Multidrug-resistant bacteria continue to be a major problem worldwide, especially among individuals with significant comorbidities and other risk factors for infection. K. pneumoniae is among the leading causes of health care-associated infections, and the organism is often resistant to multiple classes of antibiotics. A carbapenem-resistant K. pneumoniae strain known as multilocus sequence type 258 (ST258) is the predominant carbapenem-resistant Enterobacteriaceae in the health care setting in the United States. Infections caused by ST258 are often difficult to treat and new prophylactic measures and therapeutic approaches are needed. To that end, we developed a lower respiratory tract infection model in cynomolgus macaques in which to test the ability of ST258 CPS to protect against severe ST258 infection.
KEYWORDS: antibiotic resistance, capsule polysaccharide, bactericidal activity, nonhuman primate, pneumonia, vaccine, capsule, immunization
ABSTRACT
Klebsiella pneumoniae is a human gut communal organism and notorious opportunistic pathogen. The relative high burden of asymptomatic colonization by K. pneumoniae is often compounded by multidrug resistance—a potential problem for individuals with significant comorbidities or other risk factors for infection. A carbapenem-resistant K. pneumoniae strain classified as multilocus sequence type 258 (ST258) is widespread in the United States and is usually multidrug resistant. Thus, treatment of ST258 infections is often difficult. Inasmuch as new preventive and/or therapeutic measures are needed for treatment of such infections, we developed an ST258 pneumonia model in cynomolgus macaques and tested the ability of an ST258 capsule polysaccharide type 2 (CPS2) vaccine to moderate disease severity. Compared with sham-vaccinated animals, those vaccinated with ST258 CPS2 had significantly less disease as assessed by radiography 24 h after intrabronchial installation of 108 CFU of ST258. All macaques vaccinated with CPS2 ultimately developed ST258-specific antibodies that significantly enhanced serum bactericidal activity and killing of ST258 by macaque neutrophils ex vivo. Consistent with a protective immune response to CPS2, transcripts encoding inflammatory mediators were increased in infected lung tissues obtained from CPS-vaccinated animals compared with control, sham-vaccinated macaques. Taken together, our data provide support for the idea that vaccination with ST258 CPS can be used to prevent or moderate infections caused by ST258. As with studies performed decades earlier, we propose that this prime-boost vaccination approach can be extended to include multiple capsule types.
INTRODUCTION
Klebsiella pneumoniae is a Gram-negative rod-shaped bacterium often found as a part of normal human microflora in the intestines. The bacterium is also an etiological agent of pneumonia, and bloodstream, surgical site, and urinary tract infections. Such infections occur typically in individuals with severe comorbidities or significant underlying susceptibility factors (e.g., immunosuppression or mechanical ventilation). Klebsiella spp. (K. pneumoniae and K. oxytoca) were reported by Magill et al. as being among the top eight causes of health care-associated infections in the United States in 2015 (1). In this study, pneumonia was the most abundant type of infection among all patients, and Klebsiella spp. remained a significant cause of health care-associated pneumonia (1). In a separate study of 33 U.S. medical centers (across 23 states), Sader et al. reported that K. pneumoniae was the third most common microbe recovered from patients with bloodstream infections during the period of 2015 to 2017 (2). The relative high prevalence of severe health care-associated infections caused by these organisms is complicated further by antibiotic resistance.
K. pneumoniae infections have been treated historically with β-lactam antibiotics (3). Carbapenem antibiotics are a class of highly effective broad-spectrum β-lactam antibiotics that have been used widely to treat individuals with K. pneumoniae infections, especially those caused by strains that produce extended-spectrum β-lactamases (4, 5). Therefore, the emergence of carbapenem-resistant K. pneumoniae strains, especially those that are resistant to other classes of antibiotics, poses a significant problem for treatment of infections (6–8). In the United States, carbapenem-resistance in K. pneumoniae is conferred primarily by K. pneumoniae carbapenemase (KPC), which is most often encoded by blaKPC within a transposon on a plasmid (9). KPC can hydrolyze all β-lactam antibiotics (9). The first K. pneumoniae KPC-producing isolates were reported in the United States approximately twenty years ago (9), and KPC-containing strains are now widespread (8, 10). The overall mortality for infections caused by KPC-containing K. pneumoniae was estimated recently as ∼41 to 42% (10, 11).
The majority of carbapenem-resistant K. pneumoniae clinical isolates in the United States and in other regions of the world are classified as multilocus sequence type 258 (ST258) (7). ST258 is comprised of two distinct clades and their genetic differentiation is largely attributed to a region of recombination that encompasses genes, known as cps1 and cps2, involved in capsule polysaccharide (CPS) biosynthesis (12). We demonstrated previously that ST258 CPS2 contributes to evasion of the host innate immune response, including inhibition of phagocytosis and killing by human neutrophils (13, 14). Additionally, anti-CPS2 antibody produced in rabbits promotes killing of ST258 in vitro (14). Studies published in the 1980s and 1990s demonstrated that purified K. pneumoniae CPS has significant potential as a vaccine antigen (15, 16), and anti-CPS antibody (Ab) confers some protection in humans (17), but clinical trials were stopped and an immunotherapy approach for prevention and/or treatment of K. pneumoniae infections is not licensed for use in humans. Immunoprophylaxis and/or immunotherapy are viable alternatives to antibiotic therapy.
As a step toward developing an immunotherapy for carbapenem-resistant K. pneumoniae, we developed an ST258 respiratory tract infection model in cynomolgus macaques (Macaca fascicularis) and tested the efficacy of purified CPS2 as a vaccine antigen.
RESULTS
Lower respiratory tract infection in cynomolgus macaques.
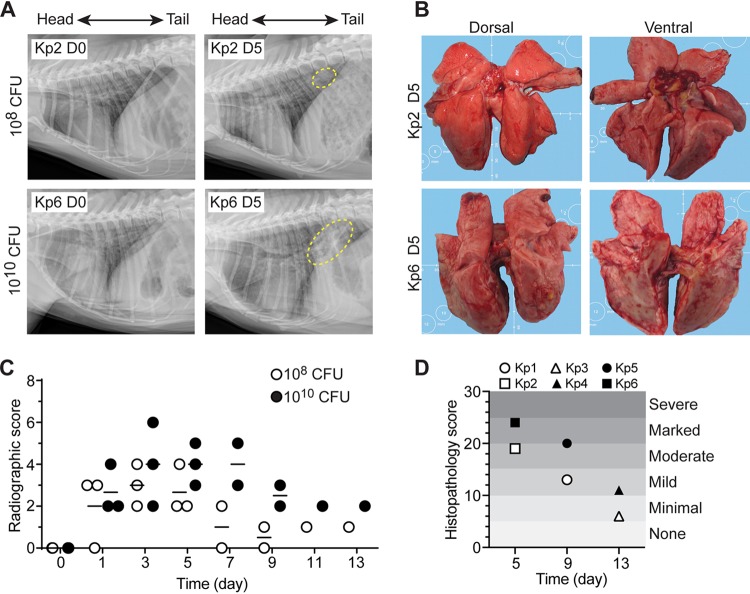
Nonhuman primates (NHPs) such as Macaca fascicularis are naturally susceptible to K. pneumoniae infections, many of which manifest as a respiratory disease (18). Moreover, the anatomy of the NHP respiratory system closely reflects that of humans (19, 20). As a first step toward testing the ability of ST258 CPS to elicit a protective immune response in the host, we developed an ST258 model of lower respiratory tract infection in cynomolgus macaques (Fig. 1). Animals (3 per group) were infected with 108 or 1010 CFU of ST258 by intrabronchial instillation, and the inoculum was divided equally between left and right lungs. Animals were monitored closely for clinical symptoms, and vital signs, SpO2, body temperature and weight, CBC and blood chemistry, and radiographs were taken every other day for 13 days (Fig. 1A, and Fig. S1 to S3 in the supplemental material). All animals had hunched posture and experienced coughing and sneezing after infection. There were patchy alveolar patterns on lung radiographs from animals infected with either inoculum starting on day 1 postinfection. At 5 days postinfection, there were regions of pulmonary consolidation located primarily in middle and caudal lobes of the lung, although the lobes affected varied among the animals in each group (Fig. 1A). The noted pulmonary consolidation remained in all infected animals until the end of the study (13 days postinfection).
FIG 1.
