Abstract
To investigate the functions of NAC-like genes, we reported the characterization and functional analysis of one Arabidopsis NAC-like gene which showed a novel function in the regulation of gibberellin biosynthesis and named as GIBBERELLIN SUPPRESSING FACTOR (GSF). GSF acts as a transcriptional activator and has transactivation capacity based on yeast transcription activity assays. YFP + GSF-TM (lacking a transmembrane domain) fusion proteins accumulated in the nuclei, while the YFP + GSF fusion proteins only accumulated in the ER membrane and were absent from the nuclei. These results revealed that GSF requires processing and release from the ER and transportation into the nucleus to perform its function. The ectopic expression of GSF-TM caused a dwarfism phenotype, which was correlated with the upregulation of the gibberellin (GA) deactivation genes GA2-oxidases 2/6 (GA2ox2/6) and the downregulation of the GA biosynthetic genes GA20-oxidases 1–4 (GA20ox1-4). The external application of GA rescued the dwarfism in the 35 S::GSF-TM plants, indicating that GSF affects GA biosynthesis, rather than the GA signaling pathway. Further analysis indicated that the upregulation of GA2ox2/6 is a key factor for the GSF function to regulate the GA level, since 35 S::GA20ox1 could not rescue the dwarfism in the 35 S::GSF-TM plants. Cold treatment induced the processing of the YFP + GSF fusion proteins from the ER membrane and their entry into the nuclei, which is correlated with the cold-induced upregulation of GA2oxs. In addition, the expression of GA2oxs was induced by drought, and the 35 S::GSF-TM plants showed drought tolerance compared to the wild-type plants. Our data suggest a role for GSF in response to abiotic stresses, such as cold and drought, by suppressing the biosynthesis of GA in Arabidopsis.
Subject terms: Gibberellins, Abiotic
Introduction
Plant hormone gibberellins (GA) have been hypothesized to play important roles in regulating various developmental processes in plants, such as stem elongation, leaf expansion, seed germination and flower development1. Mutants in the gibberellin biosynthetic genes exhibit severe GA-deficiency, such as dwarfism of the inflorescence and compact leaves, in many plant species2.
Gibberellins are comprised of tetracyclic diterpene carboxylic acids that require complex biosynthetic steps3. To date, many different gibberellins have been identified from plants4, but only a small number of gibberellins, such as GA1, GA3, GA4, and GA7, function as bioactive hormones. In Arabidopsis, the gibberellin biosynthetic genes encode such enzymes as ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS) and ent-kaurene oxidase (KO), which catalyze the early steps of GA biosynthesis. Certain GA metabolic genes, such as GA20-oxidases (GA20oxs) and GA2-oxidases (GA2oxs), are involved in the late steps in the metabolic pathway5. GA20oxs participated in the biosynthesis of the major bioactive GAs, including GA4, the primary bioactive GA form in Arabidopsis. To maintain the dynamic homeostasis of gibberellin for plant growth, the deactivation of GA by GA2oxs is important for the regulation of the concentration of bioactive gibberellin in plants1,6.
It has been reported that GA could also play a vital role in response to abiotic stresses in plants7. In general, stress may lead to the retardation of growth in plants, and a similar effect is caused by the reduction of GA to adjust its resources to resist the stress8. Additional evidence was reported that indicates that GA could respond to abiotic stresses by modulating plant growth via GA biosynthesis or signal transduction7. For example, the GA2oxs were upregulated to induce the GA deactivation in response to salt and cold stresses in Arabidopsis9–11. In contrast, the GA20oxs were reported to be downregulated in response to cold stress9,12. However, the manner in which the mechanisms control GA levels during abiotic stresses still remained to be investigated.
NAC-like genes are a group of plant-specific transcription factors that contain a conserved NAC domain (acronym of NAM of petunia, ATAF1/2 and CUC2 of Arabidopsis) in the N terminal of the proteins13,14. The NAC domain consists of approximately 150 amino acids and functions to bind DNA13. A typical NAC domain could be divided into five subdomains (A-E). Subdomain A is considered to participate in the formation of the protein dimer. The C and D subdomains were reported to bind to the binding element sequence by a positive charge, while subdomains B and E could confer the functional diversity of NAC15–17. The C-terminus of an NAC protein contains a transcriptional regulation region, which has a role to either repress or activate gene expression18. NAC transcription factors have been found to be involved in multiple functions that include the development of the shoot apical meristem (SAM)13,14,19,20, the manipulation of cell expansion in flower organs21, the regulation of lateral root formation22, senescence23,24, secondary wall biosynthesis in fibers25, xylem differentiation26,27, anther dehiscence28,29 and flower receptivity30.
A number of NAC transcription factors contain a transmembrane domain (TM) (designated NTLs) in the C-terminus31, which are anchored in the organelle membranes as a dormant form, that have been reported to be closely associated with plant responses to such environmental stresses as UV, drought, heat, cold, osmosis, high salinity, and high-light, and ER stresses, respectively32–43. However, the manner in which the mechanisms and factors are regulated by these NTLs in response to environmental stresses has not been thoroughly elucidated to date.
Thus, to further investigate the functions of more NAC-like genes is necessary. In this study, we reported the characterization and functional analysis of one Arabidopsis NAC-like gene GIBBERELLIN SUPPRESSING FACTOR (GSF). GSF, also known as RAO2/Arabidopsis NAC domain-containing protein17 (ANAC017), has been reported to be involved in mitochondrial retrograde regulation by acting as a positive regulator of AOX1a44. In addition, we identified that its function was associated with GA-related abiotic stress responses. Our results indicated that GSF could help the plants respond to cold/drought stresses through the reduction of the GA level by downregulating the GA biosynthetic genes (GA20oxs) and upregulating the GA deactivation genes (GA2oxs) in the GA metabolic pathway. Thus, our study demonstrated a linkage between the NAC-like gene and GA in the regulation of the response of plants to abiotic stresses.
Results
Isolation of GSF cDNA from Arabidopsis thaliana
One member of an Arabidopsis NAC-like gene, GIBBERELLIN SUPPRESSING FACTOR (GSF) (At1g34190), was cloned and analyzed. GSF contains three introns and four exons (Fig. S1A) and encodes a protein of 557 amino acids (Fig. S1B). The GSF protein contains a putative conserved NAC domain that consists of five subdomains (A-E) (Fig. S1B) and has been identified in the N-terminal ends of most NAC-like proteins14,45. In addition, one transmembrane motif is identified in the C-terminal end of the GSF protein31 (Fig. S1B). The GSF protein showed 82% similarity and 72% identity to the most closely related NAC-like protein, At1g34180 (Fig. S1B).
Detection of the expression for GSF by the analysis of GSF::GUS transgenic Arabidopsis
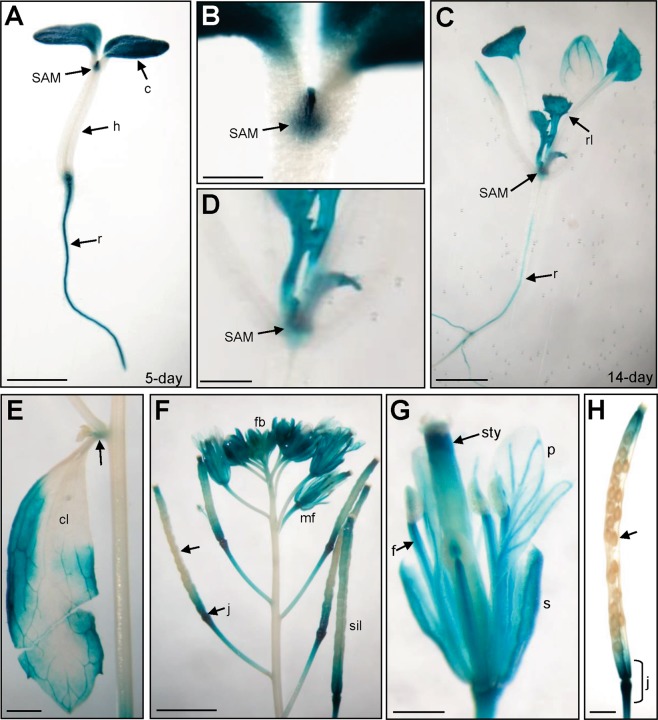
To explore the expression pattern of the GSF gene, a construct containing a 2-kb promoter region of GSF fused with GUS reporter (GSF::GUS) was constructed and transformed into Arabidopsis. In the GSF::GUS transgenic seedlings, GUS was primarily detected in the root, cotyledon and shoot apex meristem and was absent in the hypocotyl of 5-day-old (Fig. 1A,B) and 14-day-old (Fig. 1C,D) seedlings. In addition, GUS activity was detected in the rosette leaves that had emerged in 14-day-old seedlings (Fig. 1C,D). In mature plants, GUS activity was detected in the rosette and cauline leaves and the node of inflorescence (Fig. 1E). During flower development, GUS activity was primarily detected in the sepal, stamen filaments and style of carpel and weakly detected in the petals in both young and mature flowers (Fig. 1F,G). During silique development, GUS was strongly detected in the junction region of the silique and pedicel and was absent in the developing seeds (Fig. 1F,H). The pattern of GUS expression detected in this study was consistent with the Arabidopsis eFP browser data46,47 (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi).
