Abstract
Small molecule cross-linkers are invaluable for probing biomolecular interactions and for cross-linking mass spectrometry (CXMS) in addressing large protein complexes and intrinsically disordered proteins. Existing chemical cross-linkers target only a small selection of amino acid residues, limiting the number and type of cross-links, while conventional photocross-linkers target virtually all residues non-selectively, complicating data analysis. Here we report photocaged quinone methide (PQM)-based cross-linkers that are able to multitarget nine nucleophilic residues through specific Michael addition. In addition to Asp, Glu, Lys, Ser, Thr, and Tyr, PQM cross-linkers notably cross-linked Gln, Arg, and Asn hitherto untargetable by existing chemical cross-linkers, markedly increasing the number of residues targetable with a single cross-linker. Such multiplicity of cross-links will increase the abundance of cross-linked peptides for CXMS identification and afford ample constraints to facilitate structural deciphering. PQM cross-linkers were used in vitro, in E. coli, and in mammalian cells to cross-link dimeric proteins and endogenous membrane receptors. The cross-linker NHQM could directly cross-link proteins to DNA, for which few cross-linkers exist. The photoactivatable and multitargeting reactivity of these PQM cross-linkers will substantially enhance chemical cross-linking based technologies for studies of protein-protein and protein-DNA networks and for structural biology.
Keywords: quinone methide, photo-cross-linking, cross-linker, DNA-protein interaction, protein-protein interaction
Graphical Abstract
Small molecule cross-linkers have been invaluable for studying biomolecular interactions. An emerging technology for protein interaction and structural biology is the cross-linking mass spectrometry (CXMS), which analyzes proteins cross-linked by small molecule cross-linkers with tandem mass spectrometry, affording identities and distance constraints of cross-linked residues. [1] [2] It has been increasingly used to probe protein interaction networks and to derive tertiary structural information of large protein complexes and intrinsically disordered proteins, complementing X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy. Small molecule cross-linkers are also used for cross-linking DNA to proteins, which is a critical step for chromatin immunoprecipitation (ChIP), a method widely used for mapping DNA-protein interactions across eukaryotic genomes in cells, tissues, and whole organisms. [3] Chemical cross-linkers react with target residues specifically. For instance, the most commonly used cross-linkers contain homobifunctional N-hydroxysuccinimidyl (NHS) esters to react with Lys side chain or N-terminal amine. Cross-linkers targeting Cys, Glu, Asp, and His have also been developed. [4] [5] [6] [7] Expanding the repertoire of residues targetable and enabling multi-targeting ability of chemical cross-linkers would increase the number and types of constraints obtainable from CXMS. Recently, we reported a plant-and-cast strategy enabling cross-link of Lys residues with His, Ser, Thr, Tyr, and Lys side chains through sulfur-fluoride exchange reaction, showcasing the feasibility of targeting multiple residues via new chemistry. [8] However, a variety of amino acid residues remain untargetable. On the other hand, conventional photocross-linkers (such as diazirines, azides, and benzophenones) target virtually all residues non-selectively, [9] [10] but such nonspecific chemistry often results in highly complex cross-linked products, dramatically complicating MS data analysis. In addition, excessive cross-linking may artificially distort protein tertiary structures. [1] [2] For DNA-protein cross-linking, formaldehyde remains the primary reagent despite its short cross-link distance (~2 Å) and limited reactivity with few residues. [3] Therefore, new small molecule cross-linkers able to multi-target different amino acid residues, especially those inaccessible to date, with specific chemistry would be valuable for realizing the full potential of cross-linking based technologies.
We report here a new series of photocaged quinone methide (PQM)-based small molecule cross-linkers, which integrate the advantages of chemical cross-linkers (i.e., specific chemical reactivity) and conventional photocross-linkers (i.e., photo-controllability and thus potential for spatiotemporal resolution) for cross-linking biomolecules. These PQM cross-linkers were able to multitarget nine different amino acid residues through Michael addition in protein-protein cross-linking. We demonstrated their use for cross-linking proteins in vitro, in E. coli, and in mammalian cells, and for cross-linking proteins with DNA as well.
