Abstract
Aims
Fibromuscular dysplasia (FMD) is a poorly understood disease that predominantly affects women during middle-life, with features that include stenosis, aneurysm, and dissection of medium-large arteries. Recently, plasma proteomics has emerged as an important means to understand cardiovascular diseases. Our objectives were: (i) to characterize plasma proteins and determine if any exhibit differential abundance in FMD subjects vs. matched healthy controls and (ii) to leverage these protein data to conduct systems analyses to provide biologic insights on FMD, and explore if this could be developed into a blood-based FMD test.
Methods and results
Females with ‘multifocal’ FMD and matched healthy controls underwent clinical phenotyping, dermal biopsy, and blood draw. Using dual-capture proximity extension assay and nuclear magnetic resonance-spectroscopy, we evaluated plasma levels of 981 proteins and 31 lipid sub-classes, respectively. In a discovery cohort (Ncases = 90, Ncontrols = 100), we identified 105 proteins and 16 lipid sub-classes (predominantly triglycerides and fatty acids) with differential plasma abundance in FMD cases vs. controls. In an independent cohort (Ncases = 23, Ncontrols = 28), we successfully validated 37 plasma proteins and 10 lipid sub-classes with differential abundance. Among these, 5/37 proteins exhibited genetic control and Bayesian analyses identified 3 of these as potential upstream drivers of FMD. In a 3rd cohort (Ncases = 506, Ncontrols = 876) the genetic locus of one of these upstream disease drivers, CD2-associated protein (CD2AP), was independently validated as being associated with risk of having FMD (odds ratios = 1.36; P = 0.0003). Immune-fluorescence staining identified that CD2AP is expressed by the endothelium of medium-large arteries. Finally, machine learning trained on the discovery cohort was used to develop a test for FMD. When independently applied to the validation cohort, the test showed a c-statistic of 0.73 and sensitivity of 78.3%.
Conclusion
FMD exhibits a plasma proteogenomic and lipid signature that includes potential causative disease drivers, and which holds promise for developing a blood-based test for this disease.
Keywords: Plasma protein, Fibromuscular dysplasia, Proteomics, CD2AP
1. Introduction
Fibromuscular dysplasia (FMD) is a poorly understood disease that may cause aneurysm, dissection, tortuosity, stenosis, and/or occlusion of medium-large arteries.1–6 FMD most commonly affects the carotid and renal arteries, although any artery can be affected.7 The clinical manifestations reflect the arterial bed involved, with hypertension observed most often in patients with renal artery FMD, and headaches or pulsatile tinnitus in patients with cervical artery FMD.7 More severe phenotypes may result in end-organ ischaemia such as transient ischaemia attack, stroke, myocardial infarction, and/or gut ischaemia due to associated dissection and/or aneurysm rupture.2,3,8,9
In the current era, FMD is an angiographic diagnosis with a binary classification based on the presence of alternating areas of stenosis and dilation (‘multifocal’ FMD; ∼80% of patients) (Figure 1A and B), or, a single area of concentric or tubular stenosis (‘focal’ FMD; ∼20% of patients).6,7,10 For patients with multifocal FMD, mean age at diagnosis is 50–55 years and ∼90% are female.2,3 In both multifocal and focal FMD, there is often a lengthy delay from symptom onset to diagnosis. This is due in part to lack of provider knowledge and experience in caring for FMD patients, overlap with other common age-related conditions such as essential hypertension, and lack of a simple but accurate screening diagnostic test.10
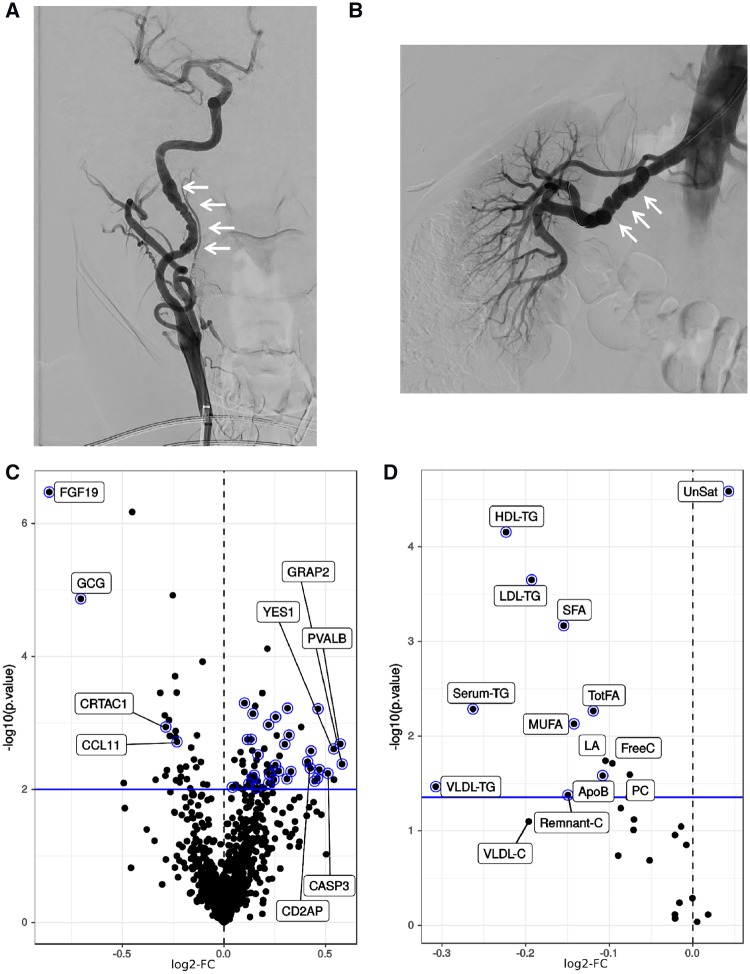
Figure 1.
Typical multifocal FMD and differential protein levels between FMD cases and matched healthy controls in the discovery cohort dataset. (A) Catheter-based angiographic image representative of the typical appearance of multifocal FMD affecting the carotid artery. (B) Typical catheter-based angiographic appearance of multifocal FMD affecting the renal artery, with so called ‘string-of-beads’ appearance. (C) Volcano plot of the differential protein level analysis comparing FMD patients to healthy controls; log2-fold change (log2-FC) on the horizontal axis, -log10 (P-value) on the vertical axis; the horizontal blue line marks the 10% FDR significance threshold. Selected proteins with large effect size or of further interest are labelled. Proteins that were subsequently validated (in the separate validation cohort) are represented with a blue halo. (D) Volcano plot of the differential lipid and lipoprotein level analysis in a fully adjusted model comparing FMD patients to healthy controls (co-variates were age, BMI, statin use, and non-statin lipid lowering medication use); log2-FC on the horizontal axis, -log10 (P-value) on the vertical axis; the horizontal blue line marks the 10% FDR significance threshold. Selected lipid and lipoprotein species with largest effect size are labelled. Lipids that were subsequently validated (in the separate validation cohort) are represented with a blue halo. ApoB, Apolipoprotein B; FreeC, free cholesterol; HDL-TG, triglycerides in HDL; LA, 18:2 linoleic acid; LDL-TG, triglycerides in LDL; MUFA, monounsaturated fatty acids 16:1, 18:1; PC, phosphatidylcholine and other cholines;. Remnant-C, remnant cholesterol (non-HDL, non-LDL-cholesterol); Serum-TG, serum total triglycerides; SFA, saturated fatty acids; TotFA, total fatty acids; UnSat, estimated degree of unsaturation of all fatty acids (the numeric value is an estimate of the average number of double bonds in the fatty acid chains); VLDL-C, total cholesterol in VLDL; VLDL-TG, triglycerides in VLDL.
There is a common misconception that FMD is a rare disease. Various studies suggest otherwise. For example, available imaging from over 3000 renal donors identified FMD in approximately 4% of cases.11 The prevalence may be even higher in symptomatic patients, with 5.8% of patients enrolled in the CORAL (Cardiovascular Outcomes in Renal Atherosclerotic Lesions) trial being found to have renal FMD on review of imaging by the angiographic core laboratory.12 This was despite the fact that FMD was an exclusion criterion for entry into that study. Among the 499 women in CORAL, 8.8% had angiographic evidence of FMD.12
Prior attempts to identify the cause or aetiology of FMD have yielded limited results. At present, only a single FMD-associated genetic locus has been identified, being the common single-nucleotide polymorphism (SNP) rs9349379 in the gene encoding Phosphatase And Actin Regulator 1 (PHACTR1).13 Although those findings have so far provided limited direct insight on the cause of FMD, they nevertheless shed light on the overall genetic basis of this disease—indicating that FMD has a complex genetic pattern of inheritance that potentially involves multiple pathobiological mechanisms.13
Circulating plasma proteins are important markers and biologic mediators of cardiovascular disease,14,15 and novel biostatistical methods which take advantage of genetic causation can now be applied to high-throughput data such as plasma proteomics to better understand disease pathobiology and causation.15–17 Moreover, as a vascular disease, affected FMD tissues are in direct contact with the bloodstream, potentially augmenting any disease-related effects on circulating plasma proteins.
Here, we undertook a high-throughput plasma proteomics study of FMD patients as compared to healthy, matched control subjects. We hypothesized that this approach might identify specific proteins that are associated with FMD, which in turn, might provide insights on the biologic basis of this disease.
