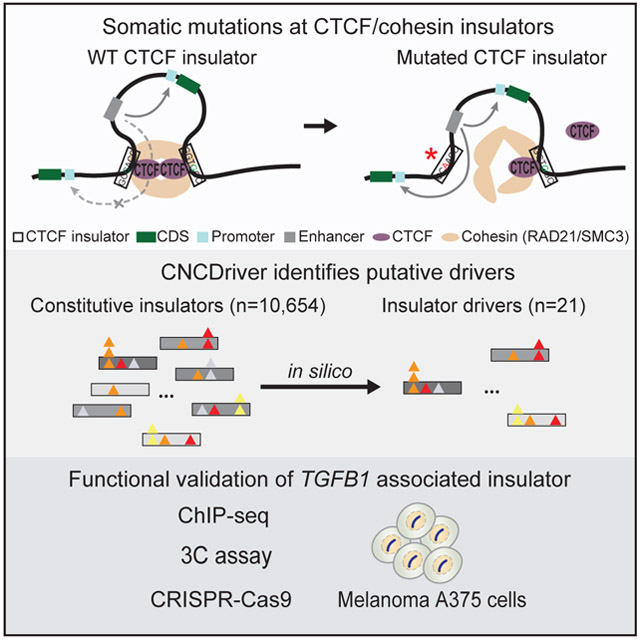
SUMMARY
Recent studies have shown that mutations at non-coding elements, such as promoters and enhancers, can act as cancer drivers. However, an important class of non-coding elements, namely CTCF insulators, has been overlooked in the previous driver analyses. We used insulator annotations from CTCF and cohesin ChIA-PET and analyzed somatic mutations in 1,962 whole-genomes from 21 cancer types. Using the heterogeneous patterns of transcription-factor-motif-disruption, functional impact and recurrence of mutations, we developed a computational method that revealed 21 insulators showing signals of positive selection. In particular, mutations in an insulator in multiple cancer types, including 16% of melanoma samples, are associated with TGFB1 up-regulation. Using CRISPR-Cas9, we find that alterations at two of the most frequently mutated regions in this insulator increase cell growth by 40-50%, supporting the role of this boundary element as a cancer driver. Thus, our study reveals several CTCF insulators as putative cancer drivers.
Graphical Abstract
In Brief
We developed a computational method that combines recurrence and functional impact of mutations to identify cancer drivers. Application of the method on 1,962 cancer whole-genomes reveals putative drivers at CTCF insulators. In particular, mutations in an insulator on chr19 are associated with TGFB1 up-regulation and may point to a novel mechanism of TGF-β signaling modulation in multiple cancer types.
INTRODUCTION
Whole-genome sequencing (WGS) of tumors has revealed that most somatic mutations occur in non-coding regions (Khurana et al., 2016). Although most of these mutations do not impact tumor growth and are called passengers, some of them can act as cancer drivers by conferring growth advantage to promote tumorigenesis. Non-coding cancer driver mutations can be identified by detecting signals of positive selection in WGS data.
Non-coding drivers at transcription factor (TF) binding sites in promoters and enhancers play a role in tumorigenesis by dysregulating gene expression. The most prominent example is the TERT promoter, which is mutated in many cancers (Vinagre et al., 2013). In other prominent examples, promoter and enhancer mutations in breast cancer can lead to FOXA1 (Rheinbay et al., 2017) and ESR1 (Bailey et al., 2016) overexpression, respectively. Most efforts to identify cis-regulatory regions under positive selection in cancer have focused on promoters and enhancers and a key functional element, namely CTCF/cohesin insulators, has been overlooked.
It is known that promoter and enhancer interactions are facilitated by partitioning of the human genome into DNA loops (Gibcus and Dekker, 2013; Gorkin et al., 2014; Rao et al., 2014). The loops that act as insulated neighborhoods preventing the interactions of promoters and enhancers across their boundaries are predominantly mediated by CCCTC-binding factors (CTCF) and cohesin (SMC1, SMC3, RAD21 and either STAG2 or STAG1) bound at the loop ends (Dowen et al., 2014; Hnisz et al., 2016; Ji et al., 2016; Tang et al., 2015). Emerging evidence from Hi-C and ChIP-seq assays suggest that other proteins, such as BRD2 (Hsu et al., 2017) and ELK4 (Mourad and Cuvier, 2018), can also co-localize with CTCF at loop anchors to affect long-range chromatin interactions. Disruption of the loop anchor regions, called CTCF/cohesin insulators (hereafter referred to as insulators), can lead to de novo enhancer-promoter interactions and the subsequent dysregulation of associated genes (Giorgio et al., 2015; Lupiáñez et al., 2015). Furthermore, smaller loops can cluster to form larger megabase-sized loops called topologically associated domains (TADs) (Bouwman and de Laat, 2015; Phillips-Cremins et al., 2013; Rao et al., 2014; Vietri Rudan et al., 2015).
It has been previously reported that CTCF/cohesin binding sites are highly mutated in several cancer types, including gastrointestinal cancers and melanoma (Guo et al., 2018; Kaiser et al., 2016; Katainen et al., 2015; Poulos et al., 2016). Although the reasons for the high mutation rates are not completely understood, these studies noted that most of these mutations are likely passengers and do not drive tumor growth. However, Hnisz et al. found the CTCF insulators of DNA loops show recurrent deletions that alter the expression of LMO2 and TALI oncogenes in T cell acute lymphoblastic leukemia (Hnisz et al., 2016). Thus, some variants at insulators may have a functional role and drive the growth of cancer cells. It is well appreciated that systematic genome-wide identification of a few drivers among tens of thousands of passengers is a challenging task. This is because computational methods to detect drivers need to account for heterogeneous mutation rates along the genome. The heterogeneous rates are a result of mutational co-variates, some of which may be specific for the type of cancer and functional element analyzed (Cuykendall et al., 2017; Khurana et al., 2016; Perera et al., 2016; Polak et al., 2015; Sabarinathan et al., 2016; Supek and Lehner, 2015). While several methods have been developed to predict non-coding drivers at promoters, enhancers, or TF binding sites in general in multiple cancer types (Araya et al., 2015; Juul et al., 2017; Lanzós et al., 2017; Lochovsky et al., 2015; Melton et al., 2015; Mularoni et al., 2016), these methods have not been developed for or applied on insulator regions.
Here, we first analyze the mutations at insulators to identify novel mutational rate covariates at these regions. We used insulator annotations obtained using ChIA-PET assays that can map long-range chromatin interactions mediated by specific proteins and therefore enable identification of chromatin loop anchor regions that are associated with CTCF and cohesin (Heidari et al., 2014; Hnisz et al., 2016; Ji et al., 2016; Tang et al., 2015). We then developed a computational method, CNCDriver (Cornell Non-Coding Driver), that incorporates the mutational rate covariates to identify insulator drivers. Out of 5,042 insulators that show recurrent mutations (i.e., present in 2 or more samples) in 1,962 whole-genomes from 21 cancer types, our method identifies 21 putative drivers. We postulate that alterations of these insulators can elicit oncogenic impact via chromatin loop rewiring. Functional validation of a predicted driver using CTCF ChIP-seq, 3C and CRISPR mutagenesis supports our computational predictions in human melanoma cells.
RESULTS
Determining mutational rate co-variates at insulators
Analysis of loop anchor regions from seven cohesin (Heidari et al., 2014; Hnisz et al., 2016) and CTCF (Li et al., 2012a; Tang et al., 2015) ChIA-PET datasets shows that the majority of them are conserved in more than one cell-type (Figure S1B, 73% insulators are conserved in more than one cell-type), as also noted by previous studies (Heidari et al., 2014; Hnisz et al., 2016; Tang et al., 2015). We only include those insulators that are conserved in at least four out of five different cell-types (GM12878, K562, Jurkat, MCF-7 and HeLa-S3) as the constitutive set in our analysis. We analyzed the patterns of somatic single nucleotide variants (SNVs) in 1,962 genomes from 21 cancer types (Table S1) at constitutive insulators. We identified the mutations predicted to disrupt CTCF binding by comparing the TF motif position weight matrix (PWM) score of the mutated vs. the reference sequence (Fu et al., 2014; Khurana et al., 2013; Mu et al., 2011) (Figure S2A and Table S14). We observe significant enrichment of CTCF motifs predicted to be disrupted due to mutations in 15 out of 21 cancer types analyzed (Figure S2B). Next, we extracted the tri-nucleotide context of the mutations predicted to disrupt CTCF motifs and compared the distributions of the 96 possible contexts with known signatures of mutational processes in human cancer from COSMIC (Alexandrov et al., 2013) as a reference control using cosine similarity as the quantitative metric (Figures S2C and S2D). We find that the tri-nucleotide distributions for CTCF motif-disrupting mutations vary considerably across cancer types and interestingly match the corresponding cancer-specific COSMIC mutational signatures in 9 cancer types (Figure S2D). As seen in Figure S2B, these 9 cancer types are also enriched for CTCF motif disruption. Thus, our results show that enrichment of CTCF motif disruption in multiple cancer types is likely due to neutral mutational processes operative in those cancers. For the remaining 6 cancer types that show enrichment of CTCF motif disruption, but do not closely match any single mutational signature identified in that cancer type, we expect the motif-disrupting mutations are likely either a combination of multiple signatures or may correspond to unknown signatures of longer sequence context (Fredriksson et al., 2017). Besides CTCF, we find that the fraction of the other 549 TFs, whose motif-disruption enrichment can be explained by known mutational processes acting in that cancer type, varies from 4% in brain low-grade glioma (LGG) to 86% in melanoma (Table S2B, Top). We provide the lists of TFs showing enriched motif disruption due to specific mutational signatures for each cancer type (Tables S2A and S2B). Our observations are consistent with those of Kaiser et al., who noted that enrichment of functional mutations at binding sites of CTCF and other TFs are likely due to neutral mutational processes, though they did not make a one-to-one comparison with the 30 mutational signatures from COSMIC (Kaiser et al., 2016). Thus, these results demonstrate that in the computational models for cancer driver detection at CTCF insulators, there is a need to balance the higher functional impact of motif-disrupting mutations and their higher frequency (Figure S3) due to background mutational processes.
Computational method to identify cancer drivers
We report a novel computational method, CNCDriver, which combines the functional impact of mutations and their recurrence across multiple cancer samples to identify the elements that show signals of positive selection. CNCDriver aims to identify the regions that show significantly more functional mutations than expected randomly. The functional impact of mutations is computed using the FunSeq2 algorithm (Methods) (Dhingra et al., 2017; Fu et al., 2014; Khurana et al., 2013). A CNCDriver score is computed for each element, which sums the functional impact and recurrence of all the mutations in the element (Figure 1A and Methods). The P value for each element is then computed by comparing its CNCDriver score to the scores in a null distribution built by repeatedly drawing the same number of mutated positions from the same element type. This framework allows us to account for previously reported covariates of mutation rates, including replication timing, mutational sequence context, DNase I hypersensitive sites (DHSs) and histone modification marks (Alexandrov et al., 2013; Polak et al., 2015). Besides insulators, it also allows identification of drivers in coding genes, promoters, enhancers and lncRNAs. To identify the insulators under positive selection, CNCDriver also incorporates the ratio of CTCF motif–disrupting to motif–preserving mutations in the null model, thus balancing the opposite effects of predicted higher functional impact of these mutations with their higher frequency (Figure S3 and Methods).