Quinone methides (QM) are efficient Michael acceptors for nucleophiles and have been versatile for chemical synthesis and chemical biology. [11] [12] [13] [14] We recently genetically encoded an unnatural amino acid FnbY containing a photocaged para-QM into proteins and showed its specific reactivity toward multiple nucleophilic residues placed in proximity. [15] The less reactive quinone has also been explored for protein cross-linking [16] [17] [18]. We therefore reasoned that integration of photocaged highly reactive QM into small molecule cross-linkers would enable the desired multi-targeting ability through specific Michael addition chemistry, as well as photocontrolled reactivity for spatiotemporal resolution.
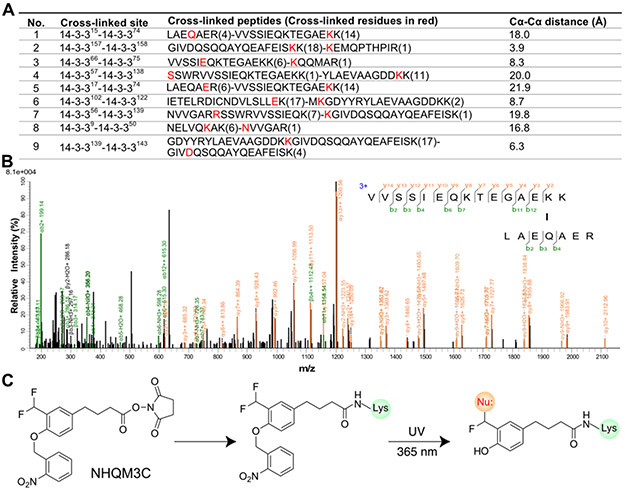
We initially designed a heterobifunctional cross-linker NHQM containing an NHS ester and a photocaged ortho-QM (o-QM) (Fig. 1A). Two fluorines were installed at the methyl group ortho to the phenolic hydroxyl, which was photocaged by an o-nitrobenzyl group. When added to proteins, the NHS ester will rapidly react with exposed Lys residues, planting the photocaged o-QM next to residues on interaction partners. UV release of o-nitrobenzyl induces elimination of a fluoride ion to generate o-QM, which will then cross-link with nearby nucleophilic side chains (Fig. 1A). To test this design, we incubated a homodimeric eukaryotic regulatory protein 14-3-3 (Fig. 1B) [19] with 1 mM NHQM followed by UV illumination (λ = 365 nm). Cross-linked dimer of 14-3-3 was detected on SDS-PAGE only with NHQM addition and UV illumination (Fig. 1C), suggesting that cross-linking was through photo-released QM. Cross-linking of 14-3-3 could be observed on SDS-PAGE as short as 1 min of UV exposure, and increased with UV exposure time (Fig. 1D, Fig S1), indicating that NHQM-mediated cross-linking could be controlled by light with temporal resolution.
Figure 1.
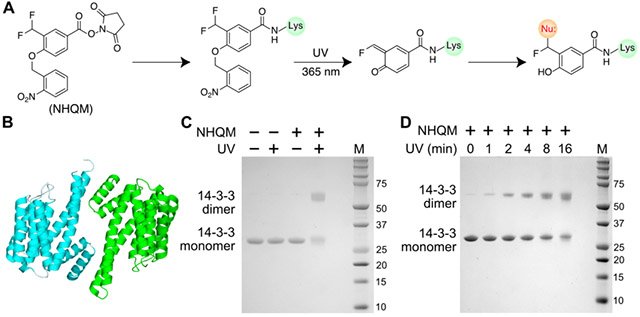
NHQM-mediated protein cross-linking in vitro. A) Scheme showing NHQM structure and photo-controlled cross-linking mechanism. Upon UV activation, a highly reactive ortho-QM was generated capable of reacting with multiple nucleophilic residues. B) Protein 14-3-3 forms a homodimer (PDB code: 4N7Y). C-D) SDS-PAGE analysis of NHQM-mediated dimeric cross-linking of 14-3-3 (C) with different UV exposure time (D).
