Abstract
Human liver microsomes (HLM) and human liver S9 fractions (HLS9) are commonly used to study drug metabolism in vitro. However, a quantitative comparison of HLM and HLS9 proteomes is lacking, resulting in the arbitrary selection of one hepatic preparation over another and in difficulties with data interpretation. In this study, we applied a label-free global absolute quantitative proteomics method to the analysis of HLS9 and the corresponding HLM prepared from 102 individual human livers. A total of 3137 proteins were absolutely quantified, and 3087 of those were determined in both HLM and HLS9. Protein concentrations were highly correlated between the two hepatic preparations (R = 0.87, P < 0.0001). We reported the concentrations of 98 drug-metabolizing enzymes (DMEs) and 51 transporters, and demonstrated significant differences between their abundances in HLM and HLS9. We also revealed the protein-protein correlations among these DMEs and transporters and the sex effect on the HLM and HLS9 proteomes. Additionally, HLM and HLS9 displayed distinct expression patterns for protein markers of cytosol and various cellular organelles. Moreover, we evaluated the interindividual variability of three housekeeping proteins, and identified five proteins with low variation across individuals that have the potential to serve as new internal controls for western blot experiments. In summary, these results will lead to better understanding of data obtained from HLM and HLS9 and assist in in vitro-in vivo extrapolations. Knowing the differences between HLM and HLS9 also allows us to make better-informed decisions when choosing between these two hepatic preparations for in vitro drug metabolism studies.
SIGNIFICANCE STATEMENT
This investigation revealed significant differences in protein concentrations of drug-metabolizing enzymes and transporters between human liver microsomes and S9 fractions. We also determined the protein-protein correlations among the drug-metabolizing enzymes and transporters and the sex effect on the proteomes of these two hepatic preparations. The results will help interpret data obtained from these two preparations and allow us to make more informed decisions when choosing between human liver microsomes and S9 fractions for in vitro drug metabolism studies.
Introduction
The liver plays a central role in the metabolism and disposition of various endogenous and foreign compounds (Meyer, 1996). Given the significant impact of hepatic metabolism and disposition on a drug’s efficacy and toxicity, preclinical investigations are routinely performed using in vitro hepatic models (Iwatsubo et al., 1997). Several in vitro models have been developed to study hepatic drug biotransformation, which include recombinant drug-metabolizing enzymes (DMEs), liver subcellular fractions (microsomes, cytosol, and S9 fractions), hepatic cell lines, primary hepatocytes, liver slices, and perfused livers (Ekins et al., 2000; Brandon et al., 2003). Among these, human liver microsomes (HLM) and human liver S9 fractions (HLS9) are the most commonly used in vitro models (Asha and Vidyavathi, 2010). Specifically, HLS9 are the supernatants of human liver homogenates after centrifugation at 9000g for 20 minutes, and consist of HLM and cytosol; HLM are the sediment separated from HLS9 by a further centrifugation at 100,000g for ∼1 hour. HLM are enriched with the membranes of the endoplasmic reticulum (ER) and other organelles, in which many DMEs and transporters are located (Fujiki et al., 1982; Ekins et al., 2000).
Hepatic DMEs and transporters are the essential elements in determining drug metabolism and disposition. The protein expression levels of hepatic DMEs and transporters greatly impact pharmacokinetics, and are of importance for drug development. Thus, characterizing protein expression profiles in HLM and HLS9 would facilitate appropriate selection of these models in the study of drug metabolism as well as improve the interpretation of results from HLM and HLS9 incubation studies. A few attempts have been made to determine protein expressions in HLM, with a focus on DMEs and transporters (Ohtsuki et al., 2012; Achour et al., 2014b, 2017a,b; Golizeh et al., 2015a; Couto et al., 2019); however, HLS9 protein profiles have been largely inconclusive. In addition, most prior studies focused on the most-studied DMEs and transporters, such as cytochrome P450s (P450s) and UDP-glucuronosyltransferases (UGTs), leaving many other important enzymes undetermined [e.g., hydrolases, sulfotransferases, N-acetyltransferases (NATs), glutathione S-transferases (GSTs), and housekeeping proteins]. In practice, HLM and HLS9 are often selected arbitrarily by investigators for in vitro metabolism studies in part due to the lack of quantitative understanding of the differences between the proteomes of these two sample types (Plant, 2004). Moreover, without knowing the differences, it is difficult for researchers to reconcile the results obtained from these distinct in vitro models. Therefore, a comparative proteomics analysis of HLM and HLS9 is needed to inform the selection of one in vitro model over another and to improve the interpretation of results generated from the two hepatic preparations.
With the rapid advance of mass spectrometry-based proteomics, absolute quantitative proteomics (AQP) has been increasingly used in both basic and clinical investigations to absolutely quantify protein levels in various biologic samples (Prasad et al., 2019). We recently developed a novel label-free global AQP method, called the data-independent acquisition (DIA)/total protein approach (TPA), using DIA and the TPA algorithm (He et al., 2019). Compared with other labeling global AQP proteomics approaches, such as metabolic labeling (e.g., stable isotope labeling using amino acid in cell culture) (Hanke et al., 2008) and chemical labeling (e.g., isobaric tags for relative and absolute quantitation) (Wiese et al., 2007), the DIA-TPA method offers an efficient and cost-effective option for absolutely measuring the whole proteome. Additionally, unlike most label-free AQP methods (Ishihama et al., 2005; Silva et al., 2006; Braisted et al., 2008; Schwanhäusser et al., 2011, 2013; Shin et al., 2013; Wiśniewski et al., 2014) that use first mass spectrometer (MS1) signals from data-dependent acquisition for quantification, the DIA-TPA method relies on the second mass spectrometer (MS2) signals from DIA, which effectively avoids the often-seen bias of data-dependent acquisition for the most intense precursors (Huang et al., 2015). Moreover, DIA-TPA employs a novel algorithm to allocate the signals of shared peptides, allowing for accurate quantification of protein isoforms. Since DMEs and transporters often consist of multiple isoforms with significant portions of shared amino acid sequences, the DIA-TPA method is ideally suited for the absolute quantification of the proteomes of HLM and HLS9.
In the present study, we performed a global absolute quantitative proteomics analysis of 102 individual HLS9 samples and the corresponding HLM preparations using the DIA-TPA method. The study revealed the proteome profiles of paired HLM and HLS9 samples and compared the levels and interindividual variabilities of DME and transporter protein concentrations between HLM and HLS9. The findings will allow us to make more informed decisions when choosing between HLM and HLS9 for in vitro drug metabolism studies, and will also lead to better understanding of the results from incubation studies involving HLM and HLS9.
Materials and Methods
Materials
Urea, ammonium bicarbonate (NH4HCO3), dl-dithiothreitol, trifluoroacetic acid, acetone, and water with 0.1% formic acid (liquid chromatography/mass spectrometry reagent) were purchased from Fisher Scientific Co. (Pittsburgh, PA). Acetonitrile with 0.1% formic acid (liquid chromatography/mass spectrometry reagent) was obtained from JT Baker Chemical Co. (Phillipsburg, NJ). Iodoacetamide was the product of Acros Organics (Morris Plains, NJ). Tosyl phenylalanyl chloromethyl ketone-treated trypsin was obtained from Worthington Biochemical Corporation (Freehold, NJ). Lysyl endopeptidase was purchased from Wako Chemicals (Richmond, VA). Water Oasis HLB columns were obtained from Waters Corporation (Milford, MA). Synthetic indexed retention time standard solution was produced by Biognosys AG (Cambridge, MA). The Pierce BCA protein assay kit and PBS were obtained from Thermo Fisher Scientific (Waltham, MA). All other chemicals and reagents were of analytical grade and commercially available.
