Abstract
Natural killer group 2D (NKG2D), an activating receptor on natural killer (NK) cells and a subset of T cells, recognizes stress-inducible proteins, including MICA and ULBP2, which are present on infected or transformed cells. Whether each NKG2D ligand (NKG2DL) has a distinct biological role is not clear. Here, using superresolution microscopy, we found that NKG2D is constitutively arranged in nanoclusters at the surface of primary human NK cells. Nanoclusters of NKG2D became smaller upon ligation with MICA but became larger upon activation by ULBP2. Additionally, ULBP2 induced the reorganization of nanoclusters of the cytokine receptor subunit for both interleukin-2 (IL-2) and IL-15 (IL-2/IL-15Rβ) such that these cytokine receptor subunits coalesced with nanoclusters of NKG2D. Functionally, the response of NK cells activated by ULBP2 was augmented by an interaction between ULBP2-bound NKG2D and IL-15R ligated by IL-15 (trans-presented by IL-15Rα–coated surfaces). These data suggest that NKG2DLs are not equivalent in their capacity to activate NKG2D and establish a previously unknown paradigm in how ligand-induced changes to the nanoscale organization of the cell surface can affect immune responses.
Introduction
Natural killer (NK) cells are lymphocytes that recognize and kill virally infected, stressed, or transformed cells and contribute to an immune response through the secretion of many cytokines and chemokines (1). Their activity is determined by the balance of signals from germ-line encoded inhibitory and activating receptors present on the cell surface (2–4). Inhibitory receptors include killer immunoglobulin (Ig)-like receptors (KIRs), which recognize self-major histocompatibility complex (MHC) class I molecules on healthy target cells and protect them from being killed by NK cells (5, 6). KIRs consist of a single polypeptide that contains immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in the cytoplasmic domain that recruit tyrosine phosphatases, such as SHP-1, upon receptor ligation (7–10).
One of the best characterized activating receptors on NK cells is the C-type, lectin-like, transmembrane receptor natural killer group 2D (NKG2D). In humans, NKG2D is a disulfide-linked homodimer present on the surface of all NK cells, most NKT cells, a subset of γδ T cells, all CD8+ T cells, and a subset of CD4+ T cells (11–13). Signaling downstream of activating receptors commonly involves phosphorylation of the immunoreceptor tyrosine-based activation motifs (ITAMs) that are either part of the receptor or present within associated adaptor proteins. However, NKG2D signals through the associated adaptor DAP10, which contains a costimulatory phosphatidylinositol-3-kinase (PI3K)–binding motif (YxNM) in its cytoplasmic domain. DAP10 recruits PI3K and the adaptor Grb2 and then Grb2 recruits the guanine nucleotide exchange factor Vav1 (14, 15).
In humans, NKG2D recognizes a set of ligands distantly related to MHC class I molecules that can be divided into two families: MHC class I–related protein A (MICA) and MICB, and UL16-binding proteins (ULBP1 to ULBP6) (11, 16–18). These ligands are rarely present on healthy cells but are induced or increase in abundance on infected or transformed cells rendering these cells susceptible to NK cell–mediated cytotoxicity (19–23). The abundance of NKG2D ligands is also increased on murine and human macrophages stimulated by a high dose of pathogen molecules, such as bacterial lipopolysaccharide (LPS) (24, 25). The different NKG2D ligands (NKG2DLs) are structurally diverse and have different patterns of expression and mechanisms of regulation. The reason for the existence of many NKG2DLs is not clear. One possibility is that they appeared during evolution of the immune system in response to selection pressures exerted by pathogens or cancer. Diversity in the ligands that activate NKG2D might make it harder for pathogens to interfere with this aspect of the immune response. It is also possible that different ligands bind to NKG2D with varying affinities; thus, this diversity might fine-tune the extent of activation, perhaps in a tissue-specific manner. A major unknown for understanding NKG2D-mediated immune responses is whether the different ligands are qualitatively and quantitatively equivalent in their capacity to trigger cellular activation.
Interleukin-15 (IL-15) is an essential cytokine for the development, maintenance, and survival of NK cells (26–29); some of its effects overlap with those of IL-2. IL-15 and IL-2 receptors have a similar structure: Both cytokine receptors form complexes that share a β-chain (IL-2/IL-15Rβ; also known as CD122), a common cytokine receptor γ-chain (γc; also known as CD132), and distinct α-chains. IL-15 increases the abundance of NKG2D and DAP10 in the NK cell surface membrane in vitro (17, 30–33). In mice, activation of the IL-15 receptor (IL-15R) results in phosphorylation of DAP10 by the kinase JNK3, thus priming DAP10 for NKG2D-dependent signaling (34). Thus, in addition to its well-established role in the survival and proliferation of NK cells, IL-15 primes NK cell cytotoxicity induced by NKG2D. However, whether direct cross-talk between NKG2D and IL-15R contributes to the activation of human NK cells or antitumor immune responses is unknown.
Here, we used dual-color direct stochastic optical reconstruction microscopy (dSTORM) to investigate how different NKG2DLs (MICA and ULBP2) affected the nanometer-scale organization of NKG2D and the organization of NKG2D in relation to the IL-15R on human primary NK (pNK) cells. We found that NKG2D was constitutively organized in nanoclusters at the surface of pNK cells and that the cluster sizes changed specifically with the activating ligand. Ligation of NKG2D by ULBP2, but not MICA, induced the reorganization of the IL-2/IL-15Rβ receptor subunit at the NK cell immune synapse where it associated with NKG2D on a nanometer scale. Functionally, degranulation of pNK cells induced by ULBP2, but not by MICA, was enhanced by IL-15 being simultaneously presented to the NK cell across the immune synapse or slide-cell contact, a process referred to as trans-presentation. At the stimulating concentrations used here, we found that ULBP2 and MICA had different capacities to activate immune responses, suggesting that the type and relative amount of NKG2DL present on tumors or virally infected cells affects the NK cell response. These results may affect the design of therapies to enhance NK cell activity.
Results
NKG2D nanoclusters at the surface of primary human NK cells reorganize upon ligation
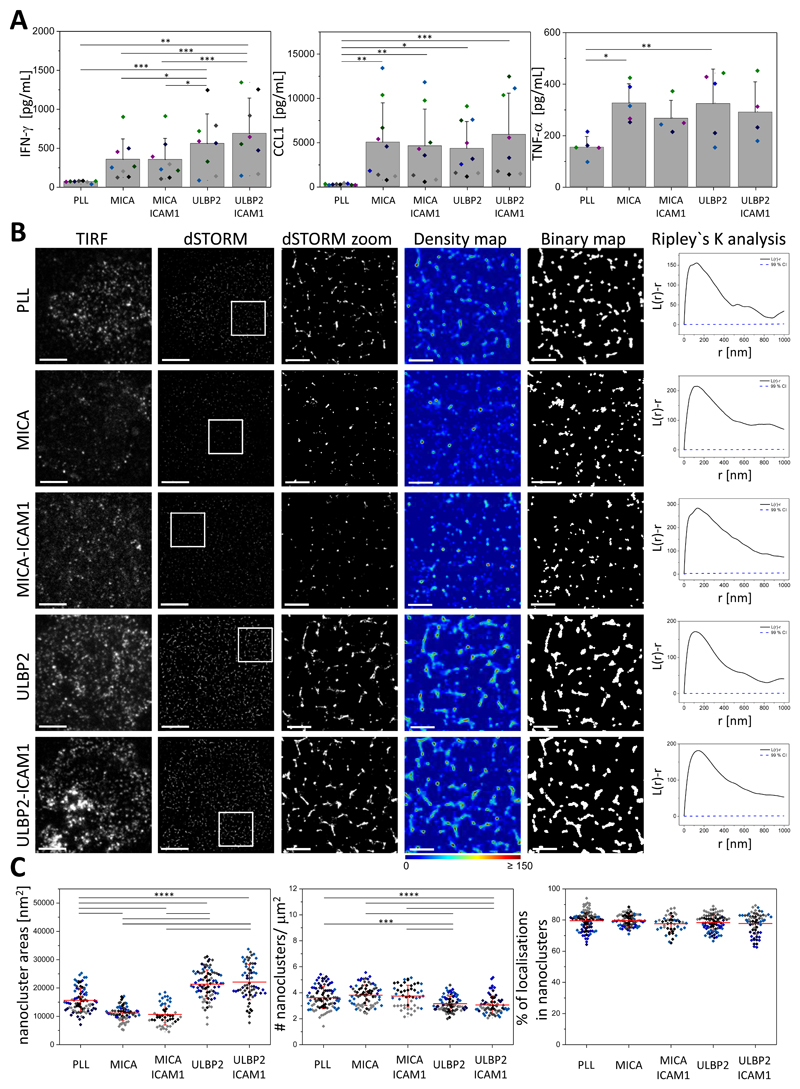
The nanoscale organization of NKG2D at the cell surface of pNK cells has not been established. Thus, we used the superresolution microscopy technique dSTORM to investigate the organization of these receptors on unstimulated cells and to test whether stimulation of NKG2D with different ligands affected its surface organization. We plated pNK cells onto poly-L-lysine–coated slides (PLL, nonactivated condition) or slides coated with the activating ligands MICA or ULBP2 in the presence or absence of the cell adhesion molecule ICAM-1. MICA and ULBP2 represent ligands for each of the two groups of NKG2DLs. We empirically determined an optimal concentration of MICA and ULBP2 for pNK cell activation based on the release of the cytokine interferon-γ (IFN-γ) and the chemokine CCL1 (fig. S1). ICAM-1 interacts with the integrin LFA-1, binding of which is necessary to elicit a full NK cell response (35, 36). At the concentrations used, activation of NKG2D with either MICA or ULBP2, with or without co-stimulation of LFA-1, stimulated secretion of IFN-γ, CCL1, and tumor necrosis factor α (TNF-α) (Fig. 1A). Both MICA and ULBP2 induced the secretion of CCL1 and TNF-α to comparable amounts, whereas ULBP2 induced greater secretion of IFN-γ than did MICA (Fig. 1A).
Fig. 1. Ligand-dependent reorganization of NKG2D nanoclusters at the surface of pNK cells.
(A) pNK cells were incubated for 24 hours in wells coated with PLL, MICA, MICA and ICAM-1, ULBP2, or ULBP2 and ICAM-1. The amounts of IFN-γ (left), CCL1 (middle), and TNF-α (right) released by the cells were assessed by ELISA. Data are means ± SD from four to nine donors. Each color represents one donor. (B) Representative TIRF and dSTORM images of NKG2D on pNK cells incubated for 10 min on slides that were coated as described in (A) and then stained with a fluorescently labelled specific monoclonal antibody (mAb) against NKG2D. Scale bars, 4 μm. Regions outlined in white are magnified (zoom) and shown with corresponding density images according to the pseudocolor scale bar, binary maps, and Ripley’s K analysis. Scale bars, 1 μm. L(r)-r, degree of clustering relative to simulated random distributions; r, radial scale. (C) Nanocluster areas (left), nanocluster density (middle), and percentage of localizations in nanoclusters (right) for NKG2D from the data shown in (B). Each symbol represents the median of several 5 μm x 5 μm regions from one cell. Lines represent means ± SD. Data are from a minimum of 40 cells from a minimum of three independent donors. Each color represents one donor. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by one-way ANOVA with Tukey’s post-hoc test. Nonsignificant differences are not indicated.