ST258 lower respiratory tract infection in cynomolgus macaques as a model nonhuman primate. (A) Representative radiographic images of lungs in uninfected animals (Kp2 D0, Kp6 D0) and then 5 days after infection with the indicated ST258 inoculum (Kp2 D5, Kp6 D5). Dashed yellow line indicates radiographic changes due to ST258 infection. (B) Gross pathology of lungs from the animals shown in panel A. (C) Quantitation of severity of radiographic scores. (D) Quantitation of severity of lung histopathology scores. Kp1 to Kp6 indicate numbers of individual animals.
ST258 lower respiratory tract infection model in cynomolgus macaques. Body weight (A) and rectal temperature (B) of animals were taken on each exam day. (C) Lung (LW) to body weight (BW) ratio of infected macaques was calculated on scheduled necropsy days. Open and closed symbols indicate animals infected with 108 (Kp1 to Kp3) and 1010 (Kp4 to Kp6) CFU of K. pneumoniae strain ST258, respectively. Download FIG S1, TIF file, 0.5 MB (541.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Lung consolidation was verified during necropsy, and there was a noted failure of infected lungs to collapse. Consistent with the radiographs, there was a high percentage of lung area visually affected in animals infected with 1010 CFU. Moreover, there were adhesions between lung lobes, adhesion of the lungs to the diaphragm, and an increased lung weight to body weight ratio. These features are consistent with severe lower respiratory tract infection (Fig. 1B, Fig. S1, and Table S1). Histopathology of lung samples correlated with subacute (day 5), chronic (day 9), and resolution (day 13) stages of the disease (Fig. 2, Fig. S4). In the most severely affected lobes, bronchioles and alveoli contained edema, organizing fibrin, aggregates of swirling and streaming fibroblasts, macrophages, and fibrin, which filled and conformed to the airway boundaries. Such pathology is consistent with bronchiolitis obliterans organizing pneumonia (BOOP) (Fig. 2). Disease was in general more severe in animals that received 1010 CFU compared to those infected with 108 CFU (Fig. 1D), a finding that underscores the idea that disease severity in the model is dose-dependent. Nonetheless, infection with either 108 or 1010 CFU of ST258 causes severe lower respiratory infection in cynomolgus macaques, and therefore 108 CFU was chosen as the inoculum for all subsequent experiments.
FIG 2.
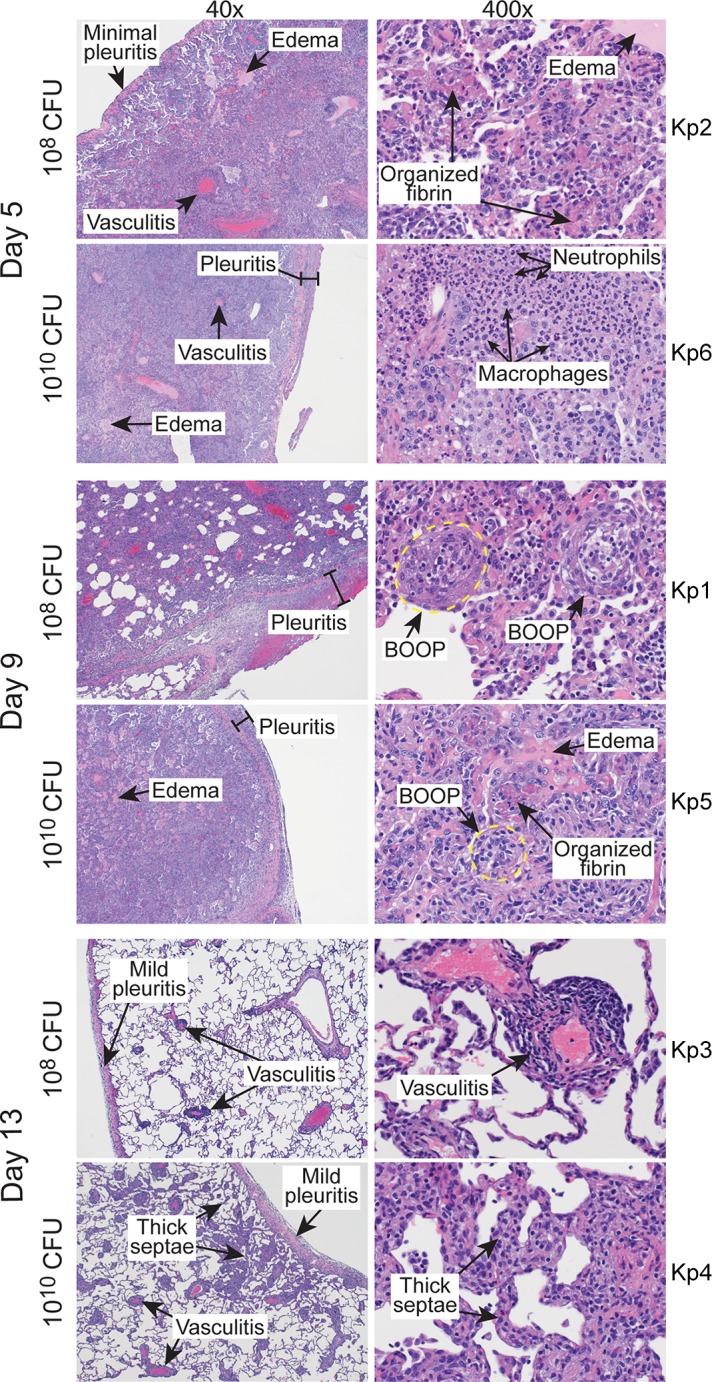
Histopathology of lungs infected with K. pneumoniae. Images represent sections of lung from cynomolgus macaques infected with 108 or 1010 CFU of K. pneumoniae strain ST258 as indicated. Tissue sections were collected during necropsy on the indicated day postinfection and stained with hematoxylin-eosin. The original magnification is 40× (left) or 400× (right). BOOP, bronchiolitis obliterans organizing pneumonia. Kp1 to Kp6 indicate numbers of individual animals.
Multiplex immunoassay analysis of selected immunomolecules in plasma samples collected from cynomolgus macaques infected with 108 (open bars) or 1010 (filled bars) CFU of K. pneumoniae strain ST258. Analysis was performed using a Multi-Analyte Profiling (MAP) technology platform (Human Inflammation Map v.1.0; Myriad RBM, Inc.). n, number of animals per group. Download FIG S2, PDF file, 0.8 MB (836KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Hematology and blood chemistry analysis of blood samples collected on exam days from cynomolgus macaques infected with 108 (open bars) or 1010 (filled bars) CFU of K. pneumoniae strain ST258. n, number of animals per group. Download FIG S3, TIF file, 2.8 MB (2.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representative images of healthy lung tissue from cynomolgus macaques. Download FIG S4, JPG file, 1.5 MB (1.6MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Percent of each lung lobe with disease as assessed by gross pathology. Download Table S1, DOCX file, 0.01 MB (15.5KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Vaccination of NHPs with CPS2 moderates severity of lower respiratory tract infection caused by ST258.
