Abstract
Eosinophilic esophagitis (EoE) is a chronic Th2 antigen driven disorder associated with tissue remodeling. Inflammation and remodeling lead to esophageal rigidity, strictures, and dysphagia. TGFβ1 drives esophageal remodeling including epithelial barrier dysfunction and subepithelial fibrosis. A functional SNP in the TGFβ1 gene that increases its transcription (C-509T) associates with elevated numbers of esophageal TGFβ1 expressing cells. We utilized esophageal biopsies and fibroblasts from TT genotype EoE children to understand if TGFβ1 influenced fibroblast and epithelial cell function in vivo. Genotype TT EoE esophageal fibroblasts had higher baseline TGFβ1, collagen1α1, periostin, and MMP-2 (p<.05) gene expression and distinct contractile properties compared with CC genotype (n=6 subjects per genotype). In vitro TGFβ1 exposure caused greater induction of target gene expression in genotype CC fibroblasts (p<.05). Esophageal biopsies from TT genotype subjects had significantly less epithelial membrane bound E-cadherin (p<.01) and wider cluster distribution at nanometer resolution. TGFβ1 treatment of stratified primary human esophageal epithelial cells and spheroids disrupted transepithelial resistance (p<.001) and E-cadherin localization (p<.0001). A TGFβ1-receptor-I inhibitor improved TGFβ1-mediated E-cadherin mislocalization. These data suggest that EoE severity can depend on genotypic differences that increase in vivo exposure to TGFβ1. TGFβ1 inhibition may be a useful therapy in subsets of EoE patients.
Introduction
Eosinophilic esophagitis (EoE) is an antigen driven chronic Th2 disease of increasing prevalence (1–3). Chronic inflammation causes esophageal remodeling with end organ rigidity, narrowing, and dysmotility with clinical dysphagia and food impaction (4–8). Despite the success of elimination diet and topical esophageal corticosteroid therapies, some patients remain unresponsive or break through standard medical interventions and progress to disease complications (9–12). The disease mechanisms that underlie therapy non-responsive and/or severe EoE are not clearly understood.
Previous studies have demonstrated that there are genetic predispositions to EoE onset. These include specific genotypes or mutations in thymic stromal lymphopoeitin and its (13) receptor(14, 15), calpain 14 (16), SPINK5/7 (17), and TGFβ1 (18, 19). In addition, genotypic variations can associate with patient phenotypes in EoE. The EoE TSLP risk allele can alter patient phenotype and associate with increased EoE food triggers (13). Children who have TGFβ1 genotype TT at the functional promoter single nucleotide polymorphism (SNP) C-509T have increased numbers of TGFβ1 expressing cells in the lamina propria and more severe histologic disease, including fibrosis, especially in the context of food sensitization (20, 21). In asthma, genotype TT at −509 can associate with asthma severity in children living close to freeways, suggesting a gene-environment interaction (22, 23).
Mechanistic studies demonstrate that TT genotype at C-509T increases TGFβ1 gene transcription either through the loss of negative regulation via AP-1 or through positive regulation by YY-1 (24). TGFβ1 positive cells are elevated in EoE subjects, especially in genotype TT, and genetic syndromes that alter the balance of TGFβ1 and its signals in vivo, such as Loeys-Dietz and Marfan’s, are associated with EoE (18–21). These findings suggest that the in vivo balance and timing of TGFβ1 expression is likely critical for proper esophageal function. Indeed, TGFβ1 has a myriad of effects in the EoE esophagus. It causes esophageal epithelial to mesenchymal transition, myofibroblast transformation, myocyte and fibroblast contraction, loss of claudin-7 with epithelial barrier dysfunction, and fibrotic gene expression (25–29). Given these effects, it is clinical impactful to understand if EoE patients who have endogenously elevated TGFβ1 have altered structural cell function. In this scenario, there could be an opportunity for personalized medicine via TGFβ1 altering therapies in subsets of patients. Herein, we utilized esophageal fibroblasts and biopsies from patients of TT and CC genotype at C-509T to understand the potential detrimental effects of increased esophageal TGFβ1 exposure in vivo.
Results
Genotype TT and CC fibroblasts are phenotypically different
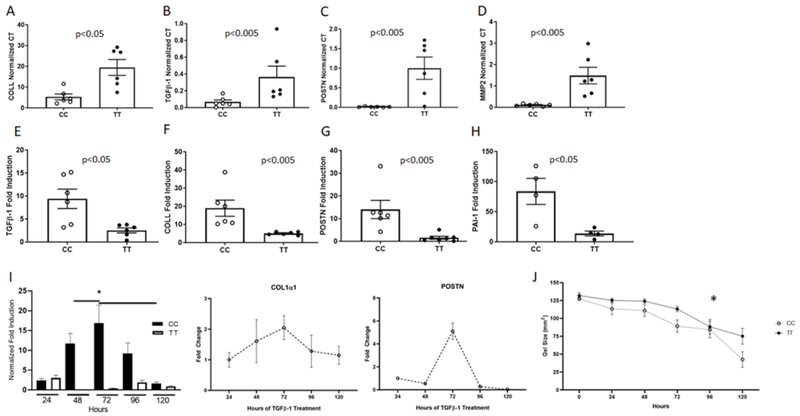
We utilized esophageal fibroblasts from 12 pediatric EoE subjects (6 genotype TT, 6 genotype CC) (see Table 1 for clinical characteristics). We assessed baseline TGFβ1 gene expression in passage and disease activity matched fibroblasts from genotype CC and TT patients. All subjects had active EoE as defined histologically at the time of fibroblast isolation. To understand if TGFβ1 gene expression was higher in genotype TT EoE patient fibroblasts, we compared baseline TGFβ1 mRNA as well as mRNA levels of TGFβ1-target genes. Genotype TT fibroblasts had significantly higher baseline TGFβ1 gene expression as compared with CC genotype fibroblasts (p<.05, Figure 1a). Genotype TT fibroblasts also had significantly higher baseline expression of the TGFβ1 target genes collagen1α1 and MMP2 as compared with genotype CC fibroblasts (p<.05, Figure 1b, c) (30). A number of additional TGFβ1 target genes trended towards significantly higher baseline expression in genotype TT fibroblasts including periostin (31), SERPINE1/Plasminogen Activator Inhibitor-1 (PAI-1) (32), and fibronectin (p=.06 for each). In addition, collagen1α1 protein was elevated in TT fibroblasts on immunoblotting of whole cell extracts (not shown). These data suggest inherent differences in fibrotic gene expression in esophageal fibroblasts from patients with CC versus TT genotype.
