Abstract
Alcohol dependence (AD) and nicotine dependence (ND) co-occur frequently (AD+ND). We integrated SNP-based, gene-based, and protein-protein interaction (PPI) network analyses to identify shared risk genes or gene subnetworks for AD+ND in African Americans (AAs, N=2094) and European Americans (EAs, N=1207). The DSM-IV criterion counts for AD and ND were modeled as two dependent variables in a multivariate linear mixed model, and analyzed separately for the two populations. The most significant SNP was rs6579845 in EAs (p<1.29×10−8) in GM2A, which encodes GM2 ganglioside activator, and is a cis-expression quantitative locus (eQTL) that affects GM2A expression in blood and brain tissues. However, this SNP was not replicated in another small sample. We identified a subnetwork of 24 genes that contributed to the AD+ND criterion counts. In the gene-set analysis for the subnetwork in an independent sample, the Study of Addiction: Genetics and Environment project (SAGE) (predominately EAs), these 24 genes as a set differed in AD+ND versus control subjects in EAs (p=0.041). Functional enrichment analysis for this subnetwork revealed that the gene enrichment involved primarily nerve growth factor pathways, and cocaine and amphetamine addiction. In conclusion, we identified a genomewide significant variant at GM2A and a gene subnetwork underlying the genetic trait of shared AD+ND. These results increase our understanding of the shared (pleiotropic) genetic risk that underlies AD+ND.
Keywords: GWAS, Network analysis, Alcohol dependence co-occurring nicotine dependence, Pleiotropy
1. INTRODUCTION
Alcohol and nicotine are often used together. Severe heavy smoking is correlated with risky drinking behaviors (John et al., 2003). Alcohol and nicotine can each moderate the physiological and psychological effects of the other (Hurley et al., 2012). Oliver et al. (Oliver et al., 2013) found that pharmacological interaction between nicotine and alcohol mutually affects craving in a complex manner. Alcohol and nicotine potentiate each other’s rewarding and antagonistic effects, and nicotine tends to alter the sedative and rewarding effects of ethanol (Rose et al., 2004, Packingham, 2014). Genetic epidemiologic studies have shown that common genetic factors influence nicotine and alcohol use (Hopfer et al., 2001, Schlaepfer et al., 2008, True et al., 1999).
Previous genome-wide association studies (GWAS) of alcohol dependence (AD) and nicotine dependence (ND) individually identified numerous significant risk loci (Gelernter et al., 2014b, Gelernter et al., 2014a, Zuo et al., 2012, Treutlein et al., 2009, Frank et al., 2012, Tobacco and Consortium, 2010, Jorgenson et al., 2017). A pooling-based GWAS for AD co-occurring with ND (AD+ND) in an Australian population identified significant markers at MARK1, DDX6, and KIAA1409 (Lind et al., 2010). Another analysis (Zuo et al., 2013) also implemented GWAS analysis in publicly-available datasets to obtain a significant region of IPO11-HIR1A, which might harbor a causal variant for AD+ND in EAs.
In this study, we sought to identify shared risk genes or gene subnetworks that contribute to shared risk for AD+ND. Our discovery GWAS analysis included individuals who met DSM-IV diagnostic criteria for both AD and ND, and alcohol- and nicotine- exposed individuals without AD or ND (total N=2094 AAs, 1207 EAs). Another 1937 subjects (1381 EAs and 556 AAs) from the SAGE study (Bierut et al., 2010, Edenberg et al., 2006) were used to validate our network findings. The analyses we carried out included (1) a multivariate linear mixed model to examine simultaneously criterion counts of AD and ND for the SNP-based analysis; (2) a dense module search within the PPI network to identify gene subnetworks enriched for AD and ND criterion count associated genes; (3) gene set analysis to assess the association of the genes involved in the identified subnetwork as a whole with AD+ND using independent case-control samples from SAGE, and (4) pathway enrichment analysis to evaluate whether genes in the subnetwork share pathway features.
2. METHODS AND MATERIALS
2.1. Subjects and diagnostic description
Yale-Penn GWAS Discovery samples
All subjects included in the discovery sample were recruited for studies of the genetics of cocaine dependence (CD), opioid dependence (OD), and AD. Subjects were recruited at five US clinical sites: Yale University School of Medicine (APT Foundation, New Haven, CT, USA), the University of Connecticut Health Center (Farmington, CT, USA), the University of Pennsylvania Perelman School of Medicine (Philadelphia, PA, USA), the Medical University of South Carolina (Charleston, SC, USA) and McLean Hospital (Belmont, MA, USA). Subjects were administered the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (Pierucci-Lagha et al., 2005) to obtain information regarding each DSM-IV (APA, 1994) lifetime criterion for AD and ND.