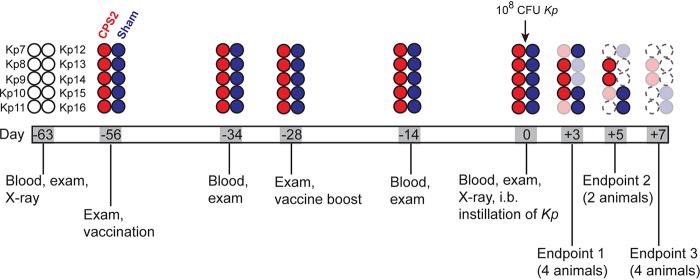
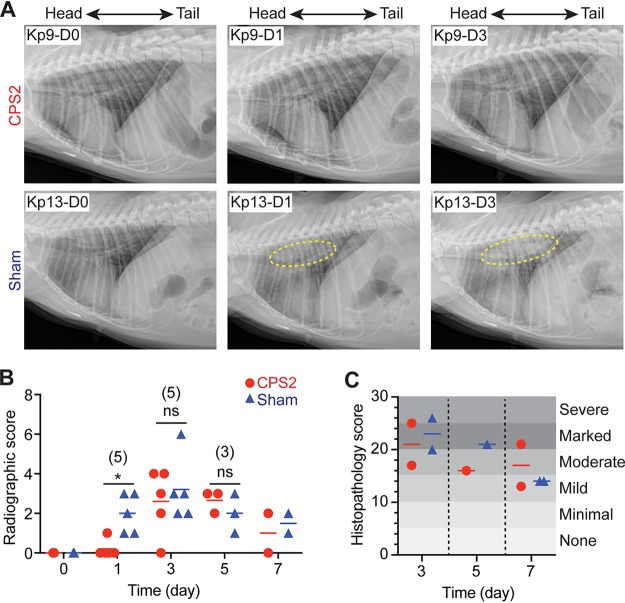
To evaluate the ability of purified CPS2 to protect NHPs against ST258 pneumonia, we infected 2 groups of 5 animals each with a 108 CFU of ST258 and then compared CPS2-vaccinated and sham-vaccinated (control) animals as outlined in Fig. 3. Within the first 24 h after infection, 3/5 animals in each group presented with a hunched posture, 2/5 animals in each group had notable sneezing, and one animal in the sham-vaccinated group presented with a cough. Although there was progressive pulmonary consolidation in all infected animals over the duration of the study, there was no radiographic change in 4/5 vaccinated animals 24 h after infection (Fig. 4A and B). Histopathology scores of lung tissues obtained from animals 3, 5, and 7 days postinfection were on average slightly reduced for CPS2-vaccinated animals compared with sham-vaccinated animals on day 3 and day 5, albeit the sample size was limited (1 to 2 animals per group were necropsied on those days) (Fig. 4C). Thus, the chest radiograph analyses indicated that CPS2-vaccinated animals had delayed disease progression compared to sham-vaccinated control animals (Fig. 4A and B). Consistent with these findings, lung tissue sections from animals vaccinated with CPS2 had decreased bacterial burden compared to those from sham-vaccinated animals, most notably at 3 and 5 days postinfection (Table 1). Collectively, these data provide clinical, radiographic, and histopathological evidence that vaccination of cynomolgus monkeys with CPS2 confers protection at least in part against severe lower respiratory tract infection caused by ST258.
FIG 3.
Schematic of the vaccination and challenge study. Individual animals are indicated with a circle. Open circles with a solid black line indicate NHPs prior to treatment (prevaccination). Red circles indicate NHPs vaccinated with CPS2. Blue circles indicate NHPs that were sham-vaccinated (control animals). Faded circles indicate the endpoint day and necropsy for that animal. Circles with dotted outlines are placeholders for animals necropsied at earlier times in the study.
FIG 4.
Vaccination with CPS2 delays the onset of ST258 pneumonia. (A) Representative radiographic images of lungs from an NHP vaccinated with CPS2 (Kp9) and from a sham-vaccinated NHP (Kp13) before infection with ST258 (Kp9-D0, Kp13-D0), 1 day postinfection (Kp9-D1, Kp13-D1), and 5 days postinfection (Kp9-D5, Kp13-D5). Dashed yellow line indicates radiographic changes due to ST258 infection. (B) Quantitation of severity of radiographic scores. (C) Quantitation of severity of lung histopathology scores. Statistical analyses for data in panel B, on day 1, day 3, and day 5, were performed using an unpaired, two-tailed t test with Welch’s correction. *, P = 0.012. The number of animals in each group (vaccinated or sham) on these days is provided in parentheses. ns, not significant at the level of P ≤ 0.05.
TABLE 1.
Bacteria in lung tissuea
| Lung lobe | CPS2 |
Sham |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Day 3 |
Day 5 |
Day 7 |
Day 3 |
Day 5 |
Day 7 |
|||||
| Kp7b | Kp11 | Kp10 | Kp8 | Kp9 | Kp13 | Kp14 | Kp12 | Kp15 | Kp16 | |
| Right upper lobe | + | ++ | − | + | − | + | +++ | + | − | − |
| Right middle lobe | ++ | − | − | − | − | + | ++ | + | − | + |
| Right lower lobe | ++ | + | − | + | − | ++ | +++ | + | + | ++ |
| Left upper lobe | ++ | − | − | − | − | + | +++ | − | − | − |
| Left middle lobe | ++ | ++ | − | + | − | − | ++ | + | − | + |
| Left lower lobe | ++ | − | − | + | − | + | ++ | + | + | − |
+, ++, +++, relative amounts of bacteria present in lung tissue samples; −, absence of bacteria in lung tissue samples.
Kp7 to Kp16 indicate numbers of individual animals.
Vaccination with CPS2 alters the host response to ST258 K. pneumoniae at the transcriptome level.
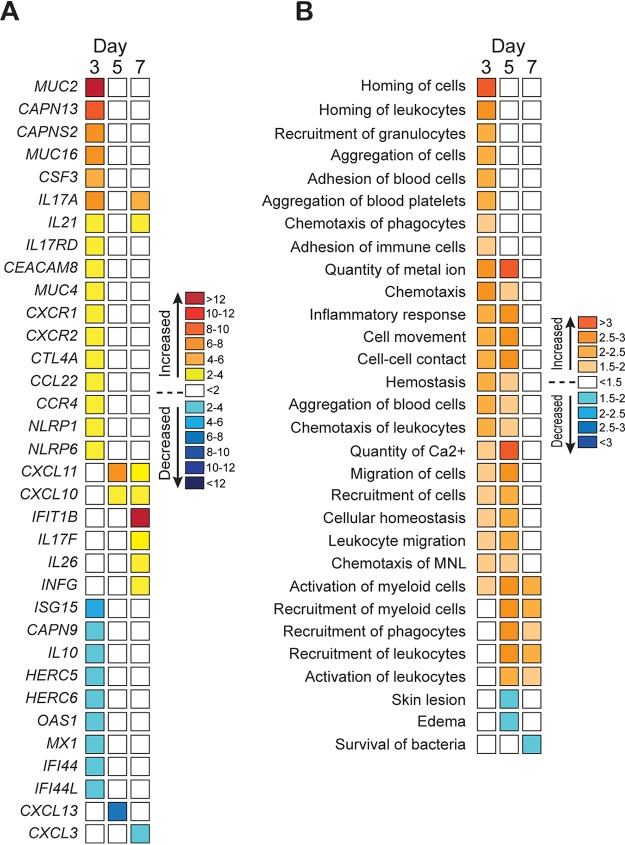
We next used an RNA sequencing (RNA-seq) approach to gain a better understanding of the host response to infection in animals vaccinated with ST258 CPS2 (Fig. 5). Lung tissue samples were obtained from 1 to 2 animals per group during necropsy on days 3, 5, and 7 postinfection and analyzed by RNA-seq. The complete list of transcripts and changes in their expression levels is compiled in Table S4. The greatest differential (between vaccinated and control animals) in host gene expression occurred on day 3 postinfection, the earliest time point tested (Fig. 5A and B). These findings are consistent with the observed delay of disease progression in CPS2-vaccinated animals (compare Fig. 4A, B and 5A, B). In general, genes encoding molecules involved in innate immune processes, such as chemotaxis and recruitment of granulocytes, were upregulated on day 3 postinfection in animals vaccinated with CPS2 (Fig. 5A and B). Compared with sham-vaccinated animals, transcripts encoding epithelial cell mucins (MUC4, MUC16) and airway-secreted mucins (MUC2, MUC5B) were upregulated in infected animals that had been vaccinated with CPS2 (Fig. 5A). Mucins play an important role in the defense of the airway against bacterial infection (21). Collectively, these data provide support for the idea that the immune response and host innate immunity is bolstered by vaccination with ST258 CPS2.
FIG 5.
Vaccination with CPS2 alters the host transcriptome during infection with ST258. (A) Three separate lung tissue samples were obtained from each animal at necropsy and processed individually for analysis by RNA-seq as described in the Methods and Materials. Results are presented as the mean fold change in transcripts from 3 to 6 separate samples from vaccinated versus sham-vaccinated animals. (B) RNA-seq data were analyzed with Ingenuity Pathway Analysis (IPA, Qiagen) to identify processes that are predicted to be increased or decreased. The intensity of orange or blue color indicates prediction strength. Increases or decreases that are ≤−2 or ≥ 2 indicate the change in predicted function is statistically significant.
Percent of each lung lobe with disease as assessed by histopathology. Download Table S2, DOCX file, 0.02 MB (17KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Histopathological changes in tissue samples obtained on scheduled necropsy days. To evaluate the degree of change for each described feature the following scale was used: 0 - no lesion/change; 1 - minimal change; 2 - mild change; 3 - moderate change; 4 - marked change; and 5 - severe change. Download Table S3, PDF file, 0.1 MB (109.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gene transcription changes in lung tissue samples of CPS2-vaccinated macaques compared to sham vaccinated, as assessed by RNA-seq. Red box indicates statistically significant upregulation (>2-fold), where green box indicates statistically significant downregulation (>2-fold) of the transcript. Download Table S4, PDF file, 2.3 MB (2.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Blood and serum obtained from vaccinated cynomolgus macaques has enhanced bactericidal activity toward ST258.