For imaging, cells were fixed and stained with a directly labelled monoclonal antibody (mAb) recognizing NKG2D. To rule out the possibility that the binding of NKG2D by MICA or ULBP2 could block accessibility for the NKG2D-specific antibody, we first pre-labeled NKL cells (an NK cell line expressing NKG2D) with MICA or ULBP2 at different concentrations. We then stained for NKG2D with the NKG2D monoclonal antibody, as used for imaging experiments, and compared whether there was a dose-dependent decrease in staining caused by the ligands. Preincubation with MICA had no effect on the staining for NKG2D (fig. S2, A and B), whereas there was a small decrease in the staining of NKG2D after incubation of the cells with ULBP2, which did not decrease further as the dose of the ligand increased (fig. S2, C and D). These data established that ligation of NKG2D with MICA or ULBP2 does not prevent accessibility to the NKG2D antibody used to mark the receptor’s location.
Our dSTORM images, as well as Ripley’s K analysis (37), showed that NKG2D is organized in nanometer-scale clusters at the surface of pNK cells in both resting and activated conditions (Fig. 1B). To gain quantitative insight into the organization of NKG2D nanoclusters, we generated probability density maps of the localizations based on univariate Getis and Franklin’s local point pattern analysis (38, 39). Density maps were then thresholded and converted to binary maps in which regions containing dense localizations of the receptor appear white (referred to as nanoclusters, Fig. 1B). Our analysis revealed that NKG2D was constitutively assembled in nanoclusters with a mean area of 15,600 ± 4,200 nm2 (Fig. 1C, left) and a mean density of 3.6 ± 0.9 nanoclusters/μm2 (Fig. 1C, middle). These became smaller upon ligation of the receptor with MICA alone or in combination with ICAM-1 (11,200 ± 2,600 nm2 and 10,700 ± 3,700 nm2, respectively, Fig. 1C, left). The density of nanoclusters did not change significantly after activation with MICA (3.8 ± 0.8 and 3.7 ± 0.8 nanoclusters/μm2, respectively, Fig. 1C, middle). In contrast to MICA, activation by ULBP2 with or without ICAM-1 increased NKG2D nanocluster sizes (21,400 ± 5,000 nm2 and 22,100 ± 6,200 nm2, respectively, Fig. 1C, left) and slightly decreased their density (3.2 ± 0.6 and 3.1 ± 0.7 nanoclusters/μm2, respectively, Fig. 1C, middle). The proportion of localizations (reflecting the proportion of receptors) in nanoclusters was very high (ranging on average from 78 to 81%), reflecting a high degree of clustering, which remained unchanged after activation with any of the ligands (Fig. 1C, right).
Label-density variation analysis is an alternative method to discriminate clustered from randomly distributed molecules and is based on experimental variation of labeling density combined with cluster analysis. An advantage to this method is that it is insensitive to artifacts generated by overcounting blinking fluorophores (40). We stained NKG2D with a range of concentrations of a directly labeled antibody and imaged by dSTORM. For each image, we calculated the relative area covered by the cluster masks (η), obtained from thresholded binary maps, and the mean density of localizations within the clusters (ρ). Clustered and random distributions can be discriminated by plotting the normalized density ρ/ρ0 (where ρ0 is the intersection of the density curves with the y axis) against η. For randomly distributed molecules, a horizontal line is observed, whereas for clustered receptors, there is an increase in ρ/ρ0. This analysis confirmed that NKG2D was clustered at the surface of pNK cells, because ρ/ρ0 increased with the concentration of the labeling antibody (fig. S3, left).
To test whether the differences in NKG2D reorganization observed after ligation with MICA versus ULBP2 were time-dependent, we assessed the organization of NKG2D at a shorter (5 min) and a longer (20 min) time point. Ligation with MICA decreased the size of NKG2D clusters over time, whereas ligation with ULBP2 induced an initial increase (within the first 10 min) in NKG2D cluster size followed by a decrease (fig. S4, A and B). At the time points tested, clusters of NKG2D were enlarged upon ligation with ULBP2, in comparison to the size of the clusters in cells stimulated with MICA (fig. S4A, B left panel). There was little, if any, change in the density of NKG2D nanoclusters, and a high fraction of localizations within nanoclusters was seen consistently across the different times tested (fig. S4B, middle and right panels).
To confirm that the observed changes in NKG2D clustering were due to the specific ligation of NKG2D by MICA or ULBP2, we blocked the activating ligands with specific antibodies before plating the cells. The pNK cells were plated for 10 min on blocked slides (slides coated with activating ligand and a blocking antibody against that ligand) in the presence of ICAM-1. The cells were then fixed and stained with a directly labeled NKG2D monoclonal antibody and imaged by dSTORM imaging. When the specific ligands were blocked, the organization of NKG2D at the cell surface remained as that seen in unstimulated cells (Fig. 2A). There was no difference in both nanocluster areas and density (Fig. 2, B left and middle) between unstimulated cells and cells with blocked ligands; however, we observed an increase in the percentage of localizations in clusters (Fig. 2B right). Thus, these data suggest that reorganization of NKG2D clusters (Fig. 1) involves binding of the receptor to the activating ligand.
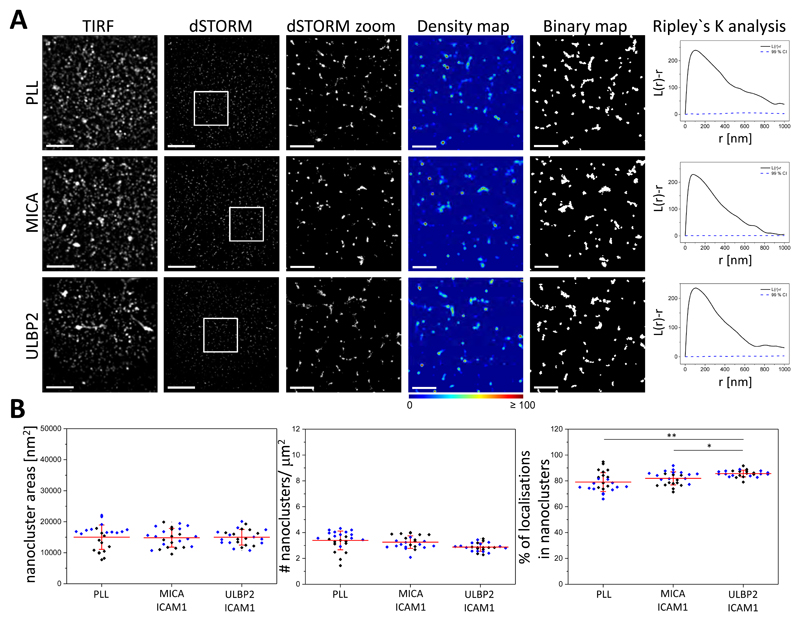
Fig. 2. Blockade of NKG2D ligands prevents the reorganization of NKG2D nanoclusters.
(A) Representative TIRF and dSTORM images of NKG2D on pNK cells incubated for 10 min on slides coated with PLL, MICA and ICAM-1, or ULBP2 and ICAM-1, blocked with specific antibodies against MICA or ULBP2 as appropriate, and then stained with fluorescently labelled specific mAb against NKG2D. Scale bars, 4 μm. Regions outlined in white are magnified and shown with corresponding density images according to the pseudocolor scale bar, thresholded binary maps, and Ripley’s K analysis. Scale bars, 1 μm. (B) Nanocluster areas (left), nanocluster density (middle), and percentage of localizations in nanoclusters (right) for NKG2D from the data shown in (A). Each symbol represents the median of several 5 μm x 5 μm regions from one cell. Lines represent means ± SD. Data are from a minimum of 12 cells from two independent donors. Each color represents one donor. *P < 0.05 and **P < 0.01 by one-way ANOVA with Tukey’s post-hoc test. Nonsignificant differences are not indicated.
There was substantial variability across cells, which is inherent to the use of primary blood samples. To account for variability in individual experiments or donors, we calculated the fold-change in nanocluster size and density by normalizing the values obtained in activating conditions from one donor to the nonactivated condition from the same donor. This showed an average increase of 1.4-fold in the size of NKG2D nanoclusters after engagement of the receptor with ULBP2 and a slight decrease after ligation with MICA (fig. S5, top). The density of nanoclusters did not change significantly after activation (fig. S5, top).
Flow cytometry showed a reduction of 76 ± 7 % and 64 ± 13 % in the amount of NKG2D at the cell surface upon activation of cells for 10 min with MICA and ULBP2, respectively (fig. S6, top). This decrease in cell surface receptor was not affected by co-ligation of LFA-1 (fig. S6. top). Thus, the decrease in NKG2D nanocluster size after ligation with MICA may relate to internalization of the receptor. However, this reduction in cell surface abundance does not easily relate to activation with ULBP2, which resulted in larger nanoclusters. The increase in nanocluster size observed with ULBP2 may involve the coalescence of individual smaller NKG2D nanoclusters or the recruitment of other proteins into the ULBP2-bound NKG2D nanoclusters. Together, these data indicate that NKG2D is organized at the cell surface in nanoscale clusters and that its organization changes upon ligand binding in a manner dependent on the ligand.
