ABSTRACT
Regeneration has fascinated scientists since well before the 20th century revolutions in genetics and molecular biology. The field of regenerative biology has grown steadily over the past decade, incorporating advances in imaging, genomics and genome editing to identify key cell types and molecules involved across many model organisms. Yet for many or most tissues, it can be difficult to predict when and how findings from these studies will advance regenerative medicine. Establishing technologies to stimulate regrowth of a lost or amputated limb with a patterned replicate, as salamanders do routinely, is one of the most challenging directives of tissue regeneration research. Here, we speculate upon what research avenues the field must explore to move closer to this capstone achievement.
Summary: This Spotlight outlines research being carried out towards achieving regrowth of a lost or amputated limb: one of the most challenging objectives of tissue regeneration research.
Limb regeneration starts with a blastema
Traditional laboratory model systems and atypical models alike have informed key concepts and mechanisms of limb regeneration. Human limb regeneration remains a dream of medical science, but it is a pressing and growing need – it has been estimated that 1.6 million people in the USA alone are living with limb loss, with the potential of that number to more than double by the mid-century point (Ziegler-Graham et al., 2008). In considering the possibility of human limb regeneration, we choose to focus our speculative piece on vertebrate models that have regeneration-competent major appendages (e.g. limbs, fins), as well as vertebrates with scant regenerative capacity resembling that of humans. The former can be interrogated for information on the molecular underpinnings of successful innate events, while the latter represent a canvas upon which manipulations, often informed by studies of the former, can push the limits of regenerative capacity and open doors to medical applications. We will primarily consider regeneration of hindlimbs, forelimbs and their equivalents. A striking example of mammalian appendage regeneration, deer antler renewal, does not restore a replicate of the original appendage and responds to hormone levels rather than to injury, and is reviewed elsewhere (Kierdorf et al., 2007). Regeneration of internal tissues such as liver and intestinal lining, and attempts to augment regeneration in poorly regenerative tissues other than limbs, have been well summarized in other reviews (Iismaa et al., 2018; Tzahor and Poss, 2017).
Teleost fish like zebrafish and urodele salamanders such as newts and axolotls (Box 1) are capable of complete regeneration after fin or limb amputation. Vertebrate appendages are complex structures composed of interdependent tissues. Thus, faithful regeneration involves not only replenishing the mass of large structures such as bone and skin, but also precise vascularization and innervation to achieve proper metabolism and function. Adult Xenopus limb amputation partially restores limb mass in the form of a cartilaginous spike, but fails to reconstitute morphology and function (Dent, 1962). These failures elevate the virtues of models like teleosts and newts, the impressive regenerative capabilities of which depend on a structure called a blastema: a transient mass of proliferative cells that forms at the site of injury and provides the source material for eventual regeneration of lost structures. The blastema has been referred to as the link between healing, a universal response to damage, and morphogenesis of a new appendage, which is the exclusive domain of a subset of species with elevated regenerative abilities (Seifert and Muneoka, 2018).
Box 1. The salamander limb: the right model at the right time – a personal case study by Maximina Yun.
While finishing my graduate studies at Cambridge University, I was invited to teach undergraduates in cell and developmental biology. This somewhat serendipitous turn took me on a journey of discoveries in the field of regeneration, and exposed me for the first time to salamanders, the vertebrates evolutionarily closest to us that are able to regenerate full limbs as adults. Ever since then I have shared with many of my scientific muses, from Thomas H. Morgan to Victor C. Twitty, a fascination for this animal and the will to uncover the molecular and cellular basis of regeneration. The latter became my primary scientific aim, driving my subsequent studies and career moves – something that, at the time, some considered a commendable yet daunting prospect. As it was bluntly put to me by a well-respected scientist during my first interview for a group leader position, ‘salamanders have been used in regeneration studies for many decades; however, significant translational progress based on this model is yet to be seen’.
Perhaps he was right then, given the ingeniously crafted yet limited tools available in the system. But a mere 5 years later, and the tide has turned. Transgenic techniques have become established, allowing crucial functional studies; advanced imaging approaches are being used for in vivo analysis of progenitor behaviors; and the giant genomes for two experimental salamander models, the axolotl and the Spanish ribbed newt, have recently become available, with all the wealth of experimental and theoretical approaches this implies (Elewa et al., 2017; Nowoshilow et al., 2018). Thus, the salamanders are finally catching up with more conventional research models such as zebrafish. And while they do so, they bring a great deal of interesting biology – besides regeneration – to the table.
And now, every day I spend in my lab I marvel at how these techniques are revolutionizing salamander studies and speeding up our understanding of the choreography of molecular and cellular events that result in the regeneration of a complex structure such as a limb. In particular, they are enabling our investigations into important issues such as the nature of the mechanisms controlling cellular plasticity in regeneration, the role of the immune-senescent cell interplay in this process and the basis for interspecies differences in regenerative capacity (Czarkwiani and Yun, 2018; Yun et al., 2015). Thus, I believe we are at an inflexion point in regeneration research, as technical advances have finally emerged to allow us to unlock the mysteries of the extreme regenerative abilities found in these captivating animals.
A wound epidermis forms after injury and acts as a signaling hub for blastema formation; without these signals, mesenchymal cells at the injury site fail to assemble the blastema (Tassava and Garling, 1979; Thornton, 1957). The structure and composition of the wound epidermis are particularly important for successful regeneration, as apico-basal polarization of wound epidermal cells is essential for proper expression and delivery of regenerative signaling factors (Chablais and Jazwinska, 2010; Chen et al., 2015; Shibata et al., 2016; Stoick-Cooper et al., 2007). Molecules distinguishing an activated regeneration epidermis have been identified through diverse experimental methods, including yeast two-hybrid assay screens, forward genetic screens and candidate gene approaches. These approaches have identified factors ranging from signaling pathway members such as Fgf20a to extracellular matrix components and regulators such as laminin β1a and newt anterior gradient protein (Chen et al., 2015; Kumar et al., 2007; Shibata et al., 2016; Whitehead et al., 2005). Yet we are far from fully comprehending the breadth, dynamics and relative importance of regeneration factors. Moving forward, recent technologies such as single cell transcriptome sequencing will help refine the genetic makeup of the wound epidermis and elaborate its crucial role as a structural and signaling center for the blastema (Aztekin et al., 2019).