A total of 102 normal individual human liver samples were obtained from XenoTech LLC (Kansas City, KS) and the Cooperative Human Tissue Network (Columbus, OH). All of the tissues were snap frozen in liquid nitrogen and stored at −80°C until use. The donors consisted of 46 males and 56 females ranging from 0.75 to 83 years of age, and included 95 Caucasians, five African Americans, one Hispanic, and one classified as other. The use of these liver samples was approved by the Institutional Review Board.
Preparation of HLS9 and HLM
HLS9 and HLM were prepared from the 102 individual human liver samples using a method described in previous publications (Wang et al., 2016; Shi et al., 2018). Briefly, ∼200 mg of individual liver tissues was homogenized in 0.5 ml PBS in 1.5 ml microcentrifuge tubes on ice using an automatic pestle (VWR International LLC, Chicago). The homogenates were then centrifuged twice at 9000g for 20 minutes at 4°C to remove the fat-containing top layers. The supernatant was collected as HLS9 and transferred to a clean tube. To prepare HLM and HLS9 were further centrifuged at 300,000g for 20 minutes at 4°C using an Optima MAX ultracentrifuge with a MLA-80 rotor (Beckman Coulter, Indianapolis, IN) (Braner et al., 2018). The pellet (HLM) formed after the centrifugation was then resuspended in PBS and transferred to a clean tube for storage. Total protein concentrations of HLS9 and HLM samples were determined using a Pierce BCA protein assay kit. Pooled HLS9 and HLM samples were prepared by mixing protein content–normalized aliquots of the given sample type from all individuals. Samples were kept on ice or at 4°C during the entire preparation process, and the prepared HLS9 and HLM samples were stored at −80°C until use. The recovery factors for HLS9 (s9 protein per gram liver) and HLM (microsomal protein per gram liver) (Wilson et al., 2003) were determined to be 107.3 ± 28.1 and 18.8 ± 6.4 mg/g liver, respectively.
Proteomics Sample Preparation
HLS9 and HLM samples were prepared for proteomics analysis according to a previously published method with some minor modifications (Shi et al., 2018). In brief, 80 μg HLS9 or HLM protein aliquot was mixed with 0.2 μg bovine serum albumin in a 1.5 ml microcentrifuge tube. One milliliter (50-fold volume) of precooled acetone was added to the mixture and stored at −20°C overnight to precipitate proteins. The samples were centrifuged at 17,000g for 15 minutes at 4°C. Following centrifugation, the supernatant was removed and the pellet was air-dried at room temperature for 5 minutes. The dried pellet was then resuspended in 100 μl freshly prepared 4 mM dl-dithiothreitol in 8 M urea solution with 100 mM NH4HCO3, followed by vortexing and sonication to ensure complete dissolution of proteins. The samples were then incubated at 37°C for 45 minutes for reduction. After cooling to room temperature, 100 μl of freshly prepared 20 mM iodoacetamide in 8 M urea solution with 100 mM NH4HCO3 was added and mixed, followed by incubation at room temperature in the dark for 30 minutes. Subsequently, the urea concentration was adjusted to 6 M by the addition of 56.6 μl 50 mM NH4HCO3. For protein digestion, samples were first incubated with lysyl endopeptidase (protein:lysyl endopeptidase = 100:1) in an orbital incubator shaker at 220 rpm and 37°C for 6 hours. Next, the urea concentration was further adjusted to 1.6 M by adding 733 μl 50 mM NH4HCO3, followed by overnight incubation with tosyl phenylalanyl chloromethyl ketone-treated trypsin (protein:trypsin = 50:1) at 220 rpm and 37°C. Digestion was terminated by adding 1 μl trifluoroacetic acid. The digested peptides were extracted and cleaned using Waters Oasis HLB columns according to the manufacturer’s instructions. The eluted peptides were dried using a SpeedVac SPD1010 vacuum concentrator (Thermo Scientific, Hudson, NH) and resuspended in 3% acetonitrile solution with 0.1% formic acid. The reconstituted peptide solution was then centrifuged at 17,000g for 10 minutes at 4°C. One-half of the supernatant was transferred to a clean autosampler vial and mixed with 1 μl indexed retention time standard solution for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS–Based Proteomics Analysis
The LC-MS/MS system consisted of a tandem quadrupole time-of-flight mass spectrometer (TripleTOF5600 plus; AB/Sciex, Framingham, MA) and an Eksigent 2D Plus LC System (Eksigent Technologies, Dublin, CA). The analysis was conducted using a previously published method with some modifications (Shi et al., 2018). Briefly, the mobile phase was composed of water with 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B). Peptides were separated through a trap-elute configuration, including a trapping column (ChromXP C18-CL, 120 Å, 5 μm, 0.3 mm catridge; Eksigent Technologies) and an analytical column (ChromXP C18-CL, 120 Å, 150 × 0.3 mm, 5 μm; Eksigent Technologies). Next, 6 μg of digested protein was injected, trapped, and cleaned on the trapping column with mobile phase A at a flow rate of 10 μl/min for 3 minutes, which was then separated on the analytical column with a gradient elution at a flow rate of 5 μl/min (Supplemental Table 1). Each sample was followed by a blank sample injection to minimize carryover. Ionization was achieved via positive ion mode with the ion spray voltage at 5500 V and source temperature at 280°C.
DIA and DIA-TPA Protein Quantification.
HLM and HLS9 samples were analyzed using a DIA method (Shi et al., 2018), which was comprised of a 250-millisecond time-of-flight mass spectrometry scan from 400 to 1250 Da, followed by tandem mass spectrometry scans from 100 to 1500 Da performed on all precursors in a cyclic manner using a 100-variable isolation window scheme. The accumulation time was 25 milliseconds per isolation window, resulting in a total cycle time of 2.8 seconds. Spectronaut Pulsar software (version 11.0; Biognosys AG, Schlieren, Switzerland) and its own standard reference spectral library “Human-Liver (fractionated)” were used to process the DIA data and obtain second mass spectrometer (MS2) signal intensities for label-free absolute protein quantification using the previously published DIA-TPA method (He et al., 2019). Quantification was performed only for proteins with at least two unique peptides. Comprehensive lists of human hydrolases and transferases were downloaded from UniProtKB/Swiss-Prot enzyme class 2 and 3, respectively.
Statistical Analysis
GraphPad Prism version 6.02 (GraphPad Software, San Diego, CA) was used for statistical analysis and generating graphs. The Venn diagram was generated using an online tool developed by the Van de Peer Laboratory (Bioinformatics & Evolutionary Genomics; http://bioinformatics.psb.ugent.be/webtools/Venn/). Since a considerable proportion of proteins had concentrations that did not follow the Gaussian distribution, nonparametric statistical analysis was performed. Specifically, the two-tailed nonparametric Spearman correlation and paired nonparametric t test (Wilcoxon matched-pairs signed rank test) were used to assess the correlations and differences between different features, respectively. A value of P ≤ 0.05 was considered statistically significant.
