Abstract
Animal and human pathogens of the genus Bordetella are not commonly considered to be intracellular pathogens, although members of the closely related classical bordetellae are known to enter and persist within macrophages in vitro and have anecdotally been reported to be intracellular in clinical samples. B. bronchiseptica, the species closest to the ancestral lineage of the classical bordetellae, infects a wide range of mammals but is known to have an alternate life cycle, persisting, replicating and disseminating with amoeba. These observations give rise to the hypothesis that the ability for intracellular survival has an ancestral origin and is common among animal-pathogenic and environmental Bordetella species. Here we analyzed the survival of B. bronchiseptica and defined its transcriptional response to internalization by murine macrophage-like cell line RAW 264.7. Although the majority of the bacteria were killed and digested by the macrophages, a consistent fraction survived and persisted inside the phagocytes. Internalization prompted the activation of a prominent stress response characterized by upregulation of genes involved in DNA repair, oxidative stress response, pH homeostasis, chaperone functions, and activation of specific metabolic pathways. Cross species genome comparisons revealed that most of these upregulated genes are highly conserved among both the classical and non-classical Bordetella species. The diverse Bordetella species also shared the ability to survive inside RAW 264.7 cells, with the single exception being the bird pathogen B. avium, which has lost several of those genes. Knock-out mutations in genes expressed intracellularly resulted in decreased persistence inside the phagocytic cells, emphasizing the importance of these genes in this environment. These data show that the ability to persist inside macrophage-like RAW 264.7 cells is shared among nearly all Bordetella species, suggesting that resisting phagocytes may be an ancient mechanism that precedes speciation in the genus and may have facilitated the adaptation of Bordetella species from environmental bacteria to mammalian respiratory pathogens.
Keywords: Bordetella, evolution, intracellular survival, macrophages, transcriptomics, stress response and adaptation
Introduction
Three closely related species of the gram-negative bacterial genus Bordetella make up the group of respiratory pathogens known as the classical bordetellae. These include the notorious human pathogen Bordetella pertussis, which is the etiological agent of pertussis or whooping cough (Mattoo and Cherry, 2005) and the closely related B. parapertussis, a species which consists of two distinct lineages that cause pertussis-like disease in humans and pneumonia in sheep, respectively (Porter et al., 1994). The third species, B. bronchiseptica, infects a wide range of mammals including many domesticated animals (Goodnow, 1980), causing a variety of pathologies ranging from chronic asymptomatic infection to acute bronchopneumonia. Multi locus sequence typing and genome comparisons revealed that B. pertussis and B. parapertussis independently arose from a B. bronchiseptica-like ancestor (Parkhill et al., 2003; Diavatopoulos et al., 2005). The classical bordetellae possess several partially characterized virulence mechanisms (Skarlupka et al., 2019) that are studied in the context of what is viewed as a completely extracellular life cycle in their mammalian hosts (Melvin et al., 2014). Yet, in vitro experiments convincingly demonstrated that the classical bordetellae can survive intracellularly within mammalian phagocytic cells (Banemann and Gross, 1997; Lamberti et al., 2010; Gorgojo et al., 2012), an ability that appears to have descended from ancestral progenitor species that lived in the environment (Hamidou Soumana et al., 2017) and acquired the ability to resist phagocytic killing by amoebae that are ubiquitous environmental predators (Taylor-Mulneix et al., 2017b). In fact, B. bronchiseptica, the species that most closely resembles the environmental ancestor of the classical bordetellae, can survive within amoeba and also disperse along with amoebic spores, highlighting a novel strategy for an environmental life cycle (Taylor-Mulneix et al., 2017a). These observations strongly suggest that intracellular survival may be an ancestral trait that might have affected the adaptation of Bordetella spp. from environmental bacteria to mammalian respiratory pathogens (Taylor-Mulneix et al., 2017b; Linz et al., 2019).
Despite not being commonly considered an intracellular pathogen, B. pertussis has repeatedly been recovered from dendritic cells and alveolar macrophages (Hellwig et al., 1999; Carbonetti et al., 2007; Paddock et al., 2008). These studies showed that B. pertussis is able to modulate human macrophages by secreting a wide range of proteins upon entry, which allows them to reside within the host cells. Interestingly, the ability to reside inside macrophages is not unique to B. pertussis, as recovery from macrophages have been confirmed for all classical bordetellae, including B. parapertussis and B. bronchiseptica (Gorgojo et al., 2012; Bendor et al., 2015).
In addition to the closely related classical bordetellae, which share about 99% sequence identity throughout their genomes, several other Bordetella species have been identified, collectively referred to as non-classical, that display much broader genetic diversity (Supplementary Figure S1). Of these, B. avium and B. hinzii cause respiratory infections in poultry and wild birds (Kersters et al., 1984; Vandamme et al., 1995). B. pseudohinzii was identified as a pathobiont in several mouse breeding colonies (Ivanov et al., 2015, 2016) and was recently shown to cause chronic ear infection in mice (Dewan et al., 2019). B. trematum is an opportunistic human pathogen that can cause severe skin disease and chronic otitis media (Vandamme et al., 1996). B. petrii was originally isolated from an anaerobic bioreactor culture enriched from river sediment (von Wintzingerode et al., 2001) and was subsequently isolated from many soil samples (Hamidou Soumana et al., 2017; Garrido-Sanz et al., 2018). Although several genomic features have changed throughout their independent evolution, including acquisition and loss of multiple virulence-associated genes (Linz et al., 2016, 2019), these Bordetella species share many characteristics that make them successful animal pathogens.
Since many of the non-classical bordetellae are animal pathogens too, we hypothesized that intracellular survival, the ability to resist digestion by phagocytic cells, may constitute an ancient environmental defense mechanism that facilitated the adaptation of Bordetella species to animals. If this were the case, then the ability to survive intracellularly would be expected to be widespread among both classical and non-classical bordetellae with shared, conserved genetic pathways. To test this hypothesis, we analyzed the transcriptome of B. bronchiseptica following internalization by macrophages and identified the induced key genes and pathways. Cross species genome comparisons revealed that most of the upregulated genes are highly conserved among the Bordetella genus. In agreement, both the classical and non-classical Bordetella species have retained the ability to survive inside murine macrophages. The only exception, B. avium – a species that has been found only among birds – has lost several of those genes and has lost the ability to survive within macrophages. Deletion of these genes in B. bronchiseptica substantially decreased its intracellular survival. These data indicate that the ability to resist phagocytic killing by host macrophages is widespread among the animal pathogenic Bordetella species and may have been an important step enabling the evolution of Bordetella species as animal pathogens.
Materials and Methods
Bacterial Strains and Growth
Bordetella bronchiseptica strain RB50, B. pseudohinzii 8-296-03, B. hinzii L60, B. petrii DSM12804, B. avium 197N and B. trematum H044680328 were grown and maintained on BG agar (Difco) supplemented with 10% defibrinated sheep’s blood (Hema Resources). Liquid cultures were grown overnight at 37°C to mid-log phase (OD ∼0.6) in Stainer Scholte (SS) liquid broth (Stainer and Scholte, 1970). Klebsiella aerogenes was grown and maintained on Luria-Bertani (LB) agar (Difco) and liquid cultures were grown at 37°C to mid-log phase in LB broth (Difco).
Intracellular Bacterial Assays
RAW 264.7 macrophages cells were grown to 80% confluency (∼1 × 105 CFU/well) in Dulbecco’s Modified Eagle Media (DMEM) supplemented with 10% FBS, glucose and glutamine in 48-well tissue-culture plates at 37°C. Bacteria were added in 10 μl PBS containing 107 CFU (MOI of 100), 106 CFU (MOI of 10) or 105 CFU (MOI of 1) as indicated. Plates were centrifuged at 250 g for 5 min at room temperature and incubated at 37°C for 1 h, after which gentamicin solution (Sigma-Aldrich) was added to a final concentration of 300 μg/ml. Plates were incubated at 37°C for an additional 1, 3, 7, or 23 h and subsequently washed with PBS. 0.1% Triton-X solution was administered, followed by 5 min incubation and vigorous pipetting to lyse the macrophages. The samples were serially diluted and plated on BG agar plates to quantify total bacteria numbers.
