Abstract
Polychlorinated biphenyls (PCBs) contaminate 19% of US superfund sites and represent a serious risk to human and environmental health. One promising strategy to remediate PCB contaminated sediments utilizes organohalide respiring bacteria (OHRB) that dechlorinate PCBs. However, functional genes that act as biomarkers for PCB dechlorination processes (i.e. reductive dehalogenase genes) are poorly understood. Here we developed anaerobic sediment microcosms that harbor an OHRB community dominated by the genus Dehalococcoides. During the 430-day microcosm incubation Dehalococcoides 16S rRNA sequences increased two orders of magnitude to 107 copies/gram of sediment, and at the same time PCB118 decreased by as much as 70%. In addition, the OHRB community dechlorinated a range of penta- and tetra-chlorinated PCB congeners including PCBs 66, 70+74+76, 95, 90+101, and PCB110 without exogenous electron donor. We quantified candidate reductive dehalogenase (RDase) genes over a 430-day incubation period and found rd14, a reductive dehalogenase that belongs to Dehalococcoides mccartyi strain CG5, was enriched to 107 copies/gram of sediment. At the same time, pcbA5 was enriched to only 105 copies/gram of sediment. A survey for additional RDase genes revealed sequences similar to strain CG5’s rd4, and rd8. In addition to demonstrating the PCB dechlorination potential of native microbial communities in contaminated freshwater sediments, our results suggest candidate functional genes with previously unexplored potential could serve as biomarkers of PCB dechlorination processes.
Keywords: PCBs, Dehalococcoides mccartyi, reductive dehalogenase genes, sediment microcosms, reductive dechlorination, polychlorinated biphenyl
Introduction
The United States banned polychlorinated biphenyl (PCB) production forty years ago; however, PCBs released into aquatic sediments before the ban persist because of their high molecular stability, tendency to sorb strongly to organic matter, and low solubility in water (Robertson &Hansen 2015). As of 2017, 257 of 1337 Superfund National Priority List sites listed PCBs as contaminants of concern (USEPA 2017). Despite the high financial cost to remove contaminated sediments, site remediation often requires dredging and excavation to mitigate PCB contamination (Gomes et al. 2013, National Research Council 2007, Perelo 2010). In addition, physical removal methods like dredging may elevate PCB concentrations in pore water, surrounding air, and increase exposure risks to humans and aquatic organisms (Martinez et al. 2017, Martinez et al. 2015, Martinez et al. 2010).
In contrast to practices that physically remove contaminated sediments, several studies suggest less intrusive methods like bioremediation can reduce PCB sediment mass in situ (Payne et al. 2013, Payne et al. 2017, Payne et al. 2011). For instance, a mesocosm study showed that bioaugmentation with PCB-dechlorinating Dehalobium chlorocoercia strain DF-1 reduced PCB congeners with five or more chlorine substituents by 58% (Payne et al. 2011). Another study revealed sediment PCB mass decreased by as much as 80% in contaminated sediments bioaugmented with multiple strains of PCB-degrading bacteria (Payne et al. 2013, Payne et al. 2017). Both studies relied on anaerobic bacteria that respire halogenated compounds to produce lesser-chlorinated congeners that are more amenable to aerobic degradation (Abraham et al. 2002). The bacteria that respire PCBs and other halogenated compounds utilize reductive dehalogenase (RDase) enzymes to catalyze the dehalogenation reaction (Hug et al. 2013), but it is unclear which RDase genes catalyze PCB dehalogenation in contaminated sediments. Further, organohalide respiration occurs naturally in the environment (Krzmarzick et al. 2012), and as a result specialized anaerobic bacteria dechlorinate PCBs without intervention at more than 20 contaminated sites (Abramowicz 1995). Because these PCB contaminated sites represent a niche for organohalide respiring bacteria (OHRB), monitored natural attenuation (MNA) may represent an additional in situ remediation strategy.
Although data on full scale MNA of PCBs is limited, MNA of chlorinated ethenes has been studied extensively (Himmelheber et al. 2007, Lebron et al. 2011, Lu et al. 2006, Wiedemeier et al. 1996). During MNA of chlorinated ethenes, the abundance of RDase genes (e.g., tceA, pceA, bvcA, and vcrA) reveals site specific dechlorination potential (Clark et al. 2018). In addition to functional gene abundance, Dehalococcoides spp. abundance is often monitored at contaminated sites because the genomes of these strict OHRB encode dozens of RDase genes (Hendrickson et al. 2002, Hug et al. 2013, van der Zaan et al. 2010). However, the abundance of Dehalococcoides spp. alone does ensure PCB specific metabolic capabilities which differ between strains (Lee et al. 2008). If bioaugmentation or MNA approaches are used to remediate PCB contaminated sediments it will be important to quantify functional genes associated with PCB dechlorination (i.e., those encoding RDases) to provide biological lines of evidence for remediation. However, RDase genes that participate in PCB dechlorination remain poorly understood. In fact, only three functional genes have been conclusively linked with PCB dechlorination processes – pcbA1, pcbA4, and pcbA5 (Wang et al. 2014), but it is unclear if the presence and abundance of these genes in contaminated sediment indicate potential for in situ PCB dechlorination.
Previously, we examined PCB contaminated sediments from a wastewater overflow lagoon in Altavista, Virginia (hereafter referred to as AVL). PCB congener analysis showed increased proportions of tri- and di-chlorinated congeners relative to Aroclor 1248, the likely contaminating PCB mixture (Mattes et al. 2018). Additionally, we sequenced partial RDase genes directly from PCB contaminated sediments, and subsequent phylogenetic analysis revealed up to 100% sequence similarity between these sequences and the RDase gene rd14 on the Dehalococcoides mccartyi strain CG5 genome (Mattes et al. 2018). Here, we aimed to confirm that the AVL sediment microbial community could actively dechlorinate the weathered PCB mixture without exogenous electron donor, identify the genera that likely respire PCBs, and conduct a survey for potential biomarker RDase genes. We also compared the abundance of rd14 with known PCB-specific RDase genes (pcbA1, pcbA4, pcbA5) in sediment microcosms to determine which genes may serve as PCB dechlorination biomarkers.
Materials and methods
PCB contaminated sediment sampling
Previously, we identified five locations within AVL where bacteria likely dechlorinated sediment PCBs (Mattes et al. 2018), and in 2016 we returned to collect sediment samples from the five identified locations (i.e., A1, C3, D2, E2, F4) (Figure S1). The PCB contaminated sediments are contained within an emergency wastewater overflow lagoon and were contaminated sometime before 1977. Additionally, PCBs are the only contaminants that have been reported in AVL sediments. We collected sediments with a hand auger from a boat and subsequently homogenized the samples in aluminum pans. Sediments were dark and sludge-like in consistency. Homogenized sediments were placed in 50 mL falcon tubes and stored at 4°C until use.
The organic carbon content of sediment samples was measured as the fraction of leachable organic carbon according to a previously described method (Mattes et al. 2018). Briefly, sediments were combined with 20 mL of deionized water and the supernatant decanted after overnight equilibration. Next, the supernatant was acidified with sulfuric acid, and total organic content was measured with a Shimadzu TOC-V analyzer operating in non-purgable organic carbon mode.
