Abstract
Receptor tyrosine kinases (RTKs) are cell surface receptors which control cell growth and differentiation, and play important roles in tumorigenesis. Despite decades of RTK research, the mechanism of RTK activation in response to their ligands is still under debate. Here we investigate the interactions that control the activation of the tropomyosin receptor kinase (Trk) family of RTKs in the plasma membrane, using a FRET-based methodology. The Trk receptors are expressed in neuronal tissues, and they guide the development of the central and peripheral nervous systems during development. We quantify the dimerization of human Trk-A, Trk-B, and Trk-C in the absence and presence of their cognate ligands: human ß-nerve growth factor (hß-NGF), human brain-derived neurotrophic factor (h-BDNF), and human neurotrophin-3 (hNT-3), respectively. We also assess conformational changes in the Trk dimers upon ligand binding. Our data support a model of Trk activation in which (1) Trks have a propensity to interact laterally and to form dimers even in the absence of ligand, (2) different Trk unliganded dimers have different stabilities, (3) ligand binding leads to Trk dimer stabilization and (4) ligand binding induces structural changes in the Trk dimers which propagate to their transmembrane and intracellular domains. This model, which we call the “transition model of RTK activation,” may hold true for many other RTKs.
INTRODUCTION
RTKs, the second largest family of membrane receptors, are known to control cell growth, differentiation, and motility via lateral dimerization in the membrane. Their dysregulation has been linked to many human diseases and disorders, including a variety of cancers (1–4). There are 58 different RTKs in humans, grouped in 20 different families, which all share the same basic architecture: an N-terminal extracellular (EC) region, a single-pass transmembrane (TM) domain, and an intracellular (IC) region containing a tyrosine kinase domain (5). The RTK ligands are polypeptides often referred to as “growth factors.” The ligands bind to the RTK EC regions and activate the kinases, via a process that involves their cross-phosphorylation on specific tyrosine residues. The activated kinases then phosphorylate additional tyrosines that serve as docking sites for adaptor proteins. The adaptors, in turn, bind cytoplasmic substrates and trigger downstream signaling pathways (1,6–10) such as the MAPK, PI3K, PKC, and STAT cascades which impact growth, development, and disease progression.
Work during the last decade has provided evidence that RTK dimerization is required, but is not sufficient, for RTK activation. Instead, there also exist structural requirements for cross-phosphorylation to occur (2,11–13). In particular, it has been proposed that a specific orientation of the kinase domains in the dimer is required, and this optimal orientation is achieved only upon ligand binding, as a result of structural rearrangements that propagate along the length of the RTK (14–16). This view is supported by experiments which show that the TM domain dimer conformation changes upon ligand binding, correlating with an increase in receptor phosphorylation (17,18). Furthermore, the rotation of the TM dimer interface has been shown to lead to periodic oscillations in kinase activity, suggesting that the TM dimer structure is sensed by the kinases (19).
An alternate view exists, however, that ligand-induced structural changes cannot be propagated along the length of the RTK because the linkers between the different domains are unstructured (20). In support of this view, there are data suggesting that the EC and IC domains can change conformations independently of each other (21,22). There is also structural evidence for a loose connection between the EC and TM domains (22,23). Within this conceptual framework, the main role of the ligand is to increase the stability of the RTK dimer (24). Thus, there is no consensus on the mechanism of RTK activation by their ligands, even after decades of RTK research.
Here, we investigate the interactions that regulate the behavior of the three RTKs from the Tropomyosin Receptor Kinase (Trk) family in the plasma membranes of mammalian cells. The Trk receptors are expressed in neuronal tissues, and they guide the development of the central and peripheral nervous systems (25–27). They also play a profound role in disease, such as cancer and neurodegeneration (28–30). The Trk receptors share a high degree of structural homology, and their extracellular domains are all composed of three leucine-rich motifs flanked by two cysteine-rich clusters at the N-terminus, followed by two Ig-like domains linked to the TM segment. The ligands of the Trk receptors are called neurotrophins. They bind to the second Ig-like domain closest to the membrane, and trigger downstream ERK, PI3K and PLC-γ signaling pathways that control differentiation, proliferation, and survival of neurons (31–34).
The biology of the Trk receptors has been studied extensively, because of their important physiological roles (25,26,35). However, the biophysical principles behind Trk receptor dimerization and activation in response to their ligands have not been elucidated. We characterize the homodimerization of the three Trk receptors in the plasma membrane, as well as their response to their cognate ligands (hß-NGF in the case of Trk-A, h-BDNF in the case of Trk-B, and hNT-3 in the case of Trk-C). We do this with the help of a FRET-based methodology that can give quantitative information about RTK dimerization, and can report on structural changes that occur in the RTK dimer upon ligand binding (36–39).
MATERIALS AND METHODS
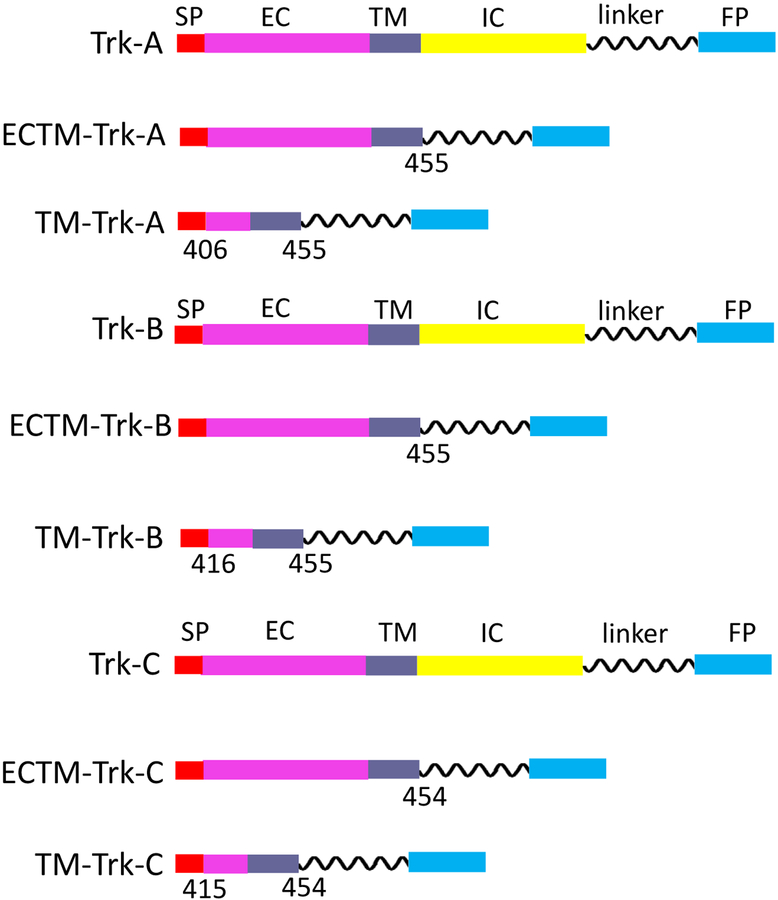
Trk-A, Trk-B and Trk-C plasmids
The pCMV5 Trk-A plasmid (40) was purchased from Addgene (# 15002). The Trk-A gene was amplified and cloned using the restriction enzyme method into a pcDNA3.1+ vector that has been previously engineered to incorporate a gene encoding for a 15 amino acid flexible (GGS)5 linker followed by either eYFP or mTurquoise (a FRET pair) (41). The ECTM-Trk-A plasmids, encoding for the Trk-A extracellular (EC) and transmembrane (TM) domains, a (GGS)5 linker, and either eYFP or mTurquoise, were similarly generated. The TM-Trk-A plasmids, containing the transmembrane (TM) domains, a (GGS)5 linker, and either eYFP or mTurquoise, were produced via Gibson assembly using the NEBuilder HiFi DNA assembly kit (New England Biolabs, E5520S), following the manufacturer’s protocols.
The pDNR-Dual Trk-B plasmid (# HsCD00022371) and pDNR-Dual Trk-C plasmid (# HsCD00022362) were purchased from the DNASU Plasmid Repository. Trk-B and Trk-C, and their ECTM and TM versions were cloned into the pcDNA 3.1+ vector, which incorporated the (GGS)5 linker and the fluorescent protein (either eYFP or mTurquoiuse). Cloning was performed via Gibson assembly using the NEBuilder HiFi DNA assembly kit (New England Biolabs, E5520S). All plasmid constructs are shown in Figure 1. The primer design for all the constructs is shown in Supplementary Table 1.
Figure 1.
Schematics of Trk-A, B, and C constructs, cloned for the FRET experiments. The numbers below the constructs indicate where the DNA sequence was cut to create the truncated constructs.
Cell culture and transfection
Human embryonic kidney cells (HEK293T) used in this work were a kind gift from Dr. D. Wirtz, Johns Hopkins University. The cells were cultured in Dublecco’s Modified Eagle Medium (DMEM, ThermoFisher Scientific, 31600034) with 10% fetal bovine serum (FBS, Hylone, SH30070.03), at 37°C in the presence of 5% CO2.