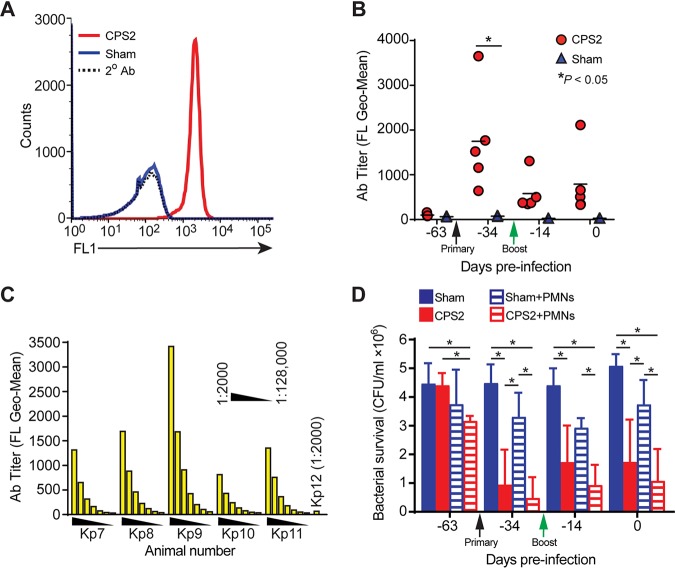
Compared with sham-vaccinated monkeys, animals vaccinated with CPS2 had increased levels of antibodies specific for ST258 (Fig. 6A and B). Antibody titers were highest at 3 weeks postvaccination and ranged from 32,000 to 64,000 as determined by flow cytometry (Fig. 6C). Although CPS2 elicited a relatively strong initial antibody response, it is noteworthy that a boost given 4 weeks after the initial vaccination failed to improve antibody titers (Fig. 6B). Moreover, antibody titers decreased within a few weeks after peak titers were measured, indicating the antibody response to CPS2 was relatively short-lived and perhaps consistent with the known immune response to purified polysaccharide alone (22). That said, the strong initial antibody response to CPS2 increased serum bactericidal activity significantly and enhanced the ability of neutrophils to kill ST258 when tested ex vivo (Fig. 6D). For example, on day −34, bacterial survival in serum from animals vaccinated with CPS2 was 21.8 ± 12.8% of that using serum from sham-vaccinated animals (P < 0.0001) (Fig. 6D). Consistent with these findings, the ability of NHP neutrophils to kill ST258 was increased significantly following vaccination with CPS2 (e.g., survival of ST258 in assays with neutrophils obtained from vaccinated NHPs was 13.2 ± 8.2% of that in assays with cells from sham-vaccinated animals, P = 0.0003) (Fig. 6D). Taken together, these results provide strong support for the idea that CPS2 can be used as a vaccine antigen for prevention or moderation of infections caused by ST258.
FIG 6.
Vaccination of cynomolgus macaques with CPS2 elicits antibodies that promote killing of ST258. (A) Antibody titers in cynomolgus macaques were determined by flow cytometry as described in the Methods and Materials. Data shown are from representative histograms from a single animal on day −34. (B) Comparison of antibody titers in cynomolgus macaques before vaccination, after primary vaccination with CPS2 (black arrow, day −56), and after a boost with CPS2 (green arrow, day −28) was determined by flow cytometry. Each symbol represents a unique animal. (C) Serum antibody titers in animals vaccinated with CPS2 (Kp7-Kp11) versus a sham-vaccinated animal (Kp12). Serum was diluted 1:2,000, 1:4,000, 1:8,000, 1:16,000, 1:32,000, 1:64,000, and 1:128,000 as indicated, and antibody binding to ST258 was determined by flow cytometry. (D) Survival of ST258 was decreased significantly by serum and neutrophils from vaccinated versus sham-vaccinated animals. ST258 (106 CFU) was incubated at 37°C with 5% serum alone (sham, CPS2) or the combination of 5% serum and neutrophils/polymorphonuclear leukocytes (PMNs) at a 1:1 CFU:PMN ratio (Sham+PMNs, CPS2+PMNs) for 1 h and bacterial survival was determined as described in the Methods and Materials. Sham, assays containing 5% serum from control animals that were sham-vaccinated; CPS2, assays containing 5% serum from animals vaccinated with ST258 CPS2. Data are presented as the mean survival ± standard error of the mean from 5 separate animals per group. *, P < 0.05 as determined by using a repeated measures one-way ANOVA and Tukey’s posttest.
DISCUSSION
Carbapenem-resistant K. pneumoniae pose a significant threat to human health and have been included as a priority pathogen by the World Health Organization since 2017 (8). Infections with ST258 are associated with high rates of morbidity and mortality, particularly among individuals with prolonged hospitalization, critically ill patients, and those exposed to indwelling devices. The rapid emergence of KPC-producing isolates worldwide presents a challenge for treatment given the expanded drug resistance profile of these strains, which has rekindled interest in development of alternative prophylactics and therapeutics such as vaccines and monoclonal antibody (MAb) therapy. In this study, we developed an ST258 respiratory tract infection model in cynomolgus macaques and evaluated the potential of purified capsule polysaccharide as an opsonophagocytic vaccine candidate for treatment of infection.
We chose NHPs to model K. pneumoniae respiratory infections due in large part to close approximation of human airway and lung anatomy, physiology and mucosal immune mechanisms. For example, primate species are distinguished by dichotomous branching of upper airways and progression to respiratory bronchioles that are not present in rodent species (19). Moreover, primate airways are highly ciliated in nature and have less abundant secretory cells than rodents (23). K. pneumoniae is part of the normal flora of nonhuman primates and is also a cause of pneumonia (18, 24). In our studies, intrabronchial administration of 108 and 1010 CFU ST258 into the right and left lungs of cynomolgus macaques resulted in development of a typical bacterial lobar pneumonia. The early stages of infection were characterized by development of radiographic signs of an alveolar pattern in the lungs by day 3. As disease progressed, pulmonary consolidation was evident in all infected NHP with animals receiving the higher inoculum showing signs of more severe disease and slower resolution.
Purified and detoxified K. pneumoniae capsular polysaccharide has been previously tested as a vaccine candidate. Vaccination with CPS was effective in reducing disease in K. pneumoniae murine infection models (25) and in a squirrel monkey model of bacteremia (26). In addition, K. pneumoniae CPS was shown to be safe in humans (27) including results of a phase I clinical trial (28). These vaccines were ultimately halted in favor of antibiotic therapy. We previously demonstrated that purified IgG specific for ST258 CPS2 promotes enhanced killing by human blood and serum, suggesting that it is a viable target of immunoprophylaxis (14). In the present study, immunization of NHPs with CPS2 resulted in a peak antibody titer at 3 weeks postvaccination (32,000 to 64,000; Fig. 6B and C). Notably, immune sera from the CPS2-vaccinated NHPs showed increased bactericidal activity and enhanced the ability of neutrophils to kill ST258 in ex vivo assays (Fig. 6D). Following the primary vaccination, NHPs received a booster at day −28 and were challenged by intrabronchiolar inoculation of 108 CFU ST258 on day 0 (Fig. 3). The severity of ST258 pneumonia was reduced and delayed in the vaccinated animals compared to the unvaccinated control group, as evidenced by clinical assessment, radiography, and pathology (Fig. 4A to C). The delay of disease in the vaccinated animals is consistent with Ingenuity Pathway Analysis (IPA) comparison of diseases and biofunctions from RNA-seq data that indicate overall upregulation of inflammatory response, activation, recruitment, and migration of immune cells compared to the sham-vaccinated control group (Fig. 5B). Upregulation of these pathways was described previously in mouse models of Pseudomonas aeruginosa (29, 30), Actinobacillus pleuropneumoniae (31), and Streptococcus pneumoniae (32) respiratory infections. Nevertheless, immunization of NHPs with CPS2 did not provide complete protection against ST258, and the induced antibody response was short-lived, as demonstrated by the ineffectiveness of the booster and the dropping antibody titer over time (Fig. 6 B). Bacterial capsular polysaccharides belong to thymus-independent (TI) antigens, are poor activators of T cells, and do not lead to generation of T and B cell memory (33). Immunogenicity and efficacy of carbohydrate vaccines can be successfully enhanced by coupling CPS to carrier proteins such as inactivated bacterial toxoids (34). To this end, Seeberger et al. recently described a semisynthetic glycoconjugate vaccine based on the structure of CPS from K. pneumoniae carbapenemase (KPC)-containing isolates conjugated to the diphtheria toxoid carrier CRM197 (35). Moreover, Diago-Navarro et al. demonstrated that treatment of mice with MAbs specific for a CPS glycoconjugate caused a reduction in the number of K. pneumoniae recovered from lungs, liver, and spleen of infected mice (36). Although the glycoconjugate successfully elicited CPS-specific antibody in mice and rabbits, the ability of the vaccine to protect against K. pneumoniae was not tested owing to the lack of an appropriate infection model for KPC strains that sufficiently approximates host comorbidities. More recently, Feldman et al. reported a K. pneumoniae CPS glycoconjugate (bioconjugate) vaccine approach directed against hypervirulent K. pneumoniae (hvKp) community strains that provided protection in a murine infection model (37). Additionally, use of MAbs specific for hvKp enhanced animal survival and decreased bacterial load in lungs, spleen, and liver in a murine infection model (38). Unlike ST258, hvKp are more virulent in animal infection models and are associated with disease in otherwise healthy individuals (39). Moving forward, it is feasible that CPS-based vaccines and/or CPS-specific MAbs will provide a viable alternative to treatment with antibiotics. For example, a CPS-based vaccine could be used for targeted prophylaxis in at-risk populations. It will be important to develop infection models that approximate ST258 infection to further test and improve current vaccine strategies.
