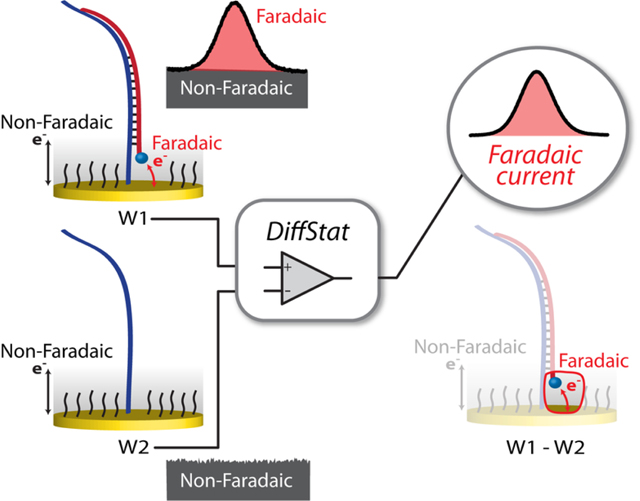
Abstract
One of the key factors limiting sensitivity in many electrochemical assays is the non-faradaic or capacitive current. This is particularly true in modern assay systems based on DNA monolayers at gold electrode surfaces, which have shown great promise for bioanalysis in complex milieu such as whole blood or serum. While various changes in analytical parameters, redox reporter molecules, DNA structures, probe coverage, and electrode surface area have been shown useful, background reduction by hardware subtraction has not yet been explored for these assays. Here, we introduce new electrochemistry hardware that considerably suppresses non-faradaic currents through real-time analog subtraction during current-to-voltage conversion in the potentiostat. This differential potentiostat (DiffStat) configuration is shown to suppress or remove capacitance currents in chronoamperometry, cyclic voltammetry, and square-wave voltammetry measurements applied to nucleic acid hybridization assays at the electrode surface. The DiffStat makes larger electrodes and higher sensitivity settings accessible to the user, providing order-of-magnitude improvements in sensitivity, and it also significantly simplifies data processing to extract faradaic currents in square-wave voltammetry (SWV). Since two working electrodes are used for differential measurements, unique arrangements are introduced such as converting signal-OFF assays to signal-ON assays, or background drift correction in 50 % human serum. Overall, this new potentiostat design should be helpful not only in improving the sensitivity of most electrochemical assays, but it should also better support adaptation of assays to the point-of-care by circumventing complex data processing.
Graphical Abstract
INTRODUCTION
Direct electrochemical analysis of living systems requires highly selective recognition and the ability to remove background interferences in matrices such as cell lysates, whole blood, or serum. Electrochemical readout has even been accomplished in vivo, although this was previously limited to enzyme-based biosensors or direct detection of endogenous electrochemically active species1. More recently, systems relying on DNA monolayers on gold electrodes for sensing or positioning of binding reagents have offered new opportunities for selective electrochemistry in complex backgrounds2–14. For example, the electrochemical DNA based (E-DNA) or aptamer-based (E-AB) biosensors introduced by the Plaxco group15 have emerged as alternatives to DNA sensors or immunoassays because of their sensitivity, affordability, selectivity, and minimal instrumentation requirements16–20. Other high-performance approaches have been devised such as nanostructured electrodes21, membrane-protected aptamers22, or antibody-oligonucleotide sensors (ECPA)23–26, all similarly relying on DNA- or RNA-based monolayers at gold electrode surfaces using standard three-electrode electrochemical cells. While these methods afford the necessary high selectivity and sensitivity for analysis of some selected analytes in living systems, there is significant room for improvement in reducing background interferences such as non-faradaic currents and signal drifts.
Optimizations of such systems have focused on instrumental parameters27, redox reporter molecules25,28, DNA structures12,29, self-assembled monolayer (SAM) coverage30,31 and chemistry32, electrode surface area21,33, and melting studies34,35. Although these are comprehensive studies, few reports have focused on reducing non-faradaic current at the electrode surface by hardware subtraction36. To our knowledge, there have been no reports exploiting hardware subtraction of non-faradaic currents with the nucleic acid-based monolayer sensors discussed above, and this presents an opportunity for further improvement in signal-to-background ratios. This non-faradaic current, which originates from the formation of a double layer at the electrode and monolayer surface, is defined by a time-dependent capacitive current in the electrochemical measurement. These currents serve as interferences (or background) to the analytical faradaic current, compromising the effectiveness of the sensors.37
One approach to reducing non-faradaic current is a working electrode surface area reduction, as the magnitude is directly proportional to surface area (SA).37 This, however, has the consequence of also reducing faradaic current, requiring higher sensitivity instrumentation. Conversely, when SA is made large, corresponding to higher currents (faradaic and non-faradaic), the instrument’s amplifiers can saturate, limiting high SA electrode use, which ultimately limits the detection sensitivity of the instrument. The non-faradaic component of the electrochemical signal can also be removed during data analysis by differential measurement techniques. Indeed, differential electrochemical measurements are commonly used for noise and background reduction.37–40 Techniques such as square-wave voltammetry (SWV) or pulse voltammetry are usually applied with nucleic acid-based sensing, and recent reports have even used clever fitting of chronoamperometric data to remove background41. However, these subtractions or corrections are carried out digitally; for example, currents from specific points in the voltammogram are subtracted by means of computer processing.40 This process affords a digital reduction in non-faradaic current, yet these currents remain in the raw data and consequently can limit usable electrode SA, detection range, and sensitivity.