To perform the FRET experiments, HEK293T cells were seeded (2.5 × 105 cells/dish) on collagen coated, glass bottom petri dishes (MatTek, P35GCOL-1.5–14-C). Upon reaching 60% confluency, cells were transfected with 2ug of DNA using Fugene HD (Promega, E2311) according to the manufacturer’s protocol. In the FRET experiments, cells were co-transfected with different ratios of Trk-mTurquoise and Trk-eYFP DNA. Control single transfection experiments were also conducted, where cells were singly transfected with either Trk-mTurquoise or Trk-eYFP DNA; the acquired spectra were used for calibration as previously described (39). Twenty four hours post transfection, cells were rinsed twice with phenol-red and serum free medium, and starved in the same media for 12 hours. In some experiments, the ligands human ß-nerve growth factor (hß-NGF, Cell Signaling Technology, 5221SC), human brain-derived neurotrophic factor (h-BDNF, Cell Signaling Technology, 3897S), or human neurotrophin-3 (hNT-3, Cell Signaling Technology, 5237SC) were added at a concentration of 5ug/mL before imaging.
Crosslinking
HEK293T cells were transfected with 2ug of Trk-eYFP DNA. A membrane impermeable crosslinker, BS3 (bis(sulfosuccinimidyl) suberate; ThermoFisher Scientific, 21580), was used in the crosslinking experiments. Twenty four hours after transfection with Trk-A-YFP DNA, cells were incubated with 2 mM BS3 for 1 hour at room temperature. The reaction was quenched with 20 mM Tris-HCl for 15 min. The cells were rinsed with ice-cold 1X PBS before lysing. In some cases, cells were incubated with the neurotrophin hß-NGF (5ug/mL) for 10–15 min before adding the crosslinker. The lysates were subjected to SDS-PAGE, and the proteins were transferred to a nitrocellulose membrane. Trk-A-eYFP was probed using anti-GFP antibody (Cell Signaling Technologies, 2555S) as the primary antibody, followed by anti-rabbit HRP conjugated antibody (Promega, W4011). The membrane was incubated with Amersham ECL Plus ™ (GE HealthCare Life Sciences, RPN2106) for 2 min and exposed to detect the chemiluminescent signal in a Chemidoc imaging system (Bio-Rad).
FRET imaging and analysis
Following published protocols (39,42,43), experiments were performed in HEK293T cells under reversible osmotic stress. The reversible osmotic swelling was necessary because the cell membrane of live cells is highly “wrinkled,” while the reversible osmotic stress eliminates these ruffles (44). Thus, the effective 3D protein concentration, determined using purified fluorescent protein standards of known concentration, can be converted into 2D receptor concentrations in the plasma membrane as previously described (39). Before imaging, the serum-free media was replaced with hypotonic swelling media (1:9 serum-free media: diH2O, 25mM HEPES) to induce reversible osmotic stress. Typically, imaging sessions started 10 min after the addition of the swelling buffer, and lasted about 2 hours. In some cases, neurotrophins were added to the swelling media prior to imaging.
Imaging was performed using a two-photon microscope equipped with the OptiMiS True Line Spectral Imaging system (Aurora Spectral Technologies, WI) (45,46). A Mai Tai laser (Spectra-Physics, Santa Clara) was used to generate femtosecond mode locked pulses, and two images were acquired for each cell: one upon excitation at 800 nm to primarily excite the donor, and a second one at 960 nm to primarily excite the acceptor (45,46). The FSI-FRET method was used to measure the FRET efficiency, the donor (Trk-mTurquoise) concentration, and the acceptor (Trk-eYFP) concentration in small membrane areas of a cell. More than 500 cells were analyzed in each case. To convert pixel level intensities of the images into concentrations, calibration solutions of purified fluorescent proteins were used as previously described (39). Soluble eYFP and mTurquoise were produced following a previously published protocol (47).
The measured FRET efficiency has contributions due to both (i) specific dimerization of the Trk receptors, ED and (ii) stochastic or “proximity FRET” that arises due to the random proximity of donors and acceptors in the two dimensional membrane, Eprox (48,49). Thus, the measured (apparent) FRET efficiency Eapp in the experiment is
| (1) |
FRET due to specific dimerization depends on the fraction of dimeric receptors, fD, and the acceptor fraction, xA, according to
| (2) |
Note that when the dimeric fraction is 100% (fD=1), equation (2) reduces to
| (3) |
The constant in equation (2) is the “Intrinsic FRET,” a structural parameter that depends only on the separation and the orientation of the two fluorescent proteins in the dimer, but does not depend on the dimerization propensity (37). The dependence of the intrinsic FRET, , on the distance between the fluorescent proteins in the dimer, d, is given by (37,50)
| (4) |
where R0 is the Forster radius for the mTurquoise-eYFP FRET pair, 54.5 Å.
Data fitting and analysis was performed as previously described (50). Briefly, we utilized a computationally-derived library of proximity FRET efficiencies that were simulated over a finite grid of equilibrium constant, K, and values, for acceptor concentrations ranging from zero to 8 × 103 acceptors/μm2, and for three different exclusion radia: 1 nm, 1.4 nm, and 2 nm. In the first step of the fitting procedure, this proximity FRET library was used to perform a gridded search for the best-fit K and Ẽ from the library, by calculating the total FRET efficiencies and comparing them to the experimentally measured ones (51). Since small changes to these parameters have little effect on the magnitude of the proximity FRET contribution, in the second step we fixed the proximity contribution and we calculated the FRET efficiencies that are due to specific dimerization, ED, using equation (1). We then fit a monomer-dimer model to the corrected ED data while varying Kdiss and , using a MATLAB non-linear least squares algorithm to find the best-fit values of Kdiss and and their 66% confidence intervals.
The stability of the dimer is related to the dissociation constant according to
| (5) |
with Kdiss reported in units of receptors per μm2, and the standard state for ΔG calculation defined as (37).
RESULTS
Full length Trk-A forms dimers in the plasma membrane, in the absence of ligand
To study Trk receptor dimerization using FRET, we attached either mTurquoise or eYFP (a FRET pair) to the C-terminus of full length Trk-A (Figure 1). Then, we co-transfected HEK 293T cells with 2 ug of DNA encoding for Trk-A-mTurquoise and Trk-A-eYFP, while varying the ratios of the two plasmids. After the receptors were trafficked to the plasma membrane, the cells were starved for 12 hours to remove all ligands, and imaged in a two photon microscope equipped with the OptiMiS spectral detection system (39,46). Images were acquired and processed using the fully quantified spectral imaging (FSI) methodology, which involves the acquisition of two complete spectra: a FRET spectrum collected upon donor excitation, and an acceptor spectrum collected upon acceptor excitation (39). As discussed in details in previous work (39), the FSI methodology yields FRET efficiencies and receptor concentrations in the plasma membrane with high precision, and can thus yield dimerization curves.
It is a challenge to measure the two-dimensional (2D) concentration of membrane proteins in the plasma membrane, because cells possess two to three times the membrane surface needed to sustain their shape and thus the plasma membrane is highly “wrinkled” or “ruffled” (52,53). While the effective 3D receptor concentrations can be determined by comparing the fluorescence intensities with standard solutions of fluorescent proteins of known concentrations (37,38), the complex membrane topology prevents their conversion into 2D receptor concentrations within the plasma membrane (39). However, cell membranes can be “un-wrinkled” in a reversible manner when the cells are subjected to controlled osmotic stress which leads to the disassembly of the caveolae (44). The application of the reversible stress does not alter the FRET efficiencies (41), indicating that membrane protein interactions are not altered in a measurable way. The reversible stress does not cause irreversible cell damage, either (42,54). In the cells under reversible osmotic stress, we analyzed membrane regions of homogenous fluorescence, about 3 μm in length, using the FSI software (39). We calculated three parameters for each of the regions: the donor concentrations, the acceptor concentrations, and the FRET efficiencies (39).
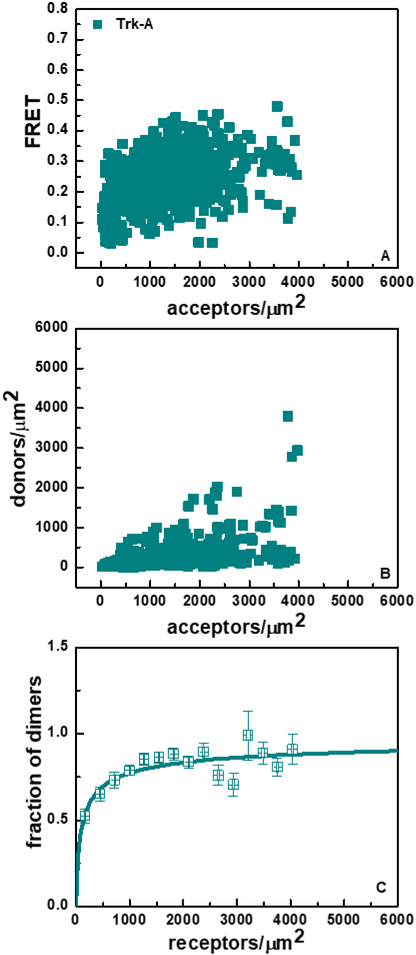
The Trk-A FRET data, for the case of the 1:3 donor-to-acceptor transfection ratio, is shown in Figure 2. Figure 2A shows the FRET efficiencies as a function of acceptor concentration. Figure 2B shows the donor versus the acceptor concentration in each region. Since each data point is derived from one cell, Figure 2B demonstrates that the analyzed cell pool exhibits a substantial variation in the donor-to-acceptor ratio and the total receptor concentration (donor + acceptor expression) despite the fact that transfection is always performed under identical conditions. This heterogeneity is embraced and exploited in the FSI-FRET method, as it enables robust fits of thermodynamic models to the FRET data (38,41,50,55,56).