Activation of NKG2D with ULBP2, but not MICA, induces the reorganization of IL-2/IL-15Rβ nanoclusters
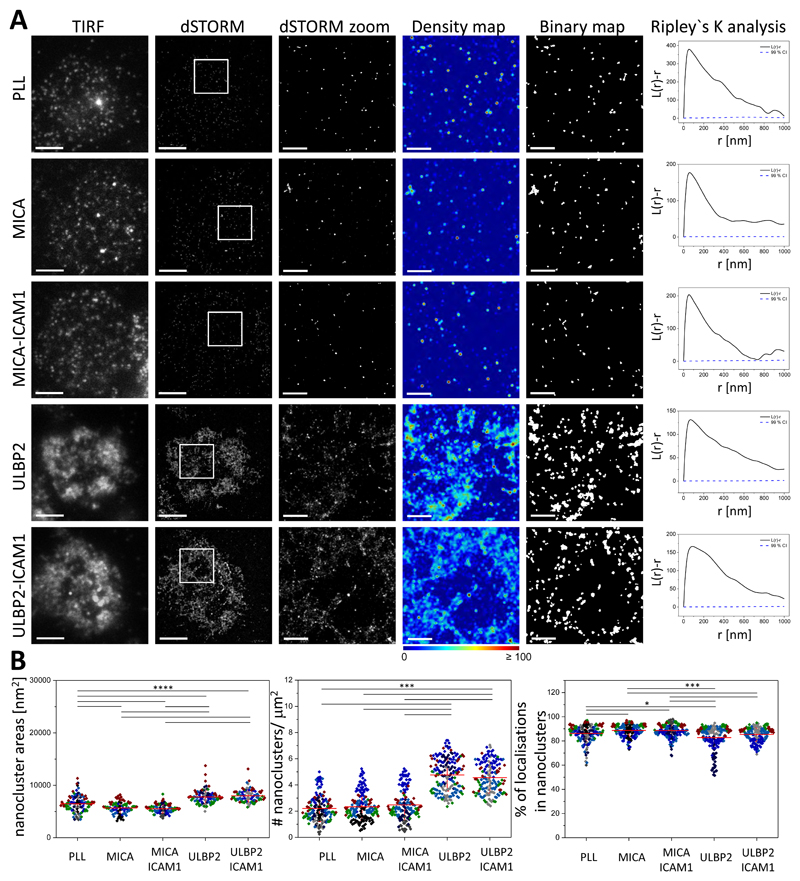
To probe for potential cross-talk between NKG2D and the IL-15R in human pNK cells and to determine whether cross-talk varies in response to activation by different NKG2D ligands, we investigated the nanometer-scale organization of the β subunit of this cytokine receptor (IL-2/IL-15Rβ) at the activating interface with and without NKG2D ligation. We plated pNK cells for 10 min onto slides coated with PLL or with the activating ligands MICA or ULBP2 with or without ICAM-1, fixed the cells, and then stained them with a directly labelled IL-2/15Rβ monoclonal antibody. dSTORM imaging followed by quantitative analysis revealed that IL-2/IL-15Rβ was constitutively arranged in nanometer-scale clusters (Fig. 3A) at the surface of pNK cells, with a mean area of 6,500 ± 1,600 nm2 (Fig. 3B, left), a mean density of 2.2 ± 1.0 nanoclusters/μm2 (Fig. 3B, middle), and a high proportion of localizations in nanoclusters (mean of 85 ± 7.0 %, Fig. 3B, right). Label-density variation analysis (40) further confirmed that IL-2/IL-15Rβ was clustered at the surface of pNK cells, because ρ/ρ0 increased when the labeling concentration was increased (fig. S3, right).
Fig. 3. Activation of NKG2D with ULBP2, but not MICA, induces the reorganization of IL-2/IL-15Rβ nanoclusters.
(A) Representative TIRF and dSTORM images of IL-2/IL-15Rβ at the surface of pNK cells incubated for 10 min on slides coated with PLL (nonactivated), MICA, MICA and ICAM-1, ULBP2, or ULBP2 and ICAM-1, and stained with a fluorescently labelled specific mAb against NKG2D. Scale bars, 4 μm. Regions outlined by the white squares are magnified and shown with corresponding density images according to the pseudocolor scale bar, thresholded binary maps, and Ripley’s K analysis. Scale bars, 1 μm. (B) Nanocluster areas (left), nanocluster density (middle), and percentage of localizations in nanoclusters (right) for IL-2/IL-15Rβ from the data shown in A were calculated by subjecting dSTORM data to spatial point-pattern analysis and thresholding. Each symbol represents the median of several 5 μm x 5 μm regions from the same cell. Horizontal lines and errors represent means ± SD. Data are from a minimum of 85 cells from a minimum of three independent donors. Each color represents one donor. *P < 0.05, ***P < 0.001, and ****P < 0.0001 by one-way ANOVA with Tukey`s post-hoc test (left) and Kruskal-Wallis with Dunn’s post-hoc test (middle and right). Nonsignificant differences are not indicated.
Activation of NKG2D with MICA, with or without co-ligation of LFA-1, did not substantially change the organization of IL-2/IL-15Rβ at the nanometer-scale (Fig. 3 and fig. S5, middle). However, ligation of NKG2D with ULBP2 induced a marked reorganization of the cytokine receptor at the activating interface (Fig. 3A). Upon ligation of NKG2D with ULBP2, nanoclusters of IL-2/IL-15Rβ increased in size (mean area of 7,800 ± 1,300 nm2, Fig. 3B, left) and in number (mean of 4.8 ± 1.2 nanoclusters/μm2, corresponding to a 2-fold increase; Fig. 3B, middle and fig. S5, middle), whereas its degree of clustering did not change (Fig. 3B, right). This reorganization of IL-2/IL-15Rβ was unaffected by co-ligation of LFA-1 (Fig. 3 and fig. S5, middle), which suggests that the effect is independent of additional signals from integrins.
The different organization of IL-2/IL-15Rβ induced by MICA versus ULBP2 persisted across a 20-min time scale (fig. S7A, B). In addition, when the specific ligands were blocked with antibodies, the organization of IL-2/IL-15Rβ at the cell surface remained as that seen in unstimulated cells (Fig. 4A), and there was no difference in any of the parameters analyzed (Fig. 4B), supporting the specificity of ULBP2-induced changes in the organization of IL-2/IL-15Rβ. Flow cytometry showed that the total surface abundance of IL-2/IL-15Rβ remained constant after ligation of NKG2D (fig. S6, bottom), indicating that the increased IL-2/IL-15Rβ density is likely due to recruitment of additional protein clusters specifically to the activating interface.
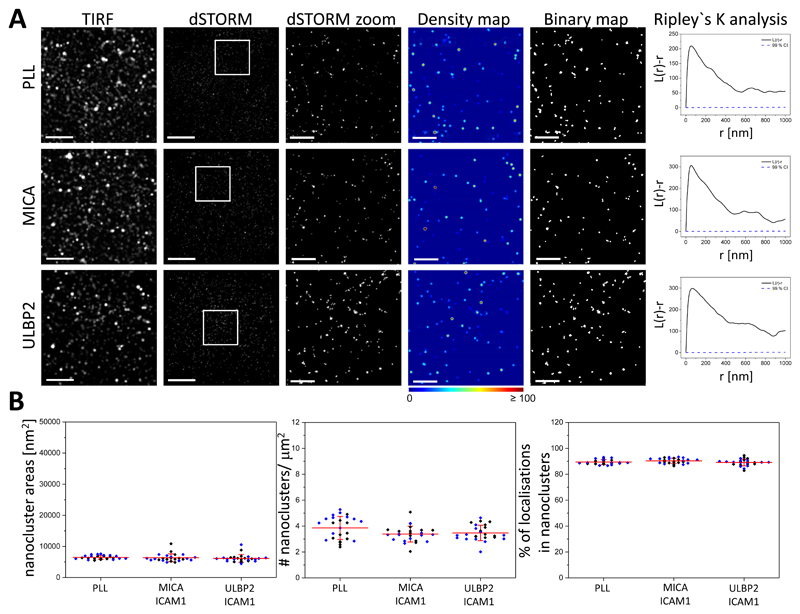
Fig. 4. Blockade of NKG2D ligands prevents the reorganization of IL-2/IL-15Rβ nanoclusters.
(A) Representative TIRF and dSTORM images of IL-2/IL-15Rβ on pNK cells incubated for 10 min on slides previously coated with PLL, MICA and ICAM-1, or ULBP2 and ICAM-1, blocked with specific antibodies against MICA or ULBP2 as appropriate, and stained with fluorescently labelled specific mAb against NKG2D. Scale bars, 4 μm. Regions outlined in white are magnified and shown with corresponding density images according to the pseudocolor scale bar, thresholded binary maps, and Ripley’s K analysis. Scale bars, 1 μm. (B) Nanocluster areas (left), nanocluster density (middle), and the percentage of localizations in nanoclusters (right) for IL-2/IL-15Rβ from the data shown in (A). Each symbol represents the median of several 5 μm x 5 μm regions from one cell. Lines represent means ± SD. Data are from a minimum of 12 cells from two independent donors. Each color represents one donor. One-way ANOVA with Tukey’s post-hoc test was used. Nosignificant differences are not indicated.
Because IL-2/IL-15Rβ is a common subunit of both the IL-2 and IL-15 receptors, we imaged the α-chain of the IL-2 receptor to establish which cytokine receptor reorganized in response to NKG2D stimulation. IL-2Rα (CD25) is specific for IL-2 and, on its own, binds to this cytokine with low affinity; IL-15Rα (CD215) is specific for IL-15 and binds to IL-15 with high affinity (41). Similar to IL-2/IL-15Rβ, IL-2Rα assembled into nanoclusters in both resting conditions and after activation of NKG2D (fig. S8A, B). However, the organization of IL-2Rα was only marginally affected after the stimulation of NKG2D with any of the ligands (fig. S5, bottom) and seemed to vary across donors rather than by stimulation conditions (fig. S8B). Together, these observations suggest that in human pNK cells, ligation of NKG2D with ULBP2, but not MICA, induces the nanometer-scale reorganization of IL-15R, but not IL-2R, at an activating interface.
IL-15 trans-presentation induces an increase in the number of IL-2/IL-15Rβ nanoclusters at the contact interface
Unlike IL-2, which is mostly secreted as a soluble factor by activated T cells and interacts with the high-affinity heterotrimeric complex IL-2Rα-IL-2/IL-15Rβ-γc (CD25-CD122-CD132), IL-15 functions mainly in a cell contact–dependent manner. IL-15 is not released as a free cytokine but is bound to IL-15Rα. Membrane-bound IL-15Rα-IL-15 complexes are presented by target cells to responding cells that have the intermediate-affinity IL-2/IL-15β-γc (CD122-CD132) receptor complex at the surface (42–45). This process of activation involving two cells that come into contact is called “trans-presentation.” Signaling by cis-presentation or through soluble complexes of IL-15Rα-IL-15 can also contribute to IL-15–induced responses, but to a much lesser extent (45–47).