If non-faradaic current is removed by hardware subtraction prior to measurement of the biosensor output, analysis can be fixated on the important faradaic components of the signal. Furthermore, larger SA electrodes should be accessible, allowing larger amplitude output signals with low background, thus higher sensitivities. This approach could also open new experimental possibilities such as continuous correction in complex matrices. While other groups have shown that sequential collection and subtraction of background and signal42–45 can provide some of these benefits with in vivo neurotransmitter detection, the measurements are fundamentally made at different times, restricting the noise rejection capabilities. A few reports have applied hardware or analog subtraction of background, such as the glucose monitoring by Deman et al.46 or high density CMOS devices36,47,48, yet these works are specialized or focused on integrated microelectrode devices. Realizing that hardware background subtraction could be very useful if applied to nucleic-acid monolayer based sensors (E-DNA, AB-DNA, ECPA, etc.), we set out to construct a differential potentiostat for this purpose, starting from an open source design49, which could be used with standard electrochemical equipment.
In this paper, a differential potentiostat (DiffStat) is constructed to utilize two working electrodes, where one electrode provides a signal and the second provides a background, which is subtracted from the first. The background subtraction is performed synchronously, during data collection from both electrodes by a differential operational amplifier configuration. The differential potentiostat is then applied to a DNA monolayer based sensor and validated against a standard potentiostat with several techniques such as chronoamperometry (CA), cyclic voltammetry (CV), and SWV. As envisioned, this instrument shows improvements in surface area accessibility, increased signal, decreased background, and decreased noise. Finally, we show unique applications such as converting a traditionally “signal-off” assay to a “signal-on” assay, as well as real-time background correction in human serum.
RESULTS AND DISCUSSION
Recent advances in bioanalytical electrochemistry—particularly those built around DNA monolayers on gold electrodes—have reinvigorated the prospect of selective electrochemistry in complex matrices2–14. However, there is no obvious consensus on how to further minimize background interferences such as non-faradaic currents and signal drifts. While various clever approaches in data analysis have been devised, one option that has yet to be explored for these nucleic acid-based monolayer sensors is to carry out hardware subtraction of non-faradaic current, i.e. the capacitive current. Capacitive current generally acts as a non-zero baseline, which can place an upper limit on signal-to-noise ratios and result in narrow detection ranges. In this work, we evaluate a novel differential potentiostat (DiffStat) design (Fig. 1B) which analog subtracts much of the capacitive current within the potentiostat circuitry, outputting a signal that is predominantly composed of faradaic current. The results herein show the DiffStat to be useful for significant background reduction and reduced noise while also supporting a wider range of electrode areas and instrument sensitivity settings.
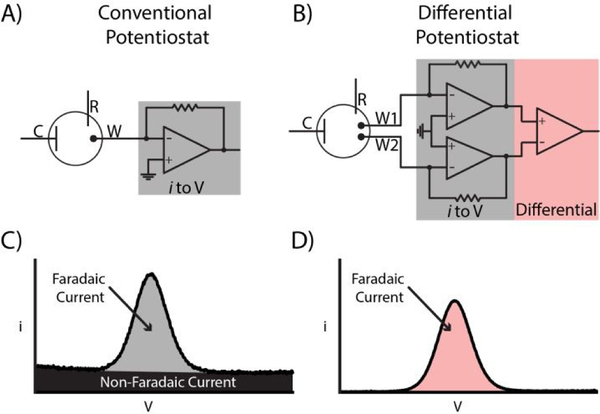
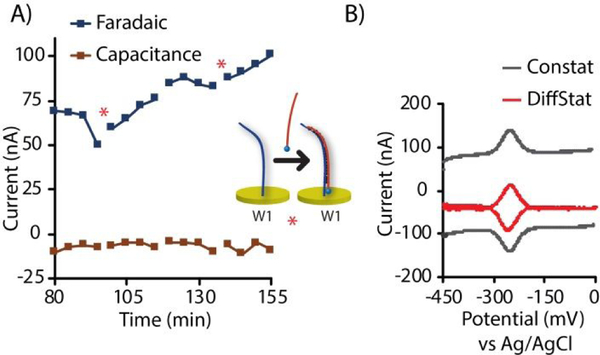
Figure 1.
