Abstract
Accurate pathologic diagnoses and molecularly informed treatment decisions for a wide variety of cancers depend on robust clinical molecular testing that uses genomic, epigenomic, and transcriptomic-based tools. Nowhere is this more essential than in the workup of brain tumors, as emphasized by the incorporation of molecular criteria into the 2016 World Health Organization classification of central nervous system tumors and the updated official guidelines of the National Comprehensive Cancer Network. Despite the medical necessity of molecular testing in brain tumors, access to and utilization of molecular diagnostics is still highly variable across institutions, and a lack of reimbursement for such testing remains a significant obstacle. The objectives of this review are (i) to identify barriers to adoption of molecular testing in brain tumors, (ii) to describe the current molecular tools recommended for the clinical evaluation of brain tumors, and (iii) to summarize how molecular data are interpreted to guide clinical care, so as to improve understanding and justification for their coverage in the routine workup of adult and pediatric brain tumor cases.
Keywords: embryona, ependymoma, glioma, meningioma, molecular
Overview and Barriers to Testing
Nervous system tumors are a unique and highly diverse group of neoplasms that develop from the central nervous system (CNS) and its coverings. This complexity makes clinical decision making and testing choices especially difficult for pathologists and treating physicians. Prognoses vary tremendously, ranging from highly malignant cancers with a median survival of less than one year despite aggressive surgery, radiation, and chemotherapy, to indolent tumors that are usually curable with surgical resection alone. Accurate diagnosis is therefore critical, yet past experience has demonstrated that, because diagnoses and classifications relying solely on histology have an element of subjectivity, their interobserver reproducibility may be low. Benign tumors can resemble lethal malignancies under the microscope, and vice versa. And, unlike most neoplasms arising elsewhere, brain tumors are often difficult to biopsy, meaning that life-altering diagnoses are frequently being rendered on extremely small samples that might not contain all the required histologic features.
In addition to their clinical and histologic complexity, brain tumors have a highly diverse set of mutations and aberrations that directly impact their biology and response to treatment. Most brain tumors have a relatively low overall mutational burden, yet mutations in key genomic drivers are linked to tumor initiation and define tumor behavior. Like many other tumor types, copy number alterations can be diagnostic and/or define key therapeutic vulnerabilities for both low- and high-grade tumors, including focal (<10 megabases) and broad chromosome-level changes (arm or whole chromosomes) included in the new World Health Organization (WHO) classification criteria.1 Gene fusions and rearrangements are important drivers in some brain tumors. Certain discrete methylation events, as well as global methylation patterns, are valuable as predictive and prognostic markers. Therefore, more than one molecular assay is usually needed to adequately evaluate a patient’s brain tumor.
Because of recent advances in our knowledge of the clinically relevant genetic underpinnings of brain tumors, and of tumors arising elsewhere in the body, there is a critical need to apply that knowledge through molecular testing that captures important diagnostic, prognostic, and predictive information. In most cases, the main purpose of molecular testing in brain tumors is to ensure that the tumor is properly classified, so as to best inform the patient and treating physician about the true prognosis of the disease and the best treatment options. Molecular testing has already been guiding brain tumor therapy for more than 10 years, as 1p/19q codeletion and O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation have influenced the use of chemotherapies like procarbazine/lomustine/vincristine (PCV) and temozolomide (TMZ), respectively. New methods are now available—such as next-generation sequencing (NGS) and copy number profiling—that can determine the 1p/19q copy number status of a tumor, as well as provide a great deal more information about other aspects of the tumor’s genome. Their use is therefore fully consistent with the goal of precision brain tumor diagnostics. The recent Food and Drug Administration approval of NGS,2 and of tumor mutation burden (TMB) as a biomarker of sensitivity to immune checkpoint inhibitors,3 provides further justification for such advanced molecular testing in all kinds of cancer, including brain tumors.
Details on the specific kinds of molecular tests required for the workup of brain tumors, including key strengths and drawbacks of each assay, are provided in the Supplementary text.
Why Molecular Testing Is Now Essential for the Workup of Brain Tumors
All the molecular panels, platforms, and assays routinely being used to improve the clinical care of brain tumor patients represent years of tremendous advancements in biotechnology, with decreasing costs to match. The first human whole-genome sequencing, officially completed in 2003, cost nearly $100 million. Now, it only costs several thousand dollars on most platforms, and targeted NGS panels are even less expensive.4 Molecular data has fundamentally and permanently transformed the diagnosis and management of patients with gliomas and embryonal tumors, with similar changes forthcoming in sellar and meningeal tumors (Table 1, Figures 1 and 2). Even though the aggregate cost of this critical component of patient management is only a small fraction of the total cost of care (including imaging, surgery, and treatment), the reimbursement of these tests has fallen short, leading to a lack of implementation or full utilization at medical centers that treat brain tumor patients. Yet molecular testing is now required to make diagnoses according to the WHO 2016 scheme, and national practice guidelines, enumerated by the National Comprehensive Cancer Network and cIMPACT-NOW, have provided evidence and recommendations for their use in patient care.5
Table 1.
Molecular testing for the accurate diagnosis and subclassification of brain tumors*
| Glioma | Embryonal Tumors | Other Tumors |
|---|---|---|
| • 1p/19q codeletion1 • ATRX mutation4 • BRAF fusion4 • BRAF mutations2 • CDK4 amplification4 • CDKN2A deletion2 • Chromosome 10 monosomy2 • Chromosome 7 gain2 • EGFR amplification2 • EGFR mutation4 • FGFR1 gain2 • FGFR1 mutation2 • FGFR3 fusions3 • H3F3A mutations1 • IDH1 and IDH2 mutation1 • Methylation profiling4 • MGMT promoter methylation3 • MYB or MYBL1 rearrangement2 • NOTCH1 mutation4 • RB mutation or deletion4 • RELA fusion1 • TERT promoter mutation2 • TP53 mutation4 |
• APC mutation1 • BRG1 mutation1 • C19MC amplification1 • CTNNB1 mutation1 • GAB1 expression1 • INI1 mutation1 • Methylation profiling4 • Monosomy 64 • MYC amplification4 • MYCN amplification4 • SHH activation1 • TP53 mutation1 • WNT activation1 • YAP1 expression1 • β-catenin expression and localization1 |
Craniopharyngioma • BRAF V600E mutation4 • CTNNB1 mutation4 • β-catenin nuclear expression4 Meningioma • BAP1 mutation or deletion4 • Methylation profiling4 • TERT promoter mutation4 |
*Each molecular marker and/or test is denoted by the main source recommending its use: 1WHO 2016 classification scheme; 2cIMPACT-NOW updates; 3treatment stratification; 4proposed with good evidence in the literature.
