Abstract
A host of hormonal-metabolic alterations take place following exposure of cardiomyocytes to hypoxia and other noxious stimuli. Here, we demonstrate that exposure of cultured rat cardiomyocytes to lipopolysaccharide (LPS) resulted in upregulation (~1.5 fold) of oxidized low-density lipoprotein receptor-1 (LOX-1). There was also a marked increase in apoptosis 12 hrs after LPS treatment with caspase-3 levels being significantly elevated (~1.3 fold) and a significant increase in LDH release at 24 hrs. Interestingly, there was a ~1.4-fold upregulation of LC-3 expression post-LPS treatment indicating development of autophagy, which probably is a compensatory response to combat cellular injury induced by LPS. Treatment with LPS also reduced the size and morphology of cardiomyocyte spheroids. In an attempt to limit LPS-induced injury, cardiomyocytes were treated with exosomes derived from mesenchymal stromal cells (MSCs). We noted a significant suppression of LOX-1 expression that in turn suppressed apoptosis as well as autophagic response and restored spheroid morphology. Mass spectrophotometric analysis of MSC exosomes revealed a cargo rich in proteins which are involved in pathways negatively modulating cell death and apoptosis while promoting cell survival. This is first report to our knowledge on the initial molecular events in MSC exosome mediated cytoprotection of stressed cardiomyocytes.
Subject terms: Apoptosis, Heart failure
Introduction
Myocardial infarction (MI) is a leading cause of mortality worldwide1. Wide-spread apoptosis occurs in the tissue areas surrounding the infarct following MI. Involvement of autophagy in acute and chronic ischemia is also well documented2,3. The importance of autophagy in limiting the extent of eventual injury has been recently demonstrated in a number of in vitro and in vivo studies. It has been reported that preceding starvation (a basic stimulant of autophagy) and intermittent reperfusion post-MI significantly reduces infarct size4,5 Reports have shown that autophagy during myocardial ischemia is a major determinant of tissue survival6,7.
Lectin-like oxidized LDL receptor-1 (LOX-1) is a type C receptor for ox-LDL which is upregulated by angiotensin II and pro-inflammatory cytokines8–11. Under physiological conditions, expression of LOX-1 in cardiomyocytes is low, but it is highly inducible when these cells are subjected to various forms of metabolic, inflammatory, ischemic, oxidative and mechanical stress. Expression of LOX-1 appears to be important in the eventual determination of extent of ischemic myocardial injury3,12–14, since abrogation of LOX-1 reduces infarct size, number of apoptotic cells, inflammatory signaling, ROS generation and cardiac function. Recent studies show that exposure of cardiomyocytes to stress also induces signals for autophagy15. The autophagy signals appear early and are short-lasting, where signals for apoptosis are relatively late in onset, but persist for a long duration3.
Attempts to exploit stem cells in hopes of regenerating/repairing cardiac muscle and improving cardiac function16–19, have so far revealed only limited degree of success20–22. A major limitation in these studies is inability of stem cells to respond and adapt favorably to the damaged microenvironment. Use of stem cell exosomes offers a promising novel approach for post-MI intervention to decrease scar formation and fibrosis, and help regenerate cardiac muscle23–27. Exosomes, a distinct class of constitutively secreted vesicles of endosomal origin, carry precious cargo of proteins and genetic material. They possess the ability to alter the transcriptome and proteome of the recipient cell, modulating pathways governing apoptosis, cell growth, proliferation and differentiation. In place of pluripotent or multipotent stem cells, which are unfavorably affected by signals from the damaged cardiac tissue microenvironment, treatment with stimulated MSC exosomes will render the ischemic heart resistant to micro-environmental stressors and help contain spread of ischemic damage28. Exosomes from various stem cell sources have been employed to provide protection against ischemic damage. However, we decided to focus our efforts on the use of MSC exosomes for the stem cells can potentially be readily harvested from MI patients without any need for differentiation using differentiating factors and also the fact that they are readily available. The primary aim of our study was to decipher molecular events in cardiomyocytes exposed to lipopolysaccharide (LPS), with particular reference to the expression of LOX-1, and signals for apoptosis and autophagy. Further, we aimed to determine if MSC exosome treatment would provide cytoprotection by blocking these early events. Lastly, we conducted proteomics to determine pathways of LPS-mediated cellular injury and their modulation by MSC exosomes.
Results
Characterization of exosomes
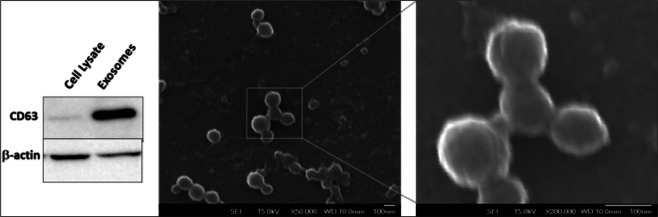
Scanning electron microscopy (SEM) analysis of suspended exosome pellet was used to confirm that the ultracentrifugation pellet isolated from MSC cultures consisted of discrete vesicles and not parts of damaged intracellular cellular organelles or membranes. Nanoparticle tracking analysis of exosomes revealed an average diameter of 118 nm (Supplement Fig. 1). Probing for exosomal marker, Western blot of equivalent quantities of proteins show enrichment of CD63 in the exosome sample compared to whole cell lysate. High magnification scanning electron microscopy micro pictographs obtained using an 80 keV electron beam showed presence of vesicles of uniform size and diameter (Fig. 1). Mass spectrometry analysis revealed the absence of several organelle markers such as endoplasmic reticulum marker calnexin, golgi marker GM130, mitochondrial marker proteins such as cytochrome C oxidase, ABCD3, ATP5A1 etc. MS analysis revealed the presence of several exosomal markers such as CD63, CD9 and CD81 (Supplementary Results). These results along with electron micropictographs indicate that the pellets isolated from conditioned media of MSC cultures by sequential centrifugation constitute a distinct set of secretory vesicles.
Figure 1.
Characterization of exosomes. Western blots show enrichment of CD63 in exosomes while it is barely detected in whole cell lysates. Exosomes were plated onto a poly-L-ornithine coated glass slide and SEM images were acquired. The exosomes show a uniform size distribution of around 80 nm. Scale bar = 100 nm.