Figure 2.
FRET data for full-length Trk-A in the absence of ligand. (A) FRET efficiencies as a function of acceptor (Trk-A-eYFP) concentration. Every data point has different donor (Trk-A-mTurquoise) concentration. (B) Donor (Trk-A-mTurquoise) concentration, plotted as a function of acceptor (Trk-A-eYFP) concentration. (C) Fraction of Trk-A dimers as a function of total Trk-A concentration. The experimentally determined dimeric fractions are binned and are shown with the symbols, along with the standard errors. The solid line indicates the best-fit dimerization curve, plotted for the best-fit Kdiss shown in Table 1.
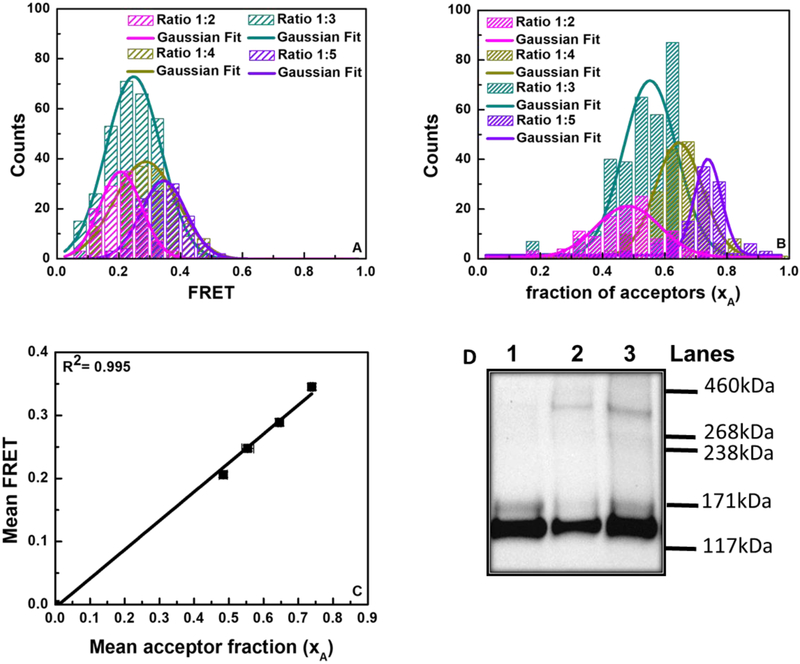
Most RTKs are known to form dimers, and we investigated if this is the case for Trk-A by examining the dependence of the FRET efficiency on the acceptor fraction. In this type of analysis, the dependence of the FRET efficiency on the acceptor fraction is known to be linear for a dimer and nonlinear for higher order oligomers (54,57–59). To investigate this dependence, we analyzed the FRET efficiencies measured for four different Trk-A-mTurquoise to Trk-A-eYFP DNA transfection ratios: 1:2, 1:3, 1:4, and 1:5 (shown in Figure 3). Histograms of FRET efficiencies and fractions of acceptor-labeled receptors are shown in Figure 3A and 3B for the four different transfection ratios. These histograms were fitted to Gaussian functions to obtain the averages and the standard errors. The average FRET efficiencies obtained from the histograms are plotted versus the average acceptor ratios in Figure 3C for the four DNA transfection ratios (symbols; standard errors are also shown but they are smaller than the symbols). The data in Figure 2 are well described by a linear function (R2=0.995), suggesting that Trk-A forms dimers in the absence of ligand.
Figure 3.
FRET and cross-linking data demonstrating full-length Trk-A dimer-formation. (A) Histograms of measured FRET efficiencies at four different donor to acceptor ratios (1:2, 1:3, 1:4, and 1:5). (B) Histograms of acceptor fractions (xA) for the four donor to acceptor ratios. (C) Average FRET efficiencies as a function of average acceptor fractions (xA). The standard errors, which are smaller than the symbols, are also shown. The linear dependence is indicative of dimer formation. (D) Trk-A Western blot in the absence and presence of a chemical cross-linker. Lane 1: Trk-A; Lane 2: Trk-A + cross-linker; Lane 3: Trk-A + hß-NGF + cross-linker. The molecular weight of monomeric, mature, fully glycosylated TrkA is ~ 140 kDa, and thus the molecular weight of fully mature Trk-A-YFP is ~170 kDa. The intense lower molecular weight band corresponds to immature, partially glycosylated TrkA found in the ER and Golgi. The fully mature Trk-A-YFP dimer molecular weight is ~340 kDa.
To confirm dimer formation, we performed cross-linking experiments with Trk-A-eYFP in the presence and absence of its ligand NGF. In these experiments, cells expressing Trk-A-eYFP were incubated with a cross-linker, lysed, and subjected to Western blotting. Since in the cross-linking experiments we do not monitor the 2D concentrations of the receptors in the membrane, no application of reversible osmotic stress was required. Trk-A bands were visualized using anti-GFP antibodies. Results, shown in Figure 3D, reveal two monomeric bands: one at molecular weight ~170 kDa, corresponding to mature, fully glycosylated Trk-A-YFP, and one at ~140 kDa, corresponding to immature, partially glycosylated Trk-A-YFP found in the ER and the Golgi (60,61). The dimer band for the fully glycosylated Trk-A-YFP is expected to be at ~340 kDa, and indeed we see such a band in the presence of the cross-linker. These results support the idea that Trk-A forms a dimer, in the absence and presence of ligand, and are consistent with the literature (62). Note that the immature band is not cross-linked because it is not localized to the plasma membrane (as the cross-linker is membrane-impermeable), and thus does not contribute to the measured FRET in Figures 2 and 3.
Next, we sought to interpret the FRET data in Figure 2 within the framework of a thermodynamic model of dimerization as described previously (50). Dimer formation can be characterized by two parameters: the two-dimensional dissociation constant, Kdiss, and the structural parameter (or “Intrinsic FRET”). The dissociation constant, Kdiss, is a measure of the dimerization propensity of Trk-A in the plasma membrane. The Intrinsic FRET is the FRET efficiency in a Trk-A dimer with one donor and one acceptor. It depends on the positioning of the fluorescent proteins within the Trk-A dimer, but not on the dimerization propensity (17,37). To interpret the FRET data, we further took into account that the measured FRET in the membrane has two contributions: one due to Trk-A dimerization and one due to the confinement of the mobile fluorophores in the two-dimensional membrane (48,50). The latter FRET occurs because of the random close approach (within 100 Å) of the donors and acceptors in the plasma membrane. This “proximity FRET” contribution is well understood, and can be corrected for as discussed in detail elsewhere (48–50). The best-fit Kdiss and the best-fit Intrinsic FRET are determined from the FRET data while accounting for proximity FRET, following a verified two-step fitting protocol that is briefly described in Materials and Methods (see also (50)).
The dimeric fraction of Trk-A as a function of the total Trk-A concentration is shown in Figure 2C. We see that Trk-A exhibits a significant dimer population in the absence of ligand. The solid line in Figure 2C shows the dimerization curve calculated for the best-fit value of Kdiss. The solid symbols show the experimentally derived dimeric fractions, which have been binned into 30 bins over the expression range of Trk-A, and then averaged within the bins.
The optimal values of Kdiss and Intrinsic FRET, as well as their 66% confidence intervals, are given in Table 1. The two dimensional dissociation constant for the Trk-A receptor, Kdiss, is 132 ± 0.37 receptors/μm2, and the corresponding dimerization free energy is −5.30 ± 0.17 kcal/mole. The Trk-A Intrinsic FRET value in the absence of ligand is 0.33 ± 0.01.
Table 1.