Microcosm Preparation
Microcosms were established in 160 mL serum bottles, and each bottle contained 100 mL of modified reduced anaerobic mineral media (Shelton &Tiedje 1984). Modifications to the media included 1 mL of the trace element solution (Table S1) and 10 mL of Vitamin Supplement (ATCC, Manassas, VA, USA). To prepare the anaerobic media, we inserted a gassing cannula into the media container and sparged with nitrogen gas for 20 minutes at a minimum flow rate of 0.2 mL/min. Next, a second gassing cannula was inserted into empty serum bottles to purge the oxygen, and the media was transferred to the serum bottles. The serum bottles were sealed with butyl rubber stoppers and aluminum crimp caps. Finally, the reducing agents L-cysteine hydrochloride (0.2 mM) and Na2S · 9 H2O (0.2 mM) were added as anaerobically prepared stock solutions. To inoculate the microcosms with PCB contaminated sediments the prepared media bottles were decrimped and unsealed in a nitrogen filled glove bag (Coy Laboratory Products, Grass Lake, MI, USA). Next, each serum bottle received 10 grams (wet weight), of the PCB contaminated AVL sediment. To obtain initial (0 days incubation) AVL sediment PCB congener concentrations, duplicate microcosms that contained media and sediment from each of the sampled locations were established and sacrificially sampled after 24 hours of equilibration. The abiotic controls contained media and sediments autoclaved at 121°C for one hour for three subsequent cycles. Duplicate microcosms were prepared for each sampled location and incubated statically in the dark at 25°C for 430 days.
DNA extraction and quantitative PCR analysis
To collect microcosm sediment samples for DNA extraction, we withdrew 3 mL of the microcosm slurry, centrifuged the samples at 16,000 × g for 8 minutes, and decanted the supernatant. Next, sediment DNA extraction was performed with the Qiagen DNeasy PowerSoil Kit according to manufacturer’s instructions. DNA extract concentrations were measured with the Qubit dsDNA high sensitivity assay kit.
We used quantitative PCR (qPCR) to amplify the 16S rRNA genes from the genera Dehalococcoides, Dehalobacter, Dehalogenimonas, and Geobacter and select RDase genes (Table S2). Each 25 μL reaction contained 12.5 μL of Power SYBR Green master mix, 1 μL of bovine serum albumin to relieve possible inhibition, and variable concentrations of primer (Table S3). The qPCR thermal cycling conditions and specific quality control methods have been described previously (Mattes et al. 2018). Briefly, for each set of primers a standard curve that ranged from 3×107 gene copies to 30 gene copies was prepared in triplicate to estimate the abundance of the target sequences. According to methods described previously, sequences amplified using AVL sediment DNA as template and transformed into clone libraries served as the standards for qPCR enumeration of all 16S sequences, CG5-rd14, bvcA, and vcrA. For primers that targeted pcbA1, pcbA4, pcbA5 and tceA g-block gene fragments (Integrated DNA Technologies, Coralville, IA, USA) served as the standards. As a quality control metric, qPCR product melt curves served as an indicator of nonspecific amplification, and melt curves were carefully screened for each reaction.
Reductive dehalogenase and 16S gene amplification and sequencing
DNA extracted from microcosm sediments after 430 days of incubation served as template in PCRs to amplify both the Dehalococcoides 16S rRNA gene and RDase gene orthologs. RDase gene orthologs were targeted with a set of previously developed degenerate primers (Hug &Edwards 2013). We attempted to amplify RDase orthologs in the template DNA with all 44 degenerate primer sets. PCR thermal cycling parameters for RDase gene orthologs were described previously (Mattes et al. 2018), and each 50 μL reaction contained approximately 100 ng of template DNA, 25 μL of TaqMan Universal PCR Master Mix, and 0.5 μL each of forward and reverse primers. Dehalococcoides 16S rRNA gene sequences were targeted with primers and thermal cycling conditions detailed elsewhere (Table S2) (Hendrickson et al. 2002, Krzmarzick et al. 2012).
Products of the RDase and Dehalococcoides 16S rRNA PCRs were screened with gel electrophoresis to determine product size. PCR products of the appropriate size were excised from the gel and purified with the QIAquick Gel Extraction Kit (Table S2). Next, a 1:1 product to vector ratio was used to clone products into pCR 2.1-TOPO vectors. Finally, the vector was transformed into TOP10 chemically competent E. coli. Successful transformations were visualized with blue and white screening on LB agar plates that contained 50 mg/L Kanamycin. White colonies were selected and grown overnight in LB broth with 50 mg/L Kanamycin. Next, plasmids were extracted with the QIAprep Spin Miniprep Kit before they were Sanger sequenced at the Iowa Institute of Human Genetics (University of Iowa, Iowa City, IA). Trimmed nucleotide sequences were translated into protein sequences, and sequence similarity was identified with NCBI Basic Local Alignment Search Tool (BLAST). Phylogenetic relationships were further analyzed with MEGA7 after alignment (Kumar et al. 2016).
PCB extraction and congener analysis
A liquid-liquid extraction based on previously described methods extracted PCBs from the media and sediment slurry (Anezaki &Nakano 2014, Marek et al. 2013). First, 0.5 mL of slurry was collected from each microcosm with a 16-gauge needle attached to a sterile syringe and placed into combusted 15 mL Pyrex tubes. Surrogate standards immediately added to the sampled slurry included 50 ng of PCB14 (3,5-dichlorobiphenyl), PCB65-d5 (2,3,5,6-tetrachlorobiphenyl-2′,3′,4′,5′,6′-d5, deuterated), and PCB166 (2,3,4,4′,5,6-hexachlorobiphenyl). Hexane was added to each sample (1:1 ratio), and the tubes were vortexed for 10 seconds before centrifugation at 3000 rpm for 10 minutes. After the organic solvent phase was decanted and transferred to a new tube, the procedure was repeated with an additional three mL of hexane. Next, two mL of concentrated sulfuric acid was added to remove lipids. After the solvent phase was decanted, the acid phase was re-extracted with three mL hexane. The combined organic solvent phase volume was concentrated to one mL and eluted with hexane through an acidified silica column prepared as described previously (Martinez &Hornbuckle 2011). The final eluent was concentrat ed to one mL and spiked with 25 ng of congeners PCB D30 (2,4,6-trichlorobiphenyl-2′,3′,4′,5′,6′-d5, deuterated), and PCB 204 (2,2′,3,4,4′,5,6,6′-octachlorobiphenyl) as internal standards. Each extracted batch of samples contained at least one laboratory blank comprised of hexane spiked with surrogate standards.
The total mass of PCBs in slurry extracts was quantified with a modified US EPA method 1668C described previously (Herkert et al. 2016, Martinez et al. 2017). All 209 PCB congeners were quantified with tandem mass spectrometry GC/MS/MS (Agilent 7000) in multiple reaction monitoring mode as 171 individual or coeluting congener peaks. The GC utilized a Supelco SBP-Octyl capillary column (30 m × 0.25 mm ID, 0.25 mm film thicknesses) with helium as a carrier gas.
As a metric of quality control and assurance, we calculated surrogate standard recoveries and quantified PCBs in method blanks to evaluate extraction procedure efficiencies and background PCB concentrations similar to methods described previously (Marek et al. 2017). A limit of quantification (LOQ) was developed using the mass of all 171 coeluting congeners quantified in each method blanks to calculate an average mass and standard deviation for each congener measured. Finally, a conservative congener specific LOQ was defined as the average plus two times the standard deviation. This LOQ was applied to all sample measurements and masses below the LOQ were treated as zero.
Statistical Analyses
PCB congener data were analyzed with an independent two-sided t-test with α = 0.05. The 16S rRNA sequence abundance and reductive dehalogenase gene abundances were analyzed with a two-factor analysis of variance (α = 0.05). Tukey’s multiple comparison tests were used to determine which means were statistically different from one another.
Nucleotide sequence accession numbers
Partial reductive dehalogenase gene sequences retrieved from sediment samples are deposited under Genbank accession numbers MK575138-MK575192. Dehalococcoides 16S rRNA gene sequences are deposited under Genbank accession numbers MK879451- MK879459.