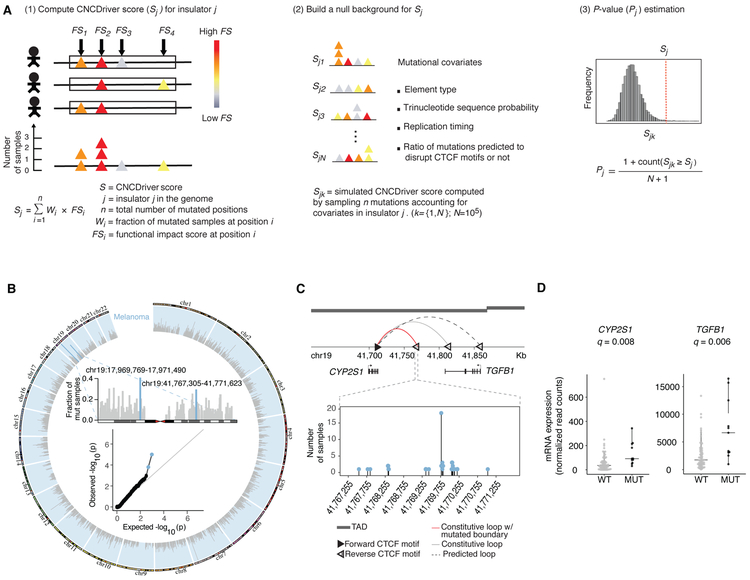
Figure 1. CNCDriver method overview and results in melanoma.
(A) CNCDriver method. CNCDriver score (Sj) combines predicted functional impact scores (FSi) and recurrence (Wi) at each mutated position in insulator j. P-value for insulator j is estimated by comparing Sj with null distribution of (Sjk). Simulated CNCDriver scores (Sjk) are obtained by sampling n mutations from other insulators accounting for mutational covariates. (B) Circos plot shows the fraction of mutated samples per insulator (light grey bars) in melanoma. Inset: (Top) Zoom-in shows the fraction of mutated samples for chr19 and the two candidate insulator drivers identified by CNCDriver in light blue bars; (Bottom): QQ-plot shows the P-value distribution and the two candidate insulator drivers in blue. (C) Predicted loop rewiring (red: original loop with significantly mutated anchor, grey: alternate constitutive loop, and dotted: predicted new loop) at the site of a candidate driver on chr19. Dark grey bars represent topological associated domains (TADs). Zoom-in needle plot shows the number of mutated samples at each position in the candidate driver. (D) Differential gene expression of TGFB1 and CYP2S1 between tumor samples with hotspot mutations (MUT) (n = 11) and those without mutations (WT) (n = 69) at candidate insulator shown in (C). See STAR Methods for CNCDriver method details. See also Figure S1-S12 and Table 1, S1 to S15.
Application of CNCDriver to coding regions demonstrates that it identifies well-known cancer genes with improved performance compared to other methods that are based solely on mutational functional bias (Mularoni et al., 2016) or burden (Lochovsky et al., 2015), demonstrating the validity of the statistical framework (Figure S4 and Table S3). Furthermore, the P values from CNCDriver follow the expected uniform distributions for all elements (Figures S4 to S8 and Tables S4 to S11), and the well-known TERT promoter is identified as a candidate in 3 cancer types (Figure S5A). We note the unique scoring scheme of CNCDriver, which includes nucleotide level impact of TF motif-disruption, allows prediction of candidate promoter drivers in melanoma where OncodriveFML gives inflated QQ plots due to hypermutated TF binding sites (Khurana et al., 2016; Mularoni et al., 2016; Perera et al., 2016; Sabarinathan et al., 2016) (Figure S5B).
Putative insulator drivers identified by CNCDriver
Using CNCDriver, we identify 21 putative insulator drivers across individual cancer types and in the joint pan-cancer analysis (Table 1 and Figure S8A). We note that only 21 insulators are predicted to be under positive selection even though many insulators show high mutational frequencies (Figure S9: grey bars in circles for each cancer type). In contrast, OncodriveFML (Mularoni et al., 2016), a method that uses only mutational functional bias to identify drivers produces high numbers of false positive hits (Figure S8B and Table S12), likely because it does not account for the higher background rates of CTCF motif-disrupting mutations associated with neutral processes. Other methods that use only high mutational burden to detect positive selection are also likely to perform poorly for CTCF insulators due to their increased mutational rates relative to flanking regions (Lanzós et al., 2017; Rheinbay et al., 2017; Weinhold et al., 2014).
Table 1. CNCDriver identified 21 putative insulator drivers, Related to Figure 1B.
Hepatocellular carcinoma (Liver), bladder urothelial carcinoma (BLCA), ovarian serous cystadenocarcinoma (Ovarian), uterine corpus endometrial carcinoma (UCEC), esophageal adenocarcinoma (ESAD), kidney cancer (Renal), colon adenocarcinoma (Colon), breast invasive carcinoma (BRCA), pancreatic adenocarcinoma (Pancreatic), and skin cutaneous melanoma (Melanoma).
| Chromosome | Insulator coordinate | Cancer Type |
|---|---|---|
| 1 | chr1:212206519-212210488 | PanCancer |
| 3 | chr3:101946566-101949238 | Liver |
| 3 | chr3:193851483-193857734 | PanCancer |
| 6 | chr6:27762081-27764969 | BLCA, PanCancer |
| 6 | chr6:28804618-28807508 | Ovarian |
| 6 | chr6:36644706-36649293 | UCEC |
| 6 | chr6:52859174-52860790 | PanCancer |
| 6 | chr6:73119843-73123728 | ESAD |
| 7 | chr7:148659158-148661781 | Renal |
| 7 | chr7:86865236-86868101 | Ovarian |
| 7 | chr7:96808154-96810356 | Colon, PanCancer |
| 8 | chr8:114448350-114451616 | ESAD |
| 12 | chr12:109830993-109832475 | BRCA |
| 14 | chr14:21076653-21082688 | Pancreatic |
| 16 | chr16:22206566-22208078 | BLCA, PanCancer |
| 16 | chr16:22307639-22310495 | Ovarian, Renal, PanCancer |
| 17 | chr17:8021414-8027560 | PanCancer |
| 19 | chr10:103602281-103604334 | UCEC |
| 19 | chr19:12901682-12905667 | Ovarian |
| 19 | chr19:17969769-17971490 | Melanoma |
| 19 | chr19:41767305-41771623 | Melanoma, PanCancer |
Among all cancer types analyzed, ovarian cancer has the maximum number of candidate drivers (i.e. four), while melanoma, esophageal, endometrial, renal, and bladder cancer follow with two candidates each (Figure S9). We find that one insulator candidate is common to both renal and ovarian cancers, while nine predicted drivers are identified in the joint pan-cancer analysis. Among these nine insulators identified in the pan-cancer analysis, four candidates are also detected to be significantly mutated in single cancer types. The remaining five candidate drivers identified via the pan-cancer analysis are likely to be important in multiple cancer types, though they do not reach statistical significance in individual cancer types due to limited cohort sizes.
To further interpret the tumorigenic role of mutations at insulator regions, we examined their clonality status. We performed integrative analysis of tumor purity, copy number alterations and read depth at mutated loci using an approach similar to the one in previous studies (Landau et al., 2013; McGranahan et al., 2015) (Figures S10A and S10B). We find that the majority of mutations in both coding and non-coding drivers tend to be clonal, likely pointing to their roles during the early stages of tumor development (Figure S10C and S10D). This result is in concordance with previous studies of coding driver genes from TCGA (The Cancer Genome Atlas) whole-exome sequencing data (McGranahan et al., 2015). However, the fraction of clonal mutations in insulators (0.60) is significantly lower than that observed in promoters (0.79) (P value = 0.049, Fisher’s exact test) suggesting the possibility that mutations at insulators may play a stronger role at later stages in cancer progression.
Predicted rewiring of chromatin loops around insulator drivers and associated genes
ChIA-PET assays provide the locations of paired loop anchors, enabling the prediction of potential loop rewiring events associated with the perturbation of the anchor regions. Perturbation of loops associated with mutations at the insulators may alter the anchor contact frequency and hence the strength of loops in the vicinity. Based on previous studies, the majority (~80%) of the loop anchors bound by CTCF and cohesin contain CTCF motifs in convergent orientations (i.e. forward-reverse) (Hnisz et al., 2016; Ji et al., 2016; Tang et al., 2015). This helps us to predict how the mutations at insulators could alter the conformations of the loops in the region leading to their rewiring. We determined the potential rewiring events by requiring that new loops: 1) contain convergent CTCF motifs at anchors within 360 kb of each other (which corresponds to the 75th percentile of CTCF-CTCF loop length distribution), and 2) if the predicted insulator driver is located within a TAD, the predicted new loops will be within the same TAD (Figure 1C and Figure S11). For example, in Figure 1C, mutations at the driver insulator with the reverse CTCF motif are predicted to weaken a constitutive loop (red), thereby strengthening an alternate constitutive loop (gray) and a predicted new loop (dotted) through the pairing of the forward CTCF motif with other reverse motifs in the vicinity.
We analyzed the genes whose expression may be altered due to the loop rewiring events. 76 genes are located within the predicted weakened or strengthened loops associated with the insulator drivers. Among these 76 genes, TGFB1, HES1, CUL1 and CDKN2A are involved in curated cancer pathways (Figures 1C, S11C, S11K and S11M) (Knijnenburg et al., 2018; Sanchez-Vega et al., 2018). Next, we asked whether these 76 genes exhibit differential expression in patients with insulator mutations vs. those without. Since RNA-seq data is not available for the esophageal cancer samples analyzed in this study (Methods), we were only able to analyze the expression of associated genes for 19 out of 21 significantly mutated insulators. We found that the expression of two neighboring genes is associated with mutations in one insulator driver candidate: TGFB1 (in melanoma and pan-cancer analysis) and CYP2S1 (in melanoma) (Figure 1D and Figure S12A). Thus, mutations in only one out of 19 insulator candidates analyzed are associated with differential expression of at least one gene (Benjamini and Hochberg method for multiple hypothesis correction, Q value ≤ 0.1). We note that this result is likely due to the small number of samples with matched WGS and RNA-seq data in the majority of cancer types. The mutational frequency of candidate drivers is also lower in other cancer types relative to melanoma, which further decreases the statistical power to detect significant differences in gene expression. For example, matched WGS and RNA-seq data is available for only 3 samples with mutations and 93 without for the candidate driver on chr12 in breast cancer, while it is available for 12 samples with mutations and 68 without for the candidate driver in melanoma.
