Abstract
The role of glycosaminoglycans (GAGs) in major biological functions are numerous and diverse, yet structural characterization of them by mass spectrometric techniques proves to be challenging. Characterization of GAG structure from tandem mass spectrometry is a tedious and time-consuming process but one that can be automated in a database-independent, high-throughput fashion through the assistance of software implementing a genetic algorithm [1]. This work presents the manner in which this data is interpreted by the software, specifically addressing the development of a scoring algorithm. The significance of glycosidic and cross-ring fragment ions and the implications that specific fragments provide for assigning the positions of modifications are discussed. The scoring algorithm is tested for statistical merit using the widely accepted expectation value as the criterion for quality. Using MS/MS data for well-characterized standards, this scoring approach is shown to assign the correct structure, with a low likelihood (1 in 1012-chance) that the assigned structure matches the data due to random chance. The integrated software that automates the structure assignment is called Glycosaminoglycan-Unambiguous Identification Technology (G-UNIT).
INTRODUCTION
Glycosaminoglycans (GAGs) are linear, polydisperse carbohydrates that are ubiquitous among living cells, and are responsible for a multitude of biological interactions including cell signaling, energy generation, protein binding conformation changes and molecular recognition [2–5]. Structurally, GAGs are composed of a repeating linear disaccharide backbone of a uronic sugar and amino sugar residue. Structural differentiation occurs based on three primary forms of modifications: O-sulfation, N-deacetylation/sulfation and uronic sugar epimerization. Recent studies suggest that patterns of sulfation has profound effect on protein binding[6, 7]. Moreover, analysis of GAGs released from proteoglycans bikunin and decorin suggest that naturally occurring glycans may contain a conserved sulfation motif that is independent of chain length [8, 9]. Modifications can be localized to specific glycan residues and further designated to specific positions with the respective use of glycosidic and cross-ring fragmentation that arise from ion activation using collisional or electron-based activation, or photodissociation [10–20].
Sulfation and other structural modifications critically influence the biological activity of GAGs, and considerable effort has been made to develop new approaches for sequencing this class of molecules. The assignment of structures for GAGs require a de novo approach in which the mass spectra must contain sufficient information to localize all the relevant features [1]. Such a method would determine GAG structure using a combination of assumed information (repeating disaccharide copolymer backbone, linkage pattern between sugars) and diagnostic and information-rich MS and MS2 fragments that can localize the position of SO3 and N-acetyl modifications. There are a number of methods that can be used to produce information-rich tandem mass spectra, using collisions, photodissociation, or electron-based activation methods. The density of fragment ions in these tandem mass spectra makes it difficult to interpret and assign structures in a high-throughput manner without assistance from specialized software. Manual interpretation of tandem MS of GAGs is possible but requires considerable time and expertise. While the interpretation of known structures can be significantly improved with mass calculation tools such as GlycoWorkBench [21], the feasibility of interpreting unknown GAG structures diminishes greatly with increasing degree-of-polymerization (dp), as the number of permutations scales as n-choose-k, where n is number of possible modification sites (up to 4 per disaccharide) and k is number of modifications.
Previous work from our laboratory proposes software for a database-independent method of automated GAG tandem MS interpretation using a genetic algorithm [1]. The approach is an effort to replicate the evolutionary cycle and determination of the most optimum structure using a survival-of-the-fittest mechanism. Theoretical structures are represented as genes and tested against the MS2 – the closeness of match between theoretical fragments and the experimental data dictates the fitness of the gene. The best genes (two or more) that best survive (i.e. structures that would have theoretical fragments that most closely match the data) breed, passing certain structural characteristics onto a new generation. Each generation has its own set of “winners”, those most accurately matching the data, and continues this process until the best candidates no longer show signs of change.
Our earlier publication provided proof-of-principle for this approach using full-length bikunin GAGs (up to dp 43 in length), but only examined glycosidic fragments [8], as enzymatic analysis has determined that there was only one type of modified disaccharide present in these samples. Most glycosaminoglycans are far more structurally diverse. Modification heterogeneity and the potential for multiple sulfo-modifications to occur on the same sugar residue dictate that we not only incorporate cross-ring fragmentation into our scoring model but also find and utilize structurally diagnostic fragments or fragment sets that can yield unambiguous assignments of modification position. The approach for automated assignment here begins at the MS1 stage, using a set of known masses for composition assignment: amino sugar, uronic sugar, delta-uronic sugar (maximum of 1) masses serve as fundamental building blocks. A network of potential masses is generated by propagation of the repeating uronic and amino sugar copolymer with variable amounts of SO3 and acetyl modifications being added. Assignment of composition is based on accurate mass, similar to an approach presented by Zaia et al. [22]. Our structural characterization technique is based on an optimization framework that generates fragments from randomly generated candidate structures in silico, and then optimizes structural possibilities with a genetic algorithm. This relies on a robust scoring algorithm to support the survival of the fittest mechanism, in which candidate structures are fragmented in silico, and the products are compared to the experimental mass spectrum. Here we explain in detail the development of the scoring method that enables this approach. This technique is considerably different from the previous work of Hu et al. [23], who have proposed a divide-and-conquer strategy that determines GAG structures by partitioning the GAG sequence into smaller parts. The approach combines and builds upon these smaller pieces of information to determine overall sequence. Our genetic algorithm approach examines a theoretical GAG structure in its entirety instead of building a structure from the ground up. The labile SO3 modifications of GAGs are prone to being lost during ion activation, thus depleting some of the fragment precursor, and can confound structure-building algorithms that attempt to assemble sequence based pieces from the whole. Also, as shown previous [1], this technique can also highlight the possibility of mixtures and/or additional GAG components in the GAGs of proteoglycans based on conflicting fragment data. Hence, our focus is more concerned with (1) does the tandem mass spectrum unambiguously explain characteristics of a particular structure on the whole (instead of its parts) and (2) is there a possible structure of appropriate composition that can more accurately match the spectrum?