2. Methods
2.1 Study design
DEFINE-FMD is a systems biology study aiming to DEFINE key disease drivers and mediators of FMD. Its forerunner pilot study, the CAUSE study (ClinicalTrials.gov Identifier: NCT01808729), enrolled its first ‘run-in’ control subject on 31 October 2012, and first FMD patient on 22 February 2013. As the CAUSE study approached target enrolment it was closed (at n = 34 subjects) and we initiated DEFINE-FMD (ClinicalTrials.gov Identifier: NCT01967511). Of the original CAUSE study subjects, four FMD cases are included in the current analysis, with the remainder of the FMD cases and all controls being from DEFINE-FMD. DEFINE-FMD continues to recruit subjects and as of July 2019 over 375 subjects have been enrolled (approximately 50% cases, 50% controls). Both the CAUSE and DEFINE-FMD studies were approved by the institutional review board of the Icahn School of Medicine at Mount Sinai, and all subjects gave written informed consent. The investigation conformed to the principles outlined in the Declaration of Helsinki.
All FMD cases in this study, in both the discovery and validation cohorts, were seen and assessed in the Mount Sinai Vascular Medicine Clinic. Inclusion criteria for entry into DEFINE-FMD include ≥18 years of age, being freely willing to participate, and fluency in English. FMD cases are required to have a clinical diagnosis of multifocal FMD that is confirmed by imaging [computed tomographic angiography (CTA), magnetic resonance angiography, or catheter-based angiography]. While DEFINE-FMD was recently expanded and is now also enrolling subjects with spontaneous coronary artery dissection (SCAD) or cervical artery dissection (CvAD) in the absence of typical multifocal FMD, these subjects with isolated SCAD or CvAD were not included in this analysis. However, for this analysis confirmed multifocal FMD cases were permitted to have SCAD and/or CvAD. In addition, per our original enrolment criteria through until early 2017, only females are included in this analysis. For healthy controls, inclusion criteria include no clinical features of FMD, CvAD, or SCAD (including no cervical or abdominal bruits, an absence of family history of sudden death or aneurysm) and absence of any major ongoing systemic disease including any condition requiring hospitalization, immune suppression, intravenous or injected medications or that result in functional impairment in the performance of activities of daily living. Healthy controls are recruited from the general population and are pre-screened by the same clinical team and matched to FMD cases according to age, sex, race/ethnicity, and body mass index (BMI). Because it would be almost impossible to identify and recruit control subjects that are of precisely the same age as every FMD case, healthy control females are recruited that broadly match the age and BMI distribution of FMD cases.
Exclusion criteria (for cases and controls) include: co-morbidities which reduce life expectancy to 1 year; any solid organ or haematological transplantation, or those in whom transplantation is considered; active autoimmune disease; illicit drug use; HIV positive; prior malignancy. In controls, an additional exclusion criteria is an early-onset family history of any form of vascular disease. Healthy controls also undergo screening clinical assessment, with specific attention paid to any history or physical examination findings suggestive of FMD or other vascular disease, by two clinical experts in FMD (J.W.O. and D.K.-D.). Notably, three patients who initially agreed to be a healthy control were diagnosed with FMD after the screening history and physical exam.
If the above entry criteria are met and following informed consent, blood draw and skin punch-biopsy (from the medial upper arm) are performed. From the skin biopsy, fibroblasts are derived using explant culture outgrowth techniques (fibroblast data are not presented here). At the blood draw, 20 mL of blood are collected: 10 mL is collected into ethylenediaminetetraacetic acid (EDTA) tubes and is reserved for deoxyribonucleic acid (DNA) extraction, while 10 mL is collected in EDTA-anticoagulated (plasma) and non-anticoagulated (serum) tubes (5 mL each) and reserved for plasma/serum preparation. Samples are transported at room temperature for processing within 15 min. To obtain plasma, EDTA-anticoagulated blood is centrifuged at 2000 g for 10 min. Blood for preparation of serum is also centrifuged at 2000 g for 10 min. Plasma and serum are then aliquoted and immediately frozen at −80°C pending batched analysis (only plasma was processed for this study). DNA is isolated from whole blood using the Puregene Blood Core kit B (cat# 158467, Qiagen, Germantown, MD, USA). DNA was aliquoted and frozen at −80°C.
2.2 Subjects
In February 2017, we identified 90 FMD cases and 100 matched controls from the CAUSE (four cases) and DEFINE datasets (86 cases and all controls) for the discovery phase of this analysis. For this analysis we imposed the additional inclusion/exclusion criteria that controls cannot be related to FMD cases, that the age ranges of cases and controls should be the same, that only persons of self-reported Caucasian race/ethnicity be included in this analysis (to reduce background genetic and proteomic variation), and that any subject taking clopidogrel, ticagrelor, or prasugrel also be excluded. The latter criterion was imposed because these medications are typically used in unstable patients or those with recent clinical events, in which acute-phase inflammatory proteins may have confounded any FMD-specific protein signature.
For the validation cohort, the same criteria were applied and in March–April 2018 an additional 23 FMD cases and 28 matched healthy controls were identified from the ongoing subjects recruited to the DEFINE-FMD study.
Importantly, while the discovery cohort was enrolled from October 2012 until February 2017, and the validation cohort was enrolled separately from March 2017 to April 2018, throughout the entire study and for both cohorts we enrolled FMD cases and controls concurrently, at the same site and by the same investigative team, and with all samples handled identically.
2.3 Power calculations and sample size estimates
For the initial discovery phase of this study, power calculations and samples size estimates were not possible, because plasma proteins had not been studied in FMD patients. However, our discovery cohort sample size was chosen to be similar to other recent successful studies of this nature.17
For the validation phase of the study, analyses and validation sample sizes were determined in advance. We first evaluated the power of a validation proteomics study based on data collected from the discovery cohort. Specifically, we considered a one-sided t-test type-1 error alpha = 0.05, with absolute effect sizes and variability estimated from the discovery cohort on the 105 FMD discovery signature proteins. Through a Monte-Carlo procedure, for validation sample sizes of n = 20, 25, and 30 per group (i.e. 40, 50, and 60 subjects in total), we estimated a median number of replicated proteins of 43, 51, and 57, respectively, out of the 105 obtained from the discovery cohort. Based on these estimates, our final validation cohort comprised 23 FMD cases and 28 matched healthy controls (51 subjects in total).
2.4 Olink protein array
For protein analysis plasma samples were shipped on dry ice and processed by Olink (Watertown, MA, USA). Analyses were performed using a high-throughput technique using all 11 available Olink panels to measure a total of 981 unique human proteins using 1012 probes. The Olink platform uses proximity extension assay (PEA) technology, where oligonucleotide-labelled antibody probe pairs are allowed to bind to their respective target present in the sample.18–20 The PEA technique has the advantage that only correctly matched antibody pairs give rise to a signal, providing high specificity. The discovery and validation cohorts were processed as two separate batches (April–May 2017 and April 2018, respectively). Data were received as normalized protein expression (NPX) values; Olink Proteomics’ arbitrary unit on log2 scale. Values below Limit Of Detection (LOD) were replaced with ‘Not a Number (NaN)’. We transformed those measures into ‘Signal-To-Noise Log-Ratio’ as follows: SNLR[i, j] = NPX[i, j] − LOD[j], where ‘i’ is the sample indicator and ‘j’ is the probe indicator. No probes or samples were excluded from the analysis after quality control.
2.5 Nightingale lipid array data
Plasma was shipped on dry ice to Nightingale Health Ltd. (Kuopio, Finland) for processing on their high-throughput nuclear magnetic resonance spectroscopy (NMR) metabolomics platform. The platform applies a single experimental setup, which allows for the simultaneous quantification of routine lipids, lipoprotein sub-classes, and individual lipids transported by these particles. Details of this platform have been published previously,21,22 and it has been widely applied in genetic and cardiovascular epidemiological studies.23–28 Due to redundancy in markers and in order to reduce multiple comparison testing, we limited our analysis to 31 primary lipid parameters classified under the categories of apolipoproteins, cholesterol, fatty acids and saturation, glycerides and phospholipids, and lipoprotein particle sizes. Notably, while in total there are 40 parameters classified into these categories by Nightingale, we removed 9 parameters that are secondary ratios derived from the primary data.
2.6 Machine learning test algorithm for FMD
We built models incorporating demographic variables plus plasma protein levels only, and demographic variables plus both plasma protein and lipid levels, to predict FMD case/control status. First, the R impute package29 was applied to fill any missing data points in the protein and lipid datasets. In detail, the R impute package employs a nearest neighbour algorithm. For each protein or lipid with missing values, the programme found the k-nearest neighbours using a Euclidean metric, where k was set to 10 (the software’s default value). Each candidate neighbour might contribute some of the missing coordinates in computing the Euclidean distance; in this case, the programme computes the distance using all the non-missing coordinates. Having found the k-nearest neighbours for a protein or lipid, the programme imputed the missing value using the average of the neighbours. For the discovery cohort plasma protein dataset there were 192 280 data points (190 subjects × 1012 Olink probes), of which 4416 were missing and were imputed (2.3%); for the discovery cohort plasma lipid dataset there were 5890 data points (190 subjects × 31 parameters) of which 210 were missing and were imputed (3.6%). For the replication cohort plasma protein dataset there were 51 612 data points (51 subjects × 1012 Olink probes), of which 1670 were missing and were imputed (3.2%); for the replication cohort plasma lipid dataset there were 1581 data points (51 subjects × 31 parameters) of which 60 were missing and were imputed (3.8%).