Results
Proteome Profile Comparison between HLM and HLS9
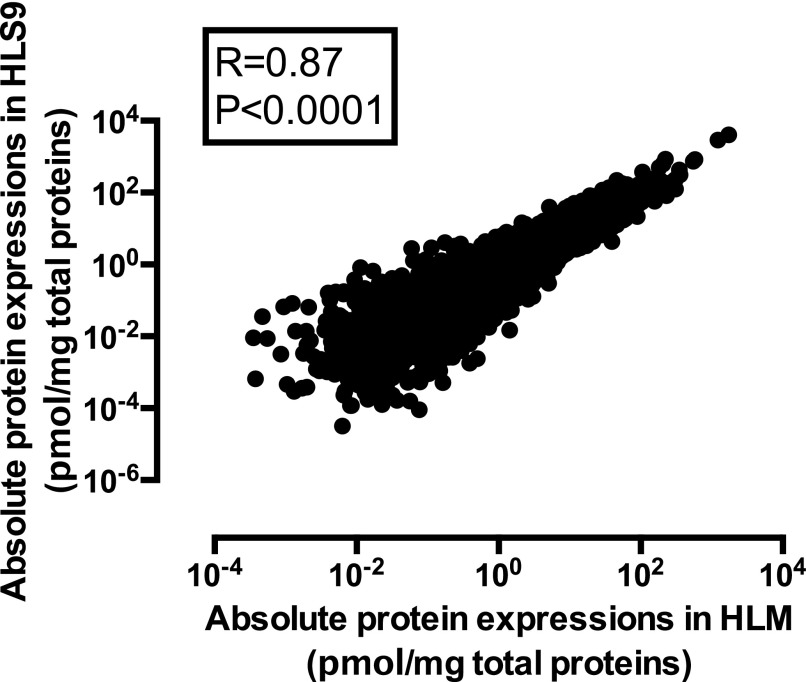
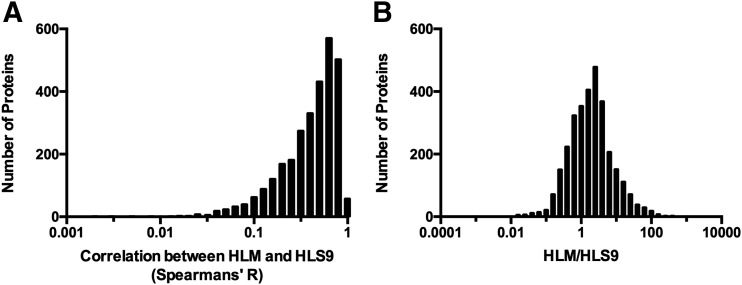
In total, 3137 proteins were absolutely quantified in the HLM and HLS9 samples. Among these, 3087 proteins (98.4%) were shared by the two hepatic preparations, and 48 and two proteins were only quantified in HLM and HLS9, respectively. In general, mean protein concentrations were highly correlated between HLM and HLS9 samples (Fig. 1, R = 0.87, P < 0.0001). Regarding correlations of specific proteins between HLM and HLS9 samples across individuals, 690 out of 3137 proteins were not correlated (R < 0.20, P > 0.05); 701 proteins showed a very weak correlation (0.20 ≤ R < 0.40, P < 0.05); 771 proteins exhibited a weak correlation (0.40 ≤ R < 0.60, P < 0.05); 742 proteins were in moderately strong correlation (0.60 ≤ R < 0.80, P < 0.05); and 233 proteins were in very strong correlation (0.80 ≤ R < 1.00, P < 0.05) (Fig. 2A). With respect to differences in protein concentrations between HLM and HLS9 samples, the concentrations of 1834 proteins were higher in HLM than in HLS9 with HLM-to-HLS9 ratios ranging from 1.11 to 826.98 (P < 0.05); meanwhile, 814 proteins were found to be more concentrated in HLS9 than HLM with HLS9-to-HLM ratios from 1.42 to 75.58 (P < 0.05) (Fig. 2B).
Fig. 1.
Correlation between the mean protein abundances in the 102 HLM and the matched HLS9 samples. Absolute protein concentrations were obtained using the label-free DIA-TPA method. Spearman’s correlation analysis was performed and P < 0.05 was considered statistically significant.
Fig. 2.
Distributions of the Spearman’s R values of the protein concentration correlations between HLM and HLS9 (A) and the HLM-to-HLS9 ratios of mean protein concentrations (B). Fifty proteins that were only quantified in one type of samples were removed from the analysis.
Phase I DMEs in HLM and HLS9
P450s.
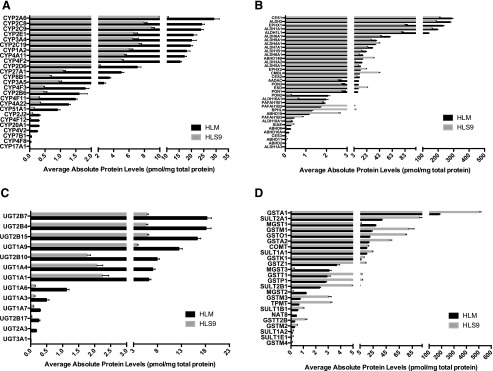
Twenty-five P450s were quantitated in both of the sample types, constituting 1.23% and 0.40% of the HLM and HLS9 proteomes, respectively. As expected, P450 protein concentrations were higher in HLM than HLS9, with HLM-to-HLS9 concentration ratios (P450(i)_HLM/P450(i)_HLS9) ranging from 2.68 to 50.64 (Supplemental Table 2). Additionally, protein levels of the 25 P450 enzymes varied significantly. CYP2A6 was the most abundant P450 enzyme in both hepatic preparations, while CYP17A1 was the least abundant in both (Fig. 3A). Meanwhile, protein concentrations between CYP2A6 and CYP17A1 differed by 5474- and 5582-fold, respectively, in HLM and HLS9 samples. Significant interindividual variability in P450 concentrations was observed across the 102 individual HLM and HLS9 samples, with CVs ranging from 41.43% to 1009.95%. CYP17A1, the least abundant P450, was found to be the most variable, with CVs of 776.11% and 1009.95% in HLM and HLS9, respectively. The substantial interindividual variability in CYP17A1 concentrations is likely in part due to less accuracy in quantifying low abundant proteins. CYP4F2 was the least variable P450 enzyme in both HLM and HLS9 with identical CVs of 41.43% (Supplemental Table 2).
Fig. 3.
Mean protein concentrations of P450s (A), hydrolases (B), UGTs (C), and other transferases (D) in the 102 matched individual HLM and HLS9 samples. Black and gray bars represent protein concentrations in HLM and HLS9, respectively, and error bars represent S.D.
Of human P450s, only about a dozen are involved in drug metabolism (Zanger and Schwab, 2013). The concentrations of these drug metabolism–related P450s ranked as follows: in HLM: CYP2A6 > CYP2C8 > CYP2C9 > CYP2E1 > CYP3A4 > CYP2C19 > CYP1A2 > CYP2D6 > CYP3A5 > CYP2B6 > CYP2J2; and in HLS9: CYP2A6 > CYP2C9 > CYP2C8 > CYP2C19 > CYP2E1 > CYP3A4 > CYP1A2 > CYP2D6 > CYP3A5 > CYP2B6 > CYP2J2 (Fig. 3A). Moreover, the protein levels of these P450s were highly correlated between HLM and HLS9 samples (Supplemental Fig. 1, R = 0.72–0.93, P < 0.0001). The HLM-to-HLS9 ratios of these P450s were within the range of 2.68 and 3.64 (Supplemental Table 2).
Hydrolases.
In total, 308 hydrolases were absolutely quantitated in the HLM and HLS9 samples, of which 36 have been reported to be involved in drug metabolism (Testa and Krämer, 2007). Among these, 11 hydrolases had significantly lower mean concentrations in HLM than HLS9 (hydrolase(i)_HLM/hydrolase(i)_HLS9: 0.22–0.85, P < 0.05), while the remaining 22 hydrolases were significantly more concentrated in HLM (hydrolase(i)_HLM/hydrolase(i)_HLS9: 1.22–26.57, P < 0.05). Carboxylesterase (CES) 1 was the most abundant hydrolase in both HLM and HLS9, with concentrations of 286.73 ± 100.28 and 205.33 ± 70.51 pmol/mg protein, respectively. Interestingly, two epoxide hydrolases [epoxide hydrolase1 (EPHX1) and epoxide hydrolase2] were distributed between HLM and HLS9 in accordance with their subcellular locations. Specifically, microsomal EPHX1 was significantly higher in HLM than HLS9 (EPHX1_HLM/EPHX1_HLS9 = 2.79), whereas cytosolic epoxide hydrolase2 had a HLM-to-HLS9 ratio of 0.57 (Fig. 3B; Supplemental Table 3).
Phase II DMEs in HLM and HLS9
UGTs.