Injurious effect of LPS on cardiomyocytes and rescue by MSC exosomes
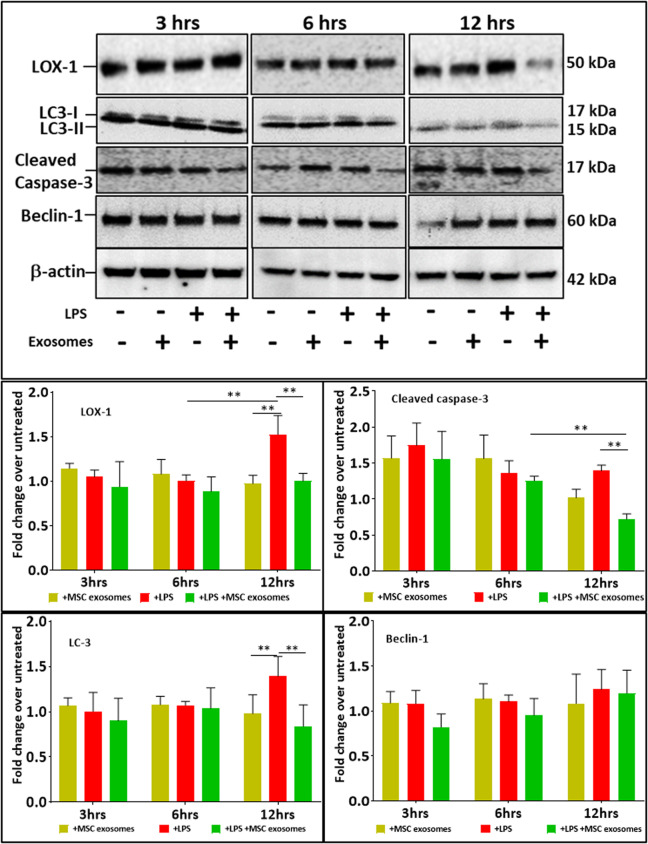
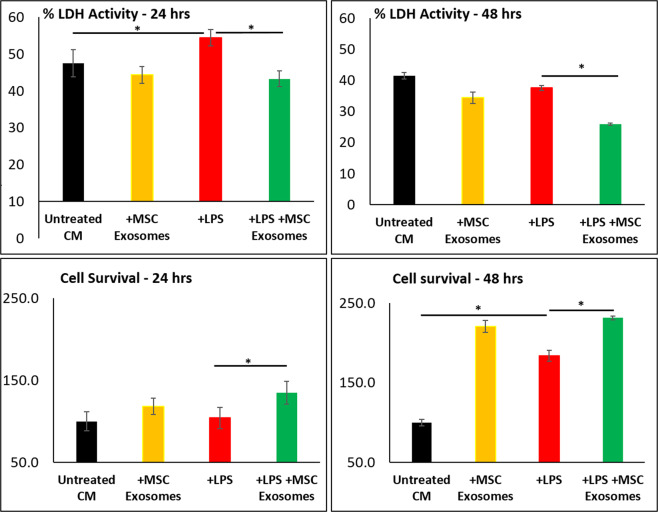
LOX-1 expression was significantly upregulated in cardiomyocytes early, at 12 hr after exposure to LPS (100 ng/ml) (Fig. 2) which correlates with our previously published data13,29. We measured occurrence of apoptosis and autophagy in response to LPS (Fig. 2). We noted a marked increase in apoptosis (expression of Caspase-3) as well as autophagy signal LC3-II and Beclin-2. Of note, while the autophagy marker LC3-II was significantly elevated (≈1.4 fold), Beclin-1 expression over the same time period also increased (≈1.2 fold), but not significantly. LDH activity as a marker of cell death slightly increased at 24 hr. Interestingly cell survival measured as MTT assay also increased under LPS stress, especially at 48 hrs (Fig. 3).
Figure 2.
Monolayer cultures of cardiomyocytes were exposed to LPS (100 ng/ml) for 1 hour and then treated with MSC exosomes. LPS induced LOX-1 expression at 12 hrs of exposure. LPS also induced apoptosis (cleaved Caspase-3) and autophagy (LC3). Treatment with MSC exosomes decreased LOX-1, cleaved caspase-3 and LC-3 levels in cardiomyocytes stressed with LPS over a period of 6–12 hrs, but had no effect on expression of Beclin-1. LOX-1 levels were decreased with MSC exosomes. Data in mean ± SD, n = 4 *p < 0.05, vs LPS treatment based on 3 independent experiments. Full-length blots are presented in Supplementary Fig. 2.
Figure 3.
Cell death (LDH release) and cell survival assays on cardiomyocytes. Cells were first treated with LPS (100 ng/ml) for 1 hr and then treated with MSC exosomes in presence of LPS. Treatment with MSC exosomes led to decrease in cell death and increase in cell survivability. Data as mean ± SD, n = 4. *p < 0.05 vs LPS treatment based on 3 independent experiments.
Most interestingly, treatment of cardiomyocytes with MSC exosomes in presence of LPS induced stress (1 hr after) rescued cells from LOX-1 expression, and occurrence of apoptosis and autophagy (Fig. 2). Treatment of cardiomyocytes with MSC exosomes also protected cardiomyocytes from death (LDH release) (P < 0.05). Notably MSC exosome treatment further increased cell survival at both 24 and 48 hr time points. Importantly, MSC exosomes alone had no effect on LOX-1 expression, occurrence of apoptosis and autophagy signals (Fig. 2). MSC exosomes, however, enhanced cardiomyocyte survival (Fig. 3).
Effect of exosomes on Cardiomyocyte spheroids
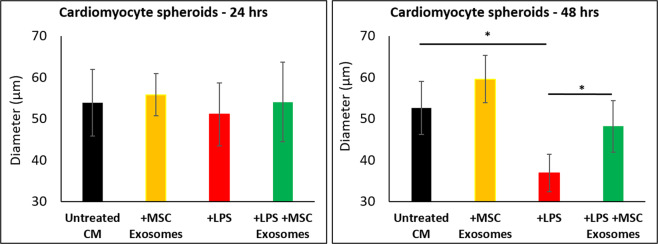
We wondered if LPS and exosomes would have any effect on tissue morphology. While monolayer cell cultures present a reductionist approach to study cellular mechanisms in response to certain stimuli, spheroid cultures accurately mimic natural physiological responses and cell-to-cell interactions30,31. Using the hanging drop method, cardiomyocytes were cultured as spheroids. Cardiomyocyte spheroids were stressed with LPS and then treated with MSC exosomes. While LPS significantly decreased cardiomyocyte spheroid diameters progressively over the 48 hr period, MSC exosomes rescued spheroid integrity over the same time point (Fig. 4). (Representative images of the spheroids are shown in Supplement Fig. 7).
Figure 4.
Cardiomyocyte spheroids were exposed to LPS (100 ng/ml) and then treated with MSC exosomes. Exposure to LPS reduced the spheroid diameter by >50% at 48 hrs. Treatment with exosomes rescued cardiomyocyte spheroid morphology in presence of LPS. *p < 0.05 vs LPS treatment based on 4 independent experiments.