EXPERIMENTAL
Mass Spectrometry
Electron detachment dissociation (EDD) experiments were performed using a 9.4T Bruker Apex Ultra QeFTMS (Billerica, MA), with an indirectly heated hollow cathode (HeatWave, Watsonville, CA) for generating EDD electrons. The solutions were ionized using flow nanoelectrospray (pulled fused silica tip FS360-75-15-N-20) Solutions were made at a concentration of 0.2 mg/mL in 50:50 methanol:H2O. Hexasaccharide solutions were injected at a rate of 25uL/h. The solutions were run in negative ion mode. Precursor ions were selected in the external quadrupole accumulated for 4 seconds in an RF only hexapole before injection into the Fourier transform-ion cyclotron resonance mass spectrometer (FT-ICR MS). Precursor isolation was refined by using in-cell isolation with coherent harmonic excitation frequency (CHEF). The isolation power of the CHEF event was 20%. For irradiation of electrons the cathode heater was set to 1.6 A and the bias was set to −19 V. The extraction lens was set to −19.2±0.2 V. Ions were irradiated for 1 s. 64 acquisitions were signal averaged per mass spectrum. Internal calibration produced a mass accuracy of 5 ppm.
Software
MS1 analysis of parent ion mass is performed using a composition assignment software module written in the MATLAB coding environment. Monoisotopic peaks and charge states are acquired from Bruker DataAnalysis (using the “FTMS” peak picking method) and deconvoluted to a neutral mass. A composition is derived from one or more neutral mass(es) by searching an automatically generated data matrix of possible chain lengths, degrees of sulfation, deacetylation, and sodium/hydrogen exchange. The user input also includes the possibility of reducing end modifications, and nonreducing ends that can terminate in unsaturated uronic acids, as is common in enzymatically produced GAG oligomers. Theoretical neutral masses from the automated data matrix are compared against user specified masses with a user-defined mass tolerance. The sequences that match are then used for performing the MS2 analysis.
For MS2 analysis, the software uses a genetic algorithm search heuristic alongside binary vector representation of glycan structures where on-bits denote an occupied site of SO3 and N-acetylation modifications. The first step generates two glycan structures at random that fit the expected composition (initialization step) and then proceeds to “breed” these structures into a new generation of candidates (crossover step). The primary three steps (crossover, mutation and fitness) are iterated until the maximum fitness value does not change after numerous cycles. The number of iterations required before termination of the algorithm can be defined by the user but is defaulted at a value of 3. The structure(s) containing the highest scores are then examined using additional data interpretation tools that assign fragment peak masses alongside their charge, intensity and mass error (in ppm).
Software is compatible for standard desktop computers, with a minimum requirement of MATLAB R2014 coding environment or newer, 2.4 GHz processor and 4GB RAM. Customization specific to GAG family (CS/DS, Hp/HS and Arixtra’s synthetic backbone and connectivity) is available using specific functions for determination of fragment masses. Additional parameters are adjustable from the command line if desired (maximum charge state of fragments, ppm mass error tolerance, maximum possible Na-H exchange, neutral loss considerations for H2O and CO2). MATLAB source code and raw mass spectral data used for testing and validation in this manuscript are available upon request.
RESULTS AND DISCUSSION
Software Architecture
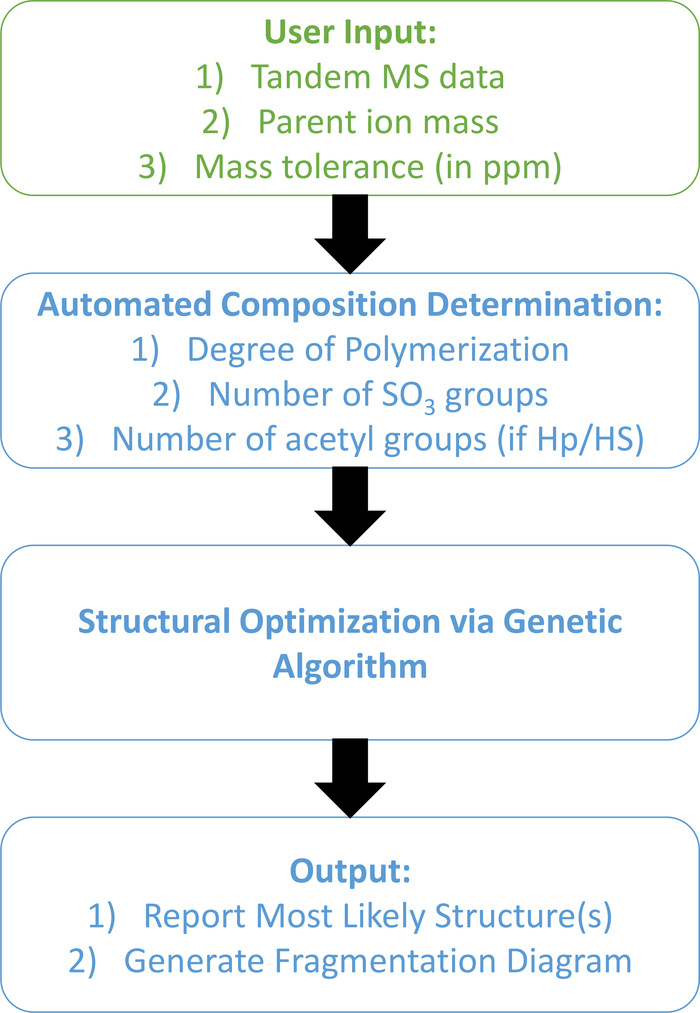
Previous work from our laboratory presented an automated and high-throughput method for characterizing GAG structure of unknown samples from tandem MS using a database independent method that generates theoretical fragments in-silico and then proceeds to optimize potential structures using a genetic algorithm [1]. The workflow is shown in Figure 1. The user provides tandem MS data in the form of a two-column comma-separate value file with masses (including isotopes) sorted ascending in column 1 and intensities (either relative or arbitrary units) in column 2 as well as additional inputs of the parent ion mass and charge of the precursor ion. An independent module calculates the composition, including degree of polymerization, number of SO3 groups, and number of acetyl groups, within a user defined ppm tolerance window. Once completed, the software automatically employs a genetic algorithm optimization model to determine the most likely structure(s) based on the data provided.
Figure 1.
The standard workflow for our current GAG identification software. The user provides the information in the green box. Blue boxes are connected module, fully automated and require no user supervision.
