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Will T, Vilcinskas A. The structural sheath protein of aphids is required for phloem feeding. Insect Biochem Mol Biol. 2015;57:34–40. 10.1016/j.ibmb.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 74.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta Proteins Proteom. 2004;1699(1–2):35–44. [DOI] [PubMed] [Google Scholar]
- 75.Habash SS, Sobczak M, Siddique S, Voigt B, Elashry A, Grundler FM. Identification and characterization of a putative protein disulfide isomerase (HsPDI) as an alleged effector of Heterodera schachtii. Sci Rep. 2017;7:13536 10.1038/s41598-017-13418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang H-J, Liu C-W, Xu H-J, Bao Y-Y, Zhang C-X. Mucin-like protein, a saliva component involved in brown planthopper virulence and host adaptation. J Insect Physiol. 2017;98:223–230. 10.1016/j.jinsphys.2017.01.012 [DOI] [PubMed] [Google Scholar]
- 77.Calderón-Cortés N, Quesada M, Watanabe H, Cano-Camacho H, Oyama K. Endogenous plant cell wall digestion: a key mechanism in insect evolution. Annu Rev Ecol Evol Syst. 2012;43:45–71. [Google Scholar]
- 78.Malgas S, van Dyk JS, Pletschke BI. A review of the enzymatic hydrolysis of mannans and synergistic interactions between β-mannanase, β-mannosidase and α-galactosidase. World J Microbiol Biotechnol. 2015;31(8):1167–1175. 10.1007/s11274-015-1878-2 [DOI] [PubMed] [Google Scholar]
- 79.Rao SA, Carolan JC, Wilkinson TL. Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PLoS One. 2013;8(2):e57413 10.1371/journal.pone.0057413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nicholson SJ, Hartson SD, Puterka GJ. Proteomic analysis of secreted saliva from Russian wheat aphid (Diuraphis noxia Kurd.) biotypes that differ in virulence to wheat. J Proteomics. 2012;75(7):2252–2268. 10.1016/j.jprot.2012.01.031 [DOI] [PubMed] [Google Scholar]
- 81.Nicholson SJ, Puterka GJ. Variation in the salivary proteomes of differentially virulent greenbug (Schizaphis graminum Rondani) biotypes. J Proteomics. 2014;105:186–203. 10.1016/j.jprot.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 82.Musser RO, Hum-Musser SM, Eichenseer H, Peiffer M, Ervin G, Murphy JB, et al. Herbivory: caterpillar saliva beats plant defences. Nat. 2002;416(6881):599–600. [DOI] [PubMed] [Google Scholar]
- 83.Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays. 2006;28(11):1091–1101. 10.1002/bies.20493 [DOI] [PubMed] [Google Scholar]
- 84.Blank L, Wolf T, Eimert K, Schröder M-B. Differential gene expression during hypersensitive response in Phylloxera-resistant rootstock ‘Börner’using custom oligonucleotide arrays. J Plant Interact. 2009;4(4):261–269. [Google Scholar]
- 85.Park C-J, Seo Y-S. Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol J. 2015;31(4):323–333. 10.5423/PPJ.RW.08.2015.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pockley AG. Heat shock proteins as regulators of the immune response. Lancet. 2003;362(9382):469–476. 10.1016/S0140-6736(03)14075-5 [DOI] [PubMed] [Google Scholar]
- 87.Kaloshian I, Walling LL. Hemipterans as plant pathogens. Annu Rev Phytopathol. 2005;43:491–521. 10.1146/annurev.phyto.43.040204.135944 [DOI] [PubMed] [Google Scholar]
- 88.Timperio AM, Egidi MG, Zolla L. Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP). J Proteomics. 2008;71(4):391–411. 10.1016/j.jprot.2008.07.005 [DOI] [PubMed] [Google Scholar]
- 89.Nabity PD. Insect‐induced plant phenotypes: Revealing mechanisms through comparative genomics of galling insects and their hosts. Am J Bot. 2016;103(6):979–981. 10.3732/ajb.1600111 [DOI] [PubMed] [Google Scholar]
- 90.Forneck A, Wöhrle A, editors. A synthetic diet for phylloxera (Daktulosphaira vitifoliae Fitch). Acta Hortic. 2001;617:129–134. [Google Scholar]