Dimerization parameters for the three full-length Trk receptors, in the absence of ligand and in the presence of 380 nM ligand. Kdiss is the two-dimensional dissociation constant, a thermodynamic parameter, which is related to the stability of the Trk dimers, ΔG, according to equation (5). is the Intrinsic FRET, which depends on the positioning of the fluorescent proteins within the Trk dimers. The average distance between the fluorescent proteins, d, is calculated from Ẽ using equation (4), under the assumption for free rotation of the fluorescent proteins. an.d.: not determined.
| Kdiss (rec.μm−2) | ΔG (kcal/mol) | Intrinsic FRET () | d(Å) | |
|---|---|---|---|---|
| Trk-A | 132 ± 37 | −5.30 ± 0.17 | 0.33 ± 0.01 | 61 ± 1 |
| Trk-A+NGF | 100% dimer | n.d.a | 0.28 ± 0.02 | 64 ± 1 |
| Trk-B | 12 ± 2 | −6.72 ± 0.10 | 0.35 ± 0.04 | 60 ± 2 |
| Trk-B+BDNF | 100% dimer | n.d. | 0.30 ± 0.02 | 63 ± 1 |
| Trk-C | 227 ± 25 | −4.98 ± 0.07 | 0.88 ± 0.02 | 39 ± 1 |
| Trk-C+NT-3 | 100% dimer | n.d. | 0.64 ± 0.03 | 50 ± 1 |
Full length Trk-B and Trk-C receptors form dimers in the plasma membrane, in the absence of ligand
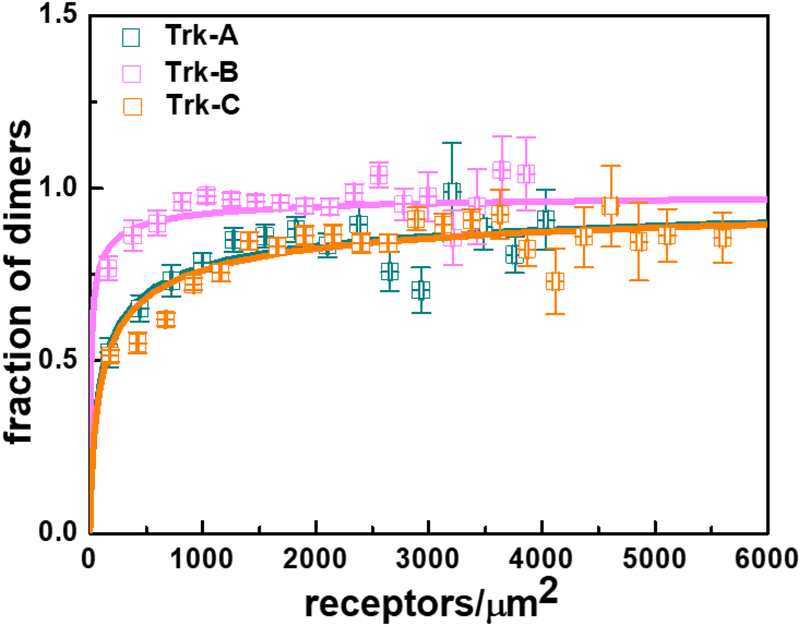
We performed similar experiments with full-length Trk-B and Trk-C, labeled with either mTurquoise or eYFP at their C-termini via flexible (GGS)5 linkers. The dimerization curves, obtained as described above, are shown in Figure 4, and are compared to the Trk-A results. The two parameter fit to the Trk-C data reveals a dimerization constant that is similar to Trk-A, but the value of the Intrinsic FRET is higher (Table 1). Trk-B, on the other hand, exhibits much higher dimerization propensity, with Kdiss = 12 ± 2 rec/μm2. Overall, the data in Figure 4 show that all three Trk receptors, and particularly Trk-B, homo-dimerize strongly in the plasma membrane, even in the absence of ligand.
Figure 4.
Dimerization curves for the three full-length Trk receptors in the absence of ligands. The best-fit dimerization parameters are shown in Table 1.
Thermodynamic contributions of Trk domains to unliganded dimerization
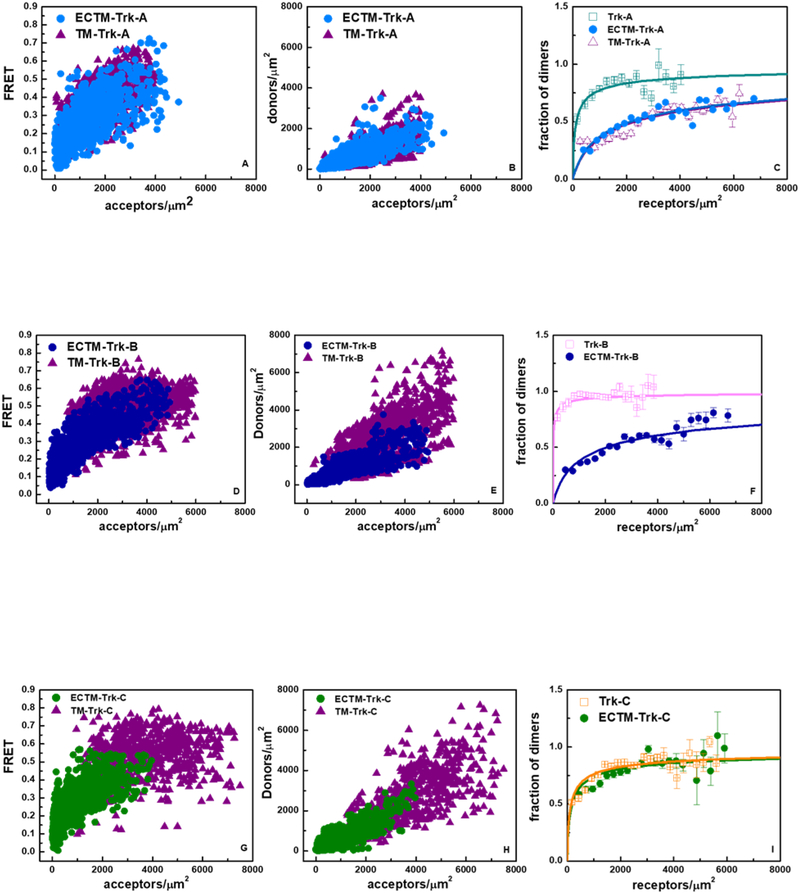
In order to gain insight into the contributions of the different domains to the stability of the unliganded Trk-A dimer, two truncated versions of the Trk-A receptor were created. First, the IC domain was removed to generate a Trk-A ECTM construct containing the EC and TM domains, with the fluorescent proteins attached to the TM domains via a 15 amino acid (GGS)5 flexible linkers (63,64) (see Figure 1). In the second truncated version of the Trk-A receptor, both the IC and EC domains were removed to generate the TM Trk-A construct, with the fluorescent proteins attached to the TM domain C-terminus by the same 15 amino acids (GGS)5 flexible linker (Figure 1). The FSI method was used to characterize the dimerization of the two truncated Trk-A constructs. Figure 5A shows FRET as a function of acceptor concentration for the ECTM and TM Trk-A constructs. Figure 5B shows the donor versus the acceptor concentration for these constructs. Figure 5C compares the dimerization curves for the ECTM and TM constructs with the dimerization curve for the full length Trk-A receptor. The calculated best-fit parameters for these constructs are reported in Table 2. According to these results, the removal of the IC domain decreases the dimer stability from −5.30 ± 0.17 to −3.65 ± 0.08 kcal/mole, indicating that contacts involving the IC domains stabilize the full-length Trk-A dimer. The removal of the EC domain, however, has no effect on dimer stability, as the ECTM and TM free energies of dimerization are the same, −3.65 ± 0.08 and −3.61 ± 0.09 kcal/mole, respectively. Thus, Trk-A dimerization in the absence of ligand occurs because of stabilizing contacts between the TM and IC domains, without a significant contribution from the EC domain.
Figure 5.
FRET data for the ECTM Trk receptors, which lack IC domains, and for the TM Trk receptors, which lack both EC and IC domains. Data are collected in the absence of ligand. (A, D, and G) Measured FRET efficiencies as a function of receptor concentrations. (B, E, and H) Donor concentrations versus acceptors concentrations. (C, F, and I) Dimeric fractions as a function of total receptor concentrations. The experimentally determined dimeric fractions are binned and are shown with the symbols, along with the standard errors. The solid lines are the dimerization curves, plotted for the optimized dimerization parameters reported in Table 2. The FRET data for the TM Trk-B and Trk-C constructs could not be fitted with a dimer model.
Table 2.
Dimerization parameters for the three truncated Trk receptors, in the absence of ligand and in the presence of 380 nM ligand.
| Kdiss (rec.μm−2) | ΔG (kcal/mol) | Intrinsic FRET () | d(Å) | |
|---|---|---|---|---|
| TM-Trk-A | 2271 ± 310 | −3.61 ± 0.09 | 0.96 ± 0.03 | 32 ± 5 |
| ECTM-Trk-A | 2127 ± 278 | −3.65 ± 0.08 | 0.76 ± 0.04 | 45 ± 2 |
| ECTM-Trk-A+NGF | 25 ± 5 | −6.28 ± 0.12 | 0.59 ± 0.01 | 51 ± 1 |
| ECTM-Trk-B | 2018 ± 134 | −3.68 ± 0.04 | 0.82 ± 0.02 | 42 ± 1 |
| ECTM-Trk B+BDNF | 23 ± 2 | −6.33 ± 0.06 | 0.50 ± 0.01 | 55 ± 1 |
| ECTM-Trk-C | 183 ± 29 | −5.10 ± 0.09 | 0.57 ± 0.01 | 52 ± 1 |
| ECTM-Trk-C+NT-3 | 15 ± 4 | −6.59 ± 0.16 | 0.64 ± 0.01 | 50 ± 1 |
We performed similar experiments with the truncated versions of Trk-B and Trk-C. Figures 5D and G show FRET as a function of acceptor concentration. Figures 5E and H show the donor versus the acceptor concentration for these constructs. Figures 5F and I compare the dimerization curves for the ECTM and full-length receptors. According to the best-fit parameters in Table 1 and Table 2, the removal of the IC domain in Trk-B decreases the dimer stability from −6.72 ± 0.10 to −3.68 ± 0.04 kcal/mole, while the IC domain in Trk-C has no significant effect on dimerization. Of note, attempts to fit the FRET data for the Trk-B and Trk-C TM constructs with a dimer model were unsuccessful, suggesting that these TM domains form oligomers in the plasma membrane.
