Abstract
Gene disruption by clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein 9 (Cas9) is highly efficient and relies on the error-prone non-homologous end-joining pathway. Conversely, precise gene editing requires homology-directed repair (HDR), which occurs at a lower frequency than non-homologous end-joining in mammalian cells. Here, by testing whether manipulation of DNA repair factors improves HDR efficacy, we show that transient ectopic co-expression of RAD52 and a dominant-negative form of tumour protein p53-binding protein 1 (dn53BP1) synergize to enable efficient HDR using a single-stranded oligonucleotide DNA donor template at multiple loci in human cells, including patient-derived induced pluripotent stem cells. Co-expression of RAD52 and dn53BP1 improves multiplexed HDR-mediated editing, whereas expression of RAD52 alone enhances HDR with Cas9 nickase. Our data show that the frequency of non-homologous end-joining-mediated double-strand break repair in the presence of these two factors is not suppressed and suggest that dn53BP1 competitively antagonizes 53BP1 to augment HDR in combination with RAD52. Importantly, co-expression of RAD52 and dn53BP1 does not alter Cas9 off-target activity. These findings support the use of RAD52 and dn53BP1 co-expression to overcome bottlenecks that limit HDR in precision genome editing.
Repurposing the type II bacterial clustered regularly interspaced short palindromic repeats (CRISPR) system as a genome-editing tool1 led to the development of a robust technology for site-directed genome editing in mammalian cells2–4, including at disease-relevant loci in primary cells5–11. Mammalian cells repair double-strand breaks (DSB) by multiple pathways, including the error-prone non-homologous end-joining (NHEJ) and high-fidelity homologous recombination (HR) pathways12. DSBs generated by CRISPR-associated protein 9 (Cas9) followed by repair through the mutagenic end-joining pathways have been exploited as a means to introduce insertions and deletions to achieve efficient gene disruption6–11. In the presence of a DNA template with sequence homology to the targeted locus and engagement of the homology-directed repair (HDR) pathway, precise gene editing enables the introduction of minor sequence modifications or larger stretches of novel DNA13–15. However, in most mammalian cell types, HDR is less frequently engaged when compared with NHEJ for DSB repair16,17. Moreover, because HDR is largely restricted to the S/G2-phases of the cell cycle18,19, engaging the HDR pathway to achieve precise genome editing in non-cycling or quiescent cells still has major limitations20. The low efficacy of HDR remains the bottleneck in clinical translation of gene editing technologies for the correction of monogenic diseases, but efforts towards augmentation of HDR utilization in clinically relevant cell types has been a topic of intensive interest, and enrichment strategies to increase the yield of gene-modified cells have recently been reported21.
Several strategies to improve HDR efficiency have been reported recently22–27, involving either NHEJ inhibition22,24,25 or augmenting HDR utilization through cell synchronization23 or with small molecules27. Transient inhibition of key NHEJ factors such as Ku70, DNA ligase IV or the DNA-dependent protein kinase catalytic sub-unit, via short hairpin RNA knockdown, small-molecule inhibition or proteolytic degradation, increased HDR in mammalian cell lines22,24,25. However, the impact that such treatments may have on Cas9 off-target activity or genome integrity remains to be investigated. Given the importance of NHEJ in genome maintenance, such strategies may have adverse consequences. Indeed, Ku70 deficiency results in growth retardation and the leaky severe combined immunodeficiency phenotype28, whereas genetic ablation of DNA ligase IV causes late embryonic lethality and impaired V(D)J recombination in mice29. In humans, DNA ligase IV mutations manifest as LIG4 syndrome in which patients exhibit immunodeficiency and developmental/growth delay30. Moreover, inhibition of NHEJ may also impose risks for quiescent cells, such as haematopoietic stem cells, as they utilize the NHEJ pathway to repair accumulated DNA damage on re-entry into cell cycle after periods of dormancy31. Alternatively, it was reported that increased HDR utilization could be achieved through the timely delivery of CRISPR–Cas9 during the S-phase of the cell cycle to cells in vitro through pharmacological means23, but such approaches may hold limitations in vivo due to potential toxicity.
Here, we hypothesized that efficient engagement of HDR may be achieved through manipulation of DNA repair pathway choice. We screened diverse factors involved in DNA repair and report that ectopic expression of RAD52 in combination with a dominant negative form of tumour protein p53-binding protein 1 (dn53BP1)32 increases HDR frequency at multiple loci in human cells, including patient-derived induced pluripotent stem (iPS) cells. Co-expression of RAD52 and dn53BP1 also improved multiplexed, HDR-mediated, precise gene editing, whereas RAD52 alone increased HDR frequency with Cas9D10A nickase. Importantly, off-target analysis using high-throughput genome-wide translocation sequencing (HTGTS)33 and targeted capture deep sequencing revealed no adverse impact on Cas9 specificity on ectopic co-expression of RAD52 and dn53BP1. Our data define a strategy to efficiently achieve precise genome editing using CRISPR–Cas9 in the context of disease modelling or to correct disease-specific mutations.
Results
Optimizing conditions for efficient HDR.
Towards our goal of improving HDR, we used an established human HEK293 reporter cell line in which HDR frequency can be assessed by repair of a broken-green fluorescent protein (GFP) cassette4. Using this cell line, we optimized transfection as well as the amount of Cas9 and guide RNA (gRNA) (Supplementary Fig. 1a–d), cell density, timing of analysis (Supplementary Fig. 1e,f), and concentration, length and orientation of the single-stranded oligonucleotide (ssODN) donor template (Supplementary Fig. 1g–i) and achieved a robust and reproducible basal HDR frequency of 15.7 ± 0.4%, representing a sevenfold increase over non-optimized conditions (Supplementary Fig. 1j). Our data are in general alignment with previously published reports23,34,35 showing that under optimized conditions, ssODN with short sequence homology to the target site can effectively be used to improve HDR.
Screening candidate factors promoting HDR.
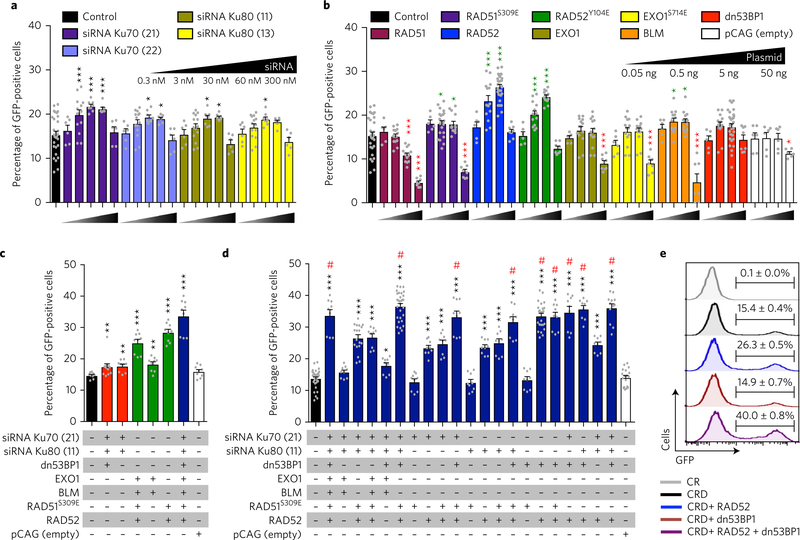
Cells lacking components of NHEJ show a propensity towards increased HDR efficiency36–38 and, consistent with this, inhibition of NHEJ components (Ku, DNA ligase IV or the DNA-dependent protein kinase catalytic subunit) via short hairpin RNA knockdown, proteolytic degradation or pharmacological inhibition has recently been shown to improve HDR frequency22,24,25. In agreement with published reports, we found that small interfering (siRNA) knockdown of Ku70 or Ku80 significantly improved HDR frequency above base-line (Fig. 1a). We reasoned that inhibition of NHEJ alone might not be sufficient to achieve maximal HDR activity and that further manipulation of factors limiting HR engagement might improve HDR. We therefore ectopically expressed factors critically involved in HDR including RAD51, RAD52, exonuclease 1 (EXO1), Bloom syndrome RecQ-like helicase (BLM), along with constitutively active versions of RAD51 (RAD51S309E)39, RAD52 (RAD52Y104E)40 and EXO1 (EXO1S714E)41. We also expressed dn53BP1, containing the tandem tudor domain, which has been reported to improve HR efficiency by counteracting the function of endogenous 53BP132. Overexpression of RAD51, EXO1, EXO1S714E or dn53BP1 did not improve HDR, while RAD51S309E (18.1 ± 0.6%, P = 0.0172) and BLM (18.7 ± 0.7%, P = 0.0102) marginally increased the HDR efficiency compared with controls (15.4 ± 0.8%) (Fig. 1b). In contrast, overexpression of either RAD52 (26.5 ± 0.5%, P < 0.0001) or RAD52Y104E (24.3 ± 0.4%, P < 0.0001) significantly improved HDR frequency (Fig. 1b).