Table 1.
Clinical characteristics of patient samples used
| Patient | Gender | Genotype | Peak Eosinophil Count | Experiments used in | |||
|---|---|---|---|---|---|---|---|
| IHC | TIRF | qPCR | Contraction Assays | ||||
| 1 | F | TT | 81 | X | X | ||
| 2 | M | TT | 100 | X | X | ||
| 3 | M | TT | 45 | X | X | ||
| 4 | M | TT | 20 | X | |||
| 5 | M | TT | 75 | X | X | ||
| 6 | M | TT | 62 | X | X | ||
| 7 | M | CC | 17 | X | |||
| 8 | M | CC | 100 | X | X | ||
| 9 | M | CC | 100 | X | X | ||
| 10 | F | CC | 30 | X | X | X | X |
| 11 | M | CC | 175 | X | X | ||
| 12 | M | CC | 50 | X | X | ||
| 13 | F | TT | 18 | X | |||
| 14 | M | TT | 61 | X | |||
| 15 | M | TT | 75 | X | |||
| 16 | F | TT | 68 | X | |||
| 17 | M | TT | 125 | X | X | ||
| 18 | M | TT | 59 | X | X | ||
| 19 | F | TT | 35 | X | |||
| 20 | F | TT | 30 | X | X | ||
| 21 | M | TT | 75 | X | |||
| 22 | M | TT | 195 | X | |||
| 23 | F | CC | 52 | X | |||
| 24 | M | CC | 66 | X | |||
| 25 | M | CC | 29 | X | |||
| 26 | M | CC | 64 | X | |||
| 27 | M | CC | 84 | X | |||
| 28 | M | CC | 31 | X | |||
| 29 | M | CC | 44 | X | |||
| 30 | M | CC | 40 | X | X | ||
| 31 | M | CC | 84 | X | X | ||
Figure 1. Genotype CC and TT fibroblasts have different transcription profiles.
Baseline transcript levels for TGFβ1 (A), collagen1α1 (COLL) (B), periostin (C), and matrix metalloproteinase 2 (MMP-2) (D) in CC and TT primary esophageal fibroblasts matched for passage number, culture conditions, and EoE disease activity. TGFβ1-induced gene expression of TGFβ1 (E), collagen1α1 (COLL) (F), periostin (POSTN) (G), and plasminogen activator inhibitor-1 (PAI-1) (H). All gene expression was normalized to the housekeeping gene ribosomal protein L13a (RPL13a) and shown as normalized Ct (A–D) or as fold induction (E-H) for the matched vehicle treated well. Each data point represents a distinct patient. Each experiment was repeated in biological triplicates on different days and in qPCR technical replicates (2–3). Chronic exposure to TGFβ1 in CC versus TT (representative assay, left panel I) and over time in CC fibroblasts (right panel, I; n=5 different individuals, bars represent SEM). *p<.01. Contraction of TT versus CC fibroblasts done in triplicate wells in esophageal fibroblasts from 10 different individuals (n=5 CC and n=5 TT) (J). *:p<.05
We hypothesized that, due to the chronic elevated local exposure to TGFβ1 in genotype TT patients and the elevated baseline gene expression of a subset of pro-fibrotic genes, esophageal fibroblasts from TT subjects would not be capable of increasing pro-fibrotic gene expression following TGFβ1 treatment to the same extent as esophageal fibroblasts derived from CC genotype subjects. Indeed, TGFβ1 induced significantly higher levels of gene expression of collagen1α1, periostin, PAI-1, and TGFβ1, itself, in genotype CC as compared with TT fibroblasts (p<.05 for all) (Figure 1d–g), suggesting that lower in vivo TGFβ1 exposure allowed for increased TGFβ1 responsivity in vitro. To further demonstrate this point, we treated genotype CC esophageal fibroblasts from EoE subjects with TGFβ1 for up to 120 hours. CC fibroblasts had collagen1α1 induction initially but after 72 hours demonstrated a decrease in their response (Figure 1i). These data are consistent with the hypothesis that tonic in vivo TGFβ1 exposure is a potential mechanism for decreased TT responsivity to exogenous TGFβ1.
In order to assess functional differences in the baseline activity of TT versus CC fibroblasts, we performed contraction and wound healing assays. TT fibroblasts had decreased contractility as compared with genotype CC fibroblasts (Figure 1j). TT fibroblasts also trended towards increased wound healing as compared to genotype CC (Supplemental Figure 1). These data demonstrate that genotype at the TGFβ1 promoter SNP −509 associates with altered fibroblast function beyond transcriptional regulation.
