Abstract
Aims
Syncope is a common condition associated with frequent hospitalization or visits to the emergency department. Family aggregation and twin studies have shown that syncope has a heritable component. We investigated whether common genetic variants predispose to syncope and collapse.
Methods and results
We used genome-wide association data on syncope on 408 961 individuals with European ancestry from the UK Biobank study. In a replication study, we used the Integrative Psychiatric Research Consortium (iPSYCH) cohort (n = 86 189), to investigate the risk of incident syncope stratified by genotype carrier status. We report on a genome-wide significant locus located on chromosome 2q32.1 [odds ratio = 1.13, 95% confidence interval (CI) 1.10–1.17, P = 5.8 × 10−15], with lead single nucleotide polymorphism rs12465214 in proximity to the gene zinc finger protein 804a (ZNF804A). This association was also shown in the iPSYCH cohort, where homozygous carriers of the C allele conferred an increased hazard ratio (1.30, 95% CI 1.15–1.46, P = 1.68 × 10−5) of incident syncope. Quantitative polymerase chain reaction analysis showed ZNF804A to be expressed most abundantly in brain tissue.
Conclusion
We identified a genome-wide significant locus (rs12465214) associated with syncope and collapse. The association was replicated in an independent cohort. This is the first genome-wide association study to associate a locus with syncope and collapse.
Keywords: Syncope, Genetic, Genome-wide association study
1. Introduction
Syncope is defined as a transient loss of consciousness due to short-term global cerebral hypoperfusion.1 It is characterized by sudden onset, short duration, and spontaneous recovery.1,2 Syncope is a common condition in the general population and accounts for approximately 1% of all visits to European emergency departments.3 It occurs more frequently in women than men, with the estimated incidence rates ranging from 2.6 to 19.5 per 1000 person-years. Highest incidence rate is seen in women aged >80.4,5 Syncope can be classified into neurally-mediated reflex syncope (neurocardiogenic), syncope due to orthostatic hypotension, and cardiac syncope. Reflex syncope (vasovagal syncope; VVS), which is a subtype of neurally-mediated reflex syncope, is the most frequent type of syncope.4 The overall mortality in relation to syncope and collapse has been reported to be 0.28% between 2000 and 2005 in the USA, and the highest risk of mortality has been observed for cardiac syncope.6
The predisposing factors for syncope comprise of a broad spectrum ranging from life-threating diseases, as well as side effects to pharmacological agents, to benign causes.2,3 Syncope occurs frequently in patients with underlying cardiac disease, in particular in genetic cardiac arrhythmias, e.g. long and short QT syndromes,7 Brugada syndrome,8 and catecholaminergic polymorph ventricular tachycardia.9 In the latter, syncope may be the only presenting symptom. Improved understanding of the pathophysiological mechanisms is therefore warranted. A recent Danish study on symptoms preceding sudden death in the young, revealed that syncope had occurred 24 h preceding death in 17% of victims.10 Despite the high use of diagnostic tests, the diagnostic yield remains low (30–48%).11
Family aggregation studies of VVS have predominantly suggested complex inheritance,12 and twin studies have shown higher concordance rates for VVS in monozygotic twin pairs compared with dizygotic twin pairs.13
Previous studies, that have associated genetic loci to syncope,14–16 have suffered from small sample sizes (n ∼ 50–150) and have failed to replicate. A genome-wide copy number variations (CNV) study of VVS, showed that CNV segments were longer in affected individuals compared with unaffected individuals.17
The UK Biobank project has extensive phenotypic and genotypic data on more than 500 000 participants from the general population, which enables researchers to study common traits. In the present study, we used UK Biobank metadata to examine the genetic basis underlying syncope and collapse.
2. Methods
2.1 Study population, UK Biobank
UK Biobank is a large, population-based, prospective cohort study with more than 500 000 participants aged between 40 and 69 years at time of recruitment (2006–2010). The biobank holds a variety of health-related information on participants, including health records, imaging, lifestyle indicators, cognitive function, biomarkers from blood and urine, and more.18 Imputed genome-wide genotype data are available for all participants, including information on population structure, relatedness, and genotype level quality. A full description on analyses of genotype data and details on quality control are provided in the UK Biobank release paper by Bycroft et al.19 In brief, several tests for marker-based quality control (QC) were performed including tests for batch effects, plate effects, deviations from Hardy–Weinberg equilibrium (HWE; P < 10−6), sex effects, array effects, and discordance across control replicates. Thereafter, sample-based QC was performed, including tests for sex mismatch and extreme heterozygosity and high missing rates (>0.05). Pre-imputation, principal component analysis (PCA) was performed in both marker and sample-based QC. In the UK Biobank pre-imputation, there were 812 428 markers before QC, and 805 426 markers left after QC. A subset of ethnically matched samples with British ancestry was identified, based on PCA.19 In this subset, syncope was defined using the International Classification of Disease (ICD)-9 code 780.2 and the ICD-10 code R55. Cases were not excluded due to other concurrent diseases.
2.2 Replication cohort, Integrative Psychiatric Research Consortium
Since 1981, all new-born babies in Denmark have had dried blood spot samples collected for the Danish Neonatal Screening Biobank (DNSB) at Statens Serum Institute (SSI). The Integrative Psychiatric Research Consortium (iPSYCH) is a population-based study selected from a birth cohort comprised of all singleton individuals born in between the years 1981 and 2005 (n = 1 472 762). In total, 86 189 individuals have been selected. Hereof, 30 000 individuals were randomly selected to represent the general population and 57 764 individuals were diagnosed with at least one of six major psychiatric diagnoses (attention deficit/hyperactivity disorder, anorexia nervosa, autism spectrum disorder, affective disorder, bipolar affective disorder, or schizophrenia). Psychiatric diseases were identified using their unique civil registration number, which enables linkage to the National Patient Registry and Psychiatric Central Research Registry in which diagnoses are coded according to the ICD 10th revision (ICD-10). Diagnoses prior to 1994 were coded according to the ICD 8th revision, and converted to equivalent ICD-10 codes.
Dried blood spots were obtained from the DNSB for genotyping of the selected patients (n = 86 189), as described previously by Pedersen et al.20 Genotyping was performed at the Broad Institute on the Infinium PsychChip v1.0 array. DNA processing, genotyping, including imputation, was successful on 77 639 samples.
Single nucleotide polymorphisms (SNPs; n = 246 369) were phased into haplotypes using SHAPEIT3 and imputed using Impute 2 with reference haplotypes from the 1000 Genomes Project phase 3, generating a total of 8 018 013 imputed variants. As a QC measure, variants with imputation quality (INFO <0.2), deviations from HWE (P < 1 × 10−6), association with imputation batch (P < 5 × 10−8), differing imputation quality between cases and controls (P < 1 × 10−6), and minor allele frequencies (MAFs) <0.01 were excluded. On a sample level, controls were made for abnormal sample heterozygosity, high levels of missing genotypes (>1%), sex concordance, inconsistencies among duplicate samples and sample relatedness. PCA and KING robust algorithm was used to identify a subset of unrelated ethnically matched individuals, as described previously by Schork et al.21 Syncope and collapse was defined by ICD-8 code 782.5 or ICD-10 code R55.