Proteomic analysis of MSC exosomes
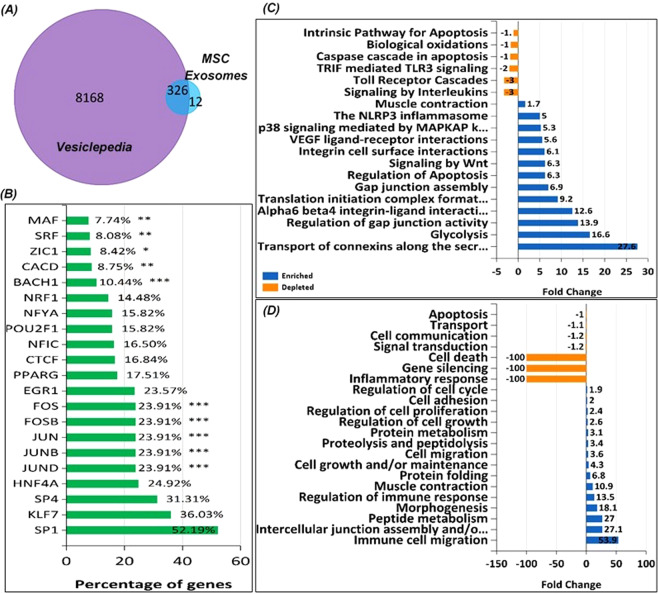
Proteomic analysis identified a host of proteins (≈338) secreted in MSC exosomes. Using FunRich32 analysis for extracellular vesicle (EV) proteins secreted by all cell types available in the database, we identified 326 common classes of proteins enriched in EV subsets (Fig. 5A) and 12 unique to the MSC exosomes. Among the entire set of proteins, we identified various transcription factors (TFs) such as SP1, KLF7, SP4, JUN, JUND, JUNB, FOSB and FOS which are involved in regulating expression of their target genes in recipient cells (Fig. 5B). Other proteins secreted via exosomes shown in Fig. 5C,D differentially modulate various pathways and are involved in processes such as apoptosis, protein metabolism, cell growth and proliferation. In addition, analysis of exosomal proteins using Integrated Pathway Analysis (IPA Qiagen Bioinformatics)33 software showed that some of the proteins are involved in upregulating pathways governing proliferation of immune cells, muscle formation and function and angiogenesis (Supplement Fig. 3A) while others are involved in downregulating processes involved in infarct size, organ degeneration, cytolysis and hypersensitive reactions (Supplement Fig. 3B).
Figure 5.
Compared to the vesicle secreted protein database, MS analysis using FunRich32 analysis tool identified a number of similar and unique proteins secreted in MSC exosomes (A). Proteomic analysis identified different transcription factors in MSC exosomes (B). FunRich analysis revealed the different pathways which the secreted proteins modulate (C) and the processes which the exosomal proteins regulate (D). IPA analysis revealed the many this exosomal cargo modulates cellular proliferation, cell survival, and in recipient cells.
Proteomic analysis of cardiomyocytes treated with LPS followed by exosomes
We conducted proteomic analysis of cardiomyocytes stimulated with LPS and then rescued with MSC exosomes. As expected, compared to untreated cardiomyocytes, LPS treatment changed the proteome (1580 identified proteins) of these cardiomyocytes (Supplement Fig. 3A) with many proteins being differentially expressed; for example, PSME4, ACADVL, FAM91A1, GNA11, STAT6 (downregulated) and NCBP1, WBP11, ACAT2, FXR2, SPART (upregulated) (Supplement Fig. 4B,C). Many of the proteins identified are related to the upregulation of apoptotic process, response to oxidative stress, regulation of cell death and myoblast migration, while proteins related to fatty acid catabolism, response to cytokine stimulus, cardiac conduction, DNA replication proof reading and regulation of cardiac conduction were downregulated. (Supplement Fig. 4D).
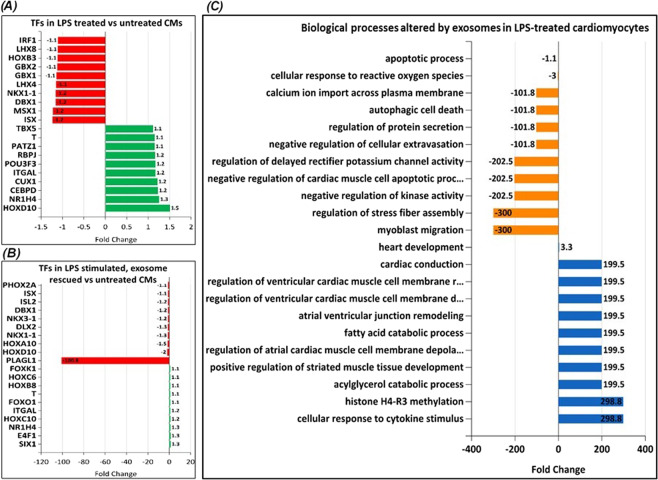
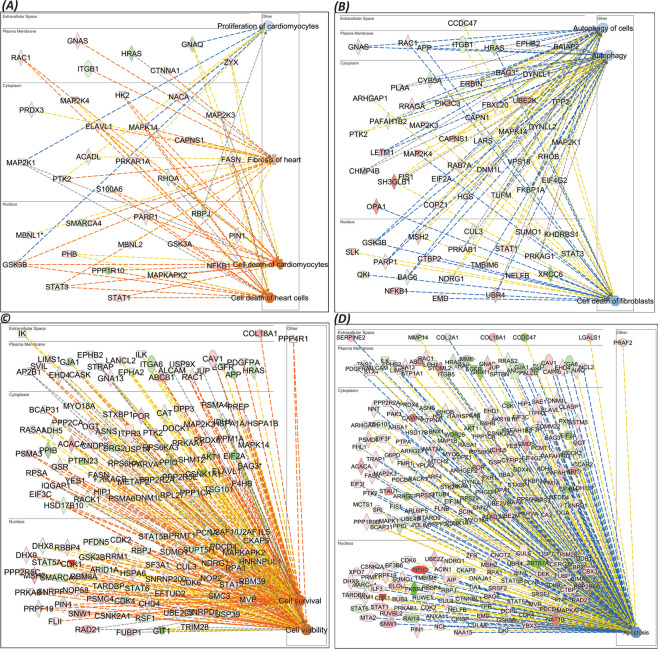
Many TFs were also found to be differentially expressed. Among the upregulated TFs were HOXD10, NR1H4, CEPBD, CUX1 and ITGAL, while ISX, MSX1, DBX1, LHX4 and GBX1 were downregulated (Fig. 6A). IPA33 network analysis revealed upregulated signaling networks leading to cardiomyocytes death and cardiac fibrosis (Fig. 7A) and suppressed pathways leading to proliferation of cardiomyocytes, autophagy and fibroblast cell death (Fig. 7B).
Figure 6.
MS analysis of cardiomyocytes (CMs) using FunRich32 analysis tool: Differences in response to exosome treatment in cells can be attributed to the functional exosomal cargo in regulating gene expression such as changes in (A) expression of transcription factors (TFs) in LPS stressed cells or (B) LPS treated cells rescued by MSC exosomes. (C) Analysis of biological processes of proteins identified by MS in exosome rescued cardiomyocytes post LPS treatment. The processes presented are in comparison against LPS stimulated cardiomyocytes.
Figure 7.
Network analysis: IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis)33 network analysis revealed upregulated signaling networks leading to cardiomyocytes death and cardiac fibrosis (A) and suppressed pathways leading to proliferation of cardiomyocytes, autophagy and fibroblast cell death (B). Exosome treatment of LPS stimulated cells upregulated signaling networks leading to increased cell viability and cell survival (C) while it decreased apoptotic signaling network (D).
Further, treatment with MSC exosomes of LPS stimulated CMs revealed expression of different sets of proteins. For example, exosome treatment of LPS stimulated CMs showed expression of 1587 identified proteins (Supplement Fig. 5A) among which GIT2, GIT1, VPS29 and DNAJA2 were downregulated, and CDK1, MACF1, ACAT2 were CAND2 were upregulated (Supplement Fig. 5B,C).