NHQM cross-linking results in a short and rigid linkage, which requires close contact of target residues for effective cross-linking. We thus tested whether this feature could be used to readily determine protein dimerization in vitro and in cell lysate. Three mutations (12LAE14 →12QQR14) of 14-3-3 disrupt the dimer interface to form a monomer, leading to a distinct function from WT 14-3-3. [20] [21] Previously the 14-3-3(QQR) mutant was isolated from cells and then characterized as a monomer using size exclusion chromatography. This in vitro process is tedious, and the results may not represent what occur in the cellular setting. We first treated purified 14-3-3 WT and QQR mutant with various amounts of NHQM and illuminated with UV light (Fig. 2A, 2B). Cross-linked dimer of WT 14-3-3 increased with higher NHQM concentration on SDS-PAGE, while 14-3-3 QQR remained a monomer (Fig. 2C, 2D). We then applied NHQM directly to lysates of E. coli cells expressing either 14-3-3 WT or QQR mutant. Western blot analysis of the cell lysates clearly showed that WT 14-3-3 was cross-linked as dimer but not the QQR mutant (Fig. 2E, 2F). These results show that NHQM-mediated cross-linking could be a facile method for determining protein interactions.
Figure 2.
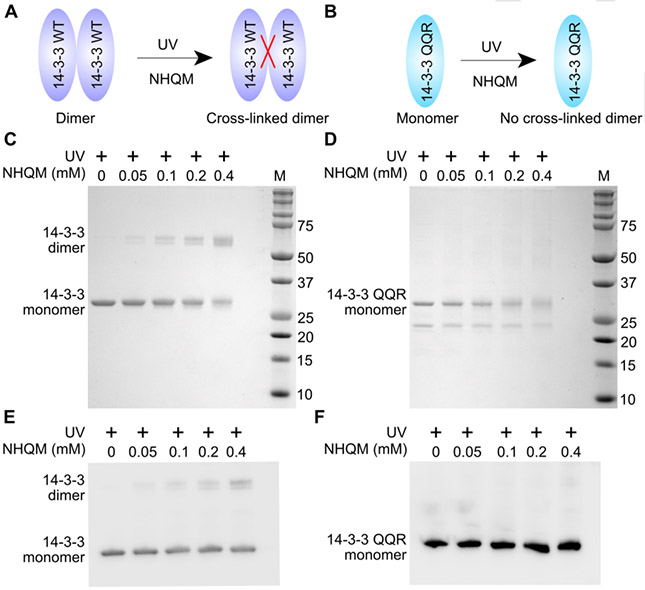
Facile differentiation of dimer and monomer via NHQM cross-linking in vitro and in cell lysate. A-B) NHQM should cross-link the WT 14-3-3 dimer into a covalent dimeric form but not the monomeric QQR mutant, which can be readily distinguished on SDS-PAGE or Western blot. C-D) SDS-PAGE analysis of NHQM-mediated in vitro cross-linking WT 14-3-3 (C) and 14-3-3(QQR) mutant (D). E-F) Western blot analysis of NHQM-mediated cross-linking of WT 14-3-3 (E) and 14-3-3(QQR) mutant (F) in E. coli cell lysate. 14-3-3 was detected by an anti-Hisx6 antibody.