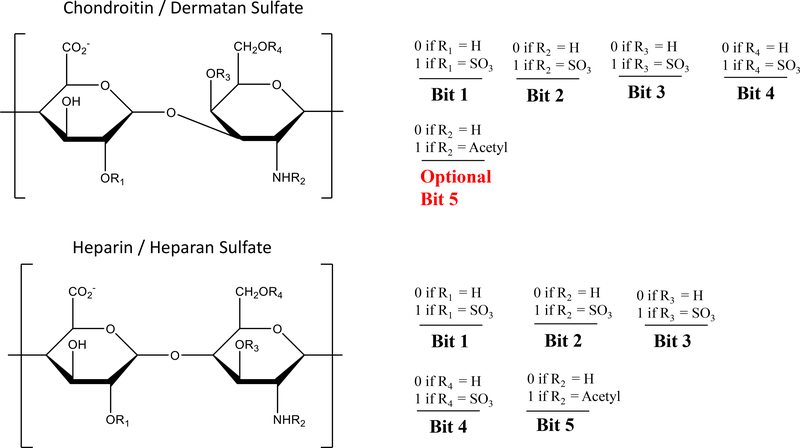
The optimization procedure generates a list of candidate structures of appropriate composition, representing them as a binary number, as shown in Figure 2. The automate procedure then determines the quality of the match of the candidates’ predicted fragmentation products with the experimental data. An essential requirement for this process is a robust scoring algorithm that can discriminate isomeric structures by their fit to the experimental data. Each iteration of the genetic algorithm attempts to improve upon the list of candidate structures while eliminating low scoring structures from consideration. This paper focuses on the development of the scoring algorithm that can be applied to any GAG family (heparin, heparan sulfate, chondroitin sulfate, dermatan sulfate) and to any chain length. The focus is on the methodology used to compare computer generated candidate structures against experimental data and to show the statistical merit of the scoring paradigm.
Figure 2.
Bit-wise matrix representation of disaccharide CS/DS and Hp/HS GAG families used in software for rapid analysis via a genetic algorithm. Multiple disaccharide units can be combined to fit the appropriate chain length and composition. In the case of CS/DS, bit 5 represents a rare and unlikely circumstance where a free-amine could exist. The default case is that bit 5 is removed, and the occupancy of bit 4 dictates either N-SO3 (when bit 4 = 1) or N-acetylation (when bit 4 = 0).
Scoring Algorithm
A fundamental feature that allows this software to operate independent of a database is that it considers all possible isomers, constrained by GAG family and by composition (degree of polymerization, or dp, degree of sulfation, and the number of acetylated amine groups). In order to allow all isomers to be considered, without having to score every possibility, the genetic algorithm optimizes the match of structures to the experimental data. Using this optimization tool allows scoring on a small subset (1% or less) of all possible permutations of a given composition while finding the correct structure. Within this optimization step is a series of comparisons between candidate structures and the experimental data – each iteration of the genetic algorithm attempts to find a more closely matched candidate structure based on its interpretation of experimental data; the fitness score of a candidate structure is the value used as a measure of closeness. Hence, the paradigm for how the software scores candidate structures must be discussed.
A 2-step system is utilized in which an initial score is given to structures based on glycosidic fragments alone and then a refined score is assigned based on the number of cross-ring fragments that can be used to unambiguously define a specific modification location. The first step determines the number and type of modifications that are located on a particular residue. For chondroitin sulfate and dermatan sulfate (CS/DS) GAG families, we consider the possibility of 2-O sulfation on the hexuronic residue and 4-O and 6-O sulfation on the N-acetyl galactosamine. Optionally, more uncommon CS modifications such as N-sulfation or a free amine group can be considered, in which case a new bit is introduced to the binary representation of the GAG candidate oligomers. As this modification is exceedingly rare for naturally occurring CS and would decrease overall software performance calculations, it is by default not used unless specified by the user. For heparin and heparan sulfate (Hp/HS) GAG families, we consider the possibility of 2-O sulfation on hexuronic residues and 3-O, 6-O and N- sulfation on glucosamine residues. Additionally, de-acetylated glucosamines are also an optional feature.
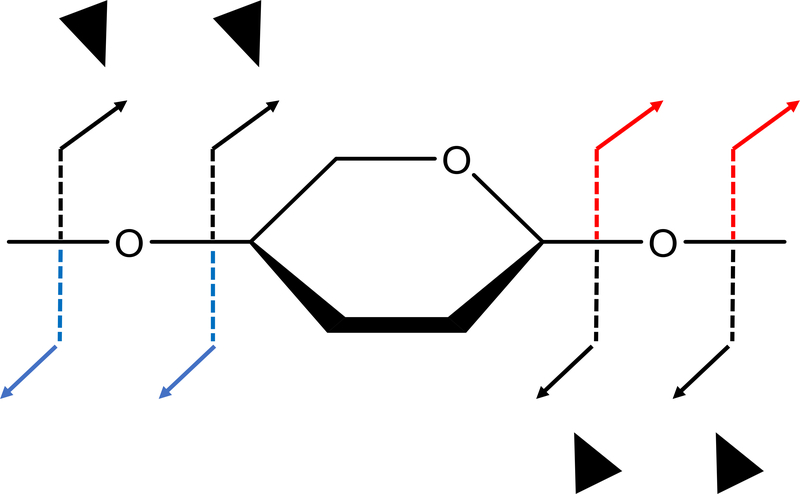
Glycosidic fragments provide the composition of each residue in the GAG oligomer, greatly reducing the search space for complete structural assignment. Each sugar residue is assigned a score of 0 to 4 (called GlycScore or SG), ranked based on the number of glycosidic fragments that surround the sugar residue as well as what information these fragments provide. Figure 3 provides the criterion for assigning a score to a residue: a GlycScore of 4 indicates that glycosidic cleavages are observed pointing directly to the specific residue and come from both the reducing and non-reducing ends. On the other hand, a low-value GlycScore indicates only a few or no fragments that are supportive of a particular residue. Assuming high quality tandem MS data with information rich spectra, one can expect that the majority of glycosidic fragments are present. When using a genetic algorithm optimization method, candidate structures that lack a large fraction (>50%) of glycosidic fragment matches are evaluated as unlikely matches and other structures with similar characteristics are unlikely to be examined.
Figure 3.
A simple paradigm for assigning GlycScore (SG) to a residue. Each residue’s SG is concerned with the set of non-reducing and reducing end fragments that point to it. For residue n, Bn, Cn, Ymax-n+1 and Zmax-n+1 (shown with black arrows) are the fragments considered to be part of SGn. Fragments in blue contribute to the SG of the residue adjacent from the non-reducing end. Likewise, red fragments contribute to the SG of the residue adjacent from the reducing end.