Subjects were selected from the larger GWASed cohort of substance use disorders (Gelernter et al., 2015, Gelernter et al., 2014a, Gelernter et al., 2014c, Kalayasiri et al., 2014) according to the inclusion criteria of extreme phenotypes for the current study. The inclusion criteria for the discovery samples (AD+ND) (1) were that individuals had to meet DSM-IV diagnostic criteria for both AD and ND. The inclusion criteria for controls were that individuals had exposure to both alcohol and nicotine, but did not meet DSM-IV criteria for dependence on either of these substances. In Stage 1 (discovery sample), we used the criterion counts as an ordinal variable in the linear mixed models with AD+ND and control samples. A total of 2094 AAs (1303 cases and 791 controls) and 1207 EAs (1079 cases and 128 controls) met the inclusion criteria (Table 1). Our prior studies (such as (Gelernter et al., 2015)) have indicated that continuous/ordinal traits have more powerful than case/control design because of greater information content and improved specificity of the dependence measure, thus considering those subjects who met criteria for dependence on both substances (APA, 1994), was then defined on the basis of criterion count. The distribution of criterion counts in the discovery samples is showed in Table 2.
Table 1.
Sample characteristics
| EA | AA | |||
|---|---|---|---|---|
| AD+ND (male %) | Control (male %) | AD+ND (male %) | Control (male %) | |
| Yale-Penn discovery sample | 1079 (61.9%) | 128 (42.9%) | 1303 (56.8%) | 791 (37.4%) |
| Yale-Penn replication sample | 637 (67.5%) | 41 (48.8%) | -- | -- |
| SAGE sample | 797 (59.1%) | 584 (31.3%) | 403 (55.1%) | 153 (40.5%) |
Table 2.
The distribution of criterion counts for AD and ND
| Symptoms | AD DSM-IV Criteria | Number of AD subjects endorsed only this criteria (EA) | Number of AD subjects endorsed only this criteria (AA) | Number of subjects endorsed both in AD and ND (EA) | Number of subjects endorsed both in AD and ND (AA) | Number of ND subjects endorsed only this criteria (EA) | Number of ND subjects endorsed only this criteria (AA) | ND DSM-IV Criteria |
|---|---|---|---|---|---|---|---|---|
| Tolerance | Tolerance means that, over time, you need more alcohol to feel the same effect | 81 | 298 | 840 | 701 | 169 | 218 | Built up a tolerance so that same amount of cigarettes had less effect than before |
| Withdrawal | As the effect of the alcohol wears off you may experience withdrawal symptoms, such as anxiety or jumpiness | 277 | 49 | 748 | 875 | 48 | 133 | Cigarette use caused emotional or psychological problems |
| Loss of Control | Drinking more than you wanted to, for longer than you intended | 106 | 156 | 954 | 1125 | 41 | 111 | Used cigarettes more often or in larger amounts than intended than intended |
| Desire to Stop- But Can’t | You have a persistent desire to cut down or stop your alcohol use, but all efforts to stop and stay stopped, have been unsuccessful | 743 | 976 | 215 | 250 | 21 | 25 | Wanted or tried to stop or cut down on your cigarette use |
| Neglecting Other Activities | You are spending less time on activities that used to be important to you (hanging out with family and friends, exercising- going to the gym, pursuing your hobbies or other interests) because of the use of alcohol | 67 | 60 | 775 | 888 | 244 | 464 | Cigarette use kept you from working, going to school, taking care of children, or engaging in recreational activities |
| Takes Up Greater Time, Energy and Focus | You spend a lot of time drinking, thinking about it, or recovering from its effects. You have few, if any, interests, social or community involvements that don’t revolve around the use of alcohol | 150 | 216 | 605 | 624 | 269 | 314 | Spent great deal of time getting, using, or getting over effects of cigarettes |
| Continued Use Despite Negative Consequences | You drink even though they know it’s causing problems. As an example, you realize that your alcohol use is interfering with your ability to do your job, is damaging your marriage, making your problems worse, or causing health problems, but you continue to drink | 314 | 475 | 754 | 708 | 28 | 91 | Cigarette use caused health problems |
AD: alcohol dependence; ND: nicotine dependence; EA: African- Americans; AA: European- Americans
Subjects gave written informed consent as approved by the institutional review boards at each clinical site, and certificates of confidentiality were obtained from the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism.
Yale-Penn GWAS EA Replication Sample
This sample included another 1,439 EA subjects, recruited from the same sites and using the same recruitment instruments as the discovery sample, Of the subjects who were exposed to alcohol or nicotine, 637 had dual diagnosis of AD+ND, and 41 had neither AD nor ND.
Independent Sample Used to Evaluate the Association of Genes Involved in the Subnetwork for AD Co-occurring ND
The network validation analyses included GWAS data from the SAGE collaboration obtained after application from dbGAP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi? study_id=phs000092.v1.p1); SAGE includes individuals from the Collaborative Study on the Genetics of Alcoholism (COGA) (Edenberg, 2002), the Family Study of Cocaine Dependence (FSCD) (Bierut et al., 2008), and the Collaborative Genetic Study of Nicotine Dependence (COGEND) (Bierut et al., 2007). Since SAGE included subjects in nuclear families and the gene-set analysis using the software SNP-set (Sequence) Kernel Association Test (SKAT) (Wu et al., 2010) allows only unrelated individuals, we randomly selected one subject from each nuclear family and combined these with the rest of the sample of unrelated individuals. Thus, we included 797 and 403 unrelated AD+ND subjects and 584 and 153 controls for EA and AA respectively, after applying the same selection criteria as for the discovery sample (Table 1).