MATERIALS AND METHODS
Ethics statement.
All animal experiments and procedures were performed in accordance with an animal study protocol (2018-026E) approved by the Animal Care and Use Committee at Rocky Mountain Laboratories (RML), National Institutes of Allergy and Infectious Diseases (NIAID), and conformed to the guidelines of the National Institutes of Health (NIH). RML is accredited fully by the Association for Assessment and Accreditation of Laboratory Animal Care International. The animals were housed according to standards detailed in the Animal Welfare Act, Public Health Service, and National Institutes of Health Animal Care and Use Policies, and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
Animals.
Female cynomolgus macaques (Macaca fascicularis) of Mauritius origin were 8 to 10 years old at the time of the study. The animals were fed a commercial nonhuman primate diet (Teklad 2055 25% Protein Primate Diet, Envigo) twice daily and had free access to water. Environmental enrichment was provided per facility enrichment plan. Macaques were pair-housed in standard nonhuman primate caging with an additional play cage attached during the vaccination period, and then housed individually following inoculation with bacteria. For installation of the inoculum, animals were anesthetized with an intramuscular (IM) injection of Telazol (3 mg/kg). Ketamine HCl (10 to 11 mg/kg, IM) was used for all other procedures and exams. Animals were euthanized by barbiturate overdose (1 ml/5 kg) administered intravenously following induction of deep anesthesia with Telazol (5 mg/kg, IM).
Bacterial strains and culture conditions.
K. pneumoniae isolate NJST258_2 (12) was cultured in Luria-Bertani (LB) broth at 37°C in a rotary shaker incubator at 225 rpm. Bacteria from overnight culture were transferred to fresh LB medium (1:200 dilution) and cultured to early exponential phase of growth (optical density at 600 nm [OD600] = 0.750). Subsequently, bacteria were harvested by centrifugation (4,200 × g for 15 min at 4°C), washed and suspended in sterile injection grade saline when used as inoculum or RPMI 1640 medium (Gibco/LifeTechnologies) containing 10 mM HEPES (RPMI/H) for use in in vitro assays.
Nonhuman primate pneumonia model.
Six female cynomolgus macaques were split randomly into 2 groups of 3 animals each. On day zero, 3 animals received 5 ml (2.5 ml to each lung) injection grade sterile saline containing 108 CFU and 3 animals received 1010 CFU of K. pneumoniae via intrabronchial instillation. Intrabronchial inoculation was performed using a bronchoscope (Pentax EB-1170K video bronchoscope). The bronchoscope was utilized to visualize the main-stem bronchi and advanced into the right and left bronchus. The inoculum was then delivered through the working channel of the bronchoscope. Following inoculation, the bronchoscope was removed. Physical exams (including body weight and body condition scoring, rectal temperature, heart rate, respiratory rate/pattern, and pulse oximetry assessment), blood collection, and thoracic radiographs were performed on day 0 prior to infection, and postinfection on day 1 and every other day afterward until day 13 (experimental endpoint). When clinically relevant dehydration was noted in at least one animal at a scheduled physical exam, all animals received 125 ml of 0.9% NaCl subcutaneously on the exam day. Animals were monitored cageside for clinical signs at least twice daily. Thoracic radiographs were taken using an IDEXX digital imaging system with PACS software (IDEXX Laboratories, Inc., Westbrook, ME). The DICOM files generated by the IDEXX system were converted to JPEG using OsiriX Lite software v10.0.2 (Pixmeo SARL, Bernex, Switzerland). Subsequently, images were analyzed and the severity of radiographic changes in each lobe was scored by clinical veterinarians in a blinded fashion, in accordance with a previously described scoring method (40). Briefly, each lung lobe was given an independent score from 0 to 3 based on the following criteria: 0, normal examination; 1, mild interstitial pulmonary infiltrates; 2, moderate interstitial infiltrates, possible partial cardiac border effacement and small areas of pulmonary consolidation; 3, progression of severity from grade 2, pulmonary consolidation. The total score per animal presented in the figures is a sum of scores assigned to all lobes. One animal from each dose group was euthanized on day 5, 9, and 13 postinfection.
Vaccine preparation and animal vaccination.
CPS2 was extracted from bacterial culture supernatant as previously described (14, 16). Residual endotoxin was inactivated by incubation of purified CPS2 with 0.1 N NaOH in 95% ethanol for 1 h at 37°C. The pH of the solution was neutralized with 2 N acetic acid and CPS2 was collected by centrifugation at 2000 × g for 10 min. The pellet was air dried and resuspended in sterile water. Subsequently, the sample was dialyzed against sterile water and concentrated using a centrifugal filter unit with a 100 kDa molecular weight cutoff (MWCO) (Amicon Ultra-15 centrifugal filter unit; MilliporeSigma, UFC910024). Endotoxin levels in purified capsule preparations were quantified with a limulus amebocyte lysate (LAL) turbidimetric endotoxin quantitation kit (Charles River Endosafe, R19000).
All animals were tested for the presence of anti-K. pneumoniae CPS2 antibody prior to vaccination with CPS2. Five animals were inoculated with up to 500 μg of purified K. pneumoniae CPS2 in 0.6 ml of injection grade saline mixed 1:1 with TiterMax Gold adjuvant (Sigma-Aldrich). Five sham-vaccinated animals received 0.6 ml of injection grade saline combined with adjuvant instead of purified CPS2. The CPS2 (or saline)/TiterMax Gold inoculum was administered as follows: 0.2 ml was injected intramuscularly into the right and left caudal thigh muscles, and 0.8 ml was injected subcutaneously in the midscapular region. Four weeks after initial vaccination, animals received a boost immunization containing ∼500 μg purified CPS2 in injection grade sterile saline (vaccinated group) or saline alone (sham-vaccinated group). The vaccination was administered by intramuscular injection (0.1 ml) into the right and left caudal thigh muscles, and by subcutaneous injection (0.4 ml) in the midscapular region. No adjuvant was used for the boost. To monitor the immune response to CPS2, blood was collected on day −63 (prior to vaccination), day −34 (3 weeks after initial vaccination), on day −14 (2 weeks after first boost), and on day 0 (day of bacteria challenge). On day 0, all animals received 5 ml (2.5 ml into each lung) of injection grade sterile saline containing 108 CFU of K. pneumoniae via intrabronchial instillation using a bronchoscope (Olympus Evis Exera BF-XP190 bronchoscope) as described above. Physical exams (including body weight and body condition scoring, rectal temperature, heart rate, respiratory rate/pattern, and pulse oximetry assessment), blood collection, and thoracic radiographs were performed as outlined in Fig. 3 until day 7 (the experimental endpoint) (Fig. S5 and S6 in the supplemental material). To evaluate histopathological changes that occurred during the course of infection, 2 animals from each group were euthanized on day 3, 1 animal from each group was euthanized on day 5, and an additional 2 animals were euthanized on day 7 postinfection (see Fig. 3 for details). Scoring of histopathological changes in the tissues samples was done in a blinded fashion by a qualified veterinary pathologist.
