Abstract
Background
Symbiotic bacteria contribute to a multitude of important biological functions such as nutrition and reproduction and affect multiple physiological factors like fitness and longevity in their insect hosts. The melon fly, Zeugodacus cucurbitae (Coquillett), is an important agricultural pest that affects a variety of cultivated plants belonging mostly to the Cucurbitaceae family. It is considered invasive and widespread in many parts of the world. Several approaches are currently being considered for the management of its populations including the environmentally friendly and effective sterile insect technique (SIT), as a component of an integrated pest management (IPM) strategy. In the present study, we examined the effect of diet and radiation on the bacterial symbiome of Z. cucurbitae flies with the use of Next Generation Sequencing technologies.
Results
Melon flies were reared on two diets at the larval stage, an artificial bran-based diet and on sweet gourd, which affected significantly the development of the bacterial profiles. Significant differentiation was also observed based on gender. The effect of radiation was mostly diet dependent, with irradiated melon flies reared on the bran diet exhibiting a significant reduction in species diversity and richness compared to their non-irradiated controls. Changes in the bacterial symbiome of the irradiated melon flies included a drastic reduction in the number of sequences affiliated with members of Citrobacter, Raoultella, and Enterobacteriaceae. At the same time, an increase was observed for members of Enterobacter, Providencia and Morganella. Interestingly, the irradiated male melon flies reared on sweet gourd showed a clear differentiation compared to their non-irradiated controls, namely a significant reduction in species richness and minor differences in the relative abundance for members of Enterobacter and Providencia.
Conclusions
The two diets in conjunction with the irradiation affected significantly the formation of the bacterial symbiome. Melon flies reared on the bran-based artificial diet displayed significant changes in the bacterial symbiome upon irradiation, in all aspects, including species richness, diversity and composition. When reared on sweet gourd, significant changes occurred to male samples due to radiation, only in terms of species richness.
Keywords: SIT, Melon fly, 16S rRNA gene, Next generation sequencing, NGS
Background
Insects harbour a variety of microbes, which are associated mostly with their reproductive and digestive tissues and range from obligate symbionts, which are crucial for the normal function of the host, to facultative symbionts, whose presence is not essential for the host [1, 2]. Depending on the type of interaction, symbionts can be further distinguished into commensals or parasites, which have either neutral or negative impact on hosts, or mutualistic which contribute to important aspects of host biology [1]. Symbiotic bacteria, especially those of the reproductive and digestive tissue, influence a variety of important physiological properties of their insect hosts. Most importantly, they provide essential nutrients, including amino acids, vitamins, carbon and nitrogen compounds, promoting development and improving host fitness, which ultimately could result in increased resistance to parasites, pathogens, pesticides and heat stress. Other aspects of host physiology include speciation, through the coevolution with their hosts, communication and reproduction, by causing embryo mortality by means of cytoplasmic incompatibility (CI), shift in the sex ratio of progeny in favour of females through feminization, parthenogenesis and male killing or increased fecundity [1, 3–19].
The genus Zeugodacus (Hendel) contains approximately 192 species [20] that are mostly distributed in regions of Asia and Oceania with a few of them occurring in eastern China and Japan. Zeugodacus (Bactrocera) cucurbitae (Coquillett) is considered an invasive species due to its introduction in many regions of Africa (East and West), in islands of the Indian Ocean and Hawaii [20–24]. The melon fly is considered an important agricultural pest affecting a variety of cultivated fruit and vegetable plants. It is mainly polyphagous, but oligophagous populations have been found in Thailand, Malaysia and France (Reunion Island, Indian Ocean) [25–28]. Its hosts were initially estimated at 81 species [23] but their number was later reduced to 45 well-recorded species that belong to 9 different families, most of them members of the Cucurbitaceae family [20].
The sterile insect technique (SIT) is an important component of an IPM strategy and is based on the release of sterile male individuals that mate with females from wild populations thus reducing the chances of producing offspring, ultimately leading to the suppression or the eradication of the target local population [29, 30]. In order to control insect pests using SIT, the production of large numbers of high quality sterile adult males is needed [29]. However, mass rearing, irradiation, handling and transport may affect the quality of the mass produced sterile insects, and this may be associated with their impact on symbiotic bacterial communities [4, 31, 32]. At the same time, insect gut bacteria can be exploited as a means of enhancing fitness and mating competitiveness of mass reared male insects [33–38].
It is therefore important to expand our knowledge of the impact of diet and radiation on the symbiotic bacterial communities of insect pest species targeted with SIT, in order to enhance its application. To that end, we used Next Generation Sequencing (NGS) technologies to examine the structure of the bacterial symbiome of irradiated and non-irradiated adult Z. cucurbitae flies that were reared on two distinct diets, one artificial and one based on a natural host.
Results
Bacterial community composition and diversity of Z. cucurbitae laboratory populations, kept on an artificial larval diet based on wheat bran and on a natural host (sweet gourd) and treated with irradiation at a 50 Gy dose, were investigated by 16S rRNA gene amplicon sequencing. In total 220,955 reads after quality filtering were used for analysis (Table 1), providing high coverage (97–98%) of the existing bacterial diversity based on the Good’s coverage index (Table 2).
Table 1.
Summary of the samples that were analyzed. The samples include irradiated male and female individuals that were raised on bran or sweet gourd and their respective non-irradiated controls
| No. | ID | Gender | Age | Fertility | Diet | No. of reads |
|---|---|---|---|---|---|---|
| 1 | F_IR_BR | Female | 15 days old | Irradiated (50 Gy) | Bran | 27,391 |
| 2 | M_IR_BR | Male | 15 days old | Irradiated (50 Gy) | Bran | 26,519 |
| 3 | F_NIR_BR | Female | 15 days old | Non-irradiated | Bran | 27,751 |
| 4 | M_NIR_BR | Male | 15 days old | Non-irradiated | Bran | 25,589 |
| 5 | F_IR_SG | Female | 15 days old | Irradiated (50 Gy) | Sweet gourd | 29,512 |
| 6 | M_IR_SG | Male | 15 days old | Irradiated (50 Gy) | Sweet gourd | 25,085 |
| 7 | F_NIR_SG | Female | 15 days old | Non-irradiated | Sweet gourd | 32,638 |
| 8 | M_NIR_SG | Male | 15 days old | Non-irradiated | Sweet gourd | 26,470 |
| Total | 220,955 |
Table 2.