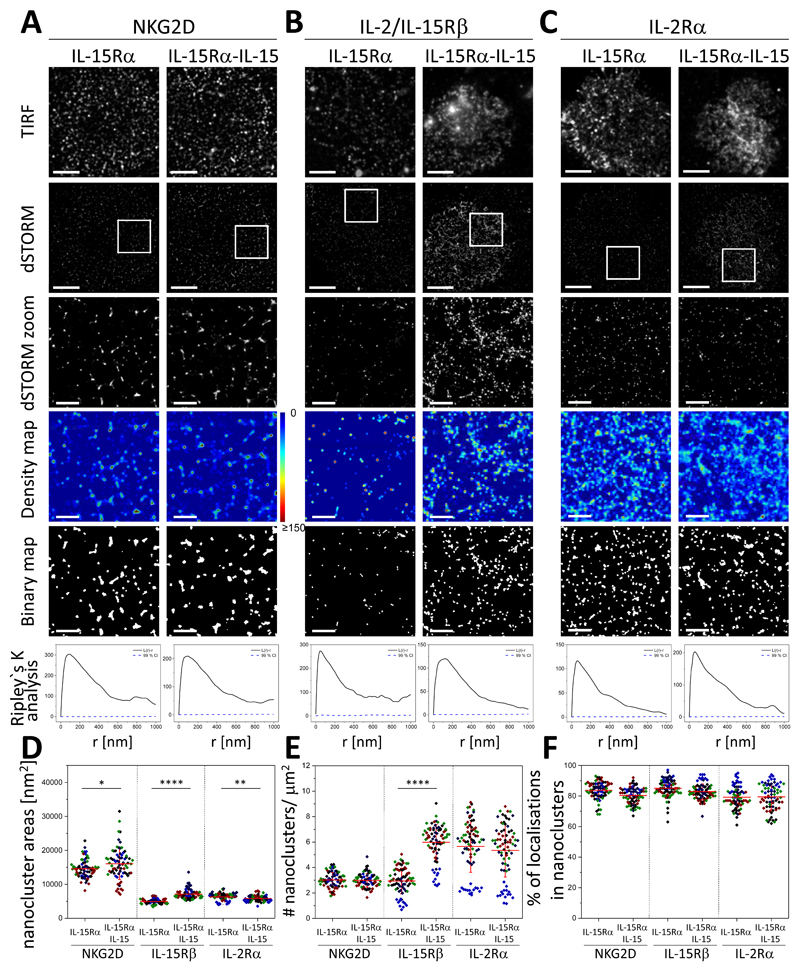
To test whether trans-presentation of IL-15Rα-IL-15 affected the nanoscale organization of NKG2D, pNK cells were plated for 10 min onto slides coated with unbound IL-15Rα, as a control, or with IL-15Rα bound to IL-15 (IL-15Rα-IL-15 complexes). Cells were then fixed, labelled, and imaged for NKG2D (Fig. 5A), IL-2/IL-15Rβ (Fig. 5B), and IL-2Rα (Fig. 5C). dSTORM images and analysis showed that ligation of IL-15 receptor by trans-presented IL-15 caused only a small change in the nanoscale organization of NKG2D (Fig. 5A). NKG2D remained organized in nanoclusters with an area of 16,040 ± 4,600 nm2, slightly increased compared to that of cells incubated on slides coated with control, unloaded IL-15Rα (14,600 ± 2,600 nm2 ; Fig. 5A, D) or in unstimulating conditions (15,600 ± 4,200 nm2; Fig. 1C and fig. S5, top). The density of nanoclusters of NKG2D (3.0 ± 0.6 nanoclusters/μm2, Fig. 5E and fig. S5, top) and their degree of clustering (~85%, Fig. 5F) was not affected when IL-15 was trans-presented in the absence of NKG2D ligation. In contrast, trans-presentation of IL-15 induced a two-fold increase in the number of IL-2/IL-15Rβ nanoclusters at the activating interface (Fig. 5, B and E, and fig. S5, middle), as well as an increase in their size (Fig. 5D and fig. S5, middle). This behavior resembles the nanoscale changes observed in IL-2/IL-15Rβ after stimulation of NKG2D with ULBP2 (Fig. 4, fig. S5). The degree of clustering of IL-2/IL-15Rβ did not change significantly after trans-presentation of IL-15 (Fig. 5F). IL-2Rα clustering remained unaltered when IL-2/IL-15Rβ was ligated with IL-15Rα-IL-15 (Fig. 5, C to F, and fig. S5, bottom), indicating that this change in organization was for IL-15R, but not IL-2R.
Fig. 5. Trans-presentation of IL-15 increases the density of IL-2/IL-15Rβ nanoclusters.
(A to C) Representative TIRF and dSTORM images of NKG2D (A), IL-2/IL-15Rβ (B), and IL-2Rα (C) at the surface of pNK cells incubated for 10 min on slides coated with IL-15Rα unloaded or loaded with IL-15 and stained with fluorescently labelled NKG2D-specific mAbs. Scale bars, 4 μm. Regions outlined by the white squares are magnified and shown with corresponding density images according to the pseudocolor scale bar, thresholded binary maps, and Ripley’s K analysis. Scale bars, 1 μm. (D to F) Nanocluster area (D), nanocluster density (E), and percentage of localizations in nanoclusters (F) for NKG2D, IL-2/IL-15Rβ, and IL-2Rα from the data shown in (A to C) were calculated by subjecting dSTORM data to spatial point-pattern analysis and thresholding. Each symbol represents the median of several 5 μm x 5 μm regions from the same cell. Horizontal lines and errors represent means ± SD. Data are from a minimum of 45 cells from a minimum of three independent donors. Each color represents one donor. *P < 0.05, **P < 0.01, and ****P < 0.0001 by two-tailed t test assuming unequal variance. Nonsignificant differences are not indicated.
ULBP2 induces the association of NKG2D with IL-2/IL-15Rβ nanoclusters
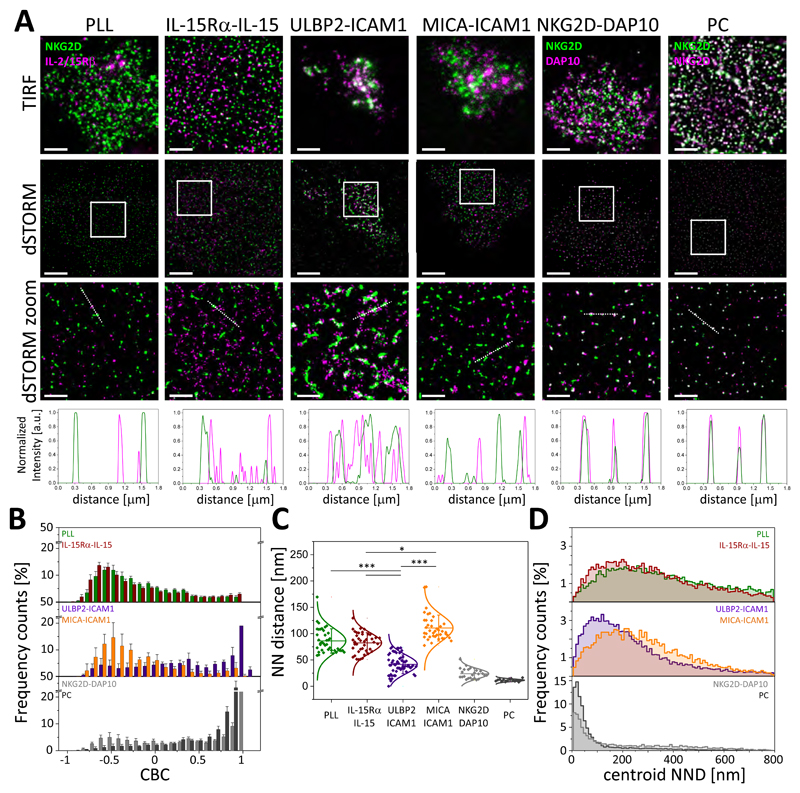
Because IL-15R primes NKG2D signaling in murine NK cells (34), the relative proximity of NKG2D and IL-2/IL-15Rβ nanoclusters may be important for signal integration. Thus, we used dual-color dSTORM to investigate the relationship between NKG2D and IL-2/IL-15Rβ at the nanometer-scale. We included two positive controls for colocalization. In one, NKG2D was stained with a primary NKG2D mAb conjugated with Atto488, which was followed by an isotype-specific secondary antibody conjugated with AF647 (Fig. 6A, column labeled PC). In the other positive control, NKG2D was labelled with an NKG2D-Atto488 mAb and its adaptor protein DAP10 (14, 48, 49) was labelled with a DAP10-AF647 mAb (Fig. 6A, column labeled NKG2D-DAP10). To investigate the organization of NKG2D and IL-2/IL-15Rβ nanoclusters, pNK cells were plated under non-activating or activating conditions for 10 min before being fixed and stained with directly labeled NKG2D-Atto488 mAb and IL-2/IL-15Rβ-AF647 mAb. Dual-color dSTORM revealed that NKG2D nanoclusters were associated with IL-2/IL-15Rβ nanoclusters at the interface when NK cells were activated by ULBP2, but not MICA (Fig. 6A).
Fig. 6. ULBP2 induces the association between NKG2D and IL-2/IL-15Rβ nanoclusters.
(A) TIRF and dSTORM images showing NKG2D and IL-2/IL-15Rβ on pNK cells incubated for 10 min on slides coated with PLL, MICA and ICAM-1, ULBP2 and ICAM-1, or IL-15Rα–IL-15 and stained with anti-NKG2D-Atto488 and anti-IL-2/IL-15Rβ-AF647 mAbs. Regions outlined in white are magnified with relative fluorescence intensity profiles along the white lines. Scale bars, 4μm, 1μm (zoom). As a positive control, cells on PLL-coated slides were stained with anti-NKG2D-Atto488 and anti-DAP10-AF647 mAbs or were stained with anti-NKG2D-Atto488 mAb followed by anti-mouse-IgG1-AF647 secondary antibody (PC). Colocalization between channels is shown in white. (B) CBC histograms of the single-molecule distributions of the colocalization parameter for NKG2D and IL-2/IL-15Rβ in cells incubated as in (A). Data are from a minimum of 10 cells from two independent donors. Bars represent means ± SD. (C) NND analysis of the data shown in (A). Data are from a minimum of 30 cells from two independent donors; two donors for the positive controls. Symbol represents the median NND of all paired single-molecule localizations from one cell. Lines/errors represent means ± SD. *P < 0.05 and ***P < 0.001 by Kruskal-Wallis (Dunn’s post-test). (D) Histogram distributions of the NND between the centroids of nanoclusters from one channel and the centroid of their nearest neighbor from the second channel from cells incubated as described in (A). Nonsignificant differences are not indicated.
The precise degree of colocalization between NKG2D and IL-2/IL-15Rβ was addressed by subjecting NKG2D and IL-2/IL-15Rβ localization lists to coordinate-based colocalization (CBC) analysis (50), which assigns a correlation coefficient to each single localization of each protein within a certain radial distance, ranging from -1 (perfectly segregated) through 0 (uncorrelated distributions) to +1 (perfectly colocalized). For CBC analysis, we chose a search radius of 50 nm based on the radius of IL-2/IL-15Rβ nanoclusters, which we calculated from the mean nanocluster area in resting conditions. After ligation of NKG2D with ULBP2, a large proportion of IL-2/IL-15Rβ associated with NKG2D; the histogram distribution of the colocalization parameter was distributed close to +1 with 69% of localizations between 0 and 1 (Fig. 6B, blue bars in middle graph). A similar trend was observed for both positive controls (Fig. 6B). In contrast, ligation of NKG2D with MICA produced a distribution of NKG2D and IL-2/IL-15Rβ similar to that of cells incubated on PLL or exposed to trans-presentation of IL-15; 70, 72, and 72% of localizations, respectively, had negative correlation coefficients (Fig. 6B, orange, green, and brown bars in middle and top graphs). As an alternative analysis, we also compared the mean nearest-neighbor distance (NND) of paired single-molecule localizations between NKG2D and IL-2/IL-15Rβ, which was 42 ± 18 nm for cells activated by ULBP2 (Fig. 6C). In contrast, in resting conditions or after ligation of NKG2D with MICA or trans-presentation of IL-15, the NND between localizations of NKG2D and IL-2/IL-15Rβ was significantly greater (86 ± 23 nm, 111 ± 25 nm and 82 ± 19 nm, respectively; Fig. 6C). A proportion of NKG2D (~20%) and IL-2/IL-15Rβ (~15%) localizations were not localized within nanoclusters (Fig. 1C and Fig. 3B, right). Because CBC analysis accounts for all localizations, we additionally tested whether nanoclusters colocalize by measuring the NND between the centroids of NKG2D and IL-2/IL-15Rβ nanoclusters. In resting conditions or after activation with MICA or trans-presentation of IL-15, the centroid NND between the two receptor nanoclusters had a mode of 205 ± 5.0 nm, 189 ± 4.5 nm, and 164 ± 4.0 nm, respectively (Fig. 6D). However, when NKG2D was activated with ULBP2, the mode for the centroid NND was markedly decreased (110 ± 3.0 nm, Fig. 6D).