Results & Discussion
The native microbial community at AVL dechlorinated a suite of penta and tetra-chlorinated PCBs
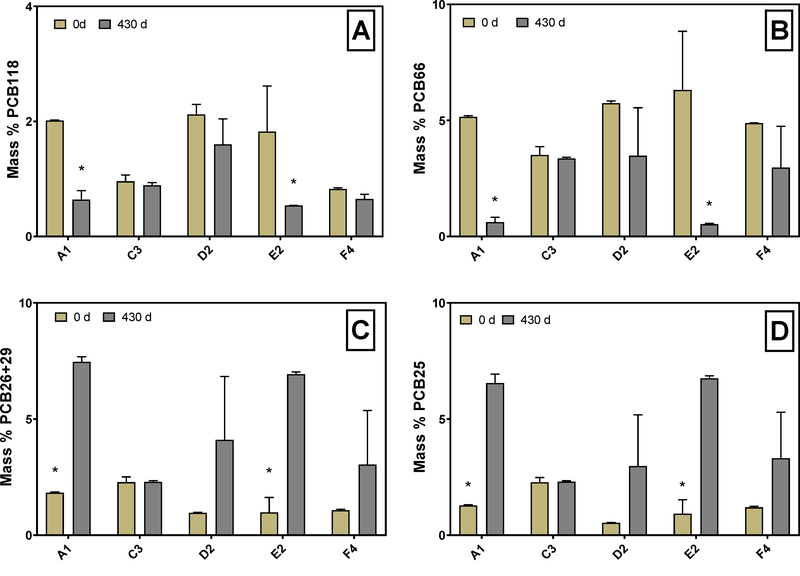
With data collected from the microcosm study, we aimed to assess if anaerobic natural attenuation could occur in the AVL PCB contaminated sediments. To examine changes in the PCB profile that result from dechlorination, we normalized each congener to the total PCB mass because the total mass did not differ significantly (p = 0.16) between the initial measurements and measurements made after 430 days (Figure S2). Previously, we determined that Aroclor 1248 is the likely contaminating PCB mixture at AVL (Mattes et al. 2018), and this mixture contains 75% penta and tetra chlorinated congeners (Frame et al. 2001). One specific congener, penta-chlorinated PCB118, represents 2% of the total PCB mass in the original A1248 mixture (Frame et al. 1996), and others have demonstrated PCB118 can be dechlorinated by sediment OHRB (Kaya et al. 2018). In addition, decreases in PCB118 are important because its coplanar dioxin-like structure poses a greater risk to exposed humans than non-dioxin like congeners (Van den Berg et al. 2006). Therefore, we compared the percentage of PCB118 in the sediment microcosms at 0 and 430 days to learn if it was dechlorinated (Figure 1A). Bacteria that respire PCBs typically remove meta- and para-position chlorines (Wiegel &Wu 2000), so we also examined the sequence of congeners that may be produced as meta- and para- position chlorines are removed from PCB118 (i.e. PCB66, 70, 74, 28, 26, and PCB25) (Figure 1 B–D). Although removal of ortho-position chlorines has been observed (Cutter et al. 1998), congeners produced by dechlorination of ortho-position chlorines did not accumulate in the sediment microcosms.
Figure 1.
Mass percentage of PCB 118 (245–34-CB) (A) and PCB 66 (24–34-CB) (B) PCB 26/29 (25–3-CB/245-CB) (C) and PCB 25 (24–3-CB) (D) at 0 days incubation compared to 430 days incubation in microcosms with variable sediment origin (A1, C3, D2, E2, F4). Asterisks indicate statistical significance determined by independent two-sample t-tests. Error bars show the standard deviation between replicates.
Initially, PCB118 comprised between 0.82 ± 0.021 and 2.1 ± 0.18 % of total PCB mass in sediment microcosms (Figure 1A). Additionally, in A1, D2, and E2 microcosms, PCB118 contributed more to total PCB mass than in C3 and F4 microcosms. The mass percent of PCB118 in A1, D2 and E2 sediments was 2.0 %, 2.1 % and 1.8% respectively, and are comparable to 2% PCB118 in Aroclor 1248. In C3 and F4 microcosms, PCB118 represented less than 1% of total PCB mass. After 430 days, PCB118 decreased significantly to 0.64 ± 0.16 and 0.54 ± 0.79 % in the sediment slurry of A1 and E2 microcosms (p = 0.05, p = 0.001). The PCB118 decrease observed in C3, D2, F4, and abiotic controls were not significant (p > 0.2) (Figure 1A, Figure S3). In contrast to PCB118, the mass percent of PCB66 ranged from 3.5 ± 0.35 to 6.3 ± 2.5 %. Similar to PCB118, PCB66 decreased over the 430-day incubation. The final contribution of PCB66 to total PCB mass ranged from 0.62 ± 0.21 and 0.53 ± 0.040 % in A1 and E2 microcosms (p = 0.02, p = 0.003). In microcosms inoculated with sediments from the C3 location, neither PCB118 nor PCB66 decreased after 430 days. Further, we observed no significant changes in any congeners that represent more than 1% of the total PCB mass in C3 microcosms (Figure S4). Unlike C3 microcosms, D2 and F4 microcosms showed decreases in PCB66 and PCB 118, but a high standard deviation prevented the decreases from being statistically significant (p > 0.2) (Figure 1A, B).
Considering only statistically significant decreases in PCB118, microcosms with A1 and E2 sediments demonstrated an average of 68% and 70% decrease in PCB118 after the 430-day incubation. Interestingly, the magnitude of PCB118 dechlorination in A1 and E2 sediments was comparable to 74% and 90% decreases observed in Grasse River (GR) and Baltimore Harbor (BH) sediments, respectively (Kaya et al. 2018). The GR and BH sediment microcosms received a fatty acid mixture (acetate, propionate, and butyrate) (Kaya et al. 2018), but the AVL sediment microcosms did not receive any exogenous electron donor. Without exogenous electron donor, the microbial community relied on the endogenous organic carbon in the sediments, the leachable fraction of which ranged from 55–232 mg/kg (Figure S5). This range of leachable organic carbon at the differing sediment collection locations did not appear to limit the extent of PCB dechlorination observed. The impact of exogenous electron donor likely influences the dechlorination lag time, as GR sediment microcosms showed 74% decrease in PCB118 after only 30 days (Kaya et al. 2018). However, we do not know the minimum time required to dechlorinate PCB118 in A1 and E2 microcosms.
To further examine dechlorination in the sediment microcosms and observe how dechlorination differs with AVL sampling location, we quantified the mass percentage of dechlorination products at 0 and 430 days in all microcosms (Figure 1C, D). PCB26 and PCB25 may form as bacteria remove meta- and para- position chlorines from PCB118 and PCB66, therefore PCB25 and PCB26 represent possible products of PCB118 dechlorination. Because PCB26 coelutes with PCB29 in our analysis methods, we reported the coelution as PCB26+29 instead of PCB26 alone. Additionally, PCB29 was not present in the original Aroclor 1248 mixture (Frame et al. 1996), so if PCB29 was present in AVL sediments it likely formed as chlorines were removed from higher chlorinated congeners. The initial mass percentage of PCB26+29 ranged from 0.98 ± 0.64 to 2.2 ± 0.23 % and PCB25 ranged from 0.53 ± 0.01 to 2.3 ± 0.21 % (Figure 1C, D). Increases in PCB26+29 and PCB25 proved statistically significant only in the A1 and E2 microcosms (p < 0.03), whereas the proportion of PCB25 and PCB26+29 did not change in C3 microcosms.