The driver candidate (chr19:41,767,305-41,771,623) identified in melanoma and in pan-cancer analysis where mutations are associated with TGFB1 up-regulation (Figures 1B, 1C, 1D and S12 and Table S13) is of particular interest. TGFB1 is involved in the transforming growth factor-β (TGF-β) signaling pathway and promotes angiogenesis and tumor cell migration in melanoma (Perrot et al., 2013). We find that the mutation frequency of this insulator is higher in metastatic (19%) relative to primary samples (12%) (Figure S12F), which is consistent with the known role of TGFB1 in melanoma metastasis (Javelaud et al., 2008; Padua and Massagué, 2009). Furthermore, this trend is even stronger when analyzing the samples with recurrent mutations (since they are likely the ones under stronger positive selection than non-recurrent mutations), with mutation frequency of 17% for metastatic and 8% for primary melanoma samples (Figure S12F). Besides melanoma, the TGF-β pathway is important in multiple cancer types and is a target for drug development (Akhurst, 2017; Seoane and Gomis, 2017). The mutations of this insulator in other cancer types (lung adenocarcinoma, endometrial carcinoma, prostate adenocarcinoma and liver hepatocellular carcinoma) and in particular the relatively high mutational frequency of 9% in colon cancer and 3% in esophageal adenocarcinoma may provide complementary mechanisms to the known genomic alterations (protein-coding mutations and copy number alterations) for modulation of TGF-β signaling, especially in gastrointestinal cancers (Korkut et al., 2018).
Functional validation of predicted driver insulator associated with differential expression of TGFB1
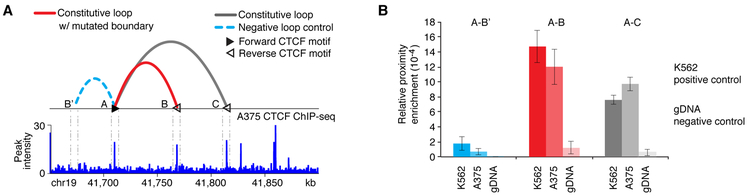
We performed functional validation for the tumorigenic role of the predicted insulator driver (chr19:41,767,305-41,771,623) in melanoma using multiple assays. We used human melanoma A375 cells and sequenced the insulator to verify the absence of mutations. We performed CTCF ChIP-Seq in A375 cells to show that the predicted driver (insulator B) is bound by CTCF (Figure 2A). Next, we performed chromosome conformation capture (3C) to confirm the presence of the relevant chromatin loop in melanoma (Figure 2B, loop A-B). We used primers on CTCF insulator ‘A’ and the insulators ‘B’ and ‘C’, which form constitutive loops in the conserved annotations, as well as a random non-insulator region B’ of similar linear distance that served as a negative control (Figure 2A). Our results show that the contact frequency of both A-B and A-C is significantly higher than the negative controls (A-B’ and naked genomic DNA) (Figure 2B) and at similar levels compared to K562 cells, one of the cell lines in which these loops were initially detected by ChIA-PET. These results validate the existence of the loop A-B in melanoma cells and support that they potentially insulate the enhancer activity from promoters outside the loops.
Figure 2. Validation of loop conformation around TGFB1.
(A) Top: Loop conformation around the TGFB1 associated insulator. Constitutive loop with the significantly mutated insulator (red), another constitutive loop (grey), and a negative control loop in chromosome conformation capture (3C) assays (light blue) are shown. CTCF motifs located at the two loop anchors are most likely to have forward-reverse orientation (faced arrows in black and white color). Bottom: CTCF ChIP–seq validation in A375 melanoma cells. The zoom-in shows the corresponding CTCF peaks at loop anchors A, B and C. (B) 3C assays confirm the existence of constitutive CTCF-CTCF loops between two pairs of CTCF anchors that are 60kb and 105 kb apart (red: loop A-B and grey: loop A-C) in A375 melanoma cells. To compensate for relative interaction efficiency based on linear distance, looping of anchor A with a random region upstream that was in closer linear distance than the downstream CTCF anchors (40kb), was also interrogated (light blue: loop A-B’) as a negative control loop. K562 leukemia cells were used as a positive control and genomic DNA that was purified, digested and ligated was used as a negative control. Relative enrichment of all samples was normalized over intra-fragment PCR. Data are represented as mean ± SD.
See also Figure S12.
Six out of 48 mutations in this insulator are located within the regions bound by CTCF (SNV4, 5, 19, 20, and 21 in Figure S12E), while 39 out of 48 mutations occur at the ChIP-seq peaks of other TFs. In particular, there are six hotspots (SNV8, 9, 11, 12, 14 and 18 in Figure S12E) where many TFs bind, including ELK4 (Table S13). Among these six SNVs, SNV8 (chr19:41,769,771) is the most recurrent mutation with a mutational frequency of 8% and is predicted to disrupt the ELK4 motif. Notably, by analysis of Hi-C data, Mourad et al. showed that besides CTCF, ELK4 is one of the TFs that show a blocking effect between long-range contacts in the genome (Mourad and Cuvier, 2018). Besides ELK4, YY1 is another prominent TF known to cooperate with CTCF for long-distance interactions (Atchison, 2014) and has been implicated for the establishment of TAD boundaries (Moore et al., 2015; Schwalie et al., 2013). Many SNVs, including recurrent mutations (SNV8, 9, 11, 12, 14 and 18) are located at an YY1 ChIP-seq peak. We find that the region spanned by SNV8 to SNV21 bound by many TFs also shows the highest mutational density in other cancer types aside from melanoma (Figure S12B). Thus, our results support the previous observation that, in addition to CTCF, other TFs may also play an important role in the maintenance of CTCF-CTCF loops.
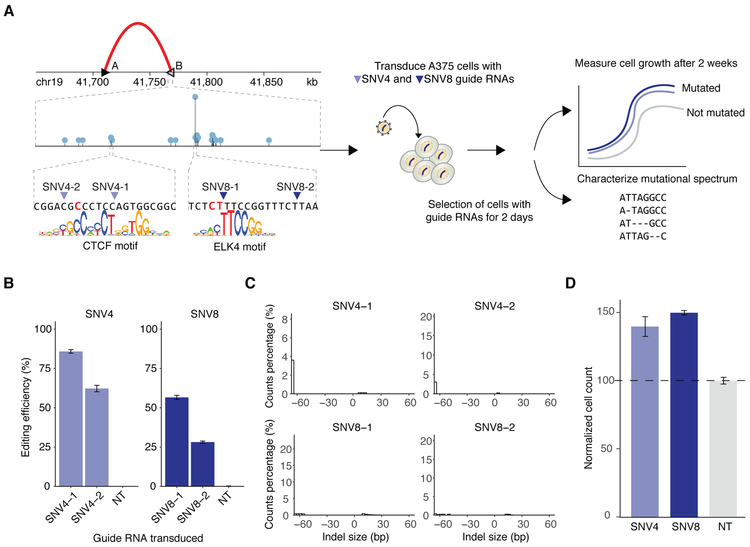
In order to experimentally test the potential role of this insulator region in tumorigenesis, we designed CRISPR-Cas9 guide RNAs to target SNV4 and SNV8 (Figure 3). We chose to edit the nucleotides at SNV4 and SNV8 positions because they constitute two of the most frequently mutated hotspots in this insulator and may represent two different mechanisms for insulator disruption by altering the binding of CTCF (SNV4) or ELK4 (SNV8). We lentivirally transduced A375 cells with 2 different guide RNAs targeting each hotspot (SNV4 and SNV8) and 2 different non-targeting (control) guide RNAs. A high rate of editing was achieved for both SNV4 and SNV8 regions (Figure 3B) and deep sequencing of the locus confirmed indels of varying lengths with the expected bias towards deletions (Figure 3C and Figure S13). We compared proliferation over a 2-week period of SNV4- and SNV8-edited cells to control cells transduced with non-targeting guide RNAs. We found increased proliferation in the SNV-edited melanoma cells — 139.8% +/− 7.3 for SNV4 and 149.8% +/− 1.7 for SNV8 — when compared to cells transduced with non-targeting guides (Figure 3D), suggesting that mutations in these regions confer a growth advantage in melanoma.
Figure 3. Functional validation of mutations in the TGFB1 associated insulator using CRISPR-Cas9 guide RNAs.
(A) Guide RNA design in the chr19 insulator region B. Needle plot shows mutations observed in B and zooms in on the sequence surrounding two of the most recurrent mutations in the region (SNV4 and SNV8). SNV4 is present in a CTCF motif and SNV8 is present in an ELK4 motif. Guide RNAs were designed to target 4 different positions (SNV4-1, SNV4-2, SNV8-1 and SNV8-2) surrounding SNV4 and SNV8. The guide RNAs are cloned into a lentiviral vector and transduced into human melanoma A375 cells. Cell proliferation is compared over a 2-week period between SNV4- and SNV8-edited cells to control cells transduced with non-targeting (NT) guide RNAs used as negative control. Deep sequencing readout characterizes the length of mutations in the selected cells. (B) Editing efficiency of each CRISPR guide RNA targeting SNV4 and SNV8. Data are represented as mean ± SEM. (C) Indel length distribution of each guide RNA that targets insulator region B. (D) Cell growth increases 40-50% in cells transduced with SNV4 and SNV8 guides. Data are represented as mean ± SEM. Cell count measurements are normalized to cells transduced with NT guide RNAs.
See also Figure S13.
DISCUSSION
This study presents a comprehensive analysis of CTCF/cohesin insulator mutations from WGS of 1,962 patients in 21 cancer types. We find that background mutational processes in different cancers lead to differential enrichment of mutations predicted to disrupt CTCF motifs; the majority of which are likely to be passengers. Using the predicted functional impact of mutations, their frequency and the patterns of CTCF motif-disruption, we developed a computational approach (CNCDriver) to identify insulator regions under positive selection. Benchmarking the statistical framework of CNCDriver on other types of known cancer drivers (coding genes and promoters) demonstrates its validity. We identify 21 candidate insulators showing signals of positive selection. Mutations in one of these 21 candidates are associated with differential gene expression that may play a role in tumorigenesis by interfering with the TGF-β pathway in melanoma and other cancers, especially gastrointestinal. Our hypothesis is supported by functional validation using CRISPR-Cas9 which shows that two of the most frequent mutations increase cell growth by 1.4- and 1.5-fold in melanoma cells. While our study clearly shows the importance of this region as cancer driver, high-throughput genome editing approaches such as the one used recently to assay the SNVs in 13 exons of BRCA1, can be used to elucidate the role of all individual mutations in tumorigenesis (Findlay et al., 2018). Thus, our study reveals several CTCF insulators as putative drivers and opens the door to multiple experimental validation and mechanistic studies of the tumorigenic impact of mutations in these elements.
With increasing numbers of cancer whole genomes sequenced and improved maps of cell-type specific annotations of insulator elements, we expect additional insulator drivers will be discovered in the future. The latest developments in genomic technology, such as the HiChIP assays, promise to reveal high-resolution maps of insulator regions (Mumbach et al., 2016). We expect that as the resolution of functional genomics assays improves, the statistical power to identify signals of positive selection in non-coding regions will also improve (Kumar and Gerstein, 2017). The framework developed in this study will guide the integration of genomic structure maps generated by the 4D Nucleome project with large-scale cancer WGS (Dekker et al., 2017).
Our study also highlights the challenge of associating CTCF-CTCF loop disruption to the affected genes in the absence of large sample sizes with matched WGS and RNA-seq data. In order to have sufficient statistical power to detect significant associations between gene expression and mutations, we estimate that we would need at least ~300 samples with matched RNA-seq data for the other cancer types in this study (Methods). Finally, while we analyzed the impact of SNVs in insulators, CTCF-CTCF loops may also be perturbed by DNA methylation, small insertions or deletions, and structural variations at insulators (Flavahan et al., 2015; Hnisz et al., 2016). Overall, identification of significantly disrupted CTCF/cohesin insulators complements the identification of other types of non-coding cancer drivers (promoters, enhancers and ncRNAs) and enables a fuller understanding of the role of non-coding alterations in tumorigenesis.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ekta Khurana (ekk2003@med.cornell.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human melanoma cells (A375) were used for CTCF ChIP-seq, chromosome conformation capture (3C) and CRISPR-Cas9 functional validation assays. Human leukemia cells (K562) were used as control in chromosome conformation capture (3C) assay. HEK293FT cells were transfected for lentiviral vector production.