We investigated the association between NKG2D and IL-2/IL-15Rβ nanoclusters at a shorter (5 min) and a longer (20 min) time of activation. At the times tested, ligation of NKG2D with MICA did not induce the association between nanoclusters of both receptors (fig. S9A), as shown by the histogram distribution of the colocalization parameter being distributed towards -1 (fig. S9B) and a mean NND of paired single-molecule localizations > 50 nm (fig. S9C). At 20 min, NKG2D and IL-2/IL-15Rβ were even more segregated than at 5 or 10 min, perhaps as a result of NKG2D internalization. After 5 min of activation with ULBP2, NKG2D and IL-2/IL-15Rβ nanoclusters were closer than in unstimulated cells (fig. S9A), although the histogram distribution of the colocalization parameter was still distributed towards -1 (fig. S9B) and the mean NND was above 50 nm (55 ± 14 nm, fig. S9C). After 20 min of activation with ULBP2, NKG2D and IL-2/IL-15Rβ nanoclusters were more segregated than at 10 min (fig. S9, A to C), perhaps due to internalization of NKG2D. Together, these data suggest that activation of NKG2D by ULBP2, but not MICA, promotes the association between NKG2D and IL-2/IL-15Rβ nanoclusters at the activating interface.
Trans-presentation of IL-15 by IL-15Rα augments the activation of NKG2D by ULBP2, but not MICA
Our analysis of how the molecular organization of the NK cell surface changes upon activation suggested that the cross-talk between NKG2D and IL-15R pathways depends on the ligand that triggers NKG2D. NK cell–mediated cytotoxicity occurs by the exocytosis of cytotoxic granules containing perforin and granzymes (51, 52). To facilitate secretion of lytic granules, the periodicity of the cortical actin mesh at the site of contact between the NK cell and its target cell (a structure called the immune synapse) changes to enable the granules to pass (53, 54). Thus, we compared the effect of MICA- and ULBP2-mediated activation of NKG2D on the remodeling of actin at an artificial immune synapse and on the degranulation capacity of pNK cells. We plated pNK cells under non-activating or activating conditions for 10 min, fixed the cells, and then labelled them with AF647-phalloidin (to stain actin) before imaging them by dSTORM. We also imaged cells by confocal microscopy to quantify the percentage of cells with a dense peripheral actin ring, a marker of NK cell activation (55).
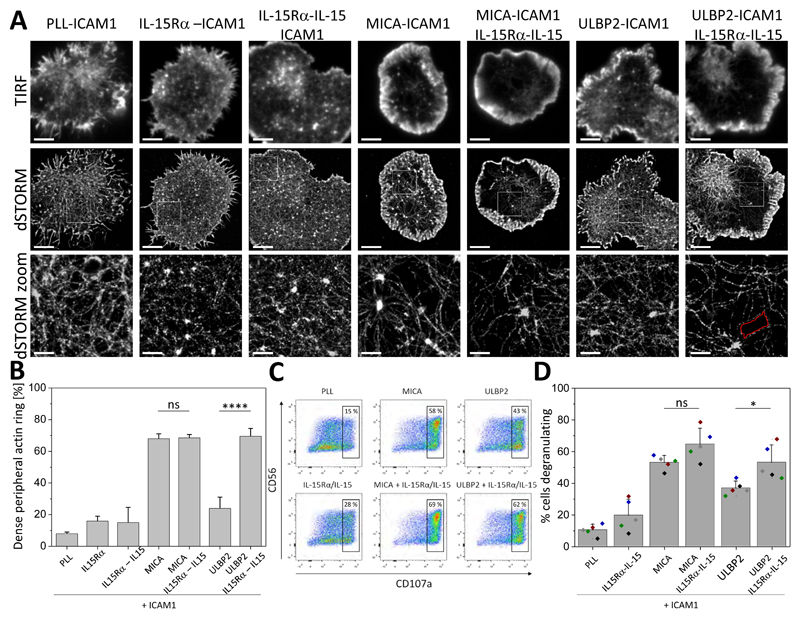
When plated onto control PLL-coated surfaces with ICAM1 or surfaces coated with IL-15Rα or IL-15Rα-IL-15 complexes with ICAM1, cells exhibited a dense cortical actin mesh at the synapse (Fig. 7A) and a low percentage of cells had dense peripheral actin rings (8 ± 1%, 16 ± 3%, and 15 ± 10%, respectively, Fig. 7B). Activation of NKG2D by MICA together with co-ligation of LFA-1 by ICAM1 induced a marked nanoscale reorganization of the synaptic actin. This was visualized by both an increase in the areas between individual actin filaments (“holes”) within the central region of the synapse (Fig. 7A) and a high percentage of cells with dense peripheral actin ring (68 ± 3%, Fig. 7B). This reorganization was not affected by the co-ligation of IL-15R with IL-15Rα-IL-15 complexes (Fig. 7, A and B). In contrast, actin remodeling was minimal when cells where stimulated by ULBP2, even with co-ligation of LFA-1 by ICAM1 (Fig. 7A), and the percentage of cells with dense peripheral actin rings was relatively low (24 ± 7%, Fig. 7B). Co-ligation of IL-15R, however, augmented the response triggered by ULBP2 such that the synaptic actin organization resembled that in cells activated with MICA, and the percentage of cells with dense peripheral actin rings increased to 70± 5% (Fig. 7, A and B).
Fig. 7. Trans-presentation of IL-15 by IL-15Rα augments the activation of NK cells by ULBP2, but not MICA.
(A) Representative TIRF and dSTORM images of membrane-proximal F-actin in pNK cells incubated for 10 min at 37°C on slides coated with PLL, unloaded IL-15Rα, IL-15–loaded IL-15Rα, or MICA or ULBP2 in the presence of ICAM-1, with or without IL15Rα-IL-15. Scale bars, 4 μm. Regions outlined by the white squares (middle row) are magnified (bottom row). Scale bars, 500 nm. An example of an actin mesh “hole” is indicated in red. (B) Histogram of the percentage of cells with dense peripheral actin rings. pNK cells were incubated as described in (A). F-actin was visualized using fluorescently labelled phalloidin, and the percentages of cells forming dense peripheral F-actin rings were quantified from confocal images. Data are from 100 to 500 cells from at least two independent donors. Bars represent means ± SD. (C) Flow cytometric analysis of CD107a surface abundance on pNK cells incubated for 5 hours at 37°C on slides coated as described in (A). Data are from one experiment and are representative of five experiments. (D) Percentage of pNK cells that degranulated as calculated from the flow cytometric data. Bars represent means ± SD from five donors. Each color represents one donor. *P < 0.05 and ****P < 0.0001 by one-way ANOVA with Tukey`s post-hoc test. ns, not significant.
To test whether degranulation of NK cells was differentially affected by the different stimuli, we analyzed the surface abundance of CD107a, a marker of degranulation (56), by flow cytometry. Stimulation of cells with MICA (with co-ligation of LFA-1) increased the amount of CD107a at the NK cell surface, compared to the basal amount, and increased the percentage of cells that degranulated from 10 ± 4% to 53 ± 4% (Fig. 7, C and D). Stimulation with ULBP2 also triggered degranulation, as evidenced by an increase in CD107a abundance at the surface (Fig. 7C) and in the percentage of cells degranulating (37 ± 4%, Fig. 7 D), and trans-presentation of IL-15 enhanced both responses significantly (53 ± 11%, Fig. 7 C and D). Together, these data suggest that costimulation by trans-presented IL-15 synergizes with the NKG2D signaling triggered by ULBP2.
Discussion
NKG2D recognizes ligands induced on infected or tumor cells, rendering these cells susceptible to NK cell–mediated killing. However, it is not known whether different ligands for NKG2D are functionally equivalent in their capacity to activate NK cells. Here, we set out to compare the ability of two NKG2D ligands, MICA and ULBP2, to activate pNK cells.
An emerging new frontier in immune cell biology is how the state of activation of cells can affect the nanoscale organization of cell surface receptors (57–61). We first investigated the nanoscale organization of NKG2D after stimulation with MICA or ULBP2. We found that NKG2D was constitutively arranged in discrete and spatially separated nanoclusters at the surface of pNK cells. After activation, receptor nanoclusters became reorganized in a way that was dependent on the activating ligand; activation by MICA induced the formation of smaller nanoclusters, whereas activation by ULBP2 induced the formation of nanoclusters that were 1.4-fold larger. We next investigated the effect of activation by either MICA or ULBP2 on the crosstalk with the IL-15R pathway. The IL-2/IL-15Rβ subunit of the receptor was also constitutively organized in nanoclusters at the surface of pNK cells. Unexpectedly, activation of NKG2D by ULBP2, but not MICA, induced an increase of two-fold in the number of IL-2/IL-15Rβ nanoclusters at the interface. Perhaps most importantly, upon activation by ULBP2, nanoclusters of NKG2D and IL-2/IL-15Rβ were associated with each other. These data imply that there is crosstalk between the two receptors, which is determined by the activating ligand for NKG2D.
The distinct nanoscale organization of NKG2D caused by ULBP2, in comparison to that caused by MICA, correlated with functional differences. At the concentrations of ligand tested, cells stimulated by ULBP2 produced more IFN-γ in comparison to cells stimulated with MICA, but degranulated less efficiently. Notably, co-ligation of IL-15R by trans-presentation of IL-15 enhanced the extent of degranulation of ULBP2-activated cells to that of MICA-activated cells. Thus, the proximity between IL-2/IL-15Rβ and NKG2D nanoclusters, observed after activation of cells by ULBP2, likely facilitates the integration of their signals to enhance NKG2D-mediated cytotoxic activity. These data suggest that there is a qualitative difference in NKG2D signaling triggered by MICA or ULBP2. There is some precedent that these ligands could signal differently. MICA is stronger than ULBP2 in inducing NKG2D downregulation, for example, and the phosphorylation of c-Cbl is more potently induced by MICA (62). The molecular basis for different NK cell responses to MICA and ULBP2 is not clear. This may result from these ligands having different affinities for NKG2D. The affinity of NKG2D for MICA is relatively low (63), but the affinity of NKG2D for ULBP2 is not known. Differences in the ways that these ligands interact might also result from differences in their different structures. ULBP2 and MICA consist of two and three Ig domains, respectively. This could feasibly alter the distance or orientation that NKG2D-MICA and NKG2D-ULBP2 spans at the intercellular contact. There is evidence that this distance is important in signal integration, because the proximity of other receptor-ligand pairs can be influenced according to whether they have similar spatial dimensions (64).