The ranking of structures and their respective step 1 score (S1) is not solely reliant on the GlycScore (SG) but also the intensities of fragment matches as shown in eq 1, where In is intensity of the 4 fragments (Bn, Cn, Ymax-n+1,Zmax-n+1) that point directly to the residue, n, in question:
| (eq.1) |
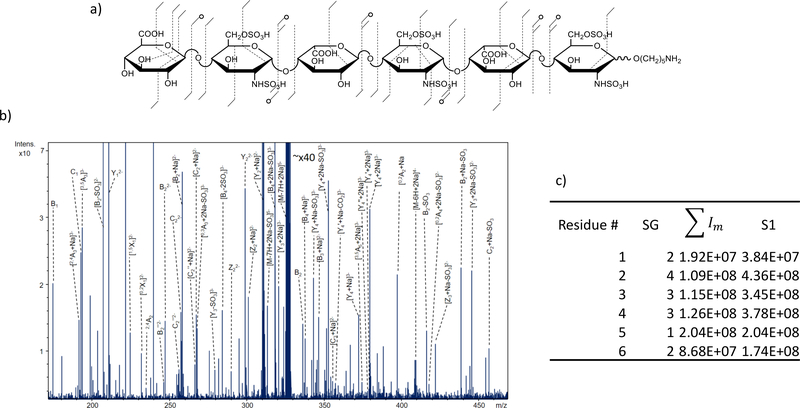
The GlycScore by itself will always yield an integer value and is prone to problematic artifacts if used as the sole measurement for ranking structures as multiple structures can have the exact same GlycScore if the number of glycosidic fragments matches are equal. The commonly occurring neutral loss of SO3 in the tandem mass spectrum exacerbates the problem, as the resulting fragments match a loss-free glycosidic fragment when the genetic algorithm presents a candidate structure that is under sulfated in a specific region compared to the true structure. The solution is to consider the intensity of matched glycosidic fragments (Im) as a scaling factor to SG. Previous work on GAG tandem MS for multiple ion activation techniques include CID, EDD, and NETD show that the intensity of SO3 loss peaks are typically lower in abundance than the non-loss peak [12, 13, 17–19]. Figure 4 shows an example of the assignment of SG and S1 for a hexasaccharide GAG with 6 sulfo-modifications.
Figure 4.
(a) Annotated structure of synthetic hexasaccharide sample with 6 SO3 groups; fragments were produced by electron detachment dissociation (EDD). (b) Annotated EDD spectrum of structure from 4a. (c) Calculated SG, Im and S1 values for each residue n. Note that in the table, the sum of all intensities for relevant fragment peaks is shown.
One challenge in assigning glycosidic fragments occurs when there are isobaric possibilities for product ions from symmetrically modified oligomers. This happens for c and z ions from a precursor with a delta-uronic acid at the non-reducing end and no derivatization of the reducing end. Isobaric fragment ions are difficult to assign either by manual interpretation or by automated glycan analysis – our software treats isobaric possibilities as both types of fragment ions. This problem is mitigated for GAG samples where the reducing end has been modified in some capacity to create an unambiguous mass difference. For example, in the case of full length glycans isolated from proteoglycans, a linker region is present on the reducing end, and this breaks the symmetry of the structure and eliminates the possibility of isobaric c/z ions [8, 9]. Some depolymerization procedures produce a non-reducing end that is not a delta-uronic acid, and these also avoid isobaric c/z fragments.
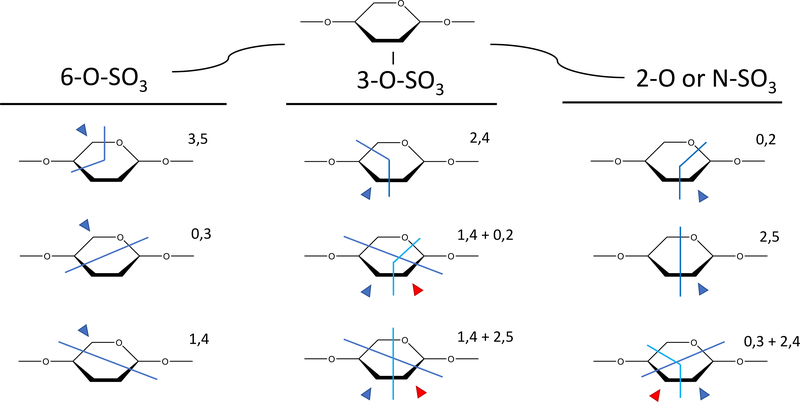
The second step of our algorithm uses a heuristic based on a priori knowledge of GAG chemical structure (Figure 4) to determine the position of SO3 modifications on sugar residues for the top scoring structures from step 1 (by default the top 3 structures). At this point in the assignment process, the structure search space is greatly constrained from the glycosidic fragment matches. At this point, it is possible to examine every glycan residue individually as there is a narrow selection of permutations based on the known biosynthetic pathways for modification. For hexuronic acids, the presence of a sulfo-modification defaults to being at the 2-O position, as no other positions are known to be modified in these residues. For amino sugars, the possible sites of modification are N, 3-O, and 6-O for heparin or heparan sulfate GAGs, or 4-O or 6-O for chondroitin or dermatan sulfate. The code looks for diagnostic cross-ring fragments or fragment combinations that would be able to unambiguously assign sulfation positions. If no cross-ring fragments are available on a residue, the code will leave the positions on that residue as ambiguous. As with all de novo sequencing methods, the mass spectra must be information-rich, that is provide enough fragmentation data to assign all the features in order to make a definitive assignment. On the other hand, a situation can arise where multiple structures are produced from conflicting cross-ring assignments. In these cases, we rank structures based on the intensities of the diagnostic cross ring fragments.
Intensity ratios between sets of fragments have been used to differentiate the presence of glucuronic versus iduronic acids in synthetic HS tetrasaccharides [24]. Furthermore, multivariate statistics have been used to identify diagnostic fragments for certain ion activation methods [12, 15, 25–27]. At the present level of knowledge, there is not sufficient knowledge to use these ratios as a definitive guide to determining all C-5 stereochemistry. The software is fully capable of applying an additional layer of analysis to determine the epimeric center once there is a fuller understanding of product ion intensities as a function of uronic acid stereochemistry. Much like the method proposed in step 2, a residue-by-residue analysis of the uronic sugars could be applied as a 3rd step without a significant increase in analysis time, and this capability is envisioned in a future release.