Species richness and diversity were estimated with the use of four indices, two in each case. The high values of the Good’s coverage index show good representation of the existing diversity in each sample. For each index, the value of the standard error is also shown
| Species richness indices | Species diversity indices | |||||
|---|---|---|---|---|---|---|
| No. | ID | Good’s coverage | Chao1 | Ace | Shannon | Simpson reciprocal |
| 1 | F_IR_BR | 0.98 | 406.27 ± 9.74a | 453.35 ± 7.74a | 2.95 ± 0.00a | 3.60 ± 0.01a |
| 2 | M_IR_BR | 0.98 | 333.04 ± 4.63b | 366.96 ± 5.54b | 3.31 ± 0.01b | 5.71 ± 0.02b |
| 3 | F_NIR_BR | 0.97 | 583.58 ± 12.45c | 594.74 ± 8.81c | 4.55 ± 0.00c | 12.38 ± 0.02c |
| 4 | M_NIR_BR | 0.98 | 471.42 ± 7.25d | 488.04 ± 6.94a | 4.41 ± 0.00d | 11.07 ± 0.02d |
| 5 | F_IR_SG | 0.97 | 622.23 ± 13.95c | 747.99 ± 13.92d | 4.55 ± 0.01c | 10.62 ± 0.02e |
| 6 | M_IR_SG | 0.97 | 522.74 ± 8.63e | 555.49 ± 5.04e | 4.90 ± 0.00e | 11.57 ± 0.04f |
| 7 | F_NIR_SG | 0.98 | 448.09 ± 13.97d | 483.93 ± 11.6a | 4.00 ± 0.00f | 6.91 ± 0.02g |
| 8 | M_NIR_SG | 0.97 | 611.78 ± 9.95c | 666.66 ± 11.69f | 4.90 ± 0.00e | 12.01 ± 0.04h |
For each diversity index, ANOVAs followed by the Tukey HSD test, (P value < 0.05 ). Significant differences are indicated by different letters
Diet and / or irradiation dependent changes
Diet and / or irradiation-dependent changes in bacterial diversity and composition were observed. Non-irradiated samples reared on the artificial bran-based diet (NIR_BR) exhibited similar species diversity and richness compared to non-irradiated controls reared on sweet gourd (NIR_SG) (t-test; df: 18; p < 1; Fig. 1). Interestingly, the non-irradiated samples kept on the bran-based diet (NIR_BR), exhibited statistically higher diversity and richness than the irradiated samples (IR_BR) (t-test; df:18; p < 0.027; Fig. 1). The non-irradiated flies that were reared on sweet gourd (NIR_SG) displayed similar richness compared to their irradiated counterparts (IR_SG) (t-test; p < 0.3; Fig. 1). Also, the irradiated samples reared on bran (IR_BR) were characterized by both lower richness and diversity compared to irradiated samples reared on sweet gourd (IR_SG) (t-test; df:18; p < 0.027; Fig. 1).
Fig. 1.
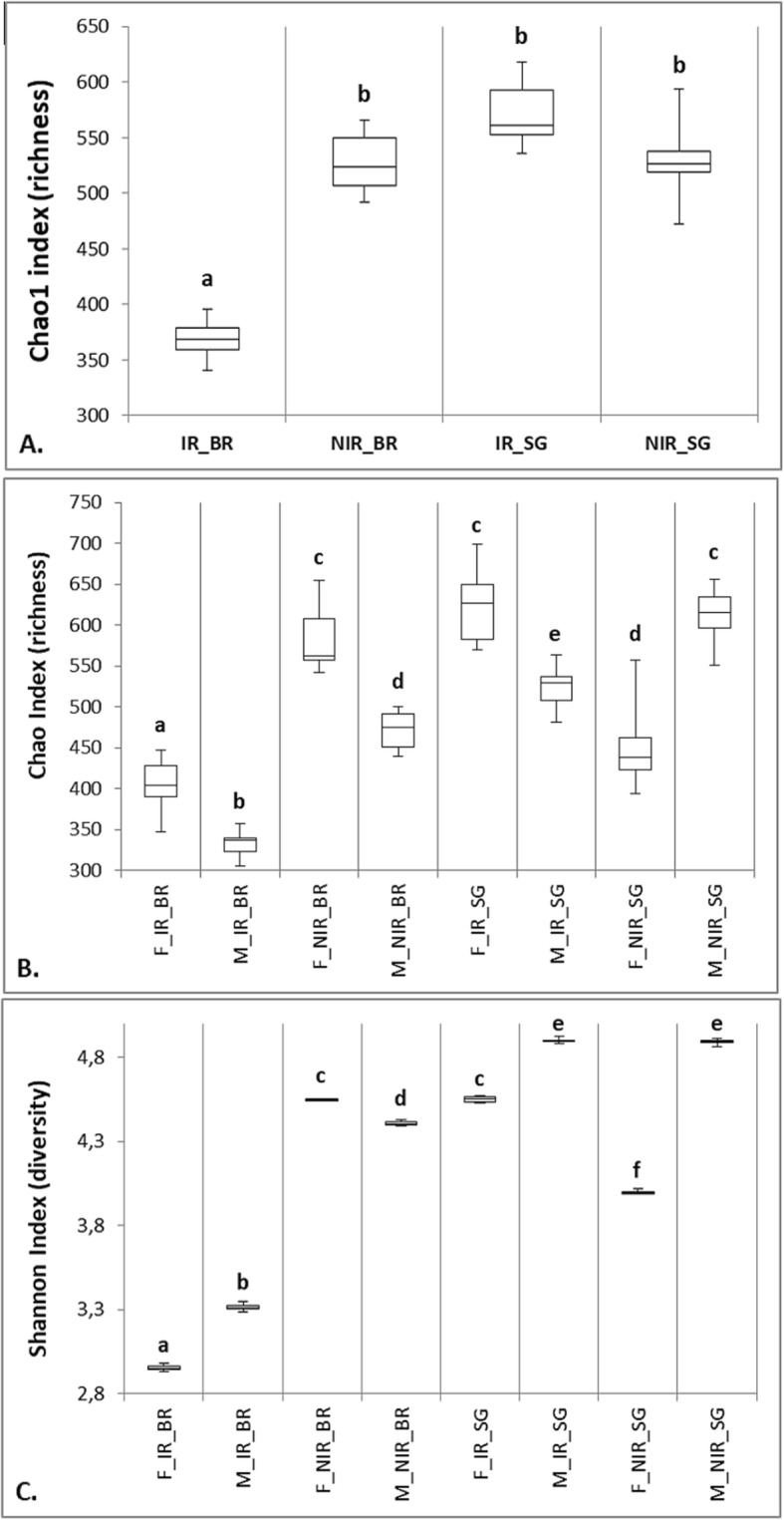
Species richness and diversity indices based on the Chao1 and Shannon indices. a Species richness of Z. cucurbitae samples reared on bran-diet, irradiated (IR_BR) and non-irradiated (NIR_BR), and samples reared on sweet gourd, irradiated (IR_SG) and non-irradiated (NIR_SG). b Species richness of female (F) and male (M) samples reared on bran-diet (BR) and sweet gourd (SG) before (NIR) and after irradiation (IR). c Diversity index of female (F) and male (M) samples reared on bran-diet (BR) and sweet gourd (SG) before (NIR) and after irradiation (IR). Boxes denote the interquartile range, the line within the box is the median, and whiskers extend to the most extreme values
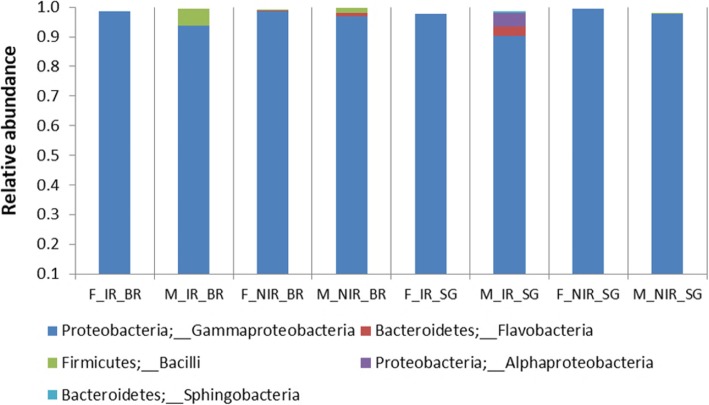
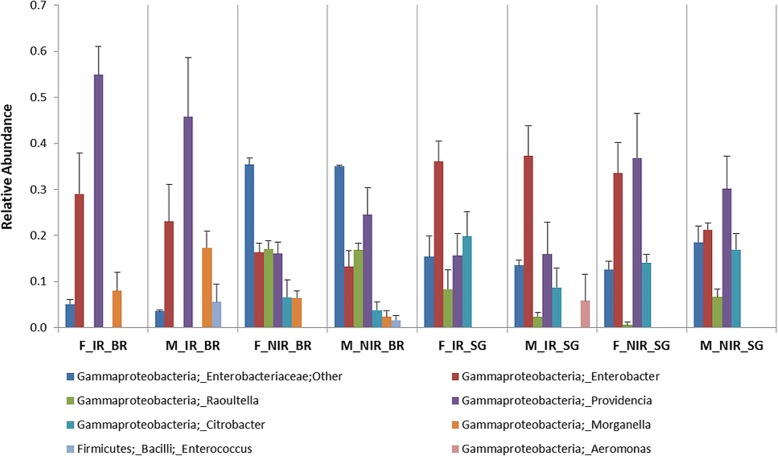