Electron Microscopy
RAW 264.7 macrophages were seeded in 6-well tissue-culture plates at a density of 1.5 × 105 cells/ml, inoculated with B. bronchiseptica RB50 at a MOI of 10:1 and centrifuged for 5 min at 250 g. Following 1 h incubation at 37°C, the macrophages were washed with PBS, and DMEM media containing 300 μg/ml gentamicin was added. After 1 h, the macrophages were washed with PBS and suspended in a final volume of 300 μl of PBS. The macrophages were then collected by centrifugation and fixed with fresh 2% glutaraldehyde for Transmission Electron Microscopy at the University of Georgia Electron Microscopy Core Facility.
Confocal Fluorescent Microscopy
Green fluorescent protein (GFP)-expressing B. bronchiseptica strain RB50 (Taylor-Mulneix et al., 2017a) was exposed to RAW 264.7 macrophages at a MOI of 100:1 for 1 h, followed by gentamycin treatment for 1 h. Live cell fluorescence microscopy was performed using a Zeiss Axio Obsever.Z1/7 microscope. Imaging was performed at 488 nm for GFP (green), and transmitted light for DIC II (white) at a magnification of 40x using an LD Plan-Neoflaur 40x/0.4 Korr M27 objective.
Z-Stack Imaging
RAW 264.7 cells were seeded in 6-well tissue-culture-treated plates with coverslips in the bottom at a density of 1.5 × 105 cells/ml in 3 ml and inoculated with B. bronchiseptica RB50 at a MOI of 10 (2 × 106 CFU) 12 h later. To synchronize the bacterial exposure to macrophages the plates were centrifuged at 300 g for 10 min. After 45 min incubation at 37°C, bacteria in the supernatant were removed by washing the macrophages three times with 1X PBS. The plates were incubated with DMEM medium containing 300 μg/ml gentamicin for 2 h to kill the remaining extracellular bacteria, and then washed 3 times with 3 ml of 1X PBS. Cells were fixed in 4% paraformaldehyde for 10 min at room temperature. The cells were then washed three times with 1X PBS and subsequently permeabilized with 0.1% Triton X-100 in 1X PBS for 20 min at room temperature. Primary antibodies derived from sera of B. bronchiseptica-infected mice were added after dilution in 1X PBS containing 2% BSA. After incubation at room temperature for 1 h, the cells were washed 3 times in 1X PBS. Then, the preparation was incubated with secondary donkey anti-mouse antibodies conjugated to FITC and with phalloidin for actin staining for 1 h. After 3 washes in 1X PBS, the coverslips were removed from the plates and fixed on glass slides with mounting medium containing DAPI. The images were taken with a Zeiss LSM 710 Confocal Laser Microscope for Z-stack imaging at 0.5 μm intervals.
Intracellular Bacterial Assay for Transcriptional Analysis
RAW 264.7 macrophages were seeded in 6-well tissue-culture plates and inoculated with B. bronchiseptica RB50 at a MOI of 100:1. A subset of the bacteria was cultured in DMEM medium without macrophages as the negative control. Following 1 h incubation at 37°C, the remaining bacteria in the supernatant were removed by washing the macrophages with PBS and followed by addition of DMEM medium containing 300 μg/ml gentamicin. After 1 h the DMEM was removed, and the macrophages were washed with PBS. The samples were suspended in 1 ml of TRIzol for RNA extraction.
RNA Isolation and Sequencing
RNA was extracted from RB50 lysates using TRIzol (Ambion) and the Bacterial RNA isolation Kit (Max Bacterial Enhancement Reagent, Ambion) with implemented PureLink DNase treatment (Invitrogen) following the manufacturer’s instructions. RNA quality was assessed using the NanoDrop 2000 (Thermo Scientific) and BioAnalyzer (Agilent). Samples were submitted for Illumina sequencing at the Molecular Research Laboratory in Shallowater, TX, United States. Ribosomal RNA was depleted from each biological replicate (n = 3) during preparation of the Illumina sequencing library.
Bioinformatic Analyses
Quality control of raw reads was performed using FASTQC and TRIMMOMATIC for filtering of low quality reads and trimming of Illumina library adapters. Filtered reads were mapped to Bordetella bronchiseptica RB50 genome assembly NC_002927.3 using “Bowtie2.” The resulting output files were used to evaluate differential gene expression between three biological replicates of intracellular B. bronchiseptica (n = 3) and controls (n = 3) using the “EdgeR” package for the statistical environment R distributed within the Bioconductor project.
Protein Similarity Analysis
Total protein sequences were extracted from the NCBI archive for: B. bronchiseptica RB50 (RefSeq assembly accession: GCF_000195675.1), B. parapertussis 12822 (GCF_000195695.1), B. pertussis Tohama I (CF_000195715.1), B. hinzii L60 (GCF_000657715.1), B. pseudohinzii 8-296-03 (GCF_000657795.2), B. avium 197N (GCF_000070465.1), B. petrii DSM12804 (GCF_000067205.1), and B. trematum H044680328 (GCF_900078695.1). Similarities between B. bronchiseptica proteins and proteins of the non-classical species were calculated in mGenomeSubtractor (Shao et al., 2010) as the H value for each protein, defined as H = i x (lm/lq). H is the highest BLASTp identity score (i), multiplied by the ratio of the matching sequence length (lm) and the query length (lq). Based on our previous work (Linz et al., 2018), proteins with an H value < 0.5 were considered absent. Pairwise tBLASTx genome comparisons in the Artemis Comparison Tool (Carver et al., 2008) validated proteins with values of H > 0.5 as true orthologs.
Quantitative Real-Time PCR
Real-time PCR analyses were performed on a QuantStudio (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems). Complementary DNA (cDNA) transcript libraries were prepared from biological triplicates of the control and of bacteria incubated with macrophages in DMEM + 10% FBS. Samples were processed for RNA extraction using TRIzol Reagent (Ambion by Life Technologies) and treated with PureLink DNase (Invitrogen). Primers were manually designed and purchased from IDT (Supplementary Table S1). The cycling parameters were as follows: 5-min preincubation at 95°C followed by 40 cycles of a 2-step PCR at 95°C and 60°C. Gene expression was calculated using the ΔΔCt method with expression of the 16S rRNA used as reference. Data were analyzed using DataAssist version 3.0 (Applied Biosystems).
Deletion Mutants
The allelic exchange vector pSS4245 (Inatsuka et al., 2010) was used for the generation of deletion mutants. Briefly, ∼1 kb of DNA flanking each end of the target gene was PCR amplified using primers provided in Supplementary Table S2, joined and inserted into the vector by PIPE cloning (Klock and Lesley, 2009). The construct was verified by sequencing, transformed into E. coli SM10λpir, and transferred into the parental B. bronchiseptica strain RB50 by mating. Colonies containing the integrated plasmid were selected and incubated on BG agar to stimulate allelic exchange by homologous recombination. Emerging colonies were screened by PCR for replacement of the wildtype by the mutant allele and confirmed by Sanger sequencing. In vitro growth curves showed that none of the deletion mutants had growth defects compared to the wildtype strain RB50 (data not shown). For complementation, the target gene was cloned into plasmid pBBR1 (Antoine and Locht, 1992).
Statistical Analysis
The mean ± standard error (error bars in figures) was determined for all appropriate data. Two-tailed, unpaired student’s t-tests were used to determine the statistical significance between two normally distributed populations. GraphPad Prism version 6.04 was used to conduct these statistical tests and to generate figures.
Results
B. bronchiseptica Entry and Persistence in Murine Macrophages
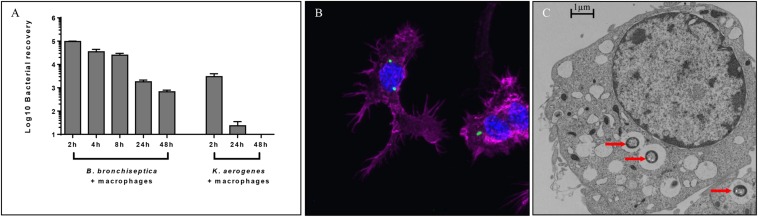
We had earlier observed using gentamicin protection assays that the prototype B. bronchiseptica strain RB50 (Bb) can enter and survive within murine macrophage-like cell line RAW 264.7 in vitro (Bendor et al., 2015). To determine the number and proportion of bacteria entering these macrophages, we performed an assay of macrophages infected with Bb at multiplicities of infection (MOI) of 100, 10 and 1 for 1 h. The percentage of recovery ranged from 0.7 to 1% of the original inoculum at all three MOIs, indicating that a relatively constant fraction of bacteria entered and resisted digestion by macrophages (Figure 1A and Supplementary Figure S2). The observation that the ratio of bacteria to macrophage did not affect survival rate suggested that this is not simply macrophages being overwhelmed or overcome by bacterial numbers. Electron microscopy (Figure 1C), confocal microscopy (Supplementary Figure S3A) and z-stack images (Figure 1B and Supplementary Figure S3B) taken after 2 h incubation confirmed the presence of bacteria within phagocytic vacuoles. Once inside the RAW 264.7 cells, bacterial numbers remained relatively stable and decreased only slowly over time. Bacterial CFUs recovered at 4 and 8 h showed no significant change in numbers for any of the MOIs used, and even at 24 and 48 h intracellular Bb were recovered in substantial numbers (Figure 1A). In contrast to Bb, Klebsiella aerogenes (Figure 1A) failed to persist in RAW 264.7 cells and was recovered at numbers over two orders of magnitude lower.