METHOD DETAILS
Data used
Somatic single nucleotide variants (SNVs) from WGS data
We collected somatic SNVs from 1,962 whole-genome sequencing (WGS) samples across 21 cancer types (Table S1). 1,332 samples are from Alexandrov et al. (Alexandrov et al., 2013), Fredriksson et al. (Fredriksson et al., 2014), and Perera et al. (Perera et al., 2016). These include hepatocellular carcinoma (liver), pancreatic adenocarcinoma (pancreatic), medulloblastoma, pilocytic astrocytoma, renal cell cancer (RECA-EU), ovarian serous cystadenocarcinoma (ovarian), colon adenocarcinoma (colon), skin cutaneous melanoma (melanoma), malignant lymphoma (MALY-DE), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), breast invasive carcinoma (BRCA), brain low grade giloma (LGG), glioblastoma multiform (GBM), head and neck squamous cell carcinoma (HNSC), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), and bladder urothelial carcinoma (BLCA). 119 esophageal adenocarcinoma samples (ESAD-UK), 150 chronic lymphocytic leukemia samples (CLLE-ES) and 124 prostate adenocarcinoma samples (PRAD-CA) were obtained from ICGC (International Cancer Genome Consortium) data portal (release 21). 183 melanoma samples (MELA-AU) were obtained from ICGC data portal (release 25). 16 of 183 melanoma patients have both primary and non-primary (9 metastasis and 7 relapse) WGS specimens. We used metastasis and relapse specimens for these 16 patients in melanoma. 57 prostate adenocarcinoma samples are from Baca et al. (Baca et al., 2013) and seven are from Berger et al (Berger et al., 2011).
A set of 226 ultra-high signal artifact regions (https://sites.google.com/site/anshulkundaje/projects/blacklists) was obtained from the ENCODE project (Dunham et al., 2012). The annotated regions are based on known sequence repeats and are characteristic of high variance in read mappability. We removed somatic mutations that overlapped with ultra-high signal artifact regions to avoid possible false mutation calls in the dataset.
Constitutive CTCF/cohesin insulator annotations
We obtained all publicly available sources of cohesin ChIA-PET data (SMC1 ChIA-PET and RAD21 ChIA-PET) and CTCF ChIA-PET data to build the constitutive CTCF/cohesin insulator annotations across five cell lines (Jurkat, K562, GM12878, MCF-7 and HeLa) (Figure S1B). The SMC1 ChIA-PET data is available in Jurkat cell line from Hnisz et al. (Hnisz et al., 2016) and the RAD21 ChIA-PET data are available in K562 and GM12878 cell lines from Heidari et al. (Heidari et al., 2014). The data in SMC1 ChIA-PET of Jurkat and RAD21 ChIA-PET data of K562 and GM12878 were processed by Mango pipeline (Phanstiel et al., 2015) at FDR threshold of 0.2. Only those anchors in the ChIA-PET loop that overlapped with corresponding cell type specific CTCF ChIP-seq peaks from ENCODE were kept as CTCF/cohesin insulators. The CTCF ChIA-PET data in K562 and MCF-7 cell line are obtained from ENCODE website and were processed by ChIA-PET v1 as described in Li et al. (Li et al., 2012a). The CTCF ChIA-PET data in GM12878 and HeLa cell line are obtained from Tang et al. (Tang et al., 2015) and were processed by ChIA-PET v2 as described in Tang et al. (Tang et al., 2015). Only those anchors of the ChIA-PET loops that co-occupied corresponding cell-type specific RAD21 and SMC3 ChIP-seq peaks from ENCODE were kept as CTCF/cohesin insulators. These CTCF/cohesin sites (loop anchors) have distinct functional role from the peaks identified by CTCF ChIP-seq or RAD21 and SMC3 ChIP-seq alone (non-loop CTCF or cohesin sites) and are highly likely to function as insulators. We intersected CTCF/cohesin insulators that are either from cohesin ChIA-PET or CTCF ChIA-PET in at least 5 out of 7 ChIA-PET data as constitutive CTCF/cohesin insulator annotations for the analysis in this study. There are 10,654 constitutive CTCF/cohesin insulators and their median length is around 2kb (Figure S1A). This length is comparable to the length of promoters (2.5kb) used by us and others for the studies of cancer drivers (Hornshøj et al., 2018; Mularoni et al., 2016; Weinhold et al., 2014).
Motifs of CTCF and other TFs and CTCF motif orientations
We used motifs for 549 TFs from the ENCODE project (http://compbio.mit.edu/encode-motifs/motifs.txt, Table S14) (Dunham et al., 2012), including TRANSFAC and JASPAR motifs, located within potentially functional regions such as DHSs or ChIP-Seq peaks. The motif models for CTCF are derived from human ChIP-seq data using curated motif models from literature and de novo motif models discovered from five motif discovery tools (AlignACE, MDscan, MEME, Trawler and Weeder) in the ENCODE project (Kheradpour and Kellis, 2014). In particular, for CTCF, we analyzed 12 CTCF motif models. We do not use RAD21 and SMC3 motif models from the ENCODE project since RAD21 and SMC3 are architectural proteins and they do not bind DNA directly.
CTCF binds DNA asymmetrically as revealed by previous study (Phillips and Corces, 2009; Rhee and Pugh, 2011). We first define the orientation of CTCF motif (MA0139.1) from the JASPAR 2018 CORE vertebrate (MA0139.1) as CTCF forward direction and the reverse-complement model as CTCF reverse direction (Figure S2A). We used FIMO to identify the location and orientation of CTCF motif at P-value threshold of 10−6. The identified locations of CTCF motif are further overlapped with locations of CTCF/cohesin insulators in 7 ChIA-PET datasets.
Annotations of coding sequence (CDS), ncRNAs, promoters and enhancers
The GENCODE v19 annotations were used to define the locations of protein-coding sequence (CDS) and lincRNAs (long intergenic non-coding RNAs). Promoters were defined as 2.5 kb upstream from the transcription starting site (TSS). Our enhancer set is obtained by the union of TF binding peaks and DHSs from ENCODE, and Segway/ChromHMM-predicted enhancers, which were defined from histone marks (H3K4me1, H3K4me2 and H3K27ac) (Fu et al., 2014). All enhancers are at least 1kb away from the TSS of the nearest gene. To associate potential regulatory targets of each enhancer, we consider all candidates genes within 1 Mb of an enhancer (Fullwood et al., 2009; Yip et al., 2012). Then, correlations between enhancer activity/inactivity signals and gene expression across multiple tissue types are computed using DNA methylation, H3K4me1, H3K27ac and RNA-seq data from the Roadmap Epigenomics project (Fu et al., 2014; Kundaje et al., 2015). H3K4me1 and H3K27ac data were considered as activity signals, and DNA methylation as inactivity signal. Significant correlation between the enhancer signal and the target gene expression was used to call enhancer-target gene pairs (see Fu et al. (Fu et al., 2014) for more details).
Replication timing data
We obtained wavelet-smoothed Repli-seq track data for 7 cell lines (HepG2, MCF-7, SK-N-SH, GM12878, BJ, K562 and IMR-90) from the University of Washington ENCODE group (http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeUwRepliSeq). We used HepG2 to represent replication timing in liver, MCF-7 for breast, GM12878 for malignant lymphoma, K562 for CLL, BJ for melanoma, IMR-90 for lung, SK-N-SH for glioblastoma, medulloblastoma and pilocytic astrocytoma. For pancreatic, kidney, ovarian, colon, esophageal, and prostate, we used the average replication timing signal from HepG2, MCF-7, GM12878, K562, BJ, IMR-90 and SK-N-SH. The replication timing tracks were further divided into 1Mb bins for each cell line. For a given functional element, we assigned a replication timing value corresponding to its overlapping 1Mb bin. If it overlapped with two bins, we took the average.
Copy number alteration data
For TCGA data, the copy number was obtained using TCGA SNP array data from Broad GDAC Firehose website (https://gdac.broadinstitute.org/). CNVKit (CNVKit version 0.9.5) was used with default parameters to get integer copy number segmentations. We used ASCAT (ASCAT version 2.4.2 ) with default parameters to identify copy number alterations, ploidy and purity for the WGS samples in the ICGC MELA-AU project.
YY1 and ELK4 ChIP-seq data
The YY1 ChIP-seq track shown in the figure (Figure S12D) is from melanoma MELAME-3M cells (Li et al., 2012b). YY1 binds the same region in MELAME-3M, K562, SKNSH and GM12878 cells (chr19:41,769,535-41,770,722). ELK4 ChIP-seq data shown in the figure (Figure S12D) is from HeLa-S3 cells from ENCODE, the same binding region is found in HEK293 cells.
Functional validation assays
CTCF ChIP-seq on A375 cells
A375 cell line was obtained from Dr. Joan Massagué (Memorial Sloan Kettering Cancer Center) and cultured in DMEM supplemented with L-glutamine (2mmol/L), penicillin (100U/ml), streptomycin (100ug/ml) and 10% heat-inactivated FBS. Cell line was cultured at 37°C with 5% CO2 and has been tested negative for mycoplasma (MycoAlert PLUS Mycoplasma Detection kit; Lonza). ChIP-Seq was performed as previously described (Chi et al., 2010). In brief, 15 million A375 cells were harvested and cross-linked with 1% paraformaldehyde for 10min at room temperature. Cross-linking was quenched with a final concentration of 0.2M glycine for 5min. Cells were washed with PBS and lysed with 0.1% SDS, 1% Triton X-100, 2mM EDTA, 150mM NaCl and 20mM Tris-HCl, pH 8.1. Chromatin was sheared to around 150bp to 600bp using Covaris E220 machine and incubated with 6ul CTCF antibody (Cell Signaling Technology; #3418) overnight at 4°C with rotation. 20ul Pierce ChIP-grade protein A/G magnetic beads was used for immunoprecipitation. The immunoprecipitated complex was washed with lithium wash buffer (0.7% sodium deoxycholate, 1% NP-40, 1mM EDTA, 500mM LiCl, 50mM HEPES-KOH, pH 7.6), reverse cross-linked and purified with Qiagen minielute column. ChIP library was constructed using KAPA HTP library prep kit and sequenced on Illumina 2500 platform for single read 50bp.
Chromosome conformation capture (3C) assays
Chromosome conformation capture (3C) was performed as described previously with some modifications (Dekker, 2006). Briefly, A375 cells and K562 cells were crosslinked in 1% formaldehyde for 10 minutes at room temperature. Cells were lysed for 20 minutes in lysis buffer (10mM Tris-HCl, 10mM NaCl, 0.2% Igepal CA630) on ice, resuspended in 0.5% SDS, quenched with Triton X-100 and the remaining nuclei were resuspended in the appropriate restriction enzyme buffer. Cells were incubated overnight at 37C and 700rpm rotation with NlaIII restriction enzyme (NEB R0125) and subsequently ligated for 4 hours at room temperature with T4 DNA ligase (NEB M0202). Crosslinks were reversed and DNA was purified using Phenol:Chloroform:Isoamylalcohol (25:24:1) purification. Human genomic DNA was digested, ligated and purified and used as a negative control. Quantification of the data was performed by qPCR using SybrGreen Supermix (Biorad 172-5270).