2.2. Genotyping and quality control
Our GWAS discovery sample was genotyped on the Illumina HumanOmni1-Quad v1.0 microarray, which included 988,306 autosomal SNPs. Genotyping was conducted at the Yale Center for Genome Analysis and at the Center for Inherited Disease Research (CIDR). Genotypes were called using the GenomeStudio software V2011.1 and genotyping module version 1.8.4 (Illumina). Samples and SNPs were excluded from analysis based on predetermined quality control (QC) criteria, as described elsewhere (Gelernter et al., 2014a). Of the discovery sample, 135 individuals with call rates <98% were excluded, and 62,076 SNPs were removed because of minor allele frequencies < 1%. After data cleaning and quality control, 5,697 individuals and 889,659 SNPs remained for imputation analysis.
In the SAGE sample, samples and SNPs were excluded either because of sample call rates ≤ 95%, SNP call rates ≤ 98%, minor allele frequencies (MAF) ≤0.01, or Hardy-Weinberg Equilibrium (HWE) P-value ≤ 10−5. 721,075 SNPs remained for analysis.
In the Yale-Penn replication sample which were genotyped using the Illumina HumanCore Exome array containing approximately 266,000 exome SNPs and 240,000 tagging SNPs for genome-wide imputation. Standard pre-imputation quality control excluded individuals with call rates < 98%, and SNPs with minor allele frequency (MAF) < 1%. The remaining SNPs were imputed into the 1000 Genomes phase 1 reference panel ( The 1000 Genomes Project Consortium, 2010).
The procedure used to control for population stratification has been described previously (Gelernter et al., 2014a). Briefly, to verify and correct misclassification of self-reported race, we compared GWAS data from all subjects included herein with genotypes from the HapMap 3 reference CEU (CEPH collection), YRI (Yoruba in Ibadan, Nigeria) and CHB (Han Chinese in Beijing, China) populations. Principal component analysis (PCA) was performed in the GWAS sample using Eigensoft software (Price et al., 2006) and 145 472 SNPs that were common to our GWAS data set and the HapMap panel (after pruning GWAS SNPs for linkage disequilibrium (LD), r2>80%) to characterize the underlying genetic architecture of the samples. The first PC score distinguished AAs and EAs; these groups were then analyzed separately. We conducted PC analyses within the two groups taken separately, and the first three PCs were used in all subsequent analyses to correct for residual population stratification.
2.3. Genotype imputation
Imputation for the discovery and replication sample was described previously (Gelernter et al., 2014c). In summary, genotypes for 37,426,733 SNPs were imputed with IMPUTE2 (Howie et al., 2009) based on 1000 Genomes reference panel released in June 2011 (http://www.1000genomes.org/) (The 1000 Genomes Project Consortium, 2010). AA and EA samples were imputed separately. Imputed SNPs with r2>0.9 were used in the following analysis.
2.4. Statistical Analysis Methods
Association Tests for the Yale-Penn GWAS sample
We used Genome-wide Efficient Mixed Model Association (GEMMA) (Zhou and Stephens, 2012), for the SNP-trait association tests, using age, sex, and the first three principle components (PCs) as covariates (Gelernter et al., 2015). GEMMA implements multivariate linear mixed models (mvLMMs) (Zhou and Stephens, 2014), which are statistical regression models that relate explanatory variables to multiple correlated outcome variables and have been widely applied in genetics because they can account for relatedness among samples (Henderson, 1984). The algorithm is based on linear algebra techniques previously used for univariate LMMs (Lippert et al., 2011) and extends them to mvLMMs. In this study, we used the mvLMMs to test associations between SNPs and criterion counts for AD and ND simultaneously. We also used quantile-quantile (QQ) plots based on p-values of genomewide SNPs to evaluate further the possibility of population stratification or other systematic biases.
Searching Modules Enriched for Genes Associated with AD+ND in the Yale-Penn GWAS sample
The module search included two steps. First, we converted the results of SNP-based association from the multivariate GWAS on AD+ND to gene-based association using VEGAS (Liu et al., 2010) to choose the smallest p-value among all SNPs mapped to each gene, i.e., based on the best p-value for each gene. Second, we used an R package, dmGWAS (Jia et al., 2010), to identify candidate subnetworks using gene-based p-value for each gene as input data and a node-weighted network analysis in the knowledge-based human protein-protein interaction (PPI) network. We used a comprehensive human brain tissue-specific PPI network downloaded from Tissue Net Database (Barshir et al., 2013) for the dense module search, which will yield subnetworks enriched with low p-value genes. The dense module searches were implemented separately for AAs and EAs.
We also searched for significant modules enriched for both AD and ND related genes by a dual-evaluation strategy, in which the modules identified in EAs were used as the discovery dataset, and the modules identified in AAs as the evaluation dataset (Jia et al., 2010). Subsequently, significant modules were extracted and merged to construct a subnetwork for further association analysis.