Physiological parameters of CPS2-vaccinated or sham-vaccinated animals. Body weight and rectal temperature of each animal were taken on each exam day. (A) Data show the mean ± standard deviation percentage change in body weight of the CPS2-vaccinated or sham-vaccinated animals compared body weight before vaccination (day −63). (B) Data indicate the mean body temperature ± standard deviation. (C) Lung (LW) to body weight (BW) ratio was calculated on each scheduled necropsy day. Download FIG S5, TIF file, 0.5 MB (510.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Antibody titers.
On designated exam days, blood from NHPs was collected in BD vacutainer tubes lacking anticoagulant. Blood was allowed to clot at room temperature for at least 20 min and serum was separated from the clot by centrifugation at 1,100 × g for 10 min. Anti-CPS2 antibody titers were determined by flow cytometry as described previously (14). Rabbit preimmune serum and rabbit anti-CPS2 antibody were used as negative and positive controls, respectively (14). Goat anti-monkey IgG conjugated with fluorescein isothiocyanate (FITC) (Abcam, product 112766) was used as a secondary antibody.
NHP neutrophil and serum bactericidal activity.
Nonhuman primate neutrophils were isolated from heparinized blood using a CD66abce MicroBead kit and magnetic-activated cell sorting (MACS) LS separation columns (Miltenyi Biotec) as described (41). Briefly, NHP blood was collected into BD vacutainer tubes containing sodium heparin (12.0 mg per 4 ml tube). Erythrocytes were lysed by incubation of blood with 7 volumes of erythrocyte lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) at room temperature for 10 min, mixing gently by inversion several times during lysis. Cells were washed with RPMI/H medium and resuspended in MACS buffer (0.5% human serum albumin [Sigma product A4327]/2 mM EDTA in Dulbecco’s phosphate-buffered saline [DPBS] without Ca++ and Mg++). CD66abce-biotin-labeled antibody was added to the cell suspension followed by addition of anti-biotin micro beads. The cell suspension was then transferred onto an LS column (Miltenyi Biotec) positioned in the magnetic stand and unlabeled cells were washed from the column using MACS buffer. Magnetically labeled cells (neutrophils) were eluted by removing the LS column from the magnetic stand and flushing it with MACS buffer. Cells were collected by centrifugation at 300 × g for 10 min at 4°C and resuspended in RPMI/H (1 × 107 cells/ml). If not used immediately, neutrophils were kept on ice. To evaluate neutrophil bactericidal activity against K. pneumoniae, purified neutrophils (106 cells) were combined with bacteria at a 1:1 bacteria: neutrophil ratio in RPMI/H containing 5% autologous serum (600 μl final volume). Assays were rotated gently (8 rpm) at 37°C for up to 60 min. Control assays without neutrophils were included in parallel, and these assays were used to evaluate serum bactericidal activity (106 CFU in 600 μl final volume). At designated times, saponin (0.1% final concentration) was added to the assay tubes and the neutrophils were lysed on ice for 15 min. Serial dilutions of assay samples were plated onto LB agar and then cultured overnight at 37°C. Bacteria were enumerated the following day. Bacterial survival was calculated with the following equations: CFU+serum_CPS-vaccinated/CFU+serum_sham-vaccinated ×100% or CFU+PMN+serum_CPS-vaccinated/CFU+PMN+serum_sham-vaccinated ×100%, where purified neutrophils are designated as polymorphonuclear leukocytes (PMN). Bactericidal activity of neutrophils purified using the positive selection method described here was compared to the bactericidal activity of neutrophils purified via gradient method and no significant difference was noted (not shown).
Histopathology analysis.
Necropsies were performed by veterinary pathologists on predetermined endpoint days. During necropsy, lungs were assessed for gross pathology and images were captured with a Nikon D300 camera. Tissue samples were also collected for histopathological analysis. Samples were fixed in 10% neutral buffered formalin (Cancer Diagnostics, Durham, NC) for a minimum of 72 h and processed using a VIP-6 Tissue Tektissue processor (Sakura Finetek, USA). Tissue samples were embedded in Ultraffin paraffin polymer (Cancer Diagnostics, Durham, NC) and sectioned serially at 5 μm. Air-dried slides were stained with hematoxylin and eosin. K. pneumoniae was visualized within tissue sections using rabbit anti-K. pneumoniae CPS2 IgG (1:500 dilution) (14) and a Discovery ULTRA System (Roche Diagnostics) and Discovery DAB Map detection kit (Roche Diagnostics) according to the manufacturer’s protocol. Images were obtained using an Olympus BX51 microscope and an Olympus DP74 camera (Olympus Corporation). Image brightness and contrast were evenly adjusted using Adobe Photoshop, and figures were created using Adobe Illustrator CC 2019 (Adobe, San Jose, CA).
Hematology, blood chemistry, multiplex immunoassays, and quantitative bacterial culture.
Hematology analysis was performed using blood containing EDTA as an anticoagulant (BD vacutainer EDTA tubes) and a ProCyte Dx hematology analyzer (IDEXX, Westbrook, ME). Blood chemistry analysis was performed with serum samples using a VetScan VS2 chemistry analyzer (Abaxis, Union City, CA). Additionally, selected serum samples were submitted for multiplex immunoassay analysis on a Multi-Analyte Profiling (MAP) technology platform (Human Inflammation Map v.1.0; Myriad RBM, Inc., Austin, TX). Samples were stored at –80°C until shipped for analysis. Results are provided in Fig. S2.
To enumerate K. pneumoniae in NHP blood, 0.1 ml of freshly collected heparinized blood was plated on LB agar and Klebsiella ChromoSelect selective agar plates (Sigma-Aldrich). Plates were evaluated for bacterial growth after incubation at 37°C for 24 h. Klebsiella pneumoniae was not detected in the blood (not shown).
RNA-seq analysis of host response during K. pneumoniae infection.
Three tissue samples (∼100 mg each) per lung/animal were taken from a visually affected section of the lung lobe during necropsy on days 3, 5, and 7 post bacterial infection. Tissue sections were immediately flash frozen in liquid nitrogen and stored at –80°C until used. To isolate total RNA, frozen samples were pulverized using a hammer, as described previously (42). Pulverized samples were added to lysis buffer containing 2-mercaptoethanol and homogenized in a FastPrep-24 sample preparation system (MP Biomedicals, Santa Ana, CA). RNA was purified initially by using a PureLink RNA minikit (Life Technologies, Carlsbad, CA). DNA was removed from purified RNA samples by treatment with TURBO DNase (Life Technologies, Grand Island, NY) for 45 min at 37°C, and RNA was purified again using an RNeasy minikit (Qiagen). Quality of purified RNA was assessed with a 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). RNA quantity was assessed by use of a Quant-iT RiboGreen RNA assay kit (Invitrogen, Inc.). To prepare samples for transcriptome sequencing (RNA-seq) analysis, 2.5 μg of purified RNA from each sample was processed in accordance with the TruSeq Stranded mRNA sample prep guide, rev. E (Illumina Inc., San Diego, CA). Each sample was uniquely indexed using the high-throughput (HT) dual-barcoding strategy as outlined in the sample prep guide. The resulting RNA libraries were fragment sized using DNA1000 Bioanalyzer chips (Agilent Technologies, Santa Clara, CA) and quantitated using a KAPA library quantification kit (Illumina) combined with Universal qPCR (quantitative PCR) mix (Kapa Biosystems, Wilmington, MA). All samples were diluted to a 2 nM working stock and multiplexed into a single pool using equal volumes. A 1.8 pM titration point was used to cluster two high-output flow cells on a NextSeq 550 Sequencing System (Illumina), which were sequenced for 75 cycles in each read direction for a total of 150 cycles each. Raw next-generation sequencing (NGS) reads were processed by first removing any Illumina adapter sequences using Cutadapt v1.12 (https://doi.org/10.14806/ej.17.1.200), and then trimmed and filtered for quality and length (min length of 35 bp using FASTX ToolKit v0.0.14, Hannon Lab, Cold Spring Harbor Laboratory). Trimmed reads for each replicate were then aligned to the Macaca fascicularis GCF_000364345.1 genome using Hisat2 v2.0.5 (43). Final read counts from mapped transcripts based on the combined replicates, normalized and then differentials for each comparison were generated using DESeq2 (https://doi.org/10.18129/B9.bioc.DESeq2) for each experimental condition. (See “Data availability” section for RNA-seq data.) Sequencing data were partially analyzed through the use of Qiagen’s Ingenuity Pathway Analysis (IPA, Qiagen Redwood City, www.qiagen.com/ingenuity). The deduced heat map analysis was regraphed for better visualization using Adobe Illustrator CC 2018 (Adobe).