The primer sequences for PCR are:
| 3C region A | CTGCTTCTCTGTATGTTACCTCATTCGATTGTCC |
| 3C region B | CCTTTGTCTGAGTAGAGATAGTGTGTGGCTTTTG |
| 3C region C | CTCCTTTATTCTGAAGGGAGTGGGCATCAAG |
| 3C region A’ | CAAGTTAGTTCCTGGTCACCTTAGATTGATGGG |
To further confirm that the 3C product indeed consisted of the assessed regions, the PCR products were run on an agarose gel to assess the predicted amplicon size, then gel-extracted and Sanger sequenced.
CRISPR guide RNA design, cloning and lentivirus production
To design the guides targeting the 3kb insulator region on chr19 (41,767,305-41,771,623) we selected regions that were most frequently mutated in human melanoma samples. We focused on two regions (termed SNV4 and SNV8). We first performed Sanger sequencing in the A375 human melanoma cell line (ATCC CRL-1619) to ensure that no mutation exists in the 3kb insulator region. We identified all possible Cas9-targetable sites on both strands in SNV4 and SNV8 and eliminated single-guide RNAs (sgRNAs) with predicted off-target binding (Benchling) which yielded 4 sgRNAs total (2 for SNV4 and 2 for SNV8). The sgRNA sequences were synthesized as single-stranded oligonucleotides (Integrated DNA Technologies).
| SNV4-1 | GCCGCCACTGGAGGGCGTCC |
| SNV4-2 | AGCCGCCACTGGAGGGCGTC |
| SNV8-1 | CTGGTGAATTCATCTCTTTC |
| SNV8-2 | ATCTCTTTCCGGTTTCTTAA |
| NT1 | CCAATACGGACCGGATTGCT |
| NT2 | GTAGCGCACGATATTAGTTC |
To clone the sgRNA guide sequences, the lentiCRISPRv2 plasmid (Addgene #52961) was digested and dephosphorylated with FastDigest BsmBI (Thermo) and FastAP (Thermo) at 37 °C for 45 minutes (Sanjana et al., 2014). The sgRNA guide sequence oligonucletodies were phosphorylated using polynucelotide kinase (New England Biolabs) at 37 °C for 30 minutes and then annealed by heating to 95 °C for 5 minutes and cooling to 25 °C at a rate of 1 °C/5 seconds. We used T7 ligase (Enzymatics) to ligate annealed oligos into purified digested vectors at 22 °C for 10 minutes. Cloned plasmids were transformed into Stbl3 (Thermo) and purified using a QIA-prep spin mini-prep kit (Qiagen).
To make lentivirus, plasmids were co-transfected with packaging plasmids pMD2.G and psPAX2 (Addgene #12259 and #12260). For each construct, a T-75 flask of 95% confluent HEK293FT cells (Thermo) in 5 ml of D10 media and transfected in OptiMEM (Thermo) using 8.3 ug of Cas9 construct, 4.6 ug of pMD2.G, and 6.6 ug of psPAX2, and 45.6 ul of PEI. D10 media consists of DMEM (Caisson Labs) supplemented with 10% FBS (Atlas Biologicals). After 4-6 hours 5 ml of D10 media with 1% bovine serum albumin (Sigma) was added to the flask to improve virus stability. After 48 hours, viral supernatant was harvested and centrifuged at 3000 rpm at 4 °C for 10 min to get rid of cell debris.
Cell culture, viral transduction and proliferation assay
For each viral construct, 1×104 A375 cells (ATCC CRL-1619) were transduced during plating with 200 ul of viral supernatant in each well of a 24 well plate in 1 ml of D10 media. Each construct was transduced in duplicate. Cells without virus were also plated in duplicate in a 24 well plate with 1 ml of D10 media as controls for puromycin selection.
At 24 hours post-transduction media was changed to D10 with 1 ug/ml puromycin (Sigma) for all wells. At 48 hours cells were fully selected, as determined by a non-transduced control well. At 2 weeks post-transduction, cells were plated in 96 well clear bottom plates (Corning) at 2,000 cells/well to measure cell viability using CellTiter Glo (Promega). Briefly, cells were equilibrated to room temperature for 30 minutes. Then, culture media was aspirated and 100 ul of CellTiter Glo reagent diluted 1:4 in PBS phosphate-buffered saline (Caisson Labs) was added to each well. Plates were then placed on orbital shaker for 2 minutes and then incubated for 10 minutes at room temperature. Luminescence was measured on an Infinite F200 Pro plate reader (Tecan) using a 1s integration time.
Deep sequencing to determine indel spectrum
To extract genomic DNA, we used QuickExtract DNA Extraction Solution (Lucigen), following the manufacturer’s protocol. To prepare samples for Illumina sequencing, a two-step PCR was performed to amplify the region of interest. For each sample, we performed 2 seperate 100 ul reactions (25 cycles each) with 250 ng of input gDNA using PfuX7 polymerase (Nørholm, 2010) and the resulting products were pooled.
The primers for the first PCR are:
F1_SNV4 TCTTGTGGAAAGGACGAAACACCGAAGACCAGCCCACCGTGTC
R1_SNV4 CCGACTCGGTGCCACTTTTTCAATGCTTTGGGTAAGGCACCCC
F1_SNV8 TCTTGTGGAAAGGACGAAACACCGATGAGCCATCGCTACCAGCTT
R1_SNV8 CCGACTCGGTGCCACTTTTTCAACTAGCCAATCAGAGCGCCGT
The second PCR was performed to attach Illumina adaptors and to barcode individual samples. The PCR was done in a 50 ul reaction with 5 ul of PCR1 product using Q5 polymerase (New England Biolabs). Amplification was carried out with 10 cycles.
The primers for the second PCR are:
F2 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT (1-9bp stagger sequence)(8bp barcode)TCTTGTGGAAAGGACGAAACACCG
R2 CAAGCAGAAGACGGCATACGAGAT (8bp barcode)GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT(1-9bp stagger)CCGACTCGGTGCCACTTTTTCAA
PCR2 products were pooled in an equimolar ratio, purified, gel extracted, and sequenced using a MiSeq 300 cycle v2 kit (Illumina). Before determining the length of indel mutations, we first filtered out primer-dimers or unrelated amplicons by removing any reads that did not contain the designed PCR1 primers and at least 5 bases beyond the 3’ end of each primer matching the intended amplicon. Reads that met this criteria were then used to measure the distance between primer pairs. This distance between primer pairs in cells transduced with sgRNAs targeting SNV4 or SNV8 regions was compared to the same distance in cells transduced with a nontargeting control sgRNA.
QUANTIFICATION AND STATISTICAL ANALYSIS
To assess the landscape of mutations at CTCF/cohesin insulators
Motif-disrupting vs. preserving mutations
To define whether a SNV disrupts a TF motif, we compute the statistical significance of the position weight matrix (PWM) score for the sequence change (alternative relative to reference). When the alternative sequence decreases the PWM score in the TF motif, we define it as a motif-disruption using a P-value threshold of 4×10−8. This approach is the same as is used in FunSeq2 (Fu et al., 2014).
Enrichment/Depletion of TF motif-disruption
To assess the enrichment of CTCF and other TF motif-disruption events within a specific cancer type, we performed a binomial test to compare the fraction of disrupted TF motifs to the corresponding proportion of the TF motifs abundance.
Deciphering the signatures of mutational processes operative in TF motif disruption
To decipher the signatures of mutational processes operative in the TF binding site regions, we used the FunSeq2 annotation of the SNVs that disrupt TF motifs and computed their normalized tri-nucleotide distributions, which determined the motif-disruption signature for each TF for each cancer type (frequency of 96 mutation types equivalent to the six substitution changes and their immediate 5’ and 3’ sequence context). To assess the similarity between motif-disruption signatures and the COSMIC signatures of mutational processes, we used a threshold of 70% cosine similarity, which is the same metric used for defining the mutational signatures by Alexandrov et al. (2013) (Alexandrov et al., 2013). We note that any previously described signature in liver cancer does not reach the 70% similarity to the CTCF motif-disrupting tri-nucleotide distribution. likely because liver cancer does not show prevalence of a single unique signature. In fact, liver cancers harbor five ubiquitous signatures (S1, S4, S5, S12, S16) with high mutation burden as has also been reported in Letouze et al. (Letouzé et al., 2017).
Estimation of observed and expected aggregated mutational rate for CTCF motif-disruption within insulators
In order to compute the rate of mutations predicted to cause CTCF motif-disruption within insulator regions, we considered the CTCF motif annotations from FunSeq2 within the flanking stretches of 2,500 nucleotides on both sides of the insulator mid-point. To exclude regions that could bias the mutation rate analyses, we filtered out any mutations annotated in coding sequences. The mutations that fall into CTCF binding sites were split into motif-disrupting or non-disrupting groups and overlapped with the insulator regions (5 kb windows). Then the 5kb windows across the genome were aligned to each other using their mid-points as reference. The aggregate mutation rate of every position within the insulator window was calculated as the total number of mutations at this position divided by the total number of nucleotides considered. In order to compute the expected CTCF motif-disruption mutation rate within insulators, we randomly introduced the same number of mutations as observed in CTCF motifs at each insulator window. The CTCF motif-disruption mutation rate of each randomly generated set of changes was computed as explained above and this procedure was repeated 100 times. Finally, the estimate of the expected rate of mutations predicted to cause CTCF motif-disruption was computed as the mean random mutation rate of every position within the windows of insulator regions. This was done for 12 cancer types that have more than 500 SNVs predicted to cause CTCF motif-disruption. The same steps were used for mutations that are not predicted to disrupt CTCF motifs.
Details of CNCDriver
Identification of mutation clusters
The clustering of mutations has been suggested to indicate signal of positive selection in cancer (Rheinbay et al., 2017; Wagner et al., 2007). We perform “density-based spatial clustering of applications with noise (DBSCAN)” algorithm to exclude mutations that are not located within mutation clusters. We use DBSCAN since the mutation clusters identified are not restricted by fixed window size or fixed number of clusters. The DBSCAN algorithm needs two values of parameters “minPts” and “ε” to detect clusters. The minPts was chosen as 2 samples or 2% of sample size, depending on which number is larger in the cohort of each cancer type. We use ε value as 50 bases by considering the length of the longest TF motif (20 bases) with 15 flanking bases in two directions.
Method framework for driver identification
To identify significantly mutated elements (CDS, promoters, enhancers, lincRNAs and CTCF insulators), we developed CNCDriver (Cornell Non-Coding Driver), which combines mutational recurrence and functional impact of variants to discern signals of positive selection.
First, CNCDriver defines a positional CNCDriverpos score for each mutated position (i) by multiplication of positional recurrence (Wi) and functional impact score (FSi).