Primary tumor isolates and tumor cell lines from different histotypes are heterogeneous with respect to the NKG2D ligands that they express and the amounts at which they are found. This can also vary with tumor progression (13, 20). For example, the melanoma cell lines Me1386 and M14 and the leukemic T cell line H9 are negative for MICA but express ULBP2 (20). Other tumor cell lines express MICA, but not ULBP2, or express both ligands at different ratios. NK cell–mediated killing correlates with the density of NKG2D ligands on tumor cells (13). In addition, some tumor cells express IL-15Rα and hence can trans-present IL-15 to NK cells. Our data suggest that this costimulatory signal may be of particular importance when the NKG2D ligand present on the tumor cell is ULBP2. In this case, trans-presentation of IL-15 could give the auxiliary signals necessary for the efficient activation of NK cell–mediated killing by ULBP2. Consistent with this, a previous study showed that IL-15 strongly enhances the immune response directed against ULBP2-expressing tumors (65). This could affect the success with which NK cells eliminate tumor cells; if tumor cells express ULBP2, but not IL-15Rα, or if IL-15 itself is not available, then the NK cells could be less efficient at eliminating them.
In summary, our data establish that activation by MICA or ULBP2 induces a different nanoscale reorganization of NKG2D at the activating interface or immunological synapse. When stimulated by MICA, NKG2D nanoclusters became smaller; in contrast, activation by ULBP2 induced the formation of larger nanoclusters. Furthermore, when activated by ULBP2, NKG2D nanoclusters colocalized with nanoclusters of IL-2/IL-15Rβ. This correlated with a functional difference between MICA and ULBP2. Trans-presentation of IL-15 augments the cytolytic response stimulate by ULBP2. There are many recent developments in using NK cells for immunotherapy (66). Our data suggest that therapies to augment NK cell activity should be tailored based on the type and amount of ligands expressed in tumors. IL-15 may play an especially important role in the immune response against tumors that express more ULBP2 than MICA.
Materials and Methods
Isolation of primary NK cells
Peripheral blood was acquired from the NHS blood service under ethics license REC 05/Q0401/108 (University of Manchester). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation (Ficoll-Paque Plus; Amersham Pharmacia Biotech). Primary human NK cells were isolated by negative magnetic selection (Miltenyi Biotec) and cultured at 1 × 106 cells/ml in clone medium [DMEM, 30% HAM`s F-12, 10% human serum, 1 mM sodium pyruvate, 1% MEM non-essential amino acids (Sigma), 2 mM L-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and 50 μM 2-mercaptoethanol (Gibco)], supplemented with 200 U/ml recombinant IL-2 (rhIL-2, Roche). For all experiments, expanded but resting NK cells were used six days after the addition of IL-2.
Flow cytometry
To assess the cell surface expression of NKG2D and IL-2/IL-15Rβ, cells were washed and blocked with phosphate-buffered saline (PBS), 2% fetal bovine serum (FBS) for 30 min at 4°C and then were stained with Zombie Aqua viability dye (Biolegend), anti-CD56-AF488 mAb (clone HCD56, Biolegend), and anti-NKG2D-APC mAb (clone 1D11, Biolegend) or anti-IL-2/IL-15Rβ-APC mAb (clone TU27, Biolegend), respectively, or isotype-matched control mAbs (mouse IgG1 isotype control, clone MOPC-21, Biolegend, conjugated with AF488 or APC) for 30 min at 4°C. Cells were then washed in PBS, 2% FBS, fixed in PBS, 2% paraformaldehyde (PFA), and analyzed with a BD FACS Canto II flow cytometer (BD Biosciences). Data were analyzed with FlowJo_V10 software.
Assay for accessibility of NKG2D/NKG2DL to staining mAb
NKL cells (a commonly used immortal human NK cell line, derived from an aggressive human leukemia) were left uncoated or coated with 30, 60 or 120 μg/ml of rhMICA-Fc or rhULBP2-Fc (R&D Systems), labeled in-house with Atto488, for 30 min at 37°C. After incubation, cells were washed in PBS, 2% FBS, fixed in PBS, 4% PFA for 15 min, and stained with anti-NKG2D-APC mAb (clone 1D11, Biolegend), or isotype-matched control mAb (mouse IgG1 isotype control, clone MOPC-21, Biolegend, conjugated with APC) for 30 min at 4°C. Cells were then washed in PBS, 2% FBS, post-fixed in PBS, 2% PFA, and analyzed with a BD FACS Canto II flow cytometer (BD Biosciences). Data were analyzed with FlowJo_V10 software.
CD107a degranulation assay
Primary NK cells were incubated on surfaces coated with antibodies or proteins used to stimulate NK cells in the presence of GolgiPlug (1/1000 dilution, BD Biosciences), monesin (1/1000 dilution, Biolegend), and anti-LAMP-1-AF647 mAb (clone H4A3, Santa Cruz Biotechnology) or isotype control mAb (mouse IgG1 isotype control, clone MOPC-21, conjugated with AF647, Biolegend) for 5 hours at 37°C. After incubation, cells were washed and stained with Zombie Aqua viability dye (Biolegend), anti-CD56-AF488 mAb (clone HCD56, Biolegend), and anti-LAMP-1-AF647, or isotype-matched control mAbs. Finally, cells were washed in PBS, 2% FBS, fixed in PBS, 2% PFA, and analyzed with a BD FACS Canto II flow cytometer (BD Biosciences). Data were analyzed with FlowJo_V10 software.
Sample preparation for imaging
Chambered glass coverslips (#1.5 Lab-Tek II, Nunc) were coated with 0.01% poly-L-lysine (PLL, Sigma) and used for the imaging of unstimulated cells. To stimulate cells, slides were additionally coated with rhMICA-Fc, rhMICA-Fc and rhICAM-1, rhULBP2-Fc, rhULBP2-Fc and rhICAM-1, or rhIL-15Rα, in PBS overnight at 4°C, as indicated in the figures. rhMICA-Fc and rhULBP2-Fc were coated at 5 μg/ml (chosen as the optimal concentration to elicit pNK cell activation; fig. S1), and rhICAM-1 and rhIL-15Rα-Fc were coated at 2.5 μg/ml (all from R&D Systems). On the next day, rhIL-15Rα was left unloaded or loaded with 50 ng/ml of IL-15 (R&D Systems) for 2 hours at room temperature. The functionality of IL-15Rα-IL-15 complexes was confirmed by Western blotting analysis of cell lysates with antibody specific for pSTAT5. Cells were then allowed to settle on the slides for 5, 10, or 20 min at 37°C, fixed with PBS, 4% PFA for 30 min at room temperature, and washed three times in PBS. Samples were blocked in PBS, 3% bovine serum albumin (BSA) at room temperature for 1 hour followed by incubation at room temperature for 1 hour with the appropriate fluorescently labelled mAbs or with AF647-labeled phalloidin (Invitrogen), for the visualization of actin, diluted in PBS, 3% BSA. Samples were then washed, post-fixed with PBS, 4% PFA for 5 min at room temperature and imaged. Primary mAbs used for microscopy were: anti-NKG2D-Atto488 (clone 1D11, Abcam), anti-IL-2/IL-15Rβ-AF647 (clone 27302, R&D Systems), and anti-IL-2Rα-AF647 (clone M-A251, BD Biosciences). All antibodies were conjugated in-house with the corresponding dye (Invitrogen) and had 6 or 7 dyes/antibody as assessed by measurements of absorption.
Assay for specificity of NKG2D ligation using a ligand blocking mAb
Chambered glass coverslips (#1.5 Lab-Tek, Nunc) were coated with 0.01% PLL or additionally with rhMICA-Fc or rhULBP2-Fc in PBS overnight at 4°C, as described earlier. On the next day, the slides were incubated with 10 μg/ml of anti-MICA or anti-ULBP2 mAbs (R&D Systems), respectively, for 2 hours at room temperature. Cells were then allowed to settle on the slides for 10 min at 37°C, fixed with PBS, 4% PFA for 30 min at room temperature, and washed three times in PBS. Samples were blocked in PBS, 3% BSA at room temperature for 1 hour, followed by incubation at room temperature for 1 hour with the appropriate fluorescently labelled mAbs diluted in PBS 3% BSA. Finally, the samples were washed, post-fixed with PBS, 4% PFA for 5 min at room temperature, and imaged.
Actin ring analysis
For actin ring analysis, chambered glass coverslips (#1.5 Lab-Tek, Nunc) were coated with 0.01% PLL or additionally with rhIL-15Rα-Fc unloaded or loaded with 50 ng/ml of IL-15, rhMICA-Fc and rhICAM-1, rhMICA-Fc and rhICAM-1 and rhIL-15Rα-IL-15 complex, rhULBP2-Fc and rhICAM-1, or rhULBP2-Fc and rhICAM-1 and rhIL-15Rα-IL-15 complex, as described earlier. Cells were allowed to settle on the slides for 10 min at 37°C, fixed in PBS, 4% PFA at room temperature for 30 min, permeabilized and blocked with PBS, 3% BSA, 0.2% Triton X-100 at room temperature for 1 hour. Actin was stained with AF647-labeled phalloidin (Invitrogen, 1/200 dilution) in PBS, 3% BSA, 0.2% Triton X-100 and imaged by confocal microscopy (Leica TCS SP8) with a 100x 1.4 N.A. oil immersion objective. Images were exported to ImageJ software (National Institutes of Health) and the percentage of cells forming peripheral actin rings was scored in a double-blinded manner to avoid the possibility of bias.