2.3 The PheWeb browser
The Michigan PheWeb browser22 is a publicly available resource generated from phenome-wide associations using UK Biobank that features 1403 in-house defined binary traits (UK Biobank application number 24460). Only variants with MAF >0.01 and info score >0.3 were used in the PheWeb browser.23
2.4 Estimation of statistical power
With an estimated disease prevalence of 0.01, ∼9000 cases and 100 000 controls in the study cohort, odds ratio (OR) of 2, and a significant P-value defined as less than 0.05/1403 (traits in PheWeb), a disease allele frequency above 0.01 is needed in order to obtain a power of 0.9. Based on this, variants with a MAF <0.01 were excluded.
2.5 Statistical analyses
In the UK Biobank cohort, a logistic mixed model was implemented in the framework of the Scalable and Accurate Implementation and Generalized mixed model (SAIGE) method.23 This allows for having related individuals in the model. SAIGE also controls for inflated Type 1 errors which otherwise can arise with imbalanced case–control ratios in logistic mixed models. SAIGE first generate relatedness estimates and a null model. Thereafter, the null model is fitted with the respective SNP, the genetic kinship matrix and adjustment for age at recruitment, sex, array batch, and principal components 1–4.
In the iPSYCH cohort subset, the underlying time scale was age. Follow-up began at age one and all individuals were followed to the event of syncope, death, emigration, or 30 April 2017, which ever occurred first. The association between genotype and risk of syncope was assessed by a Cox regression analysis adjusted for sex. Absolute risk of syncope and collapse was predicted from the Cox regression model with death as a competing risk. The absolute risk is illustrated as cumulative incidence stratified by genotype and sex. We found no violation on proportional hazard model assumptions (Supplementary Information). We also replicated significant associations using logistic regression adjusted for age, sex, and the ten first principal components.
2.6 Partitioning of the heritability
Partitioned linkage disequilibrium score regression (LDSC) was used to estimate heritability attributable to cell-type groups and H3K4Me1 cell-type annotations, as described by Finucane et al.24 The major histocompatibility region (MHC-region; chr6:25-35Mb) was excluded from analysis. We used previously computed linkage disequilibrium LD scores and allele frequencies based on data from the 1000 Genomes Project with European ancestry (see URLs).
Cell-types were grouped into: adrenal/pancreas, cardiovascular, central nervous system, connective bone, gastrointestinal, haematopoietic, kidney, liver, skeletal muscle, and other. Annotations from H3K4Me1 imputed gapped peaks were based on Roadmap Epigenomics Mapping Consortium data,25 as described by Demontis et al.26
The authors of LDSC (see URLs) state that mixed model summary statistics should not be used to estimate SNP-heritability but can be used for genetic correlation and partitioned heritability. This genome-wide association study (GWAS) is analysed with a mixed model regression. Therefore, we did not evaluate the intercept and SNP heritability.
2.7 Genetic correlations of syncope and collapse with other traits
We investigated genetic correlation (rg) of syncope and collapse with 20 other traits, using LDSC.27 Traits were selected based on availability of appropriate summary statistics, widespread prevalence, relevance for public health and if deemed to have a biological plausible correlation with syncope (Supplementary material online, Data S1). The MHC region and SNPs with MAF <0.05 were excluded from analyses and only phenotypes with a mean χ2 >1.02 were considered. Pre-computed LD scores for HapMap3 SNPs were used in the regression (see URLs). We applied Bonferroni correction and a P < 0.05/20 was considered significant.
2.8 Gene-set enrichment
We performed gene-set enrichment analyses using two independent methods, MAGMA v1.628 and MAGENTA v1.2.29 The aim was to verify enrichment in equivalent biological pathways using both methods. Only autosomal SNPs, excluding the MHC region, with a MAF >0.01 were included in the analyses. Gene-sets ranged from 10 to 1000 genes. For MAGMA, the 1000 Genomes Project Phase 3 was used as a reference panel and gene sets (n = 10 655) from MsigDB v5.2,30 with a gene window of 5 kb at each end of the gene intervals. Default settings were used for analyses using MAGENTA with a combined gene set (GO, Panther, Ingenuity, KEGG, Reactome, Biocarta; n = 10 992).
2.9 Biological and functional annotation
Functional mapping and annotation were made using FUMA v1.3.31 Genome-wide significant SNPs with the lowest P and an r2 < 0.6 from each other were defined as distinct significant SNPs and were used to define the border of the genomic risk locus.31 Lead SNPs were defined by the same conditions but with r2 < 0.1. Pairwise LD structure of SNPs was based on the 1000 Genomes Project Phase 3.32 Variant annotation for each locus was made on distinct significant SNPs and SNPs in LD with these, with a maximum distance of 250 kb and P < 0.05, referred to as candidate SNPs. Gene mapping of annotated SNPs was based on genomic position (max distance 10 kb), expression quantitative trait loci (eQTL) and chromatin interaction studies. The eQTLs were identified from GTEx v.7 with a false discovery rate (FDR) <0.05. Annotated SNPs were assigned chromatin interaction states (FDR threshold <10−6, 500 bp promoter region window) based on the most common of 15-state chromHMM across 127 cell types from Roadmap epigenomics data.25
2.10 Cross-trait association analysis
To assess pleiotropic effects of identified SNPs, we conducted a phenome-wide association study (PheWAS) on 13 different traits (syncope included) relating to arrhythmias, cardiovascular disease, mental disorders, neurological, and metabolic diseases (see Supplementary material online, Data S2; see URLs). We looked at the association between distinct significant SNPs and the abovementioned traits. Next, we investigated pleiotropy of the top ten associated lead SNPs in each of these traits. P value threshold for lead SNPs was set to P < 1 × 10−6. Lead SNPs were identified through LD clumping SNPs in PLINK v1.933 with a r2 > 0.1 and a physical distance of 1500 kb. To avoid incidental identification of the same or nearby risk loci, we used a large threshold for physical distance when clumping.
2.11 Association of gene expression and risk of syncope
We used MetaXcan34 to investigate mediating effects of gene expression levels on syncope and collapse. MetaXcan performs gene-based association analyses of predicted gene expression and phenotype risk, using summary statistics and eQTL data. We used a precomputed transcriptome prediction model based on GTEx v.7 and the 1000 Genomes Project covariance (see URLs) and default software settings. MetaXcan results were complemented with an eQTL colocalization analyses, using Sherlock.35 Sherlock uses a Bayesian framework to match GWAS associations with gene eQTL signals. A significant overlap of a locus associated with a phenotype and the eQTL of a gene implies a likely functional role for that gene in the phenotype.