The bacterial OTU composition appeared to be relatively uniform in all samples at higher taxonomic levels. The most dominant phylum was Proteobacteria exhibiting high relative abundance (94–100%), with Gammaproteobacteria being the most prevalent class (90–100%) followed by Alphaproteobacteria (4%). In some samples, Firmicutes (2–6%) and Bacteroidetes (1–4%) were also detected but to a lesser degree, with Firmicutes represented by only one class, Bacilli (2–6%), and Bacteroidetes by two, Flavobacteria (1–3%) and Sphingobacteria (1%; Fig. 2). The non-irradiated controls of the two diets displayed differences in bacterial composition. Non-irradiated samples reared on wheat bran (NIR_BR) showed higher relative abundance in sequences assigned to Raoultella and other Enterobacteriaceae and lower in Enterobacter, Providencia and Citrobacter sequences than samples reared on sweet gourd (NIR_SG), which were also characterized by the absence of Morganella (Fig. 3). The irradiated melon flies reared on wheat bran (IR_BR) exhibited significant changes in the bacterial composition from their respective non-irradiated controls (NIR_BR). These changes include a substantial increase in the relative abundance of Providencia, Enterobacter and Morganella assigned sequences (Kruskal-Wallis; p < 0.001; p < 0.02; p < 0.03), and a significant decrease in the relative abundance of members of the Enterobacteriaceae family (Kruskal-Wallis; p < 0.001) and a complete absence of sequences affiliated to Citrobacter or Raoultella (Fig. 3). On the other hand, the irradiated melon flies reared on sweet gourd (IR_SG) did not exhibit significant differences in bacterial composition from their non-irradiated controls (NIR_SG) (Fig. 3), except for a decline in the relative abundance of sequences assigned to Providencia (Fig. 3).
Fig. 2.
Relative abundance of the most dominant phyla of all samples examined
Fig. 3.
Relative abundance of the most dominant taxa among all samples examined
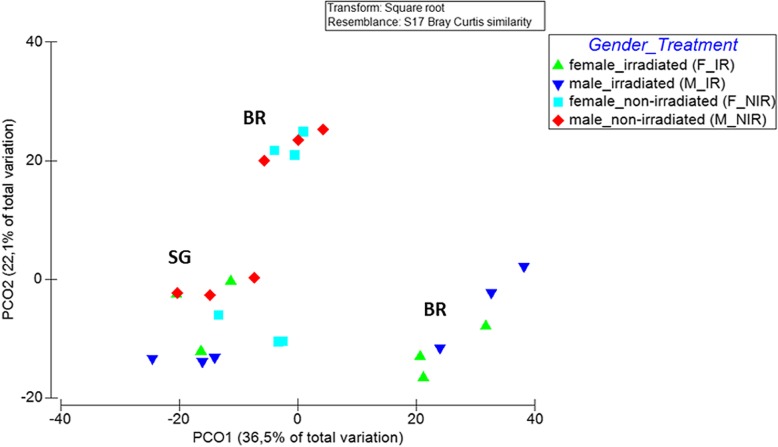
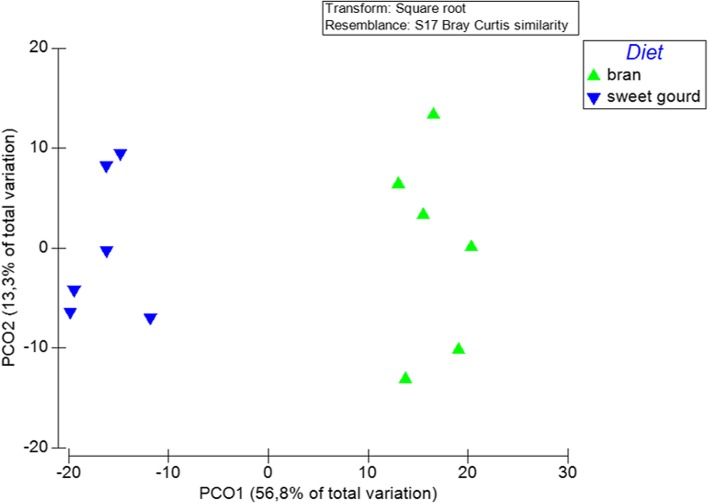
With respect to beta-diversity, bacterial communities were strongly clustered according to the larval diet and the irradiation treatment (PERMANOVA; p < 0.001; Table 3). PCoA revealed the formation of distinct clusters between the two larval diets used, wheat bran and sweet gourd (PERMANOVA; p < 0.001; Figs. 4 and 5). Furthermore, bacterial communities of samples raised on artificial wheat bran were strongly clustered according to their irradiation status, with the PCoA plot explaining 58.6% of the existing variance (Fig. 4). On the other hand, Z. cucurbitae flies that were reared on sweet gourd grouped together regardless of their irradiation status (PERMANOVA; p < 0.1; Fig. 4).
Table 3.
PERMANOVA table of results for all three factors and their combinations. Statistically significant differences (p<0.05) can be seen in bold letters in all three factors separately and in the combination of treatment and diet
| Source | df | SS | MS | Pseudo-F | P(perm) | Unique perms | (MC) |
|---|---|---|---|---|---|---|---|
| Treatment | 1 | 3109.7 | 3109.7 | 8.174 | 0.001 | 999 | 0.001 |
| Diet | 1 | 5904.4 | 5904.4 | 15.52 | 0.001 | 998 | 0.001 |
| Gender | 1 | 998.91 | 998.91 | 2.6257 | 0.03 | 999 | 0.031 |
| Treatment x Diet | 1 | 3598.6 | 3598.6 | 9.4593 | 0.001 | 997 | 0.001 |
| Treatment x Gender | 1 | 481.13 | 481.13 | 1.2647 | 0.236 | 998 | 0.273 |
| Diet x Gender | 1 | 613.49 | 613.49 | 1.6126 | 0.149 | 997 | 0.161 |
| Treat x Diet x Gender | 1 | 596.73 | 596.73 | 1.5686 | 0.169 | 998 | 0.174 |
| Res | 16 | 6087 | 380.43 | ||||
| Total | 23 | 21,390 |
Fig. 4.
Principal Coordinates Analysis (PCoA) of bacterial communities based on relative abundances of OTUs with originations from males, females, irradiated, non-irradiated, bran-based and sweet gourd samples. (BR = bran; SG = sweet gourd)
Fig. 5.