We then followed the classic machine learning work-flow and in an initial ‘training phase’ the prediction models were constructed using only the discovery cohort protein and lipid datasets (n = 190 subjects). Since we had a large number of potential predictors (i.e. protein and lipid traits) and certain predictors were inter-correlated (Figure 4), a feature selection step was applied to select a set of predictors providing non-redundant information. In detail, we applied an elastic net method which evaluates all the protein and lipid traits simultaneously, and set the regression coefficients of both the non-informative and redundant predictors as zero.30,31 Next, we selected the predictors of non-zero elastic net regression coefficients, which form a set of non-redundant predictors for FMD.30,31 To then formally construct the prediction models we applied a random forest method, specifically because it is an accurate ensemble classifier that consists of many decision trees and outputs, which can also effectively handle a large number of input variables.32 We applied the ‘randomForest’ package in R, which decorrelates the multiple trees generated though different bootstrapped procedures from the training data. Subsequently, ‘randomForest’ also reduces the variance in the trees by averaging them, which results in the final prediction models. Having finalized the predictive models and completed the ‘training phase’ using the discovery cohort datasets, we then moved into an independent ‘testing phase’. In the testing phase, the random forest models were independently applied and evaluated in the validation cohort datasets (n = 51), and we quantified prediction accuracy, receiver operating characteristic (ROC) curve, and area under the ROC curve (c-statistic). The validation cohort datasets or results were not used, in any way, to adjust or tune the predictive models.
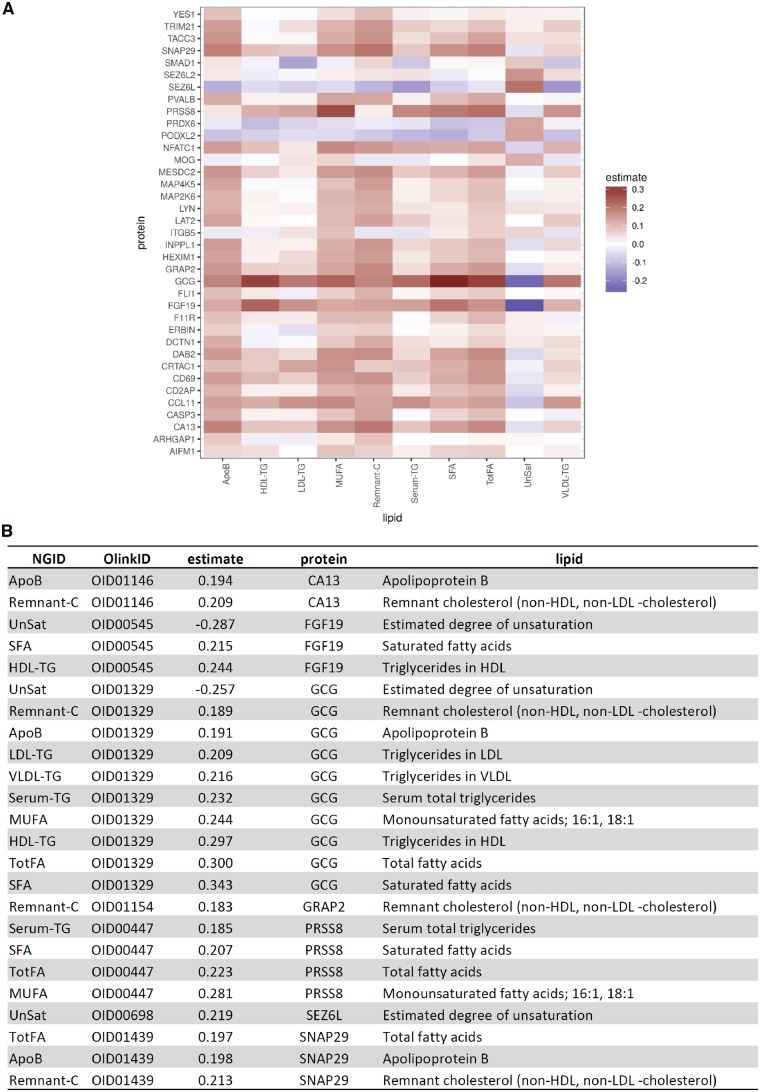
Figure 4.
Cross-correlations between validated FMD signature proteins and lipids. (A) Heatmap of Pearson cross-correlations: proteins in rows, lipids in columns. (B) Corresponding table of cross-correlations below the 10% FDR cut-off. NGID, Nightingale identification code.
2.7 Additional study details and methods
Further study details, scientific methods, and statistical methods are provided in the Supplementary material online, Appendix.
3. Results
3.1 Patient demographics
A total of 90 multifocal FMD patients and 100 healthy, age- and sex-matched controls were included in the discovery cohort of this study. All cases and controls were female and of self-reported Caucasian ethnicity (Table 1).
Table 1.
Discovery cohort demographic summary statistics
| FMD patients (n = 90) |
healthy controls (n = 100) |
|||
|---|---|---|---|---|
| Variables | Summarya | n | Summarya | n |
| Clinical | ||||
| Sex: female | 90 (100.00%) | 90 | 100 (100.00%) | 100 |
| Age at FMD diagnosis | 54 (24–73) | – | ||
| Age at study enrolment | 57 (32–74) | 90 | 50 (34.0–74.0) | 100 |
| Height (in) | 64.0 (58.0–71.0) | 90 | 65.0 (59.0–73.0) | 100 |
| Weight (lbs) | 136.5 (103.0–208.0) | 90 | 141.0 (100.0–240.0) | 100 |
| BMI | 23.1 (18.3–33.6) | 90 | 23.8 (16.6–38.7) | 100 |
| DM | 2 (2.22%) | 90 | 1 (1.00%) | 100 |
| HTN | 54 (60.00%) | 90 | 4 (4.00%) | 100 |
| Ever smoker | 18 (20.00%) | 90 | 18 (18.00%) | 100 |
| Medication use | ||||
| ACE/ARB | 31 (34.44%) | 90 | 2 (2.00%) | 100 |
| Aspirin | 72 (80.00%) | 90 | 5 (5.00%) | 100 |
| Anticoagulation | 5 (5.56%) | 90 | 0 (0.00%) | 100 |
| Beta blocker | 22 (24.44%) | 90 | 4 (4.00%) | 100 |
| Current hormone therapy | 5 (5.56%) | 90 | 10 (10.00%) | 100 |
| Statin | 33 (36.67%) | 90 | 6 (6.00%) | 100 |
| Non-statin lipid lowering | 13 (14.4%) | 90 | 1 (1.00%) | 100 |
| Thyroid replacement | 18 (20.00%) | 90 | 12 (12.00%) | 100 |
| FMD vascular features | ||||
| Aneurysmb | 30 (33.33%) | 90 | – | |
| Dissectionb | 34 (37.78%) | 90 | – | |
| Prior TIA/CVA FMD arterial disease location | 13 (14.61%) | 89 | – | |
| Cervical (carotid/vertebral) | 70 (77.78%) | 90 | – | |
| Coronaryc | 3 (3.33%) | 90 | – | |
| Iliac | 6 (6.67%) | 90 | – | |
| Intracranialc | 12 (13.33%) | 90 | – | |
| Mesenteric | 17 (18.89%) | 90 | – | |
| Renal | 66 (73.33%) | 90 | – | |
| FMD arterial bed involvement | ||||
| Total number of arterial beds involved | 2.0 (1.0–4.0) | 90 | – | |
Except where stated, all data are as at the time of study enrolment.
ACE, angiotensin converting enzyme inhibitor; ARB, angiotensin receptor blocker; DM, diabetes; HTN, hypertension; statin, HMG-CoA reductase inhibitor; TIA, transient ischaemic attack; CVA, cerebrovascular accident.
Total number of subjects with the described characteristic for yes/no features (percentage in parenthesis); median and min–max range for continuous features.
Aneurysm or dissection is considered a manifestation of FMD only if multifocal (or focal) findings are observed in a separate vascular bed.
Due to the specific features of FMD in these vascular beds, ‘coronary FMD’ implies a coronary artery dissection, while ‘intracranial FMD’ implies an intracranial aneurysm.
3.2 A plasma protein signature of FMD
The proteomics platform used for this study (PEA; Olink) allowed the evaluation of plasma levels of 981 proteins. After statistical correction for age and BMI, at 10% false discovery rate (FDR), we identified 105 proteins (104 distinct gene symbols) with differential abundance in FMD cases compared to controls (Figure 1C and Supplementary material online, Table S1). Most proteins showed moderate effect sizes with log2-fold changes (log2-FC) less than 0.5, with the most notable exceptions being fibroblast growth factor 19 (FGF19; log2-FC = −0.86) and the peptide hormone glucagon (GCG; log2-FC = −0.71) (Figure 1C).