Validation of Algorithm
In order to create a fully automated GAG interpretation software, the scoring algorithm must be 1) unsupervised and user-independent and 2) assign the correct glycan structure. Much of the glycan analysis currently published in literature uses software packages such as GlycoWorkBench [21] or similar GAG fragment calculation tools in combination with user intuition or experience to interpret the mass spectrum. Careful examination of peaks in the mass spectrum with manual supervision allows an expert to determine the likelihood of false positive while reaffirming structural features. This in-depth yet subjective form of glycan interpretation that relies on user expertise is difficult to automate and impractical for high-throughput analysis.
Assigning structure from tandem MS fragment peaks in an automated fashion requires some degree of assumption. For our software, we assume glycosidic and cross-ring fragments increase the validity of certain structural features and that enough of them will give the highest score to the most valid structure. However, the quality of the scoring system is judged not on its theoretical foundation but purely by its ability to assign the correct sequence and differentiate it from incorrect ones. We examine the quality of our scoring system with a well-accepted statistical methodology, namely by determining the likelihood that a score is given due to random chance, that is the expectation value, a statistic that has been widely applied in bioinformatics [28–30]. If x is the score of a particular spectrum S, a survival function, s(x), for a discrete score probability distribution, p(x), can be defined [31]:
| (eq.2) |
Where Pr(X > x) is defined as probability that the spectrum’s score will be higher than score x due to random matching within a defined database, D. For GAGs, the defined database is all possible permutations of the composition of the sample of spectrum S. The expectation value e(x) can be interpreted as the number of GAG structures that would be expected to have scores of at least x.
| (eq.3) |
Where n is the number of sequences scored. The expectation value can be interpreted as follows: if a score x of expectation value e(x) = y, then one would have a score of at least that value for y number of times for every replicate experiment. A lower expectation value is therefore more ideal. For example, an e(x) of 0.001 suggests that an experiment must be replicated 1 thousand times before a score of x could be obtained by random chance.
This technique has been used previously for analysis of scoring system of peptide MS [32] while using a database search engine [33]; we apply the same fundamental principles and calculations for our scoring system. To examine our GAG scoring system with this method, p(x) is determined by constructing a frequency histogram of all GAG structure scores. We take the tandem MS of a pure, single component GAG for which we know the structure (Figure 6a) and score structures of appropriate composition against the experimental data. The structures being scored against are stochastically selected and not optimized with the genetic algorithm heuristic to prevent introduction of selection bias. Among these structures, we know that only one is considered “valid” while all others are termed “stochastic”. The probability p(x) can be determined by normalizing the discrete frequency f(x) of a structure by the number of sequences scored N:
| (eq.4) |
Figure 6.
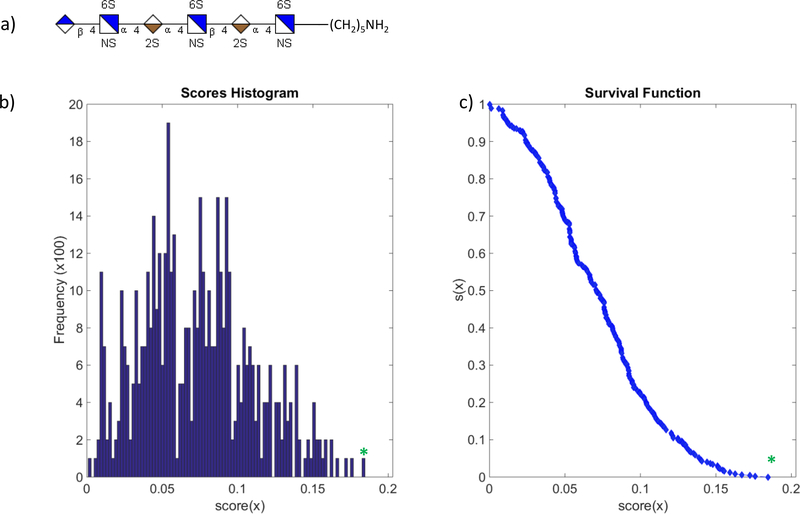
(a) The hexasaccharide structure used for calculating e(x) and s(x). Tandem MS was performed using EDD. (b) A histogram of 20,000 structures scored using the Monte Carlo method. The * represents the score that is associated with the structure shown in Figure 6a. Note that the scoring algorithm assigns the valid structure the highest score. (c) The survival function of the scoring algorithm plotted versus the score. An e(x) value of 1.96E-12 is calculated for the valid structure.
Figure 6 shows the frequency histogram of 20,000 structures scored using a Monte Carlo sampling method for a synthetically produced heparan sulfate hexasaccharide containing 8 SO3 groups. The green asterisk in the histogram represents the score associated with the valid GAG structure. Visual inspection of the histogram shows that the right-most point of the histogram contains the structure of highest score, x*, and is the valid structure for this data set. The confidence of this score increases with respect to the distance between x* and all other scores (the scores of stochastic structures). Hence, a gap between x* and the bulk majority of other scores is highly desired and, moreover, the difference between x* and the next highest scoring structure is important for evaluating the algorithms ability to discriminate similar structures. It should also be noted that the frequency of the highest score x* is the lowest in the set, implying that x* is likely a unique value observed only when matched with the valid structure.
Given how our scoring system is partially dependent on intensity while also working under some assumptions regarding the expected fragmentation results from ion-activation of GAGs, the numerical difference between two scores is not an easily interpretable measurement of the degree of difference between two structures of those respective scores. Moreover, the individual score of any structure (valid or stochastic) has little interpretability and does not serve as a good measure of fitness. A much more sophisticated estimate for confidence can be determined from the survival function (Figure 5c). Score x* for our hexasaccharide has a value of s(x*) = 5.59E-16; application of eq.3 yields an e(x*) = 1.96E-12. This expectation value indicates that an experiment would have to be repeated approximately 1012 times before a score of x* would be matched to a structure due to random chance – a figure of merit that reflects positively on our GAG scoring algorithm.
Figure 5.
Once the number of SO3 on a residue are confined to a specific residue, software searches for cross-ring fragments to determine position. In situations where a mixture might be present, structures are ranked based on the cumulative intensity of diagnostic fragments. Blue arrows indicate positions that can be assigned with a specific cross ring fragment or a combination of 2 cross-ring fragments with the assumption that they come from the same end (reducing or non-reducing). A red arrow indicates assignments that can be made if the two indicated cross-ring fragments are from different ends (i.e. an A fragment and an X fragment).