Validation of Associations Based on Genes Assigned to Subnetworks using the SAGE sample
To confirm the association of the genes involved in the gene subnetworks identified for AD+ND, a permutation test was used to obtain empirical p-values. We extracted all SNPs from the SAGE dataset that were mapped to genes in the final subnetwork. We bootstrapped the data by randomly shuffling phenotypes and genotypes while leaving the correlation of phenotype with genotype unchanged. This process was repeated 10,000 times and 10,000 results (Zn) were obtained. The empirical p value for the association of the subnetwork with AD+ND was evaluated by determining the proportion of the Zn less than or equal to the observed ones in Zn data sets.
Pathway Enrichment Analysis
We submitted the genes in the subnetwork to ConsensusPathDB (Kamburov et al., 2013), a database for annotation, for pathway enrichment analysis. A hypergeometric test implemented in ConsensusPathDB was used to compute the enrichment p-value for pathways, followed by false discovery rate (FDR) correction for multiple testing. We also used BioGPS software (Wu et al., 2009) (http://biogps.org/#goto=welcome), which implements a virtually unstructured format for integrating disparate gene annotation resources to observe gene expression.
3. RESULTS
3.1. Genomewide multivariate analysis in EAs and AAs analyzed separately
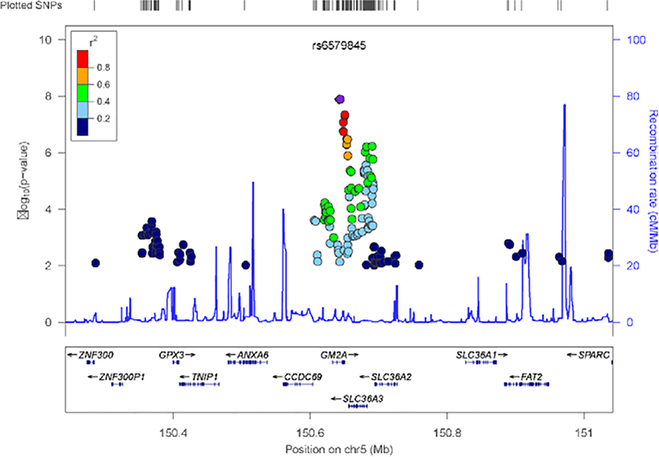
There were significant phenotypic correlations of criterion counts for AD and ND in EAs (r=0.43; p<0.001) and in AAs (r=0.78; p<0.001), as expected. In EAs, three SNPs mapped to GM2A (GM2 ganglioside activator) were significantly associated with AD+ND (Table 3), and the strongest association among the three genomewide significant (GWS) SNPs was at rs6579845 (p<1.29×10−8) (Figure 1; Supplementary Figure S1a, S1c). These three SNPs are in high linkage disequilibrium (r2 ranging 0.83~1). In AAs, no single SNP reached GWS in the analysis of the associations between SNPs and criterion counts for AD and ND simultaneously (Supplementary Figure S1b, S1d, S2).
Table 3.
Genomewide association analysis on alcohol dependence comorbid with nicotine dependence in European-Americans: Results for SNPs with P values < 5×10−7.
| chr | rs | ps | minor allele | major allele | minor allele frequency | beta_1 | beta_2 | P value | Gene |
|---|---|---|---|---|---|---|---|---|---|
| 5 | rs6579845 | 150642265 | A | G | 0.288 | 2.59E-01 | 2.45E-01 | 1.29E-08 | GM2A |
| 5 | rs12516391 | 150649006 | A | G | 0.265 | 2.53E-01 | 2.41E-01 | 4.54E-08 | GM2A |
| 5 | rs3806953 | 150648878 | C | T | 0.266 | 2.52E-01 | 2.41E-01 | 4.99E-08 | GM2A |
| 5 | rs2075783 | 150646780 | A | C | 0.279 | 2.47E-01 | 2.28E-01 | 8.45E-08 | GM2A |
| 5 | rs2075782 | 150646647 | T | C | 0.281 | 2.40E-01 | 2.25E-01 | 1.75E-07 | GM2A |
| 5 | rs6861766 | 150646736 | C | T | 0.281 | 2.40E-01 | 2.24E-01 | 1.78E-07 | GM2A |
| 5 | rs1048723 | 150647012 | A | G | 0.281 | 2.40E-01 | 2.24E-01 | 1.83E-07 | GM2A |
| 9 | rs74423531 | 94258161 | G | C | 0.012 | 8.01E-01 | 1.06E+00 | 1.83E-07 | Intergenic |
| 5 | rs1065018 | 150651775 | A | G | 0.262 | 2.39E-01 | 2.23E-01 | 3.29E-07 | Intergenic |
| 5 | rs58426811 | 150652841 | A | G | 0.262 | 2.39E-01 | 2.23E-01 | 3.42E-07 | Intergenic |
| 5 | chr5:150650679:T/C:1 | 150650679 | T | C | 0.263 | 2.39E-01 | 2.23E-01 | 3.60E-07 | Intergenic |
| 3 | rs263010 | 183497506 | A | G | 0.443 | 2.06E-01 | 1.97E-01 | 3.78E-07 | YEATS2 |
| 5 | rs35964963 | 150651584 | T | C | 0.261 | 2.37E-01 | 2.21E-01 | 4.97E-07 | Intergenic |
Chr, chromosome numbers; rs, snp ids; ps, base pair positions on the chromosome; beta_1: beta estimates for the association of SNP with AD criterion counts); beta_2: beta estimates for the association of SNP with ND criterion counts; P value was calculated for the association of SNP with criterion counts of AD and ND simultaneously.