Figure 1.
GUS staining patterns in GSF::GUS transgenic Arabidopsis. (A) GUS activity was specifically detected in the cotyledon (c), shoot apex meristem (SAM) and roots (r) of a 5-day-old seedling. GUS was absent in the hypocotyl (h). Bar = 3 mm. (B) Close-up of the shoot apex meristem (SAM) region from (A). Bar = 0.5 mm. (C) GUS activity was specifically detected in the shoot apex meristem (SAM), rosette leaves (rl) and roots (r) of 14-day-old seedlings. Bar = 3 mm. (D) Close-up of the shoot apex meristem (SAM) region from (C). Bar = 1 mm. (E) GUS activity was detected in the cauline leaf (cl) and the node of inflorescence (arrow) in the inflorescence of a mature plant. Bar = 1 mm. (F) In a mature inflorescence, GUS activity was detected in the flower organs in both flower buds (fb) and mature flowers (mf). In siliques, GUS was strongly detected in the junction region (j) of the silique and pedicel and was absent from the developing seeds (arrow). Bar = 25 mm. (G) In a mature flower from (F), GUS activity was detected in the sepal (s), stamen filaments (f) and style of the carpel (sty) and weakly detected in the petals (p). Bar = 5 mm. (H) In a mature silique from (F), GUS was strongly detected in the junction region of the silique and pedicel (j) and was absent in the developing seeds (arrow). Bar = 0.5 mm.
GSF acts as a transcriptional activator
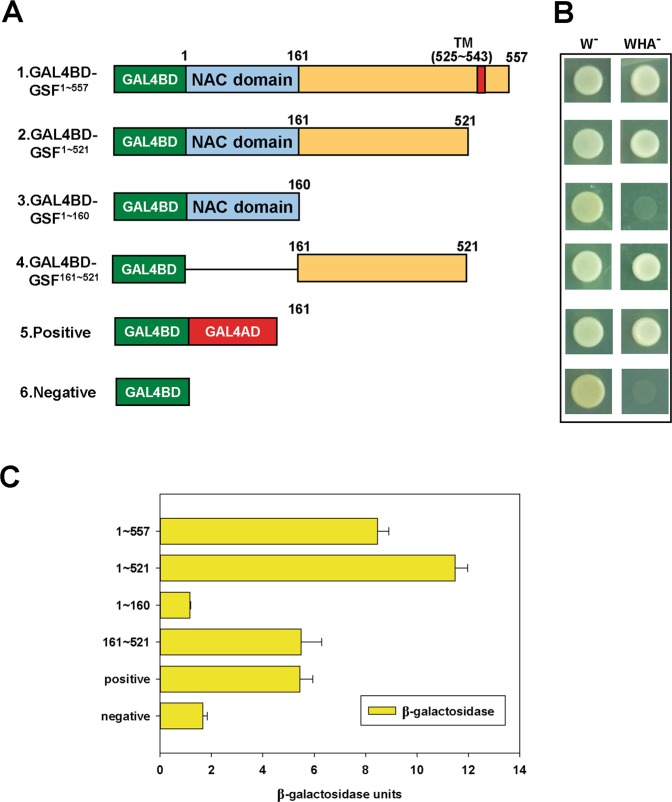
To examine whether GSF can act as a transcriptional activator, yeast transcription activity assays were performed. Different constructs containing full-length (GSF1~557, positions 1–557), truncated transmembrane (GSF1–521, positions 1–521), truncated C terminal half (GSF1–160, positions 1–160) or truncated N terminal half (GSF161–521 positions 161–521) forms of GSF (Fig. 2A) fused with the GAL4 DNA-binding domain (GAL4BD) were generated. Two constructs, the GAL4BD–GAL4 activation domain (GAL4AD) and GAL4BD, were constructed as positive and negative controls, respectively (Fig. 2A).
Figure 2.
Assay of transcriptional activation for GSF. (A) Various forms of GSF, full-length (GSF1~557, positions 1–557), truncated transmembrane (GSF1–521, positions 1–521), truncated C terminal half (GSF1–160, positions 1–160) and truncated N terminal half (GSF161–521, positions 161–521), fused with Gal4 DNA-binding domain (GAL4BD) to form four different constructs (GAL4BD-GSF1~557, GAL4BD-GSF1~521, GAL4BD-GSF1~160 and GAL4BD-GSF161~521). GAL4BD-GAL4AD is a positive control, while GAL4BD only served as a negative control. (B) The six constructs from (A) were introduced into the yeast strain AH109 and grown on SD without tryptophan (W−) or lacking tryptophan, histidine and adenine (WHA−). (C) Quantification of the β-galactosidase activity for six different transformants in yeast from (B).
All of the constructs were transformed into the yeast strain AH109 that contains several reporter genes (lacZ, His3, MEL1 and ADE2) and grew equally well on plates with histidine and adenine (W−) (Fig. 2B, left). Comparing the growth of the yeast cells on tryptophan-, histidine- and adenine-minus plates (WHA−) (Fig. 2B, right), yeast cells harboring the fusion proteins GAL4BD-GSF1~557, GAL4BD-GSF1~521 and GAL4BD-GSF161~521 grew as well as the positive controls GAL4BD-GAL4AD (Fig. 2B, right). In contrast, yeast cells that harbored the fusion protein GAL4BD-GSF1~160 did not grow as well on the WHA- plates as the negative controls GAL4BD (Fig. 2B, right). Further analysis indicated that yeasts harboring GAL4BD-GSF1~557, GAL4BD-GSF1~521 and GAL4BD-GSF161~521 have similar or even higher β–galactosidase activity with the positive controls GAL4BD-GAL4AD (Fig. 2C). Similar low β–galactosidase activity was observed in the yeasts containing GAL4BD-GSF1~160 or the negative controls GAL4BD (Fig. 2C). These results indicated that the C-terminus of GSF could activate downstream genes in plants41.
GSF truncated with a transmembrane motif could be released from the ER and enter into the nucleus
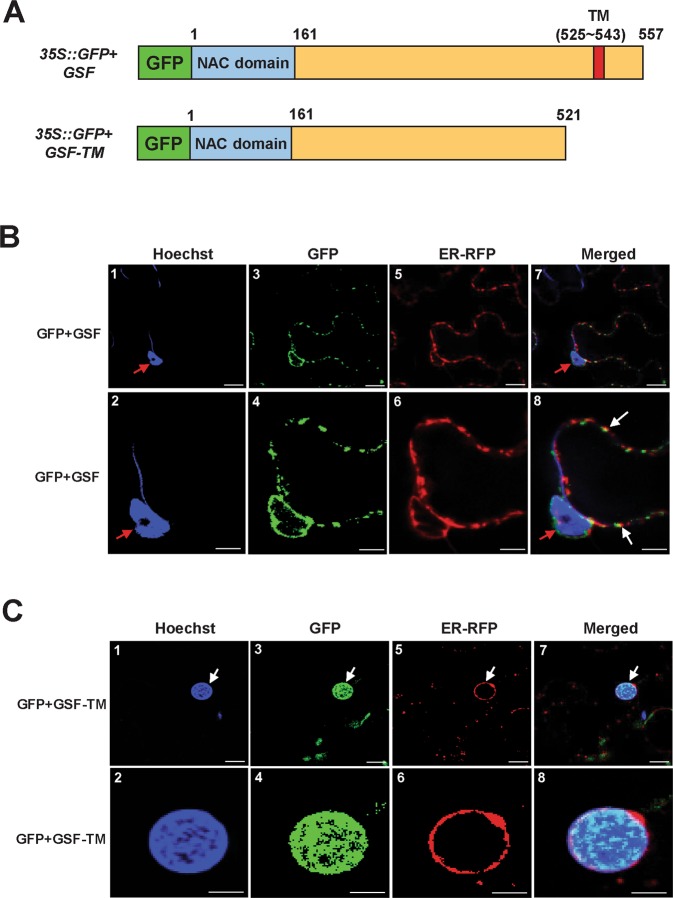
It has been reported that a group of NAC transcription factors with a transmembrane motif (designated NTLs) are translocated into the nucleus from the ER (endoplasmic reticulum) to regulate the expression of downstream genes that were activated by a proteolytic cleavage31. Since GSF contains a predicted transmembrane motif in the C-terminus that identifies them as NTL proteins31 (Figs. 3A, S1B), it is possible that GSF could be triggered and released from the ER and translocated to the nucleus to regulate the expression of the downstream genes. To examine this hypothesis, we transformed GFP fused with the full length GSF (35 S::GFP + GSF) or the GSF lacking its transmembrane motif (35 S::GFP + GSF-TM) and ER-RFP (as an ER marker) into tobacco leaves, and their fluorescence images were analyzed. The results showed that the fluorescence images of the GFP + GSF fusion proteins matched the RFP fluorescence images of ER-RFP in the ER membrane and were absent from the nuclei (Fig. 3B). In contrast, the GFP fluorescence images of the GFP + GSF-TM fusion proteins only accumulated in the nuclei of the cells, while they overlapped with the blue fluorescence of Hoechst (a dye for DNA -staining) and were absent in the ER membrane (Fig. 3C). These results indicated that GSF was localized to the ER and will be translocated into the nucleus once the transmembrane motif was deleted. This result is in agreement with the previous report44, which reported that when the N-terminal red fluorescent protein (RFP) and the C-terminal green fluorescent protein (GFP) are simultaneously tagged on the ANAC017 (GSF) protein, the N-terminal RFP can migrate to the nucleus whereas C-terminal GFP remains in the ER.