Comparison of a conventional potentiostat (ConStat) with the differential potentiostat (DiffStat) presented herein. Both instruments incorporate Counter, Reference and Working electrodes. A) In the ConStat, current generated by redox active molecules is measured and converted to a voltage proportional to the current. B) The DiffStat contains a second working electrode, W2, usually to serve as a non-faradaic “blank” or control electrode that is not exposed to the redox molecule. Both signal (faradaic) and baseline (non-faradaic) currents can be subtracted by a differential amplifier, yielding a corrected output (W1 - W2). C) Example data shows that the ConStat signal contains both faradaic and non-faradaic currents, while D) the DiffStat significantly reduces non-faradaic current yet preserves the faradaic current.
Differential Potentiostat (DiffStat) Concept.
In our attempts to address this unfavorable background at its source, we surmised that a logical way to remove the non-faradaic, capacitive currents from nucleic acid-based monolayer sensors would be to actively subtract the baseline during the measurement. One way to do this would be to build a differential circuit into the potentiostat that continuously subtracts baseline at a “blank” working electrode (W2, with no analyte) from the experimental working electrode (W1, with analyte). This would necessarily involve instrument design changes—perhaps a reason the approach has not been previously reported—yet the concept is fairly straightforward and easy to implement once the hardware changes are made. Fig. 1A shows a conventional potentiostat (ConStat) configuration using a standard cell with one working electrode connected to a current-to-voltage converter or transimpedence amplifier (TIA). By comparison, the DiffStat circuitry (Fig. 1B) utilizes a cell with two working electrodes (W1 and W2) and matching TIA circuits that feed to an additional on-board differential instrumentation amplifier. The signals from W1 and W2 are collected simultaneously, analog subtracted by the instrumentation amplifier, and quantified. By treating W1 and W2 identically, except exposing the signaling component only to W1, the capacitance devoid output can be generated. Example SWV signals from a surface hybridization based DNA sensor collected with the ConStat (Fig. 1C) and DiffStat (Fig. 1D) exemplify the non-faradaic current suppression capability of the DiffStat. While faradaic current was unchanged, non-faradaic current was greatly reduced, essentially to zero.
After initial Diffstat verification by dummy cell studies (data not shown), the DiffStat and ConStat were compared through a hybridization-based sensor for nucleic acid detection. Methylene blue (MB) appended DNA, which hybridizes to surface thiolated DNA through 40 bp, was selected as the target analyte (MB-DNA) and added to W1. The same DNA sequence in the absence of MB (CTR-DNA) was used as a control and added to W2 to carefully match the non-faradaic components between W1 and W2. To eliminate cross contamination between working electrodes, W1 and W2 were fabricated in two separate cells, and a split reference electrode and counter electrode were exploited for electrochemical contact (Fig. S-2). Comparative DiffStat and ConStat results using CA, CV, and SWV are discussed in detail below.
Chronoamperometry and Cyclic Voltammetry.
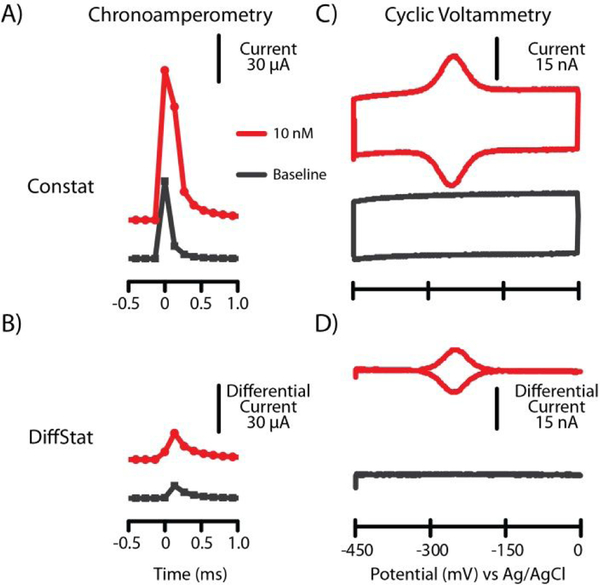
Initially, the DiffStat was tested with classic techniques such as CA and CV. In CA, the potential is quickly swept across the redox potential and held at a certain point, and current is continuously measured and plotted against the time. Fig. 2AB shows the raw chronoamperometric output of baseline (no analyte) and 10 nM MB-DNA from the ConStat and DiffStat at a collection frequency of 7.5 kHz. The DiffStat showed ~5-fold suppression in capacitance current of the baseline, and a similar trend was observed with 10 nM MB-DNA. While this experiment required a detection range setting of ± 500 μA with the ConStat, the DiffStat permitted use of the ± 50 μA range setting (10-fold higher sensitivity) due to its baseline reduction capability.
Figure 2.