2.12 Quantitative polymerase chain reaction
In order to assess gene expression in tissues, we performed quantitative polymerase chain reaction (qPCR) experiments on 12- to 15-week-old male Wistar rats from Taconic (Ejby, Denmark), euthanized by cervical dislocation. Experiments were performed in accordance with the European Union legislation for the protection of animals used for scientific purposes, and approved by the Danish National Animal Experiments Inspectorate.
RNA extraction, reverse transcription and qPCR analysis were performed as described previously.36 In brief, RNA was isolated from cerebral arteries, brain, and heart using the RNeasy Micro Kit (Qiagen, Denmark). Reverse transcription was performed as per the manufacturer’s instructions with the Nanoscript 2 kit (Primerdesign, UK). qPCR was performed with 25 ng of cDNA per well and a primer concentration of 250 nM, using Precision-iC SYBR green mastermix (Primerdesign, UK) with the CFX96 Real-Time PCR Detection System (Bio-Rad, Denmark). The most stable reference genes were determined by geNorm analysis (Biogazelle qbase+) and were found to be actin beta, topoisomerase I, and calnexin. The mean Cq values of these reference genes were used to determine the relative abundance of the zinc finger protein 804A gene (ZNF804A). Primers for ZNF804A were designed in-house and synthesized by Tag Copenhagen A/S (Denmark) with the following sequences: sense—5′-CGCTGCCCATGTTGTATCTA-3′; anti-sense—5’CCCACGATGATACCGACATAAC-3′. The following cycling conditions were used: initial activation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min, and data were collected during each cycling phase. Melt curve analysis, to ensure each primer set amplified a single, specific product, completed the protocol.
3. Results
3.1 Genome-wide significantly associated syncope risk locus
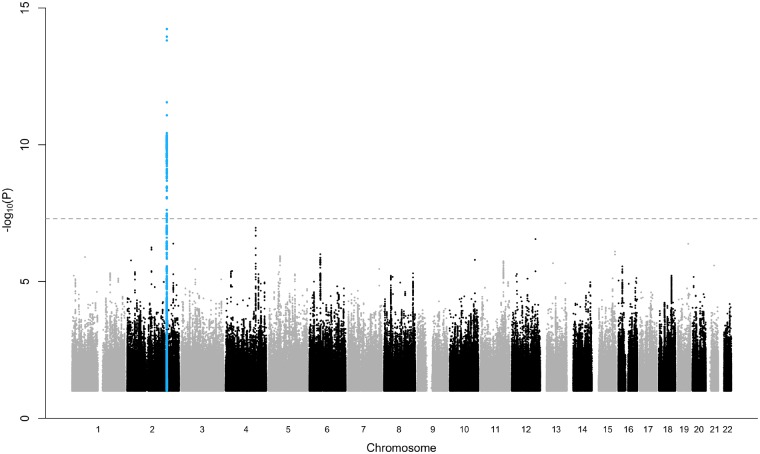
The UK Biobank subset had 408 961 ethnically matched individuals, with 9163 cases and 399 798 controls. Using logistic mixed model regression, related individuals kinship coeffcients, principal components, sex and age at recruitment were included as covariates (genomic inflation factor λ = 1.07; Figure 1, Supplementary material online, Figure S1). We identified one novel genome-wide significant locus at chromosome 2q32.1 [lead SNP rs12465214, OR = 1.13 for the risk allele C, 95% confidence interval (CI) 1.10–1.17, P = 5.8 × 10−15]. In total, we identified five distinct significant SNPs residing in the same locus (rs12465214, rs7593266, rs17582219, rs12621296, rs2219224; Table 1, Supplementary material online, Figure S2).
Figure 1.
Manhattan plot. Manhattan plot showing the association of single nucleotide polymorphisms (SNPs) with syncope and collapse in a genome-wide association study of 9163 cases and 399 798 controls. The grey dashed horizontal line marks the threshold for genome-wide significance (P = 5 × 10−8). Statistical significance of SNPs is shown on the y-axis as −log10(P) as a function of chromosomal position (x-axis). P-values are calibrated from score test with saddlepoint approximation.
Table 1.
SNPs in identified risk locus
| rsID | Chr | Position | Ref | Alt | Gnomad MAF (EUR) | P-value | nCand. SNPs |
|---|---|---|---|---|---|---|---|
| rs12465214 | 2 | 185198135 | C | A | 0.45 | 5.78E−15 | 160 |
| rs2219224 | 2 | 185211739 | G | A | 0.52 | 1.03E−09 | 41 |
| rs17582219 | 2 | 184983620 | A | G | 0.34 | 3.70E−09 | 40 |
| rs7593266 | 2 | 184965195 | T | G | 0.45 | 2.40E−08 | 149 |
| rs12621296 | 2 | 185099836 | A | G | 0.62 | 3.27E−08 | 131 |
Chr, chromosome; EUR, European (non-Finnish); MAF, minor allele frequency; nCand. SNPs, the number of unique candidate SNPs in the genomic locus.
3.2 Replication of GWAS locus
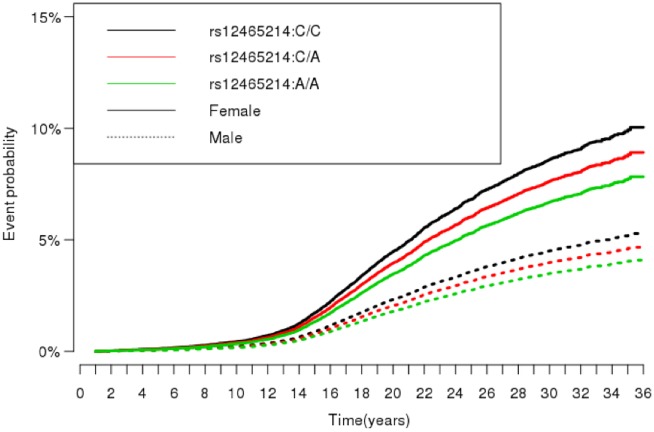
In the iPSYCH subset, we analysed 54 656 individuals. In total, 2352 syncope events occurred during a median follow-up of 24.86 years (interquartile range 19.62–29.97). The median age at syncope onset was 18.2 years. There were 51 929 controls. Sample overview can be found in Table 2. For rs12465214, we found an increased hazard ratio (HR) of syncope and collapse for the reference group (CC) compared with homozygous carriers (AA, HR = 1.30, 95% CI 1.15–1.46, P < 0.001), and compared with heterozygous carriers (CA, HR = 1.15, 95% CI 1.03–1.28, P = 0.02) (Figure 2). Women had an almost doubled hazard rate for syncope compared with men (HR = 1.93, 95% CI = 1.77–2.10, P < 0.001) (Figure 2).