Figure 3.
Transient expression of GFP + GSF, GFP + GSF-TM and ER + RFP in tobacco cells. (A) Schematic diagram for the GFP + GSF and GFP + GSF-TM constructs transiently expressed in tobacco leaves. (B) Agrobacterium-mediated transient expression of GFP + GSF and ER + RFP in the epidermal cells of N. benthamiana. The GFP + GSF fusion protein (−3, −4) accumulated in organelle-like structures that were highly similar to the ER where the RFP was localized (−5, −6). Blue DNA-staining (Hoechst) indicated the nuclei (red arrow in −1, −2, −7 and −8). A merged fluorescence image of (−1, −3, −5) in (−7); (−2, −4, −6) in (−8) showing the similar localization of GFP + GSF and ER + RFP (white arrows in −8). (−2, −4, −6 and −8) are close-up images from (−1, −3, −5 and −7), respectively. Scale bars: 10 μm in (−1, −3, −5, −7) and 5 μm in (−2, −4, −6, −8). (C) Agrobacterium-mediated transient expression of GFP + GSF-TM and ER + RFP in the epidermal cells of N. benthamiana. The GFP + GSF-TM fusion protein (−3, −4) that accumulated in the nucleus (white arrow) was highly similar to the blue DNA-staining (Hoechst) (−1, −2) and different from where ER + RFP was localized (−5, −6). A merged fluorescence image of (−1, −3, −5) in (−7); (−2, −4, −6) in (−8) showing the similar localization of GFP + GSF-TM and blue DNA-staining (Hoechst) (white arrows in −7). (−2, −4, −6 and −8) are close-up images from (−1, −3, −5 and −7), respectively. Scale bars: 10 μm in (−1, −3, −5, −7) and 5 μm in (−2, −4, −6, −8).
Ectopic expression of GSF-TM causes dwarfism of the plants
To explore the function of the GSF gene, the full length cDNA and the cDNA lacking the transmembrane motif region of the GSF gene, driven by the cauliflower mosaic virus (CaMV) 35S promoter (35S::GSF and 35S::GSF-TM), were transformed into Arabidopsis for functional analysis.
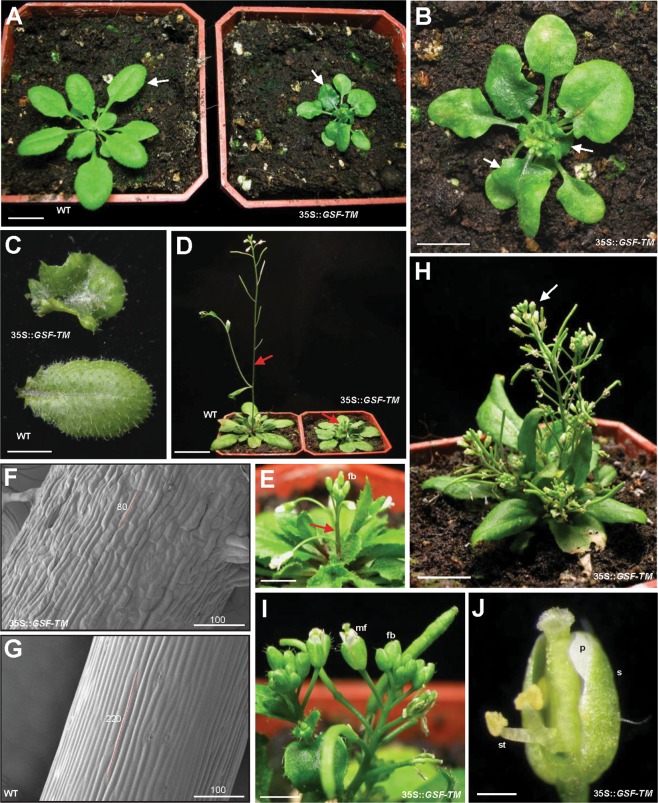
When the 35S::GSF Arabidopsis plants were analyzed, they were phenotypically indistinguishable from the wild-type plants in both vegetative and reproductive development. This result revealed that GSF could require processing and release from the ER to perform its function. Interestingly, 35S::GSF-TM transgenic Arabidopsis plants showed similar abnormal phenotypes, which were significantly different from those of the wild-type plants. These 35S::GSF-TM Arabidopsis plants exhibited phenotypic alterations, such as compact and curled rosettes leaves (Fig. 4A–C), short inflorescence internodes and severe dwarfism (Fig. 4D,E). The epidermal cells of the inflorescence in the 35S::GSF-TM transgenic plants (80 by 10 micrometers) (Fig. 4F) were significantly shorter than those in the wild type inflorescence (220 by 10 micrometers) (Fig. 4G). During the reproductive stage, flowers with short sepals and petals were observed in the compacted inflorescence of the 35S::GSF-TM plants (Fig. 4H–J). These results supported the hypothesis that GSF-TM is the functional form of GSF that regulates plant growth and development. Interestingly, a similar dwarfism phenotype was also observed in the 35S::GSF-TM + VP16 transgenic Arabidopsis (Fig. S2) in which GSF-TM was fused to the activation domain VP16-AD48,49. This result confirmed the conclusion from the yeast transcription activity assays (Fig. 2), which indicated that GSF acts as a transcriptional activator in plants.
Figure 4.
Phenotypic analysis of the 35S::GSF-TM Arabidopsis plants. (A) A 28-day-old 35S::GSF-TM plant (right) showed a severe dwarfism phenotype with small compact and curled leaves (arrow), while wild-type plants (WT, left) produced normal rosette leaves (arrow). Bar = 10 mm. (B) Close-up of the 35S::GSF-TM plant from (A). Bar = 5 mm. (C) Leaf from a severe 35S::GSF-TM transgenic plant (top) that showed a compact and curled phenotype. Leaf from a wild-type plant (WT, bottom) with a normal round shape. Bar = 5 mm. (D) A 42-day-old 35S::GSF-TM plant (right) produced a short inflorescence and internode (arrow), while wild-type plants (WT, left) produced a long inflorescence with normal internode elongation (arrow). Bar = 20 mm. (E) Close-up of the inflorescence (arrow) with the floral buds (fb) of a 35S::GSF-TM plant from (D). Bar = 2 mm. (F) Close-up of the epidermal cells (approximately 80 µm in length) in the inflorescence of a 35S::GSF-TM plant from (D). Bar = 100 µm. (G) Close-up of the epidermal cells (approximately 220 µm in length) in the inflorescence of a wild-type (WT) plant from (D). Bar = 100 µm. (H) A 56-day-old 35S::GSF-TM plant produced a short inflorescence, internode and compacted flower clusters (arrow). Bar = 10 mm. (I) Close-up of the flower buds (fb) and mature flowers (mf) with short flower organs in the inflorescence of a 35S::GSF-TM plant from (H). Bar = 2 mm. (J) Close-up of a mature flower with short sepal (s), petal (p) and stamen (st) of a 35S::GSF-TM plant from (I). Bar = 0.5 mm.