Classical electrochemical experiments. A) Chronoamperometry (CA) measurents from the ConStat showed higher capacitance, while B) the DiffStat CA data showed capacitance to be suppressed. C) Cyclic voltammetry (CV) by the ConStat gave ~25 nA of non-faradaic current, while D) CV with the DiffStat showed efficient removal of capacitance current without losing faradaic current. Data in A and B is offset for clarity.
Using CV, the current measurement is done as voltage is swept at a constant rate across the redox potential of the analyte then cycled back to the initial potential. The cyclic voltammagram outputs from the ConStat and DiffStat are shown in Fig. 2C–D. Here, the 25 nA non-faradaic current observed by the ConStat was suppressed 99.4% by the DiffStat (~160-fold). Conversely, the faradaic current generated by 10 nM MB-DNA was of similar magnitude with either potentiostat. Again, an added bonus is that the DiffStat was capable of measuring at a higher instrument sensitivity range (± 50 nA and ± 15 nA were used for ConStat and DiffStat, respectively). These data supported our hypothesis that the DiffStat circuitry could essentially negate non-faradaic current while preserving the faradaic current.
Square-Wave Voltammetry.
DNA based electrochemical sensors have been predominantly analyzed with SWV. This is a pulse voltammetry technique developed to improve the signal-to-noise ratios, where a voltage pulse is applied, and the current measurement is delayed (~ms range) until capacitance current has significantly decayed50. The approach not only gives a general increase in signal-to-background ratio, but frequency-resolved SWV has also been shown capable of discriminating target-bound probes from unbound probes51 in DNA based electrochemical sensors. It was therefore important for us to thoroughly characterize the suitability of the DiffStat for SWV readout of these sensors. We carefully evaluated the system using a wide range of electrode sizes (diameters from 0.1 to 6.0 mm). While large surface area electrodes are known to give proportionally higher faradaic signals, they also increase non-faradaic currents detrimentally, since double layer capacitance is proportional to the metal electrode’s surface area.37 Large electrodes are not commonly used for this reason, but we hypothesized that the DiffStat would make larger electrodes more accessible to SWV measurements by cancelling the capacitance currents.
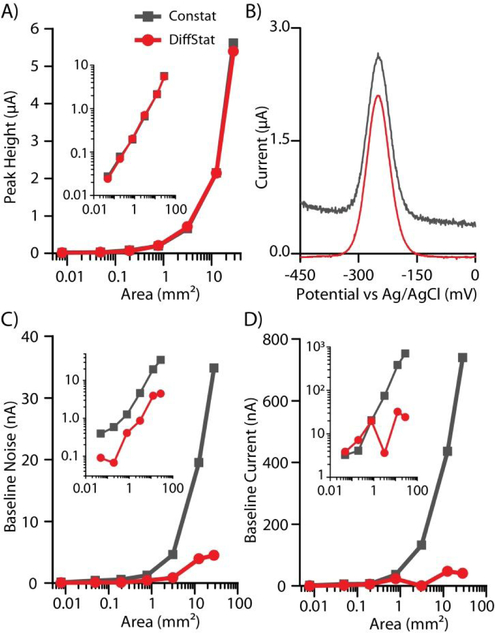
As expected, when sensing 10 nM of MB-DNA (40-bp hybridization), the increases in electrode surface area gave proportional increases in both faradaic peak current (Fig. 3A, gray) and non-faradaic baseline current (Fig. 3D, gray) with the ConStat. However, while the DiffStat’s faradaic current (Fig. 3A, red) followed the same trend as the ConStat, the baseline non-faradaic current (Fig. 3D, red) was greatly suppressed. For example, the 6 mm electrode gave ~5500 nA of faradaic peak current and ~700 nA of non-faradaic baseline when using the ConStat; while the DiffStat showed equal peak height (~5500 nA) but only ~50 nA of non-faradaic baseline. The SWV baseline was thus reduced ~14-fold using the DiffStat.
Figure 3.
Non-faradaic (capacitance) suppression across a wide range of electrode areas using square-wave voltammetry (SWV). A) Faradaic peak heights from 10 nM MB-DNA were equal using the DiffStat and ConStat, as shown in B) example unprocessed SWV data with a 4 mm diameter electrode. However, C) baseline noise and D) non-faradaic baseline current (forward SWV) was greatly reduced using the DiffStat (red) compared to the ConStat (gray). Insets show log-log plots of the same data.
Additionally, the baseline noise increased proportionally with the electrode area using the ConStat (Fig. 3C, gray), likely a result of thermal noise created by the gain resistor in the potentiostat. Since the differential instrumentation amplifier in the DiffStat combats environmental noise (including thermal) by subtracting the common mode noise (noise present on both input terminals to the amplifier) during the analysis, the overall noise floor for the DiffStat instrument was lower (Fig. 3C, red). With the 6 mm diameter electrode, an ~8-fold reduction in noise was observed with the DiffStat compared to the ConStat. Such an improvement should theoretically reduce an assay’s limit of detection (LOD) by ~8-fold if all other settings are held constant.