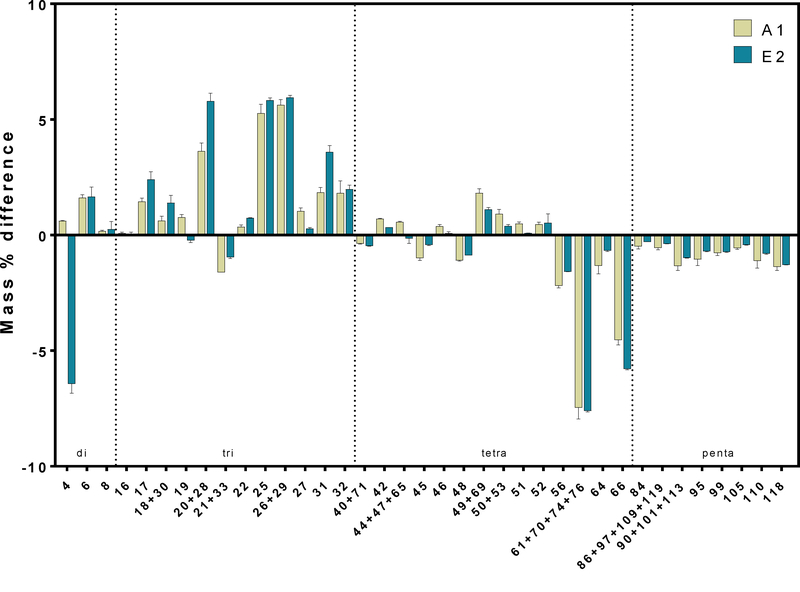
We compared PCB congener profiles before and after the 430-day incubation to determine if dechlorination activity differed between A1 and E2 sediment locations (Figure 2). Microcosms with sediments from both locations appeared to harbor bacteria that dechlorinate a range of penta-chlorinated congeners including PCB118, 110, 105, 99, 95, 84 as well as coeluting congeners PCB86+97+109+119 and PCB90+101+113. Additionally, tetra chlorinated congeners PCB66, 56, and coeluting congeners PCB61+70+74+76 decreased significantly in both A1 (p <0.03) and E2 (p < 0.003) microcosms. The most notable products that accumulated in the microcosms were tri-chlorinated congeners PCB25, PCB 26+29, and PCB20+28 (Figure 2). The same congeners that decreased in A1 microcosms also decreased in E2 microcosms and the dechlorination products appeared consistent between the sediment locations. The consistent changes in PCB congener profiles between A1 and E2 microcosms suggests that the dechlorination pattern of native microbial communities was consistent despite the different locations from which these sediments were collected.
Figure 2.
Difference between the PCB profile at 430 days and 0 days in microcosms inoculated with A1 and E2 sediments. Only congeners that represent 1% of the total mass fraction at either 0 or 430 days are shown. Error bars show the standard deviation between replicates. Positive values indicate accumulations in the specified congener over the 430 day incubation, and negative values indicate the congener decreased over time.
Dehalococcoides spp. abundance increased two orders of magnitude in cultures that exhibited significant dechlorination
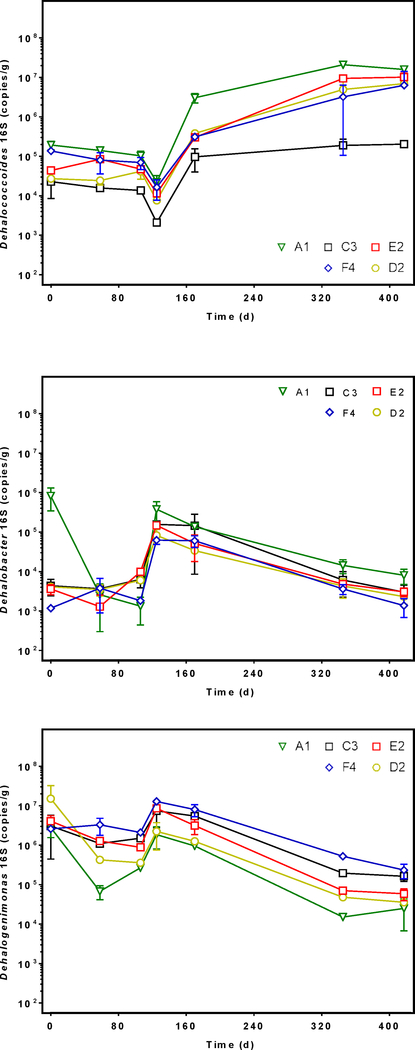
We quantified the 16S rRNA gene sequence abundance of OHRB that may dechlorinate PCBs to derive energy for growth, namely Dehalococcoides, Dehalogenimonas, and Dehalobacter (He &Bedard 2016). Additionally, we monitored the Geobacter population abundance because Geobacter activity may contribute indirectly to PCB dechlorination performed by Dehalococcoides spp. (Praveckova et al. 2016). After 417 days, the abundance of Dehalococcoides 16S rRNA gene sequences increased at least two orders of magnitude in microcosms inoculated with A1, E2, and D2 sediments from averages of 2×105 to 2×107 (A1), 4×104 to 1×107 (E2), and 3×104 to 7×106 (D2) gene copies/gram (Figure 3). The increased Dehalococcoides abundance between 0 and 417 days was significant in A1 and E2 microcosms (p < 0.0001 for both). Although D2 and F4 microcosms showed evidence of dechlorination (Figure 1), only the A1 and E2 microcosms showed statistically significant PCB dechlorination activity (Figure 2). In addition, the Dehalococcoides population in A1 was significantly greater than that of C3 (p = 0.01), and C3 microcosms showed no evidence of dechlorination (Figure S4). As expected, DNA extractions performed on the autoclaved control microcosm sediments yielded no DNA.
Figure 3.
Dehalococcoides (top), Dehalobacter (middle), and Dehalogenimonas (bottom) 16S rRNA gene sequence abundance in DNA extracted from spatially variable sediment microcosms (A1, C3, D2, E2, F4) as measured by qPCR. Error bars show the standard deviation between replicates. Each marker represents the average 16S sequence abundance at different lengths of incubation time from 0 to 417 days.
Initially the Dehalobacter populations ranged from 1×103 to 8×105 gene copies/gram, but the greatest Dehalobacter abundance at time zero was observed in A1 microcosms (Figure 4). The Dehalobacter population in A1 microcosms decreased significantly (p < 0.0001) from 8×105 to 8×103 gene copies/gram during incubation. After 417 days, the Dehalobacter populations in all microcosms were not statistically different (p = 0.06) from each other. In contrast, Dehalogenimonas abundance did not differ significantly based on sediment origin (p = 0.3). Additionally, Dehalogenimonas abundance decreased significantly over 417 days in all microcosms (p = 0.001). Finally, Geobacter populations did not change over time, but maintained relatively high abundances between 106 and 107 gene copies/gram of sediment (Figure S6).
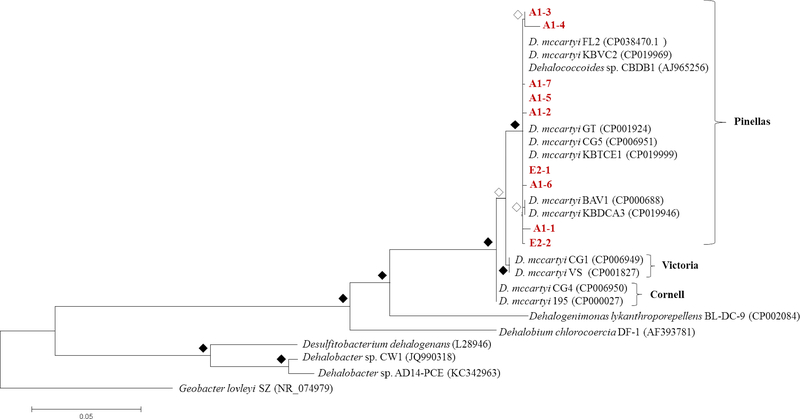
Figure 4.
Phylogenetic tree of Dehalococcoides 16S rRNA nucleotide sequences isolated from microcosms inoculated with A1 and E2 sediments. Evolutionary analysis was conducted in MEGA7 and the tree with the highest log likelihood (−4696.55) supported by a bootstrap analysis (500 replicates) is shown. The Geobacter lovleyi sequence roots the tree. Open diamonds represent bootstrap values greater than 80%, and filled diamonds represent bootstrap values below 80%. Partial Dehalococcoides 16SrRNA gene sequences obtained in this study are displayed in red.