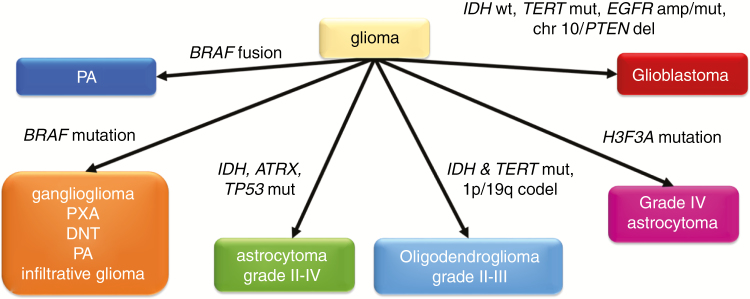
Fig. 1.
Interpretation of the most common molecular test results in a glioma or glioneuronal tumor. Molecular data must always be interpreted in the appropriate clinical and histopathologic contexts. MGMT promoter methylation testing is recommended in all grades III–IV gliomas. PXA = pleomorphic xanthoastrocytoma; DNT = dysembryoplastic neuroepithelial tumor; amp = amplification; del = deletion; codel = codeletion.
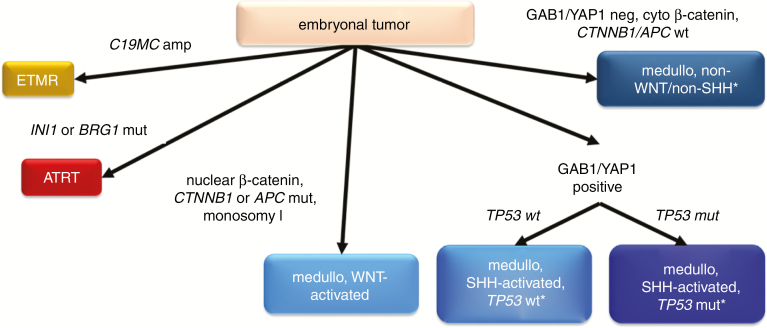
Fig. 2.
Interpretation of the most common molecular test results in an embryonal tumor. Molecular data must always be interpreted in the appropriate clinical and histopathologic contexts. medullo = medulloblastoma; cyto = cytoplasmic. *MYC/MYCN amplification testing recommended.
Glioma
Gliomas are the most common primary neoplasms arising within the brain parenchyma (meningiomas are more frequent overall but arise in the meninges covering the brain, not the brain itself).6 In clinical usage, the term “glioma” usually implies an astrocytoma or oligodendroglioma that is infiltrative and ultimately fatal. But “glioma” encompasses a heterogeneous group of neoplasms arising from glial progenitor cells that, depending on driver mutations and other cues, develop into a wide range of tumors with markedly divergent outcomes. Because these outcomes are tightly linked with specific molecular alterations (Fig. 1), testing for these alterations enhances diagnostic and prognostic accuracy well beyond the capabilities of traditional light microscopy.
Pilocytic Astrocytoma, Ganglioglioma, and Pleomorphic Xanthoastrocytoma
The most common form of glioma is astrocytoma, which is still histologically graded on a WHO scale of I to IV. Pilocytic astrocytomas (PAs) are the most frequent grade I astrocytomas, arising most often in the posterior fossa and, less commonly, the optic nerve, supratentorial region, and spinal cord, primarily in children but also sometimes in adults.7–10 The majority of PAs are indolent and curable with surgery alone. However, some can be histologically mistaken for higher-grade infiltrative gliomas, and midline PAs that are not fully resectable may require adjuvant therapy.8,9 The detection of a BRAF fusion strongly favors the diagnosis of PA, and the fusion is most characteristic of cerebellar PAs in children.10 In young adults and older patients, tumor location shifts to the supratentorium, and BRAF V600E becomes somewhat more common.10 In isolation, either a BRAF fusion or V600E mutation tends to impart an especially favorable prognosis, but if additional alterations involving cell cycle regulators are present, such as deletion of cyclin-dependent kinase inhibitor 2A (CDKN2A) and alterations of thalassemia/mental retardation syndrome X-linked (ATRX), the tumor is more likely to recur and progress to a higher grade.9,11,12 A small subset of PAs have activating alterations in fibroblast growth factor receptor (FGFR1) instead of BRAF.13 Even though both sets of alterations lead to mitogen-activated protein kinase pathway activation, PAs with FGFR1 mutations or copy number gains might behave more aggressively.12,14,15 Even so, the overall prognosis of PAs is far better than for most other gliomas. Isolated BRAF V600E is more often found in other potentially curable grade I neoplasms, like gangliogliomas and dysembryoplastic neuroepithelial tumors.16 Grades II and III pleomorphic xanthoastrocytomas typically have BRAF V600E mutations, as well as alterations in genes encoding cell cycle proteins.16,17 The Braf inhibitor, vemurafenib, is active against V600E-driven gliomas18 but does not work against gliomas with BRAF fusions.19 In this context, defining the molecular alterations surrounding BRAF can be highly informative for accurate delineation of lower-grade brain tumors, and may be impactful in their therapeutic management.
