Abstract
Polycystic ovary syndrome (PCOS) is a major reproductive disorder that is responsible for 80% of anovulatory infertility and that is associated with hyperandrogenemia, increased risk of obesity, and white adipose tissue (WAT) dysfunction. We have previously demonstrated that the combination of chronic testosterone (T) treatment and an obesogenic Western-style diet (WSD) exerts synergistic functional effects on WAT, leading to increased lipid accumulation in visceral adipocytes by an unknown mechanism. In this study, we examined the whole-genome transcriptional response in visceral WAT to T and WSD, alone and in combination. We observed a synergistic effect of T and WSD on gene expression, resulting in upregulation of lipid storage genes concomitant with adipocyte hypertrophy. Because DNA methylation is known to be associated with body fat distribution and the etiology of PCOS, we conducted whole-genome DNA methylation analysis of visceral WAT. While only a fraction of differentially expressed genes also exhibited differential DNA methylation, in silico analysis showed that differentially methylated regions were enriched in transcription factor binding motifs, suggesting a potential gene regulatory role for these regions. In summary, this study demonstrates that hyperandrogenemia alone does not induce global transcriptional and epigenetic response in young female macaques unless combined with an obesogenic diet.
Subject terms: Metabolic syndrome, DNA
Introduction
Polycystic ovary syndrome (PCOS) is a major reproductive disorder affecting 5–20% of women, depending on the diagnostic criteria employed1. The principal symptoms of PCOS include hyperandrogenemia and infertility. Additionally, women with PCOS exhibit increased rates of obesity and insulin resistance and a higher risk of developing gestational and subsequent type-2 diabetes2–5. However, the role of obesity in PCOS remains unclear. While roughly 50% of PCOS patients are obese, obesity does not appear to be required for the PCOS phenotype, and there is some evidence that the increased rate of obesity in PCOS reflects referral bias6–8. On the other hand, weight loss and insulin-sensitizing drugs are some of the most successful treatments for PCOS-associated infertility9, arguing for a more causative role of metabolic dysfunction in PCOS pathology. Studies in humans and rodent models of PCOS indicate that hyperandrogenemia is associated with the development of white adipose tissue (WAT) dysfunction, which includes increased visceral adiposity10–12 and visceral13–17 and subcutaneous10,18,19 adipocyte hypertrophy. Furthermore, both hyperandrogenemia and obesity have been found to independently reduce fertility through alterations in the hypothalamic-pituitary-ovarian axis20–22. However, whether obesity and hyperandrogenemia exert additive or synergistic effects on metabolic and reproductive functions remain unclear and challenging to study in the human population.
Animal studies have shown that prenatal programing by maternal androgens predisposes offspring to increased adiposity and other metabolic features of PCOS, as demonstrated in nonhuman primate (NHP)23,24, sheep25,26, and rodent27,28 models of PCOS. Consistent with animal models, clinical studies have demonstrated that offspring of women with PCOS develop increased body weight, reflecting the significance of the intrauterine environment and genetic factors in the etiology of PCOS29–31. Furthermore, it has been demonstrated that peripubertal obesity is associated with hyperandrogenemia and hyperinsulinemia in young girls32–34, indicating that adolescence, in the presence of obesogenic factors, is a particularly vulnerable stage for the development of PCOS. However, there is a paucity of animal studies that address the mechanisms of early-life androgen exposure on female metabolism and reproduction.
To better understand the relationship between peripubertal hyperandrogenemia and obesity, we developed a NHP model in which we could test the effects of mild hyperandrogenemia and diet on systemic metabolic and WAT-specific parameters. Prepubertal female rhesus macaques were exposed to either hyperandrogenemia (T), a high-fat, calorie-dense (per mass of diet) Western-style diet (WSD), or a T + WSD combination for 3 years35,36. T + WSD-treated animals were more insulin-resistant, and gained more fat mass over the first 3 years of treatment compared to the control (C), T, and WSD groups (Supplementary Table S1). Furthermore, the T + WSD combination induced greater alterations in WAT function compared to the other groups, including an increase in visceral adipocyte size, reduced basal lipolysis in subcutaneous and visceral (omental) WAT, and increased insulin-stimulated free fatty acid (FFA) uptake in omental WAT36. All experimental animals in this study experienced ovulatory menstrual cycles to some extent; however, mild hyperandrogenemia alone and in combination with a WSD impaired several markers of normal ovarian and uterine function37,38, which ultimately resulted in increased time to pregnancy (T and T + WSD groups), reduced pregnancy rates (WSD and T + WSD groups), and increased early pregnancy loss39. Collectively, these studies suggest a synergistic effect of T and WSD treatment on many metabolic and reproductive outcomes.
While most human studies to date have focused on subcutaneous WAT because of its relative ease of access40–43, women with PCOS display increased intra-abdominal visceral adiposity that is linked to the pathophysiology of insulin resistance and metabolic syndrome10–12. However, the mechanisms driving excess visceral adiposity in women with PCOS and in relevant animal models remain largely unknown. Previous studies suggest that DNA methylation is involved in the regulation of WAT transcriptional profiles, contributing to body fat distribution44 and modulating the pathophysiological response to obesity45. For example, studies of obese women two years after weight loss gastric bypass surgery revealed differential methylation of adipogenic genes, which may contribute to resistance to weight loss46. Furthermore, altered DNA methylation in genes involved in inflammation and glucose and lipid metabolism has been reported in peripheral and umbilical cord blood, as well as in various tissues of women with PCOS47. However, relatively few studies to date have addressed DNA methylation changes in WAT of women with PCOS40,42,43. For example, a recent genome-wide study using subcutaneous WAT of women with PCOS identified a set of genes that displayed both altered DNA methylation and mRNA levels. These genes were associated with pathways involved in inflammation, metabolism and adipogenesis42. However, no studies to date addressed a relationship between DNA methylation, gene expression, and adipocyte function in women with PCOS or in animal models.
To explore the potential mechanisms contributing to the synergistic effects of T and WSD treatment, we examined transcriptional and epigenetic changes in visceral WAT. In this study, we demonstrate that, similar to the effects of T and WSD on systemic metabolic and WAT-specific parameters described in our previous studies35,36, the combination of hyperandrogenemia and WSD induced synergistic effects on both gene expression and DNA methylation in visceral WAT of rhesus macaques. We additionally describe the relationship between differential transcription, DNA methylation, and adipocyte size, and provide in silico analysis, suggesting that T + WSD–induced differential methylation may affect transcription factor (TF) binding sites that are computationally predicted to act as the trans-regulatory elements.