The example square-wave voltammagrams in Fig. 3B, using the 4 mm diameter electrode, are representative of these findings. The ConStat data (gray) exhibited higher baseline current, higher baseline noise, and equal faradaic peak current when compared to the DiffStat data (red). These improvements given by the DiffStat contribute to better signal-to-noise ratios and effective usage of lower current ranges (higher sensitivities). Considering the standard 3σ calculation, where assay LOD is directly proportional to noise and inversely proportional to sensitivity, the DiffStat should be a useful tool for enhancing assay LODs.
Improved Detection Range or Sensitivity in SWV.
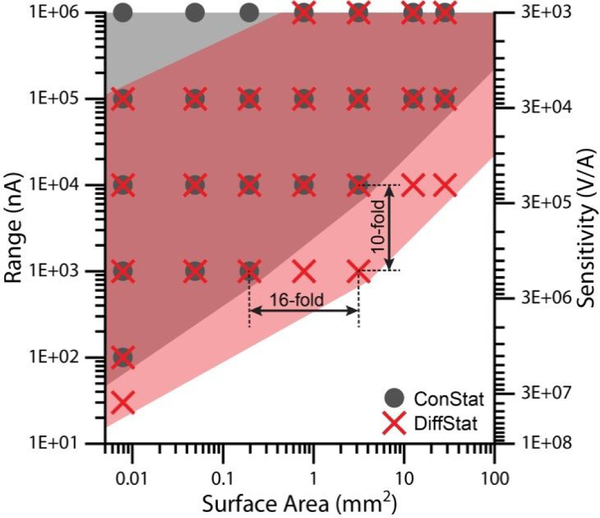
To gain a better understanding of the sensitivity improvements, we again studied the MB-DNA hybridization sensor using various electrode surface areas, where the measurements were done with 6 detection ranges (between ± 500 μA and ± 15 nA). Note that lower detection range equates to higher sensitivity in the current-to-voltage conversion. Fig. 4 shows the comparison, where the operational ranges of the DiffStat and ConStat are plotted against surface area. At all electrode areas evaluated, these data show that DiffStat enables higher sensitivity ranges (pink/red) compared to the ConStat (gray). The labeled example in Fig. 4, using 2 mm diameter electrodes (3.14 mm2 area), gave a 10-fold improvement in sensitivity compared to the ConStat. The DiffStat also allowed a 16-fold increase in accessible electrode area at the 1 μA detection range, permitting use of the 2 mm diameter electrodes under that setting. The ConStat only functioned with 0.5 mm diameter electrodes (0.196 mm2 area) in the 1 μA detection range, with signal saturating the instrument’s analog-to-digital converter when using larger electrodes. These significant improvements in SWV sensitivity or detection range result from the strong suppression of capacitive (non-faradaic) baseline currents by the DiffStat. While there were only 6 detection ranges available for testing in these instruments, generally the DiffStat was shown to be capable of detecting with at least 10-fold higher sensitivity than a conventional instrument, which should translate to lower concentrations of analytes at higher current resolution.
Figure 4.
Detection range and sensitivity can be improved significantly with the DiffStat. Larger electrodes and more sensitive amplifier settings were accessible using the DiffStat (pink shading, red X data), settings which caused instrument overload on the ConStat (gray shading, gray circles). Data points in the figure mark instrument settings which were accessible to a given instrument.
Faradaic Current Cancellation.
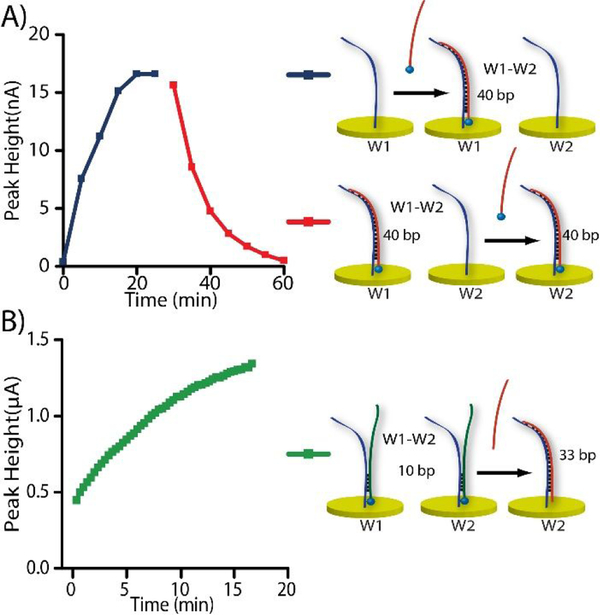
Modern methods such as E-DNA and aptasensors use redox molecule-appended DNA for quantification.52 In addition to the removal of capacitance current, if the initial “blank” faradaic current were also cancelled, the instrument could be used in a higher sensitivity range, which should help improve the assay ranges and LODs. Because the DiffStat’s fundamental property is to subtract signal between two electrodes (W1 and W2 in Fig. 1), we hypothesized that the instrument could be used to cancel initial blank currents in sensors that start with significant faradaic signals. The data in Fig. 5A supports this hypothesis. Using a DNA hybridization sensor, MB-DNA (10 nM) was added to W1, while W2 contained no faradaic signal. As expected, the DiffStat’s faradaic peak height (blue) increased over time as the MB-DNA hybridized. After it reached equilibrium (~25 min), the same MB-DNA (10 nM) was then added to W2. This introduced the same redox molecule to both W1 and W2, and the result was a suppression of faradaic current (red) in the final DiffStat output, which reached zero at ~60 min.