Table 2.
Baseline characteristics
| Syncope event | No syncope event | |
|---|---|---|
| Female | 1564 | 24 006 |
| Male | 788 | 27 923 |
| Median age (IQR) | 27.8 (8.1) | 24.7 (10.4) |
Baseline characteristics of the iPSYCH cohort at the end of follow-up, without the individuals who died during follow-up.
iPSYCH, the Integrative Psychiatric Research Consortium; IQR, interquartile range; SD, standard deviation.
Figure 2.
Cox regression model. Cumulative incidence of syncope and collapse, stratified by genotype and gender. In total, 2352 syncope and collapse events occurred in 54 656 individuals. Estimates were based on a Cox regression model taking the competing risk of death into account.
With logistic regression, the effect size of the lead SNP was comparable to the main finding in UK Biobank (risk allele C, OR = 1.13, 95% CI 1.07–1.18, P = 8.82 × 10−6).
3.3 Genetic correlation with other traits
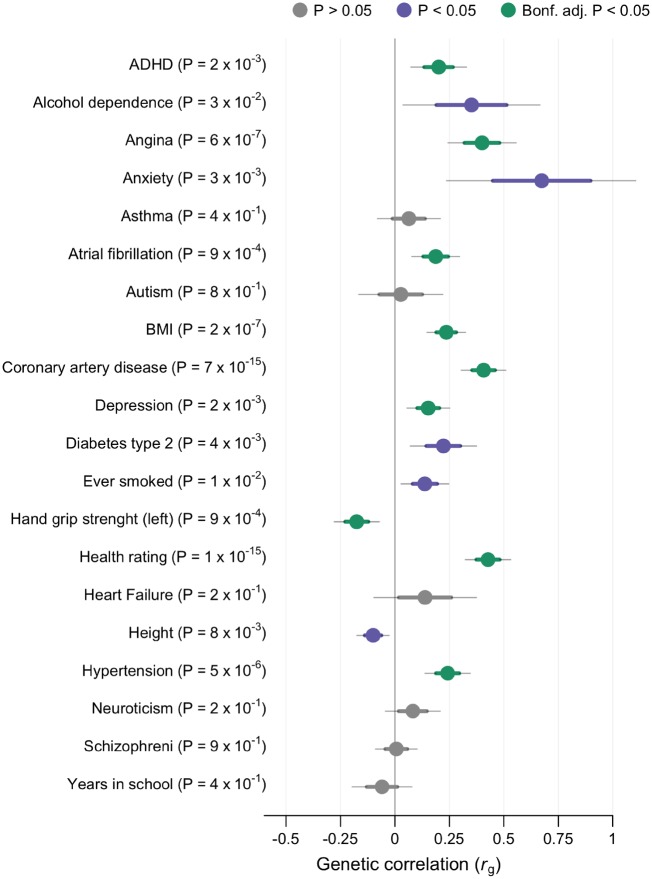
We found significant genetic correlation (rg) with nine of 20 phenotypes, and an additional five reached nominal significance (Figure 3, Supplementary material online, Data S3). Overall health rating (higher rating indicates poorer health) was the most correlated phenotype (rg = 0.43, P = 9.63 × 10−16). This was supported by a negative correlation with hand grip strength (rg = −0.18, P = 8.75 × 10−4), a proxy for overall health status.37 The genetic overlap with coronary artery disease (rg = 0.41, P = 6.99 × 10−15) was substantiated by the significant correlation with related traits such as angina, hypertension, body mass index, and atrial fibrillation (Figure 3). Anxiety was highly correlated with syncope, but with a broad mean standard error (SE) and the correlation did not reach the Bonferroni threshold (rg = 0.67, P = 2.53 × 10−3). Anxiety had a mean χ2 of 1.0298, which indicates a low polygenetic signal. The rg to anxiety is therefore rather unreliable. Two other psychiatric phenotypes, attention deficit hyperactivity disorder (rg = 0.20, P = 2.16 × 10−3) and depression (rg = 0.15, P = 2.32 × 10−3), demonstrated a significant genetic overlap with syncope. Height demonstrated a small negative correlation with syncope (rg = −0.10, P = 8.03 × 10−3). However, this association was not significant after Bonferroni adjustment.
Figure 3.
Genetic correlation. LD score regression revealing genetic correlation with syncope and collapse (9163 cases and 399 798 controls) and other phenotypes. Sample sizes of external genome-wide association studies are shown in Supplementary material online, Data S1. Phenotypes with negative log10(P) are displayed on the y-axis. X-axis show genetic correlation (rg). Dots are estimated values with thick lines indicating mean standard error (SE) and thin lines 1.96 × SE. Significant association after Bonferroni corrections is denoted with green colour. Nominal significance is denoted with violet colour and non-significant correlations with grey. Standard errors and P-values were derived from using block jackknife resampling.
3.4 Cell-type specific analyses
Annotations for ten cell types and H3K4Me1 cell-type annotations from Roadmap were used to estimate partitioned heritability of syncope and collapse into functional categories, as described by Finucane et al.24 We found a significant enrichment of SNPs located in adrenal and pancreas regulatory elements (enrichment = 7.03, SE = 2.12, P = 1 × 10−3; Supplementary material online, Figure S4, Data S4). Whereas partitioned heritability analyses based on imputed gapped peaks from H3K4Me1 did not reveal any significant H3K4Me1 cell-type annotations (Supplementary material online, Figure S5, Data S5).