Results
The combination of hyperandrogenemia and WSD elicits a greater transcriptional response than either treatment alone
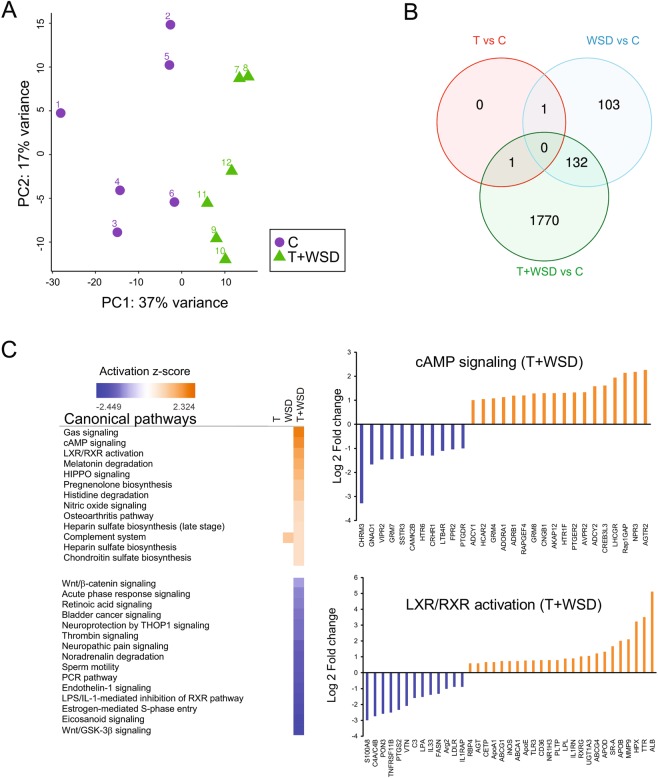
To address the potential molecular mechanisms underlying WAT dysfunction in the presence of hyperandrogenemia and/or WSD, visceral WAT biopsies were collected after a 3-year treatment period from 6 animals from each of the four experimental groups (controls (C), T, WSD, and T + WSD, see methods and Supplementary Table S1) and subjected to RNAseq analysis. Principal component analysis (PCA) showed that global patterns of gene expression were significantly different between the combined treatment (T + WSD) and control groups, with the C and T + WSD groups clearly segregating into distinct areas (Fig. 1A). The other two groups showed less separation from the C group, although they still appeared as separate groups (Supplementary Fig. S1A). To determine the relative contributions of hyperandrogenemia and WSD, we independently compared the three intervention groups to controls and identified differentially expressed genes (DEGs; FDR < 0.05; −1.5 > Fold Change > 1.5). We refer to these comparisons as T, WSD, T + WSD hereafter. As expected, we identified a substantially larger number of DEGs in the T + WSD group (n = 1,903) than in the WSD (n = 236) and T (n = 2) groups (Fig. 1B; Supplementary Tables S2–S4). These findings indicate a synergistic effect of T and WSD on the WAT transcriptome, as most differential gene expression was observed in the combination treatment. One possible limitation of our study is the relatively small sample size that might limit our power to detect smaller effects caused by WSD and T alone. Nevertheless, we did identify biologically relevant DEGs whose regulation was driven exclusively by WSD (i.e., shared between WSD and T + WSD), including GPT (glutamic-pyruvic transaminase) and MOGAT1 (monoacylglycerol O-acyltransferase 1) (Supplementary Tables S2 and S3). Altered expression of these genes has been reported in subcutaneous WAT from PCOS women42. Furthermore, HCAR2 (hydroxycarboxylic acid receptor 2) was significantly upregulated in both T and T + WSD groups, suggesting a WSD-independent effect of hyperandrogenemia (Supplementary Tables S2 and S4). Ingenuity pathway analysis (IPA) of DEGs in the T + WSD group identified activation of the G-coupled receptor Gas/cAMP, LXR/RXR, and HIPPO signaling pathways, whereas the eicosanoid, Wnt signaling, and retinol biosynthesis pathways were downregulated (Fig. 1C and Supplementary Table S5). No significantly differentially regulated pathways were identified in the WSD and T groups.
Figure 1.
Individual and combined effects of T and WSD on gene expression in omental WAT. (A) Principal component analysis (PCA) of gene expression including the 500 most variable genes among the 12 samples (n = 6 samples per each group) showing segregation of controls (C) and T + WSD. (B) Venn diagram showing the overlap between DEGs in the three independent comparisons: T (red), WSD (blue) and T + WSD (green) using a fold change cutoff of 1.5 and an FDR cutoff of 0.05. (C) Pathway analysis using IPA of DEGs in the T + WSD group. IPA assigns an activation score based on biological relevance and the number of genes in the canonical pathway. “Log Ratio” is calculated as Log2 (fold change) in gene expression compared to control. Orange, activated pathways; blue, inhibited pathways; no color, no change in activation state.
Combined T + WSD has a synergistic effect on a subset of genes
In order to identify genes for which the combined T + WSD treatment had a synergistic effect on expression, we looked at the interaction term of testosterone and WSD, utilizing the DESeq. 2 package48. Genes exceeding a significance threshold of an adjusted p-value < 0.1 in the interaction and also showing the same direction of log fold change in expression in the “T vs C” and “T + WSD vs WSD” comparisons were considered to be significantly synergistically regulated. We identified 8 up-regulated genes (CDC42BPA, ERC1, FTL, KLF8, NIN, RYR2, VPS13C, and ZNF589) and 3 down-regulated genes (CCT8, DDT and PHB2) as a result of T + WSD treatment (Table 1, Supplementary Table S6 and Supplementary Fig. S2A). We additionally performed pathway analysis including genes possibly showing a synergistic effect due to treatment, for which the unadjusted p-value was <0.1, in order to identify possible trends. Interestingly, this analysis showed that genes synergistically downregulated were enriched in cholesterol biosynthesis, glycolysis and gluconeogenesis, and SREBP signaling pathways (Supplementary Fig. S2B).
Table 1.
Synergistic regulation of gene expression by T and WSD in omental WAT.
| Gene name | Gene description | Gene ID | Log2FC | p-value |
|---|---|---|---|---|
| Upregulated genes | ||||
| RYR2 | Ryanodine Receptor 2 | ENSMMUG00000001060 | 2.22 | 0.0328 |
| FTL | Ferritin Light Chain | ENSMMUG00000003909 | 0.77 | 0.0246 |
| KLF8 | Kruppel Like Factor 8 | ENSMMUG00000014678 | 0.65 | 0.0925 |
| ZNF589 | Zinc Finger Protein 589 | ENSMMUG00000021607 | 0.61 | 0.0796 |
| NIN | Ninein | ENSMMUG00000014658 | 0.40 | 0.0992 |
| VPS13C | Vacuolar Protein Sorting 13 Homolog C | ENSMMUG00000001362 | 0.33 | 0.0662 |
| CDC42BPA | CDC42 Binding Protein Kinase Alpha | ENSMMUG00000008638 | 0.32 | 0.0942 |
| ERC1 | ELKS/RAB6-Interacting protein | ENSMMUG00000010933 | 0.31 | 0.0992 |
| Downregulated genes | ||||
| DDT | D-Dopachrome Tautomerase | ENSMMUG00000004552 | −1.07 | 0.0891 |
| PHB2 | Prohibitin | ENSMMUG00000010205 | −0.58 | 0.0548 |
| CCT8 | Chaperonin Containing TCP1 Subunit | ENSMMUG00000003023 | −0.37 | 0.0931 |
Genes surpassing a significance threshold of an adjusted p-value < 0.1 in the interaction and also showing the same direction of log fold change in expression in the individual treatment contrasts “T vs C” and “T + WSD vs WSD” were considered significant synergistic genes. Log2Ratio” is calculated as Log2 (fold change) in gene expression.
Correlation between gene expression and visceral adipocyte size
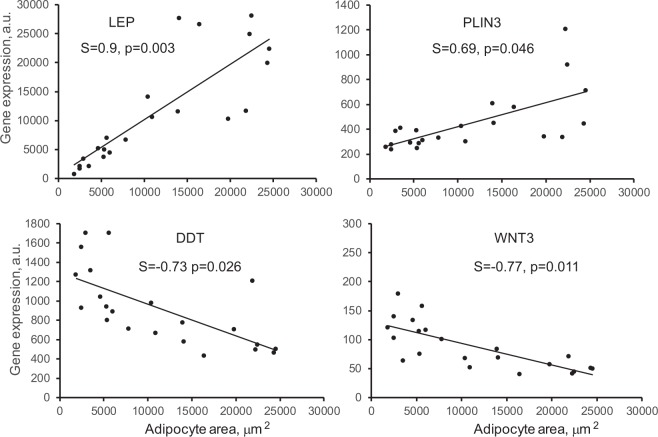
We previously demonstrated that T + WSD treatment leads to an increase in visceral adiposity and omental adipocyte size35,36. We therefore tested if the differential gene expression described in the present study is related to adipocyte size. Using a Spearman correlation analysis, we found that the expression of 235 genes exhibited significant association with adipocyte area (adjusted p < 0.05; Supplementary Table S7). Consistent with a previous report49, the expression of the leptin (LEP) gene correlated positively with omental adipocyte area (Fig. 2). Similarly, the expression of PLIN3 showed a positive association with adipocyte area. In contrast, the expression of DDT and WNT3 genes correlated negatively with adipocyte area (Fig. 2). Interestingly, DDT expression exhibited synergistic regulation by T and WSD (Table 1). Thus, it appears that the expression of a particular subset of genes is significantly associated with visceral adipocyte size.
Figure 2.
Correlation between omental adipocyte area and gene expression. Gene expression is indicated in arbitrary units (A.U.). Linear regression was determined for the combined pool of 24 samples (4 groups). Spearman correlation coefficient (S) and adjusted p-values are indicated.