Principal Coordinates Analysis (PCoA) of bacterial communities based on relative abundances of OTUs with originations from non-irradiated samples (PERMANOVA; p < 0.002)
Gender dependent changes in diversity and composition
Gender dependent differences in species diversity and composition were observed between samples. In general, female exhibited higher richness, except for the non-irradiated on sweet gourd (NIR_SG) (Fig. 1b), whereas diversity was larger in males than females, except for the non-irradiated on the bran diet (NIR_BR) (Fig. 1c). In more detail, the non-irradiated females reared on the bran diet (F_NIR_BR) exhibited higher species richness and diversity indices than the males (M_NIR_BR) (t-test; p < 0.001; Fig. 1b and c). On the other hand, samples reared on sweet gourd exhibited the opposite pattern with the non-irradiated males (M_NIR_SG), showing higher species richness and diversity indices than the females (F_NIR_SG) (t-test; p < 0.001; Fig. 1b and c). Interestingly, the bran-based male and female irradiated samples exhibited lower species richness and diversity indices than the non-irradiated males and females (t-test; df: 18; p < 0.027; Fig. 1b and c). For the samples reared on sweet gourd, the irradiated males (M_IR_SG) exhibit lower species richness (t-test;df: 18; p < 0.027) but equal diversity indices (t-test; df: 18; p < 0.9) when compared to the non-irradiated (M_NIR_SG) (Fig. 1b and c). Interestingly, the female irradiated samples kept on sweet gourd (F_IR_SG) exhibited a higher species richness and diversity when compared to the non-irradiated (F_NIR_SG) (t-test; df: 18; p < 0.027; Fig. 1b and c).
The bran-based non-irradiated female samples (F_NIR_BR) show a higher relative abundance of sequences affiliated to Citrobacter and Morganella and a lower relative abundance with sequences affiliated to Providencia when compared with the bran-based non-irradiated male samples (M_NIR_BR). For the irradiated samples we observed the presence of sequences affiliated with Enterococcus in the male samples (M_IR_BR), coupled with an increase in the relative abundance of Morganella sequences (Fig. 3). Interestingly, in male and female irradiated samples reared on bran, sequences affiliated to Citrobacter and Raoutella were not detected while members of Enterobacter and Providencia tended to increase, but with sequences assigned to Enterobacteriaceae decreasing when compared to male and female non-irradiated samples (Fig. 3). For the sweet gourd, we observed a decrease in the sequences affiliated with Raoutella in the female non-irradiated samples (F_NIR_SG) when compared with the male samples (M_NIR_SG). Interestingly, in the female irradiated samples a decrease in the relative abundance of Providencia affiliated sequences was observed coupled with an increase in the members of Raoutella when compared to the female non-irradiated samples (F_NIR_SG) (Fig. 3). For the male irradiated samples, a decrease was observed with sequences assigned to Providencia, Citrobacter, Raoutella, while an increase was observed in the relative abundance of Enterobacter and Aeromonas (Fig. 3).
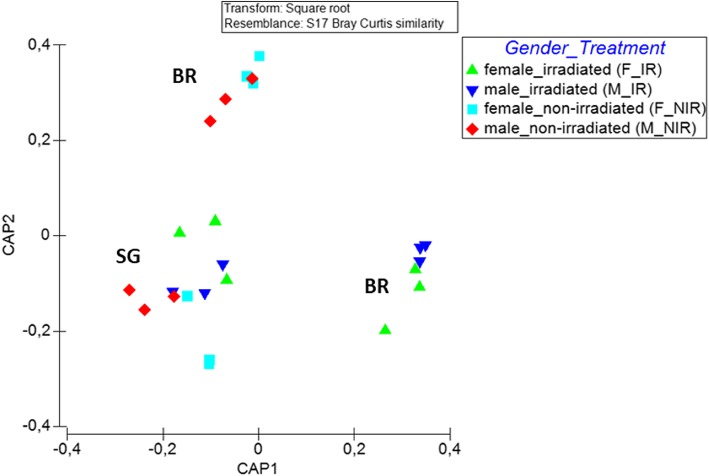
As suggested by the aforementioned differences in diversity and composition, bacterial communities seem to be affected by the sex of the melon flies (PERMANOVA; p < 0.03; Fig. 6), with non-irradiated female and male controls (NIR_F and NIR_M) forming separate but closely related clusters depending on their diet (Fig. 6). Further clustering was observed when the irradiation treatment was taken into account (PERMANOVA; p < 0.03; Fig. 6).
Fig. 6.
Canonical analysis of principal coordinates (CAP) ordinations of bacterial communities based on relative abundances of bacterial OTUs from Z. cucurbitae samples examined, based on different diets (sweet gourd (SG) and wheat bran (BR)), irradiation treatment (non-irradiated (NIR) and irradiated (IR)) and gender (male (M) and female (F)). The constrained ordinations show maximized differences among the two diets and the irradiated against the non-irradiated samples (traceQ_m’HQ_m statistics (2.52837; p < 0.001))
Discussion
The present study examined the effect of larval diet, gender and irradiation on the symbiotic bacterial communities of Z. cucurbitae laboratory flies. Our results indicate that the application of irradiation to two distinct larval diets leads to the formation of different bacterial profiles. Different bacterial profiles were also observed between samples belonging to different genders. Furthermore, statistically significant differences were observed after irradiation treatment in the two diets examined and between genders. Irradiation had a detrimental effect on the formation of bacterial communities especially in samples reared on bran whereas irradiated flies reared on sweet gourd showed mixed responses but overall managed to retain higher levels of richness and diversity, with small changes in OTU composition.
Differences in insect microbiota due to the application of distinct diets have been previously described [31, 39–47]. These studies focused on the comparison of symbiotic communities between wild populations and laboratory strains that were reared on various artificial diets or between lab populations reared on different artificial diets. In the case of wild populations of diverse Drosophila species, analysis of samples of fruit-feeding flies and samples of flower-feeders revealed the development of significantly different bacterial microbiomes between the two diets [41]. Variability among D. suzukii samples of different developmental stages, reared on fruits and artificial diet was also reported [45]. One of the most important functions of mutualistic symbionts is the provision of nutritional components from their diet to their insect-hosts, by hydrolysis, using specialized enzymes [13]. Different bacterial groups are capable of digesting different nutrients. In this way, as the composition of the symbiotic community changes, so does the ability of hosts to exploit diets with different nutritional properties. Such changes in bacterial communities have been previously described in different developmental stages of Bactrocera flies, with immature and adult flies developing different microbiota, due to different dietary habits and needs [48–50]. In these cases, larvae that mostly require diets rich in carbohydrates are characterized by different microbial communities from adults that are in need of both sugars and proteins. The development of different microbiotas between the non-irradiated samples that were reared on the artificial wheat bran diet and on sweet gourd could be due to the different bacteria that exist in each diet provided, since the majority of symbionts are usually acquired from the environment [51]. It is also possible that prevalence of certain symbionts is favoured by differences in the nutritional components of the two diets that require different types of bacteria in order to be processed.