Nearly all pure or sediment free cultures reported to dechlorinate PCBs contained Dehalococcoides strains as the dominant dechlorinators (Adrian et al. 2009, LaRoe et al. 2014, Wang et al. 2014), with the exception of Dehalobium chlorocoercia strain DF-1 (May et al. 2008). However, two dechlorinating cultures have been reported to contain either Dehalobacter or Dehalogenimonas in addition to Dehalococcoides, and all three genera likely dechlorinate PCBs in those cultures (Wang & He 2013a, b). The significant decrease (p=0.001) in Dehalogenimonas over time in all AVL microcosms suggests that they likely are not responsible for PCB dechlorination in AVL sediments. Conversely, Dehalococcoides, a genus that can survive only by respiring on halogenated compounds (Loffler et al. 2013), significantly increased in abundance in the actively dechlorinating microcosms A1 and E2. We suspect that PCBs served as electron acceptors for the increasing Dehalococcoides population, as we did not detect any chlorinated ethenes in the AVL sediment microcosms (data not shown). However, it is possible that Dehalococcoides respired on naturally-occurring organochlorines (Krzmarzick et al. 2012), but unlike PCB respiration such activity has never been explicitly demonstrated for Dehalococcoides. Finally, it is unlikely that the decreasing Dehalobacter population is responsible for PCB dechlorination in AVL microcosms. In one sediment free culture, synergy between Dehalobacter and Dehalococcoides was observed, but both populations obtained energy as they respired halogenated compounds (Wang &He 2013a). As a result, Dehalobacter and Dehalococcoides populations increased concurrently as they dechlorinated the PCB mixture Aroclor 1260 (Wang &He 2013a). In contrast, our data shows Dehalobacter populations increased sharply initially before decreasing in abundance at 125 days of incubation (Figure 4). The decrease in Dehalobacter populations as Dehalococcoides increased suggests a synergy between the two genera was not utilized to dechlorinate PCBs in AVL sediment microcosms.
At least 16 Dehalococcoides strains exist in pure culture (Zinder 2016), and highly enriched mixed cultures contain additional unique strains (Duhamel et al. 2004, Molenda et al. 2019, Schaefer et al. 2009, Wang et al. 2014). Each of these unique Dehalococcoides strains have been classified into three phylogenetic subgroups namely, Victoria, Cornell, and Pinellas (Hendrickson et al. 2002), which contain PCB-dechlorinating Dehalococcoides mccartyi strains CG1, CG4, and CG5 respectively (Wang et al. 2014). To determine the subgroup classification of the Dehalococcoides strains present in AVL microcosms, we obtained a sample of Dehalococcoides 16S rRNA gene sequences enriched after 417 days and aligned them with representative sequences from the Victoria, Cornell, and Pinellas subgroups (Table S4). Phylogenetic analysis revealed that Dehalococcoides 16S rRNA gene sequences present in the A1 and E2 AVL microcosms cluster with the Pinellas subgroup (Figure 4). In fact, most Dehalococcoides strains that dechlorinate PCBs (e.g., Dehalococcoides mccartyi strains JNA, CBDB1, and CG5) have been classified as members of the Pinellas subgroup (Adrian et al. 2009, LaRoe et al. 2014, Wang & He 2013b). In addition to Dehalococcoides that respire complex PCB mixtures, the Pinellas subgroup also contains most strains of Dehalococcoides identified as members of mixed cultures derived from contaminated sediments and enrichment culture clones (Loffler et al. 2013). One such sediment derived mixed culture that dechlorinates tetrachloroethene completely to ethene, known as KB-1, harbors at least eight distinct strains of Dehalococcoides that all classify as members of the Pinellas subgroup (Molenda et al. 2019).
Dehalococcoides CG5 rd14 abundance increased over time in microcosms
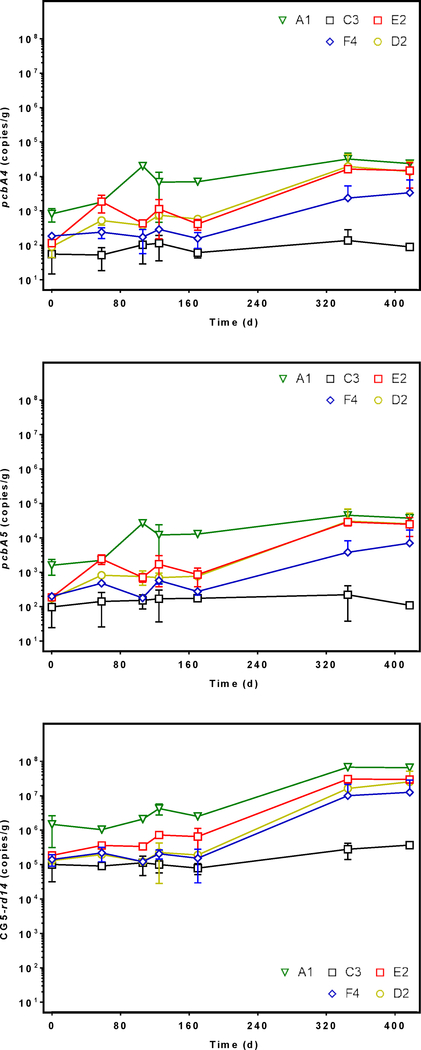
Because Dehalococcoides abundance dominated that of the other potential dechlorinators measured, we investigated which known RDase genes co-occurred and increased in abundance as the Dehalococcoides population expanded. Only three RDase genes have been linked to PCB dechlorination (pcbA1, pcbA4, pcbA5)(Wang et al. 2014), so we also targeted RDase genes involved in chlorinated ethene dechlorination (tceA, vcrA, bvcA) (Figure 5, Figure S7). The abundance of pcbA1, bvcA, and vcrA remained below detection in DNA for all time points measured, but the sequences of pcbA4, pcbA5, and tceA persisted in all microcosms. First, pcbA4 abundance only differed significantly from day zero to 417 in A1 microcosms, where it increased from 8×102 to 2×104 gene copies/gram of sediment (p = 0.001). Similar to the increase in Dehalococcoides population, the abundance of pcbA4 was greater in A1 microcosms than C3 microcosms (p = 0.01). The increase in pcbA5 sequence abundance resembled that of pcbA4 despite differing strains of Dehalococcoides known to harbor the sequences (Wang et al. 2014). The pcbA5 sequence abundance increased significantly only in A1 microcosms from 2×103 to 4×104 (p = 0.002), but pcbA5 was more abundant in A1 microcosms than in microcosms established with any other sediments. Unlike the RDase genes associated with PCB dechlorination, tceA abundance did not increase over time, and it did not differ significantly between cultures established with sediments from the six locations (Figure S7).
Figure 5.
Quantity of pcbA4 (top), pcbA5 (middle), and CG5 rd14 (bottom) gene sequence abundance in DNA extracted from spatially variable sediment microcosms (A1, C3, D2, E2, F4) as measured by quantitative real time PCR. Error bars show the standard deviation between replicates. Each marker represents the average 16S sequence abundance at different lengths of incubation time from 0 to 417 days.
We expected the rd14 gene to occur in the microcosms because we previously isolated and identified partial RDase gene sequences from AVL sediments similar to that gene (Mattes et al. 2018). We measured changes in rd14 abundance during the incubation period in all sediment microcosms (Figure 5). First, rd14 abundance was significantly greater in A1 microcosms than in microcosms with C3, D2, and F4 sediments (p = 0.008, p = 0.04, p = 0.02, respectively). Additionally, rd14 abundance in A1 microcosms increased an order of magnitude between 0 and 417 days from 1.5×106 to 6.5×107 gene copies/gram of sediment, and the observed changes were significant (p <0.0001). In E2 microcosms rd14 increased two orders of magnitude from 1.8×105 to 3×107 gene copies/gram of sediment, but rd14 abundance in E2 did not differ significantly from that of A1 microcosms (p = 0.1). Conversely, rd14 abundance remained unchanged in C3, and F4 microcosms between 0 and 417 days.