The study quantified 13 and 12 UGTs in HLM and HLS9 samples, respectively. In HLM, the protein concentrations ranged from 6.51 fmol/mg to 18.28 pmol/mg protein, versus 2.75 fmol/mg to 5.81 pmol/mg protein in HLS9 (Fig. 3C). All quantified UGTs were higher in HLM than in HLS9 with fold differences ranging from 2.75 to 60.21 (UGT(i)_HLM/UGT(i)_HLS9). The most abundant UGTs were UGT2B7 in HLM (18.28 ± 9.68 pmol/mg protein) and UGT2B15 in HLS9 (5.92 ± 2.79 pmol/mg protein). UGT3A1 had the lowest concentration in HLM (6.51 ± 0.24 fmol/mg protein), and was not detected in HLS9. Similar to other DMEs, significant interindividual variation in UGT concentrations was observed across the 102 individual human liver samples. UGT2B15 exhibited the smallest interindividual variation in both HLM (CV = 47.84%) and HLS9 (CV = 47.09%), whereas those with the greatest variation were UGT3A1 in HLM (CV = 362.19%) and UGT2A3 in HLS9 (CV = 710.72%) (Supplemental Table 4).
Other Transferases.
In addition to UGTs, 24 out of the 295 transferases quantitated in this study are also involved in the phase II reactions of various medications. Those transferases include thiopurine S-methyltransferase, catechol-O-methyltransferase, and enzymes from the GST, sufotransferase, and NAT families. The majority of the transferases had significantly higher concentrations in HLS9 than in HLM (transferase(i)_HLM/transferase(i)_HLS9: 0.15–0.85, P < 0.05), except for NAT8 and the microsomal GST1, 2, and 3) with ratios ranging from 4.21 to 14.15 (Fig. 3D; Supplemental Table 5).
Transporter Proteins in HLM and HLS9
We quantified protein concentrations of 51 transporters, including 10 ATP-binding cassette (ABC) transporters and 41 transporters from the solute carrier family (SLC). All transporters were more abundant in HLM than in HLS9. ABCB4 and six SLC transporters (SLC1A4, SLC16A1, SLC30A7, SLC39A7, SLC44A1, and SLCO1B3) were only detected in HLM, at relatively low levels. For the transporters detected in both HLM and HLS9, the differences in protein concentration between sample types ranged from 1.16- to 826.98-fold (Supplemental Table 6). ABCD3 was the most abundant ABC transporter in both HLM and HLS9 samples. The protein concentration of ABCB1/MDR1 (P-glycoprotein), the most studied efflux drug transporter, ranked ninth among the 10 ABC transporters in both HLM and HLS9 samples.
Variability of Commonly Used Housekeeping Proteins
To evaluate the variability of commonly used housekeeping proteins in human livers, we determined the protein concentrations of β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-tubulin in the 102 individual HLM and HLS9 samples. β-Actin showed comparable protein concentrations in HLM (3.32 ± 2.41 pmol/mg protein) and HLS9 (2.99 ± 1.48 pmol/mg protein); GAPDH levels were slightly higher in HLS9 (217.26 ± 38.01 pmol/mg protein) than in HLM (187.56 ± 47.20 pmol/mg protein), while β-tubulin was more abundant in HLM (10.13 ± 5.54 pmol/mg protein) compared with HLS9 (4.31 ± 2.52 pmol/mg protein). The interindividual variability of these housekeeping proteins was as follows: in HLM, β-actin (CV: 72.52%) > β-tubulin (CV: 54.68%) > GAPDH (CV: 25.16%); and in HLS9, β-tubulin (CV: 58.44%) > β-actin (CV: 49.50%) > GAPDH (CV: 17.50%). To identity other potential housekeeping proteins for the normalization of protein quantification in the liver, we also evaluated the variability of protein concentrations across the 102 individual HLM and HLS9 samples and identified five proteins with the smallest CVs (13.32%–20.76%) (Table 1). These proteins could potentially serve as new internal controls for the measurement of hepatic protein expression (e.g., in western blots).
TABLE 1.
Absolute quantifications of housekeeping proteins in HLM and HLS9
| Protein | Gene | HLM (pmol/mg protein) | HLS9 (pmol/mg protein) | ||||
|---|---|---|---|---|---|---|---|
| Mean | S.D. | CV | Mean | S.D. | CV | ||
| % | % | ||||||
| Conventional housekeeping proteins | GAPDH | 187.56 | 47.20 | 25.17 | 217.26 | 38.01 | 17.50 |
| TUBB | 10.13 | 5.54 | 54.68 | 4.31 | 2.52 | 58.44 | |
| ACTB | 3.32 | 2.41 | 72.52 | 2.99 | 1.48 | 49.50 | |
| Most stable proteins in HLM | PPIA | 63.97 | 11.21 | 17.53 | 168.24 | 35.24 | 20.94 |
| PRDX4 | 66.37 | 12.65 | 19.06 | 59.92 | 11.15 | 18.61 | |
| CAND1 | 3.51 | 0.68 | 19.41 | 2.49 | 0.68 | 27.41 | |
| APEH | 9.98 | 1.98 | 19.89 | 5.25 | 0.92 | 17.49 | |
| PAICS | 13.14 | 2.73 | 20.76 | 8.01 | 1.65 | 20.57 | |
| Most stable proteins in HLS9 | TPI1 | 35.91 | 8.55 | 23.80 | 138.19 | 18.41 | 13.32 |
| PRDX1 | 150.31 | 40.63 | 27.03 | 179.04 | 25.72 | 14.37 | |
| PGAM1 | 10.94 | 2.63 | 23.99 | 37.17 | 6.07 | 16.33 | |
| PAK1IP1 | 0.60 | 0.13 | 21.76 | 0.68 | 0.11 | 16.72 | |
| APRT | 6.46 | 1.57 | 24.29 | 19.40 | 3.25 | 16.77 | |
ACTB, β-actin; TUBB, β-tubulin.
Organelle and Cytosolic Protein Markers
The concentrations of membrane marker proteins for the endoplasmic reticulum, mitochondria, proteasome, ribosome, Golgi apparatus, and plasma membrane were all higher in HLM than in HLS9 (HLM/HLS9 > 2.00, P < 0.05). Among these organelle membrane marker proteins, plasma membrane signature protein sodium/potassium-transporting ATP1A1 showed the highest HLM-to-HLS9 ratio (7.70), followed by mitochondrial protein COX4I1 (3.76), ribosome-specific protein RPL7A (3.62), ER marker protein EPHX1 (2.79), proteasome marker protein PSMD1 (2.34), and Golgi apparatus protein formimidoyltransferase cyclodeaminase (2.24) (Table 2). As a comparison, the two cytosolic proteins TMSB4X and AKR1A1 had concentrations that were 7.68- and 4.28-fold, respectively, higher in HLS9 than in HLM (Table 2).
TABLE 2.
Absolute quantifications of cytosol and organelle membrane marker proteins in HLM and HLS9
| HLM (pmol/mg protein) | HLS9 (pmol/mg protein) | HLM/HLS9 | Localization | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | CV | Mean | S.D. | CV | ||||
| % | % | ||||||||
| Organelles protein markers | ATP1A1 | 2.08 | 1.82 | 87.74 | 0.27 | 0.23 | 84.29 | 7.70 | Plasma Membrane |
| COX4I1 | 10.46 | 6.15 | 58.84 | 2.78 | 1.65 | 59.39 | 3.76 | Mitochondria | |
| EPHX1 | 230.00 | 88.59 | 38.52 | 82.49 | 43.59 | 52.84 | 2.79 | ER | |
| FTCD | 194.81 | 67.15 | 34.47 | 87.08 | 30.88 | 35.46 | 2.24 | Golgi apparatus | |
| PSMD1 | 8.03 | 2.00 | 23.82 | 2.85 | 0.89 | 27.00 | 2.82 | Proteasome | |
| RPL7A | 19.17 | 13.94 | 72.69 | 5.29 | 3.63 | 68.65 | 3.62 | Ribosome | |
| Cytosolic protein markers | TMSB4X | 5.16 | 2.61 | 50.57 | 39.66 | 20.98 | 52.90 | 0.13 | Cytoskeleton |
| AKR1A1 | 19.23 | 6.52 | 33.91 | 82.49 | 23.42 | 28.39 | 0.23 | Cytosol | |
FTCD, formimidoyltransferase cyclodeaminase.