Advocating developmental biology.
This article is part of Development's advocacy collection – a series of review articles that make compelling arguments for the field's importance. The series is split into two: one set of articles, including this one, addresses the question ‘What are the big open questions in the field?’ We would argue that there has never been a more exciting time to get involved in developmental biology: incredible new tools mean making fundamental problems are increasingly within reach. A complementary set of articles will ask ‘What has developmental biology ever done for us?’ Together, the articles will provide a collection of case studies looking backwards to the field's achievements and forwards to its potential, and a resource for students, educators, advocates and researchers alike. To see the full collection as it grows, go to http://dev.biologists.org/content/advocating-developmental-biology.
Understanding the origins and fates of blastemal progenitor cells is a key step toward achieving limb regeneration. No vertebrate has yet been shown to engage a true pluripotent stem cell during regeneration; rather, fate-mapping experiments indicate that appendage blastemas are composed of restricted progenitor cell populations, each responsible for one or a small number of cell types (Ando et al., 2017; Fei et al., 2017; Gerber et al., 2018; Kragl et al., 2009; Lehoczky et al., 2011; Rinkevich et al., 2011; Shibata et al., 2018; Tu and Johnson, 2011). Notably, recent studies also indicate that when one progenitor population is experimentally eliminated, other progenitors can be recruited or repurposed as an ancillary source. For example, bone-forming osteoblasts normally dedifferentiate – that is, reduce their characteristic functional properties – and divide to contribute new osteoblasts in zebrafish fins, but if osteoblasts are ablated, joint-associated progenitors ramp up osteoblast production (Ando et al., 2017; Singh et al., 2012). Like osteoblasts, cardiomyocytes of zebrafish can and do re-enter the cell cycle to serve as progenitors (Knopf et al., 2011; Singh et al., 2012; Sousa et al., 2011). This strategy is also found among salamanders: satellite cells serve as progenitors in axolotl blastemas, whereas myofibers dedifferentiate and fragment in adult newt limb stumps to facilitate muscle regeneration (Sandoval-Guzmán et al., 2014; Tanaka et al., 2016). In summary, appendage regeneration does not employ a pluripotent stem cell, but system plasticity can enable multiple origins for mature tissues – a concept relevant to human limb regeneration.
Understanding how individual blastema cells behave in concert to create population-level morphogenetic changes is a technically challenging obligation of the field. Recent studies have improved the spatiotemporal resolution of regeneration by combining longitudinal imaging platforms with techniques to permanently label cells, before or during blastema formation. For example, genetic cell labeling has been used to detail the recruitment of connective tissue cells to form the blastema, as well as the extent to which these cells build different proximodistal skeletal elements (Currie et al., 2016). Real-time live imaging is being extended to follow differentiated cell populations and catalog cell shape changes, proliferation dynamics and cell death rates. Improvements in platforms that maintain animal health and stability for long periods during imaging, as well as longitudinal studies using permanent lineage tracing, have enabled capture of novel biology during appendage regeneration in fish and axolotls (Cox et al., 2018; Currie et al., 2016; Tornini et al., 2016; Xu et al., 2015). Community initiatives will be key to efficiently generate knock-in fluorescent fusion proteins, thereby allowing visualization of endogenous activity of molecular regulators. Mapping spatiotemporal changes in cell behavior in tandem with assessment of dynamic gene expression can illuminate the blueprints for appendage regeneration.
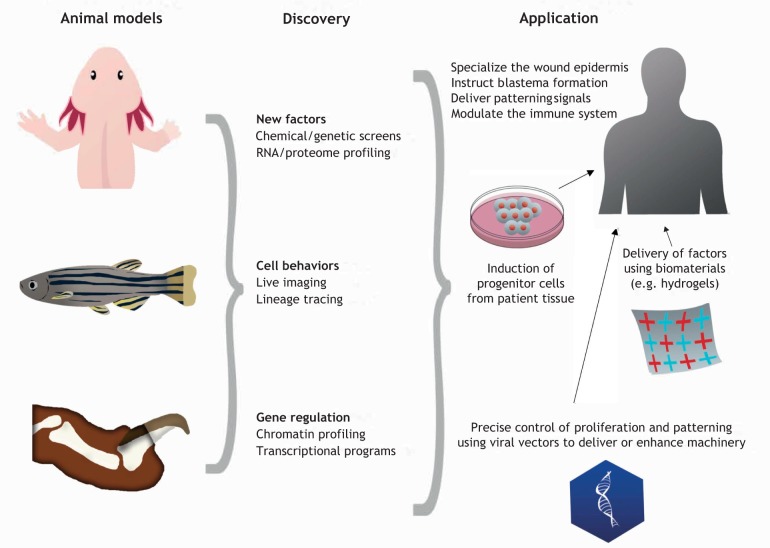
Induction of a mammalian limb blastema is a major challenge to consider at present, requiring methods to activate latent progenitor populations (if they exist) to stimulate dedifferentiation of cells in spared tissue to increase proliferative capacity or modify fate, and/or to deliver exogenous progenitor cells that will colonize the limb stump and form a functional structure. There is evidence that transplantation of embryonic limb cells enhances regeneration from adult Xenopus laevis limb stumps; thus, it is conceivable that transplantation of induced pluripotent stem cell-derived limb progenitors could facilitate limb regeneration (Lin et al., 2013). Alternatively, one can envisage clinical regeneration initiated by removing skin from the limb stump and targeting delivery of factors unique to a functional regeneration epidermis (Dawson et al., 2017; Seyedhassantehrani et al., 2017). Blastema formation could be motivated in turn by delivery of additional factors that might induce dedifferentiation and proliferation in the remaining tissue, and/or by transplantation of the proper distribution of patient-derived cells representing the precursors of all necessary cell types (Fig. 1). Given that it is possible for a blastema to form in certain mammalian contexts like mouse digit tips (discussed in the following section), the prospect of inducing blastema formation in other weakly or non-regenerative mammalian tissues seems not so far-fetched.
