Abstract
Cerebrospinal fluid (CSF) may be the best hope for minimally invasive diagnosis and treatment monitoring of central nervous system (CNS) malignancies. Discovery/validation of cell-free nucleic acid and protein biomarkers has the potential to revolutionize CNS cancer care, paving the way for presurgical evaluation, earlier detection of recurrence, and the selection of targeted therapies. While detection of mutations, changes in RNA and miRNA expression, epigenetic alterations, and elevations of protein levels have been detected in the CSF of patients with CNS tumors, most of these biomarkers remain unvalidated. In this review, we focus on the molecular changes that have been identified in a variety of CNS tumors and profile the approaches used to detect these alterations in clinical samples. We further emphasize the importance of systemic collection of CSF and the establishment of standardized collection protocols that will lead to better cross-study biomarker validation and hopefully FDA-approved clinical markers.
Keywords: biomarker, cerebrospinal fluid, circulating tumor DNA, CNS tumors, liquid biopsy
Primary malignant brain tumors of the central nervous system (CNS) are diagnosed in more than 23 000 people per year and have a dismal 5-year survival of 33%.1 Tissue sampling remains the gold standard for tumor diagnosis, but even stereotactic biopsies can carry a risk of up to 4–7% for major morbidity.2–4 Non-invasive monitoring by MRI or other advanced imaging technologies may aid in the diagnosis and monitoring of CNS tumor progression, but radiographic findings often do not depict tumor-associated changes in real time. In addition, pseudoprogression, where the tumor volume increase is not due to tumor growth but rather inflammation, can mimic the radiographic appearance of disease progression.5,6 In addition, drugs like bevacizumab may cause pseudoresponse, where imaging findings suggest tumor response but there has been no appreciable tumoricidal activity.7 Furthermore, there is no existing technology that can give insights into the molecular evolution that happens in response to therapeutic interventions.
The detection of cell-free circulating nucleic acids, including circulating free (cf)DNA and cfRNA, and proteins in cerebrospinal fluid (CSF) has the potential to revolutionize detection and clinical care for CNS tumors. Often called “liquid biopsy,” many of these tests utilize CSF to detect mutations or differentially methylated regions by next generation sequencing (NGS) or protein elevations by mass spectrometry or enzyme-linked immunosorbent assay (ELISA). The ability to obtain multiple samples at various time points may not only allow for initial cancer detection and prognostication, but also aid monitoring of treatment response and tumor evolution. Recent studies have elucidated the diverse heterogeneous molecular landscape of CNS tumors.8,9 Whereas a single biopsy is only representative of the genetic, epigenetic, and protein expression changes for one region and at one time point, a liquid biopsy may represent a snapshot of the tumor as a whole during the progression.10
Unlike other solid tumors, the use of liquid biopsy for diagnosing and monitoring CNS tumors has proven more challenging.11 The blood–brain barrier limits transport of nucleic acids and proteins into system circulation, drastically decreasing the concentration of cfDNA and tumor-associated proteins in patient sera. Liquid biopsies using CSF, however, have shown tremendous promise because the fluid interacts directly with the primary tumor11–13 (Fig. 1). In this review, we discuss the process of selecting candidate biomarkers, describe encouraging genetic, epigenetic, and protein candidates that may be amenable for liquid biopsy using CSF, and briefly discuss future directions for liquid biopsy assays in the management of patients with CNS tumors.
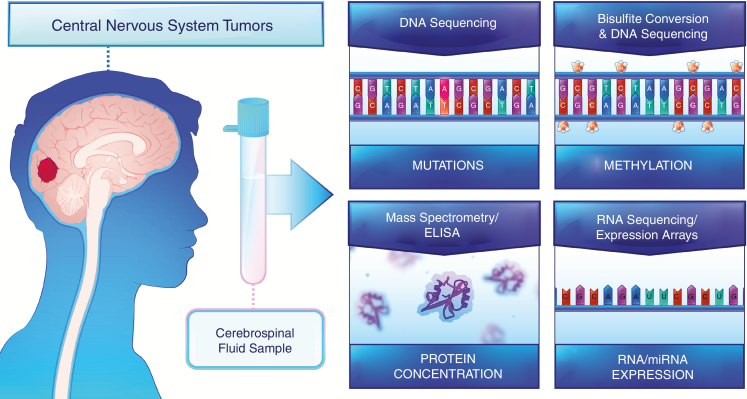
Fig. 1.
Tumors of the central nervous system interact with cerebrospinal fluid, allowing for the detection of DNA mutations, cytosine methylation alterations, and changes in protein and RNA expression.
Selection of Candidate Biomarkers
The clinical utility of liquid biopsy is rooted in the thoughtful selection of candidate biomarkers. A useful biomarker will be distinctly present in samples collected from cancer patients or at levels that exceed the background level observed in healthy individuals. While the ideal biomarker would demonstrate 100% sensitivity to detect patients with cancer and 100% specificity to exclude patients without cancer, practical selection of candidates requires trade-offs between perfect sensitivity and specificity that depend on the type of biomarker. For each of the biomarker types (mutations, gene expression, differential cytosine methylation, and protein expression), we will briefly review the biology of the biomarker, describe how the biomarker is detected, and provide examples that have shown promise.