Diffusely Infiltrative Astrocytomas
Most grades II and III astrocytomas in adults, and grade IV astrocytomas that arise from lower-grade tumors (previously known as secondary glioblastomas), are characterized by point mutations in isocitrate dehydrogenase 1 and 2 genes (hereafter referred to collectively as “IDHmut”).20 While still ultimately fatal, IDHmut astrocytomas tend to be less aggressive, and benefit more from chemotherapy (such as TMZ or PCV) than histologically similar tumors that are IDH wild-type (IDHwt).20,21 As might be expected from a mutation that causes global cytosine-phosphate-guanine hypermethylation, most IDHmut gliomas have MGMT promoter methylation.22 The majority of IDHmut astrocytomas also contain mutations in TP53 and ATRX, distinguishing them from IDHmut oligodendrogliomas (see below).23 Once an astrocytoma is identified as IDHmut, mitotic index no longer has much prognostic relevance, meaning that there is little difference in overall survival between grades II and III IDHmut astrocytomas that are diagnosed according to current histologic grading criteria.24 Of far more prognostic importance is the presence of additional molecular alterations involving the cell cycle, including homozygous deletion of CDKN2A, deletion or mutation of RB1, and CDK4 amplification, all of which are associated with shorter overall survival in grades II–III IDHmut astrocytomas.25 Because the CATNON trial showed that grade III anaplastic astrocytomas should be treated with radiotherapy (RT) and TMZ upfront,21 and the histopathologic distinction between grades II and III astrocytomas is unclear, some neuro-oncologists prefer treating both grades II and III astrocytomas with RT/TMZ immediately. The presence of necrosis and/or microvascular proliferation is still important, as IDHmut glioblastomas (GBMs) behave worse than IDHmut grades II–III astrocytomas.26 Furthermore, IDHmut is not found in any grade I glioma, so its detection essentially precludes the diagnosis of grade I tumors like ganglioglioma.27
Oligodendrogliomas
While less common than astrocytomas, oligodendrogliomas are the other major subset of diffusely infiltrative glioma. Grades II–III oligodendrogliomas (there are no grade I or IV oligodendrogliomas) are usually still lethal, but confer longer survival than even IDHmut astrocytomas. The diagnosis of an oligodendroglioma now requires the presence of IDHmut and 1p/19q codeletion.1 Interestingly, telomerase reverse transcriptase (TERT) promoter mutations, which are found in most GBM IDHwt, are also present in most oligodendrogliomas.23 Astrocytomas and oligodendrogliomas can be difficult to reliably distinguish solely based on histologic criteria. Prior to molecular diagnostics, this caused a lack of consistency among neuropathologists—a situation complicated by the varying use of the hybrid (and nebulous) term “oligoastrocytoma” for gliomas with mixed or equivocal features.28 Yet the distinction is important, because not only are oligodendrogliomas less aggressive than their astrocytic counterparts, they appear to specifically benefit from PCV chemotherapy.29,30 Because oligodendrogliomas also show increased sensitivity to TMZ, some neuro-oncologists prefer TMZ as a front-line therapy. Others prefer delaying adjuvant treatment in grade II IDHmut, 1p/19q codeleted oligodendrogliomas until the tumor shows progression to grade III. At that time, chemotherapy is sometimes given alone, thereby sparing patients the side effects of RT as long as possible.31 Such an option is available only in oligodendrogliomas because of their less aggressive nature. In current practice, ATRX, TP53, TERT, and 1p/19q screenings reliably segregate gliomas with hybrid morphology into astrocytomas or oligodendrogliomas, thus eliminating “oligoastrocytoma” as an integrated diagnosis within the WHO classification.23,32 Although IDHmut‒TERTmut‒1p/19q codeleted oligodendrogliomas do have much better prognoses overall, mutations in NOTCH are associated with more rapid progression in these tumors.33 Likewise, polysomy of 1p and 19q has been linked with shorter survival in oligodendrogliomas.34,35
Glioblastoma
The most common glioma, IDHwt de novo or primary GBM (GBM IDHwt), unfortunately is also highly malignant. If a diffusely infiltrative glioma lacks IDHmut but has other molecular alterations associated with GBM, it should probably be treated like a GBM even if necrosis and microvascular proliferation cannot be detected microscopically. The third update from cIMPACT-NOW recommended diagnostic criteria for “diffuse astrocytic gliomas, IDH-wild type, with molecular features of GBM, WHO grade IV” for such tumors; these criteria include TERT promoter mutations, gain of chromosome 7 and loss of chromosome 10, and EGFR amplification.36 Additional molecular hallmarks of GBM include amplification of other receptor tyrosine kinases like PDGFRA and MET, as well as mutations in PTEN and NF1.37 Because the vast majority of GBMs will contain at least 2 of the aforementioned alterations, great care must be taken when dealing with a lesion that has only one alteration. For example, isolated PTEN mutations and deletions are characteristic of certain nonneoplastic dysplasias, malformations, and hamartomas that might rarely be mistaken for gliomas.38
Thorough molecular characterization of a suspected GBM not only contributes to a definitive diagnosis, it also helps guide treatment decisions. Small-molecule inhibitors of epidermal growth factor receptor (EGFR), and antitumor vaccines directed against EGFR variant (v)III, have thus far failed to provide significant survival benefit in definitive clinical trials of GBM patients.39–41 However, there is renewed interest in detecting EGFR-driven GBMs, as they may be responsive to depatuxizumab mafodotin (ABT-414). This is an antibody–drug conjugate that delivers the anti-microtubule compound monomethyl auristatin F into cells with high expression of EGFR or mutant EGFRvIII.42 Another recently discovered driver of some GBM, FGFR3 fusions, may respond to FGFR inhibitors.43
Pediatric Low-Grade Gliomas
Low-grade infiltrative gliomas in children and adolescents are broadly associated with much better outcomes than in adults, even when they lack IDHmut. Some of these have BRAFV600E mutations, as discussed above. Others have FGFR1 alterations, including internal tandem duplications or mutations, or have rearrangements in MYB or MYBL1.44 Although such tumors may resemble classic astrocytomas or oligodendrogliomas, anaplastic progression is uncommon and overall survival in patients is markedly prolonged. Consequently, this subset of infiltrating gliomas warrants separate classification.44
MGMT in Gliomas
MGMT encodes O6-methylguanine-DNA methyltransferase, an enzyme that repairs the DNA damage inflicted by alkylating agents like TMZ and nitrosourea-based chemotherapies. MGMT promoter methylation is associated with much better response to TMZ.45 In several phase III trials utilizing TMZ for newly diagnosed glioblastoma, including EORTC/NCIC 26981-22981/CE3, RTOG 0525, and RTOG 0825, the median overall survival of patients with MGMT promoter-methylated GBMs ranged from 21.2 to 23.2 months, compared with 14.0–15.3 months for patients with unmethylated tumors.46–49 In conjunction with the biochemical role MGMT plays in TMZ resistance, these and other data support MGMT promoter methylation as a prognostic and predictive factor associated with favorable TMZ response in patients with previously untreated GBM.