Fig. 1 |. Ectopic expression of RAD52 and dn53BP1 increases HDR efficiency.
HDR frequency as determined by quantifying the percentage of repaired (GFP-positive) cells. a, siRNA knockdown of Ku70 (siRNA numbers 21 and 22) and Ku80 (siRNA numbers 11 and 13) (n = 4). b, Ectopic expression of RAD51, RAD51S309E, RAD52, RAD52Y104E, EXO1, EXO1S714E, BLM, dn53BP1 and a control (empty pCAG plasmid) delivered at the indicated concentrations (n = 7). Green asterisks indicate a significant increase of HDR and red asterisks indicate a significant reduction of HDR. c, Quantification of HDR frequency using combinations of factors that might inhibit NHEJ (red bars) or augment HDR (green bars), and all factors combined (blue bar) (n = 3). d, Stratification of factors to define the minimal complement of factors sufficient to achieve maximal HDR frequency (n = 3). Control cells co-transfected with Cas9, gRNA and donor template and are represented by black bars in a–d. Maximal HDR frequency is marked with by a hash symbol. e, Representative histograms showing HDR frequency (GFP-positive) in cells transfected with the indicated conditions. Statistical significance in a–d was calculated using a one-way analysis of variance with Dunnett’s multiple comparison test (*P < 0.05, **P < 0.01, ***P < 0.001). Independent experiments were performed with each condition assayed in duplicate or triplicate and pooled data are shown with individual data points as grey dots. Error bars represent s.e.m. pCAG: expression plasmid with a CAG promoter (a strong synthetic promoter consisting of the CMV early enhancer element, the promoter of the chicken β-actin gene and the splice acceptor of the rabbit β-globin gene). Numbers in parentheses against each siRNA indicate the last two digits of the respective siRNA identifier mentioned in the Methods.
Co-expression of RAD52 and dn53BP1 allows robust HDR.
Next, we took a combinatorial approach reasoning that these factors may act in a synergistic manner to further improve HDR. We tested the factors that could potentially inhibit NHEJ (siRNAs for Ku70, Ku80 and dn53BP1) or augment HDR (EXO1, BLM, RAD51S309E and RAD52), in addition to the factors combined (Fig. 1c). Although all combinations showed a significant increase in HDR, the highest frequency was achieved when all the factors were combined (33.7 ± 1.8%, P < 0.0001), suggesting that inhibition of NHEJ and augmentation of HDR may synergize to increase precise genome editing (Fig. 1c). To determine the minimal combination sufficient to maximize HDR, we further stratified the groups of factors involved in HDR and NHEJ and identified nine different conditions that showed comparably robust HDR (Fig. 1d). Strikingly, RAD52 and dn53BP1 were the only factors present in all these conditions, and indeed co-expression of RAD52 and dn53BP1 was sufficient to achieve maximal HDR using a ssODN template (33.6 ± 0.9%, P < 0.0001; Fig. 1d,e). When tested with double-stranded donor templates, co-expression of RAD52 and dn53BP1 did not improve HDR frequency (Supplementary Fig. 1k,l), indicating that ectopic expression of RAD52 and dn53BP1 is effective in improving HDR only with single-stranded donor templates. Ectopic co-expression of RAD52 and dn53BP1 did not alter cell viability or proliferation (Supplementary Fig. 2a,b).
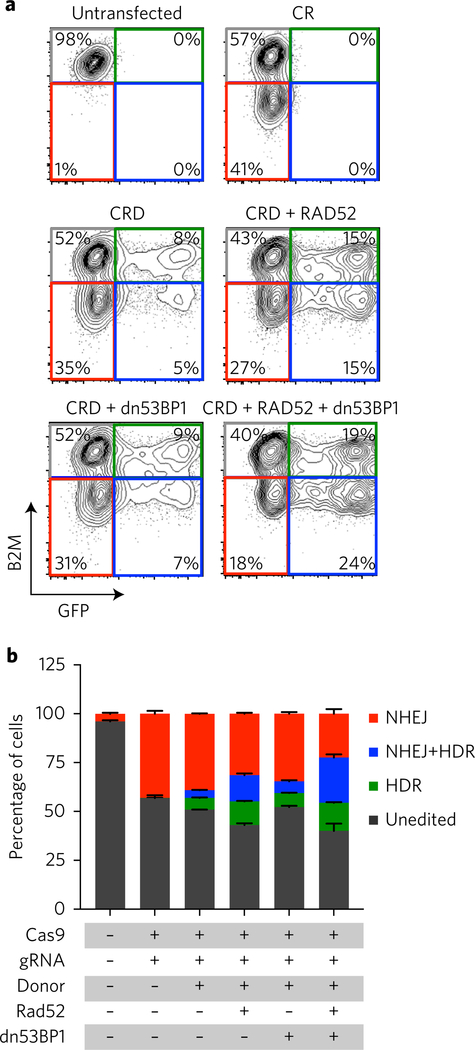
To assess the impact of RAD52 and dn53BP1 on HDR and NHEJ, we developed a system allowing simultaneous monitoring of NHEJ and HDR at different loci in which a gRNA targeting the MHC class I accessory chain β2-microglobin (B2M) provides a measure of NHEJ (ref. 9) and repair of broken-GFP reads out HDR efficacy. Upon transfection of Cas9, gRNAs targeting broken-GFP and B2M and an ssODN template for GFP, we observed exclusive loss of B2M expression in 39.1 ± 0.3% of cells indicating robust NHEJ, whereas 6.1 ± 0.3% of the cells were exclusively GFP+, indicative of HDR. A smaller fraction of cells (3.9 ± 0.2%) were GFP+ and B2M−, indicating that a minor fraction of cells had undergone both NHEJ and HDR (Fig. 2a,b). As expected, in the presence of RAD52, HDR frequency was elevated; dn53BP1 alone did not improve HDR as we had previously observed (Fig. 1e), whereas the maximal increase in HDR was observed when both proteins were co-expressed (Fig. 2a,b). Surprisingly, loss of B2M remained constant under all conditions indicating that the frequency of cells undergoing NHEJ was unaltered upon expression of RAD52 and dn53BP1, or even dn53BP1 alone (Fig. 2a,b), a result that was further confirmed when NHEJ was assessed independently of HDR (Supplementary Fig. 3a). Taken together, these data indicate that RAD52 and dn53BP1 are necessary and sufficient to significantly increase HDR utilization in HEK293 reporter cells.