Gibberellin (GA) biosynthesis genes were downregulated and gibberellin deactivation genes were upregulated in 35S::GSF-TM transgenic Arabidopsis
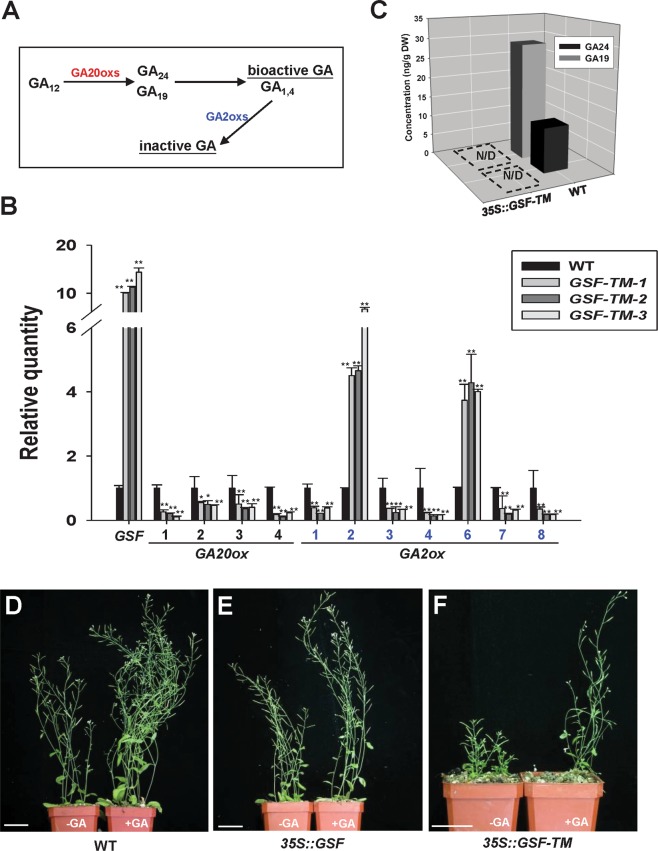
It is interesting to note that the dwarfism phenotype observed in 35S::GSF-TM transgenic plants is similar to that of the mutants deficient in GA. It was known that gibberellin metabolic genes, such as GA20-oxidases (GA20oxs) and GA2-oxidases (GA2oxs), are involved in the late steps in the metabolic pathway5. The GA20oxs participated in the biosynthesis of the major bioactive GAs, while GA2oxs is important to convert GA into inactive GAs1 (Fig. 5A).
Figure 5.
Transcript levels of GA metabolic pathway genes and the external application of GA in 35S::GSF-TM transgenic plants. (A) Schematic diagram for the gibberellin biosynthetic pathway. GA20-oxidases (GA20oxs) participate in the biosynthesis of the major bioactive GAs (GA1 and GA4) from the intermediates, such as GA19 and GA24. GA2-oxidases (GA2oxs) are involved in the late steps in the metabolic pathway to convert bioactive GAs into inactive GAs. (B) Detection of the expression for GSF, GA20oxs (1–4) and GA2oxs (1,2,3,4,6,7,8) in one wild-type (WT) plant and three severe (GSF-TM-1, 2, 3) 35S::GSF-TM transgenic Arabidopsis plants using real-time quantitative RT-PCR. The expression level relative to wild-type plants is presented. Error bars represent standard deviation. The asterisks indicate a significant difference from the wild type (WT) value (*P ≤ 0.05, **P ≤ 0.01) by Student’s T-test. (C) Detection of the concentrations of GA19 and GA24, intermediates in a bioactive GA biosynthesis pathway in wild-type (WT) and 35S::GSF-TM Arabidopsis. N/D indicates that a signal was not detected in the samples. (D–F) External supply of GA in the wild-type (D), 35S::GSF (E) and 35S::GSF-TM (F) plants. The shoot elongation was clearly observed in the wild-type (D, right) and 35S::GSF plants (E, right) after GA treatment. The significant elongation of the inflorescence (F, right) was also observed in the GA-treated 35S::GSF-TM plants. Bar = 35 mm.
To determine the profile of the gibberellin metabolic genes in 35S::GSF-TM plants, total RNA was extracted from the whole plant of the 35S::GSF-TM plants, and the expression of the genes involved in GA biosynthesis was analyzed using real-time quantitative RT-PCR analysis. As expected, the GSF mRNA was significantly upregulated in the severe-dwarf 35S::GSF-TM transgenic plants (Fig. 5B). The expression level of four GA20oxs (1–4) examined was all significantly downregulated (Fig. 5B). Among seven GA2oxs examined, GA2oxs2 and GA2oxs6 were significantly upregulated (Fig. 5B). These results strongly suggest that the similar GA-deficient phenotype in the 35S::GSF-TM transgenic plants is correlated with the altered expression of the genes that participate in GA biosynthesis.
GAs are reduced in 35S::GSF-TM Arabidopsis and dwarfism can be rescued by the external application of GA
Since the GA biosynthetic GA20ox genes were downregulated, while the GA deactivation GA2ox2/6 genes were upregulated in 35S::GSF-TM Arabidopsis, it is interesting to examine whether the levels of GAs were affected in 35S::GSF-TM Arabidopsis. To answer this question, the concentrations of GA19 and GA24, which are two intermediates in the bioactive GA1 and GA4 biosynthetic pathway (Fig. 5A), were examined in wild-type and 35S::GSF-TM Arabidopsis. The result indicated that both GA19 and GA24 can only be clearly detected in wild-type plants and are undetectable in 35S::GSF-TM Arabidopsis (Fig. 5C). Thus, the dwarfism of 35S::GSF-TM transgenic Arabidopsis is likely to be due to the reduction of the endogenous bioactive GA.
To explore whether an external supply of GA could rescue the dwarfism of the 35S::GSF-TM transgenic plants, GA was applied externally to the plants. As controls, the shoot elongation was clearly observed in the wild-type (Fig. 5D) and the 35S::GSF plants (Fig. 5E) after GA treatment. Similarly, the elongation of the inflorescence was also observed in GA-treated 35S::GSF-TM plants (Fig. 5F, right), which were different from the GA-untreated short inflorescence (Fig. 5F, left). This result indicates that the dwarfism of the 35S::GSF-TM plants was caused by the reduction of the JA level rather than the block of the signal transduction of GA and can be rescued via the external supply of GA.
Ectopic GA20ox1 expression could not rescue the dwarfism in the 35S::GSF-TM plants
Since the expression of GA20ox1, which participates in the biosynthesis of the major bioactive GAs, was repressed by 35S::GSF-TM (Fig. 5B), 35S::GA20ox1/35S::GSF-TM double transgenic Arabidopsis plants were generated, and the phenotype was analyzed to further confirm their relationship. As expected, 35S::GA20ox1 exhibited a phenotype of GA-overproduction50 by producing longer inflorescences than the wild-type Arabidopsis (Fig. S3A). The increasing length of the inflorescences was correlated with the expression level of GA20ox1 (Fig. S3B).
When 35S::GA20ox1 was introduced into 35S::GSF-TM to generate 35S::GA20ox1/35S::GSF-TM double transgenic Arabidopsis, the height and the size of the plants were significantly reduced to a level similar to those of the 35S::GSF-TM plants (Fig. 6A). As expected, the expression of GA20ox1 in these 35S::GA20ox1/35S::GSF-TM plants was higher than that of the wild-type plants (Fig. 6B). However, the expression of GA20ox2 was reduced, and GA2ox2/6 was clearly upregulated by the high level of GSF expression (Fig. 6B). This result indicated that although the ectopic expression of GA20ox1 exhibits a GA-overproduction phenotype, the high level of GSF in 35S::GA20ox1/35S::GSF-TM plants will sequentially convert the GAs into inactive ones by activating the expression of GA2ox2/6, resulting in a dwarfism phenotype. Thus, GA2ox2/6 are the ultimate key factors for the ability of GSF to regulate the GA level in plants.
Figure 6.
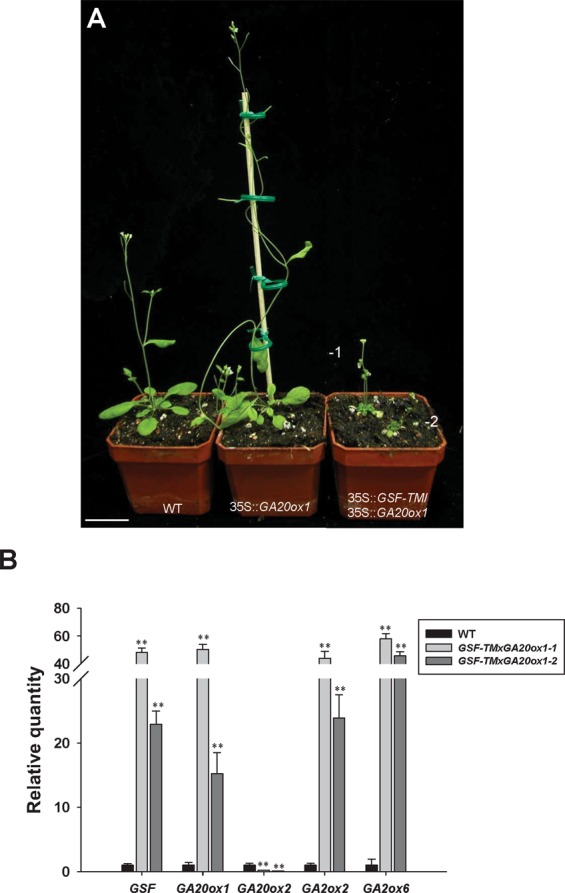
Phenotypic analysis of the 35S::GSF-TM/35S::GA20ox1 Arabidopsis plants. (A) A 32-day-old 35S::GA20ox1 Arabidopsis plant (middle) produced longer inflorescence than a wild-type (WT) (left) Arabidopsis, while two 35S::GSF-TM/35S::GA20ox1 Arabidopsis plants (lines 1 and 2) (right) showed a severe dwarfism phenotype with small compact leaves and a short inflorescence. Bar = 30 mm. (B) Detection of the expression for GSF, GA20ox1, GA20ox2, GA2ox2 and GA2ox6 in one wild-type (WT) plant and two 35S::GSF-TM/35S::GA20ox1 (lines 1 and 2) transgenic Arabidopsis plants using real-time quantitative RT-PCR. The expression level relative to wild-type plants is presented. Error bars represent standard deviation. The asterisks indicate a significant difference from the wild type (WT) value (*P ≤ 0.05, **P ≤ 0.01) by Student’s T-test.