Fig. 1.
From highly regenerative model systems to regenerative therapies. A possible pipeline for how discovery in lab model systems can influence applications for regenerative therapies of complex structures like limbs.
Instructing regeneration with pro-regenerative factors
Salamander blastemas contain all of the positional information necessary to pattern new limbs; i.e. a shoulder blastema transplanted to a limb will generate a full limb, and a transplanted wrist blastema will form distal components (Maden, 1980; Pescitelli and Stocum, 1980; Roensch et al., 2013). It is possible that a perfected therapeutic blastema will self-pattern, although it seems more likely that this striking feature would have to be replicated through additional manipulations. For example, salamander limb regeneration depends on re-innervation of axons severed during amputation; without nerves, the stump generates a blastema with reduced proliferation and differentiation capacities (Mescher and Tassava, 1975; Singer and Craven, 1948). Molecular genetic experiments have identified molecular factors accounting for the nerve dependence of regeneration in salamanders and Xenopus; hyperinnervation has been shown to have small positive effects on regeneration in adult Xenopus (Kumar et al., 2007; Zhang et al., 2018). Additionally, an ectopic blastema – and eventual limb – can be induced to sprout from an unamputated limb by creating a small skin wound, deviating a nerve to the site and applying a skin graft to provide positional information (Endo et al., 2004; Seyedhassantehrani et al., 2017). This accessory limb model has been used to model how factors encode positional information or substitute for innervation (Makanae et al., 2014; Nacu et al., 2016; Vieira et al., 2019). It has also highlighted the role of extracellular factors such as heparan sulfate in patterning (Phan et al., 2015). These discoveries offer proof of concept for the hypothesis that the environment necessary for regeneration can be recreated by manipulation of sources of signaling molecules.
Many classic developmental signaling pathways are known to play roles in patterning regenerating appendages (Currie et al., 2016; Dawson et al., 2017; Grotek et al., 2013; Kujawski et al., 2014; Whitehead et al., 2005). Broad similarities between ontogenetic and regenerative development led to the initial compilation of molecular factors that influence regeneration. The more recent generation of experiments has incorporated unbiased surveys of regenerating adult tissues or proxies for these tissues. Lineage tracing and single cell sequencing studies have highlighted that it is an oversimplification to assume that adult tissue regeneration merely recapitulates a genetic program of earlier development (Gerber et al., 2018; Tsai et al., 2019). These surveys have broadened the scope of factor discovery, e.g. implicating melanocortin 4 receptor (Mc4r) signaling, which is classically studied in energy metabolism, as a factor that can augment limb regeneration in Xenopus during stages in which it is typically defective (Zhang et al., 2018). Transcriptomic analysis of entire tissues and single cells has been employed in appendage regeneration in a variety of species to reveal candidates based on gene expression dynamics (Leigh et al., 2018; Nachtrab et al., 2013). Libraries of transcripts can in many cases be functionally assayed in cell culture; e.g. successive subfractionation of pooled cDNAs capable of inducing cell cycle entry in cultured salamander cells recently identified MARCKS-like protein as a factor with in vivo mitogenic properties in limb tissues (Sugiura et al., 2016).
Contrasting the effects of factors on poorly versus effectively regenerating tissues or species can yield insights into how genetic responses to injury may differ among vertebrates. For example, cell culture experiments have demonstrated that regeneration-competent myotubes from newts increase ERK signaling and re-enter the cell cycle upon serum stimulation, whereas mouse myotubes are unable to sustain ERK signaling or cell cycle entry, even when harboring mutations that constitutively activate the ERK pathway (Yun et al., 2014). These contrasting responses imply that there is complex genetic and epigenetic regulation of regeneration genes, which we discuss in the next section; as a result, individual molecules may be potent, but not sufficient to induce complete regeneration. Relief of cell cycle inhibition is also closely tied to regeneration competence: p53 activity is dampened in the salamander blastema (Yun et al., 2013). Likewise, the mouse Acomys cahirinus is able to regenerate hole punches in ear tissue, and cell cycle re-entry correlates with a lack of nuclear localization of the cell cycle inhibitors p21 and p27 in the ear hole blastema. By contrast, common mice (Mus musculus) maintain each of these inhibitors in muscle nuclei, and studies in cultured mouse myoblasts have found that knockdown of cell cycle regulators can modulate proliferation and indicators of regenerative capacity (Gawriluk et al., 2016; Pajcini et al., 2010; Wang et al., 2015). Thus, it is not just the presence and activity of pro-regenerative factors that determine regenerative competence, but also the attenuation of inhibitory factors. Correspondingly, a balance of triggers and breaks, produced by drug or gene therapy cocktails, is likely to be needed for consideration of human limb regeneration therapies.
The capacity of mouse limb structures to regenerate upon amputation is limited to the digits, specifically the distal half of the third phalangeal element (P3), which consists of bone surrounded by connective tissue and a nail (Seifert and Muneoka, 2018). Human fingertips also have some regenerative capacity, especially in children, so the mouse digit tip model could aid in identifying methods for augmenting limited human regeneration (Dolan et al., 2018; Illingworth, 1974). Immediately anterior to P3 in the mouse is the second phalangeal element (P2), which does not regenerate but instead truncates bone and heals skin upon amputation. Treatment with bone morphogenetic protein (Bmp) family members has been shown to induce regeneration of the P2 bone to its original length after amputation, but the regenerated region lacked hair follicles and articular cartilage, and did not initiate regeneration of the distal P3 segment (Dawson et al., 2017). Furthermore, P2 digits that were amputated and failed to regenerate could be induced to undergo modest regeneration by re-amputation and Bmp pathway activation. Similarly, while Acomys species are able to regenerate small full-thickness skin wounds, including hair follicles, common mice normally fail to regenerate hair follicles after this injury but can be induced to do so by Hedgehog signaling activation (Seifert et al., 2012). Thus, a limited number of molecules can potentially initiate an otherwise failed regenerative response, and the regeneration window can be reopened through a second injury and properly timed application of regeneration factors. The latter is an important practical consideration, as it might not be feasible to perform a clinical limb regeneration procedure immediately upon injury.