Alterations in TFs are shown in (Fig. 6B). The most prominent changes were seen in expression of SIX1, E4F1, NR1H4, HOXC10 and ITGAL(upregulated), and expression of PLAGL1, HOXD10, HOXA10, NKX1-1 and DLX2 (downregulated). Surprisingly, compared to LPS treated cells, the level of one transcription factor, PLAGL1 was dramatically modulated when LPS stimulated cells were treated with MSC exosomes (Fig. 6B). Most importantly, treatment of LPS stimulated cells with exosomes caused significant downregulation of cellular processes such as apoptosis, oxidative stress, autophagic cell death and myoblast migration and upregulation of processes such as fatty acid catabolism, response to cytokine stimulus, cardiac conduction and regulation of cardiac muscle membrane depolarization and repolarization (Fig. 6C) which is in stark contrast to those seen with LPS stimulated cells prior to treatment with exosomes (Supplement Fig. 3D). IPA network analysis showed exosome treatment of LPS stimulated cells upregulated signaling networks leading to increased cell viability and cell survival (Fig. 7C) while it decreased apoptotic signaling network (Fig. 7D).
Discussion
While various surgical and pharmacotherapeutic interventions have significantly improved patient outcomes following AMI, there is a major need for additional therapeutic options to repair or reverse damaged heart tissue post-MI. Attempts have been made to exploit stem cells by implanting them in the damaged cardiac tissue in hopes of regenerating/repairing cardiac muscle and improving cardiac function16–19. Results of these studies have so far revealed only limited degree of success20–22.
A promising novel area for recovery of various tissues from stress-induced injury is the use of stem cell exosomes23,24,27. Exosomes, a distinct class of constitutively secreted vesicles of endosomal origin, carry precious cargo of proteins and genetic material. Importantly, unlike stem cells, secreted exosomes do not respond to the damaged microenvironment but possess the ability to alter the transcriptome and proteome of the recipient cell, modulating pathways governing apoptosis, cell growth, proliferation and differentiation. Changes in exosomal cargo depend on cellular origin as well as the micro-environment in which the secreting cell finds itself in, reflecting the physiological state of the secreting cell34,35 Exosomal cargo secreted by cells plays a potent role in initiating transcription of target genes and translation of proteins which affect the phenotype of recipient cells. Importantly, unlike stem cells, secreted exosomes do not respond to the damaged microenvironment but possess the ability to alter the extracellular matrix, change the transcriptome and proteome of the recipient cell, modulating pathways governing apoptosis, cell growth, proliferation and differentiation.
Our previous work36 showed that LPS stimulates LOX-1 expression in cardiomyocytes. Increased LOX-1 expression induces oxidative stress and further drives the inflammatory pathways causing cell death. Ischemia to the heart caused by coronary artery ligation also induces LOX-1 expression37,38 The important role of LOX-1 in inducing myocardial injury became evident from studies wherein LOX-1 antibody or LOX-1 gene deletion dramatically reduced myocardial injury37,38. In other recent studies in cultured vascular smooth muscle cells, we showed that pro-inflammatory stimuli induced autophagy which was short-lasting and cellular apoptosis which was longer lasting39. In the present study, LPS indeed, significantly increased expression of LOX-1 in stimulated cardiomyocytes, which in turn led to increases in expression of proapoptotic caspase-3, and autophagy markers LC-3 and Beclin-1. However, when cardiomyocytes were first stimulated with LPS for one hour and then treated with MSC exosomes, we observed significant cytoprotective effects of MSC exosome treatment. MSC exosomes induced cytoprotective effects in cardiomyocytes by suppressing LOX-1 expression. Subsequently caspase-3 expression, and more importantly LC3 expression fell. Beclin-1 expression also increased with LPS treatment but not significantly. Previous studies have also shown discordance between LC3 and beclin-1 levels40–42. It is also possible that failure of beclin-1 to rise significantly is a type 2 error. We believe that occurrence of autophagy is a compensatory event to combat cellular injury caused by LPS3,39. These results collectively indicate that (I) LOX-1 is an early mediator of LPS-induced cardiomyocyte injury, and (II) MSC exosomes suppress LOX-1 expression and thus prevent LPS-mediated cell death. To our knowledge, these are the first set of studies showing a link between MSC exosome treatment and LOX-1 expression in cardiomyocytes which in turn affects cardiomyocyte autophagy and apoptosis.
To understand the function of exosomes, one needs to study the exosomal cargo and the influence this cargo has on recipient cells. We have previously demonstrated that changes in secreting cells’ microenvironment alters the exosomal cargo34,35. In the present investigation, we studied the proteome of MSC exosomes. Compared to all the proteins identified in secretory EVs by various tissues/cell types, we identified proteins in our samples which are unique to MSC exosomes (Fig. 5A). Among these of note were AOC1, SPARC, ARPC4-TTLL3, AFF1, THBS3, DCN and ACAN which are involved in cell-cell, cell-matrix attachment, modulation of extracellular matrix, acting polymerization while TNFRSF10D is known to inhibit TRAIL-induced apoptosis. Many of the identified exosomal proteins are involved in regulating or modulating a variety of biological pathways (Fig. 5C) in recipient cells such as downregulating intrinsic pathway for apoptosis, biological oxidation, and caspase cascade in apoptosis and toll-receptor cascades. These proteins positively regulate processes such as integrin cell surface interactions, glycolysis, MAPKAP kinase mediated p38 signaling, muscle contraction and gap junction assembly. Regulation of such pathways has a profound effect on processes such as cell death, gene silencing, inflammatory responses which are suppressed by exosomal proteins.
Further MS analysis revealed secretion of various transcription factors in exosomes. Transcription factors such as MAF, SRF, EGR1 and SP1 are involved in regulating transcription of genes which control differentiation, proliferation, metabolic pathways and cell survival. Exosomal SP1 has been implicated in providing significant protection against ischemia/reperfusion injury43. KLF7 regulates differentiation of progenitor cells and/or maintenance of phenotype cells44. Our functional assay data relative to improved cell survival and reduced cell death can be directly attributed to proteins carried in MSC exosomes.
Further, we investigated the changes in the proteome of cardiomyocytes stimulated with LPS and then rescued with MSC exosomes. We observed that MSC exosomes suppressed autophagic cell death, negatively regulated apoptotic processes, and suppressed cellular responses to reactive oxygen species, negatively regulated cellular extravasation and kinase activity. Processes such as cardiac conduction, fatty acid catabolism, regulation of striated muscle development, regulation of cardiac muscle repolarization and depolarization were positively regulated. The protective effects exhibited by MSC exosomes in LPS stimulated CMs are similar to those in cells treated with MSC exosomes alone (Supplement Fig. 6C–E). These data show the effect that MSC exosomes have on the expression of cytoprotective proteins in recipient cells.