Effect of ligands on Trk dimerization
Neurotrophins interact with the Trk receptors to initiate downstream signaling (25,31,65). Here we studied how Trk dimerization is affected by three ligands, hß-NGF, h-BDNF, and hNT-3, which are known as cognate ligands for Trk-A, Trk-B, and Trk-C, respectively. The ligand binding affinities have been characterized previously using different methods, and published values are in the pM to low nM range (26,66–70).
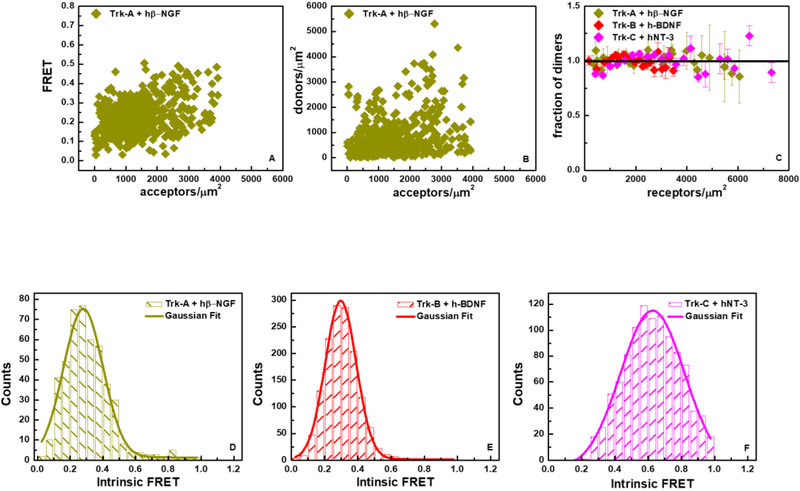
We sought to carry out FRET experiments under well-defined conditions, when all the Trk receptors are ligand-bound. We therefore performed the FRET experiments in the presence of 5 ug/ml (~ 380 nM) of each ligand, which greatly exceeds the reported dissociation constants. The FRET and concentration data for Trk-A in the presence of hß-NGF are shown in Figures 6A and 6B. The dimerization curves (dimeric fractions as a function of total Trk concentration) for the three Trk receptors are shown in Figure 6C in the presence of their respective ligands. We see that the Trk receptors are 100% dimeric over the concentration range sampled in the experiments. Thus, the two-dimensional dissociations constants are very low and could not be measured. The results demonstrate that hß-NGF, h-BDNF, and hNT-3 stabilize the Trk-A, Trk-B, and Trk-C dimers, respectively.
Figure 6.
FRET data for full-length Trk receptors in the presence of ligand at saturating concentration. (A) FRET as a function of acceptor concentration for Trk-A in the presence of hß-NGF. (B) Trk-A-mTurquoise (donor) concentration versus Trk-A-eYFP (acceptor) concentration in the presence of hß-NGF. (C) Fraction of dimers as a function of total receptor concentration for Trk-A, Trk-B and Trk-C in the presence of hß-NGF, h-BDNF and hNT-3, respectively. (D) Histogram of Trk-A Intrinsic FRET values in the presence of hß-NGF. (E) Histogram of Trk-B Intrinsic FRET values in the presence of h-BDNF. (F) Histogram of Trk-C Intrinsic FRET values in the presence of hNT-3. The dimerization parameters for the three receptors in the presence of ligands are reported in Table 1.
In the case of 100% dimers, the Intrinsic FRET is calculated for each cell using equation (3). Note that both the FRET efficiencies and the acceptor fractions, xA, are directly measured with the FSI-FRET method, and thus no fitting was required for these calculations. The Intrinsic FRET values from individual cells are shown as histograms in Figures 6D, E and F, for Trk-A, Trk-B, and Trk-C, respectively, and are fit with Gaussian functions. The Gaussian fit parameters are shown in Table 1. Comparison of the Intrinsic FRET values in Table 1 with and without ligand reveals a large change in Intrinsic FRET for Trk-C upon the addition of hNT-3, but no significant effects for Trk-A and Trk-B in response to hß-NGF and h-BDNF, respectively. As the fluorescent proteins are attached to the C-termini via flexible linkers, the differences in Intrinsic FRET for Trk-C must reflect changes in the relative positioning of the C-termini of Trk-C upon hNT-3 binding.
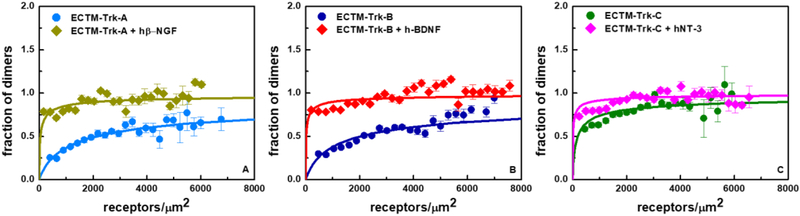
To gain additional insights into the effect of the three ligands on Trk dimerization, we performed FRET experiments with the truncated ECTM Trk constructs with deleted kinase domains, in the presence of ligands. The dimerization curves for the ECTM Trk receptors in the presence of saturating concentrations of their cognate ligands are shown in Figure 7, and are compared to the curves in the absence of ligand. The best fit dimerization parameters are shown in Table 2. Inspection of Figure 7 and Table 2 shows that the cognate ligands stabilize the ECTM Trk dimers.
Figure 7.
Dimerization curves for ECTM Trk-A, Trk-B and Trk-C in the presence and absence of ligands. (A) ECTM Trk-A in the presence and absence of hßNGF. (B) ECTM Trk-B in the presence and absence of h-BDNF. (C) ECTM Trk-C in the presence and absence of hNT-3.
As the deletion of the IC domains decreases Trk dimerization, we are able to quantify the stabilities of the ligand-bound Trk dimers and to calculate the respective dissociation constants (see Table 2). This allows us to determine the effect of the bound ligands on the thermodynamic stabilities of the dimers, by subtracting the ECTM dimer stabilities with and without ligand. We see that hß-NGF, h-BDNF, and hNT-3, at 380 nM, stabilize the Trk-A, Trk-B, and Trk-C dimers by −2.63 ± 0.14 kcal/mole, −2.65 ± 0.10 kcal/mole, and −1.49 ± 0.17 kcal/mole, respectively.
Table 2 shows the Intrinsic FRET values for the liganded truncated ECTM Trk dimers, determined from the fit of a monomer-dimer model to the measured FRET data. When we compare these values to the values in the absence of ligand, we see significant differences in the cases of Trk-A and Trk-B. As the fluorescent proteins are attached directly to the TM domains via flexible linkers, the changes in Intrinsic FRET likely reflect changes in the distance between the C-termini of the TM domains of Trk-A and Trk-B upon ligand binding (17,18,71).
Discussion
Experimental findings.
Using a FRET-based quantitative technique, we showed that the Trk receptors form dimers not only in the presence of ligand, but also in the absence of ligand. Our results are consistent with the literature (62,72,73), and yield the first measurements of Trk dimer stabilities. The Trk dissociation constants in the unliganded case, which vary from 12 to 227 rec/μm2, can be compared to the dissociation constants measured previously for the FGFRs—varying from 24 rec/μm2 (for FGFR3) to 710 rec/μm2 (for FGFR1) (17)—and for VEGFR2—35 rec/μm2 (18). Thus, the Trk receptor propensities for unliganded dimerization are similar to those of other characterized full length RTKs.
The estimates of Trk-A expression in the membrane of PC12 cells are ~20 Trk-A rec/μm2 (73,74). Since the measured Trk-A dissociation constant is 132 ± 37 rec/μm2, about 20% of the Trk-A molecules are expected to be dimeric in PC12 cells in the absence of ligand. Notably, Trk-A expression in PC12 cells is similar to the measured Trk-B dissociation constant, 12 ± 2 rec/μm2. This suggests that unliganded Trk dimerization is important in physiological contexts.