Recent innovative approaches combining molecular factors with bioengineering methodology have improved Xenopus limb regeneration beyond a cartilaginous spike. Slack and colleagues regenerated a forelimb containing multiple digits with partial mineralization by application of a fibrin patch containing larval limb progenitor cells and supplemented with additional growth factors (Lin et al., 2013). Levin and colleagues developed a wearable bioreactor that delivered progesterone in the 24 h after amputation: this induced regeneration of an appendage that lacked bone and digits but was thicker, contained more nerve bundles and vasculature than controls, and had an improved range of motion (Herrera-Rincon et al., 2018). Other engineering methods of drug and factor delivery, including the use of hydrogels, present interesting possibilities, as their protean composition could allow the creation of microenvironments for multiple cell types, thereby reproducing the different conditions amenable to formation of cartilage, bone and other tissue types over the course of regeneration (Moreira Teixeira et al., 2014). Hydrogels made from either naturally occurring molecules or synthetic polymers engineered for breakdown by naturally occurring enzymes could possibly be supplemented with regeneration-promoting factors and seeded with cells to mimic a regeneration blastema (Fig. 1). Furthermore, the immune system is a target of many current studies in tissue regeneration research, and it has become clear that there is important interplay between cells like macrophages and regulatory T cells with the regeneration machinery, ostensibly via both phagocytitic and paracrine mechanisms (Godwin et al., 2013; Hui et al., 2017; Mescher, 2017; Simkin et al., 2017). Immunomodulatory and immunoengineering approaches are thus of significant interest for the field of appendage regeneration, as they are for other fields.
Engineering and manipulation of pro-regenerative factors have been shown to significantly stimulate regeneration in multiple contexts, and CRISPR/Cas9 will likely accelerate the discovery of new pro-regenerative factors. However, the appendages regenerated in these studies typically fall short of the originals in form and function. Although a perfect replicate of a lost limb is the gold standard for regenerative therapy, this will not be achieved in the earliest cases of clinical limb regeneration. Thus, as an aside, there are likely to be many years during which patients balance the tradeoffs of mini-limbs or missing digits with prosthetics, or during which hybrid alternatives may be favored.
Regeneration is an epigenomic phenomenon
A two-step formula for creating a blastema and instructing its pattern underestimates the complexity of the epigenetic landscape for regeneration. The central questions might be: is the regenerative capacity of a tissue like the limb hard-wired, and have hundreds of millions of years of evolution permanently turned off or removed the machinery in mammals? Or, is the machinery there to be awakened? Recent work indicates that regeneration contexts are associated with large-scale changes in the expression of broad regulators of DNA and histone methylation, as well as histone acetylation (Hirose et al., 2013; Pfefferli et al., 2014; Takayama et al., 2014). These chromatin-level changes are the cause and/or consequence of changes in the expression levels of many hundreds of genes that have been cataloged in various models of appendage regeneration (Bryant et al., 2017; Johnston et al., 2016; Leigh et al., 2018; Rabinowitz et al., 2017). Disrupting the expression dynamics of individual or combinations of genes to find a cocktail capable of inducing regeneration would be a gargantuan undertaking. Alternatively, hacking into the epigenetic regulation to reboot the choreography of gene expression and interactions might represent an efficient shortcut to regeneration, with a large potential payout.
Epigenetic profiling has led to discovery of short DNA elements capable of regulating one or more genes in response to injury and/or regeneration. Studies in amputated zebrafish fins and hearts, fractured mouse bones, and injured Drosophila wing discs have identified sequences that preferentially direct gene expression upon injury and during regeneration, in some cases maintaining activated expression until regeneration is completed (Goldman et al., 2017; Guenther et al., 2015; Harris et al., 2016; Kang et al., 2016). Interestingly, a DNA region near the Drosophila wingless (wg) gene contains a silencing element that increases methylation with development and decreases induction of wg/wnt, supporting a hypothesis that regenerative capacity can be controlled through changes at gene regulatory regions (Harris et al., 2016). Developmental stage-related changes in regeneration competence have been linked to changes in epigenetic regulation of signaling factors in other contexts, such as Shh signaling in Xenopus limb regeneration (Yakushiji et al., 2007). Discovering how these elements are regulated by injury, and how their regulation changes upon recovery of regenerated structures, can ultimately reveal master upstream control mechanisms for tissue regeneration events. For example, a recent study of the acoel worm Hofstenia miamia identified regeneration-responsive chromatin regions that are enriched with binding sites for the early growth response (Egr) gene, implicating Egr as a pioneer factor that regulates gene expression during head or tail regeneration (Gehrke et al., 2019). Advances in genome assembly have paralleled epigenetic profiling and have highlighted features in the genomes of regenerative amphibians that may account for their enhanced capacity, e.g. expansion of transposable elements and genes encoding microRNAs (Elewa et al., 2017; Nowoshilow et al., 2018). These findings emphasize the importance of non-coding elements, some of them likely to be unique to highly regenerative animals, in the orchestration of large-scale changes in gene expression during regeneration.