3.5 Functional annotation
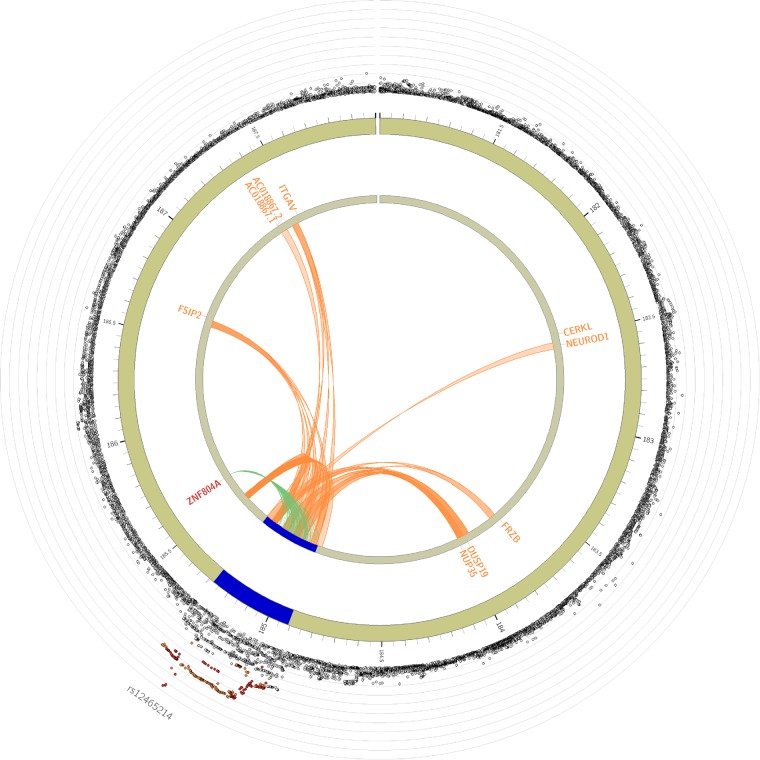
Based on the lead SNP and distinct significant SNPs in the risk locus, 244 candidate SNPs were identified and annotated (Supplementary material online, Data S6). Five SNPs (rs7349369, rs716544, rs12473165, rs12478223, rs12468883), in LD with the distinct significant SNP rs2219224 were naively mapped by position (genomic distance <10 kb) to MIR548AE1. In GTEx v.7, 134 candidate SNPs were associated with gene expression (eQTL; FDR >10−3; Figure 4, Supplementary material online, Figure S2, Data S7). The eQTLs were predominantly mapped in thyroid tissue to the nearby gene ZNF804A, with the strongest association for rs67219359 (P = 4 × 10−8, r2 with rs12465214 = 0.77). The risk locus had several significant chromatin interactions observed through Hi-C data (n = 1588; FDR <10−6), with 210 chromatin interactions mapping to regional genes (Figure 4, Supplementary material online, Data S8). In total, 26 genes mapped to the risk locus through position, eQTL and chromatin interactions (Supplementary material online, Data S9). Ten genes were protein coding, with the gene ZNF804A demonstrating strongest evidence for interaction with the risk locus.
Figure 4.
Circo plot displaying eQTL and chromatin interaction for the risk locus. The most outer layer shows a regional plot, displaying rsID of lead SNPs. The height of the y-axis indicates significance in negative log10 scale. Only SNPs with P < 0.05 are displayed. SNPs in genomic risk loci are colour-coded as a function of their maximum r2 to one of the distinct significant SNPs in the locus, as follows: red (r2 > 0.8), orange (r2 > 0.6), green (r2 > 0.4), and blue (r2 > 0.2). SNPs that are not in LD with any of the independent significant SNPs (with r2 ≤ 0.2) are grey. The second layer shows chromosome with genomic risk loci highlighted in blue. Only mapped genes by either chromatin interaction and/or eQTLs are displayed. If the gene and links are mapped only by chromatin interactions or only by eQTLs, it is coloured orange or green, respectively. When the gene is mapped by both it is coloured red.
Gene set enrichment analysis, with MAGMA and MAGENTA, did not reveal any significant gene sets.
3.6 Transcriptome-wide analyses
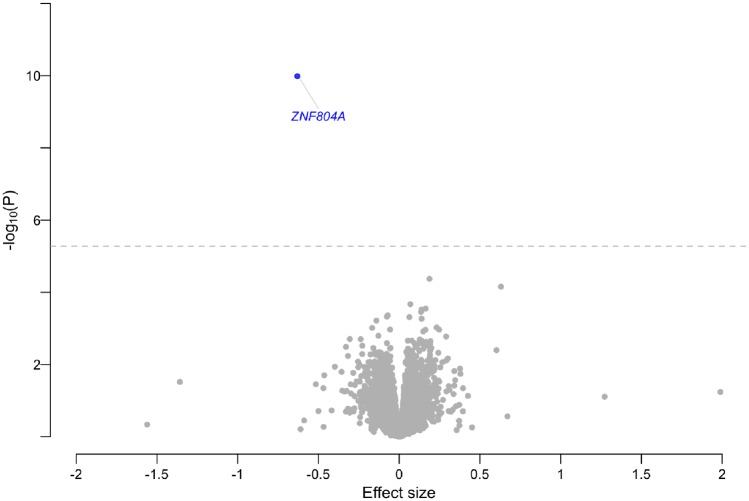
Since all but one out of the 134 candidate SNPs associated with eQTL had been identified in thyroid tissue, we used the GTEx v.7 thyroid tissue transcriptome model (n = 344; see URLs) for the MetaXcan analyses. The analyses revealed a significant association between the predicted gene expression of ZNF804A and syncope and collapse (effect size = −0.63, variance of gene expression = 0.012, P = 1.04 × 10−10; Figure 5, Supplementary material online, Data S10). The eQTL analysis with Sherlock revealed significant colocalization with the risk locus and ZNF804A (P = 1.92 × 10−5; Supplementary material online, Data S11).
Figure 5.
Volcano plot of transcriptome analysis. Plot showing results from MetaXcan transcriptome analyses based on GTEx v.7 thyroid tissue (n = 323) and syncope and collapse summary statistics (9163 cases and 399 798 controls). The effect size associated with increased or decreased expression with syncope risk is shown on the x-axis. The y-axis show −log10(P) for each tested gene. The dashed line indicates the Bonferroni threshold (P = 0.05/9504). Genes with a negative effect and significant association are demarked as blue dots. Non-significant genes are demarked as grey dots. Significance was assessed (P-values) with Wald tests.
3.7 Cross-trait associations
The PheWAS revealed a significant association between the lead SNP rs12465214 and hypertension (P = 2.12 × 10−3; Supplementary material online, Figure S6 and Data S12).
Top lead SNPs extracted from the other phenotypes did not reveal any cross-trait associations with syncope and collapse. However, with hierarchical clustering, syncope did cluster most closely to epilepsy, recurrent seizures and convulsions (Supplementary material online, Figure S7).
The gene ZNF804A has previously been associated with schizophrenia through GWAS. Therefore, we manually looked up the associations of rs12465214, rs7593266, rs17582219, rs12621296, and rs2219224 in a schizophrenia GWAS by Ripke et al.38 None of these SNPs had a nominal significant association (Supplementary material online, Data S13).