Only two other studies have examined pcbA1, pcbA4, and pcbA5 sequence abundance in PCB contaminated sediment microcosms, and the sediments were obtained from two different PCB contaminated costal waterways (Matturro et al. 2016a, Matturro et al. 2016b). Both studies reported the presence of pcbA1, pcbA4, and pcbA5 in unenriched PCB contaminated sediments with an average abundance of 105 gene copies/gram of sediment. In contrast, we did not detect pcbA1, and initially the population of Dehalococcoides in our microcosms was at least an order of magnitude lower. Therefore, the abundance of pcbA4, and pcbA5 may be useful biomarker genes for PCB dechlorination, but our work suggests rd14 is more abundant than pcbA4 and pcbA5 in environmental samples. Additionally, C3 microcosms contained the smallest Dehalococcoides population, the least pcbA4, pcbA5, and rd14 abundance, and showed no dechlorination (Figure S4). In contrast, A1 and E2 microcosms contained the greatest pcbA4, pcbA5, and rd14 abundance, in addition to the greatest Dehalococcoides population. A1 and E2 microcosms showed the most robust biological lines of evidence for PCB dechlorination and actively dechlorinated PCB118, 110, 105, 99, 95, 84, 66 and 56 among others.
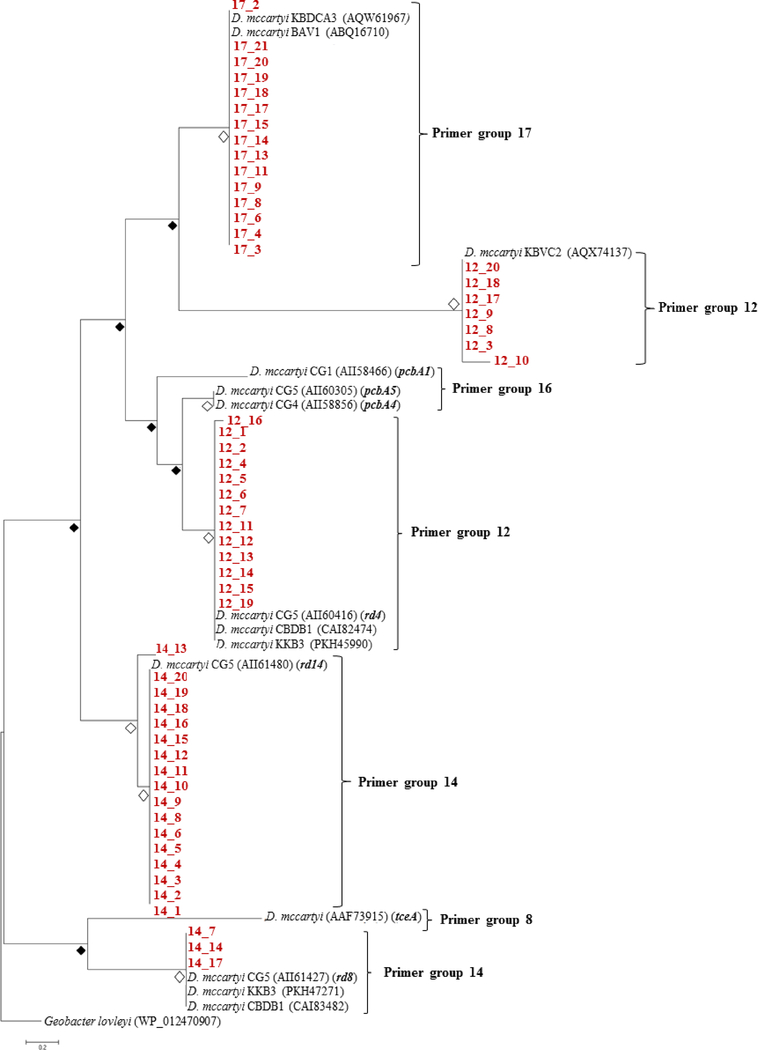
Because A1 microcosms contained the greatest Dehalococcoides populations and multiple RDase genes, we chose to perform a more comprehensive survey of RDase gene diversity on DNA extracted from A1 microcosms. The previously developed primers target orthologs of all known reductive dehalogenases (Hug &Edwards 2013), and of the 44 total primer sets we obtained PCR products from primer sets 12, 14 and 17. Primer sets 12, 14, and 17 target RDase sequences that likely belong to Dehalococcoides mccartyi, and as expected phylogenetic analysis revealed all the sequences resembled those of other D. mccartyi strains (Figure 6, Table S5). Although we detected tceA with qPCR, PCR with the primer set designed to target chlorinated ethene dehalogenases, group 8, did not produce sufficient products for cloning. Additionally, the group 16 primer set would likely amplify pcbA1, pcbA4, and pcbA5 based on sequence similarity, but we were unable to obtain sufficient products for cloning. It is important to note that qPCR was conducted with primer sets specifically designed to target smaller regions of the tceA, pcbA1, pcbA4, and pcbA5 genes with high specificity. In contrast, the primers used for the RDase survey are highly degenerate primers that produce sequences of 1000 base pairs or greater (Table S2) (Hug &Edwards 2013).
Figure 6.
Phylogenetic tree of amino acid sequences isolated from microcosms inoculated with A1 sediments. Evolutionary analysis was conducted in MEGA7 and the tree with the highest log likelihood (−482.22) supported by a bootstrap analysis (500 replicates) is shown. The WP_012470907 Geobacter lovleyi sequence roots the tree. Open diamonds represent bootstrap values greater than 80%, and filled diamonds represent bootstrap values below 80%. Partial sequences obtained in this study are displayed in red.
Phylogenetic analysis revealed the degenerate nature of the primer sets and captured diversity within the products of primer sets 14 and 12. Conversely, all of the products produced with primer set 17 appear to be part of the same clade based on the high bootstrap value (Figure 6). Unlike primer set 12 and 14 products, primer set 17 sequences did not closely resemble any sequences on the genome of Dehalococcoides strains known to dechlorinate PCBs and instead showed 95–100% sequence similarity to sequences that belong to vinyl chloride (VC) and 1,2-dichloroethane dechlorinating strains D. mccartyi BAV1 and KBDCA3 (He et al. 2003, Molenda et al. 2019). It is possible that orthologs to the BAV1 and KBDCA3 RDases participate in PCB dechlorination; however, it is also possible that these genes are not expressed in response to PCBs but reside on the genome of the PCB dechlorinating strain. Future investigations would require a transcriptomic approach to determine which RDase genes are expressed during PCB dechlorination in AVL sediment microcosms, because D. mccaryti strains are highly specialized dechlorinators and possess up to 36 different RDase genes (McMurdie et al. 2009). Additionally, different D. mccartyi strains possess identical RDase genes. For example, the CG5 rd14 sequence is identical to RDase genes on the genomes of strains JNA and CBDB1 (Adrian et al. 2009, LaRoe et al. 2014), among others.
Our previous RDase gene survey conducted on unenriched AVL sediments did not yield sequences from primer group 17, but instead only primer group 12 and 14 produced partial RDase gene sequences that were cloned and sequenced successfully (Mattes et al. 2018). The broader range of sequences obtained in this study compared to data collected for unenriched AVL sediments, suggests that the sediment microbial community became enriched in sequences targeted by primer groups 12, 14, and 17 after incubation in microcosms. The qPCR data shows rd14 became enriched significantly with time (Figure 5), so it is highly probable other reductive dehalogenase sequences were also enriched as Dehalococcoides strains increased in abundance.