Sex Effect on Protein Levels in HLS9 and HLM
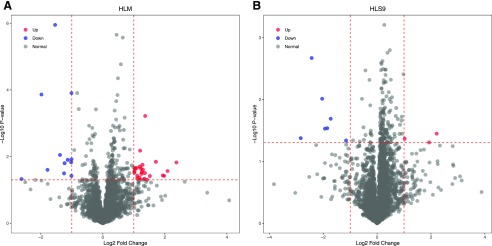
The concentrations of all quantified proteins were compared between the male (n = 46) and female (n = 56) liver samples. In total, 36 HLM proteins and 10 HLS9 proteins were significantly different between males and females (Fig. 4). Among the 36 HLM proteins, 24 proteins exhibited significantly higher levels in males than in females, whereas the remaining 12 proteins were found to have lower concentrations in males, with the male-to-female ratios of protein concentrations ranging from 0.16 to 5.18. For the 10 HLS9 proteins, the concentrations of three proteins were higher in males, while the remaining seven proteins exhibited higher levels in females. The male-to-female ratios of the concentrations of these HLS9 proteins ranged from 0.14 to 4.63 (Supplemental Table 7). Among these sex-associated proteins, UGT2B17 was found to be 2.28-fold higher in males than in females (P < 0.05), which was consistent with the previous findings (Gallagher et al., 2010).
Fig. 4.
The differences in the liver proteomes between males and females in HLM (A) and HLS9 (B). The x-axis is the log2 protein concentration ratios of males to females, with the two vertical dotted lines indicating a 2-fold difference between males and females. The y-axis indicates the −log10 (P value) with a horizontal dotted line at P = 0.05, and the higher the value of y, the more significant is the difference. The red and blue dots represent the proteins being significantly higher in males and females, respectively.
Protein-Protein Correlations in HLM and HLS9 Samples
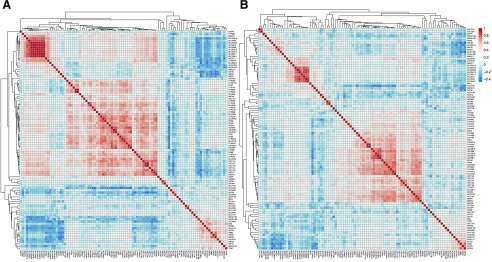
To characterize the coregulation patterns of the proteins involved in drug metabolism and disposition, we analyzed the protein-protein correlations among DMEs and transporters in HLM and HLS9 (Fig. 5). Strong protein-protein correlations were observed within several DME and transporter groups in both HLM and HLS9 samples. Nine transporters in the SLC25 family, including SLC25A1, SLC25A10, SLC25A11, SLC25A13, SLC25A15, SLC25A20, SLC25A4, SLC25A5, and SLC25A6, showed strong correlations with each other, with Spearman’s R ranging from 0.71 to 0.98 in HLM and from 0.41 to 0.90 in HLS9. In addition, four UGT1As (UGT1A4, UGT1A6, UGT1A7, and UGT1A9) and five UGT2Bs (UGT2B4, UGT2B7, UGT2B10, UGT2B15, and UGT2B17) were also strongly correlated within the corresponding subfamilies (R = 0.72–0.97 in HLM; R = 0.70–0.90 in HLS9). Moreover, intrafamily protein-protein correlations were found for the enzymes within the CES (CES1 and CES2, R = 0.62) and CYP2 families (CYP2A6, CYP2B6, CYP2C8, CYP2C9, and CYP2C19, R = 0.50–0.78) in HLM. Interestingly, significant interfamily correlations were also observed for the CES and CYP2 enzymes across the two families in HLM (R = 0.51–0.70). Similarly, the CYP2 enzymes in HLS9 were significantly correlated (R = 0.50–0.77). However, neither the CES enzymes nor the enzymes across the CES and CYP2 families were found to be significantly correlated in HLS9 (Fig. 5).
Fig. 5.
Heat map of protein-protein correlation analysis in HLM (A) and HLS9 (B): red- and blue-shaded boxes indicate the extent of positive and negative correlations, respectively; white boxes indicate no correlation.
Discussion
Although HLM and HLS9 have been widely used for the study of drug metabolism, our knowledge of their protein constitutions, especially absolute protein quantities, remains limited. Recently, in conjunction with advances in LC-MS/MS–based quantitative proteomics, there is increasing interest in studying the protein profiles of HLM and HLS9. To date, most proteomics studies have been conducted on HLM and focused on several major DMEs, such as P450s (Kawakami et al., 2011; Liu et al., 2014) and UGTs (Achour et al., 2017b), and drug transporters (Ohtsuki et al., 2012; Achour et al., 2014b, 2017a; Couto et al., 2019); only a few investigations have included both HLM and HLS9 samples. For example, Fallon et al. (2013) determined the concentrations of 14 UGT1As and UGT2Bs in individual HLM and HLS9 samples using a targeted proteomics method. Recently, a label-free AQP method, named DIA-TPA, was developed for absolute global proteome quantification (He et al., 2019). This method has been validated by comparing its quantification results of several DMEs in HLS9 with the reference values reported from previous studies based on labeled internal standard-based absolute protein quantification (He et al., 2019). Another global proteomics study compared the differences of hepatic proteins among human, rat, and mouse using pooled liver microsomes and S9 fractions and identified 1516 and 1570 proteins in HLM and HLS9, respectively, but no quantitative analysis was performed (Golizeh et al., 2015a,b). The lack of a comprehensive comparative quantitative analysis of the HLM and HLS9 proteomes contributes to uncertainty when choosing one hepatic preparation over another and leads to difficulties in interpreting the data obtained from these two different in vitro models. To fill this knowledge gap, we conducted a global quantitative proteomics analysis of 102 HLS9 samples and their corresponding HLM samples and compared the protein profiles between these two preparations. We found that HLM and HLS9 shared 3087 proteins out of a total of 3137 quantified proteins. As expected, proteins located in the cytosol or associated with the cytoskeleton were more concentrated in HLS9 relative to HLM. In contrast, membrane proteins, such as P450s, UGTs, and transporters, exhibited higher concentrations in HLM than in HLS9 (Supplemental Tables 2, 4, and 6; Table 2).
Significant interindividual variability in protein expressions has been well documented in human livers (Wilkinson, 2005; Wortham et al., 2007), and HLM and HLS9 have been widely used for the study of interindividual variability in drug metabolism (Shimada et al., 1994; Wang et al., 2016). Nevertheless, it is unclear whether the extra steps involved in preparing HLM from HLS9 would introduce additional variation on top of interindividual biologic variability in the liver. Using the 102 individual HLS9 samples and their corresponding HLM samples, we examined the interindividual variability of proteins in both preparations. Among the 3087 proteins shared by HLM and HLS9, 1205 and 1882 showed higher CVs in HLM and HLS9, respectively, suggesting that HLM sample preparation procedures did not introduce additional variation in our experiment. Interestingly, significant negative correlations were observed between CVs and the mean protein concentrations in both HLM (Supplemental Fig. 2A, R = −0.81, P < 0.0001) and HLS9 (Supplemental Fig. 2B, R = −0.88, P < 0.0001) samples. Therefore, part of the variability may be ascribable to proteomics assay errors, which tend to have a more profound impact on the quantification of low abundance proteins (e.g., CYP17A1 and UGT3A1). Demographic factors, such as sex, could contribute to the interindividual variability in the levels of some proteins. By comparing the liver proteomes between males and females, we found that UGT2B17 concentrations were significantly higher in males than in females in HLM (Supplemental Table 7, male/female = 2.28, P < 0.05). This finding was in agreement with the previous report that the protein expression and activity of UGT2B17 were about 4-fold and 1.5–3 times higher, respectively, in males than in females (Gallagher et al., 2010). However, the sex difference in UGT2B17 expression was not observed in HLS9, which is likely caused by the inaccurate protein quantification associated with the low abundance of UGT2B17 in HLS9.