We further analyzed the expression of cardiomyocyte transcription factors in response to LPS and further treatment with MSC exosomes. Compared to untreated cells, LPS alone induced expression of transcription factors such as HOX10, NR1H4, CEBPD, while suppressed expression of ISX, MSX1, DBX1, NKX1 and NOXB3 all of which control transcription of genes which negatively affect cardiac function and cell survival. Surprisingly, when LPS stimulated cardiomyocytes were treated with MSC exosomes, expression of one transcription factor, PLAGL1, was dramatically affected. PLAGL1 (pleomorphic adenoma gene-like 1) was also found to be significantly depleted in cells treated with MSC exosomes alone (Supplement Fig. 6B). PLAGL1 is a zinc-finger transcription factor whose target genes are involved in signaling, cell adhesion, extracellular matrix composition, and developmental disorders45,46.
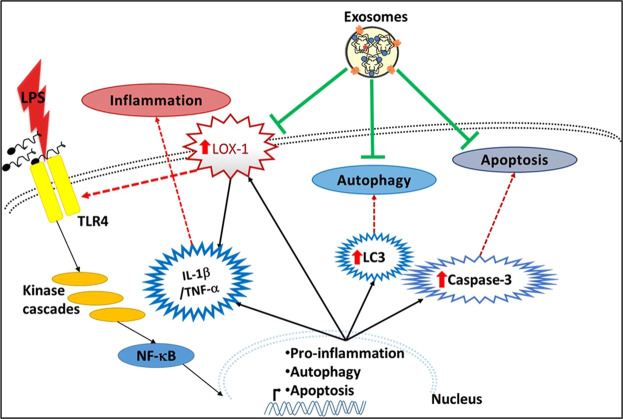
We believe that use of exosomes from MSCs would positively modulate the ischemic microenvironment of cardiac tissues and help damaged cardiomyocytes recover, repair and reverse the consequence of the inflammatory event. We have shown that MSC exosomes suppress apoptosis and autophagy in LPS stimulated cardiomyocytes as well as LOX-1 expression. These effects may be consequence of exosome cargo (Fig. 8). Although many studies have attempted to identify exosomal microRNAs as early diagnostic biomarkers for various diseases47–51, we believe that it is the physiologically active exosomal protein content which ultimately governs the role exosomes play in modulating the recipient cellular physiology. In our future studies, we will look into the secretion of many of the transcription factors via exosomes and their roles in recipient cells.
Figure 8.
MSC Exosomes provide significant cardioprotection against LPS stimulated stress responses. LPS treatment significantly increases pro-inflammatory LOX-1 expression along with cleaved caspase-3, and LC3. In spite of presence of LPS and continued stimulation of TLR4 LPS receptors, addition of MSC exosomes reverses the effects of LPS by downregulating expression of LOX-1, caspase-3 and LC3, thereby decreasing apoptosis and autophagy.
Our study will lay the ground work to utilize stimulated stem cell/cardiomyocyte exosomes, to effectuate cardioprotection in damaged heart tissues.
Methods
Cell culture and reagents
Cell culture media, as previously described34– DMEM, phosphate buffered saline (PBS), penicillin/streptomycin were obtained from Corning (Thermo Fischer Scientific, Waltham, MA), fetal bovine serum (FBS) from Atlanta Biologicals Inc (Flowery Branch, GA). LPS was purchased from Sigma Aldrich (St. Louis, MO). MTS assay regent (celltiter 96 aqueous one solution cell proliferation assay) was purchased from Promega Corporation (Madison, WI). LDH cytotoxicity assay kit was purchased from Thermo scientific (Rockford, IL). The rat myoblast cell line, H9C2 (CRL-1446) was purchased from ATCC. Mesenchymal stromal cell (MSC) line were obtained from Robert J. Griffin, Ph.D. at UAMS.
Cell culture
H9C2 cultures were maintained in complete DMEM media containing 10% FBS & 100 units/ml of penicillin/streptomycin at 37 °C in a 5% CO2 incubator. As per our previously developed protocol35, overnight cell cultures grown to 80% confluence were used for experiments. Cells cultured overnight were washed 3x with PBS and fresh serum free DMEM media was added and were then incubated for a further for indicated time points at 37 °C in a 5% CO2 incubator. For experiments, these cells were treated with LPS initially for 1 hour and then MSC exosomes were added to the culture media in presence of LPS. A small quantity of the lysates (50μg) was sued for MS analysis. For spheroid cultures, H9C2 cells were suspended in serum free DMEM medium and 20 μl of cell suspension was plated onto the covers of 6 well tissue culture plates to form hanging drops. The covers were then inverted and placed on the plates containing PBS to keep the cell suspension hydrated. Cells were allowed to form spheroids over a period of 2-3 days and then treated with exosomes alone or LPS or with LPS for 1 hour and then MSC exosomes were added without washing off the LPS.
MSCs maintained in complete DMEM media containing 10% FBS & 100 units/ml of penicillin/streptomycin at 37 °C in a 5% CO2 incubator. In brief35, cells cultured overnight were washed 3x with PBS and fresh serum free DMEM media was added and were then incubated for a further 16–18 hrs at 37 °C in a 5% CO2 incubator. Media from these cultures grown was collected to isolate exosomes. For hypoxic exosomes, MSCs were grown in an anaerobic chamber (Forma Scientific) with an atmospheric mixture of 5% CO2, 10% H2, 85% N2 producing oxygen concentrations below 0.5%.
Exosome isolation
Exosomes were isolated from conditioned media of MSC cultures incubated in serum free DMEM media for 16–18 hrs by sequential centrifugation as previously described34. In brief, media pooled from MSC cultures was subjected to series of centrifugation steps, first at 3000 g for 10 min followed by centrifugation for 30 min at 10,000 g. The pellets were discarded and followed by ultracentrifugation at 100,000 g for 3 hrs to pull down exosomes. The resulting exosome pellet was resuspended in PBS and ultracentrifuged for 2 hrs at 100,000 g. The final pellet thus obtained was resuspended in PBS and used in further experiments.
ZetaView Nanoparticle tracking analysis
Isolated exosomes were subjected to nanoparticle tracking analysis using the ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany). In brief as described previously52, he sample was diluted in 1 ml of PBS (200 fold) and loaded into the optical cell. Using two reading cycles at each position, 11 different positions inside the cells were read by the instrument. Acquisition parameters were 23 °C temp., 90 sensitivity and a frame rate of 30 frames per second (fps), a shutter speed of 100, and a laser pulse set to the shutter speed. The mean, median and mode were calculated by the software (ZetaView 8.04.02 SP2) and outliers discarded. Post-acquisition parameters were set to a minimum brightness of 25, a maximum area of 1000 pixels, and a minimum area of 10 pixels. Polystyrene particles from ThermoFisher Scientific with a known average size of 100 nm were used to calibrate the instrument prior to sample readings. Automated quality control measurements including, cell quality check and instrument alignment and focus were also performed prior to the use of the ZetaView for sample measurements.