The positional recurrence of a variant is defined as the number of mutated samples at the same position divided by the total number of samples in the cohort. For each variant, CNCDriver uses FunSeq2 to integrate functional annotations and assign a functional impact score (Fu et al., 2014; Khurana et al., 2013). The weighted functional impact-scoring scheme from FunSeq2 incorporates features such as the presence of variants in annotated regions (e.g. DHSs, histone modification marks, sensitive, ultra-sensitive, conserved, and highly occupied by transcription factor (HOT) regions) and the predicted impact of variants on TF binding. While for promoters and enhancers, a variant’s impact on TF binding is assessed for all TF motifs (549 TFs from ENCODE), only motifs for CTCF were used for variants in CTCF insulators. For all variants within an element, a CNCDriver score (Sj) is defined by summation of all positional CNCDriverpos scores within the element:
where n is the number of mutated positions within an element.
The method compares the observed CNCDriver score of an element to the null model and a P value is computed to evaluate the significance of the observed CNCDriver score. To build the null background model, a simulated CNCDriver score (Sjk) is computed by sampling n mutations from other mutations in the same element type and with similar properties, such as replication timing. The probability of drawing a mutation is determined by its tri-nucleotide sequence probability across all samples in a given cancer type. The sampling process is repeated N times (N = 105).
For CDS, promoters, enhancers and lincRNAs, replication timing threshold is chosen as the nearest 20% range between background scores and CNCDriverpos scores in the test element, so that mutations in a test element are compared with other mutations in a similar stage of the cell cycle in the Repli-seq profile (Hansen et al., 2010). For CTCF/cohesin insulators, we found the correlation (R-squared value of Pearson correlation) between CNCDriverpos score per Mb and replication timing varies substantially across cancer types. Therefore, in order to choose the best replication timing threshold for the background model of CTCF/cohesin insulators in each individual cancer type, we used the relationship between optimal threshold and R-squared value in CDS driver prediction as reference. First, when COSMIC cancer gene list was used as the gold standard for benchmarking, there is an optimal replication timing threshold in each cancer type that maximized the AUROC (area under the receiver operator characteristic curve) in the CDS driver prediction. Thus, we can learn the relationship between R-squared value and optimal replication timing threshold from CDS driver benchmarking. This relationship is then used to decide the replication timing threshold for significantly mutated insulator prediction.
For CTCF insulators, we also keep the same ratio of mutations predicted to disrupt CTCF motif or not during null background model generation.
For all elements, a pseudo-count is added to the numerator and the denominator. The P value of the tested element is determined by
where N is the number of sampling iterations, Sjk is a simulated CNCDriver score in each sampling iteration and Sj is the observed CNCDriver score of an element. We use the Benjamini and Hochberg method to correct for multiple hypothesis testing (Q value ≤ 0.1).
Filter for AID somatic hyper-mutation in CNCDriver
APOBEC and AID are cytidine deaminases, which convert cytosine to uracil, resulting in C -> T/G mutations. The APOBEC motif consists of three nucleotides and has been associated with characterized mutational signatures (Alexandrov et al., 2013). AID is expressed in several types of B cell lymphomas (Kotani et al., 2007), but in contrast to APOBEC, the AID motif consists of 4 nucleotides, [(A∣T)(A∣G)C(T∣C)] (Rogozin and Diaz, 2004) and has not been associated with a characterized signature in Alexandrov et al. (2013). Alexandrov et al. noted that signature 9, which is observed in lymphoma, does not exhibit the mutational pattern of AID likely because the AID signal is obscured by mutations caused by the error-prone polymerase η. In order to identify elements whose mutations are primarily caused by AID somatic hypermutation, we use the following formula from Roberts et al. (Roberts et al., 2013) to calculate enrichment of AID signature mutations in candidate elements,
where mutationsmotif is the number of mutations in the AID motif, mutationsC is the number of mutated cytosines, contextC is the total number of cytosines, and contextmotif is the total number of occurrences of the AID motif. We use the Fisher’s exact test to determine significance. In CNCDriver, this filter allows the user to identify whether the signals in lymphoma are predominantly due to AID somatic hypermutations.
Estimation of intra-tumor heterogeneity
We collected tumor purity estimates (p) and absolute somatic copy numbers (cn) for TCGA samples which are derived from the consensus of four methods (ABSOLUTE, ESTIMATE, LUMP and IHC) from Aran et al. (Aran et al., 2015). Following the method previously described (Landau et al., 2013; McGranahan et al., 2015), we computed the expected VAFexp given a CCFi over the entire range of CCF (cancer cell fraction) assuming a uniform prior, with CCFi ∈ [0.01, 1].
For a given mutation with alternative read counts (a), reference read counts (r) and total read depth (r + a), the probability of a given CCFi can be estimated from binomial distribution,
Then, for a given mutation with VAFobs, we computed the distribution of posterior probability P(CCFi) over 100 grid points of CCFi uniformly spanned between 0.01 to 1. The distribution over CCF was normalized by dividing them by their sum, which is the constant (C). Mutations were classified as clonal if the maximum probability of CCFi is in the top quartile of cancer cell fraction (max(P(CCFi))≥0.75); otherwise mutations were classified as subclonal.
Analysis of interactions between insulator mutations and CTCF Zn finger binding domains
Hashimoto et al. indicated CTCF Zn finger domains 3-7 (ZF3-ZF7) are critical for its binding to 15 bp core DNA motif in the CTCF-DNA binding co-crystal structure (Hashimoto et al., 2017). In addition to the core sequence, Yin et al. showed ~15% sequence in CTCF binding sites is recognized by Zn finger domains 8-11 (ZF8-ZF11) from the crystal structure, although they have 5 fold weaker binding affinity than ZF3-ZF7 (Yin et al., 2017). CTCF ZF8 servers as a spacer element with a variable length between ZF3-ZF7 and ZF9-ZF11. Schmidt et al. propose a 9 bp CTCF M2 motif to represent the consensus sequence that binds ZF9-ZF11 upstream of core motif (Schmidt et al., 2012). In this study, 293 out of 462 mutations (63%) in the 21 insulators identified to be significantly mutated locate within CTCF ChIP-seq peaks. 23 out of these 293 mutations (8%) occur in the core motif that binds ZF3-ZF7. Although we did not find CTCF M2 motif matched by FIMO (P value: 10−4) upstream of the core motif, 12 mutations are within the window that would bind other domains (ZF9-ZF11) (Supplementary Table S15).
Additionally, our major insulator candidate in melanoma has three mutated positions within the CTCF motif (SNV4, chr19:41,768,332 mutated in two patients; SNV10, chr19:41,768,799 in one patient and SNV11, chr19:41,769,800 in two patients, Figure S12E). SNV4 occurs at position 6 in the 19 bp CTCF motif (reverse orientation), overlaps with CTCF ChIP-seq peak in human melanoma A375 cells (Figure 2A), is predicted to interact with ZF4 in the CTCF protein structure and increases cell growth by 1.4 fold in melanoma cells by CRISPR experiment (Figure 3D).
Assignment of possible new CTCF-CTCF chromatin loops
Prior studies have shown that the majority (~80%) of the CTCF motifs within the two anchors of CTCF-CTCF chromatin loops are in the convergent orientation (forward-reverse) (Hnisz et al., 2016; Ji et al., 2016; Tang et al., 2015). Additionally, in the set of constitutive CTCF-CTCF chromatin loops we built from publicly available ChIA-PET datasets (Heidari et al., 2014; Hnisz et al., 2016; Li et al., 2012a; Tang et al., 2015), 75% are within the range of 360KB. We determined the number of potential rewiring events by requiring that new loops: 1) contain convergent CTCF motifs at anchors within 360 kb of each other, and (2) if the predicted insulator driver is located within a TAD, the predicted new loops will also be within the same TAD. When the predicted driver is located at a TAD boundary, we do not predict loop-rewiring events due to the possibility of formation of new TADs. Most TADs are conserved across cell-types, and we used the hESC TADs from Dixon et al (Dixon et al., 2012).
Gene expression analysis for significantly mutated insulators
We evaluated the association between the presence of somatic mutations at CTCF insulators and mRNA expression of genes within the potential new CTCF-CTCF loops. There are 76 genes that may be impacted by the change of loops associated with 21 insulator candidates predicted by CNCDriver. We used matched RNA sequencing raw counts and copy number data in ICGC data release 24 (https://dcc.icgc.org/releases/release_24) for liver, pancreatic, renal, CLL and malignant lymphoma. We used matched RNA sequencing raw counts and copy number data from TCGA (Chang et al., 2013) for LUAD, LUSC, BRCA, LGG, GBM, HNSC, THCA and UCEC. Because matched RNA-seq data were not available for medulloblastoma, pilocytic astrocytoma and ESAD, these three cancer types were not included in the mRNA expression analysis. For each cancer type, mRNA raw counts were normalized by trimmed mean of M-values (TMM) using the edgeR package (Robinson et al., 2010). In pan-cancer analysis, the mRNA expression level of a given gene and cancer type was first transformed into Z score if at least 25% tumor samples contained non-zero expression value. Then, the mRNA expression (Z score scale) from each cancer was summed up as the gene expression in pan-cancer. The Wilcoxon rank-sum test was used to test the significance of the differential gene expression between tumor samples with insulator mutations vs. those without. For gene expression analysis in pan-cancer, the Z score of a given gene in each group was summed across available cancer types followed by the Wilcoxon rank-sum test. Multiple hypothesis correction was done using the Benjamini-Hochberg method.
We have also compared the expression of genes in tumor samples with insulator mutations versus normal samples, though we note that in melanoma, there is only one normal sample with RNA-seq data available. There are only 12 cancer types in our study with RNA-seq data from normal samples. Furthermore, only 5 out of 12 cancer types with RNA-seq data from normal samples have at least one insulator candidate. Nevertheless, we find that the genes (FOXN4 and MYO1H) within the loops of insulator candidate in breast cancer (Supplementary Figure S11G) are enriched in the set of differentially expressed genes between tumor and normal samples (P value: 0.04, Fisher’s exact test).
Gene expression sample size estimation
We were able to detect significant difference in gene expression when there were 12 samples with mutations vs. 68 samples without mutations for ~2-fold change (CYP2S1) and ~3-fold change (TGFB1) in melanoma. While melanoma has mutation frequency of 16% or more in the candidate insulator drivers, the mutation frequency (2% to 4%) of candidate insulator drivers in other cancer types is lower. Thus, given a mutation frequency is 3% and assuming a minimum number of mutated samples needed with matched WGS and RNA-seq data is 10, we will likely need ~300 samples with matched RNA-seq and WGS data to detect meaningful gene expression association in other cancer types with similar fold differences as observed for CYP2S1 and TGFB1.
DATA AND SOFTWARE AVAILABILITY
Software
The code of CNCDriver is freely available online at https://github.com/khuranalab/CNCDriver.
Data Resources
The accession number for the A375 CTCF ChIP-seq data described in this study is GEO: GSE128346.
Supplementary Material
Table S1: Whole genome sequencing SNV data summary, Related to Figure 1
This study collected SNV data from 1,962 WGS samples across 21 cancer types.
Table S2: Mutational process signatures potentially associated to TF motif-disruption enrichment, Related to Figure 1
We performed a binomial test to assess the enrichment of the cancer specific fraction of TF motif-disruption with respect to the pan-cancer fraction of TF motif-disruption. We determined enrichment for a total of 549 TFs across 21 cancer types. (A) List of mutation signatures highly similar to the motif disrupting tri-nucleotide distribution (cosine similarity greater than 70%) for each TF motif-disruption enrichment per cancer type. (B) Contribution of the mutation signatures (Mut-signature) to the TF-motif disruption (TFMD) enrichment per cancer type compared to pan-cancer. The top table shows the total number of TFs with significant enrichment of motif-disruption alongside the number of TFMD signatures that match any mutational signatures and the corresponding percentage of TFMD signatures that matches any mutation signature. The bottom table shows a detailed breakdown for the contribution of each mutational signature to the matching TFMD signature. The green cells show the signatures present in each cancer type.