To confirm the important effect of GA2ox2 in GA regulation, 35S::GA2ox2 transgenic Arabidopsis plants were generated, and the phenotype analyzed. The result showed that a similar GA-deficiency dwarfism mutant phenotype, as seen in the 35S::GSF-TM plants, was observed in 35S::GA2ox2 Arabidopsis (Fig. S4A–D). The severity of the dwarfism clearly correlated with the expression level of GA2ox2 (Fig. S4E).
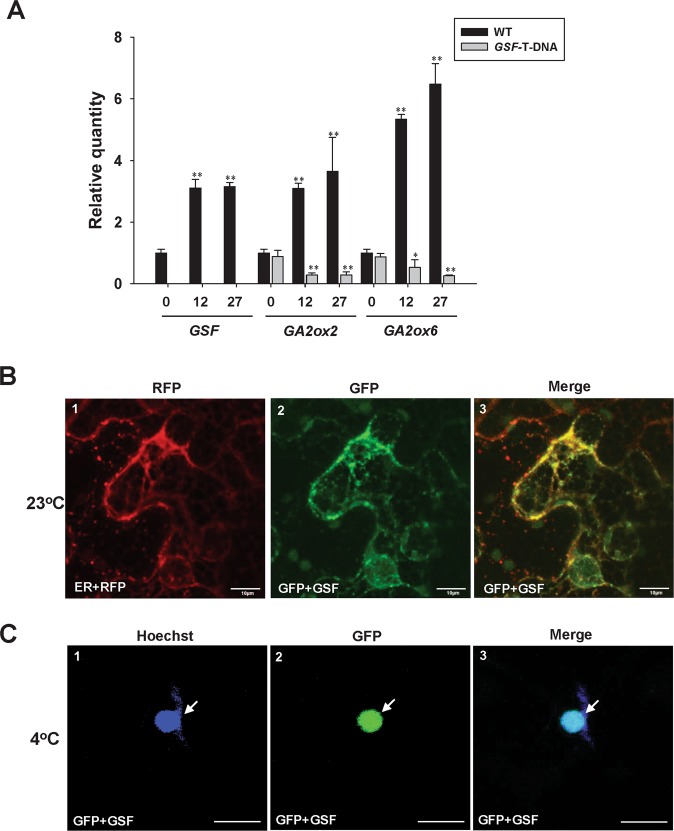
Cold stress induced the translocation of GSF from the ER to the nucleus and affected the expression of GA2oxs
It has been reported that the expression of GSF was specifically upregulated in wild-type Arabidopsis by cold stress (4 °C)32. Interestingly, it has also been demonstrated that the reduction of the GA levels retarded plant growth in response to several abiotic stresses, such as cold stress7.
To explore the relationship between cold stress and GSF-GA, 14-day-old wild-type Arabidopsis seedlings were exposed to 4 °C over a period of 12 and 27 h, respectively, and the gene expression was analyzed. The result indicated that the transcript level of GSF was gradually upregulated after 12-27-h treatment (Fig. 7A). When GA2ox2 and 6, were examined, their expression was constantly induced after cold treatment (Fig. 7A). In contrast, the expression of GA20oxs (1–4) was significantly down-regulated after 27-h treatment (Fig. S5). When the GA2ox2 and 6 were examined in the GSF T-DNA insertion mutants (SALK_022174), which showed completely abolished expression of GSF (Fig. 7A), both GA2ox2 and 6 expression was significantly down-regulated compared to that in wild-type plants (Fig. 7A) and was constantly reduced after cold treatment (Fig. 7A). These results revealed that GSF is likely to be involved in the response to cold stress by regulating the GA level primarily through the activation of GA2ox2/6 expression. Without GSF function, GA2ox2/6 expression was not induced in GSF T-DNA insertion mutants.
Figure 7.
Detection of the expression for GSF and the GA metabolic pathway genes and the cellular localization of GSF under cold treatment. (A) Detection of the expression for GSF, GA2ox2 and GA2ox6 using real-time quantitative RT-PCR for 14-day-old wild-type Arabidopsis and GSF T-DNA mutants (SALK_022174) after exposure to 4 °C over a period of 12 and 27 hours, respectively. For the detection of GSF expression in SALK_022174 mutant, the primers pair F-1 (GSF qRT for-1) and R-1 (GSF qRT rev-1) (Supplementary Table S1), which located in the two sides of the T-DNA insertion (Fig. S6), were used. The expression level relative to wild-type plants is presented. Error bars represent standard deviation. The asterisks indicate a significant difference from the untreated wild type (WT, time 0) value (*P ≤ 0.05, **P ≤ 0.01) by Student’s T-test. (B) Agrobacterium-mediated transient expression of GFP + GSF and ER + RFP in the epidermal cells of N. benthamiana at 23 °C. A GFP + GSF fusion protein (−2) accumulated in the ER where the RFP was localized (−1). A merged fluorescence image of (−1, −2) in (−3) showing the similar localization of GFP + GSF and ER + RFP. Scale bars: 10 μm. (C) Agrobacterium-mediated transient expression of GFP + GSF in the epidermal cells of N. benthamiana at 23 °C for 2 days and exposed at 4 °C (cold stress) for 6 hours. GFP + GSF fusion protein (−2) accumulated in the nucleus (arrow) where blue DNA-staining by Hoechst (arrow) was localized (−1). A merged fluorescence image of (−1, −2) in (−3) showing the similar localization (arrow) of GFP + GSF and blue DNA-staining (Hoechst). Scale bars: 10 μm.
Our data showed that GSF requires translocation from the ER to the nucleus to perform its functions (Figs. 3 and 4). To identify whether cold stress influences this GSF translocation, GFP + GSF and ER-RFP were coexpressed in tobacco leaves, and their fluorescence images were analyzed after cold (4 °C) treatment. The fluorescent images of GFP + GSF fusion proteins were localized in ER and absent in the nuclei at 23 °C (Fig. 7B). In contrast, the visualization of GFP fluorescence merged with the blue fluorescence of Hoechst (a dye for DNA staining) in some cells revealed that the GFP + GSF fusion proteins were clearly located in the nucleus during 4 °C treatment (Fig. 7C). Thus, we confirmed that the plants respond to cold stress by inducing the translocation of GSF from the ER into the nucleus, which will cause the suppression of the GA level and retard plant growth.
35S::GSF-TM transgenic plants exhibit tolerance to drought stress
It has been reported that the expression of GSF was also up-regulated in wild-type Arabidopsis by drought stress32. To explore the relationship between drought stress and GSF/GA, four-week-old wild-type Arabidopsis seedlings were exposed to drought conditions by removing the plants from the soil and exposing them directly to the air for a period of 1 and 2 h, respectively, and the gene expression was analyzed. The results indicated that the transcript level of GSF, GA2ox2 and 6 was clearly up-regulated after 1- and 2-h treatments (Fig. 8A). These results revealed that GSF is also likely to be involved in the response to drought stress by regulating the GA level primarily through the activation of GA2ox2/6 expression.
Figure 8.