Electron microscopy
Exosomes coated on glass surface were fixed overnight at 4 °C in 3% glutaraldehyde in 0.1 M cacoldylate buffer pH 7.2. Then Samples were washed three times with 0.1 M cacoldylate buffer pH 7.2 and three times with distilled water 10 min each, after that the samples were fixed in 4% osmium tetroxide in distilled water for 1 hour, and then the samples were rinsed again with 0.1 M cacoldylate buffer pH 7.2 and distilled water three times 10 min each. This was followed by dehydration process by using a graded series of ethanol with two final dehydration steps in 100% ethanol for 30 mins each. Samples were transferred to a 1:2 solution of hexamethyldisilazane (HMDS):100% ethanol for 20 minutes, then treated with a solution of 2:1 HMDS:ethanol for 20 minutes. Then the samples were submerged in 100% HMDS solution in a loosely capped container in a fume hood overnight to remove the HMDS. Finally, the samples were mounted over STUBS and then coated with carbon layers to enhance the conductivity. Images were acquired using the JEOL JSM7000F scanning electron microscope.
Cell death and viability assays
Cell viability assay was performed following the instructions published by the manufacturer of the MTS reagent. In brief, as previously performed35, H9C2 cells were seeded into 96 well plate at a density of 3000 cells per well and allowed to attach overnight (18 hrs). Fresh exosome free media was added to the cells after a wash with PBS, the cells were treated with 50 nM of lipopolysaccharide (LPS) for 1 hr and then incubated for 24 or 48 hrs with or without exosomes, in presence of LPS in a humidified, 5% CO2 atmosphere. At indicated time points, 50 μl of media from each well was collected and added into fresh wells and to the remainder 20 μl of MTS regent was added. To develop the reaction, the cells were incubated for 4 hrs after which absorbance at 570 nm was read in a 96-well plate reader. To the 50 μl of collected media in clean wells, 50 μl of LDH reagent was added and incubated for 30 mins at room temp. The reaction was quenched with addition of 50 μl of stop buffer and absorbance read at 490 and 680 nm in a 96 well plate reader.
SDS-PAGE and western blotting
Using a widely employed and previously used protocol to develop Westerns blots34,53, around 20 μg of exosomal pellets or cell lysates, determined by BCA protein estimation, were boiled with 4x Laemmle’s sample buffer containing 10% beta-mercaptoethanol and run on a 4–20% SDS-PAGE denaturing gel (Bio-Rad Laboratories, Hercules, CA). Using conventional transfer apparatus, proteins from the gel were transferred to PVDF membranes. Either 5% Bovine serum albumin (BSA) or 5% non-fat dried skimmed milk powder dissolved in Tris-buffered saline containing 0.01% tween-20 (TBS-t) was used to block the membranes for 1 hr at room temp. Blots were incubated with primary antibodies against caspase-3 (rabbit polyclonal antibody at 1:1000 dilution), LOX-1 (rabbit polyclonal antibody at1:1000 dilution), Beclin-1 (rabbit polyclonal antibody at 1:1000 dilution), LC-3 (rabbit polyclonal antibody at 1:1000 dilution), (all from Abcam, Cambridge, MA), CD63 (rabbit polyclonal antibody at 1:1000 dilution) (SBI Biosciences, Mountain View, CA) and against β-actin (mouse monoclonal antibody at 1:5000 dilution) (Abcam, Cambridge, MA)] for 1 hr at room temp. Blots were washed 3x with TBS-t and then incubated with anti-mouse or anti- rabbit antibody conjugated with horse radish peroxide (HRP) (at 1:10,000 dilution) (Santa Cruz Biotech, Dallas, TX) at room temperature for 1 hour. Blots were rinsed 3x with TBS-t and developed with a chemiluminescence developing reagent34, SuperSignal® West Femto Maximum sensitivity substrate (ThermoFisher Scientific, Rockford, IL).
Mass spectrometry
FASP Methods – Orbitrap Lumos: LC-MS/MS methods – Orbitrap Velos: Purified proteins were reduced, alkylated, and digested using filter-aided sample preparation. Tryptic peptides were then separated by reverse phase XSelect CSH C18 2.5 um resin (Waters) on an in-line 150 × 0.075 mm column using an UltiMate 3000 RSLCnano system (Thermo). Peptides were eluted using a 90 min gradient from 97:3 to 60:40 buffer A:B ratio, [Buffer A = 0.1% formic acid, 0.5% acetonitrile; buffer B = 0.1% formic acid, 99.9% acetonitrile]. Eluted peptides were ionized by electrospray (2.15 kV) followed by MS/MS analysis using higher-energy collisional dissociation (HCD) on an Orbitrap Fusion Lumos mass spectrometer (Thermo) in top-speed data-dependent mode. MS data were acquired using the FTMS analyzer in profile mode at a resolution of 240,000 over a range of 375 to 1500 m/z. Following HCD activation, MS/MS data were acquired using the ion trap analyzer in centroid mode and normal mass range with precursor mass-dependent normalized collision energy between 28.0 and 31.0. Proteins were identified by database search using MaxQuant (Max Planck Institute) with a 3 ppm parent ion tolerance and a 0.5 Da fragment ion tolerance. Peptide and protein identifications were verified using Scaffold 4 software. Protein identifications were accepted if they could be established with less than 1.0% false discovery and contained at least 2 identified peptides. Protein Prophet Algorithm54 was used to assign protein probabilities. The Mascot results were viewed and analyzed by Scaffold Q + (version Scaffold_4.8.9, Proteome Software Inc., Portland, OR). A protein threshold of 95%, minimum of 2 peptides and a 50% peptide threshold values were used as the cut-off values and resulting data exported into excel spreadsheets.
Statistical analysis
As previously described34,35,53, we used GraphPad Prism’s two way ANOVA followed by Bonferroni’s multiple comparison test to calculate statistical significance for all results, expressed as mean ± SD. Value of p < 0.05 was considered statistically significant,
Supplementary information
Acknowledgements
We would like to acknowledge the help of Ryan Porter, PhD., Dept. of Internal Medicine, UAMS, Little Rock AR 72205 in helping us with exosome characterization using ZetaView PMX 110 instrument. Supported by Grant-in-Aid from the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development, Washington, DC. Additional support was provided by Stebbins Chair in Cardiology funds to JLM. AJT acknowledges funding from the National Institutes of Health (P20GM121293 and S10OD018445) and support from the UAMS Proteomics Core Facility. RJG acknowledges the Center for Advanced Surface Engineering, under the National Science Foundation Grant No. IIA-1457888 and the Arkansas EPSCoR Program, ASSET III.
Author contributions
R.A.K. and J.L.M. conceived the study. R.A.K. performed the experiments and analyzed proteomics data. J.C.H. maintained the cardiomyocyte cell lines. R.A.K. and J.L.M. wrote the main manuscript text and prepared and edited all figures. R.N.H. wrote the scanning electron microscopy methods section and A.J.T. edited the proteomics methods section. J.C.H., R.N.H., R.J.G., A.J.T., and Z.D. reviewed the manuscript. The proteomics datasets generated and analyzed during the current study are not publicly available because they haven’t been uploaded yet, but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained an error in Figure 2, where the western blot image for cleaved Caspase-3 for the 12 hr time point was incorrect. Consequently, the Supplementary Figures file published with this Article contained an error in Supplementary Figure 2, where the full-length western blot image for cleaved Caspase-3 for the 12 hr time point was incorrect.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/12/2022
A Correction to this paper has been published: 10.1038/s41598-022-11690-y
Supplementary information
is available for this paper at 10.1038/s41598-019-55694-7.