Fig. 2 |. Co-transfection of RAD52 and dn53BP1 does not affect NHEJ frequency.
a, Representative fluorescence-activated cell sorting plots of cells transfected with the indicated conditions showing fractions of unedited (grey) and edited cells by HDR (green), NHEJ (red) or HDR and NHEJ (blue). b, Quantification of the data from a (n = 3). Error bars represent s.d. See also Supplementary Fig. 3.
Targeted genomic modification at multiple loci in HEK293 and iPS cells.
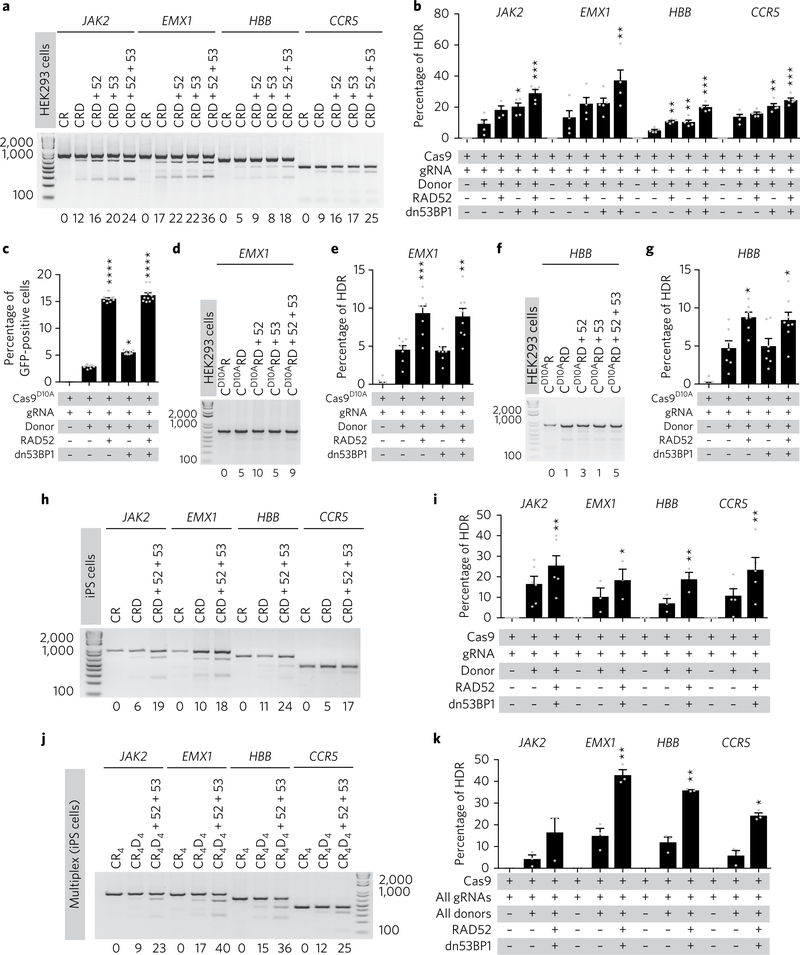
To test the robustness and broad applicability of our approach, we targeted several clinically relevant genes (JAK2, EMX1, HBB and CCR5) in HEK293 cells. To monitor HDR activity, we designed ssODN templates containing an engineered PmeI restriction site (GTTTAAAC) such that following HDR the PmeI site would be incorporated at the targeted locus and simultaneously destroy the proto-spacer sequence to prevent further cleavage by Cas9 (Supplementary Fig. 3b). To our surprise, and in contrast with repair of broken-GFP (Fig. 1b,e and Fig. 2a,b), expression of dn53BP1 alone resulted in significantly improved HDR at three of the four loci with comparable efficacy to RAD52 alone (Fig. 3a,b). Nonetheless, co-expression of RAD52 and dn53BP1 again resulted in the greatest increase in HDR frequency at all of the targeted loci (Fig. 3a,b). Consistent with our previous observations (Fig. 2a,b and Supplementary Fig. 3a), ectopic expression of dn53BP1 alone or in combination with RAD52 did not appear to alter NHEJ usage as the insertion and deletion frequency measured at JAK2 or HBB by the surveyor mismatch cleavage assay using CEL I nuclease were comparable in all the conditions tested (Supplementary Fig. 3c). Next, we tested whether co-expression of RAD52 and dn53BP1 could increase precision genome editing using a Cas9 nickase (Cas9D10A), which was devoid of any detectable off-target activity33,42. Our experiments showed that Cas9D10A-mediated HDR was significantly increased at GFP, EMX1 and HBB by ectopic expression of RAD52 alone (Fig. 3c–g). Notably, ectopic expression of dn53BP1 alone did not improve HDR efficacy, nor did it further augment HDR in the presence of RAD52 (Fig. 3c,e,g).
Fig. 3 |. Co-expression of RAD52 and dn53BP1 improves HDR frequency at multiple loci in human cells.
a,b, Representative gel electrophoresis images showing PmeI-cleaved PCR products indicative of HDR (a) and quantification of HDR frequency (b) following targeting of JAK2, EMX1, HBB and CCR5 in the presence or absence of RAD52, dn53BP1 or both, in HEK293 cells. c–g, Quantification of HDR frequency following targeting with Cas9D10A nickase of GFP (c), EMX1 (d and e) or HBB (f and g) in HEK293 cells. h,i, Representative gel electrophoresis and quantification of HDR frequency at the JAK2, EMX1, CCR5 and HBB locus in human iPS cells. j,k, Simultaneous HDR-mediated gene editing at all four loci using a multiplexed approach. CR, Cas9 + gRNA; CRD, CR + donor; CR4, Cas9 + all gRNAs; CR4D4, Cas9 + all gRNAs + all donors; 52, RAD52; 53, dn53BP1. In b, c, e and g, significance was calculated using a one-way analysis of variance with Dunnett’s multiple comparison test. In i and k, a paired two-tailed t-test was used (*P < 0.05, **P < 0.01, ***P < 0.001). Pooled data from n ≥ 3 independent experiments are shown with individual data points as grey dots. The percentage of HDR for each representative gel is shown below each figure. Error bars represent s.e.m.
Next, we tested whether our approach could be used to improve HDR-mediated gene editing in human iPS cells. Consistent with our results in HEK293 cells, HDR efficacy was significantly increased in human iPS cells upon expression of RAD52 and dn53BP1 at four independent loci (Fig. 3h,i). Cumulatively, these results indicate that the ectopic expression of RAD52 and/or dn53BP1 is effective in improving HDR mediated by Cas9 or Cas9D10A at multiple loci in human cells.
Co-expression of RAD52 and dn53BP1 allows simultaneous HDR-mediated modification of multiple genes.
CRISPR–Cas9 has been used to simultaneously target multiple loci to generate combinations of mutated alleles through multiplexed gRNA pooling2,43–46, but multiplexed HDR-mediated genome modification has not been reported. To assess the ability of CRISPR–Cas9 to mediate precise gene modification in a multiplexed fashion, we pooled gRNAs targeting JAK2, EMX1, HBB and CCR5 with ssODN donor templates containing PmeI restriction sites for each locus and co-transfected these with Cas9 in human iPS cells. Simultaneous HDR-mediated modification with multiplexed gRNA and donor templates was achieved at all of the tested loci (Fig. 3j,k) and co-expression of RAD52 and dn53BP1 consistently increased the frequency of HDR compared with levels achieved using Cas9, gRNAs and donor templates only (Fig. 3j,k). These data suggest that this approach could be used to generate cell lines harbouring multiple precisely defined genetic alterations for disease modelling or pathway discovery.
