Abstract
A single-nucleotide polymorphism in the promoter region of the Matrix Metalloproteinase-9 (MMP9) gene, rs3918242, has been shown to affect MMP9 expression in macrophages and was associated with schizophrenia by two independent groups. However, rs3918242's effects on MMP9 expression were not replicable in cell lines or brain tissue. Additionally, publically available data indicate that rs3918242 genotype is related not to MMP9 expression, but rather to expression of SLC12A5, a nearby gene coding for a K+/Cl- cotransporter, whose expression has also been related to schizophrenia. Here, we studied brain structure and function in healthy participants stratified by rs3918242 genotype using structural MRI (N = 298), functional MRI during an N-back working memory task (N = 554), and magnetoencephalography (MEG) during the same task (N = 190). We found rs3918242 was associated with gray matter volume (GMV) in the insula and dorsolateral prefrontal cortex bilaterally, closely replicated in discovery and replication samples; and with inferior parietal lobule (IPL) GMV when the samples were meta-analytically combined. Additionally, using both fMRI and MEG, rs3918242 was associated with right IPL working memory-related activation, replicated in two cohorts and across imaging modalities. These convergent results provide further impetus for examinations of the relationship of SLC12A5 with brain structure and function in neuropsychiatric disease.
Keywords: fMRI, MEG, MMP9, rs3918242, voxel-based morphometry
A single-nucleotide polymorphism (SNP) in the promoter region of the Matrix Metalloproteinase-9 (MMP9) gene, rs3918242 or C(−1562)T, has been shown to result in the loss of binding of a nuclear protein at that site, and the C-allele of this SNP has previously been linked to risk for schizophrenia in two independent association studies (Rybakowski, Skibinska, et al. 2009; Han et al. 2011). It has been suggested that this link with schizophrenia may be related to MMP9 functioning (Vafadari et al. 2016) because of the SNP’s proximity to this gene, and because rs3918242 was initially reported to be associated with increased transcription of the MMP9 gene in macrophages (Zhang et al. 1999). This proposed relationship with schizophrenia would be of potential interest because the MMP9 gene product is a component of the extracellular matrix, which is important for cell growth, neurogenesis, neuronal migration, axonal guidance, and synaptic plasticity and remodeling in the brain (Dzwonek et al. 2004; Apte and Parks 2015). However, the association of rs3918242 with MMP9 expression was not replicable in a separate study involving both macrophage cell lines and WISH amnion-derived cells (Ferrand et al. 2002) and a relationship has not been identified in brain tissue. Additionally, despite the reported link of this SNP with risk for schizophrenia, peripheral expression of MMP9 in blood was not associated with schizophrenia diagnosis in two independent studies (Niitsu et al. 2014; Rahimi et al. 2017), suggesting that another explanation for the relationship with risk is needed.
A pathway to such an alternative explanation may be provided by examination of expression quantitative trait loci in publically available resources, including the BrainSeq database (http://www.brainseq.org/eqtl) (BrainSeq AHBGC 2015) and the Common Mind Consortium (http://commonmind.org/WP) (Senthil et al. 2017). In BrainSeq, rs3918242 was not associated with MMP9 expression, but rather was related to expression of three exons in a nearby gene, SLC12A5 (puncorr = 1.23 × 10−12, pFDR = 4.8 × 10−10), and only to expression of these SLC12A5 exons. This finding was further replicated with the same exons in the Common Mind Consortium data (puncorr = 1 × 10−6). Importantly, expression of SLC12A5, which has an initiation site just 14 kb from rs3918242, has been associated with schizophrenia diagnosis in post-mortem brains (Hyde et al. 2011; Tao et al. 2012).
The SLC12A5 gene codes for a neuron-specific K+/Cl- cotransporter, KCC2, that mediates the fast hyperpolarization of GABA (Gamba 2005; Blaesse et al. 2009). It’s expression pattern rises quickly in postnatal development, and this rise is thought to mediate the GABA switch from excitatory to inhibitory actions (Rivera et al. 1999). Specific transcripts of SLC12A5 are differentially expressed in the dorsolateral prefrontal cortex (DLPFC) of schizophrenia patients (Tao et al. 2012), and ratios between the immature NKCC1 transcript and mature KCC2 transcript are altered in the hippocampi of patients with schizophrenia (Hyde et al. 2011). In view of these in vitro findings, the relationship between rs3918242 genotype and SLC12A5 expression in post-mortem brains may offer a common molecular mechanism linking the previously unrelated associations of both rs3918242 and expression of SLC12A5 with risk for schizophrenia.
Despite the evidence detailed above, associations of rs3918242 with the structure and function of the human brain in vivo have yet to be examined. Here, we tested for effects of this SNP on brain structure (measuring gray matter volume [GMV] with structural MRI) and brain function (using both fMRI and magnetoencephalography [MEG]) in healthy humans.
Materials and Methods
Participants
Study procedures were approved by the NIH Combined Neurosciences IRB, and all participants provided written informed consent. Participants were self-described Caucasians of European ancestry, non-Hispanic and were free from psychiatric, neurological, and major medical illnesses, including any substance use disorder, as determined by clinician-administered structured clinical interview for DSM-IV (SCID-IV), clinical brain MRI, routine laboratory tests, medical history, and physical examination.