To characterize this FMD plasma protein signature, we ran a Gene Set Enrichment Analysis against all collections of the MSigDB database. At 10% FDR, we found 4 enriched Gene Sets in the curated collection, 8 in the GO biological processes collection, 23 in the GO cellular components collection, and 32 in the GO molecular function collection (Supplementary material online, Table S2). Of the 8 most strongly represented terms (all <1% FDR), 4/8 were involved with chemokine and cytokine signalling.
3.3 FMD plasma protein signature is independent of medication use
Overall, FMD cases and healthy controls were well matched (Table 1). However, as might be expected, FMD cases had greater use of aspirin, ACE/ARBs (angiotensin converting enzyme inhibitors and angiotensin receptor blockers, respectively), beta blockers, and statins (Table 1). Therefore, a number of analyses were performed to determine if medication use was responsible for protein changes that were part of the FMD protein signature. First, the impact of these four medication classes on plasma protein levels was assessed by evaluating their effect within FMD patients only. There were no significant differences in protein levels with either aspirin or beta blocker use among FMD patients. ACE/ARB use was only associated with increased plasma renin levels (log2-FC = 0.93), while statin use was only associated with increased levels of PCOLCE (Procollagen C-endopeptidase enhancer 1; log2-FC = 0.93). Neither renin nor PCOLCE were among the 105 signature proteins associated with FMD. Second, we compared the impact of differing medications and FMD disease status on plasma protein levels, and found that the pattern of association of the signature proteins with FMD status was not mirrored in the medication profiles (Figure 2 and Supplementary material online, Table S3). Third, we built an alternative FMD regression model where we sequentially excluded cases and controls according to use of differing medication classes. These alternate FMD protein lists consistently overlapped with our full FMD protein signature obtained by including all subjects, with odds ratios (OR) of overlap with the full signature of between 4.4 and 10.7, and Fisher’s exact test P-values <1 × 10−31 (Supplementary material online, Table S4). These three, concordant analyses indicate that medication use is unlikely to be a major confounding factor in the FMD plasma protein signature.
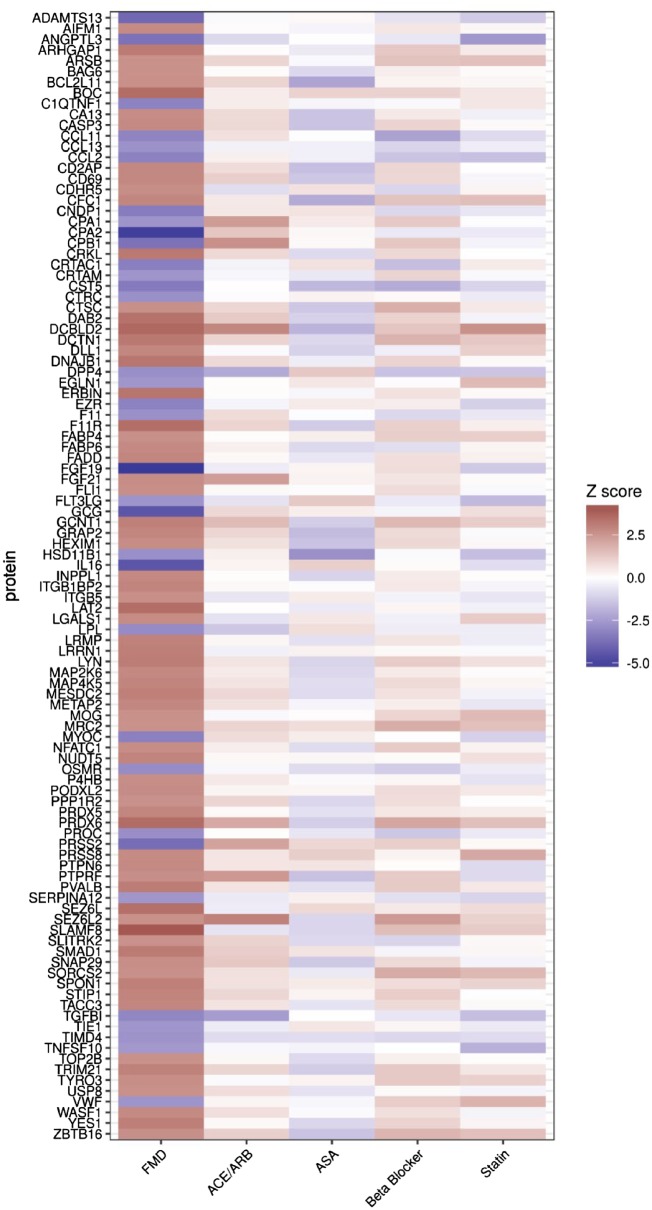
Figure 2.
The FMD protein signature is not due to medication use. FMD signature protein heatmap with the 105 signature FMD proteins from the discovery cohort presented in rows, and with z-scores of association with FMD shown in the first column, and different key medication classes in the other columns. FMD status (first column) was determined using all subjects, while medication use (other columns) was determined in FMD cases only. As expected, FMD disease status was strongly associated with each of these FMD signature proteins, while there was no association of these signature proteins with the use of differing classes of medications. z-Scores of protein associations with FMD disease status (all subjects) and differing classes of medication use (estimated within FMD patients only) corresponding to this figure are presented in Supplementary material online, Table S3.
3.4 A lipid signature of FMD
We noted that the FMD protein signature contained many proteins that regulate plasma lipid levels (Supplementary material online, Table S5). Therefore, we hypothesized that in addition to plasma proteins, FMD may also be associated with alterations in plasma lipid levels. Accordingly, we elected to profile plasma lipid and lipoprotein levels by NMR. For the data analysis, given the known associations with statin use and lipid levels, to explore the possible relationship between FMD and lipid levels we constructed a fully adjusted model that included age, BMI, statin use, and the use of non-statin lipid lowering agents. In this fully adjusted analysis, we observed differential abundance of 16 of a total of 31 lipid-related parameters (Figure 1D and Supplementary material online, Table S6). Notably, FMD cases had reduced levels of several lipid moieties; most notably of triglyceride- and fatty acid-related lipid sub-classes. Of the top three most significantly reduced lipid fractions we observed reduced LDL-triglycerides (triglycerides in LDL), reduced HDL-triglycerides (triglycerides in HDL), and reduced saturated fatty acids. We observed only a single lipid parameter that was increased in FMD cases vs. controls, which was the degree of unsaturation of all fatty acids (Figure 1D and Supplementary material online, Table S6). Alternate statistical approaches that included using a more flexible model for age as a covariate did not alter these findings (Supplementary material online, Figure S1). Furthermore, to confirm the general validity of our lipid datasets we sought and were able to replicate known lipid-associated gene loci (Supplementary material online, Figure S2). Thus, in addition to a plasma protein signature, FMD is associated with an altered lipid profile, most notably with reduced levels of triglycerides and saturated fatty acids.
3.5 Independent validation of a plasma protein and lipid signature for FMD
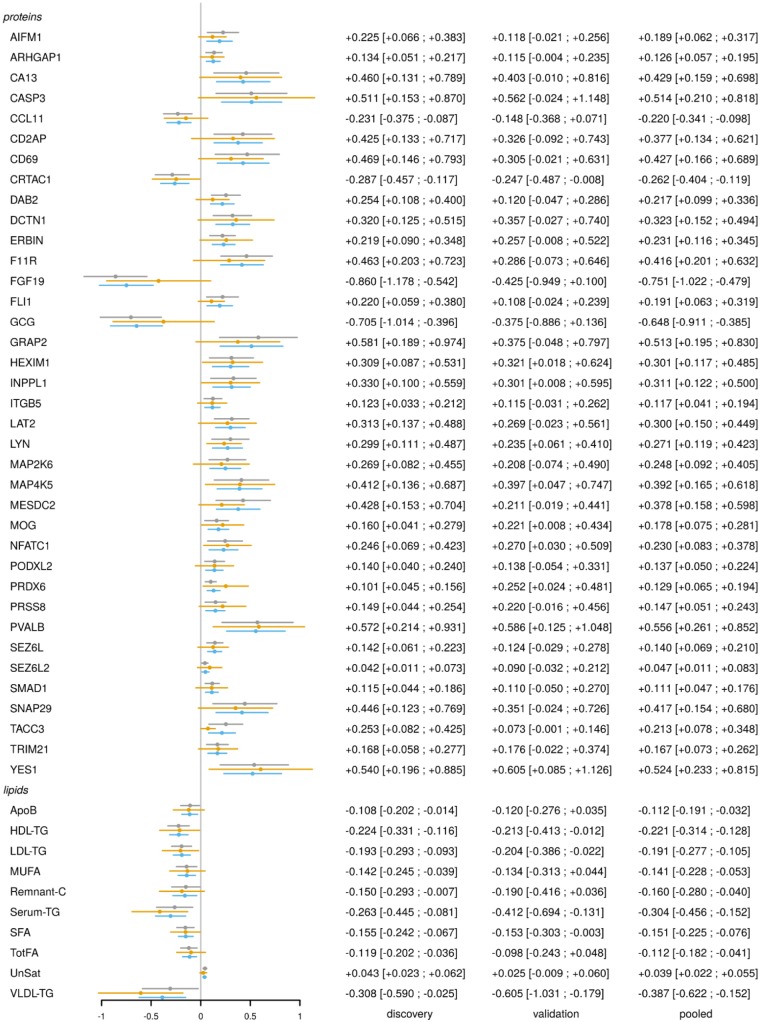
We sought to validate the FMD plasma protein and lipid signature using an independent validation cohort, which based on sample size and power estimates (see Section 2) comprised 23 FMD patients and 28 matched controls (total subjects = 51). Subject inclusion/exclusion criteria, subject recruitment, and sample processing were identical to the discovery cohort. Cases and controls in the validation cohort were well matched and the disease-specific features of the FMD cases were similar to the discovery cohort (Supplementary material online, Table S7). The criteria for validation were proteins which showed association with FMD in the validation cohort in the same direction as the discovery cohort, with a one-sided, nominal P-value <0.05. We found that 37 of the original 105 differentially abundant proteins and 10 of the original 16 differentially abundant lipids were validated in this independent cohort (Figure 1C and D—proteins with blue halo).