Highly efficient gene correction through co-expression of RAD52 and dn53BP1 in patient-derived iPS cells.
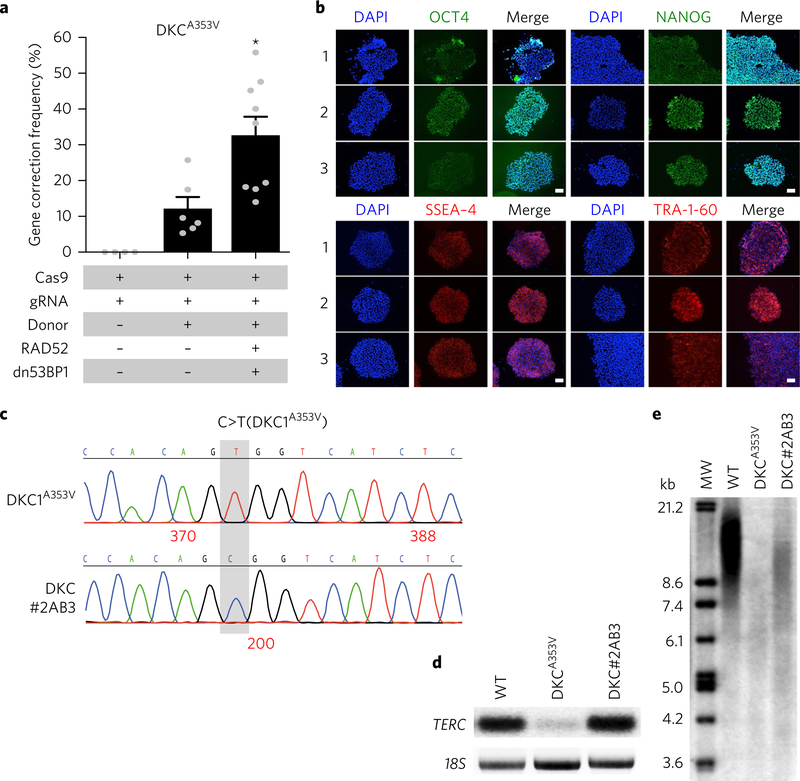
Next, we applied our approach for correcting patient-derived iPS cells bearing a mutation (c.1058 C > T (p.A353V)) in the Dyskerin (DKC1) gene underlying X-linked dyskeratosis congenita. Correction of this mutation restores an MspA1l restriction site to the locus allowing an estimation of HDR frequency upon restriction digestion analysis. Co-expression of RAD52 and dn53BP1 resulted in a correction frequency of 32.7 ± 5.2% compared with the control (12.2 ± 3.2%) (Fig. 4a) without compromising cell viability (Supplementary Fig. 4a) or expression of pluripotency markers (OCT4, NANOG, SSEA-4 and TRA-1–60) (Fig. 4b). Targeted iPS cells were clonally expanded and the correction of the mutated base was confirmed by Sanger sequencing (Fig. 4c). Corrected clones were then either spontaneously differentiated to generate embryoid bodies bearing cells of the three germ layers or, using a haematopoietic differentiation protocol47, were differentiated into CD34 + haematopoietic progenitor cells capable of giving rise to mixed lineage colonies in methylcellulose (Supplementary Fig. 4b,c). These results demonstrate that ectopic expression of RAD52 and dn53BP1 does not affect the tri-lineage differentiation potential of human iPS cells.
Fig. 4 |. Co-expression of RAD52 and dn53BP1 improves precise genomic editing in patient-specific iPS cells.
a, Gene correction frequency in patient-specific iPS cells bearing a mutation (A353V) in the DKC1 gene. b, Immunostaining of gene-corrected and -expanded iPS cells (clones 1–3) showing the expression of pluripotency markers OCT4, NANOG, SSEA-4 and TRA-1–60 (n = 3). c, Representative chromatogram of Sanger sequencing of the parental iPS cell line (DKC1A353V) and a gene-corrected iPS clone (DKC1#2AB3). d, Northern blot radiograph showing TERC, and 18S (loading control) RNA levels in wild-type (WT), DKC1A353V and gene-corrected iPS clone (DKC1#2AB3) cells. e, Southern blot telomere length analysis in WT, DKC1A353V and gene-corrected (DKC1#2AB3) iPS cells. A full blot scan is shown in Supplementary Fig. 4. Pooled data from all the independent experiments are shown with individual data points as grey dots (n = 5). Significance was calculated using paired two-tailed t-test (*P < 0.05). Error bars represent s.e.m. Scale bars: 100 μm. MW, DNA molecular weight ladder.
Dyskerin plays a crucial role in telomere maintenance by stabilization of the telomerase RNA component (TERC)48. To functionally validate DKC1 correction, we assayed dyskerin activity on a corrected clone (DKC1#2AB3) compared with wild-type and parental DKC1A353V controls by measuring TERC levels by northern blot (Fig. 4d and Supplementary Fig. 4d,e) and telomere length by Southern blot (Fig. 4e). The corrected clone showed TERC levels comparable to wild-type iPS cells (Fig. 4d) and concomitant elongation of telomere length compared with the parental cell line (Fig. 4e). These results demonstrate the utility of our approach for efficacious repair of disease-relevant mutations and functional restoration upon gene correction.
Co-expression of RAD52 and dn53BP1 does not alter Cas9 off-target activity.
To assess the impact of ectopic expression of RAD52 and dn53BP1 on CRISPR–Cas9 specificity we used two next-generation sequencing strategies to measure Cas9 off-target activity. In these experiments, we used two gRNAs targeting the chemokine receptor CCR5 (gCCR5D and gCCR5Q) that we had previously used in primary human CD34+ haematopoietic stem and progenitor cells9. Using HTGTS—an unbiased approach that has been used to monitor the off-target activity of Cas9 and other engineered nucleases33, we found that gCCR5D only exhibited on-target activity in HEK293 cells with no detectable off-target, whereas gCCR5Q exhibited significant off-target activity at seven genomic sites (Table 1). Next, we performed experiments to interrogate if ectopic expression of RAD52, dn53BP1 alone or both in combination would alter Cas9 off-target activity. These experiments revealed that Cas9 specificity was unchanged by expression of RAD52 or dn53BP1 as there were no new translocation junctions generated and sequenced other than those already associated with the guides targeting CCR5 (Tables 1 and 2) or the guide targeting the universal bait (RAG1) used in HTGTS33 (Table 2). Interestingly, analysis of the translocation junctions revealed differences in microhomology distributions at both on-target sites of gCCR5D and gCCR5Q (Fig. 5) and off-target sites of gCCR5Q and gRAG1 (RAG1B_OT1) under the various test conditions (Supplementary Fig. 5). Ectopic expression of RAD52 alone promoted increased end-joining with minimal (1 bp) microhomology over control transfected samples (Fig. 5 and Supplementary Fig. 5). In contrast, ectopic expression of dn53BP1 alone resulted in diminished direct end-joining and increased the joining of DNA ends with greater microhomology distributions (2–5 bp depending on the site) (Fig. 5 and Supplementary Fig. 5). However, upon RAD52 and dn53BP1 co-expression, the effects of RAD52 antagonized the impact of dn53BP1 and direct end-joining, and end-joining with minimal (1 bp) microhomology predominated (Fig. 5 and Supplementary Fig. 5).