Figure 1.
Regional Manhattan plot of the genomewide association analysis result for alcohol dependence co-occurring nicotine dependence in the European American sample. Significance of GWAS genotyped and imputed SNPs of the 400kb region in the chromosomal region of GM2A.
A post hoc search found that the most significant SNP, rs6579845, is a cis-expression quantitative locus (eQTL). The association between rs6579845 and GM2A expression is significant in the dorsolateral prefrontal cortex (dlPFC) from human postmortem samples (p=5.34×10−9, pFDR<0.0001) (Colantuoni et al., 2011), and also highly significant (p=9.47×10−20, pFDR<0.0001) in peripheral blood (Westra et al., 2013). Rs6579845 thus affects GM2A expression in blood and brain tissues.
However, replication for the association of rs6579845 in the Yale-Penn EA replication sample was not significant (p=0.42).
3.2. Modules Enriched for Genes Associated with AD+ND
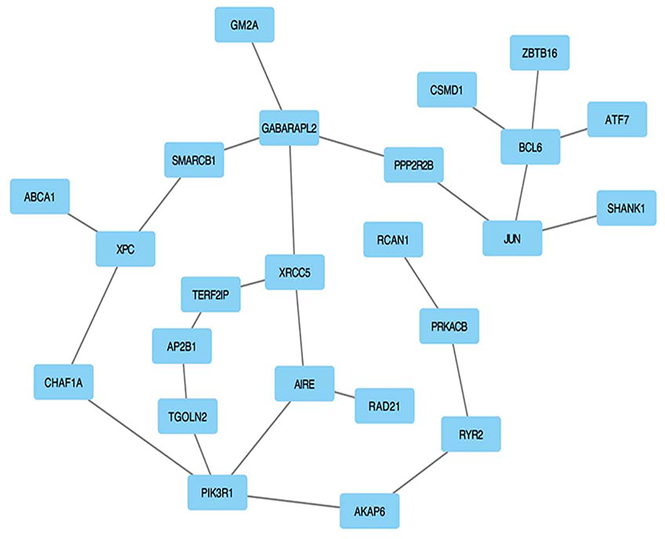
A dense module search within the PPI network was carried out for the results of gene-based analysis in AAs and EAs separately. We observed 2482 and 2804 modules in EAs and AAs, respectively, and further analyses using a dual-evaluation strategy identified five modules that were significant in the enrichment for AD+ND-associated genes for both AA and EA populations. Supplementary Table S1 shows the associated genes and statistics for each of the five modules. Out of the five modules, we removed the overlapping genes and combined all the genes to reconstruct a final subnetwork that included 24 non-redundant genes (Supplementary Table S1). Figure 2 shows the characteristics of this gene subnetwork.
Figure 2.
Gene subnetwork constructed with five significant modules generated in AAs and EAs in the AD co-occurring with ND GWAS Data Sets. All of the genes involved in each of the five significant modules were pulled together and resulted in 24 genes after the overlapping genes were removed (Table S1). Gene subnetwork of these 24 genes was generated from the human protein-protein interaction (PPI) network; we used Cytoscape software (http://cytoscape.org/) to show the relationship between these genes.
3.3. Validation of association for genes in the subnetwork in the SAGE sample
In total, we extracted 782 SNPs from the SAGE data that mapped to the 24-gene subnetwork identified in the discovery sample. We evaluated the cumulative evidence for association with AD+ND for these 782 SNPs, and observed significant enrichment of common variants for genes in the subnetwork compared to controls (p=0.041) in the EA part of the SAGE dataset, but not in the AA part.
3.4. Pathway Enrichment Analysis
This analysis revealed a number of pathways that remained significantly enriched (p<0.05) after FDR correction. The enriched pathways are related to oxytocin signaling pathway, the nerve growth factor pathway, calcium signaling in the CD4+T cell receptor (TCR) pathway, ErbB2/ErbB3 signaling events, cocaine addiction, and amphetamine addiction (Supplementary Table S2). Two of the 24 genes (JUN and PRKACB) were each involved in two of these pathways, and we found that JUN and PRKACB are highly expressed in lung and in brain occipital lobe (Supplementary Figure S3a, S3b).