- 91.Kingston K. Digestive and feeding physiology of Grape Phylloxera (Daktulosphaira vitifoliae Fitch). 2011. Dissertation of the Australian National University; Canberra: 2007. [Google Scholar]
- 92.Hewer A, Will T, van Bel AJ. Plant cues for aphid navigation in vascular tissues. J Exp Biol. 2010;213(23):4030–4042. [DOI] [PubMed] [Google Scholar]
- 93.Karley A, Ashford D, Minto L, Pritchard J, Douglas A. The significance of gut sucrase activity for osmoregulation in the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 2005;51(12):1313–1319. 10.1016/j.jinsphys.2005.08.001 [DOI] [PubMed] [Google Scholar]
- 94.Kunieda T, Fujiyuki T, Kucharski R, Foret S, Ament S, Toth A, et al. Carbohydrate metabolism genes and pathways in insects: insights from the honey bee genome. Insect Mol Biol. 2006;15(5):563–576. 10.1111/j.1365-2583.2006.00677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bayés A, de la Vega MR, Vendrell J, Aviles FX, Jongsma MA, Beekwilder J. Response of the digestive system of Helicoverpa zea to ingestion of potato carboxypeptidase inhibitor and characterization of an uninhibited carboxypeptidase B. Insect Biochem Mol Biol. 2006;36(8):654–664. 10.1016/j.ibmb.2006.05.010 [DOI] [PubMed] [Google Scholar]
- 96.Altincicek B, Vilcinskas A. Septic injury‐inducible genes in medicinal maggots of the green blow fly Lucilia sericata. Insect Mol Biol. 2009;18(1):119–125. 10.1111/j.1365-2583.2008.00856.x [DOI] [PubMed] [Google Scholar]
- 97.Mugford ST, Osbourn A. Evolution of serine carboxypeptidase-like acyltransferases in the monocots. Plant Signal Behav. 2010;5(2):193–195. 10.4161/psb.5.2.11093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sobczak M, Golinowski W. Cyst nematodes and syncytia Genomics and Molecular Genetics of Plant-Nematode Interactions: Springer Press; 2011:61–82. [Google Scholar]
- 99.Jones MG, Goto DB. Root-knot nematodes and giant cells Genomics and molecular genetics of plant-nematode interactions: Springer Press; 2011:83–100. [Google Scholar]
- 100.Schultz JC, Edger PP, Body MJ, Appel HM. A galling insect activates plant reproductive programs during gall development. bioRxivorg. 2018:383851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hassan S, Behm CA, Mathesius U. Effectors of plant parasitic nematodes that re-program root cell development. Funct Plant Biol. 2010;37(10):933–942. [Google Scholar]
- 102.Scharf I. The multifaceted effects of starvation on arthropod behaviour. Anim Behav. 2016;119:37–48. [Google Scholar]
- 103.Bansal R, Mian M, Mittapalli O, Michel AP. RNA-Seq reveals a xenobiotic stress response in the soybean aphid, Aphis glycines, when fed aphid-resistant soybean. BMC Genomics. 2014;15:972–986. 10.1186/1471-2164-15-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins IDs were based on the D. vitifoliae genome v. 3.2. Annotations and protein length were obtained by BlastP searches against the NCBI nr database (as of 12.11.2018).
(XLSX)
Proteins IDs were based on the D. vitifoliae genome v. 3.2. Annotations and protein length were obtained by BlastP searches against the NCBI nr database (as of 12.11.2018).
(XLSX)
The first table provided information about the salivary gland protein sets used for the BlastP analysis referring to the feeding category, insect species and the literature reference. The second table presented conserved DvEffectors shared with plants hoppers (green), aphids (blue), gall midges (rosa) or stink bugs (yellow). Proteins IDs were based on the D. vitifoliae genome v. 3.2. (as of 12.11.2018).
(XLSX)
Proteins IDs were based on the D. vitifoliae genome v. 3.2. The table presented the BlastP results between the identified DvEffector set against effector lists of Acyrthosiphon pisum with a cut-off e-value < e-50 and bit score > 100. Protein annotations were taken from the effector lists of Acyrthosiphon pisum [15, 16].
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.