Table 1 |.
HTGTS analysis of on- and off-target sites associated with gRNAs targeting CCR5
| Chromosome | Start | End | Target | Sequence | PAM | Total junctions |
Frequency (10,000 junctions) |
|---|---|---|---|---|---|---|---|
| chr3 | 46,411,328 | 46,420,240 | CCR5_D | TCACTATGCTGCCGCCCAGT | (GGG) | 1,619 | 307.1 |
| chr3 | 46,412,265 | 46,420,077 | CCR5_Q | GCTGTGTTTGCGTCTCTCCC | (AGG) | 1,222 | 153.4 |
| chr1 | 109,834,518 | 109,843,369 | Q_OT1 | GCTGTGTCCGCGCCTCTCCC | (CGG) | 1,932 | 242.5 |
| chr20 | 23,301,706 | 23,302,705 | Q_OT2 | GCTGTGTTTGCGCCTTTCCC | (AGG) | 58 | 7.3 |
| chr11 | 62,782,827 | 62,783,824 | Q_OT3 | GCTGTGTTTGTGCCTCCCCC | (AGG) | 11 | 1.4 |
| chr12 | 120,933,483 | 120,934,463 | Q_OT4 | GCTGTGCTAGCACCTCCCCC | (AGG) | 9 | 1.1 |
| chr10 | 77,357,580 | 77,358,098 | Q_OT5 | GCTGTGTGCTCGTCTCTCCC | (TGG) | 8 | 1 |
| chrX | 45,602,391 | 45,603,389 | Q_OT6 | TGGGTGTCTGCAGCTCTCCC | (AGG) | 8 | 1 |
| chr11 | 134,037,226 | 134,037,726 | Q_OT7 | CACGAGTGTGCGCCTCCCCC | (AGG) | 6 | 0.8 |
Pooled data from three independent experiments are shown. On-target chromosomal translocations are shown in bold. Off-target (OT) mismatches are underlined. The numbers of translocation junctions analysed were n = 52,719 for gCCR5D and n = 79,677 for gCCR5Q.
Table 2 |.
HTGTS analysis of on- and off-target sites associated with gRNAs targeting CCR5 in presence of RAD52 and/or dn53BP1
| Total junctions |
|||||
|---|---|---|---|---|---|
| Chromosome | Target | CR | CR + RAD52 | CR + dn53BP1 | CR + RAD52 + dn53BP1 |
| chr3 | CCR5_D | 905 | 1,855 | 1,091 | 1,764 |
| chr14 | RAG1B_OT1 | 508 | 990 | 581 | 643 |
| chr4 | RAG1B_OT2 | 82 | 138 | 93 | 64 |
| chr3 | CCR5_Q | 519 | 906 | 684 | 668 |
| chr1 | Q_OT1 | 723 | 1,320 | 434 | 511 |
| chr20 | Q_OT2 | 21 | 32 | 37 | 26 |
| chr11 | Q_OT3 | 4 | 7 | 10 | 19 |
| chr12 | Q_OT4 | 7 | 10 | 5 | 8 |
| chr10 | Q_OT5 | 2 | 6 | 2 | 8 |
| chrX | Q_OT6 | 4 | 10 | 4 | 12 |
| chr11 | Q_OT7 | 0 | 3 | 0 | 3 |
| chr14 | RAG1B_OT1 | 477 | 931 | 503 | 261 |
| chr4 | RAG1B_OT2 | 70 | 117 | 52 | 33 |
Pooled data from three independent experiments normalized to 22,086 total junctions are shown. On-target junctions are shown in bold. CR, Cas9 + gRNA; Off-targets (OTs) associated with universal bait RAG1B are underlined.
Fig. 5 |. Effect of RAD52 and dn53BP1 on microhomology distribution at the target site.
a,b, Microhomology distribution at the junctions with respect to RAG1 bait at the on-target sites for gCCR5D (a) and gCCR5Q (b) in the presence of RAD52 and/or dn53BP1. The number of junctions with <11-bp microhomology analysed for each condition is shown in brackets. CR, Cas9 + gRNA. Pooled data from three independent experiments normalized to 22,086 total junctions for each condition are shown. Error bars represent s.e.m. Significance was calculated using two-way analysis of variance (*P < 0.05, **P < 0.01, ***P < 0.0001).
Although our HTGTS data showed that Cas9 specificity remained unaltered in presence of RAD52 and dn53BP1, HTGTS precludes characterization of insertions and deletions. Therefore, to assess insertion and deletion activity we performed off-target analysis by targeted capture deep sequencing at algorithmically predicted off-target sites9. For this, we co-transfected Cas9, along with gRNA targeting CCR5 (gCCR5D or gCCR5Q) and respective ssODN donor templates into HEK293 cells in the presence or absence of RAD52 and/or dn53BP1 followed by targeted capture deep sequencing as previously described9 (Supplementary Table 1). As we observed with HTGTS (Tables 1 and 2), gCCR5D did not exhibit significant off-target activity at 17 predicted off-target sites in the presence or absence of RAD52 and/or dn53BP1 compared with controls (Supplementary Table 1). Off-target activity for gCCR5Q was observed, yet only at one off-target site (gCCR5Q_OT35) out of 28 predicted unique off-targets that corresponded to the predominant off-target site detected by HTGTS. Importantly, ectopic expression of RAD52 and/or dn53BP1 did not increase the insertion and deletion frequency in any setting, but rather we observed a slight yet significant decrease in the insertion and deletion frequency upon ectopic expression of dn53BP1 alone (P = 0.00343) or in combination with RAD52 (P = 0.00304) (Supplementary Table 2). Taken together, these results demonstrate that ectopic expression of RAD52 and dn53BP1 does not alter Cas9 off-target activity.
RAD52 is dispensable for basal HDR activity in human cells.
To understand the possible role of RAD52 in improving the HDR efficiency, we first pharmacologically inhibited RAD52 using a published inhibitor of RAD52 ((−)-Epigallocatechin)49 and found that RAD52 inhibition did not alter HDR frequency (Supplementary Fig. 6a). Next, we targeted exon 5 of RAD52 in the HEK293 HDR reporter cell line using CRISPR–Cas9 and established two independent RAD52-null cell lines. Consistent with our experiments using the RAD52 inhibitor, genetic loss of RAD52 did not alter basal HDR activity (Supplementary Fig. 6b). Taken together, these experiments suggest that RAD52 is dispensable for basal HDR activity in human cells.
dn53BP1 competitively antagonizes 53BP1 to improve HDR efficiency.