Exposure to hyperandrogenemia and WSD is associated with global changes in DNA methylation
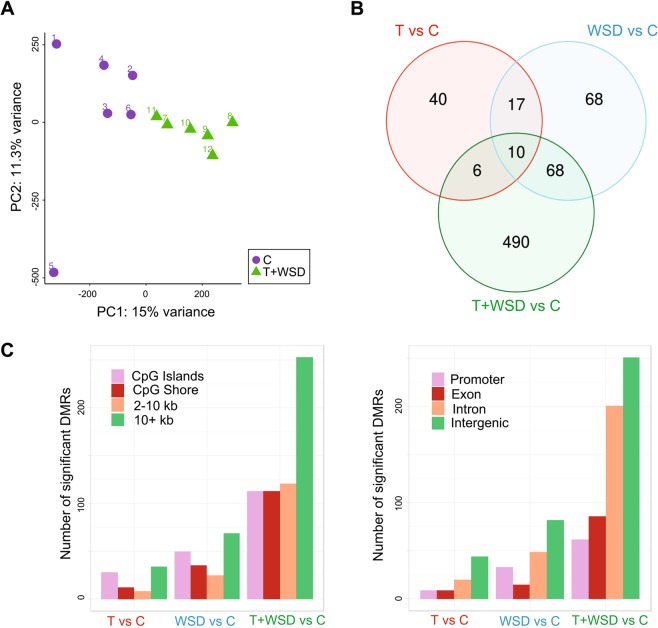
We also examined whether DNA methylation was altered in visceral WAT as a consequence of the various treatments by performing reduced-representation bisulfite sequencing (RRBS), which allows for quantitative, single-base resolution analysis of the portion of the genome enriched in genes and CpG islands50. First, we observed global differences between the T + WSD and C groups as shown by PCA (Fig. 3A). As expected, the combination of hyperandrogenemia and WSD caused more global changes in DNA methylation than either treatment alone (Supplementary Fig. S1B). Indeed, when we identified differentially methylated regions (DMRs) between each treatment group and controls, the highest number of DMRs was found in the T + WSD group (n = 574), followed by the WSD (n = 163) and T (n = 73) groups (Fig. 3B; Supplementary Tables S8–S10). In the T + WSD group, 57% of the DMRs were found to overlap with genes, with a portion of them (17%) located in promoters (Fig. 3C). Pathway analysis using GOrilla51 indicated that genes overlapping with DMRs were enriched for cellular metabolic and cAMP-signaling GO biological processes (Supplementary Table S11), indicating an overlap with pathways enriched in DEGs (Fig. 1C). There was no significant correlation between promoter methylation and adipocyte area (Spearman correlation, adjusted p < 0.05).
Figure 3.
Individual and combined effects of T and WSD on DNA methylation in omental WAT. (A) PCA of DNA methylation for C and T + WSD showing the segregation of these two groups. (B) Venn diagram representing the overlap between DMRs (10% methylation difference cutoff and an adjusted p-value cutoff of 0.05) using the same independent comparisons and color coding as for the RNAseq [T (red), WSD (blue) and T + WSD (green)]. (C) Number of significant DMRs in various genomic features (left) and genomic regions (right).
Limited correlation between differential gene expression and DNA methylation
Increased gene expression has been traditionally associated with promoter hypomethylation and gene body hypermethylation52,53. Since we observed both global differential expression and methylation, we sought to explore the relationship between these effects. We first investigated the global relationship between gene expression and promoter methylation in our study groups. To do this, we considered genes from the differential gene-expression analysis that also possessed at least one CpG with a minimum of 10X coverage in the promoter region (i.e., 3 kbp upstream from the transcription start site (TSS)). This resulted in about 11,000 genes for which we could define both an expression value and an average promoter methylation percent value. We observed a trend for an inverse correlation between gene expression and promoter methylation within each sample group (Spearman correlation value of approximately −0.2) (Supplementary Fig. S3).
We then identified genes that were both differentially expressed and differentially methylated in response to the different treatments. Specifically, we identified 63 DEGs in the T + WSD group that also overlapped or were within 5 kb of a DMR. Of these DEGs, 12 showed differential methylation at the promoter region and 9/12 displayed an inversed correlation between methylation and expression (i.e., hypomethylation with increased expression and hypermethylation with decreased expression, Table 2). Hypermethylation in gene bodies has traditionally been associated with activation of transcription. We observed that 26 of the 54 DMRs found in gene bodies were hypermethylated and 13 of those were indeed associated with increased gene expression, while 28 genes showed a “promoter-like” inverse correlation between methylation and expression (i.e., hypomethylated DMRs and increased expression, or vice versa, Table 2). The latter relationship has been reported to correspond to alternative promoters or active enhancer regions53. Using the LiftOver tool from the UCSC browser, we converted these 54 DMRs from the rhesus to the human genome (hg38), for which functional annotations are available through the ENCODE project54. We observed that DMRs in up-regulated genes corresponded more often than expected by chance to DNase I hypersensitive sites in the human genome (p-value 0.0115, Fisher’s test, two tails), suggesting that these regions might behave as transcription factor (TF) binding sites.
Table 2.
Correlations between gene expression and DNA methylation in omental WAT.
| T + WSD vs C | |||||
|---|---|---|---|---|---|
| Symbol | Region | Methyl1 | Methyl2 | Log2FC | Correlation |
| C21orf58 | P, I | −23.4 | 2.5 | Inverse | |
| MATN3 | P | −18.5 | 1.2 | Inverse | |
| CD93 | P, E | −18.6 | −17.1 | 1.0 | Inverse |
| CTXN1 | P, E | 20.6 | −1.5 | Inverse | |
| SMTNL2 | I, P | 19.6 | 32.1 | −2.0 | Inverse |
| RNF227 | P | 26.9 | −2.2 | Inverse | |
| TRIM29 | P | 19.4 | −2.6 | Inverse | |
| MISP | P | 20.3 | −3.0 | Inverse | |
| MAB21L2 | P, E | 21.1 | −6.5 | Inverse | |
| GATA5 | P | −12.8 | −1.3 | Direct | |
| SHF | P | −12.8 | −1.7 | Direct | |
| SLC6A3 | P | −14.4 | −2.0 | Direct | |
| GUCY2D | E, I | −17.7 | 1.8 | Inverse | |
| CCDC3 | I | −12.9 | 1.8 | Inverse | |
| MYH7B | I | −19.4 | 1.5 | Inverse | |
| ADCYAP1R1 | I | −16.5 | 1.4 | Inverse | |
| GALNT17 | I | −31.4 | 1.3 | Inverse | |
| EPAS1 | I | −18.6 | −16.1 | 1.0 | Inverse |
| PMEPA1 | I | −20.3 | 1.0 | Inverse | |
| APLNR | E | −17.1 | 1.0 | Inverse | |
| KIF21B | I | −14.2 | 1.0 | Inverse | |
| ADGRG1 | I | −16.1 | 1.0 | Inverse | |
| PECAM1 | I | −22.2 | 0.9 | Inverse | |
| NDST1 | I | −26.4 | 0.9 | Inverse | |
| HSPA12A | I | −19 | 0.9 | Inverse | |
| MYO9B | I | −13.2 | 0.7 | Inverse | |
| PTPRG | I | −21.3 | 0.6 | Inverse | |
| RASA3 | I | −19.2 | 0.6 | Inverse | |
| TSPAN14 | I | −23 | 0.6 | Inverse | |
| EFNA5 | I | 24.3 | 24.4 | −0.7 | Inverse |
| BAIAP2 | I | 17.9 | 18.7 | −0.7 | Inverse |
| MST1R | E, I | 18.3 | −0.7 | Inverse | |
| MCU | I | 34.3 | −0.9 | Inverse | |
| SLC9A3R1 | I | 25.2 | −1.2 | Inverse | |
| GATA6 | E, I | 23.9 | −1.3 | Inverse | |
| SSTR3 | I | 20.7 | −1.4 | Inverse | |
| C3 | I | 26.4 | −1.6 | Inverse | |
| PDZK1IP1 | E, I | 26 | −2 | Inverse | |
| LMO7 | I | 29.7 | −2.5 | Inverse | |
| KRT8 | I | 14.5 | −2.8 | Inverse | |
| IL4I1 | I | 24.9 | 2.3 | Direct | |
| ITGAX | E, I | 16.2 | 1.8 | Direct | |
| NAV1 | E, I | 18.5 | 11.9 | 1.1 | Direct |
| NEK6 | I | 10.3 | 1.0 | Direct | |
| STAB1 | I | 14.2 | 1.0 | Direct | |
| RPH3AL | I | 15.5 | 24.4 | 1.0 | Direct |
| NOS3 | E | 19.1 | 0.9 | Direct | |
| MAP7D1 | E, I | 13.5 | 0.7 | Direct | |
| GSE1 | I | 22 | 0.7 | Direct | |
| CAMKK2 | I | 18.7 | 0.7 | Direct | |
| HTRA1 | E, I | 19 | 0.6 | Direct | |
| FAM178B | I | −17.7 | −1.1 | Direct | |
| ROR2 | I | −18 | −1.2 | Direct | |
| MKX | E, I | −13.1 | −1.3 | Direct | |
| EBF4 | E, I | −11 | −1.4 | Direct | |
| GRM7 | I | −16.7 | −1.5 | Direct | |
| VIPR2 | I | −15.1 | −1.5 | Direct | |
| TNK1 | E, I | −23.6 | −2.0 | Direct | |
| WNT7B | I | −13.3 | −2 | Direct | |
| S1PR5 | E | −21.3 | −2.3 | Direct | |
| GATA4 | I | −12.5 | −2.7 | Direct | |
| WSD vs C | |||||
| HOXA10 | I | 19 | −4.0 | Inverse | |
| CCDC3 | I | −14.5 | 1.4 | Inverse | |
Genes with inverse or direct correlation between expression levels and DNA methylation changes in indicated genome regions. P, promoter; E, exon; I, intron; Methyl, differentially methylated region (DMR); Log2Ratio” is calculated as Log2 (fold change) in gene expression in “T + WSD” or “WSD” groups compared to the control (“C”). The DMRs are more than 1 bp in length and it is possible for them to overlap more than 1 gene region. For example, the DMR might span an exon/intron junction, or a promoter/1st exon junction.