Understanding the epigenetic control of regeneration may also reveal applications that reduce off-target effects of gene delivery mechanisms to induce regeneration. Gene therapy using non-integrating, non-replicative vectors like adeno-associated viruses has received recent attention as a means to deliver wild-type gene products in models or actual cases of human genetic disease (Amoasii et al., 2018; Mendell et al., 2017). This is effective in particular when the system can tolerate variable doses and tissue location of the therapeutic factor. Gene therapy for a broad application like regeneration is conceivable, although in this case potent developmental factors would be employed to reprogram tissue to a regenerative state. A major concern is therefore restricting the intended effect to the desired tissue for the appropriate length of time. This issue also exists when considering the idea of drugging limb regeneration: it is hard to imagine how one could target regenerative growth and avoid large-scale havoc wreaked by systemic application of potent developmentally interventional compounds.
Regulatory regions that activate exclusively during regeneration suggest methods to resolve this issue. For example, the activity of an enhancer linked to the zebrafish leptin b gene tracks closely with regeneration, with little or no transcriptional activation during animal development or in uninjured adult tissue, but sustained activation of gene expression in regenerating hearts or fins (Kang et al., 2016). This genetic element was used in proof of principle experiments to boost growth factor expression and local tissue regeneration in zebrafish. Conceivably, injury- and regeneration-specific enhancers could be used in adeno-associated viruses to target expression of therapeutic factors to the correct tissues and contexts when attempting to induce regeneration (Fig. 1).
Concluding remarks
Regenerating a human limb is a monumental challenge. The path to this endpoint will be dogged, and success may lie several decades away or more. Discovery science in the field of appendage regeneration has merged classic experimental methods like tissue grafting and genetic screens with modern technologies that deconstruct regeneration to context-dependent behaviors of individual cells, gene products and DNA regulatory sequences. Holistic ideas are also revealing insights into how communication between distant tissues can influence regeneration (Busse et al., 2018; Kang et al., 2013; Rodgers et al., 2014). The new mechanistic vantage points reached by contemporary studies, coupled with the evolution of delivery methods with better-than-surgical precision, will provide beacons for the journey ahead.
Acknowledgements
We thank Nutishia Lee for artwork.
Footnotes
Funding
B.D.C. was supported by a Graduate Research Fellowship (1106401) from the National Science Foundation. M.H.Y. was supported by Deutsche Forschungsgemeinschaft funds (DFG FZ111, DFG EXC168). K.D.P. was supported by a grant (R01 GM074057) from the National Institutes of Health.
References
- Amoasii L., Hildyard J. C. W., Li H., Sanchez-Ortiz E., Mireault A., Caballero D., Harron R., Stathopoulou T.-R., Massey C., Shelton J. M. et al. (2018). Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362, 86-91. 10.1126/science.aau1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K., Shibata E., Hans S., Brand M. and Kawakami A. (2017). Osteoblast production by reserved progenitor cells in zebrafish bone regeneration and maintenance. Dev. Cell 43, 643-650.e643. 10.1016/j.devcel.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Aztekin C., Hiscock T. W., Marioni J. C., Gurdon J. B., Simons B. D. and Jullien J. (2019). Identification of a regeneration-organizing cell in the Xenopus tail. Science 364, 653-658. 10.1126/science.aav9996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D. M., Sousounis K., Payzin-Dogru D., Bryant S., Sandoval A. G. W., Martinez Fernandez J., Mariano R., Oshiro R., Wong A. Y., Leigh N. D. et al. (2017). Identification of regenerative roadblocks via repeat deployment of limb regeneration in axolotls. NPJ Regener. Med. 2, 30 10.1038/s41536-017-0034-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse S. M., McMillen P. T. and Levin M. (2018). Cross-limb communication during Xenopus hindlimb regenerative response: non-local bioelectric injury signals. Development 145, dev164210 10.1242/dev.164210 [DOI] [PubMed] [Google Scholar]
- Chablais F. and Jazwinska A. (2010). IGF signaling between blastema and wound epidermis is required for fin regeneration. Development 137, 871-879. 10.1242/dev.043885 [DOI] [PubMed] [Google Scholar]
- Chen C.-H., Merriman A. F., Savage J., Willer J., Wahlig T., Katsanis N., Yin V. P. and Poss K. D. (2015). Transient laminin beta 1a induction defines the wound epidermis during zebrafish fin regeneration. PLoS Genet. 11, e1005437 10.1371/journal.pgen.1005437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B. D., De Simone A., Tornini V. A., Singh S. P., Di Talia S. and Poss K. D. (2018). In toto imaging of dynamic osteoblast behaviors in regenerating skeletal bone. Curr. Biol. 28, 3937-3947.e3934. 10.1016/j.cub.2018.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie J. D., Kawaguchi A., Traspas R. M., Schuez M., Chara O. and Tanaka E. M. (2016). Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Dev. Cell 39, 411-423. 10.1016/j.devcel.2016.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarkwiani A. and Yun M. H. (2018). Out with the old, in with the new: senescence in development. Curr. Opin. Cell Biol. 55, 74-80. 10.1016/j.ceb.2018.05.014 [DOI] [PubMed] [Google Scholar]
- Dawson L. A., Yu L., Yan M., Marrero L., Schanes P. P., Dolan C., Pela M., Petersen B., Han M. and Muneoka K. (2017). The periosteal requirement and temporal dynamics of BMP2-induced middle phalanx regeneration in the adult mouse. Regeneration 4, 140-150. 10.1002/reg2.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J. N. (1962). Limb regeneration in larvae and metamorphosing individuals of the South African clawed toad. J. Morphol. 110, 61-77. 10.1002/jmor.1051100105 [DOI] [PubMed] [Google Scholar]