4. DISCUSSION
We used a multivariate method to study the genetics of AD and ND when they co-occur, i.e. are comorbid. This was based on genetic epidemiological evidence demonstrating a shared genetic basis for these traits, and on their frequent comorbidity in clinical contexts (Hopfer et al., 2001, Schlaepfer et al., 2008, True et al., 1999). The results based on analysis of the correlated phenotypes may be due to pleiotropy, as well as shared environment and individually-relevant genetic factors (Korte et al., 2012). We observed a GWS association signal in EAs at rs6579845 (p<1.29×10−8); this maps to the GM2A (GM2 ganglioside activator) gene. The protein encoded by this gene is a small glycolipid transport protein which acts as a substrate-specific co-factor for the lysosomal enzyme β-hexosaminidase A. Along with the GM2 ganglioside activator, it catalyzes the degradation of the ganglioside GM2, as well as other molecules containing terminal N-acetyl hexosamines. Deficiency of GM2 activator proteins can cause abnormal ganglioside storage in a mouse model (Liu et al., 1997). On postnatal day 7 of brain exposure to ethanol, GM2 increased mainly in lysosomes/late endosomes of activated microglia, suggesting that GM2 may be derived from more complex gangliosides of degenerating neurons (Kolter and Sandhoff, 2006). Increased GM2 by ethanol exposure is also found in mitochondria and lysosomes in neurons (Saito et al., 2012). Abnormal lysosomes were related to chronic alcohol consumption and withdrawal, which specifically induces alterations in the medial prefrontal cortex, hippocampus, and cerebellum (Cadete-Leite et al., 1988). These findings suggested that GM2 is associated with lysosomes in neurons, and regulates the nervous system, which might influence alcohol and nicotine dependence risk. However, this SNP rs6579845 was not replicated in another small sample (the Yale-Penn sample for replication has a low sample size which might make the replication result unreliable for this complex trait of AD and ND).
Based on genes identified by the gene-based and PPI analyses, we then identified a subnetwork of 24 AD+ND-associated genes (including GM2A). Pathway enrichment analysis also revealed that oxytocin (OT) signaling may have an important role in the pathological mechanism of AD+ND. We confirmed that the gene subnetwork association as a set differed between the AD+ND versus controls in the SAGE sample. The gene subnetwork was selected based on a PPI network that might include nonlinear interactions, so we tested the association of this gene subnetwork with AD+ND using the SNP-set (Sequence) Kernel Association Test (SKAT), which applies a statistical framework that accounts for both linear and nonlinear interactions (Wu et al., 2010). There was a significant association of the gene subnetwork with AD+ND (p=0.041) in the SAGE EAs, but not AAs samples (This may be because the AA sample in SAGE is smaller than the EA sample, and provides lower power).
In the pathway enrichment analysis, the most significant pathway was oxytocin signaling. Oxytocin is mainly produced in the supraoptic, paraventricular, and accessory magnocellular nuclei of the hypothalamus (Sofroniew, 1983). Oxytocin exerts diverse central and peripheral effects, though the most known and established roles are stimulating uterine contractions during childbirth and milk production during lactation (Augustine et al., 2018). Oxytocin is also involved in cardiovascular regulation and various stress-related neuropsychiatric disorders (Vargas-Martínez et al., 2017, Iovino et al., 2018). Oxytocin peptide mRNA was significantly elevated in the prefrontal cortex of male subjects with alcohol use disorder compared to controls, using post-mortem brain tissue (Lee et al., 2017b). Other studies have also found that oxytocin was associated with nicotine dependence and withdrawal in rats (Lee et al., 2017a, Manbeck et al., 2014), specifically that prenatal ethanol and nicotine exposure affects ethanol consumption, ethanol preference, and oxytocin receptor binding in adolescent and adult rats (Williams et al., 2009).
Our study highlights several advantages of combining SNP-based and network-based analysis of GWAS. First, we applied multivariate analytical tactics to integrate criterion counts for AD and ND phenotypes. Second, by using network-based analysis, we identified 24 genes in the context of a PPI network. Third, pathway enrichment analyses of the genes in the subnetwork uncovered pathway annotations that may provide a better understanding the neurobiological mechanism of the pathway identified and thereby of AD+ND. Fourth, the significant association found in the subnetwork for AD+ND was replicated in the EA SAGE dataset, demonstrating convergent validity for the findings.
Our study has several noteworthy limitations; foremost among these is the limitation in sample size. In addition, in our gene-based analysis implementation, we mapped gene-level p values by considering the smallest p value SNP in each gene. Under this model, association signals for genes with multiple independent risk variants are not well captured.
In conclusion, through integrated SNP-based, gene-based analyses, and PPI network analysis, we identified GM2A as a risk SNP of genomewide significance, and more importantly, a specific gene subnetwork as relevant to the risk for AD+ND, which improve our understanding of one of (presumably many) shared biological mechanisms for these two traits.
Supplementary Material
Acknowledgments
Recruitment and assessment were overseen at McLean Hospital by Roger Weiss, M.D., at the Medical University of South Carolina by Kathleen Brady, M.D., Ph.D., at the University of Connecticut Health Center by Henry R. Kranzler, M.D., and at the University of Pennsylvania, initially by David Oslin, M.D. and then by Henry R. Kranzler, M.D. Genotyping services for our GWAS study were provided by the Center for Inherited Disease Research (CIDR) and Yale University (Center for Genome Analysis). We are grateful to Ann Marie Lacobelle and Christa Robinson for their technical assistance, to the SSADDA interviewers, led by Yari Nuñez and Michelle Slivinsky, to Richard Sherva, and Ryan Koesterer for their assistance with data processing and quality control, and to John Farrell for database management assistance.