FIGURE 1.
Intracellular survival of B. bronchiseptica in murine-derived macrophages. (A) Recovery of viable B. bronchiseptica RB50 and K. aerogenes post internalization by RAW 264.7 macrophages. (B) Z-stack fluorescence microscopy localizing B. bronchiseptica inside RAW 246.7 cells at 2 h post internalization (p.i.). Purple - F-actin; blue - nucleus; green - B. bronchiseptica. (C) Transmission electron microscopy of a RAW 264.7 macrophage containing B. bronchiseptica RB50 at 2 h p.i. Red arrows depict bacteria in the cell phagosomes. Scale bar: 1 μm.
B. bronchiseptica Transcriptional Response to Internalization by Macrophages
We hypothesized that to survive within professional phagocytic cells, Bb would require distinct groups of genes to be transcriptionally modulated once the bacteria reached the intracellular niche. To examine this transcriptional response, we analyzed the RNA profile of intracellular Bb at 2 h post inoculation and compared it to that of bacteria grown in vitro. Total RNA was isolated from samples collected after antibiotic treatment and sequenced on an Illumina MiSeq (RNA-Seq). On average, 8.8 × 105 reads of intracellular Bb (n = 3) and 5.6 × 106 reads of the planktonic bacterial control (n = 3) mapped to non-rRNA regions of the Bb reference genome (NC_002927.3). Those reads were used to evaluate the differential gene expression of Bb inside macrophages in comparison to that of bacteria grown in vitro.
A Principal Component Analysis (PCA) of the normalized read distribution revealed a clear difference in the global gene expression between intracellular and in vitro grown Bb (Supplementary Figure S4). The PCA plot showed clustering of the replicates and separation of the two groups along the first principal component (PC1), indicating a distinct transcriptional response to the intracellular environment. Differentially expressed genes that displayed a log2-fold change of either ≥ 1.5 (upregulated genes) or ≤−1.5 (downregulated genes) with a p-value < 0.05 were selected for further analysis, which resulted in a list of 318 upregulated and 243 downregulated genes. To validate our RNA-seq dataset we performed a quantitative real-time (qRT) PCR to assess the transcriptional changes in five highly upregulated and four strongly downregulated genes (Supplementary Figure S5), which confirmed the RNA-Seq data.
Upregulated Genes
Functional analysis of the transcriptionally upregulated genes showed major changes at the functional levels of metabolic process (102 genes), of cellular process (116 genes), regulation (24 genes) and response to stimuli (8 genes). Gene ontology evaluation revealed enrichment for genes whose products are involved in cellular processes (113 genes) including: DNA repair, protein folding and repair, oxidative stress response, and pH homeostasis, as well as enrichment for metabolic processes (102 genes) such as nutrient assimilation (24 genes) (Table 1). A list of all 318 upregulated genes can be found in Supplementary File S1.
TABLE 1.
Upregulated genes in intracellular B. bronchiseptica.
| Locus_tag | Gene | logFC | P-value | Description |
| DNA repair | ||||
| BB0180 | dksA | 3.8 | 2.4E-12 | RNA polymerase-binding transcription factor |
| BB1919 | dnaB | 1.4 | 5.4E-08 | Replicative DNA helicase |
| BB2076 | recA | 1.8 | 2.1E-08 | Recombinase RecA |
| BB2935 | dps | 2.0 | 2.0E-07 | Putative DNA-binding protein |
| Oxidative stress and pH homeostasis | ||||
| BB0020 | risA | 2.1 | 4.3E-06 | Transcriptional regulator RisA |
| BB1837 | rpoE | 3.2 | 1.7E-12 | RNA polymerase sigma factor RpoE |
| BB2275 | iscR | 4.2 | 1.7E-12 | Transcriptional regulator IscR |
| BB2276 | iscS | 2.5 | 3.3E-10 | Cysteine desulfurase |
| BB2279 | hscB | 1.6 | 1.5E-07 | HscB chaperone |
| BB2281 | fdx | 1.6 | 5.5E-04 | Ferredoxin, 2Fe–2S |
| BB3080 | slyA | 3.0 | 7.2E-06 | Transcriptional regulator SlyA |
| BB3766 | msrP | 1.6 | 8.7E-08 | Protein-methionine-sulfoxide reductase |
| BB3800 | msrB | 2.1 | 1.9E-07 | Peptide methionine sulfoxide reductase |
| BB4506 | rpoN | 3.4 | 9.5E-12 | Sigma(54) modulation protein RpoN |
| BB3942 | fur | 2.5 | 3.3E-08 | Ferric uptake regulator |
| BB4835 | rpoH | 4.5 | 2.4E-12 | RNA polymerase sigma factor RpoH |
| Protein folding | ||||
| BB0178 | hslU | 4.1 | 2.3E-12 | ATP-dependent protease |
| BB0179 | hslV | 6.0 | 1.9E-12 | ATP-dependent protease |
| BB0295 | secB | 1.6 | 1.3E-07 | Protein-export protein |
| BB0501 | htpG | 4.2 | 2.8E-10 | Chaperone protein |
| BB0962 | groEL | 4.4 | 8.4E-11 | 60 kDa chaperonin |
| BB0963 | groES | 7.8 | 6.7E-14 | 10 kDa chaperonin |
| BB2256 | 2.4 | 8.4E-11 | ATP-dependent protease | |
| BB3170 | hfq | 2.4 | 3.0E-07 | RNA-binding protein |
| BB3293 | clpB | 2.6 | 5.5E-10 | Chaperone protein |
| BB3933 | dnaJ | 1.7 | 1.1E-07 | Chaperone protein |
| BB3934 | dnaK | 3.9 | 9.7E-11 | Chaperone protein |
| BB3936 | grpE | 3.9 | 2.6E-10 | Chaperone protein |
| Metabolism | ||||
| BB0095 | glcB | 1.9 | 4.2E-06 | Malate synthase G |
| BB0096 | glcC | 1.9 | 1.5E-06 | Malate synthase G transcriptional regulator |
| BB3682 | sdhC | 2.3 | 1.0E-09 | Succinate dehydrogenase cytochrome B |
| BB3684 | mdh | 2.8 | 8.9E-10 | Malate dehydrogenase |
| BB1850 | acnB | 1.4 | 1.04.E-7 | Aconitate hydratase B |
| BB4150 | 2.4 | 1.5E-07 | Putative short-chain dehydrogenase | |
| BB3474 | ompA | 3.1 | 3.9E-11 | Outer membrane protein A |
| BB3759 | plsX | 1.6 | 1.9E-09 | Phosphate acyltransferase |
| BB3771 | pagL | 2.4 | 1.3E-08 | Lipid A deacylase |
| BB0233 | 2.9 | 2.3E-09 | Putative AMP-binding enzyme | |
| BB1556 | 4.1 | 2.4E-12 | ABC transporter, ATP-binding protein | |
| BB4592 | 4.1 | 1.7E-12 | Putative binding-protein-dependent transport | |
| BB0097 | 3.9 | 6.4E-10 | Putative dehydrogenase | |
| BB1446 | carA | 1.5 | 6.8E-09 | Carbamoyl-phosphate synthase |
| BB0235 | 5.1 | 9.5E-12 | Probable transporter | |
| BB4355 | argC | 2.6 | 5.4E-11 | N-acetyl-gamma-glutamyl-phosphate reductase |
| BB1986 | argG | 1.5 | 2.4E-08 | Argininosuccinate synthase |
| BB2000 | 2.5 | 1.3E-09 | Putative aldolase | |
| BB3179 | ndk | 2.3 | 1.9E-09 | Nucleoside diphosphate kinase |
| BB2607 | pyrH | 1.8 | 2.0E-09 | Uridylate kinase |
| BB3468 | cmk | 1.5 | 2.8E-08 | Cytidylate kinase |
| BB4376 | nrdA | 2.3 | 1.6E-09 | Ribonucleoside-diphosphate reductase |
| Virulence | ||||
| BB2992 | fimA | 2.6 | 9.7E-11 | Fimbrial protein |
| BB2994 | bvgA | 2.6 | 2.6E-09 | Transcriptional regulator of virulence |
| BB3424 | 1.7 | 3.7E-07 | Fimbrial protein | |
| BB3674 | fim2 | 6.7 | 1.7E-12 | Serotype 2 fimbrial subunit |
logFC, log2 fold change.