Mutations in cfDNA
Cancer is fundamentally a genetic disease.14 Research over the past two decades has shown that roughly 200 genes, when mutated, can drive tumor formation.15 A typical tumor contains 2 to 8 of these “driver gene” mutations, along with a range of “passenger” mutations that confer no selective advantage.14 Depending on the tumor type, a given cancer may have between 8 and 200 or more passenger mutations.
The advent of massively parallel NGS and the accompanying bioinformatic tools allows for hundreds of millions of DNA strands to be sequenced simultaneously at high coverage.16 Targeted sequencing libraries may be prepared using PCR primers that amplify loci around common oncogene (eg, KRAS, BRAF, β-catenin) and tumor suppressor (eg, TP53, APC, Rb) mutations.17–19 Alternatively, whole genome sequencing libraries may be prepared and subsequently diluted to assess for mutations,20 enriched for specific genomic loci,21,22 or simply sequenced. Regardless of the technology used to prepare sequencing libraries, the overall goal is the same: detection of rare mutations found in cfDNA.
Circulating free DNA was originally described in healthy individuals and has evolved into a remarkable diagnostic modality. While the exact cellular origins of cfDNA are unknown, cfDNA containing mutations in prototypical oncogenes and tumor suppressor genes is thought to derive from the tumor itself and is generally around 150–160 bp in length, containing exons, introns, and mitochondrial DNA.23 In addition, the relatively short half-life of less than 90 minutes makes it an ideal candidate for interrogating the dynamic changes of the tumor.24 One challenge in using cfDNA-based strategies for CNS tumors is the relative paucity of tumor-derived DNA in circulation.11,25 There appears to be greater than 10-fold enrichment in the quantity of tumor-derived DNA in the CSF (CSF-tDNA) compared with the plasma.12,26 One of the first studies by Pan et al utilized droplet digital PCR (ddPCR) and targeted amplicon sequencing to detect cancer mutations in cfDNA of CSF of primary and metastatic brain tumor patients.27 They first identified putative mutations through whole-exome sequencing of normal and primary brain tumor DNA, then interrogated the CSF samples from these patients for the same mutations. Tumor mutations were detected in 6 of 7 patients, including single nucleotide variants in NF2, AKT1, BRAF, NRAS, KRAS, and EGFR at mutant allele fractions (MAFs) ranging from 0.3% to 49.2% by ddPCR.27 In addition, the ratio of CSF to plasma mutant concentration of cfDNA correlated with burden of CNS disease.
A study by Wang et al assayed the CSF of 35 patients with CNS tumors for cfDNA mutations, including 6 medulloblastomas and 29 World Health Organization (WHO) grades I–IV gliomas.12 Similar to the study by Pan et al, the investigators utilized either targeted sequencing or whole-exome sequencing to identify mutations in TP53, IDH1, TERT, NF2, PIK3R1, PTCH1, and PTEN. The presence of these mutations was then assessed in the CSF of the same patient using a targeted sequencing approach that aimed to detect mutations with allele fractions as low as 0.01%.11,12,19 Seventy-four percent of the 35 CSF samples had detectable mutations with an average mutant allele fraction of 12.2%, ranging 0.1–77%. High-grade lesions (WHO grades III–IV) were more likely to have detectable mutations in CSF than low-grade lesions, and the levels of the mutations were also higher in high-grade lesions. However, tumor size did not appear to predict mutation detection or level. In a larger study of matched tumor, CSF, and blood from 57 patients, Pan et al detected at least one tumor-specific mutation in 82.5% of CSF-tDNA samples.27 In addition, the mutations detected in the CSF were highly concordant with the mutations detected in the primary tumor. Similar studies have supported the findings described by these groups and provide hope that the need for biopsy may be obviated for deep-seated tumors and tumors in eloquent areas, especially the brainstem, as technologies improve.25,28–31
The detection of tumor-derived DNA in CSF does depend in part on the location of the tumors. CSF-tDNA was unable to be detected in many supratentorial brain tumor cases unless it abutted the CSF space. In contrast, brainstem tumors are close to the CSF reservoir. In a recent study, Pan C et al reported that alterations were identified in the CSF circulating tumor (ct)DNA of 97.3% of brainstem glioma cases. Tumors encapsulated within the CSF, such as meningiomas and craniopharyngiomas may be attractive targets to develop CSF-based liquid biopsy approaches. This is particularly germane to craniopharyngiomas, where a subset harbor canonical BRAF V600E mutations and have demonstrated response to BRAF inhibition in early clinical trials.32 A CSF test that could obviate a biopsy could be very helpful, as the risk of surgical intervention on these tumors is not insignificant.33
The utility of CSF-tDNA as a marker of treatment response has also been explored. De Mattos-Arruda et al analyzed CSF-tDNA at various time points in 6 patients with primary or metastatic CNS tumors who also received radiographic follow-up.25 Levels of mutant CSF-tDNA decreased after surgical resection and radiographic-confirmed response to chemotherapy, whereas they increased with increased radiographic progression. They also found that patients with metastatic disease limited to the brain had positive levels of CSF-tDNA but nondetectable levels of ctDNA in the plasma. This suggests that CSF-tDNA could be used to identify new mutations that occur exclusively in brain metastases that are not found in the primary cancer. Other studies have similarly shown levels of CSF-tDNA mirror the clinical course and may predict recurrence before radiographic evidence.30,34,35 It may also be possible to ascertain sensitivity to adjuvant therapy using mutations detected in CSF-tDNA. In a study of 20 non–small cell lung cancer patients with CNS metastases, Yang et al identified 2 patients with a mutation of epidermal growth factor receptor (EGFR) not detected in the primary lung tissue.36 Both patients showed response to tyrosine kinase inhibitor treatment.