For structural MRI, there were 298 participants (an initial cohort of 220 individuals and a replication cohort of 78 individuals). For fMRI during N-back working memory, there were 554 participants (an initial cohort of 444 individuals and a replication cohort of 110). For MEG, there was a single cohort of 190 participants who performed the same N-back working memory paradigm as in the fMRI study. All participants were genotyped, and for structural MRI and fMRI, individuals were assigned to initial or replication cohorts based on chronology of genotyping. Demographic variables and MMP9 genetic distribution are shown in Table 1.
Table 1.
Sample demographics and rs3918242 genotyping
| Analysis | Sample | N | Age | Sex | rs3918242 genotype (CC/TC/TT) | HWE P-value |
|---|---|---|---|---|---|---|
| Structural | Initial | 220 | 33.9 ± 10.7 years | 116 female | 173/46/1 | P = 0.26 |
| 104 male | ||||||
| Replication | 78 | 30.0 ± 9.5 years | 41 female | 50/25/3 | P = 0.95 | |
| 37 male | ||||||
| N-back fMRI | Initial | 444 | 30.4 ± 8.8 years | 250 female | 340/97/7 | P = 0.98 |
| 194 male | ||||||
| Replication | 110 | 30.2 ± 9.4 years | 58 female | 74/33/3 | P = 0.77 | |
| 52 male | ||||||
| N-back MEG | Initial only | 190 | 32.3 ± 10.0 years | 106 female | 139/50/1 | P = 0.12 |
| 84 male |
Genotyping Procedures
Genotyping was carried out on DNA extracted from lymphoblast cell lines derived from each individual. As data from this study has been collected over time, genotyping was performed in multiple steps with an increasing number of SNPs genotyped at each step. All genotyping was done on Illumina QUAD SNP chips (ranging from 550 K to 2.5 M SNPs). Rs3918242 genotype information for each participant was obtained from an imputed genome created according to the following procedure. Pre-imputation quality control procedures were performed separately for each SNP chip, based on prior reported methods (Anderson et al. 2010). Only participants who clustered with HapMap3 CEU and TSI populations were retained for further analysis to ensure that potential results were not related to ancestry. Prior to imputation, phasing was performed on each SNP chip separately using Shapeit version 2.2 (Delaneau et al. 2013), and then imputation was performed (also on each chip separately) using IMPUTE2 (Howie et al. 2011). For the largest, densest microarray chip (the Illumina HumanOmni2.5-v1.2 chip) imputation was performed using the 1000 Genomes phase 3 data as a reference panel (Sudmant et al. 2015). For all other chips, imputation was performed using the imputed result of the HumanOmni2.5 chip as a reference panel. SNP concordance rates of the imputation were 98% for all chips, as reported by IMPUTE2. The individual chip imputations were combined to yield a final imputed genome, such that only SNPs with high quality imputation (INFO > 0.9) in all chips were retained. rs3918242 satisfied these criteria, with an INFO score = 0.990 and Certainty score = 0.997, and genotype at this SNP was extracted for further use in neuroimaging analyses. Because TT homozygotes are rare, for primary analyzes they were combined with TC heterozygotes into a T-allele carrier group, and that group was compared to CC homozygotes (Table 1). Additionally, to ensure that our results were not driven by the rare TT homozygotes, we repeated all analyses with these individuals removed.
Structural MRI Acquisition and Processing
Three-dimensional T1-weighted structural MRI scans were acquired on a 3-Tesla GE scanner (GE Medical Systems, Milwaukee, WI, USA) using a magnetization prepared rapid gradient echo (MPRAGE) sequence (repetition time 7.28 ms, echo time 2.74 ms, 120–136 slices, resolution 0.859 mm × 0.859 mm × 1.2 mm). Preprocessing of structural images included intensity nonuniformity normalization (Sled et al. 1998) and diffeomorphic registration to MNI space. Voxel-wise Jacobian-modulated gray matter maps in MNI space were calculated for each participant using SPM8 and DARTEL (Ashburner 2007), and subsequent maps were smoothed with a 6 mm FWHM kernel.
Functional MRI Acquisition and Processing
Because the SNP studied here has been associated with schizophrenia and because a large body of evidence links working memory functioning to schizophrenia, we tested for associations between rs3918242 variation and fMRI activation during an N-back working memory paradigm (with a T2-weighted echo planar BOLD sequence, repetition time = 2000 ms, echo time = 30 ms, flip angle = 90°, 3.75 mm × 3.75 mm × 6 mm voxels). The paradigm consisted of 30-second blocks of a numerical 2-back working memory task alternating with 30-second blocks of a 0-back sensorimotor control task. As described previously (Callicott et al. 2003), this paradigm reliably activates a circuit involving prefrontal cortex and the inferior parietal lobule (IPL) (Owen et al. 2005). Chance level on this four-choice paradigm is 25%, and only participants who performed with greater than 50% accuracy were included for subsequent analyses. Processing methods applied to the N-back functional data have been previously described (Callicott et al. 1998). Briefly, images were motion corrected, aligned to an MNI standard-space template, and smoothed using an 8 mm FWHM kernel. Individuals’ first-level 2-back versus 0-back contrast maps were modeled using SPM5.