Additional histograms and survival function diagrams for other GAG compositions of both experimental and synthetically generated datasets for CS/DS and Hp/HS GAG families were also calculated and and can be found in supplemental information. The expectation values for the scores of valid structures (x*) using our scoring algorithm across various chain lengths, GAG families and different degrees of modification suggests that our method can be applied to a wide variety of GAG tandem MS.
High Throughput Applications
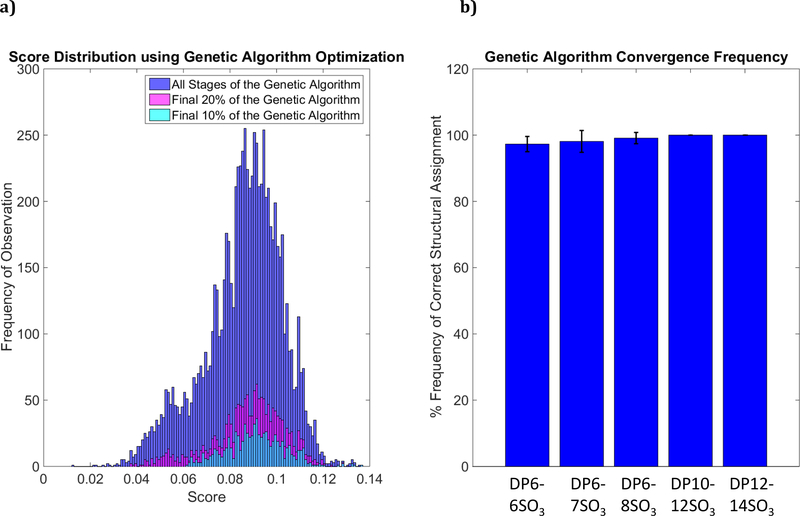
Previously published work from our laboratory proposed a high-throughput model for determining GAG structure using a genetic algorithm optimization technique [1]. While Monte Carlo simulations are performed at random and with no additional supervision, integration of our scoring method within our high-throughput platform inherently creates a bias during the optimization cycles. A genetic algorithm is modeled around a survival-of-the-fittest mechanism that mimics the characteristics of evolution. Thus, it retains information through each iteration of the algorithm – the life cycles of potential structural candidates are heavily influenced by the crossover and mutation steps that occur and create a skewed, top-heavy distribution when plotted on a histogram.
Figure 7 shows a histogram of the mean score per iteration for 8,325 optimization iterations in a genetic algorithm. Compared to the histogram in figure 6, the genetic algorithm spends most of its time refining structures that are within the upper 40th percentile of all possible structures. The magenta shaded bars in figure 7 reflect scores of structures being observed in the final 20% of the genetic algorithm, showing that the final iterations of the genetic algorithm tend to heavily bias high-scoring structural features, averaging in the 70th percentile or greater. Likewise, the cyan histogram shows the scoring range for the final 10% of the genetic algorithm. Iterations in which lower-scoring structures are observed are a byproduct of heavily mutated candidates with numerous incorrect features with frequency of observation diminishing as the algorithm progresses.
Figure 7.
(a) The distribution of mean scores per iteration for 8,235 iterations within a genetic algorithm for synthesized heparan sulfate hexasaccharide containing 8 SO3 modifications. Blue bars indicate that the majority of the genetic algorithm is optimizing structures above the 40th percentile of all possible structures. The magenta bars show the final 20% of a genetic algorithm lifecycle and is even more top-scores focused. Likewise, cyan bars show the final 10%. (b) The percent with which compositions converge upon the correct answer using our scoring system and optimized with the genetic algorithm search heuristic. Decasaccharide and dodecasaccharide data is synthetically produced in-silico and yields perfect accuracy due to lack of experimental artifacts, contaminants and noise.
These measurements are shown for 110 independent optimization cycles – that is, the genetic algorithm was applied 110 times to the same data set for the hexasaccharide structure shown in Figure 6. Among these 110 cycles, 107 iterations converged upon the correct structure, yielding a 97.3% convergence rate for this hexasaccharide (DP6). For partially stochastic methods such as the genetic algorithm (the initiation step is stochastic), it is typically best to run multiple iterations to assure convergence upon a correct answer. Time benchmarks for performance of this algorithm have been reported [1] and it is computation and temporally inexpensive to iterate multiple time to secure convergence upon the correct answer. Figure 7b shows a breakdown of percentage convergence upon the correct structure for multiple hexasaccharides as well as synthetical generated data for decasaccharide (DP10) and dodecasaccharide (DP12) structures with intensity values randomly normalized between 0.1–0.9. It should be noted that synthetically generated data has no fail rate compared to experimental data, which can be attributed to the lack of any experimental noise peaks or other potential contaminants or artifacts that could influence the structural matching elements implemented in software. This speaks to the necessity of high-quality, informationally dense mass spectra for accurate structural assignment. As noted previously [1], the genetic algorithm is by default assigned to run in multiple cycles (default of 5, adjustable by the user) with new, randomly generate starting candidates at the beginning of each cycle.
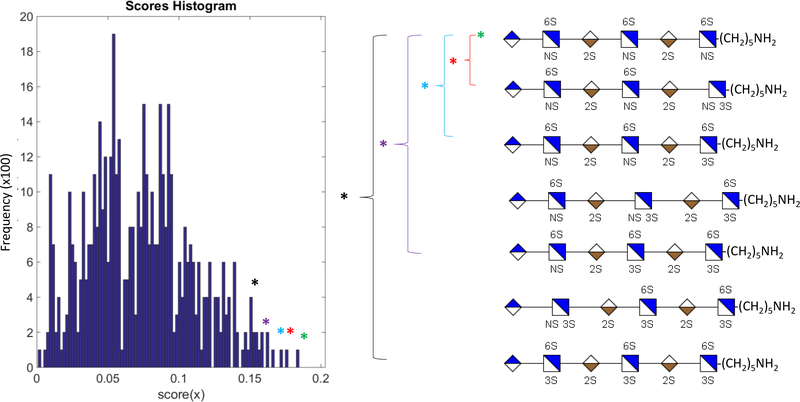
Structural ambiguity is likely to arise when structurally informative peaks within the mass spectrum is absent. When applied to a genetic algorithm or other optimization technique that involves scoring, the absence of structurally significant fragments leads to a lower maximum possible score for the experimental data. As a result, the highest-ranking structure is no longer one, unambiguous item but multiple possible elements. Figure 8 shows the increase in ambiguity when the score decreases as a result of missing pieces of structurally informative information. Reexamination of the histogram derived from Monte Carlo simulations shows that as we move lower in score, more possible structural options arise. Within our software under such circumstances, single structural convergence is no longer an option and instead, multiple structural outputs will be the final result. Tandem MS using electron activation methods such as electron detachment dissociation (EDD) [16–19, 24–26, 34–37] and negative electron transfer dissociation (NETD) [14, 20] produce an abundant series of cross-ring fragments that allow for complete or near-complete assignment of structure. Work with collision-based threshold methods [13, 26, 27, 38] are also a valid option but do not always yield all necessary cross-ring fragments for unambiguous identification, especially in longer-chain proteoglycan GAGs [8, 9]. Thus, for highly pure components where complete structural characterization is desired, the data must contain the appropriate number of structurally diagnostic peaks.