Phenotypic analysis of the 35S::GSF-TM and GSF T-DNA mutant Arabidopsis plants under drought treatment. (A) Detection of the expression for GSF, GA2ox2 and GA2ox6 using real-time quantitative RT-PCR on 21-day-old wild-type Arabidopsis after pulling the plant from the soil and exposing directly to air for 1 and 2 hours, respectively. The expression level relative to untreated control plants is presented. Error bars represent standard deviation. The asterisks indicate a significant difference from the wild type (WT) value (*P ≤ 0.05, **P ≤ 0.01) by Student’s T-test. (B) 28-day-old wild-type plant (left) showed a wilt phenotype and a more severe wilt phenotype was observed in GSF T-DNA insertion mutant (middle), while the 35S::GSF-TM plants (right) still grew normally without showing signs of wilt after growth under drought conditions (without irrigation) for 12 days. Bar = 30 mm. (C) When the plants in (B) were further irrigated for 5 days, a clearly recovery from wilt phenotype was observed for wild-type plant (left) whereas a severe wilt phenotype was still observed in GSF T-DNA insertion mutant (middle). Bar = 30 mm. (D) 40-day-old GA-treated wild-type plants (−2) and GSF T-DNA insertion mutant (−4), which showed shoot elongation, and GA-nontreated wild-type plants (−1) and GSF T-DNA insertion mutant (−3) showed a similar wilt phenotype after growth at drought condition for 12 days. 40-day-old GA-treated 35S::GSF-TM plants with shoot elongation showed a wilt phenotype (−6), while the GA-untreated 35S::GSF-TM plants (−5) still grew normally without showing signs of wilt after growth under drought conditions for 12 days. Bar = 30 mm.
We further test the relationship among GSF, GA and drought tolerance in wild-type, 35S::GSF-TM and GSF T-DNA mutant plants. When four weeks old wild-type Arabidopsis were grown at drought condition (without irrigation) for 12 days, a clearly wilt phenotype was observed (Fig. 8B, left). Interestingly, a more severe wilt phenotype was observed in GSF T-DNA insertion mutant (Fig. 8B, middle) which was indistinguishable from wild-type plants when grown in normal condition. By contrast, 35S::GSF-TM plants still grew normally without showing sign of wilt (Fig. 8B, right). When the plants in Fig. 8B were further irrigated for 5 days, a clearly recovery from wilt phenotype was observed for wild-type plant (Fig. 8C, left). In contrast, a severe wilt phenotype was still observed in GSF T-DNA insertion mutant (Fig. 8C, middle). These results revealed that GSF is likely to be involved in the tolerance to drought stress by activating the GA2ox2/6 expression. The tolerance to drought was clearly reduced in GSF T-DNA insertion mutants due to the abolishment of the GSF function.
When GA-treated wild-type plants which showed shoot elongation were grown at drought condition for 12 days, similar wilt phenotype was observed in these GA-treated (Fig. 8D–2) as in un-treated wild-type plants (Fig. 8D–1). Similar result was observed in GA-treated (Fig. 8D–4) as in un-treated GSF T-DNA insertion mutants (Fig. 8D–3). Interestingly, the tolerance to drought as seen in GA-untreated 35S::GSF-TM plants (Fig. 8D–5) was significantly altered in GA-treated 35S::GSF-TM plants (Fig. 8D–6) which showed wilt phenotype after 12 days grown at drought condition. This result supported that the low level of GA in 35S::GSF-TM plants could enhance the drought tolerance which was reduced after externally applying GA.
Discussion
Plant-specific NAC proteins are commonly identified in various plant species, suggesting that they participate in the regulation of many aspects of plant development. In this study, the NAC-like gene GSF was characterized and functionally analyzed in Arabidopsis. GSF acts as a transcriptional activator based on the activation assay in yeast. The transacting domain was found to be allocated in the C-terminal half of the GSF proteins, since GSF1–160 (truncated C terminal half) completely abolished the activation function, while the GSF161–521 (truncated N terminal half) remained as active as the full-length GSF. The nature of GSF as a transcriptional activator was further supported by the result that a similar altered dwarfism phenotype was observed in 35S::GSF-TM and 35S::GSF-TM + VP16 transgenic Arabidopsis.
It has been reported that a group of membrane-bound NAC transcription factors (NTLs) are transported into the nucleus to regulate the expression of downstream genes after their release from the ER31. Not surprisingly, we found that only GFP + GSF-TM fusion proteins accumulate in the nuclei of the cells, while the GFP + GSF fusion proteins accumulated in the ER and were absent in the nuclei. Thus, it is reasonable to propose that GSF requires processing from the ER and entry into the nucleus to perform its function. This assumption was further supported by the fact that only 35S::GSF-TM caused an altered phenotype in transgenic Arabidopsis, while 35S::GSF Arabidopsis was phenotypically indistinguishable from the wild-type plants.
To further investigate the function of GSF, the altered phenotype of 35S::GSF-TM Arabidopsis was analyzed. The ectopic expression of GSF-TM caused a dwarfism phenotype with compact and curled rosettes and short inflorescence internodes that were similar to those in the plants with GA-deficiency. Thus, this finding revealed that the function of the GSF gene is likely to be related to the modulation of GA biosynthesis or signal transduction in plants. Several lines of evidence provided additional evidence for the hypothesis that GA biosynthesis rather than the signal transduction of GA was regulated by GSF. First, when the expression of GA20oxs and GA2oxs, which are involved in late steps in the metabolic pathway5 were tested, a clear downregulation of GA20oxs (1–4) and upregulation of GA2oxs2 and GA2oxs6 were observed in 35S::GSF-TM Arabidopsis. Since the GA20oxs had participated in the biosynthesis of the major bioactive GAs, while GA2oxs functions to convert GA into inactive GAs1, the repression of GA20oxs (1–4) and activation of GA2oxs (2, 6) are expected to decrease the level of the bioactive GAs in 35S::GSF-TM Arabidopsis. Interestingly, we found that 35S::GA20ox1 could not rescue the dwarfism of the 35S::GSF-TM plants. This indicated that the products of GA-overproduction by 35S::GA20ox1 will be sequentially converted into inactive GAs by the high activity of GA2ox2/6 in the 35S::GA20ox1/35S::GSF-TM double transgenic plants. The second line of evidence results from the almost undetectable concentrations of GA19 and GA24, two intermediates in the bioactive GA biosynthesis pathway, in 35S::GSF-TM Arabidopsis. Third, an external supply of GA rescued the dwarfism of the 35S::GSF-TM transgenic plants. Thus, the dwarfism observed in 35S::GSF-TM transgenic Arabidopsis is likely to be caused by the reduction of endogenous bioactive GA due to the high level of functional GSF-TM in the nucleus.
The next question is what mechanisms GSF participated in during the regulation of GA activity. It is worth noting that the reduction of the GA levels retarded plant growth in response to several abiotic stresses7. For example, the overexpression of DDF1 causes dwarfism in Arabidopsis by upregulating the expression of GA2ox7 and reducing the contents of bioactive GA under high-salinity stress11. It has been reported that the level of several GA2oxs was upregulated, while GA20oxs was downregulated in response to cold stress9,12. However, the mechanisms of the regulation of the GA level during cold or drought stress remained to be investigated. Interestingly, GSF has also been reported32 and was demonstrated in this study to be upregulated in wild-type Arabidopsis by cold and drought stresses. Thus, GSF is likely to be a link between GSF and GA-related cold/drought response. This assumption was clearly supported by the results that GSF can be processed from the ER and enter the nucleus under cold treatment, as well as the exhibition of the tolerance to drought stress by the 35S::GSF-TM transgenic plants. All these results indicated that GSF functions to reduce the GA level in response to the cold/drought stresses in plants.
As illustrated in Fig. 9, our results reveal a possible model for the interaction of the GSF and GA in the regulation of plant growth and development. In Arabidopsis, the GSF proteins targeted to the ER membrane through the transmembrane motif (Fig. 3) when grown under normal conditions. Abiotic stresses, such as cold/drought, not only activated the GSF expression but also significantly enhanced the processing of GSF by a protease from the ER membrane and its entry into the nucleus to perform its function. This effect repressed GA20oxs and activated GA2oxs expression, eventually suppressing the biosynthesis of bioactive form of GA and retarding the plant growth in response to stress. The ectopic expression of GSF-TM continuously enhanced the reduction of the GA level during all of the stages of plant growth and resulted in a dwarfism phenotype as seen in our results. Thus, our discovery uncovered the involvement of the GSF gene in a mechanism that regulated the GA level during cold or drought stress.
Figure 9.
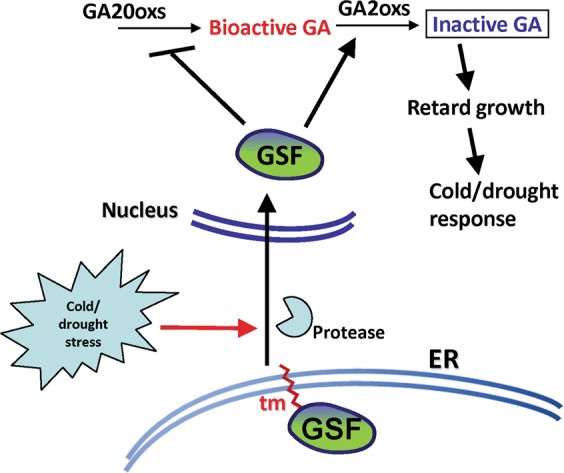
Model for the function of GSF in regulating gibberellin biosynthesis in response to cold/drought stresses in Arabidopsis. In the wild-type plants, the GSP proteins are localized at the membrane of ER through the transmembrane motif (tm) when grown under normal conditions. The cold/drought stresses activate the GSF expression and enhanced the processing of GSF by protease from the ER membrane, and it will enter the nucleus. The functional form of GSF-TM will repress GA20oxs and activate GA2oxs expression to convert the bioactive form of GA to the inactive form, resulting in the retardation of the plant growth in response to stress. 35S::GSF-TM suppressed the bioactive form of the GA level during the whole plant development and resulted in a dwarfism phenotype.