MEG Acquisition and Processing
MEG data were collected during the same N-back working memory paradigm used for fMRI with a 275-channel whole head MEG system (CTF Systems, CA) in a magnetically shielded room. Additional background noise reduction was provided by using an online synthetic third order gradiometer calculation. Data were digitized at a 600 Hz sampling rate and filtered with a 60 Hz notch filter to reduce power line noise. Head position and motion were measured using reference coils affixed to the nasion and bilateral preauricular points, which also allowed later coregistration of each individual’s MEG data to his or her structural MRI (obtained separately on a 3-Tesla GE MRI scanner with the same fiducial points marked).
Neuromagnetic data were localized to brain regions with synthetic aperture magnetometry (SAM) (Vrba and Robinson 2001), a minimum-variance beamformer method. This analysis focused on the beta band because MEG signals from this range are most often linked to fMRI BOLD activation, such that increased BOLD signal (as measured with the fMRI task here) is associated with increased MEG beta desynchronization (Singh et al. 2002). Using SAM, beta band (14–30 Hz) source power during the 400 milliseconds leading up to correct button press responses for the 2-back working memory condition was compared to the 0-back sensorimotor control condition for each individual. Log power ratios were calculated throughout the brain at 5 mm-spaced grid points (see (Rutter et al. 2009) for more mathematical details of SAM). For each individual, the resulting image was transformed into standard MNI space, and data were range-normalized by subtracting the whole-brain mean and dividing by the standard deviation using AFNI tools (Cox 1996).
Between-Genotype Statistical analyses
For structural and functional MRI, we first separately considered the data for our two independent cohorts, an initial discovery group and a replication cohort for each of these two imaging modalities. Group-level contrast maps comparing rs3918242 CC homozygotes to T-allele carriers (TC/TT genotype groups) were computed using the 3dttest++ function of AFNI (controlling for age, sex, brain volume, and the first five principal components (PCs) from a genetic principal components analysis to control for ancestry in the structural groups; and age, sex, task performance, and the first five ancestry PCs in the fMRI groups). The resulting contrast maps were thresholded at a voxel-wise P < 0.005. Clusters were reported if they were (1) at least 1000 mm3 in volume and (2) located in the same anatomical region in both cohorts.
Next, to test whether these results withstood correction for multiple comparisons, and to provide increased statistical power to uncover additional brain regions affected by genotype, statistical maps of the structural and functional MRI data for both the discovery and replication groups were combined in a meta-analytic fashion, using the Stouffer’s Z-score method to yield a combined Z statistic for each voxel (Stouffer 1949). The resulting combined maps were corrected for multiple comparisons (family-wise error correction; FWE) using the 3dClustSim function of AFNI, after estimating the smoothness of the underlying data with the ACF method (Cox et al. 2017). Using an uncorrected voxel-level threshold of P < 0.005, clusters greater than 923 voxels for the structural MRI data and clusters greater than 64 voxels for the fMRI data were significant at an FWE-corrected P < 0.05 level.
For the single MEG cohort, AFNI’s 3dttest++ was used to create group-level contrast maps of rs3918242 genotype cohorts, controlling for age, sex, task performance, and the first five ancestry PCs. Voxel-wise data were thresholded at a P < 0.005 level, and clusters greater than 1000 mm3 in volume were reported.
Results
rs3918242 Genotyping
rs3918242 SNP genotype was in Hardy-Weinberg equilibrium in all samples. Table 1 shows demographic and rs3918242 genotype information for each cohort. Genotype groups did not significantly differ by age, sex, brain volume, or task performance in any cohort (all P's > 0.2). While the results of the primary analyses that follow are based on comparing T-carriers (including both heterozygotes and rare TT homozygotes) to CC homozygotes, it should be noted that repeating all analyses after removing the TT homozygotes (1–7 per group), did not alter the location or significance of any reported clusters.
Structural MRI
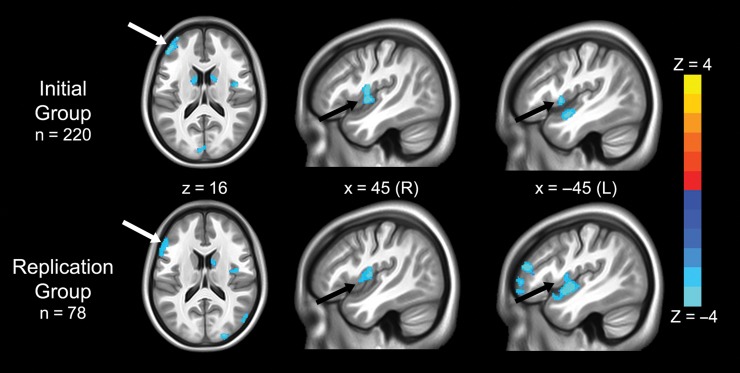
We first tested the structural MRI datasets for changes in GMV related to rs3918242 separately in the initial group of 220 participants and in the replication group of 78 participants. There was a high degree of anatomical overlap across the two groups in four regions: the bilateral posterior insula, left DLPFC, and right caudate (Fig. 1 and Table 2), and the direction of the findings was the same in both groups: in all identified regions, T-allele carriers had greater GMV than those homozygous for the C-allele.