Our study indicated that Proteobacteria were the dominant phylum in all samples examined, with Gammaproteobacteria being the most abundant class. Sequences belonging to Firmicutes and Bacteroidetes were also identified but in a lesser degree. A recent cultivation-dependent study of the midgut bacterial composition characterized from wild Z. cucurbitae flies displayed a similar picture, with Proteobacteria being the most abundant followed by Firmicutes and Actinobacteria, while Enterobacteriaceae were the most prevalent family [52]. The dominant species were from the genera Enterobacter, Klebsiella, Citrobacter, Bacillus and Providencia. All these genera, except for Bacillus, were also identified in our study showing significant relative abundance with the exception of Klebsiella, which was found only in irradiated samples reared on sweet gourd, with low relative abundance (1–2%). Interestingly, a second cultivation-dependent approach that used gut tissue from a laboratory population indicated that Firmicutes were the most abundant phylum, with species belonging to Bacillus [53]. However, female melon flies were characterized by the presence of Morganella species which were also identified in our work.
Similarly to previous studies in Ceratitis capitata [31, 32, 54], radiation altered the structure of the microbiota of Z. cucurbitae, causing reduction in bacterial symbiont richness and diversity. These alterations appeared to be much stronger in samples reared on the artificial wheat bran diet. In those samples, the relative abundance of genera like Raoultella, Citrobacter and other Enterobacteriaceae decreased, in contrast to members belonging to Providencia, Morganella and Enterobacter. Interestingly, certain strains belonging to Providencia and Morganella are known pathogens to flies or even humans [55–57]. Increase in the relative abundance could be associated with their emancipation into the hemolymph of the irradiated flies since treatment with radiation damages their gut tissue [32], although the degree to which pathogens and compounds enter the irradiated gut, and the existence of any gut dysfunction remains to be determined. Further characterization of these strains would be required to fully decipher their exact role in melon flies. Increased presence of potentially pathogenic strains, belonging to the genus Pseudomonas, was also reported in mass reared Medfly after irradiation [31]. At the same time, members of Enterobacteriaceae like Providencia, Citrobacter and Enterobacter function as attractants for both male and female Z. cucurbitae [52]. Additionally, Citrobacter, Klebsiella and Enterobacter contain species with probiotic properties to insects and arthropods [4, 7, 31, 33–38, 58]. Usually, these probiotic effects include improved fitness, longevity and increased reproductive capabilities for irradiated individuals resulting in increased competitiveness against wild populations [34, 35]. In Z. cucurbitae, enrichment of the larval diet with Enterobacter resulted in improved pupal weight, morphological indices, and adult survival rate [38] as well as increased pupal and adult productivity and faster development, particularly of males, in its closely related species C. capitata [33].
Differences based on gender in the symbiotic bacterial profiles of the non-irradiated controls varied between the two diets, with females reared on bran showing higher richness and diversity compared to males, and the exact opposite trend when reared on sweet gourd. Differences were also observed in OTU composition in both diets. Previously, the gut tissue of wild Bactrocera dorsalis females was described with lower species richness, higher diversity and differences in bacterial composition compared to males [48]. Treatment with irradiation resulted in lower richness and lower or equal diversity in male samples, in both diets, compared to their non-irradiated controls. Reduction in diversity and differences in the composition of the bacterial communities between irradiated and non-irradiated male samples was also described in studies with C. capitata, with irradiated gut samples containing the genera Salmonella, Citrobacter, Providencia, Morganella, Enterobacter, Klebsiella and Pectobacterium [31]. The majority of those genera were also identified in irradiated males in our work, as mentioned previously. On the contrary, female melon flies showed mixed trends after radiation treatment, depending on their diet. Irradiated females reared on bran followed the same trend as both irradiated males while those reared on sweet gourd the exact opposite trend, which resulted in an unusual increase in both richness and diversity compared to the non-irradiated sample. Changes in the symbiotic bacterial communities of male melon flies due to radiation could be of greater importance since males are targets for the application of the SIT. Significant alterations can be seen in irradiated male samples reared on both diets, but those on bran are affected in a larger degree. As mentioned earlier, these effects of radiation to the composition of the symbiotic bacterial communities could be possibly reversed by enriching the diet with nutrients or certain bacteria with probiotic properties.
Conclusions
Diet was found to strongly affect the structure of the microbiota in Z. cucurbitae flies. Significant differentiation in the microbiota was also observed based on the sex of the flies, but to a lesser extent. The effect of radiation was diet-dependent with sweet gourd exhibiting minor changes in the bacterial profile between irradiated and non-irradiated melon flies. A strong effect was observed on flies that were reared on an artificial bran-based diet and was characterized by a decrease in both bacterial richness and diversity, with bacterial genera like Raoultella and Citrobacter being highly reduced while sequences affiliated to members of Providencia, Morganella and Enterobacter were increased. Gender dependent radiation effects were mostly observed in terms of species richness and diversity, with males showing considerable losses and females mixed trends.
Methods
Rearing conditions and sample preparation
Zeugodacus cucurbitae flies used in the present study originated from a population collected from infested sweet gourd (Cucurbita maxima Duchesne) and have been reared for more than 500 generations at IBD, IFRB, AERE (Insect Biotechnology Division, Institute of Food and Radiation Biology, Atomic Energy Research Establishment), Ganak bari, Savar, Dhaka, Bangladesh. Larvae were reared on two distinct diets: (a) an artificial larval diet based on wheat bran which was sterilized at 80 °C for 3 days and is usually used in the artificial rearing of Z. cucurbitae ((wheat bran (26%), sugar (12%), Brewer’s yeast (3.6%), sodium benzoate (0.4%) and water (58%)) for one generation and (b) a natural host, sweet gourd, in order to examine the effect of a natural and artificial diet on the bacterial symbiome of Z. cucurbitae. Pupae were irradiated 24–48 h before adult emergence at 50 Gy, a dose capable of inducing 100% sterility in both males and females [59], with the use of a cobalt-60 gamma radiation source of IFRB. Adult flies were reared on artificial diets containing casein, yeast extract and sugar in a ratio of 1:1:2. Prior to extraction, the insects were surface sterilized.
DNA extraction, PCR amplification and sample purification
DNA extraction was performed following a simplified CTAB protocol [60]. Extracted samples were diluted in sterile deionized water and stored in − 20 °C. Each processed sample consisted of fifteen (n = 15) whole adult flies (15 days old) divided into three replicates of five flies. A fragment of approximately 460 bp belonging to the V3-V4 region of the bacterial 16S rRNA gene was amplified by PCR using the universal primer set U341F-MiSeq 5′-CCTACG GGR SGC AGC AG-3′ and 805R-MiSeq 5′-GA CTA CHV GGG TAT CTA ATC C-3′ [61]. Amplification was performed using KAPA HiFi HotStart PCR Kit (Kapa Biosystems). Each 25 μl reaction contained 5 μl of KAPA HiFi Fidelity Buffer (5X), 0.7 μl of dNTPs solution (10 mM each), 0.7 μl of each primer solution (10 μM), 0.3 μl of KAPA HiFi HotStart DNA Polymerase solution (1 U/μl), 1 μl from the template DNA solution and was finalized with 16.6 μl of sterile deionized water. The PCR protocol was comprised of an initial denaturation step at 95 °C for 3 min, followed by 30 cycles of denaturation at 98 °C for 20 s, annealing at 60 °C for 15 s and extension at 72 °C for 45 s. The reaction was terminated with a final extension step at 72 °C for 1 min. For each set of PCR reactions performed, the appropriate negative and positive controls were also prepared. From each reaction, 5 μl were loaded on a 1.5% agarose gel and separated by electrophoresis. The approximately 550 bp amplification product (size increase due to the incorporation of the 50-mer Illumina primers) was visualized in Bio-Rad’s Gel Doc™ XR+ system. Positive PCR products were purified with a 20% PEG, 2.5 M NaCl solution, centrifuged at 14.000 x g for 20 min and the precipitate was washed twice with 125 μl of a 70% v/v ethanol solution and centrifuged at 14.000 x g for 10 min as previously described [62]. The dried precipitates were suspended in 15 μl of sterile deionized water and the concentration was measured with a Quawell Q5000 micro-volume UV-Vis spectrophotometer.