While no association between differential methylation and gene expression was apparent for DEGs and DMRs from the T group, we identified two genes (HOXA1 and CCDC3) that showed an inverse correlation between expression and methylation in the WSD group, with both DMRs being intronic (Table 2). In conclusion, although we see a large global effect of T + WSD treatment on both DNA methylation and gene expression, only a portion of differentially expressed genes also exhibited differential DNA methylation.
In silico analysis of interactions between transcription factors and DMRs
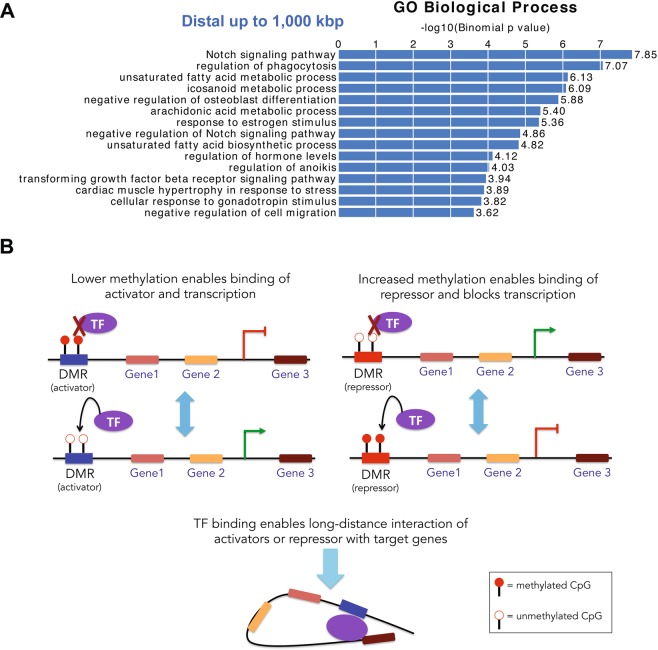
Studies show that interactions between TFs and promoter regions can occur over long genomic distances through 3D chromatin structural rearrangement55. Given the lack of a strong correlation between differential methylation and differential gene expression reported in the present study, we hypothesized that a portion of DMRs can bind TFs and act as distal regulators of gene expression. To test this hypothesis, we employed in silico analysis using the Genomic Regions Enrichment of Annotations Tool (GREAT) that associates each gene with a ‘regulatory domain’ defined as 5 kb upstream and 1 kb downstream from the TSS and an extension within 1 Mb up to the regulatory domain of the nearest upstream or downstream genes56. While no pathways were enriched in the T and WSD groups, the analysis identified several pathways of interest in the T + WSD group, including those regulating notch signaling, unsaturated fatty acid metabolism, and estrogen response (Fig. 4A). However, when we excluded long-distance regulatory domains (i.e., 1 Mbp) from the analysis, these pathways were no longer detected, suggesting that these biologically relevant processes are enriched for DMRs that act as distal regulators (Supplementary Fig. S4).
Figure 4.
DMRs distally located with respect to genes are enriched in biologically relevant pathways. (A) Pathways analysis using the Genomic Regions Enrichment of Annotations Tool (GREAT) that associates each gene with a ‘regulatory domain’ defined as a genomic region 5 kb upstream and 1 kb downstream from the TSS and an extension within 1 Mb up to the regulatory domain of the nearest upstream or downstream gene. (B) Schematic model depicting methylation-dependent modulation of TF binding to distal regulatory domains. Left diagram, hypermethylation of an activator inhibits, while hypomethylation of an activator facilitates TF binding, resulting in transcriptional repression or activation, respectively. Right diagram, particular TFs bind hypermethylated repressors, leading to transcriptional repression, while hypomethylation has the opposite effect on transcription. Bottom diagram, TF binding to a regulatory domain enables long-distance interactions with the target genes through 3D chromatin structural rearrangement.
To identify potential TF binding motifs involved in distal regulation, we performed in silico motif enrichment analysis using the Hypergeometric Optimization of Motif Enrichment (HOMER) bioinformatic tool57. We identified several significantly enriched motifs in the DMRs from the T + WSD group (Supplementary Table S12), while no significant enrichment was found for the T and WSD groups. Among the significantly enriched motifs identified in the present study, one interesting example is the Recombination Signal Binding Protein for Immunoglobulin Kappa J (RBPJ, Supplementary Table S12). Using BLASTn, we determined that the nucleotide sequence of the rhesus macaque RBPJ gene is 95% identical to human. Furthermore, BLASTp analysis showed that human and rhesus RBPJ proteins are 99% identical. This provides strong support that our RBPJ motif enrichment translates to our rhesus model. Using EnrichR58, we also determined that the promoters of DEGs in the T + WSD group were significantly enriched for the RBPJ binding motif, suggesting that the differential binding of this TF may regulate gene expression. Thus, this computational analysis suggests that DMRs may act as distal regulators through binding of biologically relevant TFs and that changes in methylation due to treatment (T + WSD) may modulate the binding of TFs, ultimately impacting gene expression (Fig. 4B).
Discussion
One of the many challenges that hinders our understanding of the pathophysiology of PCOS is dissecting out the relative contribution of hyperandrogenemia and diet in the etiology of this multifactorial disease. The role of hyperandrogenemia and diet-induced obesity as the potential co-drivers of metabolic, adipose-specific35,36, and reproductive38,39 phenotypes has been demonstrated in our recent NHP studies. These studies established that the combination of hyperandrogenemia and WSD induces greater metabolic (obesity and insulin resistance) and adipose-specific (visceral adipocyte hypertrophy, increased lipid storage, and reduced lipolysis) dysfunction than either treatment alone35,36. These and other findings32–34 raise the intriguing possibility that hyperandrogenemia alone does not induce weight gain and metabolic dysfunction in young females unless combined with an obesogenic diet.
Our previous NHP studies demonstrated that the combination of hyperandrogenemia and WSD induces intra-abdominal visceral adiposity, as evidenced by the significant enlargement of omental adipocytes36. An increase in visceral adiposity has previously been linked to the pathophysiology of insulin resistance in PCOS patients10–12, while the mechanisms remain poorly understood. One possibility is that the combination of hyperandrogenemia and WSD induces the masculinization of fat depots in females, leading to increased lipid storage in visceral WAT and decreased lipid storage in subcutaneous WAT59. The latter plays a positive role in metabolism, while the redirection of lipid stores from the subcutaneous to the visceral WAT depot is associated with metabolic syndrome60. Mechanistically, hyperandrogenemia in the presence of WSD increases insulin-stimulated FFA uptake and inhibits lipolysis in omental WAT, favoring the development of intra-abdominal obesity in NHPs36,59,61, while the mechanism remains poorly understood.