Figure 1.
Regions where gray matter volume was related to rs3918242 variation in independent groups. Blue colors indicate voxels where T-allele carriers had increased gray matter volume compared with CC homozygotes in the initial group of 220 participants (top row) and the replication group of 78 participants (bottom row). There were no regions in which gray matter volume was greater in C homozygotes. Note the high degree of spatial overlap across groups, specifically in the left dorsolateral prefrontal cortex (white arrows) and bilateral insula (black arrows). Statistical maps are thresholded at P < 0.005, uncorrected, and show clusters larger than 1000 mm3.
Table 2.
Cluster size, MNI coordinates, and peak Z-score of significant clusters identified by structural and functional brain analyses
| Location of maxima | Cluster size (mm3) | Peak voxel MNI coordinates | Peak Z-score | ||
|---|---|---|---|---|---|
| x | y | z | |||
| Gray matter volume | |||||
| Initial sample | |||||
| Left insula | 2480 | −45 | −3 | −4.5 | 4.67 |
| Left DLPFC | 2211 | −34.5 | 57 | 9 | 4.16 |
| Right insula | 1410 | 45 | 3 | 1.5 | 4.21 |
| Replication sample | |||||
| Left DLPFC | 3807 | −52.5 | 18 | 19.5 | 3.87 |
| Left insula | 1168 | −46.5 | −4.5 | 1.5 | 3.72 |
| Right insula | 1137 | 45 | −6 | 9 | 3.57 |
| Combined meta-analysis | FWE P-value | ||||
| Left insula | 7617 | −45 | −1.5 | −7.5 | ≪0.01 |
| Right insula | 6369 | 46.5 | −4.5 | 10.5 | ≪0.01 |
| Right IPL | 5991 | 57 | −66 | 12 | ≪0.01 |
| Left DLPFC | 5457 | −54 | 19.5 | 24 | ≪0.01 |
| Left fusiform | 3446 | −55.5 | −60 | −16.5 | 0.03 |
| Left IPL | 3513 | −45 | −79.5 | 30 | 0.03 |
| fMRI N-back activation | Peak Z-score | ||||
| Initial sample | |||||
| Right IPL | 1512 | 60 | −48 | 21 | 3.13 |
| Replication sample | |||||
| Right IPL | 1539 | 54 | −69 | 21 | 3.73 |
| Combined meta-analysis | FWE P-value | ||||
| Left visual cortex | 5130 | −12 | −90 | 12 | ≪0.01 |
| Right IPL | 2025 | 57 | −69 | 18 | 0.03 |
| MEG N-back beta band desynchronization | Peak Z-score | ||||
| Right IPL | 7125 | 57.5 | −47.5 | 37.5 | 3.78 |
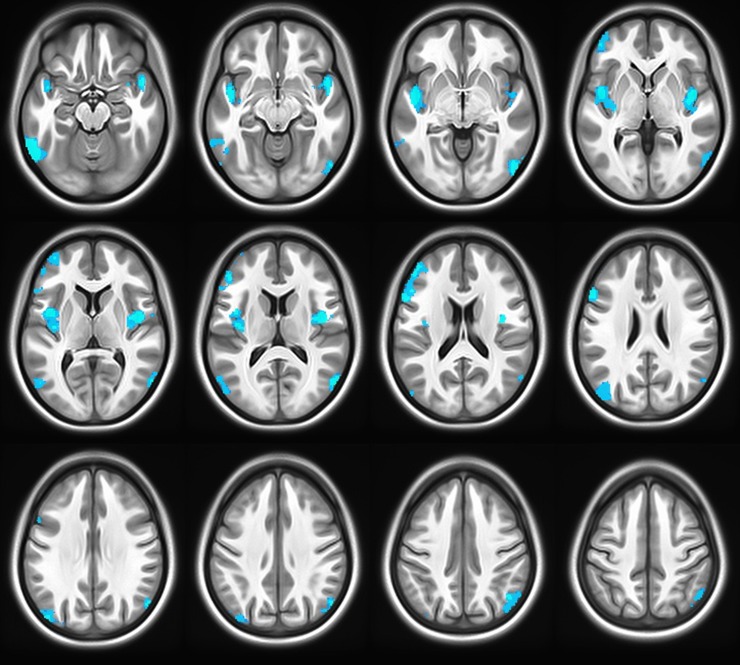
Next, when the combined voxel-wise Z-scores of the two groups were tested meta-analytically, all four of these regions were significant at a P < 0.05 threshold after FWE correction for multiple comparisons. Additionally, three other regions not observed previously in either of the individual cohorts separately also reached significance at P < 0.05 with FWE correction: the IPLs extending to occipito-temporal regions bilaterally, and a cluster in the left fusiform gyrus (Fig. 2 and Table 2).
Figure 2.