C-509T genotype and TGFβ−1 affect esophageal epithelial barrier function
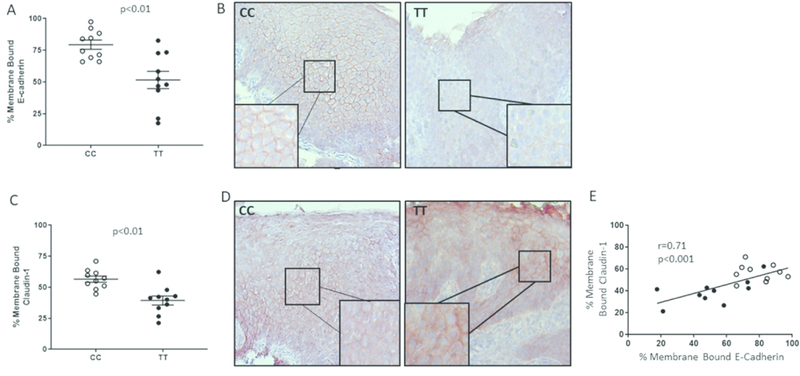
We have reported that TT genotype subjects exhibit more severe epithelial remodeling on routine histology as assessed by scoring epithelial features of basal zone hyperplasia + dilated intercellular spaces + desquamation as compared with genotype CC patients (20). For this reason, we hypothesized that esophageal epithelial barrier protein expression would be more aberrant in TT as compared to CC patients. We stained esophageal biopsies procured during active EoE from subjects of CC and TT genotype for E-cadherin or claudin-1. Quantification of membrane bound E-cadherin demonstrated that TT genotype subjects had less overall E-cadherin expression and significantly lower levels of properly localized E-cadherin as compared with CC genotype subjects (p<.01) (Figure 2a, b). Consistent with the fact that proper claudin-1 localization depends on E-cadherin, there also was significantly less membrane bound claudin-1 in genotype TT as compared with genotype CC subjects (p<.01) (Figure 2c, d). The degree of membrane-bound E-cadherin and claudin-1 expression positively correlated with one another regardless of genotype (r=0.71, p<.001), consistent with their interdependent localization patterns (Figure 2e).
Figure 2. Genotype CC and TT esophageal biopsies have distinct patterns of cell adhesion proteins.
The percentage of cells per total epithelial height that expressed membrane bound E-cadherin-1 (A) or claudin-1 (C) was quantified using immunohistochemistry (red stain) and ImageJ analysis. Representative images of CC and TT subject epithelium are shown (B, D). Each point on the graphs represents the average of 5 different epithelial regions in 10 different subject biopsies with stains repeated on 3 different days. The degree of membrane bound E-cadherin and claudin-1 correlate positively regardless of genotype (closed circles=TT, open circles=CC) (E).
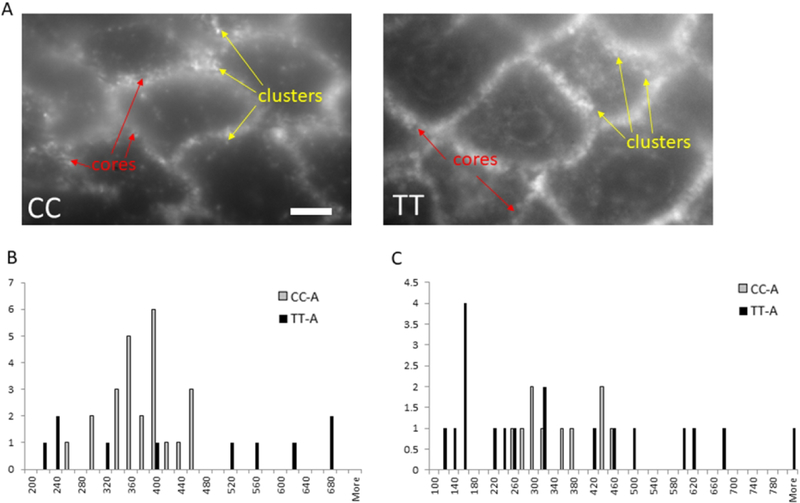
We used 6 active EoE biopsies (n= 3 CC, n= 3 TT) in high resolution TIRF microscopy to understand the distribution of E-cadherin clusters at 180 nm resolution. The TIRF imaging technique allows observation of single molecule fluorescence and determination of cluster sizes at the surfaces and boundaries of cells while minimizing background fluorescence from the rest of the tissue sample. TIRF provides near-field (evanescent wave field) excitation and only a few hundred nanometers of the specimen at the surface of the coverslip contributes to the fluorescent image. The resulting data demonstrated greater cluster size variability in TT genotype subjects as compared with CC subjects. While CC subjects had a tighter distribution of E-cadherin clusters averaging 372 ± 52 nm, TT patient biopsies had a wider distribution of E-cadherin cluster sizes that averaged 446 ± 189 nm (Figure 3a, c). In addition, the size of the central core within an E-cadherin cluster was larger and had a wider size distribution in TT as compared with CC patients (Figure 3b). Together, these data demonstrate substantial differences in E-cadherin cluster structure at the nanometer level dependent on patient genotype.
Figure 3. Total internal reflection fluorescence (TIRF) imaging shows differences in CC and TT E-cadherin.
TIRF high resolution imaging of CC and TT genotype biopsies (A) demonstrates more variability in the central core (B) and cluster (C) sizes in active genotype TT biopsies. Graphs represent quantification using TIRF imaging of core and cluster sizes and distances in at least 20 images (5 to 6 areas per tissue section that had the sharpest focus) from 6 different subjects. Representative image is shown. Scale bar = 5 microns.