With the 102 matched HLS9 and HLM samples, we were able to identify a significant positive correlation in protein concentrations between the two preparations (Fig. 1, R = 0.87, P < 0.0001). Of note, the correlation was relatively weak for proteins with low concentrations, which could be in part due to the less accurate quantifications of those low-abundance proteins. Besides the correlations of proteins between the two liver preparations, we also analyzed the protein-protein correlation within each liver preparation to explore protein expression coregulations in human livers (Wang et al., 2017). SLC transporters and CYP2 and UGT2B proteins showed significant protein-protein correlations within their own family groups in both HLM and HLS9, indicating that the proteins within these families might share certain common regulatory factors, e.g., nuclear receptors (Chen et al., 2012) and microRNAs (Ikemura et al., 2014). Interestingly, in addition to being highly correlated to each other, CES1 and CES2 were also significantly correlated with the CYP2 isoenzymes including CYP2A6, CYP2B6, CYP2C8, CYP2C9, and CYP2C19 in HLM.
To date, absolute protein quantification of HLM and HLS9 has been mostly performed using multiple reactions monitoring–based targeted proteomics methods with stable isotope internal standards, such as stable isotope-labeled with synthetic peptides (AQUA) and quantification concatamers (QconCAT) (Kawakami et al., 2011; Ohtsuki et al., 2012; Fallon et al., 2013; Achour et al., 2014b, 2017b). In comparison with label-free absolute quantitative proteomics, those stable isotope internal standards–based targeted methods are often more expensive and time consuming. More importantly, the targeted methods are limited in the number of proteins that can be simultaneously quantified (Achour et al., 2014a; Michaels and Wang, 2014; Prasad et al., 2019). Recently, a global label-free proteomics study was conducted to quantify protein concentrations in HLM, with a focus on DMEs (Achour et al., 2017a). Distinct from earlier studies, the present investigation employed a newly developed label-free absolute global quantitative proteomics method, namely DIA-TPA, to compare HLM and HLS9 proteomes using a large set of human liver samples. The DIA-TPA method allows for absolute quantification at the proteome level, and its unique algorithm is particularly suited for the quantification of protein isoforms with highly similar amino acid sequences, such as P450s and UGTs (He et al., 2019). Indeed, the present study quantified more P450s and UGTs than previous label-free absolute quantitative methods (Achour et al., 2017a; Couto et al., 2019). As an example, several CYP4F isoenzymes including CYP4F3 and CYP4F8 were successfully quantitated by the DIA-TPA method in the current study for the first time.
Recently, protein concentrations of P450s and UGTs in HLM have been intensively studied using mass spectrometer–based proteomics (Kawakami et al., 2011; Ohtsuki et al., 2012; Fallon et al., 2013; Achour et al., 2014b; Liu et al., 2014; Prasad et al., 2019); however, comparative analysis of these enzymes in HLM and HLS9 is needed. In the current study, we revealed significant differences in P450 and UGT protein concentrations between the two preparations, as well as marked interindividual variability among individuals. The concentrations of P450s and UGTs in HLM were over two times higher than in HLS9 (Supplemental Tables 2 and 4), which is consistent with both P450s and UGTs being membrane-bound proteins. Given the very low levels of UGTs in HLS9 (UGTHLM/UGTHLS9: 2.75–60.21), HLM appears to be preferable for in vitro study of UGTs. For those drug metabolism–related P450s, protein concentrations correlated well between the matched HLS9 and HLM samples (R = 0.72–0.93, P < 0.0001, Supplemental Fig. 1). We also found that the concentrations of P450s and UGTs in HLM were comparable to the values reported by previous isotope labeling–based proteomics studies (Kawakami et al., 2011; Ohtsuki et al., 2012; Achour et al., 2014b; Gröer et al., 2014; Michaels and Wang, 2014). Moreover, to further validate the protein quantification results, enzymatic activities were determined for CYP2D6 in HLS9 and UGT2B15 in HLM (Supplemental Fig. 4). We observed significant correlations between the enzymatic activities and the protein levels (Supplemental Fig. 4, CYP2D6: R = 0.6942, and UGT2B15: R = 0.6077). The strength of the correlation coefficients is comparable to that reported in previous label-based targeted proteomics studies, further supporting the validity of our label-free global AQP assay (Ohtsuki et al., 2012; Achour et al., 2014b, 2017b). In general, the P450s relevant to drug metabolism were more abundant in HLM than in HLS9. However, given that the differences were relatively small, and the protein levels of these P450s were highly correlated between the two liver preparations, either HLM or HLS9 appears appropriate for the study of these P450 enzymes in the liver.
In addition to P450s and UGTs, other phase I and II enzymes such as hydrolases and other transferases also play vital roles in catalyzing the metabolism of many drugs and toxins. However, our knowledge on the protein profiles of these enzymes in HLM and HLS9 is limited compared with P450s and UGTs. In this study, we quantified 36 hydrolases and 24 transferases in addition to UGTs in HLM and HLS9 samples (Supplemental Tables 3 and 5). In general, hydrolases were the most abundant enzyme group among the four classes of DMEs (i.e., P450s, hydrolases, UGTs, and transferases), and constituted 76.17% and 66.46% of DMEs in HLM and HLS9, respectively (Supplemental Fig. 3). CES1 was the most abundant enzyme, accounting for 15.75% and 14.31% of all DME proteins and 1.81% and 1.29% of the whole proteomes in HLM and HLS9, respectively. As expected, the hydrolases with significantly higher HLM concentrations (concentrationHLM/concentrationHLS9 > 2.0, P < 0.05) were primarily located at membranes. Interestingly, although paraoxonases (PONs) are known to be mostly excreted to the systemic circulation (Fukami and Yokoi, 2012), PON1, PON2, and PON3 exhibited significantly higher concentrations in HLM than in HLS9. Consistent with protein subcellular locations, five hydrolases located in cytosol or nucleus (CMBL, ABHD10, ESD, ABHD14B, and PAFAH1B2) showed significantly higher concentrations in HLS9 than in HLM (concentrationHLM/concentrationHLS9 < 0.50, P < 0.05), and the concentrations of hydrolases associated with membranes or the ER appeared to be significantly higher in HLM than in HLS9 (concentrationHLM/concentrationHLS9: 2.15–3.11, P < 0.05). In contrast to other classes of DMEs, the majority of transferases including the GSTA, GSTK, GSTM, GSTO, and sufotransferase families were more abundant in HLS9 than in HLM (concentrationHLM/concentrationHLS9: 0.15–0.85, P < 0.05), with the microsomal GST family and NAT8 being the only transferases that had higher concentrations in HLM (concentrationHLM/concentrationHLS9: 4.21–14.15, P < 0.05).
Transporters in human livers are another major focus in drug disposition research and have also been recognized as therapeutic targets for drug development (Leonard et al., 2002; Faber et al., 2003; Lin et al., 2015; Couto et al., 2019). In humans, ABC and SLC are the two main transporter families involved in the efflux and uptake of small molecules, resepctively. In this study, a total of 10 ABC transporters and 41 SLC transporters were quantified in HLM, all at relatively higher concentrations than those in HLS9. Of these, ABCB4 and six SLC proteins (SLC1A4, SLC16A1, SLC30A7, SLC39A7, SLC44A1, and SLCO1B3) were only detected in HLM, but not in HLS9. Thus, HLM is more suitable for transporter studies compared with HLS9 due to the extremely low concentrations of most transporters in HLS9. However, it is noted that better coverage of transporters can be achieved if utilizing a sample preparation protocol optimized for transporter protein extraction.