The present study was designed to address the potential mechanisms regulating visceral adiposity in the NHP model. We first showed that T + WSD induces a greater transcriptional response in omental WAT than either treatment alone. Interestingly, several of the DEGs that we detected in rhesus visceral WAT, including DKK2, SVEP1, NRCAM, GPT, and DMAP1, were previously reported to be differentially expressed in subcutaneous WAT of PCOS women42, confirming that our model is recapitulating some of the transcriptional changes observed in this disease, albeit in different depots. In addition, we observed that T + WSD treatment induced the upregulation of several lipid metabolism genes, including the fatty acid transporter CD36, fatty acid binding protein 4 (FABP4), triglyceride synthesis enzyme MOGAT1, and lipid droplet protein perilipin 3 (PLIN3), which collectively have a positive effect on FFA uptake and triglyceride storage in WAT. Importantly, the expression of the MOGAT1 and CD36 genes in omental WAT and circulating levels of FATP4 have been shown to be elevated in PCOS women42,62,63. In agreement with our RNAseq results, the expression of the fatty acid transporter CD36 has been shown to correlate with omental adipocyte size in women with PCOS63. Furthermore, our previous studies have demonstrated that T + WSD treatment enhances FFA uptake in omental WAT of NHPs36,61. We also observed that genes encoding the components of the LXR/RXR pathway were upregulated in T + WSD, suggesting potential changes in lipoprotein metabolism. This is consistent with a study of WAT in women with PCOS40. The increased expression of genes that regulate HDL metabolism and reverse cholesterol transport (e.g., ABCA1, ABCG1, ApoA1, APOD, CETP, PLTP, PON3, and SR-A) is compatible with other well-known effects of LXR activation, including cellular cholesterol accumulation64–66.
Our data indicate that hyperandrogenemia may attenuate cAMP-dependent lipid catabolism. The breakdown of cellular triglycerides depends on the activation of the cAMP-dependent hormone sensitive lipase (HSL), the principal lipase responsible for the β-adrenergic lipolytic response in WAT67. The HSL gene (LIPIE) has been shown to be significantly downregulated in omental WAT from T + WSD-treated NHPs61 and PCOS women63. In line with these observations, we showed that hyperandrogenemia induces lipolytic resistance in omental and subcutaneous WAT of NHPs36,61. In contrast, omental WAT from PCOS women exhibits increased catecholamine-induced lipolysis68, while subcutaneous WAT exhibits decreased catecholamine-induced lipolysis and the reduced expression of the β2-adrenergic receptor18, the regulatory II β-subunit of protein kinase A, and HSL19. Although the present study did not detect differential expression of the LIPE, PRKCA (catalytic subunit of protein kinase A), and ADRB2 (β2-adrenergic receptor) genes, several isoforms of phosphodiesterase (PDE) that mediate cAMP degradation were significantly upregulated in the T + WSD group. This finding agrees with an earlier report showing the upregulation of several PDE genes in omental WAT of PCOS women49. Furthermore, clinical studies demonstrated that the PDE4 inhibitor roflumilast added to metformin reduced fat mass in obese women with PCOS69, suggesting that hyperandrogenemia may lead to decreased cAMP levels in WAT.
There is growing evidence that body fat distribution is dynamically regulated at the transcriptional and DNA methylation levels44–46,70–74. For example, obesity induces DNA hypermethylation in the promoter of the adiponectin gene, resulting in its reduced expression and the development of insulin resistance in mice72. Similarly, DNA methylation of the perilipin-1 promoter is higher and gene expression is lower in WAT of obese subjects70. Thus, we sought to test whether hyperandrogenemia and WSD regulate the transcriptional response in WAT through the DNA methylation mechanisms. Using a genome-wide approach, we show that all treatments elicit global changes in DNA methylation, with T + WSD causing the largest effect. Although we observed a trend of global inverse correlation between methylation and transcription, only 63 genes exhibited both significant differential expression and DNA methylation. Of note, in a genome-wide study of gene expression and DNA methylation in subcutaneous WAT of PCOS women, 33 DEGs that also displayed differential DNA methylation were identified42, although none of these 33 genes were found to overlap DMRs in our study. A limited association between gene expression and DNA methylation is not entirely surprising, as the relationship between DNA methylation and gene expression is more complex than originally thought53. For instance, the functional role of DNA methylation in gene bodies is still elusive53. It is also possible that DNA methylation is regulated by WAT depot-specific mechanisms. In the original study, Xu and colleagues reported that prenatally androgenized rhesus macaques display altered genome-wide DNA methylation in visceral WAT75. In spite of significant differences in methodology and dietary regiments used by their and our groups, both studies identified 3 common DMRs associated with the CCDC3, GRM7, and NEK6 genes. Importantly, the present study showed that these genes are also differentially expressed in rhesus WAT. CCDC3 has been previously shown to be upregulated in omental WAT of obese subjects and exerts adipogenic effects in vitro and in vivo76,77. In the present study, CCDC3 was found to be upregulated and differentially methylated at the same intronic region by either WSD or T + WSD treatment, suggesting a diet-specific mechanism of gene regulation. Based on our GREAT pathway analysis, we hypothesized that a portion of the DMRs may be acting as distal regulators, rather than acting on the closest gene. Indeed, we observed that DMRs are not enriched in biologically relevant pathways, like the Notch signaling pathway, when long-distance relationships (1 Mb) were excluded in looking at cis-interactions between DMRs and genes. Moreover, our computational analysis showed that DMRs from the T + WSD group were enriched in TF binding motifs, supporting the regulatory role of DNA methylation in modulating TF binding affinities78. As an example, RBPJ has been shown to control the expression of notch signaling genes79 and to bind chromatin in a methylation-dependent fashion80,81.
Our study does have limitations. First, we did not evaluate transcriptional and epigenetic responses in isolated adipocyte vs stromal-vascular cell populations due to insufficient sample size for separation of individual cell types. However, previous analyses of human WAT also employed total tissue samples; thus, comparison of our NHP-derived data from whole WAT to these data is appropriate. Second, we did not examine transcriptional and epigenetic changes in subcutaneous WAT because this depot is insufficiently developed in young macaques to obtain enough tissue. Third, the number of animals in each group (n = 6) was relatively small. However, given a unique nature of this NHP model, the present study provides valuable translational information on the interactions between obesity and hyperandrogenemia in PCOS. Fourth, other epigenetic modifications (e.g., histone modifications and micro-RNAs82,83) may also contribute to WSD and T effects and need to be studied in order to fully understand the mechanisms underlying transcriptional changes observed in the present report.
In conclusion, this study demonstrates that the combination of hyperandrogenemia and WSD induces a synergistic transcriptional response associated with an increase in lipid anabolic capacity in the visceral WAT of NHPs36,61, similar to findings reported in women with PCOS10–12. This and other PCOS-related studies40,42 demonstrate a limited association between WAT gene expression and DNA methylation. Furthermore, the majority of genes showed no significant association with adipocyte size based on mRNA or promoter methylation levels, suggesting that hyperandrogenemia but not adipocyte hypertrophy per se is the primary etiological factor driving the transcriptional and epigenetic response in WAT. Thus, the intake of excess dietary lipids in combination with hyperandrogenemia may facilitate lipid accumulation and the development of visceral adiposity. Collectively, the results of this study support the need for future clinical trials aimed at early dietary interventions to prevent weight gain in young women with PCOS, and in developing strategies aimed at maintaining weight loss in obese women with PCOS.