To identify which residues NHQM could cross-link, we analyzed the cross-linked 14-3-3 protein with tandem mass spectrometry. Nine pairs of cross-linked peptides were identified, showing that NHQM cross-linked Lys of one peptide with Gln, Lys, Glu, Ser, Arg, Asn, and Asp of the other peptide (Fig. 3A, 3B, S2). These residues have nucleophilic side chains, consistent with the reaction mechanism of o-QM. The maximum Cα-Cα distance of NHQM cross-linker is ~22 Å considering amino acid side chain length plus the covalently connected cross-linking agent. The Cα-Cα distances for the majority of cross-linked residues fall within this range, with only one slightly exceeding the limit and measured at 26.6 Å (Fig. 3A, S3). To increase cross-linker’s flexibility, we also synthesized NHQM3C, adding three methylenes to the spacer of NHQM (Fig. 3C). When applied to the purified WT 14-3-3, NHQM3C also cross-linked 14-3-3 into covalent dimers upon UV activation (Fig. S4). Tandem mass spectrometric analysis of the cross-linked 14-3-3 identified six pairs of cross-linked peptides, with Lys cross-linked with nucleophilic residues including Gln, Glu, Thr, and Tyr (Fig. S5). The cross-linking pattern of each anchor Lys was different for NHQM and NHQM3C, and the Cα-Cα distances for NHQM3C were longer than those for NHQM, reflecting their differences in rigidity and length. Taken together, these results show that NHQM and NHQM3C expanded the repertoire of natural residues targetable with small molecule cross-linkers to a total of nine nucleophilic residues.
Figure 3.
NHQM and NHQM3C multi-target a total of nine nucleophilic residues in protein cross-linking. A) CXMS analysis of WT 14-3-3 cross-linked by NHQM. B) Representative tandem mass spectrum for cross-linked peptides showing cross-linking of Lys with Gln. Others are shown in Figures S2, S3, and S5. C) Structure of NHQM3C and its cross-linking mechanism.
We next explored potential applications of NHQM in various biological settings, starting with cross-link of interacting proteins in E. coli cell lysate. Thioredoxin (Trx) is a ubiquitous oxidoreductase regulating cellular redox through interacting with various proteins. [22] [23] We expressed His-tagged Trx in E. coli, and then applied NHQM to the cell lysate followed by UV activation. Western blot analysis of the treated cell lysates showed that many endogenous proteins were cross-linked to Trx (Fig. S6). Together with cross-linking of 14-3-3 in E. coli cell lysate (Fig. 2E), these results show that NHQM is compatible for cross-linking in E. coli cell lysate. We also attempted applying NHQM directly to E. coli cells to cross-link intracellularly expressed 14-3-3, but did not detect any 14-3-3 dimeric form on Western blot, possibly because NHQM could not enter E. coli cells efficiently.
We then tested NHQM’s cross-linking ability in mammalian cells. We incubated NHQM with live mammalian cells expressing the dimeric glutathione transferase (GST)[9] or 14-3-3, and detected cross-linking of both proteins in dimeric form in response to light, albeit in low efficiency (< 10%, Fig. S7-S8), suggesting NHQM could enter mammalian cells but at low efficiency. We thus tested using NHQM to cross-link proteins on mammalian cell surface. NHQM was applied to HEK293T cells, which were transfected with plasmids to overexpress the epidermal growth factor receptor (EGFR). EGFR dimer was detected on Western blot in response to UV and NHQM, demonstrating NHQM cross-linking of the receptor with optical control (Fig. S9). We further explored whether NHQM could probe endogenous mammalian proteins at physiologically relevant conditions. The human mammary epithelial MCF10A cells were cultured and treated with or without NHQM followed with light activation. EGFR dimerization on MCF10A cells was detected only in the presence of UV and NHQM (Fig. 4).
Figure 4.
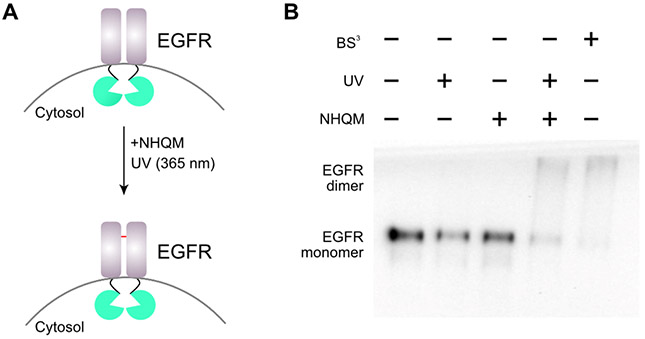
Photo-controlled dimeric cross-linking of EGFR on mammalian cell surface by NHQM. A) Scheme showing the process. B) Western blot analysis of EGFR on MCF10A cells after indicated treatment. The amine-reactive cross-linker bissulfosuccinimidyl suberate (BS3) was used as a positive control.