Enzyme-linked immunosorbent assay (ELISA)
Primary NK cells were incubated on chambered glass coverslips coated with PLL or additionally with rhMICA-Fc, rhMICA-Fc and rhICAM-1, rhULBP2-Fc, or rhULBP2-Fc and rhICAM-1, as indicated earlier, at 37°C for 24 hours. Cell supernatants were recovered and centrifuged at 350g for 10 min at room temperature to remove cell debris. IFN-γ, CCL1, and TNF-α production was quantified in the supernatants by sandwich ELISA (DuoSet ELISA, R&D Systems), according to the manufacturer’s instructions. The plates were developed with TMB ELISA substrate (Sigma) and the reaction was stopped with 1N H2SO4. Absorbance was measured at 450 nm using a 570-nm reference line to compensate for optical interference.
dSTORM imaging
dSTORM imaging (Leica SR GSD) was performed with a 160x 1.43 N.A. oil immersion objective in TIRF mode. Dual-color dSTORM imaging was performed with primary antibodies directly conjugated with AF647 and Atto488 acquired in a sequential manner. First, 642-nm laser light was used for exciting the AF647 dye and switching it to the dark state. Second, 488-nm laser light was used for exciting the Atto488 dye and switching it to the dark state. An additional 405-nm laser light was used for reactivating AF647 and Atto488 fluorescence. The emitted light from both dyes was collected by the same objective and imaged onto the EM-CCD camera at a frame rate of 10 ms per frame. A maximum of 5000 frames per condition were acquired. For each receptor, the specificity of the labeling was confirmed by staining cells with isotype-matched control antibodies (fig. S10). These controls showed a negligible amount of nonspecific binding.
dSTORM data analysis
Because dual-color dSTORM imaging is performed in sequential mode by using two different optical detection paths (the dichroic and emission filters are different), an image registration is required to generate the final two-color dSTORM image (67, 68). Therefore, fiducial markers (TetraSpek Fluorescent Microspheres, Invitrogen) of 100 nm, which were visible in both the 488- and 647-nm channels, were used to align the 488-nm imaging channel to the 647-nm channel. The images of the beads in both channels were used to calculate a polynomial transformation function that mapped the 488-nm channel onto the 647-nm channel, using the MultiStackReg plug-in of ImageJ to account for differences in magnification and rotation, for example. The transformation was applied to each frame of the 488-nm channel. dSTORM images were analyzed and rendered as previously described (69, 70) with custom written software (Insight3, kindly provided by Prof. Bo Huang, University of California, San Francisco). Briefly, peaks in single-molecule images were identified based on a threshold and then were fit to a simple Gaussian to determine the x and y positions. Only localizations with a photon count >400 photons were included, and localizations that appeared within one pixel in five consecutive frames were merged together and fitted as one localization. The final images were rendered by representing the x and y positions of the localizations as a Gaussian with a width that corresponded to the determined localization precision. Sample drift during acquisition was calculated and subtracted by reconstructing dSTORM images from subsets of frames (500 frames) and correlating these images to a reference frame (the initial time segment). Quantitative cluster analysis was based on Ripley´s K function (37) and univariate Getis and Franklin´s local point pattern analysis (38). The x and y coordinate list of localizations was used and multiple regions of 5 x 5 μm were selected for each cell giving the median value per cell. Spatial pattern analysis using Ripley´s K function was performed with SpPack (71). Quantitative color scale cluster maps based on univariate Getis and Franklin’s local point pattern analysis method were generated using a custom MATLAB script as described previously (72) with a sampling radius of 50 nm. Two dimensional pseudo-color density maps were generated by interpolating a surface plot with L(50) as the z-axis on a grid with a resolution of 5 nm. Binary maps, generated from density maps, were used to measure cluster sizes and the number of clusters per μm2 in ImageJ software with the particle analysis function. Precise values are indicative rather than definitive and analysis is effective in revealing relative differences between different receptors or conditions. Varying label density analysis was performed as described previously (40).
Coordinate-based colocalization (CBC) analysis
CBC-mediated analysis between two receptors was performed with an ImageJ plug-in (73) based on an algorithm that was described previously (50). To assess the correlation function for each localization, the x-y coordinate list from the 488-nm and 647-nm dSTORM channels was used. For each localization from the 647-nm channel, the correlation function to each localization from the 488-nm channel was calculated. This parameter can vary between -1 (perfectly segregated) through 0 (uncorrelated distributions) to +1 (perfectly colocalized). The correlation coefficients were plotted as a histogram of occurrences with a 0.1-binning. The nearest neighbor distance (NND) between each localization from the 647-nm channel and its closest localization from the 488-nm channel was measured and plotted as the median NND between localizations per cell. To assess protein cluster colocalization, the centroid NND was calculated with an ImageJ plug-in as described earlier. Dual-color dSTORM images were converted into binary maps and the x and y coordinates of cluster centroids were identified in each image with the particle analysis function in ImageJ. The NND from the centroid of a cluster in the 647-nm channel to the closest centroid of a cluster in the 488-nm channel was measured and plotted as a histogram of occurrences with a 10-nm binning. The mode of the histograms was determined by fitting the distribution to a Gaussian function.
Statistical analysis
Samples were tested for normality with a Kolmogorov-Smirnov (K-S) test. The statistical significance of differences and power analysis between two normally distributed data sets was assessed by a two-tailed t test assuming unequal variance; multiple comparisons were made with one-way ANOVA with Tukey’s post-hoc test for normally distributed data or with Kruskal-Wallis with Dunn’s post-hoc test for not normally distributed data. All statistical analysis was performed with OriginPro 9.1 (OriginLab) and with SigmaPlot 13 analysis software.
Supplementary Material
One-sentence summary.
Distinct ligands for the activating receptor NKG2D are not equivalent in their ability to stimulate natural killer cell responses.
Editor’s summary.
Distinct effects on NK cells
Natural killer (NK) cells perform immune surveillance for virally infected cells and tumor cells. The balance between the engagement of activating receptors, such as NKG2D, and inhibitory receptors on the NK cell surface determines whether the cells kill their targets. Through superresolution microscopy, Bálint et al. showed that NKG2D on the surface of unstimulated NK cells was organized into microclusters, which became differentially reorganized depending on whether the cells were exposed to MICA or ULBP2, two NKG2D ligands found on tumor cells. Furthermore, stimulation with ULBP2, but not MICA, caused NKG2D to cluster with the receptor for the cytokine IL-15, which can also be presented on tumor cells, synergizing with NKG2D to enhance NK cell responses. These data suggest that NKG2D ligands have distinct outcomes on NK cell function, which may have relevance in the use of NK cells as an immunotherapy.
Acknowledgments
We thank K. Stacey for the isolation of primary NK cells, other members of the laboratory for useful discussions, G. Howell for help with flow cytometry, and P. Paszek for help with the use of statistics.
Funding: This work was supported by the Medical Research Council (Award G1001044), a Wellcome Trust Investigator Award (110091), and the Manchester Collaborative Centre for Inflammation Research (funded by a pre-competitive open-innovation award from GSK, AstraZeneca and The University of Manchester, UK).
Footnotes
Author contributions: S.B. and F.B.L. performed all experiments and analyzed the data. S.B., F.B.L., and D.M.D. conceived the project, designed experiments, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human Nk-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115:2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 2004;23:255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiesa S, Tomasello E, Vivier E, Vély F. Coordination of activating and inhibitory signals in natural killer cells. Mol Immunol. 2005;42:477–484. doi: 10.1016/j.molimm.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 4.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ljunggren HG, Kärre K. In search of the “missing self”: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 6.Davis DM. The compatibility gene. London: Penguin; 2014. [Google Scholar]
- 7.Burshtyn DN, Scharenberg AM, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet J-P, Long EO. Recruitment of tyrosine phosphatase HCP (SHP-1) by the killer cell inhibitor receptor. Immunity. 1996;4:77–85. doi: 10.1016/s1074-7613(00)80300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olcese L, Lang P, Vély F, Cambiaggi A, Marquet D, Bléry M, Hippen KL, Biassoni R, Moretta A, Moretta L, Cambier JC, et al. Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 9.Burshtyn DN, Yang WT, Yi TL, Long EO. A novel phosphotyrosine motif with a critical amino acid at position-2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J Biol Chem. 1997;272:3066–3072. doi: 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]
- 10.Long EO. Negative signalling by inhibitory receptors: the NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 12.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 13.Nausch N, Cerwenka A. NKG2D ligands in tumour immunity. Oncogene. 2008;27:5944–5958. doi: 10.1038/onc.2008.272. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL, Phillips JH. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 15.Upshaw JL, Arneson LN, Schoon RA, Dick CJ, Billadeau DD, Leibson PJ. NKG2D-mediated signalling requires a DAP10-bound Grb2-Vav1 intermediate and phosphatidylinositol-3-kinase in human natural killer cells. Nat Immunol. 2006;7:524–532. doi: 10.1038/ni1325. [DOI] [PubMed] [Google Scholar]
- 16.Cosman D, Müllberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 17.Sutherland CL, Chalupny NJ, Schooley K, VandenBos T, Kubin M, Cosman D. UL16-binding proteins, novel MHC class I-related proteins, bind to NKG2D and activate multiple signalling pathways in primary NK cells. J Immunol. 2002;168:671–679. doi: 10.4049/jimmunol.168.2.671. [DOI] [PubMed] [Google Scholar]
- 18.Eagle RA, Traherne JA, Hair JR, Jafferji I, Trowsdale J. ULPB6/RAET1L is an additional human NKG2D ligand. Eur J Immunol. 2009;39:3207–3216. doi: 10.1002/eji.200939502. [DOI] [PubMed] [Google Scholar]
- 19.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activated lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 20.Pende D, Rivera P, Marcenaro S, Chang CC, Biassoni R, Conte R, Kubin M, Cosman D, Ferrone S, Moretta L, Moretta A. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumour cell lines of different histotypes: analysis of tumour susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62:6178–6186. [PubMed] [Google Scholar]