Methods
Animal model
All animal procedures were approved by the Oregon National Primate Research Center (ONPRC) Institutional Animal Care and Use Committee and comply with the Animal Welfare Act and the APA Guidelines for Ethical Conduct in the Care and Use of Nonhuman Animals in Research. Animal characteristics and the origin of WAT samples used in the present study have been previously described35,36. Briefly, female rhesus macaques were selected for the study and randomly assigned to one of four treatment groups (C, T, WSD and T + WSD). T-releasing capsules were prepared and implanted subcutaneously as previously described35. Animals were maintained ad libitum on either chow diet consisting of two daily meals of Fiber-balanced Monkey Diet (15% calories from fat, 27% from protein, and 59% from carbohydrates; no. 5052; Lab Diet, St. Louis, MO), supplemented with fruits and vegetables, or a WSD, containing 36% calories from fat, 18% from protein, 45% from carbohydrates (TAD Primate Diet 5LOP, 5A1F, Lab Diet). Animals had undergone 3 years of continuous treatment and were approximately 5.5 years of age and were post-pubertal at the time of WAT biopsy collected as previously described36. To perform gene expression and DNA methylation analyses, we selected 24 animals (6 representative animals from each group [C, T, WSD, and T + WSD]) that reflected the overall group findings36; i.e., increased fat mass and hyperinsulinemia at 3 years of treatment. Supplementary Table S1 provides these metabolic characteristics previously published, but for the specific subset of animals (n = 6/group) utilized in the current study.
RNA-seq libraries
Adipose biopsy procedures were previously described36. DNA and RNA were extracted from 200 mg of frozen visceral (omental) WAT homogenized using a TissueLyzer-II (QIAGEN, Hilden, Germany) with the AllPrep DNA/RNA purification kit (QIAGEN). High-quality RNA samples (RIN > 8) were used for library construction using the TruSeq Rybo-zero method and were sequenced by the Massively Parallel Sequencing Shared Resource (MPSSR) at OHSU using the Illumina HiSeq. 2500 platform, with the 100-bp, single-read protocol. RNA-seq summary statistics is shown in Supplementary Table S13.
Differential expression and synergy analysis
Sequencing reads were evaluated with FastQC (v0.11.5)84 and trimmed to remove adapters and low-quality regions using Trimmomatic (v0.36)85. After trimming, reads were aligned on the most current rhesus macaque genome assembly (rheMac8) using STAR86 with default parameters. DESeq. 2 (v1.18.1)48 was used for differential expression analysis of the count data. Pairwise comparisons were performed among the sample groups and Benjamini-Hochberg adjustment was used to adjust the p-values for multiple comparisons when performing statistical tests on thousands of genes. The lists of differentially expressed genes (FDR < 0.05; −1.5 > Fold Change > 1.5) are reported in Supplementary Tables S2–S4. Differentially expressed genes were analyzed with Ingenuity Pathway Analysis (IPA) to identify pathways enrichment. In order to find genes for which the combined T + WSD treatment might have had a synergistic effect on gene expression, we fit a 2 × 2 factorial design and looked for genes showing significance in the interaction term of T and WSD utilizing the DESeq. 2 package. This allowed us to determine genes that had a change in expression due to T treatment that was also dependent on diet condition. Genes surpassing a significance threshold of adjusted p-value < 0.1 in the interaction and also showing the same direction of log fold change in expression in the individual treatment contrasts “T vs C” and “T + WSD vs WSD” were considered significant synergistic genes. We additionally identified genes possibly showing a synergistic effect due to treatment for which the unadjusted p-value was less than 0.1 (Supplementary Table S6). To test the association between gene expression and adipocyte area, we applied a Spearman correlation analysis using ~19,000 genes which passed low count filtering to be included in our differential expression analysis pipeline.
Reduced-representation bisulfite sequencing (RRBS)
RRBS libraries were generated from ~200 ng of WAT genomic DNA following an established protocol50. Briefly, overnight digestion was performed with MspI (New England Biolabs, Ipswich, MA), which cuts the sequence CCGG and generates sticky ends, enabling every read to start with a CpG. Libraries were prepared with the NEXTflex Bisulfite-Seq Kit (Bioo Scientific Corporation, Austin, TX) and the NEBNext Methylated Adaptors (New England Biolabs). The ligated DNA was size-selected using AMPure XP magnetic beads to produce a final library size of ~350 bp. Bisulfite conversion was performed with the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA) before carrying out PCR amplification with NEBNext Multiplex Oligos (New England Biolabs) to barcode each library. The resulting libraries were normalized and multiplexed for sequencing on the Illumina NextSeq. 500 with the high-output, 75-bp cycle protocol. Sequencing reads were evaluated with FastQC84 and trimmed to remove adapters and low-quality regions with Trim Galore (v0.4.2)87 using the “RRBS” parameter. Trimmed reads were aligned to rheMac8 with Bismark (v0.16.1)88, the most widely used software for mapping bisulfite converted sequences. Summary statistics for the RRBS datasets are shown in Supplementary Table S14.
Differential methylation analysis
In order to identify differentially methylated cytosines (DMCs) in a CpG context and differentially methylated regions (DMRs) we used a multi-step approach. First, Limma (v3.34.9)89 was used to perform DMC analysis, as Limma allows modeling of more complex designs. For DMC analysis, we included CpGs with at least 10X coverage in at least 4 of the 6 replicates per group, resulting in 841,143 CpG sites. For input values, we performed an arcsine transformation on the methylation rate per CpG. Then, we used the p-values obtained from the DMC analysis as input to Comb-p90 in order to find DMRs. DMR regions are found by seeding on CpGs with corrected p-values < 0.05 and extending the region as long as it finds another CpG with a corrected p-value < 0.05 within 300 bp. All DMRs (Sidak p-value < 0.1; methylation difference > 10%) and their annotations are listed in Supplementary Tables 8–10. As many DMRs might not overlap with genes or their promoters (we annotated them as “inter-genic”) but might correspond to distal regulatory elements, we also annotated the closest transcription start site (TSS) for each inter-genic DMR. In order to identify pathways enriched in DMRs, we used the publicly available tools GREAT (http://great.stanford.edu/public/html/)56 and GOrilla (http://cbl-gorilla.cs.technion.ac.il/)51. Finally, motif enrichment was performed on the significant DMRs from the three main comparisons using HOMER (Hypergeometric Optimization of Motif EnRichment)57, specifying the use of the given size of the regions and normalizing for CpG content against the random background. Although the motif databases used by HOMER might be skewed towards human and mouse, TF binding motifs are highly conserved and therefore interchangeable between mammals and vertebrates in general57.
Supplementary information
Acknowledgements
This study was supported by NIH grants P50 HD071836 to CTR and P51 OD01192 for operation of the Oregon National Primate Research Center. Illumina sequencing was performed by the OHSU Massively Parallel Sequencing Shared Resource. RRBS library generation and data analysis was performed by the OHSU Knight Cardiovascular Institute Epigenetics consortium. We thank the OHSU ExaCloud Cluster Computational Resource and the Advanced Computing Center for performance of intensive large-scale data workflows. We also acknowledge the support of the ONPRC Bioinformatics & Biostatistics Core for help with IPA analysis.
Author contributions
L.C. designed the study (genomics), performed some of the bioinformatics analyses, and wrote the manuscript; B.A.D. analyzed the genomics data and wrote the manuscript; S.S.F. analyzed the pathway analysis data; A.W. performed tissue extraction and isolated nucleic acids; K.A.N. conducted DNA methylation experiments; D.T. coordinated sample collection; A.V. analyzed the data and edited the manuscript; C.T. analyzed the data and edited the manuscript; C.T.R. designed the study and wrote the manuscript; O.V. designed the study, collected samples, performed the statistical analyses, and wrote the manuscript.
Data availability
Gene expression and DNA methylation data are available at the NCBI Gene Expression Omnibus data repository under Accession Number GSE124709.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lucia Carbone and Brett A. Davis.
Supplementary information
is available for this paper at 10.1038/s41598-019-55291-8.