Expression of recA, dnaB, dps, and dksA, all implicated in the activation of the SOS response and DNA repair (Bearson et al., 1997; Simmons et al., 2008; Lund et al., 2014), was upregulated upon internalization by macrophages. Likewise, genes for protein folding and recycling such as molecular chaperones groES, groEL, and htpG, and protease genes hslV and hslU, were highly upregulated, as was expression of several other osmotic and heat shock response genes, including clpB, grpE, dnaK, and dnaJ.
Congruent to previous studies in other bacterial species (Buchmeier et al., 1997; Zimna et al., 2001; Clements et al., 2002; Gilberthorpe et al., 2007; Chiang and Schellhorn, 2012; Fang et al., 2016), expression of genes that promote resistance against oxidative stress and low pH was upregulated intracellularly, including transcription factor iscR and adjacent genes iscS, hscB, and fdx, and the transcription regulators slyA, risA, and fur. Additionally, RNA polymerase sigma factor genes rpoH, rpoN, and rpoE were highly upregulated (Laskos et al., 2004; Delory et al., 2006; Hanawa et al., 2013), as was expression of the RNA chaperone gene hfq, which is known to increase resistance against killing by macrophages (Bibova et al., 2013).
Increased transcription of glyoxylate cycle genes such as mdh, sdhC, glcB, glcC, and acnB, as well as of numerous ribosomal protein genes implies extensive metabolic activity in the bacterial cell in response to internalization by macrophages. Several genes of fatty acid synthesis pathways such as 3-oxoacyl-ACP reductase BB4150, long chain fatty acid Co-A ligase BB0233, outer membrane protein ompA, and ABC transport protein encoded by BB1556, were found to be strongly induced, suggesting increased membrane biosynthesis. Also, we observed an increase in expression of genes involved in amino acid biosynthesis and transport, including BB4592, carA, argC, and argG, and of de novo nucleotide biosynthesis (ndk, pyrH, cmk, and nrdA).
The fimbria encoding genes fim2, fimA and BB3424 were the only genes encoding virulence-associated factors among the 318 transcriptionally upregulated genes.
Downregulated Genes
The cya genes encoding the adenylate cyclase toxin were among the most downregulated genes in our dataset with a log2fold change of about -3 (Table 2 and Supplementary File S1), and expression of the dermonecrotic toxin gene dnt was also downregulated. In agreement with previous studies (Hausman et al., 1996; Antoine et al., 2000; Hausman and Burns, 2000), expression of the pertussis toxin operon and the associated type IV secretion system (T4SS) was barely detectable under either condition. Similarly, expression of the type III secretion system (T3SS) encoded by the bsc locus was strongly suppressed, resulting in decreased expression of both the apparatus-related and the secretion-related components (Table 2 and Supplementary File S1). In addition to toxins and secretion systems, expression of the O-Antigen–encoding wbm locus (wbmO – bplJ), was significantly downregulated.
TABLE 2.
Downregulated genes in intracellular B. bronchiseptica.
| Locus_tag | Gene | LogFC | P-value | Description |
| O-Antigen | ||||
| BB0130 | wbmO | −1.8 | 4.6E-06 | O-Antigen biosynthesis protein |
| BB0138 | wbmG | −2.2 | 1.1E-05 | Nucleotide sugar epimerase/dehydratase |
| BB0139 | wbmF | −1.8 | 9.4E-06 | Nucleotide sugar epimerase/dehydratase |
| BB0145 | bplL | −1.8 | 3.2E-06 | Lipopolysaccharide biosynthesis protein |
| BB0146 | bplJ | −1.6 | 1.2E-05 | Membrane protein |
| Adenylate cyclase toxin | ||||
| BB0325 | cyaB | −2.9 | 3.3E-09 | Cyclolysin secretion ATP-binding protein |
| BB0326 | cyaD | −3.4 | 1.0E-08 | Membrane fusion protein (MFP) family protein |
| BB0327 | cyaE | −3.3 | 5.8E-10 | Protein CyaE |
| BB0328 | cyaX | −3.9 | 1.1E-08 | Adenylate cyclase transcriptional regulator |
| Type 3 secretion system | ||||
| BB1609 | bscF | −2.1 | 6.5E-06 | Putative type III secretion protein |
| BB1623 | bcr4 | −2.6 | 6.9E-08 | Uncharacterized protein |
| BB1624 | bscI | −2.0 | 1.3E-04 | Putative type III secretion protein |
| BB1625 | bscJ | −2.3 | 4.4E-07 | Lipoprotein |
| BB1627 | bscL | −2.5 | 1.1E-04 | Type III secretion protein |
| BB1628 | bscN | −1.9 | 5.3E-06 | Type III secretion ATP synthase |
| BB1630 | bscP | −2.8 | 4.8E-07 | Type III secretion protein |
| BB1631 | bscQ | −3.0 | 4.3E-08 | Type III secretion protein |
| BB1632 | bscR | −2.1 | 6.5E-05 | Type III secretion protein |
| BB1634 | bscT | −3.0 | 3.0E-06 | Type III secretion protein |
| BB1635 | bscU | −2.8 | 7.9E-07 | Type III secretion protein |
| BB1636 | bscW | −2.8 | 9.9E-05 | Type III secretion protein |
| BB1637 | bscC | −1.9 | 3.9E-07 | Type III secretion protein |
| Electron transport | ||||
| BB3827 | −4.6 | 6.0E-10 | Putative membrane protein | |
| BB3828 | nuoN | −3.0 | 6.9E-10 | NADH-quinone oxidoreductase subunit N |
| BB3829 | nuoM | −2.3 | 2.5E-08 | NADH-ubiquinone oxidoreductase, chain M |
| BB3830 | nuoL | −2.4 | 6.9E-10 | NADH-ubiquinone oxidoreductase, chain L |
| BB3834 | nuoH | −1.7 | 1.3E-07 | NADH-quinone oxidoreductase subunit H |
| BB3835 | nuoG | −1.9 | 3.7E-08 | NADH-quinone oxidoreductase |
| BB3836 | nuoF | −2.0 | 5.7E-09 | NADH-quinone oxidoreductase subunit F |
| Cell division | ||||
| BB4188 | −2.2 | 2.4E-08 | Uncharacterized protein | |
| BB4193 | ftsZ | −2.5 | 4.3E-10 | Cell division protein FtsZ |
| BB4194 | ftsA | −3.4 | 3.3E-09 | Cell division protein FtsA |
| BB4195 | ftsQ | −3.4 | 6.0E-09 | Cell division protein FtsQ |
| BB4196 | ddl | −3.5 | 2.1E-07 | D-alanine–D-alanine ligase |
| BB4197 | murC | −3.0 | 1.1E-08 | UDP-N-acetylmuramate–L-alanine ligase |
| BB4198 | murG | −3.6 | 8.1E-10 | Undecaprenyl-PP-MurNAc-pentapeptide-UDPGlcNAc GlcNAc transferase |
| BB4199 | ftsW | −3.6 | 3.6E-08 | Cell division protein FtsW |
| BB4200 | murD | −3.0 | 5.2E-09 | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase |
| BB4201 | mraY | −2.8 | 1.3E-08 | Phospho-N-acetylmuramoyl-pentapeptide-transferase |
| BB4202 | murE | −2.1 | 1.0E-09 | Multifunctional fusion protein |
logFC, log2 fold change.