Indexing PCR and sample purification
The purified PCR products were diluted to a final concentration of 10 ng/μl and submitted to indexing PCR in order to incorporate the Illumina index primers to their sequence. During indexing PCR, each sample was amplified with a unique combination of index primers. Amplification was performed in 50 μl reactions using the KAPA HiFi HotStart PCR Kit. Each reaction contained 10 μl of KAPA HiFi Fidelity Buffer (5X), 1.5 μl of dNTPs solution (10 mM each), 5 μl of the forward index primer (10 μM), 5 μl of the reverse index primer (10 μΜ), 1 μl of KAPA HiFi HotStart DNA Polymerase (1 U/μl), 2 μl from the diluted PCR product (10 ng/μl) and 25.5 μl of sterile deionized water. The PCR program was comprised of an initial denaturation step at 95 °C for 3 min, followed by 8 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s. The reaction was terminated with a final extension step at 72 °C for 5 min. The resulting amplicons were purified using Macherey-Nagel’s NucleoMag® NGS Clean-up and Size Selection kit according to the manufacturer’s recommendations. Purified samples were suspended in 30 μl of sterile deionized water and their concentration was measured with a Quawell Q5000 micro-volume UV-Vis spectrophotometer. All samples were diluted to a final concentration of 8 nM and mixed equimolarly.
Illumina sequencing and data analysis
The library was sequenced on an Illumina MiSeq sequencing platform by IMGM SA. The sequencing results for each sample were retrieved in two FASTq files and were assembled with the use of PANDAseq v2.7 assembler [63]. Chimeric contigs were identified and removed with UCHIME [64] which is contained within the software package Qiime v1.9.1 [65]. Taxonomic assignment and alpha-diversity analysis were also performed with Qiime. Taxonomy was assigned using the SILVA 16S rRNA gene database (release 119) [66]. Species richness was estimated with Chao1 [67] and ACE indices [68] whereas species diversity was calculated with the use of Shannon’s and Simpson’s reciprocal (1/D) indices. Rarefaction analyses were performed starting with 250 sequences/sample as a minimum subsample, 5000 sequences/sample as the maximum, a 250-sequence increment/step and 10 iterations/step. Alpha-diversity comparisons were performed by t-test using Qiime including Holm-Bonferroni sequential correction. Between-sample diversity was calculated using Bray-Curtis similarity [69] on square root transformed data and principal coordinates analysis (PCoA) [70] was performed on the resulting distance matrix. Canonical analysis of principal coordinates (CAP) [71] was based on 999 permutation tests. Statistically significant differences between samples were identified with permutational multivariate analysis of variance (PERMANOVA) [72] using 999 permutations and Monte Carlo tests. Beta-diversity calculations were performed with Primer6+ [73]. OTU comparisons were performed using the non-parametric Kruskal-Wallis Rank Sum test, and the Mann-Whitney test. The obtained significance values were corrected for multiple testing using the Benjamini-Hochberg method [74]. 16S rRNA gene sequences reported in this study have been deposited in NCBI under BioProject number PRJNA487513.
Acknowledgements
Not applicable.
About this supplement
This article has been published as part of BMC Biotechnology Volume 19 Supplement 2, 2019: Proceedings of an FAO/IAEA Coordinated Research Project on Use of Symbiotic Bacteria to Reduce Mass-rearing Costs and Increase Mating Success in Selected Fruit Pests in Support of SIT Application: biotechnology. The full contents of the supplement are available online at https://bmcbiotechnol.biomedcentral.com/articles/supplements/volume-19-supplement-2.
Abbreviations
- CAP
Canonical Analysis of Principal coordinates
- CI
Cytoplasmic Incompatibility
- F_IR_BR
Female Irradiated Bran
- F_IR_SG
Female Irradiated Sweet Gourd
- F_NIR_BR
Female Non-irradiated Bran
- F_NIR_SG
Female Non-irradiated Sweet Gourd
- IPM
Integrated Pest Management
- IR_BR
Irradiated Bran
- IR_SG
Irradiated Sweet Gourd
- M_IR_BR
Male Irradiated Bran
- M_IR_SG
Male Irradiated Sweet Gourd
- M_NIR_BR
Male Non-irradiated Bran
- M_NIR_SG
Male Non-irradiated Sweet Gourd
- NGS
Next Generation Sequencing
- NIR_BR
Non-irradiated Bran
- NIR_F
Non-irradiated Female
- NIR_M
Non-irradiated Male
- NIR_SG
Non-irradiated Sweet Gourd
- OTU
Operational Taxonomic Unit
- PCoA
Principal Coordinates Analysis
- SIT
Sterile Insect Technique
Authors’ contributions
Conceived and designed the study: KB, GT. Conducted the experiments and analyzed the results: EA, MK, SP, GT. Drafted the manuscript: EA, GT. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was partially supported by the International Atomic Energy research contract no. 17011 as part of the Coordinated Research Project “Use of Symbiotic Bacteria to Reduce Mass-Rearing Costs and Increase Mating Success in Selected Fruit Pests in Support of SIT Application” and by intramural funds of the University of Patras to George Tsiamis.
Availability of data and materials
The datasets used and/or analyzed during the current study are available in NCBI.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elias D. Asimakis, Email: eliasasim@upatras.gr
Mahfuza Khan, Email: mahfuza79@gmail.com.
Panagiota Stathopoulou, Email: panayotastathopoulou@gmail.com.
Carlos Caceres, Email: C.Caceres@iaea.org.
Kostas Bourtzis, Email: K.Bourtzis@iaea.org.
George Tsiamis, Email: gtsiamis@upatras.gr.