Statistics.
Statistical analyses were performed using GraphPad Prism version 7.0e (GraphPad Software, Inc.). For comparison of 2 experimental conditions, data were analyzed with a two-tailed, unpaired t test with Welch’s correction as indicated. To compare 3 or more experimental conditions or treatments, data were analyzed with a repeated-measures one-way analysis of variance (ANOVA) and Tukey’s posttest to correct for multiple comparisons.
Data availability.
RNA-Seq data are MINSEQE compliant and have been posted online at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137135 under series number GSE137135.
Hematology and blood chemistry analysis of blood samples collected on exam days from cynomolgus macaques vaccinated with CPS2-vaccinated or sham-vaccinated and infected with 108 CFU of ST258 on day 0. Download FIG S6, TIF file, 2.2 MB (2.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
We thank Anita Mora, Ryan Kissinger, and Austin Athman (Visual Medical Arts, NIAID) for assistance with lung images.
We thank the Rocky Mountain Veterinary Branch husbandry staff for their dedicated care of the animals.
This study was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH, USA, and by a grant from NIAID, NIH (grant R01AI090155 awarded to B.N.K.).
We declare no conflictions of interest.
Footnotes
This article is a direct contribution from Frank R. DeLeo, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Franklin Lowy, Columbia University Medical Center, and Steven Projan.
Citation Malachowa N, Kobayashi SD, Porter AR, Freedman B, Hanley PW, Lovaglio J, Saturday GA, Gardner DJ, Scott DP, Griffin A, Cordova K, Long D, Rosenke R, Sturdevant DE, Bruno D, Martens C, Kreiswirth BN, DeLeo FR. 2019. Vaccine protection against multidrug-resistant Klebsiella pneumoniae in a nonhuman primate model of severe lower respiratory tract infection. mBio 10:e02994-19. https://doi.org/10.1128/mBio.02994-19.
REFERENCES
- 1.Magill SS, O'Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle J, Wilson LE, Kainer MA, Lynfield R, Greissman S, Ray SM, Beldavs Z, Gross C, Bamberg W, Sievers M, Concannon C, Buhr N, Warnke L, Maloney M, Ocampo V, Brooks J, Oyewumi T, Sharmin S, Richards K, Rainbow J, Samper M, Hancock EB, Leaptrot D, Scalise E, Badrun F, Phelps R, Edwards JR. 2018. Changes in prevalence of health care–associated infections in U.S. hospitals. N Engl J Med 379:1732–1744. doi: 10.1056/NEJMoa1801550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sader HS, Castanheira M, Streit JM, Flamm RK. 2019. Frequency of occurrence and antimicrobial susceptibility of bacteria isolated from patients hospitalized with bloodstream infections in United States medical centers (2015–2017). Diagn Microbiol Infect Dis 95:114850. doi: 10.1016/j.diagmicrobio.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Rahal JJ, Urban C, Horn D, Freeman K, Segal-Maurer S, Maurer J, Mariano N, Marks S, Burns JM, Dominick D, Lim M. 1998. Class restriction of cephalosporin use to control total cephalosporin resistance in nosocomial Klebsiella. JAMA 280:1233–1237. doi: 10.1001/jama.280.14.1233. [DOI] [PubMed] [Google Scholar]
- 4.Birnbaum J, Kahan FM, Kropp H, Macdonald JS. 1985. Carbapenems, a new class of beta-lactam antibiotics: discovery and development of imipenem/cilastatin. Am J Med 78:3–21. doi: 10.1016/0002-9343(85)90097-x. [DOI] [PubMed] [Google Scholar]
- 5.El-Gamal MI, Brahim I, Hisham N, Aladdin R, Mohammed H, Bahaaeldin A. 2017. Recent updates of carbapenem antibiotics. Eur J Med Chem 131:185–195. doi: 10.1016/j.ejmech.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasević AT, Cantón R, Carmeli Y, Friedrich AW, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Nordmann P, Poirel L, Rossolini GM, Seifert H, Vatopoulos A, Walsh T, Woodford N, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. 2017. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis 17:153–163. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 7.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. [Google Scholar]
- 9.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos-Castañeda JA, Ruano-Ravina A, Barbosa-Lorenzo R, Paillier-Gonzalez JE, Saldaña-Campos JC, Salinas DF, Lemos-Luengas EV. 2018. Mortality due to KPC carbapenemase-producing Klebsiella pneumoniae infections: systematic review and meta-analysis: mortality due to KPC Klebsiella pneumoniae infections. J Infect 76:438–448. doi: 10.1016/j.jinf.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Xu L, Sun X, Ma X. 2017. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob 16:18. doi: 10.1186/s12941-017-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLeo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi SD, Porter AR, Dorward DW, Brinkworth AJ, Chen L, Kreiswirth BN, DeLeo FR. 2016. Phagocytosis and killing of carbapenem-resistant ST258 Klebsiella pneumoniae by human neutrophils. J Infect Dis 213:1615–1622. doi: 10.1093/infdis/jiw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi SD, Porter AR, Freedman B, Pandey R, Chen L, Kreiswirth BN, DeLeo FR. 2018. Antibody-mediated killing of carbapenem-resistant ST258 Klebsiella pneumoniae by human neutrophils. mBio 9:e00297-18. doi: 10.1128/mBio.00297-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cryz SJ Jr., Fürer E, Germanier R. 1985. Safety and immunogenicity of Klebsiella pneumoniae K1 capsular polysaccharide vaccine in humans. J Infect Dis 151:665–671. doi: 10.1093/infdis/151.4.665. [DOI] [PubMed] [Google Scholar]
- 16.Cryz SJ, Fürer E, Germanier R. 1985. Purification and vaccine potential of Klebsiella capsular polysaccharides. Infect Immun 50:225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donta ST, Peduzzi P, Cross AS, Sadoff J, Haakenson C, Cryz SJ Jr., Kauffman C, Bradley S, Gafford G, Elliston D, Beam TR Jr., John JF Jr., Ribner B, Cantey R, Welsh CH, Ellison RT III, Young EJ, Hamill RJ, Leaf H, Schein RMH, Mulligan M, Johnson C, Abrutyn E, Griffiss JM, Hamadeh R, Eliasson AH, McClain JB, Melcher GP, Kelly JW, Byrne WR, Wallace M, Amundson D, Gumpert B, Slagle D. 1996. Immunoprophylaxis against Klebsiella and Pseudomonas aeruginosa infections. The federal hyperimmune immunoglobin trial study group. J Infect Dis 174:537–543. doi: 10.1093/infdis/174.3.537. [DOI] [PubMed] [Google Scholar]
- 18.Good RC, May BD. 1971. Respiratory pathogens in monkeys. Infect Immun 3:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller LA, Royer CM, Pinkerton KE, Schelegle ES. 2017. Nonhuman primate models of respiratory disease: past, present, and future. ILAR J 58:269–280. doi: 10.1093/ilar/ilx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plopper CG, Hyde DM. 2008. The non-human primate as a model for studying COPD and asthma. Pulm Pharmacol Ther 21:755–766. doi: 10.1016/j.pupt.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Weitnauer M, Mijošek V, Dalpke AH. 2016. Control of local immunity by airway epithelial cells. Mucosal Immunol 9:287. doi: 10.1038/mi.2015.126. [DOI] [PubMed] [Google Scholar]
- 22.Siegrist CA, Lambert PH. 2016. How vaccines work, p 33–42. In Bloom BR, Lambert PH (ed), The Vaccine Book, 2nd ed Academic Press, London, United Kingdom. [Google Scholar]
- 23.Whitsett JA, Alenghat T. 2015. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 16:27. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gozalo AS, Elkins WR, Lambert LE, Stock F, Thomas IIM, Woodward RA. 2016. Genetic diversity of Klebsiella pneumoniae isolates during an outbreak in a non-human primate research colony. J Med Primatol 45:312–317. doi: 10.1111/jmp.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cryz SJ Jr., Fürer E, Germanier R. 1984. Prevention of fatal experimental burn-wound sepsis due to Klebsiella pneumoniae KP1-O by immunization with homologous capsular polysaccharide. J Infect Dis 150:817–822. doi: 10.1093/infdis/150.6.817. [DOI] [PubMed] [Google Scholar]
- 26.Postal JM, Gysin J, Crenn Y. 1988. Protection against fatal Klebsiella pneumoniae sepsis in the squirrel monkey Saimiri sciureus after immunization with a capsular polysaccharide vaccine. Ann Inst Pasteur Immunol 139:401–407. doi: 10.1016/0769-2625(88)90066-9. [DOI] [PubMed] [Google Scholar]