Notably, several genes with important functions in cell structure biogenesis and proliferation were also downregulated inside macrophages, including cell division genes ftsZ, ftsA, and ftsQ and cell wall synthesis genes murC, murG, ftsW, and murD. Similarly, expression of a large gene locus (BB3827 to BB3836) encoding the oxidative respiratory chain was significantly downregulated, including NADH dehydrogenase genes nuoN, nuoM, nuoL, and nuoH.
Taken together, B. bronchiseptica responded to internalization by macrophages by rapid changes in its transcriptional profile, that were marked by suppression of growth and virulence, and strong activation of the bacterial stress response, including DNA and protein repair and pH homeostasis, and suppression of cell division, putative virulence factors and oxidative respiration.
The Non-classical Bordetellae and Intracellular Persistence
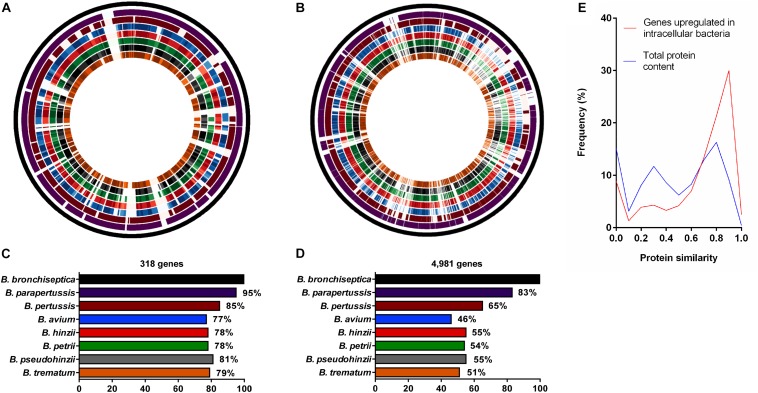
Since the human-restricted pathogens B. pertussis and B. parapertussis arose from B. bronchiseptica-like ancestors and can persist inside human macrophages, we evaluated whether the 318 upregulated genes were conserved among the three classical Bordetella species. Two hundred and seventy two intact genes (86%) were identified in the genome of B. pertussis strain Tohama I, the other genes were missing or truncated by frameshifts or premature stop codons. Similarly, 301 of the 318 genes (95%) were present in the genome of B. parapertussis strain 12822, showing conservation of most genes (Figures 2A,C).
FIGURE 2.
Comparative analysis of genes upregulated during intracellular survival and their presence/absence in non-classical Bordetella species. Analysis of protein similarity of (A) 318 B. bronchiseptica genes upregulated in macrophages and (B) 4,981 genes in the entire genome of B. bronchiseptica strain RB50 in comparison to the non-classical Bordetella species. From outside to inside: Circle 1: Virtual genome of B. bronchiseptica strain RB50. Circles 2–8: Visual representation of protein similarity between B. bronchiseptica RB50 and classical (circles 2–3) and the non-classical species (circles 4–8) represented as color shades with darker shades indicating higher protein similarity. (C) 77–81% of the genes upregulated in intracellular B. bronchiseptica were conserved among the non-classical species, (D) in contrast to only 46–55% of the 4,981 B. bronchiseptica genes in the entire genome. (E) Line plot showing the frequency of protein similarities.
While many non-classical bordetellae are also human and animal pathogens, their ability to persist inside phagocytic cells has not been evaluated. Therefore, we tested the presence or absence of the upregulated genes among the non-classical Bordetella species, including the bird pathogens B. hinzii (Vandamme et al., 1995) and B. avium (Kersters et al., 1984), the mouse pathogen B. pseudohinzii (Ivanov et al., 2016), the human opportunistic pathogen B. trematum (Vandamme et al., 1996), and the environmental species B. petrii (von Wintzingerode et al., 2001). We calculated the protein similarity (H value) of the 318 genes upregulated in B. bronchiseptica inside macrophages and their corresponding homologs in the non-classical bordetellae, with a gene considered to be present with a protein similarity value of H ≥ 0.5. An average of 77–81% of the 318 upregulated genes were present in the non-classical species (Figures 2A,C) with 95 (30%) of the genes displaying similarity values of H ≥ 0.9 (Figure 2E). In contrast, only 46–55% of the total of 4,981 evaluated B. bronchiseptica genes were identified in the genomes of the non-classical species (P < 0.0001), where only 448 (9%) of the genes reached protein similarity scores of H ≥ 0.9 (Figures 2B,D,E).
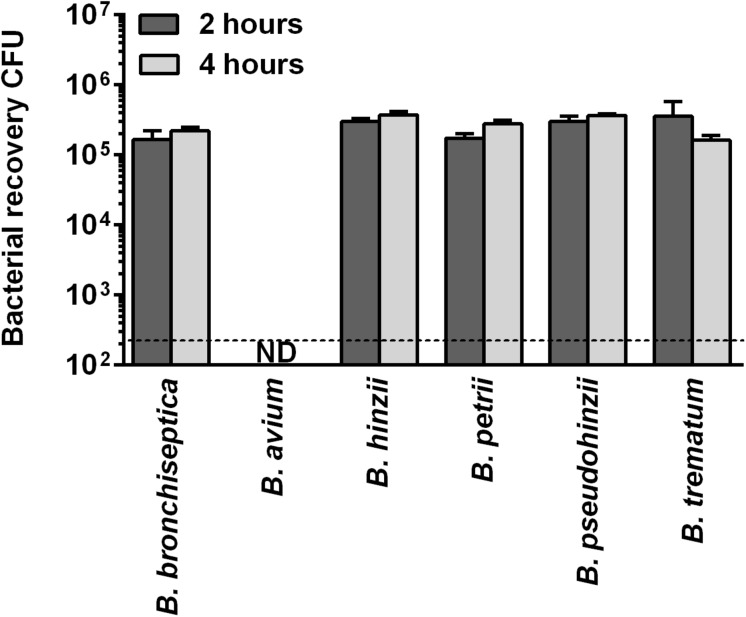
This high evolutionary conservation of genes that are upregulated in B. bronchiseptica during intracellular survival in phagocytic cells suggests that the non-classical bordetellae may be able to persist inside macrophages. To test this hypothesis, the non-classical species were assessed for intracellular survival in RAW 264.7 macrophages for 2 and 4 h. All examined Bordetella species were recovered, with the exception of B. avium (Figure 3). The inoculated B. pseudohinzii, B. hinzii, B. trematum and B. petrii bacteria survived internalization by macrophages at similar rates to B. bronchiseptica. The genomes of these species share 222 out of the 318 (70%) transcriptionally upregulated genes, which implies a critical function for intracellular persistence in mammalian phagocytic cells.
FIGURE 3.
Intracellular survival of non-classical bordetellae within macrophages. The non-classical bordetellae were recovered from macrophages in numbers similar to B. bronchiseptica. The only exception, B. avium, failed to survive internalization by macrophages.