References
- 1.Go AS, et al. Executive summary: heart disease and stroke statistics–2014 update: a report from the American Heart Association. Circulation. 2014;129:399–410. doi: 10.1161/01.cir.0000442015.53336.12. [DOI] [PubMed] [Google Scholar]
- 2.Sabbah HN. Apoptotic cell death in heart failure. Cardiovascular research. 2000;45:704–712. doi: 10.1016/S0008-6363(99)00348-X. [DOI] [PubMed] [Google Scholar]
- 3.Wang, X., Guo, Z., Ding, Z. & Mehta, J. L. Inflammation, autophagy, and apoptosis after myocardial infarction. J Am Heart Assoc7, 10.1161/jaha.117.008024 (2018). [DOI] [PMC free article] [PubMed]
- 4.Schott RJ, Rohmann S, Braun ER, Schaper W. Ischemic preconditioning reduces infarct size in swine myocardium. Circulation research. 1990;66:1133–1142. doi: 10.1161/01.RES.66.4.1133. [DOI] [PubMed] [Google Scholar]
- 5.Wagner C, Tillack D, Simonis G, Strasser RH, Weinbrenner C. Ischemic post-conditioning reduces infarct size of the in vivo rat heart: role of PI3-K, mTOR, GSK-3β, and apoptosis. Molecular and Cellular Biochemistry. 2010;339:135–147. doi: 10.1007/s11010-009-0377-x. [DOI] [PubMed] [Google Scholar]
- 6.Huang C, et al. Autophagy induced by ischemic preconditioning is essential for cardioprotection. Journal of cardiovascular translational research. 2010;3:365–373. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giricz Z, et al. Autophagosome formation is required for cardioprotection by chloramphenicol. Life sciences. 2017;186:11–16. doi: 10.1016/j.lfs.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duerrschmidt N, et al. Lectin-like oxidized low-density lipoprotein receptor-1-mediated autophagy in human granulosa cells as an alternative of programmed cell death. Endocrinology. 2006;147:3851–3860. doi: 10.1210/en.2006-0088. [DOI] [PubMed] [Google Scholar]
- 9.Kume N, et al. Inducible expression of lectin-like oxidized LDL receptor-1 in vascular endothelial cells. Circulation research. 1998;83:322–327. doi: 10.1161/01.RES.83.3.322. [DOI] [PubMed] [Google Scholar]
- 10.Li DY, Zhang YC, Philips MI, Sawamura T, Mehta JL. Upregulation of endothelial receptor for oxidized low-density lipoprotein (LOX-1) in cultured human coronary artery endothelial cells by angiotensin II type 1 receptor activation. Circulation research. 1999;84:1043–1049. doi: 10.1161/01.RES.84.9.1043. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Hermonat PL, Mehta JL, Romeo F, Novelli G. Lectin-like, oxidized low-density lipoprotein receptor-1 (LOX-1): A critical player in the development of atherosclerosis and related disorders. Cardiovascular research. 2006;69:36–45. doi: 10.1016/j.cardiores.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, et al. LOX-1 abrogation reduces cardiac hypertrophy and collagen accumulation following chronic ischemia in the mouse. Gene therapy. 2012;19:522–531. doi: 10.1038/gt.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding Z, et al. LOX-1, mtDNA damage, and NLRP3 inflammasome activation in macrophages: implications in atherogenesis. Cardiovascular research. 2014;103:619–628. doi: 10.1093/cvr/cvu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, et al. LOX-1 deletion limits cardiac angiogenesis in mice given angiotensin II. Cardiovasc Drugs Ther. 2014;28:441–446. doi: 10.1007/s10557-014-6541-x. [DOI] [PubMed] [Google Scholar]
- 15.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giri Tapan Kumar, Alexander Amit, Agrawal Mukta, Saraf Swarnalata, Saraf Shailendra, Ajazuddin Current Status of Stem Cell Therapies in Tissue Repair and Regeneration. Current Stem Cell Research & Therapy. 2019;14(2):117–126. doi: 10.2174/1574888X13666180502103831. [DOI] [PubMed] [Google Scholar]
- 17.Menasché Philippe. Cell therapy trials for heart regeneration — lessons learned and future directions. Nature Reviews Cardiology. 2018;15(11):659–671. doi: 10.1038/s41569-018-0013-0. [DOI] [PubMed] [Google Scholar]
- 18.Tang J, et al. Targeted repair of heart injury by stem cells fused with platelet nanovesicles. Nature biomedical engineering. 2018;2:17–26. doi: 10.1038/s41551-017-0182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tompkins BA, et al. Preclinical Studies of Stem Cell Therapy for Heart Disease. Circ Res. 2018;122:1006–1020. doi: 10.1161/circresaha.117.312486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Su Hyun, Cho Jang Hyun, Lee Yoon Ho, Lee Ji Hye, Kim Soo Sung, Kim Mi Yang, Lee Min Gu, Kang Won Yu, Lee Kyung Sim, Ahn Young Keun, Jeong Myung Ho, Kim Hyun Soo. Improvement in Left Ventricular Function with Intracoronary Mesenchymal Stem Cell Therapy in a Patient with Anterior Wall ST-Segment Elevation Myocardial Infarction. Cardiovascular Drugs and Therapy. 2018;32(4):329–338. doi: 10.1007/s10557-018-6804-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laguna G, et al. Effect of direct intramyocardial autologous stem cell grafting in the sub-acute phase after myocardial infarction. The Journal of cardiovascular surgery. 2018;59:259–267. doi: 10.23736/s0021-9509.17.10126-6. [DOI] [PubMed] [Google Scholar]
- 22.Lang CI, et al. Cardiac Cell Therapies for the Treatment of Acute Myocardial Infarction: A Meta-Analysis from Mouse Studies. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;42:254–268. doi: 10.1159/000477324. [DOI] [PubMed] [Google Scholar]
- 23.Adamiak M, Sahoo S. Exosomes in Myocardial Repair: Advances and Challenges in the Development of Next-Generation Therapeutics. Molecular therapy: the journal of the American Society of Gene Therapy. 2018;26:1635–1643. doi: 10.1016/j.ymthe.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagno L, Hatzistergos KE, Balkan W, Hare JM. Mesenchymal Stem Cell-Based Therapy for Cardiovascular Disease: Progress and Challenges. Molecular therapy: the journal of the American Society of Gene Therapy. 2018;26:1610–1623. doi: 10.1016/j.ymthe.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garikipati VNS, Shoja-Taheri F, Davis ME, Kishore R. Extracellular Vesicles and the Application of System Biology and Computational Modeling in Cardiac Repair. Circulation research. 2018;123:188–204. doi: 10.1161/circresaha.117.311215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gartz, M. & Strande, J. L. Examining the Paracrine Effects of Exosomes in Cardiovascular Disease and Repair. Journal of the American Heart Association7, 10.1161/jaha.117.007954 (2018). [DOI] [PMC free article] [PubMed]
- 27.Yuan Y, et al. Stem Cell-Derived Exosome in Cardiovascular Diseases: Macro Roles of Micro Particles. Frontiers in pharmacology. 2018;9:547. doi: 10.3389/fphar.2018.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezaie Jafar, Rahbarghazi Reza, Pezeshki Milad, Mazhar Mahdi, Yekani Farshid, Khaksar Majid, Shokrollahi Elhameh, Amini Hassan, Hashemzadeh Shahriar, Sokullu Sadiye Emel, Tokac Mehmet. Cardioprotective role of extracellular vesicles: A highlight on exosome beneficial effects in cardiovascular diseases. Journal of Cellular Physiology. 2019;234(12):21732–21745. doi: 10.1002/jcp.28894. [DOI] [PubMed] [Google Scholar]