References
- 1.Bourtzis K, Miller T, editors. Insect symbiosis. Florida: CRC Press, Taylor and Francis Group, LLC; 2003. [Google Scholar]
- 2.Pontes MH, Dale C. Culture and manipulation of insect facultative symbionts. Trends Microbiol. 2006;14:406–412. doi: 10.1016/j.tim.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Aharon Y, Pasternak Z, Ben Yosef M, Behar A, Lauzon C, Yuval B, et al. Phylogenetic, metabolic, and taxonomic diversities shape Mediterranean fruit fly microbiotas during ontogeny. Appl Environ Microbiol. 2013;79:303–313. doi: 10.1128/AEM.02761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behar A, Yuval B, Jurkevitch E. Gut bacterial communities in the Mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity. J Insect Physiol. 2008;54:1377–1383. doi: 10.1016/j.jinsphys.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Yosef Michael, Pasternak Zohar, Jurkevitch Edouard, Yuval Boaz. Symbiotic bacteria enable olive fly larvae to overcome host defences. Royal Society Open Science. 2015;2(7):150170. doi: 10.1098/rsos.150170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Yosef M, Pasternak Z, Jurkevitch E, Yuval B. Symbiotic bacteria enable olive flies (Bactrocera oleae) to exploit intractable sources of nitrogen. J Evol Biol. 2014;27:2695–2705. doi: 10.1111/jeb.12527. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Yosef M, Behar A, Jurkevitch E, Yuval B. Bacteria–diet interactions affect longevity in the medfly –Ceratitis capitata. J Appl Entomol. 2008;132:690–694. doi: 10.1111/j.1439-0418.2008.01330.x. [DOI] [Google Scholar]
- 8.Bourtzis K, Miller T, editors. Insect symbiosis 3. Florida: CRC Press, Taylor and Francis Group, LLC; 2009. [Google Scholar]
- 9.Bourtzis K, Miller T, editors. Insect symbiosis 2. Florida: CRC Press, Taylor and Francis Group, LLC; 2006. [Google Scholar]
- 10.Brownlie JC, Johnson KN. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009;17:348–354. doi: 10.1016/j.tim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Douglas AE. Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol. 2015;60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas AE. The microbial dimension in insect nutritional ecology. Funct Ecol. 2009;23:38–47. doi: 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- 13.Engel P, Moran NA. The gut microbiota of insects – diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 14.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors. 2013;6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montllor CB, Maxmen A, Purcell AH. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27:189–195. doi: 10.1046/j.1365-2311.2002.00393.x. [DOI] [Google Scholar]
- 16.Stouthamer R, JAJ B, GDD H. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- 17.Zchori-Fein E, Bourtzis K, editors. Manipulative tenants: bacteria associated with arthropods. Florida: CRC Press, Taylor and Francis Group, LLC; 2011. [Google Scholar]
- 18.Xie J, Vilchez I, Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS One. 2010;5:e12149. doi: 10.1371/journal.pone.0012149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng D, Guo Z, Riegler M, Xi Z, Liang G, Xu Y. Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel) Microbiome. 2017;5:13. doi: 10.1186/s40168-017-0236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Meyer Marc, Delatte Hélène, Mwatawala Maulid, Quilici Serge, Vayssieres Jean-François, Virgilio Massimiliano. A review of the current knowledge on Zeugodacus cucurbitae (Coquillett) (Diptera, Tephritidae) in Africa, with a list of species included in Zeugodacus. ZooKeys. 2015;540:539–557. doi: 10.3897/zookeys.540.9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drew R. Taxonomy. In: Drew R, Hooper G, Bateman M, editors. Economic fruit flies of the South Pacific region. Second. Department of Primary Industries Publishing, Brisbane: Australia; 1982. pp. 1–97. [Google Scholar]
- 22.Drew R. The tropical fruit flies (Diptera: Tephritidae: Dacinae) of the Australasian and Oceanian regions. Mem Qld Mus. 1989;26:521. [Google Scholar]
- 23.Dhillon MK, Singh R, Naresh JS, Sharma HC. The melon fruit fly, Bactrocera cucurbitae: a review of its biology and management. J Insect Sci. 2005;5:40. [DOI] [PMC free article] [PubMed]
- 24.Drew R, Romig MC. Tropical fruit flies (Tephritidae Dacinae) of south-East Asia: Indomalaya to north-West Australasia: Wallingford CABI; 2013.
- 25.Clarke AR, Allwood A, Chinajariyawong A, Drew RAI, Hengsawad C, Jirasurat M, et al. Seasonal abundance and host use patterns of seven Bactrocera Macquart species (Diptera: Tephritidae) in Thailand and peninsular Malaysia. Raffles Bull Zool. 2001;49:207–220. [Google Scholar]
- 26.Vayssières J, Rey J-Y, Traoré L. Distribution and host plants of Bactrocera cucurbitae in west and Central Africa. Fruits. 2007;62:391–396. doi: 10.1051/fruits:2007037. [DOI] [Google Scholar]
- 27.Vayssières J, Carel Y, Coubes M, Duyck PF. Development of immature stages and comparative demography of two cucurbit-attacking fruit flies in Reunion island: Bactrocera cucurbitae and Dacus ciliatus (Diptera Tephritidae) Environ Entomol. 2008;37:307–314. doi: 10.1093/ee/37.2.307. [DOI] [PubMed] [Google Scholar]
- 28.Hafsi A, Facon B, Ravigné V, Chiroleu F, Quilici S, Chermiti B, et al. Host plant range of a fruit fly community (Diptera: Tephritidae): does fruit composition influence larval performance? BMC Ecol. 2016;16:40. doi: 10.1186/s12898-016-0094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyck V, Hendrichs J. Robinson a, editors. Springer Netherlands: Sterile insect technique - principles and practice in area-wide; 2005. [Google Scholar]
- 30.Lees RS, Gilles JR, Hendrichs J, Vreysen MJ, Bourtzis K. Back to the future: the sterile insect technique against mosquito disease vectors. Curr Opin Insect Sci. 2015;10(Supplement C):156–162. doi: 10.1016/j.cois.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Ben Ami E, Yuval B, Jurkevitch E. Manipulation of the microbiota of mass-reared Mediterranean fruit flies Ceratitis capitata (Diptera: Tephritidae) improves sterile male sexual performance. ISME J. 2010;4:28–37. doi: 10.1038/ismej.2009.82. [DOI] [PubMed] [Google Scholar]
- 32.Lauzon CR, Potter SE. Description of the irradiated and nonirradiated midgut of Ceratitis capitata Wiedemann (Diptera: Tephritidae) and Anastrepha ludens Loew (Diptera: Tephritidae) used for sterile insect technique. J Pest Sci. 2012;85:217–226. doi: 10.1007/s10340-011-0410-1. [DOI] [Google Scholar]
- 33.Augustinos Antonios A., Kyritsis Georgios A., Papadopoulos Nikos T., Abd-Alla Adly M. M., Cáceres Carlos, Bourtzis Kostas. Exploitation of the Medfly Gut Microbiota for the Enhancement of Sterile Insect Technique: Use of Enterobacter sp. in Larval Diet-Based Probiotic Applications. PLOS ONE. 2015;10(9):e0136459. doi: 10.1371/journal.pone.0136459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavriel S, Jurkevitch E, Gazit Y, Yuval B. Bacterially enriched diet improves sexual performance of sterile male Mediterranean fruit flies. J Appl Entomol. 2011;135:564–573. doi: 10.1111/j.1439-0418.2010.01605.x. [DOI] [Google Scholar]
- 35.Hamden H, Guerfali MM, Fadhl S, Saidi M, Chevrier C. Fitness improvement of mass-reared sterile males of Ceratitis capitata (Vienna 8 strain) (Diptera: Tephritidae) after gut enrichment with probiotics. J Econ Entomol. 2013;106:641–647. doi: 10.1603/EC12362. [DOI] [PubMed] [Google Scholar]