To explore the mechanism by which dn53BP1 improves CRISPR-mediated HDR, we generated a Flag-peptide attached to a nuclear localization sequence-tagged dn53BP1 (FN-dn53BP1) and a mutant version (FN-dn53BP1D1518R) that does not bind to chromatin32. Expression of these proteins in HEK293 cells showed that FN-dn53BP1 but not FN-dn53BP1D1518R gets recruited to γ-H2AX foci positive irradiation-induced DSB sites as expected (Fig. 6a and Supplementary Fig. 7a). Co-staining for endogenous 53BP1 combined with quantitative intensity distribution revealed that 53BP1 and FN-dn53BP1 did not co-occupy γ-H2AX foci, indicating that dn53BP1 very effectively inhibited the recruitment of 53BP1 to the DSB sites, whereas FN-dn53BP1D1518R did not (Fig. 6a). Intensity correlation analysis confirmed a non-coexistence relationship between 53BP1 and FN-dn53BP1 with a maximum information coefficient score of ~0.4 (Fig. 6a). Consistent with its inability to competitively antagonize 53BP1 recruitment to DSB sites, dn53BP1D1518R was unable to augment CRISPR-mediated HDR (Fig. 6b,c). To further demonstrate that dn53BP1 acts by competitively antagonizing 53BP1, we generated a 53BP1 mutant HEK293 cell line (from the parental HDR reporter cell line with broken-GFP) that showed no 53BP1 foci formation upon irradiation, indicating a lack of recruitment to DSB sites (Supplementary Fig. 7b). This mutant line exhibited significantly higher CRISPR-mediated HDR activity at broken-GFP, JAK2 and HBB compared with the parental cell line under basal conditions (Fig. 6d–f). Importantly, ectopic expression of dn53BP1 was ineffective in improving HDR activity in this cell line (Fig. 6d–f), suggesting that the augmentation of HDR activity observed with ectopic expression of dn53BP1 occurs through competitive antagonism of endogenous 53BP1 activity at DSB sites.
Fig. 6 |. dn53BP1 competitively antagonizes 53BP1 to improve HDR efficiency.
a, Representative images (top panels) and quantified intensity correlation plots (bottom panels) showing endogenous 53BP1 and Flag-peptide attached to a nuclear localization sequence-tagged dn53BP1 (FN-dn53BP1) recruitment to the irradiation-induced DNA break sites. A mutant version of FN-dn53BP1 (FN-dn53BP1D1518R) that does not accumulate at the DSB sites is also shown. The number of data points analysed for each sample is indicated. a.u., arbitrary units. b,c, HDR activity at JAK2 (b) and HBB (c) in the presence of RAD52, dn53BP1 or dn53BP1D1518R as indicted. d–f, HDR activity in 53BP1-null cells (53BP1_KO) compared with the parental cell line at broken-GFP (d), JAK2 (e) and HBB (f). Significance was calculated using a one-way analysis of variance with Dunnett’s multiple comparison test for analyses within the same cell line and an unpaired t-test to compare the same condition in the two different cell lines (*P < 0.05, **P < 0.01, ****P < 0.0001; NS, not significant). Pooled data from all the independent experiments are shown with individual data points as grey dots (n = 3). Error bars represent s.e.m.
Discussion
CRISPR–Cas9 enables genetic manipulation of mammalian cells with unprecedented ease and efficacy, particularly for applications in which gene disruption is the desired outcome. Indeed, efficacious ablation of genes in clinically relevant primary cell types including human haematopoietic stem cells and T cells has already been achieved9,11,50–52. However, harnessing the full potential of CRISPR–Cas9 for precision gene editing, including repair of disease-causing mutations, is currently limited by infrequent utilization of HDR. Here, we show that manipulation of DNA repair pathway choice is an efficient strategy to maximize precision genome editing in human cells with CRISPR–Cas9. We found that co-expression of RAD52 and dn53BP1 enables efficient HDR that is applicable to multiple loci and multiplexing, and does not alter off-target activity. We demonstrate that this technology can be used for efficient gene correction in patient-derived iPS cells to restore functional competency.
The outcome of DSB repair depends on the engagement of various repair pathways, as well as the interplay of numerous DNA damage response effector molecules53. Previous studies have shown that genetic deficiencies in NHEJ result in channelling DSB repair to the HDR pathway36–38, and transient NHEJ inhibition can be utilized to improve HDR22,24,25. In contrast with previous approaches employed to increase HDR22,24,25, our data suggest that ectopic expression of RAD52 and dn53BP1 does not alter the insertion and deletion frequency during CRISPR-mediated editing, suggesting that NHEJ inhibition was not the primary driver of the elevated HDR. Nonetheless, the fact that microhomology distribution usage was altered by ectopic expression of these proteins indicates clear differences in end-joining activity. We observed that dn53BP1 alone resulted in the joining of DNA ends with increased microhomology distributions, which is in agreement with the implicated role of 53BP1 in protecting the end of DNA by preventing the resection54,55 and fostering fidelity of HDR56. Interestingly, however, this effect of dn53BP1 was antagonized by the introduction of RAD52, which on its own promoted end-joining with minimal microhomology usage. Importantly, using an unbiased method to examine CRISPR–Cas9 off-target activity and chromosomal translocation (HTGTS), we showed that the expression of RAD52 and/or dn53BP1 did not alter CRISPR–Cas9 specificity. Moreover, targeted capture deep sequencing analysis revealed that the insertion and deletion frequency at predicted off-targets was unaffected by the expression of RAD52 and dn53BP1.
RAD52 is a key DSB repair factor involved in homologous recombination and the single-strand annealing (SSA) pathway57–61. In yeast, loss of RAD52 is indispensable for homology-dependent DSB repair, SSA and the repair of X-ray-induced DNA damage61,62. Similarly, RAD52 deficiency leads to decreased knock-in efficacy of large double-stranded donor templates in chicken60 and mouse cells59. In contrast, RAD52 knockdown in human cell lines had a small impact on spontaneous or I-SceI endonuclease-induced HR owing to the redundant function of the breast cancer early onset-dependent pathway63,64. Consistent with these findings, our observation of unchanged HDR frequency under pharmacological inhibition of RAD52 or in RAD52-null cells indicates that RAD52 is dispensable for basal CRISPR-mediated HDR using ssODN templates in human cells. Nonetheless, ectopic expression of RAD52 improves CRISPR-mediated HDR using ssODN, presumably by favouring engagement of the SSA pathway.
The role of 53BP1 in DSB repair is complex and context dependent65,66. 53BP1 is considered to be an NHEJ promoting factor given its ability to occupy DSB sites and prevent end processing67. It has been shown that either loss of 53BP1 or expression with a dominant negative form of 53BP1 improves HR in an I-SceI system32. 53BP1 inhibits HR in Brca1-deficient cells by blocking resection of DNA breaks, which can be restored by 53BP1 deletion68. Consistent with these reports, we found that basal HDR activity was significantly enhanced by genetic ablation of 53BP1 during CRISPR-mediated editing. Our data suggest that recruitment of dn53BP1 to DSBs is required to competitively antagonize 53BP1 and augment CRISPR-mediated HDR in human cells. This is supported by our observations that dn53BP1 is ineffective in improving HDR in the absence of 53BP1. A potential role for 53BP1 in HDR has recently come to light in a study showing that silencing or exhausting the capacity of 53BP1 to bind DNA induces a switch from RAD51-dependent gene conversion to RAD52-dependent SSA56. SSA-mediated HDR is thought to be mutagenic as a result of annealing of homologous sequences flanking the DSB site69. In contrast, our HDR experiments, performed in the presence of ssODN and in combination with ectopic expression of RAD52 and/or dn53BP1, occurred without evidence of mutagenesis. This suggests that ssODN templates can be used for error-free HDR in the presence of RAD52, an interpretation consistent with studies showing that RAD52-dependent SSA is accurate and leads to error-free repair in the presence of homologous single-stranded DNA donor templates58,70. Taken together, our data suggest a two-step model in which expression of dn53BP1 prevents the recruitment of 53BP1 to Cas9-generated DSBs, facilitating RAD52-mediated HDR at the cut site using ssODNs.
Methods
Generation of CRISPR–Cas9 vectors, gRNAs and candidate gene plasmids.