The largest study to date includes 85 patients who underwent lumbar puncture postsurgery, radiation, and at least one systemic chemotherapy because they showed neurological signs or symptoms of progression.37 Using a capture-based NGS approach, Miller et al identified one or more mutations in cfDNA in the CSF of 49.4% of patients. Radiographic findings, including tumor progression, tumor burden, and spread of tumor towards the ventricular system or subarachnoid space were associated with higher levels of mutation.37 Importantly, Miller et al found that tumor evolution could be tracked through sequential biopsies of CSF. As the time between initial CSF draw and subsequent CSF collection increased, a greater diversity of mutations was observed, especially in those genes that code for growth factor signaling pathways. For example, in a patient with initial EGFR amplification and an EGFR missense mutation, subsequent CSF sampling showed amplification and mutation of platelet-derived growth factor receptor alpha (PDGFRA) and no alterations of EGFR. These findings mirror studies of sequential tumor biopsies in patients with glioma that show only 33–73% concordance of mutations over time.38–43 While more studies are clearly needed, a growing body of literature supports the use of CSF-tDNA for the diagnosis and monitoring of CNS tumors throughout the treatment course.
While most of the DNA based assays have focused on detection of somatic point mutations or small insertions/deletions, many targeted therapeutics are directed towards amplifications in genes such as EGFR.44 There are technologies, such as “personalized analysis of rearranged ends (PARE)” or whole genome sequencing, that are capable of detecting chromosomal rearrangements in a very sensitive fashion but large scale studies on CSF-tDNA are still lacking.45,46 Initial data from Mouliere et al suggest that shallow whole genome sequencing at a coverage of <0.4x was able to detect somatic copy number alterations, including EGFR amplification, in the CSF of 5/13 patients with gliomas.47 These data suggest that additional investigation is warranted.
Expression Changes in cfRNA
Circulating RNAs may also show promise as an alternative detection modality to cfDNA but are likely to be more challenging than methods that utilize cfDNA. Circulating free RNA consists of circulating messenger RNA (mRNA) and micro RNA (miRNA), likely bound to extracellular vesicles like exosomes.48–50 While mRNA codes for proteins, miRNAs are small noncoding nucleotides that target a heterogeneous population of mRNAs for degradation or translation inhibition (Fig. 2). In addition to their transient nature that is dependent upon cellular gene expression requirements, cfRNAs are likely less stable due to their single stranded topology and the presence of nucleases in bodily fluids. One benefit to RNA-based approaches is the ability to detect fusion genes, such as the KIAA:BRAF fusion in pilocytic astrocytomas, which recently has been targeted in therapeutic clinical trials.51
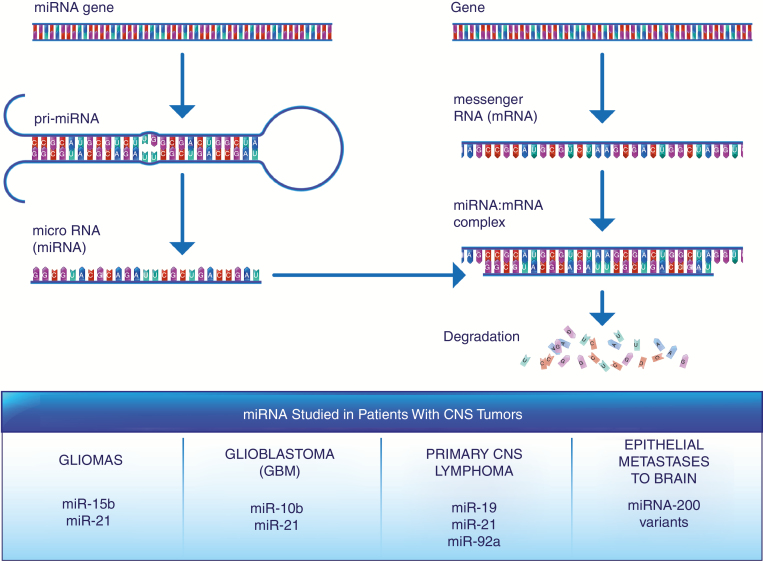
Fig. 2.
MiRNAs regulate the expression of numerous cancer-related genes, and alterations in their levels in CSF have been detected in multiple CNS tumor types.