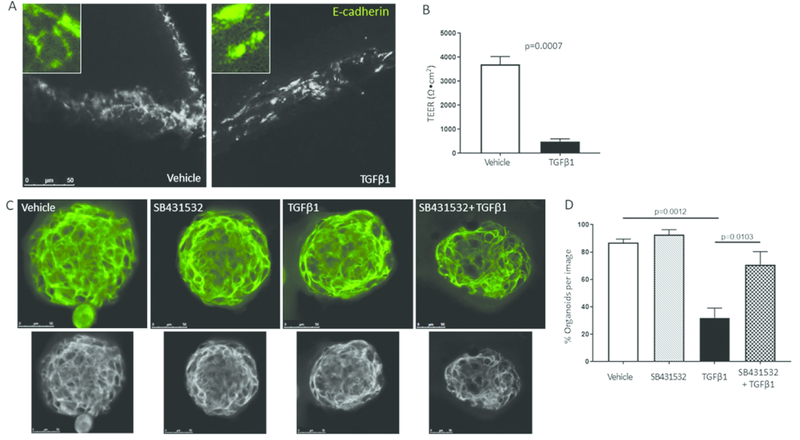
If the TT genotype causes increased local TGFβ1 exposure which in turn alters epithelial barrier protein expression, then treatment of primary esophageal epithelial cells with TGFβ1 should mimic the in vivo state and disrupt E-cadherin localization and epithelial barrier function. We isolated primary esophageal epithelial cells from normal esophagi of organ donors. We cultured these non-transformed human epithelial cells on transwells, stratified them at air-liquid interface until the generated an adequate barrier to electrical current, and then treated the cells with TGFβ1. Treatment with TGFβ1 for 2–7 days caused a significant decrease in transepithelial resistance in TGFβ1 treated versus vehicle treated wells (p<.001). E-cadherin localization pattern also was disrupted in the stratified epithelial cells such that untreated control wells had membrane localized E-cadherin while TGFβ1 treated wells had a disrupted pattern of E-cadherin localization (Figure 4a, b).
Figure 4. TGFβ1 alters epithelial resistance and E-cadherin.
Representative images of vehicle and TGFβ1 treated epithelial cells on transwells (A) and the effect on transepithelial electrical resistance (TEER) (B). Quantification of the percent of spheroids with properly localized E-cadherin during treatment with vehicle, the ALKV inhibitor SB431532, TGFβ1 or TGFβ1+SB431532 (C). Representative image of spheroids treated with vehicle, SB431532, TGFβ1 or both. Images converted to gray scale below. Each image is a single slice of a 3-dimensional confocal image (D). Spheroids were treated in 2–3 replicate wells using 2 different donors on 2 different days. TEER experiments were repeated 3 times on different days in triplicate wells using epithelial cells from 2 different donors. Bars represent means and standard error from a representative experiment.
Culture of primary esophageal epithelial cells in matrigel induces the formation of 3-dimensional esophageal epithelial cell spheroids, reminiscent of the complex structure of stratified epithelium (33, 34). To understand the effects of TGFβ1 on E-cadherin expression in epithelial cells in a 3-dimensional architecture, we treated spheroids with TGFβ1 or vehicle. TGFβ1 caused mislocalization and decreased membrane expression of E-cadherin in esophageal epithelial spheroids (p<.0001 as compared to vehicle) (Figure 4c, d). Concurrent treatment with the TGFβ1 receptor I/AlkV inhibitor SB431532 largely reversed the effect of TGFβ1 on E-cadherin localization (Figure 4c, d), suggesting that the effects of TGFβ1 were occurring via a canonical signaling pathway. Together our data suggest a genotype-influenced pathway whereby TT genotype fibroblasts and inflammatory cells (29, 35) elevate local TGFβ1 exposure and promote E-cadherin disruption and dysregulated barrier function (Figure 5).
Figure 5. Model of the effects of TT genotype at TGFβ1 SNP −509 on EoE remodeling.
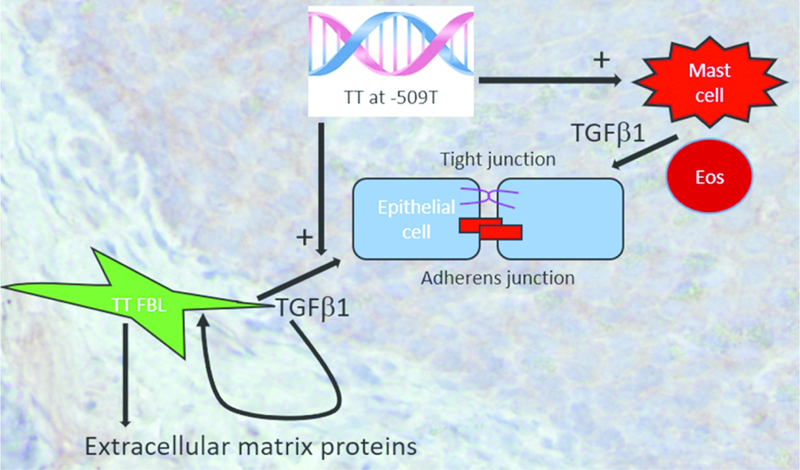
Genotype TT at −509 in the TGFβ1 promoter causes increased TGFβ1 expression from resident esophageal fibroblasts and associates with increased numbers of mast cells which, along with eosinophils, are reservoirs of TGFβ1. TGFβ1 functions in an auto- or paracrine manner on resident fibroblasts to augment production of pro-fibrotic factors such as collagen1α1, especially in genotype TT EoE patients. TGFβ1 perpetuates EoE by causing barrier breakdown of both adherens and tight junctions.