To gain insight into factors that determine the stability of the unliganded Trk dimers, we measured the thermodynamic contributions of the different Trk domains to unliganded dimerization (Figure 5). The contributions of the IC domains of Trk-A and Trk-B to dimer stabilities are favorable, since the deletion of the IC domains significantly destabilized the Trk-A and Trk-B dimers. However, the contribution of Trk-C IC domain is practically zero, suggesting that either Trk-C IC domain does not engage in contacts that stabilize the full-length Trk-C dimer, or these stabilizing contacts are balanced by repulsive ones. This finding is reminiscent of the case of the FGFRs, where the IC domains of FGFR2 and FGFR3 contribute to dimer stabilization, but the FGFR1 IC domain has no effect (17).
By measuring the dimerization of the TM Trk-A construct and comparing it to the ECTM Trk-A construct, we showed that the contribution of Trk-A EC domain to unliganded dimerization is negligible. This result is different from observations for other RTKs such as the FGFRs and VEGFR2, where removal of the EC domain increases dimer stability (17,18), thus indicating that the EC domains of those other receptors inhibit dimerization in the absence of ligand. A crystal structure of the NGF-bound isolated Trk-A EC domain shows that the ligand-bound dimer is stabilized exclusively through hß-NGF-TrkA contacts, but not through direct Trk-A—Trk-A contacts (75). Accordingly, it appears that there are no direct interactions between Trk-A EC domains that contribute to dimer stability, both in the absence and presence of the hß-NGF ligand. The Trk-A dimers are instead stabilized through contacts between the TM domains and the IC domain.
Unlike in the Trk-A case, we were not able to determine the thermodynamic contribution of Trk-B and Trk-C EC domains to unliganded dimerization, because we could not fit the FRET data for TM Trk-B and TM Trk-C with a dimer model. Thus, these TM domains likely form oligomers that are larger than dimers. Despite the propensity of these TM domains to oligomerize, it can be expected that the bulky Trk-A EC and IC domains impose constraints that do not allow three or more TM helices to interact closely, thus preventing higher order oligomerization for the full-length receptors. These observations underscore the limitations of studying RTK TM domains in isolation, outside the context of full-length RTKs. Indeed, NMR studies of isolated RTK TM domains in detergent have revealed that the TM domains can form trimers, which are likely irrelevant in biological context (76).
In this work we also sought to measure the effect of ligand binding on RTK dimer stability. It is known that bound ligands stabilize RTK dimers, but the exact contribution of ligand binding to RTK dimer stability has not been measured. Usually the stabilities of the ligand-bound RTK dimers are too high, precluding calculations of the dissociation constant, as also demonstrated here for the three full-length Trk receptors. Since the deletion of the kinase domains decreased the stabilities of the Trk dimers, however, the dissociation constants became measurable for the ligand-bound truncated ECTM Trk dimers. We were thus able to show that the contributions of the three ligands to Trk dimer stability varies from ~−1.5 to ~−2.5 kcal/mole, at 380 nM ligand. To the best of our knowledge, this is the first direct measurement of the energetics behind RTK dimer stabilization by a ligand.
Along with dimer stabilities, we quantified the Intrinsic FRET for all studied Trk dimers, both in the absence and presence of their cognate ligands. Intrinsic FRET is a structural parameter that depends on the distance between the fluorescent proteins and on their relative orientation, but does not depend on dimer stability. We used Intrinsic FRET to follow structural changes that occur on the cytoplasmic side of the receptor upon ligand binding to the extracellular domains. We characterized full-length constructs, where the fluorescent proteins were attached to the C-termini of the receptors via flexible, 15-residue (GGS)5 linkers. We also characterized truncated ECTM Trk constructs, in which the fluorescent proteins were attached to the TM domains via flexible, 15-residue (GGS)5 linkers.
We observed that the Intrinsic FRET value for full-length Trk-C changes upon ligand binding (Table 1). Since the fluorescent proteins are attached to the C-termini via flexible linkers, the measured change is indicative of a change in the relative positioning of the C-termini in response to ligand. We further observed that the Intrinsic FRET for ECTM Trk-A and Trk-B changes upon ligand binding. Since in this case the fluorescent proteins are attached to the C-termini of the TM domains via flexible linkers, differences in Intrinsic FRET are indicative of changes in the relative positioning of the TM C-termini upon ligand binding.
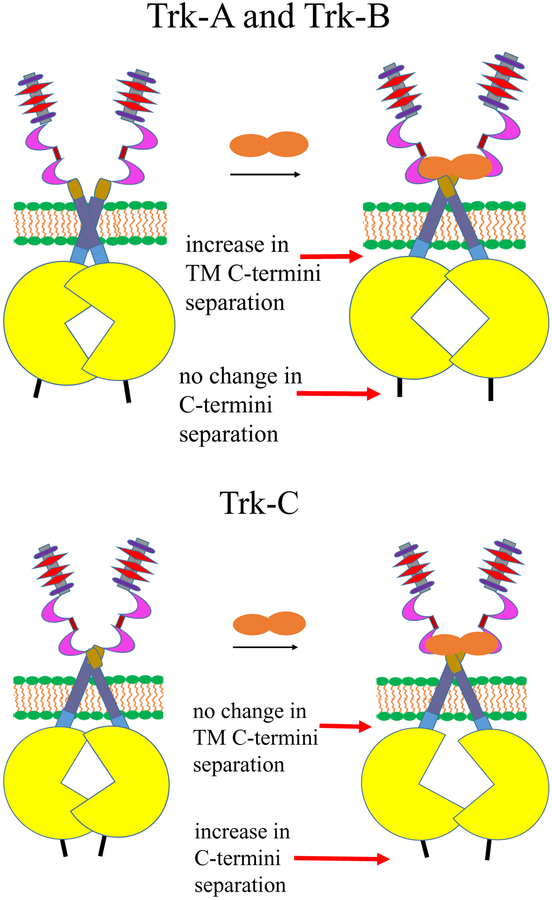
The linkers used to attach the fluorescent proteins to the Trk receptors have been shown previously to behave as random coils (63). If the orientation of the fluorescent proteins at the end of the linkers is random, the Intrinsic FRET will depend primarily on the distance between the fluorescent proteins in the Trk dimers, as shown in equation (4). In Tables 1 and 2, we calculate the distances between the fluorescent proteins under the assumption of random orientation of the fluorophores, an assumption which may not be entirely correct. Even if the exact distances are not precisely accurate, the relative changes in fluorescent protein separation due to ligand binding should still hold true. These distances allow for visualization of the conformational changes that occur in response to ligand binding to the Trk receptors. A cartoon illustrating the change in these distances is shown in Figure 8. In the top panel we depict the changes occurring in Trk-A and Trk-B in response to ligand binding. We show that the TM domain C-termini move apart upon ligand binding, while the distances between the C-termini of the receptors do not change in a significant way. This implies conformational changes in the Trk-A and Trk-B IC domain dimers upon ligand binding. In the bottom panel, we depict the Trk-C case, where the average distance between the TM domain C-termini does not change in a significant way, but the C-termini move further apart. This also implies conformational changes in the Trk-C IC domain dimer upon NT-3 binding. While the nature of the ligand-induced changes are different in the cases of Trk-A and Trk-B, on one hand, and Trk-C, on the other, the data suggest that all three Trk receptors undergo conformational changes in the IC region after their cognate ligands bind to their extracellular domains. These changes are likely critical for signal propagation across the plasma membrane.
Figure 8.
The three Trk receptors undergo conformational changes in the IC region after their cognate ligands bind to their extracellular domains. Top: Schematic depiction of the changes occurring in Trk-A and Trk-B. The TM domain C-termini move apart upon ligand binding, while the distance between the C-termini of the receptors does not change in a significant way. Bottom: Schematic depiction of the changes occurring in Trk-C in response to ligand binding. The C-termini move apart upon ligand binding, but the average distance between the TM domain C-termini does not change in a significant way.
Implications
Two RTK activation models have been widely discussed in the literature: (1) the “diffusion-based” or “canonical” model, which postulates that RTKs dimerize and activate each other only upon ligand binding (but have no propensity to interact with each other and are always monomeric in the absence of ligand) (1,5), and (2) the “pre-formed dimer model,” in which RTKs form constitutive dimers, which get activated in response to ligand binding (6,11,20,62,72,77). However, quantitative measurements of dimerization, such as the ones described here, reconcile these seemingly contradictory models. Indeed, here we find that the measured dissociation constants vary for the different Trk receptors, even though they belong to the same receptor family. Depending on the exact value of the two-dimensional dissociation constant and the expression levels, RTKs may appear to be predominantly monomeric or predominantly dimeric at a specific expression level, in accordance with the law of mass action.
An important role of the ligand in the pre-formed dimer model is to induce a structural change in the RTK dimer that reorients the catalytic domains for efficient activation (2,12,13,17,18). However, this role has been recently challenged because the linkage between the different RTK domains has been proposed to be flexible (24,78). If this is the case, the different domains are not structurally coupled, and thus structural information cannot be propagated along the length of an RTK. Resolving this controversy is challenging, as there are experimental limitations in following conformational changes in the intracellular domains of RTK dimers upon ligand binding to their EC domains, especially in the context of the full length receptors in the plasma membrane of live cells. Here, we provide evidence of conformational changes in the intracellular portions of the Trk dimers through measurements of Intrinsic FRET. The observed structural change ensures that the kinase domains adopt a signaling-competent orientation only upon ligand binding and can explain how RTKs which form very strong preformed dimers, such as Trk-B, can be activated by their ligands.