Associations between gray matter volume and rs3918242 after combining the initial and replication groups using Stouffer’s Z-score method. Significant clusters at P < 0.05, corrected for family-wise error. Blue indicates voxels in which T-allele carriers had increased gray matter volume compared with CC homozygotes. As expected, regions identified in both independent groups, left DLPFC and bilateral insulae (Fig. 1) were also highlighted here, as were several regions found only in the combined analysis. These included the inferior parietal lobules and fusiform gyrus.
N-Back Working Memory Task—fMRI
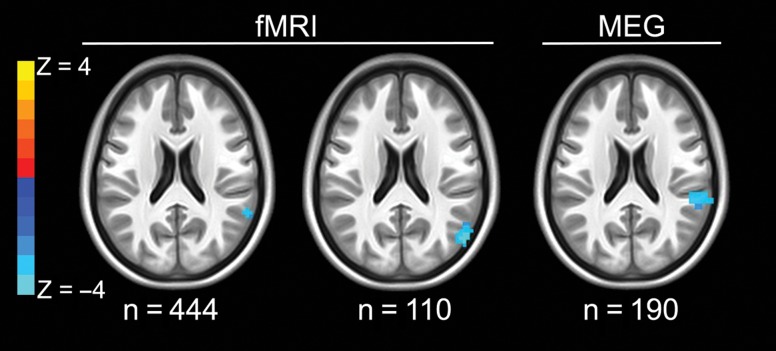
We then tested for differences in working memory-related BOLD signal as a function of rs3918242 genotype separately in the initial group of 444 and in the replication group of 110 individuals. A region in the right IPL showed differential BOLD activation as a function of genotype in both groups, such that T-allele carriers showed more robust neural recruitment than CC homozygotes (Fig. 3 and Table 2). No other regions showed differential activation in both groups.
Figure 3.
Working memory-related neurofunctional changes in the right inferior parietal lobule according to rs3918242 genotype during fMRI and MEG. Axial slices shown are at z = 21. Blue colors represent greater working memory-related activation in T-allele carriers compared with CC homozygotes, for fMRI (left: initial discovery group, n = 444; middle: replication group, n = 110) and increased beta desynchronization in T-allele carriers for the MEG data (rightmost slice: n = 190). Statistical maps are thresholded at P < 0.005 uncorrected and clusters larger than 1000 mm3.
When the combined Z-scores of the two groups were tested meta-analytically, we observed differential activation in this region of the right IPL that was significant at a threshold of P < 0.05 after FWE correction for multiple comparisons (Table 2). Moreover, this cluster, identified with functional imaging, overlapped with the right IPL cluster found in the GMV structural analysis. Additionally, the left occipital cortex was observed to have differential activation in the opposite direction (CC>T-allele carriers).
N-Back Working Memory Task—MEG
Finally, we tested for rs3918242 genotype-associated differences in brain activation as measured with MEG during the same N-back working memory paradigm. In a whole-brain analysis, we found genotype-related differential beta band activation only in a portion of the right IPL (Fig. 3 and Table 2). This region was similar to the right IPL area seen with both the combined structural MRI findings and the functional MRI findings. Moreover, the direction of the genotype differences in the MEG cohort (increased beta band desynchronization in T-allele carriers compared to CC homozygotes) was consistent with the direction of the BOLD activation findings in the fMRI cohorts (increased activation in T-allele carriers compared to CC homozygotes), reflecting the known relationship of these neurofunctional measures (Hall et al. 2014).
Discussion
Compelling pre-clinical and in vitro research has documented a role for SLC12A5 in brain development and pathology, but relevant in vivo studies of the human brain have been lacking. Here, using convergent neuroimaging methodologies—structural MRI, fMRI, and MEG—we identified brain regions where GMV and neural recruitment are affected by the functional variant rs3918242, which, in two independent databases, is related to SLC12A5 expression. We showed that the effect of this SNP on the living human brain is especially prominent in the right IPL, where T-allele carriers of rs3918242 exhibited increased GMV, increased BOLD activation during working memory as measured with fMRI, and increased beta band desynchronization as measured with MEG during the same working memory task. Additionally, this SNP was associated with GMV changes in the bilateral posterior insula and left DLPFC, findings that replicated in two independent cohorts.
The right IPL, particularly highlighted by the observed differential associations with rs3918242 variation in GMV and in two measures of working memory-related neural function, is a critical node that is reliably recruited in healthy subjects by working memory tasks such as the N-back task employed here (Berman et al. 1995; Glabus et al. 2003). Additionally, patients with IPL lesions show deficits in this cognitive domain (Berryhill and Olson 2008). The current results suggest that SLC12A5 plays a key role in this working memory-related region.
The left DLPFC, as the IPL, also showed significant differences in GMV as a function of genotype, with T-allele carriers of the rs3918242 SNP again having increased GMV compared to C-allele homozygotes. The IPL and DLPFC together form a network that is central to working memory (Salazar et al. 2012; Eriksson et al. 2015; Constantinidis and Klingberg 2016; Christophel et al. 2017). Specifically, meta-analyses have shown that these regions are preferentially activated during working memory tasks (Wager and Smith 2003; Owen et al. 2005). Furthermore, connectivity of these regions is modulated by working memory function and is diminished in patients with schizophrenia (Nielsen et al. 2017), consistent with our findings of reduced GMV in both regions and reduced working memory-related IPL activation in participants with the C-allele, the genotype previously associated with schizophrenia.