3.8 qPCR analysis
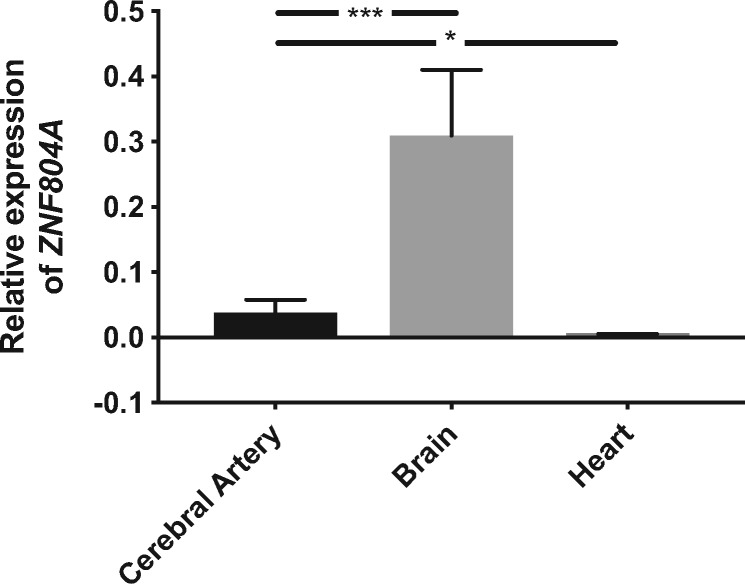
qPCR analysis revealed that ZNF804A was expressed most abundantly in brain tissue. Expression was also detected in the cerebral arteries, albeit at a lower level. The expression of ZNF804A was negligible in the heart (Figure 6).
Figure 6.
Gene expression analysis. Relative expression of ZNF804A assessed by qPCR analysis in tissue from cerebral arteries (n = 5), brain (n = 3), and heart (n = 3) from rats. Data represent the mean ± standard error of the mean (SEM). *P < 0.05, ***P < 0.001 according to unpaired t-tests.
4. Discussion
We report on the first discovery of a genome-wide significant risk locus for syncope and collapse. The lead SNP, rs12465214, residing at chromosome 2q31.1, is an intergenic variant approximately 250 kb downstream of the ZNF804A gene (Supplementary material online, Figure S2). Candidate SNPs in this locus map to 25 genes (ten protein coding) through either positional mapping, eQTLs, or chromatin interactions (Figure 4). This suggests a significant regulatory role of the risk locus on nearby genes. Transcriptome-wide analyses of eQTL suggest that the risk locus affects ZNF804A gene expression (Figure 5). ZNF804A is predominantly expressed in the brain, cerebral arteries, and endocrine tissue.39 We found no evidence of expression in heart tissue. ZNF804A gene expression in the human brain increases from embryonic to the early foetal stage and reaches a peak around the early mid-foetal stage. Eventually, the level of ZNF804A gene expression decreases and stays constant in adulthood.40 It is conceivable that abnormal blood pressure regulation or reduced cerebral perfusion are regulated at this locus, which could have an effect on the risk of syncope. However, further studies are warranted to assess such a relationship. Zinc finger protein (ZNF) genes have been associated with VVS previously. ZNF28, ZNF845, and ZNF146 genes were all significantly overexpressed in a group of children with VVS compared with a control group of healthy age-matched children.41
Partitioned heritability analyses revealed a significant enrichment of SNPs in regulatory elements of adrenal and pancreas tissue (Supplementary material online, Figure S4). This enrichment may be partly driven by the discovered risk locus, suggesting an involvement of the endocrine system in syncope pathogenesis.
The cross-trait analysis revealed an association between the lead SNP rs12465214 and hypertension (Supplementary material online, Figure S6). This suggests a potential functional link between rs12465214 and regulation of the circulatory system, which could affect the risk for syncope.
The association of rs12465214 was replicated in an independent cohort, where the SNP was significantly associated with incident syncope. We also found that women had an almost doubled HR for syncope compared with men, emphasizing the gender difference in syncope risk (Figure 2). Whether this difference is attributed to actual differences in genetic risk for syncope, or reflects sex-specific differences in healthcare-seeking behaviour is not known.
We observed a strong genetic overlap between syncope and self-reported overall health rating, and a negative correlation with hand grip strength, a proxy for health status.37 This supports the notion that syncope may be a precursor or a symptom of more severe disease. The genetic overlap with several cardiovascular diseases (CVD) reinforces this notion. Furthermore, the shared genetic component predisposing for these traits suggests similar cellular pathways to be involved in VVS and CVD.
Previously, rs1344706, in proximity to ZNF804A, has been associated with schizophrenia and bipolar disorder.42 The risk locus, rs1344706, is, however, not in LD with rs12465214 (r2 < 0.1). None of the five distinct significant SNPs that were listed in this study had a nominally significant association (P < 0.05) with schizophrenia in the GWAS by Ripke et al.38 We found no genetic correlation between syncope risk and schizophrenia (Figure 3). Alterations in the ZNF804A gene have been associated with altered functional connectivity in neurons. A knockdown of the gene in human neural stem cells or developing neurons leads to altered expression of genes related to cell adhesion, neurite outgrowth, synapse formation, synapse developing, and cytokine signalling.43 All of the mentioned functionalities have been implicated in schizophrenia.44 Also, ZNF804A has been shown to interact with the gene AXTN145 encoding the ataxin 1 protein, which has been associated with spinocerebellar ataxia characterized by Purkinje cell degeneration and loss of balance and co-ordination.46
Previous studies in relation to VVS have identified polymorphisms in the α1a-adrenergic receptor gene (ADRA1A),47 the β1-adrenergic receptor gene (ADRB1),14 the gene encoding the alpha subunit of the Gs protein (GNAS1),48 the adenosine A2a receptor gene (ADORA2A),15 and the endothelin 1 gene (EDN1).16 However, these studies were limited by small sample sizes and their results have not been replicated consistently. None of these genes were associated with syncope and collapse in the present study.
There are limitations to the present study that needs to be addressed. Firstly, phenotype definition was based on the ICD-10 code for ‘syncope and collapse’ which does not distinguish between subtypes of syncope. The use of the ICD-10 code R55 for syncope and collapse has previously been validated, with a positive predictive value of 95%.49
Secondly, drug induced prolongation of the electrocardiographic QT interval has previously been associated with increased risk of syncope and sudden unexplained death.50 Therefore, side effects of pharmaceutical agents contributing to the tendency to syncope cannot be excluded.
The ZNF804A gene and the associated pathways should be examined, both in deeply phenotyped syncope cases, arrhythmia cases, and cases with symptomatic drug interactions involving either syncope or arrhythmia.
5. Conclusions
We identified a genome-wide significant locus, rs12465214, associated with syncope and collapse (P = 5.8 × 10−15). This finding was replicated in an independent cohort. eQTL analysis pointed at the ZNF804A gene, which was found to be expressed in the brain and cerebral arteries of rats. Further genetic and functional studies are required to understand the molecular mechanism of the association between the SNP and syncope and collapse.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
URLs: Pheweb browser, http://pheweb.sph.umich.edu:5003/; Precomputed LD scores etc., https://data.broadinstitute.org/alkesgroup/LDSCORE/; LDSC, https://github.com/bulik/ldsc; MetaXcan transcriptome models, http://predictdb.org/; Phecodes, https://phewascatalog.org/phecodes.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource under the application number 24460.
Conflict of interest: none declared.