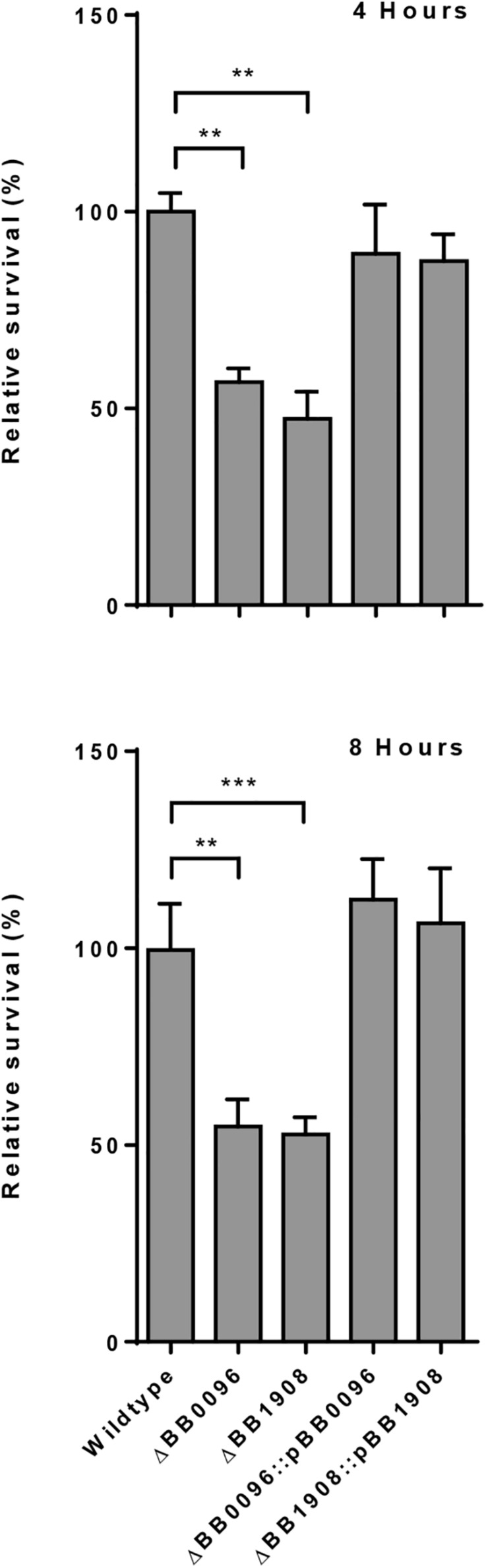
In contrast to the other analyzed Bordetella species, B. avium was severely impaired in its ability to persist inside macrophages. Only 0.001% of the inoculum was recovered after 2 h and no viable bacteria were detected after 4 h. Therefore, we performed a comparative genome analysis to identify transcriptionally upregulated genes that were only missing in B. avium, which resulted in the identification of six genes (Table 3). Deletion of two of these genes (BB0096 and BB1908) resulted in a significant reduction in intracellular survival (Figure 4 and Supplementary Figure S6). Complementation of these knock-out mutants with plasmid-borne gene copies restored the wildtype phenotype in both mutants (Figure 4), confirming that loss of malate synthase transcriptional regulator glcC (BB0096) or the tripartite tricarboxylate transporter BB1908 negatively impacts intracellular persistence in macrophages. In addition, previous studies showed important roles of transcriptionally upregulated (Table 1) risA and hfq genes in intracellular persistence of Bb and B. pertussis (Zimna et al., 2001; Bibova et al., 2013). We also assessed Bb knock-out mutants of several other transcriptionally upregulated genes (Supplementary Table S3), however, intracellular survival of none of the tested mutants was significantly different from the RB50 wildtype strain.
TABLE 3.
Genes upregulated in intracellular B. bronchiseptica and absent in B. avium.
| Locus_tag/gene | logFC | Protein |
| BB0096/glcC | 1.9 | Malate synthase transcriptional regulator |
| BB1908 | 1.6 | Tripartite tricarboxylate transporter receptor |
| BB1948 | 1.9 | Glutamate transport periplasmic receptor |
| BB1999 | 2.5 | Tripartite tricarboxylate transporter receptor |
| BB2944 | 1.6 | LysR-family transcriptional regulator |
| BB4150 | 2.4 | 3-ketoacyl-(acyl-carrier-protein) reductase |
logFC, log2 fold change.
FIGURE 4.
Assessment of B. bronchiseptica deletion mutants for intracellular survival. Deletion of malate synthase regulator gene BB0096 or tripartite tricarboxylate transporter gene BB1908 resulted in significantly reduced bacterial recovery. Plasmid-borne complementation of the gene deletion restored the wildtype phenotype. ∗∗p < 0.01; ∗∗∗p < 0.001.
Discussion
Most bacterial pathogens have specialized to either an intracellular or extracellular lifestyle, which determines the focus in studies on bacterial pathogenesis. The classical species of the genus Bordetella are broadly known as extracellular pathogens. However, an increasing number of publications have reported recovery of viable bacteria from phagocytic host cells in vitro, providing evidence for at least transient intracellular survival or persistence. There are also anecdotal reports of clinical samples harboring intracellular B. pertussis leading to speculation on the significance of this intracellular bacterial population in pathogenicity (Higgs et al., 2012).
Here we showed that intracellular survival and persistence is not restricted to the three classical bordetellae, but that the non-classical species B. hinzii, B. pseudohinzii, B. trematum, and B. petrii survived at equally high proportions, establishing the ability to survive internalization by phagocytic host cells as a common feature among the animal pathogenic bordetellae. Considering that the environmental species B. petrii did not differ from the animal pathogenic species, this common genotypic and phenotypic trait suggests the ability to survive predation by eukaryotic phagocytic cell precedes speciation in the genus.
A common ability suggests an ancestral origin and a common set of genes that are required for intracellular survival. Indeed, of the 318 B. bronchiseptica genes found to be transcriptionally upregulated during intracellular exposure, about 80% were present in the genomes of non-classical Bordetella, with 222 genes (70%) shared between all species (Figure 2A). The only exception was the bird pathogen B. avium, which was missing six out of these 222 genes and failed to persist inside macrophages (Table 3). Interestingly, B. avium has one of the smallest genomes in the Bordetella genus with only 3.73 Mb in size (Sebaihia et al., 2006) suggesting that it may have undergone genome reduction during its evolution and adaptation to a specific host (Parkhill et al., 2003; Linz et al., 2016; Taylor-Mulneix et al., 2017b).
The apparent localization of the intracellular B. bronchiseptica within phagosomes (Figure 1C) suggests that the bacteria are rapidly exposed to a variety of damaging conditions, including low pH, bactericidal factors and resource starvation. To survive such harsh conditions many pathogenic bacteria have evolved mechanisms to maintain cell homeostasis and prevent DNA damage and cell death (Simmons et al., 2008). We observed the transcriptional hallmarks of a general SOS response (Bearson et al., 1997; Lund et al., 2014), characterized by suppression of cell division via downregulation of the fts locus (Table 2) and by upregulation of DNA repair genes, of protein chaperone genes, and of B. bronchiseptica homologs (rpoH, fur, risA) of the E. coli acid tolerance genes rpoS, fur and phoP (Table 1 and Figure 5) (Simmons et al., 2008).
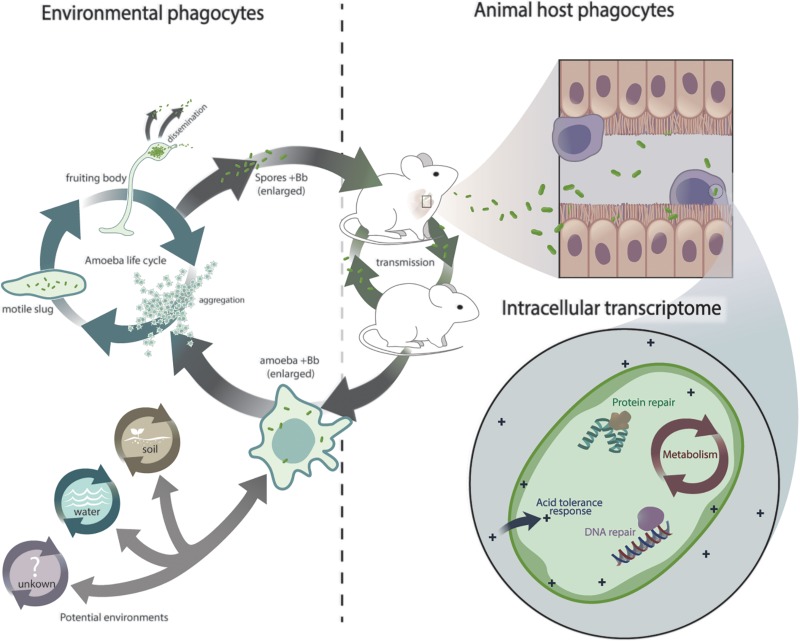
FIGURE 5.
Schematic representation of Bordetella exposure to eukaryotic phagocytes and its transcriptional response. Hypothetical scenario depicting Bordetella exposure to interconnected lifecycles and adaptation from environmental phagocytes to animal phagocytes.
Intracellular persistence was also accompanied by metabolic changes (Figure 5 and Tables 1, 2). As expected under micro-aerophilic/hypoxic conditions inside macrophages, expression of the nuoF – nuoN genes that encode the oxidative respiratory chain was strongly suppressed. In contrast, genes of the glyoxylate/TCA cycle showed elevated expression levels, including malate synthase G gene glcB and its transcriptional activator glcC, malate dehydrogenase mdh, citrate synthase gltA, and aconitase acnB. The glyoxylate cycle is important in the utilization of acetate or fatty acids as the main carbon source and may be essential to provide hexoses for nucleotide and amino acid biosynthesis under intracellular conditions (Pellicer et al., 1999; Munoz-Elias and McKinney, 2006). B. bronchiseptica genes involved in biosynthesis of nucleotides, amino acids and fatty acids were indeed upregulated, consistent with limited access to these molecules. The absence of the malate transcriptional activator glcC and tricarboxylic transporter BB1908 may explain the failure of B. avium to persist inside macrophages.