Funding and Disclosure
This work was funded by the National Institutes of Health [RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, K01 DA24758, and P50 AA12870]; the US Veterans Affairs Connecticut and Veterans Affairs Philadelphia Mental Illness Research, Education and Clinical Centers; and the VA Medical Research Service. Genotyping services at the Center for Inherited Disease Research (CIDR) at The Johns Hopkins University were supported by the National Institutes of Health [contract N01-HG-65403]. Additional funding was provided by the China Scholarship Council (Xiang).
Footnotes
Drs. Xiang, Yang, Zhou, and Gelernter declare no conflict of interest. Dr. Kranzler declares has been a consultant, advisory board member, or CME speaker for Indivior and Lundbeck. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative (ACTIVE), which in the last three years was supported by AbbVie, Alkermes, Amygdala Neurosciences, Arbor Pharmaceuticals, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, and Pfizer.
References
- Association AP (1994) Diagnostic and statistical manual of mental disorders. Forth edition. Washington, DC. [Google Scholar]
- Augustine RA, Seymour AJ, Campbell RE, Grattan DR, Brown CH (2018) Integrative neuro‐humoral regulation of oxytocin neuron activity in pregnancy and lactation. Journal of neuroendocrinology. [DOI] [PubMed] [Google Scholar]
- Barshir R, Basha O, Eluk A, Smoly IY, Lan A, Yeger-Lotem E (2013) The TissueNet database of human tissue protein–protein interactions. Nucleic acids research 41:D841–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S (2010) A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences 107:5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L (2007) Novel genes identified in a high-density genome wide association study for nicotine dependence. Human molecular genetics 16:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB (2008) Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug and alcohol dependence 95:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadete-Leite A, Alves M, Tavares M (1988) Lysosomal abnormalities in the pyramidal cells of the rat medial prefrontal cortex after chronic alcohol consumption and withdrawal. Journal of submicroscopic cytology and pathology 20:115–122. [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE (2011) Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GP (2010) A map of human genome variation from population scale sequencing. Nature 467:1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ (2002) The collaborative study on the genetics of alcoholism: an update. Alcohol Research and Health 26:214–218. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T (2006) Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Human molecular genetics 15:1539–1549. [DOI] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P (2012) Genome‐wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addiction biology 17:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler H, Sherva R, Almasy L, Koesterer R, Smith A, Anton R, Preuss U, Ridinger M, Rujescu D (2014a) Genome-wide association study of alcohol dependence: significant findings in African-and European-Americans including novel risk loci. Molecular psychiatry 19:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, Zhao H, Farrer LA (2014b) Genome-Wide Association Study of Nicotine Dependence in American Populations: Identification of Novel Risk Loci in Both African-Americans and European-Americans. Biological psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, Zhao H, Farrer LA (2015) Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biological psychiatry 77:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler H, Farrer L (2014c) Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Molecular psychiatry 19:717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CR (1984) Applications of linear models in animal breeding. Applications of linear models in animal breeding. [Google Scholar]
- Hopfer CJ, Stallings MC, Hewitt JK (2001) Common genetic and environmental vulnerability for alcohol and tobacco use in a volunteer sample of older female twins. Journal of studies on alcohol 62:717–723. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley LL, Taylor RE, Tizabi Y (2012) Positive and negative effects of alcohol and nicotine and their interactions: a mechanistic review. Neurotoxicity research 21:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovino M, Messana T, De GP, Iovino E, Dicuonzo F, Guastamacchia E, Giagulli V, Triggiani V (2018) The Role of Neurohypophyseal Hormones Vasopressin and Oxytocin in Neuropsychiatric Disorders. Endocrine, metabolic & immune disorders drug targets. [DOI] [PubMed] [Google Scholar]
- Jia P, Zheng S, Long J, Zheng W, Zhao Z (2010) dmGWAS: dense module searching for genome-wide association studies in protein–protein interaction networks. Bioinformatics 27:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John U, Meyer C, Rumpf H-J, Schumann A, Thyrian JR, Hapke U (2003) Strength of the relationship between tobacco smoking, nicotine dependence and the severity of alcohol dependence syndrome criteria in a population-based sample. Alcohol and Alcoholism 38:606–612. [DOI] [PubMed] [Google Scholar]
- Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C, Choquet H (2017) Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Molecular psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalayasiri R, Verachai V, Gelernter J, Mutirangura A, Malison RT (2014) Clinical features of methamphetamine-induced paranoia and preliminary genetic association with DBH-1021C-->T in a Thai treatment cohort. Addiction (Abingdon, England) 109:965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Stelzl U, Lehrach H, Herwig R (2013) The ConsensusPathDB interaction database: 2013 update. Nucleic acids research 41:D793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter T, Sandhoff K (2006) Sphingolipid metabolism diseases. Biochimica et Biophysica Acta (BBA)-Biomembranes 1758:2057–2079. [DOI] [PubMed] [Google Scholar]