To enable efficient cross-linking inside cells, we designed a homobifunctional cross-linker HoQM, with photocaged o-QM at both ends (Fig. 5A). We reasoned that replacing NHS ester with the photocaged QM would remove chemical reactivity of the cross-linker prior to photoactivation, thus allowing it to enter cells without causing cytotoxicity. Additionally, QM at both ends could increase the diversity of cross-link types since QM reacts with more residues than NHS ester. As expected, HoQM cross-linked 14-3-3 into dimer in vitro upon light activation with comparable efficiency as NHQM (Fig. S10). We then incubated HoQM with E. coli cells for 1 h, and found it cross-linked the intracellularly expressed 14-3-3 in dimeric form upon UV illumination (365 nm) of intact cells for 15 min (Fig. 5B), which was infeasible with NHQM. In addition, HoQM (0.6 mM) was incubated with HEK293T cells for 4 to 8 h without apparent toxicity; subsequent exposure of these cells to UV light (365 nm, 10 min) resulted in dimeric cross-linking of intracellular 14-3-3 protein (Figure 5C). These results demonstrated HoQM’s ability to cross-link proteins inside live bacterial and mammalian cells.
Figure 5.
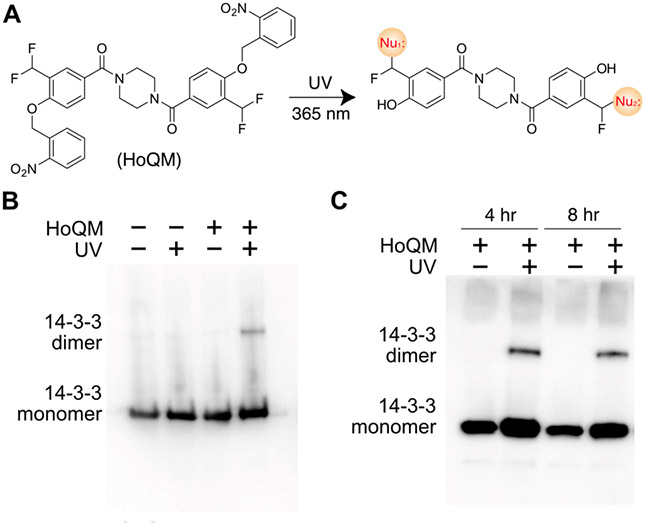
HoQM-mediated protein cross-linking in E. coli and mammalian cells. A) Scheme showing HoQM structure and cross-linking mechanism. B-C) Western blot analysis of HoQM-mediated 14-3-3 cross-linking in E. coli cells (B) and in mammalian cells (C).
Reagents to cross-link proteins with DNA remain sparse, because reactivities of most cross-linkers favor protein-protein over protein-DNA cross-linking. Formaldehyde is the primary reagent but fails to cross-link proteins not in close contact (2 Å) with DNA. [24] [25] [3] QM can alkylate deoxynucleosides efficiently. [26] We thus reasoned that NHQM should be able to cross-link protein with DNA. To test this possibility, we first incubated a single stranded DNA binding protein (SSB) [27] with a short DNA oligo of repeats 19(3x) or ATC(4x) which are reported to interact with SSB. [28] [29] The SSB-DNA complex was treated with NHQM and UV activation, and analyzed with denaturing TBE-urea gel shift assay. An upshifted band was only observed for the SSB-19(3x) or SSB-ATC(4x) complex treated with both NHQM and UV light (Figure 6A), indicating successful protein-DNA cross-linking mediated by NHQM. As expected, the NHS ester-based amine reactive BS3 cross-linker did not result in SSB-DNA cross-linking. We further investigated protein-DNA cross-linking by incubating a natural single stranded circular viral DNA M13mp18 with the SSB protein. [30] As shown in Figure 6B (also Fig. S11), the M13mp18 control DNA run as two isoforms in the denatured TBE-Urea gel due to its large size, a major form remained in the well and a minor form migrating in the gel. Only in the presence of both NHQM treatment and UV activation was the minor form of M13mp18 upshifted into the well, suggesting it was cross-linked with SSB.