- 27.Cryz SJ, Mortimer P, Cross AS, Fürer E, Germanier R. 1986. Safety and immunogenicity of a polyvalent Klebsiella capsular polysaccharide vaccine in humans. Vaccine 4:15–20. doi: 10.1016/0264-410x(86)90092-7. [DOI] [PubMed] [Google Scholar]
- 28.Edelman R, Taylor DN, Wasserman SS, McClain JB, Cross AS, Sadoff JC, Que JU, Cryz SJ. 1994. Phase 1 trial of a 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent Pseudomonas O-polysaccharide conjugate vaccine administered simultaneously. Vaccine 12:1288–1294. doi: 10.1016/s0264-410x(94)80054-4. [DOI] [PubMed] [Google Scholar]
- 29.Damron FH, Oglesby-Sherrouse AG, Wilks A, Barbier M. 2016. Dual-seq transcriptomics reveals the battle for iron during Pseudomonas aeruginosa acute murine pneumonia. Sci Rep 6:39172. doi: 10.1038/srep39172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuerth K, Lee AHY, Falsafi R, Gill EE, Hancock R. 2018. Characterization of host responses during Pseudomonas aeruginosa acute infection in the lungs and blood and after treatment with the synthetic immunomodulatory peptide IDR-1002. Infect Immun 87:e00661-18. doi: 10.1128/IAI.00661-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P, Xu Z, Sun X, Yin Y, Fan Y, Zhao J, Mao X, Huang J, Yang F, Zhu L. 2017. Transcript profiling of the immunological interactions between Actinobacillus pleuropneumoniae serotype 7 and the host by dual RNA-seq. BMC Microbiol 17:193. doi: 10.1186/s12866-017-1105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ND, Evans TJ. 2019. Dual RNA-seq in Streptococcus pneumoniae infection reveals compartmentalized neutrophil responses in lung and pleural space. mSystems 4:e00216-19. doi: 10.1128/mSystems.00216-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weintraub A. 2003. Immunology of bacterial polysaccharide antigens. Carbohydr Res 338:2539–2547. doi: 10.1016/j.carres.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Hütter J, Lepenies B. 2015. Carbohydrate-based vaccines: an overview. Methods Mol Biol 1331:1–10. doi: 10.1007/978-1-4939-2874-3_1. [DOI] [PubMed] [Google Scholar]
- 35.Seeberger PH, Pereira CL, Khan N, Xiao G, Diago-Navarro E, Reppe K, Opitz B, Fries BC, Witzenrath M. 2017. A semi-synthetic glycoconjugate vaccine candidate for carbapenem-resistant Klebsiella pneumoniae. Angew Chem Int ed Engl 56:13973–13978. doi: 10.1002/anie.201700964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diago-Navarro E, Motley MP, Ruiz-Peréz G, Yu W, Austin J, Seco BMS, Xiao G, Chikhalya A, Seeberger PH, Fries BC. 2018. Novel, broadly reactive anticapsular antibodies against carbapenem-resistant Klebsiella pneumoniae protect from infection. mBio 9:e00091-18. doi: 10.1128/mBio.01005-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldman MF, Mayer Bridwell AE, Scott NE, Vinogradov E, McKee SR, Chavez SM, Twentyman J, Stallings CL, Rosen DA, Harding CM. 2019. A promising bioconjugate vaccine against hypervirulent Klebsiella pneumoniae. Proc Natl Acad Sci U S A 201907833. doi: 10.1073/pnas.1907833116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diago-Navarro E, Calatayud-Baselga I, Sun D, Khairallah C, Mann I, Ulacia-Hernando A, Sheridan B, Shi M, Fries BC. 2017. Antibody-based immunotherapy to treat and prevent infection with hypervirulent Klebsiella pneumoniae. Clin Vaccine Immunol 24:e00456-16. doi: 10.1128/CVI.00456-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brining DL, Mattoon JS, Kercher L, LaCasse RA, Safronetz D, Feldmann H, Parnell MJ. 2010. Thoracic radiography as a refinement methodology for the study of H1N1 influenza in cynomologus macaques (Macaca fascicularis). Comp Med 60:389–395. [PMC free article] [PubMed] [Google Scholar]
- 41.Siemsen DW, Malachowa N, Schepetkin IA, Whitney AR, Kirpotina LN, Lei B, DeLeo FR, Quinn MT. 2014. Neutrophil isolation from nonhuman species. Methods Mol Biol 1124:19–37. doi: 10.1007/978-1-62703-845-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malachowa N, Kobayashi SD, Sturdevant DE, Scott DP, DeLeo FR. 2015. Insights into the Staphylococcus aureus-host interface: global changes in host and pathogen gene expression in a rabbit skin infection model. PLoS One 10:e0117713. doi: 10.1371/journal.pone.0117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ST258 lower respiratory tract infection model in cynomolgus macaques. Body weight (A) and rectal temperature (B) of animals were taken on each exam day. (C) Lung (LW) to body weight (BW) ratio of infected macaques was calculated on scheduled necropsy days. Open and closed symbols indicate animals infected with 108 (Kp1 to Kp3) and 1010 (Kp4 to Kp6) CFU of K. pneumoniae strain ST258, respectively. Download FIG S1, TIF file, 0.5 MB (541.1KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Multiplex immunoassay analysis of selected immunomolecules in plasma samples collected from cynomolgus macaques infected with 108 (open bars) or 1010 (filled bars) CFU of K. pneumoniae strain ST258. Analysis was performed using a Multi-Analyte Profiling (MAP) technology platform (Human Inflammation Map v.1.0; Myriad RBM, Inc.). n, number of animals per group. Download FIG S2, PDF file, 0.8 MB (836KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Hematology and blood chemistry analysis of blood samples collected on exam days from cynomolgus macaques infected with 108 (open bars) or 1010 (filled bars) CFU of K. pneumoniae strain ST258. n, number of animals per group. Download FIG S3, TIF file, 2.8 MB (2.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representative images of healthy lung tissue from cynomolgus macaques. Download FIG S4, JPG file, 1.5 MB (1.6MB, jpg) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Percent of each lung lobe with disease as assessed by gross pathology. Download Table S1, DOCX file, 0.01 MB (15.5KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Percent of each lung lobe with disease as assessed by histopathology. Download Table S2, DOCX file, 0.02 MB (17KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Histopathological changes in tissue samples obtained on scheduled necropsy days. To evaluate the degree of change for each described feature the following scale was used: 0 - no lesion/change; 1 - minimal change; 2 - mild change; 3 - moderate change; 4 - marked change; and 5 - severe change. Download Table S3, PDF file, 0.1 MB (109.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gene transcription changes in lung tissue samples of CPS2-vaccinated macaques compared to sham vaccinated, as assessed by RNA-seq. Red box indicates statistically significant upregulation (>2-fold), where green box indicates statistically significant downregulation (>2-fold) of the transcript. Download Table S4, PDF file, 2.3 MB (2.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Physiological parameters of CPS2-vaccinated or sham-vaccinated animals. Body weight and rectal temperature of each animal were taken on each exam day. (A) Data show the mean ± standard deviation percentage change in body weight of the CPS2-vaccinated or sham-vaccinated animals compared body weight before vaccination (day −63). (B) Data indicate the mean body temperature ± standard deviation. (C) Lung (LW) to body weight (BW) ratio was calculated on each scheduled necropsy day. Download FIG S5, TIF file, 0.5 MB (510.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Hematology and blood chemistry analysis of blood samples collected on exam days from cynomolgus macaques vaccinated with CPS2-vaccinated or sham-vaccinated and infected with 108 CFU of ST258 on day 0. Download FIG S6, TIF file, 2.2 MB (2.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
RNA-Seq data are MINSEQE compliant and have been posted online at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137135 under series number GSE137135.
Hematology and blood chemistry analysis of blood samples collected on exam days from cynomolgus macaques vaccinated with CPS2-vaccinated or sham-vaccinated and infected with 108 CFU of ST258 on day 0. Download FIG S6, TIF file, 2.2 MB (2.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.