Discussion
There is mounting evidence that EoE is a disease in which pathogenesis largely depends on intact barrier function for disease instigation and propagation.(17, 36, 37) The TT genotype at −509 in the TGFβ1 results in increased local TGFβ1 expression with increased numbers of TGFβ1 positive cells in the active EoE esophagus (24, 38, 39). Herein we demonstrate that fibroblasts from EoE children of the TT genotype transcribe more TGFβ1, that TGFβ1 alters E-cadherin localization and epithelial barrier function, and that children with the TT genotype have altered E-cadherin expression at nanometer resolution in the esophagus.
TGFβ1 has pleotropic effects on esophageal structural cells. It can disrupt esophageal epithelial cell barrier function, claudin-7 expression, and induce epithelial mesenchymal transition (25, 40). In addition, TGFβ1 induces esophageal epithelial, fibroblast, and myocyte contraction along with myofibroblast transformation and smooth muscle cell hypertrophy (27, 28, 41). Herein, we used esophageal fibroblasts derived from genotype TT children to understand intrinsic differences in fibroblast gene expression from CC genotype fibroblasts at baseline and in response to TGFβ1. TT genotype fibroblasts had elevated baseline transcription of TGFβ1 and its target genes collagen1α1 and MMP2. Consistent with the increased TGFβ1 exposure in vivo in TT subjects, CC fibroblasts had a more robust induction of TGFβ1 target gene expression following exposure to the cytokine in vitro. Genotype TT esophageal fibroblasts also demonstrated altered contractile properties, suggesting that they may be more fibrotic but less contractile than genotype CC esophageal fibroblasts. Further, we propose that, in vivo, TT fibroblasts producing more TGFβ1 would cause a more profound mislocalization of barrier proteins, resulting in a more severe disease phenotype in terms of treatment and histology (Figure 5). As such, EoE states that associate with increased TGFβ1 may predispose to a more severe disease phenotype. We have previously shown this to be the case in terms of tissue histology in food-sensitized, TT-genotype subjects. In addition, it has been demonstrated that EoE patients have mislocalized E-cadherin even while on topical corticosteroids (42). The extent of this adherens junction disruption may be dependent on local TGFβ1 exposure that can vary among patients. Our study does have the limitation that our experiments were not done using epithelial cells from genotype TT versus CC subjects. However, the numbers of epithelial cells isolated per biopsy specimen and the relatively rarity of genotype TT (10–12%) in the population limits our ability to readily complete such studies.
Not all EoE patients follow the same disease course nor do they have equal response to therapy (9, 11, 12, 43, 44). Although most patients have recurrent disease, the time to disease recrudescence can vary substantially among patients and the initial response to therapy is variable. In addition, topical corticosteroids, despite reducing eosinophilic inflammation, may not completely resolve E-cadherin localization or barrier function (42). An incompletely healed or more severely damaged epithelial barrier may predispose to harder to treat disease or EoE that is prone to recurrence. Recent data demonstrated that SPINK7 dysregulation unleashes a spontaneous alteration in esophageal barrier function with poor E-cadherin localization and expression (17). These data support a critical role of E-cadherin function in the esophageal mucosa and in EoE. Further insults to E-cadherin localization would only worsen this process in patients with dysregulated TGFβ1 expression, such as those with Loeys-Dietz, Marfans, or TT genotype at C-509T. It is interesting to suggest that a sub-phenotype of EoE patients may benefit from the concurrent use of a localized TGFβ1 inhibiting therapy. Given the ability to treat the esophagus topically, such an approach may diminish unwanted side effects of global TGFβ1 inhibition. It would be of great utility to understand if therapies that are primarily aimed at protecting epithelial barrier breakdown and normalizing E-cadherin localization are efficacious in EoE. Further, it would be of interest to understand if TGFβ1 blockade during a progressed disease state could help decrease fibrotic gene expression and its clinical sequelae. Our data suggests that such a therapeutic approach may be worth considering for clinical trials.
Methods
EoE subjects/biopsies and immunohistochemistry
Using our University of California, San Diego (UCSD)/Rady Children’s Hospital, San Diego (RCHSD) EoE database, we selected a cohort of EoE subjects who had active EoE, defined as ≥15 eosinophils per hpf. Written informed consent/assent was obtained under UCSD/RCHSD IRB approved protocol 091485. Normal esophageal epithelial cells were isolated from cadaveric/organ donor esophagi and was not considered human subjects research but approved under UCSD IRB protocol 130835.
Hematoxylin and eosin stained, formalin fixed, paraffin embedded specimens were scored by a single pathologist (R.N.) using our previously published standardized histology scoring tool (45). The numbers of epithelial eosinophils, the severity of basal zone hyperplasia (scored 0–3), the presence/absence of dilated intercellular spaces (scored 0/1), and the presence/absence of epithelial desquamation (scored 0/1) were quantified. The epithelial remodeling score is a composite index of the latter three epithelial cell features (maximum score of 5).
Paraffin embedded biopsy specimens were processed for immunohistochemistry as previously described.(46) E-cadherin (Abcam, Cambridge, MA, 1:100) and claudin-1 (Abcam, Cambridge, MA, 1:300) staining was quantified using color detection and image analysis (Image to assess cell layers with membrane bound versus cytoplasmic E-cadherin and claudin-1 relative to the total epithelial height. For any stain, 3–5 image fields were captured and the mean value for a given subject was utilized. All images were analyzed under identical light microscopic conditions, including magnification, gain, camera position, and background illumination.