Microsomes are often regarded as cellular fractions with high ER membrane content (Fouts, 1961). However, information is scarce concerning the presence of other cell organelles in HLM and HLS9 preparations. This study determined the concentrations of representative protein markers for several organelle membranes and cytosol. We found that apart from the ER marker EPHX1, protein markers for the plasma membrane and many other organelles, i.e., ribosome (RPL7A), proteasome (PSMD1), mitochondria (COX4I1), and Golgi body (formimidoyltransferase cyclodeaminase) are also more concentrated in HLM, whereas the cytosol protein marker AKR1A1 and cytoskeleton marker TMSB4X showed higher concentrations in HLS9. These observations are in accordance with the fact that HLM is mainly composed of the membranes of hepatic plasma and various organelles, while HLS9 contains both HLM and cytosol (Table 2).
For decades, housekeeping proteins have been routinely used as internal controls to normalize protein loading in western blot experiments (Sullivan-Gunn et al., 2011), with the assumption that their expression levels are constant across samples. However, it is increasingly reported that housekeeping protein expression can be affected by many physiologic and environmental factors (Ferguson et al., 2005; Rubie et al., 2005; Congiu et al., 2011), which could lead to inaccurate protein expression measurements (Sullivan-Gunn et al., 2011; Li and Shen, 2013). Hence, we analyzed the concentrations of the most widely used housekeeping proteins, including β-actin, GAPDH, and β-tublin, in the 102 matched HLM and HLS9 samples. In both the HLM and HLS9 samples, GAPDH was the most abundant housekeeping protein (ranked 22 and 17 among all proteins in HLM and HLS9, respectively), and had the least interindividual variability (CV: 25.17% in HLM and 17.50% in HLS9). To search for new housekeeping proteins suitable for use as internal controls in western blot assays of human liver samples, we identified five proteins with the smallest variability among the 102 individual HLM (CV: 17.53%–20.76%) and HLS9 (CV: 13.32%–16.77%) samples (Table 1). Further validation is warranted to evaluate the applicability of these proteins as new internal controls for western blot experiments.
In summary, the present study compared the proteome profiles of the two most commonly used hepatic preparations, HLM and HLS9, and determined the concentrations and interindividual variability of many DMEs and transporters as well as several housekeeping and subcellular marker proteins in a large set of matched HLM and HLS9 samples. The study revealed marked differences in protein concentrations between HLM and HLS9. The knowledge generated from the study may help us better interpret data obtained from the two different hepatic preparations and may assist in in vitro-in vivo extrapolations. The data also allow us to make more informed decisions when choosing between HLM and HLS9 for in vitro drug metabolism studies.
Abbreviations
- ABC
ATP-binding cassette
- AQP
absolute quantitative proteomics
- CES
carboxylesterase
- DIA
data-independent acquisition
- DME
drug-metabolism enzyme
- EPHX1
epoxide hydrolase1
- ER
endoplasmic reticulum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GST
glutathione S-transferase
- HLM
human liver microsomes
- HLS9
human liver S9 fractions
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- NAT
N-acetyltransferase
- P450
cytochrome P450
- PON
paraoxonase
- SLC
solute carrier family
- TPA
total protein approach
- UGT
UDP-glucuronosyltransferase
Authorship Contributions
Participated in research design: Wang, Shi, Zhu.
Conducted experiments: Wang, Shi.
Contributed new reagents or analytic tools: Li.
Performed data analysis: Wang, He, Shi, Zhu.
Wrote or contributed to the writing of the manuscript: Wang, Zhu.
Footnotes
This work was partially supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01HL126969 (to H.-J.Z.)], and a Michigan Institute of Clinical Health Research Pre-K Award [Grant UL1TR002240 (to X.W.)].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Achour B, Al Feteisi H, Lanucara F, Rostami-Hodjegan A, Barber J. (2017a) Global proteomic analysis of human liver microsomes: rapid characterization and quantification of hepatic drug-metabolizing enzymes. Drug Metab Dispos 45:666–675. [DOI] [PubMed] [Google Scholar]
- Achour B, Barber J, Rostami-Hodjegan A. (2014a) Expression of hepatic drug-metabolizing cytochrome P450 enzymes and their intercorrelations: a meta-analysis. Drug Metab Dispos 42:1349–1356. [DOI] [PubMed] [Google Scholar]
- Achour B, Dantonio A, Niosi M, Novak JJ, Fallon JK, Barber J, Smith PC, Rostami-Hodjegan A, Goosen TC. (2017b) Quantitative characterization of major hepatic UDP-glucuronosyltransferase enzymes in human liver microsomes: comparison of two proteomic methods and correlation with catalytic activity. Drug Metab Dispos 45:1102–1112. [DOI] [PubMed] [Google Scholar]
- Achour B, Russell MR, Barber J, Rostami-Hodjegan A. (2014b) Simultaneous quantification of the abundance of several cytochrome P450 and uridine 5′-diphospho-glucuronosyltransferase enzymes in human liver microsomes using multiplexed targeted proteomics. Drug Metab Dispos 42:500–510. [DOI] [PubMed] [Google Scholar]
- Asha S, Vidyavathi M. (2010) Role of human liver microsomes in in vitro metabolism of drugs—a review. Appl Biochem Biotechnol 160:1699–1722. [DOI] [PubMed] [Google Scholar]
- Braisted JC, Kuntumalla S, Vogel C, Marcotte EM, Rodrigues AR, Wang R, Huang ST, Ferlanti ES, Saeed AI, Fleischmann RD, et al. (2008) The APEX quantitative proteomics tool: generating protein quantitation estimates from LC-MS/MS proteomics results. BMC Bioinformatics 9:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon EF, Raap CD, Meijerman I, Beijnen JH, Schellens JH. (2003) An update on in vitro test methods in human hepatic drug biotransformation research: pros and cons. Toxicol Appl Pharmacol 189:233–246. [DOI] [PubMed] [Google Scholar]
- Braner M, Koller N, Knauer J, Herbring V, Hank S, Wieneke R, Tampé R. (2018) Optical control of the antigen translocation by synthetic photo-conditional viral inhibitors. Chem Sci (Camb) 10:2001–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tang Y, Guo C, Wang J, Boral D, Nie D. (2012) Nuclear receptors in the multidrug resistance through the regulation of drug-metabolizing enzymes and drug transporters. Biochem Pharmacol 83:1112–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congiu M, Slavin JL, Desmond PV. (2011) Expression of common housekeeping genes is affected by disease in human hepatitis C virus-infected liver. Liver Int 31:386–390. [DOI] [PubMed] [Google Scholar]
- Couto N, Al-Majdoub ZM, Achour B, Wright PC, Rostami-Hodjegan A, Barber J. (2019) Quantification of proteins involved in drug metabolism and disposition in the human liver using label-free global proteomics. Mol Pharm 16:632–647. [DOI] [PubMed] [Google Scholar]
- Ekins S, Ring BJ, Grace J, McRobie-Belle DJ, Wrighton SA. (2000) Present and future in vitro approaches for drug metabolism. J Pharmacol Toxicol Methods 44:313–324. [DOI] [PubMed] [Google Scholar]
- Faber KN, Müller M, Jansen PL. (2003) Drug transport proteins in the liver. Adv Drug Deliv Rev 55:107–124. [DOI] [PubMed] [Google Scholar]
- Fallon JK, Neubert H, Hyland R, Goosen TC, Smith PC. (2013) Targeted quantitative proteomics for the analysis of 14 UGT1As and -2Bs in human liver using NanoUPLC-MS/MS with selected reaction monitoring. J Proteome Res 12:4402–4413. [DOI] [PubMed] [Google Scholar]
- Ferguson RE, Carroll HP, Harris A, Maher ER, Selby PJ, Banks RE. (2005) Housekeeping proteins: a preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics 5:566–571. [DOI] [PubMed] [Google Scholar]