- 21.Rölle A, Mousavi-Jazi M, Eriksson M, Odeberg J, Söderberg-Nauclér C, Cosman D, Kärre K, Cerboni C. Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor on NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein. J Immunol. 2003;171:902–908. doi: 10.4049/jimmunol.171.2.902. [DOI] [PubMed] [Google Scholar]
- 22.Welte SA, Sinzger C, Lutz SZ, Singh-Jasuja H, Sampaio KL, Eknigk U, Rammensee HG, Steinle A. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur J Immunol. 2003;33:194–203. doi: 10.1002/immu.200390022. [DOI] [PubMed] [Google Scholar]
- 23.Vankayalapati R, Garg A, Porgador A, Griffith DE, Klucar P, Safi H, Girard WM, Cosman D, Spies T, Barnes PF. Role of NK cell-activating receptors and their ligands in the lysis of mononuclear phagocytes infected with an intracellular bacterium. J Immunol. 2005;175:4611–4617. doi: 10.4049/jimmunol.175.7.4611. [DOI] [PubMed] [Google Scholar]
- 24.Hamerman JA, Ogasawara K, Lanier LL. Cutting edge: Toll-like receptor signaling in macrophages indices ligands for the NKG2D receptor. J Immunol. 2004;172:2001–2005. doi: 10.4049/jimmunol.172.4.2001. [DOI] [PubMed] [Google Scholar]
- 25.Nedvetzki S, Sowinski S, Eagle RA, Harris J, Vély F, Pende D, Trowsdale J, Vivier E, Gordon S, Davis DM. Reciprocal regulation of human natural killer cells and macrophages associated with distinct immune synapses. Blood. 2007;109:3776–3785. doi: 10.1182/blood-2006-10-052977. [DOI] [PubMed] [Google Scholar]
- 26.Carson WE, Fehniger TA, Haldar S, Eckhert K, Lindemann MJ, Lai CF, Croce CM, Baumann H, Caligiuri MA. A potential role for interleukin-15 in the regulation of human natural killer cell survival. J Clin Invest. 1997;99:937–943. doi: 10.1172/JCI119258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waldmann TA, Tagaya Y. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annu Rev Immunol. 1999;17:19–49. doi: 10.1146/annurev.immunol.17.1.19. [DOI] [PubMed] [Google Scholar]
- 28.Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood. 2001;97:14–32. doi: 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- 29.Cooper MA, Bush JE, Fehniger TA, VanDeusen JB, Waite RE, Liu Y, Aquila HL, Caligiuri MA. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 2002;100:3633–3638. doi: 10.1182/blood-2001-12-0293. [DOI] [PubMed] [Google Scholar]
- 30.Roberts AI, Lee L, Schwarz E, Groh V, Spies T, Ebert EC, Jabri B. Cutting edge: NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J Immunol. 2001;167:5527–5530. doi: 10.4049/jimmunol.167.10.5527. [DOI] [PubMed] [Google Scholar]
- 31.Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, Raulet DH, Lanier LL, Groh V, Spies T, Ebert EC, et al. Coordinated induction by IL-15 of a TCR-independent NKG2D signalling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 32.Maasho K, Opoku-Anane J, Marusina AI, Coligan JE, Borrego F. NKG2D is a costimulatory receptor for human naïve CD8+ T cells. J Immunol. 2005;174:4480–4484. doi: 10.4049/jimmunol.174.8.4480. [DOI] [PubMed] [Google Scholar]
- 33.Park YP, Choi SC, Kiesler P, Gil-Krzewska A, Borrego F, Weck J, Krzewski K, Coligan JE. Complex regulation of human NKG2D-DAP10 cell surface expression: opposing roles of the γc cytokines and TGF-β1. Blood. 2011;118:3019–3027. doi: 10.1182/blood-2011-04-346825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horng T, Bezbradica JS, Medzhitov R. NKG2D signalling is coupled to the interleukin 15 receptor signalling pathway. Nat Immunol. 2007;8:1345–1352. doi: 10.1038/ni1524. [DOI] [PubMed] [Google Scholar]
- 35.Bryceson YT, Ljunggren HG, Long EO. Minimal requirements for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–2666. doi: 10.1182/blood-2009-01-201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D, Bryceson YT, Meckel T, Vasiliver-Shamis G, Dustin ML, Long EO. Integrin-dependent organisation and bidirectional vesicular traffic at cytotoxic immune synapses. Immunity. 2009;31:99–109. doi: 10.1016/j.immuni.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ripley BD. Modeling Spatial Patterns. J R Stat Soc Ser B Stat Methodol. 1977;39:172–212. [Google Scholar]
- 38.Getis A, Franklin J. Second-order neighborhood analysis of mapped point patterns. Ecology. 1987;68:473–477. [Google Scholar]
- 39.Williamson DJ, Owen DM, Rossy J, Magenau A, Wehrmann M, Gooding JJ, Gaus K. Pre-existing clusters of the adaptor Lat do not participate in signalling events. Nat Immunol. 2011;12:655–662. doi: 10.1038/ni.2049. [DOI] [PubMed] [Google Scholar]
- 40.Baumgart F, Arnold AM, Leskovar K, Staszek K, Fölser M, Weghuber J, Stockinger H, Schütz GJ. Varying label density allows artefact-free analysis of membrane-protein nanocluster. Nat Methods. 2016;13:661–664. doi: 10.1038/nmeth.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 42.Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 in trans to neighboring cells. Immunity. 2002;17:537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- 43.Koka R, Burkett PR, Chien M, Chai S, Chan F, Lodolce JP, Boone DL, Ma A. Interleukin (IL)-15Ralpha-deficient natural killer cells survive in normal but not IL-15Ralpha-deficient mice. J Exp Med. 2003;197:977–984. doi: 10.1084/jem.20021836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi H, Dubois S, Sato N, Sabzevari H, Sakai Y, Waldmann TA, Tagaya Y. Role of trans-cellular IL-15 presentation in the activation of NK cell-mediated killing, which leads to enhanced tumour immunosurveillance. Blood. 2005;105:721–727. doi: 10.1182/blood-2003-12-4187. [DOI] [PubMed] [Google Scholar]
- 45.Mortier E, Woo T, Advincula R, Gonzalo S, Ma A. IL-15Ralpha chaperones IL-15 to stable dendritic cell membrane complexes that activate NK cells via trans presentation. J Exp Med. 2008;205:1213–1225. doi: 10.1084/jem.20071913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ota N, Takase M, Uchiyama H, Olsen SK, Kanagawa O. No requirement of trans presentations of IL-15 for human CD8 T cell proliferation. J Immunol. 2010;185:6041–6048. doi: 10.4049/jimmunol.0901834. [DOI] [PubMed] [Google Scholar]
- 47.Bergamaschi C, Bear J, Rosati M, Beach RK, Alicea C, Sowder R, Chertova E, Rosenberg SA, Felber BK, Pavlakis GN. Circulating IL-15 exists as heterodimeric complex with soluble IL-15Rα in human and mouse serum. Blood. 2012;120:e1–e8. doi: 10.1182/blood-2011-10-384362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garrity D, Call ME, Feng J, Wucherpfennig KW. The activating NKG2D receptor assembles in the membrane with two signalling dimers into a hexameric structure. PNAS. 2005;102:7641–7646. doi: 10.1073/pnas.0502439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogasawara K, Lanier LL. NKG2D in NK and T cell-mediated immunity. J Clin Immunol. 2005;25:534–540. doi: 10.1007/s10875-005-8786-4. [DOI] [PubMed] [Google Scholar]
- 50.Malkusch S, Endesfelder U, Mondry J, Gelléri M, Verveer PJ, Heilemann M. Coordinate-based colocalisation analysis of single-molecule localisation microscopy data. Histochem Cell Biol. 2012;137:1–10. doi: 10.1007/s00418-011-0880-5. [DOI] [PubMed] [Google Scholar]
- 51.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 52.Dustin ML, Long EO. Cytotoxic immunological synapses. Immunol Rev. 2010;235:24–34. doi: 10.1111/j.0105-2896.2010.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rak GD, Mace EM, Banerjee PP, Svitkina T, Orange JS. Natural killer cel lytic granule secretion occurs through a pervasive actin network at the immune synapse. PLoS Bio. 2011;9:e1001151. doi: 10.1371/journal.pbio.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown ACN, Oddos S, Dobbie IM, Alakoskela J-M, Parton RM, Eissmann P, Neil MAA, Dunsby C, French PMW, Davis I, Davis DM. Remodelling of cortical actin where lytic granules dock at natural killer cell immune synapses revealed by super-resolution microscopy. PLoS Bio. 2011;9:e1001152. doi: 10.1371/journal.pbio.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Culley FJ, Johnson M, Evans JH, Kumar S, Crilly R, Casasbuenas J, Schnyder T, Mehrabi M, Deonarain MP, Ushakov DS, Braud V, et al. Natural killer cell signal integration balances synapse symmetry and migration. PLoS Bio. 2009;7:e1000159. doi: 10.1371/journal.pbio.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 57.Sherman E, Barr V, Manley S, Patterson G, Balagopalan L, Akpan I, Regan CK, Merrill RK, Sommers CL, Lippincott-Schwartz J, Samelson LE. Functional nanoscale organization of signalling molecules downstream of the T cell antigen receptor. Immunity. 2011;35:705–720. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, Batista FD. The actin and tetraspanin networks organise receptor nanoclusters to regulate B cell receptor-mediated signalling. Immunity. 2013;38:461–474. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 60.Pageon SV, Cordoba SP, Owen DM, Rothery SM, Oszmiana A, Davis DM. Superresolution microscopy reveals nanometre-scale reorganisation of inhibitory natural killer cell receptors upon activation of NKG2D. Sci Signal. 2013;6:ra62. doi: 10.1126/scisignal.2003947. [DOI] [PubMed] [Google Scholar]
- 61.Klasener K, Maity PC, Hobeika E, Yang J, Reth M. B cell activation involves nanoscale receptor reorganisations and inside-out signalling by Syk. Elife. 2014;3:e02069. doi: 10.7554/eLife.02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Molfetta R, Quatrini L, Capuano C, Gasparrini F, Zitti B, Zingoni A, Galandrini R, Santoni A, Paolini R. c-Cbl regulates MICA- but not ULBP2-induced NKG2D down-modulation in human NK cells. Eur J Immunol. 2014;44:2761–2770. doi: 10.1002/eji.201444512. [DOI] [PubMed] [Google Scholar]
- 63.Eagle RA, Trowsdale J. Promiscuity and the single receptor: NKG2D. Nat Rev Immunol. 2007;7:737–744. doi: 10.1038/nri2144. [DOI] [PubMed] [Google Scholar]
- 64.Köhler K, Xiong S, Brzostek J, Mehrabi M, Eissmann P, Harrison A, Cordoba SP, Oddos S, Miloserdov V, Gould K, Burroughs NJ, et al. Matched sizes of activating and inhibitory receptor/ligand pairs are required for optimal signal integration by human natural killer cells. PLoS One. 2010;5:e15374. doi: 10.1371/journal.pone.0015374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sutherland CL, Rabinovich B, Chalupny NJ, Brawand P, Miller R, Cosman D. ULBPs, human ligands of the NKG2D receptor, stimulate tumour immunity with enhancement by IL-15. Blood. 2006;108:1313–1319. doi: 10.1182/blood-2005-11-011320. [DOI] [PubMed] [Google Scholar]
- 66.Camille G, Huntington ND, Smyth MJ. Targeting natural killer cells I cancer immunotherapy. Nat Immunol. 2016;17:1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 67.Bates M, Dempsey GT, Chen KH, Zhuang X. Multicolor super-resolution fluorescence imaging via multi-parameter fluorophore detection. ChemPhysChem. 2012;13:99–107. doi: 10.1002/cphc.201100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balint S, Verdeny Vilanova I, Sandoval Alvarez A, Lakadamyali M. Correlative live-cell and superresolution microscopy reveals cargo transport dynamics at microtubule intersections. PNAS. 2013;110:3375–3380. doi: 10.1073/pnas.1219206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perry GLW. SpPack: spatial point pattern analysis in Excel using Visual Basic for Applications (VBA) Environ Model Softw. 2004;19:559–569. [Google Scholar]
- 72.Owen DM, Rentero C, Rossy J, Magenau A, Williamson D, Rodriguez M, Gaus K. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J Biophotonics. 2010;3:446–454. doi: 10.1002/jbio.200900089. [DOI] [PubMed] [Google Scholar]
- 73.Ovesny M, Krizek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.