Phylogenetic analysis revealed both primer sets 12 and 14 amplified two divergent groups of D. mccartyi RDase gene sequences. The two groups of primer set 12 sequences either resemble D. mccartyi CG5 rd4, a RDase gene known to be transcribed during PCB dechlorination (Wang et al. 2014), or a RDase gene that belongs to the VC-dechlorinating D. mccartyi strain KBVC2 (Edwards et al. 2018) (Figure 6). In contrast, primer set 14 sequences resembled RDase genes only on the genomes of D. mccartyi strains known to dechlorinate PCBs or structurally similar compounds, namely 1,2,3,4-tetrachlorodibenzo-p-dioxin (Dam et al. 2017, Wang et al. 2014). Although primer set 14 amplified partial RDase gene sequences related to rd14 in unenriched sediments, the microbial community enriched for PCB dechlorinators in this microcosm study also possessed sequences similar to CG5’s rd8 (Wang et al. 2014).
The genomic characterization of D. mccartyi strains CG1, CG4, and CG5 was facilitated by their dual ability to dechlorinate both chlorinated ethenes and complex PCB mixtures, and this duality is reflected in the bifunctional nature of pcbA1, pcbA4, and pcbA5 (Chen &He 2018, Wang et al. 2014). In addition to pcbA5, strain CG5 transcribed 18 other RDase genes during Aroclor 1260 respiration, and rd14 was the second most highly transcribed, followed by rd8 (Wang et al. 2014). The CG5 rd4 gene was transcribed during both PCE and Aroclor 1260 respiration, but to a lesser extent than pcbA5, rd14, and rd8. It is possible then that the bifunctional RDase pcbA5 confers less of an evolutionary advantage in an environment where chlorinated ethenes are not present, as is the case in AVL sediments. Our data suggests that rd14 may serve as a biomarker functional gene to indicate in situ PCB dechlorination potential in addition to pcbA5.
Although our AVL microcosm data highlights the potential importance of RDase gene groups 12, 14, and 17 to in situ PCB dechlorination, because OHRB possess multiple RDase genes, gene expression analysis (e.g., reverse transcription qPCR or metatranscriptomics) is required to confirm expression of these candidate biomarker genes in response to PCB mixtures during PCB dechlorination. Nonetheless, the enrichment of biomarker candidate genes that resemble Dehalococcoides mccartyi CG5’s rd14, rd8 and rd4 during PCB dechlorination in sediments suggests these genes are relevant to PCB monitored natural attenuation campaigns and perhaps even useful to monitor dechlorination at sites bioaugmented with dechlorinators.
Supplementary Material
Acknowledgements
We thank the Superfund Research Program of the National Institute of Environmental Health Sciences (Grant No. NIH P42ES013661) for funding. We thank Patrick Richards for technical assistance with organic carbon analyses. Additionally, we thank the Iowa Institute for Human Genetics Genomics Division for assistance with qPCR and DNA sequencing. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Abraham W-R, Nogales B, Golyshin PN, Pieper DH, Timmis KN (2002): Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr. Opin. Microbiol 5, 246–253 [DOI] [PubMed] [Google Scholar]
- Abramowicz DA (1995): Aerobic and anaerobic PCB biodegradation in the environment. Environ. Health Perspect 103, 97–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian L, Dudková V, Demnerová K, Bedard DL (2009): Dehalococcoides sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl. Environ. Microbiol 75, 4516–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anezaki K, Nakano T (2014): Concentration levels and congener profiles of polychlorinated biphenyls, pentachlorobenzene, and hexachlorobenzene in commercial pigments. Environ Sci Poll Res 21, 998–1009 [DOI] [PubMed] [Google Scholar]
- Chen C, He J (2018): Strategy for the rapid dechlorination of Polychlorinated Biphenyls (PCBs) by Dehalococcoides mccartyi strains. Environ. Sci. Technol 52, 13854–13862 [DOI] [PubMed] [Google Scholar]
- Clark K, Taggart DM, Baldwin BR, Ritalahti KM, Murdoch RW, Hatt JK, Loffler FE (2018): Normalized quantitative PCR measurements as predictors for ethene formation at sites impacted with chlorinated ethenes. Environ. Sci. Technol 52, 13410–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter L, Sowers KR, May HD (1998): Microbial dechlorination of 2,3,5,6-tetrachlorobiphenyl under anaerobic conditions in the absence of soil or sediment. Appl. Environ. Microbiol 64, 2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam HT, Vollmers J, Kaster A-K, Haggblom MM (2017): Reconstructed genomes of novel Dehalococcoides mccartyi strains from 1,2,3,4-tetrachlorodibenzo-p-dioxin-dechlorinating enrichment cultures reveal divergent reductive dehalogenase gene profiles. FEMS Microbiol. Ecol 93, fix151–fix151 [DOI] [PubMed] [Google Scholar]
- Duhamel M, Mo K, Edwards EAJAEM (2004): Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. 70, 5538–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EA, Molenda O, Tang S, Lomheim L (2018): Eight new genomes of organohalide-respiring Dehalococcoides mccartyi reveal evolutionary trends in reductive dehalogenase enzymes. bioRxiv, 345173 [Google Scholar]
- Frame GM, Wagner RE, Carnahan JC, Brown JF Jr, May RJ, Smullen LA, Bedard DL (1996): Comprehensive, quantitative, congener-specific analyses of eight Aroclors and complete PCB congener assignments on DB-1 capillary GC columns. Chemosphere 33, 603–623 [Google Scholar]
- Frame GM, Robertson L, Hansen L (2001): The current state-of-the-art of comprehensive, quantitative, congener-specific PCB analysis, and what we now know about the distributions of individual congeners in commercial Aroclor mixtures PCBs: Recent advances in environmental toxicology and health effects. University Press of Kentucky Lexington, KY, 3–9 pp [Google Scholar]
- Gomes HI, Dias-Ferreira C, Ribeiro AB (2013): Overview of in situ and ex situ remediation technologies for PCB-contaminated soils and sediments and obstacles for full-scale application. Sci. Total Environ 445, 237–260 [DOI] [PubMed] [Google Scholar]
- He J, Ritalahti KM, Yang K-L, Koenigsberg SS, Loffler FE (2003): Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424, 62. [DOI] [PubMed] [Google Scholar]
- He J, Bedard DL (2016): The microbiology of anaerobic PCB dechlorination, Organohalide-Respiring Bacteria. Springer, pp. 541–562 [Google Scholar]
- Hendrickson ER, Payne JA, Young RM, Starr MG, Perry MP, Fahnestock S, Ellis DE, Ebersole RC (2002): Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol 68, 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkert NJ, Martinez A, Hornbuckle KC (2016): A model using local weather data to determine the effective sampling volume for PCB congeners collected on passive air samplers. Environ. Sci. Technol 50, 6690–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelheber DW, Pennell KD, Hughes JB (2007): Natural attenuation processes during in situ capping. Environ. Sci. Technol 41, 5306–5313 [DOI] [PubMed] [Google Scholar]
- Hug L, Edwards E (2013): Diversity of reductive dehalogenase genes from environmental samples and enrichment cultures identified with degenerate primer PCR screens. Front Microbiol 4, Article 341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Maphosa F, Leys D, Loffler FE, Smidt H, Edwards EA, Adrian L (2013): Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos Trans R Soc B Biol Sci 368, 20120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya D, Imamoglu I, Sanin FD, Sowers KR (2018): A comparative evaluation of anaerobic dechlorination of PCB-118 and Aroclor 1254 in sediment microcosms from three PCB-impacted environments. J. Hazard. Mater 341, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzmarzick MJ, Crary BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SCB, Novak PJ (2012): Natural niche for organohalide-respiring Chloroflexi. Appl. Environ. Microbiol 78, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016): MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol 33, 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRoe SL, Fricker AD, Bedard DL (2014): Dehalococcoides mccartyi strain JNA in pure culture extensively dechlorinates Aroclor 1260 according to polychlorinated biphenyl (PCB) dechlorination process N. Environ. Sci. Technol 48, 9187–9196 [DOI] [PubMed] [Google Scholar]