- 29.Sun D, et al. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Molecular therapy: the journal of the American Society of Gene Therapy. 2010;18:1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell Madeline, Chabria Mamta, Figtree Gemma A., Polonchuk Liudmila, Gentile Carmine. Stem Cell Niche. New York, NY: Springer New York; 2018. Stem Cell-Derived Cardiac Spheroids as 3D In Vitro Models of the Human Heart Microenvironment; pp. 51–59. [DOI] [PubMed] [Google Scholar]
- 31.Christoffersson Jonas, Meier Florian, Kempf Henning, Schwanke Kristin, Coffee Michelle, Beilmann Mario, Zweigerdt Robert, Mandenius Carl-Fredrik. A Cardiac Cell Outgrowth Assay for Evaluating Drug Compounds Using a Cardiac Spheroid-on-a-Chip Device. Bioengineering. 2018;5(2):36. doi: 10.3390/bioengineering5020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathan Mohashin, Keerthikumar Shivakumar, Chisanga David, Alessandro Riccardo, Ang Ching-Seng, Askenase Philip, Batagov Arsen O., Benito-Martin Alberto, Camussi Giovanni, Clayton Aled, Collino Federica, Di Vizio Dolores, Falcon-Perez Juan Manuel, Fonseca Pedro, Fonseka Pamali, Fontana Simona, Gho Yong Song, Hendrix An, Hoen Esther Nolte-’t, Iraci Nunzio, Kastaniegaard Kenneth, Kislinger Thomas, Kowal Joanna, Kurochkin Igor V., Leonardi Tommaso, Liang Yaxuan, Llorente Alicia, Lunavat Taral R., Maji Sayantan, Monteleone Francesca, Øverbye Anders, Panaretakis Theocharis, Patel Tushar, Peinado Héctor, Pluchino Stefano, Principe Simona, Ronquist Goran, Royo Felix, Sahoo Susmita, Spinelli Cristiana, Stensballe Allan, Théry Clotilde, van Herwijnen Martijn J.C., Wauben Marca, Welton Joanne L., Zhao Kening, Mathivanan Suresh. A novel community driven software for functional enrichment analysis of extracellular vesicles data. Journal of Extracellular Vesicles. 2017;6(1):1321455. doi: 10.1080/20013078.2017.1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics (Oxford, England) 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kore RA, Abraham EC. Inflammatory cytokines, interleukin-1 beta and tumor necrosis factor-alpha, upregulated in glioblastoma multiforme, raise the levels of CRYAB in exosomes secreted by U373 glioma cells. Biochem Biophys Res Commun. 2014;453:326–331. doi: 10.1016/j.bbrc.2014.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kore RA, et al. Hypoxia-derived exosomes induce putative altered pathways in biosynthesis and ion regulatory channels in glioblastoma cells. Biochem Biophys Rep. 2018;14:104–113. doi: 10.1016/j.bbrep.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, et al. Cross-talk between inflammation and angiotensin II: studies based on direct transfection of cardiomyocytes with AT1R and AT2R cDNA. Experimental biology and medicine (Maywood, N.J.) 2012;237:1394–1401. doi: 10.1258/ebm.2012.012212. [DOI] [PubMed] [Google Scholar]
- 37.Li D, et al. Expression of lectin-like oxidized low-density lipoprotein receptors during ischemia-reperfusion and its role in determination of apoptosis and left ventricular dysfunction. Journal of the American College of Cardiology. 2003;41:1048–1055. doi: 10.1016/S0735-1097(02)02966-2. [DOI] [PubMed] [Google Scholar]
- 38.Hu C, et al. LOX-1 abrogation reduces myocardial ischemia-reperfusion injury in mice. Journal of molecular and cellular cardiology. 2008;44:76–83. doi: 10.1016/j.yjmcc.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Ding Z, et al. Regulation of autophagy and apoptosis in response to ox-LDL in vascular smooth muscle cells, and the modulatory effects of the microRNA hsa-let-7 g. International journal of cardiology. 2013;168:1378–1385. doi: 10.1016/j.ijcard.2012.12.045. [DOI] [PubMed] [Google Scholar]
- 40.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell death and differentiation. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 41.Tian S, et al. Beclin 1-independent autophagy induced by a Bcl-XL/Bcl-2 targeting compound, Z18. Autophagy. 2010;6:1032–1041. doi: 10.4161/auto.6.8.13336. [DOI] [PubMed] [Google Scholar]
- 42.Correa RJM, Valdes YR, Shepherd TG, DiMattia GE. Beclin-1 expression is retained in high-grade serous ovarian cancer yet is not essential for autophagy induction in vitro. Journal of Ovarian Research. 2015;8:52. doi: 10.1186/s13048-015-0182-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan X, et al. Extracellular vesicles from human-induced pluripotent stem cell-derived mesenchymal stromal cells (hiPSC-MSCs) protect against renal ischemia/reperfusion injury via delivering specificity protein (SP1) and transcriptional activating of sphingosine kinase 1 and inhibiting necroptosis. Cell Death & Disease. 2017;8:3200. doi: 10.1038/s41419-017-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laub F, et al. Developmental Expression of Mouse Krüppel-like Transcription Factor KLF7 Suggests a Potential Role in Neurogenesis. Developmental Biology. 2001;233:305–318. doi: 10.1006/dbio.2001.0243. [DOI] [PubMed] [Google Scholar]
- 45.Vega-Benedetti AF, et al. PLAGL1: an important player in diverse pathological processes. Journal of applied genetics. 2017;58:71–78. doi: 10.1007/s13353-016-0355-4. [DOI] [PubMed] [Google Scholar]
- 46.Varrault A, et al. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic acids research. 2017;45:10466–10480. doi: 10.1093/nar/gkx672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomarkers in medicine. 2013;7:769–778. doi: 10.2217/bmm.13.63. [DOI] [PubMed] [Google Scholar]
- 48.Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics. Clinical applications. 2015;9:358–367. doi: 10.1002/prca.201400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brinton LT, Sloane HS, Kester M, Kelly KA. Formation and role of exosomes in cancer. Cellular and molecular life sciences: CMLS. 2015;72:659–671. doi: 10.1007/s00018-014-1764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annual review of physiology. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 51.Lin J, et al. Exosomes: novel biomarkers for clinical diagnosis. TheScientificWorldJournal. 2015;2015:657086. doi: 10.1155/2015/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helwa I, et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PloS one. 2017;12:e0170628. doi: 10.1371/journal.pone.0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kore RA, Abraham EC. Phosphorylation negatively regulates exosome mediated secretion of cryAB in glioma cells. Biochim Biophys Acta. 2016;1863:368–377. doi: 10.1016/j.bbamcr.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.