Figure 8.
An increase in the number of possible structures for a mass spectrum occurs when the highest possible score is lower due to lack of structurally diagnostic peaks. As the score decreases, the results reported by software is no longer a single structure but multiple, equally valid structures.
CONCLUSIONS
The G-UNIT software package consist of multiple modules structured in a manner that 1) identifies GAG composition based on an accurate mass measurement within the MS1, 2) uses an in-silico fragmentation calculation tool specific to a desired GAG family (HS/Hp or CS/DS) to consider potential glycosidic and cross-ring fragments, 3) applies the multilayered scoring system specific to GAGs as discussed in this manuscript and, 4) determines the optimal structure using a genetic algorithm optimization technique using multiple iterations to minimize likelihood of converging upon local maxima. Any module or combination of modules is available to use upon request from the author. Conversion of this software platform from paid programming environments (MATLAB) to free alternatives is currently being processed.
This approach is successful both for moderately sulfated GAGs such as those examined here, or more highly sulfated GAGs such as Arixtra. The success of this automated approach requires MS/MS datasets that contain the necessary amount and type of fragments needed for structural characterization. Much like any form of automated spectral interpretation, the approach is highly reliant on the quality of the data: glycosidic fragments and structurally meaningful cross-ring fragments are necessary for complete structural characterization. The acquisition of such data can be a limiting factor in high throughput structure analysis. Fortunately, developments in activation methods for GAGs has led to a number of approaches that provide information-rich data sets (reference EDD and NETD). A lack of any necessary pieces of information both increase the possibility of structural ambiguity and simultaneously increases processing times. With this software, glycosidic fragments are particularly important as they greatly reduce search space and facilitate the assignment of cross-ring fragment peaks.
Data preprocessing methods have not been discussed in this manuscript as there are various specialized considerations that must be made for GAGs. Deconvolution of GAGs using commercialized options such as Bruker DataAnalysis or Thermo XCalibur fail to capture all relevant monoisotopic peaks. This is because isotope distributions of GAG fragments deviate a great deal from the typical proteomic averagine model, and more importantly, do not necessarily fall under a consistent chemical formula but instead change with respect to the number of sulfo modifications. Isotope deconvolution thus needs its own set of specialized rules, a point we will discuss with more detail in a separate manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful for the generous support of the National Institutes of Health, grant numbers R21HL136271, U01CA231074, and P41GM103390. The authors would also like to acknowledge Pradeep Chopra and Geert-Jan Boons (University of Georgia) for providing hexasaccharide samples and David Kilgour (Nottingham Trent University) for insight on utilizing the genetic algorithm.
REFERENCES
- 1.Duan JN, Amster IJ: An Automated, High-Throughput Method for Interpreting the Tandem Mass Spectra of Glycosaminoglycans. J Am Soc Mass Spectrom. 29, 1802–1911 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gandhi NS, Mancera RL: The Structure of Glycosaminoglycans and their Interactions with Proteins. Chemical Biology & Drug Design. 72, 455–482 (2008) [DOI] [PubMed] [Google Scholar]
- 3.Ohtsubo K, Marth JD: Glycosylation in cellular mechanisms of health and disease. Cell. 126, 855–867 (2006) [DOI] [PubMed] [Google Scholar]
- 4.Rabenstein DL: Heparin and heparan sulfate: structure and function. Natural Product Reports. 19, 312–331 (2002) [DOI] [PubMed] [Google Scholar]
- 5.Xie B, Costello CE: Carbohydrate Structure Determination by Mass Spectrometry. Carbohydrate Chemistry, Biology and Medical Applications. 29–57 (2008) [Google Scholar]
- 6.Zhao YJ, Singh A, Li LY, Linhardt RJ, Xu YM, Liu J, Woods RJ, Amster IJ: Investigating changes in the gas-phase conformation of Antithrombin III upon binding of Arixtra using traveling wave ion mobility spectrometry (TWIMS). Analyst. 14, 6980–6989 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao YJ, Singh A, Xu YM, Zong CL, Zhang FM, Boons GJ, Liu J, Linhardt RJ, Woods RJ, Amster IJ: Gas-Phase Analysis of the Complex of Fibroblast GrowthFactor 1 with Heparan Sulfate: A Traveling Wave Ion Mobility Spectrometry (TWIMS) and Molecular Modeling Study. Journal of the American Society for Mass Spectrometry. 28, 96–109 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ly M, Leach FE III, Laremore TN, Toida T, Amster IJ, Linhardt RJ: The proteoglycan bikunin has a defined sequence. Nature Chemical Biology. 7, 827–833 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu YL, Duan JN, Leach FE, Toida T, Higashi K, Zhang H, Zhang FM, Amster IJ, Linhardt RJ: Sequencing the Dermatan Sulfate Chain of Decorin. Journal of the American Chemical Society. 139, 16986–16995 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi LL, Amster J, Linhardt RJ: Mass spectrometry for the analysis of highly charged sulfated carbohydrates. Current Analytical Chemistry. 1, 223–240 (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi LL, Wolff JJ, Laremore TN, Restaino OF, Xie J, Schiraldi C, Toida T, Amster IJ, Linhardt RJ: Structural analysis of bikunin glycosaminoglycan. Journal of the American Chemical Society. 130, 2617–2625 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kailemia MJ, Li LY, Ly M, Linhardt RJ, Amster IJ: Complete Mass Spectral Characterization of a Synthetic Ultralow-Molecular-Weight Heparin Using Collision-Induced Dissociation. Analytical Chemistry. 84, 5475–5478 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kailemia MJ, Patel AB, Johnson DT, Li LY, Linhardt RJ, Amster IJ: Differentiating chondroitin sulfate glycosaminoglycans using collision-induced dissociation; uronic acid cross-ring diagnostic fragments in a single stage of tandem mass spectrometry. European Journal of Mass Spectrometry. 