In addition to our findings in IPL and DLPFC, we observed that GMV of the insula varied according to rs3918242 genotype, with T-carriers increased compared to C homozygotes in both independent samples, but we did not find co-localizing differences in brain function during working memory with either fMRI or MEG. However, the posterior portion of the insula, in contrast to the IPL, is not typically activated by working memory tasks such as the one used here (Owen et al. 2005) and, thus, our study may not have provided a sufficient “reflex hammer” for probing genotype-related neural function in this locale. Given the significant and replicable bilateral structural variation observed in the insula in both our discovery and replication cohorts, future functional imaging studies using other task probes specifically designed to activate this region may reveal differential brain function.
In contrast to the IPL, DLPFC, and insula, there was only a single region where CC homozygotes showed greater structure or function than T-allele carriers: activation of the left visual cortex during fMRI. However, it should be noted that this finding in occipital cortex was not observed in either of the independent samples individually, but, rather, only when the fMRI groups were combined in a meta-analytic fashion. Moreover, this result was not supported by the structural MRI or MEG studies, and interpretations of activation in the visual cortex during our task paradigm are difficult as this region is not differentially recruited during working memory. Thus, with the exception of this single observation, we found that T-carriers showed increased GMV and functional activation compared to individuals homozygous for the C-allele (the allele that has been associated with schizophrenia [Rybakowski, Skibinska, et al. 2009; Han et al. 2011]).
The present findings, therefore, may have implications for schizophrenia and other neuropsychiatric disorders. It is notable that all regions identified here as having genotype-related differential GMV in both of our cohorts—the DLPFC, the IPL, and the insula—have been reported to have decreased GMV in multiple meta-analyses of schizophrenia (Ellison-Wright et al. 2008; Radua et al. 2012; Ren et al. 2013). While MRI measurement of GMV may include non-neuronal factors, it is of interest that this parameter was decreased in our participants homozygous for the C-allele of the rs3918242 compared to those with the T-allele, consistent with the directionality of prior GMV findings in patients with schizophrenia, and with the fact that the C-allele, is a risk factor for schizophrenia (Rybakowski, Skibinska, et al. 2009; Han et al. 2011). However, it should be noted that though rs3918242 was associated with schizophrenia in these two case–control studies, it has not been reliably associated with schizophrenia through large-scale GWAS analyses (Schizophrenia Working Group of the Psychiatric Genomics 2014).
Further, impairments in DLPFC-IPL circuitry, along with associated working memory performance deficits (Goldman-Rakic 1994), are core features of schizophrenia. While we did not observe genotype effects on task performance in our healthy volunteers—consistent with a prior negative cognitive study of the same SNP studied here, (Rybakowski, Borkowska, et al. 2009) and with a negative study of episodic memory in healthy controls genotyped for another repeat polymorphism in the same genetic region (Vassos et al. 2008)—it is possible that, in patients, this variation interacts with additional disease-related genetic or other variables to affect cognitive performance. Thus, the constellation of findings in healthy adults reported here may provide anatomic information to guide searches for SLC12A5's effects on brain structure and function in patients.
The neural effects of SLC12A5 and the relationship of its expression with schizophrenia are putatively the result of its known involvement in GABA functioning. As mentioned above, SLC12A5 codes for a K+/Cl− cotransporter that mediates the GABA switch from excitatory to inhibitory (Rivera et al. 1999), thus offering biological plausibility for this explanation. A role for SLC12A5, rather than for MMP9, is more likely because the evidence implicating rs3918242 as being functional for MMP9 expression was only observed in macrophages (Zhang et al. 1999) and was not replicated (Ferrand et al. 2002). Moreover, a relationship between rs3918242 and MMP9 expression does not appear to exist in the human brain. Instead, two publically available expression datasets show significant association between rs3918242 and SLC12A5 expression, but not MMP9. Furthermore, the SLC12A5 gene lies just 14 kb telomeric to rs3918242 and SNPs surrounding its initiation site are in high linkage disequilibrium with rs3918242, with an R2 as high as 0.986 (per http://analysistools.nci.nih.gov/LDLink). Taken together, this evidence offers molecular plausibility for SLC12A5's role in the results reported here.
In sum, we identified brain regions where genetic variation at rs3918242 is associated with variation in brain structure and function. The results are regionally convergent across both structural and functional imaging methodologies and are reinforced by the internal replication of the study design. These data are of particular interest in view of prior associations between the SNP we tested (rs3918242) and mental illness. The implicated regions may, therefore, provide targets for future neuroimaging and molecular investigations of this association and SLC12A5's role in pathophysiology.
Notes
All authors report no competing financial interests with regard to this manuscript. The data for this work were obtained under protocol 00M0085/NCT00004571 and protocol 95M0150/NCT00001486. Some of this work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). Conflict of Interest: The authors declare no conflicts of interest related to this work.
Funding
Intramural Research Program, National Institute of Mental Health, National Institutes of Health, Bethesda, MD (ZIAMH002942).