The first step of nearly all cfRNA-related protocols involves reverse transcription of an RNA molecule to cDNA.52 Presumably, each copy of an RNA molecule is converted into a molecule of cDNA, meaning that the number of cDNA transcripts reflects the expression of a given gene. Multiple analysis methods can then be used to ascertain the expression level of one or more target genes, including real-time (RT) PCR, ddPCR, microarray, or RNA sequencing (RNA-Seq). RT-PCR and ddPCR use fluorescence-based detection of amplification products through the use of a DNA-binding dye or hybridization probe. RT-PCR requires a standard curve defined by known input quantities of RNA or DNA in order to quantify RNA expression, whereas ddPCR is able to count the absolute number of template molecules by amplifying between 1 and 5 templates per droplet.52 Both techniques are efficient, low-cost, and straightforward but are limited to analyzing 1–2 genes at a time. Microarrays and RNA-Seq, by contrast, allow for the analysis of hundreds to thousands of genes at a time. Microarray chips have a defined set of oligonucleotides precoated to a platform and allow for comparative analysis between 2 samples.52 RNA-Seq can be performed with or without enrichment for specific targets in order to better understand the global RNA profile of a sample, but it requires time-intensive and costly library preparation and NGS.52
MiRNAs are the predominant species of cfRNA that have been studied in patients with CNS tumors. One of the first studies, by Baraniskin et al, identified miRNAs characteristic of primary diffuse large B-cell lymphoma of the CNS in CSF fluid.53 A subsequent study of gliomas by the same group identified characteristic overexpression of miR-15b and miR-21.54 The combination of these 2 markers were able to differentiate patients with primary CNS lymphomas, brain metastases, or leptomeningeal carcinomas. The CSF of patients with glioblastoma (GBM) has also shown enrichment for miR-10b and miR-21.55 Circulating miRNAs may also allow clinicians to differentiate between primary CNS malignancies and tumors that have metastasized to the brain.53–57 MiR-15b and miR-21 may be specific for gliomas, while the triad of miR-19, miR-21, and miR-92a may be diagnostic for primary CNS lymphoma.53,54 In contrast, miRNA-200 variants have shown specificity for epithelial metastases to the brain.55
Methylation Changes in cfDNA
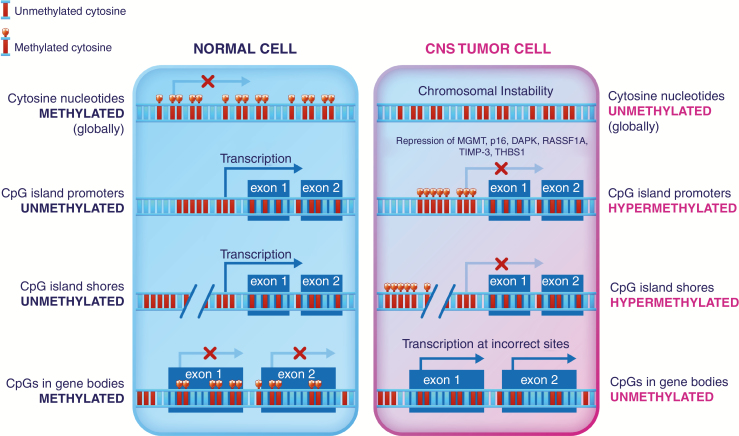
Epigenetic dysregulation is a hallmark of cancer.58 While tumor cells are globally hypomethylated compared with normal tissue, regulatory regions called cytosine-phosphate-guanine (CpG) islands and CpG shores show varying degrees of hypermethylation, especially when located near promoters or other regulatory elements of tumor suppressor genes. This increase in cytosine methylation creates regions of heterochromatin, where DNA is more tightly bound to histones and therefore inaccessible for transcription. The functional result of hypermethylation is a decrease in the protein-level expression of genes, including tumor suppressor genes (Fig. 3).58,59
Fig. 3.
CpG methylation influences the transcription of numerous target genes in cancer. CpG island promoters and CpG island shores upstream of tumor suppressor genes have shown hypermethylation in multiple studies and may serve as candidate biomarkers.
The most commonly used method to differentiate between methylated and unmethylated CpG sites utilized bisulfite treatment.60 Bisulfite treatment converts unmethylated but not methylated cytosine in DNA to uracil. Upon subsequent PCR amplification, the uracil is replaced by thymidine. Methylation-specific PCR uses 2 different sets of primers—one that binds solely to methylated sequences, and one that binds solely to unmethylated sequences—to quantify the level of DNA methylation but is primarily limited to one genomic locus.61 NGS libraries prepared by a panel of DNA strand-specific PCR primers18 or use of microarrays62 allows for many loci to be interrogated in parallel. In either case, the ratio of the signal arising from methylated versus unmethylated cytosines provides an estimate of CpG methylation.