Overall, this work supports a model of Trk activation in which (1) Trks have a propensity to interact laterally and to form dimers even in the absence of ligand, (2) different unliganded Trk dimers have different stabilities (and thus different dimer abundance at physiological concentrations), (3) ligand binding leads to Trk dimer stabilization and (4) ligand binding induces a structural changes in the Trk dimer. Thus, an increase in the expression level can cause a transition from predominantly monomeric to predominantly dimeric populations, even in the absence of ligand. When ligand binds to the unliganded dimers, it induces a transition to a structurally distinct dimeric state with higher stability. This model views RTK activation through the lens of physical chemistry and we term it the “transition model of RTK activation.”
It is tempting to speculate that all of the 58 RTKs follow the transition model of RTK activation. Alternatively, it is possible that no single model can capture the activation mechanism of all 58 RTKs. If this is the case, then the activation mechanism needs to be elucidated experimentally for every one of the 58 RTKs. The quantitative FRET methodology used in this work can help in this pursuit of basic knowledge about RTK activation.
Supplementary Material
Acknowledgements:
Supported by NIH GM068619. We thank Janet Aiyedun for help with data analysis, Michael Paul for reading the manuscript prior to publication, and Dr. Christopher King for the development of the data acquisition and analysis software.
References
- 1.Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 2.Li E, and Hristova K (2006) Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry 45, 6241–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss A, and Schlessinger J (1998) Switching signals on or off by receptor dimerization. Cell 94, 277–280 [DOI] [PubMed] [Google Scholar]
- 4.Pasquale EB (2010) Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nature Reviews Cancer 10, 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fantl WJ, Johnson DE, and Williams LT (1993) Signaling by Receptor Tyrosine Kinases. Annual Review of Biochemistry 62, 453–481 [DOI] [PubMed] [Google Scholar]
- 6.Lin CC, Melo FA, Ghosh R, Suen KM, Stagg LJ, Kirkpatrick J, Arold ST, Ahmed Z, and Ladbury JE (2012) Inhibition of Basal FGF Receptor Signaling by Dimeric Grb2. Cell 149, 1514–1524 [DOI] [PubMed] [Google Scholar]
- 7.Lemmon MA, and Schlessinger J (2010) Cell Signaling by Receptor Tyrosine Kinases. Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemmon MA, Schlessinger J, and Ferguson KM (2014) The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harbor perspectives in biology 6, a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlessinger J (2014) Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harbor perspectives in biology 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlessinger J, and Lemmon MA (2003) SH2 and PTB domains in tyrosine kinase signaling. Science’s STKE : signal transduction knowledge environment 2003, RE12. [DOI] [PubMed] [Google Scholar]
- 11.Valley CC, Lewis AK, and Sachs JN (2017) Piecing it together: Unraveling the elusive structure-function relationship in single-pass membrane receptors. Biochim Biophys Acta 1859, 1398–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriki T, Maruyama H, and Maruyama IN (2001) Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. Journal of Molecular Biology 311, 1011–1026 [DOI] [PubMed] [Google Scholar]
- 13.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang YJ, Pelton JG, Shan YB, Shaw DE, Wemmer DE, Groves JT, and Kuriyan J (2013) Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor. Cell 152, 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleishman SJ, Schlessinger J, and Ben Tal N (2002) A putative molecular-activation switch in the transmembrane domain of erbB2. Proc.Natl.Acad.Sci.U.S.A 99, 15937–15940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bocharov EV, Mayzel ML, Volynsky PE, Mineev KS, Tkach EN, Ermolyuk YS, Schulga AA, Efremov RG, and Arseniev AS (2010) Left-Handed Dimer of EphA2 Transmembrane Domain: Helix Packing Diversity among Receptor Tyrosine Kinases. Biophysical Journal 98, 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bocharov EV, Lesovoy DM, Goncharuk SA, Goncharuk MV, Hristova K, and Arseniev AS (2013) Structure of FGFR3 Transmembrane Domain Dimer: Implications for Signaling and Human Pathologies. Structure 21, 2087–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarabipour S, and Hristova K (2016) Mechanism of FGF receptor dimerization and activation. Nat.Commun 7, 10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarabipour S, Ballmer-Hofer K, and Hristova K (2016) VEGFR-2 conformational switch in response to ligand binding. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell CA, Tynan JA, Hart KC, Meyer AN, Robertson SC, and Donoghue DJ (2000) Rotational coupling of the transmembrane and kinase domains of the Neu receptor tyrosine kinase. Molecular Biology of the Cell 11, 3589–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bocharov EV, Sharonov GV, Bocharova OV, and Pavlov KV (2017) Conformational transitions and interactions underlying the function of membrane embedded receptor protein kinases. Biochim Biophys Acta 1859, 1417–1429 [DOI] [PubMed] [Google Scholar]
- 21.Lu C, Mi LZ, Schurpf T, Walz T, and Springer TA (2012) Mechanisms for kinase-mediated dimerization of the epidermal growth factor receptor. The Journal of biological chemistry 287, 38244–38253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu C, Mi LZ, Grey MJ, Zhu J, Graef E, Yokoyama S, and Springer TA (2010) Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol Cell Biol 30, 5432–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorokin A (1995) Activation of the EGF receptor by insertional mutations in its juxtamembrane regions. Oncogene 11, 1531–1540 [PubMed] [Google Scholar]
- 24.Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM, Leahy DJ, Lidke DS, and Lemmon MA (2017) EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell 171, 683–695 e618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuruvilla R, Zweifel LS, Glebova NO, Lonze BE, Valdez G, Ye HH, and Ginty DD (2004) A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell 118, 243–255 [DOI] [PubMed] [Google Scholar]
- 26.Ascano M, Richmond A, Borden P, and Kuruvilla R (2009) Axonal targeting of Trk receptors via transcytosis regulates sensitivity to neurotrophin responses. J Neurosci 29, 11674–11685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houtz J, Borden P, Ceasrine A, Minichiello L, and Kuruvilla R (2016) Neurotrophin Signaling Is Required for Glucose-Induced Insulin Secretion. Dev Cell 39, 329–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasahira T, Ueda N, Yamamoto K, Bhawal UK, Kurihara M, Kirita T, and Kuniyasu H (2013) Trks are novel oncogenes involved in the induction of neovascularization, tumor progression, and nodal metastasis in oral squamous cell carcinoma. Clinical & Experimental Metastasis 30, 165–176 [DOI] [PubMed] [Google Scholar]
- 29.Gupta VK, You YY, Gupta VB, Klistorner A, and Graham SL (2013) TrkB Receptor Signalling: Implications in Neurodegenerative, Psychiatric and Proliferative Disorders. International Journal of Molecular Sciences 14, 10122–10142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hondermarck H (2012) Neurotrophins and their receptors in breast cancer. Cytokine & Growth Factor Reviews 23, 357–365 [DOI] [PubMed] [Google Scholar]
- 31.Bothwell M (2016) Recent advances in understanding neurotrophin signaling. F1000Res 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan PK, Wang J, Littler PLH, Wong KK, Sweetnam TA, Keefe W, Nash NR, Reding EC, Piu F, Brann MR, and Schiffer HH (2007) Monitoring interactions between receptor tyrosine kinases and their downstream effector proteins in living cells using bioluminescence resonance energy transfer. Molecular Pharmacology 72, 1440–1446 [DOI] [PubMed] [Google Scholar]
- 33.Keeler AB, Suo D, Park J, and Deppmann CD (2017) Delineating neurotrophin-3 dependent signaling pathways underlying sympathetic axon growth along intermediate targets. Mol Cell Neurosci 82, 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belliveau DJ, Krivko I, Kohn J, Lachance C, Pozniak C, Rusakov D, Kaplan D, and Miller FD (1997) NGF and neurotrophin-3 both activate TrkA on sympathetic neurons but differentially regulate survival and neuritogenesis. J Cell Biol 136, 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodmer D, Ascano M, and Kuruvilla R (2011) Isoform-specific dephosphorylation of dynamin1 by calcineurin couples neurotrophin receptor endocytosis to axonal growth. Neuron 70, 1085–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Placone J, Novicky L, and Hristova K (2010) The extracellular domain of fibroblast growth factor receptor 3 inhibits ligand-independent dimerization. Science Signaling 3, ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen LR, Novicky L, Merzlyakov M, Hristov T, and Hristova K (2010) Measuring the Energetics of Membrane Protein Dimerization in Mammalian Membranes. Journal of the American Chemical Society 132, 3628–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarabipour S, Del Piccolo N, and Hristova K (2015) Characterization of Membrane Protein Interactions in Plasma Membrane Derived Vesicles with Quantitative Imaging Forster Resonance Energy Transfer. Acc.Chem.Res 48, 2262–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King C, Stoneman M, Raicu V, and Hristova K (2016) Fully quantified spectral imaging reveals in vivo membrane protein interactions. Integr.Biol.(Camb.) 8, 216–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yano H, Cong F, Birge RB, Goff SP, and Chao MV (2000) Association of the Abl tyrosine kinase with the Trk nerve growth factor receptor. J Neurosci Res 59, 356–364 [DOI] [PubMed] [Google Scholar]