- Korte A, Vilhjálmsson BJ, Segura V, Platt A, Long Q, Nordborg M (2012) A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nature genetics 44:1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Jang M, Noh J (2017a) Oxytocin attenuates aversive response to nicotine and anxiety-like behavior in adolescent rats. Neuroscience research 115:29–36. [DOI] [PubMed] [Google Scholar]
- Lee MR, Schwandt ML, Sankar V, Suchankova P, Sun H, Leggio L (2017b) Effect of alcohol use disorder on oxytocin peptide and receptor mRNA expression in human brain: A post-mortem case-control study. Psychoneuroendocrinology 85:14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, De Moor MH, Smit AB, Hottenga J-J, Richter MM, Heath AC (2010) A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin research and human genetics: the official journal of the International Society for Twin Studies 13:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D (2011) FaST linear mixed models for genome-wide association studies. Nature methods 8:833–835. [DOI] [PubMed] [Google Scholar]
- Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG (2010) A versatile gene-based test for genome-wide association studies. The American Journal of Human Genetics 87:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hoffmann A, Grinberg A, Westphal H, McDonald MP, Miller KM, Crawley JN, Sandhoff K, Suzuki K, Proia RL (1997) Mouse model of GM2 activator deficiency manifests cerebellar pathology and motor impairment. Proceedings of the National Academy of Sciences of the United States of America 94:8138–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manbeck KE, Shelley D, Schmidt CE, Harris AC (2014) Effects of oxytocin on nicotine withdrawal in rats. Pharmacology Biochemistry and Behavior 116:84–89. [DOI] [PubMed] [Google Scholar]
- Oliver JA, Blank MD, Van Rensburg KJ, MacQueen DA, Brandon TH, Drobes DJ (2013) Nicotine interactions with low-dose alcohol: pharmacological influences on smoking and drinking motivation. Journal of abnormal psychology 122:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packingham K (2014) The Effects of Cigarette Smoking During Acute Alcohol Intoxication.
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR (2005) Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA). Drug and alcohol dependence 80:303–312. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics 38:904–909. [DOI] [PubMed] [Google Scholar]
- Rose JE, Brauer LH, Behm FM, Cramblett M, Calkins K, Lawhon D (2004) Psychopharmacological interactions between nicotine and ethanol. Nicotine & Tobacco Research 6:133–144. [DOI] [PubMed] [Google Scholar]
- Saito M, Chakraborty G, Shah R, Mao RF, Kumar A, Yang DS, Dobrenis K, Saito M (2012) Elevation of GM2 ganglioside during ethanol-induced apoptotic neurodegeneration in the developing mouse brain. Journal of neurochemistry 121:649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Ehringer MA (2008) The genetic components of alcohol and nicotine co-addiction: from genes to behavior. Current drug abuse reviews 1:124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M (1983) Morphology of vasopressin and oxytocin neurones and their central and vascular projections, in Progress in brain research, Vol. 60, Progress in brain research, pp 101–114, Elsevier. [DOI] [PubMed] [Google Scholar]
- Tobacco, Consortium G (2010) Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature genetics 42:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N (2009) Genome-wide association study of alcohol dependence. Archives of general psychiatry 66:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, Eisen SA, Lyons MJ, Goldberg J, Tsuang M (1999) Common genetic vulnerability for nicotine and alcohol dependence in men. Archives of general psychiatry 56:655–661. [DOI] [PubMed] [Google Scholar]
- Vargas-Martínez F, Schanler RJ, Abrams SA, Hawthorne KM, Landers S, Guzman-Bárcenas J, Muñoz O, Henriksen T, Petersson M, Uvnäs-Moberg K (2017) Oxytocin, a main breastfeeding hormone, prevents hypertension acquired in utero: A therapeutics preview. Biochimica et Biophysica Acta (BBA)-General Subjects 1861:3071–3084. [DOI] [PubMed] [Google Scholar]
- Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, Van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, t Hoen PA, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Volker U, Perola M, Salomaa V, Brody J, Suchy-Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JB, Franke L (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nature genetics 45:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SK, Cox ET, McMurray MS, Fay EE, Jarrett TM, Walker CH, Overstreet DH, Johns JM (2009) Simultaneous prenatal ethanol and nicotine exposure affect ethanol consumption, ethanol preference and oxytocin receptor binding in adolescent and adult rats. Neurotoxicology and teratology 31:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology 10:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X (2010) Powerful SNP-set analysis for case-control genome-wide association studies. The American Journal of Human Genetics 86:929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nature genetics 44:821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M (2014) Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nature methods 11:407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li C-SR, Wang F, Zhang X-Y (2012) Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology 37:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CSR, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X (2013) Genome‐Wide significant association signals in IPO11‐HTR1A region specific for alcohol and nicotine codependence. Alcoholism: Clinical and Experimental Research 37:730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.