In gliomas, the most well-known epigenetic alteration is promoter hypermethylation of the repair gene that encodes O6-methylguanine-DNA methyltransferase (MGMT).63 Epigenetic inactivation of the MGMT gene as a result of promoter hypermethylation prevents DNA repair of damage caused by alkylating agents and is used as a positive predictor of response to temozolomide treatment.64,65 Balaña et al undertook one of the first investigations of cfDNA promoter methylation in patients with GBM.66 While this study was conducted using serum, the authors detected promoter hypermethylation of MGMT, p16, death-associated protein kinase, and Ras association domain family 1 isoform A (RASSF1A) in 39.3%, 53.6%, 34.3%, and 50% of samples, respectively. In addition, the methylation status of the cfDNA was highly concordant with that of the primary tumor. Liu et al reported close to perfect specificity in detecting aberrant promoter methylation patterns of MGMT, p16, TIMP-3, and THBS1 in matched tumor and CSF samples of patients with malignant glioma.67 Hypermethylation of MGMT and THBS1 in CSF were independent prognostic factors for progression-free survival. Subsequent reports have supported initial findings by Balaña et al and Liu et al, with sensitivities ranging from ~60% to 70%,68 and have identified other putative hypermethylation candidates such as TERT in both CSF and serum.69,70 Depending on the nature of the mutation, it may be possible to simultaneously investigate mutations in cfDNA and aberrant promoter methylation in a panel of genes in the same assay, potentially increasing the sensitivity above that of one type of biomarker alone.18
Alterations in Circulating Protein Levels
Interrogating protein levels has become an increasingly active area of research, both for biomarker identification as well as discovery of new treatment approaches. CNS tumors may secrete or shed proteins into the CSF, making them detectable by techniques such as mass spectrometry and ELISA. Most whole-protein mass spectrometry analysis requires that proteins in solution be ionized into the gas phase and accelerated through a magnetic field for analysis of their time-of-flight. The time-of-flight for the various fragments of a protein constitute a molecular fingerprint that allows sophisticated software coupled with an ever-growing body of databases to identify and quantify the proteins present in the clinical sample.71 Mass spectrometry therefore has the benefit of being able to detect a nearly unlimited number of different proteins from the same clinical specimen but requires technical know-how and is more costly than routine laboratory techniques like ELISA. ELISAs utilize antibodies to known protein targets, along with a standard curve, to quantify the amount of a given protein in solution.72 ELISAs are relatively inexpensive and straightforward to perform in a clinical laboratory but are limited to the identification of a few proteins from a given sample.
The identification of specific protein markers for CNS cancers is more challenging than for cfDNA or cfRNA changes. Elevations in the levels of proteins may be caused by disease or as a consequence of inflammation or unrelated illness. The most significant examples of specific CSF proteins that have impacted the care of CNS cancers are those related to intracranial malignant germ cell tumors.73 Germ cell tumors arise from aberrant migration and differential of various germ cell layers during embryogenesis. Intracranial germ cell tumors arise in the pineal and suprasellar regions and constitute about 3% of pediatric brain tumors. The exact constellation of proteins representative of a germ cell tumor depends on the germ cell of origin. For example, intracranial malignant germ cell tumors express beta human chorionic gonadotropin (bHCG) and alpha-fetoprotein (AFP),74 and the levels of both proteins have been shown to be markedly elevated in the CSF.75 Both bHCG and AFP have been used to diagnose malignant germ cell tumors, monitor response to treatment, and predict recurrence. While less specific, placental alkaline phosphatase and lactate dehydrogenase have also shown clinical utility.76
Researchers have also screened the CSF of glioma patients for potential protein biomarkers. In an extensive meta-analysis of the literature, Shen et al identified 19 proteins, including B2M, CA2, CA12, CALD1, DDAH1, MYCN, PPIA, SPP1, VEGFB, ALB, MAPT, SERPINA3, and SPARCL1, that were upregulated in the CSF of patients with confirmed gliomas.77 Khwaja et al were able to replicate many of these proteins in their analysis of 60 samples from WHO grades II–IV astrocytomas, metastatic brain tumors, inflammatory samples, and normal controls. Of the 103 potential tumor-specific markers, 20 were specific to high-grade astrocytomas. Similar to Shen et al, Khwaja and colleagues identified B2M, SPARCL1, and VEGFB.78 Vascular endothelial growth factor (VEGF) was further assessed in a study by Sampath et al where they assayed the CSF from 27 patients with high-grade gliomas, 39 patients with nonastrocytic CNS tumors, and 14 patients with no known CNS tumors. VEGF showed a sensitivity of 89% and a specificity of 100% for malignant astrocytoma samples and was significantly higher in high-grade astrocytomas than in nonastrocytic tumors.79 Four peptides have also been shown to distinguish GBM from controls: alpha-1-antichymotrypsin, osteopontin, transthretin, and the N-terminal residue of albumin were elevated in the CSF of GBM patients.80
Proteomic analysis may also be useful for diagnosing and monitoring medulloblastoma, the most common malignant brain tumor in children. Aiming to identify novel biomarkers in the CSF of children with diagnoses of medulloblastoma, Rajagopal et al compared the CSF proteome of 33 of these children against 25 age-matched controls.81 Unlike other studies that have noted elevations of proteins in the CSF, the level of prostaglandin D2 synthase was 6-fold less in the CSF of tumor samples compared with normal.
Limitations of Biomarkers
Cell-free nucleic acids and proteins have significant potential as biomarkers for CNS cancers. Historically, however, few biomarkers, especially proteins, discovered in the laboratory setting ever make it to clinical practice.82 Understanding the limitations behind biomarker discovery and validation can help inform good research practices and lead to the more efficient identification of these markers.
Probably the main barrier to FDA approval of biomarkers for CNS cancers is the low level of reproducibility between similar studies. Techniques for DNA, RNA, and protein isolation differ between labs, and these technical differences can lead to drastically different qualities of preparation. For example, isolation of cell-free nucleic acids from biological specimens must be carried out within hours of their collection in order to minimize the degradation caused by nucleases. Similar requirements are true for proteins. In addition, biological specimens must be properly stored and undergo no to minimal freeze-thaw cycles in order to prevent molecular degradation. Teunissen et al have suggested a protocol to standardize collection and banking of CSF samples,83 but this has not been universally adopted.