Several ongoing trials in North America and Europe are using MGMT unmethylated status as an inclusion criterion for studies that do not employ TMZ, thereby giving patients who are less likely to benefit from TMZ other options earlier in the course of disease.50 Even if a clinical trial is not an option, TMZ is often withheld from patients with low Karnofsky performance status scores if their GBMs are MGMT unmethylated, since the benefit-to-toxicity ratio may be deemed too low. Conversely, MGMT promoter hypermethylation is used as an inclusion criterion for TMZ-sensitizing strategies in the Alliance A071102 clinical trial combining adjuvant TMZ and veliparib against GBM, as well as for several planned or ongoing National Cancer Therapeutic Network trials. Elderly patients, whose tolerance for TMZ is generally lower, can still be considered candidates for TMZ if their tumors are MGMT-methylated.51–53 If a patient shows radiologic changes while on TMZ that may or may not indicate tumor progression, TMZ will usually be continued so long as the tumor has MGMT promoter methylation.
While not yet prospectively validated, the role of MGMT promoter methylation is also of growing importance in grades II–III IDHwt astrocytomas that have GBM molecular profiles. Among IDHmut grade II gliomas in the EORTC 22033 phase III trial, MGMT promoter methylation was predictive of response to TMZ, but not RT.54 It is worth noting that since the MGMT gene is located on chromosome 10q, and 10q is usually retained in grades II–III gliomas but lost in GBM, there are 2 MGMT alleles in grades II–III tumors but only 1 in GBM. MGMT promoter methylation may therefore need to be more extensive in those lower-grade tumors to have the same suppressive effect on gene expression as in GBM. Regardless, any glioma in which TMZ therapy is being considered must be tested for MGMT promoter methylation.
Histone Mutations in Gliomas
Mutations in H3F3A, encoding a histone H3 variant, are present in some diffusely infiltrative astrocytomas (mutations may also be found in the related HIST1H3B gene in the brainstem). H3-K27M is characteristic of glioma arising in the pons (known as “diffuse intrinsic pontine glioma,” or DIPG), as well as anywhere in the midline (basal ganglia, thalamus, midbrain, and brainstem) of children and adults; in contrast, H3-G34 mutations are more often found in hemispheric high-grade pediatric gliomas.55H3F3A K27M is mutually exclusive with IDHmut, rarely has MGMT promoter methylation, and is associated with aggressive behavior, such that the 2016 WHO classification scheme includes “diffuse midline gliomas with histone H3‐K27M mutation” as a grade IV diagnosis, irrespective of whether or not the tumor shows classic grade IV histologic features like necrosis and microvascular proliferation.1 Since biopsies of midline lesions tend to be small and difficult to interpret histologically, the detection of an H3F3A mutation is very helpful in supporting the clinical and pathologic suspicion of a high-grade infiltrative midline glioma. Of note, H3F3A K27M is also occasionally present in non-infiltrative gliomas and, in this context, does not automatically indicate grade IV behavior.56
Like astrocytomas and oligodendrogliomas, ependymomas arise from glial precursor cells, and hence are also classified as gliomas. Unlike astrocytomas and oligodendrogliomas, ependymomas do not contain IDHmut or 1p/19q codeletion. There are multiple molecular subtypes of ependymomas,57 but 2 that have been proposed to have unfavorable prognoses are (i) posterior fossa ependymomas group A (PF-EPN-A); and (ii) ependymomas with fusions involving v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA). The PF-EPN-A subtype mostly occurs in the cerebellum of young children, and although it does not exhibit recurrent genomic alterations, it is readily identified by a characteristic hypermethylation signature and/or an absence of H3K27me3 immunostaining in tumor cell nuclei.57,58RELA fusions cause increased signaling of nuclear factor-kappaB, are often found in supratentorial ependymomas, and may associate with much more aggressive behavior, independent of histologic grade. The latter subset of ependymomas now have their own WHO designation as “ependymoma, RELA-fusion positive.” 1,57 In addition to facilitating accurate diagnosis and guiding prognosis, molecular classification of ependymoma is beginning to influence choice of treatment, including extent of surgical resection and RT.59
Collectively, these data demonstrate that the molecular characterization of gliomas not only ensures accurate diagnoses, but also provides important prognostic information that can inform treatment decisions.
Embryonal Tumors
Brain tumors with a more primitive histologic appearance, including small nuclei with dense chromatin and sparse cytoplasm, are grouped under the category of “embryonal tumors” (Fig. 2). Embryonal tumors are rare and occur mostly in children. Some of them have only recently been codified as discrete entities by the WHO classification system, and are often very difficult to distinguish from each other histologically. Molecular studies are therefore an indispensable way to ensure that they are not misdiagnosed.
Medulloblastomas
The archetypal embryonal neoplasm is medulloblastoma, a cerebellar malignancy arising most often in children and young adults. While all medulloblastomas are grade IV, there are actually 4 major molecular subtypes, with widely varying behavior, that have warranted their own separate designations in the updated WHO classification system.1,60,61
Wingless (WNT)-driven medulloblastomas are by far the least aggressive (and the least common), and are characterized by mutations in catenin beta-1 (CTNNB1) or, less frequently, adenomatous polyposis coli (APC), as well as monosomy 6. Immunostaining for β-catenin shows nuclear localization in such tumors but can be equivocal and requires molecular confirmation.