- 36.Kyritsis GA, Augustinos AA, Cáceres C, Bourtzis K. Medfly gut microbiota and enhancement of the sterile insect technique: Similarities and differences of Klebsiella oxytoca and Enterobacter sp. AA26 probiotics during the larval and adult stages of the VIENNA 8D53+ genetic sexing strain. Front Microbiol. 2017;8:2064. [DOI] [PMC free article] [PubMed]
- 37.Niyazi N, Lauzon CR, Shelly TE. Effect of probiotic adult diets on fitness components of sterile male Mediterranean fruit flies (Diptera: Tephritidae) under laboratory and field cage conditions. J Econ Entomol. 2004;97:1570–1580. doi: 10.1603/0022-0493-97.5.1570. [DOI] [PubMed] [Google Scholar]
- 38.Yao M, Zhang H, Cai P, Gu X, Wang D, Ji Q. Enhanced fitness of a Bactrocera cucurbitae genetic sexing strain based on the addition of gut-isolated probiotics (Enterobacter spec.) to the larval diet. Entomol Exp Appl. 2017;162:197–203. doi: 10.1111/eea.12529. [DOI] [Google Scholar]
- 39.Ben-Yosef M, Aharon Y, Jurkevitch E, Yuval B. Give us the tools and we will do the job: symbiotic bacteria affect olive fly fitness in a diet-dependent fashion. Proc R Soc B-Biol Sci. 2010;277:1545–1552. doi: 10.1098/rspb.2009.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bing X, Gerlach J, Loeb G, Buchon N. Nutrient-dependent impact of microbes on Drosophila suzukii Development. mBio. 2018;9:e02199–e02117. doi: 10.1128/mBio.02199-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 2011;7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kane MD, Breznak JA. Effect of host diet on production of organic acids and methane by cockroach gut bacteria. Appl Environ Microbiol. 1991;57:2628–2634. doi: 10.1128/aem.57.9.2628-2634.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrow JL, Frommer M, Shearman DCA, Riegler M. The microbiome of field-caught and laboratory-adapted Australian Tephritid fruit fly species with different host plant use and specialisation. Microb Ecol. 2015;70:498–508. doi: 10.1007/s00248-015-0571-1. [DOI] [PubMed] [Google Scholar]
- 44.Santo Domingo JW, Kaufman MG, Klug MJ, Holben WE, Harris D, Tiedje JM. Influence of diet on the structure and function of the bacterial hindgut community of crickets. Mol Ecol. 1998;7:761–767. doi: 10.1046/j.1365-294x.1998.00390.x. [DOI] [Google Scholar]
- 45.Vacchini V, Gonella E, Crotti E, Prosdocimi EM, Mazzetto F, Chouaia B, et al. Bacterial diversity shift determined by different diets in the gut of the spotted wing fly Drosophila suzukii is primarily reflected on acetic acid bacteria. Environ Microbiol Rep. 2017;9:91–103. doi: 10.1111/1758-2229.12505. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Jin L, Zhang H. Comparison of the diversity of the bacterial communities in the intestinal tract of adult Bactrocera dorsalis from three different populations. J Appl Microbiol. 2011;110:1390–1401. doi: 10.1111/j.1365-2672.2011.05001.x. [DOI] [PubMed] [Google Scholar]
- 47.Wong AC-N, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013;7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andongma AA, Wan L, Dong Y-C, Li P, Desneux N, White JA, et al. Pyrosequencing reveals a shift in symbiotic bacteria populations across life stages of Bactrocera dorsalis. Sci Rep. 2015;5:9470. doi: 10.1038/srep09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yong H-S, Song S-L, Chua K-O, Lim P-E. High diversity of bacterial communities in developmental stages of Bactrocera carambolae (Insecta: Tephritidae) revealed by Illumina MiSeq sequencing of 16S rRNA gene. Curr Microbiol. 2017;74:1076–1082. doi: 10.1007/s00284-017-1287-x. [DOI] [PubMed] [Google Scholar]
- 50.Zhao X, Zhang X, Chen Z, Wang Z, Lu Y, Cheng D. The divergence in bacterial components associated with Bactrocera dorsalis across developmental stages. Front Microbiol. 2018;9:114. [DOI] [PMC free article] [PubMed]
- 51.Behar A, Jurkevitch E, Yuval B. Bringing back the fruit into fruit fly–bacteria interactions. Mol Ecol. 2008;17:1375–1386. doi: 10.1111/j.1365-294X.2008.03674.x. [DOI] [PubMed] [Google Scholar]
- 52.Hadapad AB, Prabhakar CS, Chandekar SC, Tripathi J, Hire RS. Diversity of bacterial communities in the midgut of Bactrocera cucurbitae (Diptera: Tephritidae) populations and their potential use as attractants. Pest Manag Sci. 2016;72:1222–1230. doi: 10.1002/ps.4102. [DOI] [PubMed] [Google Scholar]
- 53.Gujjar NR, Govindan S, Verghese A, Subramaniam S, More R. Diversity of the cultivable gut bacterial communities associated with the fruit flies Bactrocera dorsalis and Bactrocera cucurbitae (Diptera: Tephritidae). Phytoparasitica. 2017;45:453–60.
- 54.Yuval B, Ben-Ami E, Behar A, Ben-Yosef M, Jurkevitch E. The Mediterranean fruit fly and its bacteria – potential for improving sterile insect technique operations. J Appl Entomol. 2013;137:39–42. doi: 10.1111/j.1439-0418.2010.01555.x. [DOI] [Google Scholar]
- 55.Förster M, Klimpel S, Mehlhorn H, Sievert K, Messler S, Pfeffer K. Pilot study on synanthropic flies (e.g. Musca, Sarcophaga, Calliphora, Fannia, Lucilia, Stomoxys) as vectors of pathogenic microorganisms. Parasitol Res. 2007;101:243–246. doi: 10.1007/s00436-007-0522-y. [DOI] [PubMed] [Google Scholar]
- 56.Gupta AK, Nayduch D, Verma P, Shah B, Ghate HV, Patole MS, et al. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.) FEMS Microbiol Ecol. 2012;79:581–593. doi: 10.1111/j.1574-6941.2011.01248.x. [DOI] [PubMed] [Google Scholar]
- 57.Khalil S, Jacobson E, Chambers MC, Lazzaro BP. Systemic bacterial infection and immune defense phenotypes in Drosophila melanogaster. JoVE J Vis Exp. 2015;99:e52613–3. [DOI] [PMC free article] [PubMed]
- 58.Gavriel S, Gazit Y, Yuval B. Effect of diet on survival, in the laboratory and the field, of sterile male Mediterranean fruit flies. Entomol Exp Appl. 2010;135:96–104. doi: 10.1111/j.1570-7458.2010.00972.x. [DOI] [Google Scholar]
- 59.Nahar G, Howlader AJ, Rahman R. Radiation sterilization and mating competitiveness of melon fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae) male in relation to sterile insect release method. Pak J Biol Sci. 2006;9:2478–2482. doi: 10.3923/pjbs.2006.2478.2482. [DOI] [Google Scholar]
- 60.Augustinos AA, Santos-Garcia D, Dionyssopoulou E, Moreira M, Papapanagiotou A, Scarvelakis M, et al. Detection and characterization of Wolbachia infections in natural populations of aphids: is the hidden diversity fully unraveled? PLoS One. 2011;6:e28695. doi: 10.1371/journal.pone.0028695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ntougias S, Polkowska Ż, Nikolaki S, Dionyssopoulou E, Stathopoulou P, Doudoumis V, et al. Bacterial community structures in freshwater polar environments of Svalbard. Microbes Environ. 2016;31:401–409. doi: 10.1264/jsme2.ME16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chao A. Non-parametric estimation of the classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- 68.Chao A, Lee S-M. Estimating the number of classes via sample coverage. J Am Stat Assoc. 1992;87:210–217. doi: 10.1080/01621459.1992.10475194. [DOI] [Google Scholar]
- 69.Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 70.Gower JC. Principal coordinates analysis. In: Encyclopedia of biostatistics. Hoboken: John Wiley & Sons, Ltd; 2005.
- 71.Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology. 2003;84:511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2. [DOI] [Google Scholar]
- 72.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 73.Clarke K, Gorley R. PRIMER v6: user manual/tutorial. Primer-E Plymouth. 2006:192.
- 74.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available in NCBI.