A human-codon-optimized Cas9 gene with a C-terminal nuclear localization signal4 was sub-cloned into a CAG expression plasmid (an expression plasmid with a CAG promoter consisting of a CMV early enhancer element, the promoter of the chicken β-actin gene and the splice acceptor of the rabbit β-globin gene). PX459 plasmid encoding Cas9 and puromycin was purchased from Addgene. gRNA targeting the GFP sequence4 was cloned in a plasmid with the human U6 polymerase III promoter. All the other gRNA sequences published previously (Supplementary Table 3) were cloned into the pGuide plasmid using BbsI restriction sites4. The gRNA and primer sequences are listed in Supplementary Table 3. The gRNAs used in this work were selected by Surveyor assay. Plasmids encoding components of the DNA repair pathways (human RAD51, human RAD52, human EXO1, human BLM and mouse 53BP1) were obtained from the Harvard PlasmID Database. The polymerase chain reaction (PCR) products of the genes were then sub-cloned into a CAG expression plasmid and sequenced. For the generation of dn53BP1, we sub-cloned a fragment containing the tudor domain (residues 1,221 to 1,718 of mouse 53BP1). The mutant versions (RAD51S309E, RAD52Y104E and EXO1S714E) were generated by site-directed mutagenesis. siRNAs for Ku70 and Ku80 were purchased from Sigma–Aldrich (NM_021141: SASI_Hs01_00099411; SASI_Hs01_00099413 / NM_001469: SASI_Hs01_00070122; SASI_Hs01_00070121).
Cell culture.
In-house available HEK293 cells were verified by sequencing. The integration of broken-GFP cassette in the HEK293 HDR reporter cell line was confirmed by sequencing. HEK293 cells are a commonly used cell line and are easy to transfect with very high transfection efficiency. Therefore, we used these cells for various experiments. HEK293 and broken-GFP HDR reporter cell lines were maintained in Dulbecco’s Modified Eagle’s medium (Gibco) supplied with 10% foetal bovine serum (Gibco) and pencillin-streptomycin (Gibco). The human iPS cell lines (BJ RiPS) have been described previously71. The human iPS cell line DKC1A353V was derived from patient skin fibroblasts that were obtained under institutional review board-approved protocols following informed consent in accordance with the Declaration of Helsinki. DKC1A353V fibroblasts were reprogrammed and characterized using the method described in ref. 72. iPS cells were maintained onto hESC-Qualified Matrigel (BD Biosciences) in mTeSR (STEMCELL Technologies). For transfection, cells were maintained onto Matrix and Pluripro (both from Cell Guidance Systems) and enzymatic passaging was done with TrypLE (Life Technologies) and Y-27632 (10 μM; Calbiochem).
Transfection of cells.
HEK293 cells were seeded in 96-well plates (5,000 cells well−1) one day before transfection. Cells were transfected with plasmids (Cas9: 25 ng well−1; gRNA: 25 ng well−1; donor template: 4 pmol well−1; RAD52 and dn53BP1: 5 ng well−1, unless specified) mixed with Opti-MEM (Invitrogen) and 0.3 μl well−1 (optimized value) of Trans-IT 293 reagent (Mírus) according to the manufacturer’s recommendation. As the transfection efficiency of HEK293 is higher than 90% (Supplementary Fig. 1a), total cells were analysed 96 h after transfection. Human iPS cells were plated in 12-well plates the day before transfection in Pluripro (Cell Guidance Systems). After 24 h, plasmids (Cas9-puro: 250 ng well–1; gRNA: 250 ng well–1; donor template: 40 pmol well−1; RAD52 and dn53BP1: 50 ng well−1) were mixed with Opti-MEM and 3 μl well−1 Trans-IT LT1 (Mírus) according to the manufacturer’s datasheet. Concentrations for the multiplex experiment were Cas9-puro: 1,000 ng well−1; gRNA: 100 ng well−1 (of each individual plasmid); donor template: 10 pmol well−1 (of each individual single-stranded donor template); RAD52 and dn53BP1: 50 ng well−1. For antibiotics selection, 0.5–1 μg ml−1 of puromycin was added after 24 h of transfection for two days. After 96 h of transfection, DNA was extracted or iPS cells were enzymatically detached and plated onto Matrigel and mTeSR in a ratio of 5,000 cells per 60 mm dish. 10 μM of Y-27632 was added to increase single cell survival after passaging. When the colonies started to appear, each clone was manually collected and split into two wells of a 96-well plate with Matrigel and mTeSR1 (Stem Cell Technologies). One of the wells was reserved to do clonal screening by PCR and the other well was used to start clonal expansion.
Analysis of HDR by restriction digestion.
Frequencies of HDR/NHEJ were reported essentially in transfected cells. Transfection efficiency in HEK293 cells approaches 100% and in these cases analyses were done in total cells. For the iPS cells, transfected cells were selected by puromycin. The targeted region was amplified by PCR using GoTaq Hot Start Green Master Mix, 2X (Promega). The restriction digestion was performed in the PCR reaction mix supplemented with CutSmart Buffer (NEB) and two units of PmeI or MspA1I. The restriction digestion was carried out for 1.5 h at 37°C. The product was resolved on 2.5% agarose gel containing ethidium bromide. The gel picture was taken using a Carestream Gel Logic 212 Pro instrument and Molecular Imaging Software(v.5.3.2.16673, http://mi.carestreamhealth.com). The band intensity was quantitated using Image J(v.1.51m9, http://imagej.nih.gov/ij). The percentage of HDR was calculated using the equation (b + c / a + b + c) × 100, in which ‘a’ is the uncleaved band intensity and ‘b’ and ‘c’ are the cleavage products23,35.
TERC and telomere length analysis.
TERC RNA levels and telomere length were respectively analysed by northern and Southern blot, as described previously73.
Flow cytometry.
Flow cytometry analyses were performed as described previously9.
Immunostaining.
Immunostaining for pluripotency markers was performed as described previously71.
HTGTS and off-target analysis.
Off-target analysis was carried out by HTGTS as described earlier33,74. Briefly, HEK293 cells were co-transfected with expression plasmids encoding Cas9:RAG1B (20 μg), gRNA targeting CCR5 (10 μg), RAD52 and dn53BP1 (5 μg each) and GFP (5 μg) using calcium phosphate. Cells were lysed 48 h post-transfection and DNA was isolated by ethanol precipitation. 50 μg of DNA was processed for HTGTS as described33,74.
Capture sequencing and next-generation sequencing analysis.
HEK293 cells were seeded in 96-well plates (5,000 cells well−1) one day before transfection. Cells were transfected with plasmids and the donor template (Cas9: 25 ng well−1; gRNA: 25 ng well−1; donor template: 4 pmol well−1; RAD52 and dn53BP1: 5 ng well−1). Nine wells per condition were transfected with transfection mix. After five days of transfection, cells from nine wells for each condition were pooled together and treated with Proteinase K overnight at 56°C. DNA was isolated by ethanol precipitations. Capture libraries were prepared and next-generation sequencing was preformed as described previously9.
Image analysis.
The FN-dn53BP1 and endogenous 53BP1 signals colocalizing with γ-H2AX and DNA-dapi stain were detected and quantified using custom MATLAB (MathWorks, v.2017a, https://www.mathworks.com) routines that fitted the amplitudes with a two-dimensional Gaussian fitting function75 using a sigma of 1.3. False positive detections of γ-H2AX outside the cells were filtered by setting a fitted amplitude threshold at the 99th percentile of the Gaussian to the first peak of the γ-H2AX distribution. A colour-coded density scatter plot was generated using MATLAB (blue to red for low to high frequency) plotting endogenous 53BP1 versus Flag-peptide staining in the untreated sample, FN-dn53BP1 or FN-dn53BP1D1518R. Intensity correlation using maximum information coefficient, part to maximal information-based non-parametric exploration statistics76 was employed to test the dependence between endogenous 53BP1 and FN-dn53BP1 for fitted amplitudes gated above 180.