All proteins and lipids were analysed in a pooled dataset composed of all subjects from both the discovery and validation cohorts (113 FMD cases, 128 matched healthy controls). The top two protein candidates from the discovery analysis, FGF19 and the peptide hormone GCG, were both validated and in the pooled analysis these remained as the top two differentially abundant proteins. Validated proteins and lipids are shown in Figure 3, with complete results for all markers presented in Supplementary material online, Table S1 (proteins) and Table S6 (lipids).
Figure 3.
FMD protein and lipid signature. Forest plot of association between validated FMD proteins and lipids, and FMD status, in three datasets: discovery cohort (grey lines), validation cohort (gold lines), and pooled cohort (blue lines). Estimated log2-fold changes on the horizontal axis, protein, and lipid labels on the vertical axis. Point estimates and 95% confidence intervals are further reported on the right.
To gain insights on the potential mechanisms whereby upstream proteins may be associated with or regulate lipid levels in FMD patients, this pooled dataset was used to perform a cross-correlation of the 37 validated FMD signature proteins with the 10 validated plasma lipids. This revealed that levels of FGF19, GCG, PRSS8, and SNAP29 were each associated with the levels of several differing lipid sub-classes, while levels of proteins CA13, GRAP2, and SEZ6L were each associated with the levels of one or two lipid sub-classes (Figure 4). Adding validity to these findings, we note that the many of these protein-lipid associations (Figure 4) have previously been described (Supplementary material online, Table S5).
This pooled dataset was also leveraged to determine if there were any associations between plasma protein or lipid levels with FMD disease features or severity. This exploratory analysis suggested that several FMD disease features and markers of severity may be associated with specific proteins and lipids, although none of these associations reached statistical significance (Supplementary material online, Tables S8 and S9).
3.6 A proof-of-concept plasma protein test for FMD
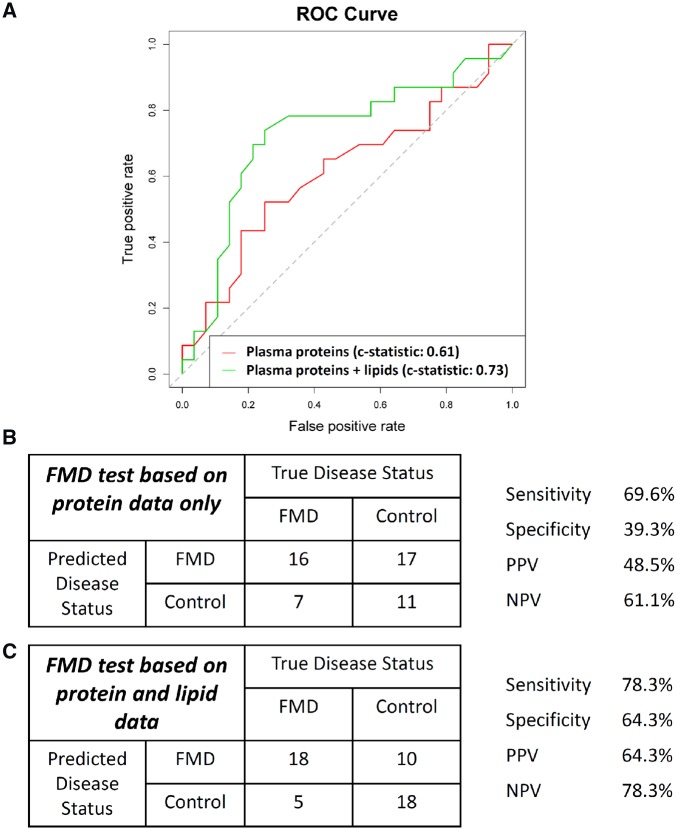
Given the diagnostic challenges posed by FMD,1,6,10 machine learning was performed to study the possibility that a specific plasma protein/lipid signature might have efficacy as a diagnostic test to predict the clinical likelihood of having FMD. We exclusively trained and developed FMD predictive models using the discovery cohort datasets, and used the validation cohort as fully independent datasets to evaluate prediction performance. An initial model incorporating 34 proteins (selected by elastic net, see Section 2) was generated using the discovery cohort datasets and independently evaluated on the validation cohort yielding a c-statistic of 0.61 (Figure 5). Of these 34 proteins, 6 were among the 37 plasma proteins that we validated as being associated with FMD (Figure 3). Next, we explored using the lipid data in conjunction with the protein and demographic data. In a model incorporating the same 34 proteins and all 31 lipid parameters, a random forest prediction model for FMD was developed and evaluated on the independent validation cohort, yielding a c-statistic of 0.73, with a sensitivity of 78.3%, specificity 64.3%, positive predictive value 64.3%, and negative predictive value 78.3% (Figure 5).
Figure 5.
A diagnostic test algorithm for FMD. Machine learning was used to generate diagnostic algorithms for predicting that a subject has FMD. Algorithms were exclusively generated using the discovery cohort datasets. Data shown in this figure represent the independent application of these algorithms to the validation cohort datasets. (A) Receiver operating characteristic (ROC) curve for a diagnostic algorithm for predicting FMD based only on plasma proteins (red), and also based on both the protein and lipid data (green). (B) Performance characteristics of the diagnostic test for FMD based only on protein data (as related to red line in A), when independently applied to the validation dataset. A total of 34 proteins were included in this model. (C) Performance characteristics of the diagnostic test for FMD based on the protein and lipid data (as related to green line in A), when independently applied to the validation dataset. A total of 34 proteins and all 31 lipid parameters were included in this model. Age and other demographic data did not enter into these models.
3.7 Identification of novel risk genes for FMD
To seek potential novel genes that are associated with the risk of having FMD, we performed protein-QTL (pQTL) mapping on confirmed Caucasian FMD patients and healthy control subjects in the discovery cohort. In brief, a pQTL is a DNA variant (SNP) that is associated with the levels of a specific plasma protein, with cis-pQTLs being local to the protein-coding gene and trans-pQTLs being at a distance. At 10% FDR, we found 193 cis- and 34 trans-pQTLs, for a total of 219 distinct probes. Of the proteins associated with these 219 pQTLs, 5 were among the 37 validated proteins that showed differential abundance between FMD cases and healthy controls (Figure 3), being CD2-associated protein (CD2AP), PODXL2, TACC3, INPPL1, and CRTAC1. Complete association statistics between all tested variants and all array probes are reported in Supplementary material online, Table S10.
We conducted a Bayesian network analysis to better understand the relationship between these five FMD signature proteins with an associated pQTL, the genotype of the distinct variants which were found to control these proteins, and FMD disease status (Table 2). Of the five analysed proteins, INPPL1 was classified as downstream of FMD (i.e. its regulation is a consequence and not a cause of FMD), CRTAC1 was classified as independent (i.e. a common latent factor simultaneously influences the protein level and disease status), while CD2AP, PODXL2, and TACC3 were classified as upstream of the disease (i.e. they may be causally related to FMD).
Table 2.
Bayesian network analysis results
| OlinkID | Gene symbol | Classification | FMD log2(FC) |
|---|---|---|---|
| OID01056 | INPPL1 | Downstream | 0.33 |
| OID01304 | CRTAC1 | Independent | −0.29 |
| OID01133 | CD2AP | Upstream | 0.43 |
| OID01389 | PODXL2 | Upstream | 0.14 |
| OID01399 | TACC3 | Upstream | 0.25 |
Bayesian network classification of the five validated FMD signature proteins which were found to be genetically regulated. In the 4th column we further report our estimated log2-fold change (log2-FC) in protein levels between FMD cases and healthy controls. By ‘classification’ according to this Bayesian analysis, ‘downstream’ implies that regulation of this protein is a consequence and not a cause of FMD, ‘independent’ implies a common latent factor simultaneously influences the protein level and disease status, while ‘upstream’ suggests that the protein may potentially be causal for FMD.