Figure 5.
Unique features of the DiffStat instrument that should prove useful in bioanalysis. A) Faradaic current cancellation was shown possible by adding labeled analyte to both W1 and W2, sequentially. The initial faradaic peak increase at W1 (blue) was cancelled by adding MB-DNA also to W2, thereby subtracting the current back to zero (red) in real time. B) A signal-OFF assay could be transformed to a signal-ON assay using a similar concept, with appropriate choice of W2.
Changing Signal-OFF to Signal-ON.
For a sensitive “signal-OFF” assay—one that decreases signal with analyte addition—the initial signal should be large. However, to measure a larger signal, the instrument must be operated at higher current range, i.e. at lower sensitivity. Since faradaic current at W1 can be corrected with an appropriately prepared W2, the DiffStat should be capable of transforming a signal-OFF assay to a signal-ON assay, thereby allowing low concentrations to be interrogated at the instrument’s highest sensitivity settings. This concept is illustrated by data in Fig. 5B, where a 33-bp competitor DNA (Com-33) was used to push pre-hybridized MB-DNA2 away from W2, thereby increasing the difference between W1 and W2. The assay which would normally be signal-OFF, was thereby transformed to a signal-ON assay. This approach should prove useful with a variety of recently developed aptasensors, E-DNA sensors, nanostructures, etc.
Measurements in human serum.
One of the many advantages of DNA-monolayer-based electrochemical sensors is their proven compliance for measurement in human serum or blood (even undiluted). Nonetheless, baseline drifts remain an issue that must be dealt with in serum or plasma. Clever approaches to overcome this drift, such as measuring multiple SWV frequencies or tagging different redox molecules to the sensor have been developed7,8, where signals are later corrected during data processing. The DiffStat, however, provides a straightforward solution to address this drift by exploiting real-time subtraction of the “blank” working electrode (W2). On engaging both working electrodes into a complex matrix, the drift should be largely cancelled. Real-time DNA detection in 50 % serum, as shown in Fig. 6A, validates the use of the DiffStat in complex matrices. In this experiment, every 20 min the W1 solution was alternated between 50 % human serum with 10 nM MB-DNA (analyte) and 50 % human serum in the absence of analyte. Faradaic current was shown to increase in the presence of analyte, while no drift in the baseline was observed with or without analyte. In Fig. 6B, CV outputs from the ConStat and DiffStat were compared in 50 % human serum. While the ConStat showed a large baseline, the baseline was absent when using the DiffStat. This characteristic of the DiffStat should make it particularly suitable for point-of-care detection, since sensitivity could be increased while post-measurement processing is minimized.
Figure 6.
Signal acquisition in 50 % human serum with the DiffStat. A) Over time, MB-DNA introduced in 50 % human serum showed increased faradaic current with stable, near-zero non-faradaic (capacitance) current. B) CV comparisons using the DiffStat and ConStat in 50 % human serum showed large non-faradaic current with the ConStat which was absent with the DiffStat, while faradaic currents were comparable.
CONCLUSIONS
A new potentiostat arrangement was introduced, in which non-faradaic current and environmental noise were suppressed or removed by analog subtraction of a “blank” working electrode. Compared to the typical digital subtraction or noise filtering, this approach of real-time hardware subtraction was shown to be both advantageous for signal enhancement and enabling for novel measurements. The DiffStat was proven useful for enhancing DNA based surface hybridization assays, enabling larger electrodes and higher sensitivity settings to be chosen. Unique arrangements of the two working electrodes allowed faradaic background subtraction that can transform signal-OFF assays to signal-ON assays or permit continuous background correction in human serum. This new instrument should also enhance the transition of SWV based assays to point-of-care settings, since faradaic currents can be directly determined without further data processing.