Funding
This work was supported by the John and Birthe Meyer Foundation, the Research Foundation of the Heart Centre, Rigshospitalet, the Research Council at Rigshospitalet, The Hallas-Møller Emerging Investigator Novo Nordisk (NNF17OC0031204), and the Arvid Nilsson Foundation. The iPSYCH study was funded by the Lundbeck Foundation Initiative for Integrative Psychiatric Research. This research has been conducted using the Danish National Biobank resource, supported by the Novo Nordisk Foundation.
Time for primary review: 53 days
Translational perspective
Our study adds new knowledge to the understanding of the pathophysiological mechanisms by which genetic variants associate with the risk of syncope and collapse. It serves to generate new hypotheses for future studies which potentially can lead to an improvement of risk stratification.
References
- 1. Brignole M, Moya A, de Lange FJ, Deharo JC, Elliott PM, Fanciulli A, Fedorowski A, Furlan R, Kenny RA, Martín A, Probst V, Reed MJ, Rice CP, Sutton R, Ungar A, van Dijk JG; ESC Scientific Document Group. 2018 ESC Guidelines for the diagnosis and management of syncope. Eur Heart J 2018;39:1883–1948. [DOI] [PubMed] [Google Scholar]
- 2. Kapoor WN. Syncope. N Engl J Med 2000;343:1856–1862. [DOI] [PubMed] [Google Scholar]
- 3. Moya A, Sutton R, Ammirati F, Blanc JJ, Brignole M, Dahm JB, Deharo JC, Gajek J, Gjesdal K, Krahn A, Massin M, Pepi M, Pezawas T, Ruiz Granell R, Sarasin F, Ungar A, van Dijk JG, Walma EP, Wieling W.. Guidelines for the diagnosis and management of syncope (version 2009). Eur Heart J 2009;30:2631–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soteriades ES, Evans JC, Larson MG, Chen MH, Chen L, Benjamin EJ, Levy D.. Incidence and prognosis of syncope. N Engl J Med 2002;347:878–885. [DOI] [PubMed] [Google Scholar]
- 5. Ganzeboom KS, Mairuhu G, Reitsma JB, Linzer M, Wieling W, van Dijk N.. Lifetime cumulative incidence of syncope in the general population: a study of 549 Dutch subjects aged 35-60 years. J Cardiovasc Electrophysiol 2006;17:1172–1176. [DOI] [PubMed] [Google Scholar]
- 6. Alshekhlee A, Shen W-K, Mackall J, Chelimsky TC.. Incidence and mortality rates of syncope in the United States. Am J Med 2009;122:181–188. [DOI] [PubMed] [Google Scholar]
- 7. Hedley PL, Jorgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M.. The genetic basis of long QT and short QT syndromes: a mutation update. Hum Mutat 2009;30:1486–1511. [DOI] [PubMed] [Google Scholar]
- 8. Hedley PL, Jørgensen P, Schlamowitz S, Moolman-Smook J, Kanters JK, Corfield VA, Christiansen M.. The genetic basis of Brugada syndrome: a mutation update. Hum Mutat 2009;30:1256–1266. [DOI] [PubMed] [Google Scholar]
- 9. Nyegaard M, Overgaard MT, Sondergaard MT, Vranas M, Behr ER, Hildebrandt LL, Lund J, Hedley PL, Camm AJ, Wettrell G, Fosdal I, Christiansen M, Borglum AD.. Mutations in calmodulin cause ventricular tachycardia and sudden cardiac death. Am J Hum Genet 2012;91:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glinge C, Jabbari R, Risgaard B, Lynge TH, Engstrom T, Albert CM, Haunso S, Winkel BG, Tfelt-Hansen J.. Symptoms before sudden arrhythmic death syndrome: a Nationwide Study Among the Young in Denmark. J Cardiovasc Electrophysiol 2015;26:761–767. [DOI] [PubMed] [Google Scholar]
- 11. Ruwald MH, Lock Hansen M, Lamberts M, Vinther M, Torp-Pedersen C, Hansen J, Gislason GH.. Unexplained syncope and diagnostic yield of tests in syncope according to the ICD-10 Discharge Diagnosis. J Clin Med Res 2013;5:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathias CJ, Deguchi K, Bleasdale-Barr K, Kimber JR.. Frequency of family history in vasovagal syncope. Lancet 1998;352:33–34. [DOI] [PubMed] [Google Scholar]
- 13. Klein KM, Xu SS, Lawrence K, Fischer A, Berkovic SF.. Evidence for genetic factors in vasovagal syncope: a twin-family study. Neurology 2012;79:561–565. [DOI] [PubMed] [Google Scholar]
- 14. Marquez MF, Hernandez-Pacheco G, Hermosillo AG, Gomez JR, Cardenas M, Vargas AG.. The Arg389Gly beta1-adrenergic receptor gene polymorphism and susceptibility to faint during head-up tilt test. Europace 2007;9:585–588. [DOI] [PubMed] [Google Scholar]
- 15. Saadjian AY, Gerolami V, Giorgi R, Mercier L, Berge-Lefranc J-L, Paganelli F, Ibrahim Z, By Y, Guéant JL, Lévy S, Guieu RP.. Head-up tilt induced syncope and adenosine A2A receptor gene polymorphism. Eur Heart J 2009;30:1510–1515. [DOI] [PubMed] [Google Scholar]
- 16. Sorrentino S, Forleo C, Iacoviello M, Guida P, D’Andria V, Favale S.. Endothelin system polymorphisms in tilt test-induced vasovagal syncope. Clin Auton Res 2009;19:347–354. [DOI] [PubMed] [Google Scholar]
- 17. Demir E, Hasdemir C, Ak H, Atay S, Aydin HH.. Genome-wide association study of copy number variations in patients with familial neurocardiogenic syncope. Biochem Genet 2016;54:487–494. [DOI] [PubMed] [Google Scholar]
- 18. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R.. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J.. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pedersen CB, Bybjerg-Grauholm J, Pedersen MG, Grove J, Agerbo E, Bækvad-Hansen M, Poulsen JB, Hansen CS, McGrath JJ, Als TD, Goldstein JI, Neale BM, Daly MJ, Hougaard DM, Mors O, Nordentoft M, Børglum AD, Werge T, Mortensen PB.. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry 2018;23:6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schork AJ, Won H, Appadurai V, Nudel R, Gandal M, Delaneau O, Hougaard D, Baekved-Hansen M, Bybjerg-Grauholm J, Pedersen MG, Pedersen CB, Neale BM, Daly MJ, Nordentoft M, Mors O, Boerglum AD, Mortensen PB, Buil A, Thompson WK, Geschwind D, Werge T.. A genome-wide association study for shared risk across major psychiatric disorders in a nation-wide birth cohort implicates fetal neurodevelopment as a key mediator. Nat Neurosci 2019;22:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.PheWeb. http://pheweb.sph.umich.edu:5003/ (5 January 2018, date last accessed).