References
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. 2010. Data quality control in genetic case-control association studies. Nat Protoc. 5:1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte SS, Parks WC. 2015. Metalloproteinases: a parade of functions in matrix biology and an outlook for the future. Matrix Biol. 44–46:1–6. [DOI] [PubMed] [Google Scholar]
- Ashburner J. 2007. A fast diffeomorphic image registration algorithm. Neuroimage. 38:95–113. [DOI] [PubMed] [Google Scholar]
- Berman KF, Ostrem JL, Randolph C, Gold J, Goldberg TE, Coppola R, Carson RE, Herscovitch P, Weinberger DR. 1995. Physiological activation of a cortical network during performance of the Wisconsin Card Sorting Test: a positron emission tomography study. Neuropsychologia. 33:1027–1046. [DOI] [PubMed] [Google Scholar]
- Berryhill ME, Olson IR. 2008. The right parietal lobe is critical for visual working memory. Neuropsychologia. 46:1767–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaesse P, Airaksinen MS, Rivera C, Kaila K. 2009. Cation-chloride cotransporters and neuronal function. Neuron. 61:820–838. [DOI] [PubMed] [Google Scholar]
- BrainSeq AHBGC 2015. BrainSeq: neurogenomics to drive novel target discovery for neuropsychiatric disorders. Neuron. 88:1078–1083. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Egan MF, Mattay VS, Bertolino A, Bone AD, Verchinksi B, Weinberger DR. 2003. Abnormal fMRI response of the dorsolateral prefrontal cortex in cognitively intact siblings of patients with schizophrenia. Am J Psychiatry. 160:709–719. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Ramsey NF, Tallent K, Bertolino A, Knable MB, Coppola R, Goldberg T, van Gelderen P, Mattay VS, Frank JA, et al. 1998. Functional magnetic resonance imaging brain mapping in psychiatry: methodological issues illustrated in a study of working memory in schizophrenia. Neuropsychopharmacology. 18:186–196. [DOI] [PubMed] [Google Scholar]
- Christophel TB, Klink PC, Spitzer B, Roelfsema PR, Haynes JD. 2017. The distributed nature of working memory. Trends Cogn Sci. 21:111–124. [DOI] [PubMed] [Google Scholar]
- Constantinidis C, Klingberg T. 2016. The neuroscience of working memory capacity and training. Nat Rev Neurosci. 17:438–449. [DOI] [PubMed] [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 29:162–173. [DOI] [PubMed] [Google Scholar]
- Cox RW, Chen G, Glen DR, Reynolds RC, Taylor PA. 2017. FMRI clustering in AFNI: false-positive rates redux. Brain Connect. 7:152–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 10:5–6. [DOI] [PubMed] [Google Scholar]
- Dzwonek J, Rylski M, Kaczmarek L. 2004. Matrix metalloproteinases and their endogenous inhibitors in neuronal physiology of the adult brain. FEBS Lett. 567:129–135. [DOI] [PubMed] [Google Scholar]
- Ellison-Wright I, Glahn DC, Laird AR, Thelen SM, Bullmore E. 2008. The anatomy of first-episode and chronic schizophrenia: an anatomical likelihood estimation meta-analysis. Am J Psychiatry. 165:1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Vogel EK, Lansner A, Bergstrom F, Nyberg L. 2015. Neurocognitive Architecture of Working Memory. Neuron. 88:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrand PE, Parry S, Sammel M, Macones GA, Kuivaniemi H, Romero R, Strauss JF 3rd. 2002. A polymorphism in the matrix metalloproteinase-9 promoter is associated with increased risk of preterm premature rupture of membranes in African Americans. Mol Hum Reprod. 8:494–501. [DOI] [PubMed] [Google Scholar]
- Gamba G. 2005. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev. 85:423–493. [DOI] [PubMed] [Google Scholar]
- Glabus MF, Horwitz B, Holt JL, Kohn PD, Gerton BK, Callicott JH, Meyer-Lindenberg A, Berman KF. 2003. Interindividual differences in functional interactions among prefrontal, parietal and parahippocampal regions during working memory. Cereb Cortex. 13:1352–1361. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS. 1994. Working memory dysfunction in schizophrenia. J Neuropsychiatry Clin Neurosci. 6:348–357. [DOI] [PubMed] [Google Scholar]
- Hall EL, Robson SE, Morris PG, Brookes MJ. 2014. The relationship between MEG and fMRI. Neuroimage. 102:80–91. [DOI] [PubMed] [Google Scholar]
- Han H, He X, Tang J, Liu W, Liu K, Zhang J, Wang X, Xu Y, Chen X. 2011. The C(-1562)T polymorphism of matrix metalloproteinase-9 gene is associated with schizophrenia in China. Psychiatry Res. 190:163–164. [DOI] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. 2011. Genotype imputation with thousands of genomes. G3. 1:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde TM, Lipska BK, Ali T, Mathew SV, Law AJ, Metitiri OE, Straub RE, Ye T, Colantuoni C, Herman MM, et al. 2011. Expression of GABA signaling molecules KCC2, NKCC1, and GAD1 in cortical development and schizophrenia. J Neurosci. 31:11088–11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JD, Madsen KH, Wang Z, Liu Z, Friston KJ, Zhou Y. 2017. Working memory modulation of frontoparietal network connectivity in first-episode schizophrenia. Cereb Cortex. 27:3832–3841. [DOI] [PubMed] [Google Scholar]