References
- 1.Azziz R, et al. Polycystic ovary syndrome. Nat Rev Dis Primers. 2016;2:16057. doi: 10.1038/nrdp.2016.57. [DOI] [PubMed] [Google Scholar]
- 2.Bellamy L, Casas JP, Hingorani AD, Williams D. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet. 2009;373:1773–1779. doi: 10.1016/S0140-6736(09)60731-5. [DOI] [PubMed] [Google Scholar]
- 3.Cefalu WT, et al. Diabetes Care. 2015. Advances in the Science, Treatment, and Prevention of the Disease of Obesity: Reflections From a Diabetes Care Editors’ Expert Forum; pp. 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao K, Bian C, Zhao X. Association of polycystic ovary syndrome with metabolic syndrome and gestational diabetes: Aggravated complication of pregnancy. Exp Ther Med. 2017;14:1271–1276. doi: 10.3892/etm.2017.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin KH, Glintborg D, Nybo M, Abrahamsen B, Andersen M. Development and risk factors of type 2 diabetes in a nationwide population of women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2017 doi: 10.1210/jc.2017-01354. [DOI] [PubMed] [Google Scholar]
- 6.Pasquali R, Pelusi C, Genghini S, Cacciari M, Gambineri A. Obesity and reproductive disorders in women. Hum Reprod Update. 2003;9:359–372. doi: 10.1093/humupd/dmg024. [DOI] [PubMed] [Google Scholar]
- 7.Ezeh U, et al. Effects of endogenous androgens and abdominal fat distribution on the interrelationship between insulin and non-insulin-mediated glucose uptake in females. J Clin Endocrinol Metab. 2013;98:1541–1548. doi: 10.1210/jc.2012-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lizneva D, et al. Phenotypes and body mass in women with polycystic ovary syndrome identified in referral versus unselected populations: systematic review and meta-analysis. Fertil Steril. 2016;106:1510–1520 e1512. doi: 10.1016/j.fertnstert.2016.07.1121. [DOI] [PubMed] [Google Scholar]
- 9.Tomic, V. & Tomic, J. Infertility Treatments in Patients with Polycystic Ovary Syndrome (PCOS). J Fertiliz In Vitro2, 10.4172/2165-7491.1000e113 (2012).
- 10.Manneras-Holm L, et al. Adipose tissue has aberrant morphology and function in PCOS: enlarged adipocytes and low serum adiponectin, but not circulating sex steroids, are strongly associated with insulin resistance. J Clin Endocrinol Metab. 2011;96:E304–311. doi: 10.1210/jc.2010-1290. [DOI] [PubMed] [Google Scholar]
- 11.Gourgari E, et al. Lipoprotein Particles in Adolescents and Young Women With PCOS Provide Insights Into Their Cardiovascular Risk. J Clin Endocrinol Metab. 2015;100:4291–4298. doi: 10.1210/jc.2015-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumesic DA, et al. Hyperandrogenism Accompanies Increased Intra-Abdominal Fat Storage in Normal Weight Polycystic Ovary Syndrome Women. J Clin Endocrinol Metab. 2016;101:4178–4188. doi: 10.1210/jc.2016-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manneras L, Jonsdottir IH, Holmang A, Lonn M, Stener-Victorin E. Low-frequency electro-acupuncture and physical exercise improve metabolic disturbances and modulate gene expression in adipose tissue in rats with dihydrotestosterone-induced polycystic ovary syndrome. Endocrinology. 2008;149:3559–3568. doi: 10.1210/en.2008-0053. [DOI] [PubMed] [Google Scholar]
- 14.Nohara K, et al. Developmental androgen excess programs sympathetic tone and adipose tissue dysfunction and predisposes to a cardiometabolic syndrome in female mice. Am J Physiol Endocrinol Metab. 2013;304:E1321–1330. doi: 10.1152/ajpendo.00620.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caldwell AS, et al. Characterization of reproductive, metabolic, and endocrine features of polycystic ovary syndrome in female hyperandrogenic mouse models. Endocrinology. 2014;155:3146–3159. doi: 10.1210/en.2014-1196. [DOI] [PubMed] [Google Scholar]
- 16.Kauffman AS, et al. A Novel Letrozole Model Recapitulates Both the Reproductive and Metabolic Phenotypes of Polycystic Ovary Syndrome in Female Mice. Biol Reprod. 2015;93:69. doi: 10.1095/biolreprod.115.131631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolic M, et al. Possible involvement of glucocorticoids in 5alpha-dihydrotestosterone-induced PCOS-like metabolic disturbances in the rat visceral adipose tissue. Mol Cell Endocrinol. 2015;399:22–31. doi: 10.1016/j.mce.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Ek I, Arner P, Bergqvist A, Carlstrom K, Wahrenberg H. Impaired adipocyte lipolysis in nonobese women with the polycystic ovary syndrome: a possible link to insulin resistance? J Clin Endocrinol Metab. 1997;82:1147–1153. doi: 10.1210/jcem.82.4.3899. [DOI] [PubMed] [Google Scholar]
- 19.Faulds G, Ryden M, Ek I, Wahrenberg H, Arner P. Mechanisms behind lipolytic catecholamine resistance of subcutaneous fat cells in the polycystic ovarian syndrome. J Clin Endocrinol Metab. 2003;88:2269–2273. doi: 10.1210/jc.2002-021573. [DOI] [PubMed] [Google Scholar]
- 20.McGee WK, et al. Elevated androgens during puberty in female rhesus monkeys lead to increased neuronal drive to the reproductive axis: a possible component of polycystic ovary syndrome. Hum Reprod. 2012;27:531–540. doi: 10.1093/humrep/der393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grulet H, et al. Roles of LH and insulin resistance in lean and obese polycystic ovary syndrome. Clin Endocrinol (Oxf) 1993;38:621–626. doi: 10.1111/j.1365-2265.1993.tb02144.x. [DOI] [PubMed] [Google Scholar]
- 22.Brewer CJ, Balen AH. The adverse effects of obesity on conception and implantation. Reproduction. 2010;140:347–364. doi: 10.1530/REP-09-0568. [DOI] [PubMed] [Google Scholar]
- 23.Eisner JR, Dumesic DA, Kemnitz JW, Colman RJ, Abbott DH. Increased adiposity in female rhesus monkeys exposed to androgen excess during early gestation. Obes Res. 2003;11:279–286. doi: 10.1038/oby.2003.42. [DOI] [PubMed] [Google Scholar]
- 24.Abbott DH, Dumesic DA, Eisner JR, Colman RJ, Kemnitz JW. Insights into the development of polycystic ovary syndrome (PCOS) from studies of prenatally androgenized female rhesus monkeys. Trends Endocrinol Metab. 1998;9:62–67. doi: 10.1016/S1043-2760(98)00019-8. [DOI] [PubMed] [Google Scholar]
- 25.Cardoso RC, Puttabyatappa M, Padmanabhan V. Steroidogenic versus Metabolic Programming of Reproductive Neuroendocrine, Ovarian and Metabolic Dysfunctions. Neuroendocrinology. 2015;102:226–237. doi: 10.1159/000381830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padmanabhan V, Veiga-Lopez A. Sheep models of polycystic ovary syndrome phenotype. Mol Cell Endocrinol. 2013;373:8–20. doi: 10.1016/j.mce.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maliqueo M, Benrick A, Stener-Victorin E. Rodent models of polycystic ovary syndrome: phenotypic presentation, pathophysiology, and the effects of different interventions. Semin Reprod Med. 2014;32:183–193. doi: 10.1055/s-0034-1371090. [DOI] [PubMed] [Google Scholar]
- 28.Roland AV, Nunemaker CS, Keller SR, Moenter SM. Prenatal androgen exposure programs metabolic dysfunction in female mice. J Endocrinol. 2010;207:213–223. doi: 10.1677/JOE-10-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang G, et al. Sex Differences in the Prenatal Programming of Adult Metabolic Syndrome by Maternal Androgens. J Clin Endocrinol Metab. 2018;103:3945–3953. doi: 10.1210/jc.2018-01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coviello AD, Sam S, Legro RS, Dunaif A. High prevalence of metabolic syndrome in first-degree male relatives of women with polycystic ovary syndrome is related to high rates of obesity. J Clin Endocrinol Metab. 2009;94:4361–4366. doi: 10.1210/jc.2009-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recabarren SE, et al. Metabolic profile in sons of women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93:1820–1826. doi: 10.1210/jc.2007-2256. [DOI] [PubMed] [Google Scholar]
- 32.McCartney CR, et al. Obesity and sex steroid changes across puberty: evidence for marked hyperandrogenemia in pre- and early pubertal obese girls. J Clin Endocrinol Metab. 2007;92:430–436. doi: 10.1210/jc.2006-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knudsen KL, et al. Hyperandrogenemia in obese peripubertal girls: correlates and potential etiological determinants. Obesity (Silver Spring) 2010;18:2118–2124. doi: 10.1038/oby.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCartney CR, et al. The association of obesity and hyperandrogenemia during the pubertal transition in girls: obesity as a potential factor in the genesis of postpubertal hyperandrogenism. J Clin Endocrinol Metab. 2006;91:1714–1722. doi: 10.1210/jc.2005-1852. [DOI] [PubMed] [Google Scholar]