- Dolan C. P., Dawson L. A. and Muneoka K. (2018). Digit tip regeneration: merging regeneration biology with regenerative medicine. Stem Cells Transl. Med. 7, 262-270. 10.1002/sctm.17-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elewa A., Wang H., Talavera-López C., Joven A., Brito G., Kumar A., Hameed L. S., Penrad-Mobayed M., Yao Z., Zamani N. et al. (2017). Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat. Commun. 8, 2286 10.1038/s41467-017-01964-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Bryant S. V. and Gardiner D. M. (2004). A stepwise model system for limb regeneration. Dev. Biol. 270, 135-145. 10.1016/j.ydbio.2004.02.016 [DOI] [PubMed] [Google Scholar]
- Fei J.-F., Schuez M., Knapp D., Taniguchi Y., Drechsel D. N. and Tanaka E. M. (2017). Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration. Proc. Natl. Acad. Sci. USA 114, 12501-12506. 10.1073/pnas.1706855114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawriluk T. R., Simkin J., Thompson K. L., Biswas S. K., Clare-Salzler Z., Kimani J. M., Kiama S. G., Smith J. J., Ezenwa V. O. and Seifert A. W. (2016). Comparative analysis of ear-hole closure identifies epimorphic regeneration as a discrete trait in mammals. Nat. Commun. 7, 11164 10.1038/ncomms11164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke A. R., Neverett E., Luo Y. J., Brandt A., Ricci L., Hulett R. E., Gompers A., Ruby J. G., Rokhsar D. S., Reddien P. W. et al. (2019). Acoel genome reveals the regulatory landscape of whole-body regeneration. Science 363, eaau6173 10.1126/science.aau6173 [DOI] [PubMed] [Google Scholar]
- Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S. et al. (2018). Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, eaaq0681 10.1126/science.aaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin J. W., Pinto A. R. and Rosenthal N. A. (2013). Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. USA 110, 9415-9420. 10.1073/pnas.1300290110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman J. A., Kuzu G., Lee N., Karasik J., Gemberling M., Foglia M. J., Karra R., Dickson A. L., Sun F., Tolstorukov M. Y. et al. (2017). Resolving heart regeneration by replacement histone profiling. Dev. Cell 40, 392-404.e395. 10.1016/j.devcel.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotek B., Wehner D. and Weidinger G. (2013). Notch signaling coordinates cellular proliferation with differentiation during zebrafish fin regeneration. Development 140, 1412-1423. 10.1242/dev.087452 [DOI] [PubMed] [Google Scholar]
- Guenther C. A., Wang Z., Li E., Tran M. C., Logan C. Y., Nusse R., Pantalena-Filho L., Yang G. P. and Kingsley D. M. (2015). A distinct regulatory region of the Bmp5 locus activates gene expression following adult bone fracture or soft tissue injury. Bone 77, 31-41. 10.1016/j.bone.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. E., Setiawan L., Saul J. and Hariharan I. K. (2016). Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. eLife 5, e11588 10.7554/eLife.11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Rincon C., Golding A. S., Moran K. M., Harrison C., Martyniuk C. J., Guay J. A., Zaltsman J., Carabello H., Kaplan D. L. and Levin M. (2018). Brief local application of progesterone via a wearable bioreactor induces long-term regenerative response in adult Xenopus hindlimb. Cell Rep. 25, 1593-1609.e1597. 10.1016/j.celrep.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K., Shimoda N. and Kikuchi Y. (2013). Transient reduction of 5-methylcytosine and 5-hydroxymethylcytosine is associated with active DNA demethylation during regeneration of zebrafish fin. Epigenetics 8, 899-906. 10.4161/epi.25653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S. P., Sheng D. Z., Sugimoto K., Gonzalez-Rajal A., Nakagawa S., Hesselson D. and Kikuchi K. (2017). Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659-672.e655. 10.1016/j.devcel.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Iismaa S. E., Kaidonis X., Nicks A. M., Bogush N., Kikuchi K., Naqvi N., Harvey R. P., Husain A. and Graham R. M. (2018). Comparative regenerative mechanisms across different mammalian tissues. NPJ Regener. Med. 3, 6 10.1038/s41536-018-0044-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C. M. (1974). Trapped fingers and amputated finger tips in children. J. Pediatr. Surg. 9, 853-858. 10.1016/S0022-3468(74)80220-4 [DOI] [PubMed] [Google Scholar]
- Johnston A. P. W., Yuzwa S. A., Carr M. J., Mahmud N., Storer M. A., Krause M. P., Jones K., Paul S., Kaplan D. R. and Miller F. D. (2016). Dedifferentiated Schwann cell precursors secreting paracrine factors are required for regeneration of the mammalian digit tip. Cell Stem Cell 19, 433-448. 10.1016/j.stem.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Kang J., Nachtrab G. and Poss K. D. (2013). Local Dkk1 crosstalk from breeding ornaments impedes regeneration of injured male zebrafish fins. Dev. Cell 27, 19-31. 10.1016/j.devcel.2013.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Hu J., Karra R., Dickson A. L., Tornini V. A., Nachtrab G., Gemberling M., Goldman J. A., Black B. L. and Poss K. D. (2016). Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201-206. 10.1038/nature17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf U., Kierdorf H. and Szuwart T. (2007). Deer antler regeneration: cells, concepts, and controversies. J. Morphol. 268, 726-738. 10.1002/jmor.10546 [DOI] [PubMed] [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C. W., Mahatma G., Fisher S., Brand M., Schulte-Merker S. et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713-724. 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Maden M., Epperlein H. H. and Tanaka E. M. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60-65. 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- Kujawski S., Lin W., Kitte F., Börmel M., Fuchs S., Arulmozhivarman G., Vogt S., Theil D., Zhang Y. and Antos C. L. (2014). Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Dev. Cell 28, 573-587. 10.1016/j.devcel.