- Lebron CA, Petrovskis E, Loeffler F, Henn K 2011: Guidance protocol: application of nucleic acid-based tools for monitoring monitored natural attenuation (MNA), biostimulation, and bioaugmentation at chlorinated solvent sites, Naval Facilities Engineering Command Port Hueneme CA Engineering Service Center [Google Scholar]
- Lee PKH, Macbeth TW, Sorenson KS, Deeb RA, Alvarez-Cohen L (2008): Quantifying genes and transcripts to assess the in situ physiology of “Dehalococcoides” spp. in a trichloroethene-contaminated groundwater site. Appl. Environ. Microbiol 74, 2728–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM (2013): Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol 63, 625–635 [DOI] [PubMed] [Google Scholar]
- Lu X, Wilson JT, Kampbell DH (2006): Relationship between Dehalococcoides DNA in ground water and rates of reductive dechlorination at field scale. Water Res. 40, 3131–3140 [DOI] [PubMed] [Google Scholar]
- Marek RF, Thorne PS, Wang K, DeWall J, Hornbuckle KC (2013): PCBs and OH-PCBs in serum from children and mothers in urban and rural U.S. communities. Environ. Sci. Technol 47, 3353–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek RF, Thorne PS, Herkert NJ, Awad AM, Hornbuckle KC (2017): Airborne PCBs and OH-PCBs inside and outside urban and rural U.S. schools. Environ. Sci. Technol 51, 7853–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Wang K, Hornbuckle KC (2010): Fate of PCB congeners in an industrial harbor of Lake Michigan. Environ. Sci. Technol. 44, 2803–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Hornbuckle KC (2011): Record of PCB congeners, sorbents and potential toxicity in core samples in Indiana Harbor and Ship Canal. Chemosphere 85, 542–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Spak SN, Petrich NT, Hu D, Carmichael GR, Hornbuckle KC (2015): Atmospheric dispersion of PCB from a contaminated Lake Michigan harbor. Atmos. Environ. 122, 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Hadnott BN, Awad AM, Herkert NJ, Tomsho K, Basra K, Scammell MK, Heiger-Bernays W, Hornbuckle KC (2017): Release of airborne polychlorinated biphenyls from New Bedford Harbor results in elevated concentrations in the surrounding air. Environ Sci Technol Lett 4, 127–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes TE, Ewald JM, Liang Y, Martinez A, Awad A, Richards P, Hornbuckle KC, Schnoor JL (2018): PCB dechlorination hotspots and reductive dehalogenase genes in sediments from a contaminated wastewater lagoon. Environ Sci Poll Res 25, 16376–16388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matturro B, Di Lenola M, Ubaldi C, Rossetti S (2016a): First evidence on the occurrence and dynamics of Dehalococcoides mccartyi PCB-dechlorinase genes in marine sediment during Aroclor1254 reductive dechlorination. Mar. Pollut. Bull 112, 189–194 [DOI] [PubMed] [Google Scholar]
- Matturro B, Ubaldi C, Rossetti S (2016b): Microbiome dynamics of a polychlorobiphenyl (PCB) historically contaminated marine sediment under conditions promoting reductive dechlorination. Front Microbiol 7, Article 1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May HD, Miller GS, Kjellerup BV, Sowers KR (2008): Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl. Environ. Microbiol 74, 2089–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Behrens SF, Müller JA, Goke J, Ritalahti KM, Wagner R, Goltsman E, Lapidus A, Holmes S, Loffler FE (2009): Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5, e1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenda O, Tang S, Lomheim L, Gautam VK, Lemak S, Yakunin AF, Maxwell KL, Edwards EA (2019): Extrachromosomal circular elements targeted by CRISPR-Cas in Dehalococcoides mccartyi are linked to mobilization of reductive dehalogenase genes. The ISME Journal 13, 24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council N (2007): Sediment dredging at Superfund megasites: Assessing the effectiveness. The National Academies Press, Washington, DC [Google Scholar]
- Payne RB, May HD, Sowers KR (2011): Enhanced reductive dechlorination of polychlorinated biphenyl impacted sediment by bioaugmentation with a dehalorespiring bacterium. Environ. Sci. Technol 45, 8772–8779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RB, Fagervold SK, May HD, Sowers KR (2013): Remediation of polychlorinated biphenyl impacted sediment by concurrent bioaugmentation with anaerobic halorespiring and aerobic degrading bacteria. Environ. Sci. Technol 47, 3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RB, Ghosh U, May HD, Marshall CW, Sowers KR (2017): Mesocosm studies on the efficacy of bioamended activated carbon for treating PCB-impacted sediment. Environ. Sci. Technol 51, 10691–10699 [DOI] [PubMed] [Google Scholar]
- Perelo LW (2010): Review: In situ and bioremediation of organic pollutants in aquatic sediments. J. Hazard. Mater. 177, 81–89 [DOI] [PubMed] [Google Scholar]
- Praveckova M, Brennerova MV, Holliger C, De Alencastro F, Rossi P (2016): Indirect Evidence Link PCB Dehalogenation with Geobacteraceae in Anaerobic Sediment-Free Microcosms. Front Microbiol 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LW, Hansen LG (2015): PCBs: recent advances in environmental toxicology and health effects. University Press of Kentucky [Google Scholar]
- Schaefer CE, Condee CW, Vainberg S, Steffan RJJC (2009): Bioaugmentation for chlorinated ethenes using Dehalococcoides sp.: Comparison between batch and column experiments. 75, 141–148 [DOI] [PubMed] [Google Scholar]
- Shelton DR, Tiedje JM (1984): General method for determining anaerobic biodegradation potential. Appl. Environ. Microbiol 47, 850–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA (2017): Search Superfund site information
- Van den Berg M, Birnbaum LS, Denison M, De Vito M, Farland W, Feeley M, Fiedler H, Hakansson H, Hanberg A, Haws L, Rose M, Safe S, Schrenk D, Tohyama C, Tritscher A, Tuomisto J, Tysklind M, Walker N, Peterson RE (2006): The 2005 World Health Organization reevaluation of human and mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol. Sci 93, 223–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zaan B, Hannes F, Hoekstra N, Rijnaarts H, de Vos WM, Smidt H, Gerritse J (2010): Correlation of Dehalococcoides 16S rRNA and chloroethene-reductive dehalogenase genes with geochemical conditions in chloroethene-contaminated groundwater. Appl. Environ. Microbiol 76, 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, He J (2013a): Dechlorination of commercial PCBs and other multiple halogenated compounds by a sediment-free culture containing Dehalococcoides and Dehalobacter. Environ. Sci. Technol 47, 10526–10534 [DOI] [PubMed] [Google Scholar]
- Wang S, He J (2013b): Phylogenetically distinct bacteria involve extensive dechlorination of Aroclor 1260 in sediment-free cultures. PLoS One 8, e59178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chng KR, Wilm A, Zhao S, Yang K-L, Nagarajan N, He J (2014): Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. PNAS 111, 12103–12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemeier TH, Wilson JT, Hansen JE, Chapelle FH, Swanson MA 1996: Technical protocol for evaluating natural attenuation of chlorinated solvents in groundwater. Revision 1, Air Force Center for Environmental Excellence Brooks AFB TX [Google Scholar]
- Wiegel J, Wu Q (2000): Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol. Ecol 32, 1–15 [DOI] [PubMed] [Google Scholar]
- Zinder SH (2016): The genus Dehalococcoides, Organohalide-respiring bacteria. Springer, pp. 107–136 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.