Many intracellular pathogens such as Burkholderia pseudomallei employ protein secretion systems to facilitate replication and spread inside their hosts (Stevens et al., 2002). Interestingly, our assays were conducted at 37°C, a temperature known to induce phosphorylation of bvgA, which in turn induces expression of virulence factors (Prugnola et al., 1995). Yet under these intracellular conditions B. bronchiseptica displayed strong suppression of other known virulence factors, including the operons encoding both T3SS and the adenylate cyclase toxin (ACT) expression, modification and secretion. While stress conditions such as low pH have been reported to induce the expression of virulence factors in many pathogenic bacteria (Rathman et al., 1996; Bearson et al., 1997), the suppression of virulence in B. bronchiseptica occurred despite their intracellular vacuolar location where similar low pH environments are expected. We had earlier reported that the avirulent stage is required for survival, persistence and replication of B. bronchiseptica within amoeba (Taylor-Mulneix et al., 2017a), which strongly suggests that repression of virulence within the intracellular environment is part of an ancient conserved stress response in the genus.
Taken together, our results show that upon internalization by macrophages a certain proportion of bordetellae are killed, but thousands of bacteria can adapt and modulate gene expression to cope with this new environment. Rapid transcriptional adaptation was marked by what can be considered a general stress response against professional phagocytes that included increased expression of genes involved in DNA and protein repair, acid tolerance and metabolism (Figure 5). Conservation of these genes throughout the genus and the demonstrated ability of non-classical species, including the environmental B. petrii, to persist inside macrophages suggests that this response to phagocytes is not confined to the commonly studied classical bordetellae. It appears to represent an ancient pathway that preceded speciation in the genus and thus likely arose from a common ancestor. The two independent but interconnected transmission cycles of B. bronchiseptica in environmental amebae and in mammalian hosts (Figure 5) lead us to speculate that early interaction with these environmental phagocytes may have played a role in the origin of this response, which subsequently facilitated the adaptation to higher animals and thus the evolution of Bordetella from environmental microbes to animal and human pathogens.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
IR, BL, and EH conceived the study. IR, BL, KD, LM, CR, DK, and EH designed the experiments. IR, BL, KD, LM, and CR performed the experiments. IR, BL, KD, LM, CR, and EH analyzed the data. IR, BL, KD, and EH wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants AI104399, GM113681, and AI142678 of the National Institutes of Health to EH. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02839/full#supplementary-material
References
- Antoine R., Locht C. (1992). Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6 1785–1799. 10.1111/j.1365-2958.1992.tb01351.x [DOI] [PubMed] [Google Scholar]
- Antoine R., Raze D., Locht C. (2000). Genomics of Bordetella pertussis toxins. Int. J. Med. Microbiol. 290 301–305. 10.1016/S1438-4221(00)80026-0 [DOI] [PubMed] [Google Scholar]
- Banemann A., Gross R. (1997). Phase variation affects long-term survival of Bordetella bronchiseptica in professional phagocytes. Infect. Immun. 65 3469–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson S., Bearson B., Foster J. W. (1997). Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147 173–180. 10.1111/j.1574-6968.1997.tb10238.x [DOI] [PubMed] [Google Scholar]
- Bendor L., Weyrich L. S., Linz B., Rolin O. Y., Taylor D. L., Goodfield L. L., et al. (2015). Type six secretion system of Bordetella bronchiseptica and adaptive immune components limit intracellular survival during infection. PloS One 10:e0140743. 10.1371/journal.pone.0140743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibova I., Skopova K., Masin J., Cerny O., Hot D., Sebo P., et al. (2013). The RNA chaperone Hfq is required for virulence of Bordetella pertussis. Infect. Immun. 81 4081–4090. 10.1128/IAI.00345-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier N., Bossie S., Chen C. Y., Fang F. C., Guiney D. G., Libby S. J. (1997). SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65 3725–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonetti N. H., Artamonova G. V., Van Rooijen N., Ayala V. I. (2007). Pertussis toxin targets airway macrophages to promote Bordetella pertussis infection of the respiratory tract. Infect. Immun. 75 1713–1720. 10.1128/IAI.01578-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T., Berriman M., Tivey A., Patel C., Bohme U., Barrell B. G., et al. (2008). Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24 2672–2676. 10.1093/bioinformatics/btn529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S. M., Schellhorn H. E. (2012). Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525 161–169. 10.1016/j.abb.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Clements M. O., Eriksson S., Thompson A., Lucchini S., Hinton J. C., Normark S., et al. (2002). Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A. 99 8784–8789. 10.1073/pnas.132047099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delory M., Hallez R., Letesson J. J., De Bolle X. (2006). An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J. Bacteriol. 188 7707–7710. 10.1128/JB.00644-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan K. K., Taylor-Mulneix D. L., Campos L. L., Skarlupka A. L., Wagner S. M., Ryman V. E., et al. (2019). A model of chronic, transmissible otitis media in mice. PLoS Pathog. 15:e1007696. 10.1371/journal.ppat.1007696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diavatopoulos D. A., Cummings C. A., Schouls L. M., Brinig M. M., Relman D. A., Mooi F. R. (2005). Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1:e45. 10.1371/journal.ppat.0010045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F. C., Frawley E. R., Tapscott T., Vazquez-Torres A. (2016). Bacterial stress responses during host infection. Cell Host Microbe 20 133–143. 10.1016/j.chom.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Sanz D., Manzano J., Martin M., Redondo-Nieto M., Rivilla R. (2018). Metagenomic analysis of a biphenyl-degrading soil bacterial consortium reveals the metabolic roles of specific populations. Front. Microbiol. 9:232. 10.3389/fmicb.2018.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilberthorpe N. J., Lee M. E., Stevanin T. M., Read R. C., Poole R. K. (2007). NsrR: a key regulator circumventing Salmonella enterica serovar typhimurium oxidative and nitrosative stress in vitro and in IFN-gamma-stimulated J774.2 macrophages. Microbiology 153(Pt 6), 1756–1771. 10.1099/mic.0.2006/003731-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow R. A. (1980). Biology of Bordetella bronchiseptica. Microbiol. Rev. 44 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgojo J., Lamberti Y., Valdez H., Harvill E. T., Rodriguez M. E. (2012). Bordetella parapertussis survives the innate interaction with human neutrophils by impairing bactericidal trafficking inside the cell through a lipid raft-dependent mechanism mediated by the lipopolysaccharide O antigen. Infect. Immun. 80 4309–4316. 10.1128/IAI.00662-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidou Soumana I., Linz B., Harvill E. T. (2017). Environmental ORigin of the Genus Bordetella. Front. Microbiol. 8:28. 10.3389/fmicb.2017.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa T., Yonezawa H., Kawakami H., Kamiya S., Armstrong S. K. (2013). Role of Bordetella pertussis RseA in the cell envelope stress response and adenylate cyclase toxin release. Pathog. Dis. 69 7–20. 10.1111/2049-632X.12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausman S. Z., Burns D. L. (2000). Use of pertussis toxin encoded by ptx genes from Bordetella bronchiseptica to model the effects of antigenic drift of pertussis toxin on antibody neutralization. Infect. Immun. 68 3763–3767. 10.1128/iai.68.6.3763-3767.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausman S. Z., Cherry J. D., Heininger U., Wirsing von Konig C. H., Burns D. L. (1996). Analysis of proteins encoded by the ptx and ptl genes of Bordetella bronchiseptica and Bordetella parapertussis. Infect. Immun. 64 4020–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig S. M., Hazenbos W. L., van de Winkel J. G., Mooi F. R. (1999). Evidence for an intracellular niche for Bordetella pertussis in broncho-alveolar lavage cells of mice. FEMS Immunol. Med. Microbiol. 26 203–207. 10.1111/j.1574-695X.1999.tb01391.x [DOI] [PubMed] [Google Scholar]