Figure 6.
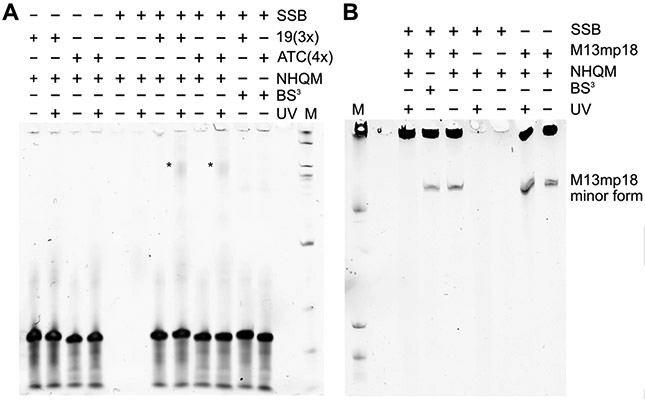
NHQM mediated protein-DNA cross-linking. A) Denaturing TBE-urea gel shift assay of NHQM-mediated cross-linking of the SSB protein with 19(3x) or ATC(4x) DNA. The upshifted cross-linked DNA band is indicated by a *. B) Denaturing TBE-urea gel shift assay of NHQM-mediated cross-linking of the SSB protein with the viral M13mp18 DNA. After cross-linking the minor form of M13mp18 was upshifted into the well and thus disappeared from the original position. Cross-linker BS3 served as a negative control.
In summary, we developed hetero- and homo-bifunctional photocaged quinone methide (PQM) cross-linkers for probing protein-protein and protein-DNA complexes. These PQM cross-linkers enable cross-linking Lys with a total of nine nucleophilic amino acid residues. In addition to Asp, Glu, Lys, Ser, Thr, and Tyr, PQM cross-linkers notably cross-linked Gln, Arg, and Asn residues hitherto untargetable by existing chemical cross-linkers, dramatically increasing the number of residues targetable with a single cross-linker [31]. Such multiplicity of cross-links will significantly increase the abundance of cross-linked peptides for CXMS identification and afford ample constraints to facilitate structural modeling of challenging large protein complexes and intrinsically disordered proteins. PQM should also react with Cys residue [11] [13] [15], although it was not identified here possibly due to poor accessibility of Cys in the tested protein. PQM cross-linkers are photo-controlled, which can be utilized to gain spatiotemporal resolution. We demonstrated their usage in vitro, as well as in E. coli and mammalian cells to cross-link dimeric proteins and endogenous membrane integral receptors. We also showed that NHQM could directly cross-link proteins to DNA, for which few cross-linkers exist. Future efforts will add sulfite or carboxyl group to increase PQM water solubility, which should further enhance its crosslinking efficiency. We expect that the photoactivatable and multitargeting reactivity of these PQM cross-linkers will be valuable for investigation of protein-protein and protein-DNA networks and structural biology through chemical cross-linking.
Supplementary Material
Acknowledgements
We thank Dr. Natalia Jura for MCF10A cells and Devan Diwanji for helpful discussion. S.R. acknowledge the support of NIH (R01GM121607); L.W. acknowledges the support of the NIH (R01GM118384, RF1MH114079).