Mutations in PTCH1, SMO, and/or SUFU characterize sonic hedgehog (SHH) medulloblastomas, which as a group have a worse prognosis compared with WNT tumors, but are usually less aggressive than non-WNT/non-SHH medulloblastomas. Immunopositivity for both growth factor receptor bound protein 2 associated–binding protein 1 (GAB1) and Yes-associated protein 1 (YAP1) is characteristic of SHH-driven medulloblastomas. SHH medulloblastomas may respond to hedgehog pathway inhibitors, such as vismodegib.62 SHH-driven tumors with TP53 mutations fare much worse than those without, as do SHH tumors with MYC or MYCN amplification and large cell/anaplastic histology.60,63
For diagnostic purposes, the last 2 molecular subtypes of medulloblastoma, Group 3 and Group 4, are combined by the WHO scheme into a single “non-WNT/non-SHH medulloblastoma” entity.1 Tumors in this group are often the most aggressive and metastatic, especially if they have large cell/anaplastic features, contain isochromosome 17q, and have amplification of MYC or MYCN.61 Such molecular data can help physicians determine whether to offer adjuvant chemotherapy after RT in patients who are otherwise considered only average risk (eg, no metastases, no histologic anaplasia).64,65
Atypical Teratoid/Rhabdoid Tumors
Another highly aggressive embryonal tumor, atypical teratoid/rhabdoid tumor, is defined by alterations of either INI1 or, rarely, BRG1.66 Loss of normal integrase interactor 1 (INI1) expression in tumor cell nuclei, demonstrable by immunohistochemistry, is a good surrogate marker of INI1 mutations.67
Primitive Neuroectodermal Tumors
For a long time, the term “primitive neuroectodermal tumor” (PNET) was used to diagnose CNS neoplasms that had embryonal features. But recent molecular studies, including methylation profiling, showed that most PNETs are actually other well-known kinds of tumor, such as GBM.68 Furthermore, new distinct entities have emerged among tumors previously grouped together as PNETs. “Embryonal tumor with abundant neuropil and true rosettes,” “ependymoblastoma,” and “medulloepithelioma” all have similarly poor prognoses, and are all characterized by amplification of the microRNA cluster C19MC. Accordingly, these terms have been merged into a new WHO entity, “embryonal tumor with multilayered rosettes” (ETMR), defined by C19MC amplification.1,69 Molecular diagnostics have defined other tumors formerly termed PNETs, including “CNS neuroblastoma with FOXR2 activation,” “CNS Ewing sarcoma family tumor with CIC alteration,” “CNS high-grade neuroepithelial tumor with MN1 alteration,” and “CNS high-grade neuroepithelial tumor with BCOR alteration.” 68
Other Tumors
Craniopharyngiomas
The sella turcica is a depression in the sphenoid bone that accommodates the pituitary gland. Pituitary adenomas are the most common sellar neoplasm, but a surprising variety of lesions and tumors can arise in this region, including craniopharyngiomas. Locally destructive and recurrent tumors that arise from embryonic pituitary tissue, craniopharyngiomas are divided into adamantinomatous and papillary subtypes, which are usually easy to distinguish from each other but sometimes histologically overlap. Although these subtypes have similar prognoses, molecular diagnostics are still very useful in their workup. Adamantinomatous craniopharyngiomas contain CTNNB1 mutations and/or nuclear localization of β-catenin, both of which help differentiate these tumors from papillary craniopharyngiomas, which are characterized by BRAF V600E.70,71 Several case reports have demonstrated that patients with BRAF-positive papillary craniopharyngiomas may respond to targeted Braf inhibitors71–75; this is currently being investigated in an Alliance cooperative group clinical trial (NCT02114767). CTNNB1 and BRAF screening also help distinguish craniopharyngioma from a nonneoplastic cystic lesion in the sella, Rathke’s cleft cyst.
Meningiomas
Meningiomas are actually more common than gliomas.6 But because they are often treatable with surgical resection and/or RT, they are widely regarded as less problematic than gliomas. Thus, they have received proportionately far less attention and research efforts. But some meningiomas repeatedly grow back, invade the underlying brain and venous sinuses, and require multiple surgical resections and high doses of RT. Such aggressive meningiomas cause serious long-term cognitive decline, and can even be lethal. Despite its relative accessibility in most cases, lack of single-cell infiltration, and nonexistent blood–brain barrier, no effective adjuvant therapy besides RT has yet been discovered for meningiomas.
While histologic grading remains the standard for prognostic stratification of meningiomas, the current WHO scheme fails to accurately predict tumor recurrence, systemic treatment options, or overall prognosis in a large proportion of cases. Massive sequencing endeavors have now identified potentially actionable targets in meningiomas, including oncogenic mutations in SMO, AKT1, and PIK3CA.76–78SMO mutations are characteristic of olfactory groove meningiomas, AKT1 mutations are seen in skull base tumors, and NF2 inactivation is most common in tumors of the cerebral convexities and is present in about half of all RT-induced meningiomas.79 Certain mutations correlate with specific subtypes of meningioma, as BAP1 alterations are often found in rhabdoid tumors, and KLF4 mutations are specific for secretory meningiomas.80,81 A national precision medicine trial is under way to explore the role of targeted therapies in meningioma (Alliance A071401; NCT02523014). TERT promoter mutation and DMD deletion suggest a worse prognosis.82,83 Although high TMB is rare in meningiomas, such tumors may respond to immune checkpoint inhibitors.84
Methylation array-based profiling of meningiomas may ultimately prove superior to light microscopy at determining which meningiomas are more likely to behave aggressively.85,86 Specific copy number alterations have also been proposed as adverse prognostic markers.87 As in gliomas, molecular testing will likely soon become necessary for optimal prognostication and treatment decisions in meningiomas.
Conclusion and Recommendations
Thorough molecular testing is now essential for the care of patients with brain tumors. Appropriate application of these testing strategies provides enhanced diagnostic and prognostic accuracy, defines targetable alterations, and improves outcomes (Fig. 3).
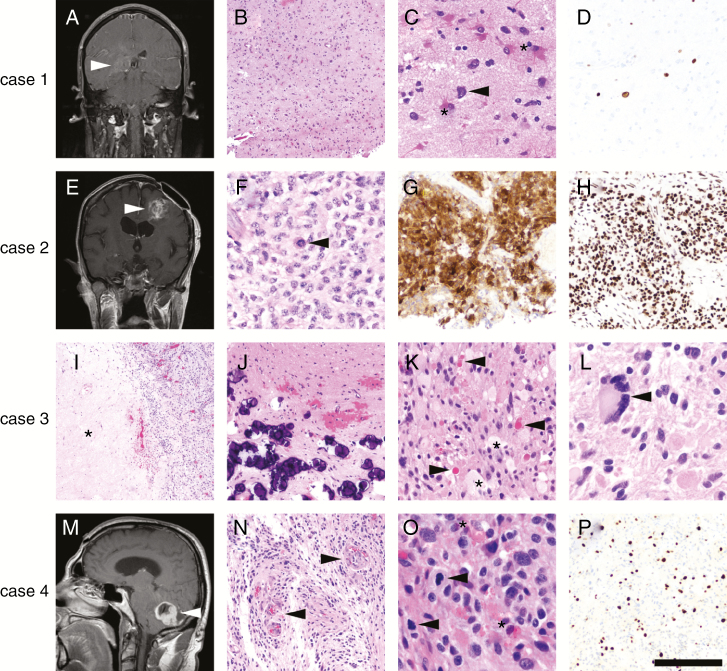
Fig. 3.