- 35.True CA, et al. Chronic combined hyperandrogenemia and western-style diet in young female rhesus macaques causes greater metabolic impairments compared to either treatment alone. Hum Reprod. 2017;32:1880–1891. doi: 10.1093/humrep/dex246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varlamov O, et al. Combined androgen excess and Western-style diet accelerates adipose tissue dysfunction in young adult, female nonhuman primates. Hum Reprod. 2017;32:1892–1902. doi: 10.1093/humrep/dex244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bishop CV, et al. Chronically elevated androgen and/or consumption of a Western-style diet impairs oocyte quality and granulosa cell function in the nonhuman primate periovulatory follicle. J Assist Reprod Genet. 2019;36:1497–1511. doi: 10.1007/s10815-019-01497-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bishop CV, et al. Chronic hyperandrogenemia in the presence and absence of a western-style diet impairs ovarian and uterine structure/function in young adult rhesus monkeys. Hum Reprod. 2018;33:128–139. doi: 10.1093/humrep/dex338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop CV, et al. Chronic hyperandrogenemia and western-style diet beginning at puberty reduces fertility and increases metabolic dysfunction during pregnancy in young adult, female macaques. Hum Reprod. 2018;33:694–705. doi: 10.1093/humrep/dey013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kokosar M, et al. A Single Bout of Electroacupuncture Remodels Epigenetic and Transcriptional Changes in Adipose Tissue in Polycystic Ovary Syndrome. Sci Rep. 2018;8:1878. doi: 10.1038/s41598-017-17919-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumesic DA, et al. Adipose Insulin Resistance in Normal-Weight Polycystic Ovary Syndrome Women. J Clin Endocrinol Metab. 2019 doi: 10.1210/jc.2018-02086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kokosar M, et al. Epigenetic and Transcriptional Alterations in Human Adipose Tissue of Polycystic Ovary Syndrome. Sci Rep. 2016;6:22883. doi: 10.1038/srep22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones MR, et al. Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity. PLoS Genet. 2015;11:e1005455. doi: 10.1371/journal.pgen.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller M, et al. Global DNA methylation levels in human adipose tissue are related to fat distribution and glucose homeostasis. Diabetologia. 2014;57:2374–2383. doi: 10.1007/s00125-014-3356-z. [DOI] [PubMed] [Google Scholar]
- 45.Arner P, et al. The epigenetic signature of systemic insulin resistance in obese women. Diabetologia. 2016;59:2393–2405. doi: 10.1007/s00125-016-4074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahlman I, et al. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. Int J Obes (Lond) 2015;39:910–919. doi: 10.1038/ijo.2015.31. [DOI] [PubMed] [Google Scholar]
- 47.ER VA-m, et al. DNA Methylation in the Pathogenesis of Polycystic Ovary Syndrome. Reproduction. 2019 doi: 10.1530/REP-18-0449. [DOI] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corton M, et al. Differential gene expression profile in omental adipose tissue in women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2007;92:328–337. doi: 10.1210/jc.2006-1665. [DOI] [PubMed] [Google Scholar]
- 50.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/S0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 53.Varley KE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 56.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varlamov O, Bethea CL, Roberts CT., Jr. Sex-specific differences in lipid and glucose metabolism. Front Endocrinol (Lausanne) 2014;5:241. doi: 10.3389/fendo.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gesta, S., Tseng, Y. H. & Kahn, C. R. Developmental origin of fat: tracking obesity to its source. Cell131, 242–256, S0092-8674(07)01272-X [pii] 10.1016/j.cell.2007.10.004 (2007). [DOI] [PubMed]
- 61.Varlamov O, et al. Ovarian cycle-specific regulation of adipose tissue lipid storage by testosterone in female nonhuman primates. Endocrinology. 2013;154:4126–4135. doi: 10.1210/en.2013-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, et al. FABP4: a novel candidate gene for polycystic ovary syndrome. Endocrine. 2009;36:392–396. doi: 10.1007/s12020-009-9228-5. [DOI] [PubMed] [Google Scholar]
- 63.Seow KM, et al. Omental adipose tissue overexpression of fatty acid transporter CD36 and decreased expression of hormone-sensitive lipase in insulin-resistant women with polycystic ovary syndrome. Hum Reprod. 2009;24:1982–1988. doi: 10.1093/humrep/dep122. [DOI] [PubMed] [Google Scholar]
- 64.Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 65.Archer A, et al. LXR activation by GW3965 alters fat tissue distribution and adipose tissue inflammation in ob/ob female mice. J Lipid Res. 2013;54:1300–1311. doi: 10.1194/jlr.M033977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kirchgessner TG, et al. Beneficial and Adverse Effects of an LXR Agonist on Human Lipid and Lipoprotein Metabolism and Circulating Neutrophils. Cell Metab. 2016;24:223–233. doi: 10.1016/j.cmet.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 67.Ryden M, et al. Comparative studies of the role of hormone-sensitive lipase and adipose triglyceride lipase in human fat cell lipolysis. Am J Physiol Endocrinol Metab. 2007;292:E1847–1855. doi: 10.1152/ajpendo.00040.2007. [DOI] [PubMed] [Google Scholar]
- 68.Ek I, et al. A unique defect in the regulation of visceral fat cell lipolysis in the polycystic ovary syndrome as an early link to insulin resistance. Diabetes. 2002;51:484–492. doi: 10.2337/diabetes.51.2.484. [DOI] [PubMed] [Google Scholar]
- 69.Jensterle M, Kocjan T, Janez A. Phosphodiesterase 4 inhibition as a potential new therapeutic target in obese women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2014;99:E1476–1481. doi: 10.1210/jc.2014-1430. [DOI] [PubMed] [Google Scholar]
- 70.Bialesova L, et al. Epigenetic Regulation of PLIN 1 in Obese Women and its Relation to Lipolysis. Sci Rep. 2017;7:10152. doi: 10.1038/s41598-017-09232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol. 2016;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim AY, et al. Obesity-induced DNA hypermethylation of the adiponectin gene mediates insulin resistance. Nat Commun. 2015;6:7585. doi: 10.1038/ncomms8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nilsson E, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63:2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 74.Ronn T, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu N, et al. Epigenetic mechanism underlying the development of polycystic ovary syndrome (PCOS)-like phenotypes in prenatally androgenized rhesus monkeys. PLoS One. 2011;6:e27286. doi: 10.1371/journal.pone.0027286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ugi S, et al. CCDC3 is specifically upregulated in omental adipose tissue in subjects with abdominal obesity. Obesity (Silver Spring) 2014;22:1070–1077. doi: 10.1002/oby.20645. [DOI] [PubMed] [Google Scholar]
- 77.Kobayashi S, et al. Fat/vessel-derived secretory protein (Favine)/CCDC3 is involved in lipid accumulation. J Biol Chem. 2015;290:7443–7451. doi: 10.1074/jbc.M114.592493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem. 2013;288:34287–34294. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 80.Rozenberg JM, Taylor JM, Mack CP. RBPJ binds to consensus and methylated cis elements within phased nucleosomes and controls gene expression in human aortic smooth muscle cells in cooperation with SRF. Nucleic Acids Res. 2018;46:8232–8244. doi: 10.1093/nar/gky562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rozenberg JM, Tesfu DB, Musunuri S, Taylor JM, Mack CP. DNA methylation of a GC repressor element in the smooth muscle myosin heavy chain promoter facilitates binding of the Notch-associated transcription factor, RBPJ/CSL1. Arterioscler Thromb Vasc Biol. 2014;34:2624–2631. doi: 10.1161/ATVBAHA.114.304634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu HL, et al. The expression of the miR-25/93/106b family of micro-RNAs in the adipose tissue of women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2014;99:E2754–2761. doi: 10.1210/jc.2013-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen YH, et al. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62:2278–2286. doi: 10.2337/db12-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andrews, S. FastQC: A quality control tool for high throughput sequence data. (2010).
- 85.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krueger, F. Trim Galore, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- 88.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Law CW, Chen Y, Shi W, Smyth G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28:2986–2988. doi: 10.1093/bioinformatics/bts545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression and DNA methylation data are available at the NCBI Gene Expression Omnibus data repository under Accession Number GSE124709.