21, 275–285 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leach FE, Riley NM, Westphall MS, Coon JJ, Amster IJ: Negative Electron Transfer Dissociation Sequencing of Increasingly Sulfated Glycosaminoglycan Oligosaccharides on an Orbitrap Mass Spectrometer. Journal of the American Society for Mass Spectrometry. 28, 1844–1854 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bin Oh H, Leach FE, Arungundram S, Al-Mafraji K, Venot A, Boons GJ, Amster IJ: Multivariate Analysis of Electron Detachment Dissociation and Infrared Multiphoton Dissociation Mass Spectra of Heparan Sulfate Tetrasaccharides Differing Only in Hexuronic acid Stereochemistry. Journal of the American Society for Mass Spectrometry. 22, 582–590 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff JJ, Amster IJ, Chi L, Linhardt RJ: Electron detachment dissociation of glycosaminoglycan tetrasaccharides. Journal of the American Society for Mass Spectrometry. 18, 234–244 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ: Electron detachment dissociation of dermatan sulfate oligosaccharides. Journal of the American Society for Mass Spectrometry. 19, 294–304 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ: Influence of charge state and sodium cationization on the electron detachment dissociation and infrared multiphoton dissociation of glycosaminoglycan oligosaccharides. Journal of the American Society for Mass Spectrometry. 19, 790–798 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolff JJ, Laremore TN, Leach FE, Linhardt RJ, Amster IJ: Electron capture dissociation, electron detachment dissociation and infrared multiphoton dissociation of sucrose octasulfate. European Journal of Mass Spectrometry. 15, 275–281 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff JJ, Leach FE, Laremore TN, Kaplan DA, Easterling ML, Linhardt RJ, Amster IJ: Negative Electron Transfer Dissociation of Glycosaminoglycans. Analytical Chemistry. 82, 3460–3466 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM: The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biological Chemistry. 393, 1357–1362 (2012) [DOI] [PubMed] [Google Scholar]
- 22.Hogan JD, Klein JA, Wu JD, Chopra P, Boons GJ, Carvalho L, Lin C, Zaia J: Software for Peak Finding and Elemental Composition Assignment for Glycosaminoglycan Tandem Mass Spectra. Mol Cell Proteomics. 17, 1448–1456 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu H, Huang Y, Mao Y, Yu X, Xu YM, Liu J, Zong CL, Boons GJ, Lin C, Xia Y, Zaia J: A Computational Framework for Heparan Sulfate Sequencing Using High-resolution Tandem Mass Spectra. Mol Cell Proteomics. 13, 2490–2502 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agyekum I, Patel AB, Zong CL, Boons GJ, Amster IJ: Assignment of hexuronic acid stereochemistry in synthetic heparan sulfate tetrasaccharides with 2-O-sulfo uronic acids using electron detachment dissociation. International Journal of Mass Spectrometry. 390, 163–169 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leach FE, Ly M, Laremore TN, Wolff JJ, Perlow J, Linhardt RJ, Amster IJ: Hexuronic Acid Stereochemistry Determination in Chondroitin Sulfate Glycosaminoglycan Oligosaccharides by Electron Detachment Dissociation. Journal of the American Society for Mass Spectrometry. 23, 1488–1497 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolff JJ, Chi LL, Linhardt RJ, Amster IJ: Distinguishing glucuronic from iduronic acid in glycosaminoglycan tetrasaccharides by using electron detachment dissociation. Analytical Chemistry. 79, 2015–2022 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaia J, Li XQ, Chan SY, Costello CE: Tandem mass spectrometric strategies for determination of sulfation positions and uronic acid epimerization in chondroitin sulfate oligosaccharides. Journal of the American Society for Mass Spectrometry. 14, 1270–1281 (2003) [DOI] [PubMed] [Google Scholar]
- 28.Karlin S, Altschul SF: Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proceedings of the National Academy of Sciences of the United States of America. 87, 2264–2268 (1990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlin S, Altschul SF: Applications and statistics for multiple high-scoring segments in molecular sequences. Proceedings of the National Academy of Sciences of the United States of America. 90, 5873–5877 (1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackey AJ, Haystead TAJ, Pearson WR: Getting more from less - Algorithms for rapid protein identification with multiple short peptide sequences. Mol Cell Proteomics. 1, 139–147 (2002) [DOI] [PubMed] [Google Scholar]
- 31.Filliben JJ, Heckert A: Exploratory Data Analysis. Engineering Statistics Handbook. [Google Scholar]
- 32.Fenyo D, Beavis RC: A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Analytical Chemistry. 75, 768–774 (2003) [DOI] [PubMed] [Google Scholar]
- 33.Field HI, Fenyo D, Beavis RC: RADARS, a bioinformatics solution that automates proteome mass spectral analysis, optimises protein identification, and archives data in a relational database. Proteomics. 2, 36–47 (2002) [PubMed] [Google Scholar]
- 34.Agyekum I, Zong CL, Boons GJ, Amster IJ: Single Stage Tandem Mass Spectrometry Assignment of the C-5 Uronic Acid Stereochemistry in Heparan Sulfate Tetrasaccharides using Electron Detachment Dissociation. Journal of the American Society for Mass Spectrometry. 28, 1741–1750 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kailemia MJ, Park M, Kaplan DA, Venot A, Boons GJ, Li LY, Linhardt RJ, Amster IJ: High-Field Asymmetric-Waveform Ion Mobility Spectrometry and Electron Detachment Dissociation of Isobaric Mixtures of Glycosaminoglycans. Journal of the American Society for Mass Spectrometry. 25, 258–268 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leach FE, Wolff JJ, Laremore TN, Linhardt RJ, Amster IJ: Evaluation of the experimental parameters which control electron detachment dissociation, and their effect on the fragmentation efficiency of glycosaminoglycan carbohydrates. International Journal of Mass Spectrometry. 276, 110–115 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leach FE, Xiao ZP, Laremore TN, Linhardt RJ, Amster IJ: Electron detachment dissociation and infrared multiphoton dissociation of heparin tetrasaccharides. International Journal of Mass Spectrometry. 308, 253–259 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaia J, Miller MJC, Seymour JL, Costello CE: The role of mobile protons in negative ion CID of oligosaccharides. Journal of the American Society for Mass Spectrometry. 18, 952–960 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.