Table S3: Area under receiver operating characteristic curve (AUROC) values for CDS results from CNCDriver, OncodriveFML and LARVA, Related to Figure 1
The COSMIC list of 547 cancer genes was further divided into cancer-type specific lists. These cancer-type specific lists were used as the gold standard to obtain AUROC values for each cancer-type using CNCDriver, OncodriveFML and LARVA.
Table S4: CNCDriver results for coding sequence (CDS), Related to Figure 1
Table S5: CNCDriver results for promoters, Related to Figure 1
Table S6: CNCDriver results for enhancers, Related to Figure 1
Table S7: CNCDriver results for lincRNAs, Related to Figure 1
Table S8: CNCDriver results for CTCF/cohesin insulators, Related to Figure 1
Table S9: CTCF/cohesin insulator results from OncoDriveFML, Related to Figure 1
Table S10: Detailed functional annotations of mutations in promoter driver candidates, Related to Figure 1
Table S11: Detailed functional annotations of mutations in enhancer driver candidates, Related to Figure 1
Table S12: Detailed functional annotations of mutations in insulator driver candidates, Related to Figure 1
Table S13: Mutation details for the significantly mutated insulator on chr 19 in melanoma and pan-cancer analysis, Related to Figure 1
Table S14: Transcription factor motif models used, Related to Figure 1
Table S15: Number of mutations located within CTCF Zinc Finger domains in insulator driver candidates, Related to Figure 1
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-CTCF | Cell Signaling Technology | #3418 (lot 1) |
| Critical Commercial Assays | ||
| KAPA HTP library prep kit | Roche | #07961901001 |
| NlaIII restriction enzyme | NEB | R0125 |
| T4 DNA ligase | NEB | M0202 |
| Sybr Green Supermix | Bio-Rad | 172-5270 |
| QIAprep Spin Miniprep Kit | Qiagen | Cat#27106 |
| CellTiter-Glo Luminescence Cell Viability Assay |
Promega | Cat#G7571 |
| QIAquick gel extraction Kit | Qiagen | Cat#28704 |
| MiSeq Reagent Kits v2 | Illumina | Cat#MS-102-2001 |
| Deposited Data | ||
| GENCODE v19 | (Harrow et al., 2012) | https://www.gencodegenes.org/releases/19.html |
| Whole genome sequencing SNV data for LUAD, LUSC, BRCA, LGG, GBM, HNSC, THCA, UCEC, and BLCA projects | (Fredriksson et al., 2014) | dbGaP: phs000178.v1.p1 |
| Whole genome sequencing SNV data for Liver, Medulloblastoma, PilocyticAstrocytoma, Renal, Ovarian, Colon, SKCM, and MalignantLymphoma projects | (Perera et al., 2016) | https://www.nature.com/articles/nature17437 |
| Whole genome sequencing SNV data for MELA-AU, CLLE-ES, ESAD-UK projects | ICGC Data Portal release v21 | https://dcc.icgc.org |
| TCGA Level 3 gene expression profiles for LUAD, LUSC, BRCA, LGG, GBM, HNSC, THCA, UCEC, and BLCA projects | The Broad Institute, Cambridge, MA | https://gdac.broadinstitute.org |
| RNA-seq profiles for ICGC LIRI-JP, PACA-AU, PACA-CA, RECA-EU, OV-AU, CLLE-ES, MALY-DE | ICGC Data Portal release v21 | https://dcc.icgc.org |
| RNA-seq profile for ICGC MELA-AU porject | European Genome-phenome Archive | https://www.ebi.ac.uk/ega/studies/EGAS00001001552 |
| Prostate adenocarcinoma whole genome sequencing SNV data | (Baca et al., 2013) | dbGaP: phs000447.v1.p1 |
| Primary prostate adenocarcinoma whole genome sequencing SNV data | (Berger et al., 2011) | dbGaP: phs000330.v1.p1 |
| Ultra-high signal artifact regions | (Dunham et al., 2012) | https://sites.google.com/site/anshulkundaje/projects/blacklists |
| Replication timing annotations | (Thurman et al., 2007) | http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeUwRepliSeq |
| CTCF/cohesin insulator annotations | (Hnisz et al., 2016) | http://science.sciencemag.org/highwire/filestream/675217/field_highwire_adjunct_files/12/aad9024_TableS8_160122.xlsx |
| TAD annotations | (Dixon et al., 2012) | https://www.encodeproject.org/comparative/chromatin/ |
| Tumor purity estimates for TCGA samples | (Aran et al., 2015) | https://media.nature.com/original/nature-assets/ncomms/2015/151204/ncomms9971/extref/ncomms9971-s2.xlsx |
| ENCODE motif models | (Kheradpour and Kellis, 2014) | http://compbio.mit.edu/encode-motifs/motifs.txt |
| A375 CTCF ChIP-seq | This paper | GEO: GSE128346 |
| Experimental Models: Cell Lines | ||
| A375, human malignant melanoma | Laboratory of Joan Massagué (Memorial Sloan Kettering Cancer Center) | N/A |
| A375, human malignant melanoma | ATCC | CRL-1619 |
| K562, human AML leukemia | ATCC | CCL-243 |
| HEK293FT | ATCC | CRL-3216 |
| Recombinant DNA | ||
| pMD2.G | Addgene | Cat#12259 |
| psPAX2 | Addgene | Cat#12260 |
| lentiCRISPRv2 plasmid | Addgene | Cat#52961 |
| Software and Algorithms | ||
| FunSeq2 | (Fu et al., 2014) | http://funseq2.gersteinlab.org/ |
| featureCounts | (Liao et al., 2014) | http://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| CNCDriver | This paper | https://github.com/khuranalab/CNCDriver |
| edgeR | (Robinson et al., 2010) | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| OncoDriveFML | (Mularoni et al., 2016) | https://bitbucket.org/bbglab/oncodrivefml.git |
| SomaticSignatures | (Gehring et al., 2015) | http://bioconductor.org/packages/release/bioc/html/SomaticSignatures.html |
| Benchling (2019) | N/A | https://benchling.com/ |
| Other | ||
| Infinite F200 Pro plate reader | Tecan | Cat#INF-MPLEX |
Highlights.
Enrichment of CTCF motif-disrupting mutations is associated with neutral signatures
Novel computational method predicts 21 insulator drivers
A predicted driver on chr19 is associated with TGFB1 up-regulation
CTCF ChIP-seq, 3C and CRISPR-Cas9 support the computational predictions
ACKNOWLEDGEMENTS
We thank TCGA (The Cancer Genome Atlas), ICGC (International Cancer Genome Consortium) as well as other groups (including Jason Wong’s group at Prince of Wales Clinical School and Lowy Cancer Research Center, Australia) who have made their data publicly available. We thank Seven Bridges Cancer Genomics Cloud for providing cloud-computing infrastructure to access TCGA data. We also thank Nils Weinhold from the Department of Radiation Oncology at Memorial Sloan Kettering Cancer Center and Martin Miller from Cancer Research UK Cambridge Institute at the University of Cambridge for assistance on data access, as well as Steven Lipkin from Weill Cornell Medicine for discussions on colorectal cancer genes and pathways. N.E.S. is supported by the NIH through NHGRI (R00HG008171, DP2-HG010099), the Sidney Kimmel Foundation, and the Melanoma Research Alliance. EK thanks the National Institutes of Health for support (R01CA218668-01A1, U24CA210989).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- Akhurst RJ (2017). Targeting TGF-β Signaling for Therapeutic Gain. Cold Spring Harb. Perspect. Biol 9, a022301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Sirota M, and Butte AJ (2015). Systematic pan-cancer analysis of tumour purity. Nat. Commun 6, 8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya CL, Cenik C, Reuter JA, Kiss G, Pande VS, Snyder MP, and Greenleaf WJ (2015). Identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations. Nat. Genet 48, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison ML (2014). Function of YY1 in Long-Distance DNA Interactions. Front. Immunol 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. (2013). Punctuated Evolution of Prostate Cancer Genomes. Cell 153, 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SD, Desai K, Kron KJ, Mazrooei P, Sinnott-Armstrong NA, Treloar AE, Dowar M, Thu KL, Cescon DW, Silvester J, et al. (2016). Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer. Nat. Genet 48, 1260–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. (2011). The genomic complexity of primary human prostate cancer. Nature 470, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman BA, and de Laat W (2015). Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol. 16, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Creighton CJ, Davis C, Donehower L, Drummond J, Wheeler D, Ally A, Balasundaram M, Birol I, Butterfield YSN, et al. (2013). The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, Fletcher JA, Dewell S, Maki RG, Zheng D, et al. (2010). ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature 467, 849–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall TN, Rubin MA, and Khurana E (2017). Non-coding genetic variation in cancer. Curr. Opin. Syst. Biol 1, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J (2006). The three “C” s of chromosome conformation capture: controls, controls, controls. Nat. Methods 3, 17–21. [DOI] [PubMed] [Google Scholar]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B, et al. (2017). The 4D nucleome project. Nature 549, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra P, Fu Y, Gerstein M, Khurana E, Dhingra P, Fu Y, Gerstein M, and Khurana E (2017). Using FunSeq2 for Coding and Non-Coding Variant Annotation and Prioritization In Current Protocols in Bioinformatics, (Hoboken, NJ, USA: John Wiley & Sons, Inc; ), p. 15.11.1–15.11.17. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. (2014). Control of Cell Identity Genes Occurs in Insulated Neighborhoods in Mammalian Chromosomes. Cell 159, 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, Janizek JD, Huang X, Starita LM, and Shendure J (2018). Accurate classification of BRCA1 variants with saturation genome editing. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvá ML, and Bernstein BE (2015). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson NJ, Ny L, Nilsson JA, and Larsson E (2014). Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet 46, 1258–1263. [DOI] [PubMed] [Google Scholar]
- Fredriksson NJ, Elliott K, Filges S, Van den Eynden J, Ståhlberg A, and Larsson E (2017). Recurrent promoter mutations in melanoma are defined by an extended context-specific mutational signature. PLOS Genet. 13, e1006773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Liu Z, Lou S, Bedford J, Mu XJ, Yip KY, Khurana E, and Gerstein M (2014). FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 15, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed Y Bin Y, Orlov YL, Velkov S, Ho A, Mei PH, et al. (2009). An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring JS, Fischer B, Lawrence M, and Huber W (2015). SomaticSignatures: inferring mutational signatures from single-nucleotide variants: Fig. 1. Bioinformatics 31, btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, and Dekker J (2013). The Hierarchy of the 3D Genome. Mol. Cell 49, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio E, Robyr D, Spielmann M, Ferrero E, Di Gregorio E, Imperiale D, Vaula G, Stamoulis G, Santoni F, Atzori C, et al. (2015). A large genomic deletion leads to enhancer adoption by the lamin B1 gene: a second path to autosomal dominant adult-onset demyelinating leukodystrophy (ADLD). Hum. Mol. Genet 24, 3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Leung D, and Ren B (2014). The 3D Genome in Transcriptional Regulation and Pluripotency. Cell Stem Cell 14, 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YA, Chang MM, Huang W, Ooi WF, Xing M, Tan P, and Skanderup AJ (2018). Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat. Commun 9, 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, and Stamatoyannopoulos JA (2010). Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci 107, 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. (2012). GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, and Cheng X (2017). Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol. Cell 66, 711–720.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, Zhang MQ, and Snyder MP (2014). Genome-wide map of regulatory interactions in the human genome. Genome Res. 24, 1905–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton A-L, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. (2016). Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science (80-. ). 351, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornshøj H, Nielsen MM, Sinnott-Armstrong NA, Świtnicki MP, Juul M, Madsen T, Sallari R, Kellis M, Ørntoft T, Hobolth A, et al. (2018). Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. Npj Genomic Med. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, Gilgenast TG, Bartman CR, Edwards CR, Stonestrom AJ, Huang P, Emerson DJ, Evans P, Werner MT, Keller CA, et al. (2017). The BET Protein BRD2 Cooperates with CTCF to Enforce Transcriptional and Architectural Boundaries. Mol. Cell 66, 102–116.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelaud D, Alexaki V-I, and Mauviel A (2008). Transforming growth factor-β in cutaneous melanoma. Pigment Cell Melanoma Res. 21, 123–132. [DOI] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, et al. (2016). 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell 18, 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juul M, Bertl J, Guo Q, Nielsen MM, Świtnicki M, Hornshøj H, Madsen T, Hobolth A, and Pedersen JS (2017). Non-coding cancer driver candidates identified with a sample- and position-specific model of the somatic mutation rate. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Taylor MS, Semple CA, Brenner S, and Guilbaud G (2016). Mutational Biases Drive Elevated Rates of Substitution at Regulatory Sites across Cancer Types. PLOS Genet. 12, e1006207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, Välimäki N, Gylfe AE, Ristolainen H, Hänninen UA, Cajuso T, et al. (2015). CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet 47, 818–821. [DOI] [PubMed] [Google Scholar]