Total Internal Reflection Fluorescence (TIRF) Microscopy
For high contrast images of fluorescently stained tissue, we used inverted Olympus IX83 microscope with a total internal reflection fluorescence objective (UAPON 100X/1.49NA, Olympus), sCMOS camera (ORCA_Flash4.0 V3, Hamamatsu), and 488nm excitation laser (Genesis MX488, Coherent). In order to ensure E-cadherin distribution was being quantified at the same region of the epithelium, tissue was evaluated initially under 10x magnification to identify mid-epithelial areas of flat tissue located close to the coverslip. We then chose sharply in focus parts of tissue areas and measured the full width at half maximum (FWHM) of E-cadherin clusters and the size of the non-fluorescent, dark core within an E-cadherin cluster.
Quantitative PCR and genotyping
Fibroblasts were collected in RNA Stat-60 (Tel Test Inc, Friendswood, Texas) for RT qPCR experiments as previously described.(28) Primer sequences were TGFβ1 forward GGTGGAAACCCA CAACGA AATC, reverse AATTCCCCTCCACGGCTCAAC; collagen I forward CAGCCGCTTCACCTACAGC, reverse TTTTGTATTCAATCACTGTCTTGCC; matrix metalloproteinase 2 (MMP2) forward AGCTCTATGGGGCCTCTCCTGA, reverse TCACCAC GGATCTGAGCGATGC; plasminogen activator inhibitor-1 (PAI-1) forward TGGTTCTGCC CAAGTTCTCCCTG, reverse TGCCACTCTCGTTCACCTCG; periostin forward GTCTTTGAGA CGCTGGAAGG, reverse AGATCCGTGAAGGTGGTTTG; fibronectin forward GGAGAATT CAAGTGTGACCCTCA, reverse TGCCACTGTTCTCCTACGTGG; RPL13A forward CATA GGAAGCTGGGAGCAAG, reverse GCCCTCCAATCAGTCTTCTG.
Genomic DNA was extracted from whole blood, peripheral blood mononuclear cells, or esophageal fibroblasts using QIAmp genomic DNA extraction kit; Genotype at −509 at the TGFβ1 promoter was determined using manufacturer’s instructions for PCR based SNP analysis (Thermo Fisher SNP ID: rs1800469).
Cell culture, epithelial cell spheroids, and treatment
Human organ transplant donor esophagi provided by the National Disease Research Interchange and the Arkansas Regional Organ Recovery Agency were processed to isolate epithelial cells. EoE biopsies were processed to isolate fibroblasts (28). Cell identity was confirmed by proper morphology and cell-relevant gene expression. All fibroblast lines were cultured in SMCM complete (ScienCell, San Diego CA) and matched for passage and disease activity state of the patients (Table 1). Fibroblasts were used at passage 7 +/− 1 passage. Cells were treated with TGFβ1 (10ng/ml) for 48 hours for qPCR experiments and for up to 120 hours with media and TGFβ1 replacement every other day for chronic exposure experiments.
Epithelial cells were isolated from human donor cadaveric esophagi, suspended in matrigel (Corning #356231), and cultured for 5–7 days in a defined medium (advanced DMEM/F12, Thermo Fisher #12634010, HEPES (20 mM, Thermo Fisher # 15630080), glutamine (Thermo Fisher # A1286001), N-2, B-27, N-Acetyl-L-cysteine (500 ng/mL, Sigma-Aldrich #A7250), r-spondin (5 ug/mL), noggin (100 ng/mL, PeproTech #250–38), EGF (500 ug/mL, Thermo Fisher # PHG0311), and rho kinase inhibitor (Y27632,1.2 ug/mL, LC Laboratories #Y-5301) replaced every other day and assessed for growth of epithelial spheroids prior to treatment with TGFβ1 (10ng/ml) and/or ALKV Inhibitor SB431532 (Invivogen # inh-sb43) for 48 hours (33). After treatment, spheroids were immunofluorescently stained for E-Cadherin (Abcam # ab1416) and fluorescent signal was amplified by tyramide signal amplification (Thermo Fisher # T20912). All images were captured using identical brightness, gain, and fluorescence microscope settings.
Contraction Assays
CC and TT patient fibroblasts were cultured in gels consisting of DMEM (Thermo Fisher Scientific # 12100061), sodium bicarbonate (Thermo Fisher Scientific # 25080094), and collagen (Advanced BioMatrix #5005–100ML) and treated with recombinant TGFβ1 (10ng/mL) over 5 days. Images were captured daily on a hemiDoc transilluminator (BioRad Laboratories) and gel area was quantified using ImageJ.
Transepithelial Resistance
Epithelial cells were plated on transwell permeable supports (12 mm insert, 0.4um polyester membrane, Costar, Cambridge, MA) coated with collagen (0.033 mg/ml), human fibronectin (0.01 mg/ml), and BSA (0.02 mg/ml) (47). Resistance was measured daily using a voltohmmeter (World Precision Instruments, Sarasota, FL) and measurements were normalized to the insert area by subtracting the resistance of a blank insert with no cells. Epithelial resistance studies were repeated 3 times on different days each in triplicate wells using esophageal epithelial cells derived from 2 different donors. Cells were treated with recombinant TGFβ1 for 2–7 days.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (San Diego, CA). Statistical analysis of data was performed by Student’s t-test or Mann-Whitney test where appropriate. A Spearman’s coefficient was calculated for correlations. A two tailed p value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank the Aceves laboratory members for technical assistance and the patients and their families for participating in the study.
Funding Sources: NIH/NIAID AI092135 (S.S.A, R.C.K.), NIH/NIAID AI135034 (S.S.A.) NIH/NCRR/NCATS UL1TR000039 (R.C.K.)