Examples of the impact of advanced molecular testing in brain tumors. Case 1 was a 44-year-old man with an ill-defined enhancing lesion in the right deep temporal lobe (A, arrowhead). A stereotactic biopsy contained moderately hypercellular brain tissue (B), composed mostly of reactive astrocytes (C, asterisks) and cells with highly atypical nuclei (C, arrowhead). No mitoses, necrosis, or microvascular proliferation was present in the biopsied material. Ki67 immunostain (D) showed only scattered positive cells. NGS detected mutations in TP53, PTEN, and the TERT promoter, leading to the diagnosis of GBM and sparing the patient a repeat surgery to obtain histologically diagnostic tissue. The patient was treated with RT/TMZ and survived less than a year after diagnosis. Case 2 was a 50-year-old woman in whom was histologically diagnosed a left frontal WHO grade II diffuse astrocytoma 10 years ago at an outside hospital (OSH), who now presented with recurrence and new enhancement (E, arrowhead). Histologically, the recurrent tumor was high grade with necrosis and microvascular proliferation (not shown), as well as mitoses (F, arrowhead) and nuclei that appeared more astrocytic than oligodendroglial. An immunostain for IDH1 R132H was positive (G), yet nuclear ATRX expression was retained (H). NGS confirmed the IDH1 R132H mutation and detected a TERT promoter mutation, as well as 1p/19q codeletion. The tumor was therefore reclassified as anaplastic oligodendroglioma, WHO grade III. The patient responded well to RT and PCV, with no recurrences 1 year later. Case 3 was a pathology consult from an OSH, consisting of a right parietal mass in a 25-year-old man (no radiology available). Histologically the tumor had necrosis (I, asterisk) and mitoses (not shown), but also had abundant mineralization (J), eosinophilic granular bodies (K, arrowheads), foamy lipidized cells (K, asterisks), and highly atypical cells, including multinucleated giant cells (L, arrowhead). The OSH diagnosed the tumor as a GBM, because the screening for BRAF V600E was negative. However, NGS detected a rare oncogenic variable lymphocyte receptor insertion between codons 506 and 507 of BRAF, previously described in Langerhans cell histiocytosis.88 No other alterations were detected, so the tumor was reclassified as anaplastic pleomorphic xanthoastrocytoma, WHO grade III. The patient had already been treated at the OSH with RT/TMZ based on the original GBM diagnosis, but was still alive 2 years later with no recurrences. Case 4 was a 50-year-old man with an enhancing, well-circumscribed cerebellar mass (M, arrowhead) containing microvascular proliferation (N, arrowheads), numerous mitoses (O, arrowheads), eosinophilic granular bodies (O, asterisks), and a high Ki67 proliferation index (P), but no necrosis or Rosenthal fibers. This tumor was originally diagnosed at the OSH as a GBM and treated with RT/TMZ, but NGS (performed several years later) detected an FGFR1 mutation and CDKN2A deletion, changing the diagnosis to an anaplastic pilocytic astrocytoma.12–15 The patient survived 7 years before the tumor recurred, and is still alive 2 years after re-resection (9 years total). Scale bar = 500 microns in I; 200 microns in B, G, H, J, N, P; 100 microns in C, D, K; 50 microns in F, L, O.
The combination of an NGS panel plus genome-wide scan for copy number variations (CNVs) is the ideal approach for the majority of gliomas and embryonal tumors, as well as for cases suspected of being metastatic tumors. Immunostains linked with specific molecular alterations are also helpful. In the next few years, the methylation 850K array and classifier might replace genome-wide CNV platforms, since methylation 850K generates genomic CNV data plus an entirely new dimension of epigenomic information on tumor subtype and tissue of origin (see Supplementary text). MGMT promoter methylation is still required for high-grade gliomas, but traditional MGMT-specific testing could be replaced with broader methylation profiling. When tissue and tumor cellularity are sparse, single-target molecular testing like fluorescence in situ hybridization (FISH), Sanger sequencing, and pyrosequencing are the next best options. Since these tissues are precious resources for patient care, the choice of tests and immunostains should be determined by highly experienced neuropathologists.
As we have demonstrated, advanced molecular diagnostics are now required to accurately guide complex, multimodal integrated brain tumor therapy. The aggregate cost of NGS and CNV or methylation arrays is less than a comparable series of older tests, including single-gene sequencing, FISH, and immunohistochemical analyses. It is also far less expensive than surgery, RT, or even a single MRI scan. Yet, while insurers routinely cover older methods of testing and all the other required elements of patient care, they are often reluctant to cover advanced molecular diagnostics. But these new molecular tools produce more accurate diagnostic and prognostic information, reduce the use of inappropriate treatment regimens, and minimize the exposure of patients to unnecessary life-altering morbidity (and sometimes even mortality) from such treatment. They are also indispensable for discovering targetable mutations and fusions, especially in uncommon variants of common CNS tumors (eg, BRAF mutation in GBM). Molecular diagnostics help stimulate new drug development, and identify patients likely to benefit from new pan-cancer clinical trials aimed at specific genetic lesions, rather than at tumor histology or tissue of origin.
Advanced molecular testing can directly lead to tangible improvements in the diagnosis, prognosis, and prediction of therapeutic response for patients with brain tumors. Leaders in the brain tumor community, including physicians and patient advocacy groups, can help by petitioning the National Cancer Institute, the Food and Drug Administration, and insurance payors to provide better coverage for these powerful new molecular diagnostic tests.
Conflict of interest statement. C.H. is a consultant for Abbvie, Eisai, and Kiyatec. K.L.L. holds patents on diagnostic methods through DFCI and is a consultant for BMS. P.B. is a consultant for Angiochem, Genentech-Roche, Lilly, and Tesaro, has received speaker’s honoraria from Genentech-Roche and Merck, and has received grant support to Massachusetts General Hospital from Merck and Pfizer. L.B.N. serves on the scientific advisory board for Abbvie, Blue Earth Diagnostics, Kiyatec, and Karyopharm, and on the data safety and monitoring board for Ziopharm. E.G. is a consultant for Oncorus, Celgene, and Genentech/Roche (honoraria to the Institution) and has received research funding from Genentech/Roche, BMS, Merck, and Tracon.
Authorship statement. C.H. and J.N.S. wrote and edited the manuscript. K.L.L., P.B., J.T.H., M.V., S.C., J.B., T.C., R.B.J., C.G., L.B.N., P.Y.W., K.J.A., R.V.L., E.G., C.G.E., and D.J.B. edited the manuscript.