Statistical analyses.
All the experiments were performed in triplicates and repeated at least three times, generating completely independent datasets. Variance remained similar among the groups that were being statistically compared. All data were pooled for analyses and statistics were performed using Graphpad Prism 6 (https://www.graphpad.com/scientific-software/prism/).
Supplementary Material
Acknowledgements
This work was supported in part by National Institutes of Health grants R01AI020047 and R01AI077595 (to F.W.A.) and RO1HL107630, HL107440, UC4DK104218 and U19HL129903 (to D.J.R.), the Translational Research Program (Boston Children’s Hospital), Pedals for Pediatrics (Dana-Farber Cancer Institute) awards (to S.A. and B.B.), The Leona M. and Harry B. Helmsley Charitable Trust (to D.J.R.) and the New York Stem Cell Foundation (to D.J.R.). The HEK293 broken-GFP reporter cell line was kindly provided by G. Church. The gRNA constructs targeting B2M were provided by C. Cowan.
Footnotes
Competing interests
D.J.R. is an academic co-founder of Intellia Therapeutics (Cambridge, MA), a biotechnology company focused on developing CRISPR–Cas9 therapies.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-017-0145-2.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability. Targeted capture deep sequence data are available from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession number SUB2968347. HTGTS data are available from the Gene Expression Omnibus GSE103241. The datasets generated and/or analysed during the current study are available from the corresponding author upon reasonable request.
References
- 1.Jinek M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu PD, Lander ES & Zhang F. Development and applications of CRISPR–Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Q. et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hruscha A. et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140, 4982–4987 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Li D. et al. Heritable gene targeting in the mouse and rat using a CRISPR–Cas system. Nat. Biotechnol 31, 681–683 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Mandal PK et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15, 643–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendel A. et al. Chemically modified guide RNAs enhance CRISPR–Cas genome editing in human primary cells. Nat. Biotechnol 33, 985–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceccaldi R, Rondinelli B. & D’Andrea AD Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell CR, Keown W, Lowe L, Kirschling D. & Kucherlapati R. Homologous recombination involving small single-stranded oligonucleotides in human cells. New Biol. 1, 223–227 (1989). [PubMed] [Google Scholar]
- 14.Igoucheva O, Alexeev V. & Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 8, 391–399 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Te Riele H, Maandag ER & Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl Acad. Sci. USA 89, 5128–5132 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao Z, Bozzella M, Seluanov A. & Gorbunova V. Comparison of non-homologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 7, 1765–1771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen NM et al. An update on targeted gene repair in mammalian cells: methods and mechanisms. J. Biomed. Sci 18, 10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karanam K, Kafri R, Loewer A. & Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell 47, 320–329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branzei D. & Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol 9, 297–308 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Genovese P. et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dever DP et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu VT et al. Increasing the efficiency of homology-directed repair for CRISPR–Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol 33, 543–548 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lin S, Staahl BT, Alla RK & Doudna JA Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maruyama T. et al. Increasing the efficiency of precise genome editing with CRISPR–Cas9 by inhibition of non-homologous end joining. Nat. Biotechnol 33, 538–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robert F, Barbeau M, Ethier S, Dostie J. & Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 7, 93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu C. et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16, 142–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu Y. et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7, 653–665 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Frank KM et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396, 173–177 (1998). [DOI] [PubMed] [Google Scholar]
- 30.O’Driscoll M. et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell 8, 1175–1185 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Beerman I, Seita J, Inlay MA, Weissman IL & Rossi DJ Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15, 37–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie A. et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell 28, 1045–1057 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frock RL et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol 33, 179–186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renaud JB et al. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR–Cas9 nucleases. Cell Rep. 14, 2263–2272 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Richardson CD, Ray GJ, DeWitt MA, Curie GL & Corn JE Enhancing homology-directed genome editing by catalytically active and inactive CRISPR–Cas9 using asymmetric donor DNA. Nat. Biotechnol 34, 339–344 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Pierce AJ, Hu P, Han M, Ellis N. & Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15, 3237–3242 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delacote F, Han M, Stamato TD, Jasin M. & Lopez BS An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 30, 3454–3463 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurosawa A. et al. DNA ligase IV and artemis act cooperatively to suppress homologous recombination in human cells: implications for DNA double-strand break repair. PLoS ONE 8, e72253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorensen CS et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol 7, 195–201 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Honda M, Okuno Y, Yoo J, Ha T. & Spies M. Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. EMBO J. 30, 3368–3382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolderson E. et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 38, 1821–1831 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ran FA et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie K, Minkenberg B. & Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl Acad. Sci. USA 112, 3570–3575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shalem O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin L. et al. Multiplex conditional mutagenesis using transgenic expression of Cas9 and sgRNAs. Genetics 200, 431–441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabadi AM, Ousterout DG, Hilton IB & Gersbach CA Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 42, e147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J. et al. mRNA-mediated glycoengineering ameliorates deficient homing of human stem cell-derived hematopoietic progenitors. J. Clin. Invest 127, 2433–2437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell JR, Wood E. & Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Hengel SR et al. Small-molecule inhibitors identify the RAD52–ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells. eLife 5, e14740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou P. et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci. Rep 5, 15577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren J. et al. Multiplex genome editing to generate universal CAR T cells resistant to PD-1 inhibition. Clin. Cancer Res 23, 2255–2266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su S. et al. CRISPR–Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep 6, 20070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Symington LS & Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet 45, 247–271 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Bothmer A. et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42, 319–329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bothmer A. et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med 207, 855–865 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ochs F. et al. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol 23, 714–721 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Bhargava R, Onyango DO & Stark JM Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32, 566–575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Storici F, Snipe JR, Chan GK, Gordenin DA & Resnick MA Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol. Cell Biol 26, 7645–7657 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rijkers T. et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell Biol 18, 6423–6429 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaguchi-Iwai Y. et al. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell Biol 18, 6430–6435 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Symington LS Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev 66, 630–670 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanamshet K, Mazina OM & Mazin AV Reappearance from obscurity: mammalian Rad52 in homologous recombination. Genes (Basel) 7, E63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng Z. et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl Acad. Sci. USA 108, 686–691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lok BH, Carley AC, Tchang B. & Powell SN RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32, 3552–3558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daley JM & Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell Biol 34, 1380–1388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kakarougkas A. et al. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 41, 9719–9731 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panier S. & Boulton SJ Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol 15, 7–18 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Bunting SF et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Finney-Manchester SP & Maheshri N. Harnessing mutagenic homologous recombination for targeted mutagenesis in vivo by TaGTEAM. Nucleic Acids Res. 41, e99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daley JM & Wilson TE Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell Biol 25, 896–906 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warren L. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park IH, Lerou PH, Zhao R, Huo H. & Daley GQ Generation of human-induced pluripotent stem cells. Nat. Protoc 3, 1180–1186 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Moon DH et al. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat. Genet 47, 1482–1488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu J. et al. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc 11, 853–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aguet F, Antonescu CN, Mettlen M, Schmid SL & Danuser G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell 26, 279–291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reshef DN et al. Detecting novel associations in large data sets. Science 334, 1518–1524 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.