Footnotes
COI: None of the authors has a relevant conflict of interest
References
- 1.Dellon ES, Liacouras CA, Molina-Infante J, Furuta GT, Spergel JM, Zevit N, et al. Updated international consensus diagnostic criteria for eosinophilic esophagitis: Proceedings of the AGREE conference. Gastroenterology. 2018. [DOI] [PMC free article] [PubMed]
- 2.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. The Journal of allergy and clinical immunology. 2011;128(1):3–20 e6; quiz 1–2. [DOI] [PubMed] [Google Scholar]
- 3.Dellon ES. Epidemiology of Eosinophilic Esophagitis. Gastroenterology clinics of North America. 2014;43(2):201–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spergel JM, Dellon ES, Liacouras CA, Hirano I, Molina-Infante J, Bredenoord AJ, et al. Summary of the updated international consensus diagnostic criteria for eosinophilic esophagitis: AGREE conference. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2018;121(3):281–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoepfer AM, Safroneeva E, Bussmann C, Kuchen T, Portmann S, Simon HU, et al. Delay in diagnosis of eosinophilic esophagitis increases risk for stricture formation in a time-dependent manner. Gastroenterology. 2013;145(6):1230–6 e1–2. [DOI] [PubMed] [Google Scholar]
- 6.Kwiatek MA, Hirano I, Kahrilas PJ, Rothe J, Luger D, Pandolfino JE. Mechanical properties of the esophagus in eosinophilic esophagitis. Gastroenterology. 2011;140(1):82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicodeme F, Hirano I, Chen J, Robinson K, Lin Z, Xiao Y, et al. Esophageal distensibility as a measure of disease severity in patients with eosinophilic esophagitis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2013;11(9):1101–7 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dellon ES, Kim HP, Sperry SL, Rybnicek DA, Woosley JT, Shaheen NJ. A phenotypic analysis shows that eosinophilic esophagitis is a progressive fibrostenotic disease. Gastrointestinal endoscopy. 2014;79(4):577–85 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greuter T, Bussmann C, Safroneeva E, Schoepfer AM, Biedermann L, Vavricka SR, et al. Long-Term Treatment of Eosinophilic Esophagitis With Swallowed Topical Corticosteroids: Development and Evaluation of a Therapeutic Concept. The American journal of gastroenterology. 2017;112(10):1527–35. [DOI] [PubMed] [Google Scholar]
- 10.Greuter T, Safroneeva E, Bussmann C, Biedermann L, Vavricka SR, Katzka DA, et al. Maintenance Treatment Of Eosinophilic Esophagitis With Swallowed Topical Steroids Alters Disease Course Over A 5-Year Follow-Up Period In Adult Patients. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2018. [DOI] [PubMed]
- 11.Rajan J, Newbury RO, Anilkumar A, Dohil R, Broide DH, Aceves SS. Long-term assessment of esophageal remodeling in patients with pediatric eosinophilic esophagitis treated with topical corticosteroids. The Journal of allergy and clinical immunology. 2016;137(1):147–56 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eluri S, Runge TM, Cotton CC, Burk CM, Wolf WA, Woosley JT, et al. The extremely narrow-caliber esophagus is a treatment-resistant subphenotype of eosinophilic esophagitis. Gastrointestinal endoscopy. 2016;83(6):1142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fahey LM, Chandramouleeswaran PM, Guan S, Benitez AJ, Furuta GT, Aceves SS, et al. Food allergen triggers are increased in children with the TSLP risk allele and eosinophilic esophagitis. Clin Transl Gastroenterol. 2018;9(3):139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nature genetics. 2010;42(4):289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nature genetics. 2014;46(8):895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis BP, Stucke EM, Khorki ME, Litosh VA, Rymer JK, Rochman M, et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 2016;1(4):e86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azouz NP, Ynga-Durand MA, Caldwell JM, Jain A, Rochman M, Fischesser DM, et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Science translational medicine. 2018;10(444). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frischmeyer-Guerrerio PA, Guerrerio AL, Oswald G, Chichester K, Myers L, Halushka MK, et al. TGFbeta receptor mutations impose a strong predisposition for human allergic disease. Science translational medicine. 2013;5(195):195ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abonia JP, Wen T, Stucke EM, Grotjan T, Griffith MS, Kemme KA, et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. The Journal of allergy and clinical immunology. 2013;132(2):378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rawson R, Anilkumar A, Newbury RO, Bafna V, Aquino M, Palmquist J, et al. The TGFbeta1 Promoter SNP C-509T and Food Sensitization Promote Esophageal Remodeling in Pediatric Eosinophilic Esophagitis. PloS one. 2015;10(12):e0144651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aceves SS, Newbury RO, Chen D, Mueller J, Dohil R, Hoffman H, et al. Resolution of remodeling in eosinophilic esophagitis correlates with epithelial response to topical corticosteroids. Allergy. 2010;65(1):109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma S, Raby BA, Hunninghake GM, Soto-Quiros M, Avila L, Murphy AJ, et al. Variants in TGFB1, dust mite exposure, and disease severity in children with asthma. American journal of respiratory and critical care medicine. 2009;179(5):356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salam MT, Gauderman WJ, McConnell R, Lin PC, Gilliland FD. Transforming growth factor- 1 C-509T polymorphism, oxidant stress, and early-onset childhood asthma. American journal of respiratory and critical care medicine. 2007;176(12):1192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, et al. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. American journal of respiratory and critical care medicine. 2004;169(2):214–9. [DOI] [PubMed] [Google Scholar]
