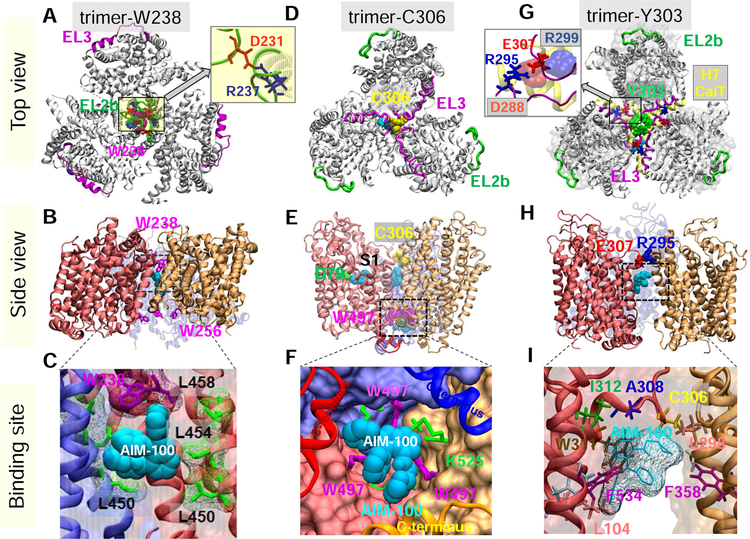
Fig. 1. Structural models for DAT trimer, and corresponding AIM-100 binding sites.
Top view, side view and AIM-100 binding site of three structural models (trimers-W38, -C306 and –Y303) are illustrated in the respective panels A-C, D-F and G-I. (A-C) Trimer-W238 is stabilized by interfacial contacts between EL2b (green), TM4 and TM9. AIM-100 (cyan van der Waals (vdW) representation) binds to the trimeric cavity with an estimated affinity of −7.6 kcal/mol. W238 and W256 (purple sticks) line the EC and IC entrances to the cavity. Inset in A displays that interfacial salt bridge R237(blue)-D231(red). (D-F) Trimer-C306 constructed from OF DAT monomer. Three C306 residues (yellow, vdW) from EL3 (magenta) cluster in the center. Inter-subunit cation-π interactions are detected between W497 and K525. Three AIM-100 binding sites are detected near: C306 (occluded state), W497 (entropically favorable; shown in panel F), and D79 at the substrate binding site (S1), all with comparable (~ −7.1 kcal/mol) binding affinities. (G-I) Trimer-Y303, homology model constructed using CaiT trimer (PDB: 2WSX) as template. In G, DAT is shown in gray ribbons and CaiT in transparent gray surface. The EL3 loop (magenta) in DAT aligns with the H7 helix (yellow ribbon) of CaiT. Three Y303 residues (green wdW balls) cluster at the center. Inset in G displays that interfacial hDAT salt bridge R295-E307, and its counterpart in CaiT, D288-R299 (semi-transparent, vdW). A high affinity AIM-100 binding site (−7.0±0.5 kcal/mol) is observed at the interface of two subunits. AIM-100 binding energies are calculated using AutoDock.