Materials and Methods
Plant materials and growth conditions
The T-DNA insertion mutants of GSF (SALK_022174) Arabidopsis seeds were obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH, USA. The seeds for Arabidopsis were sterilized and placed on agar plates containing 1/2 X MS media51 at 4 °C for 2 days. The seedlings were grown in growth chambers under long-day conditions (16-h light/8-h dark) at 22 °C for 10 days before being transplanted to the soil. The light intensity of the growth chambers was 150 μE m−2 s−1.
Cloning of Arabidopsis GSF cDNA
The Arabidopsis GSF (At1g34190), containing four exons and three introns, was identified on chromosome one. A cDNA containing an open reading frame of GSF was amplified by RT-PCR using the 5′ primer, GSF-for and the 3′ primer, GSF-rev. cDNA truncated with the transmembrane motif region of GSF-TM was amplified by RT-PCR using the 5′ primer, GSF-for, and the 3′ primer, GSF-TM-rev. All of the primers contained the generated KpnI recognition site (5′-GGTACC-3′) and the SacI recognition site (5′-GAGCTC-3′) to facilitate the cloning of the GSF cDNA. Sequences for the primers are listed in Supplementary Table S1. The amplified KpnI-SacI fragments containing either the full-length or truncated transmembrane motif region for the GSF gene was cloned into the linker region in the binary vector pBImGFP2 (CHY Lab, Taichung, Taiwan, unpublished) under the control of the cauliflower mosaic virus (CaMV) 35S-Ω promoter (35S::GSF and 35S::GSF-TM) and used for plant transformations.
Construction of the GSF-TM + VP16 construct
To generate the GSF-TM + VP16 construct, the cDNA for GSF-TM was obtained by PCR amplification using the GSF-for-KpnI and GSF-TM primers that contained the KpnI recognition sites to facilitate the cloning of the cDNA. The PCR fragment containing the GSF-TM was cloned into the pEpyon-3bK plasmid29 in front of the VP16-AD sequence and under the control of the CaMV 35S promoter and was used for plant transformation. The sequences for the primers are listed in the Supplementary Table S1.
GSF::GUS fusion construct
To generate the GSF::GUS construct, the GSF promoter (2.2 kb) was obtained by PCR amplification from the genomic DNA using the GSF-Pro-for and GSF-Pro-rev primers and cloned into the pGEMT easy vector (Promega, Madison, WI, USA). This 2.2 kb fragment for the GSF promoter was subcloned into the linker region before the β-Glucuronidase (GUS) coding region in the binary vector pBI101 (CLONTECH, Palo Alto, CA, USA). The primers contained the generated PstI (5′-CTCCAG-3′) and XbaI (5′-GGATCC-3′) recognition sites to facilitate the cloning of the promoter. The sequences for the primers are listed in Supplementary Table S1.
Construction of the 35S::GA2ox2 construct
To generate the 35S::GA2ox2 construct, the cDNA for GA2ox2 was obtained by PCR amplification using the AtGA2ox2 for PstI and AtGA2ox2 rev SalI primers that contained the generated PstI recognition site (5′-CTGCAG-3′) and SalI recognition site (5′-GTCGAC-3′) to facilitate the cloning of the cDNA. The PCR fragment containing the GA2ox2 was cloned into the linker region in the binary vector pEpyon-32K (CHY Lab, Taichung, Taiwan, unpublished) under the control of cauliflower mosaic virus (CaMV) 35S-Ω promoter (35S::GA2ox2) and used for plant transformation. The sequences for the primers are listed in the Supplementary Table S1.
Construction of the 35S::GA20ox1 construct
To generate the 35S::GA20ox1 construct, the cDNA for GA20ox1 was obtained by PCR amplification using the 5′ primer, At4g25420 (GA20ox1)-for and the 3′ primer, At4g25420 (GA20ox1)-rev primers that contained the generated KpnI recognition site (5′-GGTACC-3′) and SacI recognition site (5′-GAGCTC-3′) to facilitate the cloning of the cDNA. The PCR fragment containing the GA20ox1 was cloned into the linker region in the binary vector pEpyon-32K (CHY Lab, Taichung, Taiwan, unpublished) under the control of the cauliflower mosaic virus (CaMV) 35S-Ω promoter (35S::GA20ox1) and used for plant transformation. To generate 35S::GA20ox1/35S::GSF-TM Arabidopsis, 35S::GA20ox1 Arabidopsis (homozygote) was transformed with 35S::GSF-TM to generate 35S::GA20ox1/35S::GSF-TM Arabidopsis. The sequences for the primers are listed in the Supplementary Table S1.
GSF transcriptional activation assay in yeast
The full length and various truncated cDNA fragments for GSF were amplified using RT-PCR: GAL4BD-GSF1~557 (primer pair: GSF-for-atg-Y + GSF N-557 rev), GAL4BD-GSF1~521 (primer pair: GSF-for-atg-Y + GSF N-521 rev), GAL4BD-GSF161~521 (primer pair: GSF N-161 for + GSF N-521 rev), GAL4BD-GSF1~160 (primer pair: GSF-for-atg-Y + GSF N-160 rev) and a positive control GAL4AD (primer pair: GAL4 AD for + GAL4 AD rev). The sequences for the primers are listed in Supplementary Table S1. These amplified fragments were integrated into the NdeI–SalI site of pGBKT7 (CLONTECH, Palo Alto, CA, USA), containing the GAL4 DNA-binding domain, to produce the fusion proteins GAL4BD-GSF1~557, GAL4BD-GSF1~521, GAL4BD-GSF161~521 and GAL4BD-GSF1~160. The pGBKT7 harboring different fusion proteins was introduced into the yeast strain AH109, which contained four reporter genes (lacZ, His3, ADE2 and MEL1). The transformed yeast cells were selected using synthetic dropout (SD) plates lacking tryptophan at 30 °C for 3 days. Simultaneously, the transformed yeast cells were grown on SD plates lacking tryptophan, histidine and adenine at 30 °C for 3 days and used to quantify the β-galactosidase activity as described previously52.
Real-time PCR analysis
For real-time quantitative RT-PCR, total RNA was isolated from wild-type (WT) and transgenic plants and used as templates. The transcript levels for each genes were determined using three biological replicates and normalized against UBQ10. The RT-PCR reaction was performed on an MJ Opticon system (MJ Research, Waltham, MA) using a SYBER Green Real-time PCR Master Mix (TOYOBO Co., LTD.). The amplification condition was 95 °C for 10 minutes, followed by 40 cycles of amplification (95 °C for 15 seconds, 58 °C for 15 seconds, 72 °C for 30 seconds followed by plate reading) and melting (50–95 °C with plate readings every 1 °C). Sequences for the primers used for real-time quantitative RT-PCR for GSF, GA20ox1, GA20ox2, GA20ox3, GA20ox4, GA2ox1, GA2ox2, GA2ox3, GA2ox4, GA2ox6, GA2ox7 and GA2ox8 are listed in Supplementary Table S1. The housekeeping gene UBQ10 was used as a normalization control with the following primers: RT-UBQ10-F and RT-UBQ10-4-2. All the experiments were repeated at least twice. The data were analyzed using the Gene Expression Macro software (Version 1.1, Bio-Rad).
Plant transformation and transgenic plant analysis
Constructs generated in this study were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis plants using the floral dip method as previously described53. Transformants that survived in the media containing kanamycin (50 µg mL−1) were verified using RT-PCR analysis.
Histochemical GUS assay
Histochemical staining was performed using the standard method described previously54,55.
Scanning electron microscopy (SEM)
Scanning electron microscopy was performed as described previously52,56. Samples were fixed in 2% glutaraldehyde in 25 mM sodium phosphate buffer (pH 6.8) at 4 °C overnight. After dehydration in a graded ethanol series, the specimens were dried to their critical point in liquid CO2. The dried materials were mounted and coated with gold-palladium in a JBS sputter-coater (model 5150). Specimens were examined using a Field emission scanning electron microscope (JEOL JSM-6700F, Japan) with an accelerating voltage of 15 kV.