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Yu C, Huang L, Anal. Chem 2017, 90, 144–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sinz A, Angew. Chem. Int. Ed. Engl 2018, 57, 6390–6396. [DOI] [PubMed] [Google Scholar]
- [3].Hoffman EA, Frey BL, Smith LM, Auble DT, J. Biol. Chem 2015, 290, 26404–26411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ, Methods Enzymol. 2007, 423, 25–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Novak P, Kruppa GH, 2008, 14, 355–365. [DOI] [PubMed] [Google Scholar]
- [6].Leitner A, Joachimiak LA, Unverdorben P, Walzthoeni T, Frydman J, Förster F, Aebersold R, Proc. Natl. Acad. Sci. U. S. A 2014, 111, 9455–9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gutierrez CB, Yu C, Novitsky EJ, Huszagh AS, Rychnovsky SD, Huang L, Anal. Chem 2016, 88, 8315–8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yang B, Wu H, Schnier PD, Liu Y, Liu J, Wang N, DeGrado WF, Wang L, Proc. Natl. Acad. Sci. U. S. A 2018, 115, 11162–11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Suchanek M, Suchanek M, Radzikowska A, Radzikowska A, Thiele C, Thiele C, Nat. Methods 2005, 2, 261–267. [DOI] [PubMed] [Google Scholar]
- [10].Lössl P, Sinz A, Methods Mol. Biol 2016, 1394, 109–127. [DOI] [PubMed] [Google Scholar]
- [11].Toteva MM, Richard JP, Adv Phys Org Chem 2011, 45, 39–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bai W-J, David JG, Feng Z-G, Weaver MG, Wu K-L, Pettus TRR, Acc. Chem. Res 2014, 47, 3655–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gnaim S, Shabat D, Acc. Chem. Res 2014, 47, 2970–2984. [DOI] [PubMed] [Google Scholar]
- [14].Parra A, Tortosa M, ChemCatChem 2015, 7, 1524–1526. [Google Scholar]
- [15].Liu J, Li S, Aslam NA, Zheng F, Yang B, Cheng R, Wang N, Rozovsky S, Wang PG, Wang Q, et al. , J. Am. Chem. Soc 2019, 141, 9458–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu B, Burdine L, Kodadek T, J. Am. Chem. Soc 2006, 128, 15228–15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang J, Saggiomo V, Velders AH, Cohen Stuart MA, Kamperman M, PloS one 2016, 11, e0166490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bruins JJ, Albada B, van Delft F, Chemistry 2018, 24, 4749–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fu HA, Subramanian RR, Masters SC, Annu. Rev. Pharmacol. Toxicol 2000, 40, 617–647. [DOI] [PubMed] [Google Scholar]
- [20].Sluchanko NN, Sudnitsyna MV, Seit-Nebi AS, Antson AA, Gusev NB, Biochemistry 2011, 50, 9797–9808. [DOI] [PubMed] [Google Scholar]
- [21].Sluchanko NN, Artemova NV, Sudnitsyna MV, Safenkova IV, Antson AA, Levitsky DI, Gusev NB, Biochemistry 2012, 51, 6127–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee S, Kim SM, Lee RT, Antioxid. Redox. Signal 2013, 18, 1165–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang B, Tang S, Ma C, Li ST, Shao GC, Dang B, DeGrado WF, Dong MQ, Wang PG, Ding S, et al. , Nat. Commun 2017, 8, 2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nowak DE, Tian B, Brasier AR, BioTechniques 2005, 39, 715–725. [DOI] [PubMed] [Google Scholar]
- [25].Aoki T, Wolle D, Preger-Ben Noon E, Dai Q, Lai EC, Schedl P, Fly 2014, 8, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pande P, Shearer J, Yang J, Greenberg WA, Rokita SE, J. Am. Chem. Soc 1999, 121, 6773–6779. [Google Scholar]
- [27].Raghunathan S, Kozlov AG, Lohman TM, Waksman G, Nat. Struct. Biol 2000, 7, 648–652. [DOI] [PubMed] [Google Scholar]
- [28].Mitas M, Chock JY, Christy M, Biochem. J 1997, 324 ( Pt 3), 957–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Steen H, Petersen J, Mann M, Jensen ON, Protein Sci. 2001, 10, 1989–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Syvanen AC, Alanen M, Soderlund H, Nucleic Acids Res. 1985, 13, 2789–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].A cross-linker singly targeting Arg was reported while this work was under review: Jones AX, Cao Y, Tang Y-L, Wang J-H, Ding Y-H, Tan H, Chen Z-L, Fang R-Q, Yin J, Chen R-C, Zhu X, She Y, Huang N, Ye K, Sun R-X, He S-M, Lei X, Dong M-Q, Nat. Commun 2019, 10, 3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.