3.8 CD2AP is a novel gene that is associated with FMD
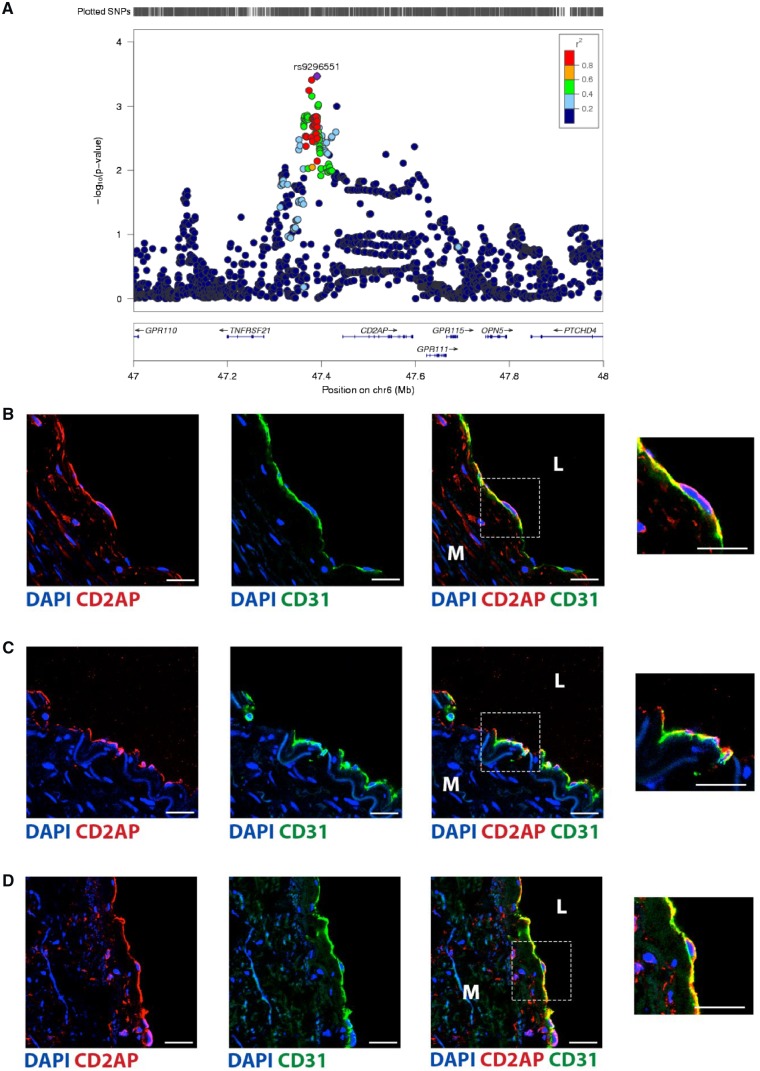
We leveraged an independent FMD genetic association study (with 506 FMD cases and 876 controls)13 and queried the associations of the genes that encode the five FMD signature proteins with an associated pQTL (CD2AP, PODXL2, TACC3, INPPL1, and CRTAC1). Consistent with the Bayesian analysis which showed they are either downstream or independent of FMD, there were no associations found for INPPL1 or CRTAC1. There were also no associations found for PODXL2 and TACC3. However, we identified a significant association with several loci that were in close linkage disequilibrium in the region upstream of CD2AP, with most of these having a P-value of ∼0.001, and with the most strongly associated being rs9296551 (OR = 1.36, P = 0.0003) (Figure 6A).
Figure 6.
CD2AP is associated with FMD and is expressed by endothelial cells. (A) Locus zoom plot showing the peak of association with FMD risk, upstream of the gene encoding CD2AP. (B–D) Immune-fluorescence staining for CD2AP was performed on adult human non-FMD samples from (B) renal artery, (C) internal mammary artery, and (D) aorta. Endothelial cells were specifically identified by staining for CD31 (green), while CD2AP is shown in red. Nuclei were stained with DAPI (blue). Scale bar represents 25 μm. Inset panels on the right, representing an enlarged view of the area in the respective dashed squares, show endothelial cells at higher magnification. M, tunica media; L, lumen. All images are representative, and consistent results were obtained from staining of the aorta, internal mammary artery, and renal artery from at least three different subjects for each sample type.
Having validated a genetic association, we sought to explore potential mechanism(s) whereby CD2AP may be causal for FMD. Although CD2AP is known to be expressed by capillary endothelial cells in the cerebral microcirculation and lung,33,34 to the best of our knowledge its expression has not been studied in medium-large arteries (as are affected by FMD). We therefore performed immune-fluorescence staining for CD2AP in the human aorta, renal artery, and internal mammary artery. These experiments revealed consistent and robust expression of CD2AP by endothelial cells in these vessels (Figure 6B–D), suggesting the possibility that changes in CD2AP may result in perturbation of endothelial cell function.
3.9 Relationship between FMD-associated SNP rs9349379 and plasma proteins/lipids
The SNP rs9349379 is associated with FMD,13 and a prior study suggested that this SNP may contribute to vascular disease pathology by modulating plasma endothelin-1 levels.13,35 Therefore, we queried potential associations with this SNP. In fully adjusted analyses, there were no plasma protein (Supplementary material online, Table S11, Figures S3 and S4) or lipid parameters (Supplementary material online, Table S12) associated with genotype at rs9349379, suggesting that this SNP is unlikely to exert any disease-relevant effects for FMD via modulation of plasma protein or lipid levels.
4. Discussion
Despite it being 60 years since the first FMD pathologic classification system was reported,36,37 remarkably little is known about the aetiology or pathologic mechanisms of this disease. Here, we present the first plasma proteomics and lipidomics analysis for FMD. Important novel findings from this study include: (i) the identification and validation of 37 proteins that comprise a unique FMD disease signature; (ii) the identification and validation of an additional 10 plasma lipid sub-fractions that comprise an FMD lipid signature; (iii) among the 37 validated signature proteins we found that 5 have an associated pQTL(s); (iv) by Bayesian analysis, 3 of these 5 proteins with an associated pQTL(s) appear as upstream in the FMD disease process and accordingly, 1 of these genes associated with a pQTL (CD2AP) was validated as being associated with FMD (P = 0.0003). It was further shown that CD2AP is expressed by endothelial cells in medium-large sized human arteries—the same vessels that are affected in FMD; and (v) as proof-of-concept, a pilot FMD test was generated by machine learning on the plasma protein and lipid signatures of the discovery cohort, that when independently applied to the validation cohort had a c-statistic of 0.73 and sensitivity of 78.3% for diagnosing FMD.
The most important finding arising from this study is the identification of a signature of plasma protein and lipid changes in patients with FMD (Figure 3). It remains to be determined which of these changes are causal for FMD, however, the most promising candidate to emerge from this study was CD2AP. In addition to showing increased abundance in FMD cases vs. controls in the discovery and validation cohorts, a link between CD2AP and FMD is suggested by the following observations: (i) CD2AP was one of only five signature proteins to exhibit genetic regulation (i.e. pQTLs); (ii) our Bayesian analysis suggests that CD2AP is an upstream driver of the FMD disease process; (iii) we validated an association between FMD and several genetic variants in the vicinity of CD2AP (P = 0.0003); (iv) CD2AP is strongly expressed in the endothelium of medium-large human arteries (as are involved in FMD) (Figure 6); and (v) based on prior literature,33,38–41 CD2AP is a strong candidate for a functional role in FMD. Specifically, while CD2AP may also be involved in other processes,42,43 polymorphism(s) in CD2AP are associated with Alzheimer’s disease.38–40 Murine studies have suggested that the mechanism of involvement in Alzheimer’s disease may be via a CD2AP-mediated perturbation in inter-cellular adhesion41 and endothelial function,33 leading to compromised vascular integrity and blood–brain barrier function.33 As a clinical correlate, one of the major FMD sub-phenotypes is arterial dissection (Table 1), where compromised vascular integrity is likely to be an underlying precipitant. Thus, there are reasonable grounds to believe that CD2AP may play a causal role in the pathogenesis of FMD.
Apart from CD2AP, other FMD signature proteins may also be involved in FMD disease causation. For example, PODXL2 (also known as endoglycan—a member of the CD34 family of proteins) is another of the three proteins identified as being potentially causative of FMD in the Bayesian analysis (Table 2). Although understudied, PODXL2 is expressed by the endothelium and acts as a ligand for vascular selectins, whereby it mediates the interaction of leucocytes with vascular surfaces through interactions with E-, P-, and L-selectins.44 Conceivably, perturbation of leucocyte–endothelial interactions in medium-large sized arteries could alter local vascular immune homeostasis, which over the longer-term might lead to unfavourable changes that culminate in FMD.
The favourable alterations in triglyceride and fatty acid levels in FMD patients were unexpected (Figures 1D and 3), but not without precedent. Interestingly, a prior study in patients with CvAD, a disease that is related to and sometimes overlaps with FMD,2 found that subjects with CvAD have a lower prevalence of hypercholesterolaemia compared to controls.45 As a possible explanation for the generally reduced lipid levels in FMD subjects, it is possible that FMD-associated changes in endothelial cells might affect endothelial lipase,46,47 or that other determinants of lipid handling by the vessel wall are perturbed in FMD. However, it is also possible that altered lipid levels are merely secondary association of FMD, or even, that altered lipid levels are upstream and causal for FMD.
At present, FMD is usually diagnosed by imaging studies. While alternatives exist, the preferred imaging modality for diagnosing FMD is CTA,1,9,48 with current data supporting one-time ‘head-to-pelvis’ CTA in all FMD patients.2,6,48,49 Screening for FMD is challenging, and asymptomatic FMD patients or those with non-specific symptoms (e.g. pulsatile tinnitus) may remain undiagnosed for many years or until they suffer a major event.1,7,10 As a possible solution, our proof-of-concept diagnostic algorithm suggests that it may be possible to develop a blood-based test for FMD. A reliable blood-based FMD test would likely have a major impact on the management of this disease, including aiding diagnosis, screening family members, facilitating family counselling, tailoring of clinical care, and other applications.