- 25.Kagalwalla AF, Akhtar N, Woodruff SA, Rea BA, Masterson JC, Mukkada V, et al. Eosinophilic esophagitis: epithelial mesenchymal transition contributes to esophageal remodeling and reverses with treatment. The Journal of allergy and clinical immunology. 2012;129(5):1387–96 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muir AB, Dods K, Henry SJ, Benitez AJ, Lee D, Whelan KA, et al. Eosinophilic Esophagitis-Associated Chemical and Mechanical Microenvironment Shapes Esophageal Fibroblast Behavior. Journal of pediatric gastroenterology and nutrition. 2016;63(2):200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muir AB, Wang JX, Nakagawa H. Epithelial-stromal crosstalk and fibrosis in eosinophilic esophagitis. Journal of gastroenterology. 2019;54(1):10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beppu LY, Anilkumar AA, Newbury RO, Dohil R, Broide DH, Aceves SS. TGF-beta1-induced phospholamban expression alters esophageal smooth muscle cell contraction in patients with eosinophilic esophagitis. The Journal of allergy and clinical immunology. 2014;134(5):1100–7 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. The Journal of allergy and clinical immunology. 2010;126(6):1198–204 e4. [DOI] [PubMed] [Google Scholar]
- 30.Beppu L, Yang T, Luk M, Newbury RO, Palmquist J, Dohil R, et al. MMPs-2 and −14 are Elevated in Eosinophilic Esophagitis and Reduced Following Topical Corticosteroid Therapy. Journal of pediatric gastroenterology and nutrition. 2014. [DOI] [PMC free article] [PubMed]
- 31.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal immunology. 2008;1(4):289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawson R, Yang T, Newbury RO, Aquino M, Doshi A, Bell B, et al. TGF-beta1-induced PAI-1 contributes to a profibrotic network in patients with eosinophilic esophagitis. The Journal of allergy and clinical immunology. 2016;138(3):791–800 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whelan KA, Muir AB, Nakagawa H. Esophageal 3D Culture Systems as Modeling Tools in Esophageal Epithelial Pathobiology and Personalized Medicine. Cell Mol Gastroenterol Hepatol. 2018;5(4):461–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeWard AD, Cramer J, Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9(2):701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. The Journal of allergy and clinical immunology. 2007;119(1):206–12. [DOI] [PubMed] [Google Scholar]
- 36.Rochman M, Azouz NP, Rothenberg ME. Epithelial origin of eosinophilic esophagitis. The Journal of allergy and clinical immunology. 2018;142(1):10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rochman M, Travers J, Miracle CE, Bedard MC, Wen T, Azouz NP, et al. Profound Loss of Esophageal Tissue Differentiation in Eosinophilic Esophagitis. The Journal of allergy and clinical immunology. 2017. [DOI] [PMC free article] [PubMed]
- 38.Che Z, Zhu X, Yao C, Liu Y, Chen Y, Cao J, et al. The association between the C-509T and T869C polymorphisms of TGF-beta1 gene and the risk of asthma: a meta-analysis. Human immunology. 2014;75(2):141–50. [DOI] [PubMed] [Google Scholar]
- 39.Khalil MS, El Nahas AM, Blakemore AI. Transforming growth factor-beta1 SNPs: genetic and phenotypic correlations in progressive kidney insufficiency. Nephron Experimental nephrology. 2005;101(2):e31–41. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen N, Fernando SD, Biette KA, Hammer JA, Capocelli KE, Kitzenberg DA, et al. TGF-beta1 alters esophageal epithelial barrier function by attenuation of claudin-7 in eosinophilic esophagitis. Mucosal immunology. 2017. [DOI] [PMC free article] [PubMed]
- 41.Muir AB, Dods K, Henry SJ, Benitez AJ, Lee D, Whelan KA, et al. Eosinophilic Esophagitis Associated Chemical and Mechanical Microenvironment Shapes Esophageal Fibroblast Behavior. Journal of pediatric gastroenterology and nutrition. 2015. [DOI] [PMC free article] [PubMed]
- 42.Simon D, Page B, Vogel M, Bussmann C, Blanchard C, Straumann A, et al. Evidence of an abnormal epithelial barrier in active, untreated and corticosteroid-treated eosinophilic esophagitis. Allergy. 2017. [DOI] [PubMed]
- 43.Eluri S, Runge TM, Hansen J, Kochar B, Reed CC, Robey BS, et al. Diminishing Effectiveness of Long-Term Maintenance Topical Steroid Therapy in PPI Non-Responsive Eosinophilic Esophagitis. Clin Transl Gastroenterol. 2017;8(6):e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf WA, Cotton CC, Green DJ, Hughes JT, Woosley JT, Shaheen NJ, et al. Predictors of response to steroid therapy for eosinophilic esophagitis and treatment of steroid-refractory patients. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2015;13(3):452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aceves SS, Newbury RO, Dohil MA, Bastian JF, Dohil R. A symptom scoring tool for identifying pediatric patients with eosinophilic esophagitis and correlating symptoms with inflammation. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2009;103(5):401–6. [DOI] [PubMed] [Google Scholar]
- 46.Doshi A, Khamishon R, Rawson R, Duong L, Dohil L, Myers SJ, et al. IL-9 Alters Epithelial Barrier and E-cadherin in Eosinophilic Esophagitis. Journal of pediatric gastroenterology and nutrition. 2018. [DOI] [PMC free article] [PubMed]
- 47.Chen X, Oshima T, Shan J, Fukui H, Watari J, Miwa H. Bile salts disrupt human esophageal squamous epithelial barrier function by modulating tight junction proteins. American journal of physiology Gastrointestinal and liver physiology. 2012;303(2):G199–208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.