Funding
This work was supported by NIH grants R01NS102669 (to C.H.), P50CA221747 (C.H. and R.V.L.), P50CA165962 (K.L.L.), and R01CA188288 (K.L.L.).
References
- 1. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Allegretti M, Fabi A, Buglioni S, et al. Tearing down the walls: FDA approves next generation sequencing (NGS) assays for actionable cancer genomic aberrations. J Exp Clin Cancer Res. 2018;37(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lemery S, Keegan P, Pazdur R. First FDA approval agnostic of cancer site—when a biomarker defines the indication. N Engl J Med. 2017;377(15):1409–1412. [DOI] [PubMed] [Google Scholar]
- 4. van Nimwegen KJ, van Soest RA, Veltman JA, et al. Is the $1000 genome as near as we think? A cost analysis of next-generation sequencing. Clin Chem. 2016;62(11):1458–1464. [DOI] [PubMed] [Google Scholar]
- 5. NCCN. Central nervous system cancers. NCCN Clinical Practice Guidelines in Oncology. 2019. https://www.nccn.org/professionals/physician_gls/pdf/cns.pdf. Accessed January 5, 2019. [Google Scholar]
- 6. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017;19(Suppl 5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horbinski C, Hamilton RL, Lovell C, Burnham J, Pollack IF. Impact of morphology, MIB-1, p53 and MGMT on outcome in pilocytic astrocytomas. Brain Pathol. 2010;20(3):581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horbinski C, Hamilton RL, Nikiforov Y, Pollack IF. Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathol. 2010;119(5):641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horbinski C, Nikiforova MN, Hagenkord JM, Hamilton RL, Pollack IF. Interplay among BRAF, p16, p53, and MIB1 in pediatric low-grade gliomas. Neuro Oncol. 2012:5:777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horbinski C. To BRAF or not to BRAF: is that even a question anymore? J Neuropathol Exp Neurol. 2013;72(1):2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodriguez FJ, Brosnan-Cashman JA, Allen SJ, et al. Alternative lengthening of telomeres, ATRX loss and H3-K27M mutations in histologically defined pilocytic astrocytoma with anaplasia. Brain Pathol. 2019;29(1):126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reinhardt A, Stichel D, Schrimpf D, et al. Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol. 2018;136(2):273–291. [DOI] [PubMed] [Google Scholar]
- 13. Jones DT, Hutter B, Jäger N, et al. ; International Cancer Genome Consortium PedBrain Tumor Project Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Becker AP, Scapulatempo-Neto C, Carloni AC, et al. KIAA1549: BRAF gene fusion and FGFR1 hotspot mutations are prognostic factors in pilocytic astrocytomas. J Neuropathol Exp Neurol. 2015;74(7):743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ballester LY, Penas-Prado M, Leeds NE, Huse JT, Fuller GN. FGFR1 tyrosine kinase domain duplication in pilocytic astrocytoma with anaplasia. Cold Spring Harb Mol Case Stud. 2018;4(2):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. [DOI] [PubMed] [Google Scholar]
- 17. Vaubel RA, Caron AA, Yamada S, et al. Recurrent copy number alterations in low-grade and anaplastic pleomorphic xanthoastrocytoma with and without BRAF V600E mutation. Brain Pathol. 2018;28(2):172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaley T, Touat M, Subbiah V, et al. BRAF inhibition in BRAF(V600)-mutant gliomas: results from the VE-BASKET study. J Clin Oncol. 2018;23:3477–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sievert AJ, Lang SS, Boucher KL, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A. 2013;110(15):5957–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van den Bent MJ, Baumert B, Erridge SC, et al. Interim results from the CATNON trial (EORTC study 26053-22054) of treatment with concurrent and adjuvant temozolomide for 1p/19q non-co-deleted anaplastic glioma: a phase 3, randomised, open-label intergroup study. Lancet. 2017;390(10103):1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horbinski C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol. 2013;125(5):621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olar A, Wani KM, Alfaro-Munoz KD, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol. 2015;129(4):585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shirahata M, Ono T, Stichel D, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018;136(1):153–166. [DOI] [PubMed] [Google Scholar]
- 26. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horbinski C, Kofler J, Yeaney G, et al. Isocitrate dehydrogenase 1 analysis differentiates gangliogliomas from infiltrative gliomas. Brain Pathol. 2011;21(5):564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fuller CE, Schmidt RE, Roth KA, et al. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/astrocytic features. J Neuropathol Exp Neurol. 2003;62(11):1118–1128. [DOI] [PubMed] [Google Scholar]
- 29. Buckner JC, Shaw EG, Pugh SL, et al. Radiation plus procarbazine, CCNU, and vincristine in low-grade glioma. N Engl J Med. 2016;374(14):1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cairncross JG, Wang M, Jenkins RB, et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol. 2014;32(8):783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas AA, Abrey LE, Terziev R, et al. Multicenter phase II study of temozolomide and myeloablative chemotherapy with autologous stem cell transplant for newly diagnosed anaplastic oligodendroglioma. Neuro Oncol. 2017;19(10):1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sahm F, Reuss D, Koelsche C, et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol. 2014;128(4):551–559. [DOI] [PubMed] [Google Scholar]
- 33. Halani SH, Yousefi S, Vega JV, et al. Multi-faceted computational assessment of risk and progression in oligodendroglioma implicates NOTCH and PI3K pathways. NPJ Precis Oncol. 2018;2(24):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen H, Thomas C, Munoz FA, et al. Polysomy is associated with poor outcome in 1p19q co-deleted oligodendroglial tumors. Neuro Oncol. 2019;29:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Snuderl M, Eichler AF, Ligon KL, et al. Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin Cancer Res. 2009;15(20):6430–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136(5):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frattini V, Trifonov V, Chan JM, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45(10):1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138(Pt 6):1613–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weller M, Butowski N, Tran DD, et al. ; ACT IV trial investigators Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18(10):1373–1385. [DOI] [PubMed] [Google Scholar]