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Kumar A., Godwin J. W., Gates P. B., Garza-Garcia A. A. and Brockes J. P. (2007). Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772-777. 10.1126/science.1147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky J. A., Robert B. and Tabin C. J. (2011). Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. USA 108, 20609-20614. 10.1073/pnas.1118017108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh N. D., Dunlap G. S., Johnson K., Mariano R., Oshiro R., Wong A. Y., Bryant D. M., Miller B. M., Ratner A., Chen A. et al. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun. 9, 5153 10.1038/s41467-018-07604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Chen Y. and Slack J. M. W. (2013). Imparting regenerative capacity to limbs by progenitor cell transplantation. Dev. Cell 24, 41-51. 10.1016/j.devcel.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M. (1980). Intercalary regeneration in the amphibian limb and the rule of distal transformation. J. Embryol. Exp. Morphol. 56, 201-209. [PubMed] [Google Scholar]
- Makanae A., Mitogawa K. and Satoh A. (2014). Co-operative Bmp- and Fgf-signaling inputs convert skin wound healing to limb formation in urodele amphibians. Dev. Biol. 396, 57-66. 10.1016/j.ydbio.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Mendell J. R., Al-Zaidy S., Shell R., Arnold W. D., Rodino-Klapac L. R., Prior T. W., Lowes L., Alfano L., Berry K., Church K. et al. (2017). Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl. J. Med. 377, 1713-1722. 10.1056/NEJMoa1706198 [DOI] [PubMed] [Google Scholar]
- Mescher A. L. (2017). Macrophages and fibroblasts during inflammation and tissue repair in models of organ regeneration. Regeneration 4, 39-53. 10.1002/reg2.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher A. L. and Tassava R. A. (1975). Denervation effects on DNA replication and mitosis during the initiation of limb regeneration in adult newts. Dev. Biol. 44, 187-197. 10.1016/0012-1606(75)90386-3 [DOI] [PubMed] [Google Scholar]
- Moreira Teixeira L. S., Patterson J. and Luyten F. P. (2014). Skeletal tissue regeneration: where can hydrogels play a role? Int. Orthop. 38, 1861-1876. 10.1007/s00264-014-2402-2 [DOI] [PubMed] [Google Scholar]
- Nachtrab G., Kikuchi K., Tornini V. A. and Poss K. D. (2013). Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development 140, 3754-3764. 10.1242/dev.098798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacu E., Gromberg E., Oliveira C. R., Drechsel D. and Tanaka E. M. (2016). FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature 533, 407-410. 10.1038/nature17972 [DOI] [PubMed] [Google Scholar]
- Nowoshilow S., Schloissnig S., Fei J.-F., Dahl A., Pang A. W. C., Pippel M., Winkler S., Hastie A. R., Young G., Roscito J. G. et al. (2018). The axolotl genome and the evolution of key tissue formation regulators. Nature 554, 50-55. 10.1038/nature25458 [DOI] [PubMed] [Google Scholar]
- Pajcini K. V., Corbel S. Y., Sage J., Pomerantz J. H. and Blau H. M. (2010). Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell 7, 198-213. 10.1016/j.stem.2010.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescitelli M. J. Jr. and Stocum D. L. (1980). The origin of skeletal structures during intercalary regeneration of larval Ambystoma limbs. Dev. Biol. 79, 255-275. 10.1016/0012-1606(80)90115-3 [DOI] [PubMed] [Google Scholar]
- Pfefferli C., Müller F., Jaźwińska A. and Wicky C. (2014). Specific NuRD components are required for fin regeneration in zebrafish. BMC Biol. 12, 30 10.1186/1741-7007-12-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. Q., Lee J., Oei M., Flath C., Hwe C., Mariano R., Vu T., Shu C., Dinh A., Simkin J. et al. (2015). Positional information in axolotl and mouse limb extracellular matrix is mediated via heparan sulfate and fibroblast growth factor during limb regeneration in the axolotl (Ambystoma mexicanum). Regeneration 2, 182-201. 10.1002/reg2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J. S., Robitaille A. M., Wang Y., Ray C. A., Thummel R., Gu H., Djukovic D., Raftery D., Berndt J. D. and Moon R. T. (2017). Transcriptomic, proteomic, and metabolomic landscape of positional memory in the caudal fin of zebrafish. Proc. Natl. Acad. Sci. USA 114, E717-E726. 10.1073/pnas.1620755114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y., Lindau P., Ueno H., Longaker M. T. and Weissman I. L. (2011). Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 476, 409-413. 10.1038/nature10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J. T., King K. Y., Brett J. O., Cromie M. J., Charville G. W., Maguire K. K., Brunson C., Mastey N., Liu L., Tsai C.-R. et al. (2014). mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510, 393-396. 10.1038/nature13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roensch K., Tazaki A., Chara O. and Tanaka E. M. (2013). Progressive specification rather than intercalation of segments during limb regeneration. Science 342, 1375-1379. 10.1126/science.1241796 [DOI] [PubMed] [Google Scholar]
- Sandoval-Guzmán T., Wang H., Khattak S., Schuez M., Roensch K., Nacu E., Tazaki A., Joven A., Tanaka E. M. and Simon A. (2014). Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell 14, 174-187. 10.1016/j.stem.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Seifert A. W. and Muneoka K. (2018). The blastema and epimorphic regeneration in mammals. Dev. Biol. 433, 190-199. 10.1016/j.ydbio.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A. W., Kiama S. G., Seifert M. G., Goheen J. R., Palmer T. M. and Maden M. (2012). Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 489, 561-565. 10.1038/nature11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedhassantehrani N., Otsuka T., Singh S. and Gardiner D. M. (2017). The axolotl limb regeneration model as a discovery tool for engineering the stem cell niche. Curr. Stem Cell Rep. 3, 156-163. 10.1007/s40778-017-0085-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata E., Yokota Y., Horita N., Kudo A., Abe G., Kawakami K. and Kawakami A. (2016). Fgf signalling controls diverse aspects of fin regeneration. Development 143, 2920-2929. 10.1242/dev.140699 [DOI] [PubMed] [Google Scholar]