- 41.Singh DR, Ahmed F, Sarabipour S, and Hristova K (2017) Intracellular Domain Contacts Contribute to Ecadherin Constitutive Dimerization in the Plasma Membrane. J Mol Biol 429, 2231–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh DR, Cao Q, King C, Salotto M, Ahmed F, Zhou XY, Pasquale EB, and Hristova K (2015) Unliganded EphA3 dimerization promoted by the SAM domain. Biochemical Journal 471, 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh DR, Pasquale EB, and Hristova K (2016) A small peptide promotes EphA2 kinase-dependent signaling by stabilizing EphA2 dimers. Biochim Biophys Acta 1860, 1922–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha B, Koster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, Morone N, Parton RG, Raposo G, Sens P, Lamaze C, and Nassoy P (2011) Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 144, 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raicu V, Stoneman MR, Fung R, Melnichuk M, Jansma DB, Pisterzi LF, Rath S, Fox M, Wells JW, and Saldin DK (2009) Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nature Photonics 3, 107–113 [Google Scholar]
- 46.Biener G, Stoneman MR, Acbas G, Holz JD, Orlova M, Komarova L, Kuchin S, and Raicu V (2014) Development and Experimental Testing of an Optical Micro-Spectroscopic Technique Incorporating True Line-Scan Excitation. International Journal of Molecular Sciences 15, 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarabipour S, King C, and Hristova K (2014) Un-induced high-yield bacterial expression of fluorescent proteins. Anal.Biochem 449, 155–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolber PK, and Hudson BS (1979) An analytic solution to the F”rster energy transfer problem in two dimensions. Biophysical Journal 28, 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.King C, Sarabipour S, Byrne P, Leahy DJ, and Hristova K (2014) The FRET signatures of non-interacting proteins in membranes: simulations and experiments. Biophysical Journal 106, 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King C, Raicu V, and Hristova K (2017) Understanding the FRET Signatures of Interacting Membrane Proteins. The Journal of biological chemistry 292, 5291–5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Press WH, Flannery BP, Teukolsky SA, and Vetterling WT (1989) Numerical Recipes. The Art of Scientific Computing, Cambridge University Press, Cambridge [Google Scholar]
- 52.Adler J, Shevchuk AI, Novak P, Korchev YE, and Parmryd I (2010) Plasma membrane topography and interpretation of single-particle tracks. Nat.Methods 7, 170–171 [DOI] [PubMed] [Google Scholar]
- 53.Parmryd I, and Onfelt B (2013) Consequences of membrane topography. FEBS.J 280, 2775–2784 [DOI] [PubMed] [Google Scholar]
- 54.Singh DR, Ahmed F, King C, Gupta N, Salotto M, Pasquale EB, and Hristova K (2015) EphA2 Receptor Unliganded Dimers Suppress EphA2 Pro-tumorigenic Signaling. Journal of Biological Chemistry 290, 27271–27279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King C, Wirth D, Workman S, and Hristova K (2017) Cooperative interactions between VEGFR2 extracellular Ig-like subdomains ensure VEGFR2 dimerization. Biochim Biophys Acta 1861, 2559–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh DR, Kanvinde P, King C, Pasquale EB, and Hristova K (2018) The EphA2 receptor is activated through induction of distinct, ligand-dependent oligomeric structures. Communications Biology 1, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adair BD, and Engelman DM (1994) Glycophorin a helical transmembrane domains dimerize in phospholipid bilayers - a resonance energy transfer study. Biochemistry 33, 5539–5544 [DOI] [PubMed] [Google Scholar]
- 58.Li M, Reddy LG, Bennett R, Silva ND, Jones LR, and Thomas DD (1999) A fluorescence energy transfer method for analyzing protein oligomeric structure: Application to phospholamban. Biophysical Journal 76, 2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schick S, Chen LR, Li E, Lin J, Koper I, and Hristova K (2010) Assembly of the M2 Tetramer Is Strongly Modulated by Lipid Chain Length. Biophysical Journal 99, 1810–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin-Zanca D, Oskam R, Mitra G, Copeland T, and Barbacid M (1989) Molecular and biochemical characterization of the human trk proto-oncogene. Mol Cell Biol 9, 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jullien J, Guili V, Reichardt LF, and Rudkin BB (2002) Molecular kinetics of nerve growth factor receptor trafficking and activation. The Journal of biological chemistry 277, 38700–38708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen JY, and Maruyama IN (2011) Nerve growth factor receptor TrkA exists as a preformed, yet inactive, dimer in living cells. Febs Letters 585, 295–299 [DOI] [PubMed] [Google Scholar]
- 63.Evers TH, van Dongen EMWM, Faesen AC, Meijer EW, and Merkx M (2006) Quantitative understanding of the energy transfer between fluorescent proteins connected via flexible peptide linkers. Biochemistry 45, 13183–13192 [DOI] [PubMed] [Google Scholar]
- 64.Sarabipour S, and Hristova K (2015) FGFR3 Unliganded Dimer Stabilization by the Juxtamembrane Domain. Journal of Molecular Biology 427, 1705–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uren RT, and Turnley AM (2014) Regulation of neurotrophin receptor (Trk) signaling: suppressor of cytokine signaling 2 (SOCS2) is a new player. Front Mol Neurosci 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soppet D, Escandon E, Maragos J, Middlemas DS, Reid SW, Blair J, Burton LE, Stanton BR, Kaplan DR, Hunter T, Nikolics K, and Parada LF (1991) The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the trkB tyrosine kinase receptor. Cell 65, 895–903 [DOI] [PubMed] [Google Scholar]
- 67.Rodriguez-Tebar A, Dechant G, Gotz R, and Barde YA (1992) Binding of neurotrophin-3 to its neuronal receptors and interactions with nerve growth factor and brain-derived neurotrophic factor. EMBO J 11, 917–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Urfer R, Tsoulfas P, O’Connell L, and Presta LG (1997) Specificity determinants in neurotrophin-3 and design of nerve growth factor-based trkC agonists by changing central beta-strand bundle residues to their neurotrophin-3 analogs. Biochemistry 36, 4775–4781 [DOI] [PubMed] [Google Scholar]
- 69.Urfer R, Tsoulfas P, Soppet D, Escandon E, Parada LF, and Presta LG (1994) The binding epitopes of neurotrophin-3 to its receptors trkC and gp75 and the design of a multifunctional human neurotrophin. EMBO J 13, 5896–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ivanisevic L, Zheng W, Woo SB, Neet KE, and Saragovi HU (2007) TrkA receptor “hot spots” for binding of NT-3 as a heterologous ligand. The Journal of biological chemistry 282, 16754–16763 [DOI] [PubMed] [Google Scholar]
- 71.Sarabipour S, and Hristova K (2016) Pathogenic Cysteine Removal Mutations in FGFR Extracellular Domains Stabilize Receptor Dimers and Perturb the TM Dimer Structure. J Mol Biol 428, 3903–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maruyama I, and Shen JY (2012) Brain-derived neurotrophic factor receptor TrkB exists as a preformed dimer in living cells. FASEB Journal 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mischel PS, Umbach JA, Eskandari S, Smith SG, Gundersen CB, and Zampighi GA (2002) Nerve growth factor signals via preexisting TrkA receptor oligomers. Biophysical Journal 83, 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clary DO, Weskamp G, Austin LR, and Reichardt LF (1994) TrkA cross-linking mimics neuronal responses to nerve growth factor. Mol Biol Cell 5, 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wehrman T, He X, Raab B, Dukipatti A, Blau H, and Garcia KC (2007) Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron 53, 25–38 [DOI] [PubMed] [Google Scholar]
- 76.Manni S, Mineev KS, Usmanova D, Lyukmanova EN, Shulepko MA, Kirpichnikov MP, Winter J, Matkovic M, Deupi X, Arseniev AS, and Ballmer-Hofer K (2014) Structural and Functional Characterization of Alternative Transmembrane Domain Conformations in VEGF Receptor 2 Activation. Structure 22, 1077–1089 [DOI] [PubMed] [Google Scholar]
- 77.He L, and Hristova K (2012) Physical-chemical principles underlying RTK activation, and their implications for human disease. Biochimica et Biophysica Acta 1818, 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bocharov EV, Mineev KS, Pavlov KV, Akimov SA, Kuznetsov AS, Efremov RG, and Arseniev AS (2017) Helix-helix interactions in membrane domains of bitopic proteins: Specificity and role of lipid environment. Biochim Biophys Acta 1859, 561–576 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.