Construction of the GFP + GSF and GFP + GSF-TM constructs
For the localization assay of the GSF protein, GFP + GSF and GFP + GSF-TM constructs were constructed. The GFP cDNA without a stop codon was amplified from a pBI-mGFP2 expression vector, while connecting 30 base pairs that encode a 5X Gly-Ala linker at the 3′ end of GFP using two step PCR57. The short oligonucleotide fragment of GFP-Gly-Ala was amplified using the 5′ primer FmGFP5L-for (KpnI) and the inner 3′ primer FmGFP5L-rve I. The full oligonucleotide fragment of GFP-Gly-Ala was amplified using the 5′ primer FmGFP5L-for (KpnI) and the inner 3′ primer FmGFP5L-rev II (HindIII). The GSF or GSF-TM cDNA with the stop codon was in frame to the 3′ end of GFP-Gly-Ala, respectively, using GSF-for-atg (HindIII) and fuse-GSF-rev for GSF, 5′ primer GSF-for-atg (HindIII) and the 3′ primer GSF-TM-rev (SacI) for GSF-TM. The sequences for the primers are listed in Supplementary Table S1. ER-RFP, which contained the ER marker fused with the RFP fluorescent protein, was obtained from the ABRC (clone name: ER-RK).
Transient expression assay of Nicotiana benthamiana
Fully expanded young leaves from 4-week-old Nicotiana benthamiana plants were infiltrated with the Agrobacterium tumefaciens strain C58C1 that contained the 35S::GSF + GFP or 35S::GSF-TM + GFP fusions as previously described58. The signal in the infiltrated leaves was observed using an Olympus FV1000 confocal microscope (Olympus FV1000, Tokyo, Japan). Hoechst 33342 (0.02 mM) that was excited at 405 nm with the laser; the GFP was excited at 488 nm, and the RFP was excited at 543 nm.
Quantification of GA intermediates level
To measure the GA intermediates at the GA19 and GA24 level, 0.5 g frozen plant sample of Arabidopsis was sent to the Plant Biotechnology Institute (PBI), National Research Council (NRC), Canada for the additional extraction, purification and quantification of the GA intermediates (https://www.nrc-cnrc.gc.ca/eng/solutions/advisory/plant_hormone.html).
Application of GA3
To determine whether the plants were responsive to gibberellin, a modified method described previously59 was applied by spraying 100 µM GA3 (Sigma) dissolved in 100% absolute ethanol to the plant grown on soil.
Supplementary information
Acknowledgements
This work was supported by grants to C-H Y from the Ministry of Science and Technology, Taiwan, ROC, grant number: MOST 103-2313-B-005-001-MY3 and MOST 106-2321-B-005-010. This work was also financially supported (in part) by the Advanced Plant Biotechnology Center from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Author contributions
C.-H.Y. developed the overall strategy, designed experiments and coordinated the project. H.-I.C. performed most of the experiments and analyzed the data. P.-F.L. performed part of gene expression experiments. C.-H.Y. prepared and revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-55429-8.
References
- 1.Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 2.Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants. Theor. Appl. Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- 3.Hedden P, Phillips AL. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/S1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 4.MacMillan J. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 2002;20:387–442. doi: 10.1007/s003440010038. [DOI] [PubMed] [Google Scholar]
- 5.Sakamoto T, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colebrook EH, Thomas SG, Phillips AL, Hedden P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014;217:67–75. doi: 10.1242/jeb.089938. [DOI] [PubMed] [Google Scholar]
- 8.Skirycz A, Inzé D. More from less: plant growth under limited water. Curr. Opin. Biotechnol. 2010;21:197–203. doi: 10.1016/j.copbio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Achard P, et al. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell. 2008;20:2117–2129. doi: 10.1105/tpc.108.058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004;37:720–729. doi: 10.1111/j.1365-313X.2003.01998.x. [DOI] [PubMed] [Google Scholar]
- 11.Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008;56:613–626. doi: 10.1111/j.1365-313X.2008.03627.x. [DOI] [PubMed] [Google Scholar]
- 12.Kilian J, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 13.Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The No Apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/S0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 14.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen MK, et al. The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochemical Journal. 2010;426:183–196. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q, Wang Q, Xiong L, Lou Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein & cell. 2011;2:55–63. doi: 10.1007/s13238-011-1010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ooka H, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 18.Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;128:1127–1135. doi: 10.1242/dev.128.7.1127. [DOI] [PubMed] [Google Scholar]
- 20.Baurle I, Laux T. Apical meristem: the plant’s fountain of youth. BioEssays. 2003;25:961–970. doi: 10.1002/bies.10341. [DOI] [PubMed] [Google Scholar]
- 21.Sablowski RWM, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/S0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 22.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes and Development. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal. 2006;46:601–612. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- 24.Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314:1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiology. 2005;138:803–818. doi: 10.1104/pp.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes and Development. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih CF, Hsu WH, Peng YJ, Yang CH. The NAC-like gene ANTHER INDEHISCENCE FACTOR acts as a repressor that controls anther dehiscence by regulating genes in jasmonate biosynthesis pathway in Arabidopsis. J. Exp. Bot. 2014;65:621–639. doi: 10.1093/jxb/ert412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Z, et al. KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis. Nature Plants. 2018 doi: 10.1038/s41477-018-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YS, et al. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. 2006;18:3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SY, et al. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic acids research. 2007;35(1):203–213. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SG, Lee AK, Yoon HK, Park CM. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 2008;55:77–88. doi: 10.1111/j.1365-313X.2008.03493.x. [DOI] [PubMed] [Google Scholar]
- 34.Tran LS, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought responsive cis-element in the EARLY RESPONSIVE TO DEHYDRATION STRESS 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He XJ, et al. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. The Plant Journal. 2005;44:903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 36.Safrany J, et al. Identification of a novel cis‐regulatory element for UV‐B‐induced transcription in Arabidopsis. Plant J. 2008;54:402–414. doi: 10.1111/j.1365-313X.2008.03435.x. [DOI] [PubMed] [Google Scholar]
- 37.Sakuraba Y, Kim YS, Han SH, Lee BD, Paek NC. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell. 2015;27:1771–1787. doi: 10.1105/tpc.15.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Seo PJ, Lee HJ, Park CM. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought induced leaf senescence in Arabidopsis. Plant J. 2012;70:831–844. doi: 10.1111/j.1365-313X.2012.04932.x. [DOI] [PubMed] [Google Scholar]
- 39.Seo PJ, et al. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010;61:661–671. doi: 10.1111/j.1365-313X.2009.04091.x. [DOI] [PubMed] [Google Scholar]
- 40.Yoon HK, Kim SG, Kim SY, Park CM. Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cell. 2008;25:438–445. [PubMed] [Google Scholar]
- 41.Morishita T, et al. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009;50:2210–2222. doi: 10.1093/pcp/pcp159. [DOI] [PubMed] [Google Scholar]
- 42.Yang ZT, et al. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014;10:e1004243. doi: 10.1371/journal.pgen.1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Frontiers in Plant Science. 2015;6:902. doi: 10.3389/fpls.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng S, et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell. 2013;25:3450–3471. doi: 10.1105/tpc.113.113985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikuchi K, et al. Molecular analysis of the NAC gene family in rice. Molecular and General Genetics. 2000;262:1047–1051. doi: 10.1007/PL00008647. [DOI] [PubMed] [Google Scholar]
- 46.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 47.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen MK, et al. The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J. 2011;68:168–185. doi: 10.1111/j.1365-313X.2011.04677.x. [DOI] [PubMed] [Google Scholar]
- 49.Peng YJ, et al. A RING-Type E3 ligase controls anther dehiscence by activating the jasmonate biosynthetic pathway gene DEFECTIVE IN ANTHER DEHISCENCE1 in Arabidopsis. Plant J. 2013;74:310–327. doi: 10.1111/tpj.12122. [DOI] [PubMed] [Google Scholar]
- 50.Huang S, et al. Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 1998;118:773–781. doi: 10.1104/pp.118.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–476. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 52.Chang YY, et al. Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 2010;152:837–853. doi: 10.1104/pp.109.147116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clough S, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 54.Jefferson RA, Kavanagh TA, Bevan M. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chou ML, Haung MD, Yang CH. EMF interact with late-flowering genes in regulating floral initiation genes during shoot development in Arabidopsis. Plant Cell Physiol. 2001;42:499–507. doi: 10.1093/pcp/pce062. [DOI] [PubMed] [Google Scholar]
- 56.Hsu HF, Yang CH. An orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant Cell Physiol. 2002;43:1198–1209. doi: 10.1093/pcp/pcf143. [DOI] [PubMed] [Google Scholar]
- 57.Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- 58.Wydro M, Kozubek E, Lehman P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotioana benthamiana. Acta Biochimica Polonica. 2006;53:289–298. doi: 10.18388/abp.2006_3341. [DOI] [PubMed] [Google Scholar]
- 59.Rieu I, et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008;53:488–504. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.