With respect to study limitations, protein levels were not separately measured by individual enzyme-linked immunosorbent assay, except endothelin 1 (Supplementary material online, Figure S3). Instead, to multiplex protein measurements we took advantage of PEA technology. Accuracy is the hallmark of PEA technology,18–20 where dual binding of antibodies must occur in close proximity to detect and measure protein abundance. Similarly, lipid parameters were measured in this study using another advanced technique—NMR spectroscopy. Recently, numerous studies have attested to the accuracy of both PEA18–20 and NMR,21–28 and rather than being a limitation, we believe that our reliance on these platforms is a potential strength of this study. Other considerations include a relatively small sample size, and the need for further mechanistic studies to understand the role of the signature FMD proteins and lipids. While promising, the proof-of-concept diagnostic FMD test also requires further development and evaluation in larger cohorts.
In conclusion, patients with multifocal FMD exhibit a plasma proteogenomic signature that includes promising candidates such as CD2AP, which merit future investigation as potential novel disease drivers. In addition, patients with FMD exhibit favourable alterations in their plasma lipid profile, most notably reduced triglyceride and fatty acid levels. While further development and evaluation will be required, our proof-of-concept analyses suggest that it may be possible to develop a blood-based test for FMD.
Supplementary Material
Acknowledgements
We thank the many patients and controls who participated in this study. We also gratefully acknowledge Pamela Mace and FMDSA for their support of this study, and Sister Christine Liegey and the people of the parish of St. Philip The Apostle in Clifton, NJ. We thank Brandy M. Haydel, CCRC, for assistance with tissue sample collections. We acknowledge support from Philippe Amouyel, Univ. Lille, Inserm, Institut Pasteur de Lille, LabEx DISTALZ-UMR1167, Lille, France. We also acknowledge the assistance and technical expertise of the Genomics and Multiscale Biology and Microscopy Core Facilities at the Icahn School of Medicine at Mount Sinai.
Conflict of interest: none declared.
Funding
This work was primarily supported by our anonymous philanthropic sponsor, whose generous gift made this study possible. K.C.M. and V.d’E. were supported by National Institutes of Health (NIH) grant T32HL007824. J.C.K. acknowledges research support from the National Institutes of Health (R01HL130423, R01HL135093). J.L.M.B. acknowledges research support from Astra-Zeneca, NIH R01HL125863, and Fondation Leducq. S.D. acknowledges research support from the Fondation Claude Pompidou. M.M. is a British Heart Foundation (BHF) Chair Holder (CH/16/3/32406) with BHF programme grant support (RG/16/14/32397). N.B.-N. is supported by a European Commission funding (ERC-2016-STG, 716628_ROSALIND).
Time for primary review: 7 days
Translational perspective
Fibromuscular dysplasia (FMD) is a poorly understood disease with no specific pharmacological therapies, which can cause stenosis, aneurysm, and dissection of medium-large arteries. At present, FMD is usually diagnosed by imaging studies, and screening for this disease can be challenging. We performed a ‘reverse-translational’ clinical study leveraging plasma and DNA samples from FMD patients and healthy matched controls to better understand this disease. We found that FMD patients exhibit a plasma proteogenomic signature that includes promising disease candidates. While further development will be required, our proof-of-concept analyses suggest that it may also be possible to develop a blood-based test for FMD.
References
- 1. Olin JW, Gornik HL, Bacharach JM, Biller J, Fine LJ, Gray BH, Gray WA, Gupta R, Hamburg NM, Katzen BT, Lookstein RA, Lumsden AB, Newburger JW, Rundek T, Sperati CJ, Stanley JC.. Fibromuscular dysplasia: state of the science and critical unanswered questions: a scientific statement from the American Heart Association. Circulation 2014;129:1048–1078. [DOI] [PubMed] [Google Scholar]
- 2. Kadian-Dodov D, Gornik HL, Gu X, Froehlich J, Bacharach JM, Chi Y-W, Gray BH, Jaff MR, Kim ESH, Mace P, Sharma A, Kline-Rogers E, White C, Olin JW.. Dissection and aneurysm in patients with fibromuscular dysplasia: findings from the U.S. registry for FMD. J Am Coll Cardiol 2016;68:176–185. [DOI] [PubMed] [Google Scholar]
- 3. Plouin P-F, Baguet J-P, Thony F, Ormezzano O, Azarine A, Silhol F, Oppenheim C, Bouhanick B, Boyer L, Persu A, Hammer F, Gosse P, Mounier-Vehier C, Le Hello C, Jeunemaitre X, Azizi M, Amar L, Chatellier G, Mousseaux E, Touzé E.. High prevalence of multiple arterial bed lesions in patients with fibromuscular dysplasia: the ARCADIA registry (Assessment of Renal and Cervical Artery Dysplasia). Hypertension 2017;70:652–658. [DOI] [PubMed] [Google Scholar]
- 4. Sethi SS, Lau JF, Godbold J, Gustavson S, Olin JW.. The S curve: a novel morphological finding in the internal carotid artery in patients with fibromuscular dysplasia. Vasc Med 2014;19:356–362. [DOI] [PubMed] [Google Scholar]
- 5. Slovut DP, Olin JW.. Fibromuscular dysplasia. N Engl J Med 2004;350:1862–1871. [DOI] [PubMed] [Google Scholar]
- 6. Gornik HL, Persu A, Adlam D, Aparicio LS, Azizi M, Boulanger M, Bruno RM, de Leeuw P, Fendrikova-Mahlay N, Froehlich J, Ganesh SK, Gray BH, Jamison C, Januszewicz A, Jeunemaitre X, Kadian-Dodov D, Kim ES, Kovacic JC, Mace P, Morganti A, Sharma A, Southerland AM, Touzé E, van der Niepen P, Wang J, Weinberg I, Wilson S, Olin JW, Plouin P-F.. First international consensus on the diagnosis and management of fibromuscular dysplasia. Vasc Med 2019;24:164–189. [DOI] [PubMed] [Google Scholar]
- 7. Olin JW, Froehlich J, Gu X, Bacharach JM, Eagle K, Gray BH, Jaff MR, Kim ESH, Mace P, Matsumoto AH, McBane RD, Kline-Rogers E, White CJ, Gornik HL.. The United States Registry for Fibromuscular Dysplasia: results in the first 447 patients. Circulation 2012;125:3182–3190. [DOI] [PubMed] [Google Scholar]
- 8. Guill CK, Benavides DC, Rees C, Fenves AZ, Burton EC.. Fatal mesenteric fibromuscular dysplasia: a case report and review of the literature. Arch Intern Med 2004;164:1148–1153. [DOI] [PubMed] [Google Scholar]
- 9. Michelis KC, Olin JW, Kadian-Dodov D, d'Escamard V, Kovacic JC.. Coronary artery manifestations of fibromuscular dysplasia. J Am Coll Cardiol 2014;64:1033–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Savard S, Steichen O, Azarine A, Azizi M, Jeunemaitre X, Plouin PF.. Association between 2 angiographic subtypes of renal artery fibromuscular dysplasia and clinical characteristics. Circulation 2012;126:3062–3069. [DOI] [PubMed] [Google Scholar]
- 11. Plouin PF, Perdu J, La Batide-Alanore A, Boutouyrie P, Gimenez-Roqueplo AP, Jeunemaitre X.. Fibromuscular dysplasia. Orphanet J Rare Dis 2007;2:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hendricks NJ, Matsumoto AH, Angle JF, Baheti A, Sabri SS, Park AW, Stone JR, Patrie JT, Dworkin L, Cooper CJ, Murphy TP, Cutlip DE.. Is fibromuscular dysplasia underdiagnosed? A comparison of the prevalence of FMD seen in CORAL trial participants versus a single institution population of renal donor candidates. Vasc Med 2014;19:363–367. [DOI] [PubMed] [Google Scholar]
- 13. Kiando SR, Tucker NR, Castro-Vega L-J, Katz A, D’Escamard V, Tréard C, Fraher D, Albuisson J, Kadian-Dodov D, Ye Z, Austin E, Yang M-L, Hunker K, Barlassina C, Cusi D, Galan P, Empana J-P, Jouven X, Gimenez-Roqueplo A-P, Bruneval P, Hyun Kim ES, Olin JW, Gornik HL, Azizi M, Plouin P-F, Ellinor PT, Kullo IJ, Milan DJ, Ganesh SK, Boutouyrie P, Kovacic JC, Jeunemaitre X, Bouatia-Naji N.. PHACTR1 is a genetic susceptibility locus for fibromuscular dysplasia supporting its complex genetic pattern of inheritance. PLoS Genet 2016;12:e1006367.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ngo D, Sinha S, Shen D, Kuhn EW, Keyes MJ, Shi X, Benson MD, O'Sullivan JF, Keshishian H, Farrell LA, Fifer MA, Vasan RS, Sabatine MS, Larson MG, Carr SA, Wang TJ, Gerszten RE.. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation 2016;134:270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benson MD, Yang Q, Ngo D, Zhu Y, Shen D, Farrell LA, Sinha S, Keyes MJ, Vasan RS, Larson MG, Smith JG, Wang TJ, Gerszten RE.. Genetic architecture of the cardiovascular risk proteome. Circulation 2018;137:1158–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Folkersen L, Fauman E, Sabater-Lleal M, Strawbridge RJ, Frånberg M, Sennblad B, Baldassarre D, Veglia F, Humphries SE, Rauramaa R, de Faire U, Smit AJ, Giral P, Kurl S, Mannarino E, Enroth S, Johansson Å, Enroth SB, Gustafsson S, Lind L, Lindgren C, Morris AP, Giedraitis V, Silveira A, Franco-Cereceda A, Tremoli E, Gyllensten U, Ingelsson E, Brunak S, Eriksson P, Ziemek D, Hamsten A, Mälarstig A.. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet 2017;13:e1006706.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Di Narzo AF, Telesco SE, Brodmerkel C, Argmann C, Peters LA, Li K, Kidd B, Dudley J, Cho J, Schadt EE, Kasarskis A, Dobrin R, Hao K.. High-throughput characterization of blood serum proteomics of IBD patients with respect to aging and genetic factors. PLoS Genet 2017;13:e1006565.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Assarsson E, Lundberg M, Holmquist G, Björkesten J, Bucht Thorsen S, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, Andersson A-C, Lindstedt P, Stenvang J, Gullberg M, Fredriksson S.. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014;9:e95192.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lind L, Arnlov J, Lindahl B, Siegbahn A, Sundstrom J, Ingelsson E.. Use of a proximity extension assay proteomics chip to discover new biomarkers for human atherosclerosis. Atherosclerosis 2015;242:205–210. [DOI] [PubMed] [Google Scholar]