Since two working electrodes are used with the DiffStat, a variety of other configurations could be devised for future studies. Care should be taken in modifying the working electrode surfaces, and the choice of using either one cell with two working electrodes or two separate cells will depend on the application. To elaborate, if the user desires to remove non-faradaic or capacitive current as shown in this work, the blank working electrode (W2) should be chosen to mimic the primary working electrode (W1) in all aspects other than the faradaic current. In other words, the non-faradaic currents between W1 and W2 should be perfectly matched in theory. In this work, we have not yet reached this idealized case, as evidenced by the non-zero capacitance in Fig. 2B and baseline current in Fig. 3B. It is feasible that hardware based, real-time adjustment could be added to further improve this match in the future. Another possibility would be to apply the DiffStat to modern aptasensors16,17,41, with W1 containing the aptamer and W2 having a mutated aptamer or scrambled sequence that does not bind the target. The DiffStat output would thus be reduced to target-binding events only. Yet another example would be to tune the system to quantify small differences in redox mediator-dependent assays5,10. In our view, these are only a few of many unique arrangements that could be devised for and enabled by the DiffStat instrument.
Supplementary Material
ACKNOWLEDGMENT
(Word Style “TD_Acknowledgments”).
Funding Sources
Support for this work was provided by the National Institutes of Health (R01 DK093810), by the National Science Foundation (CBET-1403495), and by the Department of Chemistry and Biochemistry at Auburn University.
ABBREVIATIONS
- GoG
gold on glass electrode
- PDMS
polydimethylsiloxane
- DiffStat
differential potentiostat
- ConStat
conventional potentiostat
- CA
chronoamperometry
- CV
cyclic voltammetry
- SWV
square wave voltammetry
- MB-DNA
methylene blue DNA
- CTR-DNA
control DNA
- Dstat
open source potentiostat
- TIA
transimpedance amplifier
Footnotes
Supporting Information.
This material is available free of charge via the Internet at http://pubs.acs.org.
Supporting figures and tables
The authors declare the following competing financial interest (s): The differential potentiostat and methodology presented in this article was included patent application filed in March of 2018.
REFERENCES
- (1).Wilson GS; Johnson MA Chemical reviews 2008, 108, 2462–2481. [DOI] [PubMed] [Google Scholar]
- (2).Arroyo-Curras N; Somerson J; Vieira PA; Ploense KL; Kippin TE; Plaxco KW Proceedings of the National Academy of Sciences of the United States of America 2017, 114, 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bonham AJ; Paden NG; Ricci F; Plaxco KW The Analyst 2013, 138, 5580–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ferguson BS; Hoggarth DA; Maliniak D; Ploense K; White RJ; Woodward N; Hsieh K; Bonham AJ; Eisenstein M; Kippin TE; Plaxco KW; Soh HT Science translational medicine 2013, 5, 213ra165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lam B; Das J; Holmes RD; Live L; Sage A; Sargent EH; Kelley SO Nature communications 2013, 4, 2001. [DOI] [PubMed] [Google Scholar]
- (6).Lam B; Fang Z; Sargent EH; Kelley SO Analytical chemistry 2012, 84, 21–25. [DOI] [PubMed] [Google Scholar]
- (7).Li H; Arroyo-Curras N; Kang D; Ricci F; Plaxco KW Journal of the American Chemical Society 2016, 138, 15809–15812. [DOI] [PubMed] [Google Scholar]
- (8).Li H; Dauphin-Ducharme P; Ortega G; Plaxco KW Journal of the American Chemical Society 2017, 139, 11207–11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liu J; Wagan S; Davila Morris M; Taylor J; White RJ Analytical chemistry 2014, 86, 11417–11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mahshid SS; Ricci F; Kelley SO; Vallee-Belisle A ACS sensors 2017, 2, 718–723. [DOI] [PubMed] [Google Scholar]
- (11).Schoukroun-Barnes LR; Wagan S; White RJ Analytical chemistry 2014, 86, 1131–1137. [DOI] [PubMed] [Google Scholar]
- (12).Somasundaram S; Easley CJ Journal of the American Chemical Society 2019, 141, 11721–11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Somerson J; Plaxco KW Molecules (Basel, Switzerland) 2018, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Vallee-Belisle A; Ricci F; Uzawa T; Xia F; Plaxco KW Journal of the American Chemical Society 2012, 134, 15197–15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fan C; Plaxco KW; Heeger AJ Proceedings of the National Academy of Sciences of the United States of America 2003, 100, 9134–9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Liu Y; Tuleouva N; Ramanculov E; Revzin A Analytical chemistry 2010, 82, 8131–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lubin AA; Plaxco KW Accounts of chemical research 2010, 43, 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mahshid SS; Camire S; Ricci F; Vallee-Belisle A Journal of the American Chemical Society 2015, 137, 15596–15599. [DOI] [PubMed] [Google Scholar]