Technical limitations also hamper identification of rare mutations and methylation changes in cell-free nucleic acids. While technologies like duplex sequencing and molecular barcode–assisted NGS18–20,84 have increased the signal-to-noise ratio of detecting mutations and methylation changes at low frequencies, inefficiencies in library preparation and errors generated by enzymes early in the library preparation steps obscure subtle elevations in mutations and methylation. Emerging technologies that are able to reduce this noise by sequencing both strands of a template DNA molecule may help increase the power of these technologies, but their techniques have not been widely adopted by most clinical laboratories.18,20,84
Conclusion
The collection of CSF is critical for the identification of biomarkers for CNS tumors. Due to the relative rarity of samples compared with other cancer types, it is important for investigators to bank specimens for current and future research. Low sample numbers in biomarker identification studies limit the statistical power to detect differences between malignant disease, benign disease, and normal controls, resulting in a lack of validation among studies. Standardized sample collection procedures and studies coordinated across institutions will help alleviate some of these limitations.
Compared with surgical biopsy, CSF collection is minimally invasive and may provide a picture of the genetic, epigenetic, and proteomic state of a heterogeneous CNS tumor. In addition to being useful for primary diagnosis, following the levels of biomarkers after surgery, radiation, and/or chemotherapeutic treatment may forecast response to treatment and predict recurrence before imaging modalities.
In the future, other biofluids like blood may play a role in the diagnosis and management of CNS tumors. However, the presence of the blood–brain barrier will likely limit the number and type of biomarkers identified in the blood compared with the CSF. Based on the CNS biomarkers identified so far, patients with diagnoses of medulloblastomas and high-grade gliomas may be the best population to study. These tumors will likely show the most dramatic changes in mutation number, CpG methylation status, and protein expression compared with normal controls, providing more statistical power to studies with a limited number of samples (Table 1). Studies with CSF collected at multiple time points are essential for biomarker identification. Ideally, CSF should be collected presurgery or intraoperatively before tumor resection, as the biomarker concentration will be at its highest level. Collection within a few days of surgery will help establish a baseline against which to predict recurrence. When patients return for radiographic follow-up, a separate CSF sample should be taken to understand whether a predicted biomarker can pre-date radiographic recurrence or even possibly inform clinicians about tumor evolution that suggests a benefit to using a targeted chemotherapeutic agent. A growing database of these studies conducted in a systemic manner will hopefully lead to a more effective approach to the diagnosis and monitoring of CNS tumors compared with repeated biopsy and surgery alone. Routine CSF sampling is already a part of standard of care for some CNS tumors, like medulloblastoma and primary CNS lymphoma. Expanding CSF-based diagnostics is a logical extension of existing practices but one that will require well-conducted prospective collaborative studies.
Table 1.
Benefits and considerations for various biomarkers used for the detection of CNS tumors in CSF
| Biomarker | Assay | Common Targets | Benefits | Considerations |
|---|---|---|---|---|
| cfDNA Mutations | • ddPCR • NGS |
• AKT1 • BRAF • EGFR • IDH1 • KRAS • NF2 • NRAS • PIK3R1 • PRCH1 • PTEN • TERT • TP53 |
• Short half-life enables recurrence monitoring • Both driver and passenger mutations can be used • Specific mutations may be sensitive to adjuvant therapy |
• Early stage tumors are challenging to detect • Requires whole-exome sequencing or a large targeted panel due to diversity of mutations |
| cfRNA Expression | • Real-time PCR • ddPCR • Microarray • RNA-Seq |
• miR-10b • miR-15b • miR-19 • miR-21 • miR-92a • miR-200 family |
• miRNAs may target biologically relevant pathways | • Contamination with genomic DNA can hamper interpretation of results • May be limited to assessing a few targets at a time |
| cfDNA Methylation | • Methylation- specific PCR • Microarray • NGS |
• MGMT • p16 • RASSF1A • TERT • THBS1 • TIMP-3 |
• Changes may precede mutations • Hypermethylation often occurs in tumor suppressor genes |
• Challenging to assay for hypomethylation • Design of primers and probes is challenging due to low genome complexity after bisulfite conversion |
| Protein Concentration | • ELISA • Mass spectrometry |
• AFP • B2M • bHCG • SPARCL1 • VEGF |
• Inexpensive and straightforward to implement clinically • Represents functional changes at the protein level |
• Low specificity due to confounding variables like inflammation and chronic disease |
Funding
This study was supported by the Burroughs Wellcome Career Award for Medical Scientists, NIH R37 CA230400 and U01 CA230691.
Conflict of interest statement.
C.B. is a consultant for Depuy-Synthes. The terms of this arrangement are being managed by the university in accordance with its conflict of interest policies. H.Y. is the co-founder of Genetron Health.
Acknowledgments
We thank Elizabeth Cook for her help designing and illustrating the main messages of this study.