- 20. Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S.. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res 2011;39:e102.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soininen P, Kangas AJ, Wurtz P, Suna T, Ala-Korpela M.. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet 2015;8:192–206. [DOI] [PubMed] [Google Scholar]
- 22. Soininen P, Kangas AJ, Würtz P, Tukiainen T, Tynkkynen T, Laatikainen R, Järvelin M-R, Kähönen M, Lehtimäki T, Viikari J, Raitakari OT, Savolainen MJ, Ala-Korpela M.. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst 2009;134:1781–1785. [DOI] [PubMed] [Google Scholar]
- 23. Fischer K, Kettunen J, Würtz P, Haller T, Havulinna AS, Kangas AJ, Soininen P, Esko T, Tammesoo M-L, Mägi R, Smit S, Palotie A, Ripatti S, Salomaa V, Ala-Korpela M, Perola M, Metspalu A.. Biomarker profiling by nuclear magnetic resonance spectroscopy for the prediction of all-cause mortality: an observational study of 17,345 persons. PLoS Med 2014;11:e1001606.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kettunen J, Demirkan A, Würtz P, Draisma HHM, Haller T, Rawal R, Vaarhorst A, Kangas AJ, Lyytikäinen L-P, Pirinen M, Pool R, Sarin A-P, Soininen P, Tukiainen T, Wang Q, Tiainen M, Tynkkynen T, Amin N, Zeller T, Beekman M, Deelen J, van Dijk KW, Esko T, Hottenga J-J, van Leeuwen EM, Lehtimäki T, Mihailov E, Rose RJ, de Craen AJM, Gieger C, Kähönen M, Perola M, Blankenberg S, Savolainen MJ, Verhoeven A, Viikari J, Willemsen G, Boomsma DI, van Duijn CM, Eriksson J, Jula A, Järvelin M-R, Kaprio J, Metspalu A, Raitakari O, Salomaa V, Slagboom PE, Waldenberger M, Ripatti S, Ala-Korpela M.. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun 2016;7:11122.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Q, Ferreira DLS, Nelson SM, Sattar N, Ala-Korpela M, Lawlor DA.. Metabolic characterization of menopause: cross-sectional and longitudinal evidence. BMC Med 2018;16:17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wurtz P, Havulinna AS, Soininen P, et al. Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation 2015;131:774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wurtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M.. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -omic technologies. Am J Epidemiol 2017;186:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Würtz P, Wang Q, Soininen P, Kangas AJ, Fatemifar G, Tynkkynen T, Tiainen M, Perola M, Tillin T, Hughes AD, Mäntyselkä P, Kähönen M, Lehtimäki T, Sattar N, Hingorani AD, Casas J-P, Salomaa V, Kivimäki M, Järvelin M-R, Davey Smith G, Vanhala M, Lawlor DA, Raitakari OT, Chaturvedi N, Kettunen J, Ala-Korpela M.. Metabolomic profiling of statin use and genetic inhibition of HMG-CoA reductase. J Am Coll Cardiol 2016;67:1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hastie T, Tibshirani R, Narasimhan B, Chu G. impute: Imputation for Microarray Data. R Package Version 1.54.0. 2018.
- 30. Friedman J, Hastie T, Tibshirani R.. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 31. Zou H, Hastie T.. Regularization and variable selection via the elastic net. J R Statist Soc B 2005;67(Pt. 2):301–320. [Google Scholar]
- 32. Caruana R, Karampatziakis N, Yessenalina A. Proceedings of the 25th International Conference on Machine Learning, Helsinki, Finland, 2008.
- 33. Cochran JN, Rush T, Buckingham SC, Roberson ED.. The Alzheimer's disease risk factor CD2AP maintains blood-brain barrier integrity. Hum Mol Genet 2015;24:6667–6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li C, Ruotsalainen V, Tryggvason K, Shaw AS, Miner JH.. CD2AP is expressed with nephrin in developing podocytes and is found widely in mature kidney and elsewhere. Am J Physiol Renal Physiol 2000;279:F785–792. [DOI] [PubMed] [Google Scholar]
- 35. Gupta RM, Hadaya J, Trehan A, Zekavat SM, Roselli C, Klarin D, Emdin CA, Hilvering CRE, Bianchi V, Mueller C, Khera AV, Ryan RJH, Engreitz JM, Issner R, Shoresh N, Epstein CB, de Laat W, Brown JD, Schnabel RB, Bernstein BE, Kathiresan S.. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell 2017;170:522–533.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCormack L, Harard J, Poutasse E.. Obstructive lesions of the renal artery associated with remediable hypertension. Am J Pathol 1958;34:582. [Google Scholar]
- 37. McCormack LJ, Poutasse EF, Meaney TF, Noto TJ Jr, Dustan HP.. A pathologic-arteriographic correlation of renal arterial disease. Am Heart J 1966;72:188–198. [DOI] [PubMed] [Google Scholar]
- 38. Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O'Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P.. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013;45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ERLC, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Rüther E, Schürmann B, Heun R, Kölsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Gallacher J, Hüll M, Rujescu D, Giegling I, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MMB, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues J-F, Tzourio C, Alpérovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Björnsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossù P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O'Donovan M, Amouyel P, Williams J.. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011;43:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin L-W, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD.. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet 2011;43:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang VW, Brieher WM.. FSGS3/CD2AP is a barbed-end capping protein that stabilizes actin and strengthens adherens junctions. J Cell Biol 2013;203:815–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shih NY, Li J, Karpitskii V, Nguyen A, Dustin ML, Kanagawa O, Miner JH, Shaw AS.. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science 1999;286:312–315. [DOI] [PubMed] [Google Scholar]
- 43. Raju S, Kometani K, Kurosaki T, Shaw AS, Egawa T.. The adaptor molecule CD2AP in CD4 T cells modulates differentiation of follicular helper T cells during chronic LCMV infection. PLoS Pathog 2018;14:e1007053.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fieger CB, Sassetti CM, Rosen SD.. Endoglycan, a member of the CD34 family, functions as an L-selectin ligand through modification with tyrosine sulfation and sialyl Lewis x. J Biol Chem 2003;278:27390–27398. [DOI] [PubMed] [Google Scholar]
- 45. Debette S, Metso T, Pezzini A, Abboud S, Metso A, Leys D, Bersano A, Louillet F, Caso V, Lamy C, Medeiros E, Samson Y, Grond-Ginsbach C, Engelter ST, Thijs V, Beretta S, Béjot Y, Sessa M, Lorenza Muiesan M, Amouyel P, Castellano M, Arveiler D, Tatlisumak T, Dallongeville J.. Association of vascular risk factors with cervical artery dissection and ischemic stroke in young adults. Circulation 2011;123:1537–1544. [DOI] [PubMed] [Google Scholar]
- 46. Yu JE, Han SY, Wolfson B, Zhou Q.. The role of endothelial lipase in lipid metabolism, inflammation, and cancer. Histol Histopathol 2018;33:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McCoy MG, Sun GS, Marchadier D, Maugeais C, Glick JM, Rader DJ.. Characterization of the lipolytic activity of endothelial lipase. J Lipid Res 2002;43:921–929. [PubMed] [Google Scholar]
- 48. Olin JW, Kadian-Dodov D.. Fibromuscular dysplasia: looking beyond the “string of beads”. JACC Cardiovasc Imaging 2017;10:562–564. [DOI] [PubMed] [Google Scholar]
- 49. Bolen MA, Brinza E, Renapurkar RD, Kim ESH, Gornik HL.. Screening CT angiography of the aorta, visceral branch vessels, and pelvic arteries in fibromuscular dysplasia. JACC Cardiovasc Imaging 2017;10:554–561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.