- (19).Ricci F; Plaxco KW Microchim. Acta 2008, 163, 149–155. [Google Scholar]
- (20).Xiao Y; Piorek BD; Plaxco KW; Heeger AJ Journal of the American Chemical Society 2005, 127, 17990–17991. [DOI] [PubMed] [Google Scholar]
- (21).Soleymani L; Fang Z; Sargent EH; Kelley SO Nature nanotechnology 2009, 4, 844–848. [DOI] [PubMed] [Google Scholar]
- (22).Santos-Cancel M; White RJ Analytical chemistry 2017, 89, 5598–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hu J; Wang T; Kim J; Shannon C; Easley CJ Journal of the American Chemical Society 2012, 134, 7066–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hu J; Yu Y; Brooks JC; Godwin LA; Somasundaram S; Torabinejad F; Kim J; Shannon C; Easley CJ Journal of the American Chemical Society 2014, 136, 8467–8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ren K; Wu J; Yan F; Ju H Scientific reports 2014, 4, 4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhu J; Gan H; Wu J; Ju H Analytical chemistry 2018, 90, 5503–5508. [DOI] [PubMed] [Google Scholar]
- (27).Dauphin-Ducharme P; Plaxco KW Analytical chemistry 2016, 88, 11654–11662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kang D; Ricci F; White RJ; Plaxco KW Analytical chemistry 2016, 88, 10452–10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Huang KC; White RJ Journal of the American Chemical Society 2013, 135, 12808–12817. [DOI] [PubMed] [Google Scholar]
- (30).Ricci F; Lai RY; Heeger AJ; Plaxco KW; Sumner JJ Langmuir : the ACS journal of surfaces and colloids 2007, 23, 6827–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).White RJ; Phares N; Lubin AA; Xiao Y; Plaxco KW Langmuir : the ACS journal of surfaces and colloids 2008, 24, 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li H; Dauphin-Ducharme P; Arroyo-Curras N; Tran CH; Vieira PA; Li S; Shin C; Somerson J; Kippin TE; Plaxco KW Angewandte Chemie (International ed. in English) 2017, 56, 7492–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hauke A; Kumar LSS; Kim MY; Pegan J; Khine M; Li H; Plaxco KW; Heikenfeld J Biosensors & bioelectronics 2017, 94, 438–442. [DOI] [PubMed] [Google Scholar]
- (34).Somasundaram S; Holtan MD; Easley CJ Analytical chemistry 2018, 90, 3584–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yang AH; Hsieh K; Patterson AS; Ferguson BS; Eisenstein M; Plaxco KW; Soh HT Angewandte Chemie (International ed. in English) 2014, 53, 3163–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hassibi A; Lee TH IEEE Sens. J 2006, 6, 1380–1388. [Google Scholar]
- (37).Bard AJ; Faulkner LR Electrochemical Methods: Fundamentals and Applications; Wiley, 2000. [Google Scholar]
- (38).Jacobs P; Suls J; Sansen W Sensors and Actuators B: Chemical 1994, 20, 193–198. [Google Scholar]
- (39).McGrath MJ; Iwuoha EI; Diamond D; Smyth MR Biosensors & bioelectronics 1995, 10, 937–943. [DOI] [PubMed] [Google Scholar]
- (40).Osteryoung JG; Osteryoung RA Analytical chemistry 1985, 57, 101–110. [Google Scholar]
- (41).Arroyo-Curras N; Dauphin-Ducharme P; Ortega G; Ploense KL; Kippin TE; Plaxco KW ACS sensors 2018, 3, 360–366. [DOI] [PubMed] [Google Scholar]
- (42).Dorta-Quiñones CI; Wang XY; Dokania RK; Gailey A; Lindau M; Apsel AB IEEE Transactions on Biomedical Circuits and Systems 2016, 10, 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hermans A; Keithley RB; Kita JM; Sombers LA; Wightman RM Analytical chemistry 2008, 80, 4040–4048. [DOI] [PubMed] [Google Scholar]
- (44).Howell JO; Kuhr WG; Ensman RE; Mark Wightman R Journal of Electroanalytical Chemistry and Interfacial Electrochemistry 1986, 209, 77–90. [Google Scholar]
- (45).Wang J; Dewald HD Talanta 1984, 31, 387–390. [DOI] [PubMed] [Google Scholar]
- (46).Deman P; Suls J; Sansen W Sensors and Actuators B: Chemical 1997, 44, 304–308. [Google Scholar]
- (47).Martin SM; Gebara FH; Strong TD; Brown RB IEEE Sens. J 2009, 9, 135–142. [Google Scholar]
- (48).Nazari MH; Genov R In 2009 IEEE International Symposium on Circuits and Systems, 2009, pp 2177–2180. [Google Scholar]
- (49).Dryden MD; Wheeler AR PloS one 2015, 10, e0140349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ramaley L; Krause MS Analytical chemistry 1969, 41, 1362–1365. [Google Scholar]
- (51).White RJ; Plaxco KW Analytical chemistry 2010, 82, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Kang D; Zuo X; Yang R; Xia F; Plaxco KW; White RJ Analytical chemistry 2009, 81, 9109–9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.