References
- 1. Noone AM, Howlader N, Krapcho M, et al. In: Institute NC , ed. SEER Cancer Statistics Review, 1975–2015. Bethesda, MD: NIH; 2018. [Google Scholar]
- 2. McGirt MJ, Woodworth GF, Coon AL, et al. Independent predictors of morbidity after image-guided stereotactic brain biopsy: a risk assessment of 270 cases. J Neurosurg. 2005;102(5):897–901. [DOI] [PubMed] [Google Scholar]
- 3. Kassim OO, Afolabi O, Ako-Nai KA, et al. Cytomegalovirus antibodies in breast milk and sera of mother-infant pairs. J Trop Pediatr. 1987;33(2):75–77. [DOI] [PubMed] [Google Scholar]
- 4. Quick-Weller J, Lescher S, Bruder M, et al. Stereotactic biopsy of brainstem lesions: 21 years experiences of a single center. J Neurooncol. 2016;129(2):243–250. [DOI] [PubMed] [Google Scholar]
- 5. Brandsma D, Stalpers L, Taal W, Sminia P, van den Bent MJ. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol. 2008;9(5):453–461. [DOI] [PubMed] [Google Scholar]
- 6. Cohen JV, Alomari AK, Vortmeyer AO, et al. Melanoma brain metastasis pseudoprogression after pembrolizumab treatment. Cancer Immunol Res. 2016;4(3):179–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thompson EM, Frenkel EP, Neuwelt EA. The paradoxical effect of bevacizumab in the therapy of malignant gliomas. Neurology. 2011;76(1):87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Springer S, Zhang M, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112(31):9704–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen WW, Balaj L, Liau LM, et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vogelstein B, Kinzler KW. The path to cancer –three strikes and you’re out. N Engl J Med. 2015;373(20):1895–1898. [DOI] [PubMed] [Google Scholar]
- 16. Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. [DOI] [PubMed] [Google Scholar]
- 17. Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mattox AK, Wang Y, Springer S, et al. Bisulfite-converted duplexes for the strand-specific detection and quantification of rare mutations. Proc Natl Acad Sci U S A. 2017;114(18):4733–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108(23):9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoang ML, Kinde I, Tomasetti C, et al. Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc Natl Acad Sci U S A. 2016;113(35):9846–9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen R, Im H, Snyder M. Whole-exome enrichment with the agilent sureselect human all exon platform. Cold Spring Harb Protoc. 2015;2015(7):626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. [DOI] [PubMed] [Google Scholar]
- 24. Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Mattos-Arruda L, Mayor R, Ng CKY, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panditharatna E, Kilburn LB, Aboian MS, et al. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res. 2018;24(23):5850–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61(3):514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pentsova EI, Shah RH, Tang J, et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol. 2016;34(20):2404–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swinkels DW, de Kok JB, Hanselaar A, Lamers K, Boerman RH. Early detection of leptomeningeal metastasis by PCR examination of tumor-derived K-ras DNA in cerebrospinal fluid. Clin Chem. 2000;46(1):132–133. [PubMed] [Google Scholar]
- 30. Momtaz P, Pentsova E, Abdel-Wahab O, et al. Quantification of tumor-derived cell free DNA(cfDNA) by digital PCR (DigPCR) in cerebrospinal fluid of patients with BRAFV600 mutated malignancies. Oncotarget. 2016;7(51):85430–85436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan C, Diplas BH, Chen X, et al. Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol. 2019;137(2):297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brastianos PK, Shankar GM, Gill CM, et al. Dramatic response of BRAF V600E mutant papillary craniopharyngioma to targeted therapy. J Natl Cancer Inst. 2016;108(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cavallo LM, Frank G, Cappabianca P, et al. The endoscopic endonasal approach for the management of craniopharyngiomas: a series of 103 patients. J Neurosurg. 2014;121(1):100–113. [DOI] [PubMed] [Google Scholar]
- 34. Kimoto T, Inoue M, Tokimasa S, et al. Detection of MYCN DNA in the cerebrospinal fluid for diagnosing isolated central nervous system relapse in neuroblastoma. Pediatr Blood Cancer. 2011;56(5):865–867. [DOI] [PubMed] [Google Scholar]
- 35. Li Y, Pan W, Connolly ID, et al. Tumor DNA in cerebral spinal fluid reflects clinical course in a patient with melanoma leptomeningeal brain metastases. J Neurooncol. 2016;128(1):93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang H, Cai L, Zhang Y, et al. Sensitive detection of EGFR mutations in cerebrospinal fluid from lung adenocarcinoma patients with brain metastases. J Mol Diagn. 2014;16(5):558–563. [DOI] [PubMed] [Google Scholar]
- 37. Miller AM, Shah RH, Pentsova EI, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–468. [DOI] [PubMed] [Google Scholar]
- 39. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bai H, Harmancı AS, Erson-Omay EZ, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim J, Lee IH, Cho HJ, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28(3):318–328. [DOI] [PubMed] [Google Scholar]
- 42. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee JK, Wang J, Sa JK, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49(4):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lassman AB, Aldape KD, Ansell PJ, et al. Epidermal growth factor receptor (EGFR) amplification rates observed in screening patients for randomized trials in glioblastoma. J Neurooncol. 2019;144(1):205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):20ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4(162):162ra154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mouliere F, Mair R, Chandrananda D, et al. Detection of cell-free DNA fragmentation and copy number alterations in cerebrospinal fluid from glioma patients. EMBO Mol Med. 2018;10(12):e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3(11):e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fernandez-Mercado M, Manterola L, Larrea E, et al. The circulating transcriptome as a source of non-invasive cancer biomarkers: concepts and controversies of non-coding and coding RNA in body fluids. J Cell Mol Med. 2015;19(10):2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. El-Hefnawy T, Raja S, Kelly L, et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem. 2004;50(3):564–573. [DOI] [PubMed] [Google Scholar]