- 40. Raizer JJ, Abrey LE, Lassman AB, et al. ; North American Brain Tumor Consortium A phase II trial of erlotinib in patients with recurrent malignant gliomas and nonprogressive glioblastoma multiforme postradiation therapy. Neuro Oncol. 2010;12(1):95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raizer JJ, Giglio P, Hu J, et al. ; Brain Tumor Trials Collaborative A phase II study of bevacizumab and erlotinib after radiation and temozolomide in MGMT unmethylated GBM patients. J Neurooncol. 2016;126(1):185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lassman AB, van den Bent MJ, Gan HK, et al. Safety and efficacy of depatuxizumab mafodotin + temozolomide in patients with EGFR-amplified, recurrent glioblastoma: results from an international phase I multicenter trial. Neuro Oncol. 2019;21(1):106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Di Stefano AL, Fucci A, Frattini V, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21(14):3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ellison DW, Hawkins C, Jones DTW, et al. cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600E mutation. Acta Neuropathol. 2019;137(4):683–687. [DOI] [PubMed] [Google Scholar]
- 45. Hegi ME, Genbrugge E, Gorlia T, et al. MGMT promoter methylation cutoff with safety margin for selecting glioblastoma patients into trials omitting temozolomide. a pooled analysis of four clinical trials. Clin Cancer Res. 2019;25(6):1809–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gilbert MR, Wang M, Aldape KD, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol. 2013;31(32):4085–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 48. Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 50. Hegi ME, Stupp R. Withholding temozolomide in glioblastoma patients with unmethylated MGMT promoter—still a dilemma? Neuro Oncol. 2015;17(11):1425–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perry JR, Laperriere N, O’Callaghan CJ, et al. ; Trial Investigators Short-course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med. 2017;376(11):1027–1037. [DOI] [PubMed] [Google Scholar]
- 52. Malmström A, Grønberg BH, Marosi C, et al. ; Nordic Clinical Brain Tumour Study Group (NCBTSG) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. [DOI] [PubMed] [Google Scholar]
- 53. Wick W, Platten M, Meisner C, et al. ; NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. [DOI] [PubMed] [Google Scholar]
- 54. Bady P, Kurscheid S, Delorenzi M, et al. The DNA methylome of DDR genes and benefit from RT or TMZ in IDH mutant low-grade glioma treated in EORTC 22033. Acta Neuropathol. 2018;135(4):601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 56. Louis DN, Giannini C, Capper D, et al. cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018;135(4):639–642. [DOI] [PubMed] [Google Scholar]
- 57. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Panwalkar P, Clark J, Ramaswamy V, et al. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 2017;134(5):705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zapotocky M, Beera K, Adamski J, et al. Survival and functional outcomes of molecularly defined childhood posterior fossa ependymoma: cure at a cost. Cancer. 2019;125(11):1867–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123(4):473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robinson GW, Orr BA, Wu G, et al. Vismodegib exerts targeted efficacy against recurrent sonic hedgehog-subgroup. J Clin Oncol. 2015; 33(24):2646–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhukova N, Ramaswamy V, Remke M, et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J Clin Oncol. 2013;31(23):2927–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ramaswamy V, Remke M, Adamski J, et al. Medulloblastoma subgroup-specific outcomes in irradiated children: who are the true high-risk patients? Neuro Oncol. 2016;18(2):291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ramaswamy V, Remke M, Bouffet E, et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 2016;131(6):821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Frühwald MC, Biegel JA, Bourdeaut F, Roberts CW, Chi SN. Atypical teratoid/rhabdoid tumors—current concepts, advances in biology, and potential future therapies. Neuro Oncol. 2016;18(6):764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Judkins AR. Immunohistochemistry of INI1 expression: a new tool for old challenges in CNS and soft tissue pathology. Adv Anat Pathol. 2007;14(5):335–339. [DOI] [PubMed] [Google Scholar]
- 68. Sturm D, Orr BA, Toprak UH, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Korshunov A, Sturm D, Ryzhova M, et al. Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity. Acta Neuropathol. 2014;128(2):279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Marucci G, de Biase D, Zoli M, et al. Targeted BRAF and CTNNB1 next-generation sequencing allows proper classification of nonadenomatous lesions of the sellar region in samples with limiting amounts of lesional cells. Pituitary. 2015;18(6):905–911. [DOI] [PubMed] [Google Scholar]
- 71. Brastianos PK, Taylor-Weiner A, Manley PE, et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nat Genet. 2014;46(2):161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Himes BT, Ruff MW, Van Gompel JJ, et al. Recurrent papillary craniopharyngioma with BRAF V600E mutation treated with dabrafenib: case report. J Neurosurg. 2018;1:1–5. [DOI] [PubMed] [Google Scholar]
- 73. Roque A, Odia Y. BRAF-V600E mutant papillary craniopharyngioma dramatically responds to combination BRAF and MEK inhibitors. CNS Oncol. 2017;6(2):95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Aylwin SJ, Bodi I, Beaney R. Pronounced response of papillary craniopharyngioma to treatment with vemurafenib, a BRAF inhibitor. Pituitary. 2016;19(5):544–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brastianos PK, Shankar GM, Gill CM, et al. Dramatic response of BRAF V600E mutant papillary craniopharyngioma to targeted therapy. J Natl Cancer Inst. 2016;108(2):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016;18(5):649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sahm F, Toprak UH, Hübschmann D, et al. Meningiomas induced by low-dose radiation carry structural variants of NF2 and a distinct mutational signature. Acta Neuropathol. 2017;134(1):155–158. [DOI] [PubMed] [Google Scholar]
- 80. Shankar GM, Abedalthagafi M, Vaubel RA, et al. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol. 2017;19(4):535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125(3):351–358. [DOI] [PubMed] [Google Scholar]
- 82. Sahm F, Schrimpf D, Olar A, et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. 2016;108(5):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Juratli TA, McCabe D, Nayyar N, et al. DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome. Acta Neuropathol. 2018;136(5):779–792. [DOI] [PubMed] [Google Scholar]
- 84. Dunn IF, Du Z, Touat M, et al. Mismatch repair deficiency in high-grade meningioma: a rare but recurrent event associated with dramatic immune activation and clinical response to PD-1 blockade. JCO Precis Oncol. 2018;doi: 10.1200/PO.1218.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–694. [DOI] [PubMed] [Google Scholar]
- 86. Olar A, Wani KM, Wilson CD, et al. Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 2017;133(3):431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Aizer AA, Abedalthagafi M, Bi WL, et al. A prognostic cytogenetic scoring system to guide the adjuvant management of patients with atypical meningioma. Neuro Oncol. 2016;18(2):269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Héritier S, Hélias-Rodzewicz Z, Chakraborty R, et al. New somatic BRAF splicing mutation in Langerhans cell histiocytosis. Mol Cancer. 2017;16(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]