- Shibata E., Ando K., Murase E. and Kawakami A. (2018). Heterogeneous fates and dynamic rearrangement of regenerative epidermis-derived cells during zebrafish fin regeneration. Development 145, dev162016 10.1242/dev.162016 [DOI] [PubMed] [Google Scholar]
- Simkin J., Gawriluk T. R., Gensel J. C. and Seifert A. W. (2017). Macrophages are necessary for epimorphic regeneration in African spiny mice. eLife 6, e24623 10.7554/eLife.24623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. and Craven L. (1948). The growth and morphogenesis of the regenerating forelimb of adult Triturus following denervation at various stages of development. J. Exp. Zool. 108, 279-308. 10.1002/jez.1401080207 [DOI] [PubMed] [Google Scholar]
- Singh S. P., Holdway J. E. and Poss K. D. (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell 22, 879-886. 10.1016/j.devcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S., Afonso N., Bensimon-Brito A., Fonseca M., Simoes M., Leon J., Roehl H., Cancela M. L. and Jacinto A. (2011). Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development 138, 3897-3905. 10.1242/dev.064717 [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper C. L., Weidinger G., Riehle K. J., Hubbert C., Major M. B., Fausto N. and Moon R. T. (2007). Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479-489. 10.1242/dev.001123 [DOI] [PubMed] [Google Scholar]
- Sugiura T., Wang H., Barsacchi R., Simon A. and Tanaka E. M. (2016). MARCKS-like protein is an initiating molecule in axolotl appendage regeneration. Nature 531, 237-240. 10.1038/nature16974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Shimoda N., Takanaga S., Hozumi S. and Kikuchi Y. (2014). Expression patterns of dnmt3aa, dnmt3ab, and dnmt4 during development and fin regeneration in zebrafish. Gene Expr. Patterns 14, 105-110. 10.1016/j.gep.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Tanaka H. V., Ng N. C. Y., Yang Yu Z., Casco-Robles M. M., Maruo F., Tsonis P. A. and Chiba C. (2016). A developmentally regulated switch from stem cells to dedifferentiation for limb muscle regeneration in newts. Nat. Commun. 7, 11069 10.1038/ncomms11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassava R. A. and Garling D. J. (1979). Regenerative responses in larval axolotl limbs with skin grafts over the amputation surface. J. Exp. Zool. 208, 97-110. 10.1002/jez.1402080111 [DOI] [PubMed] [Google Scholar]
- Thornton C. S. (1957). The effect of apical cap removal on limb regeneration in Amblystoma larvae. J. Exp. Zool. 134, 357-381. 10.1002/jez.1401340209 [DOI] [PubMed] [Google Scholar]
- Tornini V. A., Puliafito A., Slota L. A., Thompson J. D., Nachtrab G., Kaushik A.-L., Kapsimali M., Primo L., Di Talia S. and Poss K. D. (2016). Live monitoring of blastemal cell contributions during appendage regeneration. Curr. Biol. 26, 2981-2991. 10.1016/j.cub.2016.08.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. L., Baselga-Garriga C. and Melton D. A. (2019). Blastemal progenitors modulate immune signaling during early limb regeneration. Development 146, dev169128 10.1242/dev.169128 [DOI] [PubMed] [Google Scholar]
- Tu S. and Johnson S. L. (2011). Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 20, 725-732. 10.1016/j.devcel.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahor E. and Poss K. D. (2017). Cardiac regeneration strategies: staying young at heart. Science 356, 1035-1039. 10.1126/science.aam5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira W. A., Wells K. M., Raymond M. J., De Souza L., Garcia E. and McCusker C. D. (2019). FGF, BMP, and RA signaling are sufficient for the induction of complete limb regeneration from non-regenerating wounds on Ambystoma mexicanum limbs. Dev. Biol. 451, 146-157. 10.1016/j.ydbio.2019.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lööf S., Borg P., Nader G. A., Blau H. M. and Simon A. (2015). Turning terminally differentiated skeletal muscle cells into regenerative progenitors. Nat. Commun. 6, 7916 10.1038/ncomms8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead G. G., Makino S., Lien C. L. and Keating M. T. (2005). fgf20 is essential for initiating zebrafish fin regeneration. Science 310, 1957-1960. 10.1126/science.1117637 [DOI] [PubMed] [Google Scholar]
- Xu C., Volkery S. and Siekmann A. F. (2015). Intubation-based anesthesia for long-term time-lapse imaging of adult zebrafish. Nat. Protoc. 10, 2064-2073. 10.1038/nprot.2015.130 [DOI] [PubMed] [Google Scholar]
- Yakushiji N., Suzuki M., Satoh A., Sagai T., Shiroishi T., Kobayashi H., Sasaki H., Ide H. and Tamura K. (2007). Correlation between Shh expression and DNA methylation status of the limb-specific Shh enhancer region during limb regeneration in amphibians. Dev. Biol. 312, 171-182. 10.1016/j.ydbio.2007.09.022 [DOI] [PubMed] [Google Scholar]
- Yun M. H., Gates P. B. and Brockes J. P. (2013). Regulation of p53 is critical for vertebrate limb regeneration. Proc. Natl. Acad. Sci. USA 110, 17392-17397. 10.1073/pnas.1310519110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M. H., Gates P. B. and Brockes J. P. (2014). Sustained ERK activation underlies reprogramming in regeneration-competent salamander cells and distinguishes them from their mammalian counterparts. Stem Cell Rep. 3, 15-23. 10.1016/j.stemcr.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M. H., Davaapil H. and Brockes J. P. (2015). Recurrent turnover of senescent cells during regeneration of a complex structure. eLife 4, e05505 10.7554/eLife.05505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Chen Y., Xu H., Yang L., Yuan F., Li L., Xu Y., Chen Y., Zhang C. and Lin G. (2018). Melanocortin receptor 4 signaling regulates vertebrate limb regeneration. Dev. Cell 46, 397-409.e395. 10.1016/j.devcel.2018.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Graham K., MacKenzie E. J., Ephraim P. L., Travison T. G. and Brookmeyer R. (2008). Estimating the prevalence of limb loss in the United States: 2005 to 2050. Arch. Phys. Med. Rehabil. 89, 422-429. 10.1016/j.apmr.2007.11.005 [DOI] [PubMed] [Google Scholar]