- 51. Behling F, Schittenhelm J. Oncogenic BRAF alterations and their role in brain tumors. Cancers (Basel). 2019;11(6):794–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lovén J, Orlando DA, Sigova AA, et al. Revisiting global gene expression analysis. Cell. 2012;151(3):476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117(11):3140–3146. [DOI] [PubMed] [Google Scholar]
- 54. Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012;14(1):29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Teplyuk NM, Mollenhauer B, Gabriely G, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14(6):689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Drusco A, Bottoni A, Laganà A, et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget. 2015;6(25):20829–20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi R, Wang PY, Li XY, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6(29):26971–26981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17(5):284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–116. [DOI] [PubMed] [Google Scholar]
- 60. Wang RY, Gehrke CW, Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980;8(20):4777–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93(18):9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38(12):1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6(1):39–51. [DOI] [PubMed] [Google Scholar]
- 64. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 65. Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. [DOI] [PubMed] [Google Scholar]
- 66. Balana C, Ramirez JL, Taron M, et al. O6-methyl-guanine-DNA methyltransferase methylation in serum and tumor DNA predicts response to 1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastoma multiforme. Clin Cancer Res. 2003;9(4):1461–1468. [PubMed] [Google Scholar]
- 67. Liu BL, Cheng JX, Zhang W, et al. Quantitative detection of multiple gene promoter hypermethylation in tumor tissue, serum, and cerebrospinal fluid predicts prognosis of malignant gliomas. Neuro Oncol. 2010; 12(6):540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Z, Jiang W, Wang Y, et al. MGMT promoter methylation in serum and cerebrospinal fluid as a tumor-specific biomarker of glioma. Biomed Rep. 2015;3(4):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Juratli TA, Stasik S, Zolal A, et al. TERT promoter mutation detection in cell-free tumor-derived DNA in patients with IDH wild-type glioblastomas: a pilot prospective study. Clin Cancer Res. 2018;24(21):5282–5291. [DOI] [PubMed] [Google Scholar]
- 70. Majchrzak-Celińska A, Paluszczak J, Kleszcz R, et al. Detection of MGMT, RASSF1A, p15INK4B, and p14ARF promoter methylation in circulating tumor-derived DNA of central nervous system cancer patients. J Appl Genet. 2013;54(3):335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chait BT. Mass spectrometry in the postgenomic era. Annu Rev Biochem. 2011;80:239–246. [DOI] [PubMed] [Google Scholar]
- 72. Lequin RM. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin Chem. 2005;51(12):2415–2418. [DOI] [PubMed] [Google Scholar]
- 73. Samuel N, Remke M, Rutka JT, Raught B, Malkin D. Proteomic analyses of CSF aimed at biomarker development for pediatric brain tumors. J Neurooncol. 2014;118(2):225–238. [DOI] [PubMed] [Google Scholar]
- 74. Qaddoumi I, Sane M, Li S, et al. Diagnostic utility and correlation of tumor markers in the serum and cerebrospinal fluid of children with intracranial germ cell tumors. Childs Nerv Syst. 2012;28(7):1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nishizaki T, Kajiwara K, Adachi N, et al. Detection of craniospinal dissemination of intracranial germ cell tumours based on serum and cerebrospinal fluid levels of tumour markers. J Clin Neurosci. 2001;8(1):27–30. [DOI] [PubMed] [Google Scholar]
- 76. Watanabe S, Aihara Y, Kikuno A, et al. A highly sensitive and specific chemiluminescent enzyme immunoassay for placental alkaline phosphatase in the cerebrospinal fluid of patients with intracranial germinomas. Pediatr Neurosurg. 2012;48(3):141–145. [DOI] [PubMed] [Google Scholar]
- 77. Shen F, Zhang Y, Yao Y, et al. Proteomic analysis of cerebrospinal fluid: toward the identification of biomarkers for gliomas. Neurosurg Rev. 2014;37(3):367–380; discussion 380. [DOI] [PubMed] [Google Scholar]
- 78. Khwaja FW, Reed MS, Olson JJ, et al. Proteomic identification of biomarkers in the cerebrospinal fluid (CSF) of astrocytoma patients. J Proteome Res. 2007;6(2):559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sampath P, Weaver CE, Sungarian A, Cortez S, Alderson L, Stopa EG. Cerebrospinal fluid (vascular endothelial growth factor) and serologic (recoverin) tumor markers for malignant glioma. Cancer Control. 2004;11(3):174–180. [DOI] [PubMed] [Google Scholar]
- 80. Schuhmann MU, Zucht HD, Nassimi R, et al. Peptide screening of cerebrospinal fluid in patients with glioblastoma multiforme. Eur J Surg Oncol. 2010;36(2):201–207. [DOI] [PubMed] [Google Scholar]
- 81. Rajagopal MU, Hathout Y, MacDonald TJ, et al. Proteomic profiling of cerebrospinal fluid identifies prostaglandin D2 synthase as a putative biomarker for pediatric medulloblastoma: A pediatric brain tumor consortium study. Proteomics. 2011;11(5):935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Füzéry AK, Levin J, Chan MM, Chan DW. Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin Proteomics. 2013;10(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Teunissen CE, Petzold A, Bennett JL, et al. A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology. 2009;73(22):1914–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kennedy SR, Schmitt MW, Fox EJ, et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat Protoc. 2014;9(11):2586–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]