- Higgs R., Higgins S. C., Ross P. J., Mills K. H. (2012). Immunity to the respiratory pathogen Bordetella pertussis. Mucosal. Immunol. 5 485–500. 10.1038/mi.2012.54 [DOI] [PubMed] [Google Scholar]
- Inatsuka C. S., Xu Q., Vujkovic-Cvijin I., Wong S., Stibitz S., Miller J. F., et al. (2010). Pertactin is required for Bordetella species to resist neutrophil-mediated clearance. Infect. Immun. 78 2901–2909. 10.1128/IAI.00188-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov Y. V., Linz B., Register K. B., Newman J. D., Taylor D. L., Boschert K. R., et al. (2016). Identification and taxonomic characterization of Bordetella pseudohinzii sp. nov. isolated from laboratory-raised mice. Int. J. Syst. Evol. Microbiol. 66 5452–5459. 10.1099/ijsem.0.001540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov Y. V., Shariat N., Register K. B., Linz B., Rivera I., Hu K., et al. (2015). A newly discovered Bordetella species carries a transcriptionally active CRISPR-Cas with a small Cas9 endonuclease. BMC Genomics 16:863. 10.1186/s12864-015-2028-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersters K., Hinz K. H., Hertle A., Segers P., Lievens A., Siegmann O., et al. (1984). Bordetella avium sp. nov., isolated from the respiratory tracts of turkeys and other birds. Int. J. Syst. Bacteriol. 34 56–70. 10.1099/00207713-34-1-56 [DOI] [Google Scholar]
- Klock H. E., Lesley S. A. (2009). The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 498 91–103. 10.1007/978-1-59745-196-3_6 [DOI] [PubMed] [Google Scholar]
- Lamberti Y. A., Hayes J. A., Perez Vidakovics M. L., Harvill E. T., Rodriguez M. E. (2010). Intracellular trafficking of Bordetella pertussis in human macrophages. Infect. Immun. 78 907–913. 10.1128/IAI.01031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskos L., Ryan C. S., Fyfe J. A., Davies J. K. (2004). The RpoH-mediated stress response in Neisseria gonorrhoeae is regulated at the level of activity. J. Bacteriol. 186 8443–8452. 10.1128/JB.186.24.8443-8452.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz B., Ivanov Y. V., Preston A., Brinkac L., Parkhill J., Kim M., et al. (2016). Acquisition and loss of virulence-associated factors during genome evolution and speciation in three clades of Bordetella species. BMC Genomics 17:767. 10.1186/s12864-016-3112-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz B., Ma L., Rivera I., Harvill E. T. (2019). Genotypic and phenotypic adaptation of pathogens: lesson from the genus Bordetella. Curr. Opin. Infect. Dis. 32 223–230. 10.1097/QCO.0000000000000549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz B., Mukhtar N., Shabbir M. Z., Rivera I., Ivanov Y. V., Tahir Z., et al. (2018). Virulent epidemic pneumonia in sheep caused by the human pathogen Acinetobacter baumannii. Front. Microbiol. 9:2616. 10.3389/fmicb.2018.02616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P., Tramonti A., De Biase D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 38 1091–1125. 10.1111/1574-6976.12076 [DOI] [PubMed] [Google Scholar]
- Mattoo S., Cherry J. D. (2005). Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18 326–382. 10.1128/CMR.18.2.326-382.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin J. A., Scheller E. V., Miller J. F., Cotter P. A. (2014). Bordetella pertussis pathogenesis: current and future challenges. Nat. Rev. Microbiol. 12 274–288. 10.1038/nrmicro3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias E. J., McKinney J. D. (2006). Carbon metabolism of intracellular bacteria. Cell Microbiol. 8 10–22. 10.1111/j.1462-5822.2005.00648.x [DOI] [PubMed] [Google Scholar]
- Paddock C. D., Sanden G. N., Cherry J. D., Gal A. A., Langston C., Tatti K. M., et al. (2008). Pathology and pathogenesis of fatal Bordetella pertussis infection in infants. Clin. Infect. Dis. 47 328–338. 10.1086/589753 [DOI] [PubMed] [Google Scholar]
- Parkhill J., Sebaihia M., Preston A., Murphy L. D., Thomson N., Harris D. E., et al. (2003). Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35 32–40. [DOI] [PubMed] [Google Scholar]
- Pellicer M. T., Fernandez C., Badia J., Aguilar J., Lin E. C., Baldom L. (1999). Cross-induction of glc and ace operons of Escherichia coli attributable to pathway intersection. characterization of the glc promoter. J. Biol. Chem. 274 1745–1752. 10.1074/jbc.274.3.1745 [DOI] [PubMed] [Google Scholar]
- Porter J. F., Connor K., Donachie W. (1994). Isolation and characterization of Bordetella parapertussis-like bacteria from ovine lungs. Microbiology 140(Pt 2), 255–261. 10.1099/13500872-140-2-255 [DOI] [PubMed] [Google Scholar]
- Prugnola A., Arico B., Manetti R., Rappuoli R., Scarlato V. (1995). Response of the bvg regulon of Bordetella pertussis to different temperatures and short-term temperature shifts. Microbiology 141(Pt 10), 2529–2534. 10.1099/13500872-141-10-2529 [DOI] [PubMed] [Google Scholar]
- Rathman M., Sjaastad M. D., Falkow S. (1996). Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64 2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaihia M., Preston A., Maskell D. J., Kuzmiak H., Connell T. D., King N. D., et al. (2006). Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction. J. Bacteriol. 188 6002–6015. 10.1128/JB.01927-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., He X., Harrison E. M., Tai C., Ou H. Y., Rajakumar K., et al. (2010). mGenomeSubtractor: a web-based tool for parallel in silico subtractive hybridization analysis of multiple bacterial genomes. Nucleic Acids Res. 38 W194–W200. 10.1093/nar/gkq326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons L. A., Foti J. J., Cohen S. E., Walker G. C. (2008). The SOS regulatory network. EcoSal Plus 3 1–30. 10.1128/ecosalplus.5.4.3 [DOI] [PubMed] [Google Scholar]
- Skarlupka A. L., Linz B., Maynard J. A., Harvill E. T. (2019). “Basics of pertussis pathogenesis,” in Pertussis: Epidemiology, Immunology and Evolution, eds Rohani P., Scarpino S. V. (Oxford: Oxford University Press; ), 26–41. 10.1093/oso/9780198811879.003.0002 [DOI] [Google Scholar]
- Stainer D. W., Scholte M. J. (1970). A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63 211–220. 10.1099/00221287-63-2-211 [DOI] [PubMed] [Google Scholar]
- Stevens M. P., Wood M. W., Taylor L. A., Monaghan P., Hawes P., Jones P. W., et al. (2002). An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46 649–659. 10.1046/j.1365-2958.2002.03190.x [DOI] [PubMed] [Google Scholar]
- Taylor-Mulneix D. L., Bendor L., Linz B., Rivera I., Ryman V. E., Dewan K. K., et al. (2017a). Bordetella bronchiseptica exploits the complex life cycle of Dictyostelium discoideum as an amplifying transmission vector. PLoS Biol. 15:e2000420. 10.1371/journal.pbio.2000420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Mulneix D. L., Hamidou Soumana I., Linz B., Harvill E. T. (2017b). Evolution of bordetellae from environmental microbes to human respiratory pathogens: amoebae as a missing link. Front. Cell Infect. Microbiol. 7:510. 10.3389/fcimb.2017.00510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Heyndrickx M., Vancanneyt M., Hoste B., De Vos P., Falsen E., et al. (1996). Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans ruger and tan 1983. Int. J. Syst. Bacteriol. 46 849–858. 10.1099/00207713-46-4-849 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Hommez J., Vancanneyt M., Monsieurs M., Hoste B., Cookson B., et al. (1995). Bordetella hinzii sp. nov., isolated from poultry and humans. Int. J. Syst. Bacteriol. 45 37–45. 10.1099/00207713-45-1-37 [DOI] [PubMed] [Google Scholar]
- von Wintzingerode F., Schattke A., Siddiqui R. A., Rosick U., Gobel U. B., Gross R. (2001). Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int. J. Syst. Evol. Microbiol. 51(Pt 4), 1257–1265. 10.1099/00207713-51-4-1257 [DOI] [PubMed] [Google Scholar]
- Zimna K., Medina E., Jungnitz H., Guzman C. A. (2001). Role played by the response regulator Ris in Bordetella bronchiseptica resistance to macrophage killing. FEMS Microbiol. Lett. 201 177–180. 10.1111/j.1574-6968.2001.tb10753.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.