- Fouts JR. (1961) The metabolism of drugs by subfractions of hepatic microsomes. Biochem Biophys Res Commun 6:373–378. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol 93:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T, Yokoi T. (2012) The emerging role of human esterases. Drug Metab Pharmacokinet 27:466–477. [DOI] [PubMed] [Google Scholar]
- Gallagher CJ, Balliet RM, Sun D, Chen G, Lazarus P. (2010) Sex differences in UDP-glucuronosyltransferase 2B17 expression and activity. Drug Metab Dispos 38:2204–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golizeh M, Schneider C, Ohlund LB, Sleno L. (2015a) Dataset from proteomic analysis of rat, mouse, and human liver microsomes and S9 fractions. Data Brief 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golizeh M, Schneider C, Ohlund LB, Sleno L. (2015b) Multidimensional LC–MS/MS analysis of liver proteins in rat, mouse and human microsomal and S9 fractions. EuPA Open Proteom 6:16–27. [Google Scholar]
- Gröer C, Busch D, Patrzyk M, Beyer K, Busemann A, Heidecke CD, Drozdzik M, Siegmund W, Oswald S. (2014) Absolute protein quantification of clinically relevant cytochrome P450 enzymes and UDP-glucuronosyltransferases by mass spectrometry-based targeted proteomics. J Pharm Biomed Anal 100:393–401. [DOI] [PubMed] [Google Scholar]
- Hanke S, Besir H, Oesterhelt D, Mann M. (2008) Absolute SILAC for accurate quantitation of proteins in complex mixtures down to the attomole level. J Proteome Res 7:1118–1130. [DOI] [PubMed] [Google Scholar]
- He B, Shi J, Wang X, Jiang H, Zhu HJ. (2019) Label-free absolute protein quantification with data-independent acquisition. J Proteomics 200:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Yang L, Luo J, Guo L, Wang Z, Yang X, Jin W, Fang Y, Ye J, Shan B, et al. (2015) SWATH enables precise label-free quantification on proteome scale. Proteomics 15:1215–1223. [DOI] [PubMed] [Google Scholar]
- Ikemura K, Iwamoto T, Okuda M. (2014) MicroRNAs as regulators of drug transporters, drug-metabolizing enzymes, and tight junctions: implication for intestinal barrier function. Pharmacol Ther 143:217–224. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4:1265–1272. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Hirota N, Ooie T, Suzuki H, Shimada N, Chiba K, Ishizaki T, Green CE, Tyson CA, Sugiyama Y. (1997) Prediction of in vivo drug metabolism in the human liver from in vitro metabolism data. Pharmacol Ther 73:147–171. [DOI] [PubMed] [Google Scholar]
- Kawakami H, Ohtsuki S, Kamiie J, Suzuki T, Abe T, Terasaki T. (2011) Simultaneous absolute quantification of 11 cytochrome P450 isoforms in human liver microsomes by liquid chromatography tandem mass spectrometry with in silico target peptide selection. J Pharm Sci 100:341–352. [DOI] [PubMed] [Google Scholar]
- Leonard GD, Polgar O, Bates SE. (2002) ABC transporters and inhibitors: new targets, new agents. Curr Opin Investig Drugs 3:1652–1659. [PubMed] [Google Scholar]
- Li R, Shen Y. (2013) An old method facing a new challenge: re-visiting housekeeping proteins as internal reference control for neuroscience research. Life Sci 92:747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Yee SW, Kim RB, Giacomini KM. (2015) SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov 14:543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hu L, Ge G, Yang B, Ning J, Sun S, Yang L, Pors K, Gu J. (2014) Quantitative analysis of cytochrome P450 isoforms in human liver microsomes by the combination of proteomics and chemical probe-based assay. Proteomics 14:1943–1951. [DOI] [PubMed] [Google Scholar]
- Meyer UA. (1996) Overview of enzymes of drug metabolism. J Pharmacokinet Biopharm 24:449–459. [DOI] [PubMed] [Google Scholar]
- Michaels S, Wang MZ. (2014) The revised human liver cytochrome P450 “Pie”: absolute protein quantification of CYP4F and CYP3A enzymes using targeted quantitative proteomics. Drug Metab Dispos 42:1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, Ishiguro N, Kishimoto W, Ludwig-Schwellinger E, Ebner T, et al. (2012) Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos 40:83–92. [DOI] [PubMed] [Google Scholar]
- Plant N. (2004) Strategies for using in vitro screens in drug metabolism. Drug Discov Today 9:328–336. [DOI] [PubMed] [Google Scholar]
- Prasad B, Achour B, Artursson P, Hop CECA, Lai Y, Smith PC, Barber J, Wisniewski JR, Spellman D, Uchida Y, et al. (2019) Toward a consensus on applying quantitative liquid chromatography-tandem mass spectrometry proteomics in translational pharmacology research: a white paper. Clin Pharmacol Ther 106:525–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubie C, Kempf K, Hans J, Su T, Tilton B, Georg T, Brittner B, Ludwig B, Schilling M. (2005) Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol Cell Probes 19:101–109. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473:337–342. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. (2013) Corrigendum: global quantification of mammalian gene expression control. Nature 495:126–127. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang X, Lyu L, Jiang H, Zhu H-J. (2018) Comparison of protein expression between human livers and the hepatic cell lines HepG2, Hep3B, and Huh7 using SWATH and MRM-HR proteomics: focusing on drug-metabolizing enzymes. Drug Metab Pharmacokinet 33:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. (1994) Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270:414–423. [PubMed] [Google Scholar]
- Shin JB, Krey JF, Hassan A, Metlagel Z, Tauscher AN, Pagana JM, Sherman NE, Jeffery ED, Spinelli KJ, Zhao H, et al. (2013) Molecular architecture of the chick vestibular hair bundle. Nat Neurosci 16:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5:144–156. [DOI] [PubMed] [Google Scholar]
- Sullivan-Gunn M, Hinch E, Vaughan V, Lewandowski P. (2011) Choosing a stable housekeeping gene and protein is essential in generating valid gene and protein expression results. Br J Cancer 104:1055; author reply 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa B, Krämer SD. (2007) The biochemistry of drug metabolism—an introduction: part 3. Reactions of hydrolysis and their enzymes. Chem Biodivers 4:2031–2122. [DOI] [PubMed] [Google Scholar]
- Wang J, Huo K, Ma L, Tang L, Li D, Huang X, Yuan Y, Li C, Wang W, Guan W, et al. (2017) Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol 13:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang G, Shi J, Aa J, Comas R, Liang Y, Zhu HJ. (2016) CES1 genetic variation affects the activation of angiotensin-converting enzyme inhibitors. Pharmacogenomics J 16:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese S, Reidegeld KA, Meyer HE, Warscheid B. (2007) Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7:340–350. [DOI] [PubMed] [Google Scholar]
- Wilkinson GR. (2005) Drug metabolism and variability among patients in drug response. N Engl J Med 352:2211–2221. [DOI] [PubMed] [Google Scholar]
- Wilson ZE, Rostami-Hodjegan A, Burn JL, Tooley A, Boyle J, Ellis SW, Tucker GT. (2003) Inter-individual variability in levels of human microsomal protein and hepatocellularity per gram of liver. Br J Clin Pharmacol 56:433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Hein MY, Cox J, Mann M. (2014) A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol Cell Proteomics 13:3497–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. (2007) Expression of constitutive androstane receptor, hepatic nuclear factor 4α, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos 35:1700–1710. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Schwab M. (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. [DOI] [PubMed] [Google Scholar]