- 23. Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, LeFaive J, VandeHaar P, Gagliano SA, Gifford A, Bastarache LA, Wei W-Q, Denny JC, Lin M, Hveem K, Kang HM, Abecasis GR, Willer CJ, Lee S.. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet 2018;50:1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, Ripke S, Day FR; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium, Purcell S, Stahl E, Lindstrom S, Perry JRB, Okada Y, Raychaudhuri S, Daly MJ, Patterson N, Neale BM, Price AL.. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015;47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M.. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Baldursson G, Belliveau R, Bybjerg-Grauholm J, Bækvad-Hansen M, Cerrato F, Chambert K, Churchhouse C, Dumont A, Eriksson N, Gandal M, Goldstein JI, Grasby KL, Grove J, Gudmundsson OO, Hansen CS, Hauberg ME, Hollegaard MV, Howrigan DP, Huang H, Maller JB, Martin AR, Martin NG, Moran J, Pallesen J, Palmer DS, Pedersen CB, Pedersen MG, Poterba T, Poulsen JB, Ripke S, Robinson EB, Satterstrom FK, Stefansson H, Stevens C, Turley P, Walters GB, Won H, Wright MJ, Andreassen OA, Asherson P, Burton CL, Boomsma DI, Cormand B, Dalsgaard S, Franke B, Gelernter J, Geschwind D, Hakonarson H, Haavik J, Kranzler HR, Kuntsi J, Langley K, Lesch K-P, Middeldorp C, Reif A, Rohde LA, Roussos P, Schachar R, Sklar P, Sonuga-Barke EJS, Sullivan PF, Thapar A, Tung JY, Waldman ID, Medland SE, Stefansson K, Nordentoft M, Hougaard DM, Werge T, Mors O, Mortensen PB, Daly MJ, Faraone SV, Børglum AD, Neale BM.. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 2019;51:63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J; Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL, Neale BM.. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Leeuw CA, Mooij JM, Heskes T, Posthuma D.. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015;11:e1004219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Segrè AV, Consortium D, Investigators M, Groop L, Mootha VK, Daly MJ, Altshuler D.. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 2010;6:e1001058.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watanabe K, Taskesen E, Bochoven A V, Posthuma D.. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ.. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 2015;4:7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barbeira AN, Dickinson SP, Torres JM, Bonazzola R, Zheng J, Torstenson ES, Wheeler HE, Shah KP, Edwards T, Garcia T, Consortium G, Nicolae D, Cox NJ, Im HK.. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 2018;9:1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, Li H.. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet 2013;92:667–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jepps TA, Bentzen BH, Stott JB, Povstyan OV, Sivaloganathan K, Dalby-Brown W, Greenwood IA.. Vasorelaxant effects of novel Kv 7.4 channel enhancers ML213 and NS15370. Br J Pharmacol 2014;171:4413–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Celis-Morales CA, Welsh P, Lyall DM, Steell L, Petermann F, Anderson J, Iliodromiti S, Sillars A, Graham N, Mackay DF, Pell JP, Gill JMR, Sattar N, Gray SR.. Associations of grip strength with cardiovascular, respiratory, and cancer outcomes and all cause mortality: prospective cohort study of half a million UK Biobank participants. BMJ 2018;361:k1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ripke S, Neale BM, Corvin A, Walters JT, Farh K-H, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, Pers TH, Agartz I, Agerbo E, Albus M, Alexander M, Amin F, Bacanu SA, Begemann M, Belliveau RA, Bene J, Bergen SE, Bevilacqua E, Bigdeli TB, Black DW, Bruggeman R, Buccola NG, Buckner RL, Byerley W, Cahn W, Cai G.. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tissue expression of ZNF804A—Summary—The Human Protein Atlas. https://www.proteinatlas.org/ENSG00000170396-ZNF804A/tissue (25 February 2019, date last accessed).
- 40. Zhou Y, Dong F, Lanz TA, Reinhart V, Li M, Liu L, Zou J, Xi HS, Mao Y.. Interactome analysis reveals ZNF804A, a schizophrenia risk gene, as a novel component of protein translational machinery critical for embryonic neurodevelopment. Mol Psychiatry 2018;23:952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang Y-J, Zhou Z, Xu M, Ma Q, Yan J, Wang J, Zhang Q, Huang M, Bao L.. Alteration of gene expression profiling including GPR174 and GNG2 is associated with vasovagal syncope. Pediatr Cardiol 2015;36:475–480. [DOI] [PubMed] [Google Scholar]
- 42. O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CCA, Howie B, Leung H-T, Hartmann AM, Moller H-J, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG.. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 2008;40:1053–1055. [DOI] [PubMed] [Google Scholar]
- 43. Chang H, Xiao X, Li M.. The schizophrenia risk gene ZNF804A: clinical associations, biological mechanisms and neuronal functions. Mol Psychiatry 2017;22:944–953. [DOI] [PubMed] [Google Scholar]
- 44. Ma C, Gu C, Huo Y, Li X, Luo X-J.. The integrated landscape of causal genes and pathways in schizophrenia. Transl Psychiatry 2018;8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A.. Human protein reference database—2009 update. Nucleic Acids Res 2009;37:D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lim J, Hao T, Shaw C, Patel AJ, Szabó G, Rual J-F, Fisk CJ, Li N, Smolyar A, Hill DE, Barabási A-L, Vidal M, Zoghbi HY.. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 2006;125:801–814. [DOI] [PubMed] [Google Scholar]
- 47. Hernandez-Pacheco G, Gonzalez-Hermosillo A, Murata C, Yescas P, Espinola-Zavaleta N, Martinez M, Serrano H.. Arg347Cys polymorphism of α1a-adrenergic receptor in vasovagal syncope. Case-control study in a Mexican population. Auton Neurosci 2014;183:66–71. [DOI] [PubMed] [Google Scholar]
- 48. Lelonek M, Pietrucha T, Matyjaszczyk M, Goch JH.. Mutation T/C, Ile 131 of the gene encoding the alfa subunit of the human Gs protein and predisposition to vasovagal syncope. Circ J 2008;72:558–562. [DOI] [PubMed] [Google Scholar]
- 49. Ruwald MH, Hansen ML, Lamberts M, Kristensen SL, Wissenberg M, Olsen A-M, Christensen SB, Vinther M, Kober L, Torp-Pedersen C, Hansen J, Gislason GH.. Accuracy of the ICD-10 discharge diagnosis for syncope. Europace 2013;15:595–600. [DOI] [PubMed] [Google Scholar]
- 50. Reilly JG, Ayis SA, Ferrier IN, Jones SJ, Thomas SH.. QTc-interval abnormalities and psychotropic drug therapy in psychiatric patients. Lancet 2000;355:1048–1052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.