- Kheradpour P, and Kellis M (2014). Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42, 2976–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E, Fu Y, Colonna V, Mu XJ, Kang HM, Lappalainen T, Sboner A, Lochovsky L, Chen J, Harmanci A, et al. (2013). Integrative Annotation of Variants from 1092 Humans: Application to Cancer Genomics. Science (80-. ). 342, 1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, and Gerstein M (2016). Role of non-coding sequence variants in cancer. Nat. Rev. Genet 17, 93–108. [DOI] [PubMed] [Google Scholar]
- Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS, et al. (2018). Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 23, 239–254.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut A, Zaidi S, Kanchi RS, Rao S, Gough NR, Schultz A, Li X, Lorenzi PL, Berger AC, Robertson G, et al. (2018). A Pan-Cancer Analysis Reveals High-Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily. Cell Syst. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani A, Kakazu N, Tsuruyama T, Okazaki I. -m., Muramatsu M, Kinoshita K, Nagaoka H, Yabe D, and Honjo T (2007). Activation-induced cytidine deaminase (AID) promotes B cell lymphomagenesis in Emu-cmyc transgenic mice. Proc. Natl. Acad. Sci 104, 1616–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, and Gerstein M (2017). Cancer genomics: Less is more in the hunt for driver mutations. Nature 547, 40–41. [DOI] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. (2013). Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell 152, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzós A, Carlevaro-Fita J, Mularoni L, Reverter F, Palumbo E, Guigó R, and Johnson R (2017). Discovery of Cancer Driver Long Noncoding RNAs across 1112 Tumour Genomes: New Candidates and Distinguishing Features. Sci. Rep 7, 41544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letouzé E, Shinde J, Renault V, Couchy G, Blanc J-F, Tubacher E, Bayard Q, Bacq D, Meyer V, Semhoun J, et al. (2017). Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat. Commun 8, 1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. (2012a). Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Song JS, Bell RJA, Tran T-NT, Haq R, Liu H, Love KT, Langer R, Anderson DG, Larue L, et al. (2012b). YY1 Regulates Melanocyte Development and Function by Cooperating with MITF. PLoS Genet. 8, e1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lochovsky L, Zhang J, Fu Y, Khurana E, and Gerstein M (2015). LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations. Nucleic Acids Res. 43, 8123–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. (2015). Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 161, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, and Swanton C (2015). Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med 7, 283ra54–283ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Reuter JA, Spacek DV, and Snyder M (2015). Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet 47, 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BL, Aitken S, and Semple CA (2015). Integrative modeling reveals the principles of multi-scale chromatin boundary formation in human nuclear organization. Genome Biol. 16, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad R, and Cuvier O (2018). TAD-free analysis of architectural proteins and insulators. Nucleic Acids Res. 46, e27–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu XJ, Lu ZJ, Kong Y, Lam HYK, and Gerstein MB (2011). Analysis of genomic variation in non-coding elements using population-scale sequencing data from the 1000 Genomes Project. Nucleic Acids Res. 39, 7058–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, and López-Bigas N (2016). OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 17, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, and Chang HY (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørholm MH (2010). A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padua D, and Massagué J (2009). Roles of TGFβ in metastasis. Cell Res. 19, 89–102. [DOI] [PubMed] [Google Scholar]
- Perera D, Poulos RC, Shah A, Beck D, Pimanda JE, and Wong JWH (2016). Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature 532, 259–263. [DOI] [PubMed] [Google Scholar]
- Perrot CY, Javelaud D, and Mauviel A (2013). Insights into the Transforming Growth Factor-β Signaling Pathway in Cutaneous Melanoma. Ann. Dermatol 25, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Boyle AP, Heidari N, and Snyder MP (2015). Mango: a bias-correcting ChIA-PET analysis pipeline. Bioinformatics 31, 3092–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. (2013). Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell 153, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, and Corces VG (2009). CTCF: Master Weaver of the Genome. Cell 137, 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahoviček K, Stamatoyannopoulos JA, et al. (2015). Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos RC, Thoms JAI, Guan YF, Unnikrishnan A, Pimanda JE, and Wong JWH (2016). Functional Mutations Form at CTCF-Cohesin Binding Sites in Melanoma Due to Uneven Nucleotide Excision Repair across the Motif. Cell Rep. 17, 2865–2872. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. (2014). A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, and Pugh BF (2011). Comprehensive Genome-wide Protein-DNA Interactions Detected at Single-Nucleotide Resolution. Cell 147, 1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinbay E, Parasuraman P, Grimsby J, Tiao G, Engreitz JM, Kim J, Lawrence MS, Taylor-Weiner A, Rodriguez-Cuevas S, Rosenberg M, et al. (2017). Recurrent and functional regulatory mutations in breast cancer. Nature 547, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. (2013). An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet 45, 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, and Diaz M (2004). Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol 172, 3382–3384. [DOI] [PubMed] [Google Scholar]
- Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, and López-Bigas N (2016). Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature 532, 264–267. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, et al. (2018). Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173, 321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves Â, Kutter C, Brown GD, Marshall A, Flicek P, and Odom DT (2012). Waves of Retrotransposon Expansion Remodel Genome Organization and CTCF Binding in Multiple Mammalian Lineages. Cell 148, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalie PC, Ward MC, Cain CE, Faure AJ, Gilad Y, Odom DT, and Flicek P (2013). Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biol. 14, R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, and Gomis RR (2017). TGF-β Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb. Perspect. Biol 9, a022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, and Lehner B (2015). Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 521, 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. (2015). CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 163, 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Day N, Noble WS, and Stamatoyannopoulos JA (2007). Identification of higher-order functional domains in the human ENCODE regions. Genome Res. 17, 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, and Hadjur S (2015). Comparative Hi-C Reveals that CTCF Underlies Evolution of Chromosomal Domain Architecture. Cell Rep. 10, 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinagre J, Almeida A, Pópulo H, Batista R, Lyra J, Pinto V, Coelho R, Celestino R, Prazeres H, Lima L, et al. (2013). Frequency of TERT promoter mutations in human cancers. Nat. Commun 4, 2185. [DOI] [PubMed] [Google Scholar]
- Wagner A, Cassin RC, and Swan MS (2007). Rapid detection of positive selection in genes and genomes through variation clusters. Genetics 176, 2451–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold N, Jacobsen A, Schultz N, Sander C, and Lee W (2014). Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet 46, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Wang J, Wang M, Li X, Zhang M, Wu Q, and Wang Y (2017). Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res. 27, 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip KY, Cheng C, Bhardwaj N, Brown JB, Leng J, Kundaje A, Rozowsky J, Birney E, Bickel P, Snyder M, et al. (2012). Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 13, R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Whole genome sequencing SNV data summary, Related to Figure 1
This study collected SNV data from 1,962 WGS samples across 21 cancer types.
Table S2: Mutational process signatures potentially associated to TF motif-disruption enrichment, Related to Figure 1
We performed a binomial test to assess the enrichment of the cancer specific fraction of TF motif-disruption with respect to the pan-cancer fraction of TF motif-disruption. We determined enrichment for a total of 549 TFs across 21 cancer types. (A) List of mutation signatures highly similar to the motif disrupting tri-nucleotide distribution (cosine similarity greater than 70%) for each TF motif-disruption enrichment per cancer type. (B) Contribution of the mutation signatures (Mut-signature) to the TF-motif disruption (TFMD) enrichment per cancer type compared to pan-cancer. The top table shows the total number of TFs with significant enrichment of motif-disruption alongside the number of TFMD signatures that match any mutational signatures and the corresponding percentage of TFMD signatures that matches any mutation signature. The bottom table shows a detailed breakdown for the contribution of each mutational signature to the matching TFMD signature. The green cells show the signatures present in each cancer type.
Table S3: Area under receiver operating characteristic curve (AUROC) values for CDS results from CNCDriver, OncodriveFML and LARVA, Related to Figure 1
The COSMIC list of 547 cancer genes was further divided into cancer-type specific lists. These cancer-type specific lists were used as the gold standard to obtain AUROC values for each cancer-type using CNCDriver, OncodriveFML and LARVA.
Table S4: CNCDriver results for coding sequence (CDS), Related to Figure 1
Table S5: CNCDriver results for promoters, Related to Figure 1
Table S6: CNCDriver results for enhancers, Related to Figure 1
Table S7: CNCDriver results for lincRNAs, Related to Figure 1
Table S8: CNCDriver results for CTCF/cohesin insulators, Related to Figure 1
Table S9: CTCF/cohesin insulator results from OncoDriveFML, Related to Figure 1
Table S10: Detailed functional annotations of mutations in promoter driver candidates, Related to Figure 1
Table S11: Detailed functional annotations of mutations in enhancer driver candidates, Related to Figure 1
Table S12: Detailed functional annotations of mutations in insulator driver candidates, Related to Figure 1
Table S13: Mutation details for the significantly mutated insulator on chr 19 in melanoma and pan-cancer analysis, Related to Figure 1
Table S14: Transcription factor motif models used, Related to Figure 1
Table S15: Number of mutations located within CTCF Zinc Finger domains in insulator driver candidates, Related to Figure 1
Data Availability Statement
Software
The code of CNCDriver is freely available online at https://github.com/khuranalab/CNCDriver.
Data Resources
The accession number for the A375 CTCF ChIP-seq data described in this study is GEO: GSE128346.