- Niitsu T, Ishima T, Yoshida T, Hashimoto T, Matsuzawa D, Shirayama Y, Nakazato M, Shimizu E, Hashimoto K, Iyo M. 2014. A positive correlation between serum levels of mature brain-derived neurotrophic factor and negative symptoms in schizophrenia. Psychiatry Res. 215:268–273. [DOI] [PubMed] [Google Scholar]
- Owen AM, McMillan KM, Laird AR, Bullmore E. 2005. N-back working memory paradigm: a meta-analysis of normative functional neuroimaging studies. Hum Brain Mapp. 25:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radua J, Borgwardt S, Crescini A, Mataix-Cols D, Meyer-Lindenberg A, McGuire PK, Fusar-Poli P. 2012. Multimodal meta-analysis of structural and functional brain changes in first episode psychosis and the effects of antipsychotic medication. Neurosci Biobehav Rev. 36:2325–2333. [DOI] [PubMed] [Google Scholar]
- Rahimi S, Sayad A, Moslemi E, Ghafouri-Fard S, Taheri M. 2017. Blood assessment of the expression levels of matrix metalloproteinase 9 (MMP9) and its natural inhibitor, TIMP1 genes in Iranian schizophrenic patients. Metab Brain Dis. 32:1537–1542. [DOI] [PubMed] [Google Scholar]
- Ren W, Lui S, Deng W, Li F, Li M, Huang X, Wang Y, Li T, Sweeney JA, Gong Q. 2013. Anatomical and functional brain abnormalities in drug-naive first-episode schizophrenia. Am J Psychiatry. 170:1308–1316. [DOI] [PubMed] [Google Scholar]
- Rivera C, Voipio J, Payne JA, Ruusuvuori E, Lahtinen H, Lamsa K, Pirvola U, Saarma M, Kaila K. 1999. The K+/Cl- co-transporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature. 397:251–255. [DOI] [PubMed] [Google Scholar]
- Rutter L, Carver FW, Holroyd T, Nadar SR, Mitchell-Francis J, Apud J, Weinberger DR, Coppola R. 2009. Magnetoencephalographic gamma power reduction in patients with schizophrenia during resting condition. Hum Brain Mapp. 30:3254–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakowski JK, Borkowska A, Skibinska M, Kaczmarek L, Hauser J. 2009. The-1562 C/T polymorphism of the matrix metalloproteinase-9 gene is not associated with cognitive performance in healthy participants. Psychiatr Genet. 19:277–278. [DOI] [PubMed] [Google Scholar]
- Rybakowski JK, Skibinska M, Kapelski P, Kaczmarek L, Hauser J. 2009. Functional polymorphism of the matrix metalloproteinase-9 (MMP-9) gene in schizophrenia. Schizophr Res. 109:90–93. [DOI] [PubMed] [Google Scholar]
- Salazar RF, Dotson NM, Bressler SL, Gray CM. 2012. Content-specific fronto-parietal synchronization during visual working memory. Science. 338:1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil G, Dutka T, Bingaman L, Lehner T. 2017. Genomic resources for the study of neuropsychiatric disorders. Mol Psychiatry. 22:1659–1663. [DOI] [PubMed] [Google Scholar]
- Singh KD, Barnes GR, Hillebrand A, Forde EM, Williams AL. 2002. Task-related changes in cortical synchronization are spatially coincident with the hemodynamic response. Neuroimage. 16:103–114. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. 1998. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 17:87–97. [DOI] [PubMed] [Google Scholar]
- Stouffer SA. 1949. The American soldier. Princeton: Princeton University Press. [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature. 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Li C, Newburn EN, Ye T, Lipska BK, Herman MM, Weinberger DR, Kleinman JE, Hyde TM. 2012. Transcript-specific associations of SLC12A5 (KCC2) in human prefrontal cortex with development, schizophrenia, and affective disorders. J Neurosci. 32:5216–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafadari B, Salamian A, Kaczmarek L. 2016. MMP-9 in translation: from molecule to brain physiology, pathology, and therapy. J Neurochem. 139:91–114. [DOI] [PubMed] [Google Scholar]
- Vassos E, Ma X, Fiotti N, Wang Q, Sham PC, Liu X, Wang Y, Yan C, Meng H, Deng W, et al. 2008. The functional MMP-9 microsatellite marker is not associated with episodic memory in humans. Psychiatr Genet. 18:252. [DOI] [PubMed] [Google Scholar]
- Vrba J, Robinson SE. 2001. Signal processing in magnetoencephalography. Methods. 25:249–271. [DOI] [PubMed] [Google Scholar]
- Wager TD, Smith EE. 2003. Neuroimaging studies of working memory: a meta-analysis. Cogn Affect Behav Neurosci. 3:255–274. [DOI] [PubMed] [Google Scholar]
- Zhang B, Henney A, Eriksson P, Hamsten A, Watkins H, Ye S. 1999. Genetic variation at the matrix metalloproteinase-9 locus on chromosome 20q12.2–13.1. Hum Genet. 105:418–423. [DOI] [PubMed] [Google Scholar]